Abstract
Amid the ongoing COVID-19 pandemic, it has become increasingly important to monitor the mutations that arise in the SARS-CoV-2 virus, to prepare public health strategies and guide the further development of vaccines and therapeutics. The spike (S) protein and the proteins comprising the RNA-Dependent RNA Polymerase (RdRP) are key vaccine and drug targets, respectively, making mutation surveillance of these proteins of great importance.
Full protein sequences were downloaded from the GISAID database, aligned, and the variants identified. 437,006 unique viral genomes were analyzed. Polymorphisms in the protein sequence were investigated and examined longitudinally to identify sequence and strain variants appearing between January 5th, 2020 and January 16th, 2021. A structural analysis was also performed to investigate mutations in the receptor binding domain and the N-terminal domain of the spike protein.
Within the spike protein, there were 766 unique mutations observed in the N-terminal domain and 360 in the receptor binding domain. Four residues that directly contact ACE2 were mutated in more than 100 sequences, including positions K417, Y453, S494, and N501. Within the furin cleavage site of the spike protein, a high degree of conservation was observed, but the P681H mutation was observed in 10.47% of sequences analyzed. Within the RNA dependent RNA polymerase complex proteins, 327 unique mutations were observed in Nsp8, 166 unique mutations were observed in Nsp7, and 1157 unique mutations were observed in Nsp12. Only 4 sequences analyzed contained mutations in the 9 residues that directly interact with the therapeutic Remdesivir, suggesting limited mutations in drug interacting residues. The identification of new variants emphasizes the need for further study on the effects of the mutations and the implications of increased prevalence, particularly for vaccine or therapeutic efficacy.
Keywords: SARS-CoV-2, Computational genomics, Pathogen surveillance, Bioinformatics
1. Introduction
The global pandemic of Coronavirus Respiratory Disease 2019 (COVID-19), caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), has caused significant disruption to public health and economic activity worldwide. As of April 23, 2021, there have been over 140 million confirmed cases of COVID-19 and over 3 million deaths worldwide (John Hopkins University, 2020). Several new variants of SARS-CoV-2 have emerged and are spreading globally. The Delta variant of concern (pangolin lineage B.1.617.2, first identified in India) has much greater transmissibility (Liu and Rocklöv, 2021) relative to the original strain (Liu et al., 2020), and has been observed to be more resistant to antibody neutralization from the sera of vaccinated individuals (Edara et al., 2021). The Alpha variant of concern (B.1.1.7, first observed in the United Kingdom) has exhibited increased transmissibility (Davies et al., 2021; Graham et al., n.d.; Volz et al., 2021), the Beta variant of concern (B.1.351, first observed in South Africa) has also been observed to partially escape vaccine-induced immunity (Wu et al., 2021; Liu et al., 2021a; Garcia-Beltran et al., 2021; Madhi et al., 2021; Tada et al., 2021), and the Gamma (P.1) variant of concern first observed in Brazil has exhibited both increased transmissibility (Faria et al., 2021; Coutinho et al., 2021) and partial evasion of vaccine-induced immunity (Garcia-Beltran et al., 2021). New variants have also been discovered in California (Epsilon variant of concern, B.1.427/B.1.429 lineages) (Zhang et al., 2021; Deng et al., 2021) and New York (Iota variant of interest, B.1.526) (West et al., 2021). The Iota variant contains the E484K mutation linked to antibody escape in the Beta and Gamma variants (West et al., 2021), suggesting it may similarly decrease the effectiveness of current vaccines. The appearance of new strain variants and protein mutations have highlighted the importance of continued genomic surveillance of SARS-CoV-2 strains to understand how SARS-CoV-2 genomes are evolving over time and geographic locations.
High throughput genomic analysis of SARS-CoV-2 strains has been greatly facilitated by databases, such as the Global Initiative on Sharing all Influenza Data (GISAID) (Shu and McCauley, 2017). GISAID was initially created after the global spread of the H5N1 avian flu, to break down barriers to data sharing, enabling users to share and analyze data in a timely manner, and allowing users to access unpublished genomic data under the conditions of a data use agreement that protects the intellectual property rights of data contributors (Shu and McCauley, 2017). Many studies have leveraged the GISAID information to examine patterns in the emergence of mutations in the viral genome (Korber et al., 2020; Thomson et al., 2021; Hodcroft et al., 2020).
The genome of SARS-CoV-2 consists of 14 open reading frames (ORFs) that encode 27 proteins (Wu et al., 2020). Four of the ORFs encode structural proteins, and are named with one letter corresponding to the name of the structural protein produced: E (envelope protein), N (nucleocapsid protein), M (membrane protein), and S (spike protein) (Kim et al., 2020). The spike protein has been shown to be a key factor in viral entry, and binds to the human angiotensin-converting enzyme 2 (ACE2) (Crackower et al., 2002), resulting in the fusion with the host cell membrane (Wang et al., 2020). In addition to the four structural proteins, there are 16 non-structural proteins (Nsps) encoded by open reading frame 1ab (Wu et al., 2020). The non-structural proteins Nsp7, Nsp8, and Nsp12 have been found to form the viral RNA-dependent RNA polymerase (RdRP) complex, and each of the Nsps forming the RdRP complex must be present for the replication of viral genomic RNA to occur (Yin et al., 2020). Nsp12 contains the active site in which the antiviral drug remdesivir binds (Yin et al., 2020), making this protein of great importance for variant surveillance. Here we describe a comprehensive analysis of the amino acid mutations in the spike protein and the RdRP complex from the beginning of the pandemic to January 16th, 2021. We identify and document changes in the prevalence of mutations over time, and examine the mutations within the context of protein structures to identify patterns of mutation that may impact host-pathogen interactions, as well as vaccine and therapeutic efficacy.
To address confusion over the definitions of mutation, variant, strain, and lineage, this analysis follows the definitions outlined by Mascola et al. (2021). A mutation is defined as a change in the viral genome; for the purpose of this analysis this will specifically describe a change to the amino acid sequence transcribed from the genome (known as a non-synonymous mutation). A variant is defined as a combination of mutations that exist together in a single viral genome, and a strain is a variant confirmed to have distinct properties. The term lineage is used in the context of phylogenetic analysis and refers to variants that create new branches on a phylogenetic tree (Mascola et al., 2021).
2. Methods
2.1. Raw data download
All protein sequences submitted to GISAID by February 12th, 2021 were downloaded in a single FASTA file. The file was pre-processed to amino acid format, with one entry for each protein in every sequence. Sequence headers contained metadata including the protein, the accession ID of the sequence, the date of collection, and the geographic location. 437,006 unique viral genomes were represented in the file, though some genomes did not contain sequences for all proteins. The number of sequences included in the file for each protein is given in Table 1 . A second file containing extended metadata was also downloaded; the file was formatted as a table with one row per sequence. The reference genome used in our analysis was the Severe Acute Respiratory Syndrome Coronavirus 2 Isolate WIV04 (WIV04), sequenced in Wuhan, China on December 30th, 2019 (Zhou et al., 2020). The raw FASTA file was split by protein into 27 files using a Python script in Jupyter Notebook (version 6.1.4) (Kluyver et al., 2016), and each protein was processed separately through all subsequent steps.
Table 1.
Number of sequences remaining after each step in the analysis. The number of sequences clustered and aligned reflects the number of sequences in the cumulative analysis and structural visualizations.
| Spike | Nsp7 | Nsp8 | Nsp12 | |
|---|---|---|---|---|
| Sequences Downloaded (February 12th, 2021) | 436,506 | 435,838 | 435,848 | 435,949 |
| Sequences Passing Filter Criteria | 357,361 | 430,698 | 426,343 | 401,934 |
| Sequences Clustered and Aligned | 345,275 | 430,606 | 426,084 | 400,171 |
| Clustered Sequences Linked to Metadata | 339,387 | 423,158 | 418,772 | 393,363 |
| Metadata-Sequence Pairs Included in Time Series Analysis | 334,474 | 421,019 | 416,650 | 391,331 |
2.2. Filtering of sequences
Sequences were filtered in Python using the Biopython SeqIO module (Cock et al., 2009). In order to reduce potential incomplete sequences and lower quality sequences, all sequences that were ten or more codons shorter or longer than the reference sequence were eliminated from the analysis, along with sequences containing more than 0.1% ambiguous (“X”) codons. The number of sequences for each protein remaining after filtering is listed in Table 1.
2.3. Sequence dereplication
In order to streamline our computational pipeline, identical sequences were condensed into clusters using USEARCH (version 11.0.667) (Edgar, 2010). Clusters, representing unique sequences, were written out to a FASTA file with the ID of the cluster and the number of sequences in the cluster. Concurrently, a separate file for cluster information was created that linked the metadata for all sequences in each cluster to the cluster ID. Clusters of size one were not included in the analysis due to the low abundance and possibility of these clusters reflecting errors in sequencing rather than true variation. Each cluster represents a SARS-CoV-2 variant.
2.4. Sequence alignment
FAMSA (version 1.6.2) (Deorowicz et al., 2016) was used to align clustered sequences for comparison with the reference sequence. FAMSA was selected based on its superiority in both alignment quality and speed relative to other algorithms for large alignments (alignments with greater than 5000 sequences) (Deorowicz et al., 2016). FAMSA was ran with default settings, and the output was stored in FASTA format.
The spike alignment was then computationally edited with a Python script to correct instances where amino acids were inconsistently aligned across insertion regions. For example, -70 - H- -V75 was represented as H70 - - - -V75 in some clusters (dashes are added when insertions exist in the same region of other clusters), though the two alignments are biologically equivalent. The alignment corrections made to the spike protein alignment are given in (Supplementary Table S1).
