Abstract
Nanodiscs, which are disc-shaped entities that contain a central lipid bilayer encased by an annulus of amphipathic helices, have emerged as a leading native-like membrane mimic. The current approach for the formation of nanodiscs involves the creation of a mixed-micellar solution containing membrane scaffold protein, lipid, and detergent followed by a time consuming process (3–12 h) of dialysis and/or incubation with sorptive beads to remove the detergent molecules from the sample. In contrast, the methodology described herein provides a facile and rapid procedure for the preparation of nanodiscs in a matter of minutes (<15 min) using Sephadex® G-25 resin to remove the detergent from the sample. A panoply of biophysical techniques including analytical ultracentrifugation, dynamic light scattering, gel filtration chromatography, circular dichroism spectroscopy, and cryogenic electron microscopy were employed to unequivocally confirm that aggregates formed by this method are indeed nanodiscs. We believe that this method will be attractive for time-sensitive and high-throughput experiments.
Keywords: nanodiscs, reconstitution, circular dichroism spectroscopy, analytical ultracentrifugation, dynamic light scattering, cryogenic electron microscopy
Graphical Abstract
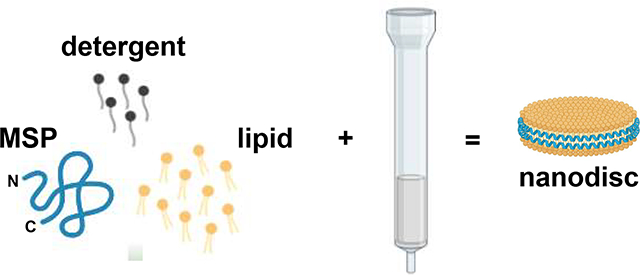
1. Introduction
Biophysical studies of membrane proteins commonly require the use of membrane mimics (e.g. vesicles, micelles, bicelles, and nanodiscs), and the ability of these mimics to recapitulate key aspects of cellular membranes is critical for maintaining the functionality of the biomolecule after reconstitution. Recently, nanodiscs pioneered by Sligar et al., have emerged as a particularly popular membrane mimic [1–11]. Nanodiscs are ovoid lipid aggregates with a diameter of 8 – 13 nm that contain a central lipid bilayer, which is encased by an annulus of amphipathic helices dubbed membrane scaffold protein (MSP) (Fig. 1). Nanodiscs are advantageous for three major reasons: 1) they possess a lipid bilayer, 2) they are amenable to a large variety of lipid compositions, and 3) they do not contain detergents.
Figure 1.
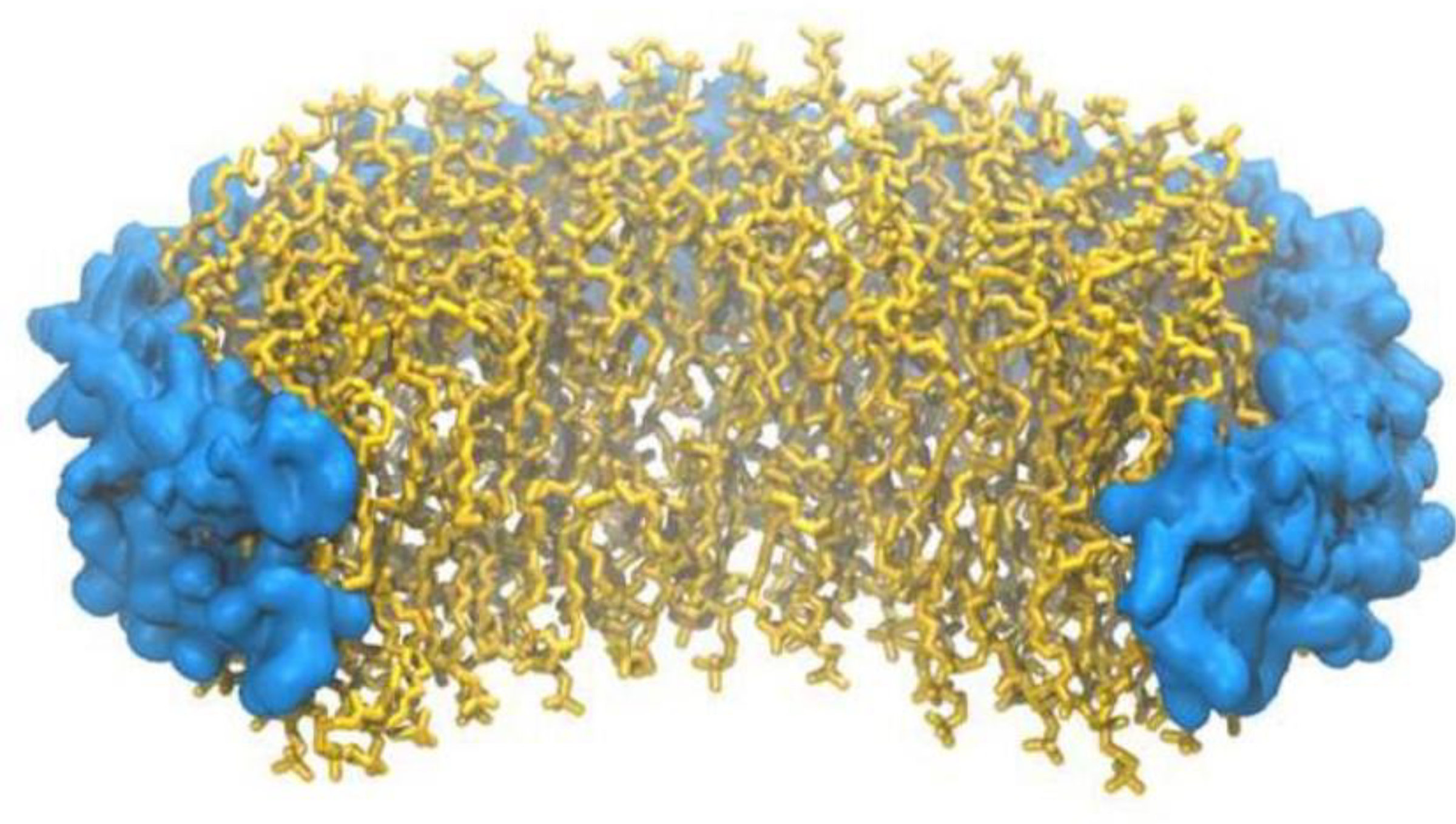
Cartoon image of a nanodisc. Membrane scaffolding protein is shown in blue. Dimyristoylphosphatidylcholine lipid is shown in yellow.
