Abstract
The marine bacterium Vibrio natriegens has recently been demonstrated to be a promising new host for molecular biology and next generation bioprocesses. V. natriegens is a Gram-negative, non-pathogenic slight-halophilic bacterium, with a high nutrient versatility and a reported doubling time of under 10 min. However, V. natriegens is not an established model organism yet, and further research is required to promote its transformation into a microbial workhorse.
In this work, the potential of V. natriegens as an amino acid producer was investigated. First, the transcription factor-based biosensor LysG, from Corynebacterium glutamicum, was adapted for expression in V. natriegens to facilitate the detection of positively charged amino acids. A set of different biosensor variants were constructed and characterized, using the expression of a fluorescent protein as sensor output. After random mutagenesis, one of the LysG-based sensors was used to screen for amino acid producer strains. Here, fluorescence-activated cell sorting enabled the selective sorting of highly fluorescent cells, i.e. potential producer cells. Using this approach, individual L-lysine, L-arginine and L-histidine producers could be obtained producing up to 1 mM of the effector amino acid, extracellularly. Genome sequencing of the producer strains provided insight into the amino acid production metabolism of V. natriegens.
This work demonstrates the successful expression and application of transcription factor-based biosensors in V. natriegens and provides insight into the underlying physiology, forming a solid basis for further development of this promising microbe.
Keywords: Transcription factor-based biosensors, Metabolic engineering, Amino acid production, High-throughput screening
Highlights
-
•
Vibrio natriegens is a promising new host for biotechnology.
-
•
Transcription factor-based biosensors were expressed in V. natriegens.
-
•
Mutagenesis and screening using FACS provided amino acid producing mutants.
-
•
Genome sequencing revealed several causal mutations leading to amino acid production.
-
•
These results will support further efforts to develop V. natriegens as a production host.
1. Introduction
Development of Vibrio natriegens as a new host for molecular biology and bioprocesses has seen a recent surge in interest. V. natriegens was first described in 1958, as a sodium (‘natrium’) requiring bacterium isolated from marsh mud (Payne, 1958). V. natriegens is a non-pathogenic, Gram-negative, slight halophile with a high nutrient versatility, which was previously classified as Pseudomonas natriegens and later Beneckea natriegens (Baumann et al., 1971; Payne et al., 1961). The most remarkable property of V. natriegens is its very short doubling time, which is below 10 min under optimal conditions (Eagon, 1962; Hoffart et al., 2017).
Recent work has highlighted the application of V. natriegens in several different areas of biotechnology (Hoff et al., 2020; Thoma and Blombach, 2021). Weinstock et al. demonstrated that V. natriegens can be used as an alternative for Escherichia coli for standard molecular biology procedures, shortening the time of standard workflows (Weinstock et al., 2016). V. natriegens high capacity for translation makes it an attractive platform for cell free protein production (Des Soye et al., 2018; Failmezger et al., 2018; Wiegand et al., 2018). Furthermore, several studies contributed to improving the available toolbox for engineering V. natriegens, describing protocols for natural (Dalia et al., 2017) and artificial (Weinstock et al., 2016) transformation, characterization of genetic parts and tools (e.g. promoters, plasmids) (Tschirhart et al., 2019), use of CRISPRi (Lee et al., 2019), which are summarized in recent review articles (Hoff et al., 2020; Thoma and Blombach, 2021).
V. natriegens was also shown to be a potential production host for biotechnology, because of its high biomass specific substrate consumption rate, which is at least two fold higher than the biomass specific substrate consumption rate of E. coli, Pseudomonas putida, Corynebacterium glutamicum and yeast, under both anaerobic and aerobic conditions (Hoffart et al., 2017). Recently, a V. natriegens platform strain was developed, which features the genomic removal of two prophage regions (Pfeifer et al., 2019). These prophages exhibited spontaneous activation under standard cultivation conditions. The prophage-free strain showed an improved tolerance to DNA-damaging conditions and hypo-osmotic conditions and outcompeted the wild type in a competitive growth experiment (Pfeifer et al., 2019). Furthermore, V. natriegens strains were successfully engineered for the production of chemicals, including L-alanine (Hoffart et al., 2017), poly-β-hydroxybutyrate (Dalia et al., 2017), 2,3-butanediol (Erian et al., 2020), melanin (Wang et al., 2020), violacein and β-carotene (Ellis et al., 2019). High natural PHB production was also described by a strain similar to V. natriegens (Chien et al., 2007). Development of V. natriegens into a production strain could be accelerated with the use of transcription factor (TF)-based biosensors. TF-based biosensors comprise a variety of proteins that enable the detection of specific cellular products (Dietrich et al., 2010; Lin et al., 2017; Mahr and Frunzke, 2016). They have multiple applications, including the dynamic regulation of production pathways (Dahl et al., 2013), monitoring of product formation in both single cells and cell cultures (Mustafi et al., 2014), and enabling screening and selection of cells with increased product formation (Chou and Keasling, 2013; Mahr et al., 2015; Raman et al., 2014). However, biosensors have not been established in V. natriegens yet.
Therefore, we aimed to establish TF-based biosensors in V. natriegens. In particular, we chose to express two biosensors from C. glutamicum, based on LysG and Lrp, which have been successfully applied in screenings and selections for amino acid production (Binder et al., 2012; Mahr et al., 2015; Schendzielorz et al., 2014). In this study, we adapted these sensors for expression in V. natriegens, and applied one of them in a mutagenesis and screening approach to obtain V. natriegens strains that produce positively charged amino acids.
2. Materials and methods
2.1. Bacterial strains and plasmids
All V. natriegens and E. coli strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used for plasmid storage and propagation (Invitrogen, Karlsruhe, Germany). All V. natriegens strains used in this work were derived from V. natriegens ATCC 14048 (Payne, 1958). All plasmids constructed in this work were based on pBR322 (Bolivar et al., 1977).
Table 1.
Bacterial strains used in this study.
| Strain/plasmid | Genotype and relevant characteristic | Reference |
|---|---|---|
| V. natriegens ATCC 14048; DSM 759 | Wild type | German Collection of Microorganisms and Cell Cultures, (Payne, 1958) |
| E. coli DH5a | supE 44 ΔlacU169 (φ80lacZDM15) hsdR17 recA1 endA1 gyrA 96 thi-1 relA1 | Invitrogen (Karlsruhe, Germany) |
| pBR322 | AmpR, TetR, | Bolivar et al. (1977) |
| pBR322-eYFP | AmpR, pBR322 derived backbone used for LysG sensor construction | This study |
| pBR322-J23100-Lrp-PbrnFE-eYFP | AmpR, pBR322 derived Lrp biosensor, Lrp expressed under the J23100 promoter | This study |
| pBR322-J23101-Lrp-PbrnFE-eYFP | AmpR, pBR322 derived Lrp biosensor, Lrp expressed under the J23101 promoter | This study |
| pBR322-J23106-Lrp-PbrnFE-eYFP | AmpR, pBR322 derived Lrp biosensor, Lrp expressed under the J23106 promoter | This study |
| pBR322-J23108-Lrp-PbrnFE-eYFP | AmpR, pBR322 derived Lrp biosensor, Lrp expressed under the J23108 promoter | This study |
| pBR322-J23110-Lrp-PbrnFE-eYFP | AmpR, pBR322 derived Lrp biosensor, Lrp expressed under the J23110 promoter | This study |
| pBR322-J23100-LysG-PlysE-eYFP | AmpR, pBR322 derived LysG biosensor, LysG expressed under the J23100 promoter | This study |
| pBR322-J23101-LysG-PlysE-eYFP | AmpR, pBR322 derived LysG biosensor, LysG expressed under the J23101 promoter | This study |
| pBR322-J23104-LysG-PlysE-eYFP | AmpR, pBR322 derived LysG biosensor, LysG expressed under the J23104 promoter | This study |
| pBR322-J23106-LysG-PlysE-eYFP | AmpR, pBR322 derived LysG biosensor, LysG expressed under the J23106 promoter | This study |
| pBR322-J23108-LysG-PlysE-eYFP | AmpR, pBR322 derived LysG biosensor, LysG expressed under the J23108 promoter | This study |
| pBR322-J23110-LysG-PlysE-eYFP | AmpR, pBR322 derived LysG biosensor, LysG expressed under the J23110 promoter | This study |
2.2. Media and culture conditions
E. coli cells were grown in liquid lysogeny broth (LB: 10 g l−1 tryptone, 5 g l−1 yeast extract, 10 g l−1 NaCl) or on LB agar plates (LB with 15 g l−1 agar [Carl Roth, Karlsruhe, Germany]) at 37 °C. Carbenicillin (50 mg l−1) was added, if not indicated otherwise.