2.5. Parsing of multiple sequence alignment
A Python script was developed in Jupyter notebook to automatically parse the aligned sequences for mutations given the ID of the cluster containing the reference sequence, which was determined by searching for “WIV04” in the cluster information file using RStudio (version 1.3.1093) (RStudio, 2020). The Python script scanned through the other clusters (Supplementary Fig. S1), comparing each codon with the corresponding codon of the reference cluster. When mutations were discovered, the program determined the type of mutation, and functions were run accordingly to store the position of the mutation relative to the reference, the ID and size of the variant cluster, and the identity of the codon in the reference and variant clusters. Based on this information, a code was computed for the mutation based on the nomenclature recommended by the Human Genome Variation Society (HGVS) (den Dunnen et al., 2016). This information was then stored for each mutation observed, as the “variant events” dataset. Deletions spanning multiple codons were recorded as a single event, and insertions at the beginning and end of the sequences were named as extensions with the format <Position of the first or last codon>ext<Identity of codon(s) inserted>. The variant events dataset was then grouped by position and the sum of the cluster sizes was taken to compute the total number of sequences with a mutation at each position (“variants by position” dataset), and a similar operation was performed to compute the total number of sequences containing each unique variant (“variants by code” dataset).
2.6. Three-dimensional visualization of frequently mutated sites
Structures of the spike protein and the RNA-dependent RNA polymerase (RdRP) complex were downloaded from the Protein Data Bank (PDB) (Berman, 2000) and visualized using PyMOL (The PyMOL Molecular Graphics System, 2020). For the spike protein, two structures were downloaded: PDB ID 6VSB (Image of 6VSB, 2020), which shows the whole spike protein with one receptor binding domain in the up conformation, and PDB ID 6M17 (Image of 6M17, 2020, (p17)), which shows the receptor binding domain of the spike protein in contact with the ACE2 receptor. For the RdRP complex, the structure PDB ID 7BV2 (Image of 7BV2, 2020) was used. The variants by position dataset was used to color each position in the structures by the frequency of variation using a log-10 scale. In a separate visualization, the spike protein and the RdRP complex were colored by domain and all positions mutated in more than 100 sequences were highlighted.
2.7. Frequency of mutations in key residues
The variants by code dataset was filtered by residue number to determine the frequency and identity of mutations in key regions of the genome, such as the receptor-binding domain (RBD), the furin cleavage site (Walls et al., 2020), and superantigen motifs (Cheng et al., 2020) in the spike protein. For the RdRP complex, the binding site of remdesivir (Yin et al., 2020) was analyzed.
2.8. Time series analysis of variants
To determine changes in the prevalence of mutations over time, the variant events dataset was linked with the extended metadata according to the process outlined in Supplementary Fig. S2. The GISAID accession ID for each sequence was extracted from the cluster information dataset using RStudio, yielding a dataframe mapping sequence accession IDs to the corresponding cluster ID. This dataframe was merged with the extended metadata on the common accession ID column, yielding a dataset listing the metadata for each sequence, along with the ID of the representative cluster. A dataframe mapping the cluster ID to the mutations observed in the cluster was created from the variant events dataset, and this dataframe was merged with the metadata with cluster IDs to yield a dataset giving the metadata for each sequence, along with the mutations observed. The number of mutations in each cluster was also computed during this step and added to the metadata for each sequence. Subsets were then taken based on the collection date of the samples: weekly time intervals beginning on January 5th, 2020 and ending on January 16th, 2021, were used. Sequences collected after January 16th were excluded from time series analysis due to the possibility of incomplete reporting of samples after this date. The frequency of each unique variant was obtained for each week and stored in a separate dataset along with the total number of sequences analyzed; variant counts were then divided by the total number of sequences to give the percentage of sequences with each unique variant by week. Subsets were also taken by continent to perform analysis by geographic region. Sequences with no defined day of collection were excluded from analysis, as well as sequences with metadata entries that could not be linked to cluster IDs. The number of sequences included in the time series dataset is given in Table 1.
2.9. Visualization of variant trends
The Python package Matplotlib (Caswell et al., 2020; Hunter, 2007) was used to visualize trends in spike and RdRP mutations over time. The top ten most common mutations were selected from the percentage table, and a line plot showing the prevalence of each mutation over time was created. To analyze trends in less common mutations, a heatmap was used: all mutations with a prevalence greater than or equal to 2% in at least one week were included. A color map for the heatmap was defined using a log-10 scale, with 0.10% as the lower bound for coloring cells. A histogram was generated to show the number of sequences represented in each week. Scatterplots plotting the number of mutations in each sample vs. the collection date of the sample were created from the merged metadata from the time series analysis. Subsets were taken by the pangolin lineage classification for each sequence, which was provided in the original metadata downloaded from GISAID. Boxplots were also created from the number of mutations in the spike protein and the proteins comprising the RdRP complex for each of the lineages associated with the variants mentioned in the introduction section.
3. Results
3.1. Spike protein
3.1.1. Common spike mutations by continent
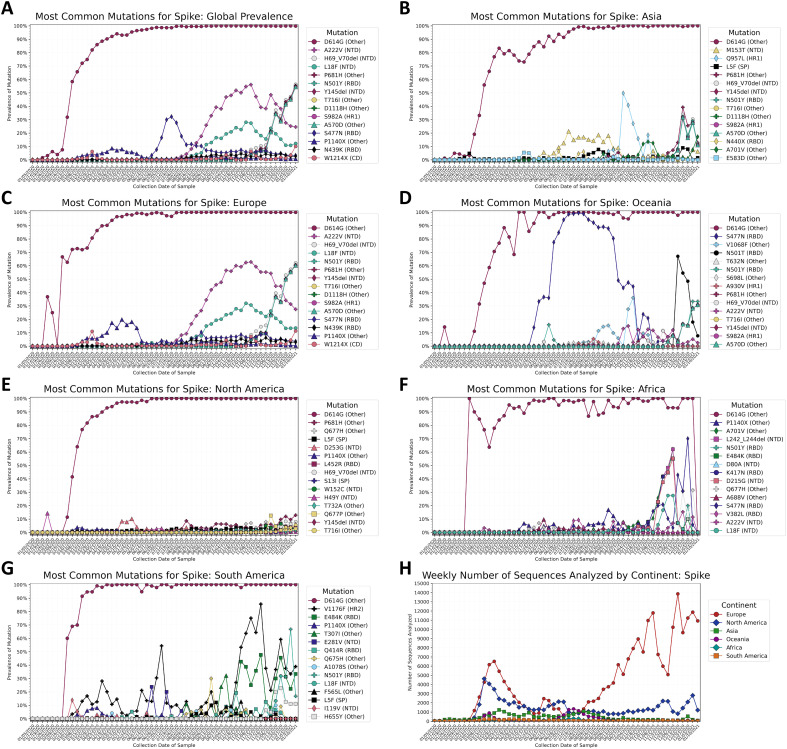
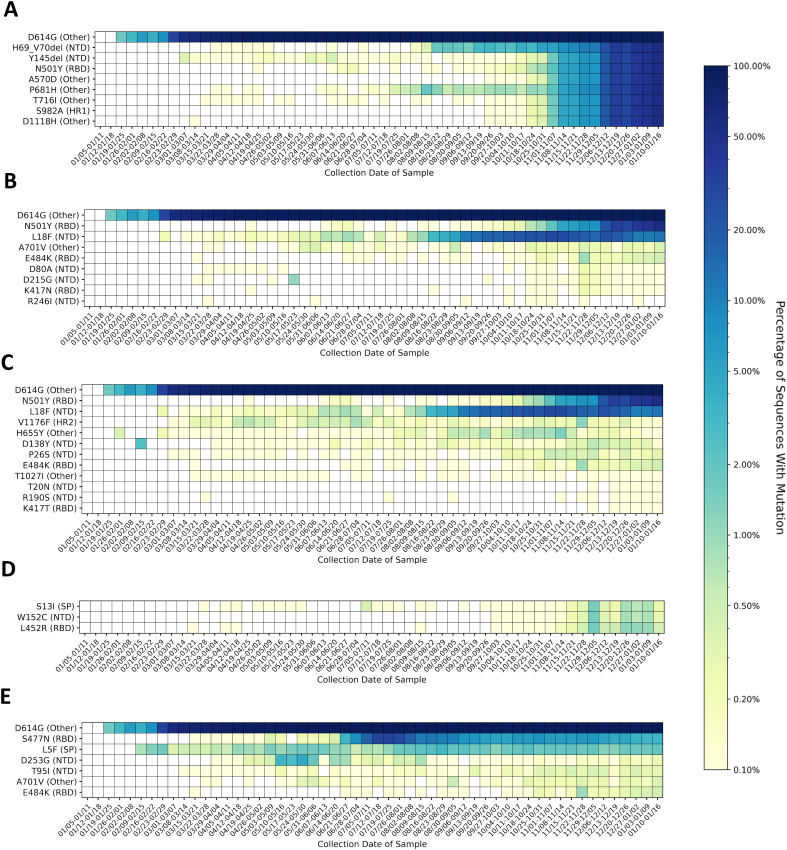
The global prevalence of the top 15 most common mutations by week of sample collection is shown in Fig. 1A, and the prevalence of the top 15 most common mutations on each continent are shown in Fig. 1B–G. The most common mutation worldwide was the substitution D614G, which quickly became prevalent after its appearance in mid-January 2020 (Fig. 1A). The mutation was observed in more than 50% of sequences collected worldwide by the week of March 1st, and in more than 90% of sequences collected by the week of April 26th. D614G quickly became the dominant variant on all continents, though its rate of establishment was much lower in Asia (Fig. 1B). D614G reached 90% prevalence in Asia during the week of June 14th.
Fig. 1.
A–H: Prevalence of the top ten most common variants for the spike protein, by collection date in one-week intervals beginning on January 5th, 2020 and ending on January 16th, 2021. Prevalence of the 15 most common mutations A) worldwide, B) in Asia, C) in Europe, D) in Oceania, E) in North America, F) in Africa, and G) in South America. Mutations that appear within the top 15 most prevalent mutations on multiple continents are given the same color and shape in every graph. The substitution D614G was the first mutation to become highly prevalent on all continents. D614G was observed in 90% of sequences analyzed globally by early May 2020. The profile of other mutations varies by continent. Several mutations are observed that are associated with variants of concern and variants of interest. These mutations may appear individually or with others associated with the variant, and some mutations are shared between variants. The mutations in Fig. 1 associated with the Alpha variant of concern first detected in the United Kingdom are H69_V70del, Y144del (identified as Y145del by alignment software), N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H (John Hopkins University, 2020). The mutations D80A, L242_L244del, K417N, E484K, N501Y, D614G, A701V, L18F, and D215G are associated with the Beta variant of concern (Liu and Rocklöv, 2021; Liu et al., 2020). L18F, E484K, N501Y, D614G, H655Y, and V1176F are associated with the Gamma variant of concern, and S13I, W152C, L452R are associated with the Epsilon variant of concern (Edara et al., 2021). Mutations in Fig. 1 associated with the Iota variant of interest are L5F, D253G, E484K, S477N, D614G, A701V) (Davies et al., 2021). H) Number of complete spike protein sequences analyzed per week, by continent. Europe has contributed the greatest number of sequences weekly since early August 2020. Lower sample sizes in regions may explain the sudden shifts in prevalence seen in (A-G).
