Abstract
RAD24 and RFC5 are required for DNA damage checkpoint control in the budding yeast Saccharomyces cerevisiae. Rad24 is structurally related to replication factor C (RFC) subunits and associates with RFC subunits Rfc2, Rfc3, Rfc4, and Rfc5. rad24Δ mutants are defective in all the G1-, S-, and G2/M-phase DNA damage checkpoints, whereas the rfc5-1 mutant is impaired only in the S-phase DNA damage checkpoint. Both the RFC subunits and Rad24 contain a consensus sequence for nucleoside triphosphate (NTP) binding. To determine whether the NTP-binding motif is important for Rad24 function, we mutated the conserved lysine115 residue in this motif. The rad24-K115E mutation, which changes lysine to glutamate, confers a complete loss-of-function phenotype, while the rad24-K115R mutation, which changes lysine to arginine, shows no apparent phenotype. Although neither rfc5-1 nor rad24-K115R single mutants are defective in the G1- and G2/M-phase DNA damage checkpoints, rfc5-1 rad24-K115R double mutants become defective in these checkpoints. Coimmunoprecipitation experiments revealed that Rad24K115R fails to interact with the RFC proteins in rfc5-1 mutants. Together, these results indicate that RFC5, like RAD24, functions in all the G1-, S- and G2/M-phase DNA damage checkpoints and suggest that the interaction of Rad24 with the RFC proteins is essential for DNA damage checkpoint control.
Eukaryotic cells employ a set of surveillance mechanisms to coordinate cell cycle events by permitting the onset of one event only after the completion of the preceding event. The mechanisms that ensure the proper ordering of cell cycle events have been termed checkpoint controls (10). DNA damage triggers the activation of checkpoint pathways that arrest the cell cycle and induce the transcription of genes that facilitate repair. Other checkpoints are activated when DNA replication is blocked. Failure to respond properly to DNA alterations may result in genomic instability, a mutagenic condition that predisposes organisms to cancer (5, 24).
The cell cycle is transiently arrested at different stages depending on the phase at which DNA damage occurs. Three responses have been characterized in the budding yeast Saccharomyces cerevisiae, known as the G1-, S- and G2/M-phase DNA damage checkpoints (16). Genetic studies have identified genes that are involved in all three checkpoints. These include RAD9, RAD17, RAD24, MEC3, DDC1, MEC1(ESR1), and RAD53 (SPK1 or MEC2) (1, 17, 18, 22, 23, 30–33, 43–45). Several lines of genetic evidence have suggested that RAD17, RAD24, MEC3, and DDC1 operate in the same checkpoint pathway, while RAD9 functions separately (17, 18, 20). Indeed, Ddc1, Mec3, and Rad17 physically interact with each other, suggesting that they function as a complex (13). RAD53 encodes a dual-specificity protein kinase (35), and Mec1 belongs to the ATM protein family (12, 28). Rad53 is phosphorylated in response to DNA damage in a MEC1-dependent manner (26, 39). DNA damage-induced Rad53 phosphorylation is also dependent on RAD9, RAD17, RAD24, MEC3, and DDC1 (21, 29, 39, 41).
Replication factor C (RFC) is required for DNA replication and repair and consists of one large and four small subunits. In S. cerevisiae, the large subunit of RFC is encoded by RFC1(CDC44), and the four small subunits are encoded by RFC2, RFC3, RFC4, and RFC5 (4). RFC is a structure-specific DNA-binding protein complex that recognizes the primer-template junction. RFC loads PCNA onto the primer terminus, and then DNA polymerases δ and ɛ bind to the DNA-RFC-PCNA complex to constitute a processive replication complex (2, 15, 42). We have demonstrated that rfc5-1 mutants are defective in the S-phase DNA damage and DNA replication block checkpoints but not in the G2/M-phase DNA damage checkpoint (36, 38). RAD24 encodes a protein structurally related to the RFC subunits (8, 19) and has an essential role in the G1-, S- and G2/M-phase DNA damage checkpoints (23, 31, 45). We isolated RAD24 in a screen for dosage-dependent suppressors of rfc5-1 and have shown that Rad24 interacts physically with Rfc2 and Rfc5 (29). Consistent with its role in DNA damage checkpoints, RAD24 overexpression suppresses the sensitivity to DNA-damaging agents and the defect in DNA damage-induced Rad53 phosphorylation in rfc5-1 mutants. Thus, the RFC proteins and Rad24 appear to form a complex that functions in the DNA damage checkpoint pathway. However, it was not known whether this complex is required for the DNA damage checkpoint only in the S phase or throughout the cell cycle.
Rad24, like the RFC subunits, contains a nucleoside triphosphate (NTP)-binding motif. In order to test if this motif is involved in Rad24 function, we created the substitution mutations rad24-K115E and rad24-K115R at the conserved lysine residue in the NTP-binding motif. From studies of cells carrying the rad24-K115R and/or rfc5-1 mutation, we show that RFC5, like RAD24, has a role in the DNA damage checkpoints not only in the S phase but also in the G1 and G2/M phases. We also show that Rad24 interacts physically with Rfc3 and Rfc4 and that in rfc5-1 mutants the Rad24K115R protein fails to associate with the RFC proteins. Our results suggest that the interaction of Rad24 with the RFC proteins is essential for DNA damage checkpoint control throughout the cell cycle.
MATERIALS AND METHODS
Strains, media, and general methods.
The yeast strains used in this study are isogenic and are listed in Table 1. Standard genetic techniques were used for manipulating yeast strains (9, 11). Synthetic complete (SC) medium containing 0.5% casamino acids and the appropriate supplements was used to maintain selection of URA3 plasmids.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype |
|---|---|
| KSC006 | MATa ade1 his2 trp1 ura3 leu2 |
| KSC835 | MATa rfc5-1::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC980 | MATa rad24Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1105 | MATa rfc5-1::LEU2 rad24Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1133 | MATa RFC1-HA::TRP1 rad24Δ::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1151 | MATa rad24-K115E::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1152 | MATa rad24-K115R::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1160 | MATa rfc5-1::LEU2 rad24-K115E::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1161 | MATa rfc5-1::LEU2 rad24-K115R::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1163 | MATa RFC3-HA::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1164 | MATa RFC4-HA::LEU2 ade1 his2 trp1 ura3 leu2 |
| KSC1168 | MATa RFC3-HA::LEU2 rad24Δ::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1170 | MATa RFC3-HA::LEU2 rfc5-1::LEU2 rad24Δ::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1172 | MATa RFC4-HA::LEU2 rad24Δ::TRP1 ade1 his2 trp1 ura3 leu2 |
| KSC1174 | MATa RFC4-HA::LEU2 rfc5-1::LEU2 rad24Δ::TRP1 ade1 his2 trp1 ura3 leu2 |
Plasmids and gene replacement.
