Summary
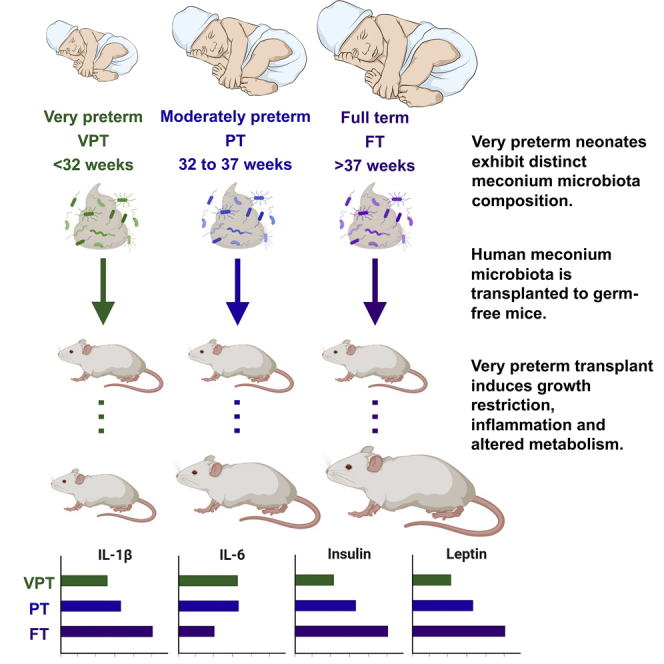
Preterm birth may result in adverse health outcomes. Very preterm infants typically exhibit postnatal growth restriction, metabolic disturbances, and exaggerated inflammatory responses. We investigated the differences in the meconium microbiota composition between very preterm (<32 weeks), moderately preterm (32–37 weeks), and term (>37 weeks) human neonates by 16S rRNA gene sequencing. Human meconium microbiota transplants to germ-free mice were conducted to investigate whether the meconium microbiota is causally related to the preterm infant phenotype in an experimental model. Our results indicate that very preterm birth is associated with a distinct meconium microbiota composition. Fecal microbiota transplant of very preterm infant meconium results in impaired growth, altered intestinal immune function, and metabolic parameters as compared to term infant meconium transplants in germ-free mice. This finding suggests that measures aiming to minimize the long-term adverse consequences of very preterm birth should be commenced during pregnancy or directly after birth.
Keywords: germ-free mice, preterm infant, gut microbiota, fecal microbiota transplant, growth, inflammation, metabolism
Graphical abstract
Highlights
-
•
Very preterm neonates exhibit a distinct meconium microbiota composition
-
•
Human meconium microbiota is transplanted to germ-free mice in this study
-
•
Preterm transplant induces growth restriction, inflammation, and altered metabolism
-
•
Initial gut microbiota may be causally related to complications of prematurity
Hiltunen et al. show that very preterm (<32 gestational weeks) birth is associated with distinct meconium microbiota composition. Very preterm infant meconium transplant to germ-free mice results in impaired growth, altered intestinal immune function, and metabolic abnormalities, suggesting a causal role for gut microbiota in the complications of prematurity.
Introduction
Preterm birth is a major global health problem affecting ∼10% of pregnancies,1 corresponding to roughly 15 million preterm neonates born every year.2 Globally, preterm birth is the leading cause of death in children younger than 5 years of age.3 In addition to increased mortality, preterm infants exhibit a high risk of both short- and long-term morbidity.4 Postnatal growth restriction is a common problem in very preterm (VPT) infants.5 While suboptimal nutritional management is an important cause of impaired postnatal growth,6 inflammation is also thought to contribute to the pathogenesis of growth failure in preterm neonates.7, 8, 9 Later in life, preterm birth is associated with significantly increased occurrence of the metabolic syndrome10 and other risk factors for metabolic and cardiovascular disease in adulthood.11
Preterm neonates exhibit aberrant gut colonization patterns.12 Reduced microbial diversity12 and delayed colonization with Bifidobacteria13 have been reported to be characteristic of the preterm gut microbiota. Interestingly, the meconium microbiota detected in preterm neonates differs not only from the gut microbiota in the same individuals later in the neonatal period and infancy,14 but also from the meconium microbiota of term neonates.15 The origin of meconium microbiota is a topic of debate, and maternal transmission has been suggested.16 It has previously been shown that defective gut microbiota maturation is concomitant with postnatal growth failure.17 According to recent reports, the gut microbiota composition at the age of 2 days may be associated with preterm infant growth during the first weeks of life,18 and specific meconium microbiota features have been linked with extrauterine growth in neonates born before 33 weeks of gestation.19 Whether the initial preterm gut microbiota causally affects early growth or the development of the inflammatory or metabolic adverse consequences of prematurity remains unknown.
In this study, we characterized the differences in the meconium microbiota in a cohort of VPT, moderately preterm (PT), and term (FT) human neonates. The causal contribution of the meconium microbiota to the growth failure, inflammatory immune state, and metabolic disturbances often encountered in VPT neonates was investigated in an experimental mouse model by performing fecal microbiota transplantation (FMT). Our results revealed significant differences in the meconium microbiota composition among the three groups. A statistically significant growth restriction, as well as an altered inflammatory response and metabolic disturbances characteristic of VPT neonates were seen in germ-free (GF) mice receiving meconium FMT from VPT neonates.
Results
The clinical characteristics of the neonates in the study are presented in Table 1. The extremely preterm infants (born at <28 weeks of gestation) and VPT infants (born from 28 to 32 weeks of gestation) in the study were combined as one group (VPT) for the statistical analyses. None of the neonates in the study exhibited symptoms or signs of perinatal asphyxia, as reflected in their Apgar scores at 5 and 15 min of age. Only mothers with symptoms and signs suggesting a high risk of spontaneous preterm delivery were recruited, and, consequently, all of the neonates included in this study had been exposed to antenatal corticosteroid treatment. It is noteworthy that the FT infants were mainly vaginally born, whereas in the preterm groups, caesarean section deliveries occurred in approximately half of the cases. The mothers were recruited during pregnancy, and the mode of delivery could not be predicted.
Table 1.
