Abstract
Telomerase is a ribonucleoprotein reverse transcriptase responsible for the maintenance of one strand of telomere terminal repeats. The key protein subunit of the telomerase complex, known as TERT, possesses reverse transcriptase-like motifs that presumably mediate catalysis. These motifs are located in the C-terminal region of the polypeptide. Hidden Markov model-based sequence analysis revealed in the N-terminal region of all TERTs the presence of four conserved motifs, named GQ, CP, QFP, and T. Point mutation analysis of conserved residues confirmed the functional importance of the GQ motif. In addition, the distinct phenotypes of the GQ mutants suggest that this motif may play at least two distinct functions in telomere maintenance. Deletion analysis indicates that even the most N-terminal nonconserved region of yeast TERT (N region) is required for telomerase function. This N region exhibits a nonspecific nucleic acid binding activity that probably reflects an important physiologic function. Expression studies of various portions of the yeast TERT in Escherichia coli suggest that the N region and the GQ motif together may constitute a stable domain. We propose that all TERTs may have a bipartite organization, with an N-GQ domain connected to the other motifs through a flexible linker.
Telomerase is a ribonucleoprotein (RNP) that is responsible for maintaining the terminal repeats of telomeres in most organisms (15). It acts as an unusual reverse transcriptase (RT), using a small segment of an integral RNA component as template for the synthesis of the dGT-rich strand of telomeres (16).
Telomerase activity has been characterized from a wide range of organisms and genes encoding both the RNA and protein components of the enzyme complex identified (for reviews, see references 2 and 41). Telomerase RNAs found in ciliated protozoa, in addition to having a short templating region, share a common secondary structure. Telomerase RNAs from yeasts and mammals are considerably larger and exhibit no evident structural conservation. The catalytic RT protein subunit (TERT), initially purified from Euplotes aediculatus as p123, was subsequently found to be homologous to Est2p, a yeast protein required for telomere maintenance (25, 26, 28). Both proteins possess RT-like motifs, alterations in which led to inactivation of telomerase activity and reduced telomere length. Subsequently, homologs of TERT were identified in Schizosaccharomyces pombe, humans, mice, and Tetrahymena, Oxytricha, and Arabidopsis spp. (3, 6, 10, 13, 22, 33, 38, 42). An evident homolog can also be discerned in the incomplete Candida albicans database (see Materials and Methods). Additional mutational analysis of the RT motifs in these latter proteins supports a role for TERT in directly mediating catalysis (20, 50). Because coexpression of TERT and telomerase RNA in vitro in the rabbit reticulocyte lysate system suffices to reconstitute enzyme activity (1, 50), these two subunits probably constitute the core of the enzyme complex.
Several groups of telomerase-associated polypeptides have been identified using either biochemical or genetic tools. First, purification of the Tetrahymena telomerase complex led to the discovery of two associated polypeptides (p80 and p95). Cloning and characterization of p80 and p95 suggest that these proteins may interact with telomerase RNA and the DNA primer, respectively (8, 12). Mouse and human homologs of p80 have also been identified and been shown to associate with the respective telomerases (19, 39). Second, a significant fraction of the human telomerase complex is apparently associated with the molecular chaperones Hsp90 and p23, which are also necessary for reconstitution of telomerase in vitro activity in the rabbit reticulocyte lysate system (21). These molecular chaperones are hypothesized to play a role in telomerase biogenesis. Third, two Sm proteins that are necessary for snRNA maturation are also components of the yeast telomerase complex, suggesting a role for these factors in telomerase RNA processing and telomerase complex assembly (47). Finally, genetic analysis of yeast identified two Est proteins (Est1p and Est3p) that act in the same pathway as telomerase RNA and TERT. Subsequent characterizations indicate that Est1p and Est3p are also subunits of the telomerase complex and that Est1p may play a role in the recruitment of telomerase core to telomere ends in vivo (9, 25, 41, 49). The precise biochemical and physiologic functions of the telomerase-associated proteins remain to be elucidated. The stoichiometry of telomerase components in the native complex is unclear. However, recent studies suggest that the yeast complex may contain more than one copy of the RNA component (43).
Previous analysis of all of the cloned TERTs revealed several salient features of their structural organization: (i) all of the RT motifs are located in the C-terminal half of the protein; (ii) a telomerase-specific motif (T motif), located just N terminal to the RT motifs, can be discerned in all TERTs; and (iii) a motif positioned further toward the N terminus (CP motif) appears to be much more highly conserved among all the ciliate telomerases and may perform a function specific to ciliate telomere formation (3). Point mutations in conserved residues of the T motif significantly impaired telomerase function in the rabbit reticulocyte lysate system (50). Other N-terminal regions of the TERT polypeptide have not been subjected to detailed molecular analysis.
To determine if the uncharacterized, N-terminal regions of TERT are functionally important and if they played conserved roles in telomere maintenance, we initiated mutagenic analysis of the yeast TERT (Est2p). As a starting point for this analysis, we performed a hidden Markov model (HMM)-based alignment of all available TERTs, including the recently identified C. albicans homolog. Several earlier applications of the HMM approach have resulted in the detection of homologies between distantly related proteins (34, 35, 37). Interestingly, such an approach in the case of TERT led to the identification of four conserved motifs in the N-terminal, non-RT region. To validate the predictions arising from comparative sequence analysis, we introduced substitution mutations into the yeast TERT (EST2) gene and investigated their effects on growth, telomere maintenance, and telomerase activity. In this paper, we report that the most N-terminal motif, called GQ, is indeed functionally important. Characterization of mutant phenotypes suggests that the GQ motif may play at least two distinct functions. Furthermore, we show that a nonconserved region located N terminal to the GQ motif (which we call the N region) is also functionally important, possibly through interacting with some nucleic acid target in the context of the native RNP.
While this work was in progress, Friedman and Cech (11) reported the identification of essential domains located in the N-terminal region of yeast TERT using a unigenic evolution approach. In this approach, the gene of interest is heavily mutagenized, and functional variants are selected. Essential and dispensable regions of the protein are then identified by statistically analyzing the distribution of missense and silent mutations. The regions identified in both their and our studies are largely concordant, and we comment on the similarities and discrepancies in the Discussion section.
MATERIALS AND METHODS
Yeast strains and plasmids.
The haploid Saccharomyces cerevisiae strain W303-a (MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1 can1-100) was used for the construction of the Δest2 strain. The disruption cassette was made by inserting ∼600 to 800 bp of the est2 gene flanking sequence upstream and downstream of the kanamycin marker of pUG6. The resulting fragment replaced amino acids 50 to 690 of Est2p open reading frame with the kanamycin resistance marker. Following transformation and selection on G418-containing plates, deletion of the est2 gene in selected isolates was confirmed by PCR.
The plasmid-borne est2 gene was derived from yeast strain JX-MH19 and contained at its C terminus both a three-Myc tag and a six-His tag. The JX-MH19 strain was constructed using the PCR recombination method described by Schneider et al. (46). The pMPY-3XMYC plasmid was amplified using the following two primers: EST2TAG1, 5′-AAGATAATATCATTCTTTTGAGAAAGGAAATTCAACACTTGCAAGCAAGGGAACAAAAGCTGG; and EST2TAG2, 5′-CCTTATCAGCATCATAAGCTGTCAGTATTTCATGTAT TATTAGTACTACTAGTGATGGTGATGGTGATGTAGGGCGAATTGGG TACC. The disruption cassette was introduced into W303-a, and transformants were selected on a Ura− plate. Subsequent selection in the presence of 5-fluoroorotic acid allowed homologous recombination of the repeated Myc tags such that the chromosomal copy of the EST2 gene became fused at its C terminus with a three-Myc tag and a six-His tag. Telomere lengths and telomerase activity in this strain (JX-MH19) are comparable to those of W303-a. To introduce this modified EST2 gene onto a plasmid, a 3.6-kb fragment encompassing the modified est2 coding region (and containing ∼500 bp of upstream region and ∼500 bp of downstream region) was amplified by PCR and inserted between the BamHI and SalI sites of pSE358 to give pSE-Est2TA. To allow for the construction of deletion mutants, the sequence surrounding the start codon of pSE-Est2TA was converted to an NdeI site by site-directed mutagenesis to give pSE-Est2Nde. Additionally, a protein A-tagged est2-containing plasmid was constructed by inserting two copies of the immunoglobulin G (IgG) binding domain of protein A (generated by PCR from pEZZ 18) between the XhoI and KpnI sites of pSE-Est2Nde to give pSE-Est2-proA.
