Deubiquitination of proteasome subunits by OTULIN regulates type I IFN production.
Abstract
OTULIN is a linear deubiquitinase that negatively regulates the nuclear factor κB (NF-κB) signaling pathway. Patients with OTULIN deficiency, termed as otulipenia or OTULIN-related autoinflammatory syndrome, present with early onset severe systemic inflammation due to increased NF-κB activation. We aimed to investigate additional disease mechanisms of OTULIN deficiency. Our study found a remarkable activation of type I interferon (IFN-I) signaling in whole blood, peripheral blood mononuclear cells, monocytes, and serum from patients with OTULIN deficiency. We observed similar immunologic findings in OTULIN-deficient cell lines generated by CRISPR. Mechanistically, we identified proteasome subunits as substrates of OTULIN deubiquitinase activity and demonstrated proteasome dysregulation in OTULIN-deficient cells as the cause of IFN-I activation. These results reveal an important role of linear ubiquitination in the regulation of proteasome function and suggest a link in the pathogenesis of proteasome-associated autoinflammatory syndromes and OTULIN deficiency.
INTRODUCTION
OTULIN [ovarian tumor (OTU) deubiquitinase with linear linkage specificity] is a methionine 1 (M1)–linked deubiquitinase that interacts with LUBAC (linear ubiquitin chain assembly complex) to remove linear ubiquitin chains generated by LUBAC (1). Binding to the LUBAC’s catalytic subunit heme-oxidized iron-responsive element-binding protein 2 (IRP2) ubiquitin ligase-1 (HOIL-1) interacting protein (HOIP) recruits OTULIN to the tumor necrosis factor receptor (TNFR) complex and restrains the nuclear factor κB (NF-κB) pathway (2–5). OTULIN also removes linear ubiquitin chains from other substrates including NF-κB essential modulator (NEMO), receptor-interacting protein kinase 1 (RIPK1), and A20 to attenuate NF-κB signaling (6). Loss of HOIP-OTULIN interaction reduces the capacity of OTULIN to restrict LUBAC-induced NF-κB activation (1). Knockin mice that express catalytically inactive OTULIN die during midgestation due to proinflammatory cell death mediated by TNFR1 and RIPK1 kinase activity (7). Phosphorylation of OTULIN regulates RIPK1 ubiquitination and promotes necroptosis (8).
In humans, biallelic loss-of-function (LoF) mutations in OTULIN cause otulipenia (6), also known as OTULIN-related autoinflammatory syndrome (ORAS) (9), an autoinflammatory disease characterized by fevers, neutrophilic dermatitis, panniculitis, arthralgia/arthritis, lymphadenopathy, hepatosplenomegaly, gastrointestinal inflammation, and failure to thrive (6). This potentially lethal disease is rare, as only seven patients have been reported in the literature to date (6, 9–11). Many clinical manifestations of otulipenia overlap with proteasome-associated autoinflammatory syndrome [PRAAS; also known as chronic atypical neutrophilic dermatosis with lipodystrophy and elevated temperature (CANDLE)] (6, 12). The first three patients with otulipenia were initially considered to have CANDLE, but all tested negative for proteasome LoF mutations (6).
The molecular pathogenesis of otulipenia is poorly understood. Defective OTULIN function leads to an up-regulation of M1 ubiquitin–mediated signaling and spontaneous NF-κB activation in myeloid cells (6, 9). Fibroblasts and monocytes from patients are sensitized to TNF-induced cell death, and increased apoptotic cells are observed in skin lesions (10). Treatment with inhibitors of TNF and interleukin-1 (IL-1) effectively reduces the inflammatory burden in OTULIN-deficient patients although disease flares have been noted (6, 9). OTULIN dysfunction in mice results in the production of type I interferons (IFN-I) in a RIPK1-dependent manner (7). However, the contribution of IFN-I to the inflammatory phenotype in patients has not yet been investigated.
In this study, we demonstrate IFN-I activation in primary cells of patients with otulipenia and identify proteasome components as substrates of OTULIN-mediated linear deubiquitination. These studies expand our understanding of the pathophysiology of otulipenia and provide an unexpected molecular link between clinically related but genetically distinct autoinflammatory diseases.
RESULTS AND DISCUSSION
Patients with otulipenia display overactivation of IFN-I signaling
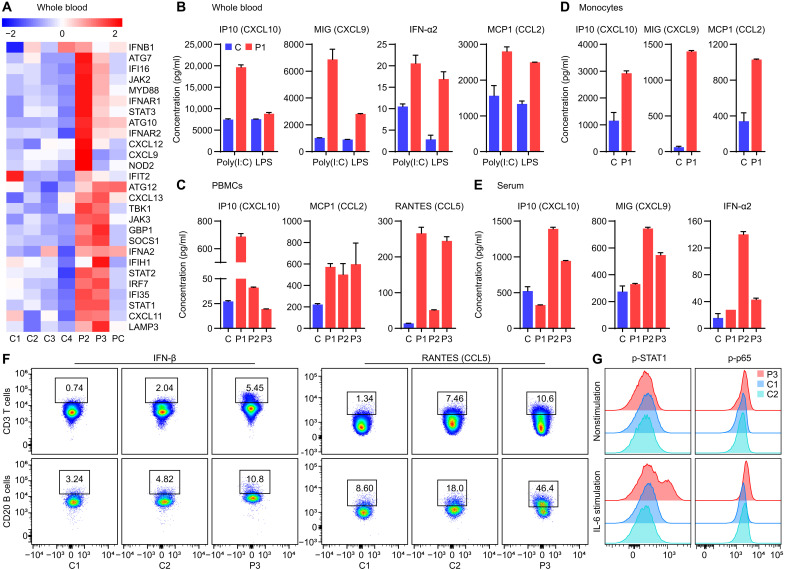
To better understand the autoinflammatory response elicited by OTULIN deficiency, we compared whole blood expression of proinflammatory mediators in patients with otulipenia (n = 2) and healthy controls (n = 4) and a patient control with type I interferonopathy using a custom-made NanoString array. In addition to activation of the NF-κB pathway (fig. S1A), we found high expression of IFN-I and IFN-inducible genes (Fig. 1A). Accordingly, whole blood, peripheral blood mononuclear cells (PBMCs), and isolated monocytes from the patients exhibited elevated production of the IFN-induced chemokines (Fig. 1, B to D). Serum measurements from patients confirmed the overproduction of IFN-α2 and IFN-induced chemokines compared to seven healthy controls (Fig. 1E). IFN-β and IFN-induced C-C motif chemokine ligand 5 (CCL5) are also up-regulated in patient T cells (CD3+) and B cells (CD20+) compared to controls (Fig. 1F). Moreover, we observed higher levels of phosphorylated signal transducer and activator of transcription 1 (p-STAT1) and NF-κB p65 in T cells (CD3+) from a patient compared to two healthy controls after IL-6 stimulation (Fig. 1G), further illustrating the contribution of IFN signaling in addition to the NF-κB pathway in the pathogenesis of otulipenia. In addition to enhanced basal IFN signaling, patient’s PBMCs exhibited increased expression of IFN-stimulated genes (ISGs) such as IFI27, IFIH1, IFIT2, IRF7, and OAS1 [by real-time polymerase chain reaction (PCR)] compared to healthy controls upon polyinosinic-polycytidylic acid [poly(I:C)] or lipopolysaccharide (LPS) stimulation (fig. S1B).
