Abstract
Alginate is a major compound of brown macroalgae and as such an important carbon and energy source for heterotrophic marine bacteria. Despite the rather simple composition of alginate only comprising mannuronate and guluronate units, these bacteria feature complex alginolytic systems that can contain up to seven alginate lyases. This reflects the necessity of large enzyme systems for the complete degradation of the abundant substrate. Numerous alginate lyases have been characterized. They belong to different polysaccharide lyase (PL) families, but only one crystal structure of a family 17 (PL17) alginate lyase has been reported to date, namely Alg17c from the gammaproteobacterium Saccharophagus degradans. Biochemical and structural characterizations are helpful to link sequence profiles to function, evolution of functions and niche-specific characteristics. Here, we combined detailed biochemical and crystallographic analysis of AlyA3, a PL17 alginate lyase from the marine flavobacteria Zobellia galactanivorans DsijT, providing the first structure of a PL17 in the Bacteroidetes phylum. AlyA3 is exo-lytic and highly specific of mannuronate stretches. As part of an “alginate utilizing locus”, its activity is complementary to that of other characterized alginate lyases from the same bacterium. Structural comparison with Alg17c highlights a common mode of action for exo-lytic cleavage of the substrate, strengthening our understanding of the PL17 catalytic mechanism. We show that unlike Alg17c, AlyA3 contains an inserted flexible loop at the entrance to the catalytic groove, likely involved in substrate recognition, processivity and turn over.
Keywords: alginate lyase, alginolytic system, bacterial metabolism, family PL17, substrate complex
Introduction
Brown macroalgae are important primary producers in coastal regions around the globe, storing a large amount of carbon from the atmosphere into their cellular constituents. In particular, alginate can account for up to 40% of the dry weight of brown macroalgae (Mabeau and Kloareg 1987). Alginate is a linear anionic polysaccharide consisting of two uronic acid monomers, α-l-guluronate (G) and β-d-mannuronate (M), that can be arranged either as homopolymeric blocks (polyG and polyM) or mixed blocks with alternating monomers (polyMG). The average molecular weight of alginate in brown algal cell wall varies depending on algal species and extraction method, with typical Mw values in the range 100–1000 kDa, corresponding to a degree of polymerization (DP) of around 500–5000 (Draget et al. 2006). The gelling properties of alginate rely on polyG blocks self-assembly in the presence of divalent cations and therefore depend on the M/G ratio and distribution of blocks along the polysaccharide chain. Brown macroalgae have long been harvested to extract alginate that has multiple applications in food, cosmetics and pharmaceutical industries. Furthermore, biological activities of alginate oligosaccharides have been demonstrated (Liu et al. 2019). Recently, alginate has also attracted attention as a potential substrate for bioengineered microorganisms in macroalgae biorefinery processes (Poblete-Castro et al. 2020), confirming the need for novel, well-characterized alginate-degrading enzymes.
Alginate lyases depolymerize alginate by catalyzing the β-elimination of the glycosidic 1→4 O-linkage between two residues, releasing unsaturated oligosaccharides with a 4-deoxy-l-erythro-hex-4-enopyranosyluronic acid motif (Δ) at the new nonreducing end (Figure 1). Based on their mode of action, alginate lyases can be classified as endolytic enzymes that cleave long alginate chains by attacking internal positions in the polymer, and exolytic oligoalginate lyases that degrade smaller oligosaccharides starting from their ends. To date, alginate lyases are classified in 12 different polysaccharide lyase (PL) families in the Carbohydrate-Active enZYmes database CAZy (Lombard et al. 2014), namely families PL5, -6, -7, -14, -15, -17, -18, -31, -32, -34, -36 and -39. The latter five families were created only recently (Helbert et al. 2019; Ji et al. 2019), showing the unexpectedly large diversity of alginate lyases in nature and consequently the importance of alginate as carbon source. To understand how the diversity of alginate lyase sequences matches diversity of alginate structures, compositions and occurrences, it is important to match the different sequences with structural and biochemical characterizations (Garron and Henrissat 2019).
Fig. 1.
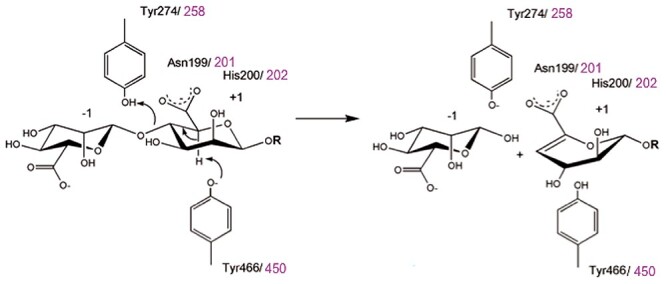
Reaction scheme for beta-elimination mechanism of alginate lyases. This catalytic mechanism is dependent on the presence of carboxylic acid groups in the substrate, which are stabilized by conserved histidine and asparagine residues in PL17 enzymes. Subsequently, a tyrosine residue abstracts a proton at C5, and finally, the formation of a double bond between C4 and C5 (beta-elimination) expels the leaving group at C4, which is protonated by a second tyrosine. Within this conserved catalytic machinery, the residue numbers are given in black for those of AlyA3, while those of Alg17c are given in magenta. This figure is available in black and white in print and in color at Glycobiology online.
The CAZy database currently contains 722 enzymes (as of May 2021) in the PL17 family, all from bacterial origin. This family is subdivided into subfamily 1 that contains endolytic hyaluronan and glucuronan lyases, and subfamily 2 (sf2) that contains exolytic oligoalginate lyases (Lombard et al. 2010). The first characterized PL17 alginate lyase was AlyII from Pseudomonas sp. OS-ALG-9 (Kraiwattanapong et al. 1999). Since then, 20 alginate lyases have been biochemically characterized in PL17 subfamily 2 (Mathieu et al. 2018), the majority originating from Gammaproteobacteria (15 enzymes) and a lower number from Bacteroidetes and Alphaproteobacteria (4 and 1 enzyme, respectively). Most of them show a substrate preference for mannuronate motifs compared with guluronate. Only one structure is available in the PL17 family, that of Alg17c from the marine gammaproteobacterium Saccharophagus degradans (Park et al. 2014). Understanding the detailed sequence–structure–function relationship of these versatile enzymes, however, necessitates numerous structural and biochemical studies (Gerlt et al. 2015). Alg17c forms a homodimer and comprises two domains: an N-terminal (α/α)6 toroid and a C-terminal β-sheet domain. The toroid domain is composed of 13 α-helices, 12 forming an open barrel structure and the additional helix maintaining its rigidity. Three layers of antiparallel β-strands form the β-sheet domain further supported by four small helices near the top layer, positioned between the two domains. Structural and mutagenesis analyses of Alg17c allowed solving its catalytic mechanism (Figure 1). The first step is the charge neutralization of the carboxylic acid group in the +1 subsite [sugar-binding subsites are numbered according to (Davies et al. 1997)] by asparagine N201 and histidine H202, followed by abstraction of the C5-proton by tyrosine Y450 acting as general base and donation of a proton to the oxygen of the glycosidic bond by tyrosine Y258 acting as a general acid. Other noncatalytic residues have been proposed to form a zinc-binding site or stabilize the binding of oligosaccharides (Park et al. 2014).
The marine bacterium Zobellia galactanivorans DsijT (phylum Bacteroidetes) has become a model organism to investigate the degradation of algal polysaccharides by members of the Flavobacteriaceae family (Barbeyron et al. 2016). It utilizes a diverse array of macroalgal substrates as sole carbon source, including agars and carrageenans from red algae and alginate, fucoidans and laminarin from brown algae (Thomas et al. 2011). Notably, Z. galactanivorans DsijT possesses a complex alginate utilization system (AUS) (Thomas et al. 2012), the expression of which is tightly controlled by the transcriptional repressor AusR (Dudek et al. 2020). Zobellia galactanivorans AUS comprises seven alginate lyases (AlyA1-AlyA7), as well as enzymes acting downstream to process unsaturated alginate monomer products into central metabolism, and membrane proteins for sugar binding/transport (Thomas et al. 2012). Most of these proteins are encoded within a single genomic region, archetypical of the polysaccharide utilization locus (PUL) organization found in Bacteroidetes (Grondin et al. 2017). The seven alginate lyases from Z. galactanivorans encompass four different CAZy families, namely PL6 (AlyA4 and AlyA6), PL7 (AlyA1, AlyA2 and AlyA5), PL14 (AlyA7) and PL17 (AlyA3). So far, activity has been validated for the four alginate lyases AlyA1, AlyA4, AlyA5 and AlyA7 (Thomas et al. 2012). The best studied enzymes are AlyA1 and AlyA5, for which detailed biochemical and structural analysis is available (Thomas et al. 2013). AlyA1 is an endolytic guluronate-specific lyase, while AlyA5 exolytically cleaves Δ, G or M residues from the nonreducing end of alginate oligosaccharides. Although both enzymes share structural similarities, additional loops obstructing the catalytic groove of AlyA5 explain its exolytic mode of action. In contrast, no biochemical or structural data are available for the predicted PL17 enzyme AlyA3. This 85.1-kDa enzyme, encoded within the alginate PUL of Z. galactanivorans DsijT, is largely conserved in other alginolytic PULs found in marine bacteria as well as in gut Bacteroidetes where it was likely acquired through horizontal gene transfer (Thomas et al. 2012; Mathieu et al. 2018). We question how the biochemical characteristics of this enzyme complete the alginolytic system in the model bacterium Z. galactanivorans, and thus, we present here the biochemical characterization and structure analysis of AlyA3. Comparison of AlyA3 native structure and in complex with an oligoalginate to that of the only other known PL17 structure of Alg17c reveals subtle differences that explain the marked preference of AlyA3 for cleaving oligomannuronate and possibly a processive mode of action.
