Abstract
Background
Preoperative determination of breast cancer molecular subtypes facilitates individualized treatment plan-making and improves patient prognosis. We aimed to develop an assembled convolutional neural network (ACNN) model for the preoperative prediction of molecular subtypes using multimodal ultrasound (US) images.
Methods
This multicentre study prospectively evaluated a dataset of greyscale US, colour Doppler flow imaging (CDFI), and shear-wave elastography (SWE) images in 807 patients with 818 breast cancers from November 2016 to February 2021. The St. Gallen molecular subtypes of breast cancer were confirmed by postoperative immunohistochemical examination. The monomodal ACNN model based on greyscale US images, the dual-modal ACNN model based on greyscale US and CDFI images, and the multimodal ACNN model based on greyscale US and CDFI as well as SWE images were constructed in the training cohort. The performances of three ACNN models in predicting four- and five-classification molecular subtypes and identifying triple negative from non-triple negative subtypes were assessed and compared. The performance of the multimodal ACNN was also compared with preoperative core needle biopsy (CNB).
Finding
The performance of the multimodal ACNN model (macroaverage area under the curve [AUC]: 0.89–0.96) was superior to that of the dual-modal ACNN model (macroaverage AUC: 0.81–0.84) and the monomodal ACNN model (macroaverage AUC: 0.73–0.75) in predicting four-classification breast cancer molecular subtypes, which was also better than that of preoperative CNB (AUC: 0.89–0.99 vs. 0.67–0.82, p < 0.05). In addition, the multimodal ACNN model outperformed the other two ACNN models in predicting five-classification molecular subtypes (AUC: 0.87–0.94 vs. 0.78-0.81 vs. 0.71–0.78) and identifying triple negative from non-triple negative breast cancers (AUC: 0.934–0.970 vs. 0.688–0.830 vs. 0.536–0.650, p < 0.05). Moreover, the multimodal ACNN model obtained satisfactory prediction performance for both T1 and non-T1 lesions (AUC: 0.957–0.958 and 0.932–0.985).
Interpretation
The multimodal US-based ACNN model is a potential noninvasive decision-making method for the management of patients with breast cancer in clinical practice.
Funding
This work was supported in part by the National Natural Science Foundation of China (Grants 81725008 and 81927801), Shanghai Municipal Health Commission (Grants 2019LJ21 and SHSLCZDZK03502), and the Science and Technology Commission of Shanghai Municipality (Grants 19441903200, 19DZ2251100, and 21Y11910800).
Keywords: Breast cancer, Molecular subtypes, Multimodal ultrasound images, Assemble convolutional neural network, Prediction
Research in context.
Evidence before this study
Preoperatively identifying molecular subtypes of breast cancer can facilitate individualized treatment plan-making and improve patient prognosis. Previous studies found that multimodal ultrasound (US) imaging manifestations were related to certain molecular subtypes of breast cancer. Convolutional neural network (CNN)-based image analysis can establish a direct link between complicated medical imaging data and disease prediction. Thus, we aimed to develop an assembled CNN (ACNN) model for the preoperative prediction of breast cancer molecular subtypes using multimodal US images.
Added value of this study
The database we built in this multicentric and prospective study was standardized multimodal US images data of breast cancers. Breast shear-wave elastography (SWE) examinations were conducted firmly according to the World Federation for Ultrasound in Medicine & Biology guidelines, which ensured the quality of SWE images. In addition, SWE image quality control was conducted independently by one senior radiologist. Based on this database, original ACNN models were designed to predict breast cancer molecular subtypes and compared their performance with preoperative core needle biopsy (CNB). The multimodal ACNN model showed satisfactory performance and robustness for predicting breast cancer molecular subtype tasks. Moreover, the multimodal ACNN model yielded similar prediction performance for T1 and non-T1 lesions. In addition, the multimodal ACNN model achieved better performance than preoperative CNB for identifying breast cancer molecular subtypes.
Implications of all the available evidence
We developed a promising ACNN model based on greyscale US, colour Doppler flow imaging (CDFI), and SWE images to predict four- and five-classification St. Gallen breast cancer molecular subtypes. The multimodal ACNN model achieved better performance than preoperative CNB. Moreover, this model also achieved superior performance in discriminating triple negative from non-triple negative breast cancers. The multimodal US image-based ACNN model is a noninvasive approach for the prediction of breast cancer molecular subtypes. Therefore, "?>this model is a potential noninvasive decision-making method for the management of patients with breast cancer in routine clinical practice.
Alt-text: Unlabelled box
1. Introduction
Breast cancer is the leading cause of cancer-related death in women worldwide and is a molecular heterogeneous disease with varying prognoses [1,2]. Determining the breast cancer molecular subtypes preoperatively can facilitate individualized treatment plan-making and thus improve the patient prognosis [3,4]. Currently, ultrasound (US)-guided core needle biopsy (CNB) is commonly performed for the preoperative pathological diagnosis of breast cancer [5]. However, some intrinsic limitations of US-guided CNB might handicap accurate determination of breast cancer molecular subtypes preoperatively [6], [7], [8], [9], [10]. The partial samples obtained by CNB might not represent the entire lesion owing to the heterogeneity of breast cancer [6].
Given the importance and challenges of preoperatively identifying breast cancer molecular subtypes, some radiology communities have explored the potential value of US images for noninvasively predicting molecular subtypes [11], [12], [13], [14]. Greyscale US, colour Doppler flow imaging (CDFI), and shear-wave elastography (SWE) examinations have been widely used to characterize breast lesions in clinical practice. Some studies found that some US image features from radiologists’ visual interpretation were related to certain molecular subtypes of breast cancer [15,16]. However, there exists high inter- and intraobserver variability in the interpretation of US images, and there is currently no practicable method to directly predict molecular subtypes of breast cancer [17].
The convolutional neural network (CNN), a newly developed type of artificial intelligence (AI), has drawn widespread attention for its excellent performance and high reproducibility in the field of medical image recognition tasks [18]. CNN-based image analysis can establish a direct link between complicated medical imaging data and disease prediction. It has been shown to achieve prominent performance in automatically recognizing breast lesions, distinguishing malignant from benign breast lesions, and predicting clinically negative axillary lymph node metastasis using US images [19], [20], [21].
Combining multimodal US images and CNN-based image analysis technology may yield a promising effect in predicting breast cancer molecular subtypes. Thus, the aim of our study was to investigate the potential for CNN to predict molecular subtypes through the use of multimodal US images of breast cancer.
2. Methods
2.1. Ethics
This multicentre study was approved by the institutional ethics committees of the three participated centres (approval number: SHSYIEC–4.1/20–120/01), and informed consent was obtained from all the patients. Prospective research of this study was registered at www.chictr.org.cn (ChiCTR2000038606).
2.2. Study population
Consecutive patients with breast cancers were prospectively enrolled from the Shanghai Tenth People's Hospital in Shanghai, China, between December 2016 and September 2020 as the training cohort. From October 2020 to February 2021, consecutive patients with breast cancers were prospectively enrolled from the Shanghai Tenth People's Hospital as the internal validation cohort, the Sun Yat-Sen University Cancer Center in Guangzhou, China, as the test cohort A, and the Ma'anshan People's Hospital in Anhui, China, as the test cohort B.
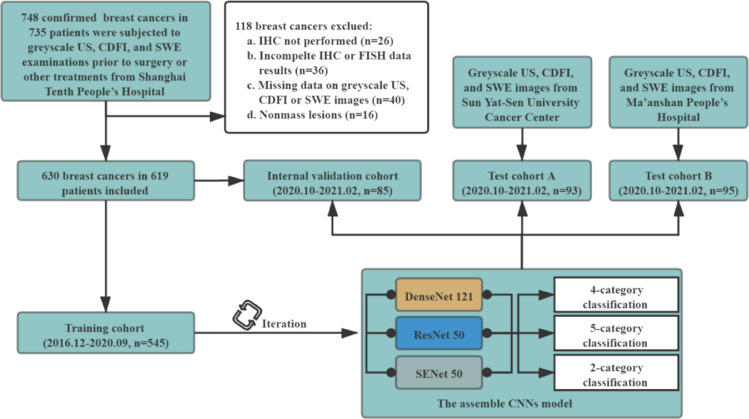
All the three centres used the same patient inclusion and exclusion criteria. The inclusion criteria were as follows: (a) patients with breast cancers underwent surgical resection; (b) greyscale US, CDFI, and SWE examinations were performed within one month before surgery and prior to any treatment, including biopsy or neoadjuvant therapies. The exclusion criteria were as follows: (a) breast cancer without immunohistochemistry (IHC) examination; (b) breast cancer with incomplete IHC data including oestrogen receptor (ER), progesterone receptor (PR), human epidermalgrowth factor receptor-2 (HER-2), or Ki-67; (c) breast cancer without fluorescence in situ hybridization (FISH) when IHC scoring 2+ in HER-2; (d) breast cancer with missing data on the greyscale US, CDFI, or SWE images; (e) nonmass lesion. A detailed flowchart of patient selection in the study is shown in Fig. 1.
Fig. 1.
Flowchart of procedures in patient enrolment and the development and evaluation of the assembled convolutional neural network (ACNN) model for automated breast cancer molecular subtypes prediction.
