Abstract
Acinetobacter baumannii is a multidrug-resistant, Gram-negative nosocomial pathogen that exhibits phenotypic heterogeneity resulting in virulent opaque (VIR-O) and avirulent translucent (AV-T) colony variants. Each variant has a distinct gene expression profile resulting in multiple phenotypic differences. Cells interconvert between the VIR-O and AV-T variants at high frequency under laboratory conditions, suggesting that the genetic mechanism underlying the phenotypic switch could be manipulated to attenuate virulence. Therefore, our group has focused on identifying and characterizing genes that regulate this switch, which led to the investigation of ABUW_1132 (1132), a highly conserved gene predicted to encode a LysR-type transcriptional regulator. ABUW_1132 was shown to be a global regulator as the expression of 74 genes was altered ≥ 2-fold in an 1132 deletion mutant. The 1132 deletion also resulted in a 16-fold decrease in VIR-O to AV-T switching, loss of 3-OH-C12-HSL secretion, and reduced surface-associated motility. Further, the deletion of 1132 in the AV-T background caused elevated capsule production, which increased colony opacity and altered the typical avirulent phenotype of translucent cells. These findings distinguish 1132 as a global regulatory gene and advance our understanding of A. baumannii’s opacity-virulence switch.
Keywords: Acinetobacter baumannii, AB5075, LysR-type transcriptional regulator, phenotypic heterogeneity, quorum sensing, motility, polysaccharide capsule, virulence
Introduction
The Gram-negative pathogen Acinetobacter baumannii poses a major threat to hospitalized patients. Cases of ventilator-associated pneumonia are among the most common A. baumannii infections, but incidences of skin and soft tissue infections, urinary tract infections, and sepsis are on the rise (Davis et al., 2005; Dijkshoorn et al., 2007; Peleg et al., 2008; Peleg and Hooper, 2010; Doyle et al., 2011; Weiner et al., 2016; Wong et al., 2017). Of primary concern is A. baumannii’s increasing resistance to treatment with antimicrobials, with 63% of infections caused by multidrug-resistant (MDR) strains (Clark et al., 2016; Lee et al., 2017; Centers for Disease Control and Prevention, 2019). In particular, A. baumannii’s rapidly growing resistance to carbapenem antibiotics and its ability to widely disseminate resistance via mobile genetic elements prompted the World Health Organization to name this organism as a critical priority for the research and development of new antimicrobial drugs in 2017 (World Health Organization, 2017). Further, its extreme resistance to desiccation and disinfectants makes it notoriously difficult to eradicate in hospital environments (Jawad et al., 1998; Chapartegui-Gonzalez et al., 2018; Rocha et al., 2018; Bravo et al., 2019; D’souza et al., 2019). In the face of such problematic phenotypes, an understanding of pathogenesis and virulence in this species is imperative. To meet this need, experiments conducted by our group have been carried out in the strain AB5075 (GenBank Accession Number CP008706.1), a highly virulent MDR clinical isolate that is genetically tractable (Jacobs et al., 2014; Gallagher et al., 2015).
Our group has sought to better understand the genetic mechanisms regulating A. baumannii virulence in light of our findings that clinical isolates of this species exhibit phenotypic heterogeneity resulting in virulent and avirulent colony opacity variants (Tipton et al., 2015; Chin et al., 2018). The virulent variant has a golden, opaque colony morphotype under oblique lighting and is termed VIR-O, while the avirulent variant is translucent and is termed AV-T ( Figure 1A ). In addition to differences in virulence, the VIR-O variant displays higher levels of capsule production, quorum sensing signal secretion, and surface-associated motility, while AV-T demonstrates greater production of biofilm and is able to utilize multiple carbon sources. Both variants switch back and forth at high frequency, with rates of approximately 4-13% conversion at 24 hours of colony growth and 20-40% at 48 hours (Chin et al., 2018). The two types have distinctly different genomic expression profiles as revealed by RNA sequencing, and a variety of gene products—including transcriptional regulators, a two-component system (OmpR/EnvZ), an efflux pump (ArpB), and a putative sRNA located upstream of a plasmid-encoded antibiotic resistance locus—appear to contribute to interconversion between VIR-O and AV-T (Tipton and Rather, 2016; Tipton et al., 2017; Chin et al., 2018; Anderson et al., 2020).
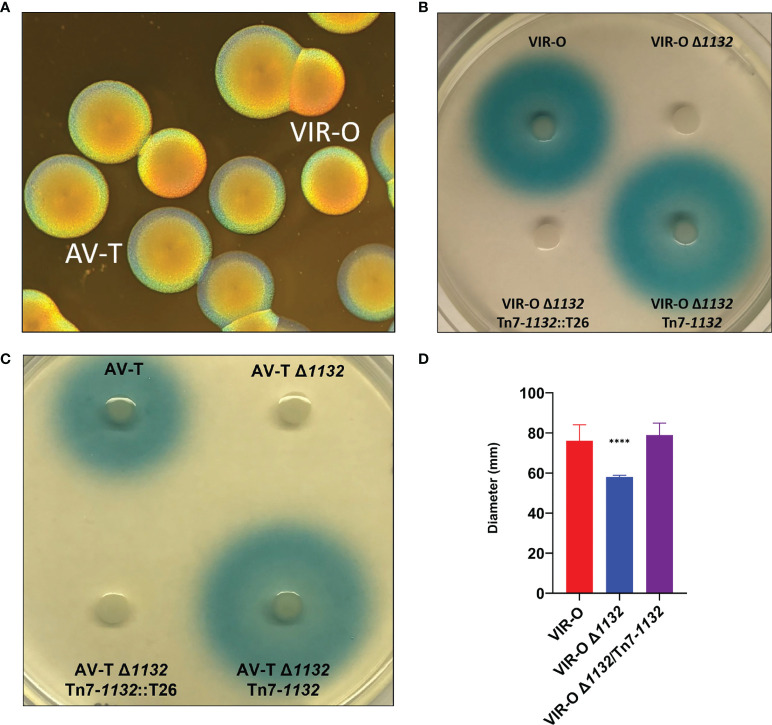
Figure 1.
(A) AB5075 Wild-type opaque (VIR-O) and translucent (AV-T) colonies, viewed under a dissecting microscope with oblique lighting from underneath. (B) Qualitative assay of AHL secretion in which cultures of the wild-type (VIR-O), the VIR-O Δ1132 mutant, the complemented mutant (VIR-O Δ1132/Tn7-1132), and a version of the complemented mutant disrupted by transposon insertion (VIR-O Δ1132/Tn7-1132::T26) were spotted onto a soft agar lawn containing X-Gal and an Agrobacterium tumefaciens traG::lacZ biosensor that reacts to the presence of exogenous AHL by cleaving X-Gal, forming a blue halo. (C) Qualitative assay of AHL secretion in the wild-type (AV-T), the AV-T Δ1132 mutant, the complemented mutant (AV-T Δ1132/Tn7-1132), and a version of the complemented mutant disrupted by transposon insertion (AV-T Δ1132/Tn7-1132::T26). Signal secretion was analyzed as in panel (B), except the amount of X-gal was increased 2-fold. (D) Surface-associated motility of wild-type (VIR-O), the VIR-O Δ1132 mutant, and the complemented mutant (VIR-O Δ1132/Tn7-1132) measured on 0.3% Eiken Agar plates. A Welch’s ANOVA (****p < 0.00005) was carried out to assess (D) error bars indicate standard deviation of the mean.
