Abstract
Objectives:
To develop and validate a deep learning algorithm to automatically detect and segment an orbital abscess depicted on computed tomography (CT).
Methods:
We retrospectively collected orbital CT scans acquired on 67 pediatric subjects with a confirmed orbital abscess in the setting of infectious orbital cellulitis. A context-aware convolutional neural network (CA-CNN) was developed and trained to automatically segment orbital abscess. To reduce the requirement for a large dataset, transfer learning was used by leveraging a pre-trained model for CT-based lung segmentation. An ophthalmologist manually delineated orbital abscesses depicted on the CT images. The classical U-Net and the CA-CNN models with and without transfer learning were trained and tested on the collected dataset using the 10-fold cross-validation method. Dice coefficient, Jaccard index, and Hausdorff distance were used as performance metrics to assess the agreement between the computerized and manual segmentations.
Results:
The context-aware U-Net with transfer learning achieved an average Dice coefficient and Jaccard index of 0.78±0.12 and 0.65±0.13, which were consistently higher than the classical U-Net or the context-aware U-Net without transfer learning (p < 0.01). The average differences of the abscess between the computerized results and the experts in terms of volume and Hausdorff distance were 0.10±0.11 mL and 1.94±1.21 mm, respectively. The context-aware U-Net detected all orbital abscess without false positives.
Conclusions:
The deep learning solution demonstrated promising performance in detecting and segmenting orbital abscesses on CT images in strong agreement with a human observer.
Keywords: orbital cellulitis, segmentation, computed tomography (CT), deep learning
I. INTRODUCTION
Pediatric orbital cellulitis is the most common sequela of acute sinusitis [1]. The public health impact of orbital cellulitis continues to be substantial. In 2009, orbital complications, such as the development of an orbital abscess (OA), accounted for 18,293 inpatient hospital stays, $10 million in hospital charges, and a shifting trend toward a higher proportion of children undergoing surgical treatment [2]. Its complications can be devastating, with the risk of blindness from orbital compartment pressure and central nervous system spread, including cavernous sinus thrombosis, meningitis, and intracranial abscess formation [3-7]. Examination of a pediatric patient with orbital cellulitis is limited by their age, acute illness, and neurological status; therefore, there is a heavy reliance on computed tomography (CT) scans to determine the site and extent of infection [8, 9]. Subperiosteal orbital abscess (SPOA), a collection of purulence between the periorbita and bone of the adjacent orbit, when presenting with visual impairment secondary to elevation in orbital pressure is an indication for emergent surgical drainage.
Currently, in clinical practice, there does not appear to be a uniform approach to assess the severity and treatment of orbital abscesses in pediatric patients [10, 11]. Clinicians typically assess the presence and severity of an orbital abscess by visual inspection and manual measurement, which is a subjective process that can lead to different treatment decisions between clinicians. An orbital abscess appears as a very low-density region depicting fluid and often surrounded by a hyperdense peripheral ring enhancement (Fig. 1), making it challenging to visually perceive and differentiate it from surrounding tissues or structures. Most retrospective studies unsurprising reported that medically treated patients had smaller orbital abscesses compared to surgically treated patients [9, 12-19]. However, size was not uniformly quantified across these studies and subjectively or objectively quantified based on a clinician’s impression [20], linear dimensions [9, 13, 15, 16], volume using a bounding box approach [12, 15, 16, 18], or volume assuming an ellipsoid [13, 14, 17, 19]. The lack of a standardized approach to quantify abscess size hinders the scientific evaluation of the impact of abscess size on the treatment approach.
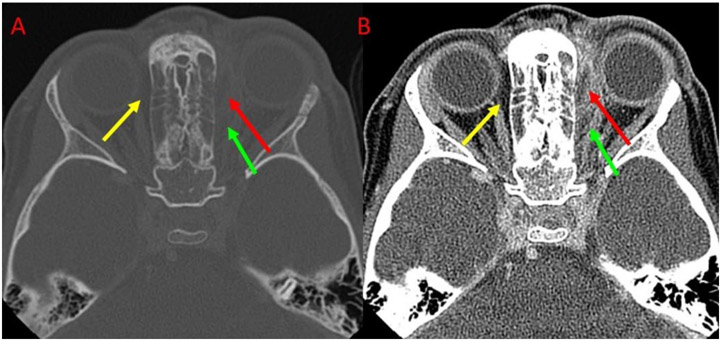
Fig 1.
An example illustrating the fuzzy appearance of an orbital abscess depicted on CT images. Red arrows indicate the orbital abscess. Green and yellow arrows indicate regions that have a similar appearance but are not orbital abscesses. (A) and (B) showed the images at different window/level values.
The use of artificial intelligence (AI) in the assessment and management of orbital abscesses would be a significant step to standardizing abscess assessment. To our knowledge, no one has investigated the application of AI to assess abscess severity or for deciding the treatment approach. An automated AI approach to detect and quantify an orbital abscess would facilitate and perhaps decrease the variability of diagnosing and treating pediatric patients with orbital abscesses. As compared with the manual delineation, the use of computerized schemes may enable a more accurate, objective, and efficient quantification of an orbital abscess and ultimately improve pediatric care. As demonstrated by other investigations [21-24], a computer approach may potentially reduce the inter- and intra-reader variability in assessing and quantifying diseases.