For deletion mutants, fragments encompassing amino acids 10 to 119, 20 to 119, 30 to 119, and 50 to 119 of Est2 were amplified by PCR and inserted between the NdeI and pflMI sites of pSE-Est2Nde to generate the desired plasmids. All point mutations were generated by using the Quick-Change protocol (Stratagene), appropriate primer oligonucleotides, and pSE-Est2Nde as template. All point mutations were confirmed by sequencing. Some of the deletion and point mutants were also made with the protein A tag by transferring the NdeI-to-NcoI fragment from the mutated pSE-Est2Nde plasmid to the pSE-Est2-proA plasmid.
To overexpress Est2p, a vector containing the triose phosphate isomerase promoter (pYX212 from Ingenious Inc.) was utilized. The NcoI site within the polylinker of pYX212 was converted to an NdeI site, and the NdeI-SalI fragments from the pSE-Est2Nde series of plasmids (containing wild-type or mutated EST2) were inserted between the NdeI and SalI sites of the resulting vector.
Sequence comparison.
All sequences used in comparative analysis, with the exception of the C. albicans TERT homolog, were obtained from GenBank. Sequence data for C. albicans was obtained from the Stanford DNA Sequencing and Technology Center website at http://www-sequence.stanford.edu/group /candida.
Determination of telomere length.
Chromosomal DNA was isolated using the Smash and Grab protocol, digested with XhoI or PstI restriction enzyme, and electrophoretically separated on a 1% agarose gel. Following capillary transfer to nylon membranes, telomere-containing fragments were detected by hybridization with a 32P-labeled poly(dG-dT) probe.
Purification of and assay for yeast telomerase.
Whole-cell extracts and active DEAE fractions were prepared as previously described (5, 29, 30). A typical telomerase reaction was carried out in 30 μl containing the following: 10 mM Tris-HCl (pH 8.0), 2 mM magnesium acetate, ∼300 mM sodium acetate (contributed by the protein fraction), 1 mM spermidine, 1 mM dithiothreitol, 5% glycerol (contributed by the protein fraction), 50 μM dTTP, 20 μCi of [α-32P]dGTP (3,000 Ci/mmol), 5 μM primer oligodeoxynucleotides (TEL66 [30]), and 15 μl of column fractions. Primer extension products were processed and analyzed by gel electrophoresis as previously described (29, 30). Signals were quantified using a PhosphorImager system (Molecular Dynamics). For quantification of activity, the signals from all labeled and RNase-sensitive products are summed.
For affinity depletion of protein A-tagged yeast telomerase, about 100 μl of DEAE fraction (in a buffer that contains ∼700 mM sodium acetate) was directly incubated with 5 μl of IgG-Sepharose beads (Pharmacia) at 4°C with gentle rocking for 2 h. The beads were pelleted by centrifugation, and the supernatant was assayed for activity.
Western analysis of protein A-tagged Est2p.
Depending on the expression level, the amount of protein A-tagged Est2p was analyzed either directly in crude extracts or following IgG-Sepharose precipitation using the ProtoBlot system (Promega). For analysis using crude extracts, ∼300 to 500 μg of total protein was electrophoresed into a sodium dodecyl sulfate (SDS)–8% polyacrylamide gel and transferred to nitrocellulose membrane. Primary anti-protein A antibody (Sigma) and secondary antibody were used at 1:1,000,000 and 1:5,000 dilutions, respectively. The high primary antibody dilution was necessary to minimize background arising from cross-reacting polypeptides. For analysis using IgG-Sepharose-purified telomerase, ∼5 mg of unfractionated extracts was treated with 40 μl of IgG-Sepharose at 4°C for 16 h. The beads were washed three times with TMG-10(600), and bound proteins were eluted with SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer. Following electrophoresis and blotting, Est2p was detected by using primary and secondary antibody at 1:20,000 and 1:5,000 dilutions, respectively.
Construction of Escherichia coli EST2 fusion protein expression plasmids and purification of fusion proteins from E. coli.
Fragments of the EST2 gene were amplified by PCR and cloned between the BamHI and PstI sites of the pMAL-cri vector (New England Biolabs). In addition, the downstream primers all possessed sequences that encode an in-frame six-His tag, allowing the fusion proteins to be purified using both nickel-affinity and amylose-affinity chromatography. The proteins expressed from these plasmids are named MBP-Est2(1–160)p, etc., with the numbers in parentheses indicating the amino acid residues of Est2p included in the fusion protein.
The plasmids carrying the maltose-binding protein (MBP)–EST2 fusion genes were transformed into BL21 cells. Transformants were inoculated into Luria-Bertani broth and grown under ampicillin selection (50 μg/ml) at 37°C overnight. A 10-ml culture of saturated cells was diluted with 1 liter of Luria-Bertani broth with ampicillin and grown at 37°C for 2.5 h. When the optical density at 600 nm of the culture reached 0.3 to 0.5, the cultures were cooled down to room temperature and induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM and growth at room temperature for 3 additional h. The cells were harvested, and extracts were prepared by sonication. Fusion proteins were purified successively over an Ni-nitrilotriacetic acid resin (Qiagen) and an amylose resin (Bio-Lab) according to the manufacturers' instructions. All preparations were found to be >90% pure by SDS-PAGE and Coomassie blue staining.
Filter binding assay.
Protein-nucleic acid binding assays were performed using a modification of a published procedure (4). Est2p fusion proteins (from 0.25 to 5 μg) were mixed with 32P-labeled nucleic acid and 50 μg of bovine serum albumin in 10 mM Tris (pH 8.0) and 10% glycerol. The reaction mixtures (50-μl total volume) were incubated on ice for 30 min and pipetted over a prewetted nitrocellulose filter (BA85; Schleicher & Schuell) sandwiched in a slot blot apparatus. The reaction mixtures were then slowly filtered through the membrane using gentle suction. The filters were washed three times with 0.5 ml of buffer (10 mM Tris, 300 mM sodium acetate, 50 μg of bovine serum albumin), air dried, and exposed to a PhosphorImager screen (Molecular Dynamics). All assays were done in duplicate, and the signals were averaged for further analysis. The difference between duplicate samples was generally <10%.
RESULTS
HMM-based analysis revealed four conserved motifs located in the N-terminal, non-RT region of TERT.
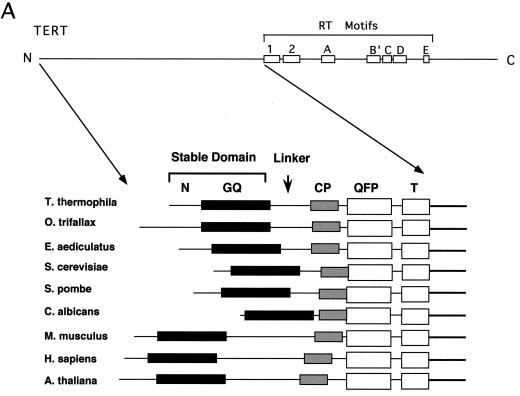
To determine if the N-terminal, non-RT regions of TERT are functionally important and if they play conserved roles in telomere maintenance, we initiated mutagenic analysis of Est2p (the yeast TERT). Several earlier applications of HMM-based sequence comparison resulted in detection of homologies between distantly related proteins (34, 35, 37). Therefore, as a starting point for our analysis, we performed an HMM-based alignment of all available TERTs, including recently identified Arabidopsis thaliana and C. albicans homologs. After obtaining an alignment of the entire amino-terminal region, we defined the motif-domain boundaries by visual inspection. The criteria used for assigning regions of the alignment as linkers included low compositional complexity, absence of clusters of conserved residues, and large numbers of gaps in the majority of sequences. Interestingly, this protocol identified four conserved motifs in the N-terminal, non-RT region of TERT (Fig. 1). We call these motifs in order from N to C terminus the GQ, CP, QFP, and T motifs, respectively. The locations of these motifs within Est2p are as follows: GQ, residues 45 to 163; CP, residues 245 to 265; QFP, residues 267 to 343; and T, residues 367 to 413. Each motif contains nearly invariant amino acid residues that are located at fixed distances from one another. The GQ and QFP motifs have not been previously recognized. In addition, the CP motif, hypothesized earlier to be ciliate specific, is shown in our analysis to possess invariant and nearly invariant residues. The identification of conserved motifs throughout the N-terminal region of TERT suggests that this region mediates a conserved function(s) in telomere synthesis. The overall comparison also revealed a particularly degenerate region that is variable in length, located between motifs GQ and CP, implying the presence of a flexible linker (Fig. 1B).