One such regulator, ABUW_1645 (1645), is a TetR-type transcriptional regulator (TTTR) whose rate of expression is 150-fold higher in the AV-T state and whose overexpression in the VIR-O background drives conversion to the AV-T state (Chin et al., 2018). Although 1645 is crucial to the maintenance of the AV-T variant and is a key regulator of the VIR-O to AV-T switch, it does not appear to act in the same pathways as other previously discovered regulators of the switch such as arpB or ompR (Tipton and Rather, 2016; Tipton et al., 2017), a fact which emphasizes the complexity of the switching mechanism and the probable functional redundancy of many of the regulatory elements.
We continued to investigate additional genes to better understand the VIR-O and AV-T variants and the processes that regulate their interconversion. This led to the characterization of ABUW_1132 (1132), a LysR-type transcriptional regulator (LTTR) that influences the VIR-O to AV-T switch. LTTRs are highly abundant in Proteobacteria (Reen et al., 2015) and are the most common type of transcriptional regulator in AB5075 at 24% (59/243) (Casella et al., 2017). They often function as global regulators and act in conjunction with a ligand molecule to repress and/or activate target genes (Maddocks and Oyston, 2008). Prototypical LTTRs are self-repressing and regulate gene(s) divergently transcribed from their own coding sequence, though these need not be the case.
A recent publication by our group detailed the identification of 1132 and its role in a relA mutant (ΔrelA), which exhibits increased quorum sensing signal secretion and hyper-motility (Perez-Varela et al., 2020). Quorum sensing in A. baumannii is carried out by a LuxI-LuxR type system composed of abaI, the autoinducer synthase, and abaR, the signal receptor and transcriptional regulator (Niu et al., 2008). We reported that the quorum sensing and motility phenotypes of ΔrelA were largely enacted through 1132, which is overexpressed 14-fold in the absence of relA (Perez-Varela et al., 2020). Specifically, 1132 overexpression results in upregulation of the autoinducer synthase abaI, resulting in a large increase to secretion of the quorum sensing signal 3-OH-C12-homoserine lactone. Expression of the abaR transcriptional regulator is also increased, resulting in strong upregulation of one of AbaR’s targets: the ABUW_3766-ABUW_3773 operon. This operon promotes production of the lipopeptide acinetin-505, which acts as a surfactant and gives rise to a hyper-motile phenotype.
This study builds on these findings and details multiple phenotypic changes resulting from the deletion of 1132; including quorum sensing signal secretion, surface-associated motility, the virulence-opacity switch, capsule expression, and virulence in a mouse pneumonia model of infection.
Results
ABUW 1132 Is a Global Regulator
To identify genes and pathways regulated by 1132, we carried out genome-wide transcriptional profiling by RNA sequencing of VIR-O Δ1132 vs. wild-type VIR-O, which revealed a total of 74 differentially regulated genes in VIR-O Δ1132 (greater than 2-fold change, p value less than 0.05) ( Supplementary Table 1 ). These results showed that 1132 impacts transcription of a variety of genes involved in regulation, metabolism, protein synthesis, and possibly the cell stress response. Genes that encode ribosomal proteins, RNA polymerase, translation initiation factors, and both transcriptional and translational elongation factors are among genes that are upregulated in VIR-O Δ1132. On the other hand, several genes that are downregulated in the absence of 1132 encode a variety of enzymes involved in oxidative stress protection including catalases, peroxidases and others that interact with glutathione.
Deletion of 1132 Impacts Quorum Sensing Signal Secretion and Motility
Deletion of 1132 results in loss of secretion of the quorum sensing signal 3-OH-C12-HSL (AHL) ( Figure 1B ). This is based on the inability to activate an Agrobacterium tumefaciens traG::lacZ fusion when grown on a soft agar lawn containing this biosensor strain and X-Gal (Niu et al., 2008; Paulk Tierney and Rather, 2019). We utilized a Tn7 transposon system (Ducas-Mowchun et al., 2019a) to provide single-copy complementation of 1132 (VIR-O Δ1132-Tn7/1132), which restored AHL secretion. This restoration is lost again if the Tn7 copy of 1132 is disrupted ( Figure 1B ). The loss of AHL secretion also occurs in the AV-T Δ1132 mutant ( Figure 1C ).
Considering our previously published findings that 1132 overexpression activates the abaI-abaR system (Perez-Varela et al., 2020), we hypothesized that loss of AHL secretion was due to downregulation of abaI when 1132 is deleted. Surprisingly, our RNA sequencing analysis indicated wild-type levels of abaI expression in VIR-O Δ1132, which we confirmed by qRT-PCR ( Supplementary Figure 1 ). This result suggested that the mutant cells synthesize AHL, but do not secrete it. To investigate this possibility, we conducted an assay utilizing the A. tumefaciens biosensor in which 10% SDS was added to a well at the center of the plate ( Supplementary Figure 2 ). Cultures of wild-type VIR-O, an abaI mutant (VIR-O abaI::T26), and VIR-O Δ1132 were then added to the plate in lines going toward the SDS-containing well. As expected, we saw that the wild-type VIR-O cells uniformly activate the biosensor. However, the VIR-O Δ1132 cells show activation of the biosensor only at the point where the VIR-O Δ1132 cells have been lysed by the SDS in the presence of non-lysed biosensor cells, which confirmed our hypothesis that AHL is synthesized but cannot exit the cell. Since there is no activation by an abaI mutant near the SDS, the activating signal in the 1132 mutant is 3-OH-C12-HSL and not a released metabolite. These results indicate a more complicated role for 1132 in quorum sensing than simple regulation of abaI, which is further considered in the Discussion section.
We previously reported that overexpression of 1132 increases surface-associated motility 3.8-fold and that this effect requires both abaI and the ABUW_3766-ABUW_3773 operon (Perez-Varela et al., 2020). As expected, both VIR-O Δ1132 and AV-T Δ1132 demonstrate a significant decrease in motility compared to their wild-type counterparts, both which are complemented by the single-copy chromosomal insertion of 1132 ( Figure 1D , only VIR-O results shown).
The 1132 deletion behaves in a manner opposite to 1132 overexpression with respect to motility, but the mechanism for this is unclear. As previously mentioned, abaI mRNA levels are unaffected by 1132 deletion, and although the RNA sequencing data shows a 1.5-fold downregulation of abaR, there are no transcriptional differences in the ABUW_3766-ABUW_3773 operon ( Supplementary Table 1 ). We also considered the possibility that AHL itself acts as a surfactant to some extent and that the loss of AHL secretion could reduce motility. To test this, we constructed a double mutant of 1132 and abaR—to control for quorum sensing-directed motility—and measured motility on 0.3% Eiken agar plates containing 1 μM synthetic 3-OH-C12-HSL. However, motility was similar between the solvent control (38.75 mm ± 0.83) and the plate with AHL added (41.0 ± 1.73) p = 0.11.
