Abstract
In this report we investigated the function of phosphoinositide-dependent protein kinase 1 (PDK1) in protein kinase B (PKB) activation and translocation to the cell surface. Wild-type and PDK1 mutants were transfected into HeLa cells, and their subcellular localization was analyzed. PDK1 was found to translocate to the plasma membrane in response to insulin, and this process did not require a functional catalytic activity, since a catalytically inactive kinase mutant (Kd) of PDK1 was capable of translocating. The PDK1 presence at the cell surface was shown to be linked to phospholipids and therefore to serum-dependent phosphatidylinositol 3-kinase activity. Using confocal microscopy in HeLa cells we found that PDK1 colocalizes with PKB at the plasma membrane. Further, after cotransfection of PKB and a PDK1 mutant (Mut) unable to translocate to the plasma membrane, PKB was prevented from moving to the cell periphery after insulin stimulation. In response to insulin, a PKB mutant with its PH domain deleted (ΔPH-PKB) retained the ability to translocate to the plasma membrane when coexpressed with PDK1. Finally, we found that ΔPH-PKB was highly active independent of insulin stimulation when cotransfected with PDK1 mutants defective in their PH domain. These findings suggest that PDK1 brings PKB to the plasma membrane upon exposure of cells to insulin and that the PH domain of PDK1 acts as a negative regulator of its enzyme activity.
Protein kinase Bα (PKBα), also known as c-Akt or RAC (related to protein kinase A and C), is composed of a N-terminal pleckstrin homology (PH) domain followed by a kinase catalytic domain which shares homology with the A and C protein kinases. Two other isoforms of PKB (termed PKBβ and PKBγ) have been identified and are expressed in ovarian, pancreatic, and breast cancer cells (10, 11). This serine/threonine kinase is rapidly activated in response to stimulation of tyrosine kinase receptors such as those for platelet-derived growth factor (PDGF), insulin, basic fibroblast growth factor, and epidermal growth factor (9, 15, 18). PKB stimulation by insulin or growth factors is thought to be dependent on phosphatidylinositol 3′-kinase (PI 3-kinase) for the following reasons: (i) it is sensitive to pharmacological inhibitors of PI 3-kinase (18), (ii) dominant-negative PI 3-kinase mutants block PKB activation (24), and (iii) constitutively active PI 3-kinase mutants stimulate PKB (9).
A model has been proposed to explain activation of PKB in response to hormones and growth factors (2). According to this model, stimulation of cell surface receptors leads to an increase in the level of phosphatidylinositol-3,4,5-trisphosphate (PtdIns-3,4,5-P3) and PtdIns-3,4-P2 via PI 3-kinase. Although it was initially reported that phospholipids could directly activate PKB by interacting with its PH domain (16), more recently it has been shown that this process most likely fulfills other and/or additional functions. Indeed, it may facilitate PKB localization to the plasma membrane. This view stems from the observation that translocation of PKB has been observed in response to interleukin-2 (1), peroxyvanadate (25), insulin-like growth factor 1 (7), and insulin (17). Further, this movement is prevented by PI 3-kinase inhibitors and by deletion of the PKB PH domain (21). In response to insulin or insulin-like growth factor 1, PKBα is phosphorylated on Thr308 and Ser473, phosphorylation on both of these residues being required for full PKB activation. A PKB kinase has been purified from muscle (5) and brain (23); it was cloned (3, 22) and found to phosphorylate PKBα on Thr308 (5), PKBβ on Thr309, and PKBγ on Thr305 (24). This 63-kDa monomeric enzyme was named 3-phosphoinositide-dependent protein kinase 1 (PDK1), since it requires PtdIns-3,4,5-P3- or PtdIns-3,4-P2-containing vesicles in order to phosphorylate PKB in vitro (3, 22). PDK1 is composed of a C-terminal PH domain and a catalytic domain similar to A, B, and C protein kinase. The fact that PDK1 possesses a PH domain that binds to phospholipids in vitro (22) points to a possible role in plasma membrane targeting and subsequent PKB activation. PDK1 in which Arg474 to Ala has been mutated in the PH domain or a PDK1 with a deletion of its PH domain shows a loss of the ability to localize at the plasma membrane (6). Whether PDK1 in fact translocates in response to cell stimulation remains controversial. Indeed, Anderson et al. (6) have shown that in PAE cells PDK1 could move to the membrane in response to PDGF in a PI 3-kinase-dependent manner, while Currie et al. have found that in 293 overexpressing cells PDK1 is constitutively located at the plasma membrane (13).
In our present work, we clarify the role of PDK1 in PKB activation and translocation. Using confocal microscopy and subcellular fractionation, we show that PDK1 moves to the plasma membrane in response to insulin. Interestingly, it appears that PDK1 promotes PKB translocation and subsequent activation. Finally, we provide evidence for the idea that the PH domain of PDK1 functions as a negative regulator of PDK1 activity in intact cells.
MATERIALS AND METHODS
Antibodies.
The antibody to Akt1 N-19 is an affinity-purified goat polyclonal antibody raised against a peptide corresponding to amino acids 3 to 21 located at the amino terminus of human Akt1 (Santa Cruz Biotechnology, Santa Cruz, Calif.). Monoclonal (12CA5) antibody to hemagglutinin (HA) was generated against a peptide (YPYDVPDYA) corresponding to the sequence of influenza virus HA and was provided by BAbCO (Richmond, Calif.). The monoclonal antibody (9E10) to the Myc epitope was from Santa Cruz Biotechnology. Monoclonal antibodies to green fluorescent protein (GFP) were obtained from Clontech (Palo Alto, Calif.). The secondary Texas red-conjugated mouse anti-sheep antibody (100-fold dilution) was provided by Amersham (Little Chalfont, Buckinghamshire, United Kingdom). The phosphorus-specific Akt (Thr308) antibody is a polyclonal antibody obtained from New England Biolabs (Beverly, Mass.). The monoclonal antibody to PDK1 was from Transduction Laboratories (Lexington, United Kingdom).
Materials.
Culture media and Geneticin were from Life Technologies, Inc. (Gaithersburg, Md.). Reagents for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were purchased from Bio-Rad (Richmond, Calif.). Enzymes for molecular biology assays were from New England Biolabs. Unless stated otherwise, all chemicals were from Sigma (St. Louis, Mo.). P81 phosphocellulose paper was purchased from Whatman (Maidstone, United Kingdom). Insulin was a kind gift from Novo-Nordisk (Copenhagen, Denmark). The Crosstide peptide was provided by Neosystem (Strasbourg, France). [γ-32P]ATP was purchased from ICN (Orsay, France). The QuickChange site-directed mutagenesis kit was from Stratagene (La Jolla, Calif.). All oligonucleotides were from Life Technologies. The T7 sequencing kit was from Pharmacia (Uppsala, Sweden), and plasmid purification kits were from Qiagen (Courtebœuf, France).
DNA constructs and expression vectors.
HA-tagged PKB in the mammalian expression vector pECE was from B. A. Hemmings (Basel, Switzerland) and has been described previously (8). Site-directed mutagenesis was performed with the QuickChange mutagenesis kit from Stratagene. The GFP-PKB fusion protein was created by cloning PKB into EcoRI and BamHI sites within the pEGFP-C2 vector (Clontech). The Myc-tagged PDK1 constructs in the mammalian expression vector pCDNA3 as well as the purified ΔPH-PKB and PDK1 proteins from baculovirus-infected Sf9 cells were kindly supplied by P. T. Hawkins (Cambridge, United Kingdom) and have been described previously (6). His-tagged PH-PDK1 was cloned in the mammalian express vector pCDNA 3.1/HISC. Myc-tagged ΔPH-PDK1 was cloned in the mammalian expression vector pCDNA 3.1/Myc-HisA.
Cell culture.
