Abstract
Diffusion-weighted magnetic resonance imaging (dMRI) is the primary method for noninvasively studying the organization of white matter in the human brain. Here we introduce QSIPrep, an integrative software platform for the processing of diffusion images that is compatible with nearly all dMRI sampling schemes. Drawing upon a diverse set of software suites to capitalize upon their complementary strengths, QSIPrep facilitates easy implementation of best practices for processing of diffusion images.
Keywords: diffusion MRI, q-space imaging, tractography, connectome, white matter, preprocessing, reconstruction
Diffusion-weighted MRI (dMRI) is the primary technique for non-invasive studies of white matter organization in humans. In recent years, dMRI methods have proliferated as the technology has advanced1–4 (see Supplementary Note 1). However, rapid progress also yielded disparate acquisition schemes, analysis approaches, and file formats that are frequently incompatible. As a result, most teams tend to use a limited set of methods, failing to capitalize upon the complementary capabilities of different tools.
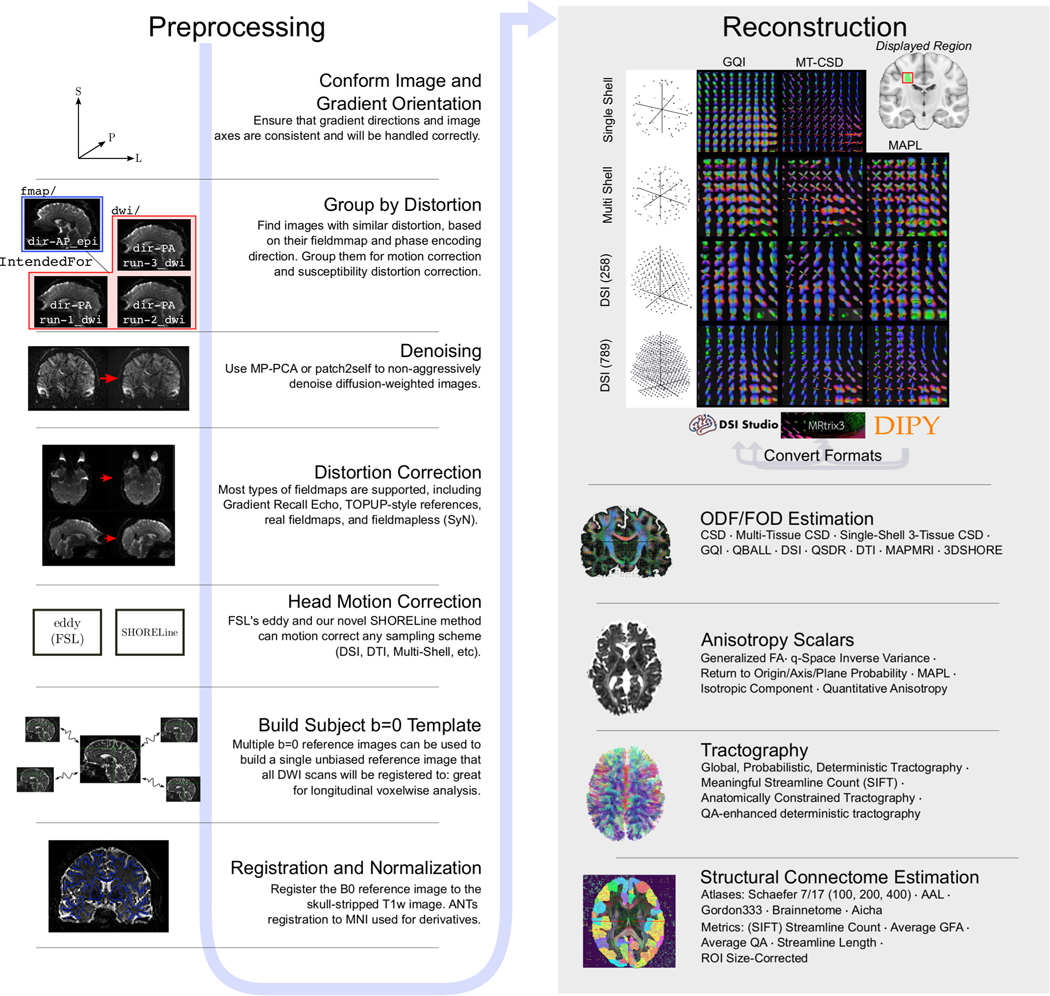
In response to these obstacles, we introduce QSIPrep, a unified and robust platform for processing and reconstructing nearly all dMRI data (Figure 1). QSIPrep leverages the metadata recorded in the Brain Imaging Data Structure (BIDS)5 to automatically configure appropriate preprocessing workflows based on the data provided. Furthermore, QSIPrep includes curated reconstruction workflows (see Extended Data Figure 3 and Online Methods) that consume the output from QSIPrep’s preprocessing pipeline and implement advanced reconstruction and tractography methods. Both preprocessing pipelines and reconstruction workflows are fully documented by animated “before vs. after” visual reports at each step, as well as standardized text that details the methods used (see supplementary figures 1 and 2 as well as Supplementary Note 2). Throughout, QSIPrep converts data to a consistent, interoperable format to capitalize upon the diverse strengths of top software packages (e.g., FSL6, DSI Studio7, DIPY8, ANTs9, and MRtrix10).
Fig 1 |. QSIPrep workflows.
QSIPrep includes preprocessing (left column) and reconstruction (right column) workflows. BIDS data enters the workflow at the top left, following the blue arrow sequentially through the possible steps. The outputs from the preprocessing pipeline are inputs for the reconstruction workflows, which includes reconstruction methods from MRtrix3, DSI Studio, and DIPY. A matrix of orientation distribution functions (ODF)s shows a fiber crossing reconstructed from multiple sampling schemes with multiple methods in QSIPrep. Gray arrows labeled “Convert Formats” indicate that a reconstruction from one software package can be converted to be used in the destination software for further processing (e.g., DIPY reconstructions can be used for tractography in MRtrix3). For further details on options for denoising workflows, see Supplementary Figure 1.
QSIPrep is distributed as both a Python package and as a Docker container that includes the necessary dependencies, ensuring that it is able to run on most computing systems11. QSIPrep has been publicly available since December 2019. Continuous integration testing, modular design and an open development environment have enabled rapid bug detection and integration of feature requests from its user base.
