The intestinal helminth Heligmosomoides polygyrus suppresses development of epithelial tuft cells, which are essential for the type 2 immune response, through secreted factors that block tuft cells both in intestinal organoids and when administered in vivo.
Abstract
Helminth parasites are adept manipulators of the immune system, using multiple strategies to evade the host type 2 response. In the intestinal niche, the epithelium is crucial for initiating type 2 immunity via tuft cells, which together with goblet cells expand dramatically in response to the type 2 cytokines IL-4 and IL-13. However, it is not known whether helminths modulate these epithelial cell populations. In vitro, using small intestinal organoids, we found that excretory/secretory products (HpES) from Heligmosomoides polygyrus blocked the effects of IL-4/13, inhibiting tuft and goblet cell gene expression and expansion, and inducing spheroid growth characteristic of fetal epithelium and homeostatic repair. Similar outcomes were seen in organoids exposed to parasite larvae. In vivo, H. polygyrus infection inhibited tuft cell responses to heterologous Nippostrongylus brasiliensis infection or succinate, and HpES also reduced succinate-stimulated tuft cell expansion. Our results demonstrate that helminth parasites reshape their intestinal environment in a novel strategy for undermining the host protective response.
Introduction
Helminth infections cause major neglected tropical diseases, afflicting >20% of the world’s population with soil-transmitted helminths alone (Jourdan et al., 2018; Pullan et al., 2014). Unlike many pathogens, parasitic helminths establish long-lasting infestations, frequently evoking minimal inflammatory reaction. This has been attributed to the release of multiple immunomodulatory molecules throughout infection, broadly referred to as excretory-secretory (ES) products (Lightowlers and Rickard, 1988; Maizels et al., 2018). Most studies have focused on how ES constituents affect immune cells, for instance the TGF-β mimic of Heligmosomoides polygyrus (TGM), which induces immunosuppressive regulatory T cells (Cook et al., 2021; Johnston et al., 2017; White et al., 2021). However, intestinal helminths localize to the intestinal lumen in close association with the epithelial layer, and the direct effects of helminths or their products on the epithelium are poorly understood.
The epithelium initially alerts the immune system to incoming parasites, producing the alarmins IL-25, IL-33, and thymic stromal lymphopoietin in response to tissue damage caused by helminths (Artis and Grencis, 2008) such as H. polygyrus, which invades the submucosa at the larval stage before returning to the lumen as an adult (Reynolds et al., 2012). Alarmins recruit innate immune cells and APCs to the infected area, and activate type 2 immune responses, including CD4+ T helper type 2 (Th2) cell effectors (Inclan-Rico and Siracusa, 2018). Parasite-secreted proteins have been identified that block epithelial alarmin signals, such as H. polygyrus alarmin release inhibitor (HpARI), which binds IL-33 to prevent its release from damaged cells (Osbourn et al., 2017). However, ES constituents with direct effects on epithelial cells themselves have yet to be identified, despite helminths inducing extensive physiological changes to the gut (McKay et al., 2017).
The intestinal epithelium is remodeled drastically during helminth infection, driven by the type 2 signature cytokines IL-4 and IL-13. These drive the “weep and sweep” response with increased epithelial mucus production and intestinal muscular contractility required for parasite expulsion, and expansion of epithelial goblet, Paneth, and tuft cells (Gerbe et al., 2016; Howitt et al., 2016; Kamal et al., 2002; Sharpe et al., 2018). Recently, tuft cells have been recognized as central orchestrators in this response, producing IL-25, leukotrienes, and other mediators that activate ILC2s to produce IL-13 (Billipp et al., 2021; McGinty et al., 2020; von Moltke et al., 2016). IL-13 stimulates epithelial cells in a positive feedback loop to increase tuft and goblet cell numbers. IL-4, which shares the receptor subunit IL-4Rα with IL-13, can also induce tuft cell hyperplasia (Gerbe et al., 2016).
To investigate the response of the intestinal epithelium to helminth ES products, we first exposed murine small intestinal organoids to H. polygyrus ES products (HpES) in combination with Th1 or Th2 cytokines, to mimic the effects of infection. RNA sequencing (RNA-seq) of these organoids revealed a down-regulation of tuft cell–associated genes by HpES, alongside inhibition of tuft cell expansion. In vivo, tuft cell induction by both succinate and the nonresident helminth Nippostrongylus brasiliensis was reduced in the presence of H. polygyrus. HpES also exerted generalized effects on organoid development, altering expression of key transcription factors and promoting a spheroid morphology. Together, these data demonstrate that H. polygyrus directly affects the development of the intestinal epithelium via ES products.
Results and discussion
Small intestinal organoid gene expression is differentially regulated by immune cytokines and HpES
To model the impact of helminth products on the intestinal epithelium, we first analyzed the transcriptional profile of mouse small intestinal organoid cultures exposed to ES molecules (HpES) released by the intestinal helminth H. polygyrus. Established organoid cultures were stimulated with IFN-γ as the main driver of type 1 responses, or a combination of IL-4 and IL-13 to induce type 2 responses, in the presence or absence of HpES. Stimulations were performed over 24 h in replicate organoids derived from four different C57BL/6 individual mice. RNA was extracted from each replicate and subjected to bulk RNA-seq analyses.
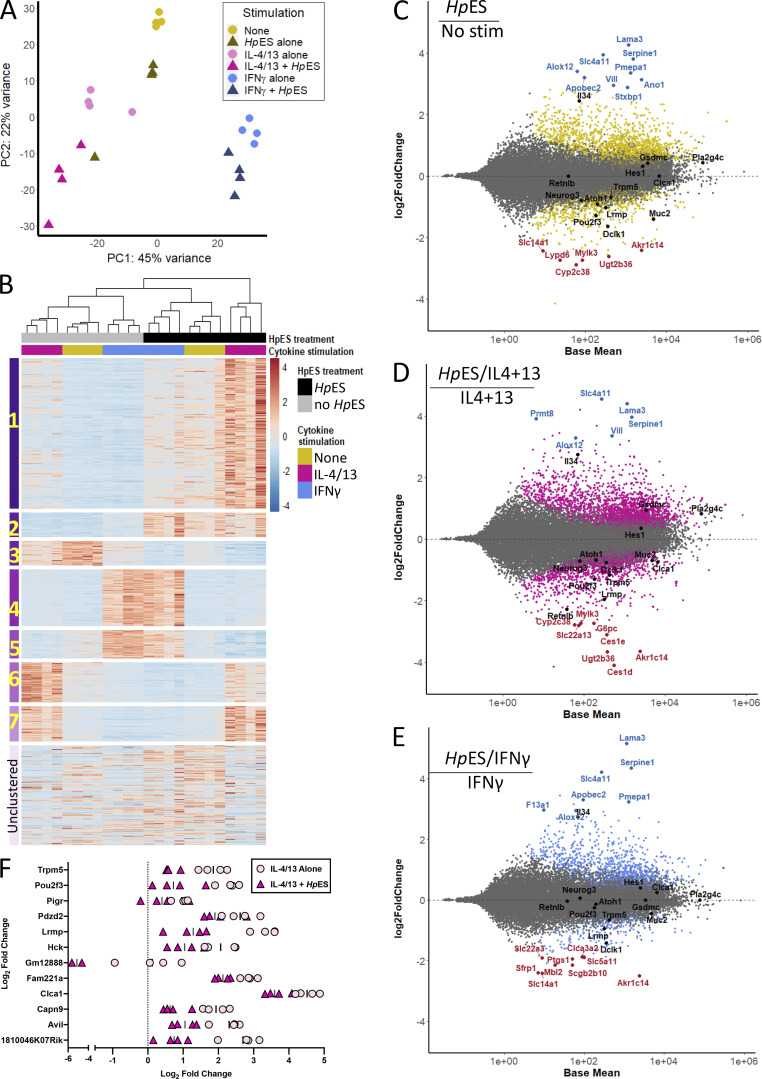
By principal component analysis, samples from the same stimulation conditions clustered together (Fig. 1 A), with the majority of variance driven by cytokine stimulation (PC1, at 45%); notably, treatment with type 1 or type 2 cytokines induced diametrically opposite effects on PC1 compared with unstimulated controls. The addition of HpES caused a consistent shift in PC2 in all groups, with the same directionality in the presence or absence of cytokines, explaining >20% of the remaining variance. These results support the hypothesis that epithelial cells are highly responsive to both type 1 and type 2 cytokines and demonstrate that these responses can be significantly affected by helminth products.
Figure 1.
Gene expression in HpES- and cytokine-treated small intestinal organoids. Organoid cultures grown from duodenal crypt cells, taken from four individual C57BL/6 mice, were stimulated in the presence or absence of the type 1 cytokine IFN-γ, the type 2 cytokines IL-4 and IL-13 (IL-4/13), and/or the ES products of adult H. polygyrus parasites (HpES). After 24 h, RNA was extracted from each replicate, and all 24 samples were subjected to parallel RNA-seq analyses. Patterns of gene expression were then analyzed. (A) Principal component analysis plot showing transcriptomes of stimulated organoids projected onto two dimensions; each condition is represented by four replicates derived from four individual mice. Principal component analysis data were produced from log-transformed normalized counts in the DESeq2 package (Love et al., 2014). (B) Heat map of the top 1,000 DEGs, organized into seven gene clusters, and a group of unclustered genes. Z-scores of normalized count values are indicated by coloring from blue (lowest) to red (highest), based on data from four replicates per group, each individually presented. (C–E) MA plots showing log2 fold change (M) plotted against log of mean normalized expression counts (A) for the comparisons of no stimulation versus HpES alone (C), IL-4/13 alone versus IL-4/13 + HpES (D), and IFN-γ alone versus IFN-γ + HpES (E). Data from four replicates per group were pooled; genes with adjusted P value < 0.05 are colored. The top 10 DEGs by log2 fold change that have known function are annotated, with those increased in the presence of HpES shown in blue and those decreased in red. Additional genes of interest are shown in black. DEGs were calculated in DESeq2. (F) Expression levels of 12 genes from cluster 6 showing the most significant (by adjusted P value) differential expression in the presence of HpES. Graphs show normalized counts/log2 fold change compared with no stimulation control from four independent biological replicates, for organoid cultures exposed to IL-4/13 alone (pink circles) or IL-4/13 + HpES (magenta triangles). Stim, stimulation.
