Highlights
-
•
LC-MS methods are used to identify intact OXA-48 like enzymes from bacterial lysates.
-
•
Individual protein variants of the OXA-48 like carbapenemases can be detected with tandem mass spectrometry.
-
•
OXA-48 like carbapenemases can be distinguished from extended spectrum β-lactamases like CTX-M-15.
-
•
The method for detection can be run in five minutes or less.
Abbreviations: MS, mass spectrometry; MS/MS, tandem mass spectrometry; LC, liquid chromatography; ESBL, extended-spectrum β-lactamase; CPO, carbapenemase-producing organism; CRE, carbapenem-resistant Enterobacterales; PCR, polymerase chain reaction; MW, molecular weight; m/z, mass-to-charge ratio; CSD, charge state distribution; MALDI, matrix-assisted laser desorption ionization; TOF, time-of-flight (mass spectrometry); ESI, electrospray ionization; CDC, Centers for Disease Control and Prevention; ATCC, American Type Culture Collection
Keywords: Antimicrobial-resistant organisms, Carbapenem-resistant Enterobacterales, Carbapenemase-producing organisms, Carbapenemase, β-Lactamase, OXA-48, OXA-48-like, CTX-M-15, Mass Spectrometry, Tandem mass spectrometry, Liquid chromatography
Abstract
Introduction
Antibiotic-resistant Gram-negative bacteria are of a growing concern globally, especially those producing enzymes conferring resistance. OXA-48-like carbapenemases hydrolyze most β-lactam antibiotics, with typically low-level hydrolysis of carbapenems, but have limited effect on broad-spectrum cephalosporins. These are frequently co-expressed with extended spectrum β-lactamases, especially CTX-M-15, which typically shows high level resistance to broad-spectrum cephalosporins, yet is carbapenem susceptible. The combined resistance profile makes the need for successful detection of these specific resistance determinants imperative for effective antibiotic therapy.
Objectives
The objective of this study is to detect and identify OXA-48-like and CTX-M-15 enzymes using mass spectrometry, and to subsequently develop a method for detection of both enzyme types in combination with liquid chromatography.
Methods
Cells grown in either broth or on agar were harvested, lysed, and, in some cases buffer-exchanged. Lysates produced from bacterial cells were separated and analyzed via liquid chromatography with mass spectrometry (LC-MS) and tandem mass spectrometry (LC-MS/MS).
Results
The intact proteins of OXA-48, OXA-181, and OXA-232 (collectively OXA-48-like herein) and CTX-M-15 were characterized and detected. Acceptance criteria based on sequence-informative fragments from each protein group were established as confirmatory markers for the presence of the protein(s). A total of 25 isolates were successfully tested for OXA-48 like (2), CTX-M-15 (3), or expression of both (7) enzymes. Thirteen isolates served as negative controls.
Conclusions
Here we present a method for the direct and independent detection of both OXA-48-like carbapenemases and CTX-M-15 β-lactamases using LC-MS/MS. The added sensitivity of MS/MS allows for simultaneous detection of at least two co-eluting, co-isolated and co-fragmented proteins from a single mass spectrum.
1. Introduction
The past several decades have seen antimicrobial-resistant organisms become one of the most significant concerns to human health across the globe [1]. Gram-negative bacteria account for a significant number of infections, with antibiotic resistance among this group increasing rapidly [2]. Bacteria that are resistant to carbapenems, a family of last-resort antibiotics [3], have garnered particular attention for their global spread over the last two decades. Carbapenem-resistant Enterobacterales (CRE) have been considered an urgent threat [4], [5], [6], [7], [8]; with significant concern for enzyme-based resistance, such as from carbapenemase-producing organisms (CPOs). The successful spread of CPOs is widely attributed to clonal expansion and horizontal gene transfer [9], [10], with the latter allowing efficient dissemination of resistance mechanisms among bacterial species.
One of the most commonly observed types of carbapenemases is the family of oxacillinase-48-like (OXA-48-like) enzymes. This family includes several variants, with OXA-48, OXA-181, and OXA-232 being among the most common globally [11]. These enzymes have been associated with particular genetic mobility attributed to the transposons on which they are encoded, having insertion sequences on either side of the gene. This has consequently played a substantial role in the global spread of these enzymes, with a significant prevalence in North Africa, the Middle East, Europe, and the Indian subcontinent [11].
Most isolates that produce OXA-48-like enzymes also co-express additional extended spectrum β-lactamases (ESBLs) [12]. Co-expression of OXA-48-like and ESBL enzymes can occur as a result of any combination of chromosomal- and/or plasmid-encoded resistance genes; however, there have been several reports of the ESBL CTX-M-15 gene being incorporated into the specific mobile genetic elements that also include OXA-48-like genes [11], such as the Tn1999.4 transposon, which harbors both resistances [13]. Although high levels of carbapenem resistance often require co-expression of ESBLs and/or porin mutations [14], [15], the specific co-expression of CTX-M-15 and similar enzymes with OXA-48-like enzymes results in a highly concerning resistance profile. Whereas OXA-48-like enzymes show carbapenemase activity, but are frequently sensitive to broad-spectrum cephalosporins, the resistance profile of CTX-M-15 enzymes provides high rates of broad-spectrum cephalosporin hydrolysis, despite being susceptible to carbapenems [16], [17], [18]. Consequently, isolates that co-express OXA-48-like carbapenemases and CTX-M ESBLs show carbapenemase activity with high levels of expanded-spectrum cephalosporin resistance [19]. The combination of specific resistance determinants adds to the complexity of detection and also illuminates the importance of identifying the specific enzymes present that contribute to the overall resistance profile of a bacterial isolate. For example, some OXA-48-like-producing isolates found not to co-express additional resistance determinants may be effectively treated with cephalosporins, such as cefepime [18], yet this therapy is often ineffective in the presence of CTX-M-15 [13]. Because of the complimentary nature of both resistance profiles of these two enyzmes, detection of one or both may not only lead to differing therapies, but may also lead to significantly different clinical outcomes.
