Abstract
ATF6, a member of the leucine zipper protein family, can constitutively induce the promoter of glucose-regulated protein (grp) genes through activation of the endoplasmic reticulum (ER) stress element (ERSE). To understand the mechanism of grp78 induction by ATF6 in cells subjected to ER calcium depletion stress mediated by thapsigargin (Tg) treatment, we discovered that ATF6 itself undergoes Tg stress-induced changes. In nonstressed cells, ATF6, which contains a putative short transmembrane domain, is primarily associated with the perinuclear region. Upon Tg stress, the ATF6 protein level dropped initially but quickly recovered with the additional appearance of a faster-migrating form. This new form of ATF6 was recovered as soluble nuclear protein by biochemical fractionation, correlating with enhanced nuclear localization of ATF6 as revealed by immunofluorescence. Optimal ATF6 stimulation requires at least two copies of the ERSE and the integrity of the tripartite structure of the ERSE. Of primary importance is a functional NF-Y complex and a high-affinity NF-Y binding site that confers selectivity among different ERSEs for ATF6 inducibility. In addition, we showed that YY1 interacts with ATF6 and in Tg-treated cells can enhance ATF6 activity. The ERSE stimulatory activity of ATF6 exhibits properties distinct from those of human Ire1p, an upstream regulator of the mammalian unfolded protein response. The requirement for a high-affinity NF-Y site for ATF6 but not human Ire1p activity suggests that they stimulate the ERSE through diverse pathways.
ATF6 was first isolated as a member of an extensive family of leucine zipper proteins able to selectively form DNA binding heterodimers (5). ATF6 is a 670-amino-acid protein; however, it exhibits an electrophoretic mobility of 90 kDa in sodium dodecyl sulfate (SDS)-polyacrylamide gels (36). The physiological role of ATF6 in the control of gene expression in mammalian cells is just emerging. Intriguingly, while ATF6 was originally isolated through probing of a cDNA expression library with a multimerized ATF site, specific DNA binding for ATF6 has not yet been demonstrated. Functional studies indicate that ATF6 can bind to the serum response factor transcriptional activation domain, and transfection experiments using antisense vector directed against ATF6 suggest that it is required for the serum induction of the c-fos promoter (36). Further, it was discovered that ATF6, rather than the serum response factor, is the target of p38 mitogen-activated protein kinase (MAPK) phosphorylation (25).
When mammalian cells are subjected to calcium depletion stress or protein glycosylation block, the transcription of a family of glucose-regulated protein (grp) genes encoding endoplasmic reticulum (ER) chaperones is induced to high levels (11, 12). This induction process, referred to as the unfolded protein response (UPR) (10), is mediated by novel promoter regulatory motifs present in multiple copies on the promoters of ER chaperone genes (20, 33). The consensus mammalian ER stress response element (ERSE) consists of a tripartite structure CCAATN9CCACG, with N9 being a strikingly GC-rich region of 9 bp. The ERSE binds multiple mammalian transcription factors including the multimeric CCAAT binding protein NF-Y (also referred to as CBF), YY1, Y-box proteins YB-1 and dbpA, and a newly discovered ER stress-inducible binding factor referred to as ERSF (14, 15, 20, 21). Using a dominant negative mutant to interfere with NF-Y function, it was demonstrated that optimal induction of ERSE requires NF-Y (35). While overexpression of YY1 has no effect on the basal activity of the grp78 promoter, it is able to stimulate the grp78 promoter activity in stressed cells (15).
The various ERSEs share similar sequence motifs and can independently confer the mammalian ER stress response to heterologous promoters (13); however, there are subtle differences which are evolutionarily conserved (3). In particular, ERSE-163 contained within the rat grp78 core (16) bears an unconventional CGAAT motif for NF-Y/CBF binding and the sequence CCAGC instead of the consensus CCACG motif found in ERSE-98, which is most proximal to the TATA element. Mutational analysis of ERSE-163 reveals that the unique CCAGC motif could be the binding site of a putative mammalian homologue of the yeast Hac1 protein (yHac1p) that binds and activates the yeast UPR and that YY1 serves as a coactivator of this factor in mammalian cells (3).
Using yeast one-hybrid screening, ATF6 was isolated as a putative ERSE binding protein (33). Overexpression of ATF6 in HeLa cells constitutively induced ERSE-mediated transcription. Subsequently, it was reported that in HeLa cells ATF6 is a transmembrane ER glycoprotein (6). Upon stress treatment, a fraction of the 90-kDa ATF6 underwent transient proteolysis, releasing an N-terminal 50-kDa protein with activating properties that could be detected as a soluble nuclear protein following biochemical fractionation (6). These important observations also raise several critical questions. For example, since in vitro-translated ATF6 failed to bind ERSE (33) and thus far direct DNA binding activity of ATF6 has not been demonstrated, how does it activate ERSE activity? Since transcription of grp genes is sustained as long as the stress inducer is present (13) whereas the appearance of p50 is transient (6, 33), are there other mechanisms to sustain ATF6 activity? Is ATF6 the predicted mammalian Hac1? The human and mouse homologues of yeast Irep, a transmembrane ER kinase with a dual endonuclease activity that splices the yeast HAC1 mRNA into a translatable form in response to ER stress, have been isolated (26, 28). Overexpression of mammalian Ire1p activates both the grp genes and CHOP-encoding genes. Further, overexpression of the murine Ire1p also leads to the development of programmed cell death in transfected cells (28). This raises the critical issue of whether ATF6 is a downstream target of the mammalian Ire1p, the putative master switch of the mammalian UPR.
In addressing these questions, we discovered that in addition to transient proteolysis, ATF6 itself undergoes specific changes in cells treated with thapsigargin (Tg), a strong inducer of the mammalian UPR through perturbance of calcium homeostasis. The stimulation of ERSE-mediated transcription activity by ATF6 requires the integrity of the tripartite structure of the ERSE, a high-affinity CCAAT binding site for NF-Y/CBF, and a functional NF-Y complex. ATF6 is an interactive partner of YY1. In Tg-treated cells, the stimulatory activity of ATF6 can be further enhanced by YY1 with a functional DNA binding domain. We showed that ATF6 exhibits properties distinct from those of human Ire1p (hIre1p) and yHac1p in activating the grp78 promoter, suggesting that they may represent diverse regulatory pathways.
MATERIALS AND METHODS
Cell culture conditions.
NIH 3T3 and Cos cells were maintained in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 2 mM glutamine, and 1% penicillin-streptomycin-neomycin at 35°C. For stress induction, cells were grown to 80% confluence and treated with 300 nM Tg for various time intervals as indicated. For treatment with genistein, 24 h following transfection, the cells were treated with 140 μM genistein for different time intervals as indicated. When the cells were treated with both genistein and Tg, genistein was added 15 min prior to Tg treatment.
Western blotting.
Whole cell lysates were prepared in radioimmunoprecipitation (RIPA) buffer as previously described (4). For ATF6 Western blotting, 50-μg aliquots of extracted proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 6 to 8% denaturing gels and transferred electrophoretically to Hybond ECL nitrocellulose membrane (Amersham, Piscataway, N.J.). For detection of the ATF6 doublet protein bands, 6% gels were run. The blotted membrane was then blocked with 5% nonfat dry milk in 1× Tris-buffered saline (20 mM Tris-HCl [pH 7.5], 500 nM NaCl) for 1 h. The primary antibody used was rabbit polyclonal ATF6 antibody directed against the c1.12 domain (36) (kindly provided by Ron Prywes, Department of Biological Science, Columbia University) at a dilution of 1:800. The secondary antibody used was horseradish peroxidase-conjugated goat anti-rabbit (Sigma), at a dilution of 1:5,000. For hemagglutinin epitope (HA)-tagged ATF6 Western blotting, 8 μg of HA-tagged pCGN-ATF6 plasmid was transfected into 2 × 106 Cos cells using SuperFect reagent (Qiagen Inc., Valencia, Calif.). Forty hours after transfection with or without specific drug treatment, the cells were lysed in RIPA buffer. Twenty micrograms of protein extract was separated by SDS-PAGE on 6% denaturing gels. The primary antibody used was mouse monoclonal HA antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) at a dilution of 1:500; the second antibody used was horseradish peroxidase-conjugated goat anti-mouse (Roche, Indianapolis, Ind.) at a dilution of 1:5,000. For the detection of GRP94, 30 μg of HeLa nuclear extract or whole cell extract was separated by SDS-PAGE on an 8% gel. The first antibody used was anti-KDEL rabbit polyclonal antibody at a 1:1,000 dilution (StressGen, Victoria, British Columbia, Canada); an anti-rabbit-horseradish peroxidase conjugate was used as secondary antibody (Santa Cruz) at a 1:5,000 dilution. For the detection of YY1, the membrane was incubated in a 1:1,000 dilution of anti-mouse monoclonal YY1 antibody (Santa Cruz) and at a 1:5,000 dilution of goat anti-mouse horseradish peroxidase-conjugated antibody (Santa Cruz). The protein bands were visualized by enhanced chemiluminescence (Amersham). The intensity of the protein bands was quantitated by scanning the autoradiographs with a Bio-Rad GS-700 densitometer.