The typical method of preparing nanodiscs is to co-dissolve the MSP and phospholipid in a solution containing detergent followed by removal of the detergent via dialysis or incubation with sorptive beads, a process that typically takes 3 – 12 h [8–14]. However, the aforementioned two processes are not the only methods to remove detergent from a mixed-micellar solution [15–17]. In particular, we desired to investigate whether detergent removal using Sephadex® G-25 resin could be used to prepare nanodiscs. If successful, this method would have the primary advantage of significant time savings which could be beneficial for time-sensitive experiments.
In this report we show, using a complementary array of biophysical techniques, that nanodiscs can indeed be formed by passage through Sephadex® G-25 resin, and that the method is compatible with a number of detergents that are commonly used for nanodisc preparations.
2. Materials and Methods
A pET-based plasmid for the over-expression of MSP MSP1E3D1 (#20066) was obtained from Addgene (Watertown, MA). Dimyristoylphosphatidylcholine (DMPC) was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Sephadex® G-25 was purchased from EMD Millipore/Sigma-Aldrich (Milwaukee, WI). High affinity Ni-charged resin was purchased from GenScript Biotech Corporation (Piscataway, NJ). Sodium trichloroacetate was purchased from Alfa Aesar (Haverhill, MA). HiLoad™ 16/600 Superdex™ 200 column was purchased from Cytiva Life Sciences (Piscataway, NJ). All other reagents were of standard laboratory grade.
Expression and purification of MSP1E3D1.
MSP1E3D1 was overexpressed in BL21(DE3) E. coli cells and purified using nickel affinity chromatography. All buffers used during the purification were supplemented with 3.0 M sodium trichloroacetate. Finally, MSP1E3D1 was concentrated to 1.4 mM using a centrifugal ultrafiltration device (Pall Microsep® Advance 3K MWCO). MSP1E3D1 concentration was determined using the Micro BCA™ Protein Assay Kit.
Preparation of mixed-micellar solutions.
20 μL of 100 mg/mL DMPC in CHCl3 was added to a 1.5 mL microcentrifuge tube and the solvent was evaporated using a stream of dry N2 gas. Next, solutions of detergent, MSP, 10× buffer, and H2O were added according to Table 1, and the sample was vortexed and briefly heated in a hot water bath (~90°C) until clear.
Table 1.
Components required for the formation of nanodiscs.
| Sample | DMPC (mg) | 200 mM detergent (μL) | 1.4 mM MSP (μL) | 10× buffer (μL) | H2O (μL) | Total Vol. (μL) | MSP: DMPC* | Detergent: DMPC* |
|---|---|---|---|---|---|---|---|---|
| Cholate | 2.0 | 30 | 13 | 30 | 227 | 300 | 1:160 | 2:1 |
| CHAPS | 2.0 | 30 | 13 | 30 | 227 | 300 | 1:160 | 2:1 |
| OG | 2.0 | 60 | 13 | 30 | 197 | 300 | 1:160 | 4:1 |
Molar ratios are based on those presented in Ritchie et al [11].
Formation of particles.
Sephadex® G-25 resin was swollen in buffer for 24 h at room temperature. Next, the slurry was poured into a column with a 1.5 cm diameter and a bed height of 4.0 cm, and a small amount of chromatography sand was gently placed onto the resin surface as protection against agitation when buffer or sample was added. After equilibrating the column with 5 column volumes of buffer, the mixed-micellar solution (300 μL) was applied to the top of the column followed by the collection of 500 μL fractions using buffer.
Quantitation of detergents.
For cholate and CHAPS samples, 50 μL of each fraction was placed into a 1.5 mL microcentrifuge tube followed by the addition of 800 μL of concentrated H2SO4. For octyl glucoside samples, 50 μL of each fraction was placed into a 1.5 mL microcentrifuge tube followed by the addition 250 μL of 5% phenol and 600 μL of concentrated H2SO4. Samples were vigorously vortexed, and then incubated at room temperature for 30 min to allow for color development. The absorbance of cholate and CHAPS samples were measured at 389 nm, while octyl glucoside samples were measured at 490 nm. This procedure was a slight modification of that presented in Urbani et. al [18].
Gel filtration chromatography.
500 μL of sample was filtered through a 0.45 μm regenerated cellulose spin filter, and injected onto a HiLoad™ 16/60 Superdex™ 200 pg (prep grade) column equilibrated with 10 mM phosphate pH 7.4, 50 mM Na2SO4. A flow rate of 1.0 mL/min was used and 1.0 mL fractions were collected.
Dynamic light scattering.
Samples (~700 μL) were filtered through a 0.22 μm polyethersulfone (PES) syringe filter, and placed in a small glass test tube (12 mm × 75 mm) for analysis. Measurements were performed in triplicate. Second order cumulant data fitting was used.
Circular Dichroism Spectroscopy.
Samples (250 μL) were filtered through a 0.45 μm regenerated cellulose spin filter, and placed in a cuvette with a 1 mm pathlength for analysis. Data was collected from 190 to 250 nm with a resolution of 1 nm, an integration time of 4 s, and a temperature of 25°C. 4 scans were averaged for each sample.
Analytical Ultracentrifugation.
All sedimentation equilibrium experiments were performed at 25°C using a Beckman XL-A analytical ultracentrifuge and a 4-hole AnTi-60 rotor. Samples at three different particle concentrations were loaded into a 6-channel charcoal-filled epon centerpiece opposite a channel containing only buffer. A volume of 120 μL of sample was loaded per channel. Equilibrium absorbance measurements (240 nm) were taken at speeds of 8,500 rpm, 12,000 rpm and 15,000 rpm using a step size of 0.003 and 64 replicates. Verification of equilibrium was determined using the match feature in the software Heteroanalysis (version 1.1.0.58, University of Connecticut, Storrs, CT). The data were fitted globally in Heteroanalysis to a single-species model using a non-linear least squares approach. During fitting, the molecular weight, baseline, and reference concentration were all allowed to float.
Cryogenic electron microscopy.
Quantifoil 2/1 grids were glow discharged using a PELCO easiGlow cleaning system. From a 2 mg/mL stock of particles, 3.5 μL of each sample was added to each grid, blotted for 3–5 s, and plunged into liquid ethane using a FEI Vitrobot mark IV. Grids were stored in liquid nitrogen until transferred to a FEI Titan Krios G3 for imaging at the Huck Institute of Life Sciences at Pennsylvania State University. Micrographs were taken at 75,000× magnification at 300 kV.
Image processing and 3D particle reconstruction.