Clinical characteristics of the study population
| Group | Intrapartum antibiotics | Gender | Mode of delivery | GA, weeks | Birth weight, g | Z score | Apgar 5 min | Apgar 15 min | Meconium analysis | FMT | Sample collection date |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Very preterm | |||||||||||
| VPT1 | yes | female | CS | 30 + 4/7 | 1,730 | 0.3 | 6 | 8 | yes | yes | 2 |
| VPT2 | yes | male | CS | 25 + 0/7 | 850 | 0.6 | 8 | N/A | yes | yes | 4 |
| VPT3 | yes | male | vaginal | 28 + 5/7 | 1,390 | 0.6 | 8 | N/A | yes | yes | 3 |
| VPT4 | yes | female | vaginal | 26 + 6/7 | 1,000 | −0.1 | 2 | 7 | yes | yes | 2 |
| VPT5 | no | female | vaginal | 30 + 4/7 | 1,420 | −0.5 | 10 | 10 | yes | no | 1 |
| Mean | – | – | – | 28 + 2/7 | 1,278 | 0.2 | – | – | – | – | – |
| Moderately preterm | |||||||||||
| PT1 | yes | female | CS | 33 + 6/7 | 3,840 | 5.4 | 8 | 9 | yes | yes | 5 |
| PT2 | yes | female | vaginal | 34 + 6/7 | 2,280 | −1 | 10 | 10 | yes | yes | 2 |
| PT3 | yes | male | vaginal | 34 + 2/7 | 1,645 | −2.7 | 9 | 9 | yes | yes | 2 |
| PT4 | yes | male | CS | 36 + 1/7 | 2,420 | −1.6 | 9 | 9 | yes | yes | 5 |
| PT5 | yes | male | CS | 34 + 2/7 | 2,295 | −0.7 | 9 | 9 | yes | yes | 3 |
| PT6 | yes | female | vaginal | 35 + 0/7 | 2,460 | −0.5 | 9 | 9 | yes | yes | 1 |
| PT7 | yes | male | vaginal | 33 + 4/7 | 2,640 | 1.3 | 8 | 8 | yes | no | 3 |
| Mean | – | – | – | 34 + 4/7 | 2,511 | 0 | – | – | – | – | – |
| Term | |||||||||||
| FT1 | no | male | vaginal | 37 + 0/7 | 3,230 | 0.1 | 9 | 9 | yes | yes | 1 |
| FT2 | yes | male | vaginal | 38 + 2/7 | 2,840 | −1.4 | 9 | 9 | yes | yes | 2 |
| FT3 | no | male | vaginal | 38 + 5/7 | 3,640 | 0.2 | 9 | 9 | yes | yes | 2 |
| FT4 | yes | female | vaginal | 38 + 6/7 | 3,190 | −0.6 | 9 | 9 | yes | yes | 2 |
| FT5 | no | female | vaginal | 41 + 5/7 | 3,400 | −0.6 | 9 | 9 | yes | yes | 2 |
| FT6 | no | female | vaginal | 40 + 6/7 | 4,440 | 1.7 | 9 | 9 | yes | yes | N/A |
| FT7 | no | female | vaginal | 38 + 4/7 | 3,580 | 0.5 | 9 | 9 | yes | yes | 1 |
| FT8 | no | female | vaginal | 38 + 4/7 | 2,820 | −1.3 | 9 | 9 | no | yes | 3 |
| Mean | – | – | – | 39 + 0 | 3,393 | −0.2 | – | – | – | – | – |
CS, caesarean section; FMT, fecal microbiota transplant; GA, gestational age.
VPT infants harbor a unique meconium microbiota
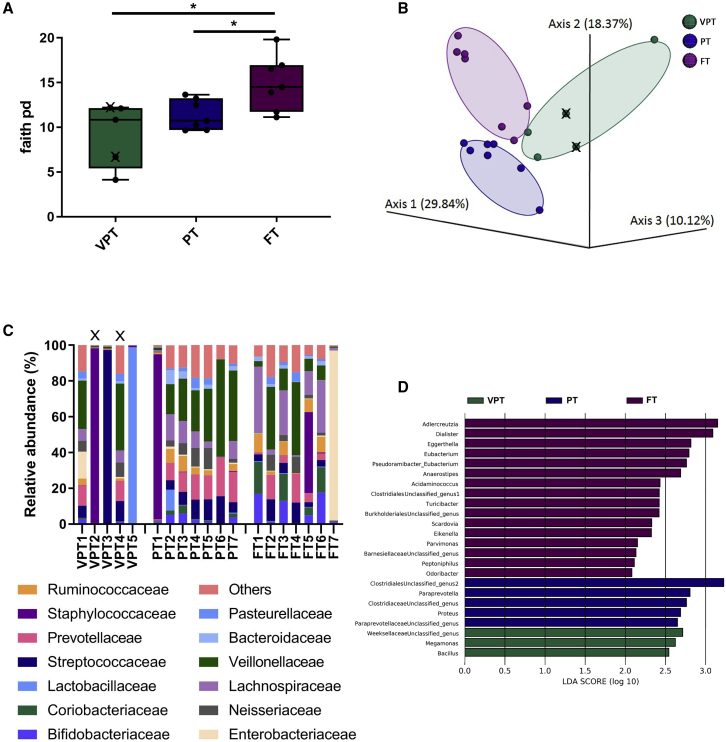
Significant differences in the meconium microbiota composition were detected between VPT, PT, and FT neonates. The meconium microbiota was analyzed by sequencing the 16S rRNA gene, and bioinformatics analyses were performed using QIIME2. Both VPT and PT neonates exhibited significantly lower alpha (within-sample) diversity as compared to FT neonates when assessed by Faith’s phylogenetic diversity (Figure 1A; p < 0.05). Moreover, significant clustering of the meconium microbiota between VPT, PT, and FT neonates was observed when beta diversity was analyzed using unweighted UniFrac (Figure 1B; q = 0.003). At the phylum level, Firmicutes were the dominant phylum in 95% of all meconium samples (highest abundance observed in the VPT neonates as compared to PT and FT neonates); the second most dominant phylum was Bacteroidetes in 50% of all meconium samples. Figure 1C summarizes the 12 most abundant bacterial families in each sample. Linear discriminant analysis effect size (LEfSe) was performed to investigate differences in the community composition between the groups, and the features differentiating the groups are summarized in Figure 1D.
Figure 1.