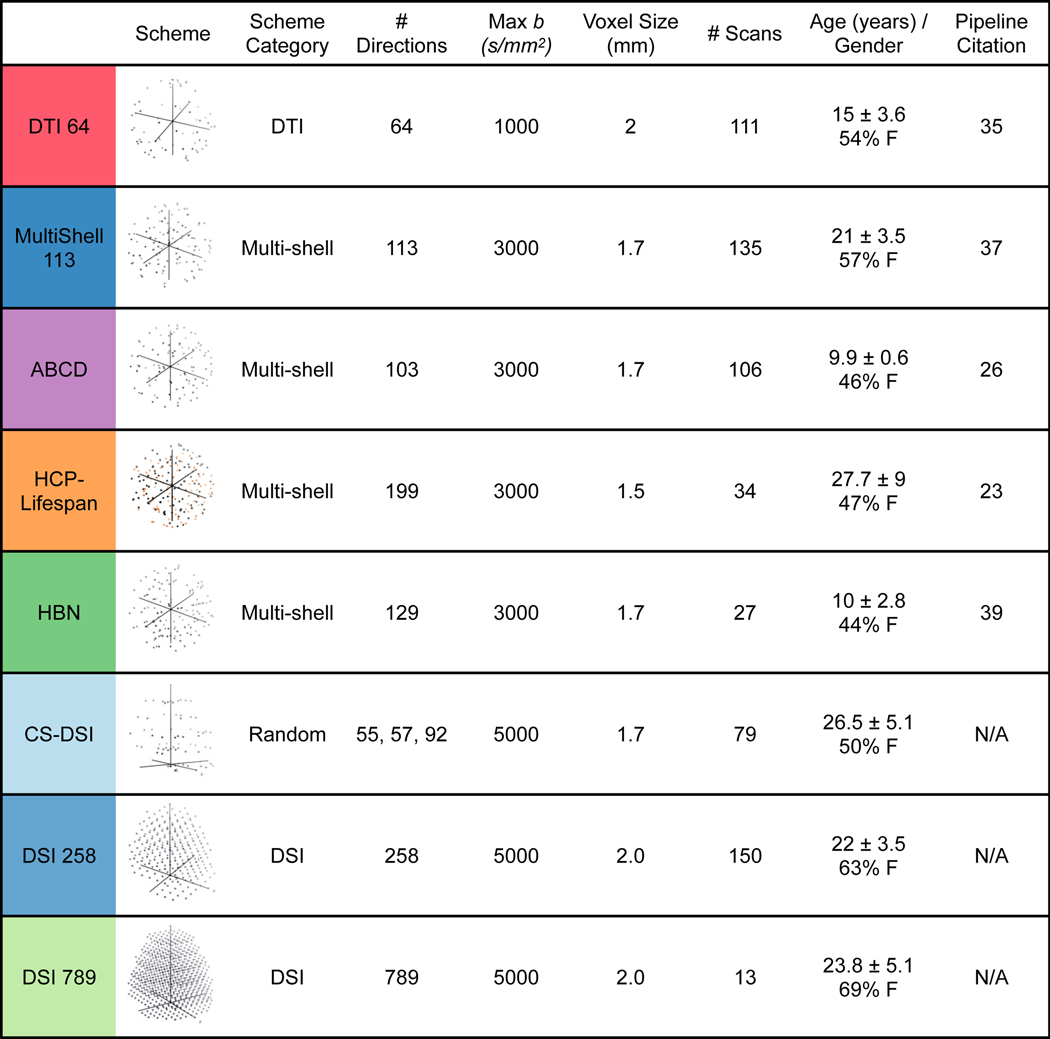
To illustrate the generalizability of QSIPrep to various static q-space sampling schemes, we processed eight different datasets acquired with a wide range of acquisition parameters and scanning platforms (n = 655 total scans). The acquisition schemes for these datasets included a standard single-shell sequence12, four different multi-shell sampling schemes13,14,15,16, two Cartesian grid diffusion spectrum images (DSI) with different sampling densities, as well as a compressed-sensing DSI (CS-DSI) sequence with random q-space sampling17 (Extended Data Fig. 1). We compared the performance of QSIPrep to published pipelines tailored for each dataset on two outcomes: image smoothness and image quality.
The spatial smoothness of the image series was characterized by the mean of the estimated full width at half maximum (FWHM) smoothness of the b=0 images. This measure is impacted by multiple interpolations and imprecise spatial resampling of images, which introduce artifactual blurriness that reduces image contrast and anatomic detail. The image quality metric we evaluated was the neighboring DWI correlation (NDC)18. NDC summarizes the pairwise spatial correlation between each pair of dMRI volumes that sample the closest points in q-space; lower values reflect reduced data quality, driven by noise and misalignment between dMRI volumes. While denoising19,20 and motion correction will increase NDC, it can also be artificially inflated by interpolation-driven spatial smoothing. Accordingly, we regressed image smoothness from the NDC values before comparing pipelines (see Supplementary Note 3).