The BamHI-HindIII fragment from YCpRAD24 (29) and an NdeI-BamHI fragment of the 5′ noncoding sequence of RAD24 were cloned into NdeI-HindIII-treated YIplac204 (6), resulting in YIpT-RAD24. To construct YIpT-RAD24-K115E and YIpT-RAD24-K115R, the 110-bp BamHI-BstBI fragment of YIpT-RAD24 was replaced by sets of complementary oligonucleotides KE-1 (5′-GATCCTACTACTGTCTGGCCCCAGTGGATGCTCTGAAAGTACGGTCATAA-3′), KE-2 (5′-GAGAGTTCTTTT ATGACCGTACTTTCAGAGCATCCACTGGGGCCAGACAGTAGTAG- 3′), ER-1 (5′-AAGAACTCTCAAAAATCTTAGTTCCTAAATACAGACAAAACAGCAACGGAACGTCCTTT-3′), and ER-2 (5′-CGAAAGGACGTTCCGTTGCCTGTTTTGTCTGTATTTAGGAACTAAGATTTTT-3′) or by KR-1 (5′-GATCCTACTACTGTCTGGCCCCAGTGGATGCTCTAGAAGTACGG TCATAA-3′), KR-2 (5′-GAGAGTTCTTTTATGACCGTACTTCTAGAGCATCCACTGGGGCCAGACAGTAGTAG-3′), ER-1, and ER-2, respectively. The 1.1-kb EcoRI-SacI fragment from YIpT-RAD24-K115R was cloned into YCpRAD24-myc (29), generating YCpRAD24-K115R-myc. The substitution at each site was confirmed by sequence analysis. To obtain rad24Δ::ura3 cells, rad24Δ::LEU2 cells were transformed with XhoI-digested pLU12 (3) and selected for Ura+ Leu−, and the resulting rad24Δ::leu2::URA3 cells were plated on medium containing 5-fluoroorotic acid to counterselect against the Ura+ marker as described before (9). To construct site-specific rad24 mutations marked with TRP1, the plasmids YIpT-RAD24-K115E and YIpT-RAD24-K115R were cleaved with ClaI, and the resulting DNA fragments were transformed into rad24Δ::ura3 cells. Correct integration of each mutant gene at the RAD24 locus was confirmed by PCR. rad24Δ cells carrying YCpRAD24-K115R-myc showed no apparent phenotype as observed for rad24-K115R::TRP1 cells with regard to sensitivity to DNA-damaging agents, such as methyl methanesulfonate (MMS) and UV light.
To construct tagged versions of RFC1, RFC3, and RFC4, sequences encoding hemagglutinin (HA) epitope tags were inserted in front of the stop codon. To construct the RFC1-HA integration plasmid YIpT-RFC1-HA, a BglII-SalI fragment from the RFC1-HA gene was subcloned into pRS304 (34). To construct the RFC3-HA and RFC4-HA integration plasmids, an MscI-SphI fragment from the RFC3-HA gene and an NcoI-XhoI fragment from the RFC4-HA gene were subcloned into YIplac128 (6), generating YIpL-RFC3-HA and YIpL-RFC4-HA, respectively. YIpT-RFC1-HA, YIpL-RFC3-HA, and YIpL-RFC4-HA were treated with SphI, KpnI, and EcoRI, respectively, and transformed into cells. The precise integration, which destroys the endogenous RFC1, RFC3, or RFC4 gene, was confirmed by PCR. These integrations did not affect the growth or DNA damage sensitivity of wild-type or rfc5-1 mutant cells. YCp-RAD53-HA was described previously (36).
UV radiation and MMS sensitivities.
The UV radiation sensitivity assay was performed as described previously (37). Cells grown at 30°C were plated on YEPD and then irradiated by UV at 254 nm. After 2 to 3 days of incubation at 30°C, the number of colonies was counted. MMS sensitivity was determined as described (37). Cells were incubated with MMS at 30°C for 30 min. Incubation was terminated by addition of sodium thiosulfate to a final concentration of 5%. Aliquots were plated on YEPD, and the number of colonies was counted after incubation at 30°C for 2 to 3 days.
UV and MMS synchrony experiments.
To analyze cell cycle delay at the G2/M transition, log-phase cultures at 30°C were prearrested with 6 μg of α-factor per ml for 120 min, washed with water, and then released into YEPD containing nocodazole (15 μg/ml) for 120 min to synchronize cells in G2/M. Cells arrested in G2/M were spread on YEPD plates and irradiated with a 254-nm UV lamp at 75 J/m2. Cells were then washed to remove nocodazole and released into fresh YEPD containing 1% dimethyl sulfoxide at 30°C. At timed intervals, cells were withdrawn and stained with 4′,6-diamidino-2-phenylindole (DAPI) for microscopic examination. An MMS synchrony experiment to monitor S-phase regulation was carried out as described elsewhere (36). To analyze cell cycle delay at the G1/S transition, log-phase cultures in YEPD were treated with α-factor (6 μg/ml) for 120 min to synchronize cells in G1. Cells arrested in G1 were spread on YEPD plates and irradiated with a 254-nm UV lamp at 75 J/m2. Cells were then washed to remove α-factor and released into fresh YEPD at 30°C. Cells were withdrawn at different times and subjected to examination as described (32).
Antibody and immunoblotting.