293 EBNA cells are human embryonic kidney cells that constitutively express the EBNA-1 protein from Epstein-Barr virus (Invitrogen, San Diego, Calif.). These cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% (vol/vol) fetal calf serum and 500 μg of Geneticin per ml. Exponentially growing cells were trypsinized, seeded at 1.25 × 105 cells/well in six-well tissue culture dishes (3.5-cm diameter), and incubated for 3 days in 2 ml of growth medium. One microgram of supercoiled DNA was mixed with 100 μl of 0.25 M CaCl2 and 100 μl of 2× BES (buffered saline containing 50 mM N,N-bis-2-hydroxyethyl-2-aminoethane sulfonic acid [pH 6.95], 280 mM NaCl, and 1.5 mM Na2HPO4). The mixture was incubated for 30 min at room temperature before being added dropwise to the cells. After incubation for 15 to 18 h at 35°C under 3% (vol/vol) CO2, the cells were moved to an incubator at 37°C and 5% (vol/vol) CO2 for 8 h before starvation in DMEM containing 0.2% (wt/vol) bovine serum albumin (BSA) for 14 h. Transfection protocols used for confocal microscopy experiments of HeLa cells were essentially identical to these used with 293 EBNA cells with the following exceptions. Cells were trypsinized and directly plated onto sterile glass coverslips at 100,000 cells/well in six-well tissue culture dishes. The next day, cells were transfected with 4 μg of DNA/well by the calcium phosphate method. Two days after transfection, cells were analyzed by confocal microscopy. COS-7 monkey kidney cells were cultured in DMEM supplemented with 10% (vol/vol) fetal calf serum and 500 μg of Geneticin per ml. They were placed in 100-ml-diameter dishes at 106 cells/dish and incubated for 3 days in 10 ml of growth medium.
Immunoprecipitation and in vitro PKB kinase assay.
After stimulation with the reagents indicated above, 293 cell extracts were prepared by lysing cells in a buffer containing 50 mM HEPES (pH 7.6), 150 mM NaCl, 10 mM EDTA, 10 mM Na4P2O7, 2 mM sodium orthovanadate, 100 mM NaF, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 100 IU of aprotinin per ml, 20 μM leupeptin, and 1% (vol/vol) Triton X-100 for 15 min at 4°C. The lysates were clarified by centrifugation at 15,000 × g for 15 min at 4°C, and PKB was immunoprecipitated with an HA (12CA5) antibody coupled to protein G-Sepharose. After washing of the immunocomplexes, kinase activity was assayed using Crosstide (12) in a reaction mixture containing 50 mM Tris, 10 mM MgCl2, 1 mM dithiothreitol, 5 μM ATP, 30 μM Crosstide, and 3.3 μCi of [γ-32P]ATP per assay. The phosphorylation reaction was allowed to proceed for 30 min at 30°C, and then was stopped by spotting 40 μl onto Whatman P81 filter papers and immersion in 1% (vol/vol) orthophosphoric acid. The papers were washed several times, rinsed in ethanol, and air dried, and the radioactivity was determined by Cerenkov counting. Background values obtained from a mixture lacking cell lysate were subtracted from all values. For detecting association between PDK1 and PKB, immunoprecipitates were washed three times in 1× phosphate-buffered saline (PBS)–1% NP-40 and twice in 10 mM Tris (pH 7.5)–100 mM NaCl–1 mM EDTA and then boiled in SDS sample buffer and resolved by SDS–10% PAGE.
In vitro phosphorylation of PDK1 and ΔPH-PKB.
Myc-PDK1 constructs were immunoprecipitated from 293 EBNA cells. Immunocomplexes were extensively washed as previously described. Myc-PDK1-immunoprecipitated proteins as well as 1 μg of PDK1 purified from baculovirus-expressing Sf9 cells was added to a mixture containing 50 mM Tris, 10 mM MgCl2, 1 mM dithiothreitol, 5 μM ATP, and 3.3 μCi of [γ-32P]ATP per assay and 1 μg of ΔPH-PKB purified from baculovirus-expressing Sf9 cells as a substrate. The phosphorylation reaction was allowed to proceed for 20 min at 30°C and then was stopped by adding Laemmli buffer. Samples were subjected to SDS-PAGE under reducing conditions. Proteins were visualized by Coomassie blue staining, and autoradiography was performed.
Immunoblot analysis.
Samples were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon; Millipore Corp.). Membranes were blocked for 30 min in a blocking buffer containing 5% (wt/vol) BSA in TBS (10 mM Tris-HCl, 140 mM NaCl [pH 7.4]) and then incubated for 1 h with the appropriate antibody diluted 1,000-fold in the same buffer. The membranes were washed extensively in TBS containing 1% (vol/vol) NP-40. Detection was performed with horseradish peroxidase-conjugated anti-rabbit, anti-mouse, or anti-goat antibody and enhanced-chemiluminescence reagents (Pierce, Bezons, France) according to the manufacturer's instructions.
Subcellular fractionation.
293 cells were washed and collected in ice-cold hypotonic buffer containing 10 mM Tris (pH 7.5), 10 mM NaF, 1 mM EDTA, 2 mM sodium orthovanadate, 0.5 mM PMSF, and 20 μM leupeptin and lysed by 20 strokes in a 1-ml syringe. Nuclei were removed by centrifugation for 10 min at 13,000 × g at 4°C. The P100 and S100 fractions were obtained by additional centrifugation at 100,000 × g for 30 min at 4°C. P100 fractions were resuspended in lysis buffer.
Fluorescent staining and confocal microscopy.
HeLa cells transfected with GFP-PKB and/or Myc-tagged PDK1 constructs and grown on coverslips were placed on ice and washed three times with ice-cold PBS prior to fixation with 4% (vol/vol) paraformaldehyde for 30 min at room temperature. Cells were then washed, treated with 50 mM ammonium chloride, and rewashed. Staining of the membrane was accomplished by incubating the cells in a humid chamber for 30 min with wheat germ agglutinin (WGA)-rhodamine (10−7 M). PDK1 constructs were visualized by incubating the cells for 1 h with a 9E10 monoclonal antibody to Myc as a primary antibody followed by another 1-h incubation with a Texas red-conjugated anti-mouse secondary antibody. Coverslips were mounted onto slides with Mowiol (Calbiochem, La Jolla, Calif.) and viewed using a Leica upright confocal microscope equipped with a Leica 63× lens objective (numerical aperture, 1.0). The molecules were excited with the 600 line of an argon-krypton laser and imaged using a 488 (GFP)-, 600 (rhodamine)-, or 568 (Texas red)-nm band-pass filter. Images were acquired with a scanning mode format of 512 by 512.
PI 3-kinase activity assays.
293 cells were washed twice with a buffer containing containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 100 mM EDTA, 10 mM Na4P2O7, and 2 mM sodium vanadate (NaVO4) and lysed in the same buffer containing 1% (vol/vol) NP-40, 20 mM leupeptin, 100 U of aprotinin per ml, and 1 mM PMSF. Cell lysates were incubated with polyclonal antibodies to IRS-1 preadsorbed to protein G-Sepharose for 3 h at 4°C. Pellets were then washed twice in PBS, pH 7.4, containing 1% (vol/vol) NP-40 and 0.2 mM NaVO4; twice in 100 mM Tris-HCl (pH 7.4) supplemented with 500 mM LiCl and 0.2 mM NaVO4; and twice in 10 mM Tris-HCl (pH 7.4) containing 100 mM NaCl, 1 mM EDTA, and 0.2 mM NaVO4. Pellets were further resuspended in 40 mM HEPES, pH 7.4, containing 20 mM MgCl2, and the kinase reaction was initiated by the addition of phosphatidylinositol (0.2 mg/ml) and 75 μM [γ-32P]ATP (7,000 Ci/mol) and performed for 20 min at room temperature. The reaction was stopped by the addition of 4 M HCl, and the phosphoinositides were extracted with a mixture of methanol and chloroform. Finally, phospholipids were analyzed by thin-layer chromatography.