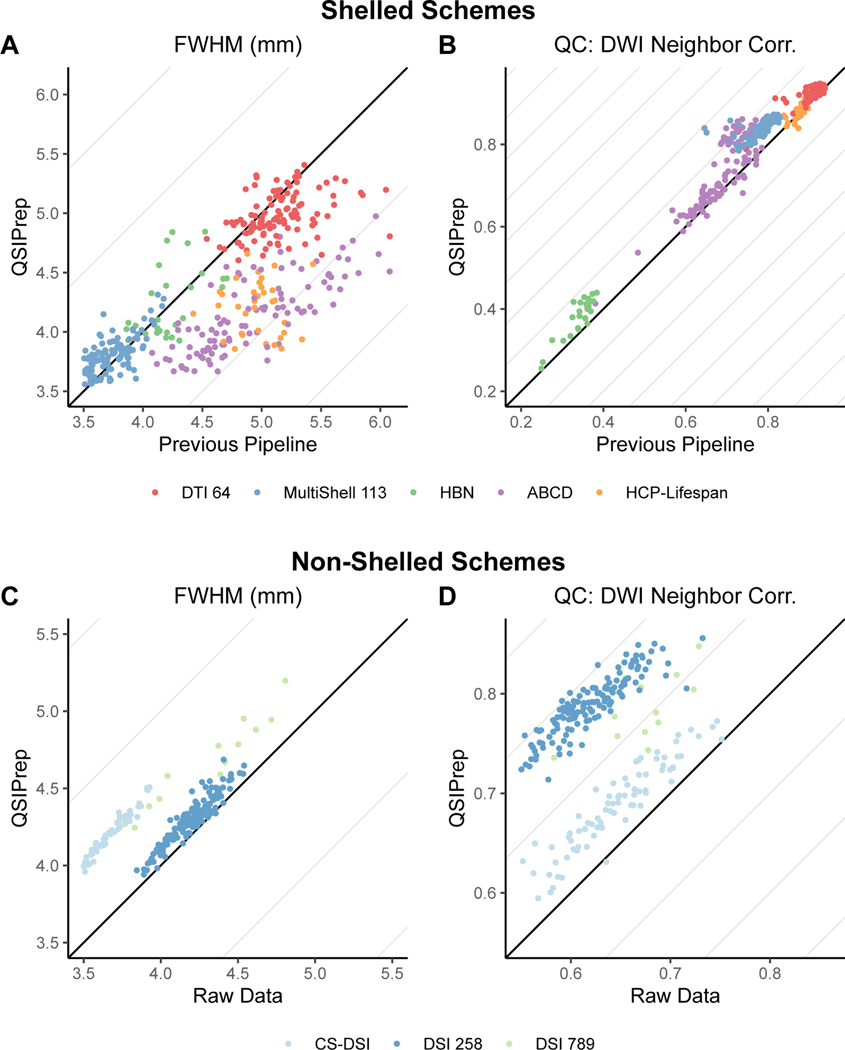
Significance in all tests reported here was determined using t-statistics from a linear mixed effects model with subject as a random intercept. Degrees of freedom and p-values were estimated using the Satterthwaite approximation. For shelled schemes, QSIPrep produced significantly less blurred images than pipelines tailored specifically for each dataset (Figure 2A and Supplementary Table 2). QSIPrep images were substantially less blurred than the custom pipelines developed for the single-shell sequence from the PNC (𝛥FWHM = −0.16mm), the multi-shell sequence from ABCD (𝛥FWHM = −0.8mm), and the multi-shell sequence from the HCP-Lifespan (𝛥FWHM = −0.75mm). Comparisons of raw and processed data further demonstrates the relatively large increase in smoothness introduced by many previously published pipelines (Extended Data Figure 2). In contrast, the smoothness of QSIPrep’s outputs was slightly higher than that produced by the pipeline developed for the NODDI-optimized MultiShell 113 sequence (𝛥FWHM = +0.09mm); no differences were seen in data from HBN.
Fig. 2 |. QSIPrep improves image quality without additional smoothing.
a,b, Comparison of image smoothness (FWHM, a) and data quality (NDC, b) produced by QSIPrep and previously published pipelines tailored for each acquisition scheme for shelled schemes. c,d, Comparison of image smoothness (c) and data quality (d) between QSIprep and raw data for nonshelled schemes (for example, Cartesian and random sampling).
Notably, QSIPrep yielded images with higher NDC than nearly all custom pipelines designed for shelled imaging sequences (Figure 2B and Supplementary Table 2). The only exception to this was the HCP pipeline, where NDC scores were not significantly different from QSIPrep. These results emphasize that QSIPrep produces images of superior (or at least noninferior) data quality compared to custom pipelines developed for a wide variety of shelled acquisition schemes.
One important advantage of QSIPrep is that in addition to shelled acquisition schemes, it can also effectively process advanced non-shelled schemes using an algorithm introduced in QSIPrep. In this case, no direct comparisons to an existing pipeline were available, so only comparisons with raw data were evaluated. Inevitably, any image processing introduces at least some increase in smoothness (Figure 2C and Supplementary Table 4). As expected, images processed with QSIPrep were slightly but significantly smoother than the raw images (𝛥FWHM = +0.53mm), similar to that seen for shelled schemes (Extended Data Figure 2). Notably, processing non-shelled sequences with QSIPrep significantly improved data quality, reflected in a large increase in NDC values (see Figure 2D and Supplementary Table 5).
Following preprocessing, QSIPrep’s set of curated reconstruction workflows provides two critical benefits to users: correct processing and a uniform derived output format. First, workflows are designed to ensure that data preprocessed by QSIPrep are handled correctly within the reconstruction workflow. Second, outputs from each reconstruction method conform to a consistent format across workflows. This emphasis on software interoperability facilitates comparisons between methods (Extended Data Figure 3). Additionally, QSIPrep allows users to apply standard processing and reconstruction methods developed for shelled sequences to advanced non-shelled sequences using a q-space-based interpolation. The ability to apply standard analytic methods to non-shelled schemes dramatically increases the accessibility of advanced non-shelled acquisition sequences.
One of the most popular applications for dMRI is to construct whole-brain structural connectomes via streamline tractography. However, file formats for storing and representing connectomes vary across software packages, thereby limiting comparisons. Furthermore, many software packages produce inconsistently sized matrices across subjects, due to some participants missing small regions from high-resolution atlases. In contrast, QSIPrep ensures that connectivity matrices are directly comparable across methods and participants. Specifically, the software checks that matrices are correctly shaped across all atlases and stores them in easily accessible HDF5 files. Finally, due to the interoperability of the component software elements, QSIPrep allows a far more diverse array of connectivity measurements to be calculated than is possible with individual software packages (see Supplementary Table 1).
Several limitations of the current version of QSIPrep should be noted. First, the software does not support double diffusion encoding q-space imaging or gradient tensor imaging. These scanning sequences are not widely used, are not currently supported by BIDS, and lack open preprocessing software. Second, it is critical to note that we do not claim that the reconstruction workflows are optimal for any given method, only that they implement current best practices. Traditionally, the question of optimality in reconstruction and tractography methods has been difficult to address, in part due to the lack of comparability of measures produced by different software packages. The interoperability provided by QSIPrep facilitates the comparison of many measures – including orientation distribution functions (ODFs), anisotropy scalars, and connectivity matrices – across reconstruction methods and sampling schemes.
Taken together, QSIPrep allows researchers to correctly apply reproducible preprocessing pipelines and advanced reconstruction methods to nearly any dMRI data: a scope currently unmatched by other current dMRI pipelines (see Supplementary Note 4). By harnessing cutting-edge techniques from individual software packages and unifying them in an interoperable framework, the widely generalizable methods provided by QSIPrep perform as well or better than existing customized solutions that can only be applied to a subset of sampling schemes. As QSIPrep’s processing workflows adapt to the characteristics of the input data, it yields an appropriate pipeline as long as the user has correctly specified their data in BIDS. This alleviates much of the burden for users who wish to follow best practices in data processing, but do not have the time or skills to learn the minutiae of multiple software packages. Critically, the adaptive pipelines configured by QSIPrep dramatically enhance accessibility and reproducibility without sacrificing quality. This is underscored by the result that pipelines automatically constructed by QSIPrep yielded results with comparable or better data quality and smoothness compared to established pipelines for multiple studies. As such, QSIPrep facilitates the adoption of fully reproducible best practices for the processing, quality assurance, and reconstruction of diffusion images.