Yeast cells were grown in synthetic complete medium selectable for URA3 plasmids. Cells were then diluted in YEPD and allowed to grow for 3 h. For cell cycle arrest, cells were treated with nocodazole (15 μg/ml) or α-factor (6 μg/ml) for 120 min and then irradiated with a 254-nm UV lamp at 150 or 200 J/m2, respectively. Cells were released into fresh YEPD containing nocodazole or α-factor and incubated for 60 min. Protein extracts for immunoblotting were prepared and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described (36). Proteins were transferred to nylon membranes, subjected to immunoblot analysis with the anti-Myc (9E10) or anti-HA (3F10 or 16B12) monoclonal antibody or anti-Rfc2 (a gift from A. Sugino) or anti-Rfc5 antibody and detected by the ECL kit (Amersham). Antibody against Rfc5 was raised by immunizing a rabbit with synthetic peptides corresponding to amino acids 22 to 42 and 119 to 134 of Rfc5 and purified with affinity chromatography. Among the RFC subunits of budding yeast, the amino acid sequences within these regions are specific to Rfc5. This antibody specifically recognized Rfc5 in immunoblots, and the signal was significantly increased when RFC5 was overexpressed (data not shown).
Immunoprecipitation.
Yeast cells were grown in SC medium appropriate to select for URA3 plasmids. Cells were then diluted in YEPD and allowed to grow for 3 h at 30°C. Cells were next harvested, washed, and resuspended in lysis buffer (36). An equal volume of glass beads was added, and the cells were lysed by vortexing. Extracts were clarified by 15 min of centrifugation at 4°C. The supernatant was diluted with lysis buffer and incubated at 4°C for 2 h with protein A-Sepharose beads bound with anti-HA (3F10) or anti-Rfc2 antibody. Protein concentrations were determined by the Bio-Rad protein assay (Bio-Rad). Immunoprecipitates were washed with lysis buffer and subsequently with a wash buffer and boiled immediately in 1× SDS-PAGE sample buffer (36). The proteins were detected after immunoblotting with antibody as described above.
RESULTS
DNA damage sensitivity of cells carrying mutations in the NTP-binding motif of Rad24.
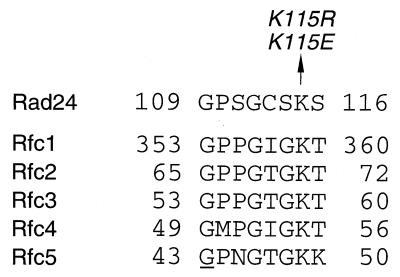
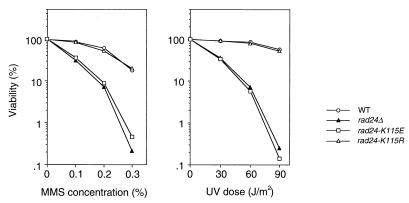
RAD24 encodes a protein structurally related to subunits of the RFC complex (8, 19). One feature of this homology is that both Rad24 and the RFC subunits contain a sequence motif characteristic of NTP-binding and -hydrolyzing proteins. The NTP-binding motif in Rad24, GXXGXXKS, deviates slightly from the classical motif, GXXGXGK(S/T) (14) (Fig. 1). The conserved lysine residue in the NTP-binding motif is involved in electrostatic interactions with the triphosphate tail of NTP, and mutation of this residue reduces NTP-binding and hydrolysis (27). To test whether this motif has a role in Rad24 function, the conserved lysine115 was changed into either glutamate or arginine, creating the rad24-K115E and rad24-K115R mutations, respectively (Fig. 1). Since the rad24Δ mutation confers sensitivity to DNA damage, we measured the sensitivity of rad24-K115E and rad24-K115R mutants to MMS treatment and UV irradiation (Fig. 2). rad24-K115E mutants showed DNA damage sensitivity very similar to that of rad24Δ mutants, while rad24-K115R mutant cells were as resistant to DNA damage as wild-type cells.
FIG. 1.
Mutations of Rad24 at the conserved lysine of the NTP-binding motif. The NTP-binding domain of Rad24 is aligned with those of all RFC subunits from S. cerevisiae. The amino acid converted by site-specific mutagenesis in the RAD24 gene is shown by an arrow with the mutation names. The amino acid underlined is the site in the rfc5-1 mutation which changes Gly to Glu at amino acid position 43.
FIG. 2.
DNA damage sensitivity in rad24 mutants. Wild-type (WT) (KSC006), rad24Δ (KSC980), rad24-K115E (KSC1151), and rad24-K115R (KSC1152) cells were grown to log phase at 30°C and treated with MMS or irradiated with UV light. Viability of cells was estimated as described in Materials and Methods.
