Abstract
Genetics and in vitro studies have shown that the direct interaction between Gal3p and Gal80p plays a central role in galactose-dependent Gal4p-mediated GAL gene expression in the yeast Saccharomyces cerevisiae. Precisely how Gal3p-Gal80p interaction effects induction is not clear. It has been assumed that Gal3p interacts with Gal80p in the nucleus upon galactose addition to release Gal80p inhibition of Gal4p. Although Gal80p has been shown to possess nuclear localization signal (NLS) peptides, the subcellular distribution of neither Gal80p nor Gal3p was previously determined. Here we report that Gal3p is located in the cytoplasm and apparently excluded from the nucleus. We show that Gal80p is located in both the cytoplasm and the nucleus. Converting Gal80p into a nucleus-localized protein (NLS-Gal80p) by exogenous NLS addition impairs GAL gene induction. The impaired induction can be partially suppressed by targeting Gal3p to the nucleus (NLS-Gal3p). We document a very rapid association between NLS-Gal3p and Gal80p in vivo in response to galactose, illustrating that the nuclear import of Gal80p is very rapid and efficient. We also demonstrate that nucleus-localized NLS-Gal80p can move out of the nucleus and shuttle between nuclei in yeast heterokaryons. These results are the first indication that the subcellular distribution dynamics of the Gal3 and Gal80 proteins play a role in regulating Gal4p-mediated GAL gene expression in vivo.
Yeast cells utilize galactose by rapidly activating expression of GAL genes (25, 26, 32, 45). Transcription of GAL genes is dependent on the DNA-binding activator Gal4p (18, 21, 27, 31). When galactose is absent, the transcription activity of the Gal4 protein is inhibited by the Gal80 protein by virtue of the direct interaction of Gal80p with a short sequence within the activation domain (amino acids 768 to 881) of the Gal4 protein (28, 33, 34, 40, 54, 62). Relief of this inhibition state in response to galactose involves the function of the GAL3 gene product (6, 7, 10, 15, 38, 55). In vitro studies have shown that Gal3p directly interacts with Gal80p in a galactose-dependent manner (8, 52, 61, 63). The capacity of Gal3p to bind to Gal80p is linked to Gal4p-mediated GAL gene activation in vivo (8) and in vitro (42). The prevailing view has been that Gal3p-Gal80p interaction gives rise to a conformational change in the Gal4p-Gal80p complex. Recently, in vitro and in vivo experiments have revealed that in the presence of galactose, Gal3p weakens the association of Gal80p with the Gal4p activation domain (50).
Interaction in vivo between Gal3p and Gal80p has been assumed to occur in the nucleus (42, 61, 63). However, the fact that the cytoplasmic galactose pathway enzyme Gal1p is 73% identical to Gal3p (3, 52) and can substitute for Gal3p in galactose-induced GAL gene transcription (5, 36) raised the question of where in the cell, the nucleus, the cytoplasm, or both, the Gal3p-Gal80p association occurs. Gal80p has been shown to possess nuclear localization signals (NLSs) (39), but the subcellular distribution of endogenous Gal80p has not been reported. No NLS is discernible in Gal3p's sequence, and its subcellular distribution has not been reported.
Here we report that Gal3p is located in the cytoplasm and is apparently excluded from the nucleus but that Gal80p is located in both the cytoplasm and the nucleus and exhibits nucleoctyoplasmic shuttling. Alterations of Gal80p and Gal3p localizations have distinct effects on GAL gene induction. These results suggest that the subcellular localization patterns and dynamics contribute to the functions of Gal3 and Gal80 proteins in GAL gene transcriptional control mechanisms in vivo.
MATERIALS AND METHODS
Strains.
Yeast strains used in this study are listed in Table 1. J. E. Hopper laboratory strains used in this study were derived from SJ21R (27), as previously described (8).
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Genotype | Reference or source |
|---|---|---|---|
| Sc722 | MATa ade1 ile leu2-3,112 ura3-52 trp1-HIII his3-Δ1 MEL1 LYS2::GAL1UAS-GAL1TATA-HIS3-URA3 | 8 | |
| Sc723 | MATa ade1 ile leu2-3,112 ura3-52 trp1-HIII his3-Δ1 MEL1 LYS2::GAL1UAS-GAL1TATA-HIS3 | 8 | |
| Sc724 | gal3Δ | MATa ade1 ile leu2-3,112 ura3-52 trp1-HIII his3-Δ1 MEL1 LYS2::GAL1UAS-GAL1TATA-HIS3 gal3Δ | 8 |
| Sc725 | gal80Δ | MATa ade1 ile leu2-3,112 ura3-52 trp1-HIII his3-Δ1 MEL1 LYS2::GAL1UAS-GAL1TATA-HIS3 gal80Δ | 8 |
| Sc726 | gal3Δ gal80Δ | MATa ade1 ile leu2-3,112 ura3-52 trp1-HIII his3-Δ1 MEL1 LYS2::GAL1UAS-GAL1TATA-HIS3 gal3Δ gal80Δ | 8 |
| Sc756 | gal3Δ gal1Δ | MATa ade1 ile leu2-3,112 ura3-52 trp1-HIII his3-Δ1 MEL1 LYS2::GAL1UAS-GAL1TATA-HIS3 gal3Δ gal1Δ | 8 |
| Sc467 | MATa/ MATα ade1/ade1 leu2-3/leu2-3 leu2-112/leu2-112 ura3-52/ura3-52 ile/ile MEL1/MEL1 | This laboratory | |
| BJ2168 | MATa leu2 trp1 ura3-52 prb1-1122 pep4-3 prc1-407gal2 | 29 | |
| MS739 | kar1-1 | MATα ura3-52 leu2-3,112 ade2-101 kar1-1 | 58 |
| JM63-1d | kar1-1 | MATαkar1-1 his4-290 ura1 | J. Haber |
| YM6141 | msn5Δ | MATα ura3-52 ADE2 his3 lys2 met gal80− msn5::HIS3 GAL1-lacZ::LEU2 | M. Johnston |
| W303 xpo1-1 | xpo1-1 | ade2-2 ura3-1 trp1-1 can1-100 his3-11,15 leu2-3,112 xpo::LEU2 pKW457 (xpo1-1 allele in a HIS3 plasmid) | 51 |
| Y1709 | cse1 | MATa ura3-52 ade2-101 his3-11 trp1-Δ901 cse1-1 | 60 |
Plasmids.
