Abstract
The eukaryotic transcript elongation factor TFIIS enables RNA polymerase II to read through blocks to elongation in vitro and interacts genetically with a variety of components of the transcription machinery in vivo. In Saccharomyces cerevisiae, the gene encoding TFIIS (PPR2) is not essential, and disruption strains exhibit only mild phenotypes and an increased sensitivity to 6-azauracil. The nonessential nature of TFIIS encouraged the use of a synthetic lethal screen to elucidate the in vivo roles of TFIIS as well as provide more information on other factors involved in the regulation of transcript elongation. Several genes were identified that are necessary for either cell survival or robust growth when the gene encoding TFIIS has been disrupted. These include UBP3, KEX2, STT4, and SWI2/SNF2. SWI1 and SNF5 disruptions were also synthetically lethal with ppr2Δ, suggesting that the reduced ability to remodel chromatin confers the synthetic phenotype. The synthetic phenotypes show marked osmosensitivity and cytoskeletal defects, including a terminal hyperelongated bud phenotype with the Swi-Snf complex. These results suggest that genes important in osmoregulation, cell membrane synthesis and integrity, and cell division may require the Swi-Snf complex and TFIIS for efficient transcription. The detection of these genetic interactions provides another functional link between the Swi-Snf complex and the elongation machinery.
TFIIS promotes the readthrough of blocks to elongation by RNA polymerase II by first stimulating the polymerase to cleave its nascent transcript and then to read through the block (62). In addition to intrinsic blocks determined by the DNA sequence, nucleic acid binding proteins can also stall the polymerase (15, 52). In the eukaryotic nucleus, the template DNA is associated with many DNA binding proteins important in both chromosome structure and regulation of gene expression. Indeed, in vitro, chromatin severely inhibits transcript elongation, and factors that allow efficient transcription of nucleosomal templates are just being identified. These include FACT, Elongator, and HMG14, all identified in vitro (18, 55–57). In addition, Spt4, Spt5, and Spt6 have been genetically associated with chromatin and transcription (5, 31, 50, 68, 69, 77). In vitro, DSIF, a human complex with Spt4 and Spt5 homologs, also can alter transcription on pure DNA templates (73). Its activity in vitro is affected by both pTEFb and a protein complex termed NELF (79). However, TFIIS itself does not facilitate efficient transcription on chromatin templates in vitro (38).
A genetic approach was taken to learn about factors required for efficient transcript elongation and the specific roles and requirements for TFIIS in vivo. To do this, synthetic lethal genetic interactions were investigated with a deletion of PPR2, the gene encoding transcript elongation factor TFIIS from Saccharomyces cerevisiae (14, 35, 40, 41). PPR2 is not an essential gene, and the disruption of the gene confers only mild phenotypes. Both the viability of the ppr2Δ strain and the presence of a TFIIS gene family in the mouse (32, 37, 39) and human (71, 72, 74) genomes suggested that TFIIS might have more than one homolog in S. cerevisiae related by sequence or function. The complete sequencing of the yeast genome (24) revealed that PPR2 is the only full TFIIS sequence homolog present in S. cerevisiae. However, the possibility that PPR2 might have a functionally overlapping protein unrelated by primary sequence was still reasonable. The possibility of a functional homolog of PPR2 was also supported by the presence of two functionally similar bacterial proteins in Escherichia coli, GreA and GreB. GreA and GreB are functionally similar to TFIIS but are unrelated to TFIIS by sequence or structure (44, 54, 65).
Several distinct complexes capable of remodeling chromatin have been identified (reviewed in reference 42). It might be expected that such activities could participate in regulating transcript elongation. However, only for the Swi-Snf complex is there evidence that it plays a role subsequent to the establishment of preinitiation complexes at the promoter (3, 6, 67). The potential involvement of the Swi-Snf complex during elongation is supported further by results presented here demonstrating synthetic lethal genetic interactions between transcript elongation factor TFIIS and several components of the Swi-Snf complex.
MATERIALS AND METHODS
Strains, genetic methods, and media.