Several methods have been employed for the detection of general β-lactamase or carbapenemase activity, such as disk diffusion, the modified carbapenem inactivation method (mCIM) [20], and assays using detection by colorimetric [21], electrochemical [22], and spectrophotometric [23] methods. Mass spectrometry (MS) has also been used to detect the presence of β-lactamase or carbapenemase activity using matrix-assisted laser desorption ionization coupled with time-of-flight (MALDI-TOF) mass spectrometry through detection of hydrolysis products of β-lactams [24] and carbapenems in Enterobacterales [25], [26], [27], Pseudomonas [28], [29] and Acinetobacter [30], [31], [32]. MALDI-TOF MS has also been used to monitor levels of growth or no growth in the presence of a carbapenem to identify resistance [33], [34].
Therapies against bacterial infections may differ depending on the type of antibiotic resistance pathways present. In the case of β-lactamase and carbapenemase-producing organisms, detection of specific enzyme types (e.g., OXA-48-like) becomes critically important for determining appropriate therapy [35]. Consequently, several different methods have been developed for direct detection of carbapenemase types, such as from immunoassays [36], [37], [38], an extension of a MALDI-TOF approach of the direct-on-target microdroplet growth assay [39], and polymerase chain reaction (PCR) assays for detection of resistance genes [40], [41], [42], [43], [44]. Other MS approaches for the detection of carbapenemases have used electrospray ionization (ESI) and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) via bottom-up proteomics techniques for the detection of peptides from trypsin-digested carbapenemases. These studies include detection of New Delhi metallo-β-lactamase (NDM) [45], Klebsiella pneumoniae carbapenemases (KPC) [46], OXA-48-like carbapenemase [47], and combinations of multiple carbapenemase enzymes [48]. Those methods using MS to identify specific proteins in targeted approaches could thereby also reveal the specific resistance mechanism(s) present. Additional advantages can be gained using high-resolution and accurate mass analysis, wherein the mass-to-charge ratio (m/z) of peptide or protein ions can be measured to within a few parts per million (ppm). This is particularly relevant for top-down proteomics, when proteolytic digestion is not incorporated, and the molecular weight (MW) and identity of intact protein ions can be determined via a combination of MS and MS/MS through direct dissociation to produce informative sequence-specific fragment ions. In line with this, we previously reported a similar study involving the direct detection of intact KPC via LC-MS and LC-MS/MS, wherein not only was the type of carbapenemase identified, the specific variants were as well [49].
Herein we present the use of MS for the direct detection of both OXA-48-like and CTX-M-15 proteins directly from Gram-negative bacterial cell lysates. This method can be applied from cells grown either in broth or on agar and, following simple sample preparation, can be used to detect OXA-48-like and/or CTX-M-15 via chromatographic separations on timescales less than five minutes. The ability to detect these enzymes can provide relevant information for therapy decisions and have significant clinical implications.
2. Methods
Bacterial isolates were obtained from American Type Culture Collection (ATCC), the Centers for Disease Control and Prevention and Food and Drug Administration Antibiotic Resistance Isolate Bank (CDC and FDA AR Isolate Bank). Antibiotic-resistant bacterial isolates used from the CDC and FDA AR Isolate Bank were from the Gram Negative Carbapenemase Detection Panel and Enterobacteriaceae Carbapenemase Diversity Panel. Information on resistant isolates, including known resistance mechanisms and minimum inhibitory concentration (MIC) values are provided by the CDC and FDA AR Isolate Bank [50].
2.1. Sample preparation
Isolates were grown either on tryptone soya agar or in trypsin soya broth for 20–24 h at 37 °C. Those grown on agar were harvested with a 10 µL loop, resuspended in 1 ml 5% acetonitrile, 0.2% formic acid in water, then transferred to bead beating tubes (MP Bio, matrix B) and were mechanically lysed using a Fisherbrand Bead Mill 4 Homogenizer (Fisher Scientific) for 120 s at 5 m/s. The tubes were then centrifuged at 12,000 g and supernatants were removed and extracted.
Isolates grown in broth were centrifuged at 4,000 g for 25 min at 10 °C. Supernatants were discarded, and the pellets were reconstituted with ice-cold 0.9% NaCl, then subjected to a second centrifugation step. Supernatants were again discarded, and the remaining pellet was reconstituted in 6 M guanidinium chloride (GuHCl), 50 mM Tris, pH 7.
Isolates prepared from either agar or broth were introduced to LC vials following centrifugal filtration with 0.2 µm spin filters (Thermo Fisher Scientific). Some isolates prepared in 6 M GuHCl, 50 mM Tris were subjected to buffer exchange into 0.2% formic acid, 5% acetonitrile in water using Amicon 10 K MWCO spin columns (Millipore Sigma), with the filtrate transferred to LC vials.
2.2. Liquid chromatography
All separations were carried out using binary gradients of Solvent A (0.2% formic acid in water) and Solvent B (0.2% formic acid in acetonitrile) using a 1 mm × 25 cm RP4H (Thermo Fisher Scientific) analytical column. Initial LC separations used cells grown in broth and were performed with Solvent B increasing from 10% to 22.5% over 10 min, during which time the flow was diverted away from the MS source, then increasing to 40% over 50 min with flow directed to the MS source, all using a flow rate of 200 µL/min. Additional separations were performed on samples in 0.2% formic acid, 5% acetonitrile using a gradient of Solvent B increasing from 5% to 28% over 1 min and continuing to 65% over an additional 6.5 min at a flow rate of 300 µL/min. These separations were performed on both a Dionex Ultimate 3000 and a Horizon Vanquish LC systems (Thermo Fisher Scientific).
An additional separation step to evaluate co-elution behavior was performed on an in-house built prototype LC system, using 4 µL/min flow rates with Solvent A (0.2% formic acid, 10% acetonitrile in water) and Solvent B (0.2% formic acid in acetonitrile) used to elute proteins off of an in-house-prepared solid-phase extraction (SPE) RP4H tip following direct introduction of lysate. This elution was performed with a binary gradient of Solvent B beginning at 10% (19% acetonitrile) and increasing to 30% (37% acetonitrile) over 4 min.
