Abstract
Objectives
Tedizolid is an oxazolidinone antimicrobial with activity against Gram-positive bacteria, including MRSA. Tedizolid resistance is uncommon and tedizolid’s capacity to select for cross-resistance to other antimicrobials is incompletely understood. The objective of this study was to further explore the phenotypic and genetic basis of tedizolid resistance in MRSA.
Methods
We selected for tedizolid resistance in an MRSA laboratory strain, N315, by serial passage until an isolate with an MIC ≥1 log2 dilution above the breakpoint for resistance (≥2 mg/L) was recovered. This isolate was subjected to WGS and susceptibility to a panel of related and unrelated antimicrobials was tested in order to determine cross-resistance. Homology modelling was performed to evaluate the potential impact of the mutation on target protein function.
Results
After 10 days of serial passage we recovered a phenotypically stable mutant with a tedizolid MIC of 4 mg/L. WGS revealed only one single nucleotide variant (A1345G) in rpoB, corresponding to amino acid substitution D449N. MICs of linezolid, chloramphenicol, retapamulin and quinupristin/dalfopristin increased by ≥2 log2 dilutions, suggesting the emergence of the so-called ‘PhLOPSa’ resistance phenotype. Susceptibility to other drugs, including rifampicin, was largely unchanged. Homology models revealed that the mutated residue of RNA polymerase would be unlikely to directly affect oxazolidinone action.
Conclusions
To the best of our knowledge, this is the first time that an rpoB mutation has been implicated in resistance to PhLOPSa antimicrobials. The mechanism of resistance remains unclear, but is likely indirect, involving σ-factor binding or other alterations in transcriptional regulation.
Introduction
Tedizolid is an oxazolidinone antimicrobial with broad-spectrum activity against Gram-positive bacteria, including MRSA.1 Resistance to tedizolid is uncommon in MRSA and, consequently, the mechanisms involved and the consequences on collateral resistance remain incompletely explored.
Like other oxazolidinones, tedizolid exerts its antibacterial effects by binding the 23S rRNA component of the 50S ribosomal subunit and thereby inhibiting protein synthesis. Mutations in the ribosomal proteins L3, L4 and L22 (encoded by rplC, rplD and rplV, respectively) and the 23S rRNA target, which also mediate the so-called ‘PhLOPSa’ (phenicol, lincosamide, oxazolidinone, pleuromutilin and streptogramin A) resistance phenotype, have been implicated as resistance mechanisms.2,3 Acquisition of the transferable rRNA methyltransferase gene, cfr, may also cause resistance to linezolid and other PhLOPSa antimicrobials, but is generally believed to be insufficient to produce tedizolid resistance on its own.4 The plasmid-borne optrA gene, which encodes an ABC transporter, has also been implicated in oxazolidinone and phenicol resistance in enterococci and staphylococci isolated from animals.5–7 In contrast, genes known to effect quinolone efflux, such as norA and mepA, or lincosamide efflux, such as mdeA, have not been reported to affect oxazolidinone activity.
Previous serial passage studies with tedizolid have seen limited success in selecting for tedizolid resistance, observing only mutations in 23S rRNA and modestly elevated MICs.8 To better understand the potential for tedizolid resistance to emerge in Staphylococcus aureus, the genetic mechanisms involved and the practical consequences of those changes on collateral antibiotic resistance, we selected for tedizolid resistance in MRSA in vitro and examined the emergence of cross-resistance to other antimicrobials in related and unrelated classes.
Methods
Serial passage and susceptibility testing
Using a well-characterized MRSA strain, N315, we selected for tedizolid resistance by serial passage similar to previously described methods.9 Briefly ∼105.5 cfu/mL of N315 was suspended in cation-adjusted Mueller–Hinton II broth (MHB; Becton, Dickinson) supplemented with 0.5× the MIC of tedizolid (Medchemexpress LLC). Once visible growth was achieved a sample of the broth was diluted 1:1000 into fresh MHB with twice the previous concentration of tedizolid and 100 μL was plated onto tryptic soy agar (TSA; Becton Dickinson) supplemented with 3 mg/L tedizolid to screen for resistance. This process was continued until growth on the resistance screening plate demonstrated an MIC of ≥4 mg/L, which is at least 1 log2 dilution above the CLSI’s breakpoint for resistance (≥2 mg/L).10 To explore the cross-resistance associated with this evolved strain we re-evaluated susceptibility to a panel of other antimicrobial agents with related and unrelated targets by broth microdilution in accordance with CLSI guidelines10 or by Etest® in the case of quinupristin/dalfopristin. These drugs, listed in Table 1, were purchased from Sigma, with the exception of daptomycin, which was purchased from its manufacturer (Merck). We also performed vancomycin population analysis profiling (PAP) to assess the emergence of hVISA as previously described.11 As always, an area under the survival: concentration curve ratio of <0.9 with the archetypal hVISA strain Mu3 (PAP AUC ratio) was considered to indicate vancomycin susceptibility. The stability of the tedizolid resistance phenotype was evaluated by passing the emergent isolate on drug-free TSA for three passages and repeated testing.