For molecular cloning procedures and for first pre-cultures, V. natriegens strains were grown in BHIN (37 g l−1 brain heart infusion [Becton, Dickinson, Franklin Lakes, NJ] with additional 15 g l−1 NaCl). When minimal media was required, V. natriegens was grown in standard VN medium (5 g l−1 (NH4)2SO4, 15 g l−1 NaCl, 1 g l−1, KH2PO4,1 g l−1 K2HPO4, 0.25 g l−1 MgSO4, 0.01 g l−1 CaCl2, 16.4 mg l−1 FeSO4·7H2O, 10 mg l−1 MnSO4·H2O, 0.3 mg l−1 CuSO4·5H2O, 1 mg l−1 ZnSO4·7H2O, 0.02 mg l−1 NiCl2·6H2O, 21 g l−1 3-(N-morpholino)propanesulfonic acid (MOPS), pH set to 7.5 by addition of NaOH), or optimized VN, VNopt (VN with in total 42 g l−1 3-(N-morpholino)propanesulfonic acid (MOPS), pH set to 8.0 by addition of NaOH).
Standard cultivations always followed the same procedure; a first preculture in BHIN medium, followed by a second preculture in VN medium, and finally the main cultivation in VN medium. If not specifically indicated, the optimized VN medium was used, supplemented with 15 g l−1 glucose. For biosensor experiments, medium was additionally supplemented with carbenicillin (50 mg l−1). When indicated, media were supplemented with specific amino acids (Sigma Aldrich, USA), or glucose was replaced with the indicated amino acid (20 mM).
Growth characterization of main cultures was done using the BioLector® microcultivation system (m2p-labs, Germany) as described previously (Heyer et al., 2012). Cultures were grown in 800 μl in 48-well FlowerPlates® (m2p-labs, Germany), at 30 °C, 80% humidity and 1500 rpm shaking frequency. When required, FlowerPlates® containing optodes for the measurement of dissolved oxygen (DO) and pH were used. Typically, cultures were inoculated to a start OD600 of 0.5, unless specified otherwise. Culture growth was measured as the backscatter of light with a wavelength of 620 nm (signal gain factor of 20). When indicated, fluorescence was measured by excitation with light with a wavelength of 510 nm and emission was measured at 532 nm (signal gain factor of 60). For measurement of dissolved oxygen and pH, standard settings were used. Measurements were taken in 5 or 10 min intervals.
When no online measurement was required, cultures were grown in 1 ml medium in 96-well deep well plates (VWR, USA)), at 30 °C and 900 rpm shaking frequency, in an HT Microtron shaker (Infors HT, Bottmingen, Switzerland).
2.3. Molecular cloning
Standard cloning methods, including PCR and DNA restriction, were carried out according to established protocols (Sambrook et al., 2001). PCR reactions were performed using Q5® High-Fidelity 2X Master Mix (New England Biolabs GmbH, Frankfurt am Main, Germany). Plasmid assembly was done using Gibson assembly (Gibson et al., 2009). Plasmid sequences were verified by Sanger sequencing, by Eurofins Genomics (Ebersberg, Germany). Primers were ordered as custom DNA oligonucleotides from Eurofins Genomics (Ebersberg, Germany). All primers are listed in Table S1.
Construction of the Lrp-based sensors was performed in one step assembly of EcoRV and NheI digested pBR322 and three inserts; a part containing lrp, a part containing eyfp and a part containing one of the Anderson promoter sequences and the lrp-brnF intergenic region with the first 30 bp of brnF. The lrp containing part was obtained by PCR from C. glutamicum genomic DNA with primers lrp_fw and lrp_rv. The eyfp containing part was obtained by PCR from plasmid pJC1-lrp-brnF′-eyfp (Mustafi et al., 2012) with primers lrp_eYFP_fw and lrp_eYFP_rv. The constitutive promoter parts were obtained by PCR from C. glutamicum genomic DNA with different P1xx_PbrnFE_fw primers and primer PbrnFE_rv.
Construction of the LysG-based sensors was performed in two steps. In the first step, the intermediate plasmid pBR322-eYFP was constructed by assembly of BamHI and NheI digested pBR322 and two PCR products, a part containing eyfp and a linker part. The eyfp containing part was obtained by PCR from plasmid pJC1-lrp-brnF′-eyfp (Mustafi et al., 2012) with primers lysG_eYFP_fw and lysG_eYFP_rv. The linker part was obtained by PCR from plasmid pDM4-vnp2-pBAD-ccdB (Pfeifer et al., 2019) with primers lysG_araC_fw and lysG_araC_rv. The LysG-based biosensor expression vectors were constructed by assembly of SmaI and BcuI digested pBR322-eYFP and three inserts, a part containing lysG, a part containing one of the Anderson promoter sequences, and a part containing the lysG-lysE intergenic region with the first 60 bp of lysE. The lysG containing part was obtained by PCR from C. glutamicum genomic DNA with primers lysG_fw and lysG_rv. The parts containing different Anderson promoter sequences were obtained by hybridization of two complementary oligonucleotides, P100_lysG_fw with P100_lysG_rv, P101_lysG_fw with P101_lysG_rv, etc. The parts containing the lysG-lysE intergenic region and an overlap sequence for assembly with a specific Anderson promoter sequence-containing part were obtained by PCR from C. glutamicum genomic DNA with different PlysE_1xx_fw primers and primer PlysE_rv.
For all biosensors plasmids, upstream of eyfp a stop codon was added followed by a ribosomal binding site (RBS) for V. natriegens (AAAGAGGAGAAA) and a 6 nucleotide spacer sequence (TAATCT), which was previously reported to enhance expression of fluorescent proteins (Lentini et al., 2013).
E. coli was transformed using the Rubidium Chloride method (Hanahan, 1983). Preparation of plasmids from E. coli was done with the QIAprep spin miniprep kit (Qiagen, Hilden, Germany). V. natriegens electroporation transformation was done following a previously established electroporation protocol (Weinstock et al., 2016).
2.4. HPLC analyses
Organic acids were quantified by high performance liquid chromatography (HPLC) using an Agilent 1100/1200 LC ChemStation combination (Agilent, Santa Clara, USA) equipped with a refractive index detector (RID). A Phenomenex rezex ROA-organic acid H+ 300 mm × 7.80 mm column (Phenomenex®, Germany) was used and isocratic elution was done with 5 mM H2SO4.
Amino acids were quantified as ortho-phthaldialdehyde derivatives by HPLC using an Agilent 1290 Infinity LC ChemStation (Agilent, Santa Clara, USA) equipped with automatic pre-column derivatization and a fluorescence detector. A Zorbax Eclipse AAA 3.5 μm 4.6 × 7.5 mm column (Agilent, Santa Clara, USA) was used and elution was performed using a gradient of Na-borate buffer (10 mM Na2HPO4; 10 mMNa2B4O7, pH 8.2) and methanol, adapted to operator's guide.
HPLC samples were prepared from cultures by centrifugation for 15 min at 4000 g and 4 °C, followed by 1:10 dilution in ddH2O, and filtration using a 0.2 μM filter. For amino acid determination, samples were prepared once cultures were at the end of the first exponential phase (after approximately 4 h of growth).