The N-terminal domain (NTD) substitution A222V and the signal peptide substitution L18F gained in prevalence globally since their appearance in late July and early August, respectively, before peaking in late October and declining in prevalence. The mutations were most common in Europe (Fig. 1C), though A222V has been observed to an increasing extent in Asia, Oceania, and Africa, and North America (Supplementary Fig. S3A), and L18F has been observed on all continents (Supplementary Fig. S3B).
The RBD substitution N439K has slowly increased in prevalence since August 2020 and has been consistently observed in 2.0–4.5% of samples worldwide since the week of August 16th. The mutation was first observed in Europe during the week of March 15th, but it has also been observed in Asia since the week of October 10th, and in North America since the week of December 13th (Supplementary Fig. S3C).
The receptor binding domain (RBD) substitution S477N appeared in June and reached peak prevalence during week of July 19th, appearing in 32.1% of sequences worldwide before decreasing in prevalence to 0.5% by the week of December 6th, 2020. An influx of sequences from Oceania was observed during this time (Fig. 1H, Supplementary Fig. 4), and S477N was observed in more than 90% sequences from Oceania between the week of July 15th and the week of August 16th (Fig. 1D). S477N has also been observed in Europe with increasing frequency, and since August it has been observed in increasing prevalence in Europe, Africa, Asia, and North America (Supplementary Fig. S3D).
Q677H has been observed in the United States in October 2020 by Hodcroft et al. (Hodcroft et al., 2021) and Tu et al. (Tu et al., 2021) The variant appears in 3.5–8.0% of samples from North America between the week of November 1st, 2020 and the week of January 10th, 2021, and it was first observed in North America during the week of March 15th, 2020 (Fig. 1E). The mutation was also observed in Europe during the same week, in Asia since the week of April 5th, in Africa since the week of May 17th, and in Oceania since the week of June 21st (Supplementary Fig. S3E). The mutation was observed in one out of 93 sequences from South America during the week of March 8th, but was not observed again on the continent until the week of July 12th, 2020.
The NTD deletion L242_L244del was observed in 62.1% of samples collected from Africa during the week of December 6th, 2020 (Fig. 1F). The deletion was first observed in Africa during the week of October 4th, 2021. L242_L244del has also been observed in Asia and Europe since the week of December 13th, 2020, in in Oceania since the week of January 3rd, 2021. The mutation has not yet been observed in North or South America (Supplementary Fig. S3F).
The RBD substitution E484K was observed in South America (Fig. 1G). This substitution is observed in the Beta (Tegally et al., 2020) and Gamma (Faria et al., 2021) variants and has been observed to decrease the neutralization efficiency of antibodies to wild-type E484 variants (Weisblum et al., 2020; Greaney et al., 2021).
Mutations associated with the Alpha variant (H69_V70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H) (Rambaut et al., 2020), have quickly increased in prevalence worldwide since November. P681H was the second most common mutation in North America, detected in 12.9% of samples from the week of January 10th 2021. This prevalence value is greater than H69_V70del and N501Y (8.0% and 5.7%, respectively), suggesting that some viral species are carrying P681H, but not other mutations associated with the Alpha variant.
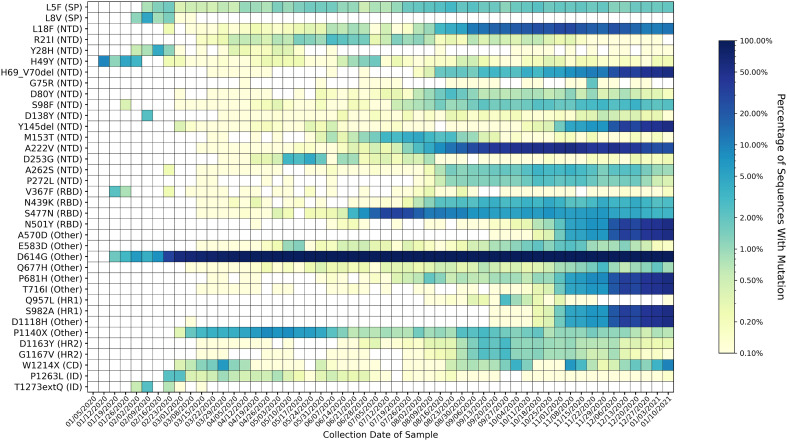
Relatively low sample sizes outside of Europe (Fig. 1H, Supplementary Fig. S4) may limit the conclusions that can be drawn about the global viral population from this data. Of the 337,474 sequences analyzed through January 16th, 2021, 226,399 (66.58%) were from Europe. 72,194 sequences (21.39%) were from North America, 22,643 (6.71%) were from Asia, 10,820 (3.21%) were from Oceania, 3610 (1.07%) were from Africa, and 3511 (1.04%) were from South America. Fig. 2 shows time series trends for all mutations present in at least 2% of samples worldwide from any given week. In addition to the mutations previously mentioned and the quickly increasing prevalence of mutations associated with the Alpha variant in the last few months of the analysis, the heatmap reveals four additional mutations that appear to be increasing in prevalence with time (L5F, S98F, A262S, and P272L). Other mutations appear to have peaked and later decreased in prevalence. Of the 36 mutations present in at least 2% of weekly samples worldwide, 15 were in the NTD, four were in the RBD, two each were in the signal peptide, the intracellular domain, heptad repeat 1, and heptad repeat 2; one was in the cytoplasmic domain, and eight were outside of a named domain. Time series analyses of variants occurring within the NTD (Supplementary Fig. S5) and the RBD (Supplementary Fig. S6) show that the N-terminal domain contains more mutations than the RBD that are consistently present and increasing in prevalence with time.
Fig. 2.
Heatmap of all mutations observed in 2% or more of genomes collected for at least one week. Variants are listed on the y-axis and sorted according to their position in the spike protein sequence. Parentheses give the domain in which each variant appears, according to the domain positions specified in Huang et al. 2020 (Graham et al., n.d.). Key for domain abbreviations: SP = signal peptide, NTD=N-terminal domain, RBD = receptor-binding domain, FP = fusion peptide, HR1 = heptad repeat 1, HR2 = heptad repeat 2, CD = cytoplasmic domain, ID = intracellular domain. The heatmap is colored based on a log-10 scale, with prevalence values of zero colored in white, and values less than or equal to 0.10% colored with the lightest shade. Time on the x-axis is categorized by week of collection date, beginning on January 5th, 2020, and ending on January 16th, 2021. Mutations associated with the Alpha variant of concern (H69_V70del, Y145del, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H) have rapidly increased in prevalence since the appearance of the variant in mid-September 2020. Varying trends are observed for other mutations; some are consistently present, while others have emerged and later disappeared. Fifteen out of the 36 variants observed in at least 2% of samples were in the N-terminal domain, four out of 33 were in the receptor-binding domain, two each were in the signal peptide, the intracellular domain, and heptad repeat 2; one was in the cytoplasmic domain, and eight were in an unspecified domain (other).
Two heptad repeat 2 residues, D1163Y and G1167V appear to increase in prevalence at the same time, near the end of August. Out of the 910 sequences containing either D1163Y, G1167V, or both, 802 contain both mutations (Supplementary Table S2). The same trend is observed for the N-terminal domain residues A262S and P272L, and these mutations occur together in 687 out of 1084 sequences containing either A262S, P272L, or both.
Some of the fluctuations in mutation prevalence may reflect the geographic trends of data deposited to GISAID. Region-specific heatmaps showing all mutations present in at least 2% of samples on each continent are shown in Supplementary Figs. S7–S16, and a complete prevalence table for all mutations is given in Supplementary Table S3 A-G.
3.1.2. Structural visualization of mutations
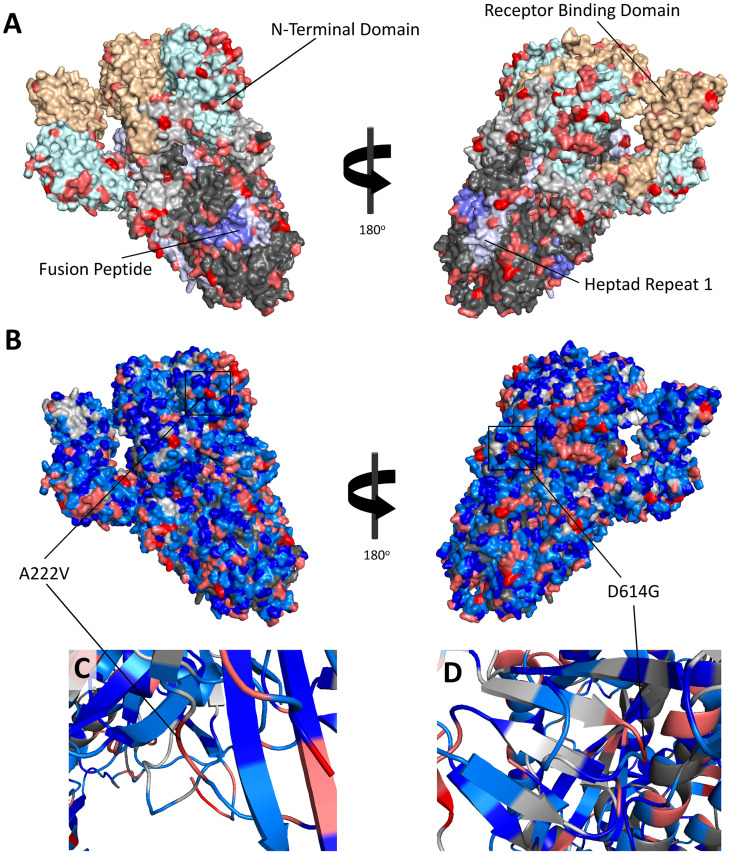
We utilized the structural visualization program PyMOL to examine the frequency of mutation at each position in the spike protein sequence and structure (Fig. 3 A–D). The N-terminal domain has many residues with more than 100 mutations, many of which are adjacent to one another (Fig. 3A). Mutations appear to be evenly distributed throughout both the S1 and S2 subunits. Of the 1735 unique mutations observed in the spike protein across all collection dates, 766 were observed in the N-terminal domain and 360 were observed in the receptor binding domain. Heptad repeats 1 and 2 contained 97 and 115 unique mutations, respectively, the intracellular domain contained 102 mutations, and the cytoplasmic domain contained 59 unique mutations (Fig. 3B). The fusion peptide contained 36 mutations. 1002 mutations were in regions of the spike protein not classified within a domain.