FIG. 1.
Identification of conserved motifs in the N-terminal, non-RT region of telomerase RT. (A) A schematic illustration of the locations of RT motifs in the telomerase RT polypeptide as determined by an earlier analysis is shown at the top (3). HMM-based analysis revealed four conserved motifs located in the N-terminal region of all TERTs that have been identified thus far (from Tetrahymena thermophila, Oxytricha trifallax, E. aediculatus, S. cerevisiae, S. pombe, C. albicans, Mus musculus, Homo sapiens, and A. thaliana). These motifs are named in order from N to C terminus the GQ, CP, QFP, and T motifs, respectively. For ease of discussion, we also designate the most N-terminal nonconserved region of TERT the N region. The segment between the GQ motif and the CP motif is particularly variable in length, consistent with the existence of a flexible linker in this region. (B) A detailed alignment of the four motifs is shown. The GQ motif is shaded yellow, and the CP, QFP, and T motifs are shaded gray. Invariant residues are italicized, and highly conserved residues are shown in red. Conserved hydrophobic residues are shown in boxes. Closed circles denote point mutations described in this study, and open triangles denote functional mutations at nonconserved positions as reported by Friedman and Cech (11). Functional amino acid substitutions at conserved positions are explicitly given at the bottom. The numbers in parentheses are the numbers of functional mutations in connecting regions. Regions I, II, III, and IV as defined by Friedman and Cech (11) are indicated at the top. Sequences are from T. thermophila, C. trifallax, E. aediculatus, S. cerevisiae, S. pombe, C. albicans, M. musculus, H. sapiens, and A. thaliana (top to bottom, respectively).
The GQ motif is required for telomere maintenance.
To validate some of the predictions arising from comparative sequence analysis, we constructed mutants of the EST2 gene bearing alanine substitutions in conserved residues in the GQ motif. The mutated EST2 gene, located on a centromeric shuttle vector under the control of its natural promoter, was transformed into an est2::Kanr strain that had been grown in the absence of Est2p for ∼25 to 50 generations. The resulting strain was then monitored for defects in growth, telomere maintenance, and telomerase activity. To facilitate future biochemical analysis, the plasmid-borne EST2 gene was fused at its 3′ end with three tandem Myc tags and a six-His tag. In some cases, an additional protein A tag (consisting of two copies of the IgG binding domain from protein A) was inserted in between the Myc and His tags to allow even more efficient affinity purification. These C-terminal modifications have no effect on telomere maintenance and telomerase activity (J. Xia, unpublished data).
A total of eight alanine substitutions at conserved GQ motif residues were constructed and tested: D66A, G85A, N104AV105A, W115A, F118AH119A, G123A, Q138AF139A, and G141A. Two additional mutants with substitutions in nonconserved residues (D93A and E154A) were also made and tested for comparison. As summarized in Table 1, three of these mutants (W115A, F118AH119A, and G123A), located quite close to one another, exhibited the most pronounced growth defects, giving rise to small and variably sized colonies suggestive of senescence (31). The senescent phenotype, characterized by progressively slower cell multiplication, has been observed in several other telomerase knockout strains (25, 48). The display of senescence suggests that the W115A, F118AH119A, and G123A mutants are quite compromised in telomerase function. In contrast, the other mutants either did not show any growth defects or grew only slightly slower (compared to a strain carrying a plasmid containing the wild-type EST2 gene) and failed to exhibit signs of senescence upon repeated restreaking.
TABLE 1.
Summary of in vivo and in vitro phenotypes of point mutants in the GQ motif
| Mutation | Senescencea | Telomeresb | Primer extension activityc |
|---|---|---|---|
| None | − | +++ | 100 |
| D66A | − | + | 70 |
| G85A | − | + | 8 |
| D93A | − | +++ | 140 |
| N104AV105A | − | + | 80 |
| W115A | + | +/− | <2 |
| F118AH119A | + | +/− | <2 |
| G123A | + | +/− | <2 |
| Q138AF139A | − | + | 5 |
| G141A | − | + | 6 |
| E154A | − | +++ | 100 |
Senescent strains (+) grew slowly on the initial transformation plate and gave rise to small colonies that are the same size as those from an Δest2 strain transformed with the pSE358 vector.
Telomere lengths were determined in Southern hybridization assays (Fig. 2) and were scored as follows: +++, wild-type telomere length; +, on average about 150 to 200 bp shorter; +/−, on average about 300 bp shorter with weak hybridization signals.
Primer extension assays were performed using TEL66 as primer under standard conditions. Each mutant fraction was assayed alongside the wild-type fraction. The signals obtained from the mutant fractions were normalized against that from the wild-type fraction, which was taken as 100.
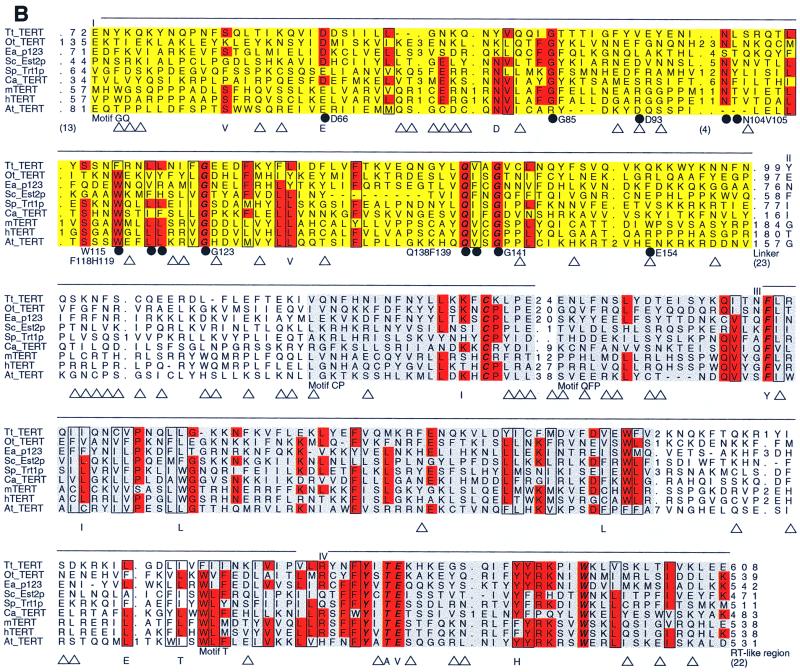
Chromosomal DNA was isolated from individual transformants following two restreaks (∼50 generations) and assayed for telomere lengths. As shown in Fig. 2, the Y′ class of telomeres in all mutants with substitutions in conserved residues shows dramatic telomere shortening of at least 150 to 200 bp (all mutants except D93A and E154A, lanes 2 to 5, 8 and 9, and 12 to 21). The senescent mutants have even shorter telomeres and gave especially weak signals for hybridization (W115A, F118AH119A, and G123A, lanes 12 to 17). Thus, there is a good correlation between growth defects and telomere repeat loss, consistent with earlier findings. In contrast to mutations in conserved residues, the two strains with mutations in nonconserved residues (D93A and E154A) failed to show any evidence of telomere shortening.
FIG. 2.