2.3. Mass spectrometry
All intact protein experiments were performed on Q-Exactive HF instrument (Thermo Fisher Scientific) with a heated electrospray ionization source (HESI) with heated metal capillary set to 325 °C. High-flow LC experiments (200–300 µL/min) used source settings of 3.7–4.0 kV for electrospray ionization voltages and 50–68% S-lens RF amplitude. Low-flow elution using a modification of an Easy-spray source was set to an electrospray voltage of 2.4 kV for ionization with the S-lens RF level set to 65. MS scans were set to a resolution of 120,000 with 5 µscans per scan. MS/MS data had resolution settings of either 120,000 for initial data acquired via long separations, or 60,000 for higher throughput data acquisition with shorter elution times. Mass isolation widths ranged from m/z 1.0 to 3.5, with the majority of data collected using a m/z 2.5 width. Intact protein ions were fragmented with nominal m/z values of 880.5, 853.9, 828.8, 805.1, 782.8 for the +32, +33, +34, +35, and +36 charge states respectively. Following MS/MS of the five charge states, most relevant data were collected from MS/MS of the +33 and +34 charge states. Mass spectra were largely interpreted manually with the assistance of Protein Prospector (http://prospector.ucsf.edu/prospector/mshome.htm), and with sequence mapping from Prosight Lite [51]. Deconvolution of intact protein mass spectra was performed using the Xtract software from the Averagine model [52], set for low sulfur content. This process considers isotopic distributions and charge states from the m/z domain and transforms them to the MW domain, based largely on the MW of an “average” amino acid.
3. Results and discussion
3.1. Detection of intact OXA-48-like carbapenemases, OXA-48, OXA-181, OXA-232
The observed full scan mass spectra in Fig. 1 were derived from lysates from bacterial isolates of the CDC and FDA AR Panels that have been determined to harbor OXA-48-like variants. Chromatographic separation of these proteins within any individual bacterial lysate led to elution times that ranged from less than 5 min to approximately 45 min, depending on the desired extent of separation. Examples of varying chromatographic conditions are described in Fig. S1. Initial detection of the specific intact protein targets from chromatographic separations was aided by using m/z values for charge states of intact OXA-48-like proteins using theoretical isotopic distributions from the MS-Isotope function, provided by Protein Prospector (http://prospector.ucsf.edu/prospector/mshome.htm, last accessed August 14, 2020). This allowed the mass spectra of the targeted protein(s) to be quickly observed from an extracted ion chromatogram (EIC, see Fig. S2). Mass spectra of intact proteins can produce overlapping charge state distributions (CSDs) in a complex sample and, even with the use of a separation method, can be challenging to interpret manually. Despite this dilemma, deconvolution techniques can be easily implemented to transform CSDs of proteins from the m/z domain into individual peaks in the MW domain. As an example, co-eluting proteins within a molecular weight range of 10,000–40,000 Da are illustrated in the deconvolution spectra and subsequent molecular weight plots shown in Fig. 1.
Fig. 1.
Mass spectra of intact OXA-48-like proteins in m/z and MW space. Spectra of intact protein elutions for m/z 650–1300 correspond to a) OXA-48, c) OXA-181, and e) OXA-232. Molecular weights of b) OXA-48, d) OXA-181, and f) OXA-232 as well as additional co-eluting proteins were calculated through deconvolution of the corresponding mass spectra. The data is from Klebsiella ozaenae AR Bank #0051 (OXA-181 and CTX-M-15), K. pneumoniae AR Bank #0066 (OXA-232), and Enterobacter aerogenes AR Bank #0074 (OXA-48) of the Gram-Negative Carbapenemase Detection Panel (CDC and FDA AR Isolate Bank).
The OXA-48-like carbapenemases OXA-48, OXA-181, and OXA-232 have very similar molecular weights around 28 kDa within an approximate mass range of 115 Da, with the specific amino acid substitutions indicated in Table 1. The experimentally-determined molecular weight of OXA-48 was 28129.3536 Da, which was 0.0707 Da from the theoretical molecular weight of 28129.2829 and corresponded to a 2.51 ppm mass difference. The observed molecular weight of OXA-181 was calculated to be 28083.2846 Da, which yielded a 0.0072 Da difference from the theoretical molecular weight of 28083.2774 and corresponded to a 0.26 ppm difference. Lastly, for OXA-232 the experimental molecular weight of 28014.2619 Da was observed, which differed from the theoretical mass of 28014.2083 Da by 0.0536 Da and corresponded to a 1.91 ppm mass difference.
Table 1.
Sequence differences between selected OXA-48-like variants. Amino acid substitution positions are based on the mature form of OXA-48-like proteins. Differences in MW correspond to the mass differences of the amino acid substitutions.
| OXA-48-like Variant | Mutation Relative to OXA-48 | Accession ID Used | Source | Δ MW |
|---|---|---|---|---|
| OXA-48 | ------- | Q6XEC0 | Uniprot | 0.0000 |
| OXA-181 | T82A, N88D, E146Q, S149A | G5CKK8 | Uniprot | −46.0055 |
| OXA-232 | T82A, N88D, E146Q, S149A, R192S | M4JTK1 | Uniprot | −115.0746 |
3.2. Fragmentation of multiple charge states of intact OXA-48
Following identification of the intact OXA-48-like protein variants with the correct MW, MS/MS was performed to determine if fragment ions specific to this family of carbapenemase could be used to confirm expression in Gram-negative pathogens. Tandem mass spectrometry allows specific precursor m/z ions to be isolated and fragmented, often providing highly informative and specific fragment ions. Fragmentation behavior can vary depending on the parameters used, such as charge state of the precursor ion, collision energy (CE), and even the nature of the background gases (i.e., He versus N2). It should be noted that the collision energy is a product of the ion charge state and the acceleration voltages, such that for a given set of voltages, the more highly charged ions will experience greater accelerations and, consequently, higher-energy collisions (i.e., an ion with a +2 charge will feel twice the acceleration from a negative potential than an ion with a +1 charge).