Plasmids used in this study are listed in Table 2. Oligonucleotides used in plasmid constructions are listed in Table 3. Details of plasmid constructions and sequence information are available upon request or through our website (http://www.collmed.psu.edu/labs/jhopper/plasmids-info.html). In brief, unique restriction sites were introduced by a PCR-based oligonucleotide-directed method. To introduce simian virus 40 (SV40) large T antigen NLS and NLS mutant (mNLS) peptides (30), oligonucleotides encoding NLS (PKKKRKVGIPQ) or mNLS (PKTKRKVGIPQ; underlining marks the site of mutation) were inserted into unique AatII sites generated in GAL80 or GAL3 genes. The green fluorescence protein (GFP) open reading frame (ORF) was amplified by PCR from pYGFP1 (14) to add appropriate restriction sites for subsequent cloning. All PCRs were carried out with high-fidelity Pfu DNA polymerase (Stratagene, La Jolla, Calif.). Recombinant junctions and PCR-amplified regions except the GFP ORF were verified by DNA sequencing. The final constructs had NLS or mNLS inserted at the protein N termini and/or GFP inserted at the protein C termini.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Reference or source |
|---|---|---|
| pRS414 | CEN6 ARS1 TRP1 | 49 |
| pRS416 | CEN6 ARS1 URA3 | 49 |
| pTEB16 | CEN ARS1 TRP1 GAL3 | 8 |
| pGP15b | CEN ARS1 URA3 GAL80GFP | This study |
| pGP15Δ | CEN ARS1 URA3 GAL80 | This study |
| pGP31 | CEN ARS1 URA3 NLS-GAL80GFP | This study |
| pGP31Δ | CEN ARS1 URA3 NLS-GAL80 | This study |
| pGP31m | CEN ARS1 URA3 mNLS-GAL80GFP | This study |
| pGP31mΔ | CEN ARS1 URA3 mNLS-GAL80 | This study |
| pGP13 | 2μm LEU2 PADH1-GAL80GFP | This study |
| pGP20c | CEN ARS1 TRP NLS-GAL3 | This study |
| pGP20GFP | CEN ARS1 TRP NLS-GAL3GFP | This study |
| pGP25 | CEN ARS1 URA3 PADH2-GAL80GFP | This study |
| pGP25-S0 | CEN ARS1 URA3 PADH2-GAL80S0-GFP | This study |
| pGP25-S1 | CEN ARS1 URA3 PADH2-GAL80S1-GFP | This study |
| pGP25-S2 | CEN ARS1 URA3 PADH2-GAL80S2-GFP | This study |
| pGP26 | CEN ARS1 TRP PADH2-NLS-GAL3 | This study |
| pGP53 | CEN ARS1 URA3 PGAL-CYC-NLS-GAL80GFP | This study |
| pGP10 | XhoI-GFP in pUC19 | This study |
PADH1 and PADH2 designate the ADH1 and the ADH2 promoters, respectively. PGAL-CYC designates the hybrid GAL-CYC promoter. Native promoters were used in all other constructs.
pGP15 has a unique AatII site at the N terminus of Gal80p that allows insertion of NLS or mNLS oligonucleotides; GFP was inserted at a unique NarI site that was previously created at the Gal80p C terminus.
NLS was inserted in pGP20 at a unique AatII site that was previously created at the N terminus of Gal3p.
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Description | Sequence |
|---|---|---|
| GANG15 | NLS, sense | CCCAAAGAAGAAGAGAAAGGTTGGTATTCCAAACGT |
| GANG16 | NLS, antisense | TTGGAATACCAACCTTTCTCTTCTTCTTTGGGACGT |
| GANG43 | mNLS, sense | CCCAAAGACTAAGAGAAAGGTTGGTATTCCAAACGT |
| GANG44 | mNLS, antisense | TTGGAATACCAACCTTTCTCTTAGTCTTTGGGACGT |
| GANG37 | GAL80, BseRI | GTCTTCTGGTCTGTTAGGGCAAG |
| GANG38 | GAL80, AatII, reverse | ACCCGACGTCCATGACGGGAGTGGAAAG |
| GANG39 | GAL80, AatII, forward | ACCCGACGTCGACTACAACAAGAGATCTTCGGTC |
| GANG40 | GAL80, EcoRI | GCTCATCTATCAGCTCTTGCTC |
| GANG7 | GFP, ClaI, forward | CCCATCGATTGATGTCTAAAGGTGAAG |
| GANG8 | GFP, ClaI, reverse | CCCATCGATATTTGTACAATTCATCCATAC |
Localization of Gal3p by indirect immunofluorescence.
Sc756 (gal3Δ gal1Δ) cells were transformed with pTEB16, which bears the gene encoding wild-type Gal3p. Transformants were grown to early log phase in medium containing 3% glycerol–2% lactic acid–0.05% glucose. Galactose was added at 2%, and the cells were incubated for 2 to 6 h. Cells were then fixed in 3.7% formaldehyde for 1 h, and indirect immunofluorescence experiments were carried out as described previously (44), with modifications previously reported (23). Rabbit anti-Gal3p serum was preabsorbed with Sc756 cells (gal3Δ gal1Δ) harboring pRS414 (49) at a 1:50 dilution in phosphate-buffered saline–1% bovine serum albumin for 4 h and then cleared by centrifugation at 50,000 rpm in a Beckman type 100 Ti rotor for 15 min, both of which were performed at 4°C. Preabsorbed and cleared anti-Gal3p antibody was used at dilutions of 1:100 to 1:400, and then fluorescein isothiocynate (FITC)-labeled anti-rabbit antibody was used at dilutions of 1:200 to 1:400. For double-labeling experiments, a 1:40,000 dilution of mouse monoclonal antibody 32D6 specific to Nsp1p (53) was simultaneously used with anti-Gal3p antibody and then with Texas red-labeled anti-mouse (1:400) antibody and FITC-labeled anti-rabbit antibody (1:400).
Localization of the Gal80GFP derivative.
Sc725 (GAL80Δ) cells were transformed with GAL80 GFP derivatives. Transformants were grown to early log phase in medium containing 3% glycerol–2% lactic acid–0.05% glucose. Galactose was added at 2%, and the cells were cultured for an additional 2 to 6 h. Cells were then spotted onto slides without further manipulation.
To costain nuclei with DAPI (4′,6′-diamidino-2-phenylindole) in living yeast cells, we used diploid strain Sc467, since Sc725 nuclei can not be readily stained by DAPI in the medium. Sc467 cells were transformed with pGP13 (PADH1-GAL80GFP). Transformants were grown to mid-exponential phase in medium containing 2% glucose. The cells were then diluted 1:10 to 1:50 in medium containing 2% glucose and 0.1 μg of DAPI per ml and cultured for about 4 to 6 h. After DAPI staining of nuclei, the cells were resuspended in medium containing glucose, galactose, or glycerol-lactic acid as the carbon sources prior to observation.
Microscopy.
Fluorescence signals were observed using a Nikon Optiphot-2 epifluorescence microscope equipped with 60× and 100× objectives. GFP was excited, and signal was detected using a Chroma Technology Corp. (Brattleboro, Vt.) filter (no. 41017). Yeast cells were observed through a Nikon differential inference contrast (DIC) attachment set. Images were acquired using a SenSys KF1400 charge-coupled device camera (Photometrics, Ltd., Tucson, Ariz.), driven by QED (Pittsburgh, Pa.) software. The final figures were prepared using Adobe Photoshop and Deneba Canvas.
Cell fractionation and nuclei enrichment.
The protease-deficient yeast strain BJ2168 (29) was grown to early exponential phase in yeast extract-peptone medium containing 3% glycerol–2% lactic acid–0.05% glucose. Galactose was added at 2%, and the cells were incubated for two additional hours. Cell fractionation and nuclei enrichment were carried out using two independent protocols, one based on a sucrose gradient method (24, 48) and the other based on a Ficoll gradient method (2, 16). Both procedures gave similar results, but a better yield for nuclei was obtained by the Ficoll gradient method.
For the cytoplasmic fraction, BJ2168 spheroplast homogenate was spun twice at 80,000 rpm in a Beckman type 100 Ti rotor for 20 min at 4°C. The supernatant was used as the cytoplasmic fraction. Proteins from the nuclear fraction and the cytoplasmic fraction were precipitated with 9 volumes of 11% trichloroacetic acid and then dissolved and boiled in 1× electrophoresis loading buffer.
Enzyme activity determination.
α-Galactosidase enzyme activities were determined by spectrometric assay as previously described (43).
Yeast heterokaryon assay.
A test of protein shuttling by a yeast heterokaryon assay was carried out essentially as described previously (17). In brief, Sc725 (gal80Δ) was transformed with pGP53 (pGAL-SVGal80GFP). For a control, Sc723 was transformed with pGAL-H2B-GFP (17). Transformants were grown to mid-exponential stage in medium containing 2% glucose, diluted, and grown overnight to a density of about 0.5 × 107 to approximately 1.0 × 107 cells/ml in medium containing 2% glycerol–3% lactic acid. Galactose was added to 2%, and the cells were grown for an additional 2 to approximately 3 h. Cells were then washed twice and resuspended in medium containing 2% glucose and grown for 3 h. For most experiments, matings were initiated by mixing 3 × 106 cells with an equal number of MS739 (a kar1-1 strain [58]) cells by concentrating the cells on a 25-mm-diameter, 0.45-μm-pore-size nitrocellulose filter and placing the filter on a yeast extract-peptone-dextrose plate. After 1 or 2 h of incubation, cells were washed off the membrane using medium containing glucose and spotted on a slide for observation. For cells to be simultaneously used for RNA analysis, approximately 108 cells were mixed with an equal number of MS739 cells and concentrated on a 82-mm-diameter filter.