Just under 34,000 (33,857) different transcripts were identified, and 15,000 were identified as differentially expressed genes (DEGs) using a likelihood ratio test across all conditions. A cluster analysis of DEGs defined seven expression patterns, and a heatmap of the 1,000 most altered genes in the study is presented in Fig. 1 B, demonstrating that HpES has profound gene-specific effects on intestinal epithelial cells. Pairwise comparisons between the different conditions revealed that while addition of IL-4/13 or IFN-γ alone modulated 1,150 and 1,673 DEGs, respectively, organoids exposed to HpES had a far greater number of DEGs than their equivalents stimulated with cytokines alone, with >6,700 (3,867 up-regulated and 2,913 down-regulated) resulting from the addition of HpES to the type 2 cytokines. The gene-specific effects of HpES are shown in organoid cultures without added cytokines (Fig. 1 C), or added IL-4/IL-13 (Fig. 1 D) and IFN-γ (Fig. 1 E), presented as MA plots of log fold-change (M) versus mean abundance (A). The more marked effect of HpES in the type 2 setting of IL-4/13 stimulation compared with type 1/IFN-γ (Fig. 1, B and D) is consistent with adaptation by this helminth parasite to modulate the mode of immunity that would lead to its expulsion (Reynolds et al., 2012).
We next analyzed gene ontology (GO) terms, which reflected induction of inflammatory responses by IFN-γ and multiple metabolic pathways by the type 2 cytokines; notably, treatment with HpES preferentially activated wound healing and angiogenesis pathways (Fig. S1 A), a finding of interest with respect to the tissue-invasive life cycle of H. polygyrus. The individual genes most up-regulated by HpES treatment of organoids included Serpine1, Lama3 (Laminin subunit α 3) and Vill (villin-like), all involved in cell adhesion, suggesting induction of structural changes in the organoids by HpES (Fig. 1 C). IL-4/13 alone up-regulated two goblet cell products, Clca1, a calcium-activated chloride channel regulator, and Retnlb, which encodes resistin-like molecule (RELM)-β, known to impair H. polygyrus feeding (Herbert et al., 2009). Both genes are down-regulated by the addition of HpES (Fig. 1 D). In agreement with investigated GO terms, we observed in all samples exposed to HpES a marked increase in arachidonate 12-lipoxygenase (Alox12), which may convert arachidonic acid into repair-promoting lipoxins and resolvins (Esser-von Bieren, 2019), as well as phospholipase A2 (Pla2g4c), which is responsible for forming arachidonic acid from phospholipid membranes.
Figure S1.
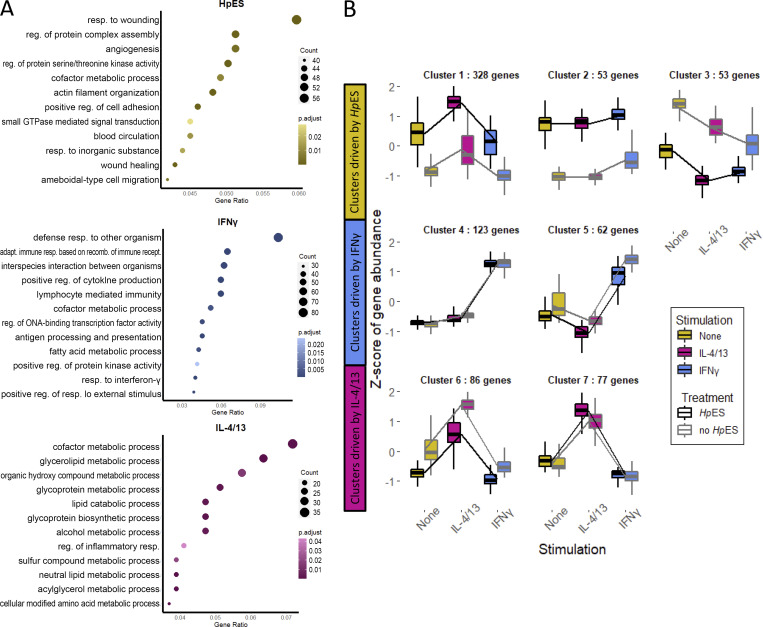
GO terms for DEGs. (A) HpES and cytokine treatment of small intestinal organoids. Top 12 GO terms, by gene ratio, for DEGs from HpES, IL-4/13, and IFN-γ individual treatments. Data are based on analyses of four biological replicates, composed of a total of 24 samples analyzed in parallel by RNA-seq. DEGs were selected as those with a P-adjusted value of <0.01. The enrichGO function from ClusterProfiler (Yu et al., 2012) was used to identify enriched GO terms, followed by ReViGO (Supek et al., 2011) to remove redundant GO terms. The top 12 GO terms were then selected using gene ratio (number of genes associated with GO term in list/total number of genes in list). (B) Responses of identified gene clusters to HpES, IL-4/13, and IFN-γ. Gene sets were split based on treatment (± HpES) and stimulation (IL-4/13, IFN-γ, or none). Clusters were identified using degPatterns, a part of the DEGreport package (Pantano et al., 2020). Adapt., adaptive; GTPase, guanosine triphosphatase; rec., reecombinant; reg., regulation; resp., response.
IL-4/13–responsive genes down-regulated by HpES include markers of intestinal tuft cells
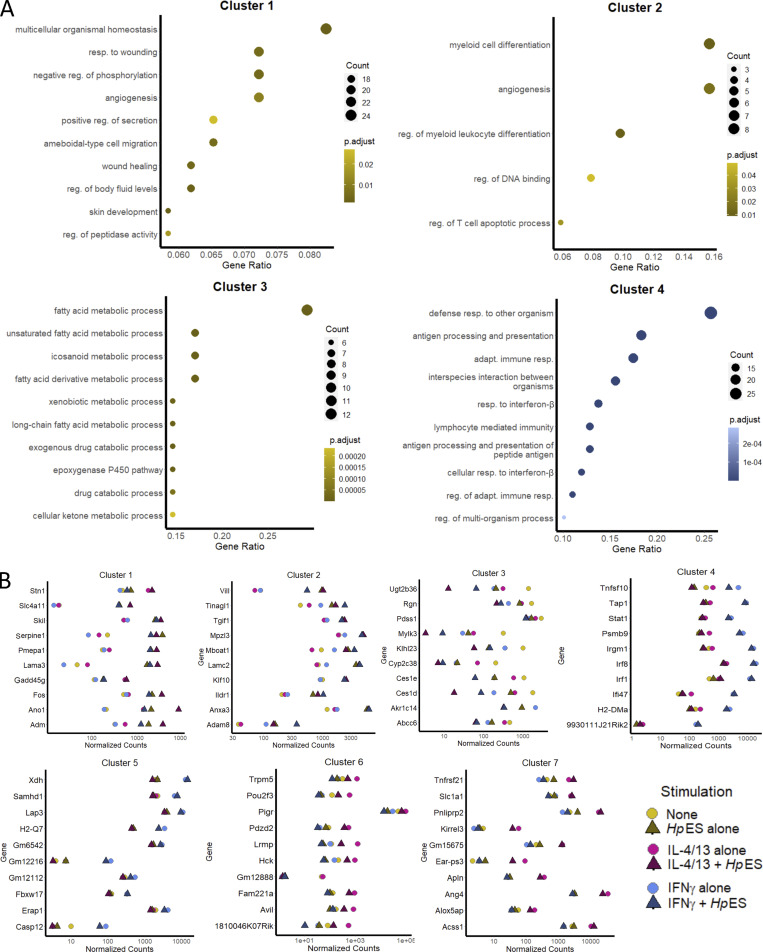
Clustering analysis on the top 1,000 DEGs (ranked by P-adjusted value), grouping genes according to their expression profile across the different stimulation, identified seven clusters with different expression profiles containing >50 genes (Fig. S1 B). The largest, cluster 1, corresponded to 328 genes that were up-regulated by both HpES and IL-4/13, many of which were represented by GO terms for homeostatic maintenance and the response to wounding (Fig. S2 A). Cluster 2 also contained genes up-regulated by HpES, irrespective of the cytokine milieu. Other clusters of interest grouped together genes down-regulated by HpES under all conditions (cluster 3), and clusters up-regulated by IFN-γ that were unaffected (cluster 4) or down-modulated (cluster 5) by HpES. The top 10 DEGs for each cluster, and their expression levels in each stimulatory condition, are shown in Fig. S2 B.
Figure S2.
Cluster analysis of HpES modulated gene expression. (A) Top 5 or 10 GO terms by gene ratio for each cluster. Only clusters 1–4 were identified as having corresponding GO terms using enrichGO from ClusterProfiler, and only five GO terms were returned for cluster 2. Redundant GO terms were removed using ReViGO before plotting. (B) Mean normalized counts for the top 10 genes from each cluster by P-adjusted values. Adapt., adaptive; reg., regulation; resp., response.