Fig. 1. Patients with otulipenia display overactivation of IFN-I signaling.
(A) NanoString analysis of IFN-I signaling in whole blood samples from two patients, four healthy controls, and a type I interferonopathy patient control (PC) with deoxyribonuclease 2 deficiency. (B) Cytokine levels in whole blood samples. The whole blood samples from P1 and his unaffected sibling were stimulated with poly(I:C) (20 μg/ml) or LPS (1 μg/ml) for 22 hours. (C) Cytokine levels in the supernatant of cultured PBMCs from three patients and one healthy control. (D) Cytokine levels in the supernatant of purified monocytes from P1 and three healthy controls. Cells used to detect IP10 and monokine induced by IFN-γ (MIG) were at basal level. Cells used to detect monocyte chemoattractant protein 1 (MCP1) were stimulated with LPS (1 μg/ml) for 48 hours. P1 was sampled twice. (E) Serum cytokine levels from three patients and seven healthy controls. (F) Intracellular cytokine staining of IFN-β and RANTES (CCL5) in T cells and B cells of P3 and two healthy controls. (G) Phosphorylation of STAT1 and p65 of patient P3 T cells compared to two healthy controls at basal level and after IL-6 (100 ng/ml) stimulation for 6 hours as determined by flow cytometry analysis. The patients carry homozygous disease-associated variants in OTULIN: P1 (Leu272Pro), P2 (Tyr244Cys), and P3 (Gly174Aspfs*2).
OTULIN knockout 293T cells display overactivation of IFN-I signaling
Considering the cross-talk between the NF-κB pathway and the IFN-I pathway, we further studied IFN signaling in OTULIN knockout (KO) 293T cells to determine whether the IFN signature observed in the patients is dependent on NF-κB pathway. We performed luciferase assay and Western blots in OTULIN KO 293T cells to confirm the observation of activated IFN-I signaling in OTULIN-deficient patients’ cells. In addition to activation of the NF-κB signaling, the induction of IFN-β was notably enhanced in OTULIN KO cell lines (KO1 and KO2) compared to wild-type (WT) 293T cells, as well as indicated by the increased levels of p-STAT2, p-TNFR-associated factor family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1), p–IFN regulatory factor 3 (IRF3) (fig. S2, A and B). In addition, IFN induced protein with tetratricopeptide repeats 3 (IFIT3), a member of the ISG family, as well as principal sensors of viral double-stranded RNA melanoma differentiation-associated protein 5 (MDA5) and retinoic acid-inducible gene I (RIG-I) were also up-regulated in OTULIN KO cell lines (fig. S2B). Consistent with the data from OTULIN-deficient patients, up-regulation of ISGs and increased production of IFN-γ induced protein 10 (IP10) were observed in OTULIN KO 293T cells (fig. S2, C and D). RNA sequencing and gene ontology enrichment analysis confirmed a strong IFN signature and antiviral response in OTULIN KO cells (fig. S2, E and F). Conversely, complementation with OTULIN expression efficiently rescued the up-regulated transcription of ISGs in OTULIN KO cells (fig. S2G). In addition, the strong IFN signature in OTULIN deficiency is independent on NF-κB signaling indicated by multiple assays, as inhibition of NF-κB was not able to suppress the enhanced IFN signaling (fig. S2, H to J). Together, these data suggested that OTULIN plays an important role in controlling autoinflammation by repressing IFN-I production in addition to restricting NF-κB signaling.
OTULIN deficiency causes proteasome dysfunction
To explore a mechanistic link between OTULIN deficiency and activation of the IFN-I pathway, we performed mass spectrometry after overexpression and immunoprecipitation of OTULIN or HOIP in 293T cells. We detected a large number of proteasome subunits that immunoprecipitated with OTULIN and HOIP (fig. S3A). These proteasome components included both 19S regulatory subunits and 20S subunits that form the catalytic core of the 26S proteasome. These results indicated that both OTULIN and LUBAC interact with the proteasome.
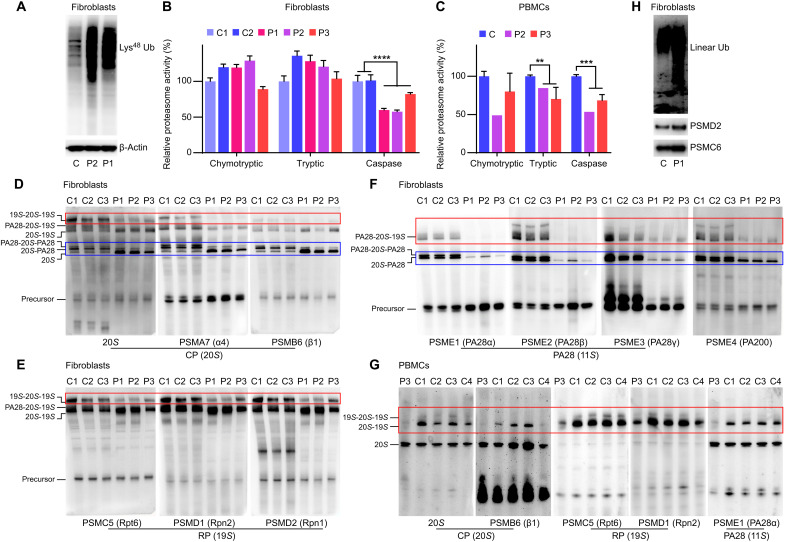
Lysine-48 (Lys48) polyubiquitin chains are the primary signal for proteasome-mediated protein degradation. Therefore, we examined Lys48 ubiquitination levels and noticed a substantial accumulation of Lys48-ubiquitinated proteins in fibroblasts from two patients with otulipenia (Fig. 2A). Similarly, we observed Lys48 ubiquitin aggregates in two OTULIN KO 293T cell lines compared to WT cells (fig. S3B). The accumulation of the proteasome-targeting Lys48-polyubiquitinated proteins suggests possible disruption of proteasome function in the setting of OTULIN deficiency.