Results
AlyA3 sequence analysis
Besides the PL17 module, AlyA3 contains a N-terminal signal peptide cleaved between residues 25 and 26 (Figure S1), suggesting that the native protein might be either located in the periplasm or secreted. Alignment with other PL17 enzymes shows a global homology with members of the sf2 subfamily (Figure S1). Notably, AlyA3 shares 42% sequence identity with Alg17c from S. degradans. Catalytic residues are well conserved, as well as residues involved in charge neutralization, substrate recognition and ion binding. One marked difference is the presence of a loop in AlyA3 inserted between helices α10 and α11 (residues 229–252). A similar loop was observed in enzymes Patl3651 and OalC17, but absent from other PL17 lyases, including Alg17c.
Overexpression and purification of AlyA3
The His-tagged recombinant AlyA3 protein was produced in soluble form in Escherichia coli BL21(DE3) and purified by nickel affinity chromatography followed by size exclusion chromatography. The yield of pure protein was ca. 100 mg L−1 of culture. The apparent molecular mass was estimated at 80 kDa on SDS-PAGE (Figure S2A), which is concordant with the theoretical mass of 83.07 kDa for the recombinant protein. Size exclusion analysis showed the protein eluted as a molecule of 163 kDa, suggesting that recombinant AlyA3 is a dimer in solution (Figure S2B).
Mode of action and end products
The action of the recombinant AlyA3 was followed by recording the absorbance at 235 nm with polymeric alginate and M-, G- and MG-blocks prepared by acid hydrolysis. M-blocks prepared this way are known to contain 5–10% of contaminating G motifs, while G-blocks typically contain 5% M motifs (Heyraud et al. 1996). MG-blocks contain mainly alternating M and G motifs, but with significant stretches of M at their extremities (Larsen et al. 1969, 1970). In all cases, biphasic kinetics were observed (Figure 2A). In the first phase, the absorbance increased, showing that AlyA3 cleaves the four substrates with a β-elimination mechanism, leading to the formation of 4,5-unsaturated 4-deoxy-l-erythro-hex-4-enepyranosyluronate (Δ) at the reducing end. This confirms the annotation of AlyA3 as an alginate lyase. The second phase consists of a decrease in the absorbance, typical for exolytic lyases which release an unsaturated monosaccharide Δ (Preiss and Ashwell 1963). This monosaccharide is known to be unstable and spontaneously converts into 4-deoxy-l-erythro-5-hexosulose uronic acid (DEH) that lacks the ability of absorbing at 235 nm due to the transfer of the unsaturation to the ketone group. The greatest activity was detected on M-blocks, while only a faint activity was detected on G-blocks. This suggests that AlyA3 is a mannuronate-specific alginate lyase and that the activity on MG-blocks is due to remaining M stretches.
Fig. 2.
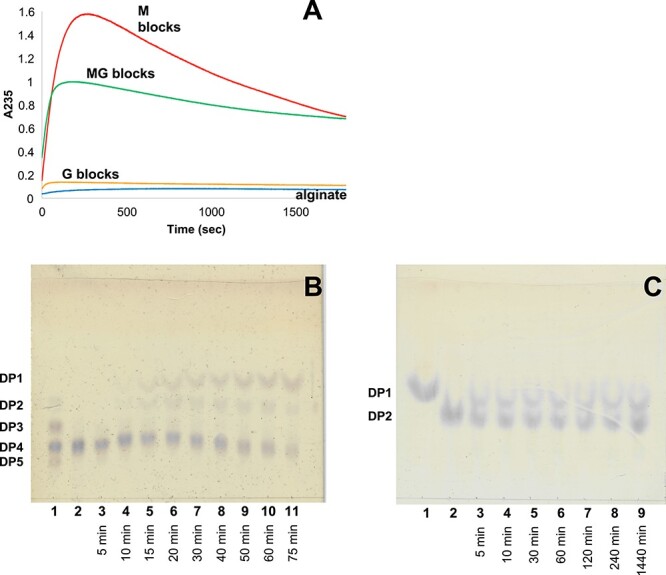
Degradation kinetics of AlyA3 on different substrates and TLC analysis of AlyA3 degradation products. (A) Degradation kinetics of M (red), MG (green), G blocks (orange) and alginate polymer (blue) by AlyA3. A235 was measured at 25°C for reaction mixtures (500 μL) composed of 100 mM Tris–HCl buffer, pH 7.0, 0.22% of substrate and 60 nM of enzyme. (B) Reaction was conducted at 30°C in 100 mM Tris–HCl buffer, pH 7.0, using sodium tetramannuronate 0.08% as substrate and 163 nM of the purified enzyme. Aliquots were sampled before adding the enzyme (lane 2) and after 5–75 min (lanes 3–11). Standard containing sodium di-, tri-, tetra- and penta-mannuronate was spotted on lane 1. (C) Reaction was conducted in the same conditions, using sodium dimannuronate as substrate. Aliquots were sampled before adding the enzyme (lane 2) and after 5–1440 min (lanes 3–9). Mannuronic acid was spotted on lane 1. This figure is available in black and white in print and in color at Glycobiology online.
TLC analysis confirmed the formation of monosaccharide products during the degradation of oligo-mannuronate from DP2 to DP6 by AlyA3 (Figure 2B and C, Figure S3). During the degradation of tetramannuronate (Figure 2B), a compound migrating above the standard DP2, attributed to DEH, was observed after only 10 min and its intensity increased during the reaction. Other bands migrating similar to DP2 were also visible and identified as minor products of the reaction. Their intensity reached a peak after ca. 30–40 min. Bands corresponding to DP3 were not observed when incubating AlyA3 with tetra- or hexa-mannuronate (Figures 2B and S3A). Incubation of AlyA3 with dimannuronate (Figure 2C) revealed the formation of the monosaccharide product within 5 min, although DP2 was still present after long-term incubation, possibly due to the presence of traces of G-containing DP2. An eluent that better resolves monosaccharide products (Figure S3B and C) allowed distinguishing two main monosaccharide products, migrating, respectively, as and above the mannuronic acid standard. Both products reacted with a keto-acid specific stain, suggesting that they correspond to DEH and a potential rearrangement product.
The presence of DEH in the reaction mixtures after action of AlyA3 was confirmed by MALDI-TOF MS (Figure S4). An ion with an m/z of 198.8 was observed before and after addition of AlyA3 on all substrates, with much greater intensity after action of the enzyme (Table I).
Table I.
Relative abundance of oligoalginates in M-blocks samples before and after degradation with AlyA3. Peak at m/z 268.17, corresponding to the buffer, was chosen as internal standard. Compounds starting with the letter “u” are oligosaccharides with an unsaturation at the nonreducing end
| m/z | M blocks w/o AlyA3 | M blocks w AlyA3 | |
|---|---|---|---|
| DEH | 198.88 | 0.428 | 4.035 |
| uDP2 | 374.92 | 0.000 | 0.648 |
| DP2 | 393.22 | 0.398 | 1.275 |
| uDP3 | 550.96 | 0.000 | 0.337 |
| uDP4 | 726.94 | 0.000 | 0.077 |
Time resolved NMR of reaction products
The activity of AlyA3 on 13C-1-enriched polyM substrate was observed using 13C time-resolved NMR analysis (Figure 3). A pseudo 2D time-resolved spectrum was recorded by acquiring 1D 13C carbon spectra every 5 min for a total of 16 h 40 min. This type of spectrum allows the observation of the change in substrate and product signals through time. Treatment of 13C-1-polyM with AlyA3 results in the release of monomers as major reaction products. This is observed through the increase of the C-1 signals of the unsaturated monomer, Δ, and subsequent increase of the C-1 signals associated with two epimers of the rearrangement product 4-deoxy-d-manno-hexulofuranosidonate (DHF) hydrate (Figure 3A), as already observed by Arntzen et al. (Arntzen et al. 2021). The conversion of Δ to DHF products proceeds as a first-order reaction that remains consistent during the entire experiment. However, the rate of formation of Δ changes and can be described in three phases. In the first phase (~0–4 h), the rate of formation of Δ is much higher than the conversion to DHF products observed as an accumulation of Δ. In the second phase (~4–6 h), Δ reaches a maximum where the rate of formation of Δ is approximately equal to the rate of conversion to DHF products. In the final phase (>8 h), Δ begins to decrease as the conversion to DHF products is greater than the formation of Δ. The rate of formation of Δ is limited by the depletion of the substrate. Additional minor signals were detected for rearrangement products of ketoaldohydrate and aldohydrate DEH hydrates (Arntzen et al. 2021). Furthermore, the recorded NMR spectra confirm that AlyA3 acts as an exo-acting lyase that processes from the nonreducing end, by the presence of an unsaturated residue at the nonreducing end of the degraded polyM substrate (Figure S5) In addition, action of AlyA3 was tested on purely alternating 13C-1-enriched-polyMG produced by in vitro epimerization, with no residual stretches of M at the extremities. It resulted in no appraisable activity detected by time-resolved NMR. This confirms the specificity of AlyA3 toward mannuronate residues.