Fig. 3.
A–D: Structure of Spike Protein (PBD ID: 6VSB) with common variants labeled. A) Spike protein domains are shown with residues mutated in 100 or more sequences highlighted. The N-terminal domain is shown in light cyan, the receptor binding domain is shown in light orange, heptad repeat 1 is shown in light blue, and the fusion peptide is shown in blue. Residues in the S1 subunit with no classified domain are shown in light grey, and unclassified residues in the S2 subunit are shown in dark grey. Residues with a variant frequency of more than 100, 1000, and 10,000 are shown in pink, red, and dark red, respectively. Variation is frequent in the N-terminal domain and is often observed in adjacent surface residues. B) All residues with variants are shown. The color-coding for variants appearing in at least 100 sequences is the same as in A) and B); light blue is used for residues with 10–100 variants, and deep blue is used for variant frequencies of 2–10. Residues with zero variants are shown based on subunit colors specified in A) and B). C) A ribbon diagram visualizing the secondary structure of the A222V variant and surrounding residues. A222V is located on a loop region, as is the case with D614G (D). Despite its distance from the receptor binding domain, D614G has been observed to alter the conformational state of the receptor binding domain by altering the conformation in the region surrounding codon 614, acting as a hinge (Volz et al., 2021). The similar secondary structure of A222 suggests that mutations of this residue may have a similar impact. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A comparison of the secondary structure of A222V to that of D614G shows that both variants occur in a loop region. A ribbon diagram of A222V (Fig. 3C), which quickly became more prevalent in Europe between mid-July and October 31st, shows that the variant occurs in a loop region, like D614G (Fig. 3D). D614G has been shown to alter the conformational state of the receptor binding domain through a hinge mechanism involving its loop structure (Yurkovetskiy et al., 2020), and it is possible that A222V may have a similar effect on the conformational state. This is supported by a stability analysis by Jacobs et al., which finds that both D614G and A222V cause rigidification in similar residues of the spike protein structure (Jacob et al., 2020).
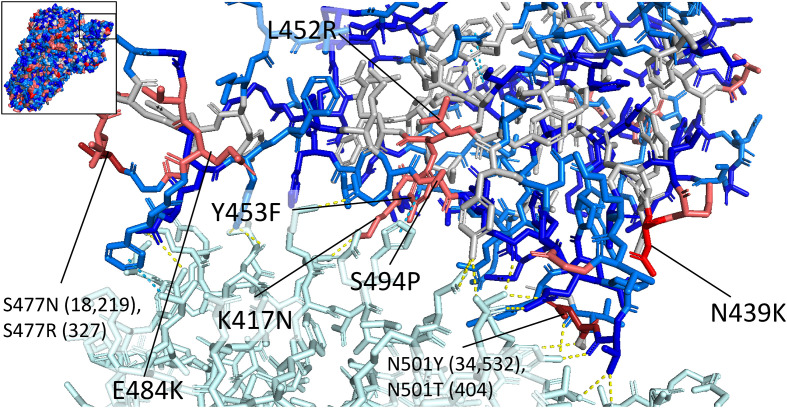
The interface between the receptor binding domain of the spike protein and ACE2 is shown in Fig. 4 (PDB ID: 6M17) (Yan et al., 2020). Four residues that directly contact ACE2 had more than 100 instances of mutation: K417, Y453, S494, and N501. The residue Y453 was shown to have pi-pi stacking interactions with the ACE2 receptor. Y453 was observed as mutating to a phenylalanine residue in 951 sequences. N501 forms two polar contacts with ACE2 residues. N501 was mutated to tyrosine in 34,532 sequences and threonine in 404 sequences. Mutation from asparagine to tyrosine at this site has been shown to increase the binding affinity to the ACE2 receptor by a factor of ten (Liu et al., 2021b).
Fig. 4.
Structure of the interface between the spike receptor binding domain (RBD) and the human ACE2 receptor (top left shows the location of the RBD on the spike protein). The ACE2 receptor is shown in light cyan, and the spike residues are colored according to the frequency of variation: red is used for residues with variants in more than 1000 sequences, pink is used for variant frequencies of 100–1000, light blue for frequencies of 10–100, blue for frequencies of 2–10, and grey for residues with no observed variants. Residues mutated in more than 100 sequences are labeled with the one-letter codes of the reference residue and the most commonly observed mutation. If multiple mutations are observed in the same residue in more than 100 sequences, each mutation is listed with the number of sequences with the mutation in parentheses. Direct contacts are shown with dotted lines: hydrogen bonds and salt bridges are shown in yellow, and pi-pi stacking interactions are shown in cyan. The residue Y453 forms pi-pi stacking interactions with the ACE2 residue H34. The mutation Y453F would result in the loss of a hydroxyl group, which may reduce steric clashing with the histidine residue. N439 does not directly contact the receptor binding domain, though it is believed that the variant lysine residue, which is positively charged, could form a salt bridge with the negatively charged ACE2 residue E329, increasing binding affinity of the RBD to ACE2 (Wu et al., 2021). The substitution L452R is observed in the Epsilon variant of concern. The mutation of a hydrophobic leucine side chain to a positively charged arginine side chain will affect electrostatic properties at the site of the mutation, which may affect antibody binding. The substitution E484K involves the replacement of a negatively charged glutamic acid residue with a positively charged lysine residue, which results in decreased efficacy of antibodies produced against wild-type E484 variants (Liu et al., 2021a; Garcia-Beltran et al., 2021). PDB structures used: PDB ID: 6M17 (RBD-ACE2 interface), PDB ID: 6VSB (whole spike protein on upper left). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
S477, N439, and L452 do not contact the ACE2 receptor directly but are in the vicinity of the binding site. S477N was observed in 18,219 sequences, and N439K was observed in 6743 sequences. L452R, which is observed in the variant of concern B.1.429 discovered in California (Zhang et al., 2021), is observed in 3.49% of the sequences from North America collected during the week of January 10th and 878 sequences total. The mutation from a hydrophobic leucine side chain to a positively charged arginine side chain will affect electrostatic properties, which may have implications for antibody binding. Mutational scanning by Starr et al. found that L452R is associated with a small increase in S protein expression (Starr et al., 2020).
3.1.3. Furin cleavage site
SARS-CoV-2 contains an insertion not found in SARS-CoV that creates an additional cleavage site for the human protease furin (Walls et al., 2020; Wrobel et al., 2020). The furin cleavage site has been shown to strongly influence host mortality rates of other RNA-based viruses such as influenza (Steinhauer, 1999), and furin cleavage sites similar to those of SARS-CoV-2 have been observed in MERS-CoV (Millet and Whittaker, 2014). The furin cleavage site occurs between residues N679 and R685 (Walls et al., 2020); these residues were analyzed for the frequency and type of variants present. A high degree of conservation was observed for residues S680, R682, and R683, which were mutated in 37, 21, and 23 sequences, respectively. P681 was found to be highly variable: P681H was observed in 36,151 sequences total (10.47%). N679 was mutated in 430 total sequences, and A684 was mutated in 146. Q677H, observed in 2146 sequences (0.62% of total sequences), occurs outside of the furin cleavage site but may affect a QTQN consensus sequence near the site (Tu et al., 2021).
3.1.4. Superantigen mimicry
Other spike protein motifs of interest include those that function as superantigens. Superantigens are proteins that stimulate the receptors of T lymphocytes (T cells) by binding at either the alpha or the beta variable chains (Saline et al., 2010), resulting in an overproduction of cytokines that leads to hyperinflammation and toxic shock syndrome (Cheng et al., 2020). The symptoms of multisystem inflammatory syndrome in children (MIS-C) infected with SARS-CoV-2 are similar to those of toxic shock syndrome (Cheng et al., 2020), suggesting that sequence patterns similar to superantigens exist in SARS-CoV-2 proteins. A sequence motif between the residues E661 and R685 was discovered that is capable of binding both the alpha and beta variable chains in T-cells, with residues S680 through R683 shown to directly contact the T-cell receptor (Cheng et al., 2020). This motif overlaps with the furin cleavage site. The sequence that directly contacts the T-cell receptor is the same sequence that is cleaved by furin. A high degree of conservation was observed for residues in the superantigen motif: all residues except for Q675, Q677, and P681 contained variants in less than 100 sequences.
A time series analysis of mutations in the superantigen motif (Supplementary Fig. S17) reveals 14 mutations out of 64 total that appear to be consistently present (P681H, Q677H, Q675H, Q677P, S673T, N679K, P681R, Q675R, P681L, T676I, T678I, A684V, Q677R, and A672V). All of these mutations except for Q675R and P681L appear to be increasing in prevalence with time, with the greatest increase observed for P681H. A deletion of the furin cleavage site (N679_S686del) was observed in 0.395% of sequences from the week of January 26th, 2020 and 0.0358% of samples from the week of May 31st, 2020. A deletion of the QTQN consensus site (N675_N679del) was observed during the week of April 12th, 2020 (0.0126% of sequences) and the weeks of May 17th, May 24th, and May 31st, 2020 (in 0.0266%, 0.1099%, and 0.0358% of sequences, respectively). The deletions were not observed during any other weeks.
