Abstract
The pH-low insertion peptide (pHLIP) and its analogs sense the microenvironmental pH variations in tumorous cells and serve as useful anticancer drug deliveries. The pHLIP binds peripherally to membranes and adopts random coil conformation at the physiological pH. The peptide switches from random coil to α-helical conformation and inserts unidirectionally into membrane bilayers when pH drops below a critical transition value that has been routinely determined by the Trp fluorescence spectroscopy. Recent high-resolution studies using solid-state NMR spectroscopy revealed the presence of thermodynamically stable intermediate states of membrane-associated pHLIP around the fluorescence-based transition pH-value. However, the molecular structural features and their mechanistic roles of these intermediate states in the pH-driven membrane insertion process of pHLIP remain largely unknown. This work utilizes solid-state NMR spectroscopy to explore 1) the mechanistic roles of key proline and arginine residues within the pHLIP sequence at intermediate pH-values, and 2) the changes in lipid dynamics at intermediate pH-values in multiple types of model bilayers with anionic phospholipid and/or cholesterol. Our results demonstrate several molecular structural and dynamics changes at around the transition pH-values, including the isomerization of proline-threonine backbone configuration, breaking of arginine-aspartic acid salt bridge and the formation of arginine-lipid interactions, and a universal decreasing of dynamics in lipid headgroups and alkyl chains. Overall, the outcomes provide important insights on the molecular interactions between pHLIP and membrane bilayers at intermediate pH-values and, therefore, prompt the understanding of pH-driven membrane insertion process of this anticancer drug-delivering peptide.
Significance
Although there have been many early successes in the developments of pH-low insertion peptide (pHLIP)-based anticancer drug delivery strategies and tumor-imaging tools, understanding of the fundamental mechanisms of pH-driven membrane insertion process of pHLIP falls behind. This creates a knowledge gap that hinders effective and rational design of pHLIP analogs for further biomedical applications. Application of high-resolution solid-state NMR spectroscopy unravels the intermediate states of membrane-associated pHLIP. This work provides insights on mechanistic roles of key residues such as the helix-breaking proline and the N-terminal arginine. These results may be utilized to guide future rational design of pHLIP sequences. Furthermore, high-resolution studies of both peptides and lipids lead to more comprehensive understanding of the membrane insertion process of pHLIP.
Introduction
The pH-low insertion peptide (pHLIP) is a 36-residue peptide (Nt-GGEQNPIYWARYADWLFTTPLLLLDLALLVDADEGT-C) that adopts pH-dependent conformational changes and membrane positioning (1,2). The pHLIP binds peripherally to membranes at physiological pH-values with random coil conformation (state II), and switches to transmembrane α-helices (state III) when pH drops below a critical value (2, 3, 4). Because the pHLIP senses microenvironmental pH changes and inserts unidirectionally into membranes, it has been utilized as a cross-membrane drug delivery tool for various types of antitumorous molecules and a carrier for imaging reagents for cancer diagnosis in the past two decades (1,5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23). Despite these early successes in biomedical applications, the mechanism that underlies such a unique pH-driven membrane insertion process of pHLIP remains unclear. The critical transition pH-value is routinely determined by Trp fluorescence spectroscopy in which both emission wavelengths and spectral intensities are sensitive to the local polarity. The nature of this approach leads to a macroscopic feature, i.e., the averaged fluorescent responses from an ensemble of individual states with different membrane positioning of Trp side chains. It was previously considered that pHLIP interacted with membranes through a two-state mechanism in which the peptide adopts a mixed state II and state III at a given intermediate pH2. Therefore, the critical transition pH is also known as pH50, meaning that 50% of peptides are in state III based on the Trp fluorescence spectral features. The presence of kinetic intermediate states of membrane-associated pHLIP were suggested based on stopped-flow fluorescence spectroscopy (4). However, molecular structural features of these intermediates remain unknown.
Recent applications of solid-state NMR (ssNMR) spectroscopy unraveled the presence of thermodynamically stable intermediate states at pH-values between state II and state III and, therefore, proposed a revised multistep membrane insertion mechanistic model for the pHLIP (24). Notably, definitive ssNMR evidence demonstrated that these intermediate states were structurally distinct from either state II or state III. For instance, residue D31 was shown to be in close proximity to the 31Ps in phospholipid headgroups at both pH 7.4 (a 6.6-Å 13C-31P internuclear distance, state II) and 5.3 (a 4.9-Å 13C-31P internuclear distance, state III) but much farther away at any intermediate pH-values (>9.0 Å at pH 6.4, 6.1, and 5.8) (24). Furthermore, inter-residue side-chain contacts between T18 and V24 were only observed at pH 6.4 but not at pH 7.4 or 5.3 (25). These results implied that membrane-associated pHLIP adopted intermediate states that could not be simplified as mixtures of state II and state III. It has been proposed that protonation of key Asp and Glu residues (i.e., titratable residues) played essential roles in the membrane insertion mechanism (26, 27, 28). Two of these residues, D14 and D25, were initially demonstrated to be critical, and their chemical modifications were shown to modulate fluorescence-based pH50-values of pHLIP (29,30). Recent ssNMR results showed that cooperative titration of the C-terminal acidic residues (D31, D33, and E34) were also required for the folding and insertion process (25). In addition to these titratable acidic groups, other key residues such as the helix-breaking proline P20 and the N-terminal arginine R11 have also been reported (31,32). Characterization of the backbone conformation of pHLIP in state III demonstrated two helical segments with a short break at T19-P20 (25). The P20G mutant of pHLIP was shown to possess higher helical contents not only in the presence of bilayers but also in water-organic solvent mixture (32) as well as increased fluorescence-based pH50 compared with the wild-type peptide (31,32). The residue R11 with a positively charged side chain was proposed to facilitate the membrane binding and/or folding processes of pHLIP through electrostatic or hydrogen bonding interactions. A recent molecular dynamic simulation supported that R11 interacted transiently with all acidic residues in state II and, more interestingly, formed stable salt bridge with D14 in a C-terminus-truncated mutant that removed the acidic residues D31, D33, and E34 (33). Experimentally, it has been shown that double mutants involving R11Q and repositioning of D14 altered the insertion pH50 of pHLIP (29).
Complementary to the structural characterizations of the pHLIP itself, explorations of lipid dynamics through 31P relaxation ssNMR spectroscopy were shown to provide mechanistic insights (24). It was shown in the bilayer containing pure zwitterionic phosphatidylcholine (PC), the interaction with pHLIP led to restricted lipid headgroup motions (increments of the nanoseconds timescale motion correlation time) in a broad pH range 5.0–7.5. Notably, the most significant changes of phosphate lipid headgroup motion occurred at pH 5.8–6.5, matching the fluorescence-based pH50-value (i.e., ∼pH 6.1) (24). Presence of other types of phospholipids has been shown to influence the interactions of membrane-active peptides (34). Previous studies demonstrated that the anionic lipids such as phosphatidylglycerol (PG) and phosphatidylserine, as well as the zwitterionic phosphatidylethanolamine, altered the value of pH50 (28,32,35,36). Incorporation of small populations (i.e., <10 mol %) of anionic lipids was shown to lead to a decrement of pH50 compared with the PC bilayer (32). However, interestingly, further increments of anionic lipid populations could either increase pH50 or had little effect based on different reports (28,35). The addition of cholesterol to PC bilayers was shown to decrease pH50, presumably by changing the bilayer thickness and/or the overall membrane fluidity (37). Applications of 31P relaxation ssNMR spectroscopy to more complex bilayer models may provide insights on the membrane insertion process of pHLIP from the aspect of membrane compositions.
In this work, we apply multiple ssNMR approaches to explore the structures and membrane interactions of key residues P20 and R11, as well as the changes of lipid dynamics in bilayer models containing anionic PG and/or cholesterol, focusing on the intermediate states at pH-values around the fluorescence-based pH50.
Materials and methods
Peptide synthesis and purification
All pHLIP peptides with and without isotope-labeled residues were synthesized manually using routine solid-phase peptide synthesis protocols with 9-fluorenylmethyloxycarbonyl chemistry. Two isotope-labeled sequences were included in this study: 1) selective 13C and 15N labeling at P20-C′ (carbonyl 13C) and T19-NH (amide 15N) respectively, and 2) uniform 13C-15N labeling at residues R11 and D14. All isotope-labeled amino acids were purchased from Cambridge Isotope (Tewksbury, MA). with preprotected 9-fluorenylmethyloxycarbonyl groups. All peptides were cleaved from resin using a cocktail mixture containing 82.5% (volume ratio, v/v) trifluoroacetic acid, 5.0% (v/v) deionized H2O, 2.5% (mass ratio, m/m) phenol, 5.0% (v/v) thioanisole, 2.5% (v/v) 1,2-ethanedithiol, and an additional 2.5% (v/v) Me2S. The crude peptides were purified by reversed phase high-performance liquid chromatography (HPLC) with a C18 column (Agilent Technologies, Santa Clara, CA) and water-acetonitrile linear gradient, and the molecular mass and purity of peptides were confirmed by liquid chromatography–mass spectrometry (LC-MS, Shimadzu, Kyoto, Japan). The peptides were stored at −20°C before usage.