Fig. 3.
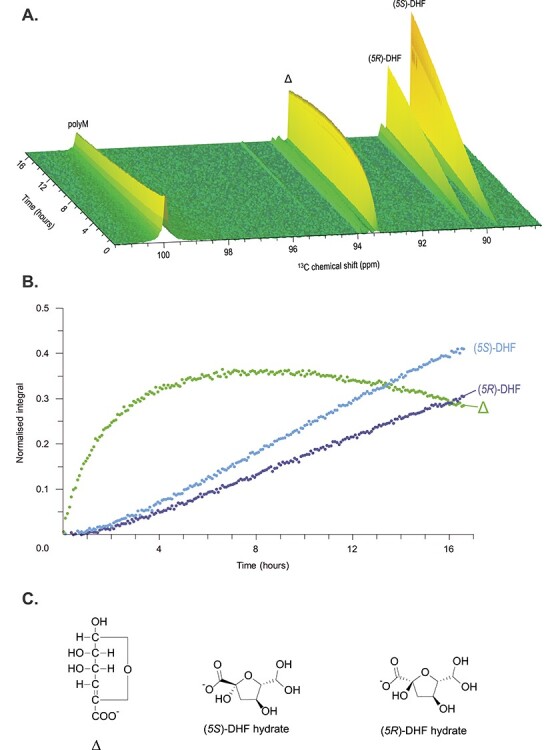
13C time-resolved spectrum of 13C-1-enriched polyM treated with AlyA3. (A) Pseudo 2D time-resolved spectra recorded by acquiring 1D carbon spectra every 5 min for a total of 16 h 40 min. Signals associated with C-1 of each component are labeled. (B) Normalized integrals of monomeric products produced during treatment of polyM with AlyA3. (C) Structures of the three main products. Δ: 4,5-unsaturated 4-deoxy-l-erythro-hex-4-enepyranosyluronate; DHF: 4-deoxy-d-manno-hexulofuranosidonate. Reaction was conducted in 3-mm NMR tube with 160 μL of substrate buffer solution (10 mg mL−1 13C-1-polyM substrate in 10-mM HEPES, pH 7.0 with 2-mM NaCl in 99.9% D2O) and 10 μL of 0.9-μM AlyA3 incubated at 25°C. This figure is available in black and white in print and in color at Glycobiology online.
Optimal activity conditions
The effects of several physical–chemical parameters on AlyA3 activity were evaluated. Activity increased linearly with temperature between 10 and 40°C and decreased rapidly until being almost abolished at 50°C (Figure 4A). Sodium chloride addition also had a strong effect on AlyA3 activity: the enzyme lost 54% of its activity after the addition of 100-mM NaCl, and only 2% of activity remained at 500-mM NaCl (Figure 4B). AlyA3 activity was higher in the pH range 6.5–8.0. Not only the pH but also the buffer had a strong impact on enzymatic activity; it was highest with HEPES (pH 8.0 and 7.5) and with Tris–HCl at pH 7.0, which was chosen as a reference due to the lower standard deviation observed with this buffer compared with HEPES. Below pH 6.5, only 20% of activity remained, and it was completely abolished at pH 9.0 and above (Figure 4C).
Fig. 4.
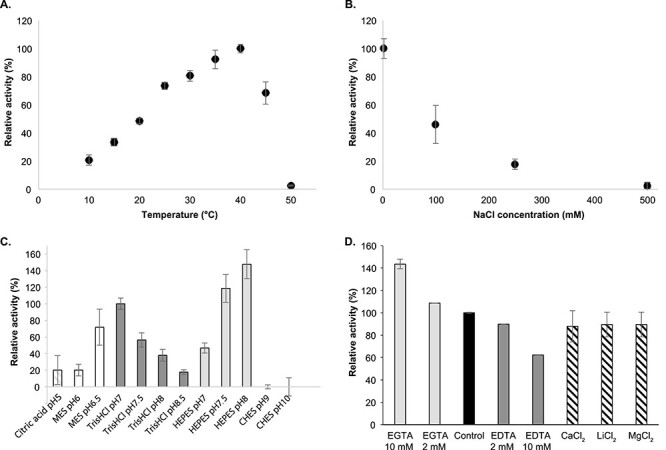
Effect of temperature (A), sodium chloride concentration (B), pH (C) and addition of chelators or cations on the activity of AlyA3. (A–D) Experiments were conducted with sodium alginate 0.22% as substrate and 6 nM of the purified enzyme. Activity was monitored by following the increase in absorbance at 235 nm during the first 2 min. Error bars indicate standard deviations of three experimental replicates. (A) Effect of temperature. Experiments were conducted in 0.1 M Tris–HCl buffer, pH 7.5. Activity at 40°C was taken as 100%. (B) Effect of NaCl concentration. Reactions were conducted in 0.1 M Tris–HCl buffer, pH 7.0 at 25°C. Activity with 2 mM NaCl was taken as 100%. (C) Effect of pH on AlyA3 activity. Experiments were performed at 25°C in 100-m buffer. Sodium citrate (stripes), MES (white), Tris–HCl (dark gray), HEPES (light gray) and CHES (black) buffers were used in the assay. Activity in Tris–HCl, pH 7.0, was taken as 100%. (D) Effect of chelators and cation chloride addition on AlyA3 activity. Reactions were conducted in 0.1 M Tris–HCl buffer, pH 7.0 at 25°C. Either CaCl2, LiCl2, MgCl2 (2 mM), EDTA (2 or 10 mM) or EGTA (2 or 10 mM) were added. Activity without any cation chloride or chelator was taken as 100%. This figure is available in black and white in print and in color at Glycobiology online.
The effects of cations on the enzymatic activity were investigated by adding various cation chloride solutions at 2 mM or by adding chelating agents, namely EDTA and EGTA at 2 or 10 mM (Figure 4D). The addition of calcium, lithium or magnesium did not affect AlyA3 activity. However, the addition of EDTA or EGTA affected its activity in a concentration-dependent manner. The presence of EDTA decreased enzymatic activity, while the presence of EGTA increased it.
Altogether, AlyA3 has optimal activity at 40°C, pH 8.0 in HEPES buffer, without addition of any other ion.
Mutagenesis analysis
Three amino-acids were selected for individual point mutations based on their conservation in other alginate lyases (Figure S1) and predicted roles as catalytic residues (Y274 and Y466) or charge neutralizer (H200). The mutations Y274A and Y466A virtually abolished the alginate lyase activity (Figure S6, 0.2 and 0.4% remaining activity compared with wild-type (WT) AlyA3, respectively). The mutation H200A strongly impaired the activity (13.9% activity remaining).
Crystal structure of AlyA3 and its complex with oligo-mannuronate substrate
The crystal structure of AlyA3 was determined at 1.42-Å resolution (Table II), using phases determined by molecular replacement and coordinates of the only other available PL17 alginate lyase (Alg17c, PDB code 4OJZ) as the starting model (Figure 5). The diffraction limit of the crystals of AlyA3_Y274A in complex with a substrate molecule was 2.16 Å. In both crystal forms, native AlyA3 and AlyA3_Y274A in complex with oligo-mannuronate, two identical polypeptide chains were present in the asymmetric unit (Figure 5A), and, for both molecules, the atomic coordinates for all residues spanning the range from 25 to 751 could be identified in the electron density. The only exception to this is the tip (residues 240–244) of the loop insertion in one of the monomers of the native structure, which does not display clear electron density for five residues, indicative of disorder due to flexibility. The dimeric interface is characteristic of a biological dimer (1088-Å2 surface area of each monomer buried at the interface) and the global arrangement was identical to that found in Alg17c (Figure 5B). The final structures are thus composed of 727 residues, two Mg2+ ions and one Ca2+, and roughly 450 water molecules, each. In the case of the native AlyA3, a malic acid and a glycerol molecule were trapped at the active site of monomer A (Figure 6B), and a malic acid molecule in monomer B, molecules identified as the most plausible interpretation of additional electron density (Figure S7) and present in the crystallization conditions. In the case of AlyA3_Y274A, the enzyme was crystallized with oligo-mannuronate DP3, and one unsaturated disaccharide identified as Δ-M, a cleavage product, was present in each of the two active sites.
Table II.
Data collection and refinement statistics for the crystal structures of the native AlyA3 and AlyA3_Y274A in complex with ΔM
| AlyA3 nat | AlyA3_Y274A + ΔM | |
|---|---|---|
| Data collection | ||
| Beamline | ESRF ID29 (03122016) | SOLEIL PX2 25012020 |
| Wavelength | 0.97 Å | 0.98 Å |
| Space group | P65 | P65 |
| Unit cell | a = b = 163.29 Å; c = 166.58 Å α = β = 90°; γ = 120° |
a = b = 163.78 Å; c = 167.92 Å α = β = 90° γ = 120° |
| Resolution range a (Å) | 43.00–1.42 (1.47–1.42) | 47.38–2.16 (2.21–2.16) |
| Total data | 2,514,603 | 2,903,416 |
| Unique data | 444,259 | 137,171 |
| Completeness (%) | 99.9 (99.9) | 99.6 (99.2) |
| CC(1/2) | 99.8 (65.6) | 99.9 (64.1) |
| Mean I/σ(I) | 4.74 (1.7) | 16.3 (1.9) |
| b R sym; cRpim(%) | 10.5 (84.0); 4.2 (18.3) | 29.5 (389); 5.4 (35.6) |
| Redundancy | 5.66 | 21.16 |
| Refinement statistics | ||
| Resolution range | 141.50–1.42 (1.46–1.42) | 47.38–2.16 (2.21–2.16) |
| Unique reflexions | 444,259 (32662) | 137,171 (7343) |
| Reflexions Rfree | 23,668 (1730) | 5319 (386) |
| R/Rfree (%) | 15.4/17.0 (21.4/23.1) | 15.4/17.8 (21.1/24.6) |
| RMSD bond lengths | 0.032 Å | 0.039 Å |
| RMSD bond angles | 2.80° | 3.02° |
| Overall B factor (Å2) | 21.88 | 29.16 |
| B factor: Molecule A (Å2) | 10.39 | 21.74 |
| B factor: Molecule B (Å2) | 10.32 | 21.31 |
| B factor: Solvant (Å2) | 25.00 | 22.67 |
| B factor: Ligands (Å2) | - | 31.56 |
aValues in parentheses concern the high-resolution shell.
b R sym = Σ | I−Iav | / Σ| I |, where the summation is over all symmetry-equivalent reflections.
c R pim = corresponds to the multiplicity weighted Rsym.