Role of 1132 in Regulation of the VIR-O to AV-T Switch
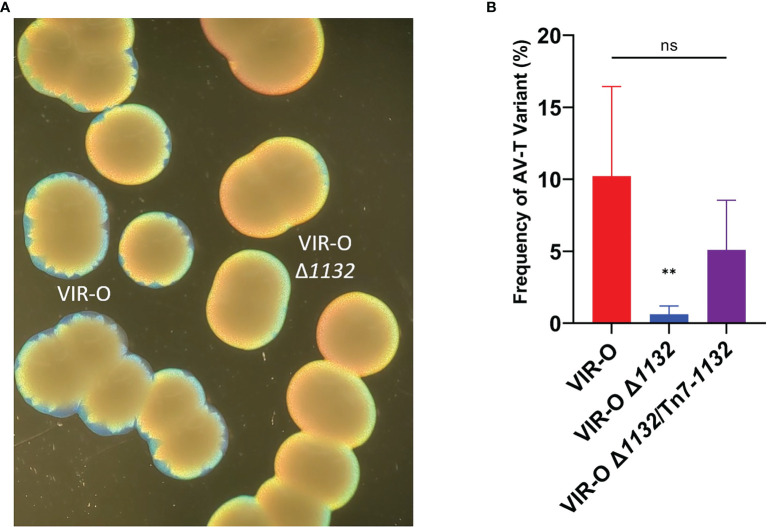
VIR-O Δ1132 exhibited lower levels of colony sectoring than observed for the wild-type VIR-O on 0.5X LB agar ( Figure 2A ). This lack of sectoring indicated a decreased rate of switching to the AV-T variant, and we subsequently quantified a 16.4-fold decrease in the switching frequency ( Figure 2B ). To confirm these effects were due to 1132 deletion, we provided single-copy complementation of 1132 using the Tn7 transposon system. This strain exhibited the wild-type phenotype of colony sectoring and restored VIR-O to AV-T switching to wild-type levels ( Figure 2B ).
Figure 2.
The Reduced Switching Phenotypes in VIR-O Δ1132. (A) Micrograph showing the loss of sectoring in VIR-O Δ1132 colonies compared to wild-type VIR-O colonies at 24 hours of growth. (B) Quantification of switching frequencies demonstrating restoration of normal switching of VIR-O Δ1132 through single copy complementation (VIR-O Δ1132/Tn7-1132). The wild-type VIR-O and VIR-O Δ1132 controls each have integrated an empty version of the pUC18T-mini-Tn7T-Apr-LAC insertion element into the attTn7 site. All micrographs were taken under a dissecting microscope illuminated from below the plate at an angle. All quantitative switching assays represent six colonies from each strain. A two-tailed Mann-Whitney test (**p < 0.005) was carried out for (B), and error bars represent the standard deviation of the mean. ns, not significant. The p value represents a comparison of both wild-type to mutant and mutant to the complemented strain.
Deletion of 1132 in the AV-T Background Increases Capsule
We further noted that AV-T Δ1132 colonies are more opaque than wild-type AV-T colonies, although AV-T Δ1132 is still translucent compared to the VIR-O Δ1132 colonies ( Figures 3A, B ). Single-copy chromosomal complementation of the AV-T Δ1132 mutant restored wild-type levels of translucence ( Figure 3A ). We first hypothesized that the increased opacity indicated hyper-switching from AV-T to VIR-O within the colony; however, the rate of switching was the same as the wild-type AV-T ( Supplementary Figure 3 ). We then considered that the AV-T Δ1132 mutant’s increased opacity may be due to increased levels of capsule.
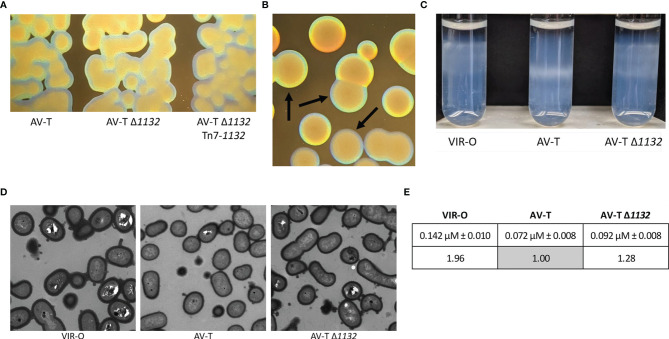
Figure 3.
Deletion of 1132 in the AV-T background alters the opacity phenotype and capsule expression. (A) Micrographs comparing representative colonies of AV-T, AV-T Δ1132 and AV-T Δ1132 Tn7-1132. (B) Micrographs comparing colonies of AV-T Δ1132, indicated by arrows, to VIR-O Δ1132. Micrographs were taken under a dissecting scope lit from underneath at an angle. (C) Wild-type VIR-O, wild-type AV-T, and AV-T Δ1132 cells were layered onto a Percoll gradient (top layer 40%, bottom layer 50%) and centrifuged for 30 minutes at 3,000 xg. AV-T Δ1132 cells migrate between the stopping points for VIR-O and AV-T wild-types, indicating intermediate capsular levels with high levels of heterogeneity in the AV-T Δ1132 mutant. Photograph shown is one of six experimental replicates. (D) Transmission electron micrographs of representative wild-type VIR-O, wild-type AV-T, and AV-T Δ1132 cells, stained with Ruthenium red. (E) Averages ± standard deviation of the mean of capsule widths of these three strains converted to ratios to the wild-type AV-T. The difference between the wild-type AV-T and AV-T Δ1132 capsule widths is significant at p < 0.0001 as determined by a Student’s two-tailed t test.
Previous work revealed that wild-type VIR-O cells exhibit a 2-fold increase in capsule compared with the wild-type AV-T (Chin et al., 2018). We utilized a Percoll density gradient, a method that was recently described as a method to separate A. baumannii strains by capsule level (Kon et al., 2020), to compare capsular polysaccharide levels in AV-T Δ1132 to wild-type VIR-O and AV-T cells. As seen in Figure 3C , the gradient is able to distinguish between capsule levels of the wild-type VIR-O and AV-T variants, with the AV-T cells migrating significantly further than the VIR-O cells. Intriguingly, AV-T Δ1132 cells exhibited a high degree of heterogeneity and occupied a space in the gradient layer that was intermediate to that of the wild-type VIR-O and AV-T. We interpret this result as indication that deletion of 1132 causes a dysregulation and increase of capsule in the AV-T state, which imparts the more opaque appearance of AV-T Δ1132.
To confirm the effect of 1132 deletion on capsule, we next carried out TEM imaging of cell samples of wild-type VIR-O, wild-type AV-T, and AV-T Δ1132 stained with Ruthenium red ( Figure 3D ). We used ImageJ to measure capsule width in 100 cells per strain with 3 measurements taken per cell, which revealed average capsule widths of 0.142 µM ± 0.010, 0.072 µM ± 0.008, and 0.092 µM ± 0.008 in VIR-O, AV-T, and AV-T Δ1132, respectively ( Figure 3E ). The resulting ratio of these values is 1.96:1.00:1.28, which demonstrates the 2-fold difference in capsule previously seen in the VIR-O vs. AV-T (Chin et al., 2018) and confirms a 28% increase in capsule in the AV-T background when 1132 is deleted. The difference between the wild-type AV-T and AV-T Δ1132 is highly significant at p < 0.0001 as determined by a Student’s two-tailed t test.
In light of these results, we considered that, in the AV-T state, 1132 may regulate the K locus genes that encode the proteins largely responsible for the biosynthesis and export of capsular polysaccharide (CPS). We assessed representative genes from the K locus—manB, galE, ABUW_3818, ABUW_3820, ABUW_3821, ABUW_3830, wza, wzb, and wzc—by qRT-PCR across three sets of samples ( Supplementary Figure 4 ). These results indicated that 1132 did not transcriptionally regulate the K locus genes.