The S. cerevisiae strains used in this study are listed in Table 1. Strains were derived from CH1305 (45), YPH499 and YPH500 (64), W303 MATa and W303 MATα, (70), and N222 and Z321 (78). Both Escherichia coli calcium-manganese-based transformations and electroporation transformations were used (29) with E. coli strain DH10B (recA1 hsdRΔ mcrA mcrBΔ mrrΔ deoR) (25). Yeast cells were transformed using lithium acetate (23). Standard yeast methods and media were used (27).
TABLE 1.
S. cerevisiae strains used
| Strain | Genotype | Source or reference |
|---|---|---|
| CH1305a | MATa ade2 ade3 ura3 leu2 lys2 | C. Holm |
| YPH499b | MATa ade2-101oc ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 lys2-801am | P. Heiter |
| YPH500b | MATα ade2-101oc ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 lys2-801am | P. Heiter |
| W303-1ac | MATa ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 | R. Rothstein |
| W303-1bc | MATα ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 | R. Rothstein |
| Z321d | MATa/MATα ade2/ade2 ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3/leu2-3 leu2-112/leu2-112 lys2Δ201/lys2Δ201 | N. Woychik |
| DM228c | MATa ade2-1 ura3-1 leu2-33,112 his3-11 trp1-1 ubp3Δ::HIS3 | D. Moazed and A. Johnson |
| FY31e | MATα ura3-52 his3Δ200 snf2Δ1::HIS3 | F. Winston |
| YBC28e | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 snf2Δ::LEU2 | F. Winston |
| CMKy1b | MATa ade2-101oc ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 lys2-801am ppr2Δ::hisG-URA3-hisG | This work |
| CMKy2b | MATa ade2-101oc ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 lys2-801am ppr2Δ::hisG | This work |
| CMKy3a | MATa ade2 ade3 ura3 leu2 lys2 ppr2Δ::hisG-URA3-hisG | This work |
| CMKy4a | MATa ade2 ade3 ura3 leu2 lys2 ppr2Δ::hisG | This work |
| CMKy5a | MATa ade2 ade3 ura3 leu2 lys2 ppr2Δ::hisG trp1Δ::hisG-URA3-hisG | This work |
| CMKy21a | MATa ade2 ade3 ura3 leu2 lys2 ppr2Δ::hisG trp1::hisG | This work |
| CMKy22a | MATα ade2 ade3 ura3 leu2 lys2 ppr2Δ::hisG trp1::hisT | This work |
| CMKy23a | MATaMATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG ppr2Δ::hisG/ ppr2::hisG/ppr2::hisG-URA3-hisG | This work |
| CMKy24a | MATa ade2 ade3 ura3 leu2 lys2 trp1::hisG ppr2Δ::hisG-URA3-hisG | This work |
| CMKy25a | MATα ade2 ade3 ura3 leu2 lys2 trp1::hisG ppr2Δ::hisG-URA3-hisG | This work |
| CMKy26a | MATα ade2 ade3 ura3 leu2 lys2 ppr2Δ::hisG-URA3-hisG | This work |
| CMKy31a | MATa ade2 ade3 ura3 leu2 lys2 trp1::hisG-URA3-hisG | This work |
| CMKy32a | MATa ade2 ade3 ura3 leu2 lys2 trp1::hisG | This work |
| CMKy80c | MATα ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 ppr2Δ::hisG-URA3-hisG | This work |
| CMKy81c | MATα ade2-1 ura3-1 leu2-3,112 his3-11 trp1-1 ppr2Δ::hisG | This work |
| CMKy19c | MATa/MATα ade2-1/ade2-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11/his3-11 trp1-1/trp1-1 PPR2/ppr2Δ::hisG-URA3-hisG UBP3/ubp3Δ::HIS3 | This work |
| CMKy20a | MATa ade2 ade3 ura3 leu2 lys2 ubp3Δ::LYS2 | This work |
| CMKy27a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 PPR2/ppr2Δ::hisG-URA3-hisG UBP3/ubp3Δ::LYS2 | This work |