ONLINE METHODS
QSIPrep’s preprocessing workflows
The preprocessing workflow is dynamically built based on data provided as BIDS input. Separate dMRI scans can be grouped and processed together depending on their acquisition parameters and user-supplied options. Image processing can include denoising, Gibbs unringing, head motion, eddy current and distortion correction, b=0 reference image creation (including an optional single-subject b=0 template), coregistration to the T1w image, spatial normalization, image resampling, and gradient rotation. Figure 1‘s left panel depicts the sequence of these steps.
The execution of the workflow is managed by Nipype21, which provides support for multi-core parallelization, algorithm input validation and interfaces to the numerous software packages used by QSIPrep. Coding style and some workflows were adapted from fMRIPrep22 version 1.2.6. The steps detailed in this section can be enabled or disabled by providing flags on the command line call to QSIPrep. We refer the user to the documentation at https://qsiprep.readthedocs.io/en/latest/usage.html#command-line-arguments.
Conform, Merge, and Denoise workflow
One of the unique challenges of dMRI preprocessing is that the q-space sampling scheme is often split into multiple separate scans. Moreover, groups of these scans may be acquired with opposite phase encoding directions so that their b=0 images can be used for Susceptibility Distortion Correction (SDC). The heuristic used by QSIPrep is to divide the scans into “warped groups” that share the same susceptibility distortions. The warped groups are sent to the conform, merge, and denoise workflow.
All spatial transformation operations in QSIPrep (excluding TOPUP/eddy) are performed using ANTs9. ANTs internally uses an LPS+ coordinate system. The FSL-style bvec format required by BIDS specifies gradient directions with respect to the image axis, not world coordinates. By conforming all images and bvecs to LPS+ image orientation, ANTs can be used directly for registration and transformation on both the images and the gradient vectors. The conform step enforces this orientation and checks that the images have matching qform/sform mappings.
Next, warped groups undergo image-based artifact corrections (using MP-PCA23 or Patch2Self20, Gibbs unringing24, bias correction25, and b=0 image-based intensity normalization), and concatenated if multiple runs are present. This step can be done as concatenate-then-denoise or denoise-then-concatenate (default), depending on the user’s preference. If images are concatenated before denoising, there will be more data for MP-PCA/patch2self to include in denoising. However, if the concatenated scans are very far out of alignment with one another, the performance of MP-PCA may be sub-optimal. The other denoising methods are not affected by when data is concatenated. The user can select the concatenate-then-denoise order using a command line flag. A visual description of these workflows is presented in Supplementary Figure 3.
Head Motion, Eddy Current and Susceptibility Distortion Correction workflow
We combined head motion correction (HMC), eddy current correction (ECC), and susceptibility distortion (SDC) into a single workflow due to the interdependence of the TOPUP and eddy tools. This workflow is split into special cases for shelled sampling schemes (multi-shell or single-shell) and all other sampling schemes.
Shelled sampling schemes.
If a reverse-phase encoding direction image is available in the fmap/ or dwi/ directories, a fieldmap is calculated using TOPUP and sent to eddy to be applied in addition to HMC and ECC. In all other cases the fieldmap is calculated using workflows adapted from fMRIPrep and applied to the motion-corrected and imputed output from eddy.
Cartesian and random sampling schemes.
These schemes are processed using the QSIPrep’s novel SHORELine algorithm (https://qsiprep.readthedocs.io/en/latest/preprocessing.html?#head-motion-estimation-shoreline) before being processed using the distortion correction workflows.
Regardless of the sampling schemes, SDC requires a careful selection of representative b=0 images from each DWI scan. QSIPrep selects up to three (depending on availability) b=0 images evenly spaced in time from each group of phase encoding directions. Using a representative subset of all b=0 images is required to limit the run time of TOPUP. The details of which images are used for SDC are included in the HTML report. Newer versions of QSIPrep implement the “best b=0” workflow used by the developmental HCP pipelines15, where b=0 images are selected that have the highest average spatial correlation to the other b=0 images in the same warped group.
b=0 template workflow
The reference image for each DWI series is created by extracting the b=0 images from the series after HMC, ECC, and SDC. They are combined using a normalized average as implemented in ANTs and undergo a histogram equalization as implemented in DIPY. A visual report is generated showing the b=0 template before and after histogram equalization.
Intramodal template workflow
In cases where there are multiple sessions or multiple separate DWI scans that should not be merged, there will be multiple b=0 reference images. Each can be affected by errors in SDC or intermodal co-registration to the T1w image. QSIPrep provides the option to create an “intramodal template” using ANTs template construction26 on the set of b=0 reference images. The intramodal template is co-registered to the T1w image instead of each individual b=0 reference image. The transform to the intramodal template as well as the intramodal template’s transform to the T1w image are added to the stack of transforms that get combined and applied to each DWI (avoiding an additional interpolation).
Co-registration and resampling workflow
Coregistration between the b=0 template image (or the intramodal b=0 template) is performed using antsRegistration. If the user requests a T1w-based spatial normalization to a template, this is also performed using the antsRegistration-based workflow adapted from fMRIPrep. Similar to the HCP Pipelines27 and the ABCD MMPS pipeline13, QSIPrep uses a rigid transformation to register the skull-stripped T1w image to AC-PC alignment. Unlike these other pipelines, QSIPrep combines all spatial transformations so that only a single resampling can be applied. However, the eddy-based workflow uses eddy’s interpolation along with a separate final interpolation to AC-PC alignment. The final resampling uses a Lanczos-windowed Sinc interpolation if the requested output resolution is close to the resolution of the input data. If more than a 10% increase in spatial resolution is requested, then a BSpline interpolation is performed to prevent ringing artifact. The final resampling can at most include the affine head motion correction, the polynomial eddy current correction, the nonlinear susceptibility distortion correction, the nonlinear registration to the b=0 template, the coregistration to the T1w image, and the realignment to AC-PC orientation. Combining these into a single shot interpolation helps preserve high frequency spatial features and precision of the final transform. Any T1w-based steps can be skipped using the --dwi-only flag.