Meconium composition in the very preterm (VPT), moderately preterm (PT), and term (FT) neonates
(A) Alpha diversity among the VPT, PT, and FT neonates using Faith’s phylogenetic diversity index, compared using t tests (p < 0.05). Data are means ± standard errors of the mean (SEMs).
(B) Principal coordinate analysis (PCoA) of unweighted UniFrac distances between the microbiota of the 3 groups (PERMANOVA, q = 0.003).
(C) Relative abundance at the family level.
(D) The greatest differences in taxa between the 3 groups are presented according to the linear discriminant analysis (LDA) scores (log10), as determined using the LEfSe method.
∗p < 0.05; VPT, very preterm (n = 5); PT, moderately preterm (n = 7); FT, term (n = 7). X denotes extremely preterm neonates (gestational age <28 weeks) within the VPT group.
Differences in weight gain, inflammatory cytokine gene expression, and metabolic hormone levels in mice after FMT
FMT with meconium from VPT, PT, or FT neonates was performed to 18 eight-week-old GF mice to establish whether the altered meconium microbiota in VPT neonates may play a role in the development of the preterm infant phenotype. Mouse stool samples were collected at 7 and 35 days post-FMT. At 7 days, Akkermansia and Faecalibacterium were found to be overrepresented in mice receiving FMT from VPT infants (LEfSe; Figure S1). An unclassified Clostridiales was overexpressed in mice that received FMT from PT infants 35 days after the transplant (Figure S2).
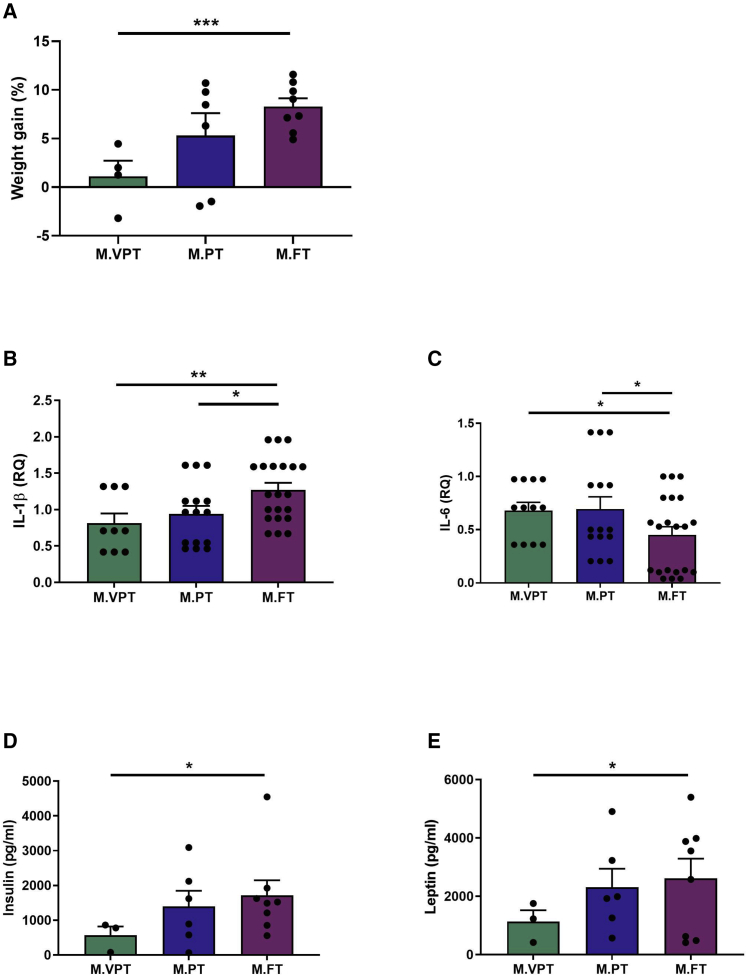
To assess the association between the meconium microbiota and growth, mouse weight was monitored, and weight fold change was calculated in the mice as a measure of growth after FMT. After 35 days, the mice that received the meconium microbiota of VPT neonates had gained significantly less weight compared to the mice that received the meconium microbiota of FT neonates (Figure 2A; p < 0.001), while the mice receiving FMT from PT neonates exhibited intermediate growth. These results suggest that the distinct microbiota in VPT infant meconium is associated with growth failure in mice.
Figure 2.
The effect of FMT using meconium on growth, metabolic hormones, and inflammatory cytokines in germ-free mice
(A) Percent weight gain (M.VPT n = 4, M.PT n = 6, M.FT n = 8).
(B and C) Expression of inflammatory cytokines in mice terminal ileum, determined by quantitative PCR analysis.
(B) Expression of IL-1β (M.VPT n = 3, M.PT n = 5, M.FT n = 7).
(C) Expression of IL-6 (M.VPT n = 3, M.PT n = 5, M.FT n = 7).
(D and E) Concentration of metabolic hormones in mouse blood determined by multiplex assay; .
(D) Concentration of insulin (M.VPT n = 3, M.PT n = 6, M.FT n = 8).
(E) Concentration of leptin (M.VPT n = 3, M.PT n = 6, M.FT n = 8).
Data represent the means ± SEMs. Regarding mice microbiota results, see Figures S1 and S2. FMT, fecal microbiota transplantation; M.VPT, mouse very preterm; M.PT, mouse moderately preterm; M.FT, mouse term. ∗∗∗p < 0.001; ∗∗∗p < 0.01; ∗p < 0.05 (t test).
We next investigated the impact of FMT from VPT, PT, and FT neonates on intestinal immune activation. The expression of mRNA encoding the inflammatory cytokines interleukin-1β (IL-1β) and interleukin-6 (IL-6) was measured from the mouse terminal ileum samples collected after sacrificing the mice on day 35 of the experiment. The mice receiving the microbiota from FT neonates had significantly higher levels of IL-1β compared to mice transplanted from VPT (p < 0.01) and PT (p < 0.05) neonates (Figure 2B). The mice receiving meconium microbiota from VPT and PT neonates displayed significantly higher intestinal inflammatory activation, as measured by IL-6 mRNA (Figure 2C; p < 0.05) levels as compared to the mice receiving the meconium microbiota of FT neonates. These data indicate that mice receiving FMT with very premature meconium exhibit altered intestinal inflammatory activation.