CryoSPARC was used for motion correction and contrast transfer function estimation of 1380, 804, and 900 micrographs of cholate, CHAPS, and octyl glucoside nanodiscs, respectively [19]. Initial 2D classification templates were formed from manually picking ~300 particles across 20 micrographs for each sample. Automated particle picking was performed across all micrographs using the initial templates. From these, new templates were generated and selected to exclude artifacts. CryoSPARC Ab-initio reconstruction and heterogeneous refinement was performed to generate 3–5 3D models from the selected templates. For each sample, the largest model was chosen for 3D visualization, which were constructed from 75193, 32961, and 29197 particles for cholate, CHAPS, and octyl glucoside nanodiscs, respectively. 3D models were visualized using UCSF Chimera [20].
Results and Discussion
Preparation of particles via Sephadex® G-25 resin.
For our studies we employed the ubiquitous MSP, MSP1E3D1, at a molar ratio of 1 : 160 MSP1E3D1 to DMPC (dimyristoylphosphatidylcholine), which is the established stoichiometry for preparing nanodiscs using this particular MSP [11]. However, it is important to note that this stoichiometry translates into 2 MSP1E3D1 molecules and 320 DMPC molecules per nanodisc (twice this ratio) [10,11,21,22]. First, the mixed-micellar solution is prepared by creating a homogeneous solution containing MSP1E3D1, DMPC, and detergent (Table 1). This results in a mixed micelle solution containing final sodium trichloroacetate, DMPC, and detergent concentrations of 61 mM, 10 mM and 20 – 40 mM, respectively. Three detergents commonly used for nanodisc preparations were chosen: sodium cholate, CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate), and octyl glucoside at DMPC to detergent ratios of 1:2, 1:2, and 1:4, respectively. Next, the mixed-micellar solution was applied to a short gravity column containing 7.0 mL of Sephadex® G-25 resin. Our hypothesis was that as the sample percolates through the resin, the monomers of the detergent that are in equilibrium with the mixed-micelle will be trapped in the pores of the media, resulting in the gentle and efficient removal of detergent followed by the spontaneous formation of nanodiscs. Fig. 2 shows our hypothesis graphically.
Figure 2.
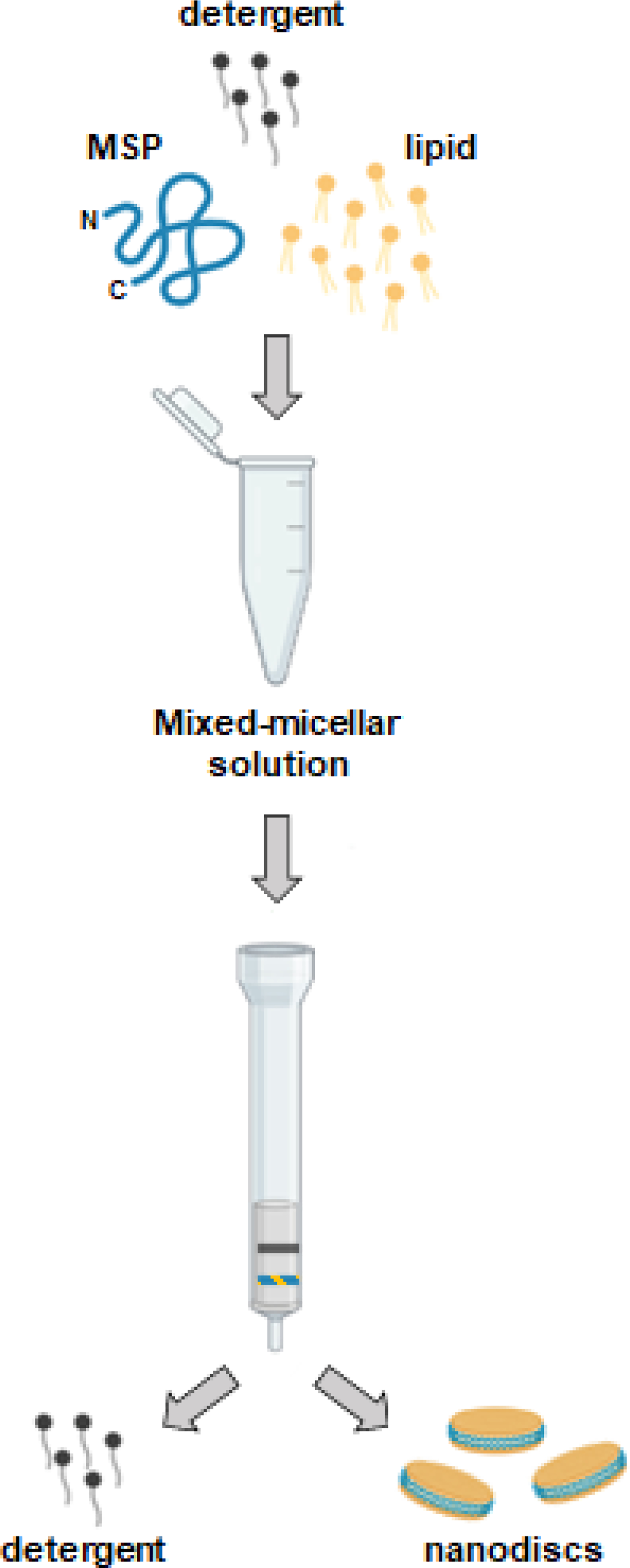
Pictorial representation of hypothetical nanodisc formation using Sephadex® G-25 resin.
When this procedure was performed, a species with a strong absorbance at 280 nm eluted in the void of the column (Fig. 3, black trace and Fig. S1), which indicated that it was much larger than the pores of the media. Importantly, if the voided species was an aggregate that did not contain MSP, which has 3 tryptophan and 9 tyrosine residues, it would not absorb at 280 nm. In addition, visually, the fractions containing the voided species were optically clear and did not show any turbidity, revealing that the 280 absorbance was not due to light scattering, and is indeed a MSP-containing species. For all 3 detergents, 99.6% of this species eluted in 2.0 mL (Fig. 3, grayed area). Next, all fractions were assayed for the concentration of detergent which revealed that in each case (Fig. 3, red trace), the detergent is effectively removed from the voided species (>99.9%).
Figure 3.