Fig. 2. OTULIN deficiency causes proteasome dysfunction.
(A) The Lys48 ubiquitination (Ub) levels in patients’ and one healthy control’s fibroblasts. (B) Proteasome chymotryptic, tryptic, and caspase-like activity in fibroblasts from three patients and two healthy controls. (C) Proteasome chymotryptic, tryptic, and caspase-like activity in PBMCs from two patients and six healthy controls. P3 was sampled twice. (D to F) Native gel analysis of the proteasome assembly in fibroblasts from three patients and three healthy controls. The assembly of different parts of the proteasome was illustrated using antibodies of different subunits correspondingly. CP, core particle; RP, regulatory particle. (G) Native gel analysis of the proteasome assembly in PBMCs from the patient P3 and four healthy controls. (H) Western blots analysis of linear ubiquitination levels on purified proteasome from patient P1’s and one healthy control’s fibroblasts. Data in (B) and (C) are shown as means ± SEM from four repeated technical analyses. P values were determined by unpaired two-tailed t test. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Quantification of caspase-like activity confirmed a significantly reduced proteasome activity in fibroblasts from patients with otulipenia compared to healthy controls (Fig. 2B). Reconstitution with a plasmid encoding WT OTULIN showed increased proteasome caspase-like activity in OTULIN KO 293T cells, while expressing the pathogenic OTULIN variant Leu272Pro or catalytically inactive variant Cys129Ala failed to increase proteasome activity (fig. S3C). In addition to the defect in caspase-like activity, trypsin activities were also reduced in patient’s PBMCs (Fig. 2C). These results further demonstrate that the deficiency of OTULIN is associated with impaired proteasome function.
Next, we performed native polyacrylamide gel electrophoresis to analyze the assembly of proteasome complex in patients’ fibroblasts. The 26S proteasome consists of 20S core component; and the regulatory complexes, including the 19S, the PA28α/β, or the PA28γ complex, attached to one or both ends of 20S. Immunoblotting using antibodies to 20S and the subunits PSMA7 and PSMB6 showed an accumulation of the 20S proteasome but decreased levels of 20S with PA28 in fibroblasts from patients compared to controls (Fig. 2D). The levels of immunoprecipitated 26S with the 19S were also reduced in patients’ cells. The reduced association of the 20S with 19S was further demonstrated by immunoblotting with antibodies to 19S regulatory proteasome subunits PSMC5, PSMD1, and PSMD2 (Fig. 2E). The association of 20S with PA28α, PA28β, PA28γ, and PA200 was also attenuated as indicated by immunoblotting of PSME1, PSME2, PSME3, and PSME4 (Fig. 2F). Moreover, we observed parallel 26S proteasome assembly defects in PBMCs from a patient with otulipenia compared to healthy controls using five different antibodies for subunits of 20S, 19S, and 11S (Fig. 2G). Analysis of OTULIN KO 293T cells further supported the decreased association of 20S with PA28 compared to WT cells (fig. S3D), although the difference was less notable compared to fibroblasts from patients.
To further investigate how OTULIN interacts with the proteasome, we purified proteasomes and performed Western blot for linear ubiquitination. We observed substantially elevated levels of linear ubiquitination in purified proteasomes from patient fibroblasts and OTULIN KO 293T cells (Fig. 2H and fig. S3E). Together, these results illustrate that linear deubiquitination of the proteasome by OTULIN is required for proper proteasome assembly and function.
OTULIN deubiquitinate proteasome subunits to maintain proteasome assembly and function
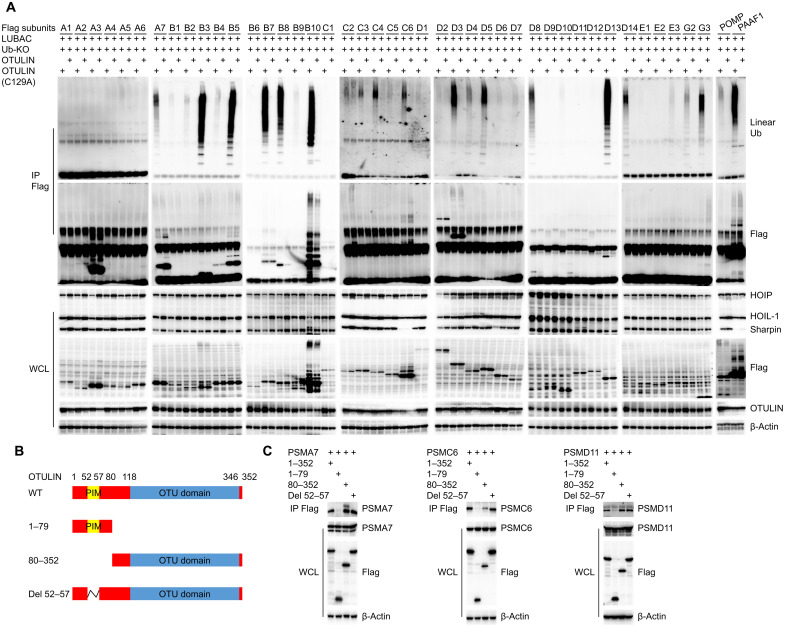
To identify the specific proteasome subunit(s) targeted by OTULIN, we screened each proteasome subunit by overexpression of LUBAC, WT ubiquitin, or ubiquitin-KO, together with Flag-tagged proteasome subunits with or without OTULIN, and then performed immunoprecipitation to Flag. We confirmed that multiple proteasome and immunoproteasome subunits are subjected to linear deubiquitination by OTULIN. For the 20S proteasome, PSMA7 and PSMB7 were markedly linear ubiquitinated by LUBAC, and OTULIN effectively removed the linear ubiquitin chains from these two subunits (fig. S4A). The other subunits, including PSMB1 to PSMB6, were also modified by linear ubiquitination (fig. S4A). All three catalytic immunoproteasome subunits (PSMB8/β5i, PSMB9/β1i, and PSMB10/β2i) were subjected to strong linear ubiquitination by LUBAC, which can be cleaved by OTULIN (Fig. 3A and fig. S4A). Linear ubiquitination can be branched with Lys63 ubiquitination, and it is likely that most proteasome subunits are modified by branched ubiquitin chains and some subunits can be linear ubiquitination only when WT ubiquitin is co-overexpressed. However, the immunoproteasome subunits have accumulated pure linear ubiquitin chains. In addition, multiple 19S proteasome subunits, including PSMC2, PSMC4, PSMC6, PSMD3, PSMD13, and PSMD14, were also identified as substrates of OTULIN (Fig. 3A and fig. S4B). Moreover, proteasome assembly factors including PSMG2, PSMG3, and PAAF1 were also subjected to linear ubiquitination modification (Fig. 3A).