RESULTS
PDK1 catalytic activity is not required for its insulin-induced translocation.
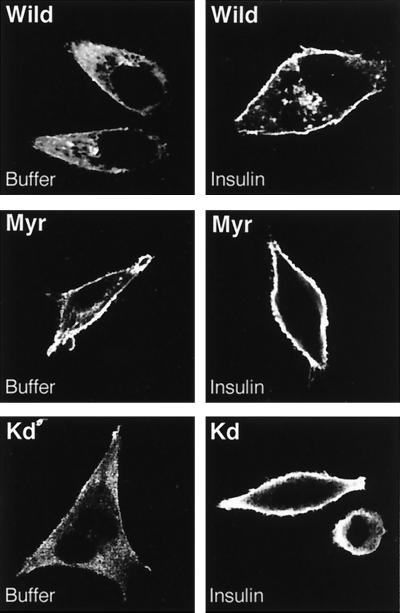
In response to PDGF, PDK1 has been shown to move to the plasma membrane (6). However, more recent studies have not supported this initial report. Indeed, PDK1 was not found to translocate to the membrane in response to either PDGF or insulin, while it is to a small extent constitutively present at the membrane (13). In an attempt to clarify these contradictory results, we transfected HeLa cells with expression vectors coding for a 63-kDa PDK1 tagged with a Myc epitope at its amino terminus. A PDK1 variant (Myr), modified by the addition of a myristoylation-palmitoylation sequence to the amino-terminal end and containing a Myc tag at the carboxy-terminal end, was used as a control to show constitutive localization of PDK1 at the plasma membrane. A third construct used consisted of a PDK1 catalytically inactive due to K110 substitution to Q (designated Kd for kinase deficient). Cells were transfected with these constructs and serum starved for 12 h prior to stimulation with insulin. As shown in Fig. 1, in nonstimulated cells PDK1 is partially located at the plasma membrane, but in response to insulin there is an increased amount at the membrane. Myr-PDK1 was present for the greater part at the plasma membrane either with or without insulin treatment of the cells. Finally, we found that the kinase-deficient PDK1 mutant retained its ability to translocate to the plasma membrane in response to insulin. This indicates that the catalytic activity of PDK1 is not required for the hormone-induced translocation process. Taken together, our results confirm that PDK1 translocation to the plasma membrane occurs in response to insulin similar to what we have seen for PKB in the same cells (21).
FIG. 1.
Effect of insulin on localization of PDK1 mutants. HeLa cells were transfected with 4 μg of either wild-type (Wild), myristoylated (Myr), or kinase dead (Kd) PDK1 as described in Materials and Methods. Forty-eight hours later, HeLa cells were incubated for 5 min with buffer or insulin (10−6 M). Cells were washed and fixed with paraformaldehyde (4%) prior to incubation with an antibody to Myc followed by incubation with a Texas red-linked mouse antibody to label PDK1 mutants. Slides were mounted and analyzed by confocal microscopy. Images represent the center section of the x-y plane.
Expression of a kinase-deficient PDK1 mutant permits normal PKB activation by insulin while basal PKB activity is reduced.
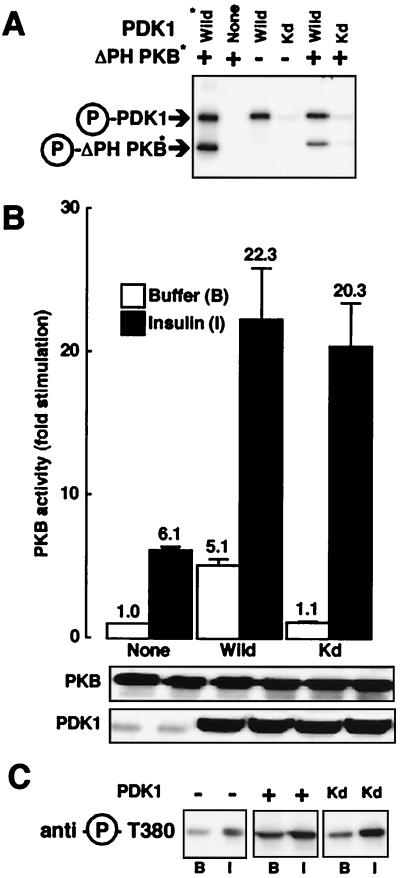
Since catalytically inactive PDK1 still translocates to the plasma membrane in response to insulin, we determined its effect on PKB activity. It has been previously shown that ΔPH-PKB can serve as an efficient in vitro substrate for PDK1 even in the absence of phospholipids (4, 13, 20). First, we performed an in vitro kinase assay to confirm that Kd-PDK1 had completely lost its ability to autophosphorylate and to phosphorylate ΔPH-PKB. As shown in Fig. 2A, although wild-type PDK1 (purified from PDK1-expressing baculovirus-infected Sf9 cells or immunoprecipitated from 293 cells) remains phosphorylated and able to phosphorylate ΔPH-PKB in vitro, the kinase-deficient PDK1 is essentially not phosphorylated under basal conditions and is unable to phosphorylate ΔPH-PKB. Next, 293 cells were transfected with PKB and the different PDK1 constructs. In response to insulin there was a 6.1-fold stimulation of PKB (Fig. 2B). When PDK1 was coexpressed in the absence of insulin, there was a 5.1-fold stimulation of PKB. A similar observation has been made by others (6) and was interpreted to mean that PDK1 is a constitutively active kinase, the overexpression of which leads to potentiation of PKB activity even in the absence of insulin. This view is supported by the observation that coexpression of Kd-PDK1 and PKB does not change basal PKB activity. Surprisingly, there was no significant difference in insulin-induced PKB activation when Kd-PDK1 is expressed compared to when wild-type PDK1 is expressed. Western blot analysis showed that the increase in PKB activity was not due to enhanced protein levels. Experiments using an antibody to phospho-Thr308 of PKB were also performed in transfected HeLa cells (Fig. 2C). We see an increase in PKB Thr308 phosphorylation in response to insulin compared to basal conditions when PKB was transfected alone, and a higher phosphorylation under basal conditions when PKB is expressed jointly with PDK1. Together these observations confirm the results obtained in 293 cells. Further, we found an increase in the level of PKB phosphorylation in response to insulin when coexpressed with Kd-PDK1. This indicates that at least in these two cell types Kd-PDK1 does not act as a dominant-negative molecule on endogenous PDK1.
FIG. 2.
Effect of kinase-dead PDK1 on PKB activation. (A) Myc-PDK1 constructs (Wild and Kd) were expressed separately in 293 cells. Immunoprecipitates and PDK1 purified from baculovirus were incubated with [γ-32P]ATP (20 min at 30°C) and ΔPH-PKB purified from baculovirus-expressing Sf9 cells. Samples were subjected to SDS–10% PAGE under reducing conditions followed by autoradiography. Arrows point to the positions of Myc-PDK1, PDK1, and ΔPH-PKB. Asterisks indicate proteins purified from baculovirus-expressing Sf9 cells. (B) 293 cells were transfected with wild-type PKB and, where indicated, with PDK1 constructs. Serum-starved cells were incubated for 5 min in the absence (white bars) or presence (black bars) of insulin (10−6 M). After incubation, cells were lysed, PKB was immunoprecipitated, and its kinase activity was determined using Crosstide as described in Materials and Methods. PKB activity is expressed as fold stimulation compared to basal levels, and the corresponding value is indicated above each column. Values shown are representative of at least four independent experiments performed in triplicate. The levels of the expressed enzymes measured by immunoblotting are shown below the bar graph. (C) HeLa cells were transfected with wild-type PKB and, where indicated, with PDK1 constructs. Serum-starved cells were incubated for 5 min with buffer (B) or insulin (I). After incubation, cells were lysed and 50 μg of the lysates was subjected to SDS-PAGE. PKB phosphorylation was determined by immunoblot analysis with an antibody specifically recognizing phospho-Thr308.