| CMKy29a | MATa ade2 ade3 ura3 leu2 lys2 ppr2Δ::hisG-URA3-hisG ubp3Δ::LYS2 | This work |
| CMKy35a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG PPR2/ppr2Δ::hisG-URA3-hisG | This work |
| CMKy36a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG PPR2/ppr2Δ::hisG-URA3-hisG STT4/stt4Δ::kanr | This work |
| CMKy38a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG PPR2/ppr2Δ::hisG-URA3-hisG SNF2/snf2Δ::kanr | This work |
| CMKy39a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG PPR2/ppr2Δ::hisG-URA3-hisG KEX2/kex2Δ::kanr | This work |
| CMKy41a | MATa ade2 ade3 ura3 leu2 lys2 snf2Δ::kanr | This work |
| CMKy42a | MATα ade2 ade3 ura3 leu2 lys2 snf2Δ::kanr | This work |
| CMKy43a | MATa ade2 ade3 ura3 leu2 lys2 trp1::hisG snf2Δ::kanr | This work |
| CMKy44a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG PPR2/ppr2Δ::hisG-URA3-hisG SNF2/snf2ΔHind::kanr | This work |
| CMKy45a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG ppr2Δ::hisG/ppr2Δ::hisG-URA3-hisG | This work |
| CMKy46a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG ppr2Δ::hisG/ppr2::hisG-URA3-hisG SNF2/snf2Δ::kanr | This work |
| CMKy47d | MATa/MATα ade2/ade2 ura3-52/ura3-52 his3-Δ200/his3-Δ200 leu2-3/leu2-3 leu2-112/leu2-112 lys2Δ201/lys2Δ201 SNF2/snf2Δ::kanr | This work |
| CMKy48a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG-URA3-hisG | This work |
| CMKy49a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG -URA3-hisG SNF2/snf2Δ::kanr | This work |
| CMKy60a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG PPR2/ppr2Δ::hisG-URA3-hisG SWI1/swi1Δ::kanr | This work |
| CMKy61a | MATa ade2 ade3 ura3 leu2 lys2 swi1Δ::kanr | This work |
| CMKy62a | MATa ade2 ade3 ura3 leu2 lys2 trp1::hisG swi1Δ::kanr | This work |
| CMKy63a | MATα ade2 ade3 ura3 leu2 lys2 swi1Δ::kanr | This work |
| CMKy64a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG PPR2/ppr2Δ::hisG-URA3-hisG SNF5/snf5Δ::kanr | This work |
| CMKy65a | MATa ade2 ade3 ura3 leu2 lys2 snf5 Δ::kanr | This work |
| CMKy66a | MATα ade2 ade3 ura3 leu2 lys2 snf5 Δ::kanr | This work |
| CMKy67a | MATa ade2 ade3 ura3 leu2 lys2 trp1::hisG snf5Δ::kanr | This work |
| CMKy73a | MATa/MATα ade2/ade2 ade3/ade3 ura3/ura3 leu2/leu2 lys2/lys2 TRP1/trp1::hisG SNF2/snf2Δ::kanrHTA1-HTB1/(hta1-htb1)Δ::LEU2 | This work |
| CMKy76b | MATa/MATα ade2-101oc/ade2-101oc ura3-52/ura3-53 leu2-Δ1/leu2-Δ1 his3-Δ200/his3-Δ200 trp1-Δ63/trp1-Δ63 lys2-801am/lys2-801am PPR2/ppr2Δ::hisG-URA3-hisG | This work |
| CMKy77b | MATa/MATα ade2-101oc/ade2-101oc ura3-52/ura3-53 leu2-Δ1/leu2-Δ1 his3-Δ200/his3-Δ200 trp1-Δ63/trp1-Δ63 lys2-801am/lys2-801am PPR2/ppr2Δ::hisG-URA3-hisG SNF2/snf2Δ::kanr | This work |
| CMKy78c | MATa/MATα ade2-1/ade2-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11/his3-11 trp1-1/trp1-1 PPR2/ppr2Δ::hisG-URA3-hisG | This work |
| CMKy79c | MATa/MATα ade2-1/ade2-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11/his3-11 trp1-1/trp1-1 PPR2/ppr2Δ::hisG-URA3-hisG SNF2/snf2Δ::kanr | This work |
CH1305 background.