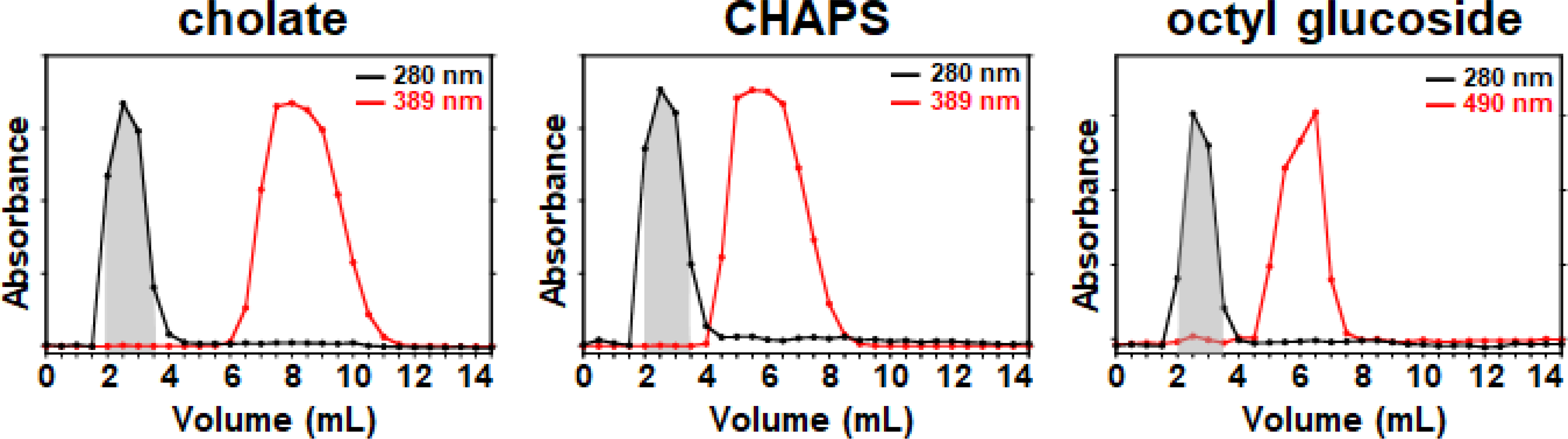
Elution profiles of mixed micellar solutions passed over Sephadex® G-25 resin. Spherical data points represent elution fractions. Red traces indicate the elution profile of detergent. Absorbances are normalized for facile viewing. 10 mM KH2PO4 pH 7.4, 50 mM Na2SO4 was used to equilibrate the column and collect fractions.
In all, processing the mixed micelle solution on the Sephadex® G-25 column takes less than 15 minutes from start to finish.
Determination of particle homogeneity by gel filtration chromatography.
Next, the particles produced using the SephadexⓇ G-25 column were subjected to analysis by gel filtration chromatography to assess the overall homogeneity of the sample. A HiLoad® 16/60 Superdex® 200 pg (prep grade) column was utilized as it is a common choice for researchers analyzing nanodisc samples [11,23–25]. Fig. 4 shows that for each detergent, a single major species was formed, and that processing of the mixed-micellar solution on the Sephadex® G-25 column does not result in an extremely inhomogeneous mixture of products. In addition, the elution volume of the major peak was nearly identical for all three detergents which strongly suggests that the same species is being formed in each case.
Figure 4.
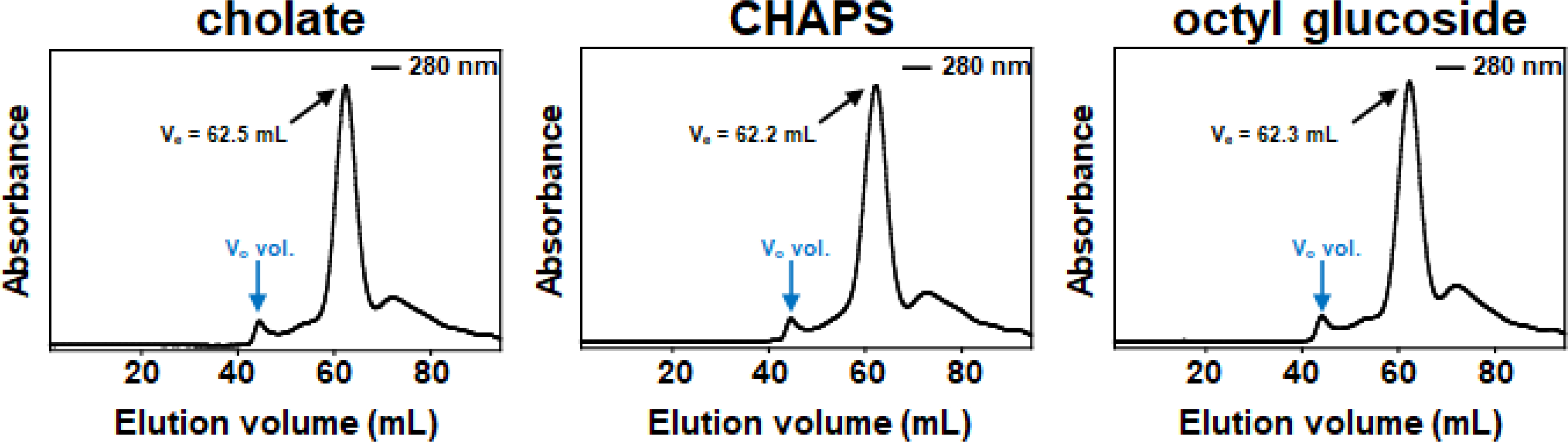
Elution profiles of particles on a HiLoad™ 16/60 Superdex™ 200 column. vo is void volume and ve is elution volume. 10 mM KH2PO4 pH 7.4, 50 mM Na2SO4 was used to equilibrate the column and collect fractions.
Determination of particle size by dynamic light scattering.
To characterize the dimensions of the particles produced, dynamic light scattering was employed. In a dynamic light scattering experiment, one obtains the hydrodynamic radius, Rh, of a hypothetical sphere that diffuses as fast as the particles being analyzed. Therefore, to investigate a particle such as a nanodisc that is discoidal and not spherical, the Rh of its equivalent sphere must be determined. This can be done using Eqn. 1 developed by Mazer et al. where t and r are the thickness and radius of the disc, respectively [26]. In our case, the thickness is taken to be 5.0 nm which is that of a fully-hydrated DMPC bilayer and the radius is taken to be 6.05 nm which is the well-established literature value for the radius of a MSP1E3D1 nanodisc [11,26,27].
| Eqn. 1 |
When this is done, the expected Rh for MSP1E3D1 nanodiscs is 5.53 nm. Fig. 5 shows the dynamic light scattering data obtained for each detergent. Second-order cumulant fitting was applied to the autocorrelation function which yielded the values of Rh and . is often called the polydispersity index and it can be used as indication of sample homogeneity. Furthermore, it is widely accepted that a sample can be considered reasonably homogeneous if the value of this parameter is less than 0.3 [28,29].