PDK1 subcellular localization and PKB basal activity depend on IRS-1-associated PI 3-kinase activity.
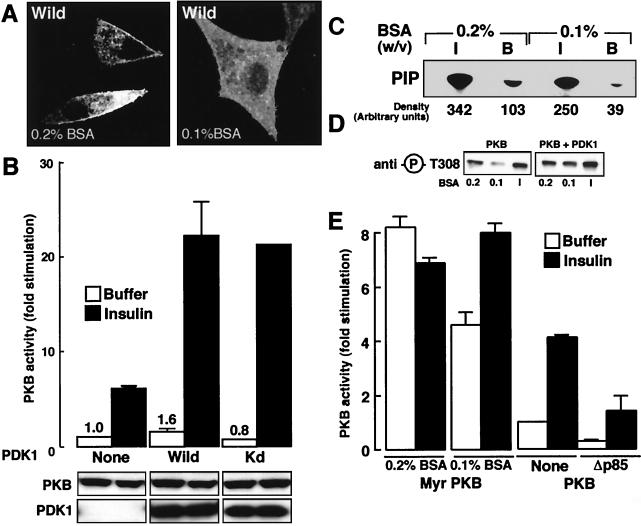
We have shown in Fig. 1 that without insulin stimulation a small fraction of PDK1 was found close to the cell membrane. This localization could explain the basal PKB activity (5.1-fold stimulation) seen after PDK1 coexpression. To further test this hypothesis, we serum starved cells in 0.1% (wt/vol) BSA instead of 0.2% (wt/vol) BSA and for 24 h instead of 12 h. Then confocal microscopy was performed on PDK1-transfected cells in the absence of insulin stimulation. As can be seen in Fig. 3A, when cells are serum starved in the presence of 0.2% (wt/vol) BSA, PDK1 is found at the plasma membrane. However, after a longer serum starvation period in 0.1% (wt/vol) BSA, PDK1 was no longer found to be cell membrane associated. This disappearance of plasma membrane association of PDK1 seen in 0.1% (wt/vol) BSA-treated cells is accompanied by a loss in basal PKB stimulation compared to results seen in Fig. 2, with “basal” being defined as prior to addition of growth factors or hormones (Fig. 3B). However, this treatment did not modify the effect of insulin, since a 20-fold stimulation of PKB activity is maintained when PKB and PDK1 are coexpressed. Upon coexpression of Kd-PDK1, no significant decrease in PKB activity was observed (0.8-fold stimulation) in the absence of insulin, and a normal level of activation following insulin stimulation (20-fold) is found. To determine whether the reduction in membrane association of PDK1 was linked to the basal level of phospholipids, we measured PI 3-kinase activity associated with IRS-1 immunopurified from 293 cells cultured in different BSA concentrations. We observed a decrease in the estimated PI 3P production in the presence of 0.1% (wt/vol) BSA compared to 0.2% (wt/vol) BSA (Fig. 3C). However, with insulin no significant difference is seen in PI 3P production. This decrease in IRS-1-associated PI 3-kinase activity observed with 0.1% (wt/vol) BSA compared to that found with 0.2% (wt/vol) BSA could explain both the basal localization of PDK1 to the cell surface and the PKB activity observed with PDK1 cotransfected compared to PKB transfected alone. According to our hypothesis, 24-h starvation reduces the basal PI 3-kinase activity compared to a 12-h-long treatment. If this view is correct, fewer PKB molecules should be phosphorylated on Thr308 after 24 h of serum starvation. To check this, we transfected HeLa cells with PKB either alone or together with PDK1 as described in the legend to Fig. 3D. When PKB activity was monitored using the phospho-Thr308 antibody, we saw a clear reduction in basal PKB phosphorylation after depletion in 0.1% BSA for 24 h compared to a 0.2% BSA treatment for 12 h. This was also the case for PKB cotransfected with PDK1, which correlated with the results obtained with 293 cells in Fig. 3B. Finally, to further illustrate the involvement of phospholipids in PDK1 activation, we transfected 293 cells with constitutively active Myr-PKB. This PKB mutant has been reported to be attached to the cell membrane and to be highly active in the absence of growth factor stimulation. After a 24-h starvation in 0.1% BSA, Myr-PKB was 40% less active than after serum depletion for 12 h in 0.2% BSA (Fig. 3E). Further, after cotransfecting a dominant-negative form of PI 3-kinase (Δp85), we saw a decrease in basal PKB activation in the absence of insulin stimulation and, as expected, a decrease after insulin stimulation, similar to previous reports (9).
FIG. 3.
Role of phospholipids in PDK1 localization and PKB basal activation. (A) Wild-type PDK1 (Wild) was expressed in HeLa cells and after 24 h, cells were serum starved with either 0.2% (wt/vol) BSA for 12 h or 0.1% (wt/vol) BSA in DMEM for 24 h. Cells were washed and fixed with paraformaldehyde (4%) prior to incubation with an antibody to Myc followed by incubation with a Texas red-linked mouse antibody to label PDK1. Slides were mounted and analyzed by confocal microscopy. Images represent the center section of the x-y plane. (B) 293 cells were transfected with wild-type PKB and PDK1 mutants. Cells were serum starved with 0.1% (wt/vol) BSA and incubated for 5 min in buffer (white bars) or insulin (10−6 M) (black bars). Then, PKB activity was determined as described in Materials and Methods. PKB activity is expressed as fold stimulation compared to basal levels, and the corresponding value is indicated above each column. Values shown are representative of three independent experiments performed in triplicate. The levels of the expressed enzymes measured by immunoblotting are shown below the bar graph. (C) 293 cells were serum starved in either 0.2% (wt/vol) or 0.1% (wt/vol) BSA for 24 h prior to exposure for 5 min to insulin (I) (10−6 M) or to buffer. Total cellular protein lysates were subjected to immunoprecipitation with an antibody to IRS-1, and associated PI 3-kinase activity was measured with PtdIns or PtdIns-4-P plus PtdIns-4,5-P2 as substrates. The position of 32P-labeled Pi (PIP) is indicated. Radioactivity associated with each spot was quantified using phosphorimaging and is indicated below the autoradiograph. (D) HeLa cells were transfected with wild-type PKB and where indicated with PDK1 constructs. Cells were serum starved with either 0.2% (wt/vol) BSA for 12 h or 0.1% (wt/vol) BSA for 24 h and incubated for 5 min with buffer or insulin (I). After incubation, cells were lysed and 50 μg of the lysates was subjected to SDS-PAGE. PKB phosphorylation was revealed by immunoblotting with an antibody specific for phospho-Thr308 peptide. (E) 293 cells were transfected with Myr-PKB or wild-type PKB and, where indicated, with Δp85. Cells were serum starved and lysed, PKB was immunoprecipitated, and its activity was determined as described in Materials and Methods.
PDK1 affects PKB subcellular localization.