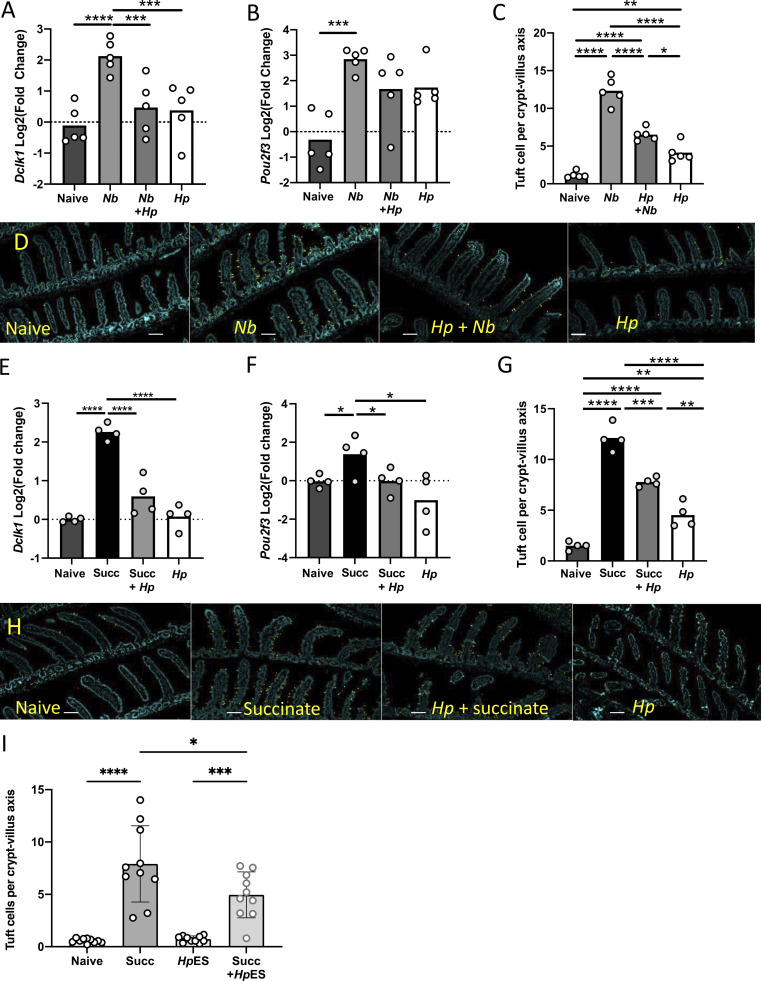
We then focused on cluster 6, containing 86 genes up-regulated by IL-4/13 treatment but repressed by HpES, including Lrmp, Pou2f3, and Trpm5 (Fig. 1 F), which in the intestinal epithelium are specific for tuft cells, a chemosensory cell type that differentiates in response to IL-13 (Gerbe et al., 2016; von Moltke et al., 2016). Mice lacking either Pou2f3 (Gerbe et al., 2016) or Trpm5 (Howitt et al., 2016) are defective in their ability to expel helminth parasites. The goblet cell marker Clca1 (Leverkoehne and Gruber, 2002) is also present in this set of DEGs. The down-regulation of these markers suggests that H. polygyrus may interfere with intestinal development to inhibit cell types involved in helminth elimination.
HpES and L3 larvae repress tuft cell expansion in small intestinal organoids
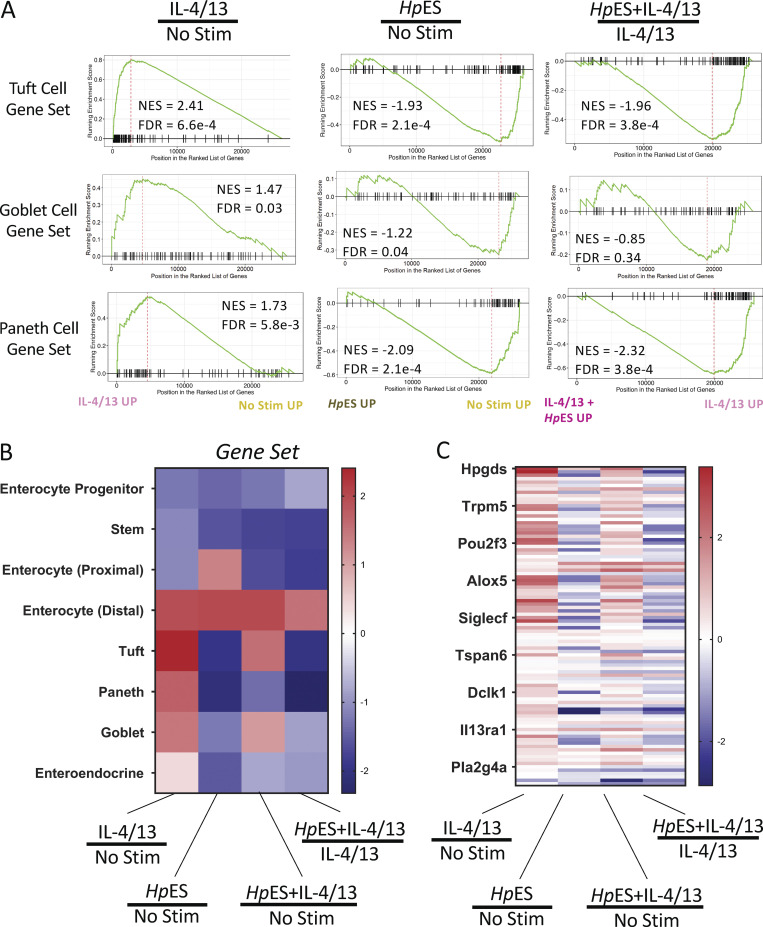
To search more systematically for potential cell-specific inhibitory effects, we then applied gene set enrichment analysis (GSEA) on the differentially regulated genes. Using gene sets identified from epithelial cell types in single-cell RNA-seq analyses of the intestinal epithelium (Haber et al., 2017), we found that HpES treatment caused a marked reduction in normalized enrichment scores (NESs) for goblet, Paneth, and tuft cell–associated gene sets, both alone and in the presence of IL-4/13 (Fig. 2, A and B). Reduced scores for each of these gene sets suggest regulation of intestinal cell differentiation by components of HpES, as all three are secretory cell types (Chiacchiera, 2019).
Figure 2.
GSEA of HpES modulated gene expression. (A) GSEA of the genes expressed by organoids exposed to IL-4/13, HpES, or neither, for gene sets of tuft cells, goblet cells, and Paneth cells as described by Haber et al. (2017). Graphs depict the enrichment score (y axis, green line) with positive values where gene sets are induced, and negative values where they are inhibited. Each vertical bar on the x axis represents an individual gene within the gene set for the stated cell type, and its relative ranking against all genes analyzed. NES and false discovery rate (FDR) are indicated on each graph. Gene expression data are pooled from four independent biological replicates, which were analyzed in parallel by RNA-seq. (B) Heat map of NES for cell type gene sets from Haber et al. (2017) in organoids treated with combinations of IL-4/13, HpES, or neither. (C) Heat map of tuft cell gene set expression, with key genes indicated on the y axis, showing log2 fold change of genes in organoids treated with combinations of IL-4/13, HpES, or neither. Stim, stimulation.
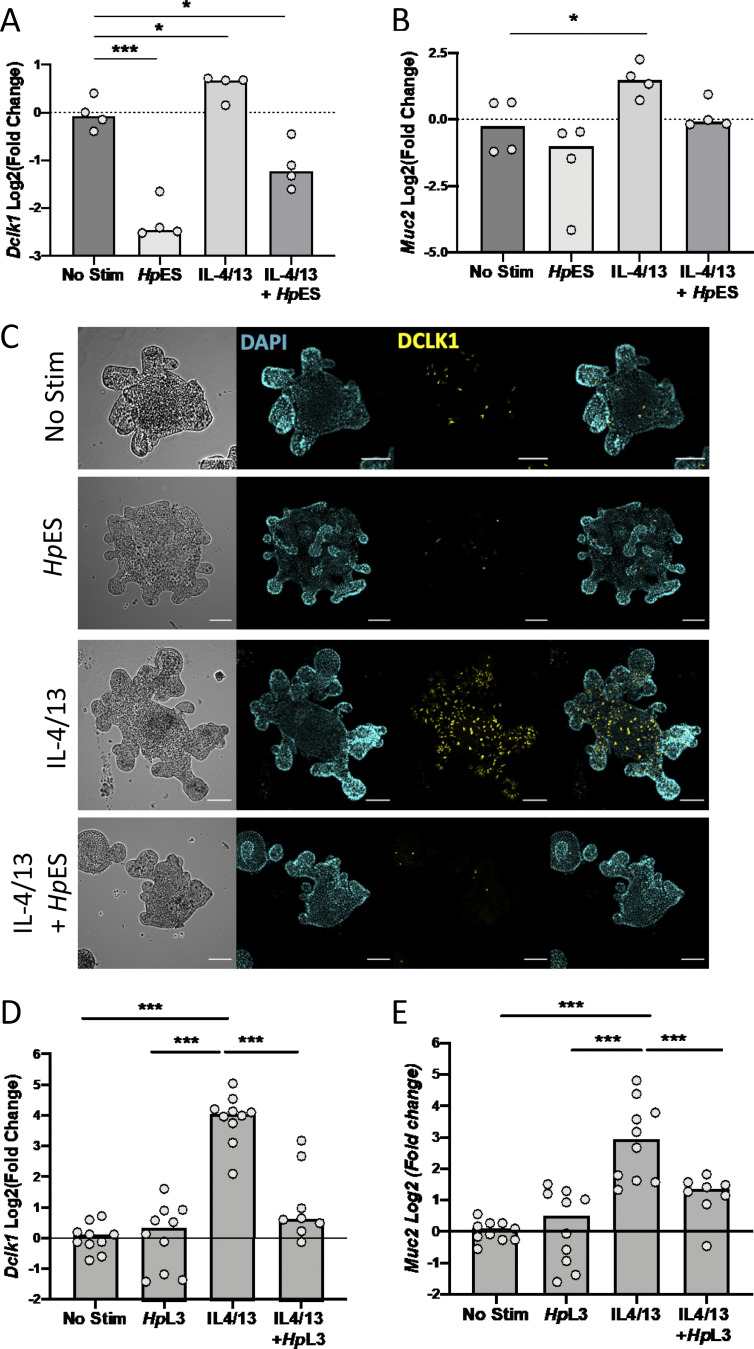
Within the tuft cell gene set, multiple canonical genes such as Dclk1, Pou2f3, and Trpm5 showed similar patterns of induction by IL-4/13, but were suppressed by HpES treatment, even in the presence of type 2 cytokines (Fig. 2 C). Loss of Dclk1 and Pou2f3 induction was also confirmed by quantitative RT-PCR (qRT-PCR) analysis of cytokine- and HpES-treated organoids (Fig. 3 A and Fig. S3 A). Futhermore, HpES suppressed the induction of Muc2, a major gene product of goblet cells (Fig. 3 B). In addition, organoids stained with anti-Dclk1 antibody showed that HpES completely blocked tuft cell induction by IL-4/13, and even reduced baseline tuft cell differentiation in unstimulated organoids (Fig. 3 C).
Figure 3.