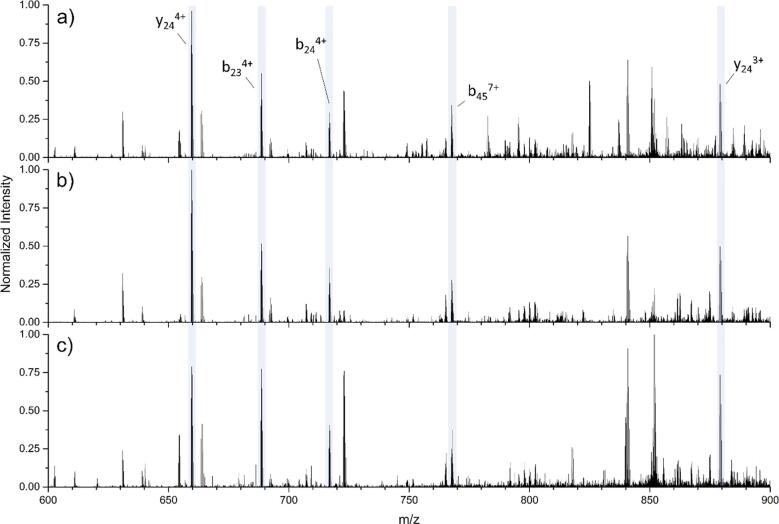
Fig. 2 illustrates the fragmentation spectra of five separate charge states of OXA-48, ranging from +32 to +36, all with CE set to 20 eV, within an m/z range of 600–900. A selection of five sequence-informative fragment ions, detailed in Table 2, are highlighted in blue and annotated in the top panel (Fig. 2a). An illustration of the residue positions of backbone peptide cleavages produced from these fragment ions are presented in Fig. 3. Although similar fragments are observed following dissociation of each of the selected charge states, the most extensive production of informative fragment ions, based on a ProSight Lite search, was observed for the +33 charge state, followed by the +32 and +34 charge states. It is worth noting that most fragments correspond to cleavages approximately 20–25 residues from either terminus.
Fig. 2.
Fragmentation of a) +32, b) +33, c) +34, d) +35, and e) +36 charge states of OXA-48, with selected shared fragments highlighted in blue. Data from E. aerogenes AR Bank #0074.
Table 2.
Mass accuracy of selected fragments from Fig. 2 relative to theoretical m/z. Calculations are based on the theoretically most abundant isotope for each fragment.
| Precursor, z | y24+3 |
y24+4 |
b23+4 |
b24+4 |
b45+7 |
|---|---|---|---|---|---|
| 879.1788 | 659.6359 | 688.592 | 716.863 | 767.8187 | |
| 880.58, +32 | −0.34 | 0.45 | 0.29 | −0.70 | 1.04 |
| 853.93, +33 | 1.36 | 0.76 | 0.44 | 0.42 | 0.65 |
| 828.84, +34 | −0.11 | 0.45 | 0.58 | 0.42 | 1.69 |
| 805.12, +35 | 1.25 | 1.36 | 1.45 | 2.09 | 1.17 |
| 782.85, +36 | 0.45 | 0.91 | 0.73 | 2.51 | 0.26 |
Fig. 3.
Sequence information provided by fragment ions for each charge state with set acceleration voltages for charges +32 through +36. Each charge state has a unique symbols, with blue representing b-ions and red representing y-ions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Fragmentation comparisons of OXA-48, OXA-181, and OXA-232
Following comparisons between the dissociation of different charge states of OXA-48, the +33 and +34 charge states corresponding to m/z 852.2 and 827.5 were selected for subsequent MS/MS experiments for the OXA-48-like variants. Using similar dissociation conditions associated with the experiments in Fig. 2, the +33 and +34 precursor ions of OXA-48, OXA-181, and OXA-232 were fragmented for evaluation. Comparisons of those fragmentation spectra are presented in Fig. 4. The same selection of sequence-informative fragment ions produced from OXA-48 are also present for OXA-181 and OXA-232, and are highlighted in blue. These ions are identical between the three OXA-48-like proteins as a result of the sequence homology they share.
Fig. 4.
Comparison of fragmentation spectra of the +33 charge state between a) OXA-48 (E. aerogenes AR Bank #0074), b) OXA-181 (K. ozaenae AR Bank #0051) and c) OXA-232 (K. pneumoniae AR Bank #0066), with selected fragment ions highlighted in blue.
Bacterial cell lysates consist of many different proteins at varying concentration levels, making individual evaluations of targeted proteins challenging without the introduction of a separation step prior to mass analysis. In the absence of separation, the individual components subjected to ESI are competing for a finite amount of charge, such that only those components with the most favorable characteristics (e.g., concentration, gas phase basicity, droplet surface activity) will be ionized. Liquid chromatography is inherently compatible with ESI, effectively reducing the number of multiple proteins co-eluting and competing for charge at any given time. Under appropriate conditions, the use of chromatography can often offer the possibility of separating proteins well enough from each other to be able to identify individual charge state distributions. As demonstrated above, this inherent complexity requires high-resolution and accurate mass analysis for both detection of the general OXA-48-like family, as well as the individual protein variants.
Intact protein MWs may be suggestive, but are typically not confirmatory, for protein identity. Likewise, fragment ions on their own may be suggestive, but only in connection to the specific precursor ions. Successful determination of protein identity often involves combining the intact protein MW information of MS with the sequence and/or structural information gained from fragments generated via MS/MS. As an example, OXA-163 and OXA-405 share 98% sequence homology to OXA-48, yet they have significantly reduced carbapenemase activity with increased cephalosporinase activity. This sequence homology would lead to the production of identical fragment ions to OXA-48; however, the precursor m/z for OXA-163 and OXA-405 at the +33 charge state would be 839.8 and 838.8, respectively; both well-removed from the 853.8 apex of OXA-48 for the same charge state.
3.4. Detection and fragmentation of CTX-M-15
One important example when considering potential interferences in the analysis of OXA-48-like carbapenemases is that of the extended spectrum β-lactamase CTX-M-15 (Uniprot.org accession Q2PUH3, accessed August 14, 2020). This ESBL is endemic in many Gram-negative pathogens, with one of the most prominent being Klebsiella pneumoniae. Although it is found in isolates without any carbapenemase genes, it is frequently observed co-expressed with OXA-48-like proteins. Detection of CTX-M-15 was performed here in a similar manner to OXA-48-like proteins, based on intact protein mass spectra with CSDs corresponding to a protein with a MW of approximately 28 kDa. The theoretical MW of CTX-M-15 is 28090.5440 Da, which differs from OXA-48 by approximately 39 Da, OXA-181 by 7 Da, and OXA-232 by 76 Da. The intact protein mass spectrum for CTX-M-15 is displayed in Fig. 5a, with the corresponding MW of proteins detected within a range of 10,000–40,000 Da illustrated in the inset.
Fig. 5.