Figure 5.
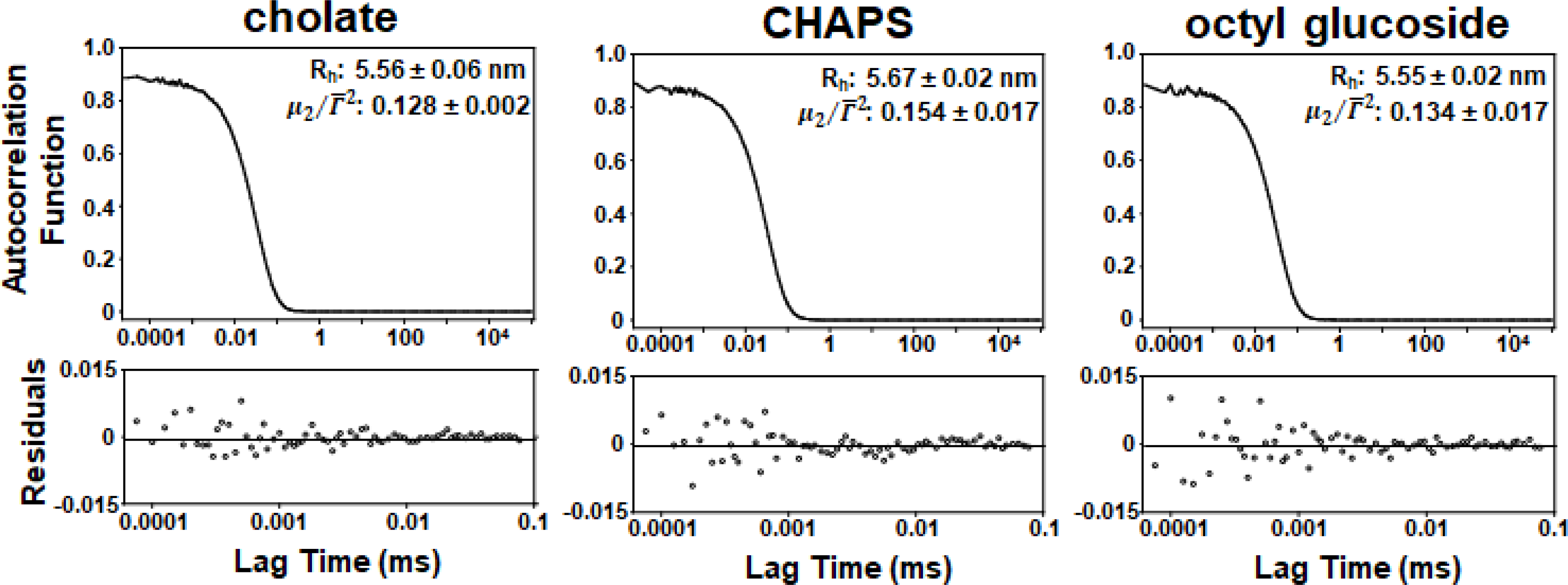
DLS autocorrelation functions, Rh, and values of the two highest intensity fractions from the Superdex™ 200 elution profile of particles produced on the Sephadex® G-25 column using 1× phosphate-buffered saline (PBS). Data was collected for 5 mins at 25° C using values of viscosity and refractive index of 1.0200 cP and 1.335 respectively for PBS.[35]
Clearly, the Rh values of 5.56 nm, 5.67 nm, and 5.55 nm obtained for each detergent match very well with the expected Rh for a nanodisc of 5.53 nm. In addition, the values of for each detergent were well below 0.3 indicating that the population of particles was quite homogeneous. Therefore, these results provide clear evidence that the particles produced were likely nanodiscs.
Determination of particle molecular weight by analytical ultracentrifugation.
Next, the particles were subjected to analysis by analytical ultracentrifugation using the method of sedimentation equilibrium. Sedimentation equilibrium is a powerful technique that yields molecular weight information that is independent of particle shape. Based on well-established nanodisc stoichiometry, there are two MSPs and 320 DMPC molecules per nanodisc [10,11,21,22]. From this ratio, the molecular weight of a nanodisc can be calculated using the formula below.
where Mnanodisc, MMSP, and MDMPC are the molecular weights of the nanodisc, MSP1E3D1, and DMPC, respectively. Using this formula, the molecular weight of a nanodisc is 282 kD. Another parameter that is needed for the experiment is the partial specific volume of the nanodisc which can be calculated as a weighted average of the partial specific volumes of DMPC (0.963 mL/g) [30] and MSP1E3D1 (0.732 mL/g; SEDENTERP software). Using this methodology, the calculated partial specific volume is 0.909 mL/g. Fig. 6 shows the sedimentation curves at 3 speeds and 3 concentrations for particles prepared with all three detergents.
Figure 6.