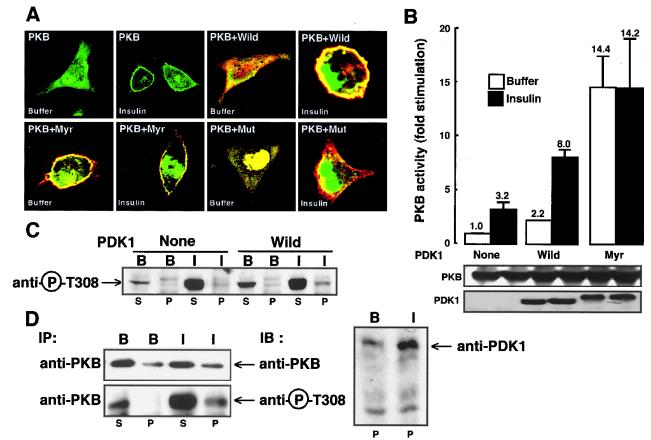
HeLa cells were transfected with plasmids coding for GFP-PKB and different Myc-tagged PDK1 constructs. As previously described, cells expressing GFP-PKB exhibit both cytosolic staining and a labeled nucleus, whereas following insulin stimulation a clear cell membrane staining appears, reflecting PKB translocation (14). To test the hypothesis that PDK1 could modify PKB movement, HeLa cells were cotransfected with PKB and various PDK1 mutants. In the presence of myristoylated PDK1 (Myr), PKB clearly localized to the plasma membrane, which was visualized as a yellow ring due to the colocalization of green GFP-PKB (PKB) and red myrPDK1 (Myr) (Fig. 4A). Next, we used a PDK1 mutant which lacks the ability to translocate to the plasma membrane due to the replacement of arginine 474 by alanine (Mut) (6). When GFP-PKB and this translocation-deficient PDK1 mutant were cotransfected, no membrane localization of PKB was seen after insulin stimulation. We determined then the effect of Myr-PDK1 on PKB activity after their cotransfection in HeLa cells. As shown in Fig. 4B the activity of PKB obtained from cells transfected with Myr-PDK1 is higher than that seen in cells transfected with PDK1 under basal conditions (14.4-fold in the case of Myr-PDK1 compared to 2.2-fold stimulation in the case of PDK1). Further, no increase in PKB activity compared to basal levels is seen upon insulin stimulation when cells are cotransfected with Myr-PDK1 (14.2-fold stimulation). In contrast, when wild-type PDK1 is transfected into cells, increased basal PKB activity is observed (2.2-fold stimulation) as well as an increased response to insulin compared to the control (8-fold stimulation). This is expected since Myr-PDK1, which is constitutively present at the plasma membrane, will of course not translocate upon insulin stimulation. To see whether PDK1-induced PKB plasma membrane localization can account for the increased PKB activity, we performed fractionation studies in transfected 293 cells and COS cells. PKB activity was monitored using the phospho-Thr308 antibody. Upon coexpression of PDK1 and PKB in 293 cells, PKB activity in the particulate fraction was higher under basal conditions (Fig. 4C). Upon insulin stimulation, we saw an increase in PKB phosphorylation when 293 cells were cotransfected with PDK1 compared to PKB activity when the cells were transfected with PKB alone. Next, we looked at endogenous PKB and PDK1 localization in COS cells, which we previously found to express measurable levels of endogenous PKB (14). Fractionation studies showed that upon insulin stimulation the occurrence of PKB and its phospho-Thr308 form are increased in the particulate fraction (Fig. 4D). However, most of the phospho-Thr308 PKB was seen in the cytosolic fraction. For endogenous PDK1 as for PKB, we found an increased occurrence in the particulate fraction with insulin stimulation. However, as for PKB, most of the protein was localized in the cytosolic fraction (data not shown).
FIG. 4.
Effect of PDK1 mutants on PKB subcellular localization and activation. (A) GFP-PKB was coexpressed in HeLa cells together with wild-type PDK1 (Wild), myristoylated PDK1 (Myr), or PDK1 point mutated in its PH domain (Mut). Forty-eight hours later, HeLa cells were exposed for 5 min to buffer or to insulin (10−6 M), washed, and fixed with paraformaldehyde (4%) prior to incubation with an antibody to Myc, followed by an incubation with a Texas red-linked mouse antibody to label PDK1 constructs. Slides were mounted and analyzed by confocal microscopy. Images represent the center section of the x-y plane. (B) HeLa cells were transfected with wild-type PKB and wild-type PDK1 (Wild) or Myr-PDK1 (Myr). Cells were serum starved in 0.2% (wt/vol) BSA and incubated for 5 min in the presence (black bars) or absence (white bars) of insulin (10−6 M). Then, PKB activity was determined as described in Materials and Methods. PKB activity is expressed as fold stimulation compared to buffer conditions, and the corresponding value is indicated above each column. Values shown are representative of at least three independent experiments performed in triplicate. The levels of the expressed enzymes measured by immunoblotting are shown below the bar graph. (C) 293 cells were transfected with HA-PKB or both Myc-PDK1 and HA-PKB. The cytosolic (S100) and particulate (P100) fractions were prepared as described in Materials and Methods. The PKB phosphorylation state was revealed by immunoblotting with an antibody specific for phospho-Thr308 peptide. (D) COS-7 cells were serum starved in 0.2% (wt/vol) BSA and incubated for 5 min with insulin (I) (10−6 M) or buffer (B). Then, cytosolic (S100) and particulate (P100) fractions were prepared as described in Materials and Methods. PKB was immunoprecipitated (IP) from 500 μg of the cytosolic or particulate fraction, and changes in PKB and PDK1 distributions were detected by immunoblotting (IB) with antibodies to PKB or PDK1. PKB phosphorylation was revealed by immunoblotting with an antibody specific for phospho-Thr308 peptide.
In response to insulin, PDK1 promotes ΔPH-PKB translocation to the plasma membrane.
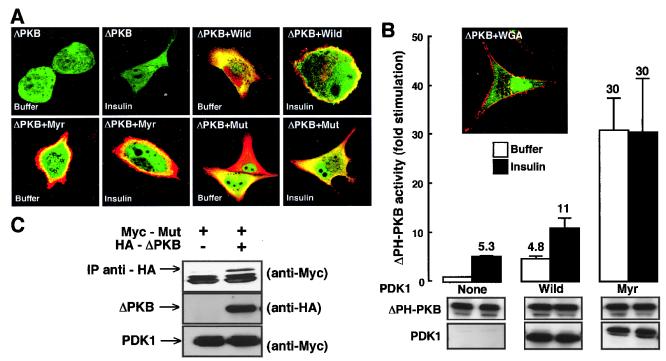
To further investigate the effect of PDK1 on PKB movement, we transfected HeLa cells with a PKB mutant devoid of its PH domain (ΔPH-PKB). We showed earlier that this construct was not capable of translocating to the plasma membrane in response to insulin (Fig. 5A) (21). When GFP–ΔPH-PKB is coexpressed with Myr-PDK1, the cells show the following features: (i) red staining around their cell membranes, corresponding to Myr-PDK1; (ii) green staining in the cytosol, corresponding to GFP–ΔPH-PKB; and (iii) yellow staining just under the plasma membrane, corresponding to colocalization of ΔPH-PKB and Myr-PDK1. The third colocalization is visible without or with insulin treatment. Taken together, these results support the idea that PDK1 translocates jointly with PKB but more importantly support the idea that PDK1 brings PKB to the cell surface. To ascertain that the yellow ring was not an artifact due to protein overexpression, we performed a negative control experiment for colocalization by staining the membrane in red (WGA-rhodamine) and transfecting green ΔPH-PKB. The absence of a yellow ring in the insert of Fig. 5B illustrates that after insulin stimulation ΔPH-PKB is not found around the cell membrane. Next we looked at ΔPH-PKB activity in the presence of the wild type or Myr-PDK1. The activity of ΔPH-PKB was found to be much higher in the presence of Myr-PDK1 than in the presence of wild-type PDK1, even without insulin stimulation (Fig. 5B). This result is in agreement with what we have shown in Fig. 4B, i.e., that PKB basal activity in the presence of Myr-PDK1 is higher than that seen in cells transfected with PDK1. As ΔPH-PKB is more active when cotransfected with Mut-PDK1 (Fig. 6B), we thought that the interaction between those two proteins could be sufficiently strong to allow its detection. Indeed, as seen in Fig. 5C, Mut-PDK1 coimmunoprecipitates with ΔPH-PKB. In contrast, we have been unable to visualize coimmunoprecipitation between wild-type PKB and the PDK1 mutants (data not shown).
FIG. 5.