3.1.5. Emergent SARS-CoV-2 strains
The time series trends of mutations specific to the Alpha, Beta, and Gamma variants first detected in the UK, South Africa, and Brazil, respectively, as well as the Delta variant, are shown in Fig. 5 A-E. The ten mutations associated with the Alpha variant (Fig. 5A) have quickly gained prevalence worldwide since their appearance in mid-September, appearing in more than half of sequences collected worldwide during the week of January 17th. Mutations associated with the Alpha variant appear to be propagating at a greater rate than those in the other variants/strains. The RBD mutation N484K is observed in the Beta (Fig. 5B) and Gamma (Fig. 5C) variants, and N501K is observed in the Alpha, Beta, and Gamma variants. Both lineages associated with the Epsilon variant (B.1.427 and B.1.429, Fig. 5D) share the same set of mutations on the spike protein (S13I, W152C, L452R) (Zhang et al., 2021; Deng et al., 2021), and have similar effect on transmissibility and antibody evasion (Deng et al., 2021). The three mutations associated with these variants are first observed together during the week of September 27th, 2020. S13I and L452R were detected separately in previous sequences, but W152C was not. The mutations associated with the variant of interest Iota (Fig. 5E), first detected in New York (L5F, T95I, D253G; E484K or S477N; D614G, and A701V) (West et al., 2021), appear together consistently after the week of October 4th, 2020, though the fact that many of these mutations are shared with other phylogenetic lineages makes it difficult to conclude that the variant originated at this time point. Individual mutations associated with the Delta variant were also observed in the data (Supplementary Fig. S18), but they do not appear together in the same sequence, suggesting that sequences containing all the mutations in the Delta variant as currently defined did not yet appear in the GISAID database as of January 16th, 2021.
Fig. 5.
A–E: Heatmap of variants observed in the A) Alpha, B) Beta, C) Gamma, D) Epsilon, variants of concern and the E) Iota variant of interest. Variants are listed on the y-axis, and time is categorized by week of collection date on the x-axis, beginning on January 5th, 2020, and ending on January 16th, 2021. Parentheses give the domain in which each variant appears, according to the domain ranges specified in Huang et al. 2020. Key for domain abbreviations: SP = signal peptide, NTD=N-terminal domain, RBD = receptor-binding domain, FP = fusion peptide, HR1 = heptad repeat 1, HR2 = heptad repeat 2, CD = cytoplasmic domain, ID = intracellular domain. The heatmap is colored based on a log-10 scale, with prevalence values of zero colored in white, and values less than or equal to 0.10% colored with the lightest shade. A) The ten mutations associated with the Alpha variant of concern have quickly increased in prevalence after appearing together in mid-September 2020 and are now observed in more than 50% of sequences worldwide. Some mutations associated with the Beta variant of concern (H69_V70del, Y145del, N501Y, D614G, P681H, and T716I) were present separately before the emergence of the variant in mid-September 2020, while others (A570D, S982A, and D1118H) appeared together at this point. Y144del sometimes appears as Y145del due to ambiguities in the alignment software with identical adjacent amino acids. B) The mutations associated with the Gamma variant of concern appeared together in mid-October 2020 and have increased in prevalence over time in samples worldwide, but to a lesser extent than the variants in the Alpha variant. C) The mutations associated with the Gamma variant of concern appeared together in mid-December 2020; all mutations except for T20N and K417T were present separately before the emergence of the variant. D) The mutations associated with the Epsilon variant of concern appeared together in mid-September 2020. The substitutions L452R and S13I appeared separately before the emergence of the variant, while W152C did not. E) The mutations associated with the Iota variant of interest consistently appear together after early October 2020. All mutations associated with this variant appeared separately before its emergence. Some mutations are present in multiple variants: the RBD substitution E484K is also observed in the Beta, Gamma, and Iota variants, and the RBD substitution N501Y is also observed in the Alpha, Beta, and Gamma variants.
A plot of the number of spike protein amino acid polymorphisms in individual samples by collection date and variant is given in Supplementary Fig. S19A. The plot shows that the rate of change in the number of spike protein polymorphisms relative to the reference changes over time. The rate decreases after mid-March 2020 and remains close to zero and increases again after mid-August 2020. Samples identified as the Alpha, Beta, and Gamma variants exhibit a gap between the number of polymorphisms for these samples relative to other variants, suggesting that the mutations related to these variants appeared all at once. This is consistent with observations of mutations related to these variants appearing in longitudinal samples from immunocompromised patients (Choi et al., 2020). Samples from the Alpha, Beta, and Gamma variants have more amino acid polymorphisms on average than the Epsilon or the Iota variants (Supplementary Fig. S19B).
3.2. RNA-dependent RNA polymerase (RdRP) complex
3.2.1. Common RdRP mutations by continent
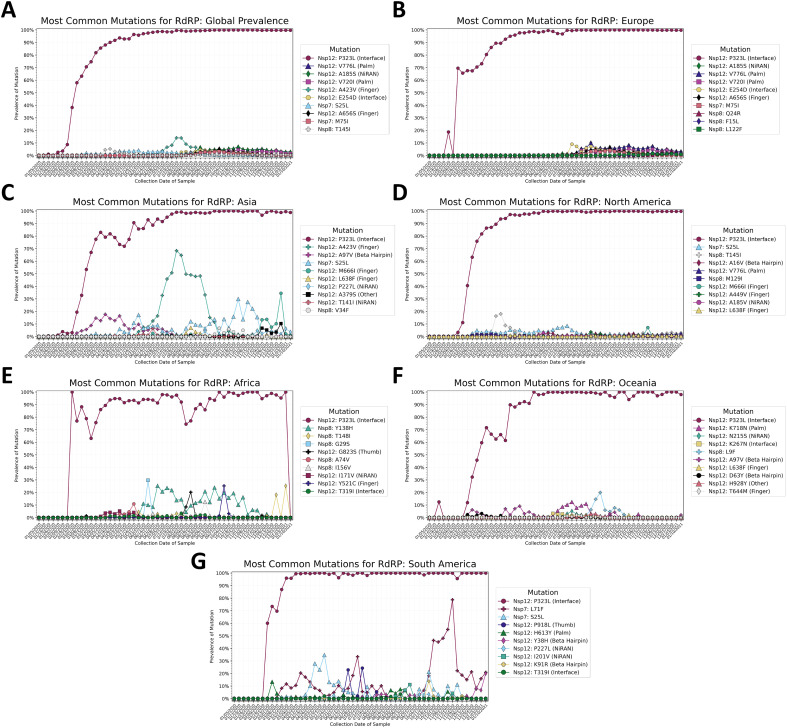
The Nsp12: P323L mutation appeared in late January 2020 and was present in 50% of the sequences by early March, and 90% of the sequences by late April (Fig. 6A). The trend of predominance for Nsp12: P323L was observed on all continents (Fig. 6B-G), but the rate of establishment was lower in Asia than in other continents, as with spike: D614G. Other mutations have emerged and are relatively rare but consistently present, plateauing at 3.5–4.0% of samples collected weekly since August 2020. These mutations include the Nsp12 mutations E254D, A423V, A656S, V720I, and V776L; the Nsp7 mutations S25L and M75I; and the Nsp8 mutation T145I. Time series trends in Europe (Fig. 6B) match global trends, except for the Nsp12 mutation A424V, which is seen in the global data but not in Europe.
Fig. 6.
A–G: Prevalence of the top ten most common mutations for the constituent proteins of the RNA-dependent RNA polymerase (RdRP) complex, by collection date in one-week intervals beginning on January 5th, 2020 and ending on January 16th, 2021. The top ten mutations A) worldwide, B) in Europe, C) in Asia, D) in North America, E) in Africa, F) in Oceania, and G) in South America, are shown. Mutations that appear within the top ten most prevalent mutations on multiple continents are given the same color and shape in every graph. The combination and prevalence of mutations present varies by continent, with the Nsp12 substitution P323L being most common on all continents, and most other mutations present in less than 20% of samples from each continent. Exceptions include the Nsp12 finger subdomain substitution A423V, which was common in Asia during July and August 2020, and the Nsp7 substitution L71F, which was common in South America during October and November 2020. Nearly all mutations besides the Nsp12 substitution P323L have either peaked and subsequently decreased in prevalence, or have plateaued at low prevalence values of 5–10%.
The spike in A424V in the global data can be attributed to the high prevalence of the mutation in Asia during July 2020 (Fig. 6C). The mutation peaked in prevalence in Asia at 63.36% during the week of July 26th, and then decreased in prevalence. The mutation was not observed in any sequences during and after the week of November 22nd. Similar trends are observed for the Nsp12 mutations A97V and M666I and the Nsp7 mutation S25L. The Nsp12 mutation A97V peaked at 17.7% prevalence during the week of April 12th, 2020, the Nsp7 mutation S25L peaked during the week of October 25th, 2020 at 29.9%, and the Nsp12 mutation M666I peaked at 34.4% prevalence during the week of December 27th, 2020.
The Nsp7 mutation S25L was also observed in North America (Fig. 6D). The mutation was first observed during the week of March 1st, and peaked in prevalence during the week of July 26th, 2020 at 8.44%. The mutation appears to have decreased in prevalence after August 2020 and is observed in 0.25% of sequences in North America as of January 10th, 2021. As of January 10th, 2021, mutations in the polymerase complex besides the Nsp12 mutation P323L were rare but their presence is stable. The Nsp8 mutations T145I and Q24R along with the Nsp12 mutation V776L were present in 2.0–2.5% of sequences during the week of January 10th, 2021, and the Nsp12 mutations P227L, V354L, and V605 were present in 1.0–2.0% of sequences.
In Africa (Fig. 6E), the Nsp8 mutation Y138H appears consistently in 5–30% of weekly sequences between late May 2020 and early November. Mutations observed in more than 10% of weekly samples include the Nsp8 mutations A47V, T148I, and I156V; and the Nsp12 mutations Y521C and G823S; but these mutations are not consistently observed above this threshold.