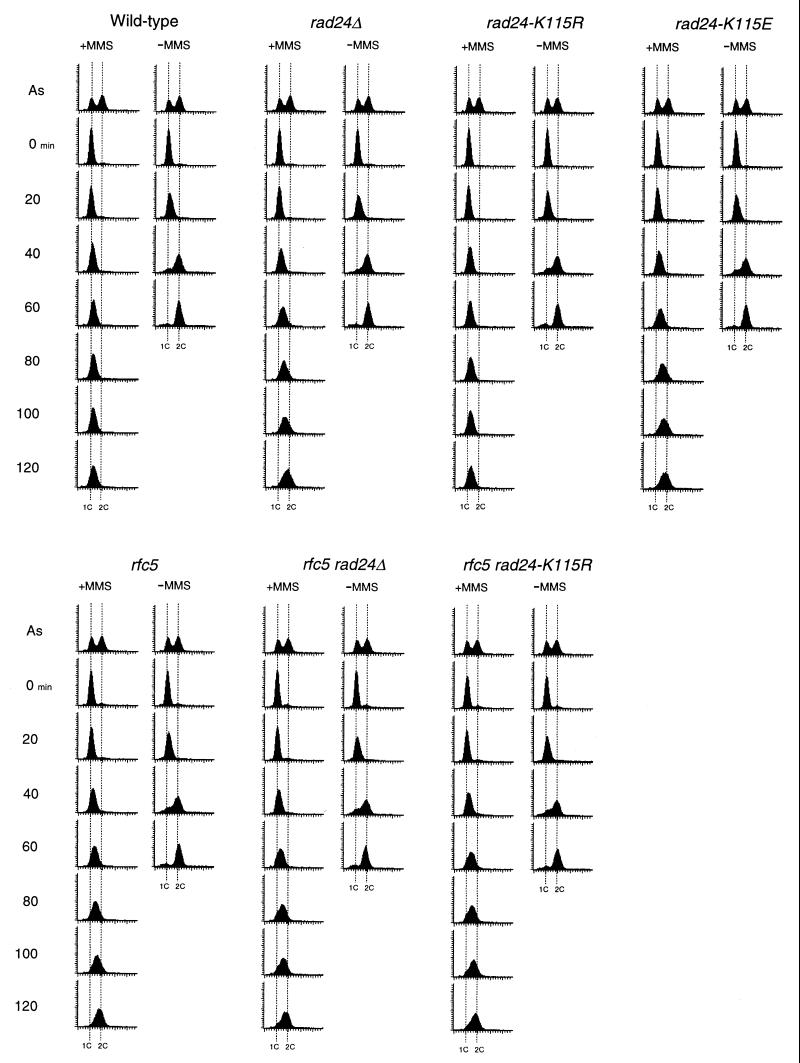
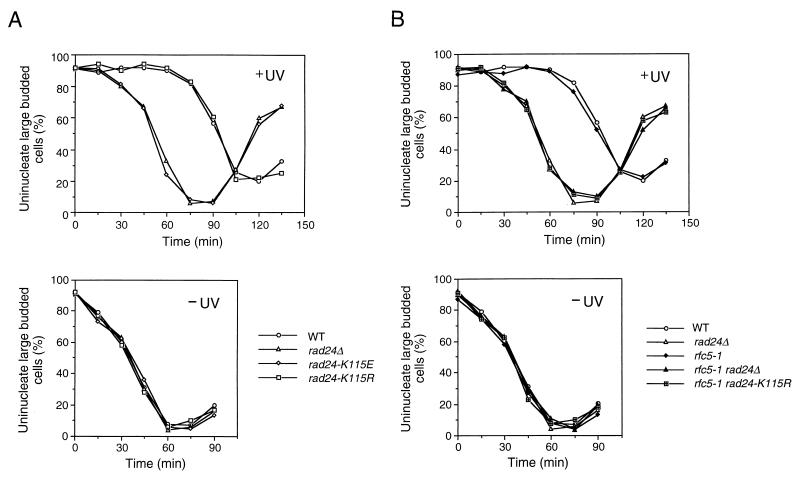
To further investigate the properties of these rad24 mutations, we evaluated DNA damage checkpoints in the corresponding mutant cells. It has been shown that RAD24 is required for the G1-, S- and G2/M-phase DNA damage checkpoints (23, 31, 45). We first examined the S-phase checkpoint by monitoring the DNA content of cells experiencing DNA damage after release from a G1 block (Fig. 3). When released from α-factor arrest and exposed to MMS, wild-type cells exhibited lower rates of DNA synthesis. In contrast, rad24Δ mutants showed some delay but progressed through the S phase faster than wild-type cells. The partial defect of rad24Δ mutants in the S-phase DNA damage checkpoint was reported previously (23). Under the same conditions, rad24-K115R cells proceeded through the S phase as slowly as wild-type cells, whereas rad24-K115E mutant cells completed the S phase as fast as rad24Δ cells. We next examined the G2/M-phase DNA damage checkpoint by monitoring mitotic division following UV irradiation (Fig. 4A). When cell cultures were released from nocodazole arrest after UV irradiation, rad24-K115R cells delayed nuclear division similar to wild-type cells, while rad24Δ and rad24-K115E cells went through mitosis much faster than wild-type cells. We further analyzed the G1-phase DNA damage checkpoint in the rad24 mutants by monitoring the appearance of budded cells after release from α-factor arrest (Fig. 5A). When cell cultures were released from α-factor arrest after UV irradiation, rad24-K115R cells were delayed in bud emergence, similar to the wild-type cells. This delay at the G1/S progression was equally reduced in rad24Δ and rad24-K115E cells. Thus, rad24-K115E appears to be a complete loss-of-function mutation, whereas rad24-K115R appears to be functionally equivalent to the wild-type gene. However, we show below that the rad24-K115R mutation confers a DNA damage checkpoint defect when combined with the rfc5-1 mutation (see below). These results suggest that the NTP-binding motif is important for Rad24 function.
FIG. 3.
S-phase DNA damage checkpoint in rad24 and rfc5-1 rad24 mutants. Wild-type (KSC006), rad24Δ (KSC980), rad24-K115E (KSC1151), rad24-K115R (KSC1152), rfc5-1 (KSC835), rfc5-1 rad24Δ (KSC1105), and rfc5-1 rad24-K115R (KSC1161) cells were synchronized with α-factor in G1 and released in either the presence (+) or the absence (−) of 0.05% MMS at 30°C as described in Materials and Methods. Aliquots of cells were collected at the indicated times after release from α-factor treatment and examined for DNA content by flow cytometry. Dotted lines indicate the DNA content of 1C and 2C cells. The top panels represent asynchronous (As) cells not treated with MMS at 30°C and are included as a reference.
FIG. 4.
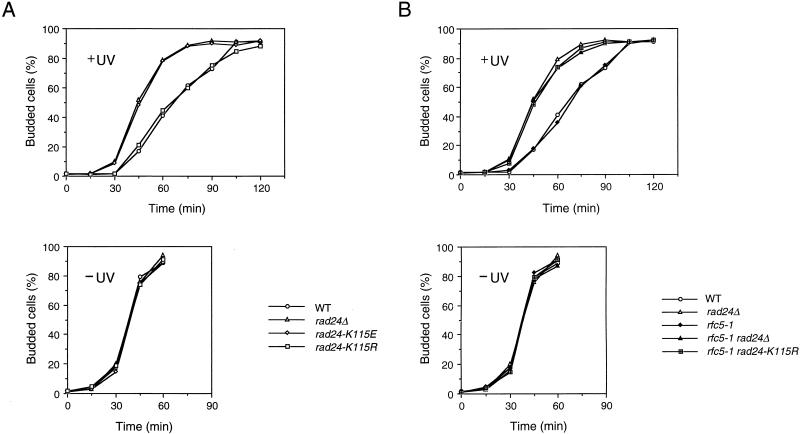
G2/M-phase DNA damage checkpoint in rad24 and rfc5-1 rad24 mutants. Cells were grown at 30°C, arrested with nocodazole, and irradiated or not irradiated with UV light. At the indicated times after release of UV-irradiated (+UV) and unirradiated (−UV) cultures from nocodazole, the percentage of uninucleate large budded cells was scored by DAPI staining. (A) Wild-type (WT) (KSC006), rad24Δ (KSC980), rad24-K115E (KSC1151), and rad24-K115R (KSC1152) cells; (B) wild-type (KSC006), rfc5-1 (KSC835), rad24Δ (KSC980), rfc5-1 rad24Δ (KSC1105), and rfc5-1 rad24-K115R (KSC1161) cells.