The Δ1132 Mutation Increases Virulence in the AV-T Background
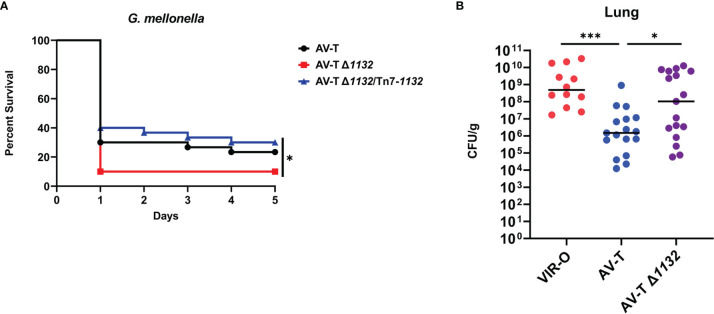
Capsule is a known virulence factor in A. baumannii, and presumably contributes to virulent phenotype of the VIR-O variant relative to AV-T (Russo et al., 2010; Chin et al., 2018; Singh et al., 2018; Tipton et al., 2018; Talyansky et al., 2021). The intermediate capsular levels of AV-T Δ1132 therefore suggested that this deletion may increase virulence. Before initiating virulence studies, we first tested the growth rates of the AV-T and AV-T Δ1132 strains and found no significant differences under laboratory conditions ( Supplementary Figure 5A . Using the Galleria mellonella (waxworm) model of infection, a modest, but statistically significant increase in AV-T Δ1132 virulence was observed ( Figure 4A ). Waxworms injected with the wild-type AV-T control showed a 23.3% survival rate after five days, while only 10% of those injected with AV-T Δ1132 survived. Complementation of AV-T Δ1132 with the single-copy chromosomal insertion of 1132 (AV-T Δ1132/Tn7-1132) reversed the increase in virulence, bringing the rate of survival back up to 30%.
Figure 4.
Deletion of 1132 in AV-T background increases virulence. (A) Galleria mellonella infected with AV-T Δ1132 are killed at higher rates compared with wild-type AV-T or the complemented mutant (AV-T Δ1132/Tn7-1132). (B) Bacterial CFU/g recovered from mice lungs 24 hours after intranasal inoculation with pure cultures of wild-type VIR-O, wild-type AV-T, or AV-T Δ1132. A Log-Rank (Mantel-Cox) test (*p < 0.05) was carried out for (A) and a two-tailed Mann-Whitney test (*p < 0.05; ***p < 0.0005) was carried out for (B).
We next examined the virulence of AV-T Δ1132 in a mouse pneumonia model of infection. In three experiments, mice were intranasally inoculated with 1 x 108 CFU/mL of VIR-O (n=10), AV-T (n=17), or AV-T Δ1132 (n=17). At 24 hours post-inoculation, lungs were harvested, and the CFU/g for each tissue was calculated where a significant increase in CFU/g in AV-T Δ1132 was observed compared to wild-type AV-T in the lungs ( Figure 4B ). Both strains exhibited a similar number of VIR-O variants recovered from the lungs (approximately 0.1%). The highly virulent VIR-O variant was unaffected by the Δ1132 mutation in both a Galleria mellonella waxworm model and in a mouse pneumonia model ( Supplementary Figures 5B, C ) and both strains exhibited similar growth rates in-vitro ( Supplementary Figure 5A ).
Discussion
This study confirms 1132 as a global transcriptional regulator that impacts multiple pathways: surface-associated motility, AHL secretion, regulation of the opacity-virulence switch, and capsule expression. Most importantly, our work demonstrates that an 1132 deletion, and possibly its natural downregulation, has the potential to increase virulence of the typically avirulent translucent colony variant in the clinical isolate AB5075. The 1132 gene was highly conserved in all completed A. baumannii genome sequences (n = 330), where at least 98.7% nucleotide homology was observed. The gene was also conserved in A. nosocomialis (91.7% or greater nucleotide identity), A. seifertii (89.8% or greater identity) and A. pittii (83.7% or greater identity). Given the high conservation of 1132 among A. baumannii strains, it is likely that the 1132-associated phenotypes we have reported here and in a previous publication (Perez-Varela et al., 2020) will also be conserved in other strains.
Some questions remain regarding the mechanisms through which 1132 acts in the described phenotypes. As previously mentioned, in our earlier publication we determined that overexpression of 1132 causes large increases in AHL secretion and surface-associated motility. This occurs through activation of the abaI-abaR system and follows a well-defined downstream pathway ending with overproduction of the surfactant acinetin-505 (Perez-Varela et al., 2020) Perplexingly, although the deletion of 1132 results in phenotypes opposite to that of 1132 overexpression—loss of both AHL secretion and motility—our experiments show that neither of these phenotypes are due to transcriptional downregulation of the abaI-abaR system or the ABUW_3766-ABUW_3773 operon. A post-transcriptional effect of 1132 on abaI also seems unlikely, as VIR-O Δ1132 cells lysed in the presence of the AHL-detecting biosensor are able to activate the traG::lacZ fusion, indicating a functional AbaI protein ( Supplementary Figure 2 ). It is possible that the length of the N-acyl chain of the AHL molecule is altered due to the global changes that occur when 1132 is deleted. Both shortened and elongated chains could affect the hydrophobicity of the AHL molecule and therefore have the potential to disrupt diffusion or export of the autoinducer across the membrane. Due to the trapping of signal within the cell, it is possible that the quorum sensing response is altered, possibly being activated earlier in cell density, or being constitutive. With respect to capsule, the effect of 1132 on capsule thickness did not involve transcriptional changes in representative genes within the capsule locus. Therefore, the 1132 mutation may alter biosynthetic pathways that impact precursors for capsule synthesis in a manner that increases their abundance and results in enhanced capsule thickness.
This work revealed that 1132 positively regulated the VIR-O to AV-T switch and experiments to address the mechanism for this increase are ongoing. It is possible that one or more of several regulatory genes revealed by the RNA sequencing as differentially regulated by 1132 is involved. However, it is intriguing that other known regulators of the switch, including the OmpR/EnvZ two-component system and the ArpB efflux pump, were not regulated by 1132. A possibility is that the 1132 deletion creates a cellular metabolome that withholds ligands or other molecules required by other known regulators of the switch, such as the TetR regulator ABUW_1645 (Chin et al., 2018). It is important to note that the lack of AHL secretion is not expected to impact the switch, as deletion of abaI does not cause alteration of switching frequency (Tipton et al., 2015).
Our work here has important implications for the characterizations of VIR-O cells as virulent and AV-T cells as avirulent. While the increase in virulence in the AV-T variant caused by 1132 deletion was modest, this finding demonstrates that stochastic downregulation of 1132—potentially mediated by RelA’s repression of 1132—may allow an AV-T cell to be moderately virulent in a mammalian host without necessitating a switch to the VIR-O state. We have previously hypothesized that AV-T cells possess an advantage in natural environments due to their versatility in utilization of carbon sources and their improved formation of biofilm (Chin et al., 2018). Combined, these observations suggest that AB5075, and possibly other A. baumannii strains, could survive in a natural environment while existing in a virulent state.
Materials and Methods
Bacterial Strains and Growth Conditions
All experiments were conducted using the clinical isolate AB5075. For all experiments, cultures were started from frozen glycerol stocks containing at least 99.5% of the desired colony variant (VIR-O or AV-T). Cultures were grown in either broth or on solid media containing LB (10 g tryptone, 5 g yeast extract, and 5 g NaCl per liter) and 1.5% agar (Difco) (“1X” LB agar) or 0.8% agar (“0.5X” LB agar).