Effect of PDK1 mutants on ΔPH-PKB subcellular localization and activation. (A) GFP–ΔPH-PKB was coexpressed in HeLa cells together with wild-type PDK1 (Wild), myristoylated PDK1 (Myr), or PDK1 point mutated in its PH domain (Mut). Forty-eight hours later, HeLa cells were exposed for 5 min to buffer or to insulin (10−6 M), washed, and fixed with paraformaldehyde (4%) prior to incubation with an antibody to Myc, which was followed by an incubation with a Texas red-linked mouse antibody to label PDK1 constructs. Slides were mounted and analyzed by confocal microscopy. Images represent the center section of the x-y plane. (B) 293 cells were transfected with ΔPH-PKB and wild-type PDK1 (Wild) or Myr-PDK1 (Myr). Cells were serum starved in 0.2% (wt/vol) BSA and incubated for 5 min in the presence (black bars) or absence (white bars) of insulin (10−6 M). Then, PKB activity was determined as described in Materials and Methods. PKB activity is expressed as fold stimulation compared to buffer conditions, and the corresponding value is indicated above each column. Values shown are representative of at least three independent experiments performed in triplicate. HeLa cells were transfected with 8 μg of ΔPH-PKB construct as described in Materials and Methods. Forty-eight hours later, cells were stimulated for 5 min with insulin (10−6 M). Cells were washed and fixed with paraformaldehyde (4%) prior to incubation with WGA-rhodamine (red) to label the plasma membrane. Slides were mounted and analyzed by confocal microscopy. Images represent the center section of the x-y plane. Membrane staining is in red, and GFP is in green. Areas of colocalization appear as yellow. The levels of the expressed enzymes measured by immunoblotting are shown below the bar graph. (C) 293 cells were transfected with ΔPH-PKB with or without Mut-PDK1. Anti-HA (ΔPH-PKB) immunoprecipitates were resolved by SDS-PAGE followed by Myc (Mut-PDK1) immunoblotting. Whole-cell lysates (100 μg) were resolved; Mut-PDK1 and ΔPH-PKB expression are detected using the Myc and HA antibodies, respectively.
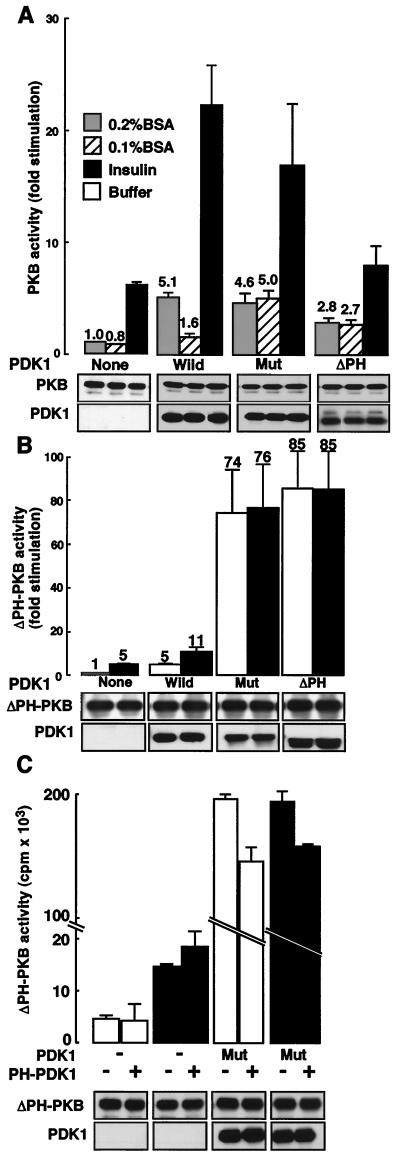
FIG. 6.
Role of the PDK1 PH domain in PKB activation. (A) PKB and PDK1 proteins (Wild, Mut, or ΔPH) were expressed in 293 cells. Cells were serum starved in 0.2% (wt/vol) (white bars) or 0.1% (wt/vol) (dashed bars) BSA and stimulated for 5 min with insulin (10−6 M) (black bars), and PKB kinase activity was determined as described in Materials and Methods. Kinase activity is expressed as fold increase compared to that of nonstimulated wild-type cells. The levels of the expressed enzymes measured by immunoblotting are shown below the bar graph. (B) ΔPH-PKB and PDK1 constructs (Wild, Mut, or ΔPH) were expressed in 293 cells. Prior to stimulation for 5 min with insulin (10−6 M) (black bars) cells were serum starved in 0.2% (wt/vol) BSA, and ΔPH-PKB kinase activity was determined as described in Materials and Methods. Kinase activity is expressed as fold increase compared to that of ΔPH-PKB nonstimulated cells. Values shown are representative of at least three independent experiments performed in triplicate. The levels of the expressed enzymes measured by immunoblotting are shown below the bar graph. (C) 293 cells were transfected with ΔPH-PKB and with PDK1 and PH-PDK1 as indicated. ΔPH-PKB activity was determined as described in Materials and Methods. The levels of the expressed enzymes measured by immunoblotting are shown below the bar graph.
PDK1 activity is negatively regulated by its PH domain.
Next, we examined the role of the PDK1 PH domain in PKB activation. It has been shown by Anderson et al. (6) that a point mutation in the PH domain (R474A) of PDK1 (Mut) results in a loss of its ability to localize to the plasma membrane. A similar phenomenon has been found for PKB since the PKB-PH R25C mutant has lost both its ability to move to the cell surface and most of its activity. Further, as for PKB, a PDK1 with its PH domain deleted fails to localize to the cell surface (data not shown). The ability of ΔPH-PDK1 and Mut-PDK1 to induce PKB activation was tested (Fig. 6A). Stimulation of PKB activity is seen after coexpression of either Mut-PDK1 (4.6-fold) or ΔPH-PDK1 (2.8-fold) even in the absence of insulin. This is not significantly different from the stimulation seen upon cotransfection of the wild-type PDK1 construct (5.1-fold stimulation). In contrast, insulin-stimulated PKB activity is significantly diminished upon coexpression of the PDK1 PH domain mutants. This reduced stimulation was not surprising, since PDK1 cannot translocate to the cell surface, and consequently PKB will not either. Note that the activity seen was comparable to that found with PKB transfected alone. This reflects very likely the action of endogenous PDK1, which can bring PKB to the cell surface, leading to its phosphorylation on Thr308 and on Ser474 by PDK2 activity. In this case basal activity can be linked to a small quantity of PDK1 going to the membrane, as seen for the wild-type PDK1 construct in 0.2% (wt/vol) BSA. To confirm this hypothesis, cells were depleted in 0.1% (wt/vol) BSA. This led to a return to basal activity values for PKB cotransfected with wild-type PDK1, but increased PKB activity was maintained with the PDK1 mutants. To further approach this phenomenon, a similar experiment with ΔPH-PKB was performed. As seen in Fig. 6B, ΔPH-PKB is robustly activated by Mut- and ΔPH-PDK1 mutants. Indeed, a 75- to 85-fold stimulation was seen, which we have never found for PKB in these cells and under these conditions. We interpret these findings to mean that the PH domain of PDK1 acts as a negative modulator of PDK1 enzyme activity. If this hypothesis is correct, we should be able to obtain a decrease in ΔPH-PKB activity when Mut-PDK1 is transfected together with its nonmutated wild-type PDK1 PH domain alone. This was indeed the case, since ΔPH-PKB is 25% less active when transfected together with Mut-PDK1 and the PH domain of PDK1 compared to ΔPH-PKB activity seen after transfection of Mut-PDK1 alone (Fig. 6C). As a control, we have transfected ΔPH-PKB only with the PH domain of PDK1 to be sure that the PH domain impacts on PDK1 activity and not ΔPH-PKB activity (Fig. 6C).