2.3. The greyscale US, CDFI, and SWE examination protocol
The greyscale US, CDFI, and SWE examinations were performed by one of six experienced radiologists in breast US examination with the same US system (Aixplorer; SuperSonic Imagine, Aixen Provence, France) with a 15–4 MHz linear transducer following the standard protocol.
The greyscale US, CDFI, and SWE images of each breast lesion at the largest long axis cross-section and the largest transverse cross-section were routinely recorded. Two greyscale US images, including a nonmarked image and an image with enclosing callipers at the edge of the lesion for measurement, were obtained at each plane. During CDFI examination, a default equipment setting was implemented for all lesions: a scale of 4 cm/s, medium wall filter, and pulse repetition frequency of 700 Hz. One CDFI image was obtained at each plane. Then, SWE image acquisition was immediately performed. Breast SWE examinations were conducted according to the World Federation for Ultrasound in Medicine & Biology guidelines for performing US elastography of the breast [22]. Briefly, the probe was vertically placed on the skin without consciously applying any vibration/compression, lightly touching the skin, and trying not to apply pressure. The stabilized SWE images of the lesion were saved after a few seconds of immobilization. The tissue elasticity was represented as a colour-coded map in kilopascals (kPa) at each pixel and a colour scale ranging from 0 (dark blue, soft) to 180 kPa (red, hard). A total of 12 US images (one nonmarked greyscale US image, one greyscale US image with callipers, one CDFI image, and three SWE images per plane) were obtained from each breast lesion. Greyscale US images with callipers were used to assist nonmarked greyscale US image resizing. The greyscale US, CDFI, and SWE images were stored on the Supersonic imagine system platform in DICOM format and then exported in JPEG format for subsequent analysis.
2.4. Image quality control
One radiologist (T. Ren) with 10 years of experience in performing breast US examinations independently conducted image quality control. One SWE image per plane was selected for the ACNN models. Finally, eight US images (one nonmarked greyscale US image, one greyscale US image with callipers, one CDFI image, and one SWE image per plane) were obtained from each breast lesion for further analysis.
2.5. Data annotation
According to IHC results of ER, PR, HER-2, Ki-67, and FISH status, all breast cancers were classified into four and five types of St. Gallen molecular subtypes (luminal A, luminal B [including luminal B-HER-2 negative and luminal B-HER-2 positive], HER-2 positive, and triple negative) [23].
Briefly, the breast cancers were identified as ① luminal A: ER (+) and/or PR (+), HER-2 (-) and Ki-67 (< 14%); ② luminal B (-): ER (+) and/or PR (+), HER-2 (-) and Ki-67 (> 14%); ③ luminal B (+): ER (+) and/or PR (+), HER-2 (+) and any Ki-67; ④ HER-2 positive: ER (-) and PR (-), HER-2 (+); ⑤ triple negative: ER (-) and PR (-), HER-2 (-).
ER and PR were deemed positive if the percentage of stained cells and intensity were over 1%. For HER-2, IHC scoring 0 or 1+ was considered negative, and those scoring 3+ were considered positive. IHC scoring 2+ was further tested with FISH, and HER-2 was considered positive if (a) the ratio of the HER-2 gene signal to the chromosome 17 probe signal (HER-2/CEP17 ratio) ≥ 2.0; (b) HER-2/CEP17 ratio < 2.0, while average HER-2 signals/cell ≥ 6.0.
2.6. Data preprocessing
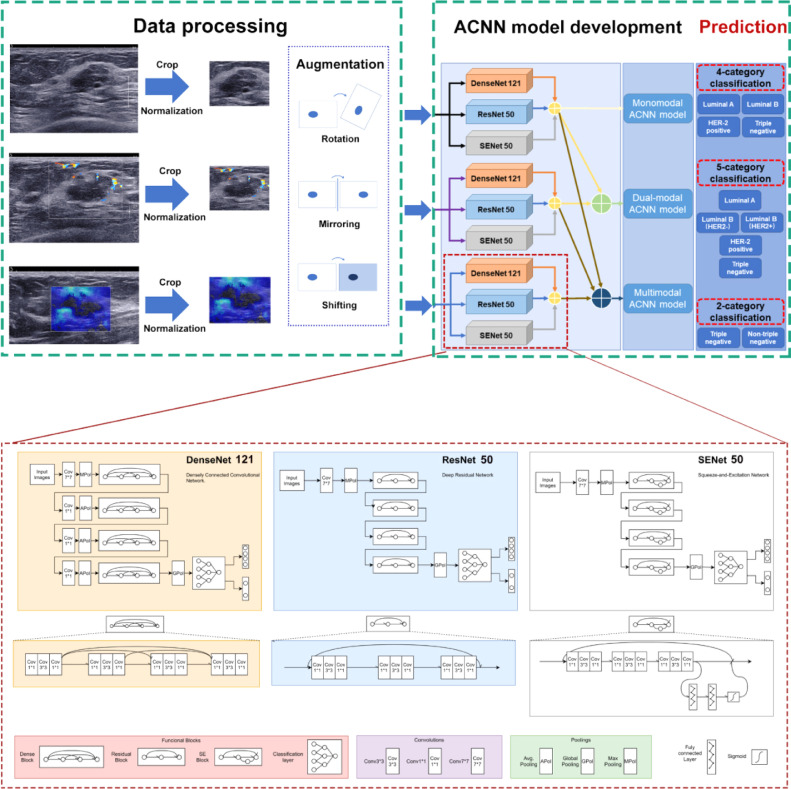
A Faster Region-based Convolutional Neural Network (FR-CNN) detection algorithm was used to obtain the regions of interest (ROIs) of multimodal US images [24]. Greyscale US images with callipers were used to assist nonmarked greyscale US images resizing. Four callipers at the edge of the lesion were first detected by FR-CNN in greyscale US images with callipers. Then, a rectangular ROI was generated by extending 64 pixels from enclosing callipers. Third, the rectangle ROI was duplicated to the corresponding greyscale US image without callipers at the same location. FR-CNN was used to detect and extract the embedded ROI in CDFI and SWE images. Finally, all extracted multimodal US images in JPEG format were normalized to 448 × 448 pixels to standardize the distance scale. This procedure facilitates the unification of the input size of the model. To avoid overfitting, we applied offline data augmentation techniques for each image during training. By means of random geometric image transformations (rotation, mirroring, and shifting), it can artificially increase the training image dataset up to 88 times its original size. This strategy can mimic the data diversity observed in reality, which results in better generalization performance of the model. Detailed information on the data preprocessing is provided in Appendix S1. All preprocessing steps were conducted in Python (Version 3.6.8; Python Software Foundation, Wilmington, Del) by using PyTorch Transform (https://pytorch.org/docs/stable/torchvision/transforms.html) and Dataloader (https://pytorch.org/docs/stable/data.html?highlight=dataloader#torch.utils.data.DataLoader). Image augmentation was not performed for the internal validation and test cohorts.
2.7. ACNN model development
An ACNN model architecture was designed to predict breast cancer molecular subtypes. It is a combination architecture of three CNN networks (DenseNet 121, ResNet 50, and SENet 50). ResNet uses residual learning blocks to increase the ability of function layers for learning semantic information at the same time, and preserves the information flow over a long range via skip connections. However, ResNet has obvious redundancy, where each layer with a relatively simple structure extracts only a few features [25]. DenseNet allows every layer block to have access to all proceeding layer blocks, which significantly improves the data flow between layers. As a result, it reduces the number of parameters to prevent overfitting or gradient vanishing or exploding. This method makes up for the redundancy of ResNet. However, as the number of dense connections grows, the volume of DenseNet increases. This results in the increasing training time of DenseNet [26]. Compared to DenseNet, SENet could selectively emphasize informative features and suppress less useful features by means of attention and gating mechanisms, which would achieve better results [27]. Therefore, these three basic CNN networks were chosen and then parallelly assembled via a majority vote algorithm that has been proven to have prominent and stable performance in image recognition tasks [28]. To optimize the hyperparameter configurations, three CNN networks were pretrained on ImageNet. Appendix S2 shows the detailed information about development of the ACNN model architecture based on three CNN networks. All programs were run in Python version 3.6.8. The monomodal ACNN model applied to greyscale US images only was trained and tested. The dual-modal ACNN model applied to both greyscale US and CDFI images was trained and tested. The multimodal ACNN model applied to greyscale US and CDFI as well as SWE images was trained and tested. The workflow of the ACNN model development is presented in Fig. 2.
Fig. 2.
Workflow in the development of three ACNN models for automated breast cancer molecular subtypes prediction and the structure of each CNN network.
2.8. Heat map generation
To better interpret the network predictions, the class activation mapping method was utilized to produce heat maps. Heat maps can visualize the most indicative areas of the image to interpret the predictive mechanism of the ACNN model, which reflects the contribution of each pixel in the three modal US images for prediction of breast cancer molecular subtypes. All heat maps were produced by applying the packages of OpenCV (opencv-python 4.1.0.25, https://opencv.org/releases.html) and Matplotlib (https://pypi.org/project/matplotlib/).