Methods to Distinguish Colony Variants
Cultures were plated or streaked on 0.5X LB agar and viewed under a dissecting microscope, illuminated from underneath at an oblique angle. Colonies must be viewed at high colony density (at least 100 colonies per plate) to effectively distinguish VIR-O and AV-T colonies.
Detection of 3-OH-C12-HSL
Plates containing the Agrobacterium tumefaciens traG::lacZ biosensor and X-Gal were prepared as described in Paulk Tierney and Rather (2019). AB5075 cell cultures were grown in 2 mL LB to OD600 ~ 0.3, and 1 µL was spotted onto the soft agar lawn. Plates were incubated at 28°C overnight.
Quantification of VIR-O and AV-T Switching
Dilutions were plated from frozen pure stocks of the strains to be assessed, and plates were incubated at 37°C for a set number of growth hours. High density plates were used to verify the purity of the stock. Isolated colonies were extracted from the plate by cutting out a section of agar underlying the colony and then resuspended and dilutions plated. The percent variant was then determined for each set of resuspended colonies from each strain. All experiments were performed twice with three colonies each for a total of six colonies.
Electroporation
Cell cultures for competent cells were grown in 2 mL LB to OD600 ~ 0.5 (mid-log phase) at 37°C, shaking from frozen stocks. Cultures were pelleted and washed twice in sterile dH2O, then resuspended to accommodate a volume of approximately 50 µL of cell per electroporation. Plasmid minipreps or ligation products mixed with cells were added to 2 mm cuvettes and electroporated at 2.5 kV. Cells were recovered in 1 mL LB at 37°C, shaking for one hour, then plated onto media containing selective antibiotics.
Construction of Deletion Mutations
In-frame deletion of ABUW_1132 was generated using sucrose counterselection and a suicide vector containing the sacB marker (pEX18Tc) using methods previously described (Hoang et al., 1998). Briefly, PCR amplification (Phusion polymerase, Thermo-Fisher Scientific) was used to amplify the 2-kb regions upstream and downstream of the gene to be deleted, gel purified (UltraClean 15 DNA Purification Kit, MO BIO Laboratories), and ligated (Fast-Link DNA Ligation Kit, Epicentre Biotechnologies). The 4-kb ligation product was PCR amplified, gel purified, and ligated into the pEX18Tc vector MCS. The product vector was verified by sequencing and then transformed into competent E. coli Transformax EC100D cells (Epicentre Biotechnologies) by electroporation. The resulting suicide vector was confirmed by PCR and transformed by electroporation into AB5075, grown to OD600 of 0.5 in 2-mL LB and washed twice in 10% glycerol. Transformants were plated on 1X LB agar plus Tetracycline (5-µg/mL) to yield single-crossover mutants. Counterselection was carried out at room temperature on 1X LB plates with 10% sucrose and no NaCl, and colonies were screened by PCR for the deletion. Primer sequences are recorded in Supplementary Table 2 .
To construct the Δ1132, abaR::T26 double mutant, genomic DNA was purified from an abaR::T26 mutant obtained from the University of Washington AB5075 transposon mutant library. Genomic DNA was then electroporated into VIR-O Δ1132. Transformants were selected on 1X LB plus Tetracycline (5-µg/mL) and confirmed by PCR.
Construction of Single-Copy Complementation in Deletion Mutants
We utilized a Tn7-based single-copy insertion element system (Ducas-Mowchun et al., 2019a; Ducas-Mowchun et al., 2019b) to reintroduce 1132 into the VIR-O Δ1132 and AV-T Δ1132 deletion mutants. We opted to use a segment of 1132 that includes a large portion of the up and downstream regions (GenBank Accession NZ_CP008706.1:1152309-1156236). This is the portion of the genome contained within the original 1132-containing fragment isolated from a high-copy AB5075 chromosomal library during the screen that led to our first discovery of 1132 (Perez-Varela et al., 2020), and previous experiments showed that this fragment allows for optimal expression of 1132. A modified version of this construct containing a T26 transposon (tetracycline) in 1132 had also been made to confirm loss of the phenotypes presumably caused by 1132 overexpression. This was made by digestion of the plasmid with PmlI, which cuts once near the beginning of the 1132 ORF, and re-ligation with a PCR-amplified T26 transposon. The original 1132-containing fragment and the modified version with 1132::T26 were excised from these plasmids by digestion with XbaI, which flanks the site into which the library was cloned, and gel purified to be used in construction of the suicide vector.
The pUC18T-mini-Tn7T-Apr-LAC construct was digested with SpeI and ligated with the 1132- or 1132::T26-containing fragment (Fast-Link DNA Ligation Kit, Epicentre Biotechnologies). The ligation product was transformed into competent E. coli Transformax EC100D cells (Epicentre Biotechnologies) by electroporation. The resulting constructs—pUC18T-mini-Tn7T-Apr-LAC/1132 and pUC18T-mini-Tn7T-Apr-LAC/1132::T26—were confirmed by PCR and sequencing then transformed into pure cultures of VIR-O Δ1132 and AV-T Δ1132 by electroporation along with the helper plasmid pTNS2. Transformant colonies were screened by PCR for insertion into the attTn7 site and those containing the correct insertion were again verified by sequencing. Wild-type Tn7 and Δ1132-Tn7 control strains were also made in both VIR-O and AV-T backgrounds using the same methods with an empty pUC18T-mini-Tn7T-Apr-LAC construct. Primer sequences are recorded in Supplementary Table 2 .
RNA Preparation and qRT-PCR Analysis
Strains for qRT-PCR analysis in Supplementary Figure 1 were prepared for RNA isolation by growing them from pure frozen stocks to OD600 ~ 0.15 then plating 100 µL on a 1X LB plate. Plates were incubated at 37°C for 6 hours, then pooled with 3 mL ice cold LB. Resuspensions were normalized to within OD600 of 0.01, then 1 mL of the resuspension was pelleted, flash frozen in an ethanol-dry ice bath and stored at -80°C. The resuspension cultures were streaked on 0.5X LB to ensure sample purity.
Strains for qRT-PCR analysis in Supplementary Figure 4 were grown up to match the growth conditions of the cells prepared for electron microscopy (see below). Samples were grown from pure frozen stocks in overnight cultures of 2 mL LB and were then shaken at 37°C to the same OD600 of 0.3. 20 µL of each culture was then plated as a line onto a 0.5X plate, allowed to incubate at 37°C for 4 hours, and then harvested off the plate. Scraped cells were resuspended to an OD600 of 0.6 in 2 mL LB. 1 mL of cells was pelleted, flash frozen in an ethanol-dry ice bath, then stored at -80°C. The resuspension cultures were streaked on 0.5X LB to ensure sample purity.
RNA was prepared using the Epicentre MasterPure RNA Purification kit according to the manufacturer’s protocols. The resulting nucleic acid product was purified to remove DNA contamination using the Invitrogen TURBO DNA-free kit according to the manufacturer’s instructions. Following quantification of RNA concentration using a NanoDrop ND-1000 spectrophotometer, cDNA was prepared from 1 µg of RNA using the High-Capacity cDNA Reverse Transcription Kit by Applied Biosystems and subsequently diluted 1:10 in nuclease-free water. qRT-PCR experiments were carried out on a Bio-Rad CFX Connect Real-Time PCR Detection System using iQ SYBR Green Supermix reverse transcriptase from Bio-Rad. RNA purity was confirmed by qRT-PCR through the inclusion of template controls made without reverse transcriptase. qRT-PCR data was analyzed by the delta-delta Ct method (2–∆∆Ct) with comparison to 16S as an internal control. This method was carried out for three biological replicates. Each biological replicate had three technical replicates for each primer set. Primer sequences are recorded in Supplementary Table 2 .