Mass spectra of CTX-M-15, with a) the charge state distribution of the intact protein, with inset showing the deconvoluted MW of all proteins present in the spectrum from 10,000 to 40,000 Da, and b) the fragmentation spectrum of the +33 charge state, with selected fragment ions highlighted in red. It should be noted that the intensity scale for b) has been set to 0.5 from 1.0, wherein the y637+ and y63+6 ion intensities correspond to 1.0 (100%) and 0.92 (92%), respectively. Data from K. ozaenae AR Bank #0051. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For the MS/MS of CTX-M-15, the +33 and +34 charge states are used for dissociation, as was done for the OXA-48-like suite of variants discussed above. At these charge states, the m/z difference between the proteins is approximately 0.2–3.5, which consequently allows a single precursor window to be used for all 4 proteins. Therefore, similar precursor charge states used for OXA-48-like (z = +33 and z = +34, m/z windows centered on 852.20 and 827.50, respectively) were used to dissociate similar charge states of CTX-M-15. Fig. 5b illustrates the fragmentation spectrum of CTX-M-15 from the +33 charge precursor ion, which is further illustrated in the supplemental data section in Fig. S3. Selected sequence-informative CTX-M-15-specific fragments are highlighted in red, with measurement accuracies presented in Table 3.
Table 3.
Mass accuracy (in ppm) of selected CTX-M-15 fragments from Fig. 5 relative to theoretical m/z. Calculations are based on the theoretically most abundant isotope for each fragment.
| Precursor m/z, z | fragment: | y42+6 | b81+10 | y112+11 | y63+7 | y63+6 |
|---|---|---|---|---|---|---|
| m/z: | 773.6040 | 893.1694 | 1077.2110 | 958.2281 | 1117.7650 | |
| 852.20, +33 | ppm: | −2.59 | −2.91 | −3.71 | −3.34 | −3.85 |
3.5. Distinguishing between OXA-48-like from CTX-M-15 intact proteins
The three OXA-48-like proteins and CTX-M-15 have similar MWs that span an m/z range of 3.5 for the +33 charge state. Fig. 6a illustrates the +33 charge states of OXA-48, OXA-181, OXA-232 and CTX-M-15 within the m/z range of 850–854.5, using separate spectra overlaid on a single plot. OXA-181 and CTX-M-15 show the smallest difference of the four proteins, with a MW difference of approximately 7 Da, corresponding to an m/z of 0.22 for the +33 charge state. Fig. 6b illustrates a closer view of the isotopic overlap for the +33 charge states of the two proteins within m/z 852.2–853.2.
Fig. 6.

Isotopic distribution and overlap of the +33 charge states of OXA-48-like variants with CTX-M-15, from separate bacterial isolates. Data from K. ozaenae AR Bank #0051 (OXA-181 and CTX-M-15), K. pneumoniae AR Bank #0066 (OXA-232), and E. aerogenes AR Bank #0074 (OXA-48).
3.5.1. Distinguishing OXA-48-like from CTX-M-15 using chromatography
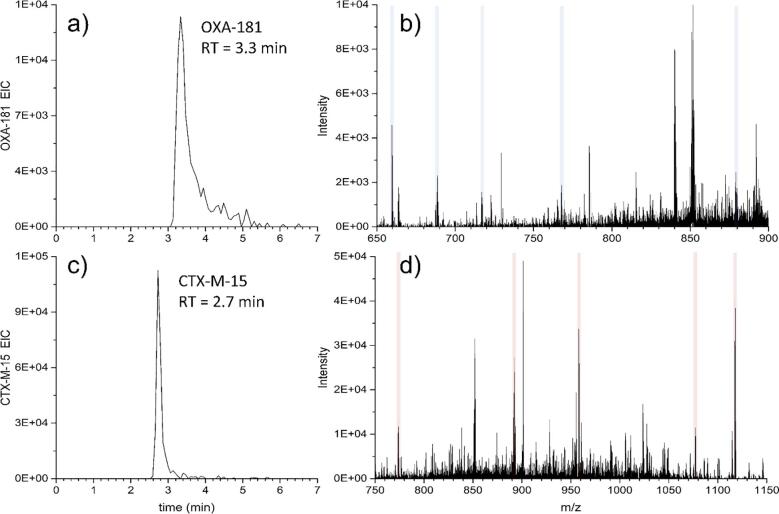
Reducing the complexity of a protein mixture to view only a small portion of an overall sample will increase detection efficiency as well as provide the opportunity to search for multiple targets. Whereas OXA-181 and CTX-M-15 have an increasingly high percentage of isotopic overlap as charge state increases, Fig. 7 clearly demonstrates separation of both proteins in less than 5 min, as shown with the EIC of each protein. In these data, the retention times of CTX-M-15 and OXA-181 were 2.7 and 3.3 min, respectively. The fragment ions used in Table 2 and Table 3 are highlighted in blue for OXA-181 fragments, and red for CTX-M-15 fragments. Additionally, the combination of LC-MS/MS significantly improves the detection capability for OXA-181. Although the MS of intact CTX-M-15 is detected only at a low level, intact OXA-181 is not detected (Fig. S4). However, fragment ions from both proteins are detected with sufficient signal-to-noise using MS/MS. This phenomenon is commonly observed in complex protein samples, wherein the m/z space may be too crowded for sensitive measurements to be employed, yet the simplification through the use of mass-selective filtering, dissociation, and detection of fragment ions, as performed through MS/MS, allows much more sensitive measurements to be made.
Fig. 7.
Detection of both OXA-181 and CTX-M-15 using LC-MS/MS. a) Extracted ion chromatogram (EIC) of OXA-181, with b) MS/MS spectrum of OXA-181; c) EIC of CTX-M-15 with d) MS/MS spectrum. Selected OXA-181 fragments are highlighted in blue, while CTX-M-15 fragments are highlighted in red. Data from K. ozaenae AR Bank #0051.
3.5.2. Distinguishing co-fragmented OXA-48-like from CTX-M-15 using MS/MS
The use of MS/MS to clearly determine protein identity based on both precursor and fragment ions can be applied directly to distinguish OXA-48-like and CTX-M-15 under conditions in which chromatography is unable to separate the two proteins. Fig. 8 shows the MS/MS spectrum of both proteins being co-isolated and co-fragmented. Sequence-informative fragments corresponding to OXA-181 are highlighted in blue, while the sequence-informative fragments corresponding to CTX-M-15 are highlighted in red. A more-detailed view of the three diagnostic fragments from each protein is presented in the lower portion of Fig. 8, (for additional examples see Fig. S5). In combination with restricting the range of intact protein masses with the precursor m/z window, knowing the specific fragmentation behavior of the individual targets can make possible the detection of both proteins using sequence-specific fragment ions as markers.