2.9. Statistical analysis
All statistical analyses were performed using SPSS (Version 22.0, IBM Corporation, Armonk, USA) and R software (Version 3.4.1, R Foundation for Statistical Computing, Vienna, Austria). Comparisons of the differences in clinical factors among the different molecular subtypes of breast cancer were analysed using the chi-square test or t test. To evaluate the prediction performance of the three ACNN models, receiver operating characteristic (ROC) curves were constructed. The area under the ROC curve (AUC) with 95% confidence intervals (CIs), sensitivity, specificity, and accuracy were investigated. In addition, the F1-score was also calculated (F1 = ). Delong's test was performed to compare the AUCs. A confusion matrix was applied to illustrate the specific examples of molecular subtype classes where the prediction results of the ACNN model were discordant with the ground truth from the IHC results. A p value less than 0.05 was considered statistically significant.
2.10. Role of the funding source
The funders had no role in study design, data collection, data analyses, interpretation, or writing of the manuscript. The corresponding authors (C. Zhao and H. Xu) had full access to all of the data and the final responsibility for the decision to submit for publication.
3. Results
3.1. Patient characteristics and pathological features of breast cancers
A total of 807 women (mean age, 59.30 years ± 12.75; range, 20–95 years) with 818 breast cancers were prospectively enrolled for analysis. There were 534 women (mean age, 59.3 years ± 13.04; range, 26–95 years) with 545 breast cancers in the training cohort and 85 women (mean age, 59.55 years ± 11.31; range, 35–81 years) with 85 breast cancers in the internal validation cohort from the principal hospital. In addition, 93 women (mean age, 56.33 years ± 13.78; range, 20–88 years) with 93 breast cancers were enrolled in test cohort A from the Sun Yat-Sen University Cancer Center. There were 95 women (mean age, 55.97 years ± 14.56; range, 26–87 years) with 95 breast cancers in test cohort B from Ma'anshan People's Hospital.
A total of 306 breast cancers presented in T1 stage and 239 non-T1 cancers in the training cohort. There were 39, 44, and 39 cancers in the T1 subgroup and 46, 49, and 56 cancers in the non-T1 subgroup in the internal validation cohort, test cohort A, and test cohort B, respectively. Other baseline characteristics of all patients and breast cancers (including age, sex, pathological findings, and US Breast Imaging Reporting and Data System [BI-RADS] category) are presented in Table 1. As reported in Table 1, the distribution of each breast cancer molecular subtype was similar in the training cohort, internal validation cohort, and two independent test cohorts.
Table 1.
The basic characteristics of patients and breast cancers.
| Characteristics | Training cohort | Internal validation cohort | Test cohort A | Test cohort B |
|---|---|---|---|---|
| No. patients | 534 | 85 | 93 | 95 |
| Age (y) | ||||
| Mean ± SD | 59.30 ± 13.04 | 59.55 ± 11.31 | 56.33 ± 13.78 | 55.97 ± 14.56 |
| Median (range) | 60 (26-95) | 62 (35-81) | 57 (20-88) | 56 (26-87) |
| No. breast cancers | 545 | 85 | 93 | 95 |
| Tumor size | ||||
| T1 (< 2 cm) | 306 (56.1) | 39 (45.9) | 44 (47.3) | 39 (41.1) |
| T2 (2-5 cm) | 223 (40.9) | 42 (49.4) | 47 (50.5) | 54 (56.8) |
| T3 (> 5 cm) | 16 (3.0) | 4 (4.7) | 2 (2.2) | 2 (2.1) |
| Location | ||||
| Left | 280 (51.4) | 46 (54.1) | 46 (49.4) | 47 (49.5) |
| Right | 265 (48.6) | 39 (45.9) | 47 (50.6) | 48 (50.5) |
| Histologic type | ||||
| Invasive ductal cancer | 482 (88.4) | 78 (91.8) | 87 (93.5) | 86 (90.5) |
| Invasive lobular cancer | 12 (2.2) | 1 (1.2) | 4 (4.3) | 3 (3.2) |
| Mixed | 7 (1.3) | 0 (0.0) | 1 (1.1) | 0 (0.0) |
| Other | 44 (8.1) | 6 (7.0) | 1 (1.1) | 6 (6.3) |
| Histologic grade | ||||
| I | 46 (8.5) | 7 (8.2) | 9 (9.7) | 8 (8.4) |
| II | 271 (49.7) | 43 (50.6) | 46(49.5) | 47 (49.5) |
| III | 228 (41.8) | 35 (41.2) | 39 (40.8) | 40 (42.1) |
| BI-RADS | ||||
| 3 | 2 (0.4) | 1 (1.2) | 0 (0.0) | 0 (0.0) |
| 4a | 84 (15.4) | 9 (10.6) | 10 (10.8) | 12 (12.6) |
| 4b | 154 (28.3) | 17 (20.0) | 19 (20.4) | 20 (21.1) |
| 4c | 239 (43.8) | 41 (48.2)) | 41 (44.1) | 48 (50.5) |
| 5 | 66 (12.1) | 17 (20.0) | 23 (24.7) | 15 (15.8) |
| ER status | ||||
| Positive | 395 (72.5) | 66 (77.6) | 68 (73.1) | 67 (70.5) |
| Negative | 150 (27.5) | 19 (22.4) | 25 (26.9) | 28 (29.5) |
| PR status | ||||
| Positive | 349 (64.0) | 50 (58.8) | 55 (59.1) | 59 (62.1) |
| Negative | 196 (36.0) | 35 (41.2) | 38 (40.9) | 36 (37.9) |
| HER-2 status | ||||
| Positive | 135 (24.8) | 21 (25.0) | 28 (30.1) | 29 (30.5) |
| Negative | 410 (75.2) | 64 (75.0) | 65 (69.9) | 66 (69.5) |
| Ki-67 | ||||
| ≤ 14% | 118 (21.7) | 18 (24.7) | 16 (17.2) | 22 (23.2) |
| > 14% | 427 (78.3) | 67 (75.3) | 77 (82.8) | 73 (76.8) |
| Molecular subtypes | ||||
| Luminal A | 93 (17.1) | 15 (17.6) | 14 (15.1) | 16 (16.8) |
| Luminal B | 307 (56.3) | 52 (61.2) | 54 (58.1) | 51 (53.7) |
| Luminal B (-) | 240 (44.0) | 41 (48.3) | 42 (45.2) | 40 (42.1) |
| Luminal B (+) | 67 (12.3) | 11 (12.9) | 12 (12.9) | 11 (11.6) |
| HER-2 positive | 72 (13.2) | 7 (8.3) | 12 (12.9) | 9 (9.5) |
| Triple negative | 73 (13.4) | 11 (12.9) | 13 (13.9) | 19 (20.0) |
Note. —Unless otherwise specified, data in parentheses are percentages.
SD, standard deviation; BI-RADS, breast imaging reporting and data system; ER, oestrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor-2.
The clinical factors (including age, location, size, and BI-RADS category) were not relevant to breast cancer molecular subtypes (p = 0.808, 0.851, 0.940, and 0.969, respectively [chi-square test and t test]) (Table S5). Therefore, clinical factors were not included in AI model. Additionally, the aim of this study was to predict breast cancer molecular subtypes preoperatively. Thus, pathological factors were also not used in AI model development.
3.2. Performance of the three ACNN models in predicting four-classification St. Gallen molecular subtypes
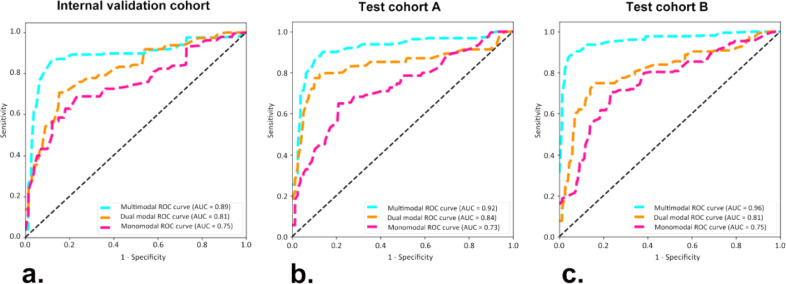
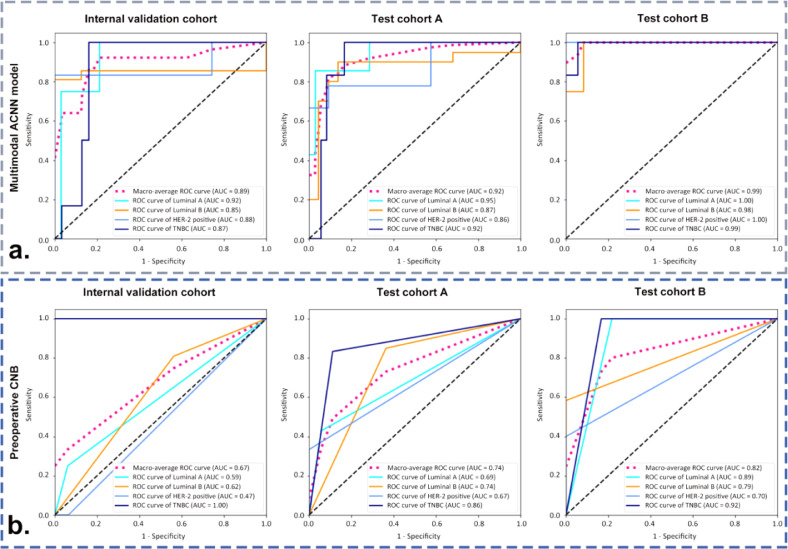
In comparison with the monomodal and dual-modal ACNN models, the multimodal ACNN model showed the best performance in predicting four-classification St. Gallen molecular subtypes (luminal A, luminal B, HER-2 positive, and triple negative). This was measured in terms of macroaverage AUC (0.89 vs. 0.81 vs. 0.75 for the internal validation cohort, 0.92 vs. 0.84 vs. 0.73 for test cohort A, and 0.96 vs. 0.81 vs. 0.75 for test cohort B) (Fig. S1 and Fig. 3). The sensitivity, specificity, accuracy, and F1 score of the ACNN models are presented in Table 2.