Finally, we assessed the impact of FMT from VPT, PT, and FT neonates on mouse metabolism. Plasma levels of the metabolic hormones insulin and leptin were measured from blood samples collected after sacrificing the mice on day 35 of the experiment. The mice receiving the meconium of VPT neonates exhibited statistically significant differences in the metabolic state as demonstrated by significantly lower plasma levels of both insulin and leptin, as compared to the mice that received meconium from FT neonates (Figures 2D and 2E; p < 0.05). The mice receiving FMT from PT neonates exhibited intermediate levels of metabolic hormones.
Discussion
Our results demonstrate that the common VPT infant phenotype, which includes growth restriction, altered immune responsiveness, and metabolism, can be transferred to GF mice by meconium microbiota transplant. The meconium microbiota composition of VPT neonates was significantly distinct from that of FT neonates and exhibited lower alpha diversity. It is well established that the gut microbiota in preterm infants during the first weeks of life is markedly different from that of term infants. This may be caused, at least in part, by exposures known to perturb normal gut colonization, including caesarean section delivery20 and antibiotic exposure,21,22 as well as reduced breastfeeding and skin-to-skin contact, which often coexist in preterm neonates. However, we have previously reported that in addition to these exposures, prematurity per se affects early gut colonization patterns.23 According to a detailed study by Korpela and colleagues,24 the gut microbiota of VPT and PT neonates is initially dominated by only a few bacterial taxa, but the gut microbiota composition over the first postnatal weeks gradually evolves to be primarily composed of Bifidobacteria and thus resembles that of FT infants. Interestingly, while the overall microbiota composition appeared to be strongly correlated with postnatal age in the study, postmenstrual age was the strongest determinant of gut microbiota composition. In the present study, significant differences in gut microbiota composition between VPT, PT, and FT neonates were already detected in meconium. These results are consistent with a previous study indicating that the meconium microbiota in preterm neonates differs from the meconium microbiota in FT neonates, but also from the gut microbiota in the same individuals later in the neonatal period and infancy.14 Even though postnatal influences cannot completely be ruled out, these data may be interpreted to indicate that gestational age and prenatal exposures may influence the initial gut microbiota. It has been suggested that the altered meconium microbiota in preterm neonates may be pro-inflammatory and causally linked to the maternal and fetal inflammatory responses associated with preterm delivery.15 The present study endeavored to systematically assess the consequences of aberrant initial gut colonization.
Postnatal growth restriction is a substantial clinical problem in very low birth weight preterm infants.25 Undoubtedly, suboptimal nutritional management often contributes to the growth failure.26 Interestingly, it has been reported that early gut microbiota composition assessed at 2 and 10 days of age is associated with weight gain in preterm neonates born between 28 and 33 weeks of gestation.18 According to a recent report, defective gut microbiota maturation was observed in extremely preterm infants with growth failure as compared to those exhibiting normal growth.17 It is of particular interest that lower intestinal microbiota diversity preceded the development of impaired growth and was detectable already during the first week of life. The results of the present study provide evidence for a possible causal relationship between aberrant initial gut microbiota composition and growth failure in preterm neonates. We demonstrate that mice receiving VPT meconium microbiota transplants exhibit statistically significantly restricted growth as compared to mice receiving FMT of meconium from FT neonates, despite uniform nutritional management of the experimental animals. The association between gut microbiota composition and growth is well established by clinical and experimental studies in the context of overweight and obesity in later life.27 A recent report shows an association between the first stool and obesity at the age of 3 years.28 Intestinal microbes are known to contribute to energy harvest from the diet and modulate host metabolism associated with growth and energy storage. Defective gut microbiota maturation has previously been reported to be associated with growth failure in undernourished children.29 The potential causal role of aberrant initial gut colonization in subsequent growth restriction observed in this study may be explained by the inflammatory activation observed in both the VPT neonates and the experimental animals receiving the FMT.
Inflammatory activation is associated with preterm birth and many of its adverse consequences. According to a report by Schreurs and colleagues,30 the fetal gut harbors naive CD4+ T cells, which support mucosal development. Dysregulation of this process by preterm birth and premature exposure to antigens was suggested to contribute to intestinal inflammation in the study. In the present study, an intestinal inflammatory tone reflected by the modest but statistically significant increase in baseline IL-6 expression was seen in the mice receiving FMT from VPT infants but not in those receiving FMT of PT or FT meconium. We also observed a decrease in intestinal IL-1β in mice receiving VPT neonate meconium microbiota. Despite its proinflammatory roles, this cytokine has been reported to also play a role in intestinal repair.31 It is well established that a propensity to exaggerated intestinal inflammatory responses in preterm neonates plays a role in the pathogenesis of necrotizing enterocolitis (NEC), an inflammatory intestinal disease afflicting preterm neonates.32 Based on experiments using a gnotobiotic mouse model, Yu et al.33 have reported that FMT with early gut microbiota from preterm infants exhibiting poor growth resulted in defective intestinal maturation as compared to those receiving FMT from preterm infants with normal weight gain. These data suggest that aberrant initial gut microbiota composition may impair growth and increase NEC risk by inducing inflammation and perturbing gut maturation. In addition, inflammation has been linked to adverse neurological and growth outcomes in VPT infants.7, 8, 9 Interventions aiming to modulate gut colonization and intestinal immune responses, including early human milk feeding34,35 and probiotics,36 have been shown to reduce both intestinal inflammation and adverse clinical outcomes,37, 38, 39 including NEC.40 Our present data may be interpreted to suggest that preventive approaches involving interventions aiming to influence early gut colonization could be initiated prenatally.
Metabolic alterations are common in preterm infants in the neonatal period and beyond. According to epidemiological and cohort studies, preterm birth is associated with an increased incidence of the metabolic syndrome and a significantly increased occurrence of risk factors for later metabolic and cardiovascular disease, including lower lean body mass, higher body fat content, and impaired glucose metabolism.10,11 The cord blood concentration of leptin has been reported to be lower in preterm as compared to term neonates.41,42 Insulin and leptin both function as fetal growth factors,41,43 and fetal and neonatal exposure to leptin has been suggested to protect from the development of adverse metabolic outcomes in later life.44 It has recently been suggested that the aberrant gut microbiota observed in extremely preterm infants suffering from growth retardation may contribute to a persistent metabolic state resembling fasting characterized by defective anabolic glucose metabolism.17 In the present study, mice receiving FMT from VPT infants exhibited significantly lower plasma concentrations of insulin and leptin, which may explain the growth restriction observed in the mice receiving FMT from VPT infants. This demonstrates a causal connection between initial gut colonization patterns and the adverse metabolic consequences of very preterm birth.