Fig. 8.
Fragmentation spectrum of both OXA-181 and CTX-M-15 following co-isolation and co-fragmentation. Fragments corresponding to OXA-181 are highlighted in blue, while those corresponding to CTX-M-15 are highlighted in red. Isotopic distributions of individual selected fragments are shown in the lower panel. Data from K. pneumoniae AR Bank #0140. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Further evaluations for OXA-48-like and CTX-M-15 proteins from bacterial isolates
Determining the presence or absence of these proteins is based on both MS and MS/MS data. Although intact protein MS data may be suggestive of protein identity, only MS/MS data will be confirmatory; consequently, more significance is given to MS/MS data. Criteria for determination of the presence of a protein from this approach may vary depending on the stringency or leniency and associated error tolerance. Those criteria set herein define diagnostic fragment ions necessary for identification as having 1) the correct precursor m/z for the intact protein ion; 2) m/z accuracies within ±10 ppm of the theoretical values; 3) correct charge state, as determined by isotopic m/z spacing; 4) signal-to-noise ratio greater than three (S/N > 3); 5) at least 2 contiguous isotopes; and 6) similar retention time (RT) from a chromatographic elution, with data corresponding to either a single scan or a sum of up to 3 scans. A minimum of three diagnostic ions (i.e., those fragments that meet all criteria listed above) should be observed, as illustrated in Fig. 8. Additionally, by alternating between MS and MS/MS, detection of the intact protein is achieved in tandem with production of fragment ions; however, under some conditions of high protein ion flux, the intact protein might not always be observed. Based on these criteria, Table 4 presents results from analysis of multiple bacterial isolates for the detection of OXA-48-like and CTX-M-15 proteins.
Table 4.
Isolates evaluated for presence of OXA-48-like and CTX-M-15 proteins.
| Identifier | Species | Expected | OXA-48-like | CTX-M-15 |
|---|---|---|---|---|
| ATCC 11775 | Escherichia coli | ---- | none | none |
| ATCC 13883 | Klebsiella pneumoniae | ---- | none | none |
| ATCC 700721 | Klebsiella pneumoniae | ---- | none | none |
| DH5α | Escherichia coli | ---- | none | none |
| AR Bank #0039 | Klebsiella pneumoniae | OXA-181, CTX-M-15 | Detected | Detected |
| AR Bank #0042 | Klebsiella pneumoniae | CTX-M-15 | none | Detected |
| AR Bank #0043 | Klebsiella pneumoniae | ---- | none | none |
| AR Bank #0044 | Klebsiella pneumoniae | CTX-M-15 | none | Detected |
| AR Bank #0047 | Klebsiella pneumoniae | ---- | none | none |
| AR Bank #0051 | Klebsiella ozaenae | OXA-181, CTX-M-15 | Detected | Detected |
| AR Bank #0060 | Enterobacter cloacae | ---- | none | none |
| AR Bank #0065 | Enterobacter cloacae | ---- | none | none |
| AR Bank #0066 | Klebsiella pneumoniae | OXA-232, CTX-M-15 | Detected | Detected |
| AR Bank #0072 | Enterobacter cloacae | ---- | none | none |
| AR Bank #0073 | Enterobacter cloacae | ---- | none | none |
| AR Bank #0074 | Enterobacter aerogenes | OXA-48 | Detected | none |
| AR Bank #0075 | Klebsiella pneumoniae | OXA-232, CTX-M-15 | Detected | Detected |
| AR Bank #0077 | Escherichia coli | ---- | none | none |
| AR Bank #0087 | Klebsiella pneumoniae | ---- | none | none |
| AR Bank #0089 | Escherichia coli | ---- | none | none |
| AR Bank #0140 | Klebsiella pneumoniae | OXA-181, CTX-M-15 | Detected | Detected |
| AR Bank #0141 | Klebsiella pneumoniae | OXA-181, CTX-M-15 | Detected | Detected |
| AR Bank #0142 | Klebsiella pneumoniae | OXA-181, CTX-M-15 | Detected | Detected |
| AR Bank #0160 | Klebsiella pneumoniae | OXA-48 | Detected | none |
| AR Bank #0163 | Enterobacter cloacae | CTX-M-15 | none | Detected |
4. Conclusion
Mass spectrometry has been shown to be an invaluable tool for the detection and identification of proteins, especially of those conferring antibiotic resistance. Clinical laboratories could immediately benefit from the ability to detect carbapenemases such as OXA-48-like and ESBLs (such as CTX-M-15) through the detection of any combination of these enzymes. This approach allows detection of specific intact proteins directly from cell lysates, with optional cleanup steps, and with the ability for LC-MS/MS analysis time to be less than five minutes.
There are both benefits and limitations to the detection of specific proteins using MS. In the case of OXA-48-like carbapenemases, they are frequently co-expressed with ESBLs, often belonging to SHV, TEM, or CTX-M families. The breadth of diversity among the three groups is beyond the scope of the current detection approach, and consequently CTX-M-15 is used as a representative ESBL. Whereas susceptibility detection offers insight into which antibiotics are likely ineffective against particular bacterial isolates, resistance detection—the act of searching for specific resistance determinants—provides information about the presence or absence of a particular resistance mechanism, but does not provide a comprehensive view of the overall resistance profile for a bacterial isolate. Consequently, there is a trade-off between broad susceptibility testing and focusing on detection of specific resistance mechanisms, in which the former offers broad information but might not be specific enough (e.g., identifying carbapenem resistance without identifying carbapenemase activity, or identifying carbapenemase activity but without identifying the specific mechanism(s)); on the other hand, the latter can provide extremely specific information but might miss relevant resistance(s). Until resistance detection methods can thoroughly detect the full extent of relevant resistance mechanisms present, the information provided through susceptibility testing is likely also needed. Despite these limitations, MS provides a digital platform that allows multiple targets to be analyzed over the course of a chromatographic timeframe, illustrated herein using separate charge states that could otherwise be easily adjusted for detection of multiple proteins.