Fig. 3. Proteasome subunits as substrates of OTULIN deubiquitinase activity.
(A) Identification of proteasome subunits that are regulated by OTULIN-mediated deubiquitination of linear ubiquitin using immunoprecipitation (IP). LUBAC complex consists of the catalytic subunit HOIP and two accessory proteins: HOIL-1 and Sharpin. WCL, whole-cell lysate. Ub-KO, ubiquitin mutant with all lysines mutated to arginines, which only forms linear polyubiquitin chains. (B) Schematic illustration of the different OTULIN deletion constructs that are used in experiments shown in (C). (C) Immunoprecipitation of 293T cells transfected with the indicated OTULIN constructs and various proteasome subunits provides evidence that the interaction between OTULIN and proteasome subunits relies on the OTU domain.
To confirm the interaction of these proteasome subunits with OTULIN, we also performed immunoprecipitation in 293T cells by expressing the proteasome subunits plasmids with OTULIN plasmid (fig. S4C). These experiments confirmed the interaction of various proteasome subunits with OTULIN, although the strength of interaction varied among the different subunits. To identify the domain of interaction between OTULIN and proteasome subunits, we constructed plasmids that express truncated forms of OTULIN, including amino acids 1 to 79 of the PUB (peptide:N-glycanase and UBA or UBX-containing proteins)-interacting motif (PIM) domain, amino acids 80 to 352 of the OTU domain, and the construct with amino acid deletions 52 to 57 in the PIM domain (Fig. 3B). When coexpressed with different proteasome subunits (PSMA7, PSMC6, and PSMD11), only the amino acids 1 to 79 OTULIN construct could not establish these interactions (Fig. 3C), indicating that the OTU domain is required for OTULIN-mediated deubiquitination of proteasome subunits. The same interaction with endogenous proteasome subunits was also observed (fig. S4D).
Last, we explored the impact of linear ubiquitin modification on proteasome function using 293T cells. Proteasome peptidase activity was measured in two OTULIN KO 293T cell lines (KO1 and KO2) and WT 293T cells after lysis and fractionation by glycerol density gradient centrifugation. The peak of 26S proteasome peptidase activity was shifted in both OTULIN KO cell lines compared to WT (fig. S3F). Proteasome subunits shifted to higher glycerol density fraction, indicating a potential change in proteasome configuration due to the retained linear ubiquitination in OTULIN-deficient cells. Supporting the association of proteasome dysregulation and aberrant ubiquitination, aggregates of linear ubiquitination were especially prominent in fractions that constitute the 26S proteasome (fractions 20 to 28) in OTULIN KO cells (fig. S3G). The peak of chymotrypsin activity and the shift of proteasome subunits were rescued by restoring WT OTULIN expression in OTULIN KO cells (fig. S3F).
OTU-deubiquitinating enzymes such as OTUD6B and OTUD7A have been implicated in the regulation of proteasome function, and patients with deficiency of OTUD6B or OTUD7A had reduced proteasome assembly, decreased proteasome activity, and accumulation of ubiquitin-protein conjugates (13, 14). These observations support that OTULIN, as a deubiquitinase, could function to regulate proteasome activity.
In addition, multiple protein ubiquitination enzymes interact with the proteasome, and many subunits of the 26S proteasome can be lysine-63–ubiquitinated (15, 16). The in situ ubiquitination of the 26S proteasome regulates its own activity. Ubiquitination of proteasome subunits significantly impairs the ability of 26S proteasome to bind, deubiquitinate, and degrade ubiquitinated proteins (15, 16). When the increase in proteasome autoubiquitination is associated with a partial loss of proteasomal capacity, proteasomal polyubiquitin chain–trimming enzymes with deubiquitinating activities will exert opposite function to antagonize ubiquitination of the proteasome. Therefore, OTU-deubiquitinating enzymes are critical for proteasome function, but the direct role of linear ubiquitination in regulating proteasome function has never been reported.
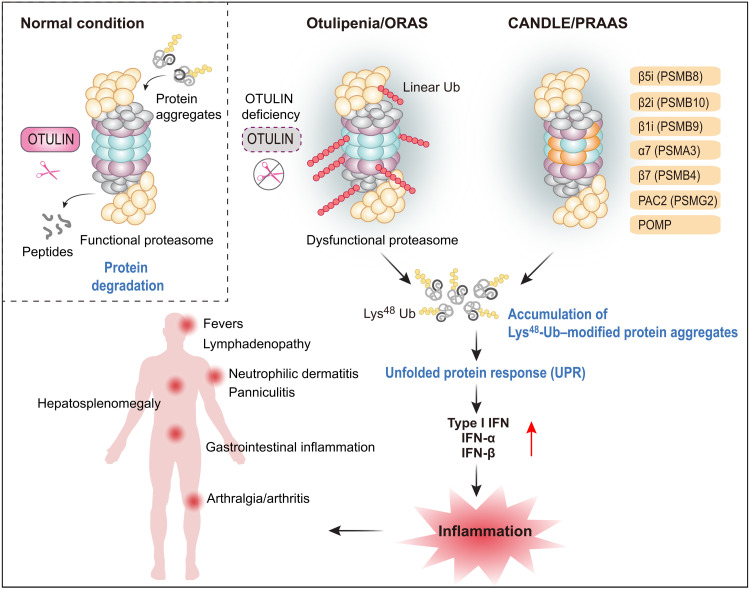
Our data collectively reveal an important role of linear ubiquitination in the maintenance of proteasome function. We found that the dysregulation in linear deubiquitination of the proteasome complex leads to up-regulation in IFN-I signaling in OTULIN-deficient cells. These findings also establish a link between otulipenia and PRAAS/CANDLE, a primary interferonopathy caused by mutations that disrupt proteasome assembly and function. Clinically, patients with otulipenia and PRAAS/CANDLE share features of severe multiorgan systemic inflammation, in particular, skin findings of lipodystrophy and panniculitis (Fig. 4).