Northern blotting.
A Northern blot analysis was carried out according to standard protocols (11). Total RNA yeast samples were prepared from cells subjected to induced, repressed, or mating conditions as described above under “Yeast heterokaryon assay.” A 700-bp XhoI fragment from pGP10 containing the GFP ORF and a 1.1-kb BamHI fragment from pGEM-ACT1 (57) containing the ACT1 ORF were radiolabeled with [α-32P]dCTP by the random-priming method using a NEBlot kit (New England Biolabs, Beverly, Mass.) and used as probes.
RESULTS
Gal3p is a cytoplasmic protein.
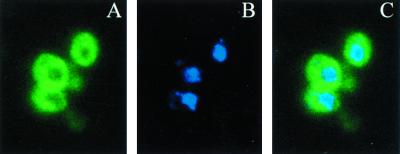
We employed indirect immunofluorescence to determine the subcellular location of Gal3p. The anti-Gal3p antiserum detects only Gal3p, as cells with Gal3p deleted (Sc756 [gal3Δ gal1Δ] cells harboring the vector pRS414 alone) did not yield an immunofluorescence signal (data not shown). In contrast, galactose-induced Sc756 cells carrying GAL3 expressed from the native GAL3 promoter on a single-copy plasmid (pTEB16) had a distinct Gal3p signal (Fig. 1). Surprisingly, all detectable Gal3p was located in the cytoplasm (Fig. 1A). Nuclear regions defined by DAPI staining of DNA (Fig. 1B) or by use of the Nsp1p-specific monoclonal antibody (53) (not shown) were devoid of the Gal3p signal (Fig. 1C). The results indicate that Gal3p is located in the cytoplasm and that it may be excluded from the nucleus.
FIG. 1.
Gal3p is located in the cytoplasm and apparently excluded from the nucleus. Sc756 (gal3Δ gal1Δ) cells carrying plasmid pTEB16 (GAL3) were induced with 2% galactose for 3 h and prepared for immunofluorescence. Cells were stained with anti-Gal3p antibodies followed by FITC anti-rabbit antibody (A), anti-Nsp1p monoclonal antibody 32D6, Texas red anti-mouse antibody (not shown), and DAPI (B). A converged image is shown in panel C.
Gal80GFP is located in both the nucleus and the cytoplasm.
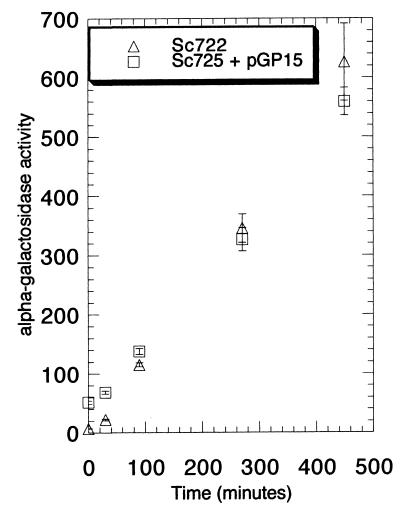
Although Gal80 peptides were previously shown to localize β-galactosidase to the nucleus (39), no localization studies of functional Gal80p have been carried out. We investigated the location of Gal80p in the living cells by tagging Gal80p with GFP (14). The Gal80GFP was determined to be fully functional by a histidine reporter plate assay (8; see details below) and a MEL1 (α-galactosidase) induction kinetics assay (Fig. 2).
FIG. 2.
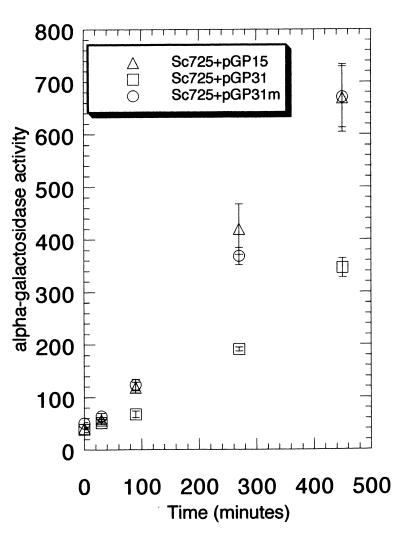
Cells carrying Gal80GFP exhibit normal MEL1 gene (α-galactosidase) induction kinetics in response to galactose. Sc722 cells or Sc725 (gal80Δ) cells carrying pGP15 (Gal80GFP) were grown in glycerol-lactic acid medium to an optical density at 600 nm of ∼0.2. Galactose was then added to a final concentration of 2%. At the indicated time points, an aliquot of cells was removed and processed for α-galactosidase enzyme activity assay. Activities are expressed as picomoles of p-nitrophenyl-α-d-galactopyranoside released per milligram of protein per minute at 30°C. Error bars show standard deviations. To take into account errors generated from the protein concentration estimation and the enzymatic activity assay, the standard deviations were calculated based on a linear approximation method for error transmission in a nonlinear function of several variables (9).
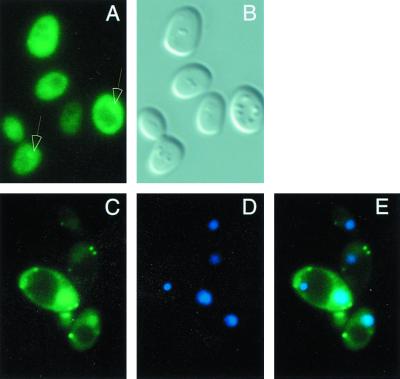
Sc725 (gal80Δ) cells carrying GAL80GFP expressed from the native GAL80 promoter on a single-copy plasmid (pGP15) were grown in medium containing glycerol–lactic acid to early log phase and then induced with galactose for 3 h. Direct fluorescence microscopy showed Gal80GFP as a diffuse signal throughout the cell, with the nuclear signal (Fig. 3A) being slightly more intense than the surrounding cytoplasm signal in some cells (Fig. 3A and B). We also expressed Gal80GFP from a truncated 410-bp ADH1 promoter (4, 47, 56) on a 2μm plasmid (pGP13). This truncated promoter yielded moderately higher levels of Gal80GFP than those from the native GAL80 promoter. In a number of cells, the level of Gal80GFP was higher than in others. The nuclear signal was distinct from the cytoplasmic signal (Fig. 3C to E) when Gal80GFP was overproduced in these cells (this stronger nuclear accumulation might have been due to the overexpression of Gal80GFP and, possibly, relative to the levels of Gal3p). We conclude that the functional Gal80p derivative Gal80GFP is located in both the cytoplasm and the nucleus.
FIG. 3.
Gal80GFP is located in both the cytoplasm and the nucleus. Sc725 (gal80Δ) cells carrying pGP15 (Gal80GFP) were induced with 2% galactose for 3 h (A and B), or Sc467 cells carrying pGP13 (PADH1-Gal80GFP) were grown in medium containing 2% glucose and stained with DAPI (C to E). Cells were then spotted on the slide and observed directly through a GFP (A and C) or DIC (B) filter set. (D) DAPI staining; (E) converged image. Arrows point to the fluorescent nuclear signal.
Gal80p fractionates as both a cytoplasmic and a nuclear protein, whereas Gal3p fractionates as a cytoplasmic protein.