Fig. 5.
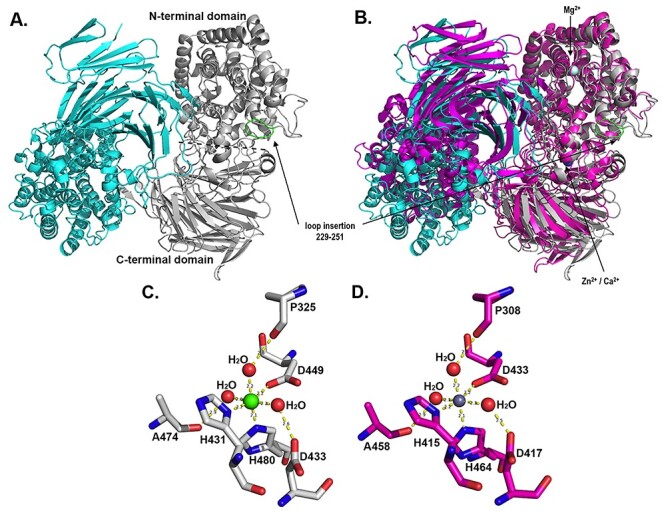
Crystal structure representation of the AlyA3 biological dimer. (A) Cartoon representation of the 2-fold dimeric quaternary arrangement of AlyA3. Monomer A is colored in cyan and monomer B in light gray. In the gray monomer, the N-terminal (α/α)6 toroid and a C-terminal β-sheet domain are separated by a dotted line. The loop insertion found in AlyA3 but absent in Alg17c (see Figure 1) is colored in green and indicated by an arrow. (B) Superimposition of the dimer of AlyA3 (cyan and gray) with the dimer of Alg17c (colored in magenta, pdb code 4OJZ). The positions of ions in AlyA3 are indicated by an arrow. (C) Stick representation of the conserved residues together with the water molecules forming the octahedral coordination sphere of the Ca2+ ion in AlyA3. (D) Stick representation of the conserved residues together with the water molecules forming the octahedral coordination sphere of the Zn2+ ion in Alg17c. The figure was produced using PyMOL (The PyMOL Molecular Graphic Systems 2002). This figure is available in black and white in print and in color at Glycobiology online.
Fig. 6.
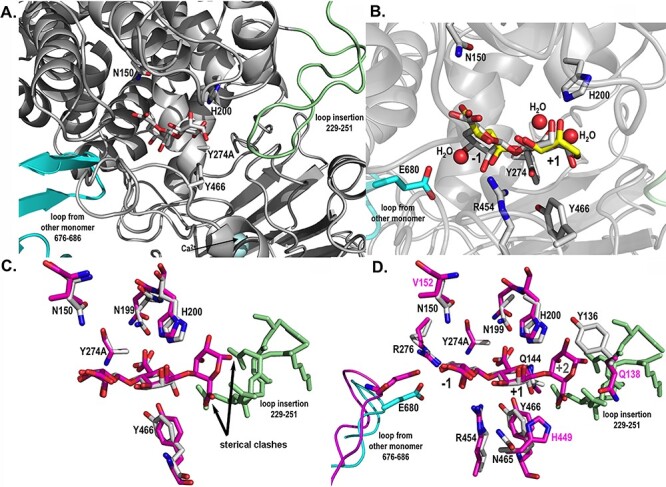
Catalytic active site structures of AlyA3 with bound molecules. (A) Close up view into the active site of AlyA3_Y274A, showing the relative position of a bound Δ-M substrate molecule with respect to the two loops blocking the entrance to the groove. The β-hairpin loop coming from the neighboring monomer B is colored in cyan, and the loop insertion, characteristic of AlyA3, is colored in light green. (B) Close up view into the active site of native AlyA3, showing the position of solvent molecules, malic acid and glycerol (carbons in yellow) that together with several water molecules (red spheres) occupy positions of the hydroxyl and acidic groups of the bound DP2 (Δ-M) substrate molecule (carbons in white) positioned here by superimposition with the AlyA3_Y274A complex with DP2 (panel A). Conserved and catalytic active residues are labeled. (C) Close up view into the active site of AlyA3_Y274A with Δ-M (carbons in white) superimposed with the DP3 molecule Δ-M-G bound in Alg17c (carbons in magenta). The figure highlights the sterical clashes, pointed out by the black arrows, of any additional guluronate unit at the potential +2 binding site with the inserted loop of AlyA3 (colored in light green). The atoms of the +2 guluronate unit would come to lie at 0.1 and 1.3 Å from atoms of the amino acids within the loop insertion. (D) Close up view into the active site of AlyA3_Y274A with Δ-M (carbons in white) superimposed with the DP3 molecule bound in Alg17c (carbons in magenta). The figure additionally displays the residues surrounding the substrate molecule and that differ between AlyA3 and Alg17c. Residue numbers in black label the residues of AlyA3, and labels in magenta highlight the residues that differ between the two enzymes, in close proximity of the catalytic site. This figure is available in black and white in print and in color at Glycobiology online.
The overall fold of AlyA3 belongs to the multidomain (α/α)6 toroid structural class. It is globally similar to that of the homologous Alg17c and organized in two domains, an N-terminal (α/α)6 barrel fused to a C-terminal β-sheet domain (Garron and Cygler 2010). The root mean square deviation (rmsd) calculated between monomers of the two alginate lyases is 1.65 Å, and the superimposition of dimers of AlyA3 to Alg17c leads to an rmsd of 2.76 Å (Figure 5B). Comparable to Alg17c, AlyA3 contains a roughly 16 Å long antiparallel β-turn, part of the C-terminal β-sheet, which extends from the globular monomer and points into the catalytic active site of the second monomer of the dimer (Figure 5A). This loop appears to block the “entrance” of the substrate-binding site, next to the −1 binding site (Figures 5A and6A). In AlyA3, a Ca2+ ion is coordinated by highly conserved residues (His431, His480 and Asp449) with perfect octahedral geometry, at the interface of the N- and C-terminal domains (Figure 5B and C). This contrasts with Alg17c, where the divalent cation was modeled as zinc (Figure 5D). Since our data diffracted to higher resolution and the difference in electrons between Ca2+ and Zn2+ is significant (18 vs. 28, respectively), we have verified the chemical nature of this ion in AlyA3, by refining either with a Ca or Zn ion at this position. While electron density is well explained by Ca2+, refinement in the presence of Zn2+ reveals a negative peak in the Fo-Fc map (see Figure S8A and B). While the octahedral coordination sphere is more in favor of the presence of Ca, the nature of the chelating residues (especially the histidine residues) are much more frequently found to coordinate zinc ions (Harding 2004). Although we cannot exclude that a mixture of both ions is present, our structural refinement is in favor of the ion being a Ca2+ in AlyA3. Besides, AlyA3 seems to contain additional ions that were refined as Mg2+ (Figure 5B). Again, it is difficult to distinguish Mg2+ from H2O in the crystal structure since they have the same number of electrons; but since the coordination sphere is octahedral and the enzyme activity appears to be Mg2+dependant, we refined these additional ions as Mg2+.
Despite the overall structural similarity between the two PL17 alginate lyases, some crucial differences can be highlighted in AlyA3, with respect to Alg17c. Indeed, AlyA3 contains a substantial loop-insertion, from residues 229 to 251, the tip of which comes to lie close to the +2 substrate binding site of the catalytic groove in the enzyme (Figures S1,5 and 6). This loop blocks a potential +2 substrate-binding subsite that is occupied by a mannuronate unit in the substrate complex of Alg17c and absent in AlyA3. It is interesting to note that, while the extended antiparallel β-turn that connects the two monomers, both in Alg17c and AlyA3, displays low-temperature factors, the loop insertion of AlyA3 has on average higher temperature factors than the core protein, indicating that this loop might be rather flexible (Figure S8C). Moreover, in the native AlyA3 structure, the tip of this loop (residues 240–243) in one of the monomers (monomer B) did not reveal clear, unique electron density, which is a typical sign of disorder and indirectly linked to flexibility.
Another interesting difference concerns some of the amino acids surrounding binding subsites of the active site. Indeed, Gln138, Val152 and His449 of Alg17c are replaced by Tyr136, Asn150 and Asn465 in AlyA3, respectively (Figure 6). Besides these three residues, the amino acids interacting with the bound substrate and/or important for the catalytic cleavage are perfectly conserved in sequence and structural position.