Telomere length determination in strains that contain mutations in the conserved and nonconserved residues within the GQ motif. The Δest2 strain that had been grown for ∼25 to 50 generations was transformed with plasmids bearing either wild-type (W.T.) or mutated EST2. The transformants were restreaked twice, and single colonies were picked for growth in liquid culture. Chromosomal DNAs were isolated from the cultures, digested with PstI, electrophoresed into a 1% agarose gel, transferred to a nylon membrane, and probed with 32P-labeled poly(dG-dT). The locations of the Y′ telomeres and an X telomere are indicated by a vertical bar and an arrowhead, respectively. The mobilities of several molecular size standards are indicated on the left.
Two mutations impaired telomere maintenance without affecting telomerase activity.
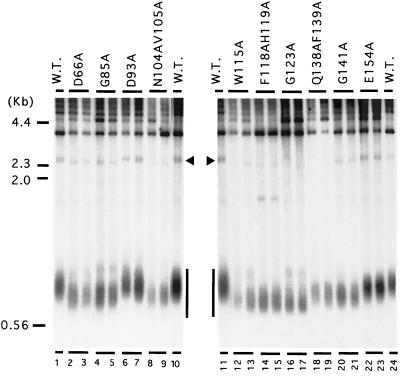
Because several studies point to the existence of mutations that uncouple in vitro telomerase activity from in vivo telomere maintenance (e.g., est1 and est3 mutants [27]), we were interested in determining if any of the GQ mutations had similar properties. Extracts were prepared from each of the mutant strains, and telomerase activity was partially purified over DEAE columns and tested. Earlier studies indicated that, under standard reaction conditions, this chromatographic fraction yields labeled products that are almost entirely due to TLC1 and Est2p (5; N. Lue, unpublished data). As shown in Fig. 3A, almost the entire primer extension signal in this preparation is sensitive to RNase A pretreatment. In addition, when the Est2p in the strain is tagged with two copies of the IgG binding domain from protein A, >80% of the activity in the DEAE fraction can be specifically trapped on IgG-Sepharose beads and depleted from supernatant, indicating that this fraction is largely free of other contaminating activities (Fig. 3A).
FIG. 3.
Telomerase primer extension assays for wild-type and mutated RNPs. (A) The DEAE fraction was prepared from strains whose Est2p was either tagged or untagged with two copies of the IgG binding domain from protein A. The fractions were incubated either in the absence (−) or in the presence (+) of IgG-Sepharose beads. The supernatants were then recovered, incubated in the absence (−) or the presence (+) of RNase A, and assayed for telomerase activity using TEL66 (TAGGGTAGTAGTAGGG) as the primer oligonucleotide. The position of the primer +3 products is marked to the left of the panel. (B) DEAE fractions were prepared from strains bearing wild-type and mutated Est2p. Each mutant fraction was tested alongside the wild-type fraction using equal amounts of total protein. Each panel presents results from a single set of assays. The identities of the mutations are indicated at the top of each panel, and RNase-sensitive signals derived from telomerase are indicated by vertical bars to the left of each panel. An RNase-insensitive band (indicated by an asterisk) is occasionally observed in some assays.
Each mutant telomerase was tested side by side with the wild-type fraction using equal amounts of total protein (Fig. 3B). As expected, the control mutants that exhibited no telomere shortening had in vitro activities that were comparable to those of the wild-type enzyme (D93A and E154A, lanes 5 and 15). This result further demonstrates the reproducibility of our protocol. Six of the eight mutants with substitutions in conserved residues exhibited greatly reduced telomerase activity, ranging from ∼12-fold reduction for G85A (lane 3) to more than 50-fold reduction for the senescent mutants (W115A and G123A, lanes 9 and 10). Interestingly, two mutants, D66A and N104AV105A (lanes 2 and 7), appear to uncouple telomerase activity in vitro from telomere maintenance in vivo; though the telomeres in these two strains are greatly shortened, the mutant enzymes exhibited nearly wild-type levels of activity. Other strains that had similar telomere length defects such as G85 and Q138AF139A (lanes 3 and 12) suffered a ∼12- to 20-fold reduction in telomerase activity (Table 1). All mutant fractions were assayed at least twice, and where a significant reduction in activity relative to that of the wild-type fraction was observed, the mutant activity as a percentage of the wild-type activity varied by less than 5%.
The extreme N-terminal region of Est2p (N region) is also required for telomere maintenance in vivo and telomerase activity in vitro.
Our comparative analysis suggests that the extreme N-terminal 50 amino acids (termed N region) of yeast TERT could not be reliably aligned with its homologs from other species. To test the importance of this N region, we constructed N-terminal truncation mutants of the EST2 gene and tested their ability to support telomere length maintenance and telomerase activity using the previously described system.
Four deletion mutants missing amino acids 2 to 10, 2 to 20, 2 to 30, and 2 to 50 (abbreviated as N-10, N-20, N-30, and N-50, respectively) were tested in this system. As shown in Fig. 4A and Table 2, the N-10 and N-20 mutants exhibited significantly reduced growth rates in minimal medium on agar plates. However, they showed no evidence of senescence upon repeated restreaking (data not shown). In contrast, the N-30 and N-50 mutants gave rise to heterogeneously sized colonies suggestive of senescence. Chromosomal DNA from two independent clones of each strain was isolated following two restreaks and analyzed for telomere length by Southern hybridization. Consistent with the growth defects, all deletion mutants, including the smallest one (N-10), possessed significantly shortened telomeres relative to the control strain (Fig. 4B, compare lanes 1 and 8 with lanes 2 to 7). Both Y′-type telomeres (marked by a vertical bar) and X-type telomeres (marked by arrows) in the mutant strains were shortened, consistent with a general defect in telomere maintenance.
FIG. 4.
N-terminal deletions result in defective Est2p function. (A) Transformants bearing either wild-type or N-terminally deleted Est2p were restreaked twice and monitored for growth defects on minimal plates. The photographs show colonies from the second restreak after 2 days of growth. The identities of the clones are indicated at the left of each panel. (B) Transformants bearing either wild-type or N-terminally deleted Est2p were restreaked twice, and single colonies were picked for growth in liquid cultures. Chromosomal DNAs were isolated from the cultures, digested with XhoI electrophoresed into a 1% agarose gel, transferred to a nylon membrane, and probed with 32P-labeled poly(dG-dT). The locations of the Y′ telomeres and X telomeres are indicated by a vertical bar and several arrowheads to the right of the panel, respectively. (C) DEAE fractions were prepared from strains bearing wild-type and N-terminally deleted Est2p and tested for telomerase primer extension activity using TEL66 as the primer oligonucleotide.
TABLE 2.
Summary of in vivo and in vitro phenotypes of N-terminal deletions of Est2p
| Residues deleted | Senescencea | Telomeresb | Primer extension activityc |
|---|---|---|---|
| None | − | +++ | 100 |
| 2–10 | − | +/− | ∼2 |
| 2–20 | − | +/− | ∼2 |
| 2–30 | + | +/− | ND |
| 2–50 | + | +/− | ND |
Senescent strains (+) grew slowly on the initial transformation plate and gave rise to small colonies that are the same size as those from an Δest2 strain transformed with the pSE358 vector.
Telomere lengths were determined in Southern hybridization assays (Fig. 2) and were scored as follows: +++, wild-type telomere length; +/−, on average about 300 bp shorter with weak hybridization signals.
Primer extension assays were performed using TEL66 as primer under standard conditions. Each mutant fraction was assayed alongside the wild-type fraction (Fig. 3). The signals obtained from the mutant fractions were normalized against that from the wild-type fraction, which was taken as 100. ND, not determined.
The deletion mutants were also tested for defects in telomerase activity using the primer extension assay. As shown in Fig. 4C, fractions derived from the N-10 and N-20 strains exhibited significantly reduced telomerase activity compared to those from the control strain. Quantification by PhosphorImager analysis indicates that the reduction is approximately 40- to 50-fold. The N-30 and N-50 strains were not tested for in vitro activity. However, based on the growth defects, it is likely that these latter strains would have exhibited the same or less telomerase activity. We conclude that the N region of Est2p is required for full telomerase primer extension activity as measured in vitro.
Loss of telomerase activity in the mutants cannot be accounted for by loss of protein expression-stability alone.