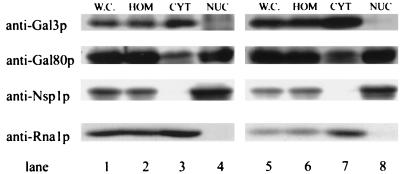
Cell fractionation was carried out to independently assess the subcellular localization of endogenous Gal3 and Gal80 proteins. The protease-deficient stain BJ2168 (29) was used to minimize protein degradation during the cell fractionation process. The nucleus-specific antibody 32D6 (a mouse monoclonal antibody against Nsp1p [53], a component of the nuclear pore complex [46]) and the cytoplasm-specific anti-Rna1p antibody (23) were used to monitor the cross-contamination between the nuclear and the cytoplasmic fractions. Representative results are shown in Fig. 4. Gal3p and Gal80p were present in the whole-cell extracts (lanes 1 and 5) and the spheroplast homogenate (lanes 2 and 6). Gal3p was enriched in the cytoplasmic fraction (lanes 3 and 7) but was not present in the nuclear fraction (lanes 4 and 8). In contrast, Gal80p was present in both the cytoplasmic and nuclear fractions (lanes 3 and 7 and lanes 4 and 8). The amounts of Gal80p were comparable between the spheroplast homogenate and the nuclear fractions, but the amount was somewhat lower in the cytoplasmic fraction. Since the cytoplasmic fraction was the supernatant from ultracentrifugation, the lower level of Gal80p in this fraction might reflect some loss in the pellet. Alternatively, there may be less Gal80p in the cytoplasm than in the nucleus. These subcellular fractionation results closely mirror the microscopy results and point to a marked difference in the subcellular distributions of the Gal80 and Gal3 proteins.
FIG. 4.
Endogenous Gal80p fractionates as both a cytoplasmic and a nuclear protein, whereas Gal3p fractionates as a cytoplasmic protein. Lanes 1 to 4 each received 50 μg of protein from cells grown in glycerol-lactic acid medium; lanes 5 to 8 each received 25 μg of protein from cells induced with galactose for 2 h. Exposure times for different blots were different. W.C., whole-cell extract; HOM, homogenate; CYT, cytoplasm fraction; NUC, enriched-nucleus fraction.
Targeting Gal80p to the nucleus compromises GAL gene induction.
Since Gal80p is both nuclear and cytoplasmic and Gal3p is cytoplasmic, we addressed whether these distinct subcellular distributions have physiological relevance. To do so, we altered the normal distribution profile of Ga80 and/or Gal3 proteins and determined whether altered cellular distribution affects GAL gene induction. We took advantage of the well-characterized SV40 large T antigen NLS (30). This NLS works efficiently in yeast, and a single amino acid mutation abolishes its NLS function (for an example, see reference 37). Insertion of this NLS causes Gal80GFP (NLS-Gal80GFP) to be localized in the nucleus (Fig. 5A to C), while Gal80GFP with the mutant form of this NLS (mNLS-Gal80GFP) shows a signal throughout the cell (Fig. 5D to F) similar to that of the wild-type Gal80GFP (Fig. 3A to C).
FIG. 5.
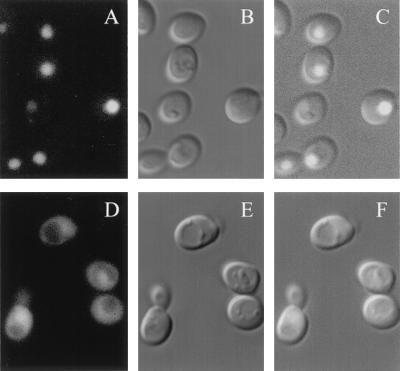
NLS-Gal80GFP is located in the nucleus, while Gal80GFP with the mutant form of the NLS is located in both the cytoplasm and the nucleus. Sc725 (gal80Δ) cells carrying either pGP31 (NLS-Gal80GFP) (A to C) or pGP31m (mNLS-Gal80GFP) (D to F) were grown to late exponential phase in medium containing glucose and then streaked on plates containing galactose-glycerol-lactic acid. Plates were incubated at 30°C for 2 days. Cells were then transferred to liquid medium containing galactose, spotted on slides, and observed through a GFP or DIC filter set.
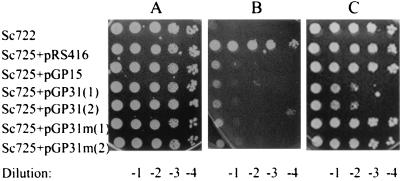
Two methods were used to assess the effects of NLS-altered Gal80GFP on GAL gene induction. First, we evaluated the gene expression of the galactose-inducible HIS3 reporter (having four GAL upstream activation sequence [UASGAL] sites on its promoter) by a plate-based colony growth assay (8). Sc725 (gal80Δ) has a chromosomal insertion of this PGAL1-HIS3 reporter that allows HIS3 transcription to be regulated by the GAL gene induction mechanism. Growth of Sc725 cells carrying different Gal80p derivatives on synthetic complete (SC) medium containing galactose but lacking histidine allowed us to assess the function of the Gal80p derivatives (Fig. 6). Sc725 harboring NLS-Gal80GFP showed markedly slower growth than the cells carrying either Gal80GFP or mNLS-Gal80GFP (Fig. 6C). Levels of growth of these cells on SC medium containing histidine were comparable (Fig. 6A and data not shown). Results indistinguishable from these were obtained in experiments using GAL80 constructs without GFP tags (data not shown). Second, we determined the induction kinetics for the expression of the galactose-inducible MEL1 gene (which has a single low-affinity UASGAL site on its promoter) by performing α-galactosidase enzyme activity assays (43). Cells harboring Gal80GFP or mNLS-Gal80GFP exhibited similar levels of MEL1 induction, whereas cells harboring NLS-Gal80GFP exhibited twofold lower induction (Fig. 7).
FIG. 6.
NLS-Gal80p causes slower cell growth in a HIS3 reporter plate assay. A plasmid containing either no Gal80p (pRS416), wild-type Gal80GFP (pGP15), NLS-Gal80GFP (pGP31), or Gal80GFP with the mutant form of the NLS insertion (pGP31m) was transformed into Sc725 (gal80Δ) cells which have the PGAL1-HIS3 reporter cassette integrated in the genome. Sc722 is the parental strain for Sc725 and was used as the wild-type GAL80 control. Transformants and Sc722 cells were grown to late log phase in SC medium lacking uracil containing 2% glucose. The cell density was determined and adjusted to 5 × 107 cells/ml. Serial 10-fold dilutions were made with SC medium lacking uracil and histidine. Eight-microliter samples of 100, 10−1, 10−2, 10−3, and 10−4 dilutions of each cell culture were then spotted onto solid growth media: SC medium lacking uracil plus 2% glucose (A), SC medium lacking uracil and histidine plus glycerol-lactic acid plus 10 mM 3-AT (3-amino-1,2,4-triazole) (B), and SC medium lacking uracil and histidine plus 2% galactose-glycerol-lactic acid plus 10 mM 3-AT (C). These plates were then incubated at 30°C for 3 to 6 days. The result on day 6 is shown here. Cells were also spotted on plates with SC medium lacking uracil plus glycerol-lactic acid. No growth differences were observed on those plates (data not shown).
FIG. 7.
NLS-Gal80p markedly reduces galactose-inducible MEL1 gene expression. Induction kinetics for Sc725 (gal80Δ) cells carrying pGP15, pGP31, or pGP31m was determined as described for Fig. 2. Two independent experiments were carried out. The values from these two independent experiments varied by <10%.
Compromised GAL gene induction conferred by NLS-Gal80p can be partially suppressed by NLS-Gal3p.