Table 1.
MICs of various antimicrobials for the parent strain (N315) and the tedizolid-passaged rpoB mutant (N315-TDZ4), and log2 fold change in MIC (i.e. number of doubling dilutions) and antimicrobial target
| Drug | MIC (mg/L) |
Target | ||
|---|---|---|---|---|
| N315 | N315-TDZ4 | log2 fold change | ||
| Chloramphenicola | 8 | 128 | 4 | 50S ribosome |
| Daptomycin | 0.25 | 0.25 | 0 | phosphatidylglycerol of the cell membrane |
| Doxycycline | 0.125 | 0.125 | 0 | 30S ribosome |
| Linezolida | 2 | 8 | 2 | 50S ribosome |
| Moxifloxacin | 0.0625 | 0.0625 | 0 | DNA gyrase/topoisomerase |
| Quinupristin/dalfopristina | 0.38 | 2 | ≥2 | 50S ribosome |
| Retapamulina | 0.0625 | 16 | 8 | 50S ribosome |
| Rifampicin | 0.001 | 0.001 | 0 | RNAP |
| Tedizolida | 0.25 | 4 | 4 | 50S ribosome |
| Vancomycin | 0.5 | 1 | 1 | cell-wall synthesis |
| PAP AUC ratio with Mu3 | 0.44 | 0.85 | 0.95 | |
PhLOPSa antimicrobials.
WGS and homology modelling
The recovered isolate with reduced tedizolid susceptibility (N315-TDZ4) was subjected to WGS using the MiSeq platform (Illumina, San Diego, CA, USA) as previously described12 to an average read depth of at least 50×. Sequence data from this study are freely available through the NCBI Sequence Read Archive (PRJNA578164). Using a strategy of combined de novo genome assembly and short-read re-mapping,12 we catalogued all forms of genetic variation in the sequenced isolate, ranging from single nucleotide variants (SNVs) and small insertions and deletions (indels) to large-scale structural changes. We constructed homology models of any mutated proteins to elucidate the potential mechanistic basis for changes in drug susceptibility using I-TASSER.13 While homology modelling is necessarily imprecise, I-TASSER is among the most reliable algorithms available14 and the wealth of structural information about bacterial RNA polymerases (RNAPs) lends confidence to our predictions. In the case of RpoB, the estimated template modelling score was 0.67 (values >0.5 indicate successful prediction of protein topology) and the estimated root-mean-square deviation between the homology model and the true structure was 10.1 Å.
Results
After 10 days of serial passage we recovered an isolate (N315-TDZ4) with a tedizolid MIC of 4 mg/L, i.e. 16× the MIC (4 log2 dilutions) for the parent strain, N315. We found that some degree of cross-resistance (increases in MIC of ≥2 log2 dilutions) was evident for chloramphenicol, linezolid, retapamulin and quinupristin/dalfopristin, but susceptibility to other drugs tested was relatively unchanged (Table 1). The vancomycin MIC increased from 0.5 mg/L for N315 to 1 mg/L for N315-TDZ4 and the vancomycin PAP AUC ratio with Mu3 increased from 0.44 to 0.85, indicating continued vancomycin susceptibility (PAP AUC ratio ≥0.9 indicates hVISA). Passage on drug-free TSA indicated that the tedizolid resistance phenotype was stable for at least three passages.