Liposome preparation
The phospholipids, including 1-palmitoyl-2-oleoyl-sn-glycerol-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG), and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL). The phospholipids and cholesterol were dissolved in chloroform to make 25-mg/mL stock solutions and stored at −20°C before usage. To make liposome samples, appropriate volumes of phospholipids and cholesterol stock solutions were mixed and dried under a stream of N2 air before drying overnight under high vacuum. The lipid films were then resuspended and vortexed in 10 mM phosphate buffer (pH 8.0, containing 0.01% w/v NaN3) to the total lipid concentration of 1.5 mM. After eight freeze-thaw cycles using liquid N2 and a 41°C water bath, the liposomes were extruded for 31 cycles using a 200-nm pore-size filter membrane. All liposomes were prepared freshly before the fluorescence measurements and ssNMR sample preparation.
Tryptophan fluorescence assay
Lyophilized pHLIP peptides were firstly dissolved in dimethylformamide and then mixed with liposome solution in phosphate buffer at pH 8.0. The final concentrations of pHLIP and total lipids were kept at 5 μM and 1.5 mM, respectively, to obtain a peptide/lipid molar ratio 1:300. Liposomes in the absence of pHLIP were used as controls. The pHLIP-liposomes were incubated at 4°C for overnight to allow absorption of peptides to membranes and then divided into 200-μL aliquots. The pH was then adjusted using concentrated 50 mM phosphate/acetate buffer solutions to different values in a range 4.0–8.0. The samples were then allowed to equilibrate for 1–2 h at ambient temperature before their tryptophan fluorescence spectra were recorded.
Tryptophan emission spectra were recorded on a PerkinElmer LS-55 spectrometer (Waltham, MA) with the excitation wavelength at 285 nm and the emission wavelength range of 310–400 nm. The excitation and emission slits were both set to 5 nm. For background subtraction, liposome-only controls were prepared with pH-values 4.0, 5.0, 6.0, 7.0, and 8.0. The controls at different pH-values gave similar fluorescence emissions and thus the averaged spectra from multiple pH-values were used as background for subtraction.
To obtain the pH-dependent transition curves, the fluorescence spectral center of mass (CM) was calculated as:
| (1) |
where Ii represents the spectral intensity at the wavelength λi. The quantitative analysis of CM was reported to provide information about the entire spectral range and was more sensitive to both the shapes and widths of emission spectra (38, 39, 40). The pH-dependent transition curves were fitted to the Boltzmann equation below:
| (2) |
where the parameters A1, A2, x0, and dx represent the minimal and maximal wavelengths, the transition pH (i.e., pH50), and the slope of transition, respectively.
The ssNMR spectroscopy
All ssNMR samples were prepared following the same protocols as described for the fluorescence spectroscopy, except that the pHLIP/total lipid molar ratio was kept at 1:75 instead of 1:300. We previously showed that this higher peptide/lipid ratio did not affect the pH-dependent fluorescence and circular dichroism spectral features of pHLIP (41). All ssNMR spectra were collected on a 600 MHz Bruker Avance III spectrometer (Billerica, MA) equipped with a 2.5-mm TriGamma magic angle spinning (MAS) probe, which was tuned to 1H, 13C, or 15N for 13C-15N rotational-echo double-resonance (REDOR) experiments and 1H, 31P, or 13C for 13C-31P REDOR and 31P relaxation measurements.
The following spectrometer parameters were utilized for the 13C-15N REDOR experiment applied to the samples with selectively isotope-labeled T19 and P20: a 60-kHz 1H π/2 pulse, a 1.5-ms cross-polarization between 1H and 13C with 50 kHz 1H field and 57 kHz 13C field with a linear ramp, 50 kHz 13C/15N π-pulse trains for the heteronuclear dipolar recoupling, and a pulsed-spin locking (42) acquisition algorithm with 90 kHz 1H decoupling. For the 13C-15N REDOR applied to samples with uniformly labeled R11 and D14, 1.0-ms frequency-selective Gaussian pulses (43) were applied to both 13C and 15N channels in the middle of dipolar recoupling periods. The 13C and 15N carrier frequencies for the Gaussian pulses were set to the D14-Cγ (i.e., ∼181 ppm) and R11-Nε (i.e., ∼90 ppm), respectively. For the 13C-31P REDOR, the 33 kHz 31P π-pulse train was applied and the frequency-selective pulse was applied only to the 13C channel with the carrier frequency at R11-Cε (i.e., ∼165–170 ppm). All REDOR spectra were collected with 8000 ± 2 Hz MAS frequency. Samples were kept at 280 K by monitoring the 1H signal in H2O before and after each set of measurements.
The T1 (spin-lattice) and T2 (spin-spin) relaxation rates (R1 and R2, respectively) were obtained using routine inversion-recovery (π-τ-π/2) and Hartmann-Hahn Echo (π/2-τ-π-τ) pulse sequences with 55 kHz 31P pulses and ∼100 kHz 1H two-pulse phase modulation (TPPM) decoupling field. The sample temperatures were precisely determined from the H2O 1H chemical shifts. The experimental temperatures were chosen to be above the transition temperature of POPC to keep the samples in liquid phase (the addition of POPG does not alter the transition temperature of liposomes). The MAS frequency was set at 9000 ± 2 Hz, and all samples were fully hydrated throughout the measurements.
Results and discussion
The pH-dependent interaction of R11 with D14 and phospholipids
Figs. 1 and 2 show the representative 13C-15N and 13C-31P REDOR spectra, which probe the internuclear distances between R11-Nε and D14-Cγ and between R11-Cε and 31P in lipid headgroups, respectively. Spectral were collected on samples with pH 6.4, 6.1, 5.8, and 5.3. These pH-values were chosen based on our previous works that characterized the protonation states of individual titratable residues (e.g., Asp and Glu) in pHLIP (24,25). Experimental REDOR dephasing were determined as the differences between normalized 13C peak volumes in the full (S0, with 15N pulses off) and reduced (S1, with 15N pulses on) spectra, and were plotted in Fig. 3 with the simulated curves for different 13C-15N and 13C-31P internuclear distances.
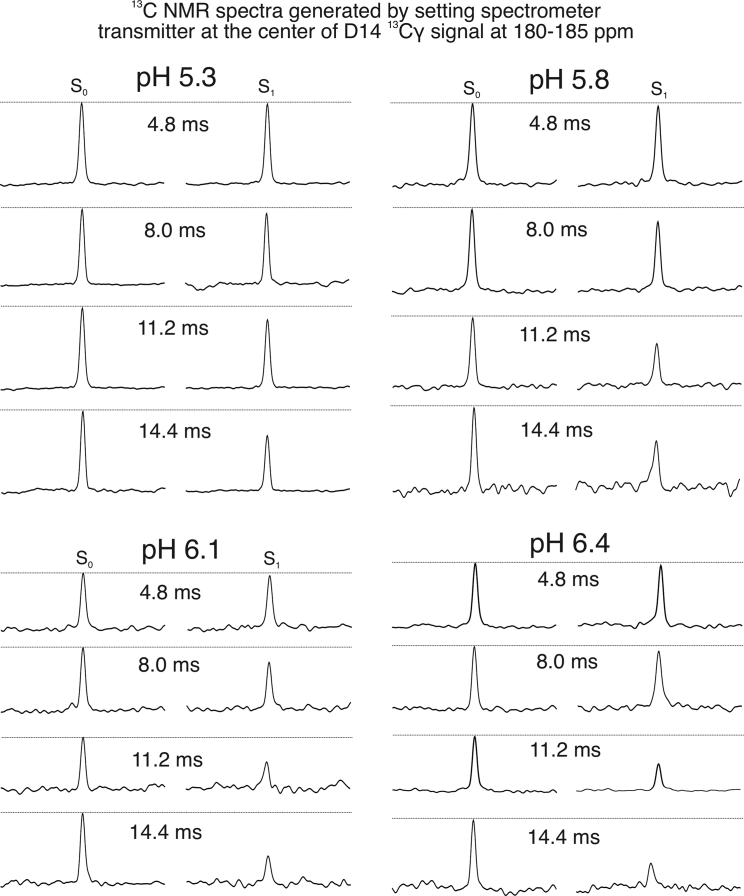
Figure 1.