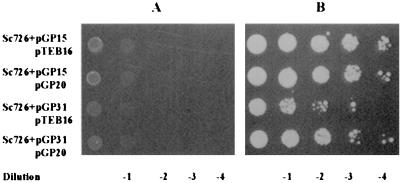
If the poor induction phenotype conferred by NLS-Gal80p is due to the physical separation of the now nucleus-localized NLS-Gal80p and the cytoplasm-localized Gal3p, we ought to be able to enhance induction by targeting Gal3p to the nucleus. To test this prediction, we constructed an NLS-Gal3p derivative that performed like wild-type Gal3p in the HIS3 reporter plate assay (data not shown; see below also). NLS-Gal3p is located in the nucleus (Fig. 8). We found repeatedly that the presence of both NLS-Gal3p and NLS-Gal80p partially restored the growth rate of cells, as evaluated by the PGAL1-HIS3 reporter plate assay (Fig. 9).
FIG. 8.
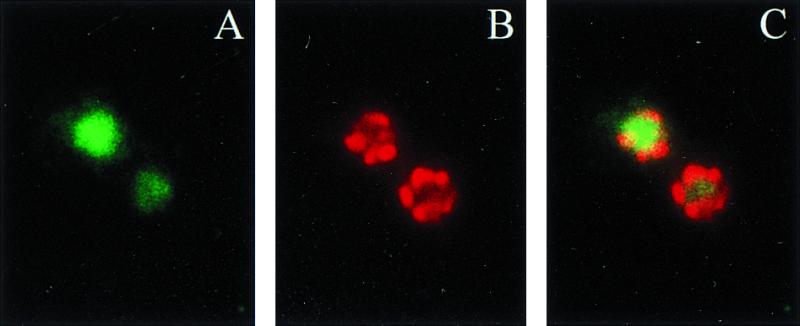
NLS-Gal3p is located in the nucleus. Sc756 (gal3Δ gal1Δ) cells carrying plasmid pGP20 (NLS-Gal3p) were induced with 2% galactose for 3 h and prepared for immunofluorescence, as described for Fig. 1. Cells were stained with anti-Gal3p antibodies, followed by consecutive staining with FITC anti-rabbit antibody (A), anti-Nsp1p monoclonal antibody 32D6, Texas red anti-mouse antibody (B), and then DAPI (not shown). A converged image is shown in panel C. A 100× objective was used.
FIG. 9.
NLS-Gal3p partially suppresses the poor induction caused by NLS-Gal80p. Sc726 cells (gal3Δ gal80Δ) carrying either wild-type Gal80GFP (pGP15) and wild-type GAL3 (pTEB16), pGP15 and NLS-Gal3p (pGP20), NLS-Gal80p (pGP31) and pTEB16, or pGP31 and pGP20 were assayed for HIS3 reporter gene expression as described for Fig. 6, except SC medium lacking uracil, tryptophan, and histidine was used. The result on day 4 is shown. Cells spotted on plates containing SC medium lacking uracil and tryptophan plus 2% glucose plates did not have any growth differences (data not shown). Minus galactose in the medium; (B) plus galactose in the medium.
Gal80p and NLS-Gal3p associate rapidly in vivo in response to galactose, and Gal80p is able to quickly enter the nucleus.
The preceding results indicate that Gal3p must have access to Gal80p for normal induction and that Gal80p-Gal3p interaction may normally occur in the cytoplasm. Since the association of Gal80p with Gal4p at the UASGAL sites within the GAL promoters presumably is the ultimate target for induction, there must then be a mechanism(s) to transmit the effect of the Gal3p-Gal80p interaction into the nucleus. The mechanism might involve the dual cytoplasmic and nuclear location of Gal80p.
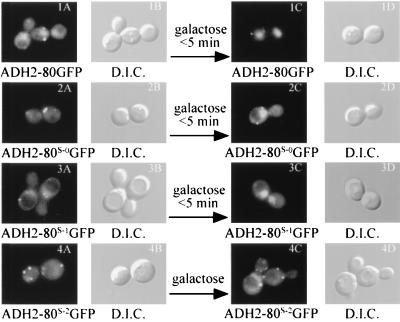
To address this issue, we analyzed the Gal80p subcellular distribution in more detail in the living cells. We utilized the Gal80GFP and NLS-Gal3p derivatives. Both were expressed under the control of the ADH2 promoter. Sc726 (gal3Δ gal80Δ) cells carrying both derivatives were cultured in glycerol-lactic acid medium, in which case the cell nucleus was loaded with NLS-Gal3p (not probed for in the experiment whose results are shown in Fig. 10, but documented in Fig. 8), and Gal80GFP was found to be in both the nucleus and the cytoplasm (Fig. 10, panels 1A and B). In less than 5 min after galactose addition, all detectable Gal80GFP was located in the nucleus (Fig. 10, panels 1C and D). In contrast, the GFP derivative of the GAL80S2 mutant protein, known to have little or no interaction with Gal3p (61), did not shift location (Fig. 10, panels 4A to D). The GFP derivatives of two other GAL80S mutant proteins, which have been shown to interact with Gal3p, but at reduced levels compared to that of wild-type Gal80p (M. P. Woods, A. K. Sil, and J. E. Hopper, unpublished data), showed translocation to the nucleus, albeit to a lesser degree than did Gal80GFP (Fig. 10, panels 2C and D and panels 3C and D). These results suggest that the nuclear import of Gal80p may be very fast and is apparently efficient. In addition, these results illustrate for the first time in living cells the rapid association of Gal3p and Gal80p in response to galactose.
FIG. 10.
The Gal80GFP derivatives can undergo rapid translocation in the cell. Sc726 cells (gal3Δ gal180Δ) carrying pGP26 and either pGP25 (1A to D), pGP25-S0 (2A to D), pGP25-S1 (3A to D), or pGP25-S2 (4A to D) were grown to early exponential phase in SC medium lacking tryptophan and uracil, containing glycerol-lactic acid. For each culture, 180 μl was withdrawn and added to 20 μl of 20% galactose. After being quickly mixed by pipetting, cells were spotted on slides and observed through a GFP (A and C) or DIC (B and D) filter set. For Sc726 with pGP26 and pGP25-S2, translocation was not observed even after >60 min of incubation in medium containing galactose. ADH2 designates the ADH2 promoter.
In these experiments, stronger nuclear accumulation of Gal80GFP was apparent in the glycerol-lactic acid-grown cells (Fig. 10, panels 1A and B). We attribute this to the natural intrinsic low-level interaction between Gal80p and Gal3p in the absence of galactose that has been documented elsewhere (52, 61). Consistent with this, none of the three GAL80S mutant GFP derivatives gave rise to stronger nuclear-signal accumulation in the absence of galactose.
NLS-Gal80p shuttles between nuclei in yeast heterokaryons.
Rapid movement of Gal80p from the cytoplasm to the nucleus indicates that the two peptides, which target the β-galactosidase fusion protein to the nucleus (39), function as efficient NLSs in native Gal80p conformation. This notion, taken together with our result showing the dual nuclear and cytoplasmic location of Gal80p, led us to test whether Gal80p can be exported out of the nucleus.