Plasmids.
The construction of grp promoter-chloramphenicol acetyltransferase (CAT) fusion genes −457, −154, −130, −104, and −85 have been described elsewhere (29). The construction of (−169/−135)MCAT, (−159/−110)MCAT, and (−109/−74)MCAT has been described elsewhere (13). Mutants of (−109/−74)MCAT [CCAAT(m), CCACG(m1), and GGC(m)] were previously described (20). The (−169/−135)MCAT mutant CGAAT(m) was constructed similarly. The ERSE-131 mutant CCAAG(m1) was constructed using a Quikchange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The mutated bases in all constructs were confirmed by DNA sequencing. The mammalian expression vector pCGN-ATF6 containing HA-tagged full-length ATF6 driven by the cytomegalovirus (CMV) promoter was provided by Ron Prywes (Department of Biological Science, Columbia University); its construction has been described elsewhere (36). Expression vectors CMV-YY1 and the DNA binding site deletion mutant CMV-YY1Δ were gifts from Y. Shi (Harvard Medical School); their construction has been described previously (24). The expression vector for hIre1p (26) was a gift from R. Kaufman (University of Michigan). The construction of pMCX-Hac1, the mammalian expression vector for the 230-amino-acid yHac1 protein, has been described elsewhere (3). The expression vector for NF-YA29 has been described elsewhere (18).
Transfection conditions.
NIH 3T3 cells were seeded in six-well plates and grown to 60 to 80% confluence. One microgram of the reporter plasmid was cotransfected with 1 μg of pCH110, an expression vector for β-galactosidase under the control of the simian virus 40 (SV40) promoter, along with 1 μg of pCGN-ATF6 or CMV empty vector, using SuperFect reagent (Qiagen). In some experiments, 1 μg of CMV-YY1 or CMV-YY1Δ was also added. Transfections with hIre1p and yHac1p expression vectors were performed identically to that of ATF6. For stress induction, 24 h after transfection the cells were treated with 300 nM Tg for 16 h prior to harvesting. Preparation of the cell lysates for CAT assays and the quantitation of the CAT assays have been described elsewhere (15). Cell extracts corresponding to equal β-galactosidase units were used. Each transfection was repeated three to five times.
Immunofluorescence staining.
NIH 3T3 cells were grown to 80% confluence in chamber slides (Nalge Nunc International, Naperville, Ill.), washed with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde in PBS for 10 min. For the detection of ATF6, the cells were stained with anti-ATF6 (c1.12) antibody (1:100 dilution) as primary antibody and fluorescein-labeled anti-rabbit immunoglobulin G (IgG; 1:100 dilution; Vector Laboratories, Inc., Burlingame, Calif.) as secondary antibody. For the detection of YY1, cells were stained with anti-YY1 monoclonal antibody (1:100 dilution; Santa Cruz) as primary antibody and Texas red anti-mouse IgG (1:100 dilution) as secondary antibody.
Biochemical fractionation.
NIH 3T3 cells grown in duplicate in 150-mm-diameter dishes to 70% confluence were either nontreated or treated with Tg for 6 h. The cells were trypsinized, washed with PBS twice, then resuspended in 600 μl of hypotonic buffer (10 mM HEPES [pH 7.9], 0.1 mM EDTA, 1 mM dithiothreitol [DTT], 10 mM KCl, protease inhibitor cocktail [Roche]), and incubated at 4°C for 10 min. The cells were lysed by the addition of 3 μl of NP-40 and vortexed for 30 s. After confirmation of cell lysis by Trypan blue uptake assay, the free nuclei were pelleted from the cytoplasmic fraction by centrifugation at 5,000 rpm and then washed twice in hypotonic buffer. For the preparation of soluble nuclear protein, the nuclei were incubated successively in 300 μl of hypotonic buffer containing 150, 250, or 500 mM NaCl and 10% glycerol at 4°C for 10 min. The supernatant from each wash was separated from the nuclei by centrifugation at 5,000 rpm. Aliquots of these supernatants were concentrated 2.5-fold by Microcon-10 centrifugal filter devices (Millipore Corporation, Bedford, Mass.). For preparation of the membrane-bound protein, the final nucleus pellet was incubated at room temperature with 300 μl of hypotonic buffer containing 10% glycerol and 1% SDS. Following low-speed shaking for 1 h, the sample was centrifuged at 15,000 rpm for 30 min and the supernatant was collected. For evaluation of protein concentration and integrity, equal volumes of the protein samples were incubated in Laemmli buffer at 95°C for 10 min, loaded on an SDS–8% polyacrylamide gel, and run at 50 V for 20 min and then 140 V for 45 min. The gel was incubated in Coomassie blue staining solution for 1 h at room temperature and then destained overnight. Preparation of the highly concentrated HeLa nuclear extract from control cells and cells treated with Tg for 6 h has been described elsewhere (22).
EMSA.
Conditions for the electrophoretic mobility shift assays (EMSAs) using HeLa nuclear extract and synthetic ERSE as probe have been described elsewhere (20, 21).
GST pull-down assays.
Glutathione S-transferase (GST) was used alone or as a GST-YY1 or GST-Ras fusion. In vitro-translated 35S-labeled ATF6 was prepared with the TNT coupled reticulocyte system from Promega (Madison, Wis.). Five microliters of the labeled ATF6 was mixed with bacterially expressed GST proteins, and the pull-down assays were performed as previously described (31).
Coimmunoprecipitation assays.
A total of 2 × 106 Cos cells were transfected with 8 μg of plasmid pCGN-ATF6 or pCGN using SuperFect reagent. The cells with or without Tg treatment were harvested 48 h after transfection and lysed in 300 μl of NP-40 buffer (0.5% NP-40, 50 mM HEPES [pH 7.5], 150 mM sodium chloride). Protein extract (500 μg) from each sample was pretreated with 50 μl of protein A-Sepharose beads (Sigma) for 1 h at 4°C prior to incubation with 3 μl of anti-YY1 monoclonal antibody (Santa Cruz), 2 μl of normal rabbit serum, or 2 μl of NF-Y/CBF rabbit polyclonal antibody (kindly provided by Sankar Maity, University of Texas) for 30 min. Following the incubation period, 50 μl of protein A-Sepharose beads was added, and the mixtures were rotated at 4°C overnight. The beads were then washed successively once with buffer I (PBS containing 0.5% NP-40 and 1 mM DTT), buffer II (100 mM Tris-HCl [pH 7.4], 500 mM LiCl, 1 mM DTT), and buffer III (10 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM DTT). The immunoprecipitate was released from the washed beads by the addition of 30 μl of 1 × SDS loading buffer (50 mM Tris-HCl [pH 6.8], 100 mM DTT, 2% SDS, 0.1% bromophenol blue, 10% glycerol), followed by heating at 100°C for 5 min. The supernatant obtained after centrifugation was resolved by SDS-PAGE on an 8% gel and subjected to Western blot analysis to detect the coimmunoprecipitated protein.
RESULTS
Tg stress-induced changes of ATF6.
The human cDNA sequence of ATF6 predicts a protein of 670 amino acids (Fig. 1A). As previously noted, ATF6 contains a serine-rich region at its N terminus and a central basic region spanning from amino acids 303 to 330 followed by a leucine zipper sequence (the b-ZIP region) (36). The transactivation domain of ATF6 is contained within the first 273 amino acids (25). Computer analysis of the primary sequence of ATF6 further predicts the location of several putative nuclear localization signals around amino acids 170, 330, and 380 and, strikingly, a short transmembrane region from amino acids 378 to 398 flanked by highly hydrophilic domains (7).
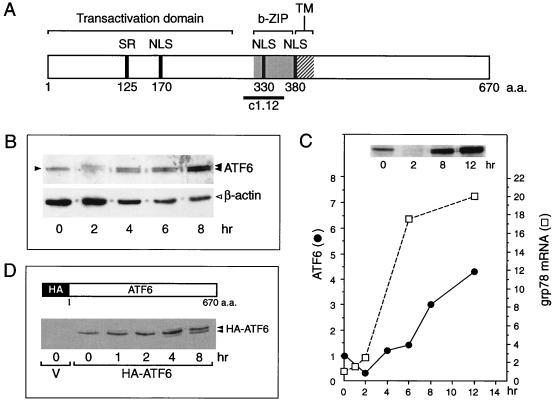
FIG. 1.