2.5. MNNG mutagenesis
For mutagenesis, methyl nitronitrosoguanidine (MNNG) was dissolved in DMSO to a concentration of 3 g l−1. From that, working solutions were prepared from 3 g l−1 to 1 g l−1 in 0.5 stepwise increments. Cell cultures were grown in BHIN supplemented with 50 mg l−1 carbenicillin in 500 ml shake flasks for 4 h to a final OD600 of 4.6 and left in stationary phase for 30 min prior to mutagenesis. Upon addition of 250 μl MNNG solution (or DMSO as control) to 5 ml of culture volume, cell cultures were incubated for 15 min in 50 ml centrifuge tubes in a 37 °C rotary shaker, including handling time. Immediately afterwards, 45 ml 0.9% NaCl was added before culture harvest by centrifugation was performed (20 °C, 4000 rpm, 7 min, Heraeus X3R, Thermo Scientific, Waltham, USA). Cells were washed twice with 0.9% NaCl and transferred to BHIN, supplemented with 50 mg l−1 carbenicillin, for 1 h regeneration at 37 °C.
Afterwards, for each different MNNG concentration-culture, 160 cells were sorted on BHIN agar plates supplemented with 50 mg l−1 carbenicillin, using a FACSAria Fusion flow cytometer (Becton Dickinson, San Jose, USA). After one day, viability of the cultures was determined by counting the number of clones on each plates.
2.6. Flow cytometry analysis and sorting
Characterization of sensor performance was done by flow cytometry using a MACSQuant® (Miltenyi Biotec, Germany) equipped with a 488 nm excitation laser and a 525/50 nm band-pass detection filter. Cultures were grown for one day, diluted 1:100 into sterile VNopt medium and incubated for 30 min on room temperature before measurement.
Cell sorting experiments by flow cytometry were performed on a FACSAria Fusion flow cytometer (Becton Dickinson, San Jose, USA) equipped with a 488 nm excitation laser and a 85 μm nozzle. Forward scatter was filtered using a 1.5 neutral density (ND) filter, side scatter was measured using a 488/10 nm band-pass filter. Fluorescence (eYFP) was measured using a 530/30 nm band-pass filter. FACS-Diva software version 8.0.2 was used to record and export the measurements. For cell sorting, the single-cell precision mode was used. Prior to flow cytometry analysis, cultures were diluted 500 times in VNopt medium without glucose, filtered using a 30 μm filter (Sysmex, Goerlitz, Germany) and incubated for 30 min on room temperature to allow for eYFP protein maturation. A hierarchical gating strategy was used for sorting. Non-bacterial particles were excluded using forward scatter and side scatter values, and doublet discrimination (avoiding sorting of multiple cells in one droplet) was done using side scatter and forward scatter area and width values. A rectangular gate was used to sort the top 5% most fluorescent cells. For bulk sorting experiments, cells were collected in a 15 ml tube prefilled with 5 ml sterile filtered VNopt medium without glucose. Afterwards, tubes were centrifuged at 1500 rpm for 5 min, excess medium was removed and the remainder was resuspended in VNopt medium to a total volume of 800 μl.
Cultures were grown until stationary phase overnight. In the morning, a 10x dilution in fresh medium was done and the FACS sorting procedure was started once cultures were at the end of the first exponential phase (after approximately 4 h of growth).
2.7. Data analysis and visualization
Analysis and visualization of growth, HPLC and fluorescence data was done using R v3.6.3 (R Core Team, 2020) and tidyverse (Wickham et al., 2019). Fluorescence data was parsed using flowcore (Ellis et al., 2021), hill curve fitting for dose response curves was done using drc (Ritz et al., 2015).
2.8. Whole genome sequencing
V. natriegens genomic DNA was purified using a NucleoSpin Microbial DNA Mini kit (MACHEREY-NAGEL, Dueren, Germany) from an overnight BHIN culture. DNA concentration was measured using the Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, USA). Afterwards, 1 μg of DNA was used for library preparation using the NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (NEB, Frankfurt am Main, Germany). Library evaluation was done by qPCR using the KAPA library quantification kit (Peqlab, Erlangen, Germany). Afterwards, normalization for pooling was done and paired-end sequencing was performed using a Miseq (Illumina, San Diego, CA), with a read length of 2 × 150 bases. Sequencing output (base calls) were stored as demultiplexed fastq files. Processing of the data (e.g. trimming, mapping, coverage extraction) was done with the CLC Genomic Workbench software (Qiagen Aarhus A/S, Aarhus, Denmark). Reads were mapped against the V. natriegens ATCC 14048 genome (RefSeq replicon entries NZ_CP009977.1 and NZ_CP009978.1). The relevance of identified mutations was assessed manually. Only SNPs with a frequency higher than 50% were considered.
3. Results and discussion
3.1. Optimizing a defined medium for repetitive-batch cultivation
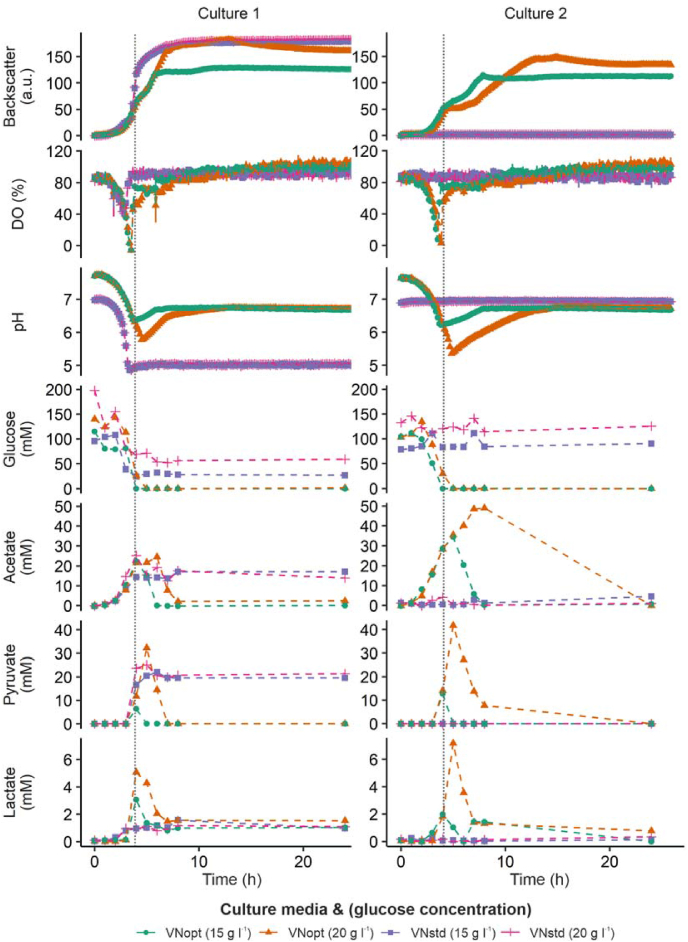
A V. natriegens medium that supports repetitive-batch cultivations is required for iterative screening experiments, such as fluorescence activated cell sorting (FACS) based enrichments and adaptive laboratory evolution (ALE) experiments. Therefore, initial microtiter growth cultivations were performed with V. natriegens to find a medium that supports repetitive-batch cultivations (i.e., in which iterative cultivations show nearly identical growth behavior). Several different growth media have been described for V. natriegens, which are generally adapted media originally used to grow other bacteria (e.g., E. coli or C. glutamicum), usually with increased sodium chloride concentrations; examples are M9-based (Weinstock et al., 2016) and VN (CGXII-based) (Hoffart et al., 2017) media. When V. natriegens was grown in VN medium with 20 g l−1 glucose using the BioLector® cultivation system, growth was severely hampered when repetitive cultivations where performed (Fig. 1). It was hypothesized that a low pH, reached during stationary phase in the first culture, could be the reason, as it was previously shown that V. natriegens converts glucose to organic acids such as acetate, pyruvate and lactate during batch cultivations in VN medium (Hoffart et al., 2017; Payne et al., 1961).
Fig. 1.