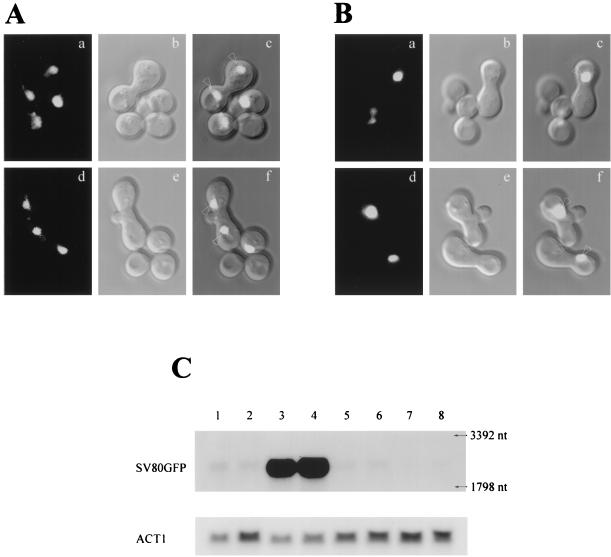
We performed a kar1-1 cross (13) to determine if nucleus-localized Gal80p molecules are able to move out of one nucleus and into another via the cytoplasm in yeast heterokaryons as described before for nucleocytoplasmic shuttling proteins (17). We placed NLS-GAL80GFP under the control of a GAL-CYC promoter (20) (pGP53) to distinguish nucleus-localized Gal80p and to regulate NLS-Gal80GFP synthesis during the experimental procedure. To test for shuttling, Sc725 (gal80Δ) was transformed with pGP53. Nuclei of the transformants were loaded with NLS-Gal80GFP by galactose induction followed by glucose repression. The cells were then mated with MS739, a kar1-1 strain (58), on a yeast extract-peptone-dextrose plate. Formation of heterokaryons was assessed by their characteristic morphology (13). With the protocol and the strains (Sc725 × MS739) used in this study, heterokaryons started to appear about 1 h after mixing of the mating parents (Fig. 11A, images a to c, and 11B, panels a to c). Zygotic buds were formed in some heterokaryons at approximately 2 h after mixing (Fig. 11A, images d to f, and 11B, panels d to f).
FIG. 11.
NLS-Gal80GFP shuttles between nuclei in yeast heterokaryons. (A) Sc725 cells (gal80Δ) harboring pGP53 (PGAL-CYC-NLS-Gal80GFP) were mated with MS739 cells (kar1-1) (panel a to c) 1 h after mixing of parents cells; (panel d to f) 2 h after mixing of parents cells; (panels a and d) GFP filter set; (panels b and e) DIC filter set; (panels c and f) converged images. Arrows indicate heterokaryon nuclei. (B) Sc723 cells harboring pGAL-H2B-GFP were mated with MS739 cells. Other conditions were the same as those described for panel A. (C) Northern blot analysis for the NLS-GAL80GFP mRNA expression time course during the yeast heterokaryon assay. SV80GFP designates the NLS-GAL80GFP transcripts; ACT1 designates the ACT1 transcripts. Total RNA was prepared from Sc725 cells harboring pGP53 grown in media containing glycerol-lactic acid (lane 1), containing glucose (lane 2), induced with 2% galactose for 1.5 h (lane 3), induced with galactose for 3 h (lane 4), repressed with 2% glucose for 1.5 h (lane 5), repressed with glucose for 3 h (lane 6), mated with MS739 cells for 1 h (lane 7), and mated with MS739 cells for 2 h (lane 8). Thirty micrograms of total RNA was loaded in each lane. Yeast rRNAs served as size markers. After being probed for NLS-GAL80GFP transcripts, the membrane was stripped and probed for ACT1 transcripts. nt, nucleotide.
NLS-Gal80GFP molecules originally present in Sc725 nuclei (see below) were distributed in both nuclei in Sc725 × MS739 heterokaryons (Fig. 11A), indicating that NLS-Gal80GFP moved out of the original nuclei and into the other via the cytoplasm of the heterokaryon. The experiments were repeated three times. In over 100 heterokaryons with distinct GFP signals observed, NLS-Gal80GFP was distributed in both nuclei. There were two exceptions. A cell with the morphology of a heterokaryon had a GFP signal in one nucleus, perhaps due to a low nuclear-fusion rate (∼1% for the MS739 cross [58]) or to the cell being in the stage prior to cytoplasmic fusion (both cells were in the early mating stage, as assessed by their morphology). An independent kar1-1 strain (JM63-1b, gift from J. Haber) was also used and yielded a similar result (not shown). To ensure that the appearance of NLS-Gal80GFP in both heterokaryon nuclei was not due to transient nuclear fusion or leaky nuclei, we conducted parallel experiments using pGAL-H2B-GFP. This plasmid bears the genes that encodes a GFP-tagged histone, H2B, which should not exhibit shuttling (17). We observed no instances of shuttling in these parallel experiments (Fig. 11B).
To confirm that the NLS-Gal80GFP detected in heterokaryons in Fig. 11A originated from nucleus-localized NLS-Gal80GFP synthesized during the galactose induction in Sc725 cells, rather than during the mating in heterokaryons, we determined NLS-GAL80GFP mRNA levels through the time course of the heterokaryon assay (Fig. 11C). NLS-GAL80GFP transcripts were present at a very low level when cells were grown in media containing glycerol-lactic acid or glucose (Fig. 11C, lanes 1 and 2), and GFP fluorescence was not observable with the low levels of NLS-GAL80GFP mRNA in these cells (data not shown). Galactose induction resulted in high levels of NLS-GAL80GFP mRNA in 1.5 to 3 h (Fig. 11C, lanes 3 and 4), and distinct GFP fluorescence was observed as a signal restricted to the nucleus (data not shown, but results were similar to those shown in Fig. 5A to C). By 1.5 and 3 h after galactose removal and glucose repression, NLS-GAL80GFP transcripts were down to very low levels (Fig. 11C, lanes 5 and 6) and GFP florescence was restricted to the nucleus without a significant decrease in signal strength (data not shown). The kinetics of NLS-GAL80GFP transcript decay during glucose repression was consistent with the apparent half-life of GAL80 mRNA determined by a RNA polymerase II temperature shutoff procedure (the half-life is about 16 min; see http://web.wi.mit.edu/young/expression/halflife.html [22]). Finally, NLS-GAL80GFP transcripts were maintained at very low levels in cells during mating (Fig. 11C, lanes 7 and 8).
DISCUSSION
The signal transduction pathway and gene transcription control mechanisms of galactose-induced GAL gene expression have been studied intensively. Now that it is established that Gal3p and Gal80p interaction plays a vital role in GAL gene induction (8, 52, 59, 61, 63), the immediate question is how Gal3p-Gal80p interaction triggers the relief of Gal80p inhibition of Gal4p. In this study, we have addressed this question from a cell biology perspective.
The indirect immunofluorescence and cell fractionation experiments presented here demonstrate that Gal3p is located in the cytoplasm and apparently excluded from the nucleus. This perhaps could have been anticipated. Gal3p has strikingly high similarity to Gal1p (3, 52), the galactose kinase of the yeast galactose pathway, presumably a cytoplasmic protein. Gal1p can substitute for Gal3p inducing function (5, 36), and overproduction of either Gal3p or Gal1p can cause GAL gene expression in the absence of galactose (6).
The nucleus has been implicated as the location where the effective Gal3p and Gal80p interaction occurs (42, 52, 61). Our data showing cytoplasmic localization of Gal3p, however, point out that this may not be the case. Based on these data, we cannot exclude the formal possibility that a small undetectable fraction of Gal3p or Gal1p resides in the nucleus. Nevertheless, it seems unlikely that a small fraction of Gal3p or Gal1p should be sufficient to effect Gal4p activation. It has been shown in vitro that a 20- to 30-fold molar excess of Gal3p over the amount of Gal80p is required to alleviate Gal80p's inhibition on Gal4p (42, 59). And yet, simply increasing the level of Gal3p in the nucleus, as we did by targeting Gal3p to the nucleus by NLS addition, did not alter the GAL gene expression regulation in cells containing native Gal80p. In addition, our preliminary data showed that the subcellular location of the GAL3C mutant proteins (8) was indistinguishable from that of the wild-type Gal3p (data not shown). Therefore, it seems unlikely that Gal3p's distribution alone is the critical feature in the GAL gene induction mechanism. Rather, the significance of Gal3p subcellular distribution appears to derive from its relation to the subcellular distribution and dynamics of Gal80p.