Fig. 3.
Receiver operating characteristic (ROC) curves of three ACNN models in predicting four-classification breast cancer molecular subtypes for (a) the internal validation cohort from the Shanghai Tenth People's Hospital, (b) test cohort A from the Sun Yat-Sen University Cancer Center, and (c) test cohort B from the Ma'anshan People's Hospital. Numbers in parentheses are areas under the receiver operating characteristic curves (AUCs).
Table 2.
Comparison of performance in three ACNN models for predicting four-classification molecular subtypes of breast cancers.
| ACNN models | Datasets | AUC | Sensitivity (%) | Specificity (%) | Accuracy (%) | F1-score |
|---|---|---|---|---|---|---|
| Monomodal ACNN model | Internal validation cohort (n=85) | 0.75 | 66.67 | 53.33 | 51.76 | 0.53 |
| Test cohort A (n=93) | 0.73 | 64.29 | 64.29 | 53.76 | 0.59 | |
| Test cohort B (n=95) | 0.75 | 52.94 | 56.25 | 54.73 | 0.55 | |
| Dual-modal ACNN model | Internal validation cohort (n=85) | 0.81 | 71.43 | 66.67 | 62.35 | 0.64 |
| Test cohort A (n=93) | 0.84 | 75.00 | 64.28 | 74.19 | 0.69 | |
| Test cohort B (n=95) | 0.81 | 61.11 | 68.75 | 68.42 | 0.69 | |
| Multimodal ACNN model | Internal validation cohort (n=85) | 0.89 | 92.31 | 80.00 | 84.71 | 0.82 |
| Test cohort A (n=93) | 0.92 | 91.67 | 78.57 | 81.72 | 0.80 | |
| Test cohort B (n=95) | 0.96 | 87.50 | 87.50 | 82.11 | 0.85 |
Note. —The monomodal ACNN model was trained and tested with greyscale US images.
The dual-modal ACNN model was trained and tested with greyscale US and CDFI images.
The multimodal ACNN model was trained and tested with greyscale US and CDFI as well as SWE images.
ACNN, assembled convolutional neural network; AUC, area under the receiver operating characteristic curve.
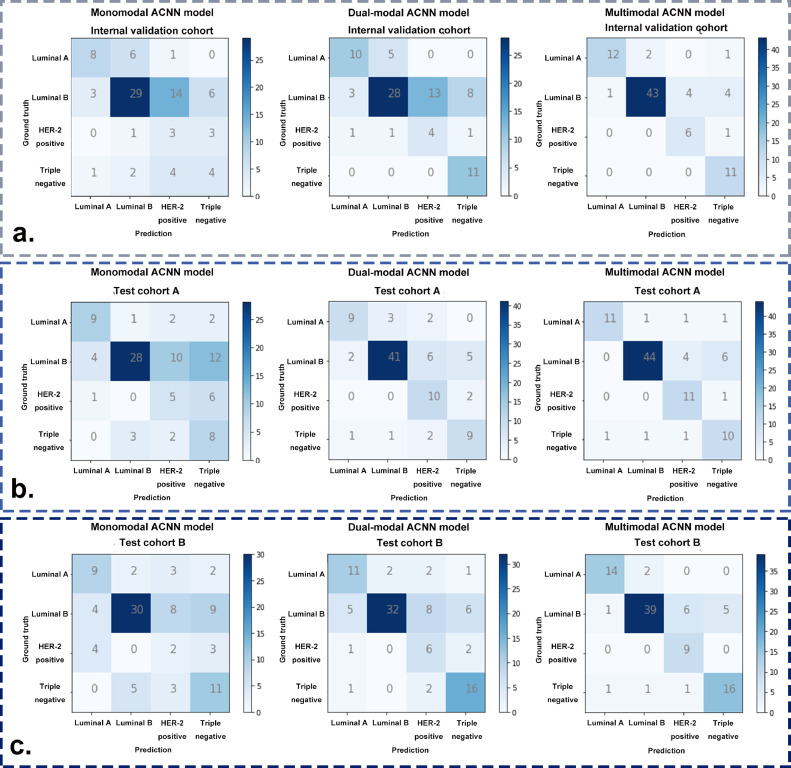
Additionally, the detailed confusion matrices of the three ACNN models revealed particular examples of breast cancer molecular subtypes where the ACNN prediction was in accordance with the postoperative IHC results. After adding CDFI to develop the dual-modal ACNN model, an additional two luminal A, seven triple negative, and one HER-2 positive cancers were correctly predicted in the internal validation cohort, an additional thirteen luminal B and five HER-2 positive cancers were correctly predicted in test cohort A, an additional two luminal A, two luminal B cancers, four HER-2 positive, and five triple negative cancers were correctly predicted in test cohort B. The dual-modal ACNN model had a better performance based on its more accurate prediction of luminal B, HER-2 positive, and triple negative cancers (Fig. 4). Compared with the dual-modal ACNN model, the multimodal ACNN model made an additional two, fifteen, and two correct predictions in luminal A, luminal B and HER-2 positive cancers, respectively, in the internal validation cohort; an additional two, three, and one correct predictions in luminal A, luminal B, and HER-2 positive, respectively, in test cohort A; and an additional three, seven, and three correct predictions in luminal A, luminal B and HER-2 positive cancers, respectively, in test cohort B. The multimodal ACNN model rendered a higher performance than the dual-modal ACNN model by predicting more correct cases in luminal A, luminal B and HER-2 positive cancers (Fig. 4).
Fig. 4.
The confusion matrices of three ACNN models for predicting four-classification breast cancer molecular subtypes in (a) the internal validation cohort, (b) test cohort A, and (c) test cohort B.
3.3. Comparison of the performance between the multimodal ACNN model and preoperative CNB in predicting four-classification St. Gallen molecular subtypes
Preoperative CNBs were performed in 82.4% (70/85) breast cancers in internal validation cohort, 84.9% (79/93) breast cancers in test cohort A, and 53.7% (51/95) breast cancers in test cohort B. In addition, there were 37 (43.5%, 37/85), 42 (45.2%, 42/93), and 24 (25.3%, 24/95) breast cancers that underwent preoperative CNB and had complete preoperative IHC results in the internal validation cohort, test cohort A, and test cohort B, respectively. The κ-values of ER, PR, HER-2, and Ki-67 between CNB and surgical excision specimens ranged from 0.21 to 0.67. The detailed IHC results of breast cancers in CNB and postoperative pathological examinations are presented in Table S6.
The multimodal ACNN model achieved better performance than preoperative CNB for predicting breast cancer molecular subtypes in the validation and test cohorts. In the internal validation cohort, the performance of the multimodal ACNN model was superior to that of preoperative CNB (AUC: 0.89 vs. 0.67, p < 0.05 [Delong's test]). The multimodal ACNN model made an additional seven correct predictions than preoperative CNB (31 vs. 24 correct predictions in 37 breast cancers). There were 4, 17, 5, and 11 breast cancers classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by the multimodal ACNN model, respectively; 3, 26, 2, and 6 cancers were classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by preoperative CNB, respectively; 4, 21, 6, and 6 cancers were classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by postoperative IHC, respectively (Tables 3, S7, and Fig. 5).
Table 3.
Comparison of performance in the multimodal ACNN model and preoperative CNB for predicting four-classification molecular subtypes of breast cancers.
| Methods | Datasets | AUC | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|
| Multimodal ACNN model | Internal validation cohort (n=37) | 0.89 | 84.87 | 82.47 | 83.73 |
| Test cohort A (n=42) | 0.92 | 83.00 | 81.35 | 83.33 | |
| Test cohort B (n=24) | 0.99 | 89.62 | 76.27 | 83.33 | |
| Preoperative CNB | Internal validation cohort (n=37) | 0.67 | 51.56 | 49.70 | 64.86 |
| Test cohort A (n=42) | 0.74 | 61.11 | 70.93 | 66.67 | |
| Test cohort B (n=24) | 0.82 | 49.66 | 63.54 | 62.50 |
Note. —The multimodal ACNN model was trained and tested with greyscale US and CDFI as well as SWE images.
ACNN, assembled convolutional neural network; AUC, area under the receiver operating characteristic curve; CNB, core needle biopsy.
Fig. 5.
ROC curves of (a) the multimodal ACNN model and (b) preoperative CNB in classifying four-classification breast cancer molecular subtypes in the internal validation cohort from the Shanghai Tenth People's Hospital, test cohort A from the Sun Yat-Sen University Cancer Center, and test cohort B from the Ma'anshan People's Hospital. Numbers in parentheses are AUCs.
In test cohort A, the performance of the multimodal ACNN model was better than preoperative CNB (AUC: 0.92 vs. 0.74, p < 0.05 [Delong's test]). The multimodal ACNN model made an additional seven correct predictions than preoperative CNB (35 vs. 28 correct predictions in 42 breast cancers). There were 7, 19, 8, and 8 breast cancers classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by the multimodal ACNN model, respectively; 5, 25, 3, and 9 cancers were classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by preoperative CNB, respectively; and 7, 20, 9, and 6 cancers were classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by postoperative IHC, respectively (Tables 3, S7, and Fig. 5).