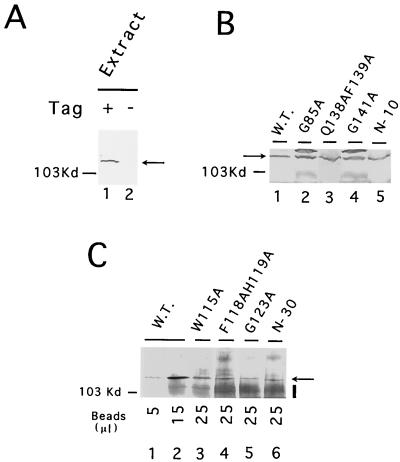
To determine if reduced telomerase activity of the deletion and point mutants was due to reduced expression-stability of Est2p, we determined the amount of protein A-tagged Est2p in the mutant strains by Western analysis. To minimize potential variations introduced by IgG-Sepharose binding, we first directly analyzed unfractionated extracts for the presence of Est2p. An immunoreactive species of ∼115 kDa can be detected in extracts from the tagged strain, but not from the untagged strain, supporting the specificity of our assay (Fig. 5A). As expected, the two functionally defective mutants that exhibited wild-type levels of in vitro telomerase activity (D66A and N104AV105A) had wild-type levels of Est2p (data not shown). Four mutants with reduced telomerase activity in vitro (G85A, Q138AF139A, G141A, and N-10) also exhibited levels of Est2p comparable to that of the wild-type strain (Fig. 5B). The 12-fold or greater loss of in vitro activity of these four mutants was therefore not due to reduced Est2p expression-stability. Interestingly, the four senescent mutants (W115A, F118AH119A, G123A, and N-30) did manifest a significant reduction in Est2p level such that it was not possible to detect these polypeptides unequivocally in unfractionated extracts (data not shown). However, following IgG-Sepharose purification, even these mutant proteins can be clearly visualized in Western analysis (Fig. 5C). The increased background in these latter assays (marked by a vertical bar to the right of the panel) came from IgG that was released by heating of the IgG-Sepharose beads in SDS and that reacted with the secondary antibody. Using signals derived from different amounts of Sepharose beads carrying wild-type Est2p as standards, these four mutant polypeptides appear to be present at approximately one-third to one-fifth of the wild-type protein level (compare lanes 3 to 6 with lane 1 and lane 2). The N-30 mutant polypeptide exhibited a slightly increased mobility by SDS-PAGE, further confirming the authenticity of our signal (Fig. 5C, lane 6). Given that telomerase activity was reduced by 50-fold or more in these senescent mutants, it appears that the mild reduction in Est2p level cannot solely account for the enzymatic defect.
FIG. 5.
Analysis of mutant protein expression. (A) Whole-cell extracts were prepared from strains whose Est2p was either tagged (+) or untagged (−) with the IgG binding domain of protein A and subjected to Western analysis. For each extract, 300 μg of total protein was examined. The position of the protein A-tagged Est2p is indicated by an arrow. (B) Est2p levels in whole-cell extracts from wild-type (W.T.) or mutant strains were analyzed by Western blotting. For each extract, 500 μg of total protein was examined. The position of the protein A-tagged Est2p is indicated by an arrow. (C) Est2p levels in whole-cell extracts from wild-type (W.T.) or mutant strains were assessed by affinity precipitation followed by Western analysis. For each extract, 4 mg of total proteins was incubated with 40 μl of IgG-Sepharose resin with gentle agitation at 4°C for 16 h. The beads were washed extensively, and the indicated amount of Sepharose was boiled in SDS-PAGE loading buffer. The eluted proteins were then subjected to immunoblotting. The position of the protein A-tagged Est2p is indicated by an arrow. The increased background (marked by a vertical bar) was due to IgG that eluted from the beads and that cross-reacted with the secondary antibody.
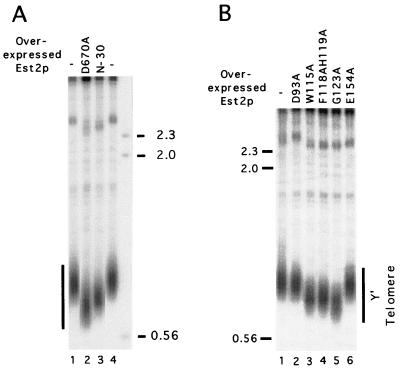
If the senescent mutant proteins were indeed defective in function (as opposed to defective only in expression-stability), then they might act in a dominant-negative fashion when overexpressed in the presence of wild-type protein. This was found to be indeed the case. Four mutants that caused senescence (W115A, F118AH119A, G123A, and N-30) were placed downstream of a strong constitutive promoter, and the resulting plasmids were introduced into a wild-type strain (W303). Following three restreaks (∼75 generations), chromosomal DNAs were isolated from the transformants and analyzed for telomere length alteration (Fig. 6). Consistent with a defect in function, all four mutants caused significant telomere shortening in the host strain (Fig. 6A, compare lane 3 with lanes 1 and 4; Fig. 6B, compare lanes 3 to 5 with lane 1). As expected, two mutant proteins that supported normal telomere maintenance (D93A and E154A) had no effect when overproduced (Fig. 6B, lanes 2 and 6), and an Est2p with an RT active site mutation (D670A) caused the most severe telomere shortening (Fig. 6A, lane 2). The other nonsenescent GQ motif mutants caused at most a slight shortening of telomeres (∼50 bp) when overexpressed in W303, possibly because they retain a significant level of function (data not shown).
FIG. 6.
Several Est2p mutants can cause telomere shortening when overexpressed in the presence of wild-type protein. (A) W303 was transformed with a pYX212 plasmid expressing mutated Est2p. After restreaking of the transformants three times, chromosomal DNAs were isolated from the strains, digested with PstI, and analyzed for telomere lengths. DNAs were also isolated from untransformed W303 and tested for comparison. The source of the DNA is indicated at the top of the panel. (B) DNAs from strains overexpressing different Est2p mutant proteins were isolated and digested as for panel A and tested for telomere lengths. The source of the DNA is indicated at the top of the panel. The mobilities of several molecular size standards (in kilobases) are indicated at the sides.
Identification of a protease-resistant stable domain in the N-terminal region of Est2p.
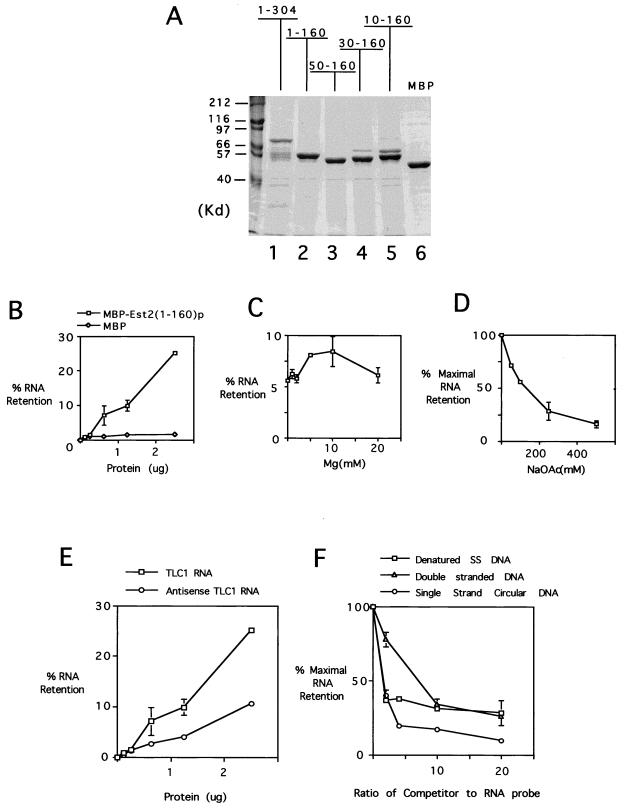
To begin to biochemically dissect the TERT polypeptide, we attempted to express recombinantly the N-terminal region of Est2p in Escherichia coli as MBP (maltose-binding protein) fusion proteins. To facilitate purification, the proteins were also fused to a six-His tag at its C terminus. Three fragments, 1–304, 1–270, and 1–160, were chosen for initial characterization as parts of the MBP fusion protein [designated MBP-Est(1–304)p, MBP-Est2(1–270)p, and MBP-Est2(1–160)p, respectively]. Interestingly, the two larger fragments appear to be sensitive to proteolysis in E. coli. For example, following affinity purification over a maltose column, fractions derived from the MBP-Est2(1–304)p-overproducing strain contained not only the full-length fusion polypeptide but also several smaller fragments (Fig. 7A, lane 1). These smaller fragments most likely resulted from proteolysis of the Est2p segment in vivo, because they still retained the MBP domain, and because the MBP domain on its own was stably expressed in E. coli (Fig. 7A, lane 6). Based on their size, the proteolyzed fragments appear to retain ∼160 amino acids of Est2p (Fig. 7A, compare lanes 1 and 2). Interestingly, the MBP-Est2(1–160)p can be easily overproduced and purified as a single polypeptide from E. coli, again suggesting that this segment of Est2p, encompassing the N region and GQ motif, can form a stable domain in vivo that is resistant to proteolysis.