Tg stress-induced changes of ATF6. (A) Schematic drawing of the primary structure of ATF6, a 670-amino-acid (a.a.) protein. The positions of the transactivation domain, the b-ZIP domain, and a putative transmembrane (TM) domain are in brackets. The locations of the serine-rich (SR) region, putative nuclear localization signal (NLS), and the subfragment c1.12 used to generate antibody against the basic region of ATF6 are also indicated. (B) Total cell lysates were prepared from NIH 3T3 cells treated with Tg for the indicated time and analyzed by immunoblotting using anti-ATF6 antibody. Twenty micrograms of each protein sample was applied to each lane. The single 90-kDa ATF6 band (at time zero) is indicated by a single closed arrowhead and the ATF6 doublet band in the later time points is marked by the double arrowheads. The β-actin protein band (open arrowhead) in the same Western blot served as the protein loading control. (C) The level of ATF6 following Tg stress was quantitated by densitometry and plotted against the kinetics of accumulation of the grp78 mRNA. The measurement of grp78 mRNA levels in Tg-treated NIH 3T3 cells has been described elsewhere (13). A Western blot of ATF6 protein level after 0, 2, 8, and 12 h of Tg treatment is shown in the inset. Fifty micrograms of each protein sample was applied to each lane. (D) Schematic drawing of HA-tagged full-length ATF6 (36). Shown below is the Western blot with anti-HA antibody performed on total cell lysate prepared from Cos cells transfected with empty vector (V) or with HA-ATF6 expression vector and treated with Tg for the indicated time. The position of the HA-ATF6 doublet is indicated.
To detect ATF6 expression, Western blotting of total NIH 3T3 cell lysate was performed, using an antibody with established specificity against ATF6 (36). In nonstressed NIH 3T3 cells, ATF6 was constitutively expressed and migrated as a single 90-kDa protein band in SDS-PAGE (Fig. 1B). Upon Tg treatment, a reduction in endogenous ATF6 was observed at 2 h, suggesting proteolysis as previously reported (6, 33). By 4 h, the basal level of ATF6 was restored; at the same time, a new and slightly faster-migrating band was detected (Fig. 1B). This doublet band pattern persisted while the level of ATF6 started to increase. By 12 h of Tg treatment, there was a four- to fivefold increase in the steady-state level of ATF6 compared to nonstressed cells (Fig. 1C). We noted that the doublet ATF6 band pattern was best resolved with low protein amount loaded on a 6% gel; when the protein amount was high, as shown in the inset of Fig. 1C, the doublet pattern merged into one band. The kinetics of accumulation of p90 ATF6 as related to the increase in grp78 transcript level is summarized in Fig. 1C.
To confirm that the new band was derived from ATF6, Cos cells were transfected with an expression vector of HA-tagged p90 ATF6. Total cell lysate was prepared from cells treated with Tg for various times and subjected to Western blot analysis with the anti-HA antibody (Fig. 1D). The anti-HA antibody was highly specific for the exogenous ATF6 since the HA-ATF6 protein band was detected in cell lysate transfected with the ATF6 expression vector but not with the empty vector. As in the case of the endogenous ATF6 protein, a doublet was observed starting at 4 h after Tg treatment. In contrast to the endogenous protein, no decrease in the HA-ATF6 protein level was detected following Tg treatment (Fig. 1D). For exogenously expressed ATF6, we could not detect any 50-kDa protein of HA-ATF6 in either control or Tg-treated cells even when large amounts of protein lysates were analyzed (data not shown). We also had difficulty detecting the endogenous p50 ATF6 band in a consistent manner using the anti-ATF6 antibody.
Enhanced nuclear localization of p90 ATF6 after Tg stress.
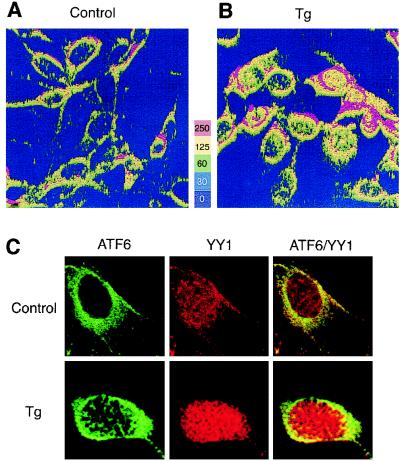
To investigate the subcellular localization of endogenous ATF6 in control and Tg-treated cells, immunofluorescence studies were performed using anti-ATF6 antibody. As shown by the relative fluorescence intensity compiled by three-dimensional rendering of the z-sectioning data, ATF6 was detected predominantly in the perinuclear region in nonstressed control cells (Fig. 2A), in agreement with a putative transmembrane domain in ATF6 (Fig. 1A). YY1, a soluble nuclear transcription factor, was primarily localized in the nucleus, while minor overlap of YY1 and ATF6 immunofluorescence in the cytoplasm was also observed in control cells (Fig. 2C). Upon Tg treatment for 8 h correlating with the rapid increase in grp78 transcript level (Fig. 1C), both ATF6 and YY1 showed increased nuclear localization while ATF6 retained perinuclear staining (Fig. 2B and C). Further, through confocal microscopy, the appearance of ATF6 in close proximity with YY1 inside the nucleus was detected in Tg-treated but not control cells (Fig. 2C).
FIG. 2.
Tg-induced changes in subcellular localization of ATF6. NIH 3T3 cells were grown to 80% confluence in chamber slides and not treated (A) or treated with 300 nM Tg for 8 h (B). The cells were stained with anti-ATF6 antibody and viewed with a 40× oil immersion lens, yielding a magnification of ×400 using a Zeiss dual-photon confocal microscope with LSM 510 imaging software. Z sectioning was evaluated at 0.1-μm intervals yielding 23 sections. The z-sectioning data were used for three-dimensional rendering of relative fluorescence intensity represented in panels A and B as a five-color, 250-unit scale. (C) Control or Tg-treated NIH 3T3 cells were reacted with antibodies against ATF6 and YY1 and subjected to confocal microscopy. ATF6 is shown as green and YY1 is shown as red fluorescence, while yellow suggests colocalization.
The subcellular localization of p90 ATF6 was further investigated through biochemical fractionation of NIH 3T3 cell extracts. Endogenous ATF6 was detected by immunoblotting as shown in Fig. 3A. Cell lysates were prepared from control cells and cell treated with Tg for 6 h when the doublet form of p90 ATF6 was prominent (Fig. 3B, lanes 1 and 2). Free nuclei were isolated from the cytoplasm and subjected to successive salt washes for the elution of soluble nuclear protein, and the membrane-bound protein was released from the nuclei pellet following the final wash by resuspension in a buffer containing 1% SDS. The protein samples were subjected to SDS-PAGE (6% gel) and Western blotted with anti-ATF6 antibody. In agreement with the immunofluorescence staining (Fig. 2), p90 ATF6 from control cells was predominantly associated with the membrane fraction in control cells (Fig. 3B, lane 11). For Tg-treated cells, p90 ATF6 was detected also in the 0.25 and 0.5 M NaCl wash fractions (Fig. 3B, lanes 8 and 10). Further, it was the faster-migrating form of ATF6 induced by Tg treatment that was preferentially recovered as the soluble nuclear protein. The enrichment of the soluble nuclear protein in the salt washes was confirmed by rehybridization of the same Western blot with an antibody directed against TFII-I/BAP-135, a soluble nuclear protein (32) (data not shown).
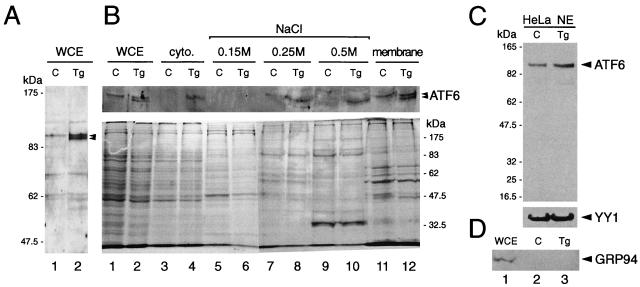
FIG. 3.