QSIPrep’s reconstruction workflows
A major challenge in comparing reconstruction methods is that many dMRI software packages have their own file formats, coordinate systems, orientation conventions, and visualization tools (see Supplementary Note 1). This diversity is compounded by the large number of possible dMRI acquisition schemes, many of which only meet the requirements of a subset of reconstruction methods. QSIPrep’s set of curated reconstruction workflows provides two critical benefits to users: correct processing and a uniform derived output format. First, workflows are designed to ensure that data preprocessed by QSIPrep are handled correctly within the reconstruction workflow. Second, outputs from each reconstruction method conform to a consistent format across workflows.
This emphasis on software interoperability facilitates comparisons between methods. For example, Extended Data Figure 3 displays the results from a number of reconstruction workflows, depicting disparate sampling schemes reconstructed using popular methods from MRtrix3, DSI Studio, and the Laplacian-regularized MAPMRI (MAPL)3 implementation from DIPY. The visual similarity of the reconstructed ODFs and Fiber Orientation Distributions (FODs) suggests that many of these methods share important features like peak directions. All reconstruction outputs are produced in the native file format of each software package used and also consistently provided in a DSI Studio (fib format) file.
Capitalizing upon the interoperability described above, QSIPrep also allows users to apply standard processing and reconstruction methods developed for shelled sequences to advanced non-shelled sequences. To do this, QSIPrep converts non-shelled sampling schemes to a multi-shell scheme using a 3dSHORE-based q-space interpolation. This conversion allows, for example, the use of multi-shell multi-tissue reconstruction and MRtrix3 tractography methods on any non-shelled sampling scheme. The ability to apply standard analytic methods to non-shelled schemes dramatically increases the accessibility of these advanced acquisition sequences.
QSIPrep’s curated reconstruction workflows apply developer-recommended postprocessing and reconstruction steps, storing the results in both the software-native and DSI Studio formats. The pipelines were chosen from the most popular open-source diffusion imaging software packages such that there is at least one workflow for each q-space sampling scheme. A comparison of pipelines is shown in Supplementary Table 1 and their implementation details are described below grouped by software. While the included workflows use fixed parameters, users can download and edit workflow configuration files to change the workflow’s behavior.
MRtrix3
There are a number of MRtrix3-based workflows that share the same initial steps but differ in how the FOD estimation is performed. In each MRtrix3-based workflow the fiber response function is estimated using dwi2response dhollander28 with a brain mask based on the T1w. The main differences are the MRtrix3 workflows are in 1) the CSD algorithm used to estimate WM FODs and GM/CSF compartments (either multi-shell multi-tissue CSD, MSMTCSD; or single-shell 3-tissue28,29 CSD, SS3T-CSD) and 2) whether a T1w-based tissue segmentation is used during tractography. The *_noACT versions of the pipelines must be used if SDC was not performed during preprocessing. ACT requires SDC to align the T1w-based segmentation for accurate use during tractography. Otherwise, cropping is performed at the T1w-based GM/WM interface along with backtracking. In all MRtrix3 pipelines, tractography is performed using tckgen, which employs the iFOD2 probabilistic tracking method to generate 107 streamlines with a maximum length of 250mm, minimum length of 30mm, FOD power of 0.33. Weights for each streamline are calculated using SIFT2, which is then used to estimate the structural connectivity matrix.
mrtrix_multishell_msmt.
This workflow uses the dwi2fod msmt_csd algorithm28 to estimate FODs for white matter, gray matter and cerebrospinal fluid using multi-shell acquisitions. The white matter FODs are used for tractography and the T1w segmentation is used for anatomical constraints30. mrtrix_multishell_msmt_noACT is identical except that no T1w-based anatomical constraints are used in tractography. mrtrix_singleshell_ss3t is optimized for single-shell acquisitions and also estimates multi-tissue FODs for white matter, gray matter and cerebrospinal fluid using the ss3t_csd_beta1 (SS3T-CSD) algorithm28,29, provided via the MRtrix3Tissue fork of MRtrix3. The white matter FODs are used for tractography and the T1w segmentation is used for anatomical constraints30. mrtrix_singleshell_ss3t_noACT removes the anatomical constraints from tractography.
DSI Studio
dsi_studio_gqi runs the standard GQI reconstruction31 followed by deterministic tractography32. GQI works on almost any sampling scheme. GQI models the diffusion ODF. Diffusion ODFs exhibit smaller peaks than are commonly seen with CSD, but robustly detect fiber crossings33. Although GQI technically works on DTI scans, with spherical sampling on a single-shell around b=1000 s/mm2, its performance markedly improves when more q-space samples are available. The tractography performed in this pipeline ensures that 5 million streamlines are created with a maximum length of 250mm, a minimum length of 30mm, random seeding, a step size of 1mm, and an automatically calculated Quantitative Anisotropy32 (QA) threshold. Additionally, a number of anisotropy scalar images are produced such as quantitative anisotropy (QA)31, generalized fractional anisotropy (GFA), and the isotropic component of the ODF.
DIPY
dipy_mapmri.
Mean Apparent Propagator MRI (MAPMRI) is a recently proposed reconstruction method34 that can estimate ensemble average diffusion propagators (EAPs) and ODFs analytically using multi-shell, Cartesian, or random q-space sampling schemes. This method produces EAP-derived scalars like return to origin probability (RTOP), return to axis probability (RTAP), return to plane probability (RTPP), q-space inverse variance (QIV), and mean squared displacement (MSD). The ODFs are saved in DSI Studio format and optionally as spherical harmonics coefficients in the MRtrix3 format. dipy_3dshore. The 3D Simple Harmonic Oscillator-based Reconstruction and Estimation (3dSHORE)35 method also uses a closed-form solution to estimate EAPs and ODFs from q-space data. This workflow uses the BrainSuite 3dSHORE basis in a DIPY reconstruction. Much like dipy_mapmri, EAP-related scalars such as RTOP, RTAP, RTPP, and MSD are estimated. For both of these reconstruction pipelines, tractography is run identically to the dsi_studio_gqi.
Experimental DSI scheme-converting reconstruction
csdsi_3dshore
This pipeline is for DSI or compressed-sensing DSI. The first step is an L2-regularized 3dSHORE reconstruction3 of the ensemble average propagator in each voxel. These EAPs are then used to 1) calculate ODFs, which are then sent to DSI Studio for tractography and 2) impute signal for a multi-shell (specifically HCP) sampling scheme, which is run through the mrtrix_multishell_msmt pipeline. The resampling is similar to a previouslydescribed GQI-based method36 but uses the 3dSHORE basis set to estimate out-of-sample images.