Kinetic parameters and substrate specificity
AlyA3 kinetic parameters were estimated on M-rich alginate, M blocks and oligomannuronates with varying degrees of polymer lengths (DPs) (Table III). Reactions followed Michaelis–Menten kinetics with all the substrates tested. AlyA3 affinity, turnover rate and catalytic efficiency were low for polymeric alginate with M/G = 2.0 (Km = 4.4 mM, kcat = 4.7 s−1 and kcat/Km = 1.1 mM−1 s−1, respectively). These parameters were more favorable on structures with lower DP, showing that AlyA3 is an oligo-alginate lyase. We further compared kinetic parameters on purified oligomannuronates with increasing length, from DP2 to DP6. AlyA3 had the strongest affinity for penta- and hexa-mannuronate (Km = 414 and 428 μM, respectively). Similarly, these substrates yielded the highest catalytic efficiencies (kcat/Km = 93.1 and 100.4 mM−1 s−1, respectively). In contrast, the catalytic efficiency was 4-fold lower on di-mannuronate and intermediate on tri- and tetra-mannuronate.
Table III.
Kinetic parameters of AlyA3 on different substrates
| Substrate | K m (μM) | Vm (μM min−1) | k cat (s−1) | k cat/Km (mM−1 s−1) |
|---|---|---|---|---|
| Alginate M/G = 2 | 4355 ± 4158 | 1.7 ± 1.0 | 4.7 ± 2.9 | 1.1 ± 0.9 |
| M-blocks | 1250 ± 520 | 14.2 ± 2.6 | 39.5 ± 7.2 | 31.6 ± 8.2 |
| M-DP2 | 603 ± 208 | 4.8 ± 0.6 | 13.4 ± 1.6 | 22.2 ± 3.7 |
| M-DP3 | 781 ± 245 | 16.0 ± 1.9 | 44.5 ± 5.3 | 57.0 ± 9.5 |
| M-DP4 | 786 ± 221 | 13.8 ± 1.5 | 38.3 ± 4.1 | 48.8 ± 7.3 |
| M-DP5 | 414 ± 172 | 13.9 ± 1.8 | 38.6 ± 4.9 | 93.1 ± 16.8 |
| M-DP6 | 428 ± 199 | 15.5 ± 2.4 | 43.1 ± 6.6 | 100.4 ± 21.9 |
Experiments were performed at 35°C, in a Tris–HCl buffer (100 mM, pH 7.0). Substrate concentration range was 0.025–0.5 g L−1. Values are derived from independent triplicate experiments.
Discussion
Despite the apparent simplicity of its chemical structure (composed of G and M units only), and in analogy to the abundant cellulosic component of plant cell walls, alginate necessitates the synergy of several enzymes to be broken down into monosaccharide components (Wong et al. 2000). In general, this is also reflected by the presence of four to seven alginate lyases in microorganisms that utilize this substrate as a carbon source (Thomas et al. 2012). The heterotrophic marine model Bacteroidetes, Z. galactanivorans, possesses seven alginate lyases. Two of them, AlyA1 and AlyA5 belonging to family PL7, have been biochemically characterized in the past (Thomas et al. 2013). AlyA1 was identified as an endo-acting enzyme highly specific of G-blocks, requiring a minimal stretch of four consecutive G-units to be active. Complementary to this, AlyA5 was shown to be exo-active, displaying activity on degradation products of AlyA1, indicating activity toward oligosaccharides containing an unsaturated sugar at the nonreducing end, but no difference in activity, whether the pattern was Δ-M or Δ-G. Altogether, these two enzymes are able to fully convert commercial alginate into monosaccharides (Thomas et al. 2013). In contrast, AlyA3 that we describe here has no activity on guluronate homopolymers or alternating M/G regions. This alginate lyase belonging to family PL17 is exo-lytic and highly specific of M stretches, with highest activity on oligosaccharides ranging from DP3 to DP6. Oligo-mannuronate of DP2 is only partially degraded within the time scale of our experiments (1440 min). Thus, within the alginolytic system of Z. galactanivorans, the activity of AlyA3 is complementary to both AlyA1 and AlyA5, which may also represent an alternative degradation mode as a function of the chemical composition of the alginate substrate. Furthermore, its preference for short oligosaccharides and low-salt tolerance matches its predicted localization in the periplasm, while AlyA1 is secreted to the external medium and AlyA5 attached to the outer membrane (Thomas et al. 2012).
The lower activity of AlyA3 on DP2 compared with longer oligosaccharides corroborates previous observations on Alg17c (Kim et al. 2012) and suggests that at least, three sugar residues are recognized for optimal activity. In this context, the absence of detection of DP3 when incubating AlyA3 with longer oligosaccharides (Figure 2B) supports the hypothesis of a processive mode of action. Possibly, the enzymes attach to medium-sized oligosaccharides present in the periplasm that are processed through the active site with the help of the two flexible loops until DP2 is reached. This could also explain the trapping of a DP2 in the crystal structure of AlyA3_Y274A (see below).
AlyA3 kinetic parameters on M-blocks are in the same range of magnitude of other exolytic PL17 enzymes (Table SII). Interestingly, although, to date, all characterized PL17 alginate lyases are exolytic and most display higher activity on polyM containing oligosaccharides (Mathieu et al. 2018), the complete inactivity of AlyA3 on G-containing oligo-alginates seems to date to be unique. For example and in contrast, Alg17c from S. degradans was active on both polyM and polyG substrates (Kim et al. 2012). However, it is important to point out that in general, alginate-oligosaccharides or alginate block-structures are difficult to obtain as 100% pure, with perfectly defined composition. Consequently, activity observed on polyG or MG blocks of the various PL17 alginate lyases might result from M stretches present as impurities in those substrates. Conversely, the M-blocks we used for the determination of kinetic parameters were contaminated with 13% G motifs (estimated by NMR, data not shown). This might explain why kcat/Km was lower on M-blocks compared with the activity on DP5/6 oligo-mannuronates.
The crystal structure of AlyA3 is globally similar to that described for Alg17c (PDB code 4NEI), having the same fold and quaternary arrangement, and structural comparison of this family to PL15 enzymes has been described previously (Park et al. 2014). Interestingly, domain movements, displacing the relative positions of the C- and N-terminal domains, have been observed for native Alg17c with respect to the enzyme in complex with a substrate molecule. While these global movements are not evidenced by the crystals structures of AlyA3 compared with AlyA3_Y274A in complex with Δ-M, several features do also point toward possible mobility and domain movements for AlyA3. First, the superimposition of monomers of AlyA3 to Alg17c gives lower rmsd values than superposing the dimer, although the global dimeric arrangement of the two enzymes is highly similar and involves identical regions. This indicates that the N- and C-terminal domain positions within the dimer of AlyA3 in our crystal structure are not exactly in the same conformation as the one of Alg17c. In addition, normal mode analysis of the AlyA3 dimeric structure also supports domain movements, highlighting a closing and opening of the N-terminal domains (that contain the active site) with respect to each other (Figure S8D), while the C-terminal antiparallel β-sheet domain appears to play the role of a base that stabilizes the biological dimer. Second, the tip of loop 229–252 is disordered in one of the monomers (monomer B) of native AlyA3, while it is perfectly defined by electron density in the complexed structure. This is further supported when analyzing the B-factor distribution (Figure S8C), since the tip of this loop has on average higher B-factors than the rest of the enzyme molecule. Moreover, since AlyA3 is perfectly active on longer oligosaccharides, DP3–6 and poly-M blocks, the loops on either side of the catalytic active site necessarily change conformation to open access to the catalytic machinery.
The active site of AlyA3 reveals two preferential binding sites, since only two sugar-units are bound, even though the inactivated enzyme was crystallized in the presence of DP3 (MMM). Furthermore, any additional binding site would necessitate that the loops boarding the active site move away. Several explanations can be proposed: either a very low-residual activity of AlyA3_Y274A has led to cleavage producing the Δ-M found in the active site; or the commercial DP3 substrate contained traces of Δ-M and crystallization has selected for the most tightly bound molecule. These hypotheses both correlate with the fact that AlyA3 degrades DP2 slower than longer oligosaccharides. Taken together, this might implicate that constrains apply to longer oligosaccharides upon binding, possibly through binding with the flexible loops, which are relaxed when DP2 is reached. Indeed, as for example in cellulases, substrate distortions have been described to be important for the enzymatic activity of glycosidases (Sulzenbacher et al. 1997; Davies et al. 1998), since they are part of the driving force lowering the energy barrier for the reaction to take place.
The change of activities revealed by the experiments when adding chelating agents, such as EDTA or EGTA, points toward some influence of divalent ions on enzymatic reactivity. EDTA had the more drastic effect, and being more specific toward magnesium than EGTA, it suggests that the preferred divalent cations required for AlyA3 activity would rather be magnesium. On the other hand, adding Ca2+, Li2+ or Mg2+ ions to the reaction solutions did not enhance activity, showing that the presence (or absence) of these ions only have subtle effects on activity. Indeed, alginate lyase activity has regularly been shown to depend on divalent cations; it is believed that these ions compensate the negative charges of the carboxylate groups of the substrate and help extracting the single alginate chains from gels that also contain large amounts of cations (Wong et al. 2000). Our crystal structure of AlyA3 revealed two potential Mg2+ sites, which are rather distant (roughly 11 and 16 Å, respectively) to the active site. Nevertheless, they are bound to residues that are part of the loops forming the substrate binding subsites. Potentially, the presence (or absence) of these ions could influence the flexibility of the structural loops and thus the kinetics of the catalytic activity.