Fig. 4. Graphical abstract of the pathogenic mechanism of otulipenia.
OTULIN deficiency leads to the excessive linear ubiquitination on proteasome subunits, especially on the immunoproteasome subunit, which disrupts immunoproteasome assembly and function. The defected proteasome will cause accumulation of unfolded or ubiquitinated protein, leading to a strong IFN-I response. Clinically, patients with otulipenia usually suffer from severe multiorgan systemic inflammation.
Although multiple proteasome subunits are susceptible to linear ubiquitination, this modification is more prominent on immunoproteasome subunits. The excessive linear ubiquitination due to OTULIN deficiency disrupts immunoproteasome assembly and function. Immunoproteasome plays an important role in the pathogenicity of autoinflammation, and pathogenic mutations in all three immunoproteasome subunits have been identified in patients with PRAAS/CANDLE (12, 17), which further implicated the tight link between linear ubiquitination defects associated with proteasome dysfunction and dysregulation of IFN-I signaling.
The coexistence of NF-κB and IFN-I inflammatory signatures in patients with otulipenia is clinically informative for the management of patients who only respond partially to TNF inhibitors. This phenomenon has been observed in patients with other severe autoinflammatory disorders (18). Suppression of IFN-I signaling by Janus kinase (JAK) inhibitors may be therapeutically indicated in patients with the OTULIN deficiency, and they are now frequently used to treat CANDLE/PRAAS and other interferonopathies (19). Initial experiments showed that patient’s fibroblasts responded to JAK inhibitor treatment (fig. S1, C to F). Further studies are needed to elucidate the molecular mechanism of IFN-I induction in otulipenia and PRAAS/CANDLE. Enhanced endoplasmic reticulum stress and activation of the unfolded protein response due to proteasome defects have been linked to increased IFN-I production (12, 20, 21). This mechanism may also explain the notable IFN-I activation in primary cells of patients with otulipenia.
A previous study indicated that IFN-I is triggered by RIPK1 activation and increased necroptosis in OTULIN-deficient mice (7). We and others recently described cleavage-resistant RIPK1-induced autoinflammatory (CRIA) syndrome due to gain-of-function mutations in RIPK1 that renders the protein uncleavable by caspase-8, leading to increased RIPK1 activation and enhanced necroptosis. Distinct from the features of otulipenia, patients with CRIA present with recurrent fevers and lymphadenopathy (22, 23). Therefore, how much RIPK1 activation contributes to IFN-I production in human OTULIN deficiency remains unknown.
In summary, our work identified proteasome subunits as substrates of the OTULIN deubiquitinase activity, adding linear ubiquitination to the repertoire of proteasome posttranslation modifications that includes phosphorylation and lysine-63 ubiquitination. We demonstrated that proteasome dysfunction due to dysregulation in linear ubiquitination directly contributes to IFN-I production in the setting of OTULIN deficiency.
MATERIALS AND METHODS
Study design
Patients with OTULIN deficiency, termed as otulipenia or ORAS, present with early onset severe systemic inflammation. However, the molecular pathogenesis of otulipenia is poorly understood. The aim of this study is to investigate the pathophysiology of OTULIN deficiency. For this purpose, we performed luciferase assay, RNA sequencing, NanoString assay, flow cytometry, quantitative PCR (qPCR), and cytokine measurements in cells from patients and OTULIN-deficient cell lines. We used mass spectrometry, proteasome activity assay, glycerol gradient ultracentrifugation, native polyacrylamide gel electrophoresis, Western blotting, and immunoprecipitation to study proteasome activity, assembly, and linear ubiquitination in OTULIN deficiency. For human primary materials based on experiments, multiple age- and gender-matched healthy controls used in the experiment were randomly selected. Patient samples were taken multiple times and used for independent biological replicate experiments. For cell-based experiments, biological triplicates were performed in each single experiment, in general, unless otherwise stated. No statistical methods were used to predetermine the sample sizes. No data were excluded from analysis. Blinding was not possible, as the authors who performed the experiment also analyzed the data.
Study approval
Patients enrolled in this study were evaluated at the Hacettepe University, Turkey and the National Institutes of Health, USA under protocols approved by the Institutional Review Boards of their respective institutions and provided written informed consents.
Cell preparation, culture, and stimulation
PBMCs were separated by lymphocyte separation medium and SepMate tubes (STEMCELL Technologies) according to the manufacturer’s instructions. Monocytes were purified from PBMCs by negative selection (Monocyte Isolation Kit II, Miltenyi Biotec). The human embryonic kidney (HEK) 293T cell line was acquired from the American Type Culture Collection. Fibroblasts were derived from skin biopsies of patients and control donors. Fibroblasts and HEK293T cells were grown in Dulbecco’s modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (ExCell Bio) and penicillin/streptomycin (HyClone). PBMCs and monocytes were grown in RPMI 1640 (Gibco) supplemented with 10% FBS and penicillin/streptomycin. All cell lines tested negative for mycoplasma contamination.
Poly(I:C) (Sigma-Aldrich, P1530) (20 μg/ml) was used to stimulate whole blood samples and PBMCs for the indicated amount of time. LPS (Sigma-Aldrich, L6529) (1 μg/ml) was used to stimulate monocytes and PBMCs for the indicated amount of time. IL-6 (PeproTech, 200-06) (100 ng/ml) was used to stimulate PBMCs for the indicated amount of time.
CRISPR-Cas9–mediated OTULIN KO 293T cells
Guide RNA sequences (ATTAAGCGTAGCTCCTGAAA) targeting exon 3 of OTULIN were cloned into the plasmid pX330. Constructs were transfected into 293T followed by selecting with puromycin (1 μg/ml) to obtain transformed cells, and single colonies were obtained by serial dilution and amplification. Gene deletion was assessed by Sanger sequencing and immunoblotting.
Luciferase assay
NF-κB/IFN stimulation response element/IFN-β firefly reporter plasmid and Renilla luciferase expression plasmid were used to cotransfect WT and OTULIN KO 293T cells. Cells were processed with the indicated treatment after 24 hours of transfection, and luciferase activity was measured using a multimode plate reader (BioTek). Data were calculated as fold induction by normalizing firefly luciferase activity to Renilla luciferase activity.