RNA Sequencing and Analysis
Three independent sets of RNA were prepared from each of AB5075 strains VIR-O and VIR-O Δ1132. Cultures were grown in 2 mL LB from pure frozen stocks to OD600 ~ 0.5 (shaking, 37°C) and were streaked on 0.5X LB to ensure purity of samples. 1 mL of cells was pelleted and flash frozen in a dry ice bath before being stored at -80°C. RNA was prepared using the Epicentre MasterPure RNA Purification kit according to the manufacturer’s protocols.
RNA quality control, sequencing, and analysis was carried out by the Yerkes Non-Human Primate Genomics Core at Emory University. RNA quantity and quality assessments were carried out using a Thermo Nanodrop2000 and Agilent 2100 Bioanalyzer, respectively. RNA sequencing was carried out using an Illumina HiSeq 3000, and reads were normalized and mapped using Cufflinks software. The RNA-Seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE185730 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185730).
Percoll Density Gradient
Method slightly modified from Kon et al. (2020). Cell cultures of VIR-O, AV-T, and AV-T Δ1132 were grown in 2 mL LB from pure frozen stocks to OD600 ~ 0.6 (shaking, 37°C). For each strain, 600 µL of cells was pelleted and washed in PBS 1X, then resuspended in 100 µL PBS 1X. Percoll solutions were prepared by first mixing 9 parts Percoll (Sigma Aldrich Cat. P4937) and 1 part 1.5 M NaCl, which was further diluted using 0.15 M NaCl to make 40% and 50% Percoll solutions. For each cell sample to be tested, 1 mL of 40% Percoll was added to a 12x75 mm glass tube (Fisherbrand 14958C), and 1 mL 50% Percoll was gently layered underneath using a 1cc syringe and a 1.5 in. 23G needle (BD 305194). 100 µL of cells were gently layered at the top of the gradient. Samples were spun at 3,000 xg for 30 minutes at room temperature and then visually assessed. This protocol was repeated 6 times.
Electron Microscopy
Ruthenium red-lysine fixation and cell staining and subsequent transmission electron microscopy were performed by the Robert P. Apkarian Integrated Electron Microscopy Core at Emory University using previously described techniques (Fassel et al., 1997; Fassel et al.,1998; Beaussart et al., 2014). Samples were provided to the EM Core as follows: cells grown from pure frozen stocks in overnight cultures were shaken at 37°C until OD ~ 0.30, and 20 µL of each culture was plated as a line onto the same 0.5X LB plate. Plates were then incubated at 37°C for 4 hours and stored overnight at 4°C before being transported to the EM Core.
Capsule widths in the electron micrographs were measured by ImageJ 1.53e (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, https://imagej.nih.gov/ij/, 1997-2018). Three measurements were taken for 100 cells of each strain at the same magnification, which were averaged and then converted to micrometers.
Galleria mellonella Larvae (Waxworm) Virulence Assays
Waxworms were purchased from Speedy Worm (www.speedyworm.com). Cultures of AV-T, AV-T Δ1132, and AV-T Δ1132/Tn7-1132 were grown in 2 mL LB from pure frozen stocks to OD600 ~ 0.5 (shaking, 37°C). 5 µL of a LB control, AV-T, AV-T Δ1132, or AV-T Δ1132/Tn7-1132 was injected into the hemolymph of a larvae of mass 150-200 mg (n=30 per condition; approximately 1.2 x 106 CFU were administered per strain). Larvae were then housed in petri dishes in a humidified incubator at 37°C for 5 days. Each day, dead larvae were removed and surviving larvae counted.
Mouse Virulence Assays
Approximately 1 × 108 CFU were administered per mouse for infections to quantify the bacterial load. For mouse infections, overnight standing bacterial cultures at room temperature were sub-cultured in LB broth and grown at 37°C with shaking to an OD600 ~0.15, washed and re-suspended in PBS. Each mouse was inoculated intranasally with 50 µL of bacteria. Mice were anesthetized with isoflurane immediately prior to intranasal inoculation. At 24 hours, the mice were sacrificed and the lungs were harvested, homogenized, and ten-fold serial dilutions plated for CFU on 0.5X LB plates. The mouse strain used was C57BL/6J (females at 8-10 weeks of age) from Jackson Laboratories (JAX stock #000664). Experiments were carried out under the Institutional Animal Care and Use Committee guidelines.
Statistics
Statistics were performed using GraphPad Prism 9.2.0 software for Windows (GraphPad Software, San Diego, California USA, www.graphpad.com). The following statistical tests were utilized: (1) Mann-Whitney test for the mouse experiments (two-tailed), two-sample switching assays (two-tailed), motility assays (two-tailed) and qRT-PCR experiments (two-tailed or multiple, as indicated); (2) Log-Rank (Mantel-Cox) test for the Galleria mellonella experiments; (3) Welch’s ANOVA analysis for three-sample switching assays; and (4) Student’s two-tailed t test for capsule width measurements from the TEM micrographs. The Mann-Whitney test was utilized instead of a Student’s t test to allow for the possibility or reality of a non-Gaussian distribution. Welch’s ANOVA was chosen over standard ANOVA to allow for unequal variance between samples.
Data Availability Statement
The RNA-Seq data presented in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE185730 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185730).
Ethics Statement
The animal study was reviewed and approved by Emory University Institutional Animal Care and Use Committee.
Author Contributions
PR and AT conceptualized and designed the study and experiments. AT carried out all of the experiments except the RNA sequencing, mouse experiments, and electron microscopy. CC performed the mouse experiments and subsequent data analysis. AT analysed all other data with guidance from PR. AT and CC wrote the original manuscript. All authors contributed to the article and approved the submitted version.
Funding
Research in the PNR Laboratory is supported by T32 AI106699 to AT and NIH R01 AI72219 and R21 AI115183 and Department of Veterans Affairs awards I01 BX001725 and IK6BX004470 to PR. Research in the DSW Laboratory is supported by a Department of Veteran’s Affairs award BX002788. DW is also supported by a Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Disease award. The content expressed herein is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Department of Veterans Affairs.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dr. M. Pérez-Varela for critical reading of this manuscript and Drs. K.A. Tipton, S.E. Anderson, J.M. Colquhoun, and M. Pérez-Varela for their valuable input. We are grateful to Dr. Ayush Kumar at the University of Manitoba for providing pUC18T-mini-Tn7T-Apr-LAC. This work was supported in part by the Robert P. Apkarian Integrated Electron Microscopy Core at Emory University, and we especially thank Jeannette Taylor and Dr. Ricardo C. Guerrero for conducting the electron microscopy.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.778331/full#supplementary-material
Deletion of 1132 does not affect abaI expression. qRT-PCR experiments comparing wild-type VIR-O and VIR-O Δ1132 indicate no significant difference in the expression of abaI, using 16S as an internal control (two-tailed Mann-Whitney test, ns, not significant). Results are the average of three biological replicates.