In addition, in both crystal structures of PL17 members, a conserved metal ion site, located at 11 Å from Tyr466 in AlyA3, was identified and seems to be present in all PL17 enzymes, since the amino acids binding this ion are globally conserved. The nature of this ion, however, is still open to debate; in our crystal structure, we presume that the nature of the ion is calcium, while it was modelled as zinc in Alg17c. The octahedral coordination observed in PL17 enzymes is more frequent for calcium binding sites, while zinc is more generally found in tetrahedral coordination. On the other hand, histidines are rather rare ligands for binding of calcium ions, whereas they are frequently found binding zinc ions. Nevertheless, the position of this conserved binding site at the interface of the N- and C-terminal domains of AlyA3 or Alg17c points more toward a structural role of this ion. This is also the interpretation provided by Park et al. (2014), who have replaced one of the coordinating His residues by mutagenesis and have shown a negative effect on kcat/Km in Alg17c, which was nearly 1000-fold lower than that of the WT. Having said this, one needs again to consider that domain movements could be influenced or moderated by the presence of this structuring ion, and thus indirectly influence the catalytic activity.
Analyses of AlyA3 activity on various substrates seem to indicate that the enzyme dissociates from the substrate as soon as a G-unit enters the catalytic active site. Nevertheless, it is difficult to identify crucial features that define this specificity, when superimposing with the structure of the PL15 alginate lyase Atu3025 (3AFL, data not shown) that has a similar fold and is G-specific. Interestingly, the inserted loop in AlyA3 and the loop coming from the symmetric monomer in PL17 alginate lyases appear to be structural features that distinguish PL17 from PL15. These loops delimit binding subsites and might be involved in specific recognition of M-stretches. As illustrated in Figure 6C, when superimposing with the Δ-M-G molecule crystallized in the active site of Alg17c, the G-unit would be in steric clash to the unique loop of AlyA3, whereas an additional M-unit could be compatible with binding in the active site of AlyA3. This feature could therefore be a plausible explanation for the particular tight substrate specificity toward mannuronate containing oligosaccharides that we observe for AlyA3. In addition, and together with domain mobility within the dimer and flexibility of the inserted loop that displaces the relative positions of these loops with respect to the catalytic machinery, these features might as well be involved in the processive mechanism, positioning or pushing the M-stretches in the correct sites for catalysis. This opens the question of whether the presence of the C-terminal domain and the dimerization of this enzyme family are important for substrate turnover and/or processivity.
Material and methods
Substrate materials
Unless otherwise stated, all chemicals were from Sigma (Darmstadt, Germany). Sodium alginate with M/G ratio 2.0 was kindly provided by Danisco. Homopolymeric M and G blocks and heteropolymeric MG blocks were prepared from sodium alginate by partial acid hydrolysis as described previously (Haug et al. 1974). The composition of blocks prepared this way was reported elsewhere. M and G blocks typically contain 5–10% of contaminating residues from the other type (Heyraud et al. 1996). MG blocks mainly consist of alternating mannuronate and guluronate residues, with remaining stretches of M at their extremities (Larsen et al. 1969, 1970). Di-, tri-, tetra- and hexa-mannuronic acid sodium salts were purchased from Carbosynth (Compton, UK). 13C-1-enriched poly-mannuronic acid (polyM) substrate (fraction of G motif FG = 0.0, Mw ~ 6–10 kDa) was produced and purified from an epimerase negative AlgG mutant of Pseudomonas fluorescens as previously described (Gimmestad et al. 2003) using 13C-1-fructose (Cambridge Isotope Laboratories, Tewksbury, USA) as the sole carbon source. Alternating poly-mannuronic acid/guluronic acid polyMG (FG = 0.46, fraction of G diads FGG = 0.0, Mw ~ 6–8 kDa) was made using in vitro epimerization of poly-M with AlgE4 as previously reported (Donati et al. 2005).
Sequence analysis
Characterized members of the PL17 family were retrieved from the CAZy database (Lombard et al. 2014) and a literature survey. Their protein sequences were aligned using MAFFT (Katoh et al. 2019) with the L-INS-I algorithm and the scoring matrix BLOSUM62, using a structural alignment of Alg17C (PDB accession 4NEI) and AlyA3 (this study) as a constraint. The alignment was visualized using ESPript 3.0 (Robert and Gouet 2014).
Cloning procedures
The alyA3 gene (locus identifier: zgal_2624) from Z. galactanivorans DsijT encoding a putative alginate lyase PL17_2 protein was cloned as per Groisillier and coworkers (Groisillier et al. 2010). Briefly, a pair of primers was designed (see ORL692 and ORL693 in Table SI) to PCR-amplify the coding region corresponding to the full-length protein deleted for its signal peptide from Z. galactanivorans genomic DNA. After digestion with the restriction enzymes BamHI and PstI, the purified PCR product was ligated using the T4 DNA ligase into the expression vector pFO4 (Groisillier et al. 2010) linearized with BamHI and NsiI, resulting in a recombinant chimeric protein with an N-terminal hexa-histidine tag. This plasmid, named pZG227, was subsequently transformed into E. coli DH5α strain for storage and in E. coli BL21(DE3) strain for protein expression after being confirmed by sequencing.
Site-directed mutagenesis of alyA3
Three different residues, namely H200, Y274 and Y466, were targeted for independent site-directed mutagenesis and replaced by alanines to further characterize the enzymatic activity of AlyA3. These mutageneses were performed using the QuickChange II XL site-directed mutagenesis kit (Stratagene, Bellingham, USA) by manipulating the WT overexpression plasmid pZG227 for the template with the primer pairs, ORL1112-ORL1124, ORL1113-ORL1125 and ORL1114-ORL1126 (Table SI), to yield the mutated plasmid versions pZG353, pZG354 and pZG355 bearing the above point mutations, respectively. Every mutation was confirmed by sequencing.
Protein production and purification procedures
For biochemical assays and crystallography, recombinant E. coli BL21(DE3) cells were precultured overnight at 37°C in LB medium containing 100 μg mL−1 ampicillin. One liter of autoinducible ZYP5052 medium (Studier 2005) with ampicillin was inoculated with 1 mL of preculture and incubated for 72 h at 20°C, 200 rpm. Cells were harvested by centrifugation (20 min, 5000 rpm, 4°C) and pellets were stored at −20°C. Cells were resuspended in buffer A (25 mM Tris–HCl pH 7.5, 200 mM NaCl, 20 mM imidazole) containing DNase (5 U μL−1) and a protease inhibitor cocktail (Complete EDTA-free, Roche Applied Science, Penzberg, Germany). Cells were lysed either with a French Press or by sonication. The cell lysate was clarified by centrifugation (20,000 g, 30 min, 4°C). The supernatant was loaded at 1 mL min−1 on a HisTrap FF 5-mL column (GE Healthcare) preloaded with nickel and equilibrated in buffer A. Proteins were eluted with a linear gradient between buffer A and buffer B (25-mM Tris–HCl pH 7.5, 200-mM NaCl, 1-M imidazole) in 50 mL at 5 mL min−1 and collected in fractions of 1.5 mL. Recombinant proteins were further purified on a size exclusion chromatography column (Superdex 200 HiLoad 16/60 prep grade, GE Healthcare, Chicago, USA) in buffer C (25-mM Tris–HCl pH 7.5, 200-mM NaCl) at 1 mL min−1. Fractions showing the presence of the pure proteins on 12% SDS-PAGE were pooled and concentrated by ultrafiltration on membranes with a 10-kDa molecular weight cut-off (Amicon Ultra-15 10 K, Merck Millipore, Darmstadt, Germany).
To test the effect of point mutations, recombinant E. coli BL21(DE3) cells expressing WT and mutant AlyA3 were grown for 3 days at 20°C in 5-mL ZYP5052 medium containing 100-μg mL−1 ampicillin. After centrifugation, pellets were resuspended in 500-μL ice-cold lysis buffer (20-mM Tris–HCl pH 8.0, 500-mM NaCl, 1-mM EDTA, 0.1% Triton X-100, 0.1-mg mL−1 DNase) and cells were lysed by sonication. After centrifugation (20,000 g, 30 min, 4°C), recombinant proteins were purified from the supernatant using His SpinTrap columns (GE Healthcare, Chicago, USA) and eluted with 20-mM sodium phosphate buffer pH 7.4, 500-mM NaCl, 500-mM imidazole.