We developed and validated an artificial intelligence (AI) approach, namely context-aware convolutional neural network (CA-CNN), to automate the process of segmenting and quantifying orbital abscess features. The underlying idea is motivated by the fact that an orbital abscess typically occurs in the regions between the periorbita and bone of the adjacent orbit. To leverage this knowledge, we combined two sets of image patches with different resolutions to identify an orbital abscess. One with a lower image resolution but a wider field-of-view was used to emphasize and learn the context regions of the orbital abscess. The other one with a higher image resolution but a smaller field-of-view was used to characterize the details of the orbital abscess. In addition, a pre-trained CNN model for lung segmentation was used for transfer learning to alleviate the large data requirement of the deep learning technology. The feasibility and performance of this scheme were evaluated on a dataset of orbital CT scans acquired on 67 pediatric subjects using the 10-fold cross-validation method.
II. MATERIALS AND METHODS
A. The dataset
The dataset included 67 pediatric subjects who were diagnosed with an orbital abscess, underwent CT examinations, and treated at the Children’s Hospital of the University of Pittsburgh Medical Center (UPMC) from 2005 to 2016 (Table 1). None of the subjects had bilateral orbital abscesses. Four had facial trauma or fractures. This study was approved by the University of Pittsburgh Institutional Review Board (STUDY20030023). Written informed consent was waived because all data were de-identified by an honest broker prior to dissemination to the investigators.
Table 1:
Subject demographics and subgroups (n=67)
| Age, year (SD) | 6.0±3.98 |
| Male, n (%) | 54 (80.6) |
| Laterality | |
| Right, n (%) | 32 (47.8) |
| Left, n (%) | 35 (52.2) |
| Bilateral, n (%) | 0 (0) |
| Abscess location (AL medial) (%) | 63 (94.0) |
| Facial trauma / fractures, n (%) | 4 (6.0) |
| Hospitalization, days | 4.0±1.44 |
Abbreviations: SD – standard deviation
The orbital CT examinations were acquired on a CT scanner (LightSpeed VCT, GE Healthcare, Waukesha, WI, USA) with the use of radiopaque contrast. Scans were acquired using a helical technique at the following parameters: 32×0.625 mm detector configuration, 0.531 pitch, 120 kVp tube energy, and 105~140 mA tube current. Images were reconstructed to encompass the orbit field in a 512×512 pixel matrix. The slice thickness ranged from 0.625 to 1.25 mm, and the in-plane pixel dimensions ranged from 0.292 to 0.323 mm.
A board-certified, experienced ophthalmologist specializing in orbital surgery reviewed the CT scans and manually outlined the orbital abscesses depicted on the CT images in the cohort. The ophthalmologist viewed and outlined the entire abscesses on a standard personal computer and monitor. The user was able to adjust the magnification and window/level settings as necessary. After outlining the abscess on CT images, the outlines were saved using in-house software. The user was blind to any information related to the subject (e.g., medical intervention or surgical outcome).
B. context-aware convolutional neural network (CA-CNN)
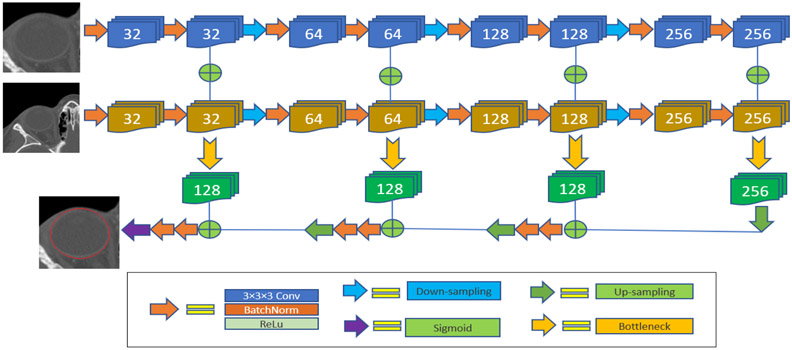
On the basis of the classical U-Net [25, 26], a four-stage context-aware deep learning architecture was developed for image segmentation (Fig. 2). This architecture is formed by two encoder pathways with progressive down-samplings and one decoder pathway with progressive up-sampling. The first encoder pathway accepts a three-dimensional (3-D) sub-volume image patch with a higher resolution but smaller field-of-view (FOV). The second encoder pathway accepts a sub-volume image patch with a lower resolution but larger FOV to leverage the knowledge that an orbital abscess typically occurs in the regions between the periorbita and bone of the adjacent orbit. Each stage of the encoder path was composed of two consecutive 3×3×3 convolution operations with batch normalization and rectified linear unit (ReLu) activation. The neighboring stages were connected by a 2×2×2 down-sampling operation. Each stage of the decoder path was composed of two consecutive convolutions on the output of the bottleneck layers, and the neighboring stages were connected by 2×2×2 up-sampling. The initial number of the convolutional filters was set at 32 and increased by two times progressively in the following stages. The output layer was activated by a sigmoid function to obtain the final predictions of the ROI and its background. Adam optimizer was used to update network weights during the training. The soft Dice coefficient was used as the loss function (Eq. (1)).
| (1) |
where N is the total number of voxels, pi is the predicted probability of a voxel to be part of the regions-of-interest, and gi is the binary value of a voxel that belongs to the manual segmentation. The soft Dice coefficient was originally proposed by Milletari et al. [27] to establish the balance between foreground and background voxels.