Structural connectivity matrices
Tractography resulting in connectivity matrices are conformed to a standard HDF5-based output format so as to be directly comparable across methods and software packages. A set of commonly used parcellation schemes are included with QSIPrep, such as the Schaefer atlases in the 100, 200, and 400 parcel resolutions, the brainnetome atlas (264 regions), AICHA (384 regions), Gordon (333 regions), the AAL (116 regions), and the Power atlas (264 regions). Furthermore, users can easily add their own custom atlases as required.
Evaluation Data
Data were gathered from a number of independent studies from multiple institutions. Each study obtained informed consent from participants and was approved by their institutions’ ethics committee. These samples were selected to test a variety of q-space sampling schemes and evaluate if QSIPrep handles each one correctly. An overview of the acquisition parameters is provided in Extended Data Figure 1. QSIPrep was run on the raw data from each study. Spatial smoothness and neighboring DWI correlation18 were calculated for the QSIPrep-preprocessed data and for the data processed using a pipeline specifically designed for that sampling scheme. In the case of non-shelled schemes, QSIPrep was compared to unprocessed data.
Single-shell, DTI.
The single shell data was collected as part of the Philadelphia Neurodevelopmental Cohort (PNC)12 and processed according to the methods described by Roalf et al.37. This pipeline is similar to QSIPrep, utilizing eddy (from FSL5) and custom code for applying distortion correction. The QSIPrep pipeline differs in that it adds MP-PCA, Gibbs unringing, FSL6, and resampling using ANTs. A total of 111 subjects were randomly selected from the available PNC dMRI data.
Multi-shell, NODDI-optimized.
This sampling scheme was designed with the goal of fitting microstructural models such as NODDI38. The data were published Pines et al.14, where the preprocessing scheme used FSL5’s TOPUP and eddy with outlier replacement enabled. The QSIPrep pipeline differed in that it added MP-PCA denoising, Gibbs unringing, FSL6, and resampling using ANTs. We evaluated a sample of 136 participants from the cohort used by Pines et al.
Multi-shell, ABCD.
A total of 106 datasets were downloaded from the NIMH Data Archive (NDA) repository as raw NIfTIs and following minimal preprocessing. The ABCD dMRI preprocessing pipeline13 does not use any of the same software as QSIPrep but performs similar steps. The ABCD pipeline includes gradient nonlinearity correction and uses in-house code for performing Eddy current and distortion correction. QSIPrep adds MP-PCA, Gibbs unringing, ECC, SDC using FSL6, and resampling using ANTs.
Multi-shell, HCP-Lifespan.
A total of 34 subjects were scanned using the HCP-Lifespan imaging protocol15 and processed using both the official HCP diffusion pipelines39 (v4.0.0-alpha.5) and QSIPrep. The HCP diffusion pipeline included motion and eddy current correction, distortion correction, across-scan intensity normalization, coregistration to the T1w image, gradient unwarping and image pair averaging. QSIPrep was upgraded as part of 0.9.0beta1 to include the image pair averaging so that QC measures could be compared directly between the QSIPrep and HCP pipeline outputs. QSIPrep was adjusted to use a quadratic first-level model in eddy to match the HCP diffusion pipeline.
Multi-shell, HBN.
A total of 27 HBN16 subjects were processed using both an early prototype version of dMRIPrep (https://github.com/nipy/dmriprep) and QSIPrep. Both dMRIPrep and QSIPrep use TOPUP and eddy for distortion, eddy current, and motion correction, but dMRIPrep did not include Gibbs unringing or MP-PCA.
Cartesian grid (DSI) schemes.
Prior to QSIPrep there was no publicly available software for applying head motion correction to DSI or CS-DSI acquisitions. Therefore, the QSIPrep-preprocessed images were compared directly to the NDC calculated on the raw images. DSI258 was a repeated measures study with eight repeated scans per subject. CS-DSI acquired four different random schemes per subject. DSI 789 scans acquired 789 unique coordinates on a Cartesian grid in q-space.
Extended Data
Extended Data Fig. 1. Diffusion imaging data used in QSIPrep development and evaluation.
Cartesian (DSI), random (CS-DSI), and shelled (single-shell DTI and multi-shell) sequences were used to test the preprocessing and reconstruction workflows in QSIPrep. Sequences varied widely in their maximum b-value (1000–5000 s/mm2), number of q-space samples (64–789) and voxel size (1.5–2.3 mm). The row colors represent these schemes across all figures. The colors in the HCP-Lifespan image indicate that these samples came from different scans, grouped by phase-encoding direction.
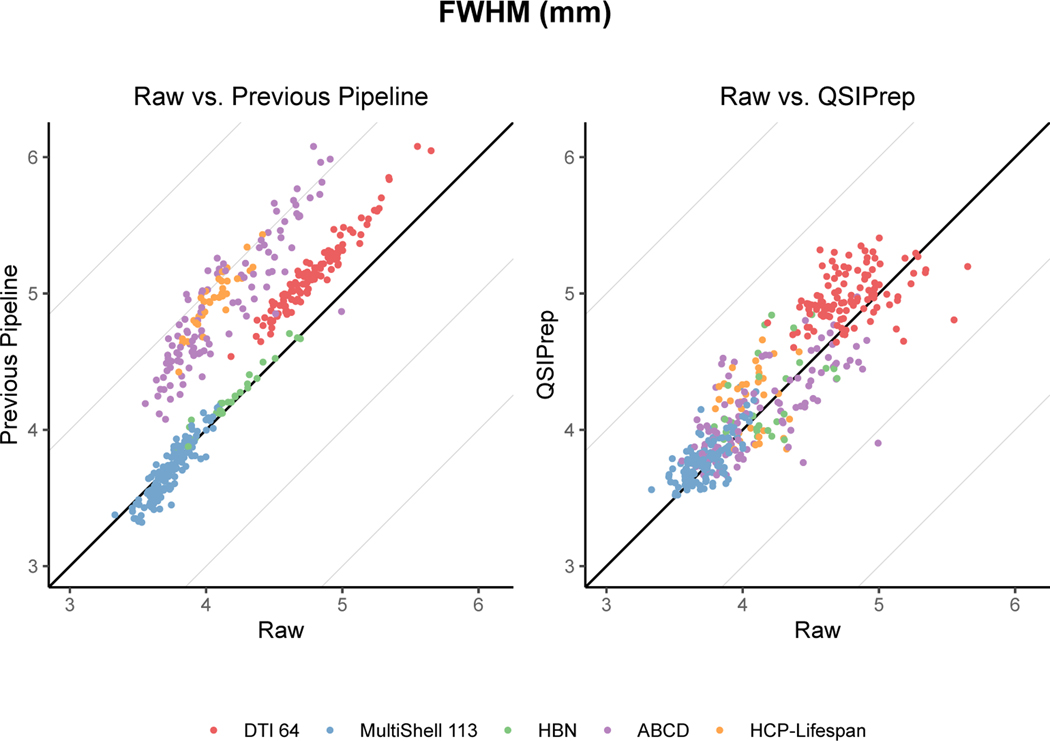
Extended Data Fig. 2. Comparing added smoothness from QSIPrep and previous pipelines.