In test cohort B, the multimodal ACNN model also performed better in comparison to preoperative CNB (AUC: 0.99 vs. 0.82, p < 0.05 [Delong's test]), and made an additional five correct predictions compared to preoperative CNB (20 vs. 15 correct predictions in 24 breast cancers). There were 2, 9, 7, and 6 breast cancers classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by the multimodal ACNN model, respectively; 5, 8, 2, and 9 cancers were classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by preoperative CNB, respectively; and 1, 12, 5, and 6 cancers were classified as luminal A, luminal B, HER-2 positive, and triple negative cancers by postoperative IHC, respectively (Tables 3, S7, and Fig. 5).
3.4. Performance of the three ACNN models in predicting five-classification St. Gallen molecular subtypes
Comparing the monomodal and dual-modal ACNN models, the multimodal ACNN model showed the best performance in predicting five-classification St. Gallen molecular subtypes (luminal A, luminal B (-), luminal B (+), HER-2 positive, and triple negative). This was measured in terms of macroaverage AUC (0.88 vs. 0.78 vs. 0.71 for the internal validation cohort, 0.94 vs. 0.81 vs. 0.78 for test cohort A, and 0.87 vs. 0.79 vs. 0.75 for test cohort B) (Fig. S2). The sensitivity, specificity, accuracy, and F1 score of the ACNN models could be found in Table S8.
As shown in Fig. S3, the multimodal ACNN model could correctly generate an additional 29 and 15 predictions in comparison with the monomodal ACNN and the dual-modal ACNN models in the internal validation cohort, respectively. In test cohort A, an additional 30 and 14 correct predictions were made by the multimodal ACNN model in comparison with the monomodal ACNN and the dual-modal ACNN models, respectively. In test cohort B, an additional 28 and 17 correct predictions were yielded by the multimodal ACNN model in comparison with the monomodal ACNN and the dual-modal ACNN models, respectively.
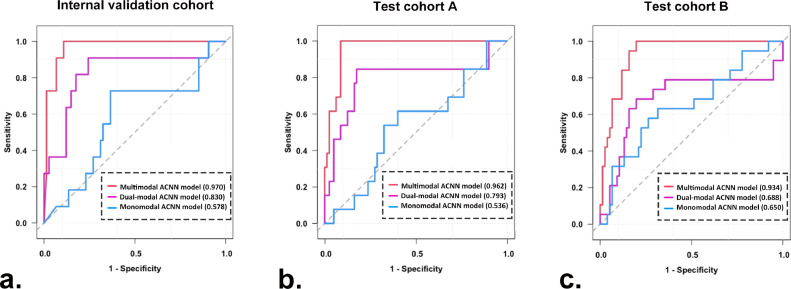
3.5. Performance of the three ACNN models in predicting triple negative from non-triple negative breast cancers
The multimodal ACNN model had better performance in comparison with the dual-modal and monomodal ACNN models in predicting triple negative from non-triple negative breast cancers. For the internal validation cohort, the AUCs were 0.970 (95% CI: 0.936, 1.000) for the multimodal ACNN model, 0.830 (95% CI: 0.672, 0.989) for the dual-modal ACNN model, and 0.578 (95% CI: 0.386, 0.770) for the monomodal ACNN model (p < 0.05 for the multimodal ACNN model vs. dual-modal ACNN model, multimodal ACNN model vs. monomodal ACNN model, and dual-modal ACNN model vs. monomodal ACNN model [Delong's test]). For test cohort A, the AUCs were 0.962 (95% CI: 0.926, 0.997) for the multimodal ACNN model, 0.793 (95% CI: 0.618, 0.968) for the dual-modal ACNN model, and 0.536 (95% CI: 0.364, 0.707) for the monomodal ACNN model (p < 0.05 for multimodal ACNN model vs. dual-modal ACNN model, multimodal ACNN model vs. monomodal ACNN model, and dual-modal ACNN model vs. monomodal ACNN model [Delong's test]). For test cohort B, the AUCs were 0.934 (95% CI: 0.886, 0.981) for the multimodal ACNN model, 0.688 (95% CI: 0.519, 0.858) for the dual-modal ACNN model, and 0.650 (95% CI: 0.503, 0.796) for the monomodal ACNN model (p < 0.05 for multimodal ACNN model vs. dual-modal ACNN model, multimodal ACNN model vs. monomodal ACNN model; p = 0.46 for dual-modal ACNN model vs. monomodal ACNN model [Delong's test]) (Fig. 6). The performance of the multimodal ACNN model was also measured in terms of specificity, sensitivity, accuracy, and F1 score. The detailed results of prediction performance appear in Table 4, and the classification confusion matrices for the ACNN models are shown in Fig. S4.
Fig. 6.
ROC curves of three ACNN models for predicting triple negative from non-triple negative breast cancers in (a) the internal validation cohort, (b) test cohort A, and (c) test cohort B. Numbers in parentheses are AUCs.
Table 4.
The performance of three ACNN models for predicting triple negative from non-triple negative breast cancers.
| ACNN models | Datasets | AUC | Sensitivity (%) | Specificity (%) | Accuracy (%) | F1-score |
|---|---|---|---|---|---|---|
| Monomodal ACNN model | Internal validation cohort (n=85) | 0.578 (0.386-0.770) | 45.45 (21.25-72.01) | 68.92 (56.23-77.17) | 64.71 (54.09-74.05) | 0.66 |
| Test cohort A (n=93) | 0.536 (0.364-0.707) | 61.54 (35.41-82.40) | 57.50 (46.56-67.75) | 58.06 (47.91-67.58) | 0.58 | |
| Test cohort B (n=95) | 0.650 (0.503-0.796) | 57.89 (36.24-76.89) | 69.74 (58.62-78.95) | 67.37 (57.40-75.98) | 0.69 | |
| Dual-modal ACNN model | Internal validation cohort (n=85) | 0.830 (0.672-0.989) | 81.82 (51.15-96.01) | 75.68 (66.16-85.22) | 77.65 (67.63-85.27) | 0.77 |
| Test cohort A (n=93) | 0.793 (0.618-0.968) | 84.62 (56.54-96.90) | 71.25 (60.49-80.06) | 73.12 (63.28-81.12) | 0.72 | |
| Test cohort B (n=95) | 0.688 (0.519-0.858) | 68.42 (45.80-84.84) | 71.05 (59.99-80.09) | 70.53 (60.67-78.79) | 0.71 | |
| Multimodal ACNN model | Internal validation cohort (n=85) | 0.970 (0.936-1.000) | 100 (69.98-100) | 83.78 (79.85-94.66) | 90.59 (82.28-95.38) | 0.90 |
| Test cohort A (n=93) | 0.962 (0.926-0.997) | 100 (73.41-100) | 91.14 (81.25-95.08) | 91.40 (83.72-95.80) | 0.91 | |
| Test cohort B (n=95) | 0.934 (0.886-0.981) | 94.74 (73.52-100) | 84.21 (74.24-90.89) | 86.32 (77.85-91.96) | 0.85 |
Note. —95% confidence intervals are included in brackets.
ACNN, assembled convolutional neural network; AUC, area under the receiver operating characteristic curve.
Additionally, the multimodal ACNN model obtained satisfactory prediction performance for both T1 and non-T1 lesions. This was measured in terms of the AUC for the T1 subgroup and non-T1 subgroup (Fig. S5): 0.957 (95% CI: 0.891, 1.000) and 0.985 (95% CI: 0.958, 1.000) for the internal validation cohort, 0.958 (95% CI: 0.897, 1.000) and 0.961 (95% CI: 0.903, 1.000) for the test cohort A, and 0.957 (95% CI: 0.889, 1.000) and 0.932 (95% CI: 0.868, 0.996) for the test cohort B. The sensitivity, specificity, accuracy, and F1 score for two subgroups are presented in Table 5.
Table 5.
The performance of the multimodal ACNN model for predicting triple negative from non-triple negative breast cancers in two subgroups with different T stages.
| Datasets | Subgroups | AUC | Sensitivity (%) | Specificity (%) | Accuracy (%) | F1-score |
|---|---|---|---|---|---|---|
| Internal validation cohort | T1 stage (n=39) | 0.957 (0.891-1.000) | 100 (45.41-100) | 88.57 (73.45-96.06) | 89.74 (75.85-96.51) | 0.89 |
| non-T1 stage (n=46) | 0.985 (0.958-1.000) | 100 (59.56-100) | 89.74 (75.85-96.51) | 91.30 (79.14-97.10) | 0.91 | |
| Test cohort A | T1 stage (n=44) | 0.958 (0.897-1.000) | 100 (59.56-100) | 94.59 (81.37-99.43) | 95.45 (84.04-99.58) | 0.95 |
| non-T1 stage (n=49) | 0.961 (0.903-1.000) | 100 (55.72-100) | 86.05 (72.36-93.82) | 87.76 (75.39-94.64) | 0.87 | |
| Test cohort B | T1 stage (n=39) | 0.957 (0.889-1.000) | 100 (45.41-100) | 82.86 (66.94-92.28) | 84.62 (69.89-93.14) | 0.85 |
| non-T1 stage (n=56) | 0.932 (0.868-0.996) | 93.33 (68.16-100) | 85.37 (71.17-93.50) | 87.50 (76.07-94.12) | 0.84 |
Note. —95% confidence intervals are included in brackets.