Representative 13C-15N REDOR spectra for measuring the internuclear distance between D14-Cγ and R11-Nε. The 13C carrier frequency was set to ∼180–185 ppm (D14-Cγ, specific for individual samples) for both the Gaussian selective pulse and the pulsed-spin locking acquisition period (as described in the experimental section). Thus, all peaks centered at 0 ppm in the outcoming spectra, and the x axes were therefore not labeled. All spectra were processed with 20 Hz Gaussian line broadening.
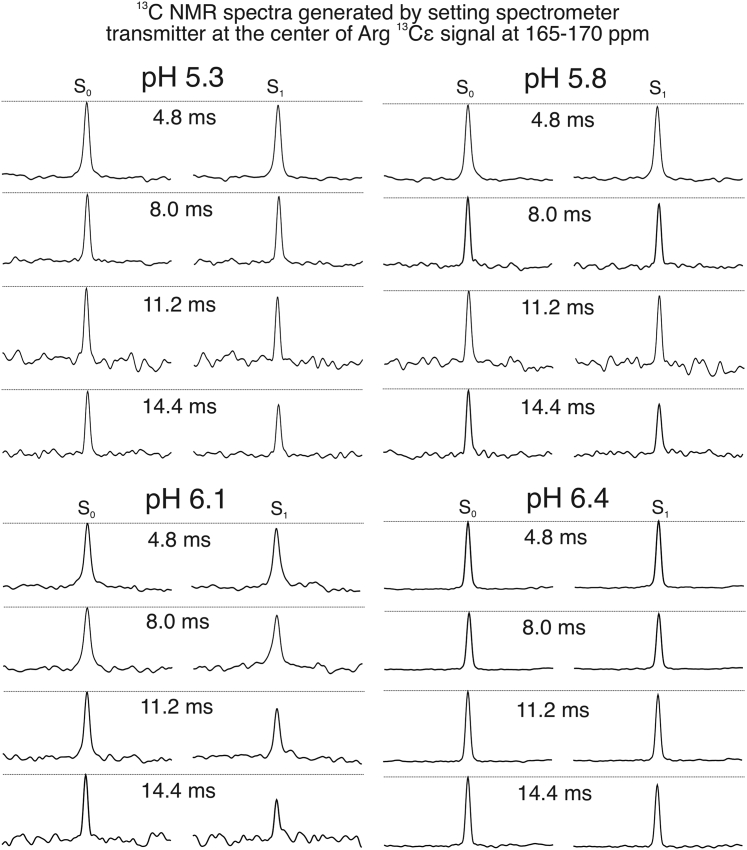
Figure 2.
Representative 13C-31P REDOR spectra for measuring the internuclear distance between R11-Cε and the lipid phosphate headgroup 31Ps. The 13C carrier frequency was set to ∼165–170 ppm (R11-Cε, specific for individual samples) for both the Gaussian selective pulse and the pulsed-spin locking acquisition period (as described in the experimental section). Thus, all peaks centered at 0 ppm in the outcoming spectra, and the x axes were therefore not labeled. All spectra were processed with 20 Hz Gaussian line broadening.
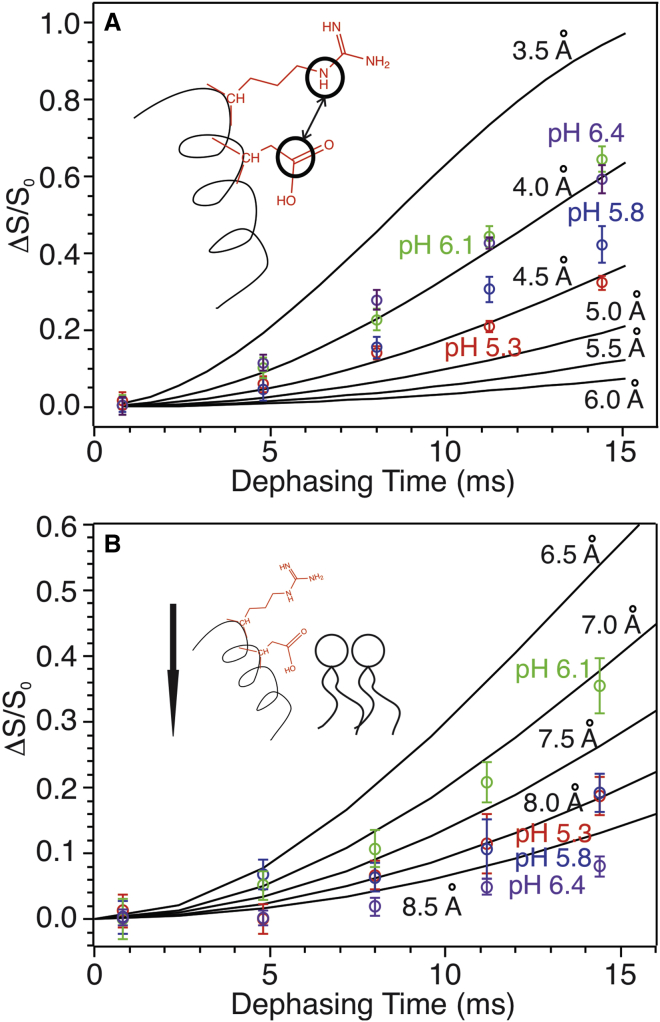
Figure 3.
(A) 13C-15N and (B) 13C-31P REDOR buildup curves obtained by analyzing the corresponding spectra (Figs. 1 and 2, respectively). Experimental dephasing (ΔS/S0) data were shown in open symbols with color coding: red, pH 5.3; blue, pH 5.8; green, pH 6.1; and purple, pH 6.4. The solid lines show simulated 13C-15N and 13C-31P buildup curves for the two-spin pairs with different internuclear distances. Cartoon models show the measured distances (R11 and D14 side chains) and the relative geometries in pHLIP and lipid bilayer. Error bars represent the spectral noises.
At pH 6.4 and 6.1, the internuclear distances between D14-Cγ and R11-Nε fitted well to the simulation curve with 4.0 Å internuclear distance, indicating the formation of salt bridge between the two side chains (44). Interestingly, the R11-D14 13C-15N internuclear distance became longer when pH dropped below 6.1. We showed previously that the side-chain carboxylic group of D14 underwent protonation between pH 5.6–6.1 (25), which might remove the negative charge on D14 and thus disfavored the electrostatic interaction between D14 and R11. Meanwhile, the internuclear distance between R11-Cε and lipid headgroup 31Ps became shorter at pH 6.1 (∼7.0 Å) compared with pH 6.4 (>8.5 Å), and remained at 8.0 Å at pH 5.8 and 5.3. These results suggest that positively charged R11 side chain may interact with phospholipid headgroups when the intrapeptide electrostatic interaction between D14 and R11 is weakened because of the protonation of D14 upon pH dropping below 6.1. Previous molecular dynamic simulation suggested that R11 formed transient salt bridge with D14 in state II (physiological pH ∼7.4), which could be stabilized by the removal of C-terminal D/E residues (33). These ssNMR data support the simulation results because C-terminal D/E residues were shown to protonate at higher pH-values. Therefore, decreasing of the environmental pH is likely to eliminate C-terminal negative charges, which has similar effects as the removal of D/E residues.