Preprocessing generally increases the spatial smoothness of images relative to the raw images. Here the raw image smoothness (x-axis) is compared to the same images after being processed by the published pipeline for each dataset (left) and QSIPrep (right). The direct comparison between QSIPrep and the Previous Pipeline is presented in Fig. 2.
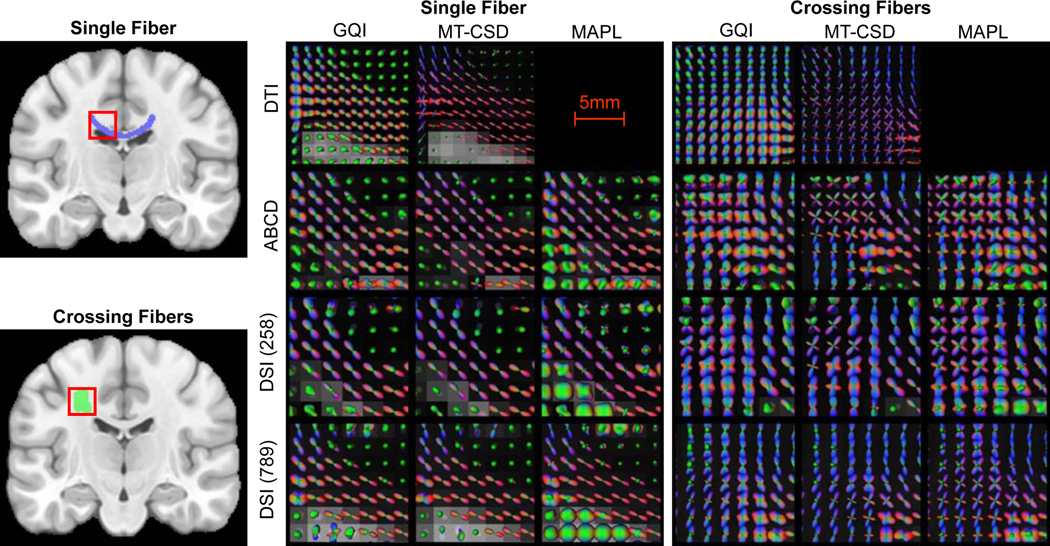
Extended Data Fig. 3. QSIPrep reconstruction workflows produce comparable output across diverse sampling schemes and reconstruction methods.
Four sampling schemes each reconstructed using four methods: GQI from DSI Studio, multi-tissue CSD from MRtrix, and MAPL from Dipy. ODF fields are shown in two white matter regions (left), a single fiber area in the corpus callosum (top) and a crossing fiber region in the centrum semiovale (bottom). The middle panel shows ODFs reconstructed in the single fiber region, and the right panel shows ODFs reconstructed in the crossing fiber region for the four sampling schemes (rows) and the three reconstruction methods (columns).
Supplementary Material
ACKNOWLEDGEMENTS
R01 MH111886 and R01 MH11886-02 for DJO, NINDS R01-NS099348-01 for XH and DSB, UL1TR001878 for JD and MK, 1 U01 EY025864-01 to GKA, MH080243 and Staunton Farm Foundation for BSL, T32 MH 018951 for LMC, R01MH113550, RF1MH116920, R01MH120482 for TDS, CBICA Software Seed Grants for MC and AA, R01-EB027585-01 for EG, SF, AR, AHR AK, W911NF-16-1-0474 from the Army Research Office and by the Institute for Collaborative Biotechnologies under Cooperative Agreement W911NF-19-2-0026 with the Army Research Office to STG and JMV, RF1AG054409 to CD. BL was supported by T32MH014654. Support for the collection of the data for Philadelphia Neurodevelopment Cohort (PNC) was provided by grant RC2MH089983 awarded to REG. Data used in the preparation of this article were obtained from the Adolescent Brain Cognitive Development (ABCD) Study (https://abcdstudy.org), held in the NIMH Data Archive (NDA). This is a multisite, longitudinal study designed to recruit more than 10,000 children age 9–10 and follow them over 10 years into early adulthood. The ABCD Study is supported by the National Institutes of Health and additional federal partners under award numbers U01DA041048, U01DA050989, U01DA051016, U01DA041022, U01DA051018, U01DA051037, U01DA050987, U01DA041174, U01DA041106, U01DA041117, U01DA041028, U01DA041134, U01DA050988, U01DA051039, U01DA041156, U01DA041025, U01DA041120, U01DA051038, U01DA041148, U01DA041093, U01DA041089, U24DA041123, U24DA041147. A full list of supporters is available at https://abcdstudy.org/federal-partners.html. A listing of participating sites and a complete listing of the study investigators can be found at https://abcdstudy.org/consortium_members/. ABCD consortium investigators designed and implemented the study and/or provided data but did not necessarily participate in analysis or writing of this report. This manuscript reflects the views of the authors and may not reflect the opinions or views of the NIH or ABCD consortium investigators.
Footnotes
CODE AVAILABILITY
All code used to perform the statistical tests are available at:
https://pennlinc.github.io/qsiprep_paper/ (DOI: 10.5281/zenodo.4014341) and the QSIPrep source code is available at github.com/pennbbl/qsiprep. Docker images for the versions of QSIPrep used in this manuscript are available on DockerHub at version 0.8.0 and 0.9.0beta1. Newer versions are available.
DATA AVAILABILITY
Source data for Figure 2 and Supplementary Tables 1–4 are available at https://pennlinc.github.io/qsiprep_paper/ (DOI: 10.5281/zenodo.4667846). Imaging data is available with restrictions depending on the original source of the data. PNC data is available on dbGAP [https://www.ncbi.nlm.nih.gov/projects/gap/cgibin/study.cgi?study_id=phs000607.v3.p2]. ABCD data is publicly available in both raw BIDS (https://nda.nih.gov/edit_collection.html?id=3165) and preprocessed (https://nda.nih.gov/edit_collection.html?id=2573) states. HBN data is available on the NeuroImaging Tools and Resources Collaboratory (NITRC [http://fcon_1000.projects.nitrc.org/indi/cmi_healthy_brain_network/sharing_neuro.html]) under a data use agreement. HCP-Lifespan imaging data is available upon request to GKA. DSI 258 is available upon request to JMV. DSI 756 is available upon request to DSB. CS-DSI, DTI 64 and MultiShell 113 are available upon request to TDS.