Biochemical fractionation of ATF6 in control and Tg-treated cells. (A) Representative immunoblot of whole cell extract (WCE) prepared from control NIH 3T3 cells (C) and cells treated with Tg for 16 h (Tg) with the anti-ATF6 antibody. Positions of the molecular size marker are indicated on the left, and the position of the ATF6 doublet is indicated by arrowheads. (B) Control cells and cells treated with Tg for 6 h were fractionated into cytoplasmic and nuclear fractions after successive washes with hypotonic buffer containing 0.15, 0.25, and 0.5 M NaCl. The membrane-bound protein was released from the final nucleus pellet with buffer containing 1% SDS. WCE was prepared by lysing an aliquot of the cells directly in RIPA buffer. Equal volumes of the samples from control and Tg-treated cells were applied onto SDS–6% polyacrylamide gels. For the NaCl wash fractions, the supernatants were concentrated 2.5-fold prior to loading onto the gel. The upper panel shows an immunoblot with anti-ATF6 antibody. The lower panel shows Coomassie blue staining pattern of 5 μl each of WCE and 30 μl each of the other samples as indicated above. (C) Western blot of ATF6 using 30 μg of HeLa nuclear extract (NE) from control and Tg-treated cells applied onto an SDS–8% polyacrylamide gel. The same blot was reacted with anti-YY1 antibody. In panels B and C, positions of the molecular size marker are indicated. (D) Thirty-microgram aliquots of HeLa WCE or NE from control and Tg-treated cells were subjected to Western blotting for detection of GRP94.
Localization of p90 ATF6 inside the nucleus is not confined to NIH 3T3 cells. In nonstressed HeLa cells, nuclear staining of ATF6 in addition to perinuclear staining was observed by immunofluorescence (6). In highly concentrated HeLa nuclear extract containing both soluble and membrane-bound nuclear protein prepared by the method of Shapiro et al. (22), p90 ATF6 could be detected along with YY1 from both control cells and cells treated with Tg for 6 h (Fig. 3C). Since the gel electrophoresis was performed for a shorter time, the doublet form of ATF6 was not resolved under these conditions. These nuclear extracts were free of contamination from ER proteins such as GRP94 (19), which was readily detectable from the HeLa whole cell extract (Fig. 3D, lane 1) but was absent from the nuclear protein preparations (lanes 2 and 3).
Selective activation of ERSEs by ATF6.
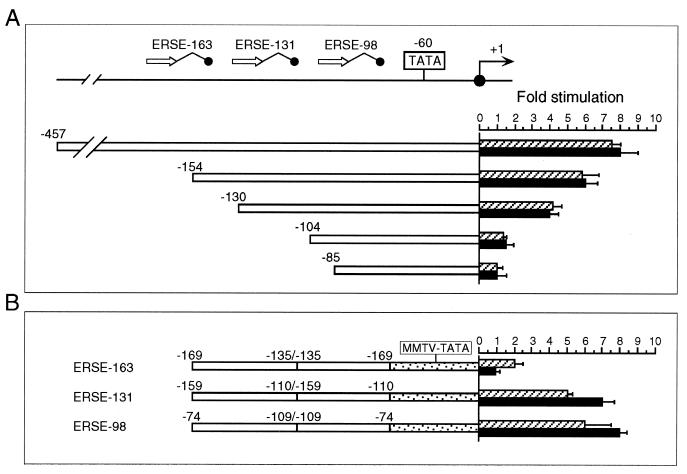
To map the ATF6 target sites within the grp78 promoter, ATF6 expression vector was cotransfected with a series of 5′ deletion mutants of the rat grp78 promoter linked to the CAT reporter gene. In every construct examined, the ability of ATF6 to stimulate the grp78 promoter directly correlated with that of Tg stress. Thus, we observed eight- and sixfold activation by ATF6 for the −457 and −154 constructs and fourfold stimulation for the −130 construct but no stimulation with the −104 or −85 construct (Fig. 4A). The failure to stimulate the −104 construct showed that ATF6 activation of the rat grp78 promoter requires at least two copies of ERSE, and the failure to stimulate the −85 construct ruled out the possibility that ATF6 acts through the TATA element of the grp78 promoter. When the ERSEs were examined individually, ERSE-98 [contained in duplicate within (−109/−74) MCAT)] and ERSE-131 [contained in duplicate within (−159/−110) MCAT] were efficiently about five- to eightfold) activated by ATF6 and Tg (Fig. 4B). In contrast, ATF6 was not able to activate ERSE-163 [contained in duplicate within (−169/−135)MCAT], which responded to Tg stress with a twofold induction. These results show that ATF6 induction of the individual ERSE is selective, with ERSE-98 as the most efficient target.
FIG. 4.
Comparison of the sequence requirement for ATF6 and Tg stimulation of the rat grp78 promoter. (A) Summary of the effect of 5′ deletion (−457, −154, −130, −104, and −85) of the rat grp78 promoter on ATF6 stimulation. The locations of the three ERSEs in the rat grp78 promoter with respect to the TATA sequence motifs are indicated, and (+1) represents the site of transcription initiation. The consensus ERSE unit is comprised of a CCAAT( ) and a CCACG (●) sequence separated by a 9-bp (∧) GC-rich motif. The grp-CAT constructs were transiently transfected into NIH 3T3 cells with either ATF6 expression vector or empty CMV vector. The fold stimulation by ATF6 overexpression (black bar) was compared to Tg treatment (striped bar), with standard deviations as indicated. (B) Selective stimulation of the ERSEs by ATF6. Schematic drawings of the CAT genes driven by grp78 promoter subfragments containing the respective ERSE linked in duplicate copies to the minimal mouse mammary tumor virus promoter are shown. The fold stimulation by ATF6 overexpression (black bar) is compared to Tg treatment (striped bar), with standard deviations as indicated.
) and a CCACG (●) sequence separated by a 9-bp (∧) GC-rich motif. The grp-CAT constructs were transiently transfected into NIH 3T3 cells with either ATF6 expression vector or empty CMV vector. The fold stimulation by ATF6 overexpression (black bar) was compared to Tg treatment (striped bar), with standard deviations as indicated. (B) Selective stimulation of the ERSEs by ATF6. Schematic drawings of the CAT genes driven by grp78 promoter subfragments containing the respective ERSE linked in duplicate copies to the minimal mouse mammary tumor virus promoter are shown. The fold stimulation by ATF6 overexpression (black bar) is compared to Tg treatment (striped bar), with standard deviations as indicated.
ATF6 activation of the ERSE requires a strong NF-Y binding site and a functional NF-Y complex.
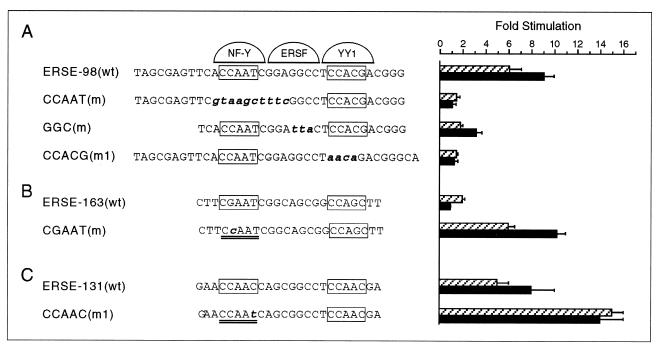
To map the sequence within wild-type (wt) ERSE-98 required for ATF6 stimulation, ERSE-98 with a mutated binding site for either NF-Y/CBF, ERSF, or YY1 was tested for ATF6 inducibility (Fig. 5A). As confirmed in EMSAs using HeLa nuclear extract, ERSE-98 formed three protein complexes with the above three factors (20) (Fig. 6A). The ability to compete for YY1 and NF-Y/CBF binding was lost upon mutation of the CCACG and CCAAT sequences, respectively (Fig. 6B, lanes 3 and 4). These same mutations also eliminated the stimulation of ERSE-98 by ATF6, whereas mutation in the GGC motif which abolished ERSF binding (20) reduced the stimulation by ATF6 to about half (Fig. 5A). Thus, optimal stimulation by ATF6 requires the sequence integrity of the tripartite structure of ERSE.
FIG. 5.
Target sites for ATF6 and Tg stimulation within ERSE. (A) Summary of effect of mutation of the NF-Y binding site [CCAAT(m)], ERSF site [GGC(m)], and YY1 site [CCACG(m1)] within ERSE-98 on ATF6 and Tg stimulation. The locations of these sites within ERSE-98 are shown, with the mutated bases highlighted in bold lowercase. Transient transfections into NIH 3T3 cells were performed. The fold stimulation by ATF6 overexpression (black bar) is compared to Tg treatment (striped bar), with standard deviations as indicated. (B) Effect of mutation of the NF-Y binding site on ERSE-163 on ATF6 and Tg stimulation. The single base change within (−169/−135)MCAT is as indicated. (C) Effect of mutation of the NF-Y binding site on ERSE-131 on ATF6 and Tg stimulation. The single base change within (−159/−110)MCAT is indicated.