The pH-dependent backbone conformational change of P20 determined by ssNMR
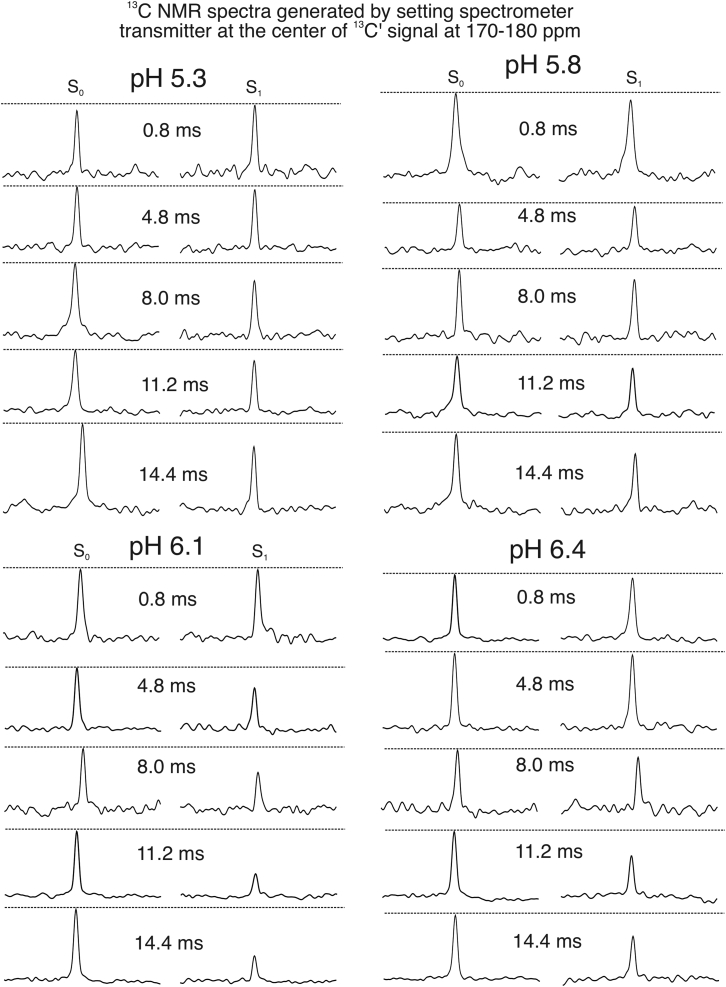
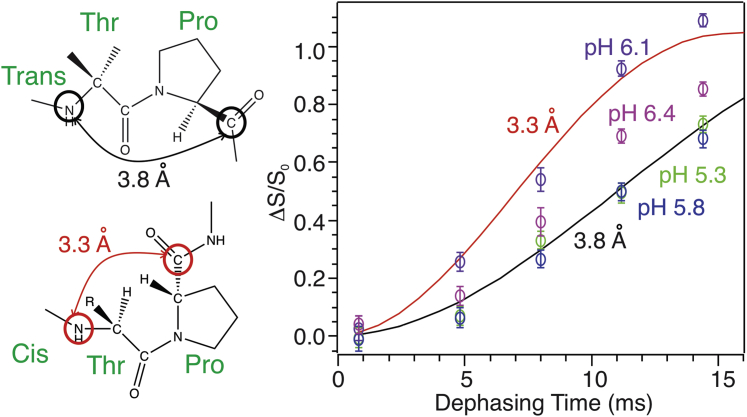
As a known breaker for helical conformations, the single proline residue P20 that locates within the helix-forming segment of pHLIP is believed to play important roles in its membrane insertion process. Fig. 4 shows the 13C-15N REDOR spectra that specifically probe the internuclear distance between T19-NH and P20-C′, which is sensitive to the backbone dihedral angles around the T19-P20 peptide bond. These internuclear distances are expected to be 3.3 and 3.8 Å when P20 adopts “cis”- and “trans”-backbone conformations respectively, which correspond to 85 and 55 Hz 13C-15N dipolar coupling strengths and distinct REDOR buildup curves (Fig. 5). The experimental REDOR dephasing was corrected for natural abundance contributions from unlabeled amino acids and lipids (see Supporting materials and methods) and the corrected dephasing at different pH-values were plotted in Fig. 5.
Figure 4.
The 13C-15N REDOR spectra for measuring the internuclear distance between P20-C′ and R11-Nε. The 13C carrier frequency was set to ∼170–180 ppm (the general C′ region) for the pulsed-spin locking acquisition period. Thus, all peaks centered at 0 ppm in the outcoming spectra, and the x axes were therefore not labeled. All spectra were processed with 20 Hz Gaussian line broadening.
Figure 5.
(Left) Illustration of the 13C-15N internuclear distances between P20-C′ and T19-NH (circled in the chemical structures) in the Thr-Pro segment with “trans”- and “cis”-backbone configurations. (Right) Shown are corrected REDOR dephasing data (ΔS/S0, open symbols, obtained from experimental REDOR spectra shown in Fig. 4 and processed with natural abundance corrections) with color coding for different pH-values: green, pH 5.3; blue, pH 5.8; purple, pH 6.1; and magenta, pH 6.4. Simulated curves are shown for two-spin pairs with internuclear distances 3.8 Å (black) and 3.3 Å (red). Error bars represent spectral noises in S0 and S1 spectra.
Notably, the fitting of 13C-15N REDOR data support a backbone conformational switch at P20 from “cis” to “trans” around pH 6.1. It remains unclear which factors might drive this dramatic backbone conformational switch. The activation energy for proline cis- or trans-conformational change was reported to be 17–19 kcal/mol at room temperature (45,46). These and previous ssNMR results implied that two processes might compensate energies. First, the N-terminal segments A10-T18 might undergo conformational changes from partially random coils to α-helices, which might release ∼0.12 kcal/mol energy per residue and ∼1 kcal/mol in total (41,47). Second, the repositioning of W15 and/or W9 toward more hydrophobic bilayer interior might compensate a couple of kcal/mol free energy based on the Whimley-White hydrophobic scale of amino acids (48). However apparently, the magnitude of these free energy changes was not comparable with the activation energy of proline backbone conformational switch. Interestingly at pH 6.4, REDOR dephasing curve of P20-C′ showed a mixture of both “cis”- and “trans”-conformations. At pH 6.4, the pHLIP segment from L21 to C-terminus adopted α-helical conformation, whereas the N-terminal segment adopted a mixture of nonhelical and α-helical conformations (41). It is possible that the helical population of pHLIP possesses a “cis”-P20 and peptide favors a bundled helical conformation at pH 6.4 and 6.1 when sinking across the polar lipid headgroup region, which requires a “cis”-configuration of P20. This hypothesis is supported by our previous ssNMR result that the side chains of T18 and L24 were in close proximity and both residues adopted α-helical conformation (25). However, such a bundled helix only extends ∼20 Å and, therefore, is insufficient for extending across the bilayers. Hypothetically, this may unfavorably create increased volumes only to the outer leaflet of membrane bilayer or place certain polar side chains within the hydrophobic bilayer interiors, which may eventually drive the formation of “trans”-configuration of P20 and more extended α-helices.