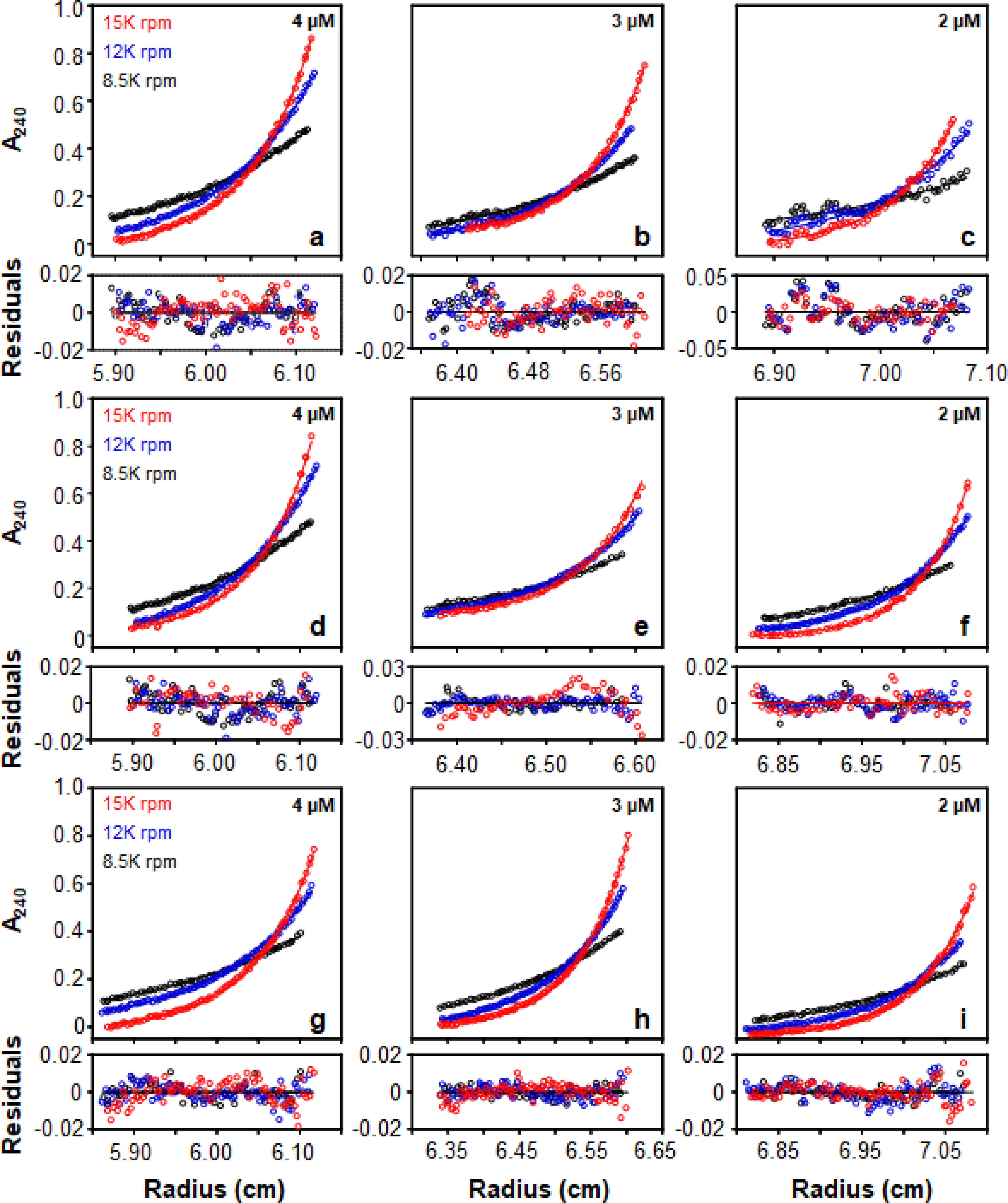
Sedimentation equilibrium profiles of particles produced on the Sephadex® G-25 column using sodium cholate (a-c), CHAPS (d-f), and octyl glucoside (g-i) in 10 mM KH2PO4 pH 7.4, 50 mM Na2SO4 at 25°C. Residuals are displayed underneath each equilibrium profile.
When this data was fit to the Lamm equation using non-linear regression and an ideal single species model, a molecular weight of 281 ± 3, 281 ± 2, and 289 ± 1 kD was obtained for cholate, CHAPS, and OG, respectively. All three of these values were very close to the ideal molecular weight of a nanodisc which is 282 kD. Therefore, the analytical ultracentrifugation data also supported that the particles obtained were likely nanodiscs.
Determination of particle secondary structure by circular dichroism spectroscopy.
Next, the particles were investigated by circular dichroism spectroscopy. Circular dichroism (CD) spectroscopy is a powerful tool that probes protein secondary structure.
Fig. 7 shows that the particles produced by all three detergents possessed the hallmark signature of a strongly ⍺-helical protein, which is minima at 208 and 222 nm along with a maximum at 192 nm. In addition, analysis of the spectrum using the K2D algorithm developed by Andrade et al. confirmed that the overall ⍺-helicity was greater than 60% for all 3 preparations [31–34]. To be certain that the DMPC molecules are not influencing the spectrum of the particles, a spectrum of DMPC in a micellar solution at a concentration identical to the DMPC concentration in the particles was acquired. The DMPC lipids did not produce any significant CD signal (Fig. S2); therefore, the observed CD spectrum of the particles was that of the MSP. This result was congruent with the established secondary structure of MSPs in nanodiscs as they are a series of amphipathic ⍺-helices that wrap around the lipid bilayer [23].
Figure 7.
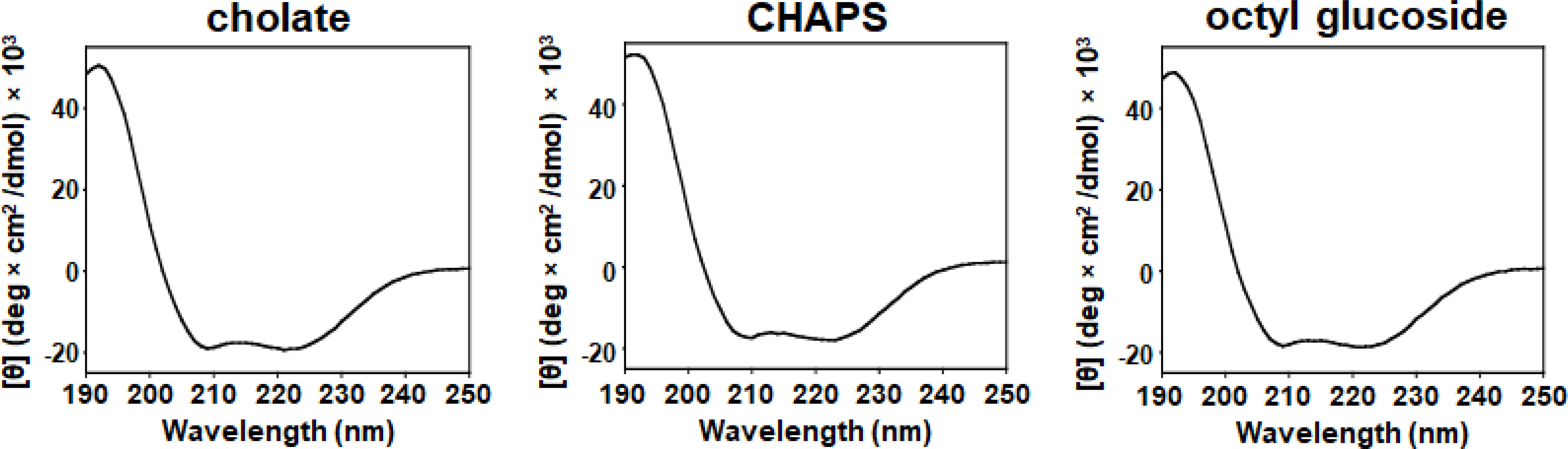
Circular dichroism spectra of particles in 10 mM KH2PO4 pH 7.4, 50 mM Na2SO4 at 25°C. [Θ] is mean residue ellipticity.
Evaluation of particle shape and size by cryogenic electron microscopy.