Enzymatic assays
Protein concentration was determined using the Bradford assay and by measuring the absorbance at 280 nm, assuming a theoretical molar extinction coefficient of 101,205 M−1 cm−1 as estimated by Expasy (https://web.expasy.org/protparam/). Alginate lyase activity was assayed by following the increase in absorbance at 235 nm due to the release of unsaturated uronates, in a 1-cm quartz cuvette containing 0.22% sodium alginate M/G = 2 as substrate and 6-nM purified enzyme in 500-μL reaction volume, using a thermo-regulated spectrophotometer (UV-2401PC, Shimadzu, Kyoto, Japan). Activity was measured as the initial velocity during the first 2 min. All tests were performed in triplicates. The effect of temperature was tested in 100-mM Tris–HCl pH 7.5. The effect of pH was tested at 25°C using various buffers at 100 mM: citrate (pH 5.0), MES (pH 6.0–6.5), Tris–HCl (pH 7.0–8.5), HEPES (pH 7.0–8.0) and CHES (pH 9.0–10.0). The effects of NaCl (2–500 mM), cations (CaCl2, LiCl2 or MgCl2, 2 mM) and chelating agents (EDTA or EGTA, 2 and 10 mM) were tested in 100-mM Tris pH 7.0 at 25°C. Kinetic parameters were determined at 35°C in 100-mM Tris–HCl pH 7.0, 2-mM NaCl using sodium alginate with M/G = 2.0, defined M-blocks and purified oligomannuronates (DP2-DP6) as substrates, using concentrations ranging from 0.025 to 0.5 g L−1. Kinetic parameters were estimated by fitting a hyperbolic regression with function nls in R v3.5.0 using substrate concentrations derived from the molecular mass of a monosaccharide (194 g mol−1) and a molar extinction coefficient for reaction products ε = 8500 L mol−1 cm−1 (Preiss and Ashwell 1962).
Thin layer chromatography
Two microliters of reaction mixture were spotted on silica 60 gel plates. The eluent was either formic acid/n-butanol/water (6:4:1, v/v/v) or ethanol/butanol/water (5:5:3, v/v/v). Staining was performed by immersing plates either in the general carbohydrate stain ethanol/sulfuric acid (9:1, v/v) containing 0.1% orcinol, or the keto-acid specific stain 2% phenylediamine in 80% ethanol, followed by incubation at 100°C for 5 min.
Mass spectrometry analysis
Reaction mixtures were mixed (1:1, v/v) with a matrix composed of 2,5-dihydroxybenzoic acid 10 mg mL−1 in tetrahydrofuran, then spotted (1 μL) on a stainless steel target and air-dried. Mass spectra were recorded using an Autoflex III SmartBeam (Bruker Daltonics, Billerica, MA) equipped with a 337-nm laser, in positive mode and analyzed using Spectrus Processor (Advanced Chemistry Development, Inc. Toronto, Canada).
Time-resolved analysis with NMR spectroscopy
For the time-resolved NMR analysis of lyase reactions, a stock solution of 10 mg mL−1 of substrate in 10-mM HEPES, pH 7.0 with 2-mM NaCl in 99.9% D2O was prepared (Gimmestad et al. 2003; Donati et al. 2005). For each reaction, 160 μL of substrate buffer solution was added to 3-mm NMR tube and preheated to a temperature of 25°C in the NMR instrument. 1D carbon spectrum was recorded to ensure that the sample had not undergone any degradation or contamination prior to the time-resolved NMR experiment. The reaction was started by adding 10 μL of 0.9-μM AlyA3 to preheated substrate and mixed by inverting the sample several times. The sample was hereafter inserted into the preheated NMR instrument and the experiment was started. The recorded spectrum is a pseudo-2D type experiment recording a 1D carbon NMR spectrum every 5 min with a total of 200 time points (total experiment time 16 h 40 min). The recorded 1D carbon spectrum (using inverse gated proton decoupling) contains 32 K data points and has a spectral width of 200 ppm, 32 scans with a 30° flip angle and relaxation delay of 2.1 s (total recording time of 81 s). After completion of the time-resolved experiment, a 13C-1H HSQC spectrum was recorded.
All NMR spectra were recorded on a BRUKER AVIIIHD 800-MHz spectrometer (Bruker BioSpin AG, Fällanden, Switzerland) equipped with 5-mm cryogenic TCI probe. The spectra were recorded, processed and analyzed using TopSpin 3.6pl7 and TopSpin 4.0.7 software (Bruker BioSpin AG).
Protein crystallization and 3D crystal structure determination
Protein crystallization was initiated by screening with JCSG+ and PACT screens from Qiagen (Hilden, Germany) in 96-well Corning plates (no. 3551), which makes a total of 192 conditions. These experiments for both the AlyA3 native and AlyA3_Y274A mutant samples were carried out at 292 K with the sitting-drop vapor diffusion method. A crystallization robot (Dunn Labortechnik, Crystal Gryphon, Asbach, Germany) was used for dispensing the drops that contained 200-nL protein solution that were mixed with 100 nL of reservoir solution. After visual identification of initial crystallization conditions, these were further optimized for AlyA3 native crystals in 24-well Linbro plates by the hanging-drop vapor diffusion method. For AlyA3, optimal conditions were obtained for a protein solution of 11.3 mg mL−1 concentration. Drops of 2-μL volume of this protein solution were mixed with 1 μL of crystallization solution that contained 2.1-M DL-Malic acid pH 7.0 and equilibrated against a reservoir containing 500 μL. For AlyA3_Y274A in complex with oligo-mannuronate, the optimized conditions were directly obtained from the screening solution. A volume of 0.1 μL of protein solution at 13.4 mg mL−1, supplemented with 1.6 mM of oligo-mannuronate of DP3, was mixed with 0.2 μL of crystallization solution that contained 2.4-M sodium malonate (dibasic monohydrate) and equilibrated against a reservoir containing 100 μL. The native and complexed crystals were flash-frozen at 100 K in a crystallization solution supplemented with 10% glycerol. Data were collected on ID29 at the ESRF (Grenoble) to 1.42-Å resolution for AlyA3 and on Proxima-2 (SOLEIL, St Aubin, France) at 2.16-Å resolution for AlyA3_Y274A in complex with oligo-mannuronate. All crystals belonged to space group P65 with unit cell parameters as given in Table II, and all structures had two molecules in the crystallographic asymmetric unit. All data sets were treated and converted with the program suite XDS (Kabsch 2010) and scaled using aimless (Winn et al. 2011). The 3D structure of AlyA3 was solved by the molecular replacement method using PHASER (McCoy 2007) with Alg17c from S. degradans (PDB 4OJZ) as initial model. The complexed structure of AlyA3_Y274A was obtained starting from the coordinates of native AlyA3, after deleting all ligands, ions and water molecules. The starting phases were optimized by alternating cycles of model building in COOT (Emsley and Cowtan 2004) and refinement using the program REFMAC 5 (Murshudov et al. 2011) as part of the CCP4 suite (Collaborative Computational Project, Number 4 1994). The overall refinement was carried out with REFMAC 5, and water molecules were added automatically with REFMAC-ARP/wARP (Perrakis et al. 2001) and visually verified. In all cases, the stereochemistry of the final structure was evaluated using MOLPROBITY (Chen et al. 2010). All further data collection and refinement statistics are summarized in Table II.
Supplementary Material
Acknowledgements
We are grateful to Nelly Kervarec from the Mass Spectrometry platform at the Université de Bretagne Occidentale for recording the MALDI-TOF mass spectra and to Dr. Lionel Cladière for assistance with the calibration of the size exclusion column. We thank Martin Sevko and William Shepard for their precious local contact support on beamline Proxima2 at SOLEIL (Paris, France) and ID29 beamline staff for their help at ESRF (European Synchrotron Research Facilities, Grenoble, France). The authors appreciate the access to the CristalO platform (FR2424, Station Biologique de Roscoff), which is part of the Biogenouest core facility network. We thank Håvard Sletta from the Department of Biotechnology and Nanomedicine (SINTEF Industry) in Trondheim, Norway, who provided purified alginate samples for the NMR experiments.
Contributor Information
Diane Jouanneau, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Leesa J Klau, Norwegian Biopolymer Laboratory (NOBIPOL), Department of Biotechnology and Food Science, NTNU Norwegian University of Science and Technology, Sem Sælands vei 6/8, N-7491 Trondheim, Norway.
Robert Larocque, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Agathe Jaffrennou, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Ghislain Duval, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Nolwen Le Duff, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Thomas Roret, Sorbonne Université, CNRS, FR 2424, Station Biologique de Roscoff, 29680 Roscoff, France.
Alexandra Jeudy, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Finn L Aachmann, Norwegian Biopolymer Laboratory (NOBIPOL), Department of Biotechnology and Food Science, NTNU Norwegian University of Science and Technology, Sem Sælands vei 6/8, N-7491 Trondheim, Norway.
Mirjam Czjzek, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
François Thomas, Sorbonne Université, CNRS, Integrative Biology of Marine Models (LBI2M), Station Biologique de Roscoff (SBR), 29680 Roscoff, France.
Data availability
The coordinates of AlyA3 and AlyA3-Y274A in complex with oligo-mannuronate were deposited at the Protein Data Bank (http://wwwpdb.org/), with the accession codes 7BJT and 7BM6, respectively.
Authors’ contributions
Contributions are as follows, following the CRediT taxonomy. Conceptualization: FT, MC; Investigation: all authors; Visualization: DJ, MC, FLA and FT. Writing—original draft: DJ, MC and FT. Writing—review and editing: DJ, LJK, FLA, MC and FT.
Funding
EU for its support with regards to the GENIALG Program (FP7-NMP, project 604530, to M.C. and D.J.); ANR for its support with regards to the investment expenditure program IDEALG (http://www.idealg.ueb.eu/, grant agreement No. ANR-10-BTBR-04, to M.C. and T.R.); ANR for its support with regards to project ALGAVOR (ANR-18-CE02–0001-01 to F.T. and N.L.D.); Research Council of Norway through grants 294946 (The Norwegian Seaweed Biorefinery Platform) and Norwegian NMR platform (NNP) 226244 to L.K. and F.L.A.