HpES and H. polygyrus larvae repress tuft cell expansion in small intestinal organoids. (A and B) Expression of canonical tuft cell and goblet cell genes in organoids treated with combinations of IL-4/13, HpES, or neither. Log2 fold change of qRT-PCR values for Dclk1 and Muc2 compared with nonstimulated control in four independent biological replicates analyzed by RNA-seq. Statistical analysis was by ordinary one-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; ***, P < 0.001. (C) Representative images of organoids stained for tuft cells (anti-DCLK1), shown in yellow, in in organoids treated with combinations of IL-4/13, HpES, or neither. Nuclear staining (DAPI) shown in cyan. Scale bar is 100 µm. (D and E) Expression of Dclk1 and Muc2 in H. polygyrus L3 larvae exposed organoids, Log2 fold change of qRT-PCR values shown compared with nonstimulated control. Data are pooled from four experiments each with two to four replicates, total n = 8–10 per group. Statistical analysis was by ordinary one-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; ***, P < 0.001. Stim, stimulation.
Figure S3.
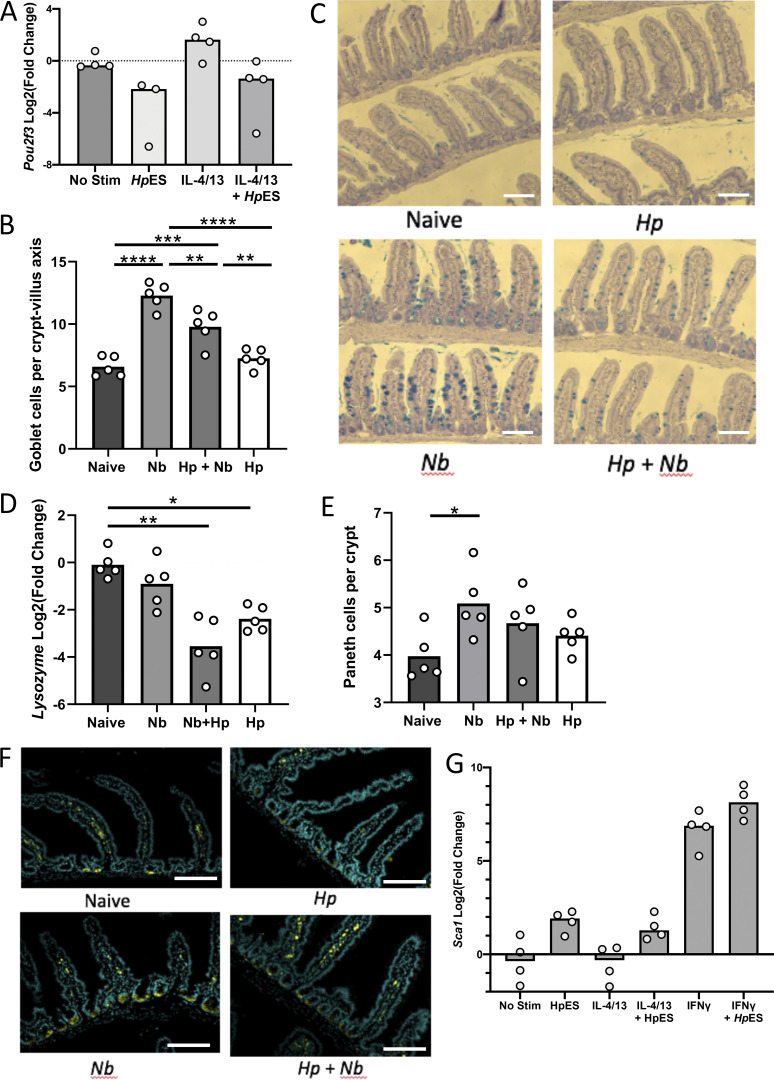
Cell type–specific inhibition by HpES and H. polygyrus. (A) Pou2f3 gene expression in HpES and IL-4/13 treated organoids. Log2 fold change shown compared with nonstimulated control in four separate biological repeats analyzed in parallel by RNA-seq. (B) Goblet cell counts in mice singly or coinfected with H. polygyrus and/or N. brasiliensis, counted villus tip to tip after periodic acid–Schiff staining; 25 counts villus-crypt units were analyzed per mouse. Data are from one experiment with four mice per group, representative of three similar experiments. Kruskal–Wallis test with Dunn’s multiple comparisons test was used; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Representative images of goblet cell staining using periodic acid–Schiff staining. Scale bar is 100 µm. (D) Lysozyme gene expression in intestinal samples. Data are from one experiment with four mice per group, representative of three similar experiments. Ordinary one-way ANOVA with Tukey’s multiple comparisons test was used; *, P < 0.05; **, P < 0.01. (E) Paneth cell counts, number of cells per crypt after staining with anti-lysozyme. Data are from one experiment with four mice per group, representative of three similar experiments. Kruskal–Wallis test with Dunn’s multiple comparisons test was used; *, P < 0.05. (F) Representative images of Paneth cell staining with anti-lysozyme. Scale bar is 100 µm. (G) Expression of Sca1 from organoid cultures under the indicated conditions. Change shown compared with nonstimulated control in four independent biological replicates analyzed in parallel by RNA-seq. Hp, H. polygyrus; Nb, N. brasiliensis; Stim, stimulation.
HpES is derived from adult worms collected from the lumen of the intestine. We were interested in whether infective L3 larvae of H. polygyrus, which burrow through the epithelium and establish in submucosal tissue, had a similar effect. Adding L3 larvae to the organoids similarly inhibited the induction of Dclk1 in response to IL-4/13 (Fig. 3 D), suggesting H. polygyrus is capable of inhibiting tuft cells at different stages of its life cycle within the host. Furthermore, L3 larvae recapitulated the effect of HpES in suppressing Muc2 expression in organoids (Fig. 3 E).
H. polygyrus inhibits tuft cell expansion in vivo
To investigate whether inhibition of tuft cell expansion also occurs in vivo, we performed coinfection experiments with the nematode N. brasiliensis, a natural rat parasite that is rapidly expelled by mice through strong tuft cell–dependent type 2 immune responses (Gerbe et al., 2016). We first infected mice with H. polygyrus, allowing chronic infection to develop, with adults in the intestinal lumen, over 28 d. We then infected with N. brasiliensis for 7 d. Expression of the tuft cell markers Dclk1 and Pou2f3 was substantially increased above naive controls in N. brasiliensis–infected mice, as expected. However, in H. polygyrus infection alone or coinfected mice, increases were muted and were not significantly different from naive controls (Fig. 4, A and B). Dclk1+ tuft cell numbers increased sharply in N. brasiliensis infections, but significantly less so in H. polygyrus– and co-infected mice (Fig. 4, C and D). In a similar fashion, H. polygyrus infection also reduced the number of goblet cells induced by N. brasiliensis (Fig. S3, B and C), suggesting either that tuft cell inhibition reduces the IL-13–dependent stimulation of goblet cells, or that HpES constituents more broadly affect differentiation of secretory lineage cells. Expression of lysozyme, a Paneth cell marker, was also reduced by H. polygyrus infection, though no reduction in Paneth cell number was seen (Fig. S3, D–F).
Figure 4.
H. polygyrus inhibits tuft cell expansion in vivo. Mice were infected with 200 H. polygyrus L3 for 28 d before infection with N. brasiliensis for 7 d (A–D) or treatment with 100 mM succinate in drinking water for 7 d. Intestinal tissues were taken at day 7 for mRNA isolation and immunohistological analysis. (A and B) Expression of canonical tuft cell genes Dclk1 (A) and Pou2f3 (B) measured by qRT-PCR in intestinal samples taken from the singly and coinfected mice, presented as log2 fold change compared with uninfected controls. Statistical analysis was by ordinary one-way ANOVA with Tukey’s multiple comparisons test. Data are from one of three replicate experiments, each with four or five mice per group. ***, P < 0.001; ****, P < 0.0001. (C and D) Tuft cell counts (C) and representative images of tuft cell staining (D) in singly and coinfected mice. Scale bar is 100 µm. Tuft cell counts from ≥20 villus/crypt units were averaged per mouse, and the means for each of five mice per group are presented. Experiments were performed three times with similar results, and data from one representative experiment are shown. Kruskal–Wallis test with Dunn’s multiple comparisons test was used as the discrete data gathered will not be normally (Gaussian) distributed. *, P < 0.05; ****, P < 0.0001. (E and F) Expression of canonical tuft cell genes Dclk1 (E) and Pou2f3 (F) measured by qRT-PCR in intestinal samples taken from succinate and H. polygyrus–infected mice, presented as log2 fold change compared with untreated and uninfected controls. Statistical analysis was by ordinary one-way ANOVA with Tukey’s multiple comparisons test. Data are from one of two replicate experiments, each with four mice per group. *, P < 0.05; ****, P < 0.0001. (G and H) Tuft cell counts (G) and representative images of tuft cell staining (H) from succinate-treated mice. Scale bar is 100 µm. Tuft cell counts are given as mean for each of four mice per group. Data are from one of two replicate experiments, each with four mice per group. Kruskal–Wallis test with Dunn’s multiple comparisons test was used. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (I) Tuft cell counts from mice administered with succinate and/or HpES; 5 µg HpES was given at days −1, 0, +1, +2, +3 and +4, i.p. Results are pooled from two independent experiments each with five mice per group. Kruskal–Wallis test with Dunn’s multiple comparisons test was used. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. Hp, H. polygyrus; Nb, N. brasiliensis; Succ, succinate.
We also wanted to investigate whether H. polygyrus infection could prevent the induction of tuft cells by the metabolite succinate, which can activate tuft cells through the succinate receptor SUCNR1, inducing a type 2 immune response, including proliferation of tuft cells (Nadjsombati et al., 2018; Schneider et al., 2018). Mice carrying a 28-d H. polygyrus infection were treated with 100 mM succinate in drinking water for 7 d. Succinate treatment alone caused increased transcription of the tuft cell markers Dclk1 (Fig. 4 E) and Pou2f3 (Fig. 4 F), whereas succinate treatment in H. polygyrus–infected mice failed to induce these genes above naive levels. Immunohistochemistry of intestinal tissue samples stained with anti-Dclk1 showed a similar suppression of tuft cell responses in infected mice (Fig. 4, G and H).
We also tested whether HpES, administered in vivo, could recapitulate the suppressive effects of live infection. Mice receiving 5 µg of HpES i.p. daily during succinate treatment showed a modest but significant reduction in tuft cell numbers (Fig. 4 I). Together these results show that H. polygyrus infection and parasite products can inhibit tuft cell expansion induced both by other helminth species and the metabolite succinate.