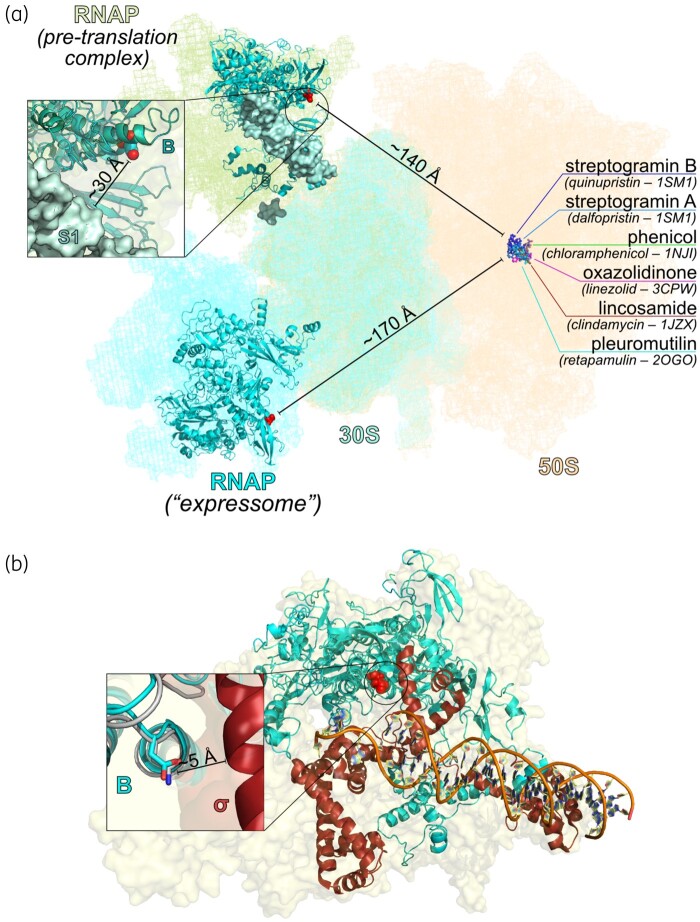
Although short-read WGS cannot confidently recover variants occurring in repetitive or structurally complex genomic regions,15whole-genome analysis of the strain pair revealed only an SNV in the rpoB gene (A-1345-G), corresponding to amino acid substitution Asn-449-Asp in the β-subunit of RNAP. This mutation lies outside of the rifampicin resistance-determining regions, which span nucleotides 1384–1464 (amino acids 462–488) and 1543–1590 (amino acids 462–488),16,17 and we correspondingly did not observe any change in rifampicin susceptibility (Table 1). No other variants were identified in the evolved strain, N315-TDZ4, showing that it is inherently isogenic to the parent. Homology modelling predicted that the mutation in rpoB is located on a helix-turn-helix motif that makes up part of the σ-factor binding interface and may also interact with protein S1 of the 30S subunit of the ribosome. However, in modelled RNAP/ribosome complexes this mutation lies ≥140 Å from the binding site of PhLOPSa drugs on the 50S ribosomal subunit and would therefore be unlikely to interact directly with their target sites (Figure 1).
Figure 1.
(a) Homology model of S. aureus RpoB N449D docked into available models of RNAP/ribosome interactions, reflecting either a pre-translational RNAP : 30S subunit complex (PDB ID 6AWB)18 or an active transcribing-translating RNAP/70S ribosome complex (the ‘expressome’, PDB ID 5MY1).19 In either case, the mutated residue in RNAP is distant from the binding site of PHLOPSa drugs on the 50S ribosomal subunit. Coordinates for the 50S subunit are from PDB IDP 5U9F, while drug-binding sites are inferred from the PDB IDs indicated in parentheses.23–27 The inset shows that the shortest distance between the site of the mutation and any part of the ribosome is to protein S1. (b) Homology model of S. aureus RpoB aligned with a crystal structure of the RNAP transcription initiation complex from Thermus aquaticus (PDB ID 4XLP28), showing that the site of mutation comprises part of the σ-factor binding surface. In the inset, cyan represents the S. aureus homology model, while grey and burgundy, respectively, represent the β-subunit and σ-factor from the crystal structure. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.