We used cryogenic electron microscopy to directly image the particles produced on the Sephadex® G-25 column.
As shown in Fig. 8, the particles had a discoidal shape and average size of 10.06 ± 2.12 nm, 10.27 ± 1.80 nm, and 10.74 ± 2.08 nm for the cholate, CHAPS, and octyl glucoside preparations, respectively. Therefore, the overall shape and size of the particles produced were in congruence with the formation of nanodiscs using the MSP1E3D1 scaffold protein, which have been previously reported to have a diameter of 12.1 nm [11]. A variety of nanodisc orientations were apparent in the micrographs, which allowed for particle reconstruction analysis. Reconstruction of the entities produced from each detergent preparation further supports the formation of nanodiscs as the electron density of the 2 MSP1E3D1 scaffolding proteins along the circumference of particles is clearly visible (Fig. 9).
Figure 8.
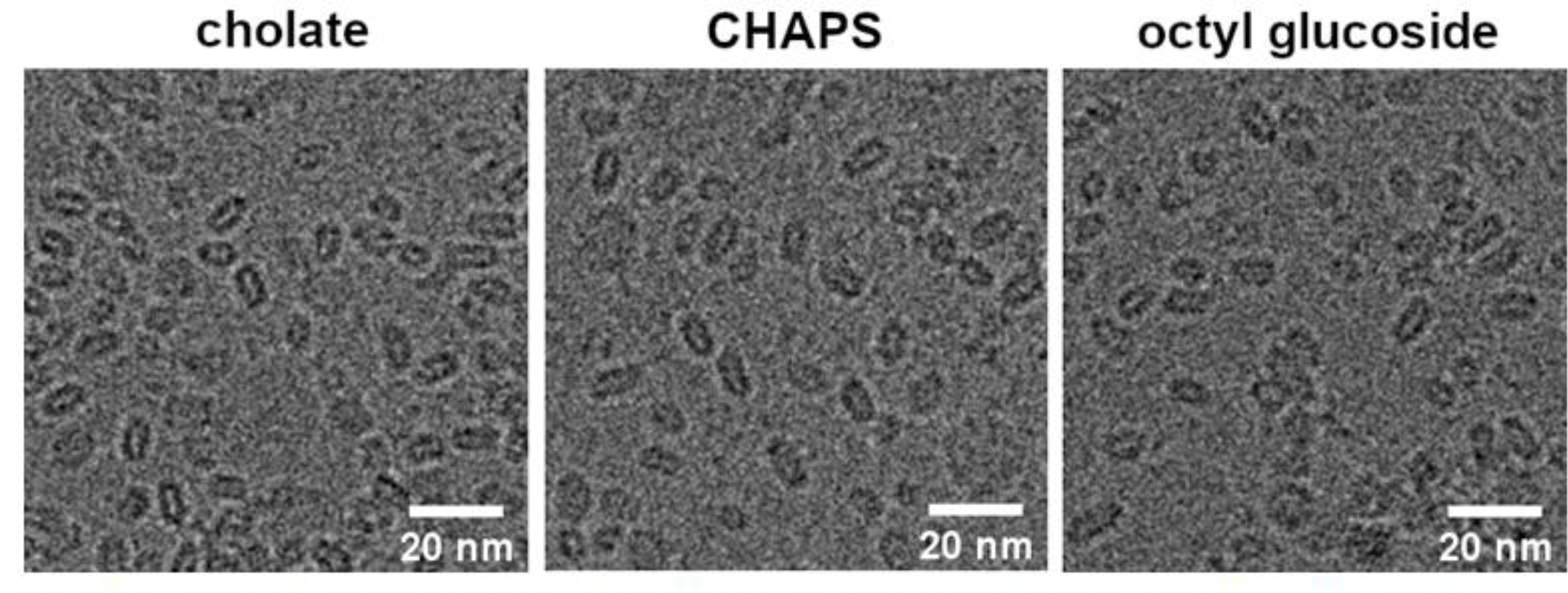
Cryogenic electron microscopy images of particles produced using Sephadex® G-25 resin with 10 mM KH2PO4 pH 7.4, 50 mM Na2SO4.
Figure 9.

Particle reconstruction images of particles produced using Sephadex® G-25 resin with 10 mM KH2PO4 pH 7.4, 50 mM Na2SO4.
The methodology detailed herein represents a rapid and attractive alternative to produce nanodiscs. Future studies will explore the potential of this method to facilitate the rapid reconstitution of membrane proteins in nanodiscs. This could be potentially beneficial for labile proteins as it dramatically limits the time the protein spends in detergent. In addition, this significant decrease in time also introduces the possibility of high-throughput screens of nanodisc reconstitution. Lastly, we believe that the membrane protein community will benefit from having the option of another methodology for the preparation of nanodiscs.
Supplementary Material
Acknowledgements
We thank Thomas Perone and Marvens Jean for laboratory assistance and Soohyung Park for preparing Figure 1. We thank Jennie Cawley for assistance in processing electron microscopy images in ImageJ. The authors sincerely thank Dr. Hyunwook Lee for useful advice and expertise concerning the cryo-EM experiments. Figure 2 was created using BioRender.com. This work was supported by NIH R15 GM141606-01 awarded to K.J.G. J.D.M., E.W.G., and E.D.G. acknowledge support from the Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the US Department of Energy, Office of Science, Basic Energy Sciences under award no. DE-SC0001090.
Footnotes
Conflicts of interests.