The method presented allows identification of OXA-48-like and CTX-M-15, two clinically-relevant enzymes whose combined resistance profile can render β-lactam and carbapenem antibiotics ineffective. Consequently, because clinical therapies may differ based on the detection of one enzyme, both, or neither, the information produced from this resistance detection method could be used to provide valuable insight that may help to support critical clinical decisions. We anticipate that similar methods can be developed and adapted to detect additional carbapenemase resistance mechanisms in the future.
Acknowledgments
Acknowledgements
The authors would like to thank Mary Ann Silvius and Dr. Nathan A. Ledeboer for helpful conversations.
Conflicts of Interest
All authors are employees of Thermo Fisher Scientific. Several of the authors are associated with intellectual property development that is relevant to this research.
-
(1)
WO2018222345 – Automated Determination of Mass Spectrometer Collision Energy Ping F. Yip, Helene Cardasis, James L. Stephenson, Jr. International filing date 07.05.2018.
-
(2)
WO2018067772 – System and Method for Real-Time Isotope Identification Scott R. Kronewitter, James L. Stephenson, Jr., Ping F. Yip International Filing Date 05.10.2017.
-
(3)
US20140051113 – Apparatus and Methods for Microbial Identification by Mass Spectrometry James L. Stephenson, Jr., Oksana Gvozdyak, Roger Grist, Clay Campbell, Ian D, Jardine, Iain C. Mylchreest Publication Date 20.02.2014.
-
(4)
WO2017127733 – Rapid Mass Spectrometry Methods for Antimicrobial Susceptibility Testing Using Top-Down Mass Spectrometry James Stephenson, Jr., Roger Grist, William McGee, Jason Neil, David Sarracino International Filing Date 20.01.2017.
-
(5)
WO2016145331 – Methods for Data-Dependent Mass Spectrometry of Mixed Biomolecular Analytes Ping F. Yip, James L. Stephenson, Jr., Oksana Gvozdyak International Filing Date 11.03.2016.
-
(6)
WO2016011355 – Methods for Mass Spectrometry of Mixtures of Proteins of Polypeptides using Proton Transfer Reaction James L. Stephenson, Jr., John E.P. Syka, August A. Specht International Filing Date 17.07.2015.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmsacl.2021.05.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Seale A.C., et al. AMR Surveillance in low and middle-income settings - A roadmap for participation in the Global Antimicrobial Surveillance System (GLASS) Wellcome Open Res. 2017;2:92. doi: 10.12688/wellcomeopenres.12527.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho J., Tambyah P.A., Paterson D.L. Multiresistant Gram-negative infections: a global perspective. Curr Opin Infect Dis. 2010;23(6):546–553. doi: 10.1097/QCO.0b013e32833f0d3e. [DOI] [PubMed] [Google Scholar]
- 3.Papp-Wallace K.M., Endimiani A., Taracila M.A., Bonomo R.A. Carbapenems: past, present, and future. Antimicrob. Agents Chemother. 2011;55(11):4943–4960. doi: 10.1128/AAC.00296-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzouvelekis L.S., et al. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 2012;25(4):682–707. doi: 10.1128/CMR.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(CDC), C.f.D.C.a.P., Antibiotic Resistance Threats. http://www.cdc.gov/drugresistance/threat-report-2013/, 2013.
- 6.Michael C.A., Dominey-Howes D., Labbate M. The antimicrobial resistance crisis: causes, consequences, and management. Front. Public Health. 2014;2:145. doi: 10.3389/fpubh.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventola, The antibiotic resistance crisis. Pharm. Ther. 40(4): 277–283. [PMC free article] [PubMed]
- 8.Roca I., et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 2015;6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Duin D., Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. 2017;8(4):460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arana D.M., et al. Concurrent interspecies and clonal dissemination of OXA-48 carbapenemase. Clin. Microbiol. Infect. 2015;21(2):148.e1–148.e4. doi: 10.1016/j.cmi.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Pitout J.D.D., et al. The global ascendency of OXA-48-type carbapenemases. Clin. Microbiol. Rev. 2019;33(1) doi: 10.1128/CMR.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A. Potron, al., Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro. Surveill., 2013. 18. [DOI] [PubMed]
- 13.Potron A., et al. A mosaic transposon encoding OXA-48 and CTX-M-15: towards pan-resistance. J. Antimicrob. Chemother. 2013;68(2):476–477. doi: 10.1093/jac/dks397. [DOI] [PubMed] [Google Scholar]
- 14.Nordmann P., Naas T., Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2011;17(10):1791–1800. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyos-Mallecot Y., et al. OXA-244-producing Escherichia coli isolates, a challenge for clinical microbiology laboratories. Antimicrob. Agents Chemother. 2017;61(9) doi: 10.1128/AAC.00818-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel L., et al. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004;48(1):15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L., Potron A., Nordmann P. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 2012;67(7):1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 18.Escola-Verge L., et al. Infections by OXA-48-like-producing Klebsiella pneumoniae non-co-producing extended-spectrum beta-lactamase: Can they be successfully treated with cephalosporins? J Glob. Antimicrob. Resist. 2019;19:28–31. doi: 10.1016/j.jgar.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 19.Tomic Paradzik M., et al. Hidden carbapenem resistance in OXA-48 and extended-spectrum beta-lactamase-positive Escherichia coli. Microb. Drug Resist. 2019;25(5):696–702. doi: 10.1089/mdr.2018.0309. [DOI] [PubMed] [Google Scholar]
- 20.Pierce V.M., et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among enterobacteriaceae. J. Clin. Microbiol. 2017;55(8):2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nordmann P., Poirel L., Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2012;18(9):1503–1700. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogaerts P., et al. Evaluation of the BYG carba test, a new electrochemical assay for rapid laboratory detection of carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 2016;54(2):349–358. doi: 10.1128/JCM.02404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernabeu S., Poirel L., Nordmann P. Spectrophotometry-based detection of carbapenemase producers among Enterobacteriaceae. Diagn. Microbiol. Infect. Dis. 2012;74(1):88–90. doi: 10.1016/j.diagmicrobio.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 24.Hooff G.P., et al. Characterization of beta-lactamase enzyme activity in bacterial lysates using MALDI-mass spectrometry. J. Proteome Res. 2012;11(1):79–84. doi: 10.1021/pr200858r. [DOI] [PubMed] [Google Scholar]