Fig. 2:
The architecture of the developed context-aware U-Net.
The developed multi-scale architecture enables the network to learn the features at different scales and utilize the contextual information to improve the segmentation. However, this dual-pathway architecture requires a high computational cost in both memory and efficiency. To alleviate this problem, bottleneck layers were used to reduce the number of feature parameters.
The training and internal validation subgroups were used to train the CNN architecture by performing the following procedures:
(1) An entire CT scan cannot be processed by the GPU for deep learning due to the memory limit of the graphical processing unit (GPU). As a solution, the volumetric CT scans were subdivided into a number of three-dimensional (3D) patches (e.g., 96×96×96 voxels). The CT images were made isotropic with each voxel having a uniform dimension of 0.5 mm3. In the implementation, we randomly sampled the 3D patches on the CT images. The patches with low and high resolutions were centered on the same locations. Since the patches are randomly sampled from the 3D CT images, the patches with low resolution do not always fully enclose the regions between the periorbita and bone of the adjacent orbit. This typically happens when the patches with high resolution are located around the abscess.
(2) A pre-trained model for CT-based lung segmentation was used for transfer learning. This pre-trained model was based on the developed context-aware U-Net and trained on a dataset consisting of more than 100 chest CT scans acquired using different image protocols [28]. Transfer learning can improve the learning efficiency and reduce the requirement for a large and diverse dataset [29-31].
(3) The orbital CT images were augmented on the fly by randomly performing various affine transformation and image filtering operations alone and in combination. The affine transformations included rotation, translation, flip, and scaling. The image filtering included image smoothing, Gaussian noise addition, and intensity shifts. These image augmentations increased the diversity of the images by generating a large number of variations of the original images. The usage of the data augmentation can not only avoid overfitting but also improve the robustness of the trained models [32, 33]. The on-the-fly data augmentation will automatically assure the training performance. If the performance of a network can be improved further via data augmentation, this improvement will be reflected in a decreasing validation loss in the following epochs. A detailed description and its impact on the performance of the trained models can be found in a survey [34].
(4) During the training, 64 volumetric patches were randomly from the foreground and the background of each CT scan in one training step. The initial learning rate was set at 0.001. If the validation performance did not increase in two epochs, the learning rate would be reduced by a factor of 0.5. The training would stop if the validation performance of the current epoch did not improve compared to the previous 10 epochs.
C. Performance evaluation
The 10-fold cross-validation method was used to train and evaluate the performance of the developed models. We firstly shuffled the dataset (n=67) and then split them into 10 folds, among which eight folds had seven cases, one fold had six cases, and one fold had 5 cases. When training the models, we chose eight folds for training, one fold for the interval validation, and one fold for the independent test. We repeated this procedure ten times so that each sample was observed in the training and testing.
The performance of the CNN models to identify orbital abscesses was compared to the manually outlined regions using the independent test set. A threshold of 0.5 was used to binarize the computerized results. The regular Dice coefficient (Eq. 2), Jaccard index (JI) (Eq. 3), and the Hausdorff distance (Eq. 4) were computed as the performance metrics to assess the agreement between the computer and the human observer.
| (2) |
| (3) |
| (4) |
where A and B are two different regions (i.e., the outlines by the computer and the human observer).
We evaluated the impact of transfer learning by training the context-aware U-Net architecture with and without the pre-trained model and comparing their performance on the same test sets. The segmentation performance of the context-aware U-Net architecture was also compared to the classical U-Net architecture. For a fair comparison, we trained the three models using the same set of parameters (e.g., learning rate and optimizer). A paired-samples t-test was performed to assess the performance of the segmentation approaches. A p-value of less than 0.05 was considered statistically significant. IBM SPSS v14 was used for the statistical analyses.
III. RESULTS
The agreement for orbital abscesses segmentation between manual segmentation and the computer methods was significantly stronger for the context-aware U-Net with transfer learning compared to the context-aware without transfer learning or the classical U-Net (Table 2 and Fig. 3). Several examples were randomly selected and presented in Fig.3 and Fig. 4 to visually demonstrate the segmentation performance of the developed CNN models. Visual inspection revealed a reasonable agreement between the computer and manual segmentation (Figs. 3-4). The average Dice coefficient, Jaccard index, and Hausdorff distance were 0.78±0.12, 0.65±0.13, and 1.94±1.21 mm, respectively, for comparing the context-aware U-Net algorithm segmentation to the manual segmentation. Among the three models, the context-aware U-Net with the transfer learning demonstrated the best performance (p<0.001), while the performance of the context-aware U-Net without transfer learning was better than the classical U-Net (p=0.036) (Table 2). The context-aware U-Net detected all orbital abscesses regardless of the utilization of transfer learning or not. In contrast, the classical U-Net missed the detection of a small orbital abscess (Fig. 5).
Table 2.