FIG. 5.
G1-phase DNA damage checkpoint in rad24 and rfc5-1 rad24 mutants. Cells were grown at 30°C, arrested with α-factor, and irradiated or not irradiated with UV light. The percentage of budded cells was scored at the indicated times after release of UV-irradiated (+UV) and unirradiated (−UV) cultures from α-factor. (A) Wild-type (WT) (KSC006), rad24Δ (KSC980), rad24-K115E (KSC1151), and rad24-K115R (KSC1152) cells; (B) wild-type (KSC006), rfc5-1 (KSC835), rad24Δ (KSC980), rfc5-1 rad24Δ (KSC1105), and rfc5-1 rad24-K115R (KSC1161) cells.
DNA damage checkpoints in rfc5-1 rad24-K115R double mutants.
We have shown that Rad24 interacts physically with the RFC subunit Rfc5 (29). If these proteins function as a complex, its complex activity should be dependent on the properties of the proteins and abolished by loss of either protein. Consistently, rfc5-1 rad24Δ double mutants were defective in DNA damage checkpoints, similar to rad24Δ single mutants (Fig. 3, 4B, and 5B). Since the rad24-K115R mutation confers no apparent phenotype, we examined the genetic interaction between the rad24-K115R and rfc5-1 mutations in the DNA damage checkpoints. We first evaluated the S-phase DNA damage checkpoint of the rfc5-1 and rfc5-1 rad24-K115R double mutants. As observed previously (36), rfc5-1 single mutant cells progressed through S phase faster than wild-type cells in the presence of MMS. The defect of rfc5-1 single mutants in the S-phase checkpoint was similar to that of rad24Δ single and rfc5-1 rad24-K115R double mutants (Fig. 3). We next analyzed the G2/M-phase DNA damage checkpoint in rfc5-1 and rfc5-1 rad24-K115R double mutants. Although neither rfc5-1 nor rad24-K115R single mutants were defective in the G2/M-phase DNA damage checkpoint, rfc5-1 rad24-K115R double mutants became defective in this checkpoint; these double mutants failed to delay mitosis, similar to rad24Δ mutants (Fig. 4B). We further examined the G1-phase DNA damage checkpoint in rfc5-1 and rfc5-1 rad24 double mutants. Although neither rfc5-1 nor rad24-K115R cells were defective, rfc5-1 rad24-K115R double mutants were as defective as rad24Δ mutants in the G1-phase DNA damage checkpoint (Fig. 5B). These results show that RFC5 is involved in DNA damage checkpoint control not only in the S phase but also in the G1 and G2/M phases.
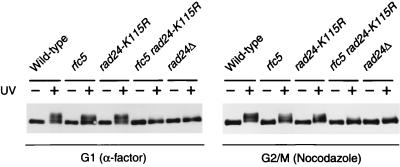
Rad53 phosphorylation in α-factor- or nocodazole-arrested rfc5-1 rad24-K115R double mutants.
Rad53 is required for DNA damage checkpoint control and is hyperphosphorylated in response to DNA damage. Consistent with the role of Rad24 in the DNA damage checkpoints, RAD24 is required for DNA damage-induced Rad53 phosphorylation (29, 41). Since rfc5-1 rad24-K115R double mutants become as defective as rad24Δ cells in the G1- and G2/M-phase DNA damage checkpoints, we expected that rfc5-1 rad24-K115R cells in the G1 or G2/M phase would be defective in DNA damage-induced Rad53 phosphorylation. To test this hypothesis, Rad53 phosphorylation following UV irradiation was examined by immunoblot analysis in cells arrested in the G1 phase with α-factor or in the G2/M phase with nocodazole (Fig. 6). Under these conditions, Rad53 hyperphosphorylation occurred in wild-type, rfc5-1, and rad24-K115R single mutant cells. However, Rad53 phosphorylation in rfc5-1 rad24-K115R double mutants was significantly reduced, similar to rad24Δ mutants in the G1 and G2/M phases. These results are consistent with the finding that rfc5-1 rad24-K115R double mutants are defective in the G1- and G2/M-phase DNA damage checkpoints and further support the idea that RFC5 functions in the G1- and G2/M-phase checkpoints.
FIG. 6.
DNA damage-induced Rad53 modification in G1- and G2/M-arrested rfc5-1 rad24-K115R mutants. Wild-type (KSC006), rfc5-1 (KSC835), rad24-K115R (KSC1152), rfc5-1 rad24-K115R (KSC1161), and rad24Δ (KSC980) cells carrying YCpRAD53-HA were grown at 30°C, arrested in G1 with α-factor or in G2/M with nocodazole, and unirradiated (−) or irradiated with UV light (+). Cells were then incubated at 30°C, maintaining arrest in medium containing α-factor or nocodazole for 60 min, and subjected to immunoblotting analysis as described in Materials and Methods.
Effect of the rfc5-1 mutation on the interaction between Rad24K115R and the RFC proteins.