In Oceania (Fig. 6F), there are no stable mutations in the RdRP complex besides the Nsp12 mutation P323L. Several mutations, such as the Nsp12 mutations A97V, N215S, K718N, and N911S; and the Nsp8 mutation L9F, emerged, peaked in prevalence between 5 and 25%, and later decreased to less than 4% prevalence.
In South America (Fig. 6G), the Nsp7 mutation L71F was observed in 78.7% of sequences from the week of November 22nd, 2020, and in 15–25% of samples from the following weeks. The Nsp7 mutation S25L was also observed in the region, peaking at 34.7% prevalence during the week of May 17th, 2020, and peaking again at 21.2% during the week of October 18th, 2020. The Nsp12 thumb subdomain mutation P918L was observed in 22.8% of samples collected during the week of June 21st and 24.2% of sequences collected during the week of July 12th. This amino acid residue is near M924, which is involved in RNA binding.
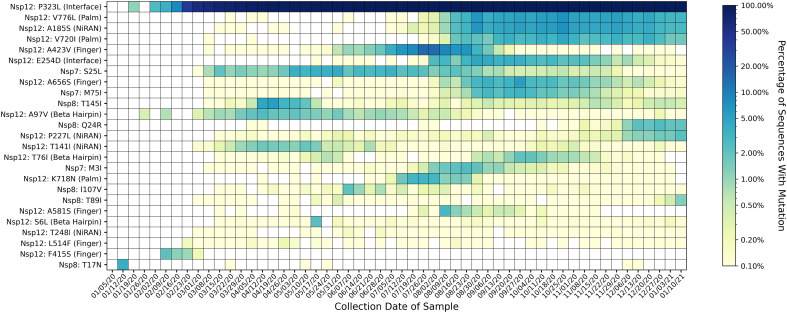
A heatmap of time series data for mutations in the RdRP complex is shown in Fig. 7. The Nsp12 substitutions P323L, V776L, A185S, and V720I appear to be stable or increasing in prevalence, while other mutations have peaked in prevalence and later disappeared. The Nsp12 substitutions A185S and V776L appear to increase in prevalence together, suggesting a correlation between these variants. This is supported by the table of variant combinations for each cluster (Supplementary Table S4, Supplementary Table S5, Supplementary Table S6), which shows V776L occurring together with A185 in 10,273 sequences, and V776L and A185S occurring separately in 84 and 379 sequences, respectively.
Fig. 7.
Heatmap of all mutations observed in 2% or more of genomes collected for at least one week. Mutations are listed on the y-axis, and parentheses give the domain in which each mutation appears, according to the domain ranges specified in Yin et al. 2020 (Madhi et al., 2021). The heatmap is colored based on a log-10 scale, with prevalence values of zero colored in white, and values less than or equal to 0.10% colored with the lightest shade. Time on the x-axis is categorized by week of collection date, beginning on January 5th, 2020, and ending on January 16th, 2021. Most mutations in the polymerase complex do not appear to persist, except for the Nsp12 substitutions P323L, V776L, A185S, and V720I. Of the 25 mutations observed in at least 2% of sequences in any week, 17 are in Nsp12, five are in Nsp8, and three are in Nsp7. Of the 17 Nsp12 mutations, five are in the finger subdomain, four are in the NiRAN domain; three each are in the palm subdomain and the Beta Hairpin; two are in the interface domain, and zero are in the thumb subdomain.
Relatively low sample sizes on some continents (Supplementary Fig. 20A–F) limit the conclusions that can be drawn from the time series data. Supplementary Figs. S21–S27 show regional heatmaps of all variants present in at least 2% of samples on each continent, and Supplementary Table S7 A–G shows complete prevalence data of all variants.
3.2.2. Structural visualization of variants in RdRP complex
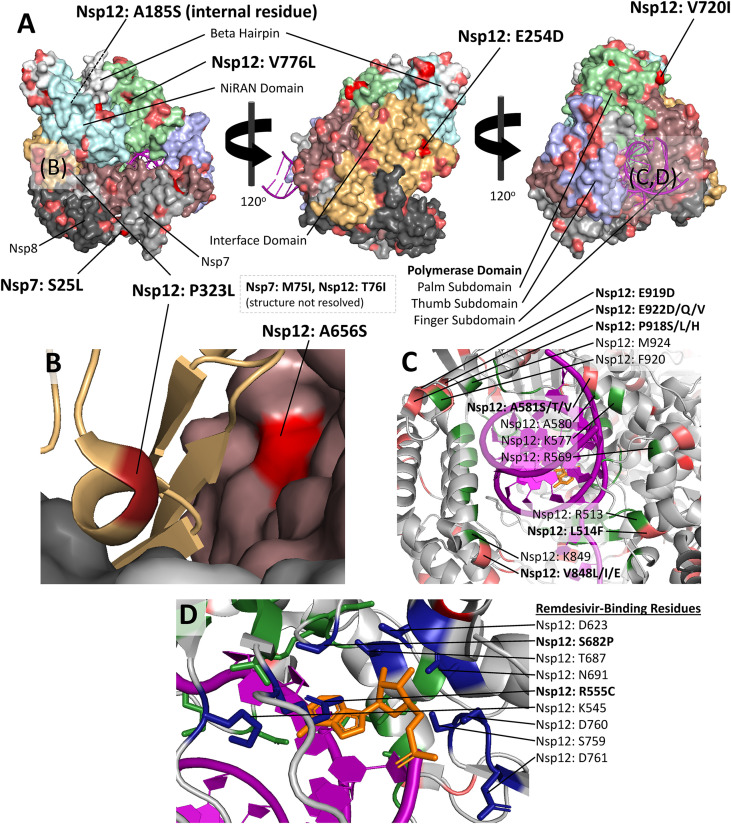
327 unique variants were observed in Nsp8, 166 unique variants were observed in Nsp7, and 1157 unique variants were observed in Nsp12 (Fig. 8A). Of the 1157 variants in Nsp12, 242 were observed in the finger subdomain, 190 were observed in the beta hairpin, 210 were observed in the palm subdomain, 202 were observed in the NiRAN domain, 151 were observed in the interface, and 115 were observed in the thumb subdomain. Forty-seven variants occurred in an uncharacterized region of Nsp12.
Fig. 8.
A–D: Structural visualization of variants in the RNA-Dependent RNA Polymerase (RdRP) complex. A) RdRP complex colored by domain, with high-frequency variants highlighted. Nsp7 and Nsp8 are colored in grey and dark grey, respectively. Nsp12 is colored and labeled by domain. Residues with more than 100 variants are highlighted according to the number of sequences with a variant at that position (worldwide, all collection dates). dark red is used for residues with variants in 10,000+ sequences, red is used for variant frequencies of 1000–10,000, and pink is used for variant frequencies of 100–1000. Nsp7 has few variants compared with Nsp8 and Nsp12, and the interface and NiRAN domain of Nsp12 also appear to have relatively few variants compared with other domains. The common variant Nsp7: S25L exists at the interface between Nsp7 and Nsp8. B) A ribbon diagram of the highly prevalent Nsp12: P323L substitution. The substitution exists within a small alpha helix surrounded by loop regions. Proline helps maintain a tight helical geometry, and substitution of this variant may disrupt the secondary structure. The common Nsp12 substitution A656S is also shown. C) Ribbon diagram of RNA-binding residues of Nsp12 (shown in green). Remdesivir is shown in orange, and mutations are colored according to the scheme in (A). There are no RNA-binding residues with variation in more than 100 sequences, though mutations are observed in residues within four alpha-helix regions containing RNA-binding residues (labeled, with mutations in bold with the substituted residue(s) observed). D) Ribbon diagram of remdesivir-binding site with side chains of remdesivir-binding residues (dark blue). Variation was observed in two remdesivir-binding residues: R555, which is mutated to cysteine in two sequences and S862, which is mutated to proline in two sequences. PDB structures used: PDB ID: 7BV2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The Nsp12 residue P323 occurs in a small alpha helix region within a loop secondary structure (Fig. 8B). This residue is not known to directly bind residues in neighboring domains, though it is near residue A656, which contacts the interface domain and is mutated to a serine in 3631 sequences across all time points (0.907%). A656 has also been observed to mutate to a threonine in 57 sequences, and to a valine in 37 sequences. A656S peaked in prevalence in mid-September and was observed in a decreasing proportion of sequences afterward, while A656T and A656V do not appear to become more prevalent with time.
3.2.3. RNA-binding residues
RNA-binding residues of in the finger, thumb, and palm subdomains were highly conserved (Fig. 8C). Out of the 41 residues of Nsp12 known to bind RNA (Yin et al., 2020), only ten were mutated in at least two sequences and no residues were mutated in more than nine sequences (0.0022% of all sequences analyzed). Several residues occupying the same secondary structure as RNA-binding residues were mutated in more than 100 sequences. The residue L514, which is adjacent to the RNA-binding residue R513, was mutated to phenylalanine in 143 sequences. A581, which is in the alpha helix containing RNA-binding residues A580, K577, and R569, was mutated to serine in 480 sequences, threonine in 29 sequences, and valine in 16 sequences. V848, adjacent to the RNA-binding residues K849, was mutated to leucine in 146 sequences, isoleucine in 25 sequences, and glutamate in 2 sequences. The mutations P918S/L/H (161, 85, and 3 sequences, respectively), E919D (126 sequences), and E922D/Q/V (201, 2, and 2 sequences, respectively) are observed in the alpha helix containing the RNA binding residues F920 and M924.
3.2.4. Variation in the binding site for Remdesivir
Remdesivir blocks viral replication by binding to the following residues in Nsp12: K545, S682, R555, T687, S759, N691, D623, D760, and D761 (Yin et al., 2020). Remdesivir-binding residues are shown in Fig. 8D. Of these nine amino acid residues, variants were observed for only two: S862, which mutated to proline in two sequences worldwide, and R555, which mutated to cysteine in two sequences.
Scatterplots of the number of Nsp7, Nsp8, and Nsp12 polymorphisms in individual samples by collection date is given in Supplementary Fig. S28A–C. The scatterplots show a much lower rate of increase in the number of polymorphisms over time relative to the spike protein. The rate of increase is greater for Nsp12 than for Nsp7 and Nsp8. Boxplots of the number of polymorphisms by variant (Supplementary Fig. S29A–C) show that all the analyzed variants of concern have a median of zero polymorphisms in Nsp7 and Nsp8, and a median of one polymorphism in Nsp12.