YPH499 background.
W303 background.
Z321 background.
S288C background.
For sporulation medium, the auxotrophic requirements of the diploids were supplemented by adding the appropriate amino acids at 25% of the concentration used for synthetic complete (SC) medium. The 5-fluoroorotic acid plates were made as previously described (4). Sucrose and raffinose plates included 2% of the appropriate sugar added to SC medium without dextrose. YPGal was made as described for YPD, with 2% galactose instead of dextrose. G418 (Gibco-BRL) medium (YPD with 200 μg of G418 per ml) was made as previously described (26). Sorbitol medium contained 1 M sorbitol in YPD.
Medium lacking inositol was made as described (63). 6-Azauracil (Aldrich) was added to SC-Ura medium from a 5-mg/ml stock solution dissolved in water to final concentrations of 25, 50, 75, or 100 μg of 6-azauracil per ml. Mycophenolic acid (Sigma) medium was made by adding an appropriate amount of a 5-mg/ml stock dissolved in methanol to 10, 25, 50, or 100 μg/ml in SC medium.
Plasmids.
All plasmids used and their cloning strategies are listed in Table 2. The plasmid for the synthetic lethal screen was based on pDS1, derived from pDK221 (43) by replacement of the LEU2 gene with Ampr. Vent polymerase (NEB) was used for all PCR-generated subclones.
TABLE 2.
Plasmids used in this study
| Plasmid | Construction | Reference(s) |
|---|---|---|
| pJD3 | XbaI (blunted)-EcoRI hisG-URA3-hisG fragment from pUC19 inserted into MunI (blunted)- and EcoRI-digested pKC8 | 12 |
| pJD4 | BamHI genomic fragment of PPR2 inserted into BamHI site of pDS1 | 43 |
| pJD5 | XhoI-NsiI PPR2 fragment from pKC3 inserted into XhoI-PstI-digested pRS315 | 12, 64 |
| pLPUBP3 | Complementing library plasmid for synthetic lethal mutant 70B | |
| pLPKEX2 | Complementing library plasmid for synthetic lethal mutant 44B | |
| pLPSTT4 | Complementing library plasmid for synthetic lethal mutant 56B | |
| pLPSNF2 | Complementing library plasmid for synthetic lethal mutant 28A | |
| pJD7 | 4.2-kb HindIII fragment from pLPUBP3 inserted into HindIII site of pRS315 | |
| pJD8 | 3.7-kb HindIII fragment from pLPUBP3 inserted into HindIII site of pRS315 | |
| pJD9 | PCR-generated BamHI-XbaI fragment (using pLPUBP3 as template) inserted into BamHI-XbaI-digested pRS315 | |
| pJD10 | PCR-generated BamHI-PstI fragment (using pLPUBP3 as template) inserted into BamHI-PstI-digested pRS315 | |
| pJD11 | PCR-generated SpeI-PstI fragment (using pLPUBP3 as template) inserted into SpeI-PstI-digested pRS315 | |
| pJD12 | PCR-generated SpeI-SacII fragment (using pLPUBP3 as template) inserted into SpeI-SacII-digested pRS315 | |
| pJD17 | PCR-generated BamHI-PstI fragment (using pLPSTT4 as template) inserted into BamHI-PstI-digested pRS315 | |
| pJD19 | NcoI digest and religation of pLPSNF2 (disrupts SNF2 open reading frame) | |
| pJD27 | PPR2-containing PvuII fragment from pKC(1–309) inserted into PvuII-digested pRS315 | 12 |
| pJD28 | PPR2-containing PvuII fragment from pKC(131–309) inserted into PvuII-digested pRS315 | 12 |
Yeast whole-colony PCR.