Gal80p has been shown to possess an NLS (39). Yet, here we show with living yeast cells that a fully functional Gal80 GFP derivative is located in both the cytoplasm and the nucleus. Subcellular fractionation experiments confirm that native Gal80p is present in both the cytoplasm and the nucleus. That this steady-state distribution of Gal80p stems simply from the relatively small size of Gal80p (48 kDa) seems unlikely. In principle, proteins smaller than about 40 kDa can passively cross the nuclear envelope through the aqueous channels formed by the nuclear pore complex in yeast. In practice, however, very few proteins do so (12, 35). Moreover, the fully functional Gal80GFP we have utilized in this study is too large (about 70 kDa) for passive diffusion to occur.
The subcellular distribution of Gal80p to both the nucleus and the cytoplasm, as we observed here, is very likely a result of nucleocytoplasmic shuttling processes. The nuclear import of Gal80p is very rapid and seemingly very efficient (Fig. 10). This indicates that NLS peptides in the Gal80p sequence (39) are functional in the native Gal80p conformation. Consistently, nucleus-localized Gal80p molecules can shuttle between nuclei in yeast heterokaryons (Fig. 11), indicating that Gal80p can be exported out of the nucleus. Taken together, these data suggest that Gal80p is a nucleocytoplasmic shuttling protein. We have tested three known protein nucleocytoplasmic export factors, Msn5p, Cse1p, and Crm1p (1, 19, 35, 41), for their effects on the distribution of Gal80p. Our preliminary results indicated that apparently none alters Gal80p localization (data not shown). The mechanism(s) for Gal80p shuttling requires further investigation.
Our data showing that NLS-Gal80p impairs GAL induction indicate that the subcellular distribution dynamics of Gal80p is an intrinsic feature of GAL gene induction mechanisms. Alteration of Gal80p subcellular distribution dynamics had a marked effect on transcription of a GAL gene having either high-affinity (in the PGAL-HIS3 reporter) or low-affinity (in the MEL1 gene) Gal4p binding sites. More remarkably, the cellular level of NLS-Gal80p was determined by Western blotting to be about two- to approximately threefold lower than that of native Gal80p (data not shown). This indicates that NLS-Gal80p is a more potent inhibitor of GAL gene transcription than wild-type Gal80p, presumably due to the predominant nuclear location of NLS-Gal80p. These data strengthen the notion that the location of endogenous Gal80p affects GAL gene transcription control. The fact that NLS-Gal80p did not completely prevent induction does not distract us from this notion, since NLS addition most likely serves only to alter rather than to eliminate the dynamics of Gal80p subcellular distribution. That is, NLS-Gal80p is not completely inaccessible to cytoplasm-located Gal3p. Indeed, we demonstrated that NLS-Gal80GFP molecules were able to move out of the nucleus and access the cytoplasm in yeast heterokaryons.
The impaired GAL gene induction caused by NLS-Gal80p may be partially suppressed by targeting Gal3p to the nucleus. This result suggests that the subcellular distribution and dynamics of Gal80p and Gal3p contribute to their function in the GAL gene induction mechanism. That the suppression was only partial leads us to hypothesize that a cytoplasmic localization for Gal3p is required for it to most effectively perform its function relative to that of Gal80p.
In summary, these findings implicate subcellular localization patterns and dynamics of Gal3 and Gal80 proteins as intrinsic properties of the GAL gene induction mechanisms in vivo. To integrate these new properties to explain how Gal3p-Gal80p interaction affects Gal80p-Gal4p complexes in vivo, three possibilities are currently being considered. One is that a small fraction of cellular Gal3p enters the nucleus and destabilizes the Gal80p-Gal4p association in the presence of galactose (nuclear entry). Another possibility is that in response to galactose, Gal80p interacts with Gal3p in the cytoplasm, causing accumulation of Gal80p in the cytoplasm (cytoplasmic trapping). A third possibility is that cytoplasm-located Gal3p interacts with Gal80p in the presence of galactose, resulting in modified Gal80p having an altered affinity for Gal4p (cytoplasmic modification). This third possibility takes into consideration the recent discovery that KlGal80p, the S. cerevisiae Gal80p ortholog and functional counterpart in the yeast Kluyveromyces lactis, is subjected to KlGal1p-dependent, carbon source-regulated dephosphorylation (64). Further investigation will address the mechanisms for Gal80p shuttling, to what extent the shuttling of Gal80p affects GAL gene induction, whether Gal80p is a modified protein, and whether Gal3p exerts its induction function solely in the cytoplasm.
ACKNOWLEDGMENTS
We thank S. Alam, J. P. Aris, B. P. Cormack, T. Fukasawa, J. Haber, P. Heiter, A. K. Hopper, M. Johnston, E. W. Jones, M. Rose, and P. Silver for providing the plasmids, strains, and antisera used in this work. We thank M. G. Fried, A. K. Hopper, R. Shiman, members of A. K. Hopper's laboratory, and members of J. E. Hopper's laboratory for stimulating discussions and encouragement. We thank A. K. Hopper, S. Sarkar, and A. K. Sil for critical evaluation of the manuscript.
This work is supported by Public Health Service grant GM27975 to James E. Hopper.
REFERENCES
- 1.Adam S A. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr Opin Cell Biol. 1999;11:402–406. doi: 10.1016/S0955-0674(99)80056-8. [DOI] [PubMed] [Google Scholar]
- 2.Aris J P, Blobel G. Isolation of yeast nuclei. Methods Enzymol. 1991;194:735–749. doi: 10.1016/0076-6879(91)94056-i. [DOI] [PubMed] [Google Scholar]
- 3.Bajwa W, Torchia T E, Hopper J E. Yeast regulatory gene GAL3: carbon regulation; UASgal elements in common with GAL1, GAL2, GAL7, GAL10, GAL80, and MEL1; and encoded protein strikingly similar to yeast and Escherichia coli galactokinases. Mol Cell Biol. 1988;8:3439–3447. doi: 10.1128/mcb.8.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beier D R, Young E T. Characterization of a regulatory region upstream of the ADR2 locus of S. cerevisiae. Nature. 1982;300:724–728. doi: 10.1038/300724a0. [DOI] [PubMed] [Google Scholar]
- 5.Bhat P J, Hopper J E. The mechanism of inducer formation in gal3 mutants of the yeast galactose system is independent of normal galactose metabolism and mitochondrial respiratory function. Genetics. 1991;128:233–239. doi: 10.1093/genetics/128.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhat P J, Hopper J E. Overproduction of the GAL1 or GAL3 protein causes galactose-independent activation of the GAL4 protein: evidence for a new model of induction for the yeast GAL/MEL regulon. Mol Cell Biol. 1992;12:2701–2707. doi: 10.1128/mcb.12.6.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat P J, Oh D, Hopper J E. Analysis of the GAL3 signal transduction pathway activating GAL4 protein-dependent transcription in Saccharomyces cerevisiae. Genetics. 1990;125:281–291. doi: 10.1093/genetics/125.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blank T E, Woods M P, Lebo C M, Xin P, Hopper J E. Novel Gal3 proteins showing altered Gal80p binding cause constitutive transcription of Gal4p-activated genes in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2566–2575. doi: 10.1128/mcb.17.5.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Box G E P, Hunter W G, Hunter J S. Transmission of error. In: Bradley R A, Kendall D G, Hunter J S, Watson G S, editors. Statistics for experimenters. New York, N.Y: John Wiley & Sons; 1978. pp. 563–570. [Google Scholar]
- 10.Broach J R. Galactose regulation in Saccharomyces cerevisiae. The enzymes encoded by the GAL7, 10, 1 cluster are co-ordinately controlled and separately translated. J Mol Biol. 1979;131:41–53. doi: 10.1016/0022-2836(79)90300-0. [DOI] [PubMed] [Google Scholar]
- 11.Brown T, Mackey K. Analysis of RNA by Northern and slot blot hybridization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman D D J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1994. pp. 4.9.1–4.9.16. [Google Scholar]
- 12.Cole C N, Hammell C M. Nucleocytoplasmic transport: driving and directing transport. Curr Biol. 1998;8:R368–R372. doi: 10.1016/s0960-9822(98)70239-8. [DOI] [PubMed] [Google Scholar]