Our results suggest that the prenatal events leading to very preterm birth and their impact on initial gut colonization may play an important role in the development of the adverse consequences of preterm birth. Experimental and clinical studies aiming to further elucidate the exposures and events modulating the meconium microbiota may provide a basis for means to improve the long-term health of individuals born preterm.
Limitations of the study
Our study has obvious limitations. We included a relatively small number of subjects and, consequently, neonates born as extremely preterm infants (before 28 weeks of gestation) and very preterm (born between 28 and 32 weeks of gestation) were analyzed as a single group. The meconium samples were collected from diapers and therefore potentially exposed to environmental microbes. However, uniform sample collection protocol and tools were used to avoid systemic bias. The meconium samples collection time had some variation, since they were collected 0–5 days after birth. Nonetheless, our results regarding meconium microbiota composition are significant and consistent with previously published larger studies. We therefore believe the samples to be representative. There was variation in the groups regarding the mode of birth. Due to the study design, the mode of birth could not be predicted. In future research, the caesarean section rate differences should be minimized. A further limitation of our study is the fact that the weight gain, inflammatory responsiveness, or metabolic outcomes of the infants participating in this study were not reported due to the small sample size and poor availability of data. Instead, we relied on previously published data, which indicate that postnatal growth failure is a frequently encountered problem in very preterm infants, and that adverse metabolic consequences are common. Despite these limitations, we believe that our data reliably demonstrates an association between the aberrant meconium microbiota and the characteristic preterm infant phenotype, including growth failure, inflammatory immune tone, and metabolic disturbances associated with impaired growth and metabolic disease. Furthermore, our results from the FMT experiments in GF mice suggest that a causal relationship may underlie these associations. Although assuming interspecies transfer of pathology is prone to criticism and the results should be interpreted with caution,45 FMT from individuals with and without pathology into GF rodents, followed by a comparative analysis of pathological phenotypes in the recipient animals, regardless of actual microbiota composition, remains the most commonly used model to make causal inferences regarding gut microbiota aberrancies and pathogenesis of disease.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Newborn fecal samples | Turku University Hospital, Turku, Finland | N/A |
| Critical commercial assays | ||
| PureLink Microbiome DNA Purification Kit | Invitrogen, ThermoFisher Scientific | CAT# A29790 |
| AMPure XP for PCR clean-up (magnetic beads) | Beckman Coulter | CAT# A63882 |
| Quant-iT PicoGreen dsDNA quantitation kit | Invitrogen, ThermoFisher Scientific | CAT# P7589 |
| MO BIO’s PowerSoil DNA Isolation Kit Handbook | QIAGEN | CAT# 12855 |
| 5x All-in-One RT MasterMix | abm | CAT# G592 |
| Fast SYBR Green Master mix | Applied Biosystems | CAT# 43-856-12 |
| Total RNA Purification Kit | Norgen Biotek Corp | CAT# 17200 |
| Deposited data | ||
| Sequence data | This paper | ENA:PRJEB46240; DDBJ:ERP130461 |
| Experimental models: organisms/strains | ||
| Mouse: Swiss Webster | Azrieli Medical Faculty Animal Facility | WT – established colony in the faculty |
| Software and algorithms | ||
| QIIME2 | Bolyen et al., 201946 | https://qiime2.org/ |
| DADA2 | Callahan et al., 201647 | https://benjjneb.github.io/dada2/tutorial.html |
| Other | ||
| Mouse Chow: Maintenance Diet | Altromin International | Diet 1324 |
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact, Samuli Rautava (samulirautava@gmail.com).
Materials Availability
The study did not generate new unique reagents.
Experimental model and subject details
Ethics statement
The children were born at the Turku University Hospital, Turku, Finland. The study was approved by the Ethics Committee of the Hospital District of Southwest Finland. Oral and written informed consent was obtained from the caregivers. All mouse experimental protocols were approved by the Bar Ilan Ethics Committee.
Human cohort
The study was based on samples collected from 20 neonates participating in an observational study at the Turku University Hospital in Turku, Finland. Pregnant women with symptoms or signs suggesting high risk of spontaneous preterm delivery were included. No randomization was conducted. The recruited mothers and their infants were grouped by gestational age at delivery. Altogether, 5 neonates were born VPT (before 320/7 weeks of gestation), 7 neonates were born PT (320/7-366/7 weeks of gestation) and 8 neonates were born FT (after 370/7 weeks of gestation). It is of note that both neonates born before 28 weeks of gestation and those born between 28-32 weeks of gestation were included in the VPT group.
Mice
8-10 weeks old germ-free Swiss Webster female mice were used for this study. The mice were housed in isolated cages and maintained on a 12-h light/dark cycle. All mice were handled uniformly and fed the same diet (Maintenance Diet 1324, Altromin International, Germany). All mouse experimental protocols were approved by the Bar Ilan Ethics Committee.
Method details
Meconium microbiota
Meconium samples (the first stool passed by the neonate after birth) were collected from the study subjects. To minimize environmental contamination, standardized protocols were used in sample collection. Meconium samples were frozen immediately after collection and kept at −70°C until analysis. DNA was extracted from all meconium samples together with appropriate positive and negative controls using the PureLink Microbiome DNA Purification Kit (Invitrogen). The V4 region of the bacterial 16S rRNA gene was amplified by polymerase chain reaction (PCR) using the 515F (AATGATACGGCGACCACCGAGATCTACACGCT) barcoded and 806R (TATGGTAATTGTGTGYCAGCMGCCGCGGTAA) primers. PCR were carried out by 35 cycles of denaturation (95°C), annealing (55°C) and extension (72°C), with final elongation at 72°C. PCR products were purified using AMPure magnetic beads (Beckman Coulter) and quantified using Quant-iT PicoGreen dsDNA quantitation kit (Invitrogen). Samples (30 ng), were loaded on 2% agarose E-Gel (Thermo Fisher), purified and sequenced using the Illumina MiSeq platform (Genomics Center, Azrieli Faculty of Medicine, BIU, Israel). A meconium sample from one subject was excluded from the analyses because the isolated DNA was not PCR amplifiable. The analyses were therefore performed on samples from 5 very preterm, 7 moderately preterm, and 7 term neonates (n = 19). In addition to positive and negative controls included throughout the pipeline, in a subset of the samples, real-time PCR (qPCR) of the 16S rRNA region was performed to confirm ample bacterial load for further analysis.