RNA sequencing
RNA libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) and then sequenced on Illumina NovaSeq to get 150–base pair paired-end reads. featureCounts was used to count the reads numbers mapped to each gene. Differential expression analysis was performed using the DESeq2 R package.
NanoString assay
One hundred nanograms of total RNA was used for NanoString assay, and gene expression analysis was conducted using the nCounter Analysis System (NanoString Technologies) with a code set designed to target 594 immunologically related genes. NanoString assay and data analysis were performed as previously described (6).
Intracellular cytokine staining
Intracellular cytokine staining for IFN-β and CCL5 were measured in PBMCs at baseline as previously described (22). Cells were stained by antibodies CD3 (BD Biosciences, 557943), CD20 (BD Biosciences, 740333), IFN-β (Invitrogen, BMS1044FI), and CCL5 (BioLegend, 515507). All events were acquired on BD LSRFortessa (BD Biosciences) and analyzed by FlowJo (TreeStar).
Flow cytometry analysis of phosphorylation
For phosflow staining, isolated PBMCs were treated with IL-6 for 6 hours at 37°C with 5% CO2 and then permeabilized with Perm Buffer III according to the manufacturer’s instructions (BD Biosciences). Cells were stained by antibodies CD3 (BD Biosciences, 557851), p-p65 (pS529) (BD Biosciences, 565446), and p-STAT1 (pY701) (BD Biosciences, 612564). All events were acquired on a BD LSRFortessa (BD Biosciences) and analyzed with FlowJo (TreeStar).
Quantitative reverse transcription PCR assay
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN, 74104). cDNA was generated by the PrimeScript RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, RR047A). qPCR was performed using TB Green Premix Ex Taq II (Tli RNase H Plus) (Takara, RR820A) and TaqMan Gene Expression Master Mix (Applied Biosystems) on Roche 480II and ABI 7500, respectively. Relative mRNA expression was normalized to ACTB or GAPDH and analyzed by the ΔΔCt method.
Antibodies and expression plasmids
The following antibodies were from Cell Signaling Technology: β-Actin (4970), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 5174), p-STAT2 (88410), STAT2 (72604), p-IRF3 (4947), IRF3 (4302), p-TBK1 (5483), TBK1 (3504), MDA5 (5321), Rig-I (3743), SHARPIN (12541), Lys48 polyubiquitin (8081), and OTULIN (14127). The following antibodies were from ABclonal Technology: PSMD2 (A1999), PSMA7 (A4052), PSMB6 (A4053), PSMC5 (A1538), PSMD1 (A16420), PSME1 (A5358), PSME2 (A5562), PSME3 (A12697), PSME4 (A13815), and PSMD11 (A15306). IFIT3 (ab76818), PSMC6 (ab22639), and 20S (ab22673) were purchased from Abcam. PSMB9 (sc28809) and HOIL-1 (sc365523) were purchased from Santa Cruz Biotechnology. HOIP (MAB8039) was purchased from R&D. Flag (F3165) was purchased from Sigma-Aldrich. Linear polyubiquitin (1F11/3F5/Y102L) was from Genentech.
The plasmids for HOIP (#50015), HOIL-1 (#50016), SHARPIN (#50014), and green fluorescent protein (GFP)–ubiquitin (#11928) were from Addgene. Ubiquitin-WT plasmid was constructed by removing the GFP tag from GFP-ubiquitin plasmid. Ubiquitin-KO plasmid was constructed by site-direct mutagenesis of all lysines mutated to arginines, which only forms linear polyubiquitin chains. The plasmid for OTULIN (sc317556) was from OriGene.
Western blotting and immunoprecipitation
Cells were lysed in cell lysis buffer supplemented with complete protease inhibitors. Immunoprecipitation and immunoblotting were conducted as described previously with specific antibodies (22).
Cytokine detection
The concentrations of cytokines in the supernatants of stimulated and nonstimulated whole blood and monocytes and in the serum were determined using Bio-Plex Pro Human Cytokine 27-plex and 21-plex immunoassay kits (Bio-Rad Hercules). The concentrations of cytokines in the supernatants of PBMCs were determined using ProcartaPlex 13-plex (Affymetrix). The concentrations of cytokines in the supernatants of 293T cells were measured by enzyme-linked immunosorbent assay kits (R&D).
Purification of the human 26S proteasome
Proteasome was purified by the Rapid 26S Proteasome Purification Kit (UBPBio, J4310). This approach uses the N-terminal ubiquitin-like (Ubl) domain of human RAD23B as the affinity bait, which allows the rapid and gentle isolation of endogenous 26S proteasomes. The bound 26S proteasome is subsequently eluted using the C-terminal two ubiquitin-interacting motifs of human S5a that bind Ubl.
Proteasome activity assay
Proteasome activities in cultured cells were measured with Proteasome-Glo cell–based assay kit (Promega, G8532). Chymotryptic-like, tryptic-like, and caspase-like activities were measured separately following the company’s recommended protocols.
Glycerol gradient ultracentrifugation
Glycerol gradient ultracentrifugation was based on standard procedures using a 12 to 40% gradient (12 ml) generated by the BR-188 Density Gradient Fractionation System (Brandel). Whole-cell lysates of 293T WT and OTULIN KO cells were centrifuged at 100,000g for 20 hours at 4°C, and the fractions of proteasome subunits were separated on the basis of their density. One hundred microliters of fractions were collected for analysis of activity and Western blots. Proteasome activity was measured by Suc-Leu-Leu-Val-Tyr-aminomethylcoumarin for chymotrypsin-like activity.
Native polyacrylamide gel electrophoresis
TSDG buffer–lysed protein [10 mM tris-HCl, 10 mM NaCl, 1.1 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 2 mM adenosine 5′-triphosphate, and 10% glycerol (pH 7.2)] was separated on native polyacrylamide gel electrophoresis (3 to 12%; Invitrogen) and immunoblotted for proteasome subunits as indicated.
Mass spectrometry
Flag-OTULIN and Flag-HOIP plasmids were transfected into 293T cells for 24 hours. Cells were lysed followed by immunoprecipitation. Immunoprecipitation protein was resolved on a homemade SDS–polyacrylamide gel electrophoresis gel. The gel was then stained with Coomassie blue, and target bands were excised and digested with trypsin.