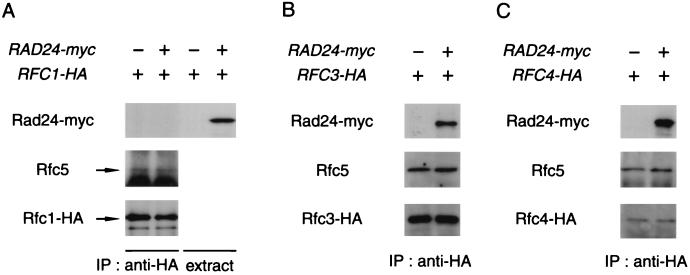
We have shown previously that Rad24 associates with Rfc2 and Rfc5 (29). Next, we examined whether Rad24 interacts physically with the other RFC proteins Rfc1, Rfc3, and Rfc4. To detect these RFC proteins, we constructed cells containing HA-tagged versions of the RFC1, RFC3, and RFC4 genes. A low-copy-number plasmid carrying RAD24-myc (YCpRad24-myc) or vector alone was transformed into cells containing the HA-tagged RFC1 (RFC1-HA), RFC3 (RFC3-HA), or RFC4 (RFC4-HA) gene. Cell extracts were prepared and subjected to immunoprecipitation using anti-HA antibody. The immunocomplexes were analyzed by immunoblotting with anti-Rfc5 and anti-Myc antibodies. Immunoaffinity-purified anti-Rfc5 antibody recognized Rfc5 in immunocomplexes from RFC1-HA cells, but anti-Myc antibody failed to detect Rad24-myc (Fig. 7A). In contrast, Rad24-myc was detected in immunocomplexes from cells expressing Rad24-myc together with Rfc3-HA or Rfc4-HA (Fig. 7B and 7C). These and our previous observations show that Rad24 interacts physically with Rfc2, Rfc3, Rfc4, and Rfc5 but not with Rfc1. During preparation of the manuscript, Green et al. (7) presented similar results.
FIG. 7.
Physical interaction of Rad24 with RFC proteins. Cells containing RFC1-HA (KSC1133) (A), RFC3-HA (KSC1163) (B), or RFC4-HA (KSC1164) (C) were transformed with YCpRAD24-myc or empty vector. Extracts prepared from the transformants were subjected to immunoprecipitation (IP) with anti-HA antibody. The immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-Myc, anti-Rfc5, or anti-HA antibody. Whole extracts were immunoblotted with anti-Myc antibody.
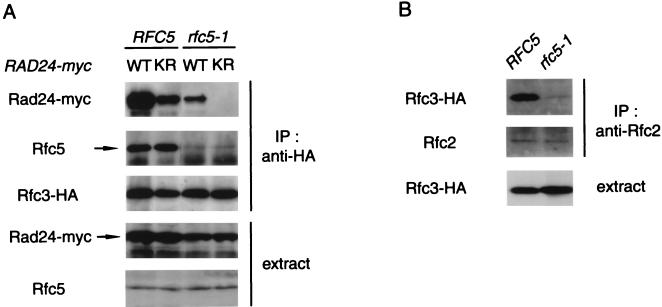
To understand the phenotype of rfc5-1 rad24-K115R double mutants, we next examined the interaction between the Rad24K115R and RFC proteins in wild-type and rfc5-1 cells. RFC5 RFC3-HA rad24Δ and rfc5-1 RFC3-HA rad24Δ cells were transformed with YCpRAD24-myc or YCpRAD24-K115R-myc, and extracts prepared from the cells were examined by immunoblotting analysis with anti-Myc and anti-Rfc5 antibodies. The expression levels of the Rfc5 and Rad24 proteins were not significantly altered in either RFC5 or rfc5-1 cells (Fig. 8A). Cell extracts were also subjected to immunoprecipitation with anti-HA antibody followed by immunoblotting analysis with anti-Myc and anti-Rfc5 antibodies to detect coprecipitation of the Rad24-myc and Rfc5 proteins (Fig. 8A). In RFC5 cells, the interaction of Rfc3-HA with Rad24K115R-myc was slightly decreased compared to its interaction with Rad24-myc. In rfc5-1 mutants, coprecipitation of Rad24-myc with Rfc3-HA was reduced compared to wild-type cells and strikingly, coprecipitation of Rad24K115R-myc was undetectable. In rfc5-1 mutants, the interaction between Rfc3-HA and Rfc5 was also decreased, suggesting that interactions among the other RFC proteins may also be affected. To address this possibility, we examined the interaction between Rfc2 and Rfc3 in rfc5-1 mutants. Cell extracts were prepared from RFC5 RFC3-HA and rfc5-1 RFC3-HA cells and subjected to immunoprecipitation with anti-Rfc2 antibody. The immunoprecipitates were then analyzed by immunoblotting analysis with anti-Rfc2 and anti-HA antibodies. The interaction of Rfc2 with Rfc3-HA was decreased in rfc5-1 mutants compared to wild-type cells, although the expression level of Rfc3-HA was not altered (Fig. 8B). Furthermore, in wild-type cells, the interaction of Rfc4-HA with Rad24K115R-myc was reduced compared to its interaction with Rad24-myc, while in rfc5-1 mutants no interaction of Rfc4-HA with Rad24K115R-myc was detected (data not shown). These results indicate that the rfc5-1 mutation causes a defect in the interaction between Rad24K115R and the RFC proteins. Together with the genetic observations provided above, these results suggest that the interaction of Rad24 with the RFC proteins is essential for DNA damage checkpoint control throughout the cell cycle.
FIG. 8.
Effect of the rfc5-1 mutation on the interaction of Rfc3 with Rad24K115R and Rfc2. (A) Interaction of Rad24K115R with Rfc3 in RFC5 and rfc5-1 mutant cells. RFC5 RFC3-HA rad24Δ (KSC1168) and rfc5-1 RFC3-HA rad24Δ (KSC1170) cells were transformed with YCpRAD24-myc (WT) or YCpRAD24-K115R-myc (KR). Extracts prepared from the transformants were subjected to immunoprecipitation (IP) with anti-HA antibody. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with anti-Myc, anti-Rfc5 and anti-Myc, anti-Rfc5, or anti-HA antibody. (B) Interaction of Rfc3 with Rfc2 in RFC5 and rfc5-1 mutant cells. Extracts from RFC5 RFC3-HA rad24Δ (KSC1168) and rfc5-1 RFC3-HA rad24Δ (KSC1170) cells carrying YCpRAD24-myc were subjected to immunoprecipitation (IP) with anti-Rfc2 antibody. The extracts and immunocomplexes were separated by SDS-PAGE and immunoblotted with the corresponding antibody.