Conflict of interest statements
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Collaborative Computational Project, Number 4 . 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 50:760–763. [DOI] [PubMed] [Google Scholar]
- Arntzen MØ, Pedersen B, Klau LJ, Stokke R, Oftebro M, Antonsen SG, Fredriksen L, Sletta H, Aarstad OA, Aachmann FL, et al. 2021. Insights into alginate degradation through the characterization of a thermophilic exolytic alginate lyase. Appl Environ Microbiol. 87:e02399–e02320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeyron T, Thomas F, Barbe V, Teeling H, Schenowitz C, Dossat C, Goesmann A, Leblanc C, Oliver Glöckner F, Czjzek M, et al. 2016. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: example of the model algae-associated bacteriumZobellia galactanivoransDsijT. Environ Microbiol. 18:4610–4627. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies GJ, Mackenzie L, Varrot A, Dauter M, Brzozowski AM, Schülein M, Withers SG. 1998. Snapshots along an enzymatic reaction coordinate: Analysis of a retaining β-glycoside hydrolase. Biochemistry. 37:11707–11713. [DOI] [PubMed] [Google Scholar]
- Davies GJ, Wilson KS, Henrissat B. 1997. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J Lett. 321:557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati I, Holtan S, Mørch YA, Borgogna M, Dentini M, Skjåk-Bræk G. 2005. New hypothesis on the role of alternating sequences in calcium-alginate gels. Biomacromolecules. 6:1031–1040. [DOI] [PubMed] [Google Scholar]
- Draget KI, Moe ST, Skjåk-Bræk G, Smidsrød O. 2006. Alginates. In: Stephen AM, Phillips GO, Williams PA, editors. Food polysaccharides and their applications: Boca Raton, USA, CRC Press. p. 289–334. [Google Scholar]
- Dudek M, Dieudonné A, Jouanneau D, Rochat T, Michel G, Sarels B, Thomas F. 2020. Regulation of alginate catabolism involves a GntR family repressor in the marine flavobacterium Zobellia galactanivorans DsijT. Nucleic Acids Res. 48:7786–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- Garron M-L, Cygler M. 2010. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 20:1547–1573. [DOI] [PubMed] [Google Scholar]
- Garron ML, Henrissat B. 2019. The continuing expansion of CAZymes and their families. Curr Opin Chem Biol. 53:82–87. [DOI] [PubMed] [Google Scholar]
- Gerlt J, Bouvier J, Davidson D, Imker H, Sadkhin B, Slater D, Whalen K. 2015. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 1854:1019–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimmestad M, Sletta H, Ertesvåg H, Bakkevig K, Jain S, Suh S j, Skjåk-Bræk G, Ellingsen TE, Ohman DE, Valla S. 2003. The Pseudomonas fluorescens AlgG protein, but not its mannuronan C-5-epimerase activity, is needed for alginate polymer formation. J Bacteriol. 185:3515–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisillier A, Hervé C, Jeudy A, Rebuffet E, Pluchon P, Chevolot Y, Flament D, Geslin C, Morgado IM, Power D, et al. 2010. MARINE-EXPRESS: taking advantage of high throughput cloning and expression strategies for the post-genomic analysis of marine organisms. Microb Cell Fact. 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. 2017. Polysaccharide utilization loci: Fuelling microbial communities. J Bacteriol. 199:e00860–e00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding MM. 2004. The architecture of metal coordination groups in proteins. Acta Crystallogr Sect D Biol Crystallogr. 60:849–859. [DOI] [PubMed] [Google Scholar]
- Haug A, Larsen B, Smidsrød O. 1974. Uronic acid sequence in alginate from different sources. Carbohydr Res. 32:217–225. [Google Scholar]
- Helbert W, Poulet L, Drouillard S, Mathieu S, Loiodice M, Couturier M, Lombard V, Terrapon N, Turchetto J, Vincentelli R, et al. 2019. Discovery of novel carbohydrate-active enzymes through the rational exploration of the protein sequences space. Proc Natl Acad Sci U S A. 116:6063–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyraud A, Gey C, Leonard C, Rochas C, Girond S, Kloareg B. 1996. NMR spectroscopy analysis of oligoguluronates and oligomannuronates prepared by acid or enzymatic hydrolysis of homopolymeric blocks of alginic acid. Application to the determination of the substrate specificity of Haliotis tuberculata alginate lyase. Carbohydr Res. 289:11–23. [DOI] [PubMed] [Google Scholar]
- Ji S, Dix SR, Aziz AA, Sedelnikova SE, Baker PJ, Rafferty JB, Bullough PA, Tzokov SB, Agirre J, Li FL, et al. 2019. The molecular basis of endolytic activity of a multidomain alginate lyase from Defluviitalea phaphyphila, a representative of a new lyase family, PL39. J Biol Chem. 294:18077–18091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr. 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Chung JH, Wang D, Lee J, Woo HC, Choi IG, Kim KH. 2012. Depolymerization of alginate into a monomeric sugar acid using Alg17C, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Appl Microbiol Biotechnol. 93:2233–2239. [DOI] [PubMed] [Google Scholar]
- Kraiwattanapong J, Motomura K, Ooi T, Kinoshita S. 1999. Characterization of alginate lyase (ALYII) from Pseudomonas sp. OS-ALG-9 expressed in recombinant Escherichia coli. World J Microbiol Biotechnol. 15:105–109. [Google Scholar]
- Larsen B, Smidsrod O, Haug A, Painter T. 1969. Determination by a kinetic method of the nearest-neighbour frequencies in a fragment of alginic acid. Acta Chem Scand. 23:2375–2388. [DOI] [PubMed] [Google Scholar]
- Larsen B, Smidsrød O, Painter T, Haug A, Rasmussen SE, Sunde E, Sørensen NA. 1970. Calculation of the nearest-neighbour frequencies in fragments of alginate from the yields of free monomers after partial hydrolysis. Acta Chem Scand. 24:726–728. [Google Scholar]
- Liu J, Yang S, Li X, Yan Q, Reaney MJT, Jiang Z. 2019. Alginate oligosaccharides: production, biological activities, and potential applications. Compr Rev Food Sci Food Saf. 18:1859–1881. [DOI] [PubMed] [Google Scholar]
- Lombard V, Bernard T, Rancurel C, Brumer H, Coutinho PM, Henrissat B. 2010. A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem J. 432:437–444. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabeau S, Kloareg B. 1987. Isolation and analysis of the cell walls of brown algae: Fucus spiralis, F. ceranoides, F. vesiculosus, F. serratus, Bifurcaria bifurcata and Laminaria digitata. J Exp Bot. 38:1573–1580. [Google Scholar]
- Mathieu S, Touvrey-Loiodice M, Poulet L, Drouillard S, Vincentelli R, Henrissat B, Skjåk-Bræk G, Helbert W. 2018. Ancient acquisition of “alginate utilization loci” by human gut microbiota. Sci Rep. 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. 2007. Solving structures of protein complexes by molecular replacement with phaser. Acta Crystallogr Sect D Biol Crystallogr. 63:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr Sect D Biol Crystallogr. 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Jagtap S, Nair SK. 2014. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J Biol Chem. 289:8645–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. 2001. ARP/wARP and molecular replacement. Acta Crystallogr - Sect D Biol Crystallogr. 57:1445–1450. [DOI] [PubMed] [Google Scholar]
- Poblete-Castro I, Hoffmann SL, Becker J, Wittmann C. 2020. Cascaded valorization of seaweed using microbial cell factories. Curr Opin Biotechnol. 65:102–113. [DOI] [PubMed] [Google Scholar]
- Preiss J, Ashwell G. 1962. Alginic acid metabolism in bacteria. I. Enzymatic formation of unsaturated oligosaccharides and 4-deoxy-L-erythro-5-hexoseulose uronic acid. J Biol Chem. 237:309–316. [PubMed] [Google Scholar]
- Preiss J, Ashwell G. 1963. Polygalacturonic acid metabolism in bacteria: I. Enzymatic formation of 4-deoxy-L-threo-5-hexoseulose uronic acid. J Biol Chem. 238:1571–1576. [PubMed] [Google Scholar]
- Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42:320–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 41:207–234. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher G, Schülein M, Davies GJ. 1997. Structure of the endoglucanase I from Fusarium oxysporum: Native, cellobiose, and 3,4-epoxybutyl β-D-cellobioside-inhibited forms, at 2.3 Å resolution. Biochemistry. 36:5902–5911. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphic Systems . 2002.
- Thomas F, Barbeyron T, Michel G. 2011. Evaluation of reference genes for real-time quantitative PCR in the marine flavobacterium Zobellia galactanivorans. J Microbiol Methods. 84:61–66. [DOI] [PubMed] [Google Scholar]
- Thomas F, Barbeyron T, Tonon T, Génicot S, Czjzek M, Michel G. 2012. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ Microbiol. 14:2379–2394. [DOI] [PubMed] [Google Scholar]
- Thomas F, Lundqvist LCE, Jam M, Jeudy A, Barbeyron T, Sandström C, Michel G, Czjzek M. 2013. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J Biol Chem. 288:23021–23037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr Sect D Biol Crystallogr. 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T, Preston L, Schiller N. 2000. Alginate lyase: Review of major sources and enzyme characteristics, structure-function analysis, biological roles, and applications. Annu Rev Microbiol. 54:289–340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of AlyA3 and AlyA3-Y274A in complex with oligo-mannuronate were deposited at the Protein Data Bank (http://wwwpdb.org/), with the accession codes 7BJT and 7BM6, respectively.