Discussion
We report for the first time (to the best of our knowledge) a novel mechanism of resistance to oxazolidinones, phenicols, pleuromutilins and streptogramins that was selected for using escalating concentrations of tedizolid. The mechanism requires only a single nucleotide substitution in rpoB, which encodes the β-subunit of RNAP. The parent strain N315 is resistant to clindamycin, so lincosamide cross-resistance could not be evaluated in this study, but we otherwise observed susceptibility changes for all of the so-called PhLOPSa antimicrobials that were conferred by this mutation. Although tedizolid resistance remains uncommon in MRSA and the clinical relevance of the rpoB mutation is still unknown, our results suggest an evolutionarily efficient mechanism of MDR via a novel, indirect pathway.
RNAP binds the 30S ribosomal subunit in at least two different modes, one thought to promote ribosome assembly and translation initiation, and the other facilitating the tight linkage between transcription and translation in bacteria18,19(Figure 1a), while no significant interactions between the 50S subunit and RNAP have been detected. Despite the absence of a direct link between RNAP and the 50S subunit, we observed cross-resistance only to agents that target 50S and observed no changes in susceptibility to 30S-targeting agents (doxycycline) or non-ribosome-targeting agents (daptomycin, moxifloxacin and rifampicin). This suggests a relatively specific effect of the SNV rather than a broader modulation of translation or protein synthesis. While macrolides are not considered part of the PhLOPSa group, they also target the 50S subunit; however, macrolide susceptibility was unevaluable in this study since N315 is resistant to erythromycin at baseline. Consequently, we could not determine whether there was even broader resistance to all 50S-targeting agents. Based on the absence of altered susceptibility to other intracellularly active agents, including moxifloxacin, doxycycline and rifampicin, it seems unlikely that this variant somehow facilitates multidrug efflux.
Previous studies have reported an association between vancomycin or daptomycin susceptibility and certain RpoB mutations, including Ala-621-Glu, Ala-477-Asp and His-481-Tyr.20–22 We did not observe any change in daptomycin MIC for our mutant, but we did see a consistent 1 log2 dilution increase in vancomycin MIC. While this difference is commonly thought to be within the error of the assay, we also performed vancomycin PAP and found a substantial, albeit also subclinical, change in vancomycin PAP AUC ratio with Mu3 from 0.44 to 0.85. Vancomycin non-susceptibility rarely emerges from only one SNV, often requiring multiple, step-wise mutations in multiple genes. Consequently, it remains plausible that the rpoB variant in N315-TDZ4 could contribute to the VISA phenotype, especially in other strains with higher vancomycin MICs at baseline or other mutations that predispose an isolate towards glycopeptide resistance. While the change in vancomycin susceptibility is small, to the best of our knowledge, we believe that this is the first report that suggests that some degree of oxazolidinone cross-resistance with glycopeptides is possible.
For now, tedizolid resistance remains uncommon clinically and it is unknown whether this unique mutation could be selected for by other PhLOPSa drugs or would be likely to emerge after clinical exposure to tedizolid. The precise molecular mechanism by which this mutation mediates this resistance phenotype is still unclear, but may involve transcriptional modulation by altered σ-factor binding. Under other circumstances it may be necessary to perform allelic exchange experiments to attribute this phenotype to the rpoB mutation that we identified. However, since this was the only variant detected in the entire genome, including non-coding regions, we feel confident that this SNV is responsible for the changes in antimicrobial susceptibility that we observed. Given the apparent role of rpoB in manifesting many antimicrobial resistance phenotypes, additional work to systematically characterize the pathways affected by all functional regions of RNAP in multiple species and with consideration to disparate drug classes seems warranted.
Acknowledgements
This study was initially presented in part at IDWeek 2018, San Francisco, CA, USA.
Funding
This work was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (1R21AI132994, 1R01AI136979), the Cystic Fibrosis Foundation (SINGH19R0) and by the University of Washington.
Transparency declarations
B.J.W. has received research grants from commercial sources, including Shionogi Inc. All other authors: none to declare.