The performance of the CNN models in identifying orbital abscess depicted on CT images based on the 10-fold cross-validation method.
| Case | Dice Coefficient | Jaccard Index | Hausdorff distance | Volume difference |
|---|---|---|---|---|
| M-1 | 0.78±0.12 | 0.65±0.13 | 1.94±1.21 mm | 0.10±0.11 mL |
| M-2 | 0.63±0.19 | 0.47±0.18 | 3.11±1.40 mm | 0.15±0.15 mL |
| M-3 | 0.61±0.21 | 0.48±0.19 | 3.21±1.66 mm | 0.17±0.18 mL |
M-1: the context-aware U-Net with transfer learning
M-2: the context-aware U-Net without transfer learning
M-3: the classical U-Net. (The missed orbital abscess was not included in the computations)
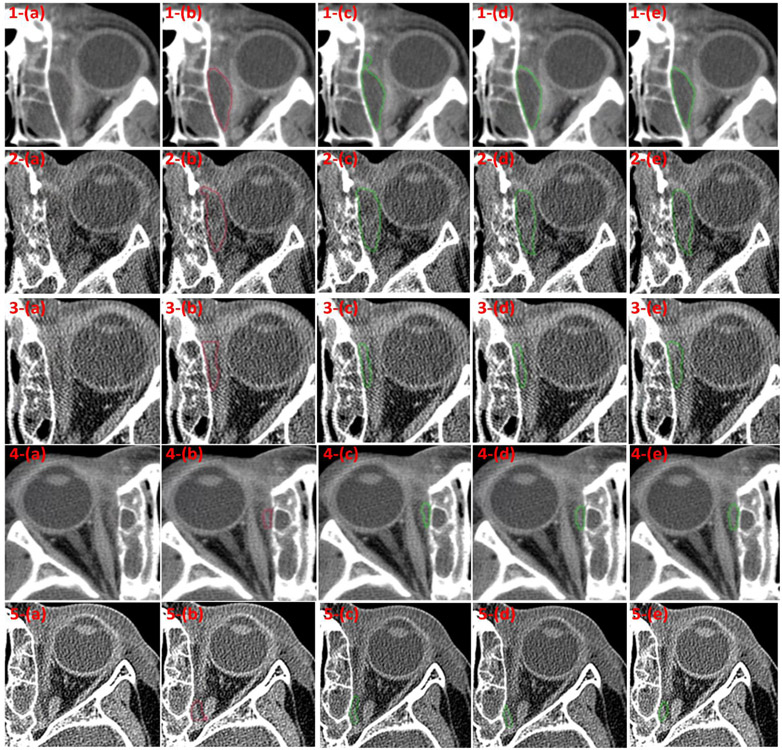
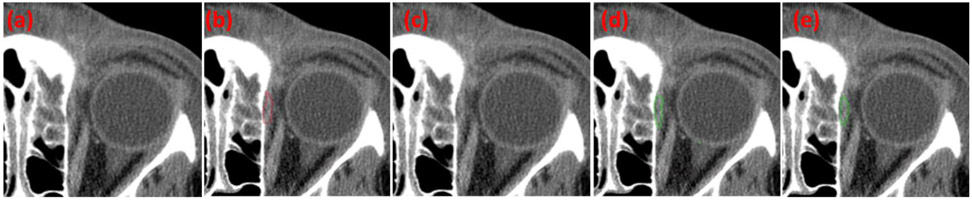
Fig. 3.
Some examples with various sizes of orbital abscesses showing good agreement between the manual outlines and the results obtained by the context-aware U-Net model with transfer learning. (a) the original CT images, (b) the manual outlines by the ophthalmologist, (c) the segmentations by the classical U-Net model, (d) the segmentations by the context-aware U-Net model without transfer learning, and (e) the segmentations by the context-aware U-Net model with transfer learning.
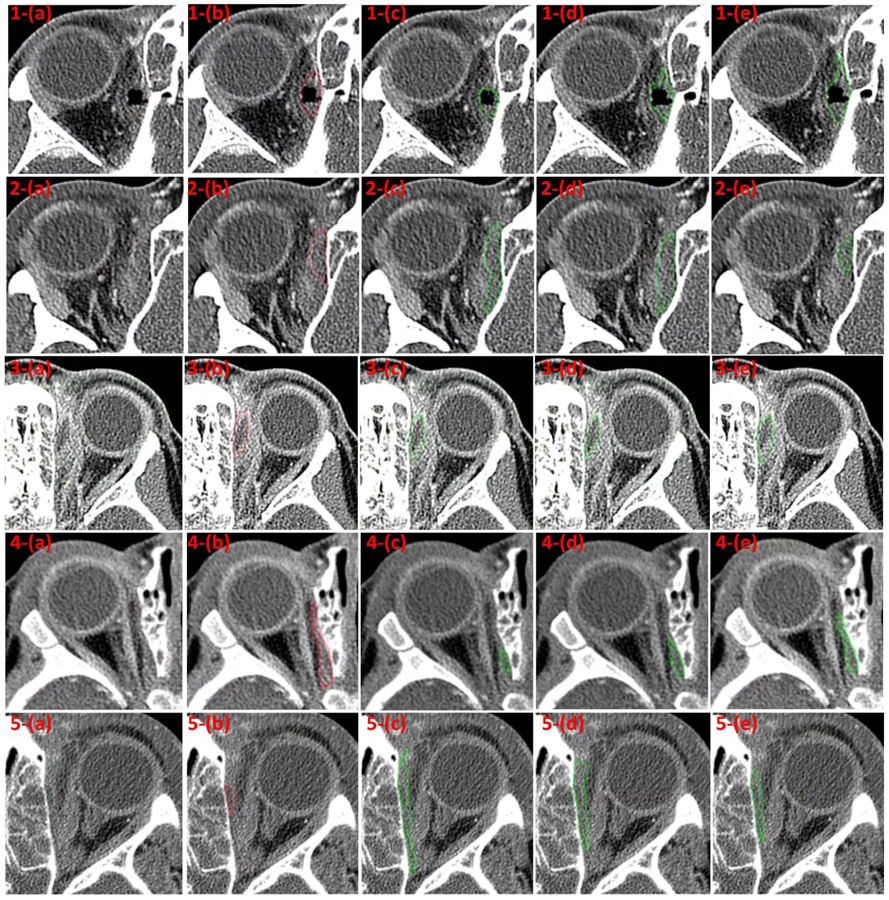
Fig. 4.