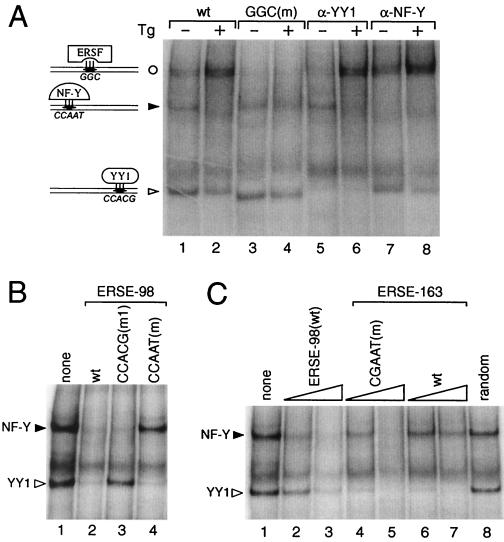
FIG. 6.
Effect of sequence mutations on factor binding affinity. (A) In the EMSA reactions, radiolabeled ERSE-98 (wt) was mixed with HeLa nuclear extract prepared from control (−) or Tg-treated cells (+) (lanes 1 and 2); the GGC(m) oligomer was used as radiolabeled probe (lanes 3 and 4); anti-YY1 antibody was added to the EMSA reaction with the wt probe (lanes 5 and 6), and anti-NF-Y antibody was added to the EMSA reaction with the wt probe (lanes 7 and 8). (B) The NF-Y and YY1 complexes formed were subjected to competition by unlabeled oligomers. No competitor (lane 1) or a 50-fold molar excess of wt ERSE-98 (lane 2) or its mutated forms (lanes 3 and 4) as indicated at the top was added. (C) The NF-Y and YY1 complexes were subjected to competition in EMSA. Lanes contained no competitor (lane 1), 10- and 50-fold molar excess of wt ERSE-98 (lanes 2 and 3), CGAAT mutant (lanes 4 and 5), or wt ERSE-163 (lanes 6 and 7), and 50-fold molar excess of a random synthetic oligomer of equal length (lane 8). The sequences of the wt and mutated forms of ERSE-98 and ERSE-163 are shown in Fig. 5. Positions of the ERSF, NF-Y, and YY1 complexes are indicated.
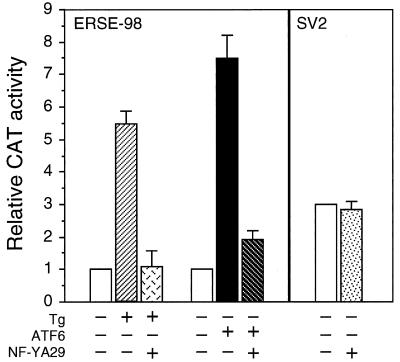
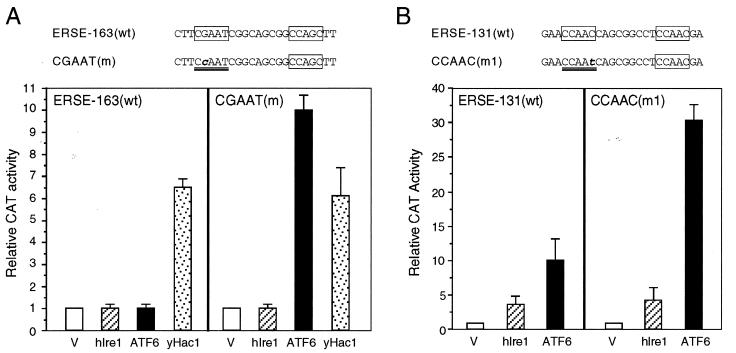
The importance of a strong NF-Y binding site for ATF6 stimulation was further demonstrated by mutation analysis of ERSE-163, which cannot be stimulated by ATF6. ERSE-163 differs from ERSE-98 in that it contains a weak NF-Y binding site but a stronger YY1 binding site (15). Thus, ERSE-163 contains a CGAAT sequence for NF-Y binding instead of the consensus CCAAT contained within ERSE-98 (Fig. 5B). This resulted in lower binding affinity for NF-Y, as confirmed by the weaker ability of ERSE-163 than of ERSE-98 to compete for NF-Y binding in EMSA (Fig. 6C, compare lanes 2 and 3 to lanes 6 and 7). Mutation of this weak CGAAT site to the consensus CCAAT site resulted in equivalent binding affinity for NF-Y as ERSE-98 (Fig. 6C, compare lanes 2 and 3 to lanes 4 and 5). Correspondingly, the inability of ATF6 to activate ERSE-163 was completely reversed when the CGAAT motif of ERSE-163 was mutated to conform to the consensus CCAAT motifs among the ERSEs (Fig. 5B). Similarly, conversion of the first CCAAC motif of ERSE-131 to CCAAT further increased its ability to respond to ATF6 and Tg induction (Fig. 5C). These collective results establish high-affinity NF-Y/CBF binding as critical for ATF6 stimulation of the ERSE. Further, the requirement of a functional complex of NF-Y/CBF for ATF6 stimulation was tested through the use of NF-YA29, a dominant negative mutant of NF-Y (18). NF-Y/CBF is a heteromeric transcription factor consisting of at least three subunits (17). NF-YA29, in which three amino acids in the DNA binding domain of NF-YA have been mutated, forms a complex with NF-YB, rendering it functionally inactive as a transcription activator. Expression of NF-YA29 suppressed ATF6 as well as Tg stimulation of ERSE-98 (Fig. 7). The same amount of NF-YA29 was without effect on the SV40 promoter-driven CAT activity (Fig. 7).
FIG. 7.
Effect of coexpression of NF-YA29 on promoter activities. For ERSE-98-mediated promoter activity, NIH 3T3 cells were transfected with (−109/74)MCAT as the reporter gene. The cells were either nontreated, treated with Tg, or cotransfected with empty CMV vector, pCGN-ATF6, or NF-YA29 as indicated into NIH 3T3 cells. The CAT activity in nonstressed cells transfected with the empty CMV vector was set at 1. pSV2CAT, used as the reporter gene for SV40-mediated promoter activity, was cotransfected with either empty vector or NF-YA29. The relative promoter activities are shown.
YY1 is an interactive partner and a coactivator of ATF6 in Tg-stressed cells.
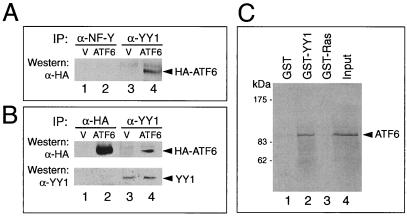
The lack of demonstrable binding of ATF6 to the ERSE and the functional dependence of ATF6 on an intact NF-Y and YY1 binding site on ERSE-98 prompted us to determine whether ATF6 activation is mediated by protein-protein interaction with either NF-Y, YY1, or both. To test for their in vivo association, Cos cells were transfected with either the empty vector or the expression vector for the HA-tagged ATF6. Total cell lysates prepared from nontreated or Tg-treated cells were subjected to immunoprecipitation with either anti-NF-Y, anti-YY1, or anti-HA antibody, with normal rabbit serum or preimmune serum as a negative control. The immunoprecipitates were then Western blotted with the antibodies to detect coimmunoprecipitation of the proteins. Examples for these reactions are shown in Fig. 8. While we could not detect interaction between HA-ATF6 with NF-Y, YY1 was able to interact with HA-ATF6 in both nonstressed (Fig. 8A) and Tg-stressed (Fig. 8B) extracts. To test for direct interaction between ATF6 and YY1, in vitro-translated p90 ATF6 was mixed with GST-YY1, GST-Ras, or the GST vector alone. Our results showed that ATF6 could associate with recombinant YY1 in vitro but showed no affinity for either GST alone or GST-Ras (Fig. 8C).
FIG. 8.