H. polygyrus alters differentiation of secretory-lineage cell types
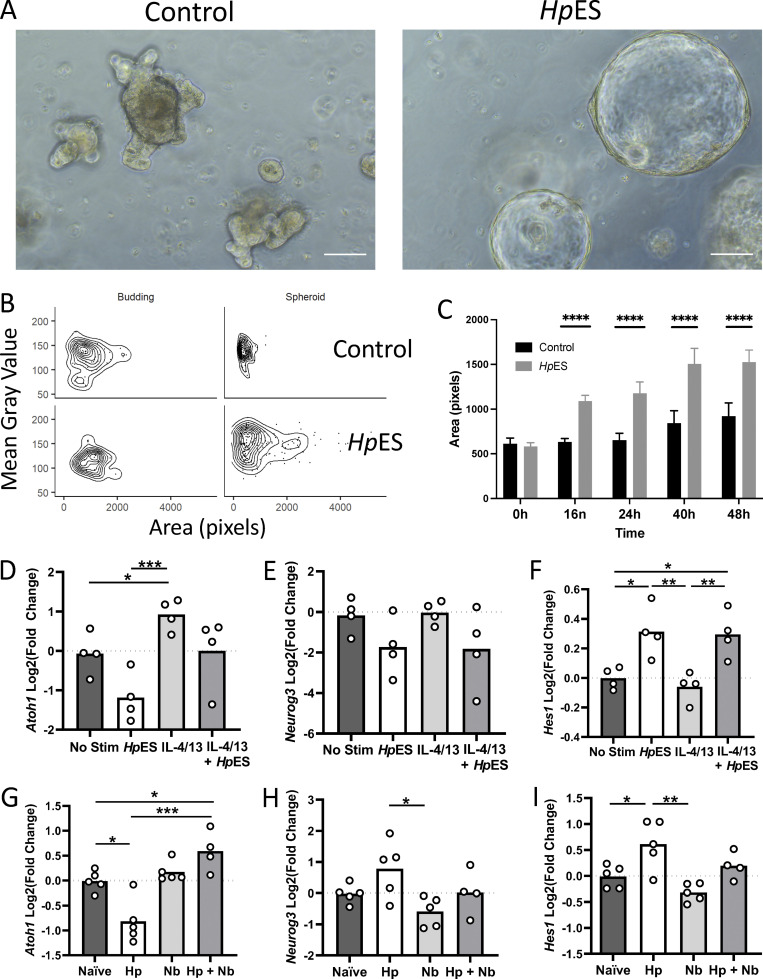
The inhibition of tuft cell expansion by H. polygyrus, as well as inhibition of gene sets for other defense-related cell types by HpES, suggested a more global effect of this nematode on the intestinal epithelium than compromising individual cell lineages. In this context, we noted marked effects on the morphology of organoids in the presence of HpES, inducing large spheroid structures with minimal crypt budding when exposed shortly after splitting (Fig. 5 A). Using an automated image analysis pipeline, we were able to categorize organoids as having the immature “spheroid” or mature “budding” morphology (Lindholm et al., 2020 Preprint). These results confirmed that HpES skews organoid morphology toward the enlarged spheroid type (Fig. 5, B and C).
Figure 5.
H. polygyrus infection alters organoid morphology and the intestinal stem cell niche. Developmental changes in the intestinal epithelium were investigated by analysis of organoid morphology and key developmental gene expression following HpES exposure, and expression of the same key genes in intestinal tissue following infection with H. polygyrus. (A) Images 16 h after culture of control organoids and organoids incubated with HpES. Scale bar is 100 µm. (B and C) Quantification of organoid architecture after addition of HpES, showing in the distribution of organoids classed as budding or spheroid in the control (top) and HpES (bottom) treatment conditions (B); and quantification by the area of organoid images over 48 h (C). Unpaired t tests were used for statistical analysis; ****, P < 0.0001. (D–F) Expression of intestinal development–related genes, Atoh1, Neurog3, and Hes1, from organoid cultures under the indicated conditions. Change shown compared with nonstimulated control in four independent biological replicates analyzed in parallel by RNA-seq. One-way ANOVA with Tukey’s multiple comparisons test was used; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (G–I) qRT-PCR on in vivo intestinal samples for the same intestinal development–related genes. Data shown are from five individual mice in one of three replicate experiments. One-way ANOVA with Tukey’s multiple comparisons test was used; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Hp, H. polygyrus; Nb, N. brasiliensis; Stim, stimulation.
Spheroid organoids have also been observed when grown from stem cells from mice infected with the same parasite, H. polygyrus (Nusse et al., 2018); these authors suggested that infection could induce IFN-γ signaling within the epithelium, leading to a fetal-like phenotype of stem cells characterized by Sca-1 expression that restrains differentiation of secretory cells, while maintaining proliferation (Nusse et al., 2018). Although Sca-1 expression was highly induced by IFN-γ treatment in our RNA-seq dataset, HpES treatment had only a minor effect (Fig. S3 G), indicating that H. polygyrus modulates intestinal stem cell fate through an IFN-γ– and Sca-1–independent pathway.
Due to the changes in morphology induced by HpES, and the inhibition of diverse secretory lineage cells, we next investigated expression of key developmental genes in the intestinal epithelium. Atoh1, a basic helix-loop-helix (bHLH) transcription factor, is considered to be the master regulator of the secretory cell lineage in the intestinal epithelium, and its expression drives differentiation into the secretory cell types (Yang et al., 2001). Notably, IL-4/13 treatment increases Atoh1, but this is ablated in the presence of HpES (Fig. 5 D), and indeed, HpES alone decreases Atoh1 gene expression below levels seen with no stimulation. Genes downstream of Atoh1, such as Neurog3, which specifies enteroendocrine cells (Jenny et al., 2002), also showed a trend toward suppression by HpES (Fig. 5 E). Atoh1 is known to be repressed by another bHLH transcription factor, Hes1, which is expressed in proliferating cells and preserves the status of stem cells and transit amplifying cells (Jensen et al., 2000; Yang et al., 2001). While Hes1 expression is unaffected by IL-4/13, the presence of HpES treatment markedly increases the transcription of this factor (Fig. 5 F), suggesting a pathway through which Atoh1 suppression is achieved and secretory cell differentiation inhibited. qRT-PCR from H. polygyrus–infected tissues confirmed repression of Atoh1 expression (Fig. 5 G) and up-regulation of Hes1 (Fig. 5 I).
Taken together, our results indicate that H. polygyrus may redirect the cellular makeup of the intestinal epithelium both by promoting epithelial proliferation (and potentially, repair), as well as reprogramming developmental pathways to favor enterocyte differentiation at the expense of specialized secretory cells that produce mediators detrimental to parasite survival.
It is broadly recognized that helminth parasites modify their environment to optimize their survival, through direct manipulation of host cell signals and responses (Brehm and Koziol, 2017; Yap and Gause, 2018). Previously, attention has mostly focused on modulation of immune system cells and disruption of protective immunity (Gazzinelli-Guimaraes and Nutman, 2018; Maizels et al., 2018; Ryan et al., 2020). However, it is increasingly evident that epithelial cells, specifically stem cells, are affected by Th2 cytokines induced by helminth infection (Biton et al., 2018; Lindholm et al., 2020 Preprint). Here, we show that the helminth H. polygyrus, and its secreted products, can act directly on the intestinal epithelium that forms its natural niche in vivo. Targeting the epithelium has many advantages for the parasite; it is this tissue that first responds to helminth presence, releasing signals such as thymic stromal lymphopoietin, IL-25, and IL-33. Molecules within HpES have already been identified that interfere with the IL-33 pathway (Osbourn et al., 2017; Vacca et al., 2020). One of these, HpARI, acts on lung epithelial cells to prevent IL-33 release (Osbourn et al., 2017), and may serve this role in the intestine. In parallel, the suppression of tuft cells will reduce IL-25 in the intestinal milieu, required to trigger ILC2 activation and IL-13 production for type 2 immunity (Gerbe et al., 2016). Indeed, both IL-25–deficient (Neill et al., 2010; Smith et al., 2018) and tuft cell–deficient (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016) mice are highly susceptible to intestinal helminth infection.
Tuft cell expansion can be induced by two separate pathways, one defined by the action of succinate on its receptor, SUCNR1, and the second through unknown stimuli emanating from parasitic helminths acting in a SUCNRI-independent fashion (Lei et al., 2018; Nadjsombati et al., 2018; Schneider et al., 2018). The ability of H. polygyrus to inhibit tuft cell differentiation in vivo both when exogenous succinate is administered and when a different helminth, N. brasiliensis, is introduced, points to a hypothesis that H. polygyrus inhibits the development, rather than the function, of tuft cells. Furthermore, replicating the inhibitory effect in vitro in small intestinal organoid cultures confirms that there is a direct helminth–epithelial interaction to dictate cellular differentiation that gives rise across the secretory cell lineages.
Our finding that genes involved in cell fate decisions such as Atoh1 and Hes1 are affected by HpES confirms that this helminth is capable of inducing changes in development. Atoh1 and Hes1 modulate the decision of Lgr5+ stem cells to follow the secretory cell type lineage, composed of goblet, Paneth, tuft, and enteroendocrine cells (Chiacchiera, 2019). We found down-regulation of all these gene sets in our RNA-seq data, consistent with the role of other specialized cells in helminth defense (Coakley and Harris, 2020; Sorobetea et al., 2018). Goblet cells are well known for their role in the “weep and sweep” response in helminth defense, producing large quantities of mucins as well as RELM-β, which prevents the worm from locating its food source (Herbert et al., 2009). Paneth cells undergo hyperplasia in response to helminth infection (Kamal et al., 2002), and although primarily associated with anti-microbial immunity, are essential in fostering the Lgr5+ stem cell niche (Sato et al., 2011). Similarly, although enteroendocrine cells have no clear role in anti-helminth immunity, they alter in response to helminth infection (Haber et al., 2017; Manocha et al., 2013; Worthington et al., 2018; Worthington et al., 2013), producing serotonin to promote intestinal contractility and the “sweep” part of immunity to helminth infection (Coakley and Harris, 2020; Wang et al., 2018; Zhao et al., 2006). By targeting the whole secretory cell lineage, H. polygyrus may coordinately neutralize each of these functions required for effective helminth defense.