ACNN, assembled convolutional neural network; AUC, area under the receiver operating characteristic curve.
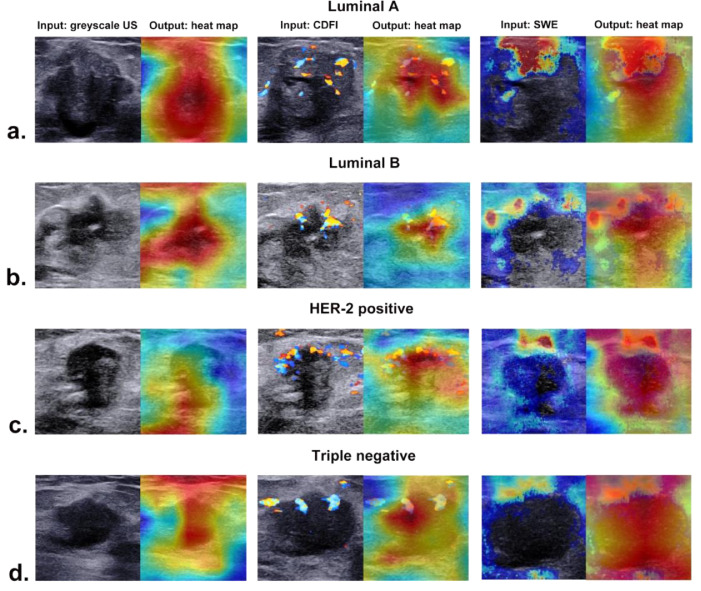
3.6. Visual interpretation of the ACNN model
The corresponding heat maps of greyscale US, CDFI, and SWE images of different molecular subtypes are presented in Fig. 7, which could produce a crude localization highlighting the import regions during the prediction process. Different colour distributions reflected that the ACNN model focused on the most predictive areas and image features in different phenotypes. The red parts of the heat map indicated that those parts provided more informative features during the network's predictive process.
Fig. 7.
The corresponding heat maps of greyscale US, CDFI, and SWE images in four molecular subtypes of breast cancer from (a) a 74-year-old woman with luminal A invasive ductal carcinoma, (b) a 52-year-old woman with luminal B invasive ductal carcinoma, (c) a 48-year-old woman with HER-2 positive invasive ductal carcinoma, and (d) a 57-year-old woman with triple negative invasive ductal carcinoma.
4. Discussion
In this multicentre study, we successfully developed a prominent multimodal US image-based ACNN model for predicting breast cancer molecular subtypes. It yielded satisfactory performance in predicting four- and five-classification St. Gallen breast cancer molecular subtypes, with AUCs of 0.89–0.96 and 0.87–0.94 in the validation and test cohorts, respectively. The multimodal ACNN model outperformed preoperative CNB in identifying four-classification molecular subtypes in the validation and test cohorts (AUC: 0.89–0.99 vs. 0.67–0.82). Moreover, this multimodal ACNN model achieved the best performance in discriminating triple negative from non-triple negative cancers compared with the monomodal and the dual-modal ACNN models. In addition, the multimodal ACNN model obtained satisfactory prediction performance for both T1 and non-T1 lesions. These promising results in the study were attributed to the advantages of standardized US image acquisition criteria, a large-scale sample size from a multicentre database, and the ACNN model architecture. To the best of our knowledge, this is the first multicentre study to apply the ACNN approach to decode multimodal US images for predicting breast cancer molecular subtypes.
Breast cancer molecular subtypes can greatly influence the initial selection of treatment options. The presence of ER or PR in luminal cancers indicates the possibility of benefit from preoperative neoadjuvant endocrine therapy (NET) [29]. Preoperative NET could decrease Ki-67 in the majority of patients while low Ki-67 scores at week 2 are associated with a longer time to recurrence [30]. HER-2 positive cancers can benefit from targeted anti-HER-2 antibody therapy [31]. The combination of chemotherapy and preoperative targeted therapy could significantly increase pathological complete response (pCR) and tail pCR [32]. Triple negative cancers with unfavourable prognostic features do not benefit from conventional endocrine and targeted therapies. Chemotherapy is the only established therapeutic option until the appearance of immunotherapy [33,34]. The KEYNOTE-522 study showed that the addition of pembrolizumab to preoperative chemotherapy increased pCR [35].
Prior studies found that some greyscale US image features were associated with certain breast cancer molecular subtypes [[11], [12], [13], [14],36,37]. Echogenic halo and posterior acoustic shadowing are predictive features of luminal A cancer [13]. The absence of an echogenic halo is a specific characteristic of luminal B cancer, and calcifications are highly correlated with luminal B cancer [12]. Luminal cancer usually shows irregular shapes, while HER-2 positive cancer and triple negative cancer appear oval or round with posterior acoustic enhancement [11,13,14]. Calcifications, echogenic halo, and posterior acoustic enhancement are more commonly observed in HER-2 positive cancer [13,36,37]. Ko et al. reported that triple negative cancer was more likely to exhibit circumscribed and markedly hypoechoic shadowing but less likely to have posterior acoustic shadowing on greyscale US [38]. Furthermore, some studies have shown that blood vessel distribution and lesion stiffness varied among different molecular subtypes [11,16,39]. HER-2 positive cancers are more likely to have internal vessels, while luminal cancers are internal vessel poor with prominent external vessels [11]. Shear wave velocity shows significant differences in different subtypes of breast cancer [16,39]. These findings supported a strong link between intrinsic biological properties and imaging manifestations.
Recently, some efforts have been made to apply AI methods to US images for the assessment of breast cancer molecular subtypes [12, 40]. A preliminary study by Zhang et al. established an ensemble decision method based on US features collected from radiologists’ interpretations to predict breast cancer molecular subtypes and achieved accuracies from 77.4% to 92.7% [12]. However, high inter- and intrareader variability in the interpretation of breast lesions on US will undoubtedly lessen the actual value of the traditional machine learning approach. Unlike traditional machine learning approach, which requires radiologists to design specific image characteristics beforehand, CNNs can automatically learn characteristics from medical images [18,41]. A retrospective study attempted to apply a CNN model to process greyscale US images for predicting four-classification breast cancer molecular subtypes [40]. However, their predictive results simply comprised outputs of four separate binary tasks (including luminal A and non-luminal A set, luminal B and non-luminal B set, HER-2 positive and non- HER-2 positive set, and triple negative and non-triple negative set). Similarly, another study also only evaluated the performance of retrospectively collected greyscale US images for prediction of binary classification breast cancer molecular subtypes using a CNN approach [42]. Additionally, previous studies used radiomics, machine learning, or deep learning methods to decipher breast cancer molecular subtypes seen on magnetic resonance image (MRI) [43], [44], [45], [46]. Nevertheless, in comparison to those studies using MRI, US is a much more widely available and cheaper option. The ACNN models could realize the overall predictive performance for four subtypes simultaneously by using a macroaverage statistical method. Furthermore, their monostructured CNN model was constructed based on the greyscale US image alone. When analysing the molecular heterogeneity of cancer, the vascular architecture and tissue stiffness should be taken into consideration. CDFI could provide blood information on breast lesions, and SWE could assess the mechanical properties of the tumour tissue elasticity. Our results verified that CDFI and SWE could provide added value in improving the predictive performance of the ACNN model. Biological changes in tumour tissues can affect the radiologic manifestations on US images that can be encoded by the CNN algorithm. By complementing the advantages and disadvantages of DenseNet, ResNet, and SENet, the ACNN model can integrate informative features in different modal US images. Thus, the multimodal strategy based on greyscale US, CDFI, and SWE images is more effective and valuable for predicting breast cancer molecular subtypes than monomodal (greyscale US image) and dual-modal (greyscale US and CDFI images).
The application in medical imaging of CNN models is mostly dubbed the “black box” medical algorithm [47]. The method of visualization with a heat map by highlighting the import regions of the image can solve the problem of how the algorithm identifies input images and establishes links with output labels [48]. The ACNN model focused on the US features of breast cancer associated with molecular subtype instead of the nonrelevant regions of the image and exhibited the strongest activation regions corresponding to areas with certain features. Our study indicated that there were usually two locations valuable for predicting molecular subtype, including the boundary of the tumour and the region inside the tumour for greyscale US. For CDFI, the distribution of supplying vessels was mostly noticed in our study, while we also found that the relatively rigid region of SWE was related to molecular subtype features.