4. Discussion
This study analyzed viral genomic data made available through GISAID and used structural information to identify trends in mutations over time and geographic location and to examine the structural features of mutations. The study validated previous findings of the predominance of spike: D614G and Nsp12: P323L in the population (Korber et al., 2020) while uncovering recent increases in the prevalence of new mutations in Nsp12 and the spike protein. Trends in the emergence of new variants vary by geographic area.
The very high prevalence of the D614G mutant in the sequences sampled reflects studies documenting increased viral counts in-vitro (Korber et al., 2020; Hou et al., 2020) as well as higher viral loads in infected individuals (Korber et al., 2020). D614G has also been observed to co-occur with Nsp12: P323L (Korber et al., 2020), the most common mutation observed in Nsp12. For both the spike protein and Nsp12, new mutations appeared in late July to early August and have since steadily increased in prevalence. Most mutations generally have neutral or deleterious effects on protein function (Soskine and Tawfik, 2010); these mutations would not be expected to increase in prevalence over time unless they occur with a variant that confers an increase in fitness, replication, or transmission potential. The increasing prevalence of new mutations in the population suggests that these provide an evolutionary advantage, though follow-up studies in-vitro or in-vivo are required to definitively determine the effects of these variants.
Recent studies suggest the emergence of mutations associated with higher case fatality rates of COVID-19. The Nsp7 mutation L71F has been linked to higher mortality rates in several studies (Farkas et al., 2020; Nagy et al., 2021; Fang et al., 2021), though these studies have yet to enter peer review. The spike protein mutation V1176F results in increased stability of the spike protein (Farkas et al., 2020) and has also been associated with higher mortality rates (Farkas et al., 2020; Nagy et al., 2021). The exact extent to which case fatality rates increase is not yet known, though these studies suggest that mutations that increase disease severity are beginning to spread.
The high conservation observed in the RNA-binding residues of the RdRP indicates that there is no evidence of remdesivir resistant mutations as of January 10th, 2021. Continued surveillance of contact residues for remdesivir (Yin et al., 2020) and other drug candidates targeting the RdRP (Sada et al., 2020) is necessary to effectively respond to potential drug resistance in the future.
The N-terminal domain and the receptor binding domain (RBD) of the spike protein were observed to be variable, which may have implications for monoclonal antibody treatments that target these domains (Chi et al., 2020; Premkumar et al., 2020; Rogers et al., 2020). The RBD variant N439K, which has been shown to result in antibody evasion (Thomson et al., 2021; Starr et al., 2020), is becoming more common over time. Several variants observed in the N-terminal domain and the signal peptide (L18F, Y144del, and D253G) have also been shown to decrease the effectiveness of antibodies (McCallum et al., 2021). In addition to single mutations, combinations of mutations are of concern for antibody effectiveness. Combinations of mutations in the Alpha strain have been shown to have a compound effect on antibody escape, even when the individual mutations involved have no effect by themselves (Tada et al., 2021). The combination of variants involved in the Beta strain has affected some of the antibodies used in the Regeneron treatment: REGN10987 was not affected, but in at least one study REGN10933 was shown to be 773 times less effective against the Beta strain (Zhou et al., 2021). This finding highlights the need to continue sequencing of viral genomes worldwide to ensure the continued effectiveness of existing therapeutics and vaccines against SARS-CoV-2, and to guide the development of new treatments.
The analysis in this study is limited in some respects, since the files used and downloaded for this study consist only of amino acid sequences, without the accompanying nucleotide information. Amino acid data can robustly capture information on non-synonymous mutations, but analyses such as codon bias (Plotkin and Dushoff, 2003) cannot be performed without nucleotide information. The lack of nucleotide information also limits the ability of alignment programs to distinguish between two possible deletions in regions where two or more identical amino acids are adjacent to one another; this causes the spike variant Y144del to be identified as Y145del in some sequences of our analysis. In addition, the identity of ambiguous “X” codons cannot be fully elucidated without nucleotide data. Despite stringent filtering of sequences, ambiguous codons were still observed in some sequences.
Due to the lack of accompanying clinical data posted to the GISAID platform, it is possible that some samples may represent longitudinal samples taken from the same patient across different time points. If this is the case, rare variants in these samples may be over-represented. The time series data may also be influenced by reporting bias. A sudden influx of samples from a region where a variant is present may over-represent that variant, and variants may appear to become less common due to decreasing reporting of sequences from regions where they are common. European variants are likely over-represented in the time series data from August onward due to the very high number of samples collected in this region relative to others in the same time interval. Without extensive sequencing of samples worldwide, it is likely that there are functionally significant variants present that have not yet been discovered.
Future directions for study include segmenting the data by location as well as time to characterize regional trends in variation. The analyses performed in this study can be repeated for other SARS-CoV-2 proteins, and differing trends in variation by protein could be determined by normalizing variant frequencies to the length of each protein. Also, correlation analysis of variants can be performed to see which mutations happen concurrently, verified by identifying variants that co-occur on specific viral protein sequences. This would allow for preemptive identification of new strains consisting of multiple variants, which can be evaluated in follow-up studies for functional implications.
The following are the supplementary data related to this article.
Supplementary Table S1: Alignment corrections made to the spike protein alignment.
Supplementary Table S2: Table of all variants observed for the spike protein. The "Variant" column gives the combination of amino acid polymorphisms separated by commas, and the "Number_of_Sequences" column states the number of clustered sequences globally and accross all time points with the mutation combination. The variant "None" indicates sequences with no changes in the spike protein relative to the reference sequence WIV04.
Supplementary Table S3: Complete prevalence table for all spike protein variants over time, beginning on January 5th, 2020. Each week is indicated at the top of the columns on the x-axis, and amino acid variants are indicated on the rows of the y-axis. The global and region-specific prevalence is shown in each of tables S3A-S3G.
Supplementary Table S4: Table of all variants observed for Nsp7. The "Variant" column gives the combination of amino acid polymorphisms separated by commas, and the "Number_of_Sequences" column states the number of clustered sequences globally and accross all time points with the mutation combination. The variant "None" indicates sequences with no changes in Nsp7 relative to the reference sequence WIV04..
Supplementary Table S5: Table of all variants observed for Nsp8. The "Variant" column gives the combination of amino acid polymorphisms separated by commas, and the "Number_of_Sequences" column states the number of clustered sequences globally and accross all time points with the mutation combination. The variant "None" indicates sequences with no changes in Nsp8 relative to the reference sequence WIV04.
Supplementary Table S6: Table of all variants observed for Nsp12. The "Variant" column gives the combination of amino acid polymorphisms separated by commas, and the "Number_of_Sequences" column states the number of clustered sequences globally and accross all time points with the mutation combination. The variant "None" indicates sequences with no changes in Nsp12 relative to the reference sequence WIV04.
Supplementary Table S7: Complete prevalence table for all RNA-dependent RNA polymerase (RdRP) complex protein variants over time, beginning on January 5th, 2020. Each week is indicated at the top of the columns on the x-axis, and amino acid variations are indicated on the rows of the y-axis. The global and region-specific prevalence is shown in each of tables S7A-S7G.
Supplementary Table S8: List of Authors from the Originating laboratories responsible for obtaining the specimens, as well as the Submitting laboratories where the genome data were generated and shared via GISAID, on which this research is based. All Submitters of data may be contacted directly via www.gisaid.org. Authors are sorted alphabetically.
Supplementary Figs. S1-S29
Acknowledgments
Acknowledgements
We gratefully acknowledge the authors of the submitting laboratories to the GISAID portal, as well as the authors from the originating laboratories who obtained the sequences. The entire analysis is based on data from the GISAID portal (Supplementary Fig. S8). Many thanks to our colleagues, Cody Glickman, Elaine Epperson, Nabeeh Hasan, and Jo Hendrix, in the Strong Laboratory for their input and feedback. WS thanks the countless faculty and peers that have served as role models and furthered his learning. MS was supported by a Colorado Advanced Industries grant.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
None.
References
- Berman H.M. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell TA, Droettboom M, Lee A, Hunter J, Firing E, Andrade ESD, Hoffmann T, Stansby D, Klymak J, Varoquaux N, et al. matplotlib/matplotlib: REL: v3.2.2. Zenodo; 2020. https://zenodo.org/record/3898017. doi: 10.5281/ZENODO.3898017. [DOI]
- Cheng M.H., Zhang S., Porritt R.A., Noval Rivas M., Paschold L., Willscher E., Binder M., Arditi M., Bahar I. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc. Natl. Acad. Sci. 2020;117(41):25254. doi: 10.1073/pnas.2010722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., Solomon I.H., Kuo H.-H., Boucau J., Bowman K., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383(23):2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho R.M., Marquitti F.M.D., Ferreira L.S., Borges M.E., Paixão da Silva R.L., Canton O., Portella T.P., Poloni S., Franco C., Plucinski M.M., et al. Model-based estimation of transmissibility and reinfection of SARS-CoV-2 P.1 variant. Infect. Dis. 2021 doi: 10.1101/2021.03.03.21252706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Sarao R., Oliveira-dos-Santos A.J., Chappell M.C., Backx P.H., Yagil Y. Vol. 417. 2002. Angiotensin-Converting Enzyme 2 is an Essential Regulator of Heart Function; p. 7. [DOI] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538) doi: 10.1126/science.abg3055. (eabg3055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.-F., Smith T., Antonarakis S.E., Taschner P.E.M., et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum. Mutat. 2016;37(6):564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-González A., Glasner D.R., Reyes K.R., Gliwa A.S., et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021 doi: 10.1016/j.cell.2021.04.025. (Apr:S0092867421005055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deorowicz S., Debudaj-Grabysz A., Gudyś A. FAMSA: fast and accurate multiple sequence alignment of huge protein families. Sci. Rep. 2016;6(1):33964. doi: 10.1038/srep33964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edara V.-V., Lai L., Sahoo M.K., Floyd K., Sibai M., Solis D., Flowers M.W., Hussaini L., Ciric C.R., Bechnack S., et al. Infection and Vaccine-Induced Neutralizing-Antibody Responses to the SARS-CoV-2 B.1.617 Variants. N Engl J Med. 2021;385(7):664–666. doi: 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fang S., Liu S., Shen J., Lu A.Z., Zhang Y., Li K., Liu J., Yang L., Hu C.-D., Wan J. Updated SARS-CoV-2 single nucleotide variants and mortality association. Health Inform. 2021 doi: 10.1101/2021.01.29.21250757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., da Candido D.S., Mishra S., MAE Crispim, Sales F.C., Hawryluk I., JT McCrone, et al. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. Epidemiology. 2021 doi: 10.1101/2021.02.26.21252554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas C., Mella A., Haigh J.J. 2020. Large-scale population analysis of SARS-CoV-2 whole genome sequences reveals host-mediated viral evolution with emergence of mutations in the viral Spike protein associated with elevated mortality rates. [DOI] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St. Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021 doi: 10.1016/j.cell.2021.03.013. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MS, Sudre CH, May A, Antonelli M, Murray B, Varsavsky T, Kläser K, Canas LS, Molteni E, Modat M, et al. The effect of SARS-CoV-2 variant B.1.1.7 on symptomatology, re-infection and transmissibility:31.