FIG. 7.
The Est2(1–160)p fragment exhibits a nonspecific nucleic acid binding activity. (A) Several MBP-Est2p fusion proteins were expressed and purified from E. coli using both nickel-affinity and maltose-affinity columns. The resulting preparations were analyzed in an SDS–10% polyacrylamide gel. The identities of the fusion proteins are indicated at the top of the panel. All fusion proteins (lanes 1 to 5) contain ∼40 kDa of an MBP domain besides the Est2p fragments. The MBP (lane 6) derived from the original cloning vector contains an extra protein fragment beyond the polylinker and is therefore ∼48 kDa in size. (B) Increasing amounts of MBP and MBP-Est2(1–160)p were incubated with 20 ng of in vitro-transcribed, labeled TLC1 RNA. The resulting mixtures were passed through a nitrocellulose filter, and the percentage of probe retained on the filter was plotted against the amount of protein used. Assays were performed in duplicate, and the averages and spreads are indicated. (C) Filter binding assays were performed using 1.3 μg of MBP-Est2(1–160)p and 20 ng of TLC1 RNA in the presence of increasing Mg concentrations. The percentage of probe retained on the filter was plotted against the Mg concentration. (D) Filter binding assays were performed using 1.3 μg of MBP-Est2(1–160)p and 20 ng of TLC1 RNA in the presence of increasing sodium acetate concentrations. The percentage of probe retained on the filter was plotted against the sodium acetate concentration (taking the amount retained in the presence of no salt as 100%). (E) Filter binding assays were performed using increasing amounts of MBP-Est2(1–160)p and 20 ng of either TLC1 RNA or antisense TLC1 RNA probe. The percentage of probe retained on the filter was plotted against the amount of protein used. (F) Filter binding assays were performed using 1.3 μg of MBP-Est2(1–160)p and 10 ng of labeled TLC1 RNA in the presence of increasing amounts of three different unlabeled DNA competitors: denatured salmon sperm DNA, double-stranded linear DNA, and single-stranded (SS) circular DNA. The percentage of probe retained on the filter was plotted against the ratio of competitor to probe (taking the amount retained in the presence of no competitor as 100%).
The N-terminal domain of Est2p possesses a nucleic acid binding activity.
One potential function for the N-terminal domain is involvement in protein-RNA interactions. To test this idea, we assayed the N-terminal fusion protein for RNA binding activity using a filter retention assay. Initial studies employed MBP-Est2(1–160)p and 32P-labeled full-length TLC1 RNA. As shown in Fig. 7B, the amount of RNA retained on the filter increased with increasing protein concentrations. At the highest protein concentration used (0.05 μg/μl), ∼25% of the input RNA was retained on the filter. The apparent dissociation constant was about 5 μM. The amount of RNA retained by MBP was substantially less than that by the fusion protein, indicating that RNA binding was mediated by the N-terminal Est2p fragment.
We investigated several reaction parameters and found that both salt and Mg2+ concentrations significantly affected the efficiency of binding. RNA retention as a function of Mg2+ concentration is a bell-shaped curve, with a peak at ∼5 to 10 mM (Fig. 7C). The protein-RNA interaction was favored at low salt concentrations; the binding efficiency was reduced by ∼75% at 250 mM sodium acetate relative to no salt (Fig. 7D).
To determine the sequence specificity of binding, we generated both sense and antisense TLC1 RNA probes by in vitro transcription and compared their abilities to interact with the fusion protein. As shown in Fig. 7E, the extent of binding is only twofold higher for the sense probe, suggesting that the N-terminal domain, at least by itself, does not recognize RNA with significant sequence specificity. We also tested the effect of RNA length on the efficiency of binding using TLC1 RNA missing increasing numbers of 3′-end residues. The results indicate that the fusion protein has similar affinities for RNAs ranging from 450 to 1,300 nucleotides long (data not shown).
We next tested binding of the fusion protein to DNAs of different structure using a competition filter-binding assay. A fixed amount of fusion protein [MBP-Est2(1–160)p, 1.3 μg] was mixed with both a fixed amount of labeled TLC1 RNA (10 ng) and variable amounts of unlabeled DNA competitors. The resulting mixture was then subjected to filtration through a nitrocellulose membrane as previously described. Single-stranded circular φX174 virion DNA, sheared and denatured salmon sperm DNA, and double-stranded linear DNA were used as the competitors. As shown in Fig. 7F, at an RNA/DNA ratio of 2:1, both the φX174 DNA and denatured salmon sperm DNA reduced the binding efficiency by 60%. Comparable inhibition of RNA binding by double-stranded linear DNA was achieved at a ratio of ∼8:1. We also tested short single-stranded telomere oligonucleotides (the G-rich strand) in the competition assay, and the same amount of these oligonucleotides (by weight) was no more effective than the other single-stranded DNAs in reducing the binding signal (data not shown). Thus, the Est2p(1–160) fragment can also bind DNA in a non-sequence-specific fashion, with a slight preference for single-stranded DNA over double-stranded DNA.
The first 50 amino acid residues of Est2p are required for the nucleic acid binding activity.
To further define the nucleic acid binding domain, we constructed a series of plasmids for expressing subfragments of the Est2p N-terminal domain. As before, these subfragments were fused to both an MBP and a six-His tag. The fusion proteins were expressed in and purified from E. coli as previously described (Fig. 7A) and used in filter binding assays.
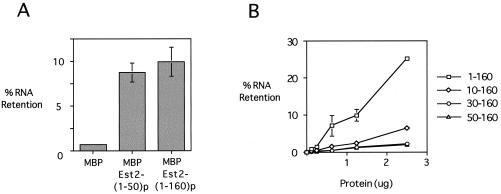
As shown in Fig. 8A, a large deletion from the C-terminal end of the Est2p fragment had little effect on RNA binding. Even a fusion protein with only the first 50 amino acid residues of Est2p [MBP-Est2(1–50)p] showed binding to RNA comparable to that of MBP-Est2(1–160)p. In contrast, a 10-amino-acid deletion from the N terminus significantly reduced the RNA binding activity, by approximately fivefold. When 30 or 50 amino acids were deleted from the N terminus, the RNA binding activity was reduced to the background level (Fig. 8B; also see the MBP plot in Fig. 7B). A similar series of assays using a double-stranded linear DNA as the probe gave essentially identical results (data not shown). These observations suggest that the N region (first 50 amino acids) of Est2p is largely responsible for the nonspecific nucleic acid binding activity exhibited by the 1–160 fragment.
FIG. 8.
The first 50 amino acid residues of Est2p are largely responsible for the nucleic acid binding activity exhibited by the N-GQ fragment. (A) Filter binding assays were performed using 1.3 μg of MBP, MBP-Est2(1–50)p, and MBP-Est2(1–160)p and 20 ng of labeled TLC1 RNA. The percentage of probe retained on the filter for each protein was plotted. (B) Filter binding assays were performed using increasing amounts of N-terminally truncated fusion proteins and 20 ng of labeled TLC1 RNA. The percentage of probe retained on the filter was plotted against the amount of protein used.