In our study, ACNN models were initially developed for predicting four-classification breast cancer molecular subtypes. However, the luminal B subtype is the dominant subtype and could be further classified as luminal B (-) and luminal B (+). These two subtypes are clinically different diseases and treated with different systemic modalities [11]. Therefore, we investigated the feasibility and performance of the ACNN model in a five-classification molecular subtype task. Our results indicated that the multimodal ACNN model was capable of processing a five-classification task and outperformed the other two ACNN models. In addition, among all breast cancer molecular subtypes, triple negative cancer is a special type of breast cancer due to a lack of targeted molecular therapies and poor prognosis [33,34]. The ACNN models were able to yield a prediction of triple negative cancer in four and five-classification tasks, whereas the prediction performance of triple negative cancer was affected by non-triple negative cancer. Therefore, we further optimized our ACNN models to specifically identify triple negative from non-triple negative cancers. Our results demonstrated that the multimodal ACNN model also performed best in comparison with the monomodal ACNN model and the dual-modal ACNN model. Moreover, the multimodal ACNN model obtained similar and satisfactory prediction performance for T1 and non-T1 subgroups. The T stage of breast cancer has been considered an essential and critical determinant of clinical outcome, while even T1 triple negative cancer has the tendency to behave aggressively [49,50]. This result indicated that multimodal ACNN has the potential to help clinicians in the management of breast cancer, especially for early-stage triple negative cancers.
A multimodal ACNN algorithm for the classification of breast cancer molecular subtypes could make a difference in clinical practice. US-guided breast biopsy is the standard method for preoperative diagnosis of breast cancer molecular subtypes in the current clinical practice. It is often used for clinicians in pre-treatment plan-making of breast cancer patients. However, high variability in ER, PR, HER-2, and Ki-67 between partial samples obtained by CNB and surgical excision specimens was found in our study (κ-value from 0.21 to 0.67), which was similar to previous studies (κ-value from 0.195 to 0.522) [8,9]. The diagnostic performance of preoperative CNB might be lessened owing to the heterogeneity of breast cancer. Deep learning is well known for its capability to learn high-level representations of data through multiple functional layers and to represent characteristic biological processes of different subtypes, which has the potential to better dissect molecular heterogeneity [51]. In addition, different modalities of US images contain different information on breast cancers [15,16]. Thus, the multimodal US image-based ACNN model achieved better performance than preoperative CNB in this study. If the result of the multimodal ACNN model was not in accordance with US-guided CNB, clinicians would consider an additional biopsy because inadequate tissue might result in inaccurate IHC results, especially for small cancers. On the other hand, if presented with the same results, it can increase the clinicians’ confidence in making the optimum treatment strategy. In addition, the ACNN model can convert obscure and inexplicably derived image features into intelligible heat maps. In actual clinical applications, heat maps can be a visual tool that provides highly informative regions on US images. Our study provided a method to supplement US-guided CNB instead of obviating the need for which. With the help of generated heat map, the ACNN model has the potential to assist clinicians in determining the sampling area for the lesion during US-guided CNB procedures. Therefore, our study provided a noninvasive decision-making method for the preoperative prediction of breast cancer molecular subtypes in clinical practice, especially in remote areas.
Our study had some limitations. First, the breast cancer molecular subtype classification was based on IHC phenotypes in our study rather than genetic subtypes. However, IHC is still recommended for clinical application in current clinical practice guidelines because it is easily accessible and inexpensive [52]. Second, although multimodal US images were analysed in our study, other imaging modalities (for example, mammography and MRI) were not studied. Accordingly, further efforts will focus on assessing predictions that combine different imaging methods of breast cancer. Third, the sample size of the validation set in our study was limited due to the prospective design and relatively short study period. Therefore, further studies with long-term periods are needed to effectively validate the performance of our ACNN model. Finally, although our proposed ACNN model had prominent performances for predicting four-, five-, and two-classification breast cancer molecular subtypes, other newly developed algorithms (for example, transformer and transfer learning) and well-performed CNNs (for example, InceptionNet and NASNet) were not involved in this study. Thus, we suggest that the continually updated AI algorithms should be explored for decoding the molecular subtypes of breast cancer seen on multimodal US images in future studies.
In conclusion, we developed an original multimodal ACNN that combined greyscale US and CDFI as well as SWE images to predict molecular subtypes of breast cancer and outperforms ACNN models applied to dual-modal or monomodal US images. The multimodal US-based ACNN model is therefore a potential noninvasive decision-making method for the management of patients with breast cancer in routine clinical practice.
Contributors
Conception and design: H. Xu. and C. Zhao. Data verification: B. Zhou, L. Wang, T. Ren, C. Peng, L. Sun, C. Zhao, and H. Xu. Data acquisition: B. Zhou, L. Wang, T. Ren, and C. Peng. Data analysis/interpretation: B. Zhou, L. Wang, and C. Zhao. Data analysis/interpretation in revision: B. Zhou, L. Wang, H. Yin, and L. Sun. Algorithm development: B. Zhou, C. Zhao, and D. Li. Literature research: B. Zhou, C. Zhao, L. Wang, T. Ren, C. Peng, and H. Shi. Statistical analysis: B. Zhou, T. Wu, and C. Zhao. Writing, review, and/or revision of the manuscript: B. Zhou, C. Zhao, and H. Xu. All authors read and approved the final version of the manuscript.
Data sharing statement
As the study involved human participants, the data cannot be made freely available in the manuscript, nor a public repository because of ethical restrictions. However, the data are available from Shanghai Tenth People's Hospital for researchers who meet the criteria for access to confidential data. Interested researchers can send data access requests to the corresponding author (H.Xu. and C.Zhao).
Declaration of Competing Interest
T. Wu is an employee of GE Healthcare. D. Li is an employee of XiaoBaiShiJi Network Technical Co., Ltd. They do not hold any other financial interests in these companies. These companies had no role in the study design, data collection, analysis, patient recruitment or writing of the manuscript, the decision to submit the manuscript for publication or any aspect pertinent to the study. No products from this company were used in this study. The other authors declare that they have no conflicts of interest.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (Grants 81725008 and 81927801), Shanghai Municipal Health Commission (Grants 2019LJ21 and SHSLCZDZK03502), and the Science and Technology Commission of Shanghai Municipality (Grants 19441903200, 19DZ2251100, and 21Y11910800).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103684.
Contributor Information
Chong-Ke Zhao, Email: zhaochongke123@163.com.
Hui-Xiong Xu, Email: xuhuixiong@126.com.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Markham M.J., Wachter K., Agarwal N., et al. Clinical cancer advances 2020: annual report on progress against cancer from the american society of clinical oncology. J Clin Oncol. 2020;38(10):1081. doi: 10.1200/JCO.19.03141. [DOI] [PubMed] [Google Scholar]
- 3.Andre F., Ismaila N., Henry N.L., et al. Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: ASCO clinical practice guideline update-integration of results from TAILORx. J Clin Oncol. 2019;37(22):1956–1964. doi: 10.1200/JCO.19.00945. [DOI] [PubMed] [Google Scholar]
- 4.Henry N.L., Somerfield M.R., Abramson V.G., et al. Role of patient and disease factors in adjuvant systemic therapy decision making for early-stage, operable breast cancer: update of the ASCO endorsement of the cancer care ontario guideline. J Clin Oncol. 2019;37(22):1965–1977. doi: 10.1200/JCO.19.00948. [DOI] [PubMed] [Google Scholar]
- 5.Wang M., He X., Chang Y., Sun G., Thabane L. A sensitivity and specificity comparison of fine needle aspiration cytology and core needle biopsy in evaluation of suspicious breast lesions: a systematic review and meta-analysis. Breast. 2017;31:157–166. doi: 10.1016/j.breast.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Yeo S.K., Guan J.L. Breast cancer: multiple subtypes within a tumor? Trends Cancer. 2017;3(11):753–760. doi: 10.1016/j.trecan.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singer G., Stadlmann S. Pathology of breast tissue obtained in minimally invasive biopsy procedures. Recent Results Cancer Res. 2009;173:137–147. doi: 10.1007/978-3-540-31611-4_8. [DOI] [PubMed] [Google Scholar]
- 8.Agosto-Arroyo E., Tahmasbi M., Al Diffalha S., et al. Invasive breast carcinoma tumor size on core needle biopsy: analysis of practice patterns and effect on final pathologic tumor stage. Clin Breast Cancer. 2018;18(5):e1027–e1030. doi: 10.1016/j.clbc.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Prendeville S., Feeley L., Bennett M.W., O'Connell F., Browne T.J. Reflex repeat HER2 testing of grade 3 breast carcinoma at excision using immunohistochemistry and in situ analysis: frequency of HER2 discordance and utility of core needle biopsy parameters to refine case selection. Am J Clin Pathol. 2016;145(1):75–80. doi: 10.1093/ajcp/aqv018. [DOI] [PubMed] [Google Scholar]
- 10.Chen J., Wang Z., Lv Q., et al. Comparison of core needle biopsy and excision specimens for the accurate evaluation of breast cancer molecular markers: a report of 1003 cases. Pathol Oncol Res. 2017;23(4):769–775. doi: 10.1007/s12253-017-0187-5. [DOI] [PubMed] [Google Scholar]