Fecal microbiota transplant (FMT) experiments
Germ free status of mice was verified using 16S rRNA PCR on fecal samples as described above for meconium samples. FMT of meconium from neonates were introduced to GF Swiss Webster female mice (8-10 weeks old), defining day 0 of the experiment. Each meconium sample collected in the study was separately suspended in 800 μl sterile phosphate-buffered saline (PBS) and dissociated by vortex for 1 min 200 μl from each suspension was gavaged orally to each GF mouse. A total of 18 meconium specimens were transferred according to the three groups: VPT (n = 4), PT (n = 6) and FT (n = 8). Mouse stool samples were collected on day 7 and 35. Samples were also collected on days 14, 21, and 28; the results were generally in-line with those presented and are therefore not presented herein. DNA from stool samples was extracted using the Mobio Powersoil DNA extraction kit (Mobio). The rest of the analysis steps were similar to those for the meconium samples. Mice were weighed at day 35 of the experiment, and terminal ileum specimens were collected from the mice after sacrifice by CO2 inhalation. A 1-cm section of terminal ileum was immediately removed using sterile scissors. The section was immediately placed in a tube containing RNAlater and stored at −80°C. Total RNA was extracted from tissues using Total RNA Purification Kit (Norgen Biotek Corp). In brief, terminal ileum tissue samples were homogenized in 2 mL cold RL buffer (Norgen Biotek Corp) using the Bio-Gen PRO200 tissue homogenizer (PRO Scientific). Homogenates were centrifuged, and the supernatant was taken for purification according to the manufacturer’s protocol (Total RNA Purification Kit, Norgen Biotek Corp). RNA quantity was determined on a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific) and then reverse transcribed using 5x All-in-One RT MasterMix (Abm). In brief, 200 μg of total RNA were added into a 10 μl reaction volume, and then the mixture was incubated at 25°C for 10 min, followed by synthesis at 42°C for 15 min, and a termination reaction at 85°C for 5 min. Next, 190 μL nuclease-free water was added, and the cDNA stored at −80°C until needed for qPCR. Real-time PCR amplification was performed to determine expression of IL-1β and IL-6 (primers in Table S1) with cDNA in a reaction containing with a final concentration of 0.1 μM for each primer and 5 μL Fast SYBR Green Master mix (Applied Biosystems) with 80 ng cDNA in 10 μL total volume. Reactions were run in a ViiA 7 Real-Time PCR System (Applied Biosystems). Cycle condition were: 95°C for 20 min, 40 cycles at 95°C for 1 min, and 60°C for 20 min. The results were analyzed using the ViiA 7 software and exported into Microsoft Excel for further analysis. Every sample was processed in technical triplicates. In all, 15 samples were analyzed (M.VPT n = 3; M.PT n = 5; M.FT n = 7); one terminal ileum from each group was not included due to technical problems. Blood samples were taken from the heart by performing thoracotomy on day 35. Blood was separated by centrifugation and the plasma was stored at −80°C. Plasma samples were used for measuring metabolic hormones (insulin, leptin) using the MILLIPLEX MAGPIX system (Merck Millipore). We analyzed 17 samples (M.VPT n = 3, M.PT n = 6, M.FT n = 8); one blood sample was not included in the analyses due to due to technical problems. Independent sample t tests were used to identify the differences between fold change in mouse weight at day 35 and between study groups. T Ttests were performed in order to examine the differences between blood leptin and insulin levels, and between expression of the inflammatory cytokines IL-1β and IL-6 in the mouse terminal ileum.
Quantification and statistical analysis
Data analysis was performed using QIIME2.46 Sequence reads were demultiplexed by per-sample barcodes and Illumina-sequenced amplicon read errors were corrected by Divisive Amplicon Denoising Algorithm (DADA2).47 A phylogenetic tree was generated. Alpha and beta diversity analysis was performed based on a feature table (species) containing features observed in at least 40% samples of one or more groups and rarefied at 40,000 sequences. Alpha diversity (within sample diversity) parameters, were calculated using Faith’s Phylogenetic Diversity.48 Beta diversity (between sample diversity) was analyzed using UniFrac distances.49 Principal Coordinate Analysis (PCoA) was performed based on unweighted UniFrac distance matrices.
Linear discriminant analysis Effect Size (LEfSe) was also performed,50 to identify the features that significantly differed between samples according to relative abundances. The statistical analyses regarding each test have been described in the figure legends. No assumptions were tested.
Acknowledgments
We would like to thank our research nurses for their assistance. The study was funded by the Finnish Society for Pediatric Research and the Päivikki and Sakari Sohlberg Foundation. The study sponsors had no role in study design; the collection, analysis, and interpretation of data; the writing of the report; or the decision to submit the paper for publication.
Author contributions
The clinical part of the study was designed by H. Hiltunen, R.L., E.I., S.S., and S.R. The fecal microbiological analyses and animal experiments were designed and performed by H. Hanani, S.T., O.Z., and O.K. All of the authors participated in analyzing and interpreting the data. The first draft of the manuscript was written by H. Hiltunen, H. Hanani, O.K., and S.R. All of the authors contributed to writing and revising the manuscript, have seen and approved the submission of the final version of the manuscript, and take full responsibility for the content. The births took place and samples and clinical data were collected at the Turku University Hospital, Finland. The fecal sample analysis and mouse experiments were conducted at the Azrieli Faculty of Medicine, Safed, Israel. No honorarium, grant, or other form of payment was given to anyone to produce the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100447.
Contributor Information
Omry Koren, Email: omry.koren@biu.ac.il.
Samuli Rautava, Email: samulirautava@gmail.com.