Interaction of ATF6 with YY1 in coimmunoprecipitation assays. (A) Cell lysates in NP-40 buffer prepared from Cos cells transfected with either the empty vector (V) or pCGN-ATF6 (ATF6) were immunoprecipitated (IP) with 2 μl of anti-NF-Y (lanes 1 and 2) or anti-YY1 (lanes 3 and 4) antibody. The immunoprecipitates were applied to SDS–8% denaturing polyacrylamide gels and Western blotted with anti-HA antibody. The position of HA-ATF6 is indicated. (B) Cos cells transfected either with the empty vector (V) or HA-ATF6 (ATF6) were subjected to Tg treatment. The lysate was immunoprecipitated with anti-HA (lanes 1 and 2) or anti-YY1 (lanes 3 and 4) antibody and Western blotted with anti-HA antibody. The position of HA-ATF6 is indicated. The same blot (lanes 1 through 4) was washed and Western blotted with anti-YY1 antibody. The position of YY1 is indicated. (C) GST pull-down assays. The reactions were performed using in vitro-translated ATF6 and GST (lane 1), GST-YY1 (lane 2), and GST-Ras (lane 3). Lane 4 represents 20% of input radiolabeled ATF6. The protein bound onto the beads was eluted, applied to an SDS–8% polyacrylamide gel, and detected by autoradiography. Positions of the protein size markers are indicated.
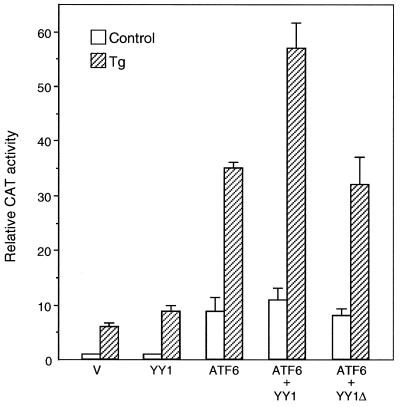
The functional relationship between ATF6 and YY1 was examined by cotransfection of expression vectors for these two proteins into NIH 3T3 cells (Fig. 9). We observed that while ATF6 itself was a strong inducer of ERSE-98 in nonstressed cells, coexpression of YY1 minimally increased the ATF6 activity, and YY1 deleted of its zinc finger DNA binding domain (YY1Δ) was without effect (Fig. 9). In contrast, under the Tg-stressed conditions, YY1 enhanced ATF6 stimulation of ERSE-98. Although the magnitude of stimulation was only in the range of 1.5- to 2-fold, probably due to the high abundance of endogenous YY1, YY1Δ was not able to stimulate ATF6 activity. We noted in these experiments that addition of Tg to the ATF6-overexpressing cells further increased the promoter activity from 10- to 35-fold (Fig. 9). This additional increase in promoter activity following Tg treatment was consistently observed even in transfectants with maximum dosage for ATF6 overexpression (data not shown), suggesting that additional mechanisms were activated to maximize grp78 promoter activity under Tg stress conditions.
FIG. 9.
Effect of overexpression of ATF6, YY1, and YY1Δ on ERSE-98-mediated CAT activity. The construct (−109/−74)MCAT, used as the reporter gene, was cotransfected with either the empty CMV vector (V) or expression vector for ATF6, YY1, or YY1Δ, alone and in combinations as indicated, into NIH 3T3 cells. The transfected cells were either grown under normal culture conditions (control) or treated with Tg. The CAT activity in nonstressed cells transfected with the empty CMV vector was set at 1. Relative promoter activities are shown with standard deviations.
Distinct activating properties of hIre1p and ATF6.
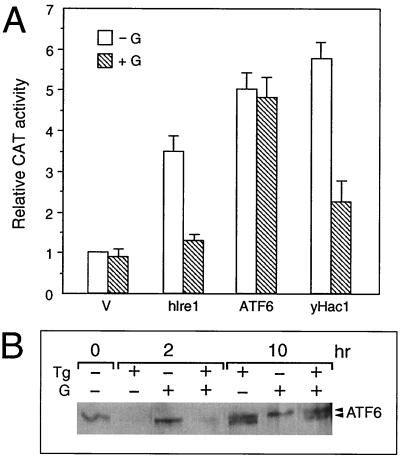
With the identification of Ire1p as an upstream activator of the mammalian UPR (9, 26, 28), we tested whether ATF6 is a downstream target of hIre1p. Also, since yHac1p is an activator of mammalian grp78 and grp94 promoter activities (3), we asked whether ATF6 is the mammalian counterpart of yHac1 by comparing the activating properties of ATF6 and yHac1. The stress induction of the grp78 promoter has been shown to be sensitive to the tyrosine kinase inhibitor genistein, which has no effect on the basal grp78 promoter activity (1, 35). All three proteins, hIre1p, ATF6, and yHac1p, were able to stimulate the grp78 promoter three- to sixfold in the absence of ER stress (Fig. 10A). If the three proteins are functionally equivalent, their levels of sensitivity to genistein may be similar. Our results showed that the induction of the grp78 promoter activity by both hIre1p and yHac1p was inhibited by genistein. In contrast, ATF6-mediated stimulation of the grp78 promoter activity was resistant to genistein treatment. Further, genistein exhibited no effect on the Tg stress-induced changes of ATF6 (Fig. 10B). These results indicate either that ATF6 is downstream of the genistein inhibitory step which suppresses hIre1p action or that ATF6 and hIre1p could act through diverse mechanisms.
FIG. 10.
Effect of genistein on ATF6 activity and protein level. (A) Effect of genistein on hIre1p, ATF6, and yHac1p stimulation of the grp78 promoter. NIH 3T3 cells were transiently transfected with −154CAT and either the empty CMV vector (V) or expression vector for hIre1p, ATF6, or yHac1p as indicated. The cells were either nontreated (−G) or treated with 140 μM genistein (+G). CAT activity in the nontreated cells transfected with the empty CMV vector was set at 1. Relative promoter activities are shown with standard deviations. (B) Effect of genistein on ATF6 protein level. Total cell lysates were prepared from NIH 3T3 cells with no drug treatment or Tg treated for 2 or 10 h, in the presence or absence of genistein (G), as indicated at the top; 30 μg of cell lysate from each sample was applied to an SDS–6% polyacrylamide gel and Western blotted with anti-ATF6 antibody.
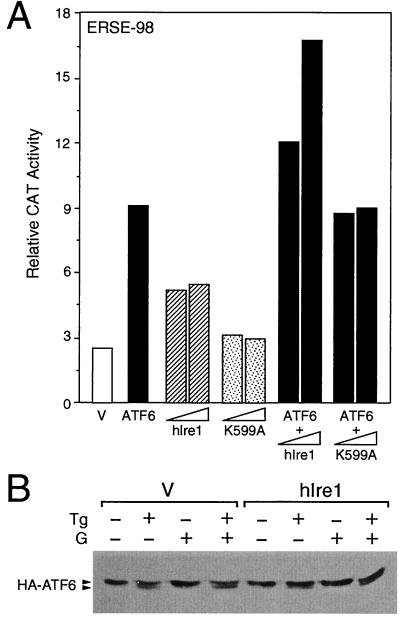
Next we examined the effect of coexpression of hIre1p and ATF6. When NIH 3T3 cells were transfected with ATF6 or hIre1p individually, we detected a 9- or 5-fold, respectively, induction of ERSE-98 activity (Fig. 11A). Coexpression of hIre1p with ATF6 resulted in a dosage-dependent increase in ERSE-98 activity up to 17-fold. A kinase-defective form of hIre1p, referred to as hIre1p(K599A), was constructed by substituting the conserved lysine at residue 599 in the putative ATP binding domain with alanine (26). When hIre1p(K599A) was coexpressed with ATF6, no additive effect was observed (Fig. 11A). The stimulatory effect of hIre1p on ATF6 was clearly not due to increased proteolytic cleavage of p90 ATF6. In cells cotransfected with hIre1p, the levels of ATF6 in control, Tg-treated, genistein-treated, or Tg- and genistein-cotreated cells were similar or slightly higher than those transfected with the empty vector (Fig. 11B).
FIG. 11.
Effect of hIre1p coexpression on ATF6. (A) NIH 3T3 cells were transiently transfected with (−109/−74)MCAT and either the empty CMV vector (V), ATF6 expression vector, or hIre1p and its dominant negative mutant K599A, alone and in combination, as indicated. The relative promoter activities are shown with standard deviations. (B) Plasmid pCGN-ATF6 was cotransfected into Cos cells with the empty CMV vector (V) (lanes 1 to 4) or expression vector for hIre1p (lanes 5 to 8). The transfected cells were either nontreated (lanes 1 and 5) or treated with Tg (lanes 2 and 6), genistein (lanes 3 and 7), or genistein and Tg (lanes 4 and 8). Equal amounts of cell lysate from each sample were Western blotted with anti-HA antibody. The position of HA-ATF6 is indicated.