Liquid chromatography–tandem mass spectrometry (MS/MS) analysis was performed on a Q Exactive mass spectrometer (Thermo Fisher Scientific) that was coupled to EASY-nLC (Thermo Fisher Scientific) for 2 hours. MS/MS spectra were searched using Mascot engine (version 2.2; Matrix Science, London, UK) against a nonredundant International Protein Index Arabidopsis sequence database v3.85 (released at September 2011; 39,679 sequences) from the European Bioinformatics Institute (www.ebi.ac.uk). For protein identification, the following options were used: peptide mass tolerance = 20 parts per million, MS/MS tolerance = 0.1 Da, enzyme = trypsin, missed cleavage = 2, fixed modification: carbamidomethyl (C), and variable modification: oxidation (M).
Statistical analysis
No statistical methods were used to predetermine sample size. For cell-based experiments, biological triplicates were performed in each single experiment, in general, unless otherwise stated. All values were expressed as means ± SEM and calculated from the average of at least three independent biological replicates unless specifically stated. Statistical analysis was performed using GraphPad Prism 8 software (GraphPad Software Inc.). For comparisons between two groups, the Student’s t test (unpaired and two-tailed) was applied. In all tests, a 95% confidence interval was used, for which P < 0.05 was considered a significant difference. Statistical analysis of RNA sequencing was performed using R software (R version 3.5.2).
Acknowledgments
We thank the patients and the unaffected controls for support during this research. We thank the Life Sciences Institute core facilities, Zhejiang University for technical assistance. We thank P. Xu at Zhejiang University for providing IFN-β reporter plasmid. We thank X. Guo at Zhejiang University for help.
Funding: This work was supported by the National Key R&D Program of China (grant no. 2018YFC1004903 to Q.Z.), the National Natural Science Foundation of China (grant nos. 31771548 and 81971528 to Q.Z.), and the Zhejiang Provincial Natural Science Foundation of China (grant no. LR19H100001 to Q.Z.).
Author contributions: Q.Z. and X.Y. designed the study and directed and supervised the research. P.T., S.W., J.Z., H.H., Z.Y., R.F., and W.L.T. performed experiments. P.T., P.Y.L., J.W., and X.Y. analyzed the data. S.O., H.Y., E.S., R.T., and I.A. enrolled the patients and collected and interpreted clinical information. Q.Z., P.T., P.Y.L., and I.A. wrote the manuscript with input from others. All authors contributed to the review and approval of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
REFERENCES AND NOTES
- 1.Elliott P. R., Nielsen S. V., Marco-Casanova P., Fiil B. K., Keusekotten K., Mailand N., Freund S. M., Gyrd-Hansen M., Komander D., Molecular basis and regulation of OTULIN-LUBAC interaction. Mol. Cell 54, 335–348 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keusekotten K., Elliott P. R., Glockner L., Fiil B. K., Damgaard R. B., Kulathu Y., Wauer T., Hospenthal M. K., Gyrd-Hansen M., Krappmann D., Hofmann K., Komander D., OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153, 1312–1326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiil B. K., Damgaard R. B., Wagner S. A., Keusekotten K., Fritsch M., Bekker-Jensen S., Mailand N., Choudhary C., Komander D., Gyrd-Hansen M., OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol. Cell 50, 818–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaeffer V., Akutsu M., Olma M. H., Gomes L. C., Kawasaki M., Dikic I., Binding of OTULIN to the PUB domain of HOIP controls NF-κB signaling. Mol. Cell 54, 349–361 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Rivkin E., Almeida S. M., Ceccarelli D. F., Juang Y.-C., MacLean T. A., Srikumar T., Huang H., Dunham W. H., Fukumura R., Xie G., Gondo Y., Raught B., Gingras A.-C., Sicheri F., Cordes S. P., The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498, 318–324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q., Yu X., Demirkaya E., Deuitch N., Stone D., Tsai W. L., Kuehn H. S., Wang H., Yang D., Park Y. H., Ombrello A. K., Blake M., Romeo T., Remmers E. F., Chae J. J., Mullikin J. C., Güzel F., Milner J. D., Boehm M., Rosenzweig S. D., Gadina M., Welch S. B., Özen S., Topaloglu R., Abinun M., Kastner D. L., Aksentijevich I., Biallelic hypomorphic mutations in a linear deubiquitinase define otulipenia, an early-onset autoinflammatory disease. Proc. Natl. Acad. Sci. U.S.A. 113, 10127–10132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heger K., Wickliffe K. E., Ndoja A., Zhang J., Murthy A., Dugger D. L., Maltzman A., de Sousa E. M. F., Hung J., Zeng Y., Verschueren E., Kirkpatrick D. S., Vucic D., Lee W. P., Roose-Girma M., Newman R. J., Warming S., Hsiao Y. C., Kőműves L. G., Webster J. D., Newton K., Dixit V. M., OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature 559, 120–124 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Douglas T., Saleh M., Post-translational modification of OTULIN regulates ubiquitin dynamics and cell death. Cell Rep. 29, 3652–3663.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Damgaard R. B., Walker J. A., Marco-Casanova P., Morgan N. V., Titheradge H. L., Elliott P. R., McHale D., Maher E. R., McKenzie A. N. J., Komander D., The deubiquitinase OTULIN is an essential negative regulator of inflammation and autoimmunity. Cell 166, 1215–1230.e20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damgaard R. B., Elliott P. R., Swatek K. N., Maher E. R., Stepensky P., Elpeleg O., Komander D., Berkun Y., OTULIN deficiency in ORAS causes cell type-specific LUBAC degradation, dysregulated TNF signalling and cell death. EMBO Mol. Med. 11, e9324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nabavi M., Shahrooei M., Rokni-Zadeh H., Vrancken J., Changi-Ashtiani M., Darabi K., Manian M., Seif F., Meyts I., Voet A., Moens L., Bossuyt X., Auto-inflammation in a patient with a novel homozygous OTULIN mutation. J. Clin. Immunol. 