- 25.Johansson Å., et al. The detection and verification of carbapenemases using ertapenem and Matrix Assisted Laser Desorption Ionization-Time of Flight. BMC Microbiol. 2014;14(89) doi: 10.1186/1471-2180-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasserre C., et al. Efficient detection of carbapenemase activity in enterobacteriaceae by matrix-assisted laser desorption ionization-time of flight mass spectrometry in less than 30 minutes. J. Clin. Microbiol. 2015;53(7):2163–2171. doi: 10.1128/JCM.03467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sparbier K., et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J. Clin. Microbiol. 2012;50(3):927–937. doi: 10.1128/JCM.05737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hrabák J., et al. Carbapenemase activity detection by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2011;49(9):3222–3227. doi: 10.1128/JCM.00984-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burckhardt I., Zimmermann S. Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 2011;49(9):3321–3324. doi: 10.1128/JCM.00287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrabak J., et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2012;50(7):2441–2443. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carvalhaes C.G., et al. Detection of SPM-1-producing Pseudomonas aeruginosa and class D beta-lactamase-producing Acinetobacter baumannii isolates by use of liquid chromatography-mass spectrometry and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 2013;51(1):287–290. doi: 10.1128/JCM.02365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.M. Kempf, et al., Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One, 2012; 7(2): e31676. [DOI] [PMC free article] [PubMed]
- 33.Lange C., et al. Quantitative matrix-assisted laser desorption ionization-time of flight mass spectrometry for rapid resistance detection. J. Clin. Microbiol. 2014;52(12):4155–4162. doi: 10.1128/JCM.01872-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Idelevich E.A., et al. Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin. Microbiol. Infect. 2018;24(7):738–743. doi: 10.1016/j.cmi.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 35.Banerjee R., Humphries R. Clinical and laboratory considerations for the rapid detection of carbapenem-resistant Enterobacteriaceae. Virulence. 2017;8(4):427–439. doi: 10.1080/21505594.2016.1185577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.H. Boutal, et al., A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother., 2018; 73(4): 90. [DOI] [PMC free article] [PubMed]
- 37.Rosner S., et al. Evaluation of a novel immunochromatographic lateral flow assay for rapid detection of OXA-48, NDM, KPC and VIM carbapenemases in multidrug-resistant Enterobacteriaceae. J. Med. Microbiol. 2019;68(3):379–381. doi: 10.1099/jmm.0.000925. [DOI] [PubMed] [Google Scholar]
- 38.Cointe A., et al. Detection of carbapenemase-producing Enterobacteriaceae in positive blood culture using an immunochromatographic RESIST-4 O.K.N.V. assay. Antimicrob. Agents Chemother. 2018;62(12) doi: 10.1128/AAC.01828-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Correa-Martínez C.L., et al. Development of a MALDI-TOF MS-based screening panel for accelerated differential detection of carbapenemases in Enterobacterales using the direct-on-target microdroplet growth assay. Sci. Rep. 2020;10(1):4988. doi: 10.1038/s41598-020-61890-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Smith, et al., Rapid and accurate detection of carbapenemase genes in Enterobacteriaceae with the Cepheid Xpert Carba-R assay. J. Med. Microbiol., 2016; 65(9): 951–953. [DOI] [PubMed]
- 41.Lund M., et al. Rapid real-time PCR for the detection of IMP, NDM, VIM, KPC and OXA-48 carbapenemase genes in isolates and spiked stool samples. Diagn. Microbiol. Infect. Dis. 2018;92(1):8–12. doi: 10.1016/j.diagmicrobio.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Bordin A., et al. Evaluation of the SpeeDx Carba (beta) multiplex real-time PCR assay for detection of NDM, KPC, OXA-48-like, IMP-4-like and VIM carbapenemase genes. BMC Infect. Dis. 2019;19(1):571. doi: 10.1186/s12879-019-4176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traczewski M.M., Carretto E., Canton R., Moore N.M. Multicenter evaluation of the Xpert Carba-R assay for detection of carbapenemase genes in GRAM-negative isolates. J. Clin. Microbiol. 2018;56(8) doi: 10.1128/JCM.00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mentasti M., et al. Rapid detection of IMP, NDM, VIM, KPC and OXA-48-like carbapenemases from Enterobacteriales and Gram-negative non-fermenter bacteria by real-time PCR and melt-curve analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2019;38(11):2029–2036. doi: 10.1007/s10096-019-03637-5. [DOI] [PubMed] [Google Scholar]
- 45.Cecchini T., et al. Deciphering multifactorial resistance phenotypes in Acinetobacter baumannii by genomics and targeted label-free proteomics. Mol. Cell. Proteomics. 2018;17(3):442–456. doi: 10.1074/mcp.RA117.000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H., et al. Peptide markers for rapid detection of KPC carbapenemase by LC-MS/MS. Sci. Rep. 2017;7(1):2531. doi: 10.1038/s41598-017-02749-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strich J.R., et al. Identification of the OXA-48 carbapenemase family by use of tryptic peptides and liquid chromatography-tandem mass spectrometry. J. Clin. Microbiol. 2019;57(5) doi: 10.1128/JCM.01240-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foudraine D.E., et al. Accurate detection of the four most prevalent carbapenemases in E. coli and K. pneumoniae by high-resolution mass spectrometry. Front. Microbiol. 2019;10:2760. doi: 10.3389/fmicb.2019.02760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGee W.M., et al. Direct detection of intact Klebsiella pneumoniae carbapenemase variants from cell lysates: Identification, characterization and clinical implications. Clin. Mass Spectrom. 2020;17:12–21. doi: 10.1016/j.clinms.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.CDC & FDA Antibiotic Resistance Isolate Bank. April 2021, CDC: Atlanta, GA.
- 51.Fellers R.T., Greer J.B., Early B.P., Yu X., LeDuc R.D., Kelleher N.L., et al. ProSight Lite: Graphical software to analyze top-down mass spectrometry data. Proteomics. 2015;15(7):1235–1238. doi: 10.1002/pmic.201570050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Senko M.W., Beu S.C., McLaffertycor F.W. Determination of monoisotopic masses and ion populations for large biomolecules from resolved isotopic distributions. J. Am. Soc. Mass Spectrom. 1995;6(4):229–233. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.