DISCUSSION
RAD24 and RFC5 are required for DNA damage checkpoint control in the budding yeast S. cerevisiae. The budding yeast RFC is composed of one large subunit, Rfc1, and four small subunits (Rfc2, Rfc3, Rfc4, and Rfc5). The RFC subunits are related to one another in their primary amino acid sequence. Rad24 and the RFC subunits also have sequence homology, including the presence of an NTP-binding motif. To evaluate the role of the NTP-binding motif in Rad24 function, we created two different mutations in the motif. One mutation, rad24-K115E, changes the conserved lysine to glutamic acid, converting a basic residue to an acidic residue. The other mutation, rad24-K115R, changes the conserved lysine to arginine, a similar basic amino acid. The phenotype of rad24-K115E mutants is identical to that of rad24 null mutants, suggesting that the NTP-binding motif has an essential role in Rad24 function. It is noted, however, that such an extreme amino acid change may alter the conformation of the entire Rad24 protein. In contrast, rad24-K115R mutants show no apparent phenotype with respect to DNA damage sensitivity or DNA damage checkpoint control. The fission yeast Rad17 protein, a structural and functional homolog of Rad24, also contains an NTP-binding motif in the corresponding region. Previously, Griffiths et al. (8) mutated the conserved lysine118 to arginine or glutamate in the Rad17 protein and obtained results essentially the same as ours; rad17.K118R cells showed no phenotype, whereas rad17.K118E cells were phenotypically similar to null mutants. These results are consistent with the current view that the function of checkpoint genes is highly conserved in eukaryotes.
RAD24 has an essential role in all the G1-, S- and G2/M-phase DNA damage checkpoints. We previously demonstrated that rfc5-1 mutants are defective for the S-phase DNA damage checkpoint. Rad24 and Rfc5 interact physically and appear to function together in regulating the response to DNA damage. However, there was no evidence to suggest that RFC5 has a role in the G1- and/or G2/M-phase DNA damage checkpoints. To address this possibility, we examined the G1- and G2/M-phase checkpoints in rfc5-1 rad24-K115R double mutants. Although neither rfc5-1 nor rad24-K115R single mutants are defective in the G1- and G2/M-phase DNA damage checkpoints, rfc5-1 rad24-K115R double mutants become as defective in these checkpoints as rad24Δ mutants. Consistent with the idea that Rad24 and Rfc5 function as a complex that controls DNA damage checkpoints, the rfc5-1 and rad24Δ mutations are genetically nonadditive with respect to the checkpoint defects in the G1 and G2/M phases. We also showed that rfc5-1 rad24-K115R and rfc5-1 rad24Δ double mutants are as defective as rad24Δ mutants in the S-phase DNA damage checkpoint. Rad53 is phosphorylated in response to DNA damage, and its phosphorylation correlates with the activation of the checkpoint pathway. Although rad24-K115R and rfc5-1 single mutants are not defective in DNA damage-induced Rad53 phosphorylation in the G2/M phase, rfc5-1 rad24-K115R double mutants are defective. Thus, the rfc5-1 mutation in the presence of the wild-type RAD24 gene is defective only in the S-phase DNA damage checkpoint, while the rfc5-1 rad24-K115R double mutation becomes defective in all the G1-, S- and G2/M-phase DNA damage checkpoints. These observations suggest that RFC5, like RAD24, has a role in the DNA damage checkpoints throughout the cell cycle.
To further understand the phenotypes of rfc5-1 and rfc5-1 rad24-K115R double mutants, we examined the physical interaction between the Rad24 and RFC proteins in wild-type and rfc5-1 mutant cells. Coimmunoprecipitation experiments revealed that the interaction of Rad24K115R with Rfc3 or Rfc4 is decreased in RFC5 cells, despite the fact that the rad24-K115R mutation does not appear to affect the DNA damage responses. In rfc5-1 mutants, the interaction between Rad24 and the RFC proteins is decreased compared to wild-type cells, and the interactions among the RFC proteins are also impaired. The rfc5-1 mutation changes glycine to glutamate at amino acid position 43 in the Rfc5 NTP-binding motif (Fig. 1). Involvement of the NTP-binding motif in complex formation may be a common feature of Rad24 and RFC proteins. It was reported that mutation of the NTP-binding motif in the p140, p40, or p36 subunit of the human RFC complex results in decreased complex assembly and/or stability (25). One explanation why rfc5-1 mutants are defective only in the S-phase checkpoint could be that the interaction between Rad24 and the RFC proteins is decreased specifically in the S phase. However, this possibility is unlikely because the interaction between Rad24 and Rfc3 in rfc5-1 mutants was constant during the cell cycle (data not shown). It is, rather, possible that the S-phase DNA damage checkpoint is more sensitive to the level of the interaction between Rad24 and the RFC proteins than the G1- and G2/M-phase DNA damage checkpoints.
Alternatively, the rfc5-1 defect may result from alterations in Rfc5 function and/or impaired interactions among the RFC proteins. For example, DNA damage may be processed differently in the S phase and Rfc5 may be more specifically involved in recognition of this damage processing. The significance of the Rad24-RFC protein interaction in the DNA damage checkpoints is demonstrated from studies of rfc5-1 mutants expressing Rad24K115R. When expressed in rfc5-1 mutants, Rad24K115R shows no detectable association with the RFC proteins. Accordingly, rfc5-1 rad24-K115R double mutants are defective in all the G1-, S- and G2/M-phase DNA damage checkpoints. Importantly, these rfc5-1 rad24-K115R double mutants are as defective as rad24Δ mutants in the DNA damage checkpoints. Thus, the interaction between Rad24 and the RFC proteins appears to be critical for the DNA damage checkpoints.
RFC is composed of one large subunit (Rfc1) and four small subunits (Rfc2, Rfc3, Rfc4, and Rfc5). RFC has an established role in recognizing the primer-template junction and loading PCNA onto the primer terminus. We have shown that Rad24 interacts physically with the small RFC subunits Rfc2, Rfc3, Rfc4, and Rfc5 but not with the large RFC subunit Rfc1. Recently, Green et al. (7) purified Rad24 to homogeneity and found that Rfc2 and Rfc3 copurify with Rad24. They also performed coimmunoprecipitation studies with Rad24 and the RFC subunits and obtained results similar to ours; Rad24 associates with Rfc2, Rfc3, Rfc4, and Rfc5 but not with Rfc1. They further showed that Rad24 does not cofractionate with Rfc1. These results suggest that Rad24 forms a complex closely related to but distinct from RFC and that the Rad24 complex functions in the DNA damage checkpoints. Genetic analysis has suggested that RAD24 is involved in the same checkpoint pathway as RAD17, MEC3, and DDC1 (7, 17, 18, 20). Rad17 has been suggested to share structural similarity with PCNA (40) and to function in a complex with Mec3 and Ddc1 (13). Interestingly, overexpression of DDC1 suppresses the rad24Δ mutant phenotype (13). These observations raise the possibility that the Rad24-RFC proteins complex is required for loading the Rad17-Mec3-Ddc1 complex onto specific structures on damaged DNA.