Optimization of a V. natriegens growth medium suitable for repetitive-batch cultivation.V. natriegens growth in standard VN (VNstd) with 15 g l−1 glucose or 20 g l−1 glucose, and optimized VN (VNopt) with 15 g l−1 glucose or 20 g l−1 glucose. For two repetitive cultivations, backscatter (indicates biomass), dissolved oxygen and pH were measured online (n = 3, mean and standard deviation are shown), and samples were taken for measurement of glucose, acetate, pyruvate and lactate concentrations (n = 1 representative culture). Vertical grey lines indicate end of first exponential phase of culture in VNopt with 15 g l−1 glucose.
To enable stable repetitive growth in VN medium, the initial pH was increased from 7.5 to 8.0, and the amount of buffer (MOPS) was doubled to 42 g l−1. V. natriegens was grown in the standard VN medium and in the new, optimized medium, with different glucose concentrations, and extracellular concentrations of glucose, acetate, pyruvate and lactate were measured over time by HPLC (Fig. 1).
When grown in the standard medium with normal (20 g l−1) or lowered (15 g l−1) glucose concentrations, only the first culture showed growth. In the second culture no change in any of the measured parameters was observed. In contrast, for cultivations in the optimized medium with both glucose concentrations, growth was observed in both the first and second cultivation. Thus, in standard VN medium the V. natriegens cultures seemed to lose their viability during the first cultivation. This was likely due to the low pH values that were reached after approximately 4 h, due to production of the organic acids acetate, pyruvate and lactate. After reaching a pH value of 5, no more decrease in glucose concentration was observed and the stable dissolved oxygen (DO) values indicated no respiratory activity. The backscatter measurement did not represent worse growth in standard VN medium, compared to optimized VN medium, but this was likely an artefact of the backscatter measurement; final optical density values (OD600) were 10.0 ± 0.2 (20 g l−1 glucose) and 10.3 ± 0.1 (15 g l−1 glucose) for standard VN medium, and 20.6 ± 0.0 (20 g l−1 glucose) and 18.0 ± 0.2 (15 g l−1 glucose) for optimized VN medium. These values support the hypothesis that the V. natriegens cultures were inhibited by low pH values, which led to growth arrest. This effect was also found in a previous study that reported a negative effect on growth when pH values dropped below 7, when V. natriegens was grown on trypticase-based medium (Payne et al., 1961).
When V. natriegens was grown in optimized VN medium, two distinct growth phases were observed. First, an exponential growth phase in which all glucose was consumed and acetate, pyruvate and lactate were produced (Fig. 1). During this phase the DO values and pH rapidly decreased. Then, a second growth phase started, in which the concentrations of acetate, pyruvate and lactate decreased to below the detection limit, and the pH value increased. The lowered DO values during this phase indicate respiration of organic acids. The second cultivation in optimized VN medium showed the same trend as the first one, but especially for the higher glucose concentration (20 g l−1), more acetate, pyruvate and lactate were produced and a lower pH value was reached, compared to the first cultivation. Previous studies also reported the production of organic acids, especially under anaerobic conditions (Hoffart et al., 2017; Long et al., 2017; Payne et al., 1961; Thiele et al., 2021).
Repetitive cultivations, showing a similar growth profile, could be done in optimized VN medium with both 20 g l−1 glucose and 15 g l−1 glucose for at least 8 cultivations (data not shown). Because of the faster cultivation time and lower organic acid production, all remaining experiments were carried out using VN medium with a pH initially set to 8.0, with 42 g l−1 MOPS and 15 g l−1 glucose.
3.2. Orthogonal transfer of TF-based biosensors to V. natriegens
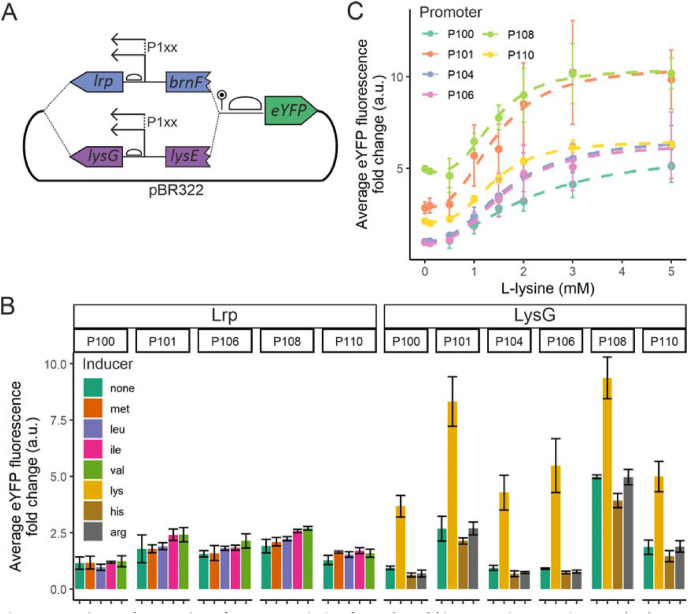
To establish biosensors in V. natriegens, two transcription factor-based biosensors previously used in C. glutamicum were adapted for expression in V. natriegens; LysG and Lrp. Both are used for the detection of amino acids: Lrp for L-methionine, L-leucine, L-isoleucine and L-valine; LysG for L-histidine, L-arginine and L-threonine (Binder et al., 2012; Mustafi et al., 2012).
For both Lrp and LysG, a set of sensor variants were designed, following a similar design principle (Fig. 2A). The fluorescent protein eYFP was chosen as the sensor reporter. The Lrp sensor consists of the C. glutamicum lrp gene, the lrp-brnF intergenic region and the first part of brnF (Mustafi et al., 2012). The LysG sensor consists of the C. glutamicum lysG gene, the lysG-lysE intergenic region and the first part of lysE (Binder et al., 2012). The first part of brnF or lysG were added, because these genes are transcribed as a leaderless transcripts (Pfeifer-Sancar et al., 2013). Therefore, the first parts of brnF or lysG probably have regulatory functions and are important for Lrp or LysG binding. For each sensor, an in-frame stop codon, RBS and linker fragment was added upstream of eyfp. For both sensors, a small sensor library was constructed by expressing the lrp or lysG gene under different promoters from the Anderson promoter library, which was previously shown to be functional in V. natriegens (Anderson, 2006; Stukenberg et al., 2021; Tschirhart et al., 2019).
Fig. 2.
Design and expression of two transcription factor-based biosensors in V. natriegens A) Schematic overview of LysG and Lrp biosensor design for expression in V. natriegens. Different sensor variants were created using promoters from the Anderson promoter library to express lrp or lysG. The first part of brnF or lysG were added to the respective construct, because these genes are transcribed as a leaderless transcripts. After the first part of brnF/lysE, a stop codon (pin) and an additional RBS (bubble) were added (for further details, see supplementary data 2 and 3). B) Sensor response of the Lrp and LysG based biosensor variants. Strains were induced with 3 mM of the indicated inducer, or no inducer was added. C) Dose-reponse signal of the LysG based biosensor variants and L-lysine. A hill-curve was fitted to the measured values. For all fluorescence measurements, fluorescence per cell of 10,000 cells per culture were measured and averaged, and fold change over background was calculated. Average and standard deviation of three cultures are shown (n = 3). Promoters from the Anderson promoter library are shown as abbreviations (P100 = J23100, etc.).
Functionality of each sensor set was tested with each sensor inducer, by measuring the reporter output in presence and absence of inducer (Fig. 2B). The sensor variants with different promoters showed different outputs, both in the presence or absence of inducers. Interestingly, the relative maximum output did not necessarily correspond to the promoter strength measured in previous studies (Stukenberg et al., 2021; Tschirhart et al., 2019), but this could be due to the different reporter or media used. Cultivation media were shown to have an influence on promoter functioning in V. natriegens (Wu et al., 2020). Another explanation could be the lack of promoter insulation, since the Anderson promoter sequences are relatively short (35bp), and only a short RBS was added (21bp). Lack of promoter insulation and different RBSs have been shown to influence expression levels (Mutalik et al., 2013). Only for the LysG sensor induced with L-lysine, a clear difference between background and induced expression was observed, with all sensor variants (up to 5-fold for P106). However, some constructs exhibited a high background level in the absence of the inducer (P101 and P108), which would have a negative implication for their application in screening procedures. To rule out that low induction levels for the other sensor-inducer combinations were due to inhibited uptake of amino acids, V. natriegens was grown on amino acids as sole carbon and energy source. Growth was possible on L-leucine, L-histidine and L-arginine (data not shown), growth on L-histidine and L-arginine was also shown in previous studies (Baumann et al., 1971; Ellis et al., 2019), ruling out uptake problems for these amino acids. When the LysG-based biosensor was used in C. glutamicum, the fluorescence-output was higher with addition of L-arginine and L-histidine than with L-lysine (Binder et al., 2012). Thus, the reason for the low response with these biosensor-inducer combinations in V. natriegens cannot be explained. A LysG variant that is not responsive to L-lysine anymore was recently described, and could be an interesting alternative for use in V. natriegens for the specific isolation of histidine producers (Della Corte et al., 2020).