Some examples with moderate (row: 1-3) and poor (row: 4-5) agreement between the manual outlines and the results by the context-aware U-Net model with transfer learning. (a) the original CT image, (b) the manual outlines by the ophthalmologist, (c) the segmentations by the classical U-Net model, (d) the segmentations by the context-aware U-Net model without transfer learning, and (e) the segmentations by the context-aware U-Net model with transfer learning.
Fig. 5.
The case with a small orbital abscess missed by the classical U-Net model but well segmented by the context-aware U-Net. (a) the original CT image, (b) the manual outline by the ophthalmologist, (c) the segmentation (no result) by the classical U-Net model, (d) the segmentation by context-aware U-Net model without transfer learning, and (e) the segmentation by the context-aware U-Net model with transfer learning.
The average orbital abscess volumes computed from the manual outlines were 0.65±0.49 mL with a range from 0.05 mL to 1.4 mL. The average orbital abscess volume computed by the context-aware U-Net with context-aware learning was 0.59±0.51 mL with a range from 0.05 mL to 2.25 mL, as compared with 0.61±0.52 mL with a range from 0.01 mL to 2.2 mL for the CA-CNN without transfer learning and 0.52±0.47 mL with a range from 0.01 mL to 2.2 mL for the classical U-Net. The absolute volume differences between the manual outlines and the CA-CNN algorithm were 0.10±0.11 mL. These differences were smaller than the CA-CNN without transfer learning (0.15±0.15 mL), and the classical U-Net (0.17±0.18 mL).
The training time when transfer learning was implemented was reduced from ~15 hours to ~9 hours compared to without transfer learning on our workstation (Intel Xeon CPU 3.60GHz, Nvidia Titan Xp 12GB). It took less than 30 seconds to process an orbital CT scan and identify the orbital abscesses.
IV. DISCUSSION
Pediatric patients presenting with signs and symptoms of orbital cellulitis need an immediate and thorough evaluation. Once a diagnosis of an orbital abscess is confirmed, the decision to treat the patient with a medical or surgical approach is critical and extremely time-sensitive. These patients often require continuous monitoring, especially if medical treatment is initiated. To improve and possibly expedite the treatment decision, investigators have recommended including abscess size in the decision-making process. However, there are challenges and variations related to timely abscess measurement. Manually outlining the abscess on all the images depicting the abscess can be extremely time-consuming and variable across clinicians. To our knowledge, no investigative effort has been made to automate this process.
Our AI approach to automatically segment and quantify an orbital abscess in pediatric patients demonstrated very promising results. The results suggested that deep learning technology can be utilized to address the challenging problem. The unique characteristic of our approach is the utilization of contextual information and transfer learning. The former explores the contextual information to improve the segmentation, and the latter significantly reduces the requirement of a large dataset and improves training efficiency. This AI approach makes it relatively straightforward to quantify the features (e.g., volume and density) of an orbital abscess. To our knowledge, this is the first investigation that used AI to automate the segmentation and quantification of an orbital abscess depicted on CT images.
In our cohort (n=67), the orbital abscess volumes measured by the manual outline ranged from 0.045 mL to 2.69 mL with an average 0.64±0.63 mL. In the independent test set, the difference between the manual outline and the best performing model was 0.10±0.11 mL. The average Dice coefficient, Jaccard index, and Hausdorff distance were 0.78±0.12, 0.65±0.13, and 1.94±1.21 mm, respectively. As demonstrated by the average Hausdorff distance, which measures how far two outlines are from each other, there was a reasonable agreement between the computerized outlines and the manual outlines. Notably, the relatively small dimension of an orbital abscess could result in lower than expected Dice coefficients and Jaccard indices, because a small difference between the computer and manual segmentations may lead to obviously low values in the Dice coefficient and the Jaccard index. Given the fuzzy appearance and small size of an orbital abscess, we believe that the underlying differences between the manual and automated segmentation are limited and most likely will not significantly affect the clinical implementation of the developed algorithm.