DISCUSSION
We have approached here the mechanism of PKB activation by PDK1 using immunocytochemistry and biochemical analysis. One of the key observations of our work is that in HeLa cells PDK1 translocates to the plasma membrane in response to insulin. This translocation does not require the catalytic activity of PDK1. Our results are consistent with those published by Anderson et al. (6) showing PDK1 translocation in PAE cells exposed to PDGF. However, another recent study from Currie et al. is at variance with this view (13). Indeed, these authors found that in 293 cells PDK1 is constitutively localized at the plasma membrane and that no increase in its membrane presence is observed after either insulin or PDGF stimulation. Here we provide evidence which helps to explain the contradictory results obtained by the two groups. Indeed, it is possible that the discrepancy could be due at least in part to differences in the basal level of the 3′-phospholipid products of PI 3-kinase in resting cells. We found that a proportion of PDK1 was at the plasma membrane under basal conditions; this occurred with a modest amount of PI 3-kinase associated with IRS-1. The use of more-stringent deprivation conditions, i.e., 0.1% BSA, was sufficient to (i) reduce basal PI 3-kinase activity, (ii) decrease basal phosphorylation of PKB Thr308, and (iii) reduce the corresponding PDK1 plasma membrane localization. Since the affinity of PDK1 for PI-3,4,5-P3 has been shown to be 22-fold higher than that of PKB (13) (Kd = 1.6 nM for PDK1 versus 35 nM for PKB), it is reasonable to assume that the basal levels of 3′ phospholipids are sufficient to promote the localization of PDK1 to the plasma membrane whereas they are not high enough to result in PKB plasma membrane recruitment.
We next examined the effects of coexpression of different mutants of PDK1 with PKB on the PKB kinase activity. Coexpression of PKB with wild-type PDK1 resulted in an increment in activity in the absence of insulin as compared to that seen without coexpression of PDK1. Expression of PKB together with kinase-inactive PDK1 did not result in an increase in the basal PKB kinase activity, indicating that the kinase activity of PDK1 is essential for PKB stimulation under these conditions. Ectopic expression of wild-type PDK1 also resulted in a significant enhancement of the insulin-induced PKB activation. Surprisingly, coexpression of the Kd-PDK1 resulted in a similar potentiation of the insulin-induced PKB activation. Those results illustrate that Kd-PDK1 does not act as a dominant-negative form in response to insulin as it was shown by Pullen et al. (20) with p70S6k activation by PDK1 in insulin-stimulated 293 cells. Indeed, cell surface translocation and phospholipids are not required for p70S6k phosphorylation and activation (3), while they are for PKB. The activation mechanism of PKB differs from the one occurring for p70S6k at least for those two major aspects. Therefore, it is not surprising that Kd-PDK1 differently affects PKB activity and that of p70S6k.
At first glance our data would seem at variance with the widely accepted model in which PDK1 is one of the major kinases responsible for the phosphorylation and activation of PKB in response to growth factor stimulation. However, these apparent contradictions can be explained if the key role of PDK1 is in fact to cause translocation of PKB to the plasma membrane and if this is the rate-limiting step in PKB activation. As we have shown in this report, kinase-deficient PDK1 is still able to be transported to the cell surface in response to insulin. Therefore, ectopic expression of Kd-PDK1 with PKB could increase the efficiency of PKB translocation to the plasma membrane where it can be activated by endogenous PDK1 and a putative PDK2.
To verify that PDK1 is involved in promoting the intracellular movement of PKB, we localized these two kinases using confocal microscopy in HeLa cells and subcellular fractionation in 293 cells. For confocal microscopy analysis, HeLa cells were used instead of 293 cells, since we found that 293 cells are difficult cells on which to perform imaging. While preliminary data indicated that the distribution of PKB in 293 cells was similar to that seen in HeLa cells, these data are not conclusive at this point in time. Cotransfection of GFP-PKB along with different PDK1 constructs clearly showed that PDK1 was able to promote PKB translocation to the plasma membrane. In addition, a PDK1 mutant defective in its PH domain was unable to induce PKB translocation, demonstrating the role of the PDK1 PH domain in PKB translocation. However, the PH domain of PKB does not appear to be essential for intracellular movement, since ectopic expression of PDK1 resulted in translocation of a PKB mutant lacking its PH domain.
In a previous report we investigated the role of the PKB PH domain in activation of the enzyme and found that it was required for PKB translocation to the cell surface (21). This membrane targeting likely resulted in a change in PKB conformation allowing its phosphorylation and subsequent activation by PDK1 and PDK2. In the present report, we analyzed the role of the PDK1 PH domain. We have shown, as previously reported (13), that PH domain-defective PDK1s are unable to translocate to the plasma membrane. However, it is not known whether binding to phospholipids is the only important function of the PDK1 PH domain. To determine if the PH domain of PDK1 fulfills other functions as well, we cotransfected with PKB or ΔPH-PKB different PDK1 constructs mutated in or devoid of their PH domain. An intriguing result was obtained when ΔPH-PKB was cotransfected with either ΔPH-PDK1 or Mut-PDK1. Under these conditions, ΔPH-PKB was active to the same extent without or with stimulation by insulin, demonstrating that generation of 3′ phospholipids by PI 3-kinase does not contribute to this activation process. This can be explained if the PH domain of PDK1 is somehow acting in an inhibitory fashion to prevent PKB activation by PDK1 and if phospholipid binding to the PH domain removes this negative constraint. However, when we performed in vitro phosphorylation of ΔPH-PKB by ΔPH-PDK1 versus that by PDK1, we did not see a much higher activation of ΔPH-PKB by ΔPH-PDK1 than by PDK1 (data not shown). This could be explained if in intact cells another protein is required to produce increased ΔPH-PDK1-mediated activation of ΔPH-PKB. PDK1 appears to be a constitutively active kinase that does not require phosphorylation by another kinase for its activation, since its autophosphorylation seems to be activating. Several recent reports, including our own, support the view that both kinases may be functionally regulated by PtdIns-3,4,5-P3. This evidence can be summarized as follows: (i) PDK1-induced phosphorylation of a PH domain-deleted mutant of PKB (ΔPH-PKB) in vitro is enhanced by PtdIns-3,4,5-P3 (23); (ii) although a fraction of PDK1 is constitutively associated with the plasma membrane, more molecules become associated via a PH domain-dependent mechanism following PDGF stimulation (6, 13); (iii) suboptimal doses of PDGF and insulin synergize with PDK1 to activate PKB (6); (iv) phosphorylation, but not membrane association, of myristoylated PKB is partially inhibited by wortmannin or by depletion of transfected 293 cells in 0.1% BSA for 24 h (19).
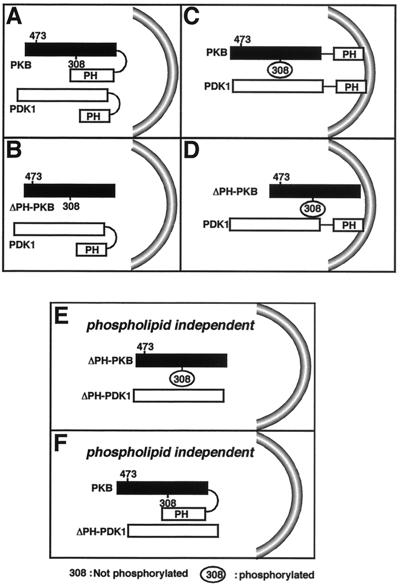
Taking our results as a whole, we propose the following model to explain the activation of PKB by PDK1 and the requirement for phospholipids. When PKB resides in the cytosol, its conformation is such that its PH domain blocks access to residue Thr308, which therefore cannot be phosphorylated by PDK1 (Fig. 7A). After stimulation by insulin, 3′ phospholipids which promote PDK1 translocation to the membrane together with PKB are produced. There, PKB is phosphorylated by PDK1 and probably by PDK2 (Fig. 7C). At that point, both kinases are in the proper conformation and in the adequate compartment for optimal activation. When PKB is in the favorable conformation but located in the cytosol, such as is the case with ΔPH-PKB, PDK1 is not in a functionally active configuration due to its folded PH domain (Fig. 7B). When both PKB and PDK1 PH domains are removed, negative constraints are relieved and the molecules are in configurations allowing maximal activity independent of phospholipids (Fig. 7E). Finally, PKB in the presence of ΔPH-PDK1 is slightly activated, but because PKB is not in an optimal conformation, the extent of activation is less than that seen with ΔPH-PKB (Fig. 7F). This is the case even after insulin stimulation, since translocation is not possible due to the lack of the PDK1 PH domain. Elevated basal PKB activation (4.6-fold stimulation in the case of mutPDK1 and 2.8-fold in the case of ΔPH-PDK1) under these conditions is independent of the presence of phospholipids.