In summary, our results suggest that the Rad24-RFC proteins complex functions in all the G1-, S- and G2/M-phase DNA damage checkpoints. It has been proposed that there may be one DNA damage surveillance system that functions throughout the cell cycle, as opposed to multiple distinct mechanisms that operate at different checkpoints (24). Future experiments will be necessary to determine whether the Rad24-RFC protein complex functions as an RFC-related complex, which might recognize DNA damage and recruit other checkpoint proteins.
ACKNOWLEDGMENTS
We thank A. Sugino for materials, M. Lamphier for critical readings of the manuscript, M. Mayer for discussion, and C. Green and N. Lowndes for communicating results prior to publication. K.S. acknowledges H. Mishima, Y. Miyake, H. Takahashi and H. Terasaki for suggestions and encouragement.
T.K. is a recipient of a JSPS predoctoral fellowship. This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas and General Research from the Ministry of Education, Science, Sports and Culture of Japan (K.M. and K.S.).
REFERENCES
- 1.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 2.Burgers P M J. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerase δ and ɛ. J Biol Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 3.Cross F R. Marker swap plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 4.Cullmann G, Fien K, Kobayashi R, Stillman B. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 6.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 7.Green C M, Erdjument-Bromage H, Tempst P, Lowndes N F. A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr Biol. 2000;10:39–42. doi: 10.1016/s0960-9822(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths D J F, Barbet N C, McCready S, Lehmann A R, Carr A M. Fission yeast rad17; a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 1995;14:5812–5823. doi: 10.1002/j.1460-2075.1995.tb00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guthrie C, Fink G R. Guide to yeast genetics and molecular biology. New York, N.Y: Academic Press; 1991. [Google Scholar]
- 10.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:229–234. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 11.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 12.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair, and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin E V. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 15.Lee S-H, Kwong A D, Pan Z-Q, Hurwitz J. Studies on the activator 1 protein complex, an accessory factor for proliferating cell nuclear antigen-dependent DNA polymerase δ. J Biol Chem. 1991;266:594–602. [PubMed] [Google Scholar]
- 16.Longhese M P, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Longhese M P, Fraschini R, Plevani P, Lucchini G. Yeast pip3/mec3 mutants fail to delay entry into S phase and to slow down DNA replication in response to DNA damage, and they define a functional link between Mec3 and DNA primase. Mol Cell Biol. 1996;16:3235–3244. doi: 10.1128/mcb.16.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longhese M P, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 1997;17:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lydall D, Weinert T. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol Gen Genet. 1997;256:638–651. doi: 10.1007/s004380050612. [DOI] [PubMed] [Google Scholar]
- 20.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 21.Paciotti V, Lucchini G, Plevani P, Longhese M P. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 23.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulovich A G, Toczyski D P, Harwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 25.Podust V N, Tiwari N, Ott R, Fanning E. Functional interactions among the subunits of replication factor C potentiate and modulate its ATPase activity. J Biol Chem. 1998;273:12935–12942. doi: 10.1074/jbc.273.21.12935. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinase MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 27.Saraste M, Sibbald P R, Wittinghofer A. The P-loop—a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 28.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanajali S R, Simmons A, Clines G A, Sartiel A, Gatti R A, Chessa L, Sanal O, Lavin M F, Jaspers N G J, Taylor A M R, Arlett C F, Miki T, Weissman S M, Lovett M, Collins F S, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 29.Shimomura T, Ando S, Matsumoto K, Sugimoto K. Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol Cell Biol. 1998;18:5485–5491. doi: 10.1128/mcb.18.9.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siede W, Allen J B, Elledge S J, Friedberg E C. The Saccharomyces cerevisiae MEC1 gene, which encodes a homolog of the human ATM gene product, is required for G1 arrest following radiation treatment. J Bacteriol. 1996;178:5841–5843. doi: 10.1128/jb.178.19.5841-5843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siede W, Friedberg A S, Dianova I, Friedberg E C. Characterization of G1 checkpoint control in the yeast Saccharomyces cerevisiae following exposure to DNA-damaging agent. Genetics. 1994;138:271–281. doi: 10.1093/genetics/138.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siede W, Friedberg A S, Friedberg E C. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siede W, Nusspaumer G, Portillo V, Rodriguez R, Friedberg E C. Cloning and characterization of RAD17, a gene controlling cell cycle responses to DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:1669–1675. doi: 10.1093/nar/24.9.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stern D F, Zheng P, Beidler D R, Zerillo C. Spk1, a new kinase from Saccharomyces cerevisiae, phosphorylates proteins on serine, threonine, and tyrosine. Mol Cell Biol. 1991;13:3744–3755. doi: 10.1128/mcb.11.2.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimoto K, Ando S, Shimomura T, Matsumoto K. Rfc5, replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol Cell Biol. 1997;17:5905–5914. doi: 10.1128/mcb.17.10.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugimoto K, Sakamoto Y, Takahashi O, Matsumoto K. HYS2, an essential gene required for DNA replication in Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:3493–3500. doi: 10.1093/nar/23.17.3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugimoto K, Shimomura T, Hashimoto K, Araki H, Sugino A, Matsumoto K. Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitosis in budding yeast. Proc Natl Acad Sci USA. 1996;93:7048–7052. doi: 10.1073/pnas.93.14.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 40.Thelen M P, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;19:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 41.Torre-Ruiz M-A, Green C M, Lowndes N F. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;17:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsurimoto T, Stillman B. Functions of replication factor C and proliferating-cell nuclear antigen: functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci USA. 1990;87:1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinert T A, Hartwell L H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 45.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]