DISCUSSION
We have investigated the function of the N-terminal region of Est2p (yeast TERT) by a combination of HMM-based sequence comparison, mutagenesis, and biochemical studies. In this report, we describe the identification of four phylogenetically conserved non-RT motifs (named GQ, CP, QFP, and T) located in the N-terminal region of TERTs. The locations of these motifs within Est2p are as follows: GQ, residues 45 to 163; CP, residues 245 to 265; QFP, residues 267 to 343; and T, residues 367 to 413. Alanine substitutions of conserved GQ motif residues confirmed the functional importance of this motif. Indeed, the identification of two phenotypic classes of mutations within this motif suggests that it may mediate at least two distinct biochemical functions. Finally, we showed that the extreme N-terminal, nonconserved region (N region) of TERT possesses a nonspecific nucleic acid binding activity when analyzed in isolation. Furthermore, the N region proved to be important for telomere maintenance and telomerase activity in the context of the native telomerase RNP.
Comparison of the unigenic evolution approach and the HMM-based alignment approach in identifying functionally important TERT regions.
While our work was in progress, Friedman and Cech (11) reported the identification of essential yeast TERT N-terminal domains using a unigenic evolution approach. In this approach, the gene of interest is heavily mutagenized, and functional variants are selected. Essential and dispensable regions of the protein are then identified by statistically analyzing the distribution of missense and silent mutations. This analysis led to the identification of four essential regions in the Est2p N-terminal portion: region I, residues 31 to 163; region II, 214 to 265; region III, 285 to 374; and region IV, 378 to 432. A casual comparison between these regions and our motifs immediately indicates that they are largely concordant, with the GQ motif corresponding to region I, the CP motif corresponding to region II, and the QFP and T motifs corresponding to regions III and IV, respectively (Fig. 1B).
Because the unigenic evolution approach is aimed at identifying all essential regions, one would predict that the conserved motifs should constitute subparts of these regions. This is indeed the case for both the GQ and CP motifs. However, in the case of QFP and T motifs, there are two evident discrepancies that are worth noting. First, the QFP motif appears to encompass more residues at its N-terminal boundary than does region III. Nevertheless, mutations that are tolerated in the spacer between region II and region III do not affect conserved residues. Second, the boundary between region III and region IV and that between motif QFP and motif do not correspond. However, as noted by Friedman and Cech (11), because of the close proximity of these two regions, the choice of boundary is somewhat arbitrary.
While we did not present mutagenesis of conserved CP, QFP, and T motif residues in this report, preliminary studies indicate that conserved residues within these motifs are also required for normal telomere maintenance (Y. Peng and N. Lue, unpublished data). Furthermore, close inspection of tolerated mutations reported by Friedman and Cech (11) lends further credence to the validity of the alignment. Out of the 166 tolerated missense mutations, only 14 affect conserved or nearly conserved residues, and out of the 14 mutations, only 5 (N80→D, A360→E, T384→A, T384→I, and E385→V) change a conserved residue to a chemically dissimilar one. The identification of physiologically important conserved motifs that account for much of the N-terminal portion of TERTs suggests that this region shares a common structure and mediates conserved functions in telomere maintenance.
The function of the GQ motif.
While mutagenesis indicates that the GQ motif mediates an important function(s), its precise mechanisms are not understood. As described earlier, two mutations in nonconserved residues failed to affect telomerase function in vitro and in vivo. In contrast, eight out of eight mutants with changes in conserved residues show defective telomerase function in vivo. These mutations can be further classified according to the level of telomerase primer extension activity in vitro: one class of mutants (called class A, six of eight mutants) has 1/12 or less the wild-type levels of activity, while the other class (called class B, D66A and N104AV105A) has nearly wild-type levels of activity. The identification of phenotypically distinct mutants suggests that the GQ motif may play at least two functions in telomere maintenance. Potential defects for the two classes of mutants are discussed separately below.
For class A mutants, there is a good correlation between telomere shortening and loss of telomerase activity. For example, the G85A, Q138AF139A, and G141A mutants, which exhibit ∼1/12 to 1/20 of the wild-type activity, have telomeres that are on average ∼150 bp shorter and do not exhibit signs of senescence. The senescent strains (W115A, F118AH119A, and G123A) have nearly undetectable levels of telomerase activity (>50-fold reduction) and even shorter telomeres. Taken together, these results are consistent with the prevailing model that the equilibrium telomere length is established by both lengthening and shortening mechanisms, with the level of telomerase activity being a key component of the lengthening mechanism (32).
Since class A mutant proteins are present at normal or slightly reduced levels, their defects in telomere maintenance and telomerase activity cannot be explained by loss of expression or stability. Two possibilities can be considered. First, the assembly of Est2p into the telomerase complex may be affected. Consistent with this hypothesis, Friedman and Cech (11) found that two of their alanine substitution mutants (Ala-4, which changes residues 110 to 119 all to alanine, and Ala-5, which changes residues 145 to 154 all to alanine) exhibited reduced binding to TLC1 RNA. In this regard, it is also interesting to note that the region of hTERT (1 to 200) hypothesized to bind Hsp90 and P23 contains the GQ motif (21). Binding to Hsp90 and P23 has been suggested to play a role in the assembly of human telomerase RNP. It is possible that some of the class A mutations may disrupt Est2p interaction with Hsp90 and/or P23, thereby causing a reduction in telomerase activity. However, it should be noted that the ability of the four senescent mutants to act in a dominant-negative fashion suggests that the mutant proteins are capable of interacting with some component of the telomere pathway and interfering with its action. Second, it is possible that some of the class A residues participate directly in telomerase primer extension. Interestingly, secondary structure predictions suggest that the residues mutated in the senescent strains (W115A, F118AH119A, and G123A) may lie on the same face of an alpha helix and may therefore mediate the same function (I. Saira Mian, unpublished data).
Because the class B mutants (D66A and N104AV105A) have wild-type levels of telomerase activity, the synthesis and assembly of the RNP are probably not affected. Instead, functional interaction between Est2p and other factors that regulate telomerase in vivo may be disrupted. Three groups of factors are potential candidates for this interaction based on epistasis analysis: Est1p and Est3p, the Rad50-Mre11-Xrs2 complex, and factors in the Tel1p-Tel2p pathway. Strains that contain mutations in both telomerase core components (Est2p and TLC1) and factors in one of these three groups do not exhibit more severe growth defects or telomere shortening than do strains that carry single mutations (24, 40, 44). Thus, these factors may functionally interact with telomerase core components. Est1p and Est3p are presumed components of the telomerase holoenzyme but neither is required for in vitro activity. While the function of Est3p is not known, recent studies suggest that the Est1p may be involved in the recruitment of the core telomerase to chromosomal ends (9). The Rad50-Mre11-Xrs2 complex participates in nonhomologous double-stranded DNA break end-joining repair and possesses a 5′-to-3′ exonuclease activity (17). Tel1p is homologous to DNA-dependent protein kinases and other kinases involved in cell cycle checkpoint control (14, 36). Tel2p is an essential protein that exhibits a telomere sequence-specific DNA and RNA binding activity in vitro (23, 45). The mechanisms of these non-Est factors in telomere maintenance are poorly understood. Regardless of the precise interaction partner for the Est2p GQ motif, the fact that disruption of interaction can be caused by mutations in conserved residues suggests that the interaction may also be evolutionarily conserved.
The function of the N region.
Our results suggest that the first 50 amino acid residues of Est2p (N region) are required for telomere maintenance in vivo and telomerase activity in vitro. These 50 amino acid residues, when separated from the rest of the RNP, are also capable of mediating nucleic acid binding in vitro. Deleting the first 10 amino acids of Est2p results in significant telomere shortening and an ∼50-fold reduction in telomerase activity. The same deletion reduced the binding activity of the 1–160 fragment by fivefold. Deleting the first 30 or 50 amino acids had more severe effects both on the function of the full-length protein and on the binding activity of the 1–160 fragment. The correlation between function of the full-length protein and the binding activity of the N-terminal fragment suggests that the binding activity reflects an important physiologic function. We have been unable to express and purify a significant amount of full-length Est2p. Thus, it is not possible to compare the binding properties of N-terminal domain with those of the full-length protein. However, given the localization of the RT motifs in the C terminus of Est2p, the N-terminal domain is unlikely to account completely for the binding activity of the full-length protein.