VIR-O Δ1132 cells contain quorum sensing signal (AHL), but do not secrete it. Cultures of wild-type VIR-O, VIR-O Δ1132, and VIR-O ΔabaI were grown to the same OD600 and plated as a line onto a soft-agar lawn containing Agrobacterium tumefaciens (traG::lacZ) and X-Gal. 10% SDS was added to a well at the center of the plate, lysing the cells.
AV-T Δ1132 switches to the VIR-O variant at the same frequency as wild-type AV-T. Bars represent averages of six colonies assayed for switching frequency at 18 hours of growth. Error bars indicate standard deviation of the mean, and there is no statistical significance as assessed by Welch’s ANOVA.
K locus genes for capsule polysaccharide synthesis and export are not transcriptionally regulated by 1132. Bars for AV-T Δ1132 and AV-T Δ1132/Tn7-1132 represent averages of three biological replicates. Error bars indicate standard deviation of the mean, and there is no statistical significance in expression across strains as assessed by multiple Mann-Whitney tests.
(A) Growth curves for VIR-O, AV-T, VIR-O Δ1132, and AV-T Δ1132 in LB media. Data represents two biological replicates, and error bars indicate standard deviation of the mean. (B) Galleria mellonella infected with VIR-O Δ1132 show no significant difference in mortality compared with wild-type VIR-O (n=30 per strain) as assessed by a log-rank (Mantel-Cox) test. (C) Mice were infected intranasally with either VIR-O or VIR-O Δ1132. At 24 hours post-inoculation, bacteria were recovered from the lungs, spleen, and liver tissues and CFU/g quantified. There is no significant difference between the two strains in all tissues as assessed by a Mann-Whitney test. ns, not significant.
References
- Anderson S. E., Chin C. Y., Weiss D. S., Rather P. N. (2020). Copy Number of an Integron-Encoded Antibiotic Resistance Locus Regulates a Virulence and Opacity Switch in Acinetobacter Baumannii AB5075. mBio 11. doi: 10.1128/mBio.02338-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaussart A., Péchoux C., Trieu-Cuot P., Hols P., Mistou M. Y., Dufrêne Y. F. (2014). Molecular Mapping of the Cell Wall Polysaccharides of the Human Pathogen Streptococcus Agalactiae. Nanoscale 6, 14820–14827. doi: 10.1039/C4NR05280C [DOI] [PubMed] [Google Scholar]
- Bravo Z., Orruno M., Navascues T., Ogayar E., Ramos-Vivas J., Kaberdin V. R., et al. (2019). Analysis of Acinetobacter Baumannii Survival in Liquid Media and on Solid Matrices as Well as Effect of Disinfectants. J. Hosp. Infect. 103, e42–e52. doi: 10.1016/j.jhin.2019.04.009 [DOI] [PubMed] [Google Scholar]
- Casella L. G., Weiss A., Pérez-Rueda E., Antonio Ibarra J., Shaw L. N. (2017). Towards the Complete Proteinaceous Regulome of Acinetobacter Baumannii. Microb. Genom. 3, mgen000107. doi: 10.1099/mgen.0.000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2019). Antibiotic Resistance Threats in the United States, 2019 (Atlanta, GA: ). Available at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (Accessed August 2 2021). [Google Scholar]
- Chapartegui-Gonzalez I., Lazaro-Diez M., Bravo Z., Navas J., Icardo J. M., Ramos-Vivas J. (2018). Acinetobacter Baumannii Maintains its Virulence After Long-Time Starvation. PloS One 13, e0201961. doi: 10.1371/journal.pone.0201961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. Y., Tipton K. A., Farokhyfar M., Burd E. M., Weiss D. S., Rather P. N. (2018). A High-Frequency Phenotypic Switch Links Bacterial Virulence and Environmental Survival in Acinetobacter Baumannii. Nat. Microbiol. 3, 563–569. doi: 10.1038/s41564-018-0151-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark N. M., Zhanel G. G., Lynch J. P., 3rd (2016). Emergence of Antimicrobial Resistance Among Acinetobacter Species: A Global Threat. Curr. Opin. Crit. Care 22, 491–499. doi: 10.1097/MCC.0000000000000337 [DOI] [PubMed] [Google Scholar]
- D'souza A. W., Potter R. F., Wallace M., Shupe A., Patel S., Sun X., et al. (2019). Spatiotemporal Dynamics of Multidrug Resistant Bacteria on Intensive Care Unit Surfaces. Nat. Commun. 10, 4569. doi: 10.1038/s41467-019-12563-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K. A., Moran K. A., Mcallister C. K., Gray P. J. (2005). Multidrug-Resistant Acinetobacter Extremity Infections in Soldiers. Emerg. Infect. Dis. 11, 1218–1224. doi: 10.3201/1108.050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkshoorn L., Nemec A., Seifert H. (2007). An Increasing Threat in Hospitals: Multidrug-Resistant Acinetobacter Baumannii. Nat. Rev. Microbiol. 5, 939–951. doi: 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- Doyle J. S., Buising K. L., Thursky K. A., Worth L. J., Richards M. J. (2011). Epidemiology of Infections Acquired in Intensive Care Units. Semin. Respir. Crit. Care Med. 32, 115–138. doi: 10.1055/s-0031-1275525 [DOI] [PubMed] [Google Scholar]
- Ducas-Mowchun K., De Silva P. M., Crisostomo L., Fernando D. M., Chao T. C., Pelka P., et al. (2019. a). Next Generation of Tn7-Based Single-Copy Insertion Elements for Use in Multi- and Pan-Drug-Resistant Strains of Acinetobacter Baumannii. Appl. Environ. Microbiol. 85. doi: 10.1128/AEM.00066-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducas-Mowchun K., De Silva P. M., Patidar R., Schweizer H. P., Kumar A. (2019. b). Tn7-Based Single-Copy Insertion Vectors for Acinetobacter Baumannii. Methods Mol. Biol. 1946, 135–150. doi: 10.1007/978-1-4939-9118-1_13 [DOI] [PubMed] [Google Scholar]
- Fassel T. A., Mozdziak P. E., Sanger J. R., Edmiston C. E. (1997). Paraformaldehyde Effect on Ruthenium Red and Lysine Preservation and Staining of the Staphylococcal Glycocalyx. Microsc. Res. Tech. 36, 422–427. doi: [DOI] [PubMed] [Google Scholar]
- Fassel T. A., Mozdziak P. E., Sanger J. R., Edmiston C. E. (1998). Superior Preservation of the Staphylococcal Glycocalyx With Aldehyde-Ruthenium Red and Select Lysine Salts Using Extended Fixation Times. Microsc. Res. Tech. 41, 291–297. doi: [DOI] [PubMed] [Google Scholar]
- Gallagher L. A., Ramage E., Weiss E. J., Radey M., Hayden H. S., Held K. G., et al. (2015). Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter Baumannii. J. Bacteriol. 197, 2027–2035. doi: 10.1128/JB.00131-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998). A Broad-Host-Range Flp-FRT Recombination System for Site-Specific Excision of Chromosomally-Located DNA Sequences: Application for Isolation of Unmarked Pseudomonas Aeruginosa Mutants. Gene 212, 77–86. doi: 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Jacobs A. C., Thompson M. G., Black C. C., Kessler J. L., Clark L. P., Mcqueary C. N., et al. (2014). AB5075, a Highly Virulent Isolate of Acinetobacter Baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. mBio 5, e01076–e01014. doi: 10.1128/mBio.01076-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawad A., Seifert H., Snelling A. M., Heritage J., Hawkey P. M. (1998). Survival of Acinetobacter Baumannii on Dry Surfaces: Comparison of Outbreak and Sporadic Isolates. J. Clin. Microbiol. 36, 1938–1941. doi: 10.1128/JCM.36.7.1938-1941.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon H., Schwartz D., Temkin E., Carmeli Y., Lellouche J. (2020). Rapid Identification of Capsulated Acinetobacter Baumannii Using a Density-Dependent Gradient Test. BMC Microbiol. 20, 285. doi: 10.1186/s12866-020-01971-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. R., Lee J. H., Park M., Park K. S., Bae I. K., Kim Y. B., et al. (2017). Biology of Acinetobacter Baumannii: Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front. Cell Infect. Microbiol. 7, 55. doi: 10.3389/fcimb.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks S. E., Oyston P. C. F. (2008). Structure and Function of the LysR-Type Transcriptional Regulator (LTTR) Family Proteins. Microbiology 154, 3609–3623. doi: 10.1099/mic.0.2008/022772-0 [DOI] [PubMed] [Google Scholar]