Quantitative analysis of 31P relaxation ssNMR reveals pHLIP-induced changes in lipid dynamics
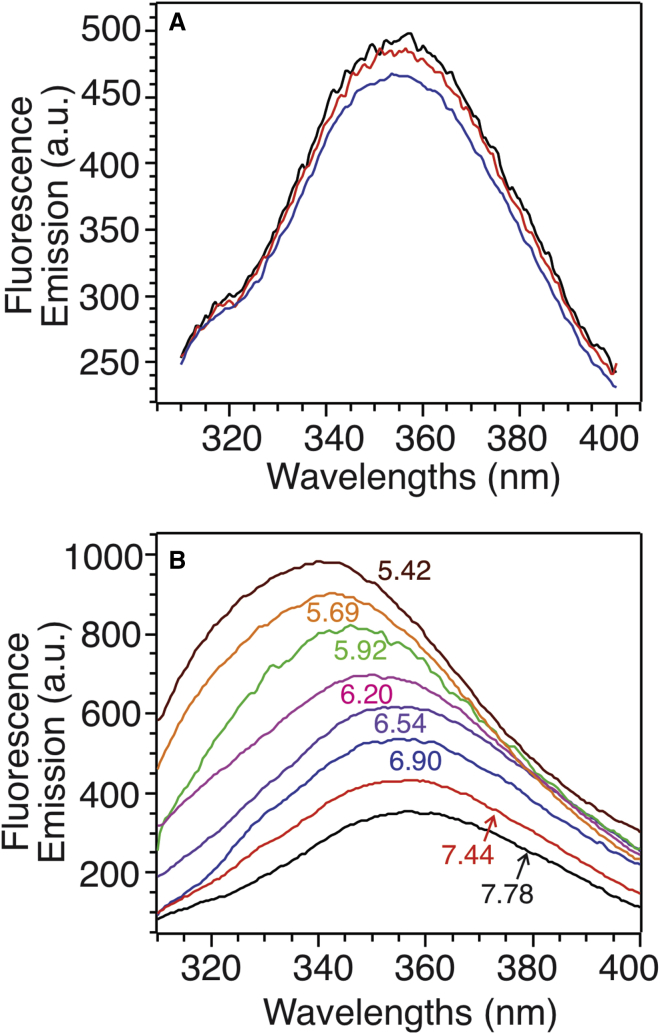
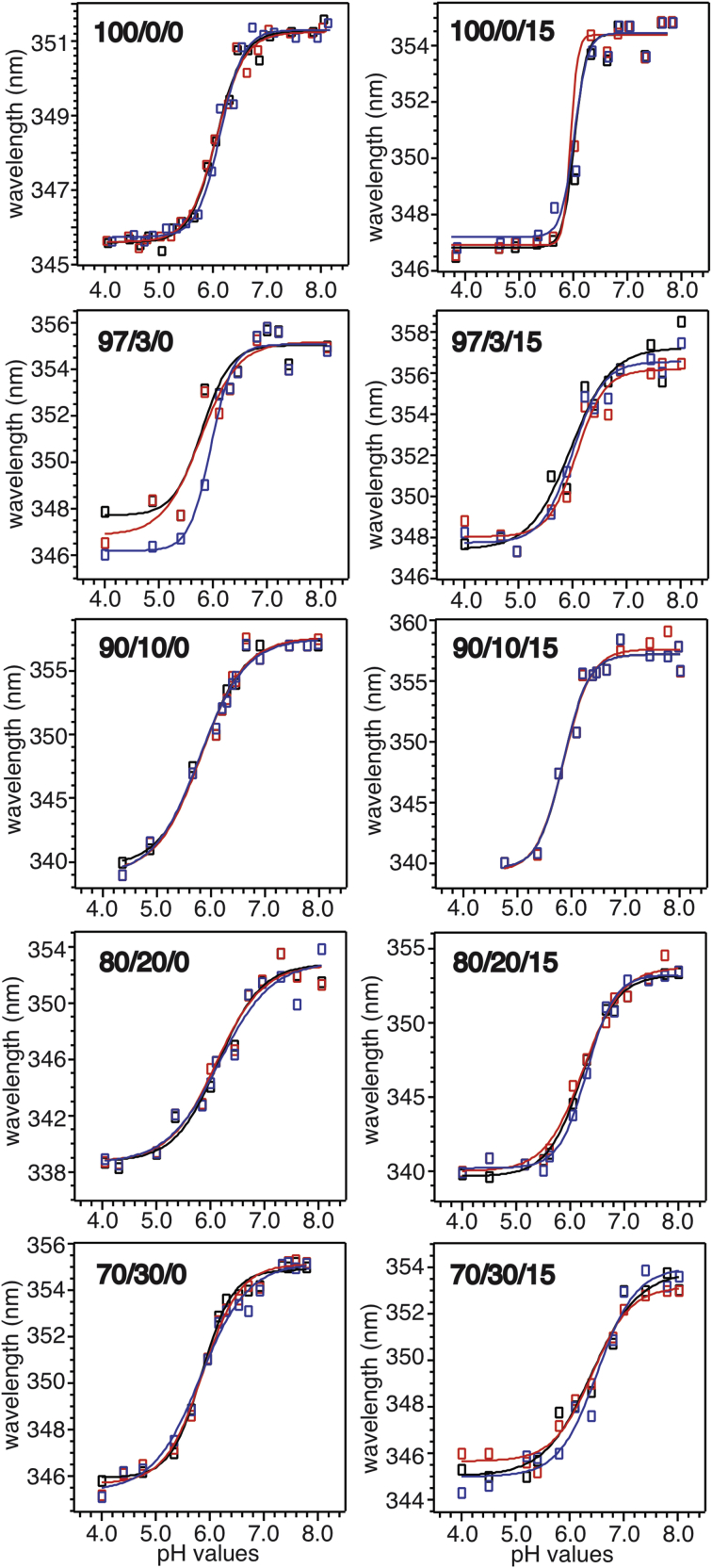
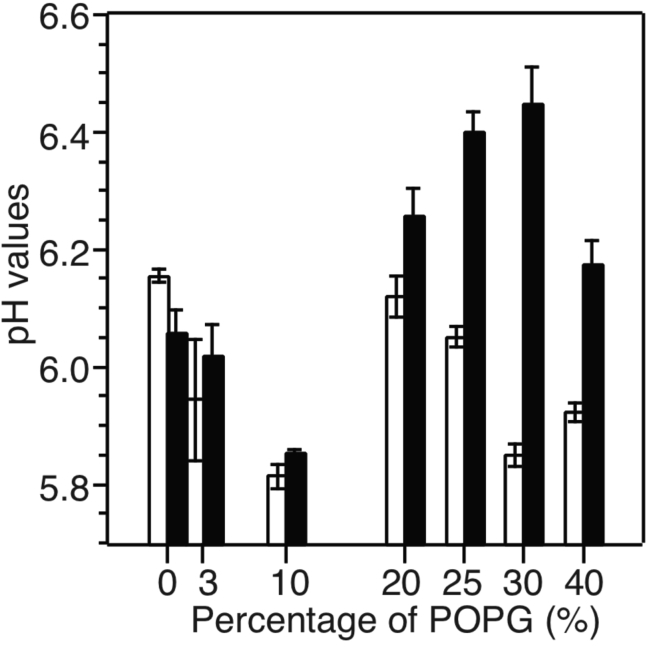
Trp fluorescence spectroscopy was utilized to monitor the pH-dependent membrane insertion of pHLIP in POPC bilayers with different molar percentages of anionic POPG and the presence or absence of cholesterol (representative fluorescence spectra shown in Fig. 6). The CM wavelengths were calculated using Eq. 1 and their pH dependence (Fig. 7) were fitted to sigmoidal curves to obtain pH50-values (Fig. 8). The addition of ∼3–∼10 mol % POPG led to a considerable decrement of pH50 in bilayer models both with and without cholesterol, which was consistent with a previous report on the effects of an anionic phosphatidylserine (POPS) (35). Interestingly, further increments in POPG population showed distinct influences on pH50 in the presence or absence of cholesterol. In general, pHLIP had higher pH50-values in model bilayers with cholesterol, and the largest difference occurring at 30 mol % POPG (∼0.5 pH unit difference).
Figure 6.
Representative tryptophan fluorescence emission spectra. (A) Three repetitions for the model bilayer with 3 mol % POPG (denoted 97/3/0) and the pHLIP peptides at pH ∼7.4. (B) The pH-dependent tryptophan fluorescence traces for the model bilayer with 3 mol % PG and 15 mol % cholesterol and the pHLIP peptides (denoted 97/3/15).
Figure 7.
Plots of pH-dependent CM wavelengths from the Trp fluorescence spectra of membrane-associated pHLIP peptides in different bilayer models (the molar percentages of POPC, POPG, and cholesterol are shown in black for each plot). Three repetitions are shown for each sample with experimental data in open symbols and fittings in solid curves.
Figure 8.
Plots of the pH50in the samples with different molar percentage of POPG. The open and solid columns indicate the samples without and with 15 mol % cholesterol, respectively. The error bars are derived from three repetitions of fluorescence spectra shown in Fig. 6.
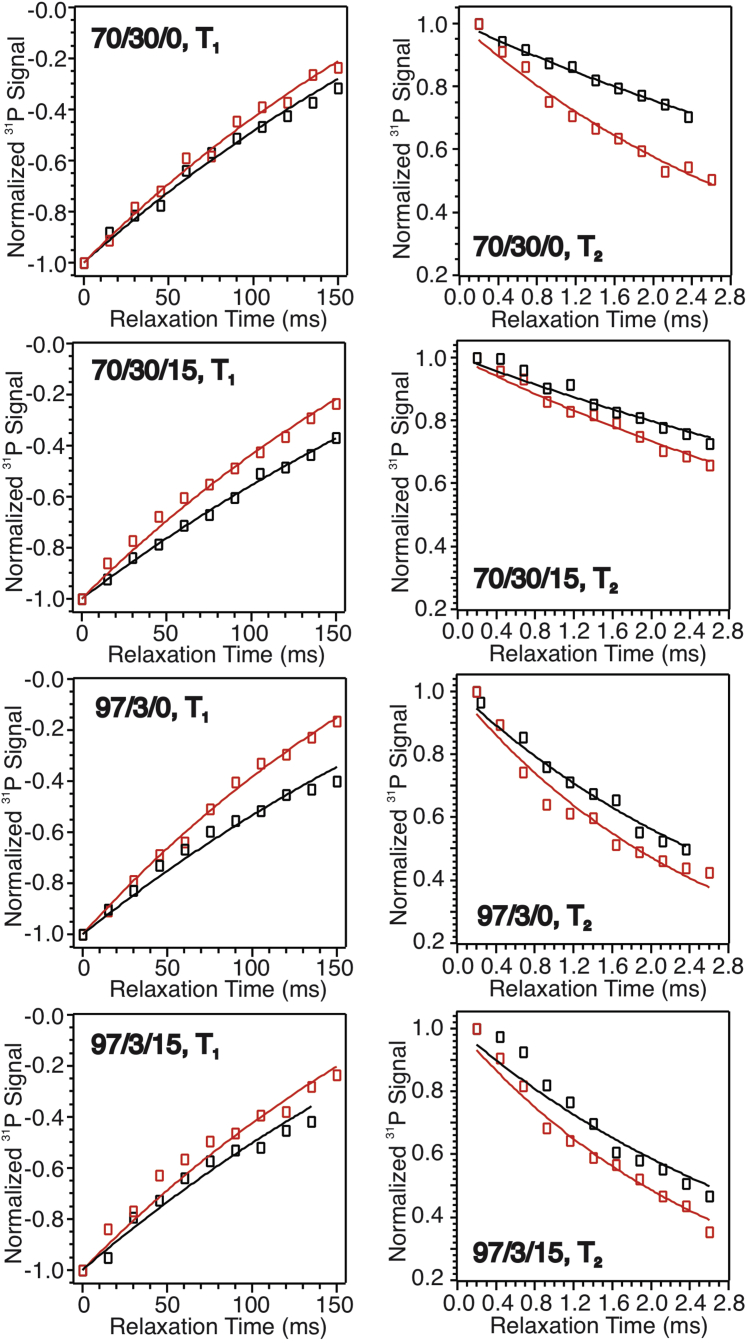
We then applied 31P ssNMR relaxation spectroscopy to explore the quantitative changes in phospholipid dynamics, which were shown to be sensitive to the molecular interactions between pHLIP and lipid bilayers in our previous work (24). In this work, we selected four different bilayer models: the bilayers with 3 mol % POPG in POPC, both in the absence and presence of additional 15 mol % cholesterol (named as 97/3/0 and 97/3/15, respectively), and the ones with 30 mol % POPG without and with 15 mol % cholesterol (70/30/0 and 70/30/15, respectively). The POPG molar percentages were chosen based on this Trp fluorescence result (Fig. 8) in which the greatest cholesterol-dependent change in pH50 occurred at 30 mol % POPG, and a previous report showing that small populations of anionic lipids (i.e., <5 mol %) led to significant decrease of pH50 (31). Fig. 9 shows the representative spin-lattice (T1) and spin-spin (T2) 31P relaxation curves in different model bilayers at 296 K. Qualitatively, the addition of pHLIP in bilayers led to more rapid T1 and T2 decay compared with the non-pHLIP controls, which indicated stronger dipolar coupling between the phosphate headgroups and therefore more restricted lipid dynamics.