The physiologic binding target for the Est2p N region is not clear. It seems unlikely that this fragment is binding both RNA and DNA nonspecifically in the context of the entire telomerase complex. Possibly, it has a specific binding target in vivo, but that it requires other domains of Est2p or other components of telomerase to achieve specific binding. One attractive target is the telomerase RNA. The N-terminal fragment may, for example, be involved in the formation of a stable RNP. This idea would be consistent with its important role in telomerase activity and telomere maintenance. Another attractive target is the telomeric DNA. TERT has been reported to possess a second primer-binding site away from the reverse transcription catalytic site (also known as the anchor site [7, 18]). The anchor site has been postulated to be important for processive elongation by ciliate telomerases. Earlier studies also suggest that anchor site interactions may be important for high-affinity primer-telomerase complex formation (29). Further studies will be necessary to define the precise target of the Est2p N region.
The N-GQ fragment may constitute a stable domain of TERT.
Sequence analysis suggests that the conserved GQ motif is followed in the alignment by a presumed flexible spacer, which exhibits little conservation and is variable in length. This conjecture is supported by our expression studies with E. coli. As described in the Results section, while the 1–160 fragment can be easily overproduced and purified from E. coli, larger fragments are quite susceptible to proteolysis. In addition, unigenic evolution analysis indicates that this linker region may be largely dispensable for function. The sequence alignment, mutagenesis, and biochemical studies, taken together, suggest that all TERTs may have at least a bipartite domain organization, with conserved N-terminal (N-GQ fragment) and C-terminal (CP-QFP-T-RT fragment) domains connected through a flexible spacer. The identification of a stable domain of TERT should open the way toward detailed structural analysis of at least an important part of telomerase. Given that telomerase has been shown to be an attractive anticancer drug target, such a structural study will no doubt be interesting both from a biochemical and from a pharmacological perspective.
ACKNOWLEDGMENTS
We thank B. Futcher, B. Schneider, and B. Schwer for strains and plasmids and B. Schwer for comments on the manuscript.
This work was supported by the American Cancer Society, the Concert for the Cure, and the AMDeC Foundation (N. F. Lue) and by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under contract no. DE-AC03-76SF00098 (I.S. Mian). Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
J.X. and Y.P. contributed equally to the work.
REFERENCES
- 1.Beattie T L, Zhou W, Robinson M O, Harrington L. Reconstitution of human telomerase activity in vitro. Curr Biol. 1998;8:177–180. doi: 10.1016/s0960-9822(98)70067-3. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E H. Telomerases. Annu Rev Biochem. 1992;61:113–129. doi: 10.1146/annurev.bi.61.070192.000553. [DOI] [PubMed] [Google Scholar]
- 3.Bryan T M, Sperger J M, Chapman K B, Cech T R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc Natl Acad Sci USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchman A R, Lue N F, Kornberg R D. Connections between transcriptional activators, silencers, and telomeres revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988;8:5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohn M, Blackburn E H. Telomerase in yeast. Science. 1995;269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 6.Collins K, Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc Natl Acad Sci USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins K, Greider C W. Tetrahymena telomerase catalyzes nucleolytic cleavage and nonprocessive elongation. Genes Dev. 1993;7:1364–1376. doi: 10.1101/gad.7.7b.1364. [DOI] [PubMed] [Google Scholar]
- 8.Collins K, Kobayashi R, Greider C W. Purification of Tetrahymena telomerase and cloning of genes encoding the two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- 9.Evans S K, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald M S, Riha K, Gao F, Ren S, McKnight T D, Shippen D E. Disruption of the catalytic telomerase subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman K L, Cech T R. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gandhi L, Collins K. Interaction of recombinant Tetrahymena telomerase proteins p80 and p95 with telomerase RNA and telomeric DNA substrates. Genes Dev. 1998;12:721–733. doi: 10.1101/gad.12.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenberg R A, Allsopp R C, Chin L, Morin G B, DePinho R A. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 14.Greenwell P W, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 15.Greider C W, Blackburn E H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 16.Greider C W, Blackburn E H. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 17.Haber J E. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 18.Hammond P W, Lively T N, Cech T R. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Program A E, Bass M B, Arruda I, Robinson M O. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- 20.Harrington L, Zhou W, McPhail T, Oulton R, Yeung D S K, Mar V, Bass M B, Robinson M O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holt S E, Aisner D L, Bauer J, Tesmer V M, Dy M, Ouellette M, Trager J B, Morin G B, Toft D O, Shay J W, Wright W E, White M A. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 1999;13:817–826. doi: 10.1101/gad.13.7.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilian A, Bowtell D D, Abud H E, Hime G R, Venter D J, Keese P K, Duncan E R, Reddel R R, Jefferson R A. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 23.Kota R S, Runge K W. The yeast telomere length regulator TEL2 encodes a protein that binds to telomeric DNA. Nucleic Acids Res. 1998;26:1528–1535. doi: 10.1093/nar/26.6.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le S, Moore J K, Haber J E, Greider C W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner J, Cech T R. Purification of telomerase from Euplotes aediculatis: requirement of a primer 3′ overhang. Proc Natl Acad Sci USA. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lingner J, Cech T R, Hughes T R, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 29.Lue N F, Peng Y. Negative regulation of yeast telomerase activity through an interaction with an upstream region of the DNA primer. Nucleic Acids Res. 1998;26:1487–1494. doi: 10.1093/nar/26.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lue N F, Xia J. Species-specific and sequence-specific recognition of the dG-rich strand of telomeres by yeast telomerase. Nucleic Acids Res. 1998;26:1495–1502. doi: 10.1093/nar/26.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundblad V, Szostak J W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 32.Marcand S, Brevet V, Gilson E. Progressive cis-inhibition of telomerase upon telomere elongation. EMBO J. 1999;18:3509–3519. doi: 10.1093/emboj/18.12.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyerson M, Counter C M, Eaton E N, Ellisen L W, Steiner P, Caddle S D, Ziaugra L, Beijersbergen R L, Davidoof M J, Liu Q E A. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 34.Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mian I S, Moser M J, Holley W R, Chatterjee A. Statistical modelling and phylogenetic analysis of a deaminase domain. J Comput Biol. 1998;5:57–72. doi: 10.1089/cmb.1998.5.57. [DOI] [PubMed] [Google Scholar]
- 36.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 37.Moser M J, Holley W R, Chatterjee A, Mian I S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura T M, Morin G B, Chapman K B, Weinrich S L, Andrews W H, Lingner J, Harley C B, Cech T R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama J-I, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: a gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- 40.Nugent C I, Bosco G, Ross L O, Evans S K, Salinger A P, Moore J K, Haber J E, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 41.Nugent C I, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 42.Oguchi K, Liu H, Tamura K, Takahashi H. Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana. FEBS Lett. 1999;457:465–469. doi: 10.1016/s0014-5793(99)01083-2. [DOI] [PubMed] [Google Scholar]
- 43.Prescott J, Blackburn E H. Functionally interacting telomerase RNAs in the yeast telomerase complex. Genes Dev. 1997;11:2790–2800. doi: 10.1101/gad.11.21.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ritchie K B, Mallory J C, Petes T D. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Runge K W, Zakian V A. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider B L, Seufert W, Steiner B, Yang Q H, Futcher A B. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- 47.Seto A G, Zaug A J, Sobel S G, Wolin S L, Cech T R. Saccharomyces cerevisiae telomerase is an Sm sn-RNP. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 48.Singer M S, Gottschling D E. TLC1:template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 49.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 50.Weinrich S L, Pruzan R, Ma L, Ouellette M, Tesmer V M, Holt S E, Bodnar A G, Lichtsteiner S, Kim N W, Trager J B, Taylor R D, Carlos R, Andrews W H, Wright W E, Shay J W, Harley C B, Morin G B. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]