For the Lrp-based sensor, none of the added inducers resulted a measurable difference in output. It is therefore likely that correct expression or functioning of this protein was not possible in V. natriegens.
To further characterize the LysG sensor induced with L-lysine, the dose-response relationship was measured (Fig. 2C). All sensor variants could be used to detect differences in the lower millimolar ranges, the different promoters resulted in differences in the background output and dynamic range of the sensors.
3.3. Mutagenesis and FACS-based enrichment
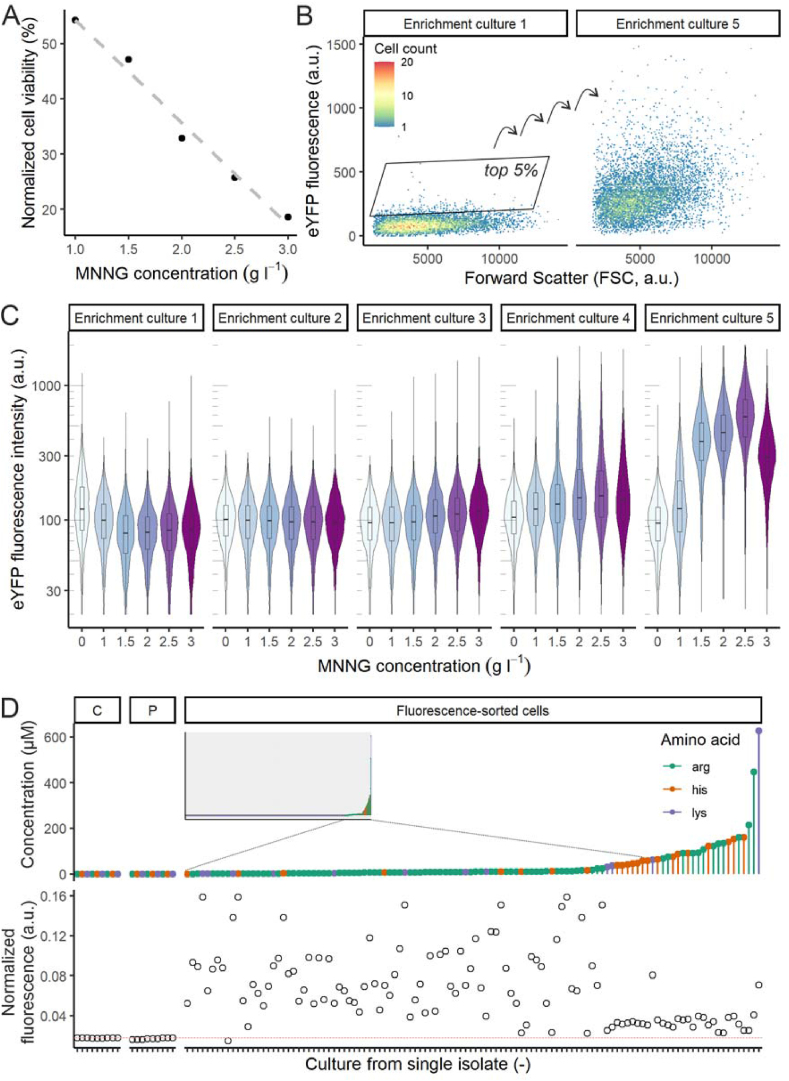
To demonstrate the application of transcription factor-based biosensors for the development of V. natriegens, the LysG-based sensor strain was used to screen for amino acid producer strains. Production of positively charged amino acids has not been shown for V. natriegens, and the WT strain does not produce measurable quantities of these components (Fig. 3). Using fluorescence-activated cell sorting (FACS), mutant cells with increased production can be selected, based on the higher fluorescence output (Binder et al., 2012; Mahr et al., 2015). The sensor harboring promoter P106 (J23106) was used for this experiment, because it has a low basal fluorescence output in combination with a good dynamic range (Fig. 2C) reducing the selection of false positive cells (i.e. fluorescent cells that are not producer cells).
Fig. 3.
Mutagenesis and FACS-based screening of V. natriegens harboring a LysG-based biosensor to isolate amino acid producing strains. A) Viability of V. natriegens cells after treatment with different MNNG concentrations. Values are normalized based on the viability of a control that was not subjected to MNNG. B) FACS scatterplots showing distribution of single cells treated with 3.0 g l−1 MNNG for the first and final enrichment culture (n = 10,000). C) Fluorescence per cell distribution per enrichment step (n = 10,000). Boxplots show median and first and third quartiles (lower and upper hinges). For the MNNG treated strains, the 300,000 cells of the top 5% fluorescent cells were sorted after each cultivation. For the non-MNNG treated control, 300,000 cells of the main population (no selection) were sorted after each cultivation. D) Overview of amino acid production and fluorescence of single isolates. C denotes control strain (non-MNNG treated, main population sorted), P denotes parental strain. Small frame shows amino acid concentrations of all fluorescence-sorted cells, selection shows strains for which more than 10 μM extracellular amino acid was measured. The concentration of each amino acid is shown separately for each culture (i.e. up to 3 amino acids concentrations for each). Fluorescence values are based on normalized bulk fluorescence measurements of the culture, red line indicates average fluoresce value of parental and control strains. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To generate genotypic diversity, a culture carrying the P106-LysG-biosensor was subjected to the mutagen MNNG (N-methyl-N′-nitro-N-nitrosoguanidine) (Adelberg et al., 1965). Because the effect of MNNG on V. natriegens has not been described yet, the cultures were subjected to different concentrations (1.0, 1.5, 2.0, 2.5 and 3.0 g l−1). The viability was calculated after MNNG treatment, which ranged from 55% to 20% (Fig. 3A). Enrichment of potential producer strains was done for each mutagenized culture, by repetitive sorting of 300,000 of the top 5% fluorescent strains followed by an overnight cultivation in optimized VN medium described in section 3.1 (Fig. 3B). Each MNNG treated culture showed an upward shift in fluorescence after three to four enrichment steps (Fig. 3C). After the fourth enrichment step, single cells were sorted on plates for the top 5% fluorescent cells, from each MNNG treated culture. Cells that formed colonies were grown in microtiter plates and the concentration of extracellular positively charged amino acids was quantified by HPLC measurement (Fig. 3D). From 245 measured strains, 37 (15%) produced more than 10 μM extracellular amino acids, and producers were isolated from cultures treated with each different MNNG concentration. Single producers of each amino acid were obtained, as well as strains that produced two or all three positively charged amino acids. In C. glutamicum, the LysG-based biosensor has been used in FACS-based screenings to isolate L-lysine and L-arginine producers, but not L-histidine producers (Binder et al., 2012; Schendzielorz et al., 2014). As a control experiment, we isolated eight clones from a gate covering the whole population; none of them produced any of the effector amino acids. As a further control, we randomly picked 89 MNNG-treated clones from plates. Among these, a total of only 3 clones produced more than 10 μM of the LysG effector amino acids (one clone produced 128 μM L-arginine and two clones produced 70 and 22 μM L-lysine, respectively). These controls confirm the functionality of the biosensor-based FACS-screening approach.