Figure 9.
Plots of T1 (left column) and T2 (right column) relaxation curves for different samples in the absence (black) and presence (red) of pHLIP. For each sample, the experimental data were shown in open symbols, and the fitting to exponential functions were shown in solid curves.
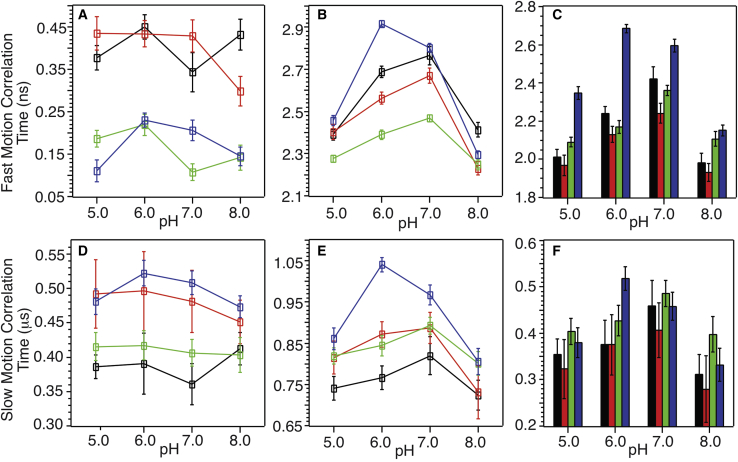
Quantitative analysis of 31P relaxation rates led to the microsecond- and nanosecond-timescale correlation times (denoted as τs and τf, respectively) (see Supporting materials and methods for derivations of the relationship between τs and τf and the relaxation rate constants). The τs and τf are dominated by the lateral diffusive motion and the headgroup uniaxial rotation/wobbling motions of phospholipids respectively (49). Fig. 10 plots the correlation times in all bilayer models with the absence or presence of pHLIP at 296 K. In the bilayers without pHLIP, values of τf and τs showed correlations with membrane compositions, but not with pH-values. In general, τf were influenced more significantly by POPG and τs were affected more by cholesterol. The addition of 30 mol % POPG led to more rapid lipid headgroup motions, presumably due to the formation of more defective charge networks (between the negatively charged phosphate groups and the positively charged choline groups in POPC). The addition of 15 mol % cholesterol showed restrictive effect to the diffusive motion. Both τf and τs increased significantly with the addition to pHLIP (compared with the changes in τf and τs in the absence of pHLIP), implying generally stronger pHLIP-lipid interactions. The panels C and F plot the differences in τf and τs due to the addition of pHLIP, respectively. Notably, the values of τf, which represented the lipid headgroup motions, were influenced significantly by both the membrane compositions and the pH-values. Quantitatively, the lipid headgroup motions were more restricted at intermediate pH-values (6.0 and 7.0) compared with terminal values for any membrane compositions. This was in parallel with the known mechanistic scenes of pHLIP insertion where the peptides showed strongest interactions with the headgroup regions of bilayers in this pH range.
Figure 10.
Plots of the pH-dependent (A and B) fast-motion and (D and E) slow-motion correlation times derived from 31P ssNMR relaxation spectroscopy for different bilayer models. (A) and (D) and (B) and (E) show the correlation times without and with the addition of pHLIP, respectively. Colors represent different membrane compositions: black, 97/3/0; red, 97/3/15; green, 70/30/0; and blue, 70/30/15. (C and F) show the differences in fast and slow-motion correlation times upon addition of pHLIP (subtraction of the correlation times in A and D from the ones in B and E, respectively). The same color coding applied for individual membrane compositions. Error bars are determined using the algorithms in Supporting materials and methods)
Effects of PG and cholesterol on lipid dynamics and the correlations with the fluorescence-based pH50
Comparing with the analysis on the POPC-only bilayer model, it is important to note that introduction of POPG and/or cholesterol led to significantly different variations in pHLIP-induced lipid dynamics changes, which may correlate with the fluorescence-based pH50-values (24). These measurements of pH50 in the presence of POPG agree with a previous study on 10 and 25 mol % POPG in POPC bilayer (28). In addition, previous works using POPS led to similar conclusions that the incorporation of low percentages of anionic lipid induced decrease of pH50 (32,35). Considering the trend of fast-motion correlation times τf in the absence and presence of POPG (e.g., ∼1.0–2.0 ns in 100% POPC (24), ∼2.4–2.8 ns with 3 mol % POPG, and ∼2.2–2.5 ns with 30 mol % POPG), there seems to be stronger peptide-lipid interactions with small percentage of POPG that restrict the headgroup motions. This may correlate with the decrease of pH50 with the addition of anionic lipids because stronger peptide-lipid-headgroup interactions may stabilize a peripherally bound state II.
Cholesterol is an essential membrane composition that is known to alternate the physicochemical properties of bilayers such as the fluidity/rigidity and chain orientation homogeneity (50,51). Our measurements on the fluorescence-based pH50 showed that the addition of 15 mol % cholesterol into 100% POPC bilayer led to ∼0.1 pH unit decrement, which was consistent with a previous report using similar bilayer models (31). It has been suggested that cholesterol affected the pH50 through the modulation of bilayer fluidity (31,37), which is supported by our results on the slow diffusive motion correlation time τs. As shown in Fig. 10 D, the addition of cholesterol caused increment in τs in bilayers with both 3 and 30 mol % POPG. Interestingly, when comparing the fast-motion correlation times between different membrane compositions, it was found that the lipid motions were most significantly restricted by pHLIP in membranes containing 30 mol % POPG and an additional 15 mol % cholesterol (Fig. 10 C, blue columns comparing with black, red, and green columns at pH 5.0, 6.0, and 7.0). This observation implies that pHLIP may have strongest interactions with the lipid headgroups in this membrane environment, which may correlate with the most significant increment in pH50 in this bilayer model.
Insights on pHLIP-membrane insertion mechanistic models
We previously proposed two steps, namely “sinking” and “extending,” at intermediate pH-values between states II and III defined by tryptophan fluorescence, based on measurements of residue-specific 13C-31P distances in the POPC bilayer (24). In this work, 13C-15N and 13C-31P REDOR data on R11 suggest that this residue may contribute to the “sinking” step through changes of electrostatic interactions. Table 1 below summarizes the residue-specific 13C-31P distances within the segment A10-D14 from this and previous works (24,41). Notably, the residues that are in close proximities to 31P (i.e., phospholipid headgroups) changes from pH 6.4 to 6.1 and lower. Although D14-A13 locates near 31P at the higher pH, R11-A10 becomes closer to the phosphate headgroups when pH drops. This corresponds to a movement of half-to-full helical cycle toward the interior of lipid bilayer, or in other words, a “sinking” in terms of the membrane positioning of this segment. Within the same pH range, the electrostatic interaction between R11 and D14 is weakened, which releases the intrapeptide salt bridge (Fig. 3 A). Therefore, the protonation of D14 between pH 5.6–6.1 may have two contributions to the positioning change of pHLIP: 1) it decreases the polarity of the side chain of D14 and makes the nearby peptide segment preferably inserts more deeply to bilayer center, and 2) it frees the positively charged R11 side chain, which potentially strengthens the electrostatic interaction to lipid headgroups.