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Crawford K.H.D., Bloom J.D., Veesler D., Vaughan T.G., Comas I., Candelas F.G., SeqCOVID-SPAIN Consortium, et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Epidemiology. 2020 doi: 10.1101/2020.10.25.20219063. [DOI] [Google Scholar]
- Hodcroft E.B., Domman D.B., Snyder D.J., Oguntuyo K.Y., Van Diest M., Densmore K.H., Schwalm K.C., Femling J., Carroll J.L., Scott R.S., et al. Emergence in late 2020 of multiple lineages of SARS-CoV-2 spike protein variants affecting amino acid position 677. Infect. Dis. 2021 doi: 10.1101/2021.02.12.21251658. [DOI] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9(3):90–95. doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
- Image of 6M17 (Yan, R. et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448 (2020)) created with PyMOL (The PyMOL Molecular Graphics System) Schrödinger, LLC; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Image of 6VSB (Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020)) created with PyMOL (The PyMOL Molecular Graphics System) Schrödinger, LLC; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Image of 7BV2 (Yin, W. et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368, 1499–1504 (2020)) created with PyMOL (The PyMOL Molecular Graphics System) Schrödinger, LLC; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J.J., Vasudevan K., Pragasam A.K., Gunasekaran K., Kang G., Veeraraghavan B., Mutreja A. Evolutionary tracking of SARS-CoV-2 genetic variants highlights intricate balance of stabilizing and destabilizing mutations. Genomics. 2020 doi: 10.1101/2020.12.22.423920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Hopkins University . Johns Hopkins Coronavirus Resource Center; 2020. COVID-19 Dashboard.https://coronavirus.jhu.edu/map.html [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluyver T., Ragan-Kelley B., Pérez F., Bussonnier M., Frederic J., Hamrick J., Grout J., Corlay S., Ivanov P., Abdalla S., et al. In: Positioning and Power in Academic Publishing: Players, Agents and Agendas. Loizides F., Schmidt B., editors. IOS Press; 2016. Jupyter notebooks—a publishing format for reproducible computational workflows; pp. 87–90. [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Rocklöv J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J. Travel Med. 2021;28(7) doi: 10.1093/jtm/taab124. (taab124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gayle A.A., Wilder-Smith A., Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J. Travel Med. 2020;27(2) doi: 10.1093/jtm/taaa021. (taaa021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;3 doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang Q., Wei P., Chen Z., Aviszus K., Yang J., Downing W., Peterson S., Jiang C., Liang B., et al. The basis of a more contagious 501Y.V1 variant of SARS-COV-2. Biochemistry. 2021 doi: 10.1101/2021.02.02.428884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. Efficacy of the ChAdOx1 nCoV-19 Covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2102214. Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola J.R., Graham B.S., Fauci A.S. SARS-CoV-2 viral variants—tackling a moving target. JAMA. 2021 doi: 10.1001/jama.2021.2088. https://jamanetwork.com/journals/jama/fullarticle/2776542 Feb 11. [DOI] [PubMed] [Google Scholar]
- McCallum M., Marco A.D., Lempp F., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Immunology. 2021 doi: 10.1101/2021.01.14.426475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc. Natl. Acad. Sci. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Á., Pongor S., Győrffy B. Different mutations in SARS-CoV-2 associate with severe and mild outcome. Int. J. Antimicrob. Agents. 2021;57(2) doi: 10.1016/j.ijantimicag.2020.106272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J.B., Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza a virus. Proc. Natl. Acad. Sci. 2003;100(12):7152–7157. doi: 10.1073/pnas.1132114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y., et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5(48) doi: 10.1126/sciimmunol.abc8413. (eabc8413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D., Volz E. COVID-19 Genomics Consortium UK. 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations - SARS-CoV-2 coronavirus / nCoV-2019 Genomic Epidemiology.https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369(6506):956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio . RStudio, PBC; Boston, MA: 2020. Integrated Development Environment for R.http://www.rstudio.com/ [Google Scholar]
- Sada M., Saraya T., Ishii H., Okayama K., Hayashi Y., Tsugawa T., Nishina A., Murakami K., Kuroda M., Ryo A., et al. Detailed molecular interactions of Favipiravir with SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza virus polymerases in silico. Microorganisms. 2020;8(10):1610. doi: 10.3390/microorganisms8101610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saline M., Rödström K.E.J., Fischer G., Orekhov Vyu, Karlsson B.G., Lindkvist-Petersson K. The structure of superantigen complexed with TCR and MHC reveals novel insights into superantigenic T cell activation. Nat. Commun. 2010;1(1) doi: 10.1038/ncomms1117. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance. 2017;22(13):30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soskine M., Tawfik D.S. Mutational effects and the evolution of new protein functions. Nat. Rev. Genet. 2010;11(8):572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer D.A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258(1):1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- Tada T., Dcosta B.M., Samanovic-Golden M., Herati R.S., Cornelius A., Mulligan M.J., Landau N.R. Convalescent-Phase Sera and Vaccine-Elicited Antibodies Largely Maintain Neutralizing Titer against Global SARS-CoV-2 Variant Spikes. mBio. 2021;12(3):e0069621. doi: 10.1128/mBio.00696-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. Epidemiology. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- The PyMOL Molecular Graphics System. Schrödinger, LLC; 2020. https://pymol.org/2/ [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021 doi: 10.1016/j.cell.2021.01.037. https://linkinghub.elsevier.com/retrieve/pii/S0092867421000805 Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H., Avenarius M.R., Kubatko L., Hunt M., Pan X., Ru P., Garee J., Thomas K., Mohler P., Pancholi P., et al. Distinct patterns of emergence of SARS-CoV-2 spike variants including N501Y in clinical samples in Columbus Ohio. Genomics. 2021 doi: 10.1101/2021.01.12.426407. [DOI] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034. Jan 1. [DOI] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.-Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181(4):894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Barnes C.O., Yang Z., Bjorkman P.J. SARS-CoV-2 lineage B.1.526 emerging in the New York region detected by software utility created to query the spike mutational landscape. Bioinformatics. 2021 doi: 10.1101/2021.02.14.431043. [DOI] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27(8):763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Werner A.P., Moliva J.I., Koch M., Choi A., Stewart-Jones G.B.E., Bennett H., Boyoglu-Barnum S., Shi W., Graham B.S., et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.01.25.427948. Jan 1. [DOI] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.-D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021 doi: 10.1001/jama.2021.1612. https://jamanetwork.com/journals/jama/fullarticle/2776543 Feb 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021 doi: 10.1016/j.cell.2021.02.037. (Feb:S0092867421002269) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Alignment corrections made to the spike protein alignment.
Supplementary Table S2: Table of all variants observed for the spike protein. The "Variant" column gives the combination of amino acid polymorphisms separated by commas, and the "Number_of_Sequences" column states the number of clustered sequences globally and accross all time points with the mutation combination. The variant "None" indicates sequences with no changes in the spike protein relative to the reference sequence WIV04.
Supplementary Table S3: Complete prevalence table for all spike protein variants over time, beginning on January 5th, 2020. Each week is indicated at the top of the columns on the x-axis, and amino acid variants are indicated on the rows of the y-axis. The global and region-specific prevalence is shown in each of tables S3A-S3G.
Supplementary Table S4: Table of all variants observed for Nsp7. The "Variant" column gives the combination of amino acid polymorphisms separated by commas, and the "Number_of_Sequences" column states the number of clustered sequences globally and accross all time points with the mutation combination. The variant "None" indicates sequences with no changes in Nsp7 relative to the reference sequence WIV04..
Supplementary Table S5: Table of all variants observed for Nsp8. The "Variant" column gives the combination of amino acid polymorphisms separated by commas, and the "Number_of_Sequences" column states the number of clustered sequences globally and accross all time points with the mutation combination. The variant "None" indicates sequences with no changes in Nsp8 relative to the reference sequence WIV04.
Supplementary Table S6: Table of all variants observed for Nsp12. The "Variant" column gives the combination of amino acid polymorphisms separated by commas, and the "Number_of_Sequences" column states the number of clustered sequences globally and accross all time points with the mutation combination. The variant "None" indicates sequences with no changes in Nsp12 relative to the reference sequence WIV04.
Supplementary Table S7: Complete prevalence table for all RNA-dependent RNA polymerase (RdRP) complex protein variants over time, beginning on January 5th, 2020. Each week is indicated at the top of the columns on the x-axis, and amino acid variations are indicated on the rows of the y-axis. The global and region-specific prevalence is shown in each of tables S7A-S7G.
Supplementary Table S8: List of Authors from the Originating laboratories responsible for obtaining the specimens, as well as the Submitting laboratories where the genome data were generated and shared via GISAID, on which this research is based. All Submitters of data may be contacted directly via www.gisaid.org. Authors are sorted alphabetically.
Supplementary Figs. S1-S29