Since NF-Y is a critical component for ATF6 stimulation of ERSE-directed transcription, we next examined the effect of a high-affinity NF-Y binding site on the stimulatory activities of hIre1p, ATF6, and yHac1. Using wt ERSE-163/CAT and mutated ERSE-163/CAT as the test genes, we observed that only yHac1p was able to stimulate the wt ERSE-163/CAT (Fig. 12A). The induction level was about sixfold. In contrast, both hIre1p and ATF6 were without effect. By converting the weak NF-Y binding site CGAAT within ERSE-163 to a CCAAT consensus motif, ATF6 was able to stimulate this promoter about 10-fold. The same mutation did not rescue hIre1p activity, nor did it further enhance the yHac1p activity. Similarly, ATF6 stimulation of ERSE-131/CAT was increased from 10- to 30-fold when the CCAAC motif was mutated to the consensus CCAAT motif; however, this same mutation had minimal effect on hIre1p activity (Fig. 12B). Collectively, these experiments established that ATF6 stimulation of the grp78 promoter is enhanced by a high-affinity NF-Y binding site, whereas the activity of the downstream target of hIre1p that mediates the activation of the grp78 promoter is not dependent on a strong NF-Y binding site. We further observed that in contrast to hIre1p and ATF6, yHac1p can neither bind nor stimulate ERSE-98 activity (3).
FIG. 12.
Distinct activating properties of ATF6, hIre1p, and yHac1. (A) Effect of mutation of ERSE-163 on hIre1p, ATF6, and yHac1p stimulation. NIH 3T3 cells were transiently transfected with the wt construct (−169/−135)MCAT or the construct bearing the CGAAT(m) mutation and either the empty CMV vector (V) or expression vector for hIre1p, ATF6, or yHac1p as indicated. CAT activity in cells transfected with the empty CMV vector was set at 1. Relative promoter activities are shown with standard deviations. (B) Effect of mutation of ERSE-131 on hIre1p and ATF6 stimulation. NIH 3T3 cells were transiently transfected with the wt construct (−159/−110)MCAT or the construct bearing the CCAAC(m1) mutation and either the empty CMV vector (V) or expression vector for hIre1p or ATF6 as indicated. Relative promoter activities are shown with standard deviations.
DISCUSSION
The recent identification of ATF6 as a transcription factor that can constitutively induce the grp promoter in an ERSE-dependent manner (33) raises the question of how this stimulation can be achieved. One proposed mechanism is that ER stress induced proteolysis of the membrane-bound p90 ATF6, releasing a soluble p50ATF6 and leading to induced transcription in the nucleus (6). This is similar to the proteolytic maturation cascade of steroid regulatory element binding protein (SREBP) to regulate cholesterol and fatty acid biosynthesis, a process which also takes place in the ER (27, 30). However, whereas the proteolytic cleavage product of SREBP is abundant and stable, p50 ATF6 was observed only transiently and in very low amounts (33). Further, the ability of ATF6 to bind DNA has not yet been established.
We report here that ATF6 undergoes multiple forms of change upon treatment of cells with Tg which perturbs calcium homeostasis and propose a mechanism for its activation of the grp78 promoter through physical and functional interaction with previously established DNA binding components of the ERSE (Fig. 13). ATF6 is constitutively expressed as a 90-kDa protein in nonstressed cells. Within 2 h of Tg treatment, there was an initial drop of endogenous ATF6 protein level, in agreement with proteolytic cleavage as reported elsewhere (33). For exogenously expressed p90 ATF6 that is highly potent as a transactivator for ERSE, this drop in protein level was not observed. Rather, for both endogenous and exogenously expressed ATF6, at 4 h following Tg stress, and an additional, faster-migrating form was detected. With prolonged Tg treatment, as the grp78 transcript accumulated to high levels, the total amount of ATF6 also increased. The new form of ATF6 can be recovered as a soluble nuclear protein, in support of enhanced nuclear localization of ATF6 as detected by immunofluorescence. How might the new form of ATF6 be generated? Since this new form is also readily detected with exogenous expression of p90 ATF6 tagged at its N terminus with the HA epitope, it must have been generated from p90 ATF6 through either alternative splicing or posttranslational modification. The former mechanism is particularly attractive since splicing of the short transmembrane domain from ATF6 will be able to convert it from a predominantly membrane-associated form to a soluble form. For posttranslational modification, it is possible Tg treatment triggers phosphorylation of ATF6, which is a known target for the p38 MAPK (25), or other kinases are involved. The faster-migrating form could also arise from alteration of glycosylation, acetylation, or methylation status of ATF6 triggered by Tg stress. However, the ATF6 doublet appeared to migrate slower than the nonglycosylated form of ATF6 generated by tunicamycin treatment of the cells (data not shown). An additional mechanism is through proteolytic cleavage. Since the HA tag at the N terminus was retained in the new form, proteolytic trimming involving the immediate N terminus of the protein is unlikely. Future investigations are needed to resolve this.
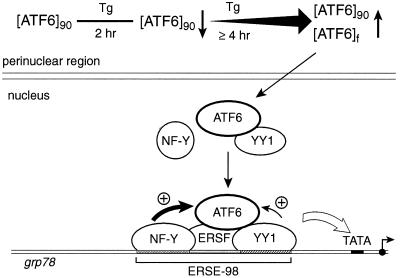
FIG. 13.
Model for ATF6 induction of grp78 following Tg stress. The 90-kDa ATF6 is primarily associated with the perinuclear region in nonstressed cells. Upon Tg treatment, the ATF6 level initially drops but quickly recovers with the additional appearance of a faster-migrating form ([ATF6]f) and an increase in the amount of ATF6. A fraction of ATF6 enters the nucleus. Through interaction with YY1, ATF6 becomes part of a multiprotein complex binding onto the ERSE of the grp78 promoter. YY1 also enhances ATF6 activity. Other factors that bind to ERSE include an ERSF and the CCAAT binding protein NF-Y. The stimulatory activity of ATF6 on the grp78 promoter depends on the integrity of the ERSE structure. In addition, a high-affinity NF-Y binding site and a functional NF-Y complex are required for optimal stimulation by ATF6. Other Tg-induced modifications of the transcription factors may also occur. This multiprotein complex, acting in concert with the basal transcription machinery, stimulates grp78 transcription.
Considering that ATF6 either has no or extremely weak DNA binding affinity, it may require interaction with other proteins to activate the ERSE. Likely candidates for the ATF6 interactive partners in achieving ERSE activation include NF-Y, YY1, and ERSF, proteins known to bind to ERSE (20). Mutation analysis to define the sites on ERSE-98 required for ATF6 stimulation revealed that binding sites for all three proteins are important. Of primary importance is NF-Y binding to the CCAAT sequence motif within all ERSEs. NF-Y is a multimeric CCAAT binding protein and requires at least three subunits to bind to its DNA site (17). Here we showed that mutation of the CCAAT binding site within the ERSE abolished ATF6 stimulation. Conversely, by changing the low-affinity NF-Y binding site to the consensus CCAAT binding site within ERSE-163, ATF6 inducibility was restored. Similar effects were observed for ERSE-131. We further showed that a functional NF-Y complex is required for optimal ATF6 stimulation. Interestingly, NF-Y, acting synergistically with SREBP, is also found to be critical for the transcriptional activation of cholesterol biosynthesis genes (2, 8).
YY1 is a ubiquitous transcription factor unique to higher eukaryotes (23). Unconventional YY1 binding sites are located within the ERSEs (3, 20). Previous studies have shown that YY1 regulates the c-fos promoter through direct interaction with ATF/CREB, such that the C-terminal zinc finger domain of YY1 is necessary and sufficient for binding with the b-ZIP region of ATF (34). Here we showed that YY1 can interact with ATF6 both in vitro and in vivo. We further observed that overexpression of YY1 also enhanced ATF6 stimulation of ERSE-98, suggesting that YY1 can act as a coactivator of ATF6. Recently, we showed that YY1 can stimulate yHac1p-mediated activation of the grp78 promoter (3). Like ATF6, yHac1p is a b-ZIP protein and is an activator of the grp promoters in nonstressed cells. However, ATF6 and yHac1p appear to act on different target sites of the grp78 promoter, with yHac1p having the unique ability to stimulate ERSE-163 bearing the weak NF-Y site (3). While the magnitude of activation by YY1 is modest due to the abundance of endogenous YY1, its stimulatory effect for both yHac1 and ATF6 is dependent on the zinc finger DNA binding domain. This suggests that YY1 could enhance ATF6 action through anchoring of the protein complex onto the ERSE, or it could involve protein-protein interactions, leading to maximal stimulation. It is also possible that the YY1 binding site within the ERSE also binds other, yet unidentified proteins capable of interacting with ATF6 and modifying ATF6 activity. Since Tg treatment of cells already expressing high levels of ATF6 results in substantial further enhancement of the grp response, additional mechanisms specifically activated by Tg may contribute to maximal Tg stress induction of the grp promoter. Such mechanisms may include modification of ATF6 to convert it to a more potent form, as well as the activation of YY1 and other cofactors.