Table 1.
Summary of 13C-31P distances in pHLIP segment A10-D14
The backbone conformational change of P20 at around pH 6.1 agrees well with the proposed “extending” mechanistic step and may be important for pHLIP to achieve the final transmembrane configuration. However currently, it remains unclear about the overall structural change for pHLIP at this critical pH (i.e., close to the fluorescence-based pH50 in POPC bilayer). Nevertheless, it seems to be reasonable to hypothesize that pHLIP may not tightly associate with lipids in this state because the backbone conformational change of P20 essentially leads to a nearly 180° reorientation in the C-terminal half of peptide. Such a hypothesis is supported by our previous 13C-31P REDOR data in which none of the Asp-Glu residues is close to the lipid headgroups.
Finally, the 31P relaxation data provide insights on how the pHLIP insertion mechanisms may change with other lipids and/or cholesterol. It was shown that pHLIP adopted a different peripherally bound state II in bilayer models with more complicated lipid compositions (i.e., “shallow” state II) compared with pure POPC bilayer (32). The comparison of fast-motion correlation times τf between pure POPC bilayer and more complicated bilayer models at physiological pH-values provides supporting evidence: in POPC bilayer, τf is ∼1.0 ns at pH 7.4 with the addition of pHLIP. In bilayers with POPG and cholesterol, τf is 2.3–2.5 ns at pH 8.0 and 2.5–2.9 at pH 7.0. The significant increments of τf indicate stronger pHLIP-lipid-headgroup interactions in the more complex bilayer models before the insertion of pHLIP (i.e., state II), which is consistent with the “shallow” peripheral binding model where the peptide locates in the phosphate-aqueous interface. In addition, stronger peptide-lipid interaction in the presence of cholesterol implies that pHLIP might preferentially bind to membrane regions such as rafts, which are rich in cholesterol. Future REDOR spectroscopy will be applied to probe the changes of R11-phospholipid interactions in more complex bilayers. Based on our results in POPC bilayer and the hypothesis of stronger pHLIP-lipid interactions in the “shallow” state II model, we expect that the 13C-31P distances may become closer in these bilayers. Furthermore, it will be useful to explore whether the backbone conformational change at P20 is a universal mechanistic step for pHLIP insertion in more complex bilayers because several studies have shown that the mutation of P20 altered the peptide helicity and pH50 and, possibly, the insertion process (31,32).
Conclusions
In summary, we showed in this work that the intermediate states of pHLIP-membrane system possess unique features in peptide conformational changes, peptide-lipid interactions as well as the lipid dynamics, which can be quantitatively determined using ssNMR spectroscopy. Major conformational changes, including the backbone conformational change from cis- to trans-configuration at P20 and the weakening the electrostatic interactions between R11 and D14 side chains, were demonstrated at pH-values around 6.1, which provided experimental evidence for the previously proposed “sinking” and “extending” steps in mechanisms of pHLIP-membrane insertion. Quantitative analysis of 31P relaxation data revealed significant restriction of lipid headgroup motions in bilayers upon the addition of pHLIP. Importantly, the increments of correlation time, which quantitatively reported the changes in lipid motion, were considerably larger at intermediate pH-values (i.e., 6.0–7.0 vs. 5.0 or 8.0), supporting stronger peptide-lipid interactions at intermediate states. Lastly, lipid dynamics data suggested that the strongest pHLIP-lipid interactions occurred in bilayer model containing 30 mol % POPG and an additional 15 mol % cholesterol compared with other tested compositions, which correlated with the most significant changes in fluorescence-based pH50.
Author contributions
S.A.O. performed the experiments, collected and analyzed the data, and wrote the article. W.Q. designed the project, analyzed the data, and wrote the article.
Acknowledgments
This work is supported by the National Institutes of Health (R01-GM125853) and the National Science Foundation Major Research Instrumentation Grant (NSF-0922815).
Editor: Charles Deber.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.10.001.
Supporting material
References
- 1.Andreev O.A., Dupuy A.D., et al. Reshetnyak Y.K. Mechanism and uses of a membrane peptide that targets tumors and other acidic tissues in vivo. Proc. Natl. Acad. Sci. USA. 2007;104:7893–7898. doi: 10.1073/pnas.0702439104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reshetnyak Y.K., Segala M., et al. Engelman D.M. A monomeric membrane peptide that lives in three worlds: in solution, attached to, and inserted across lipid bilayers. Biophys. J. 2007;93:2363–2372. doi: 10.1529/biophysj.107.109967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reshetnyak Y.K., Andreev O.A., et al. Engelman D.M. Energetics of peptide (pHLIP) binding to and folding across a lipid bilayer membrane. Proc. Natl. Acad. Sci. USA. 2008;105:15340–15345. doi: 10.1073/pnas.0804746105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreev O.A., Karabadzhak A.G., et al. Reshetnyak Y.K. pH (low) insertion peptide (pHLIP) inserts across a lipid bilayer as a helix and exits by a different path. Proc. Natl. Acad. Sci. USA. 2010;107:4081–4086. doi: 10.1073/pnas.0914330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andreev O.A., Engelman D.M., Reshetnyak Y.K. Targeting acidic diseased tissue: new technology based on use of the pH (low) insertion peptide (pHLIP) Chim. Oggi. 2009;27:34–37. [PMC free article] [PubMed] [Google Scholar]
- 6.Burns K.E., Robinson M.K., Thévenin D. Inhibition of cancer cell proliferation and breast tumor targeting of pHLIP-monomethyl auristatin E conjugates. Mol. Pharm. 2015;12:1250–1258. doi: 10.1021/mp500779k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wyatt L.C., Lewis J.S., et al. Engelman D.M. Applications of pHLIP technology for cancer imaging and therapy. Trends Biotechnol. 2017;35:653–664. doi: 10.1016/j.tibtech.2017.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng C.J., Bahal R., et al. Slack F.J. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Z., Li C., et al. Yan B. pH low insertion peptide mediated cell division cycle-associated protein 1 -siRNA transportation for prostatic cancer therapy targeted to the tumor microenvironment. Biochem. Biophys. Res. Commun. 2018;503:1761–1767. doi: 10.1016/j.bbrc.2018.07.110. [DOI] [PubMed] [Google Scholar]
- 10.Wyatt L.C., Moshnikova A., et al. Reshetnyak Y.K. Peptides of pHLIP family for targeted intracellular and extracellular delivery of cargo molecules to tumors. Proc. Natl. Acad. Sci. USA. 2018;115:E2811–E2818. doi: 10.1073/pnas.1715350115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ai F., Wang N., et al. Zhu G. An upconversion nanoplatform with extracellular pH-driven tumor-targeting ability for improved photodynamic therapy. Nanoscale. 2018;10:4432–4441. doi: 10.1039/c7nr06874c. [DOI] [PubMed] [Google Scholar]
- 12.Gerhart J., Thévenin A.F., et al. Thévenin D. Inhibiting epidermal growth factor receptor dimerization and signaling through targeted delivery of a juxtamembrane domain peptide mimic. ACS Chem. Biol. 2018;13:2623–2632. doi: 10.1021/acschembio.8b00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brito J., Golijanin B., et al. Golijanin D. Ex-vivo imaging of upper tract urothelial carcinoma using novel pH low insertion peptide (variant 3), a molecular imaging probe. Urology. 2020;139:134–140. doi: 10.1016/j.urology.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu W., Deacon J., et al. Glazer P. Tumor-targeted pH-low insertion peptide delivery of theranostic gadolinium nanoparticles for image-guided nanoparticle-enhanced radiation therapy. Transl. Oncol. 2020;13:100839. doi: 10.1016/j.tranon.2020.100839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H.-J., Zhao X., et al. Yan X.-P. pH-driven targeting nanoprobe with dual-responsive drug release for persistent luminescence imaging and chemotherapy of tumor. Anal. Chem. 2020;92:1179–1188. doi: 10.1021/acs.analchem.9b04318. [DOI] [PubMed] [Google Scholar]