During growth of the cultures from the isolates, the sensor output was also measured as bulk culture fluorescence. The strongest producer strains did not show the highest fluorescence (Fig. 3D). This only confirms that the sensor output cannot directly provide information about the efficiency of amino acid secretion. While the TF-based biosensor detects the intracellular amino acid concentration, the extracellular concentration is measured by HPLC in the secondary screening. This means that, for example, a strain with very good secretion properties may feature a low intracellular amino acid level. This was also observed previously when a similar analysis was performed on isolated C. glutamicum L-lysine producers (Binder et al., 2012). There, the authors identified two clusters of isolates based on L-lysine production and fluorescence output: one cluster of low-fluorescent, high L-lysine producing strains, and one cluster of highly fluorescent, low-producing strains (false-positives). We observe a similar result, with a high variation in fluorescence for the non-producing isolates, and a low variation in fluorescence for the producer-isolates. This effect could be due to mutations in amino acid exporters, or due to mutations in the biosensors. Sequencing of the sensor region of two false-positive strains revealed mutations in lysG (E76K and S270F), which likely increased the fluorescence output without requiring an increase in amino acid production. In conclusion, the LysG sensor could be successfully applied in a high-throughput screening to isolate positively charged amino acid producing V. natriegens strains.
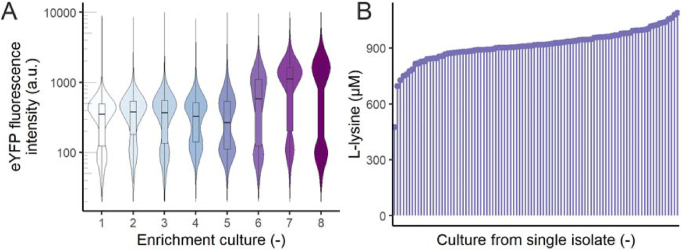
3.4. FACS-based enrichment for improved L-lysine production
Analysis of the cultures from single isolates after the FACS enrichment revealed that the strongest producers did not show the highest fluorescence output. This indicates that the production in these mutants did not lead to maximum sensor induction. Therefore, the best producer, which produced more than 600 μM L-lysine, was subjected to another series of FACS-based enrichments. No additional mutagen was applied. After five new enrichments, an increase in fluorescence was observed (Fig. 4A). During the FACS-based enrichment, a population of low-fluorescent cells was observed at each sorting step (Fig. 4A), which was not observable during the first enrichment (Fig. 3D). A possible explanation for this observation is plasmid instability, as one study reported a low plasmid maintenance for the pBR322 backbone in V. natriegens (Tschirhart et al., 2019).
Fig. 4.
FACS-based enrichment of V. natriegens L-lysine producers harboring a LysG-based biosensor. A) Fluorescence per cell distribution per enrichment step (n = 10,000). Boxplots show median and first and third quartiles (lower and upper hinges); 300,000 cells of the top 5% fluorescent cells were sorted after each cultivation. B) Extracellular L-lysine concentration of 96 cultures from sorted single isolates.
After the seventh enrichment, single cells of the top 5% fluorescent population were sorted. Cultures from 96 colonies were grown in microtiter plates and the concentration of extracellular L-lysine was quantified by HPLC measurement (Fig. 4B). Almost all cultures showed similar levels of L-lysine, in higher amounts than the parental strain (max. 1.1 mM). Therefore, it is likely that this approach resulted in the efficient enrichment of the most competitive clones under the chosen selection regime, suggesting a low genetic diversity of the population at the end of the enrichment workflow.
3.5. Identification of causal mutations in producer strains
Whole genome sequencing of the parental strain and the single top L-lysine, L-histidine and L-arginine producers after the first 5-round FACS enrichment (Fig. 3) was performed to identify causal mutations for the producer phenotype (Table 2, Suppl. Data 1). Overall, 81 single nucleotide polymorphisms (SNPs) could be identified, of which 68 (84%) were in coding regions, and 50 (62%) resulted in amino acid exchanges. Furthermore, all but one mutations were G to A (32, 40%) or C to T (48, 59%) mutations, which was expected since MNNG is known to predominantly induce G to A conversions. For the L-histidine producer, 23 SNPs were found, of which three could be related to L-histidine metabolism based on gene annotations and predicted metabolic networks. The first, a mutation in a gene encoding a serine ammonia-lyase (V395I) which catalyzes the conversion of serine to pyruvate, could result in an indirect increase of L-histidine. A lower activity of this enzyme would increase the reaction of serine to glycine. This reaction yields methyl tetrahydrofolate (mTHF) as a cofactor, which is needed to remove the toxic intermediate 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) in the L-histidine synthesis pathway (Schwentner et al., 2019). Two other mutations were found, one in a gene encoding IMP dehydrogenase (L433F) and one in a carA (P358L), encoding carbamoyl phosphate synthetase, which are both involved in nucleotide biosynthesis pathways, which are linked to L-histidine biosynthesis.
Table 2.
Relevant mutations of isolated V. natriegens amino acid producer strains. Total SNPs for each strain are indicated in brackets.
| # | Phenotype | Relevant mutations |
|---|---|---|
| H1 | L-histidine producer (23 SNPs) | serine ammonia-lyase (V395I) |
| IMP dehydrogenase (L433F) | ||
| carA, carbamoyl phosphate synthetase (P358L) | ||
| A1 | L-arginine producer (35 SNPs) | agmatine deiminase (P77L) |
| potD, ABC transporter substrate-binding protein (L15F) | ||
| acetolactate synthase large subunit (Y296C and W486*) | ||
| threonine synthase (A186V) | ||
| upstream of glutamate synthase (G to A and G to A) | ||
| L1 | L-lysine producer (23 SNPs) | dapA, 4-hydroxy-tetrahydrodipicolinate synthase (H56Y) |
| DUF1338 domain-containing protein (V92I) | ||
| L2 | L-lysine improved producer (2 SNPs) | All mutations of L1 |
| DNA topoisomerase I (S500P) |
For the L-arginine producer, 35 SNPs were identified, seven of which could be related to positively charged amino acid metabolism. Two mutations could reduce the conversion of L-arginine to other compounds, thereby increasing the L-arginine concentration. The first of these mutations was found in the gene encoding an agmatine deiminase (P77L), which catalyzes a step in an L-arginine degradation pathway. The other mutation was found in potD, encoding an ABC transporter substrate-binding protein (L15F) that transports spermidine and putrescine, which are compounds that are produced from L-arginine. A possible decrease in transport could increase their concentration, leading to an increase in L-arginine concentrations due to lower conversion requirements. Another set of mutations were all involved in metabolism of other amino acids, which are connected to L-arginine biosynthesis by biochemical pathways. Two mutations were found in a gene encoding the large subunit of acetolactate synthase (Y296C and W486*), and one mutation was found in a gene encoding threonine synthase (A186V). Both enzymes are involved in L-threonine and branched chain amino acid biosynthesis. Furthermore, two mutations were found upstream of the gene encoding glutamate synthase (two G to A mutations), which could alter the expression of this gene. The multiple mutations in genes involved in amino acids biosynthesis and degradation found in the L-arginine producers could indicate an intricate rewiring of metabolism to increase the production of L-arginine.
A total of 23 SNPs were found in the L-lysine producer isolated after the first round of FACS enrichment, of which two could be related to L-lysine metabolism. The first is a mutation in gapA (H56Y), which encodes 4-hydroxy-tetrahydrodipicolinate synthase. This is the third enzyme in the L-lysine biosynthesis pathway, which is known to be feedback-inhibited by L-lysine, and mutation could result in a lower inhibition leading to an increased L-lysine production. Another mutation was found in a gene encoding an enzyme active in a putative L-lysine degradation pathway, identified as a DUF1338 domain-containing protein (V92I). In P. putida, a homologue of this protein catalyzes the conversion of 2-oxoadipate to 2-hydroxyglutarate, in a pathway that leads to the conversion of L-lysine to 2-ketoglutarate (Thompson et al., 2019). While this pathway has not been described in V. natriegens, this mutation could result in a lower L-lysine degradation rate. Furthermore, SNPs in several ABC transporters were found. While these could influence amino acid transport, their specific function still need to be elucidated.