In defining the relationship between ATF6 and hIre1p, we discovered different properties for the two proteins in activating the grp78 promoter. First, while overexpression of both proteins can stimulate the grp78 promoter in nonstressed cells, the enhancement mediated by hIre1p, but not ATF6, is sensitive to the tyrosine kinase inhibitor genistein, previously shown to suppress the grp stress response (1, 35). Second, ATF6 stimulation requires a strong NF-Y binding site, whereas the activity of the downstream target of hIre1p that mediates the activation of the grp78 promoter does not. Last, overexpression of hIre1p does not promote proteolysis of ATF6, nor does it substantially increase its protein level. While the critical issue of whether ATF6 is a downstream target of Ire1p requires further investigations, the difference in their activating properties suggests diverse regulatory pathways.
ACKNOWLEDGMENTS
We are greatly indebted to Ron Prywes, Yang Shi, Randy Kaufman, Sankar Maity, and Peter Edwards for the ATF6, YY1, hIre1p, and NF-Y/CBF reagents; we thank Ebrahim Zandi, Ramachandra Reddy, and Xinke Chen for helpful discussions.
We thank David Hinton and Ernesto Barron for helpful suggestions and assistance in microscopy, which was performed at the Electron Microscopy Core Facility at the Doheny Eye Institute, USC, supported by NEI/NIH core grant EY03040 and the Norris Cancer Center. This work was supported by Public Health Service grant CA27607 from the NCI, National Institutes of Health, to A.S.L.
REFERENCES
- 1.Cao X, Zhou Y, Lee A S. Requirement of tyrosine- and serine/threonine kinases in the transcriptional activation of the mammalian grp78/BiP promoter by thapsigargin. J Biol Chem. 1995;270:494–502. doi: 10.1074/jbc.270.1.494. [DOI] [PubMed] [Google Scholar]
- 2.Dooley K A, Millinder S, Osborne T F. Sterol regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene through a direct interaction between sterol regulatory element binding protein and the trimeric CCAAT-binding factor/nuclear factor Y. J Biol Chem. 1998;273:1349–1356. doi: 10.1074/jbc.273.3.1349. [DOI] [PubMed] [Google Scholar]
- 3.Foti D M, Welihinda A, Kaufman R J, Lee A S. Conservation and divergence of the yeast and mammalian unfolded protein response. Activation of specific mammalian endoplasmic reticulum stress element of the grp78/BiP promoter by yeast Hac1. J Biol Chem. 1999;274:30402–30409. doi: 10.1074/jbc.274.43.30402. [DOI] [PubMed] [Google Scholar]
- 4.Gazit G, Lu J, Lee A S. De-regulation of GRP stress protein expression in human breast cancer cell lines. Breast Cancer Res Treat. 1999;54:135–146. doi: 10.1023/a:1006102411439. [DOI] [PubMed] [Google Scholar]
- 5.Hai T W, Liu F, Coukos W J, Green M R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 6.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmann K, Stoffel W. TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 8.Jackson S M, Ericsson J, Mantovani R, Edwards P A. Synergistic activation of transcription by nuclear factor Y and sterol regulatory element binding protein. J Lipid Res. 1998;39:1712–1721. [PubMed] [Google Scholar]
- 9.Kaufman R J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 10.Kozutsumi Y, Segal M, Normington K, Gething M J, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 11.Lee A S. Coordinated regulation of a set of genes by glucose and calcium ionophores in mammalian cells. Trends Biochem Sci. 1987;12:20–23. [Google Scholar]
- 12.Lee A S. Mammalian stress response: induction of the glucose-regulated protein family. Curr Opin Cell Biol. 1992;4:267–273. doi: 10.1016/0955-0674(92)90042-b. [DOI] [PubMed] [Google Scholar]
- 13.Li W W, Alexandre S, Cao X, Lee A S. Transactivation of the grp78 promoter by Ca2+ depletion. A comparative analysis with A23187 and the endoplasmic reticulum Ca2+-ATPase inhibitor thapsigargin. J Biol Chem. 1993;268:12003–12009. [PubMed] [Google Scholar]
- 14.Li W W, Hsiung Y, Wong V, Galvin K, Zhou Y, Shi Y, Lee A S. Suppression of grp78 core promoter element-mediated stress induction by the dbpA and dbpB (YB-1) cold shock domain proteins. Mol Cell Biol. 1997;17:61–68. doi: 10.1128/mcb.17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W W, Hsiung Y, Zhou Y, Roy B, Lee A S. Induction of the mammalian GRP78/BiP gene by Ca2+ depletion and formation of aberrant proteins: activation of the conserved stress-inducible grp core promoter element by the human nuclear factor YY1. Mol Cell Biol. 1997;17:54–60. doi: 10.1128/mcb.17.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W W, Sistonen L, Morimoto R I, Lee A S. Stress induction of the mammalian GRP78/BiP protein gene: in vivo genomic footprinting and identification of p70CORE from human nuclear extract as a DNA-binding component specific to the stress regulatory element. Mol Cell Biol. 1994;14:5533–5546. doi: 10.1128/mcb.14.8.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maity S N, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 18.Mantovani R, Li X Y, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 19.Reddy R K, Lu J, Lee A S. The endoplasmic reticulum chaperone glycoprotein GRP94 with Ca2+-binding and antiapoptotic properties is a novel proteolytic target of calpain during etoposide-induced apoptosis. J Biol Chem. 1999;274:28476–28483. doi: 10.1074/jbc.274.40.28476. [DOI] [PubMed] [Google Scholar]
- 20.Roy B, Lee A S. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 1999;27:1437–1443. doi: 10.1093/nar/27.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy B, Li W W, Lee A S. Calcium-sensitive transcriptional activation of the proximal CCAAT regulatory element of the grp78/BiP promoter by the human nuclear factor CBF/NF-Y. J Biol Chem. 1996;271:28995–29002. doi: 10.1074/jbc.271.46.28995. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro D J, Sharp P A, Wahli W W, Keller M J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 23.Shi Y, Lee J-S, Galvin K M. Everything you have ever wanted to know about Yin Yang 1 … . Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Seto E, Chang L S, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 25.Thuerauf D J, Arnold N D, Zechner D, Hanford D S, DeMartin K M, McDonough P M, Prywes R, Glembotski C C. p38 Mitogen-activated protein kinase mediates the transcriptional induction of the atrial natriuretic factor gene through a serum response element. A potential role for the transcription factor ATF6. J Biol Chem. 1998;273:20636–20643. doi: 10.1074/jbc.273.32.20636. [DOI] [PubMed] [Google Scholar]
- 26.Tirasophon W, Welihinda A A, Kaufman R J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Sato R, Brown M S, Hua X, Goldstein J L. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 28.Wang X Z, Harding H P, Zhang Y, Jolicoeur E M, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wooden S K, Li L J, Navarro D, Qadri I, Pereira L, Lee A S. Transactivation of the grp78 promoter by malfolded proteins, glycosylation block, and calcium ionophore is mediated through a proximal region containing a CCAAT motif which interacts with CTF/NF-I. Mol Cell Biol. 1991;11:5612–5623. doi: 10.1128/mcb.11.11.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worgall T S, Sturley S L, Seo T, Osborne T F, Deckelbaum R J. Polyunsaturated fatty acids decrease expression of promoters with sterol regulatory elements by decreasing levels of mature sterol regulatory element-binding protein. J Biol Chem. 1998;273:25537–25540. doi: 10.1074/jbc.273.40.25537. [DOI] [PubMed] [Google Scholar]
- 31.Wu F, Lee A S. Identification of AP-2 as an interactive target of Rb and a regulator of the G1/S control element of the hamster histone H3.2 promoter. Nucleic Acids Res. 1998;26:4837–4845. doi: 10.1093/nar/26.21.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang W, Desiderio S. BAP-135, a target for Bruton's tyrosine kinase in response to B cell receptor engagement. Proc Natl Acad Sci USA. 1997;94:604–609. doi: 10.1073/pnas.94.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q, Gedrich R W, Engel D A. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J Virol. 1995;69:4323–4330. doi: 10.1128/jvi.69.7.4323-4330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou Y, Lee A S. Mechanism for the suppression of the mammalian stress response by genistein, an anticancer phytoestrogen from soy. J Natl Cancer Inst. 1998;90:381–388. doi: 10.1093/jnci/90.5.381. [DOI] [PubMed] [Google Scholar]
- 36.Zhu C, Johansen F E, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]