Other helminth parasites are known to influence the epithelium of the organ they inhabit, primarily in pro-tumorigenic settings thought to arise as side effects of metabolite release, induction of inflammation, or alterations to commensal bacteria populations (Brindley and Loukas, 2017; Scholte et al., 2018). Interestingly, the human liver fluke Opisthorchis viverrini, which can cause cholangiocarcinoma, secretes a granulin-like growth factor, Ov-GRN-1, that induces proliferation of host cells (Smout et al., 2009; Smout et al., 2015). Our findings therefore build on these earlier reports that helminth interactions with epithelial tissues are central to the processes of invasion and pathogenesis in parasite infections. We are now endeavoring to identify the H. polygyrus–derived factor(s) that mediate the recasting of intestinal epithelial cell fate, to shed greater insight into the host pathways exploited by the parasite, and potentially to discover new molecular tools that can be used to modulate intestinal cell differentiation.
Accompanying the suppression of the secretory cell lineage, we also observed that organoid cultures exposed to HpES formed a greater proportion of spheroids, strikingly similar to the morphology of organoids produced from stem cells collected from H. polygyrus–infected mice (Nusse et al., 2018); in this case, the authors proposed that IFN-γ–induced transcriptional changes, reflecting an absence of Lgr5+ stem cells, replaced by Sca1+ cells in the infected tissues, led to a fetal reversion of the stem cells. However, our ability to create spheroids in vitro by HpES exposure argues that host IFN-γ is not required, and that developmental changes are due to the direct effects of secreted factors from the parasite. More broadly, it is interesting to speculate that the spheroid phenotype represents a pro-proliferative tissue repair process that may be essential to recover from parasite migration and epithelial disruption during this infection, raising the possibility that H. polygyrus has evolved to mitigate damage and prolong survival of the host in its own interest. Taken together, our findings illustrate the complex relationship between immunity and development at the epithelium, which are manipulated by a sophisticated parasitic helminth to its own advantage and perhaps also that of its host.
Materials and methods
Mice and parasites
8–12-wk-old female C57BL/6 mice bred in-house or purchased from Envigo UK housed in individually ventilated cages were used throughout this study. All animal studies were performed under UK Home Office Licence and approved by the University of Glasgow Ethical Review Board.
Infections employed H. polygyrus and N. brasiliensis, both maintained as previously described (Camberis et al., 2003). HpES was collected as previously described (Johnston et al., 2015). For succinate experiments, sodium succinate dibasic hexahydrate (Sigma-Aldrich) was dissolved in autoclaved tap water to 100 mM, filter-sterilized, and given to mice as their drinking water. To administer HpES in vivo, 5 µg in 100 µl PBS was given i.p. daily from day −1 to day +4 relative to addition of succinate to drinking water.
Organoid culture
Organoids were cultured from crypts isolated from the proximal 10 cm of small intestine (duodenum). Briefly, the duodenum was cut into 2-mm pieces and washed three times with cold PBS before incubation with 2 mM EDTA in PBS for 30 min at 4°C. After removal of EDTA solution, cold PBS was added and crypts isolated from basal membrane by pipetting. This procedure was repeated with more vigorous pipetting to create six fractions. Fractions with enriched crypts were identified using microscopy and pooled through a 70-µm cell strainer (Greiner), then spun at 300 ×g for 3 min at 4°C to pellet crypts. Crypts were resuspended and spun at 100 ×g for 3 min at 4°C to remove single cells. The pellet was then resuspended in 10 ml basic medium (Advanced DMEM/F12, supplemented with 1% PenStrep, 1% L-glutamine, and 10 mM Hepes, all from Gibco) and crypt numbers counted by microscopy. 500 crypts in 40 µl Matrigel (Corning) were seeded into wells of a 24-well flat-bottomed plate (Corning). After incubation for 10 min at 37°C, 400 µl of complete crypt medium was added (basic medium plus 1× N2 supplement [Life Technologies], 1× B27 supplement [Life Technologies], 50 ng/ml murine epidermal growth factor [Invitrogen], 100 ng/ml murine Noggin [Peprotech], and 500 ng/ml murine R-spondin-1 [R&D]; 3 µM CHIR99021 [Miltenyi] was added only for initial plating). Crypts were then cultured at 37°C in 5% CO2, medium was changed every 2–3 d, and organoids were passaged once a week by dissociation and washing in cold basic medium, reseeding at 500 crypts per well in 40 µl Matrigel. Organoids were passaged at least three times before use. Stimulations were as follows: HpES at 10 µg/ml, IL-4 (Miltenyi) at 400 ng/ml, IL-13 (Peprotech) at 400 ng/ml, and IFN-γ at 1 ng/ml, or 500 H. polygyrus L3 stage larvae. Organoids were stimulated for 24 h before collection for RNA-seq.
RNA extraction
Tissues were taken and stored in either 400 µl TRIzol (Invitrogen; for organoids) or 1 ml RNAlater (Qiagen; for intestine samples) until RNA extraction using the RNeasy mini kit (Qiagen) could be performed. For intestinal samples, tissue was disrupted using a TissueLyser (Qiagen) in 600 µl lysis buffer (RLT, Qiagen) for 2 min at 25 Hz before continuing according to kit instructions. For organoid samples, after spinning at 12,000 ×g for 2 min to remove debris, 100 µl chloroform was added to the supernatant before mixing well. After incubation at room temperature for 3 min, samples were centrifuged at 12,000 ×g for 15 min at 4°C. The upper aqueous phase was transferred to a new tube and 1.5 volumes of 100% ethanol added before transfer to a RNeasy spin column and purification as per kit instructions. An on-column DNA digestion step was included in RNA purification using a RNase free DNase (Qiagen). RNA concentration was determined using a Nanodrop 2000 (Thermo Fisher Scientific).
RNA-seq
Library preparation and whole transcriptome profiling was performed by Glasgow Polyomics. PolyA selection library preparation was performed using the TruSeq stranded mRNA kit (Illumina) before single-end sequencing was performed with 30 million reads per sample, 1 x75 nt read length, on the NextSeq 500 platform (Illumina), with results returned as fastq files. Trim Galore! was used to cut adapters (using Cutadapt) and run quality control (using FastQC), with MultiQC used to inspect all samples together. HISAT2 was used to align reads to the mouse genome, followed by featureCounts to aggregate mapped reads. Count data were then analyzed using DESeq2 to identify differential gene expression analysis (Love et al., 2014).
The software degPatterns, within the DEGreport package (Pantano et al., 2020), was used to identify clusters within DEGs, and GO enrichment analysis was performed using enrichGO from clusterProfiler, followed by removal of redundant GO terms using ReViGO (Supek et al., 2011). Heatmaps were created with the aid of the Pheatmap package (Kolde, 2015). UpSet plots were generated using the UpSetR package (Conway et al., 2017). ggplot2 was used for generation of graphs (Wickham, 2016). The R package clusterProfiler was used for GSEA analysis with 10,000 permutations and otherwise default settings.
All RNA-seq data have been submitted to the European Nucleotide Archive with the ArrayExpress accession no. E-MTAB-11118.
qRT-PCR
Samples used for qRT-PCR were reverse-transcribed using the qScript cDNA synthesis kit (Quanta Bio) according to the kit instructions. cDNA was then diluted 1/10 for use in 12.5 µl qRT-PCR reactions with PerfeCTa SYBR Green Supermix (Quanta Bio) and appropriate primers as previously published (Aronson et al., 2014; Guo et al., 2018; Nusse et al., 2018), detailed in Table 1. Reactions were performed in a 384-well plate (Applied Biosystems) on the QuantStudio 7 Flex real-time PCR system (Applied Biosystems). Analysis was performed using the delta-delta Ct method, with target gene expression normalized against two reference genes. The best reference genes were determined from a selection using the NormFinder plugin for Microsoft Excel (Andersen et al., 2004).
Table 1. Primers used for qRT-PCR reactions.
| Name | Use | Sequence |
|---|---|---|
| GAPDH | Reference ER | F 5′-ATGACATCATCAAGAAGGTGGTG-3′ |
| R 5′-CATACCAGGAAATGAGCTTG-3′ | ||
| HPRT | Reference | F 5′-TTTACTGGCAACATCAACAG-3′ |
| R 5′-CAGATCCCACATACTCACTG-3′ | ||
| RPS29 | Reference | F 5′-ACGGTCTGATCCGCAAATAC-3′ |
| R 5′-CATGATCGGTTCCACTTGGT-3′ | ||
| Pou2f3 | Tuft cell gene | F 5′-AGAGAATCAACTGCCCCGTG-3′ |
| R 5′-GGAAGGCACGACTCTCTTCC-3′ | ||
| DCLK1 | Tuft cell gene | F 5′-CAGCCTGGACGAGCTGGTGG-3′ |
| R 5′-TGACCAGTTGGGGTTCACAT-3′ | ||
| Muc2 | Goblet cell gene | F 5′-CAGTTTATTCCTGTGTGCCCAAGG-3′ |
| R 5′-GGCTTCAGAATAATGTACTGCTGC-3′ | ||
| Lysozyme | Paneth cell gene | F 5′-GGAATGGATGGCTACCGTGG-3′ |
| R 5′-CATGCCACCCATGCTCGAAT-3′ | ||
| Sca1 | Stem cell gene | F 5′-GATGGACACTTCTCACACTACA-3′ |
| R 5′-GCAGGTAATTGATGGGCAAGA-3′ | ||
| Lgr5 | Stem cell gene | F 5′-CTCCAACCTCAGCGTCTTC-3′ |
| R 5′-GTCAAAGCATTTCCAGCAAGA-3′ | ||
| Atoh1 | Cell fate gene | F 5′-AGCTTCCTCTGGGGGTTACT-3′ |
| R 5′-TTCTGTGCCATCATCGCTGT-3′ | ||
| Neurog3 | Cell fate gene | F 5′-CTCAGCAAACAGCGAAGAAG-3′ |
| R 5′-GGGAAGGTGGGCAGGAC-3′ | ||
| Hes1 | Cell fate gene | F 5′-AGTGTCACCTTCCAGTGGCT-3′ |
| R 5′-TGGGCTAGGGACTTTACGGG-3′ |
Organoid immunofluorescent staining
Organoids were seeded out in 4- or 8-well chamber slides (Thermo Fisher Scientific) before use in stimulations and microscopy. Organoids were stimulated for 48 h before use. For immunofluorescence staining, cultured organoids were washed in PBS twice and then fixed in 4% paraformaldehyde for 20 min at room temperature. Organoids were then permeabilized with PBS containing 0.5% Triton X-100 for 10 min at 4°C, followed by rinsing three times with PBS containing 100 mM glycine. Organoids were blocked with IF buffer (PBS containing 0.1% BSA, 0.2% Triton X-100, 0.05% Tween-20, and 10% FCS) for 1 h at room temperature before incubation with 1/1,000 anti-mouse Dclk1 (Abcam) in antibody diluent (Invitrogen) at 4°C overnight. After rinsing three times with IF buffer, slides were incubated with anti-rabbit FITC (Dako) for 1 h at room temperature. Slides were then washed with IF buffer followed by three rinses with PBS before being mounted with Vectashield mounting media containing DAPI (Vector Laboratories). Slides were imaged using a Leica DMi8 inverted microscope and Leica Application Suite (LAS) X software (Leica Microsystems). Resulting image files were analyzed using ImageJ/Fiji (Schindelin et al., 2012).