The models based on the context-aware U-Net successfully identified all of the orbital abscesses without any false positive detection. In contrast, the classical U-Net failed to detect a small orbital abscess, which appeared as a region with relatively low contrast on the CT images (Fig. 5). Hence, the context-aware architecture can somewhat improve the performance of the CNN-based segmentation. As our experiments demonstrated, the utilization of the transfer learning technology significantly improved the performance of the context-aware U-Net, especially when the size of the dataset for machine learning is limited. Notably, the utilization of transfer learning cannot guarantee the best performance of a CNN model. Since CNN-based deep learning is data-hungry, we believe that the segmentation performance can be further improved when additional data is available.
In transfer learning, a pre-trained model from a different task is used as the starting point of the training for the problem under investigation. Typically, the more related the tasks, the easier it is to transfer or cross-utilize the knowledge. If the network architecture of a pre-trained model is the same as the network architecture of the current problem, the pre-trained model can be directly applied as the starting point of the training on the new dataset. Otherwise, we may need to extract the first layers in the pre-trained model and keep the trained weight and then add additional layers specific to the new CNN model. If the network architectures for two tasks are completely different, it is unreasonable to use transfer. Since the weights of many layers in the pre-trained model are learned from a previous task and already available, the training performance, including the accuracy and efficiency, can be improved significantly even when the target task has limited data for machine learning. In our implementation, we trained the developed context-aware U-Net using a dataset from a different task described in [28]. The pre-trained model was used directly as the starting point of the training for segmenting abscess.
We are aware of the limitations with this study. First, the orbital CT scans were retrospectively collected from a single institution and acquired using similar CT protocols. The extensive image augmentation implemented during the training process may have addressed some of the differences in image acquisition protocols. Second, the dataset was small for both training and validation. At this moment, it is difficult to collect a large dataset for this specific problem. Hence, we used transfer learning to improve the performance and the 10-fold cross-validation method to evaluate and compare the performance of the trained models. We believe that our preliminary results demonstrate the potential of deep learning technology to assess important clinical features of an orbital abscess. Third, it is always desirable to develop novel CNN architectures to maximize the performance with a limited dataset. Since no investigative effort has been made to automatically identify the orbital abscess depicted on CT images, it is prudent to first demonstrate the feasibility in this regard, especially given the very fuzzy appearance of an orbital abscess. Finally, we did not assess the clinical impact of the difference in orbital abscess measurements between the manual and computer segmentation. At this time, we did not evaluate intra- and inter-reader agreement because we are in the development stage and trained on the output of a single reader. Our primary focus in this study was to develop and validate the potential of the deep learning algorithm for automated segmentation of orbital abscesses in a small dataset.
V. CONCLUSION
We presented an artificial intelligence approach to automate the segmentation and quantification of orbital abscess in pediatric patients depicted on CT images. We used a pre-trained lung segmentation model to implement transfer learning in the CNN architecture. Our abscess segmentation results demonstrated the feasibility and advantages of the proposed CNN architecture for providing a clinically useful assessment of patients presenting with an orbital abscess.
ACKNOWLEDGEMENT
This work is supported by the National Institutes of Health (NIH) (Grant No. R01CA237277) and the UPMC Hillman Developmental Pilot Program. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Nvidia Titan Xp GPU for this research.
Footnotes
Disclosure Statement:
The authors have no conflicts of interest to declare.
REFERENCES
- 1.Wald ER, Applegate KE, Bordley C, Darrow DH, Glode MP, Marcy SM, Nelson CE, Rosenfeld RM, Shaikh N, Smith MJ, Williams PV, Weinberg ST, American Academy of P. Clinical practice guideline for the diagnosis and management of acute bacterial sinusitis in children aged 1 to 18 years. Pediatrics. 2013;132(1):e262–80. Epub 2013/06/26. doi: 10.1542/peds.2013-1071. PubMed PMID: 23796742. [DOI] [PubMed] [Google Scholar]