References
- 1. Kisgen JJ, Mansour H, Unger NR. et al. Tedizolid: a new oxazolidinone antimicrobial. Am J Health Syst Pharm 2014; 71: 621–33. [DOI] [PubMed] [Google Scholar]
- 2. Shaw KJ, Poppe S, Schaadt R. et al. In vitro activity of TR-700, the antibacterial moiety of the prodrug TR-701, against linezolid-resistant strains. Antimicrob Agents Chemother 2008; 52: 4442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Freitas AR, Dilek AR, Peixe L. et al. Dissemination of Staphylococcus epidermidis ST22 with stable, high-level resistance to linezolid and tedizolid in the Greek-Turkish region (2008-2016). Infect Control Hosp Epidemiol 2018; 39: 492–4. [DOI] [PubMed] [Google Scholar]
- 4. Zhanel GG, Love R, Adam H. et al. Tedizolid: a novel oxazolidinone with potent activity against multidrug-resistant gram-positive pathogens. Drugs 2015; 75: 253–70. [DOI] [PubMed] [Google Scholar]
- 5. Wang Y, Lv Y, Cai J. et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 2015; 70: 2182–90. [DOI] [PubMed] [Google Scholar]
- 6. Liu B, Sun H, Pan Y. et al. Prevalence, resistance pattern, and molecular characterization of Staphylococcus aureus isolates from healthy animals and sick populations in Henan Province, China. Gut Pathog 2018; 10: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fan R, Li D, Wang Y. et al. Presence of the optrA gene in methicillin-resistant Staphylococcus sciuri of porcine origin. Antimicrob Agents Chemother 2016; 60: 7200–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Locke JB, Hilgers M, Shaw KJ.. Novel ribosomal mutations in Staphylococcus aureus strains identified through selection with the oxazolidinones linezolid and torezolid (TR-700). Antimicrob Agents Chemother 2009; 53: 5265–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silverman JA, Oliver N, Andrew T. et al. Resistance studies with daptomycin. Antimicrob Agents Chemother 2001; 45: 1799–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Edition: M100. 2017.
- 11. Wootton M, Howe RA, Hillman R. et al. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother 2001; 47: 399–403. [DOI] [PubMed] [Google Scholar]
- 12. Roach DJ, Burton JN, Lee C. et al. A year of infection in the intensive care unit: prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet 2015; 11: e1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang J, Zhang Y.. Protein structure and function prediction using I-TASSER. Curr Protoc Bioinformatics 2015; 52: 5.8.1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kryshtafovych A, Monastyrskyy B, Fidelis K. et al. Evaluation of the template-based modeling in CASP12. Proteins 2018; 86 Suppl 1: 321–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berbers B, Saltykova A, Garcia-Graells C. et al. Combining short and long read sequencing to characterize antimicrobial resistance genes on plasmids applied to an unauthorized genetically modified Bacillus. Sci Rep 2020; 10: 4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alifano P, Palumbo C, Pasanisi D. et al. Rifampicin-resistance, rpoB polymorphism and RNA polymerase genetic engineering. J Biotechnol 2015; 202: 60–77. [DOI] [PubMed] [Google Scholar]
- 17. Zhou W, Shan W, Ma X. et al. Molecular characterization of rifampicin-resistant Staphylococcus aureus isolates in a Chinese teaching hospital from Anhui, China. BMC Microbiol 2012; 12: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Demo G, Rasouly A, Vasilyev N. et al. Structure of RNA polymerase bound to ribosomal 30S subunit. eLife 2017; 6: e28560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kohler R, Mooney RA, Mills DJ. et al. Architecture of a transcribing-translating expressome. Science 2017; 356: 194–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cui L, Isii T, Fukuda M. et al. An RpoB mutation confers dual heteroresistance to daptomycin and vancomycin in Staphylococcus aureus. Antimicrob Agents Chemother 2010; 54: 5222–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bæk KT, Thøgersen L, Mogenssen RG. et al. Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob Agents Chemother 2015; 59: 6983–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watanabe Y, Cui L, Katayama Y. et al. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J Clin Microbiol 2011; 49: 2680–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harms JM, Schlunzen F, Fucini P. et al. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol 2004; 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hansen JL, Moore PB, Steitz TA.. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol 2003; 330: 1061–75. [DOI] [PubMed] [Google Scholar]
- 25. Ippolito JA, Kanyo ZF, Wang D. et al. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J Med Chem 2008; 51: 3353–6. [DOI] [PubMed] [Google Scholar]
- 26. Schlünzen F, Zarivach R, Harms J. et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 2001; 413: 814–21. [DOI] [PubMed] [Google Scholar]
- 27. Davidovich C, Bashan A, Auerbach-Nevo T. et al. Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity. Proc Natl Acad Sci USA 2007; 104: 4291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bae B, Feklistov A, Lass-Napiorkowska A. et al. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife 2015; 4: e08504. [DOI] [PMC free article] [PubMed] [Google Scholar]