Finally, the genome of the improved L-lysine production strain, isolated after the second FACS enrichment, was analyzed and compared to the parental L-lysine producer. Only two additional SNPs were found, this low number is likely because no additional mutagenesis was done on this strain. The first mutation was in a gene encoding DNA topoisomerase I (S500P), the second one was a C to G mutation in the intergenic region between a hypothetical protein and phosphate ABC transporter substrate-binding protein. It is possible that the mutation in the DNA topoisomerase has pleiotropic effects that potentially affect the expression of genes involved in L-lysine biosynthesis.
4. Conclusion
V. natriegens is a promising host for next-generation bioprocesses. A recent study showed a high volumentric productivity of engineered V. natriegens for L-alanine production (0.56 ± 0.10 g L-alanine liter−1 min−1) using resting cells under anaerobic conditions (Hoffart et al., 2017). These results impressively emphasize V. natriegens as a potential production host for amino acid production.
In this work, we describe the first expression of transcription factor-based biosensors in V. natriegens, and use them in a high-throughput screening to isolate producer strains. Individual L-lysine, L-arginine and L-histidine were obtained, but all showed relatively low productivity in the high micro molar range. This is in stark contrast to the results of screenings with the same sensor in the established amino acid producer C. glutamicum. Here, FACS screenings resulted in the isolation of clones secreting up to 40 mM L-lysine (Binder et al., 2012; Schendzielorz et al., 2014), but no L-histidine producers were obtained. These clones carried mutations leading to a loss of feedback inhibition of the aspartate kinase – a key target for engineering L-lysine production in these strains. Nevertheless, the successful isolation of V. natriegens clones secreting low amounts of LysG effector amino acids demonstrates the functionality of this approach. FACS screening enables the rapid isolation of positive clones from a large library (Dietrich et al., 2010). The high rate of false-positive clones - a typical characteristic of such an approach - naturally requires the examination of the isolates with regard to their production properties by means of HPLC. Results shown in Fig. 3D indicate that the initial FACS screening provides qualitative information, but may not provide quantitative information on the overall productivity of the strain, since the sensor acts intracellularly. Here, mutation of the sensor construct itself or mutation in exporters may drastically affect the sensor output. The high-throughput character of the overall approach does, nevertheless, enable the identification of potential causal mutations, demonstrating the power of this approach. The mutated genes provide information about the V. natriegens amino acid metabolism and regulation and could be targeted in future engineering work to further improve V. natriegens as a host for amino acid production. Multiplexed re-engineering of the identified genetic variants, e.g. by using the MuGent approach (Dalia et al., 2017), could identify novel and non-intuitive beneficial mutations and provide insights in epistatic interactions.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement no 638718) and the Helmholtz Association (grant number W2/W3-096). The authors would like to thank Tino Polen for help with the analysis of sequencing data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2021.e00187.
Author contributions
Roberto Giuseppe Stella: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data Curation, Writing – Original Draft, Visualization, Supervision, Project administration. Philipp Baumann: Conceptualization, Methodology, Investigation, Writing – Review & Editing. Sophia Lorke: Investigation. Felix Münstermann: Investigation. Astrid Wirtz: Methodology. Jan Marienhagen: Conceptualization, Resources, Writing – Review & Editing, Funding acquisition. Julia Frunzke: Conceptualization, Resources, Writing – Review & Editing, Supervision, Project administration, Funding acquisition.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adelberg E.A., Mandel M., Ching Chen G.C. Optimal conditions for mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine in Escherichia coli K12. Biochem. Biophys. Res. Commun. 1965;18:788–795. doi: 10.1016/0006-291X(65)90855-7. [DOI] [Google Scholar]
- Anderson J.C. 2006. http://parts.igem.org/Promoters/Catalog/Anderson Anderson promoter library [WWW Document]. URL.
- Baumann P., Baumann L., Mandel M. Taxonomy of marine bacteria: the genus Beneckea. J. Bacteriol. 1971;107:268–294. doi: 10.1128/jb.107.1.268-294.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder S., Schendzielorz G., Stäbler N., Krumbach K., Hoffmann K., Bott M., Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R.L., Greene P.J., Betlach M.C., Heyneker H.L., Boyer H.W., Crosa J.H., Falkow S. Construction and characterization of new cloning vehicle. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- Chien C.-C., Chen C.-C., Choi M.-H., Kung S.-S., Wei Y.-H. Production of poly-β-hydroxybutyrate (PHB) by Vibrio spp. isolated from marine environment. J. Biotechnol. 2007;132:259–263. doi: 10.1016/j.jbiotec.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Chou H.H., Keasling J.D. Programming adaptive control to evolve increased metabolite production. Nat. Commun. 2013;4:1–8. doi: 10.1038/ncomms3595. [DOI] [PubMed] [Google Scholar]
- Dahl R.H., Zhang F., Alonso-Gutierrez J., Baidoo E., Batth T.S., Redding-Johanson A.M., Petzold C.J., Mukhopadhyay A., Lee T.S., Adams P.D., Keasling J.D. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- Dalia T.N., Hayes C.A., Stolyar S., Marx C.J., McKinlay J.B., Dalia A.B. Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. ACS Synth. Biol. 2017;6:1650–1655. doi: 10.1021/acssynbio.7b00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Corte D., van Beek H.L., Syberg F., Schallmey M., Tobola F., Cormann K.U., Schlicker C., Baumann P.T., Krumbach K., Sokolowsky S., Morris C.J., Grünberger A., Hofmann E., Schröder G.F., Marienhagen J. Engineering and application of a biosensor with focused ligand specificity. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-18400-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Soye B.J., Davidson S.R., Weinstock M.T., Gibson D.G., Jewett M.C. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 2018;7:2245–2255. doi: 10.1021/acssynbio.8b00252. [DOI] [PubMed] [Google Scholar]
- Dietrich J.A., McKee A.E., Keasling J.D. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu. Rev. Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- Eagon R.G. Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J. Bacteriol. 1962;83:736–737. doi: 10.1128/jb.83.4.736-737.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis B., Haaland P., Hahne F., Le Meur N., Gopalakrishnan N., Spidlen J., Jiang M., Finak G. flowCore: flowCore: basic structures for flow cytometry data. R Packag. version. 2021 2017. 2.4.0. [Google Scholar]
- Ellis G.A., Tschirhart T., Spangler J., Walper S.A., Medintz I.L., Vora G.J. Exploiting the feedstock flexibility of the emergent synthetic biology chassis Vibrio natriegens for engineered natural product production. Mar. Drugs. 2019;17:1–21. doi: 10.3390/md17120679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erian A.M., Freitag P., Gibisch M., Pflügl S. High rate 2,3-butanediol production with Vibrio natriegens. Bioresour. Technol. Reports. 2020;10 doi: 10.1016/j.biteb.2020.100408. [DOI] [Google Scholar]
- Failmezger J., Scholz S., Blombach B., Siemann-Herzberg M. Cell-free protein synthesis from fast-growing Vibrio natriegens. Front. Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Heyer A., Gätgens C., Hentschel E., Kalinowski J., Bott M., Frunzke J. The two-component system ChrSA is crucial for haem tolerance and interferes with HrrSA in haem-dependent gene regulation in Corynebacterium glutamicum. Microbiology. 2012;158:3020–3031. doi: 10.1099/mic.0.062638-0. [DOI] [PubMed] [Google Scholar]