- Niu C., Clemmer K. M., Bonomo R. A., Rather P. N. (2008). Isolation and Characterization of an Autoinducer Synthase From Acinetobacter Baumannii. J. Bacteriol. 190, 3386–3392. doi: 10.1128/JB.01929-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulk Tierney A. R., Rather P. N. (2019). Methods for Detecting N-Acyl Homoserine Lactone Production in Acinetobacter Baumannii. Methods Mol. Biol. 1946, 253–258. doi: 10.1007/978-1-4939-9118-1_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Hooper D. C. (2010). Hospital-Acquired Infections Due to Gram-Negative Bacteria. N. Engl. J. Med. 362, 1804–1813. doi: 10.1056/NEJMra0904124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter Baumannii: Emergence of a Successful Pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Varela M., Tierney A. R. P., Kim J., Vazquez-Torres A., Rather P. N. (2020). Characterization of RelA in Acinetobacter Baumannii. J. Bacteriol. doi: 10.1128/JB.00045-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reen F. J., Romano S., Dobson A. D., O'gara F. (2015). The Sound of Silence: Activating Silent Biosynthetic Gene Clusters in Marine Microorganisms. Mar. Drugs 13, 4754–4783. doi: 10.3390/md13084754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha I. V., Xavier D. E., Almeida K. R. H., Oliveira S. R., Leal N. C. (2018). Multidrug-Resistant Acinetobacter Baumannii Clones Persist on Hospital Inanimate Surfaces. Braz. J. Infect. Dis. 22, 438–441. doi: 10.1016/j.bjid.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo T. A., Luke N. R., Beanan J. M., Olson R., Sauberan S. L., Macdonald U., et al. (2010). The K1 Capsular Polysaccharide of Acinetobacter Baumannii Strain 307-0294 Is a Major Virulence Factor. Infect. Immun. 78, 3993–4000. doi: 10.1128/IAI.00366-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J. K., Adams F. G., Brown M. H. (2018). Diversity and Function of Capsular Polysaccharide in Acinetobacter Baumannii. Front. Microbiol. 9, 3301. doi: 10.3389/fmicb.2018.03301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talyansky Y., Nielsen T. B., Yan J., Carlino-Macdonald U., Di Venanzio G., Chakravorty S., et al. (2021). Capsule Carbohydrate Structure Determines Virulence in Acinetobacter Baumannii. PloS Pathog. 17, e1009291. doi: 10.1371/journal.ppat.1009291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton K. A., Chin C. Y., Farokhyfar M., Weiss D. S., Rather P. N. (2018). Role of Capsule in Resistance to Disinfectants, Host Antimicrobials, and Desiccation in Acinetobacter Baumannii. Antimicrob. Agents Chemother. 62. doi: 10.1128/AAC.01188-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton K. A., Dimitrova D., Rather P. N. (2015). Phase-Variable Control of Multiple Phenotypes in Acinetobacter Baumannii Strain Ab5075. J. Bacteriol. 197, 2593–2599. doi: 10.1128/JB.00188-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton K. A., Farokhyfar M., Rather P. N. (2017). Multiple Roles for a Novel RND-Type Efflux System in Acinetobacter Baumannii AB5075. Microbiologyopen 6. doi: 10.1002/mbo3.418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton K. A., Rather P. N. (2016). An Ompr/envZ Two-Component System Ortholog Regulates Phase Variation, Osmotic Tolerance, Motility, and Virulence in Acinetobacter Baumannii Strain AB5075. J. Bacteriol. doi: 10.1128/JB.00188-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner L. M., Webb A. K., Limbago B., Dudeck M. A., Patel J., Kallen A. J., et al. (2016). Antimicrobial-Resistant Pathogens Associated With Healthcare-Associated Infections: Summary of Data Reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention 2011-2014. Infect. Control Hosp. Epidemiol. 37, 1288–1301. doi: 10.1017/ice.2016.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong D., Nielsen T. B., Bonomo R. A., Pantapalangkoor P., Luna B., Spellberg B. (2017). Clinical and Pathophysiological Overview of Acinetobacter Infections: A Century of Challenges. Clin. Microbiol. Rev. 30, 409–447. doi: 10.1128/CMR.00058-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2017) Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available at: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (Accessed August 2 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Deletion of 1132 does not affect abaI expression. qRT-PCR experiments comparing wild-type VIR-O and VIR-O Δ1132 indicate no significant difference in the expression of abaI, using 16S as an internal control (two-tailed Mann-Whitney test, ns, not significant). Results are the average of three biological replicates.
VIR-O Δ1132 cells contain quorum sensing signal (AHL), but do not secrete it. Cultures of wild-type VIR-O, VIR-O Δ1132, and VIR-O ΔabaI were grown to the same OD600 and plated as a line onto a soft-agar lawn containing Agrobacterium tumefaciens (traG::lacZ) and X-Gal. 10% SDS was added to a well at the center of the plate, lysing the cells.
AV-T Δ1132 switches to the VIR-O variant at the same frequency as wild-type AV-T. Bars represent averages of six colonies assayed for switching frequency at 18 hours of growth. Error bars indicate standard deviation of the mean, and there is no statistical significance as assessed by Welch’s ANOVA.
K locus genes for capsule polysaccharide synthesis and export are not transcriptionally regulated by 1132. Bars for AV-T Δ1132 and AV-T Δ1132/Tn7-1132 represent averages of three biological replicates. Error bars indicate standard deviation of the mean, and there is no statistical significance in expression across strains as assessed by multiple Mann-Whitney tests.
(A) Growth curves for VIR-O, AV-T, VIR-O Δ1132, and AV-T Δ1132 in LB media. Data represents two biological replicates, and error bars indicate standard deviation of the mean. (B) Galleria mellonella infected with VIR-O Δ1132 show no significant difference in mortality compared with wild-type VIR-O (n=30 per strain) as assessed by a log-rank (Mantel-Cox) test. (C) Mice were infected intranasally with either VIR-O or VIR-O Δ1132. At 24 hours post-inoculation, bacteria were recovered from the lungs, spleen, and liver tissues and CFU/g quantified. There is no significant difference between the two strains in all tissues as assessed by a Mann-Whitney test. ns, not significant.
Data Availability Statement
The RNA-Seq data presented in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE185730 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185730).