Supplemental information
Data and code availability
-
•
The sequence data have been deposited to ENA and DDBJ and are publicly available as of the date of publication. The project numbers are provided in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Goldenberg R.L., Culhane J.F., Iams J.D., Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blencowe H., Cousens S., Chou D., Oestergaard M., Say L., Moller A.B., Kinney M., Lawn J., Born Too Soon Preterm Birth Action Group Born too soon: the global epidemiology of 15 million preterm births. Reprod. Health. 2013;10(Suppl 1):S2. doi: 10.1186/1742-4755-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L., Oza S., Hogan D., Perin J., Rudan I., Lawn J.E., Cousens S., Mathers C., Black R.E. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385:430–440. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 4.Patel R.M. Short- and Long-Term Outcomes for Extremely Preterm Infants. Am. J. Perinatol. 2016;33:318–328. doi: 10.1055/s-0035-1571202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrenkranz R.A., Younes N., Lemons J.A., Fanaroff A.A., Donovan E.F., Wright L.L., Katsikiotis V., Tyson J.E., Oh W., Shankaran S., et al. Longitudinal growth of hospitalized very low birth weight infants. Pediatrics. 1999;104:280–289. doi: 10.1542/peds.104.2.280. [DOI] [PubMed] [Google Scholar]
- 6.Brennan A.M., Murphy B.P., Kiely M.E. Optimising preterm nutrition: present and future. Proc. Nutr. Soc. 2016;75:154–161. doi: 10.1017/S0029665116000136. [DOI] [PubMed] [Google Scholar]
- 7.Cuestas E., Aguilera B., Cerutti M., Rizzotti A. Sustained Neonatal Inflammation Is Associated with Poor Growth in Infants Born Very Preterm during the First Year of Life. J. Pediatr. 2019;205:91–97. doi: 10.1016/j.jpeds.2018.09.032. [DOI] [PubMed] [Google Scholar]
- 8.Trevisanuto D., Peruzzetto C., Cavallin F., Vedovato S., Cosmi E., Visentin S., Chiarelli S., Zanardo V. Fetal placental inflammation is associated with poor neonatal growth of preterm infants: a case-control study. J. Matern. Fetal Neonatal Med. 2013;26:1484–1490. doi: 10.3109/14767058.2013.789849. [DOI] [PubMed] [Google Scholar]
- 9.Mestan K., Yu Y., Matoba N., Cerda S., Demmin B., Pearson C., Ortiz K., Wang X. Placental inflammatory response is associated with poor neonatal growth: preterm birth cohort study. Pediatrics. 2010;125:e891–e898. doi: 10.1542/peds.2009-0313. [DOI] [PubMed] [Google Scholar]
- 10.Heidemann L.A., Procianoy R.S., Silveira R.C. Prevalence of metabolic syndrome-like in the follow-up of very low birth weight preterm infants and associated factors. J. Pediatr. (Rio J.) 2019;95:291–297. doi: 10.1016/j.jped.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Sipola-Leppänen M., Vääräsmäki M., Tikanmäki M., Matinolli H.M., Miettola S., Hovi P., Wehkalampi K., Ruokonen A., Sundvall J., Pouta A., et al. Cardiometabolic risk factors in young adults who were born preterm. Am. J. Epidemiol. 2015;181:861–873. doi: 10.1093/aje/kwu443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Underwood M.A., Sohn K. The Microbiota of the Extremely Preterm Infant. Clin. Perinatol. 2017;44:407–427. doi: 10.1016/j.clp.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drell T., Lutsar I., Stšepetova J., Parm U., Metsvaht T., Ilmoja M.L., Simm J., Sepp E. The development of gut microbiota in critically ill extremely low birth weight infants assessed with 16S rRNA gene based sequencing. Gut Microbes. 2014;5:304–312. doi: 10.4161/gmic.28849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gómez M., Moles L., Espinosa-Martos I., Bustos G., de Vos W.M., Fernández L., Rodríguez J.M., Fuentes S., Jiménez E. Bacteriological and immunological profiling of meconium and fecal samples from preterm infants: a two-year follow-up study. Nutrients. 2017;9:1–19. doi: 10.3390/nu9121293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ardissone A.N., de la Cruz D.M., Davis-Richardson A.G., Rechcigl K.T., Li N., Drew J.C., Murgas-Torrazza R., Sharma R., Hudak M.L., Triplett E.W., Neu J. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS ONE. 2014;9:e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He Q., Kwok L.Y., Xi X., Zhong Z., Ma T., Xu H., Meng H., Zhao F., Zhang H. The meconium microbiota shares more features with the amniotic fluid microbiota than the maternal fecal and vaginal microbiota. Gut Microbes. 2020;12:1794266. doi: 10.1080/19490976.2020.1794266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Younge N.E., Newgard C.B., Cotten C.M., Goldberg R.N., Muehlbauer M.J., Bain J.R., Stevens R.D., O’Connell T.M., Rawls J.F., Seed P.C., Ashley P.L. Disrupted Maturation of the Microbiota and Metabolome among Extremely Preterm Infants with Postnatal Growth Failure. Sci. Rep. 2019;9:8167. doi: 10.1038/s41598-019-44547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arboleya S., Martinez-Camblor P., Solís G., Suárez M., Fernández N., de Los Reyes-Gavilán C.G., Gueimonde M. Intestinal microbiota and weight-gain in preterm neonates. Front. Microbiol. 2017;8:183. doi: 10.3389/fmicb.2017.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terrazzan A.C., Procianoy R.S., Roesch L.F.W., Corso A.L., Dobbler P.T., Silveira R.C. Meconium microbiome and its relation to neonatal growth and head circumference catch-up in preterm infants. PLoS ONE. 2020;15:e0238632. doi: 10.1371/journal.pone.0238632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stokholm J., Thorsen J., Chawes B.L., Schjørring S., Krogfelt K.A., Bønnelykke K., Bisgaard H. Cesarean section changes neonatal gut colonization. J. Allergy Clin. Immunol. 2016;138:881–889.e2. doi: 10.1016/j.jaci.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 21.Arboleya S., Sánchez B., Milani C., Duranti S., Solís G., Fernández N., de los Reyes-Gavilán C.G., Ventura M., Margolles A., Gueimonde M. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J. Pediatr. 2015;166:538–544. doi: 10.1016/j.jpeds.2014.09.041. [DOI] [PubMed] [Google Scholar]