- 2.Capra G, Liming B, Boseley ME, Brigger MT. Trends in orbital complications of pediatric rhinosinusitis in the United States. JAMA Otolaryngol Head Neck Surg. 2015;141(1):12–7. Epub 2014/10/24. doi: 10.1001/jamaoto.2014.2626. PubMed PMID: 25340955. [DOI] [PubMed] [Google Scholar]
- 3.Wald ER, Pang D, Milmoe GJ, Schramm VL Jr., Sinusitis and its complications in the pediatric patient. Pediatr Clin North Am. 1981;28(4):777–96. Epub 1981/11/01. doi: 10.1016/s0031-3955(16)34065-2. PubMed PMID: 7312451. [DOI] [PubMed] [Google Scholar]
- 4.Davis JP, Stearns MP. Orbital complications of sinusitis: avoid delays in diagnosis. Postgrad Med J. 1994;70(820):108–10. Epub 1994/02/01. doi: 10.1136/pgmj.70.820.108. PubMed PMID: 8170879; PMCID: PMC2397650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagenmann M, Naclerio RM. Complications of sinusitis. J Allergy Clin Immunol. 1992;90(3 Pt 2):552–4. Epub 1992/09/01. doi: 10.1016/0091-6749(92)90184-4. PubMed PMID: 1527349. [DOI] [PubMed] [Google Scholar]
- 6.Moloney JR, Badham NJ, McRae A. The acute orbit. Preseptal (periorbital) cellulitis, subperiosteal abscess and orbital cellulitis due to sinusitis. J Laryngol Otol Suppl. 1987;12:1–18. Epub 1987/01/01. PubMed PMID: 3471826. [PubMed] [Google Scholar]
- 7.Swift AC, Charlton G. Sinusitis and the acute orbit in children. J Laryngol Otol. 1990;104(3):213–6. Epub 1990/03/01. doi: 10.1017/s0022215100112319. PubMed PMID: 2187942. [DOI] [PubMed] [Google Scholar]
- 8.Rudloe TF, Harper MB, Prabhu SP, Rahbar R, Vanderveen D, Kimia AA. Acute periorbital infections: who needs emergent imaging? Pediatrics. 2010;125(4):e719–26. Epub 2010/03/03. doi: 10.1542/peds.2009-1709. PubMed PMID: 20194288. [DOI] [PubMed] [Google Scholar]
- 9.Ryan JT, Preciado DA, Bauman N, Pena M, Bose S, Zalzal GH, Choi S. Management of pediatric orbital cellulitis in patients with radiographic findings of subperiosteal abscess. Otolaryngol Head Neck Surg. 2009;140(6):907–11. Epub 2009/05/27. doi: 10.1016/j.otohns.2009.02.014. PubMed PMID: 19467413. [DOI] [PubMed] [Google Scholar]
- 10.Adil EA, Muir ME, Kawai K, Dombrowski ND, Cunningham MJ. Pediatric Subperiosteal Abscess Secondary to Acute Sinusitis: A Systematic Review and Meta-analysis. Laryngoscope. 2020;130(12):2906–12. Epub 2020/02/18. doi: 10.1002/lary.28570. PubMed PMID: 32065412. [DOI] [PubMed] [Google Scholar]
- 11.Appachi S, Bedwell JR. Predictors of surgical management for paediatric orbital subperiosteal abscesses. Curr Opin Otolaryngol Head Neck Surg. 2020;28(6):430–4. Epub 2020/10/08. doi: 10.1097/MOO.0000000000000668. PubMed PMID: 33027139. [DOI] [PubMed] [Google Scholar]
- 12.Todman MS, Enzer YR. Medical management versus surgical intervention of pediatric orbital cellulitis: the importance of subperiosteal abscess volume as a new criterion. Ophthalmic Plast Reconstr Surg. 2011;27(4):255–9. Epub 2011/03/19. doi: 10.1097/IOP.0b013e3182082b17. PubMed PMID: 21415801. [DOI] [PubMed] [Google Scholar]
- 13.Gavriel H, Yeheskeli E, Aviram E, Yehoshua L, Eviatar E. Dimension of subperiosteal orbital abscess as an indication for surgical management in children. Otolaryngol Head Neck Surg. 2011;145(5):823–7. Epub 2011/07/23. doi: 10.1177/0194599811416559. PubMed PMID: 21778515. [DOI] [PubMed] [Google Scholar]
- 14.Le TD, Liu ES, Adatia FA, Buncic JR, Blaser S. The effect of adding orbital computed tomography findings to the Chandler criteria for classifying pediatric orbital cellulitis in predicting which patients will require surgical intervention. J AAPOS. 2014;18(3):271–7. Epub 2014/06/14. doi: 10.1016/j.jaapos.2014.01.015. PubMed PMID: 24924283. [DOI] [PubMed] [Google Scholar]
- 15.Nation J, Lopez A, Grover N, Carvalho D, Vinocur D, Jiang W. Management of Large-Volume Subperiosteal Abscesses of the Orbit: Medical vs Surgical Outcomes. Otolaryngol Head Neck Surg. 2017;157(5):891–7. Epub 2017/08/30. doi: 10.1177/0194599817728490. PubMed PMID: 28849710. [DOI] [PubMed] [Google Scholar]
- 16.Oxford LE, McClay J. Medical and surgical management of subperiosteal orbital abscess secondary to acute sinusitis in children. Int J Pediatr Otorhinolaryngol. 2006;70(11):1853–61. Epub 2006/08/15. doi: 10.1016/j.ijporl.2006.05.012. PubMed PMID: 16905200. [DOI] [PubMed] [Google Scholar]
- 17.Quintanilla-Dieck L, Chinnadurai S, Goudy SL, Virgin FW. Characteristics of superior orbital subperiosteal abscesses in children. Laryngoscope. 2017;127(3):735–40. Epub 2016/06/14. doi: 10.1002/lary.26082. PubMed PMID: 27291943. [DOI] [PubMed] [Google Scholar]