Immunohistochemistry
The small intestine was taken and prepared for processing and embedding in paraffin using the Swiss-rolling technique (Bialkowska et al., 2016), which enables many villi to be investigated in the same cut section. Transverse sections were made using a microtome through the gut rolls at a thickness of 5 µm before mounting on glass slides. Sections were deparaffinized by immersing slides in xylene, then hydrated through 100%, 90%, and 70% ethanol successively. Heat-induced epitope retrieval was performed in citrate buffer (Thermo Fisher Scientific), and then sections were blocked using 2.5% normal horse serum blocking solution (Vector Laboratories) for 1 h at room temperature. Slides were then incubated overnight at 4°C with rabbit anti-mouse Dclk1 (Abcam) or rabbit anti-mouse lysozyme (Abcam) at 1:1,000 in 2.5% normal horse serum blocking solution. Polyclonal rabbit IgGs (Abcam) were used as an isotype control. After washing, sections were incubated with swine anti-rabbit-IgGs/FITC (Agilent Dako), washed, and mounted using Vectashield Vibrance antifade mounting medium with DAPI (Vactor Laboratories). Slides were imaged using a Leica DMi8 inverted microscope and Leica Application Suite (LAS) X software (Leica Microsystems). The resulting image files were analyzed using ImageJ/Fiji.
Periodic acid–Schiff staining
Slides were prepared, deparaffinized, and hydrated as above before undergoing periodic acid–Schiff staining, following the instructions in the kit (Atom Scientific).
Statistics
All statistical analysis was performed using Prism 8 (Graphpad Software Inc.). Error bars on graphs display the mean and SEM. Ordinary one-way ANOVA with Tukey’s multiple comparisons test was used. In the case of tuft cell count data, a Kruskal–Wallis test with Dunn’s multiple comparisons test was used as the discrete data gathered is not normally (Gaussian) distributed; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1, Fig. S2, and Fig. S3 present additional data on the GO terms most affected by HpES and cytokines, and the seven clusters of DEGs (Fig. S1); the GO terms most altered within each cluster, and the individual genes most associated with each cluster (Fig. S2); and changes in tuft cell, Paneth cell, and goblet cell differentiation in singly and coinfected mice (Fig. S3).
Acknowledgments
We thank Claire Ciancia and Anne-Marie Donachie for excellent technical assistance. The authors gratefully acknowledge the Flow Core Facility and Glasgow Imaging Facility, University of Glasgow, for their support and assistance.
This work was supported by the Wellcome Trust through an Investigator Award to R.M. Maizels (ref. 1219530), a Collaborative Grant (ref. 211814), and the Wellcome Trust core-funded Wellcome Centre for Integrative Parasitology (ref. 104111).
Author contributions: C. Drurey, G. Coakley, P. Jay, N. Harris, and R.M. Maizels conceived and planned the research aims and experiments. C. Drurey, G. Coakley, M.C. Poveda, S. Löser, and R. Doolan carried out the experiments. C. Drurey, H.T. Lindholm, and M.J. Oudhoff performed bioinformatic and imaging analyses; F. Gerbe, P. Jay, N. Harris, M.J. Oudhoff, and R.M. Maizels provided methodological advice and contributed to the interpretation of the results. C. Drurey and R.M. Maizels organized the data and wrote the first draft of the manuscript. All authors provided critical feedback and helped shape the research, analysis, and manuscript.
References
- Andersen, C.L., Jensen J.L., and Ørntoft T.F.. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64:5245–5250. 10.1158/0008-5472.CAN-04-0496 [DOI] [PubMed] [Google Scholar]
- Aronson, B.E., Stapleton K.A., Vissers L.A., Stokhuijzen E., Bruijnzeel H., and Krasinski S.D.. 2014. Spdef deletion rescues the crypt cell proliferation defect in conditional Gata6 null mouse small intestine. BMC Mol. Biol. 15:3. 10.1186/1471-2199-15-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis, D., and Grencis R.K.. 2008. The intestinal epithelium: sensors to effectors in nematode infection. Mucosal Immunol. 1:252–264. 10.1038/mi.2008.21 [DOI] [PubMed] [Google Scholar]
- Bialkowska, A.B., Ghaleb A.M., Nandan M.O., and Yang V.W.. 2016. Improved Swiss-rolling technique for intestinal tissue preparation for immunohistochemical and immunofluorescent analyses. J. Vis. Exp. (113):54161. 10.3791/54161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billipp, T.E., Nadjsombati M.S., and von Moltke J.. 2021. Tuning tuft cells: new ligands and effector functions reveal tissue-specific function. Curr. Opin. Immunol. 68:98–106. 10.1016/j.coi.2020.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton, M., Haber A.L., Rogel N., Burgin G., Beyaz S., Schnell A., Ashenberg O., Su C.W., Smillie C., Shekhar K., et al. 2018. T helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell. 175:1307–1320.e22. 10.1016/j.cell.2018.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm, K., and Koziol U.. 2017. Echinococcus-host interactions at cellular and molecular levels. Adv. Parasitol. 95:147–212. 10.1016/bs.apar.2016.09.001 [DOI] [PubMed] [Google Scholar]
- Brindley, P.J., and Loukas A.. 2017. Helminth infection-induced malignancy. PLoS Pathog. 13:e1006393. 10.1371/journal.ppat.1006393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberis, M., Le Gros G., and Urban J. Jr. 2003. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. In Current Protocols in Immunology. R. Coico, editor. John Wiley and Sons, Inc. 19.12.11–19.12.27. 10.1002/0471142735.im1912s55 [DOI] [PubMed] [Google Scholar]
- Chiacchiera, F. 2019. Transcriptional and epigenetic mechanisms controlling intestinal cell fate. In Epigenetics and Regeneration. Palacios D., editor. Academic Press, Elsevier; 10.1016/B978-0-12-814879-2.00011-X [DOI] [Google Scholar]
- Coakley, G., and Harris N.L.. 2020. The intestinal epithelium at the forefront of host-helminth interactions. Trends Parasitol. 36:761–772. 10.1016/j.pt.2020.07.002 [DOI] [PubMed] [Google Scholar]
- Conway, J.R., Lex A., and Gehlenborg N.. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 33:2938–2940. 10.1093/bioinformatics/btx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, L., Reid K.T., Häkkinen E., de Bie B., Tanaka S., Smyth D.J., White M.P., Wong M.Q., Huang Q., Gillies J.K., et al. 2021. Induction of stable human FOXP3+ Tregs by a parasite-derived TGF-β mimic. Immunol. Cell Biol. 99:833–847. 10.1111/imcb.12475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser-von Bieren, J. 2019. Eicosanoids in tissue repair. Immunol. Cell Biol. 97:279–288. 10.1111/imcb.12226 [DOI] [PubMed] [Google Scholar]
- Gazzinelli-Guimaraes, P.H., and Nutman T.B.. 2018. Helminth parasites and immune regulation. F1000 Res. 7:1685. 10.12688/f1000research.15596.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe, F., Sidot E., Smyth D.J., Ohmoto M., Matsumoto I., Dardalhon V., Cesses P., Garnier L., Pouzolles M., Brulin B., et al. 2016. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 529:226–230. 10.1038/nature16527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X.K., Ou J., Liang S., Zhou X., and Hu X.. 2018. Epithelial Hes1 maintains gut homeostasis by preventing microbial dysbiosis. Mucosal Immunol. 11:716–726. 10.1038/mi.2017.111 [DOI] [PubMed] [Google Scholar]
- Haber, A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. 2017. A single-cell survey of the small intestinal epithelium. Nature. 551:333–339. 10.1038/nature24489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, D.R., Yang J.-Q., Hogan S.P., Groschwitz K., Khodoun M., Munitz A., Orekov T., Perkins C., Wang Q., Brombacher F., et al. 2009. Intestinal epithelial cell secretion of RELM-β protects against gastrointestinal worm infection. J. Exp. Med. 206:2947–2957. 10.1084/jem.20091268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt, M.R., Lavoie S., Michaud M., Blum A.M., Tran S.V., Weinstock J.V., Gallini C.A., Redding K., Margolskee R.F., Osborne L.C., et al. 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 351:1329–1333. 10.1126/science.aaf1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inclan-Rico, J.M., and Siracusa M.C.. 2018. First Responders: Innate Immunity to Helminths. Trends Parasitol. 34:861–880. 10.1016/j.pt.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny, M., Uhl C., Roche C., Duluc I., Guillermin V., Guillemot F., Jensen J., Kedinger M., and Gradwohl G.. 2002. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 21:6338–6347. 10.1093/emboj/cdf649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, J., Pedersen E.E., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., and Madsen O.D.. 2000. Control of endodermal endocrine development by Hes-1. Nat. Genet. 24:36–44. 10.1038/71657 [DOI] [PubMed] [Google Scholar]
- Johnston, C.J., Robertson E., Harcus Y., Grainger J.R., Coakley G., Smyth D.J., McSorley H.J., and Maizels R.. 2015. Cultivation of Heligmosomoides polygyrus: an immunomodulatory nematode parasite and its secreted products. J. Vis. Exp. (98):e52412. 10.3791/52412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, C.J.C., Smyth D.J., Kodali R.B., White M.P.J., Harcus Y., Filbey K.J., Hewitson J.P., Hinck C.S., Ivens A., Kemter A.M., et al. 2017. A structurally distinct TGF-β mimic from an intestinal helminth parasite potently induces regulatory T cells. Nat. Commun. 8:1741. 10.1038/s41467-017-01886-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan, P.M., Lamberton P.H.L., Fenwick A., and Addiss D.G.. 2018. Soil-transmitted helminth infections. Lancet. 391:252–265. 10.1016/S0140-6736(17)31930-X [DOI] [PubMed] [Google Scholar]