ETHICS OVERSIGHT
No new data was collected specifically for this study. All other data was acquired with IRB approval at their original institutions and informed consent from participants. The University of Pennsylvania IRB approved DTI64 (810336), HCP-Lifespan (819931), MultiShell113 (822831), CS-DSI (829744), DSI789 (828062). DSI258 was approved by the Institutional Review Board at the University of California Santa Barbara (23-17-0532). ABCD and HBN datasets are publicly available, de-identified data resources collected as part of large consortia.
COMPETING INTERESTS
The authors have no competing interests to declare.
REFERENCES
- 1.Wedeen VJ, Hagmann P, Tseng W-YI, Reese TG & Weisskoff RM Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. Magn. Reson. Med. 54, 1377–1386 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Alexander DC A general framework for experiment design in diffusion MRI and its application in measuring direct tissue-microstructure features. Magn. Reson. Med. 60, 439–448 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Fick RHJ, Wassermann D, Caruyer E. & Deriche R. MAPL: Tissue microstructure estimation using Laplacian-regularized MAP-MRI and its application to HCP data. Neuroimage 134, 365–385 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Yeh CH, Smith RE, Liang X, Calamante F. & Connelly A. Correction for diffusion MRI fibre tracking biases: The consequences for structural connectomic metrics. Neuroimage 142, 150–162 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Gorgolewski KJ et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson JLR & Sotiropoulos SN An integrated approach to correction for offresonance effects and subject movement in diffusion MR imaging. Neuroimage 125, 1063–1078 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeh F-C & Tseng W-YI NTU-90: A high angular resolution brain atlas constructed by q-space diffeomorphic reconstruction. Neuroimage 58, 91–99 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Garyfallidis E. et al. Dipy, a library for the analysis of diffusion MRI data. Front. Neuroinform. 8, 8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avants BB, Epstein CL, Grossman M. & Gee JC Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tournier JD et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage vol. 202 116137 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Gorgolewski KJ et al. BIDS apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLOS Comput. Biol. 13, e1005209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satterthwaite TD et al. Neuroimaging of the Philadelphia Neurodevelopmental Cohort. NeuroImage vol. 86 544–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagler DJ et al. Image processing and analysis methods for the Adolescent Brain Cognitive Development Study. Neuroimage 202, 116091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pines AR et al. Leveraging multi-shell diffusion for studies of brain development in youth and young adulthood. Dev. Cogn. Neurosci. 43, 100788 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harms MP et al. Extending the Human Connectome Project across ages: Imaging protocols for the Lifespan Development and Aging projects. Neuroimage 183, 972–984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Connor D. et al. The Healthy Brain Network Serial Scanning Initiative: a resource for evaluating inter-individual differences and their reliabilities across scan conditions and sessions. Gigascience 6, giw011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paquette M, Merlet S, Gilbert G, Deriche R. & Descoteaux M. Comparison of sampling strategies and sparsifying transforms to improve compressed sensing diffusion spectrum imaging. Magn. Reson. Med. 73, 401–416 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Yeh FC et al. Differential tractography as a track-based biomarker for neuronal injury. Neuroimage 202, 116131 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veraart J. et al. Denoising of diffusion MRI using random matrix theory. Neuroimage 142, 394–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fadnavis S, Batson J. & Garyfallidis E. Patch2Self: Denoising Diffusion MRI with Self-Supervised Learning. NeurIPS 1–11 (2020). [Google Scholar]
- 21.Gorgolewski K. et al. Nipype: A flexible, lightweight and extensible neuroimaging data processing framework in Python. Front. Neuroinform. 5, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteban O. et al. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16, 111–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veraart J, Sijbers J, Sunaert S, Leemans A. & Jeurissen B. Weighted linear least squares estimation of diffusion MRI parameters: Strengths, limitations, and pitfalls. Neuroimage 81, 335–346 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Kellner E, Dhital B, Kiselev VG & Reisert M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magn. Reson. Med. 76, 1574–1581 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Tustison NJ et al. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med. Imaging 29, 1310–1320 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avants BB et al. The optimal template effect in hippocampus studies of diseased populations. Neuroimage 49, 2457–2466 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sotiropoulos SN et al. Advances in diffusion MRI acquisition and processing in the Human Connectome Project. Neuroimage 80, 125–143 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhollander T, Raffelt D. & Connelly A. Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion MR data without a co-registered T1 image. in ISMRM Workshop on Breaking the Barriers of Diffusion MRI; vol. 5 5 (2016). [Google Scholar]
- 29.MRtrix3Tissue | MRtrix3Tissue is a fork of MRtrix3. https://3tissue.github.io/.
- 30.Smith RE, Tournier JD, Calamante F. & Connelly A. Anatomically-constrained tractography: Improved diffusion MRI streamlines tractography through effective use of anatomical information. Neuroimage 62, 1924–1938 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Yeh F-C, Wedeen VJ & Tseng W-YI Generalized q-Sampling Imaging. IEEE Trans. Med. Imaging 29, 1626–1635 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Yeh FC, Verstynen TD, Wang Y, Fernández-Miranda JC & Tseng WYI. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS One 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh FC, Wedeen VJ & Tseng WYI Practical crossing fiber imaging with combined DTI datasets and generalized reconstruction algorithm. Proc Intl Soc Mag Reson Med (2009). [Google Scholar]
- 34.Özarslan E. et al. Mean apparent propagator (MAP) MRI: A novel diffusion imaging method for mapping tissue microstructure. Neuroimage 78, 16–32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Özarslan E, Koay CG & Basser PJ Simple harmonic oscillator based reconstruction and estimation for one-dimensional Q-space magnetic resonance (1D-SHORE). in Applied and Numerical Harmonic Analysis 373–399 (Springer International Publishing, 2013). doi: 10.1007/978-0-8176-8379-5_19. [DOI] [Google Scholar]
- 36.Yeh F-C & Verstynen TD Converting Multi-Shell and Diffusion Spectrum Imaging to High Angular Resolution Diffusion Imaging. Front. Neurosci. 10, 418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roalf DR et al. The impact of quality assurance assessment on diffusion tensor imaging outcomes in a large-scale population-based cohort. Neuroimage 125, 903–919 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H, Schneider T, Wheeler-Kingshott CA & Alexander DC NODDI: Practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Glasser MF et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.