39, 138–141 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Brehm A., Liu Y., Sheikh A., Marrero B., Omoyinmi E., Zhou Q., Montealegre G., Biancotto A., Reinhardt A., de Jesus A. A., Pelletier M., Tsai W. L., Remmers E. F., Kardava L., Hill S., Kim H., Lachmann H. J., Megarbane A., Chae J. J., Brady J., Castillo R. D., Brown D., Casano A. V., Gao L., Chapelle D., Huang Y., Stone D., Chen Y., Sotzny F., Lee C. C., Kastner D. L., Torrelo A., Zlotogorski A., Moir S., Gadina M., McCoy P., Wesley R., Rother K. I., Hildebrand P. W., Brogan P., Kruger E., Aksentijevich I., Goldbach-Mansky R., Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J. Clin. Invest. 126, 795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santiago-Sim T., Burrage L. C., Ebstein F., Tokita M. J., Miller M., Bi W., Braxton A. A., Rosenfeld J. A., Shahrour M., Lehmann A., Cogne B., Kury S., Besnard T., Isidor B., Bezieau S., Hazart I., Nagakura H., Immken L. L., Littlejohn R. O., Roeder E.; EuroEpinomics Res Consortium Autosomal Recessive working group, Kara B., Hardies K., Weckhuysen S., May P., Lemke J. R., Elpeleg O., Abu-Libdeh B., James K. N., Silhavy J. L., Issa M. Y., Zaki M. S., Gleeson J. G., Seavitt J. R., Dickinson M. E., Ljungberg M. C., Wells S., Johnson S. J., Teboul L., Eng C. M., Yang Y., Kloetzel P. M., Heaney J. D., Walkiewicz M. A., Biallelic variants in OTUD6B cause an intellectual disability syndrome associated with seizures and dysmorphic features. Am. J. Hum. Genet. 100, 676–688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garret P., Ebstein F., Delplancq G., Dozieres-Puyravel B., Boughalem A., Auvin S., Duffourd Y., Klafack S., Zieba B. A., Mahmoudi S., Singh K. K., Duplomb L., Thauvin-Robinet C., Costa J. M., Kruger E., Trost D., Verloes A., Faivre L., Vitobello A., Report of the first patient with a homozygous OTUD7A variant responsible for epileptic encephalopathy and related proteasome dysfunction. Clin. Genet. 97, 567–575 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Jacobson A. D., MacFadden A., Wu Z., Peng J., Liu C. W., Autoregulation of the 26S proteasome by in situ ubiquitination. Mol. Biol. Cell 25, 1824–1835 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besche H. C., Sha Z., Kukushkin N. V., Peth A., Hock E. M., Kim W., Gygi S., Gutierrez J. A., Liao H., Dick L., Goldberg A. L., Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J. 33, 1159–1176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarrabay G., Mechin D., Salhi A., Boursier G., Rittore C., Crow Y., Rice G., Tran T. A., Cezar R., Duffy D., Bondet V., Boudhane L., Broca C., Kant B. P., VanGijn M., Grandemange S., Richard E., Apparailly F., Touitou I., PSMB10, the last immunoproteasome gene missing for PRAAS. J. Allergy Clin. Immunol. 145, 1015–1017.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Schwartz D. M., Blackstone S. A., Sampaio-Moura N., Rosenzweig S., Burma A. M., Stone D., Hoffmann P., Jones A., Romeo T., Barron K. S., Waldman M. A., Aksentijevich I., Kastner D. L., Milner J. D., Ombrello A. K., Type I interferon signature predicts response to JAK inhibition in haploinsufficiency of A20. Ann. Rheum. Dis. 79, 429–431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez G. A. M., Reinhardt A., Ramsey S., Wittkowski H., Hashkes P. J., Berkun Y., Schalm S., Murias S., Dare J. A., Brown D., Stone D. L., Gao L., Klausmeier T., Foell D., de Jesus A. A., Chapelle D. C., Kim H., Dill S., Colbert R. A., Failla L., Kost B., O’Brien M., Reynolds J. C., Folio L. R., Calvo K. R., Paul S. M., Weir N., Brofferio A., Soldatos A., Biancotto A., Cowen E. W., Digiovanna J. J., Gadina M., Lipton A. J., Hadigan C., Holland S. M., Fontana J., Alawad A. S., Brown R. J., Rother K. I., Heller T., Brooks K. M., Kumar P., Brooks S. R., Waldman M., Singh H. K., Nickeleit V., Silk M., Prakash A., Janes J. M., Ozen S., Wakim P. G., Brogan P. A., Macias W. L., Goldbach-Mansky R., JAK1/2 inhibition with baricitinib in the treatment of autoinflammatory interferonopathies. J. Clin. Invest. 128, 3041–3052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poli M. C., Ebstein F., Nicholas S. K., de Guzman M. M., Forbes L. R., Chinn I. K., Mace E. M., Vogel T. P., Carisey A. F., Benavides F., Coban-Akdemir Z. H., Gibbs R. A., Jhangiani S. N., Muzny D. M., Carvalho C. M. B., Schady D. A., Jain M., Rosenfeld J. A., Emrick L., Lewis R. A., Lee B.; Undiagnosed Diseases Network, Zieba B. A., Kury S., Kruger E., Lupski J. R., Bostwick B. L., Orange J. S., Heterozygous truncating variants in POMP escape nonsense-mediated decay and cause a unique immune dysregulatory syndrome. Am. J. Hum. Genet. 102, 1126–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebstein F., Poli Harlowe M. C., Studencka-Turski M., Kruger E., Contribution of the Unfolded Protein Response (UPR) to the pathogenesis of proteasome-associated autoinflammatory syndromes (PRAAS). Front. Immunol. 10, 2756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao P., Sun J., Wu Z., Wang S., Wang J., Li W., Pan H., Bai R., Zhang J., Wang Y., Lee P. Y., Ying W., Zhou Q., Hou J., Wang W., Sun B., Yang M., Liu D., Fang R., Han H., Yang Z., Huang X., Li H., Deuitch N., Zhang Y., Dissanayake D., Haude K., McWalter K., Roadhouse C., MacKenzie J. J., Laxer R. M., Aksentijevich I., Yu X., Wang X., Yuan J., Zhou Q., A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature 577, 109–114 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Lalaoui N., Boyden S. E., Oda H., Wood G. M., Stone D. L., Chau D., Liu L., Stoffels M., Kratina T., Lawlor K. E., Zaal K. J. M., Hoffmann P. M., Etemadi N., Shield-Artin K., Biben C., Tsai W. L., Blake M. D., Kuehn H. S., Yang D., Anderton H., Silke N., Wachsmuth L., Zheng L., Moura N. S., Beck D. B., Gutierrez-Cruz G., Ombrello A. K., Pinto-Patarroyo G. P., Kueh A. J., Herold M. J., Hall C., Wang H., Chae J. J., Dmitrieva N. I., McKenzie M., Light A., Barham B. K., Jones A., Romeo T. M., Zhou Q., Aksentijevich I., Mullikin J. C., Gross A. J., Shum A. K., Hawkins E. D., Masters S. L., Lenardo M. J., Boehm M., Rosenzweig S. D., Pasparakis M., Voss A. K., Gadina M., Kastner D. L., Silke J., Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 577, 103–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4