- Kamal, M., Dehlawi M.S., Brunet L.R., and Wakelin D.. 2002. Paneth and intermediate cell hyperplasia induced in mice by helminth infections. Parasitology. 125:275–281. 10.1017/S0031182002002068 [DOI] [PubMed] [Google Scholar]
- Kolde, R. 2015. pheatmap: Pretty Heatmaps. https://mran.microsoft.com/snapshot/2017-09-01/web/packages/pheatmap/pheatmap.pdf
- Lei, W., Ren W., Ohmoto M., Urban J.F. Jr., Matsumoto I., Margolskee R.F., and Jiang P.. 2018. Activation of intestinal tuft cell-expressed Sucnr1 triggers type 2 immunity in the mouse small intestine. Proc. Natl. Acad. Sci. USA. 115:5552–5557. 10.1073/pnas.1720758115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverkoehne, I., and Gruber A.D.. 2002. The murine mCLCA3 (alias gob-5) protein is located in the mucin granule membranes of intestinal, respiratory, and uterine goblet cells. J. Histochem. Cytochem. 50:829–838. 10.1177/002215540205000609 [DOI] [PubMed] [Google Scholar]
- Lightowlers, M.W., and Rickard M.D.. 1988. Excretory-secretory products of helminth parasites: effects on host immune responses. Parasitology. 96(S1, Suppl):S123–S166. 10.1017/S0031182000086017 [DOI] [PubMed] [Google Scholar]
- Lindholm, H.T., Parmar N., Drurey C., Ostrop J., Diez-Sanchez A., Maizels R.M., and Oudhoff M.J.. 2020. Developmental pathways regulate cytokine-driven effector and feedback responses in the intestinal epithelium. bioRxiv. (Preprint posted June 20, 2020). 10.1101/2020.06.19.160747 [DOI]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels, R.M., Smits H.H., and McSorley H.J.. 2018. Modulation of host immunity by helminths: the expanding repertoire of parasite effector molecules. Immunity. 49:801–818. 10.1016/j.immuni.2018.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manocha, M., Shajib M.S., Rahman M.M., Wang H., Rengasamy P., Bogunovic M., Jordana M., Mayer L., and Khan W.I.. 2013. IL-13-mediated immunological control of enterochromaffin cell hyperplasia and serotonin production in the gut. Mucosal Immunol. 6:146–155. 10.1038/mi.2012.58 [DOI] [PubMed] [Google Scholar]
- McGinty, J.W., Ting H.A., Billipp T.E., Nadjsombati M.S., Khan D.M., Barrett N.A., Liang H.E., Matsumoto I., and von Moltke J.. 2020. Tuft-cell-derived leukotrienes drive rapid anti-helminth immunity in the small intestine but are dispensable for anti-protist immunity. Immunity. 52:528–541.e7. 10.1016/j.immuni.2020.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay, D.M., Shute A., and Lopes F.. 2017. Helminths and intestinal barrier function. Tissue Barriers. 5:e1283385. 10.1080/21688370.2017.1283385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadjsombati, M.S., McGinty J.W., Lyons-Cohen M.R., Jaffe J.B., DiPeso L., Schneider C., Miller C.N., Pollack J.L., Nagana Gowda G.A., Fontana M.F., et al. 2018. Detection of succinate by intestinal tuft cells triggers a Type 2 innate immune circuit. Immunity. 49:33–41.e7. 10.1016/j.immuni.2018.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill, D.R., Wong S.H., Bellosi A., Flynn R.J., Daly M., Langford T.K.A., Bucks C., Kane C.M., Fallon P.G., Pannell R., et al. 2010. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 464:1367–1370. 10.1038/nature08900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse, Y.M., Savage A.K., Marangoni P., Rosendahl-Huber A.K.M., Landman T.A., de Sauvage F.J., Locksley R.M., and Klein O.D.. 2018. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature. 559:109–113. 10.1038/s41586-018-0257-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn, M., Soares D.C., Vacca F., Cohen E.S., Scott I.C., Gregory W.F., Smyth D.J., Toivakka M., Kemter A.M., le Bihan T., et al. 2017. HpARI protein secreted by a helminth parasite suppresses interleukin-33. Immunity. 47:739–751.e5. 10.1016/j.immuni.2017.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantano, L., Hutchinson J., Barrera V., Piper M., Khetani R., Dally K., Perumarl T.M., Kirchner R., and Steinbaugh M.. 2020. DEGreport: Report of DEG analysis. R package version 1.26.0. http://lpantano.github.io/DEGreport/.
- Pullan, R.L., Smith J.L., Jasrasaria R., and Brooker S.J.. 2014. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit. Vectors. 7:37. 10.1186/1756-3305-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds, L.A., Filbey K.J., and Maizels R.M.. 2012. Immunity to the model intestinal helminth parasite Heligmosomoides polygyrus. Semin. Immunopathol. 34:829–846. 10.1007/s00281-012-0347-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, S., Shiels J., Taggart C.C., Dalton J.P., and Weldon S.. 2020. Fasciola hepatica-derived molecules as regulators of the host immune response. Front. Immunol. 11:2182. 10.3389/fimmu.2020.02182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., and Clevers H.. 2011. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 469:415–418. 10.1038/nature09637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C., O’Leary C.E., von Moltke J., Liang H.E., Ang Q.Y., Turnbaugh P.J., Radhakrishnan S., Pellizzon M., Ma A., and Locksley R.M.. 2018. A metabolite-triggered tuft cell-ILC2 circuit drives small intestinal remodeling. Cell. 174:271–284.e14. 10.1016/j.cell.2018.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholte, L.L.S., Pascoal-Xavier M.A., and Nahum L.A.. 2018. Helminths and cancers from the evolutionary perspective. Front. Med. (Lausanne). 5:90. 10.3389/fmed.2018.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe, C., Thornton D.J., and Grencis R.K.. 2018. A sticky end for gastrointestinal helminths; the role of the mucus barrier. Parasite Immunol. 40:e12517. 10.1111/pim.12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K.A., Löser S., Varyani F., Harcus Y., McSorley H.J., McKenzie A.N.J., and Maizels R.M.. 2018. Concerted IL-25R and IL-4Rα signaling drive innate type 2 effector immunity for optimal helminth expulsion. eLife. 7:e38269. 10.7554/eLife.38269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smout, M.J., Laha T., Mulvenna J., Sripa B., Suttiprapa S., Jones A., Brindley P.J., and Loukas A.. 2009. A granulin-like growth factor secreted by the carcinogenic liver fluke, Opisthorchis viverrini, promotes proliferation of host cells. PLoS Pathog. 5:e1000611. 10.1371/journal.ppat.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smout, M.J., Sotillo J., Laha T., Papatpremsiri A., Rinaldi G., Pimenta R.N., Chan L.Y., Johnson M.S., Turnbull L., Whitchurch C.B., et al. 2015. Carcinogenic parasite secretes growth factor that accelerates wound healing and potentially promotes neoplasia. PLoS Pathog. 11:e1005209. 10.1371/journal.ppat.1005209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorobetea, D., Svensson-Frej M., and Grencis R.. 2018. Immunity to gastrointestinal nematode infections. Mucosal Immunol. 11:304–315. 10.1038/mi.2017.113 [DOI] [PubMed] [Google Scholar]
- Supek, F., Bošnjak M., Škunca N., and Šmuc T.. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 6:e21800. 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca, F., Chauché C., Jamwal A., Hinchy E.C., Heieis G., Webster H., Ogunkanbi A., Sekne Z., Gregory W.F., Wear M., et al. 2020. A helminth-derived suppressor of ST2 blocks allergic responses. eLife. 9:e54017. 10.7554/eLife.54017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke, J., Ji M., Liang H.E., and Locksley R.M.. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 529:221–225. 10.1038/nature16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.J., Sharkey K.A., and McKay D.M.. 2018. Modulation of the immune response by helminths: a role for serotonin? Biosci. Rep. 38:BSR20180027. 10.1042/BSR20180027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, M.P.J., Smyth D.J., Cook L., Ziegler S.F., Levings M.K., and Maizels R.M.. 2021. The parasite cytokine mimic Hp-TGM potently replicates the regulatory effects of TGF-β on murine CD4+ T cells. Immunol. Cell Biol. 99:848–864. 10.1111/imcb.12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. 2016. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York. 10.1007/978-3-319-24277-4 [DOI] [Google Scholar]
- Worthington, J.J., Samuelson L.C., Grencis R.K., and McLaughlin J.T.. 2013. Adaptive immunity alters distinct host feeding pathways during nematode induced inflammation, a novel mechanism in parasite expulsion. PLoS Pathog. 9:e1003122. 10.1371/journal.ppat.1003122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington, J.J., Reimann F., and Gribble F.M.. 2018. Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 11:3–20. 10.1038/mi.2017.73 [DOI] [PubMed] [Google Scholar]
- Yang, Q., Bermingham N.A., Finegold M.J., and Zoghbi H.Y.. 2001. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 294:2155–2158. 10.1126/science.1065718 [DOI] [PubMed] [Google Scholar]
- Yap, G.S., and Gause W.C.. 2018. Helminth Infections Induce Tissue Tolerance Mitigating Immunopathology but Enhancing Microbial Pathogen Susceptibility. Front. Immunol. 9:2135. 10.3389/fimmu.2018.02135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., Han Y., and He Q.Y.. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, A., Urban J.F. Jr., Morimoto M., Elfrey J.E., Madden K.B., Finkelman F.D., and Shea-Donohue T.. 2006. Contribution of 5-HT2A receptor in nematode infection-induced murine intestinal smooth muscle hypercontractility. Gastroenterology. 131:568–578. 10.1053/j.gastro.2006.05.013 [DOI] [PubMed] [Google Scholar]