- Hoff J., Daniel B., Stukenberg D., Thuronyi B.W., Waldminghaus T., Fritz G. Vibrio natriegens: an ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ. Microbiol. 2020;22:4394–4408. doi: 10.1111/1462-2920.15128. [DOI] [PubMed] [Google Scholar]
- Hoffart E., Grenz S., Lange J., Nitschel R., Müller F., Schwentner A., Feith A., Lenfers-Lücker M., Takors R., Blombach B. High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.01614-17. e01614–e01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.H., Ostrov N., Wong B.G., Gold M.A., Khalil A.S., Church G.M. Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nat. Microbiol. 2019;4:1105–1113. doi: 10.1038/s41564-019-0423-8. [DOI] [PubMed] [Google Scholar]
- Lentini R., Forlin M., Martini L., Del Bianco C., Spencer A.C., Torino D., Mansy S.S. Fluorescent proteins and in vitro genetic organization for cell-free synthetic biology. ACS Synth. Biol. 2013;2:482–489. doi: 10.1021/sb400003y. [DOI] [PubMed] [Google Scholar]
- Lin J.L., Wagner J.M., Alper H.S. Enabling tools for high-throughput detection of metabolites: metabolic engineering and directed evolution applications. Biotechnol. Adv. 2017;35:950–970. doi: 10.1016/j.biotechadv.2017.07.005. [DOI] [PubMed] [Google Scholar]
- Long C.P., Gonzalez J.E., Cipolla R.M., Antoniewicz M.R. Metabolism of the fast-growing bacterium Vibrio natriegens elucidated by13C metabolic flux analysis. Metab. Eng. 2017;44:191–197. doi: 10.1016/j.ymben.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr R., Frunzke J. Transcription factor-based biosensors in biotechnology: current state and future prospects. Appl. Microbiol. Biotechnol. 2016;100:79–90. doi: 10.1007/s00253-015-7090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr R., Gätgens C., Gätgens J., Polen T., Kalinowski J., Frunzke J. Biosensor-driven adaptive laboratory evolution of L-valine production in Corynebacterium glutamicum. Metab. Eng. 2015;32:184–194. doi: 10.1016/j.ymben.2015.09.017. [DOI] [PubMed] [Google Scholar]
- Mustafi N., Grünberger A., Kohlheyer D., Bott M., Frunzke J. The development and application of a single-cell biosensor for the detection of L-methionine and branched-chain amino acids. Metab. Eng. 2012;14:449–457. doi: 10.1016/j.ymben.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Mustafi N., Grünberger A., Mahr R., Helfrich S., Nöh K., Blombach B., Kohlheyer D., Frunzke J. Application of a genetically encoded biosensor for live cell imaging of L-valine production in pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum strains. PLoS One. 2014;9 doi: 10.1371/journal.pone.0085731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalik V.K., Guimaraes J.C., Cambray G., Mai Q.-A., Christoffersen M.J., Martin L., Yu A., Lam C., Rodriguez C., Bennett G. Quantitative estimation of activity and quality for collections of functional genetic elements. Nat. Methods. 2013;10:347–353. doi: 10.1038/nmeth.2403. [DOI] [PubMed] [Google Scholar]
- Payne W.J. Studies on bacterial utilization of uronic acids. III. Induction of oxidative enzymes in a marine isolate. J. Bacteriol. 1958;76:301–307. doi: 10.1128/jb.76.3.301-307.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne W.J., Eagon R.G., Williams A.K. Some observations on the physiology of Psuedomonas natriegens nov. Spec. Antonie Van Leeuwenhoek. 1961;27:121–128. doi: 10.1007/BF02538432. [DOI] [PubMed] [Google Scholar]
- Pfeifer-Sancar K., Mentz A., Rückert C., Kalinowski J. Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNAseq technique. BMC Genom. 2013;14:1–23. doi: 10.1186/1471-2164-14-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer E., Michniewski S., Gätgens C., Münch E., Müller F., Polen T., Millard A., Blombach B., Frunzke J. Generation of a prophage-free variant of the fast-growing bacterium Vibrio natriegens. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/aem.00853-19. e00853-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Found. Stat. Comput.; Vienna, Austria: 2020. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- Raman S., Rogers J.K., Taylor N.D., Church G.M. Evolution-guided optimization of biosynthetic pathways. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:17803–17808. doi: 10.1073/PNAS.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz C., Baty F., Streibig J.C., Gerhard D. Dose-response analysis using R. PLoS One. 2015;10 doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., MacCallum P., Russell D.W. Cold Spring Harbor Laboratory, Cold Spring Harbor; NY: 2001. Molecular Cloning: a Laboratory Manual. [Google Scholar]
- Schendzielorz G., Dippong M., Grünberger A., Kohlheyer D., Yoshida A., Binder S., Nishiyama C., Nishiyama M., Bott M., Eggeling L. Taking control over control: use of product sensing in single cells to remove flux control at key enzymes in biosynthesis pathways. ACS Synth. Biol. 2014;3:21–29. doi: 10.1021/sb400059y. [DOI] [PubMed] [Google Scholar]
- Schwentner A., Feith A., Münch E., Stiefelmaier J., Lauer I., Favilli L., Massner C., Öhrlein J., Grund B., Hüser A., Takors R., Blombach B. Modular systems metabolic engineering enables balancing of relevant pathways for l-histidine production with Corynebacterium glutamicum. Biotechnol. Biofuels. 2019;12:1–21. doi: 10.1186/s13068-019-1410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg D., Hensel T., Hoff J., Daniel B., Inckemann R., Tedeschi J.N., Nousch F., Fritz G. The marburg collection: a golden gate DNA assembly framework for synthetic biology applications in Vibrio natriegens. bioRxiv. 2021;26:437105. doi: 10.1021/acssynbio.1c00126. 2021.03. [DOI] [PubMed] [Google Scholar]
- Thiele I., Gutschmann B., Aulich L., Girard M., Neubauer P., Riedel S.L. High-cell-density fed-batch cultivations of Vibrio natriegens. Biotechnol. Lett. 2021 doi: 10.1007/s10529-021-03147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Blombach B. Metabolic engineering of Vibrio natriegens. Essays Biochem. 2021;14048:1–12. doi: 10.1042/ebc20200135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.G., Blake-Hedges J.M., Cruz-Morales P., Barajas J.F., Curran S.C., Eiben C.B., Harris N.C., Benites V.T., Gin J.W., Sharpless W.A., Twigg F.F., Skyrud W., Krishna R.N., Pereira J.H., Baidoo E.E.K., Petzold C.J., Adams P.D., Arkin A.P., Deutschbauer A.M., Keasling J.D. Massively parallel fitness profiling reveals multiple novel enzymes in Pseudomonas putida lysine metabolism. mBio. 2019;10 doi: 10.1128/mBio.02577-18. e02577-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschirhart T., Shukla V., Kelly E.E., Schultzhaus Z., Newringeisen E., Erickson J.S., Wang Z., Garcia W., Curl E., Egbert R.G., Yeung E., Vora G.J. Synthetic biology tools for the fast-growing marine bacterium Vibrio natriegens. ACS Synth. Biol. 2019;8:2069–2079. doi: 10.1021/acssynbio.9b00176. [DOI] [PubMed] [Google Scholar]
- Wang Z., Tschirhart T., Schultzhaus Z., Kelly E.E., Chen A., Oh E., Nag O., Glaser E.R., Kim E., Lloyd P.F., Charles P.T., Li W., Leary D., Compton J., Phillips D.A., Dhinojwala A., Payne G.F., Vora G.J. Melanin produced by the fast-growing marine bacterium Vibrio natriegens through heterologous biosynthesis: characterization and application. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/AEM.02749-19. e02749-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock M.T., Hesek E.D., Wilson C.M., Gibson D.G. Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods. 2016;13:849–851. doi: 10.1038/nmeth.3970. [DOI] [PubMed] [Google Scholar]
- Wickham H., Averick M., Bryan J., Chang W., McGowan L.D., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the tidyverse. J. Open Source Softw. 2019;4:1686. [Google Scholar]
- Wiegand D.J., Lee H.H., Ostrov N., Church G.M. Establishing a cell-free Vibrio natriegens expression system. ACS Synth. Biol. 2018;7:2475–2479. doi: 10.1021/acssynbio.8b00222. [DOI] [PubMed] [Google Scholar]
- Wu F., Chen W., Peng Y., Tu R., Lin Y., Xing J., Wang Q. Design and reconstruction of regulatory parts for fast-frowing Vibrio natriegens synthetic biology. ACS Synth. Biol. 2020;9:2399–2409. doi: 10.1021/acssynbio.0c00158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.