- 22.Gibson M.K., Wang B., Ahmadi S., Burnham C.A., Tarr P.I., Warner B.B., Dantas G. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 2016;1:16024. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forsgren M., Isolauri E., Salminen S., Rautava S. Late preterm birth has direct and indirect effects on infant gut microbiota development during the first six months of life. Acta Paediatr. 2017;106:1103–1109. doi: 10.1111/apa.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korpela K., Blakstad E.W., Moltu S.J., Strømmen K., Nakstad B., Rønnestad A.E., Brække K., Iversen P.O., Drevon C.A., de Vos W. Intestinal microbiota development and gestational age in preterm neonates. Sci. Rep. 2018;8:2453. doi: 10.1038/s41598-018-20827-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fanaroff A.A., Stoll B.J., Wright L.L., Carlo W.A., Ehrenkranz R.A., Stark A.R., Bauer C.R., Donovan E.F., Korones S.B., Laptook A.R., et al. NICHD Neonatal Research Network Trends in neonatal morbidity and mortality for very low birthweight infants. Am. J. Obstet. Gynecol. 2007;196:147.e1–147.e8. doi: 10.1016/j.ajog.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 26.Lapointe M., Barrington K.J., Savaria M., Janvier A. Preventing postnatal growth restriction in infants with birthweight less than 1300 g. Acta Paediatr. 2016;105:e54–e59. doi: 10.1111/apa.13237. [DOI] [PubMed] [Google Scholar]
- 27.Tremaroli V., Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 28.Korpela K., Renko M., Vänni P., Paalanne N., Salo J., Tejesvi M.V., Koivusaari P., Ojaniemi M., Pokka T., Kaukola T., et al. Microbiome of the first stool and overweight at age 3 years: a prospective cohort study. Pediatr. Obes. 2020;15:e12680. doi: 10.1111/ijpo.12680. [DOI] [PubMed] [Google Scholar]
- 29.Blanton L.V., Charbonneau M.R., Salih T., Barratt M.J., Venkatesh S., Ilkaveya O., Subramanian S., Manary M.J., Trehan I., Jorgensen J.M., et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351 doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreurs R.R.C.E., Baumdick M.E., Sagebiel A.F., Kaufmann M., Mokry M., Klarenbeek P.L., et al. Human Fetal TNF-α-Cytokine-Producing CD4+ Effector Memory T Cells Promote Intestinal Development and Mediate Inflammation Early in Life. Immunity. 2019;50:462–476.e8. doi: 10.1016/j.immuni.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Voronov E., Apte R.N. IL-1 in Colon Inflammation, Colon Carcinogenesis and Invasiveness of Colon Cancer. Cancer Microenviron. 2015;8:187–200. doi: 10.1007/s12307-015-0177-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neu J., Walker W.A. Necrotizing enterocolitis. N. Engl. J. Med. 2011;364:255–264. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y., Lu L., Sun J., Petrof E.O., Claud E.C. Preterm infant gut microbiota affects intestinal epithelial development in a humanized microbiome gnotobiotic mouse model. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;311:G521–G532. doi: 10.1152/ajpgi.00022.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis E.D., Richard C., Larsen B.M., Field C.J. The Importance of Human Milk for Immunity in Preterm Infants. Clin. Perinatol. 2017;44:23–47. doi: 10.1016/j.clp.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Walker W.A., Iyengar R.S. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr. Res. 2015;77:220–228. doi: 10.1038/pr.2014.160. [DOI] [PubMed] [Google Scholar]
- 36.Sohn K., Underwood M.A. Prenatal and postnatal administration of prebiotics and probiotics. Semin. Fetal Neonatal Med. 2017;22:284–289. doi: 10.1016/j.siny.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia J. Human milk and the premature infant. Ann. Nutr. Metab. 2013;62(Suppl 3):8–14. doi: 10.1159/000351537. [DOI] [PubMed] [Google Scholar]
- 38.Lechner B.E., Vohr B.R. Neurodevelopmental Outcomes of Preterm Infants Fed Human Milk: A Systematic Review. Clin. Perinatol. 2017;44:69–83. doi: 10.1016/j.clp.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Dermyshi E., Wang Y., Yan C., Hong W., Qiu G., Gong X., Zhang T. The “Golden Age” of Probiotics: A Systematic Review and Meta-Analysis of Randomized and Observational Studies in Preterm Infants. Neonatology. 2017;112:9–23. doi: 10.1159/000454668. [DOI] [PubMed] [Google Scholar]
- 40.Maffei D., Schanler R.J. Human milk is the feeding strategy to prevent necrotizing enterocolitis! Semin. Perinatol. 2017;41:36–40. doi: 10.1053/j.semperi.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 41.Özdemir Z.C., Akşit M.A. The association of ghrelin, leptin, and insulin levels in umbilical cord blood with fetal anthropometric measurements and glucose levels at birth. J. Matern. Fetal Neonatal Med. 2020;33:1486–1491. doi: 10.1080/14767058.2018.1520828. [DOI] [PubMed] [Google Scholar]
- 42.Matsuda J., Yokota I., Iida M., Murakami T., Yamada M., Saijo T., Naito E., Ito M., Shima K., Kuroda Y. Dynamic changes in serum leptin concentrations during the fetal and neonatal periods. Pediatr. Res. 1999;45:71–75. doi: 10.1203/00006450-199901000-00012. [DOI] [PubMed] [Google Scholar]
- 43.Fant M.E., Weisoly D. Insulin and insulin-like growth factors in human development: implications for the perinatal period. Semin. Perinatol. 2001;25:426–435. doi: 10.1053/sper.2001.29036. [DOI] [PubMed] [Google Scholar]
- 44.Stocker C.J., Cawthorne M.A. The influence of leptin on early life programming of obesity. Trends Biotechnol. 2008;26:545–551. doi: 10.1016/j.tibtech.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Walter J., Armet A.M., Finlay B.B., Shanahan F. Establishing or Exaggerating Causality for the Gut Microbiome: Lessons from Human Microbiota-Associated Rodents. Cell. 2020;180:221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 46.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faith D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992;61:1–10. [Google Scholar]
- 49.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The sequence data have been deposited to ENA and DDBJ and are publicly available as of the date of publication. The project numbers are provided in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.