- 18.Rahbar R, Robson CD, Petersen RA, DiCanzio J, Rosbe KW, McGill TJ, Healy GB. Management of orbital subperiosteal abscess in children. Arch Otolaryngol Head Neck Surg. 2001;127(3):281–6. Epub 2001/03/20. doi: 10.1001/archotol.127.3.281. PubMed PMID: 11255472. [DOI] [PubMed] [Google Scholar]
- 19.Tabarino F, Elmaleh-Berges M, Quesnel S, Lorrot M, Van Den Abbeele T, Teissier N. Subperiosteal orbital abscess: volumetric criteria for surgical drainage. Int J Pediatr Otorhinolaryngol. 2015;79(2):131–5. Epub 2014/12/23. doi: 10.1016/j.ijporl.2014.11.021. PubMed PMID: 25529362. [DOI] [PubMed] [Google Scholar]
- 20.Garcia GH, Harris GJ. Criteria for nonsurgical management of subperiosteal abscess of the orbit: analysis of outcomes 1988-1998. Ophthalmology. 2000;107(8):1454–8. Epub 2000/08/02. doi: 10.1016/s0161-6420(00)00242-6. PubMed PMID: 10919887. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Yu J, Zhu Q, Li S, Zhao Z, Yang B, Pu J. Potential of deep learning in assessing pneumoconiosis depicted on digital chest radiography. Occup Environ Med. 2020;77(9):597–602. Epub 2020/05/31. doi: 10.1136/oemed-2019-106386. PubMed PMID: 32471837. [DOI] [PubMed] [Google Scholar]
- 22.Zhen Y, Chen H, Zhang X, Meng X, Zhang J, Pu J. Assessment of Central Serous Chorioretinopathy Depicted on Color Fundus Photographs Using Deep Learning. Retina. 2020;40(8):1558–64. Epub 2019/07/10. doi: 10.1097/IAE.0000000000002621. PubMed PMID: 31283737. [DOI] [PubMed] [Google Scholar]
- 23.Tibrewala R, Ozhinsky E, Shah R, Flament I, Crossley K, Srinivasan R, Souza R, Link TM, Pedoia V, Majumdar S. Computer-Aided Detection AI Reduces Interreader Variability in Grading Hip Abnormalities With MRI. J Magn Reson Imaging. 2020;52(4):1163–72. Epub 2020/04/16. doi: 10.1002/jmri.27164. PubMed PMID: 32293775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heijman E, Aben JP, Penners C, Niessen P, Guillaume R, van Eys G, Nicolay K, Strijkers GJ. Evaluation of manual and automatic segmentation of the mouse heart from CINE MR images. J Magn Reson Imaging. 2008;27(1):86–93. Epub 2007/12/01. doi: 10.1002/jmri.21236. PubMed PMID: 18050352. [DOI] [PubMed] [Google Scholar]
- 25.Ronneberger O, Fischer P, Brox T, editors. U-Net: Convolutional Networks for Biomedical Image Segmentation. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015; 2015. 2015//; Cham: Springer International Publishing. [Google Scholar]
- 26.Falk T, Mai D, Bensch R, Cicek O, Abdulkadir A, Marrakchi Y, Bohm A, Deubner J, Jackel Z, Seiwald K, Dovzhenko A, Tietz O, Dal Bosco C, Walsh S, Saltukoglu D, Tay TL, Prinz M, Palme K, Simons M, Diester I, Brox T, Ronneberger O. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16(1):67–70. Epub 2018/12/19. doi: 10.1038/s41592-018-0261-2. PubMed PMID: 30559429. [DOI] [PubMed] [Google Scholar]
- 27.Milletari F, Navab N, Ahmadi S-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation 2016. 565–71 p. [Google Scholar]
- 28.Pu J, Leader JK, Bandos A, Ke S, Wang J, Shi J, Du P, Guo Y, Wenzel SE, Fuhrman CR, Wilson DO, Sciurba FC, Jin C. Automated quantification of COVID-19 severity and progression using chest CT images. Eur Radiol. 2020. Epub 2020/08/14. doi: 10.1007/s00330-020-07156-2. PubMed PMID: 32789756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Opbroek A, Ikram MA, Vernooij MW, de Bruijne M. Transfer learning improves supervised image segmentation across imaging protocols. IEEE Trans Med Imaging. 2015;34(5):1018–30. Epub 2014/11/07. doi: 10.1109/TMI.2014.2366792. PubMed PMID: 25376036. [DOI] [PubMed] [Google Scholar]
- 30.Van Opbroek A, Achterberg HC, Vernooij MW, De Bruijne M. Transfer Learning for Image Segmentation by Combining Image Weighting and Kernel Learning. IEEE Trans Med Imaging. 2019;38(1):213–24. Epub 2018/07/27. doi: 10.1109/TMI.2018.2859478. PubMed PMID: 30047874. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, MacKinnon T. Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Clin Radiol. 2018;73(5):439–45. Epub 2017/12/23. doi: 10.1016/j.crad.2017.11.015. PubMed PMID: 29269036. [DOI] [PubMed] [Google Scholar]
- 32.Sandfort V, Yan K, Pickhardt PJ, Summers RM. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci Rep. 2019;9(1):16884. Epub 2019/11/16. doi: 10.1038/s41598-019-52737-x. PubMed PMID: 31729403; PMCID: PMC6858365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hussain Z, Gimenez F, Yi D, Rubin D. Differential Data Augmentation Techniques for Medical Imaging Classification Tasks. AMIA Annu Symp Proc. 2017;2017:979–84. Epub 2018/06/02. PubMed PMID: 29854165; PMCID: PMC5977656. [PMC free article] [PubMed] [Google Scholar]
- 34.Shorten C, Khoshgoftaar TM. A survey on Image Data Augmentation for Deep Learning. Journal of Big Data. 2019;6(1):60. doi: 10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]