FIG. 7.
Working hypothesis for activation of PKB by PDK1. (A and B) Activation without insulin; (C and D) activation with insulin; (E and F) activation with or without insulin.
Considered together, our results strengthen the concept that certain PH domains can act as specific membrane recruitment devices, regulating the translocation of soluble proteins such as PKB. Moreover, we demonstrate that some PH domains can exert functions other than just binding to phospholipids in that they also appear to control the activity level of the kinase. Indeed, our data indicate that the PH domain of PDK1 appears to modulate the enzyme conformation, which permits it to modify the kinase activity depending on intracellular localization.
ACKNOWLEDGMENTS
Our research was supported by the Institut National de la Santé et de la Recherche Médicale, the Association pour la Recherche sur le Cancer, the Université de Nice-Sophia Antipolis, the Ligue contre le Cancer, Groupe LIPHA-Merck (Lyon, France), and the European Community (grant QLG1-CT-1999-00674). C.S. was a recipient of a Poste Vert from INSERM.
We thank P. T. Hawkins for the generous gift of Myc-tagged PDK1 constructs and proteins purified from baculovirus-expressing Sf9 cells.
REFERENCES
- 1.Ahmed N N, Grimes H L, Bellacosa A, Chan T O, Tsichlis P N. Transduction of interleukin-2 antiapoptotic and proliferative signals via Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:3627–3632. doi: 10.1073/pnas.94.8.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alessi D, Cohen P. Mechanism of activation and function of protein kinase B. Curr Opin Genet Dev. 1998;8:55–62. doi: 10.1016/s0959-437x(98)80062-2. [DOI] [PubMed] [Google Scholar]
- 3.Alessi D, Kozlowski M T, Weng Q-P, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1997;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 4.Alessi D R, Deak M, Casamayor A, Caudwell F B, Morrice N, Norman D G, Gaffney P, Reese C B, MacDougall C N, Harbison D, Ashworth A, Bownes M. 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr Biol. 1997;7:776–789. doi: 10.1016/s0960-9822(06)00336-8. [DOI] [PubMed] [Google Scholar]
- 5.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R J, Reese C B, Cohen P. Characterization of a 3-phosphoinositide-dependent kinase which phosphorylates and activates protein kinase Bα. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 6.Anderson K E, Coadwell J, Stephens L R, Hawkins P T. Translocation of PDK-1 to the plasma membrane is important in allowing PDK-1 to activate protein kinase B. Curr Biol. 1998;8:684–691. doi: 10.1016/s0960-9822(98)70274-x. [DOI] [PubMed] [Google Scholar]
- 7.Andjelkovic M, Alessi D, Meier R, Fernandez A, Lamb N, Frech M, Cron P, Cohen P, Lucocq J, Hemmings B. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 8.Andjelkovic M, Jakubowicz T, Cron P, Ming X-F, Han J-W, Hemmings B A. Activation and phosphorylation of a pleckstrin holology domain containing protein kinase (RAC-PK/PKB) promoted by serum and protein phosphatase inhibitors. Proc Natl Acad Sci USA. 1996;93:5699–5704. doi: 10.1073/pnas.93.12.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgering T, Boudewijn M, Coffer P J. Protein kinase B (c-Akt) in phosphotidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 10.Cheng J Q, Godwin A K, Bellacosa A, Taguchi T, Franke T F, Hamilton T C, Tsichlis P N, Testa J R. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases is amplified in human ovarian carcinomas. Proc Natl Acad Sci USA. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng J Q, Ruggeri B, Klein W M, Sonoda G, Altomare D A, Watson D K, Testa J R. Amplification of AKT2 in human pancreatic-cancer cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci USA. 1996;93:3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross D A E, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthetase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 13.Currie R A, Walker K S, Gray A, Deak M, Casamayor A, Downes C P, Cohen P, Alessi D R, Lucocq J. Role of phosphatidylinositol 3,4,5-trisphosphate in regulating the activity and localization of 3-phosphoinositide-dependent protein kinase-1. Biochem J. 1999;337:575–583. [PMC free article] [PubMed] [Google Scholar]
- 14.Filippa N, Sable C L, Filloux C, Hemmings B A, Van Obberghen E. Mechanism of protein kinase B activation by cyclic AMP-dependent protein kinase. Mol Cell Biol. 1999;19:4989–5000. doi: 10.1128/mcb.19.7.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franke T F, Yang S-I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinosital 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 16.Frech M, Andjelkovic M, Ingley E, Reddy K K, Falck J R, Hemmings B A. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J Biol Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 17.Göransson O, Wijkander J, Manganiello V, Degerman E. Insulin-induced translocation of protein kinase B to the plasma membrane in rat adipocytes. Biophys Biochem Res Commun. 1998;246:249–254. doi: 10.1006/bbrc.1998.8602. [DOI] [PubMed] [Google Scholar]
- 18.Kohn A D, Kovacina K S, Roth R A. Insulin stimulates the kinase activity of RAC-PK, a pleckstrin homology domain containing Ser/thr kinase. EMBO J. 1995;14:4288–4295. doi: 10.1002/j.1460-2075.1995.tb00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Wahl M, Eguinoa A, Stephens L, Hawkins P, Witte O. Proc. Natl Acad Sci USA. 1997;94:13820–13825. doi: 10.1073/pnas.94.25.13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pullen N, Dennis P, Andjelkovic M, Dufner A, Kozma S, Hemmings B, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 21.Sable C L, Filippa N, Filloux C, Hemmings B A, Van Obberghen E. Involvement of the pleckstrin homology domain in the insulin-stimulated activation of protein kinase B. J Biol Chem. 1998;273:29600–29606. doi: 10.1074/jbc.273.45.29600. [DOI] [PubMed] [Google Scholar]
- 22.Stephens L, Anderson K, Stokoe D, Erdjument-Bromage H, Painter G, Holmes A, Gaffney P, Reese C, McCormick F, Tempst P, Coadwell J, Hawkins P. Protein kinase B kinase that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 23.Stokoe D, Stephens L R, Copeland T, Gaffney P R J, Reese C B, Painter G F, Holmes A B, McCormick F, Hawkins P T. Duel role of phosphatidylinositol-3,4,5,-trisphosphate in the activation of protein kinase B. Science. 1997;277:567–570. doi: 10.1126/science.277.5325.567. [DOI] [PubMed] [Google Scholar]
- 24.Walker K, Deak M, Paterson A, Hudson K, Cohen P, Alessi D. Activation of protein kinase B beta and gamma isoforms by insulin and by 3-phosphoinositide-dependent protein kinase-1 in vitro: comparison with protein kinase B. Biochem J. 1998;331:299–308. doi: 10.1042/bj3310299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wijkander J, Stenson-Holst L, Rahn T, Resjo S, Castan I, Manganiello V, Belfrage P, Degerman E. Regulation of protein kinase B in rat adipocytes by insulin, vanadate, and peroxyvanadate. J Biol Chem. 1997;272:21520–21526. doi: 10.1074/jbc.272.34.21520. [DOI] [PubMed] [Google Scholar]