- 13.Conde J, Fink G R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 15.Douglas H, Pelroy G. A gene controlling inducibility of the galactose pathway enzymes in Saccharomyces. Biochim Biophys Acta. 1963;68:155–156. [Google Scholar]
- 16.Dove J E, Brockenbrough J S, Aris J P. Isolation of nuclei and nucleoli from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 1998;53:33–46. doi: 10.1016/s0091-679x(08)60873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins D A, Silver P A. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giniger E, Varnum S M, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- 19.Gorlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto H, Kikuchi Y, Nogi Y, Fukasawa T. Regulation of expression of the galactose gene cluster in Saccharomyces cerevisiae. Isolation and characterization of the regulatory gene GAL4. Mol Gen Genet. 1983;191:31–38. doi: 10.1007/BF00330886. [DOI] [PubMed] [Google Scholar]
- 22.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 23.Hopper A K, Traglia H M, Dunst R W. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurt E C, McDowall A, Schimmang T. Nucleolar and nuclear envelope proteins of the yeast Saccharomyces cerevisiae. Eur J Cell Biol. 1988;46:554–563. [PubMed] [Google Scholar]
- 25.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. II. Plainview, N.Y: Cold Spring Harbor Press; 1992. [Google Scholar]
- 27.Johnston S A, Hopper J E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci USA. 1982;79:6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston S A, Salmeron J M, Jr, Dincher S S. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987;50:143–146. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- 29.Jones E W. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 30.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 31.Laughon A, Gesteland R F. Isolation and preliminary characterization of the GAL4 gene, a positive regulator of transcription in yeast. Proc Natl Acad Sci USA. 1982;79:6827–6831. doi: 10.1073/pnas.79.22.6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 33.Lue N F, Chasman D I, Buchman A R, Kornberg R D. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol Cell Biol. 1987;7:3446–3451. doi: 10.1128/mcb.7.10.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–142. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- 35.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 36.Meyer J, Walker-Jonah A, Hollenberg C P. Galactokinase encoded by GAL1 is a bifunctional protein required for induction of the GAL genes in Kluyveromyces lactis and is able to suppress the gal3 phenotype in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5454–5461. doi: 10.1128/mcb.11.11.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson M, Silver P. Context affects nuclear protein localization in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:384–389. doi: 10.1128/mcb.9.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nogi Y. The GAL3 gene product is required for maintenance of the induced state of the GAL cluster genes in Saccharomyces cerevisiae. J Bacteriol. 1986;165:101–106. doi: 10.1128/jb.165.1.101-106.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nogi Y, Fukasawa T. Functional domains of a negative regulatory protein, GAL80, of Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3009–3017. doi: 10.1128/mcb.9.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nogi Y, Matsumoto K, Toh-e A, Oshima Y. Interaction of super-repressible and dominant constitutive mutations for the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae. Mol Gen Genet. 1977;152:137–144. doi: 10.1007/BF00268810. [DOI] [PubMed] [Google Scholar]
- 41.Pemberton L F, Blobel G, Rosenblum J S. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 42.Platt A, Reece R J. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 1998;17:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Post-Beittenmiller M A, Hamilton R W, Hopper J E. Regulation of basal and induced levels of the MEL1 transcript in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1238–1245. doi: 10.1128/mcb.4.7.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pringle J R, Adams A E, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 45.Reece R J, Platt A. Signaling activation and repression of RNA polymerase II transcription in yeast. Bioessays. 1997;19:1001–1010. doi: 10.1002/bies.950191110. [DOI] [PubMed] [Google Scholar]
- 46.Rout M P, Aitchison J D, Suprapto A, Hjertaas K, Zhao Y, Chait B T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruohonen L, Aalto M K, Keranen S. Modifications to the ADH1 promoter of Saccharomyces cerevisiae for efficient production of heterologous proteins. J Biotechnol. 1995;39:193–203. doi: 10.1016/0168-1656(95)00024-k. [DOI] [PubMed] [Google Scholar]
- 48.Shen W C, Selvakumar D, Stanford D R, Hopper A K. The Saccharomyces cerevisiae LOS1 gene involved in pre-tRNA splicing encodes a nuclear protein that behaves as a component of the nuclear matrix. J Biol Chem. 1993;268:19436–19444. [PubMed] [Google Scholar]
- 49.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sil A K, Alam S, Xin P, Ma L, Morgan M, Lebo C M, Woods M P, Hopper J E. The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p-Gal4p complex in response to galactose and ATP. Mol Cell Biol. 1999;19:7828–7840. doi: 10.1128/mcb.19.11.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki-Fujimoto T, Fukuma M, Yano K I, Sakurai H, Vonika A, Johnston S A, Fukasawa T. Analysis of the galactose signal transduction pathway in Saccharomyces cerevisiae: interaction between Gal3p and Gal80p. Mol Cell Biol. 1996;16:2504–2508. doi: 10.1128/mcb.16.5.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tolerico L H, Benko A L, Aris J P, Stanford D R, Martin N C, Hopper A K. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics. 1999;151:57–75. doi: 10.1093/genetics/151.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Torchia T E, Hamilton R W, Cano C L, Hopper J E. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: effects on carbon-controlled regulation of the galactose/melibiose pathway genes. Mol Cell Biol. 1984;4:1521–1527. doi: 10.1128/mcb.4.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torchia T E, Hopper J E. Genetic and molecular analysis of the GAL3 gene in the expression of the galactose/melibiose regulon of Saccharomyces cerevisiae. Genetics. 1986;113:229–246. doi: 10.1093/genetics/113.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tornow J, Santangelo G M. Efficient expression of the Saccharomyces cerevisiae glycolytic gene ADH1 is dependent upon a cis-acting regulatory element (UASRPG) found initially in genes encoding ribosomal proteins. Gene. 1990;90:79–85. doi: 10.1016/0378-1119(90)90441-s. [DOI] [PubMed] [Google Scholar]
- 57.Vaduva G, Martin N C, Hopper A K. Actin-binding verprolin is a polarity development protein required for the morphogenesis and function of the yeast actin cytoskeleton. J Cell Biol. 1997;139:1821–1833. doi: 10.1083/jcb.139.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallen E A, Hiller M A, Scherson T Y, Rose M D. Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J Cell Biol. 1992;117:1277–1287. doi: 10.1083/jcb.117.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vollenbroich V, Meyer J, Engels R, Cardinali G, Menezes R A, Hollenberg C P. Galactose induction in yeast involves association of Gal80p with Gal1p or Gal3p. Mol Gen Genet. 1999;261:495–507. doi: 10.1007/s004380050993. [DOI] [PubMed] [Google Scholar]
- 60.Xiao Z, McGrew J T, Schroeder A J, Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yano K, Fukasawa T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:1721–1726. doi: 10.1073/pnas.94.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yocum R R, Johnston M. Molecular cloning of the GAL80 gene from Saccharomyces cerevisiae and characterization of a gal80 deletion. Gene. 1984;32:75–82. doi: 10.1016/0378-1119(84)90034-9. [DOI] [PubMed] [Google Scholar]
- 63.Zenke F T, Engles R, Vollenbroich V, Meyer J, Hollenberg C P, Breunig K D. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science. 1996;272:1662–1665. doi: 10.1126/science.272.5268.1662. . (Erratum, 273:417, 1996.) [DOI] [PubMed] [Google Scholar]
- 64.Zenke F T, Kapp L, Breunig K D. Regulated phosphorylation of the Gal4p inhibitor Gal80p of Kluyveromyces lactis revealed by mutational analysis. Biol Chem. 1999;380:419–430. doi: 10.1515/BC.1999.056. [DOI] [PubMed] [Google Scholar]