- 11.Dogan B.E., Menezes G.L.G., Butler R.S., et al. Optoacoustic imaging and gray-scale US features of breast cancers: correlation with molecular subtypes. Radiology. 2019;292(3):564–572. doi: 10.1148/radiol.2019182071. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L., Li J., Xiao Y., et al. Identifying ultrasound and clinical features of breast cancer molecular subtypes by ensemble decision. Sci Rep. 2015;5:11085. doi: 10.1038/srep11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng F.Y., Lu Q., Huang B.J., et al. Imaging features of automated breast volume scanner: correlation with molecular subtypes of breast cancer. Eur J Radiol. 2017;86:267–275. doi: 10.1016/j.ejrad.2016.11.032. [DOI] [PubMed] [Google Scholar]
- 14.Wu T., Li J., Wang D., et al. Identification of a correlation between the sonographic appearance and molecular subtype of invasive breast cancer: a review of 311 cases. Clin Imaging. 2019;53:179–185. doi: 10.1016/j.clinimag.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 15.Sigrist R.M.S., Liau J., Kaffas A.E., Chammas M.C., Willmann J.K. Ultrasound elastography: review of techniques and clinical applications. Theranostics. 2017;7(5):1303–1329. doi: 10.7150/thno.18650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu H., Wan J., Xu G., et al. Conventional US and 2-D shear wave elastography of virtual touch tissue imaging quantification: correlation with immunohistochemical subtypes of breast cancer. Ultrasound Med Biol. 2019;45(10):2612–2622. doi: 10.1016/j.ultrasmedbio.2019.06.421. [DOI] [PubMed] [Google Scholar]
- 17.Yang S., Gao X., Liu L., et al. Performance and reading time of automated breast US with or without computer-aided detection. Radiology. 2019;292(3):540–549. doi: 10.1148/radiol.2019181816. [DOI] [PubMed] [Google Scholar]
- 18.Hosny A., Parmar C., Quackenbush J., Schwartz L.H., Aerts H. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18(8):500–510. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liao W.X., He P., Hao J., et al. Automatic identification of breast ultrasound image based on supervised block-based region segmentation algorithm and features combination migration deep learning model. IEEE J Biomed Health Inform. 2020;24(4):984–993. doi: 10.1109/JBHI.2019.2960821. [DOI] [PubMed] [Google Scholar]
- 20.Ciritsis A., Rossi C., Eberhard M., et al. Automatic classification of ultrasound breast lesions using a deep convolutional neural network mimicking human decision-making. Eur Radiol. 2019;29(10):5458–5468. doi: 10.1007/s00330-019-06118-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhou L.Q., Wu X.L., Huang S.Y., et al. Lymph node metastasis prediction from primary breast cancer US images using deep learning. Radiology. 2020;294(1):19–28. doi: 10.1148/radiol.2019190372. [DOI] [PubMed] [Google Scholar]
- 22.Barr R.G., Nakashima K., Amy D., et al. WFUMB guidelines and recommendations for clinical use of ultrasound elastography: Part 2: breast. Ultrasound Med Biol. 2015;41(5):1148–1160. doi: 10.1016/j.ultrasmedbio.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 23.Goldhirsch A., Winer E.P., Coates A.S., et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen international expert consensus on the primary therapy of early breast cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren S., He K., Girshick R., Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017;39(6):1137–1149. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 25.He F., Liu T., Tao D. Why ResNet works? Residuals generalize. IEEE Trans Neural Netw Learn Syst. 2020;31(12):5349–5362. doi: 10.1109/TNNLS.2020.2966319. [DOI] [PubMed] [Google Scholar]
- 26.G. Huang, Z. Liu, G. Pleiss, L. Van Der Maaten, K. Weinberger Convolutional networks with dense connectivity. IEEE Trans Pattern Anal Mach Intell 2019. Doi: 10.1109/TPAMI.2019.2918284. [DOI] [PubMed]
- 27.Hu J., Shen L., Albanie S., Sun G., Wu E. Squeeze-and-excitation networks. IEEE Trans Pattern Anal Mach Intell. 2020;42(8):2011–2023. doi: 10.1109/TPAMI.2019.2913372. [DOI] [PubMed] [Google Scholar]
- 28.Li X., Zhang S., Zhang Q., et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: a retrospective, multicohort, diagnostic study. Lancet Oncol. 2019;20(2):193–201. doi: 10.1016/S1470-2045(18)30762-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran B., Bedard PL. Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res. 2011;13(6):221. doi: 10.1186/bcr2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith I., Robertson J., Kilburn L., et al. Long-term outcome and prognostic value of Ki67 after perioperative endocrine therapy in postmenopausal women with hormone-sensitive early breast cancer (POETIC): an open-label, multicentre, parallel-group, randomised, phase 3 trial. Lancet Oncol. 2020;21(11):1443–1454. doi: 10.1016/S1470-2045(20)30458-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu B., Yan M., Ma F., et al. Pyrotinib plus capecitabine versus lapatinib plus capecitabine for the treatment of HER2-positive metastatic breast cancer (PHOEBE): a multicentre, open-label, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22(3):351–360. doi: 10.1016/S1470-2045(20)30702-6. [DOI] [PubMed] [Google Scholar]
- 32.Ma W., Zhao F., Zhou C., et al. Targeted neoadjuvant therapy in the HER-2-positive breast cancer patients: a systematic review and meta-analysis. Oncol Targets Ther. 2019;12:379–390. doi: 10.2147/OTT.S183304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Xie X., Wang X., et al. Locoregional and distant recurrences after breast conserving therapy in patients with triple-negative breast cancer: a meta-analysis. Surg Oncol. 2013;22(4):247–255. doi: 10.1016/j.suronc.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Adams S., Schmid P., Rugo H.S., et al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann Oncol. 2019;30(3):397–404. doi: 10.1093/annonc/mdy517. [DOI] [PubMed] [Google Scholar]
- 35.Schmid P., Cortes J., Pusztai L., et al. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. 2020;382(9):810–821. doi: 10.1056/NEJMoa1910549. [DOI] [PubMed] [Google Scholar]
- 36.Seo B.K., Pisano E.D., Kuzimak C.M., et al. Correlation of HER-2/neu overexpression with mammography and age distribution in primary breast carcinomas. Acad Radiol. 2006;13(10):1211–1218. doi: 10.1016/j.acra.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Kim S.H., Seo B.K., Lee J., et al. Correlation of ultrasound findings with histology, tumor grade, and biological markers in breast cancer. Acta Oncol. 2008;47(8):1531–1538. doi: 10.1080/02841860801971413. [DOI] [PubMed] [Google Scholar]
- 38.Ko E.S., Lee B.H., Kim H.A., et al. Triple-negative breast cancer: correlation between imaging and pathological findings. Eur Radiol. 2010;20(5):1111–1117. doi: 10.1007/s00330-009-1656-3. [DOI] [PubMed] [Google Scholar]
- 39.Song E.J., Sohn Y.M., Seo M. Tumor stiffness measured by quantitative and qualitative shear wave elastography of breast cancer. Br J Radiol. 2018;91(1086) doi: 10.1259/bjr.20170830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang M., Zhang D., Tang S.C., et al. Deep learning with convolutional neural network in the assessment of breast cancer molecular subtypes based on US images: a multicenter retrospective study. Eur Radiol. 2021;31(6):3673–3682. doi: 10.1007/s00330-020-07544-8. [DOI] [PubMed] [Google Scholar]
- 41.Zhao C.K., Ren T.T., Yin Y.F., et al. A comparative analysis of two machine learning-based diagnostic patterns with thyroid imaging reporting and data system for thyroid nodules: diagnostic performance and unnecessary biopsy rate. Thyroid. 2020;31(3):470–481. doi: 10.1089/thy.2020.0305. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., Li H, Wang C., et al. Evaluating the accuracy of breast cancer and molecular subtype diagnosis by ultrasound image deep learning model. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.623506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng W., Sun Y., Qian H., et al. Computer-aided diagnosis evaluation of the correlation between magnetic resonance imaging with molecular subtypes in breast cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.693339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leithner D., Mayerhoefer M.E., Martinez D.F., et al. Non-invasive assessment of breast cancer molecular subtypes with multiparametric magnetic resonance imaging radiomics. J Clin Med. 2020;9(6):1853. doi: 10.3390/jcm9061853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leithner D., Bernard-Davila B., Martinez D.F., et al. Radiomic signatures derived from diffusion-weighted imaging for the assessment of breast cancer receptor status and molecular subtypes. Mol Imaging Biol. 2020;22(2):453–461. doi: 10.1007/s11307-019-01383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leithner D., Horvat J.V., Marino M.A., et al. Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: initial results. Breast Cancer Res. 2019;21(1):106. doi: 10.1186/s13058-019-1187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price WN. Big data and black-box medical algorithms. Sci Transl Med. 2018;10(471):eaao5333. doi: 10.1126/scitranslmed.aao5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zurowietz M., Nattkemper TW. An interactive visualization for feature localization in deep neural networks. Front Artif Intell. 2020;3:49. doi: 10.3389/frai.2020.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foulkes W.D., Reis-Filho J.S., Narod SA. Tumor size and survival in breast cancer–a reappraisal. Nat Rev Clin Oncol. 2010;7(6):348–353. doi: 10.1038/nrclinonc.2010.39. [DOI] [PubMed] [Google Scholar]
- 50.Sopik V., Narod SA. The relationship between tumour size, nodal status and distant metastases: on the origins of breast cancer. Breast Cancer Res Treat. 2018;170(3):647–656. doi: 10.1007/s10549-018-4796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao F., Wang W., Tan M., et al. DeepCC: a novel deep learning-based framework for cancer molecular subtype classification. Oncogenesis. 2019;8(9):44. doi: 10.1038/s41389-019-0157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolff A.C., Hammond M.E.H., Allison K.H., et al. Human epidermal growth factor receptor 2 testing in breast cancer: american society of clinical oncology/college of american pathologists clinical practice guideline focused update. J Clin Oncol. 2018;36(20):2105–2122. doi: 10.1200/JCO.2018.77.8738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.