- 16.Vāvere A.L., Biddlecombe G.B., et al. Lewis J.S. A novel technology for the imaging of acidic prostate tumors by positron emission tomography. Cancer Res. 2009;69:4510–4516. doi: 10.1158/0008-5472.CAN-08-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An M., Wijesinghe D., et al. Engelman D.M. pH-(low)-insertion-peptide (pHLIP) translocation of membrane impermeable phalloidin toxin inhibits cancer cell proliferation. Proc. Natl. Acad. Sci. USA. 2010;107:20246–20250. doi: 10.1073/pnas.1014403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wijesinghe D., Engelman D.M., et al. Reshetnyak Y.K. Tuning a polar molecule for selective cytoplasmic delivery by a pH (Low) insertion peptide. Biochemistry. 2011;50:10215–10222. doi: 10.1021/bi2009773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies A., Lewis D.J., et al. Pikramenou Z. pH-controlled delivery of luminescent europium coated nanoparticles into platelets. Proc. Natl. Acad. Sci. USA. 2012;109:1862–1867. doi: 10.1073/pnas.1112132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao L., Daniels J., et al. Reshetnyak Y.K. pHLIP®-mediated delivery of PEGylated liposomes to cancer cells. J. Control. Release. 2013;167:228–237. doi: 10.1016/j.jconrel.2013.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N., Yin L., et al. Engelward B.P. Peptide targeting and imaging of damaged lung tissue in influenza-infected mice. Future Microbiol. 2013;8:257–269. doi: 10.2217/fmb.12.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adochite R.-C., Moshnikova A., et al. Reshetnyak Y.K. Targeting breast tumors with pH (low) insertion peptides. Mol. Pharm. 2014;11:2896–2905. doi: 10.1021/mp5002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reshetnyak Y.K. Imaging tumor acidity: pH-low insertion peptide probe for optoacoustic tomography. Clin. Cancer Res. 2015;21:4502–4504. doi: 10.1158/1078-0432.CCR-15-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otieno S.A., Hanz S.Z., et al. Qiang W. pH-dependent thermodynamic intermediates of pHLIP membrane insertion determined by solid-state NMR spectroscopy. Proc. Natl. Acad. Sci. USA. 2018;115:12194–12199. doi: 10.1073/pnas.1809190115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanz S.Z., Shu N.S., et al. Qiang W. Protonation-driven membrane insertion of a pH-low insertion peptide. Angew. Chem. Int.Engl. 2016;55:12376–12381. doi: 10.1002/anie.201605203. [DOI] [PubMed] [Google Scholar]
- 26.Barrera F.N., Weerakkody D., et al. Engelman D.M. Roles of carboxyl groups in the transmembrane insertion of peptides. J. Mol. Biol. 2011;413:359–371. doi: 10.1016/j.jmb.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreev O.A., Engelman D.M., Reshetnyak Y.K. pH-sensitive membrane peptides (pHLIPs) as a novel class of delivery agents. Mol. Membr. Biol. 2010;27:341–352. doi: 10.3109/09687688.2010.509285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyrychenko A., Vasquez-Montes V., et al. Ladokhin A.S. Lipid headgroups modulate membrane insertion of pHLIP peptide. Biophys. J. 2015;108:791–794. doi: 10.1016/j.bpj.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fendos J., Barrera F.N., Engelman D.M. Aspartate embedding depth affects pHLIP’s insertion pKa. Biochemistry. 2013;52:4595–4604. doi: 10.1021/bi400252k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onyango J.O., Chung M.S., et al. An M. Noncanonical amino acids to improve the pH response of pHLIP insertion at tumor acidity. Angew. Chem. Int.Engl. 2015;54:3658–3663. doi: 10.1002/anie.201409770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrera F.N., Fendos J., Engelman D.M. Membrane physical properties influence transmembrane helix formation. Proc. Natl. Acad. Sci. USA. 2012;109:14422–14427. doi: 10.1073/pnas.1212665109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasquez-Montes V., Gerhart J., et al. Ladokhin A.S. Comparison of lipid-dependent bilayer insertion of pHLIP and its P20G variant. Biochim. Biophys. Acta Biomembr. 2018;1860:534–543. doi: 10.1016/j.bbamem.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta C., Ren Y., Mertz B. Cooperative nonbonded forces control membrane binding of the pH-low insertion peptide pHLIP. Biophys. J. 2018;115:2403–2412. doi: 10.1016/j.bpj.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santana H.J.A., Caseli L. A bactericide peptide changing the static and dilatational surface elasticity properties of zwitterionic lipids at the air-water interface: relationship with the thermodynamic, structural and morphological properties. Biophys. Chem. 2021;277:106638. doi: 10.1016/j.bpc.2021.106638. [DOI] [PubMed] [Google Scholar]
- 35.Scott H.L., Nguyen V.P., et al. Barrera F.N. The negative charge of the membrane has opposite effects on the membrane entry and exit of pH-low insertion peptide. Biochemistry. 2015;54:1709–1712. doi: 10.1021/acs.biochem.5b00069. [DOI] [PubMed] [Google Scholar]
- 36.Scott H.L., Heberle F.A., et al. Barrera F.N. Phosphatidylserine asymmetry promotes the membrane insertion of a transmembrane helix. Biophys. J. 2019;116:1495–1506. doi: 10.1016/j.bpj.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karabadzhak A.G., Weerakkody D., et al. Engelman D.M. Bilayer thickness and curvature influence binding and insertion of a pHLIP peptide. Biophys. J. 2018;114:2107–2115. doi: 10.1016/j.bpj.2018.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrera F.N., Poveda J.A., et al. Neira J.L. Binding of the C-terminal sterile alpha motif (SAM) domain of human p73 to lipid membranes. J. Biol. Chem. 2003;278:46878–46885. doi: 10.1074/jbc.M307846200. [DOI] [PubMed] [Google Scholar]
- 39.Royer C.A., Scarlata S.F. Fluorescence approaches to quantifying biomolecular interactions. Methods Enzymol. 2008;450:79–106. doi: 10.1016/S0076-6879(08)03405-8. [DOI] [PubMed] [Google Scholar]
- 40.Scott H.L., Westerfield J.M., Barrera F.N. Determination of the membrane translocation pK of the pH-low insertion peptide. Biophys. J. 2017;113:869–879. doi: 10.1016/j.bpj.2017.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shu N.S., Chung M.S., et al. Qiang W. Residue-specific structures and membrane locations of pH-low insertion peptide by solid-state nuclear magnetic resonance. Nat. Commun. 2015;6:7787. doi: 10.1038/ncomms8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petkova A.T., Tycko R. Sensitivity enhancement in structural measurements by solid state NMR through pulsed spin locking. J. Magn. Reson. 2002;155:293–299. doi: 10.1006/jmre.2002.2519. [DOI] [PubMed] [Google Scholar]
- 43.Jaroniec C.P., Tounge B.A., et al. Griffin R.G. Frequency selective heteronuclear dipolar recoupling in rotating solids: accurate (13)C-(15)N distance measurements in uniformly (13)C,(15)N-labeled peptides. J. Am. Chem. Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 44.Ban X., Lahiri P., et al. Kaustubh B. Evolutionary stability of salt bridges hints its contribution to stability of proteins. Comput. Struct. Biotechnol. J. 2019;17:895–903. doi: 10.1016/j.csbj.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joseph A.P., Srinivasan N., de Brevern A.G. Cis-trans peptide variations in structurally similar proteins. Amino Acids. 2012;43:1369–1381. doi: 10.1007/s00726-011-1211-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masiero A., Nelly L., et al. Catherine P. The impact of proline isomerization on antigen binding and the analytical profile of a trispecific anti-HIV antibody. MAbs. 2020;12:1698128. doi: 10.1080/19420862.2019.1698128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wieprecht T., Beyermann M., Seelig J. Thermodynamics of the coil-alpha-helix transition of amphipathic peptides in a membrane environment: the role of vesicle curvature. Biophys. Chem. 2002;96:191–201. doi: 10.1016/s0301-4622(02)00025-x. [DOI] [PubMed] [Google Scholar]
- 48.Wimley W.C., White S.H. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 49.Yang Y., Yao H., Hong M. Distinguishing bicontinuous lipid cubic phases from isotropic membrane morphologies using (31)P solid-state NMR spectroscopy. J. Phys. Chem. B. 2015;119:4993–5001. doi: 10.1021/acs.jpcb.5b01001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Favela-Rosales F., Galván-Hernández A., et al. Ortega-Blake I. A molecular dynamics study proposing the existence of statistical structural heterogeneity due to chain orientation in the POPC-cholesterol bilayer. Biophys. Chem. 2020;257:106275. doi: 10.1016/j.bpc.2019.106275. [DOI] [PubMed] [Google Scholar]
- 51.Surmeier G., Paulus M., et al. Nase J. Cholesterol modulates the pressure response of DMPC membranes. Biophys. Chem. 2019;252:106210. doi: 10.1016/j.bpc.2019.106210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.