The authors declare that there are no competing conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Denisov IG, Sligar SG, Nanodiscs in Membrane Biochemistry and Biophysics, Chem. Rev. 117 (2017) 4669–4713. 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Boldog T, Li M, Hazelbauer GL, [14] - Using Nanodiscs to Create Water-Soluble Transmembrane Chemoreceptors Inserted in Lipid Bilayers, in: Simon MI, Crane BR, Crane A (Eds.), Meth. Enzymol, Academic Press, 2007: pp. 317–335. 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- [3].Yokogawa M, Fukuda M, Osawa M, Nanodiscs for Structural Biology in a Membranous Environment, Chem. Pharm. Bull. (Tokyo). 67 (2019) 321–326. 10.1248/cpb.c18-00941. [DOI] [PubMed] [Google Scholar]
- [4].Sun C, Gennis RB, Single-particle cryo-EM studies of transmembrane proteins in SMA copolymer nanodiscs, Chem. Phys. Lipids. 221 (2019) 114–119. 10.1016/j.chemphyslip.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Raschle T, Hiller S, Yu T-Y, Rice AJ, Walz T, Wagner G, Structural and Functional Characterization of the Integral Membrane Protein VDAC-1 in Lipid Bilayer Nanodiscs, J. Am. Chem. Soc. 131 (2009) 17777–17779. 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Matthies D, Bae C, Toombes GE, Fox T, Bartesaghi A, Subramaniam S, Swartz KJ, Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs, Elife. 7 (2018). 10.7554/eLife.37558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li MJ, Atkins WM, McClary WD, Preparation of Lipid Nanodiscs with Lipid Mixtures, Curr. Protoc. Protein Sci. 98 (2019) e100. 10.1002/cpps.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Denisov IG, Grinkova YV, Lazarides AA, Sligar SG, Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size, J. Am. Chem. Soc. 126 (2004) 3477–3487. 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- [9].Denisov IG, Sligar SG, Nanodiscs for structural and functional studies of membrane proteins, Nat. Struct. Mol. Biol. 23 (2016) 481–486. 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bayburt TH, Sligar SG, Membrane protein assembly into Nanodiscs, FEBS Lett. 584 (2010) 1721–1727. 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG, Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs, Meth. Enzymol. 464 (2009) 211–231. 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nath A, Atkins WM, Sligar SG, Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins, Biochem. 46 (2007) 2059–2069. 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- [13].Bayburt TH, Grinkova YV, Sligar SG, Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins, Nano Lett. 2 (2002) 853–856. 10.1021/nl025623k. [DOI] [Google Scholar]
- [14].Ivanova L, Denisov IG, Grinkova YV, Sligar SG, Fæste CK, Biotransformation of the Mycotoxin Enniatin B1 by CYP P450 3A4 and Potential for Drug-Drug Interactions, Metabolites. 9 (2019). 10.3390/metabo9080158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brunner J, Skrabal P, Hausser H, Single bilayer vesicles prepared without sonication physico-chemical properties, Biochim. Biophys. Acta Biomembr. 455 (1976) 322–331. 10.1016/0005-2736(76)90308-4. [DOI] [PubMed] [Google Scholar]
- [16].Allen TM, Romans AY, Kercret H, Segrest JP, Detergent removal during membrane reconstitution, Biochim. Biophys. Acta Biomembr. 601 (1980) 328–342. 10.1016/0005-2736(80)90537-4. [DOI] [PubMed] [Google Scholar]
- [17].Ollivon M, Lesieur S, Grabielle-Madelmont C, Paternostre M, Vesicle reconstitution from lipid–detergent mixed micelles, Biochim. Biophys. Acta Biomembr. 1508 (2000) 34–50. 10.1016/S0304-4157(00)00006-X. [DOI] [PubMed] [Google Scholar]
- [18].Urbani A, Warne T, A colorimetric determination for glycosidic and bile salt-based detergents: applications in membrane protein research, Anal. Biochem. 336 (2005) 117–124. 10.1016/j.ab.2004.09.040. [DOI] [PubMed] [Google Scholar]
- [19].Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination, Nat. Methods. 14 (2017) 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- [20].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera—A visualization system for exploratory research and analysis, J. Comput. Chem. 25 (2004) 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- [21].Puthenveetil R, Vinogradova O, Optimization of the Design and Preparation of Nanoscale Phospholipid Bilayers for its Application to Solution NMR, Proteins. 81 (2013) 1222–1231. 10.1002/prot.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Inagaki S, Ghirlando R, Grisshammer R, Biophysical characterization of membrane proteins in nanodiscs, Methods. 59 (2013) 287–300. 10.1016/j.ymeth.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Johansen NT, Tidemand FG, Nguyen TTTN, Rand KD, Pedersen MC, Arleth L, Circularized and solubility-enhanced MSPs facilitate simple and high-yield production of stable nanodiscs for studies of membrane proteins in solution, The FEBS J. 286 (2019) 1734–1751. 10.1111/febs.14766. [DOI] [PubMed] [Google Scholar]
- [24].Yeh V, Lee T-Y, Chen C-W, Kuo P-C, Shiue J, Chu L-K, Yu T-Y, Highly Efficient Transfer of 7TM Membrane Protein from Native Membrane to Covalently Circularized Nanodisc, Sci. Rep. 8 (2018) 13501. 10.1038/s41598-018-31925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lindhoud S, Carvalho V, Pronk JW, Aubin-Tam M-E, SMA-SH: Modified Styrene–Maleic Acid Copolymer for Functionalization of Lipid Nanodiscs, Biomacromolecules. 17 (2016) 1516–1522. 10.1021/acs.biomac.6b00140. [DOI] [PubMed] [Google Scholar]
- [26].Mazer NA, Benedek GB, Carey MC, Quasielastic light-scattering studies of aqueous biliary lipid systems. Mixed micelle formation in bile salt-lecithin solutions, Biochem. 19 (1980) 601–615. [DOI] [PubMed] [Google Scholar]
- [27].Small DM, Phase equilibria and structure of dry and hydrated egg lecithin, J. Lipid Res. 8 (1967) 551–557. [PubMed] [Google Scholar]
- [28].Sadeghi R, Etemad S.Gh., Keshavarzi E, Haghshenasfard M, Investigation of alumina nanofluid stability by UV–vis spectrum, Microfluid Nanofluid. 18 (2015) 1023–1030. 10.1007/s10404-014-1491-y. [DOI] [Google Scholar]
- [29].Danaei M, Dehghankhold M, Ataei S, Hasanzadeh Davarani F, Javanmard R, Dokhani A, Khorasani S, Mozafari MR, Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems, Pharmaceutics. 10 (2018). 10.3390/pharmaceutics10020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aune KC, Gallagher JG, Gotto AM, Morrisett JD, Physical properties of the dimyristoylphosphatidylcholine vesicle and of complexes formed by its interaction with apolipoprotein C-III, Biochem. 16 (1977) 2151–2156. 10.1021/bi00629a017. [DOI] [PubMed] [Google Scholar]
- [31].Whitmore L, Wallace BA, Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases, Biopolymers. 89 (2008) 392–400. 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- [32].Whitmore L, Wallace BA, DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data, Nucleic Acids Res. 32 (2004) W668–673. 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lobley A, Whitmore L, Wallace B, DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra RID C-5651-2008 RID C-3753-2008, J. Bioinform. 18 (2002) 211–212. 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- [34].Andrade MA, Chacón P, Merelo JJ, Morán F, Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network, Protein Eng. 6 (1993) 383–390. 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- [35].Barchanski A, Laser-Generated Functional Nanoparticle Bioconjugates: Design for Application in Biomedical Science and Reproductive Biology, Springer, 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