A procedure for direct whole-colony PCR was modified for this work (36). The reaction buffer contained 12.5 μl of 10 mM Tris-HCl (pH 8.3)–50 mM KCl–2.5 mM MgCl2–170 μg of bovine serum albumin (BSA) per ml–200 μM each of the four deoxynucleoside triphosphates–1.0 μM each primer–50 U of AmpliTaq (Perkin-Elmer) per ml. A yeast colony was added to a 12.5-μl reaction, and the reaction mix was overlaid with mineral oil. The PCR was initiated with a 5-min 92°C denaturation step, followed by 35 cycles of 92°C for 1 min, primer annealing at the appropriate temperature for 2 min, and 72°C for 2 min. All primer annealing temperatures were above 50°C.
Synthetic lethal screen.
The red-white colony sectoring assay has been described previously (43) and subsequently applied to a synthetic lethal approach (45). An ADE3 plasmid (confers a red color when placed in an ade2 ade3 background) containing a nonessential gene is lost at a high frequency in the absence of selective pressure due to a mutant ARS/CEN sequence, leading to the appearance of sectored colonies. An ade2 ade3 ppr2Δ strain (CMKy4) was transformed with this ADE3 plasmid also containing PPR2 and URA3 (pJD4).
Ten independent cultures were grown in SC-Ura medium overnight to an optical density at 600 nm (OD600) of approximately 1.0. Plating cell density was 10,000 per 150-mm 4% YPD plate, in which the 4% glucose enhanced the appearance of sectors. The plates were irradiated with 65 J of UV light, providing a viability after irradiation of ∼15%. Plates were incubated for 5 days at 30°C. The initial 24 h of incubation were in the absence of light to avoid light-induced repair. Individual solid red colonies were selected and streaked on a new 4% YPD plate. Only colonies that produced >95% nonsectored colonies on the second plate were studied further.
Plasmid dependence of the nonsectoring phenotype was confirmed by growth on SC-Ura medium. Dominant and recessive mutations were identified by mating each mutant to CMKy22. If the mutation was recessive, the diploid regained the ability to sector. Dominant mutations or integration of the ADE3 URA3 PPR2 plasmid into the genome produced a nonsectoring phenotype in the diploid. To determine if the synthetic lethality was due to PPR2 or one of the other genes on the plasmid, a plasmid with PPR2 and LEU2, pJD5, was shuffled into each mutant, replacing pJD4. A growth requirement for leucine and a sectoring phenotype on YPD indicated that only PPR2 was required, as the ADE3-containing plasmid could now be lost. To determine if a single mutation was responsible, 12 tetrads were dissected for each mutant following mating to CMKy22 to determine if the sectoring phenotype segregated as one gene. The tetrads were also used to determine if any growth defect associated with the synthetic lethal mutation segregated with the sectoring phenotype.
Subsequent to these procedures, 49 recessive mutations were recovered and placed into 10 complementation groups. The rapid appearance of apparent suppressors was noted for 28 of the mutants.
Cloning of synthetic lethal genes.
A LEU2 CEN/ARS genomic library (22) was used for the complementation screens. High-efficiency transformation conditions were optimized for each mutant. Between 5,000 and 10,000 transformants were screened for each mutant to ensure genomic coverage, and plasmids were recovered from transformants that sectored reproducibly. Each plasmid was used to transform the original mutant to confirm that it restored sectoring. Sequencing identified the genomic fragment responsible for restoring sectoring, and the specific gene was isolated after subclones of each library plasmid were tested.
Gene disruptions.
Disruptions of PPR2 were made by replacing codons 40 to 284 of PPR2 with the hisG-URA3-hisG cassette (2). The disruption was confirmed by Southern blot analysis on 6-azauracil-sensitive transformants. These strains were plated onto 5-fluoroorotic acid medium to select for Ura− recombinants (2).
The TRP1 disruption was made by inserting the hisG-URA3-hisG cassette into the middle of the TRP1 gene using pNKY1009 (2).
A PCR strategy was used to disrupt the UBP3 gene in haploid CH1305. PCR primers were designed that contained 45 to 50 bp of UBP3 flanking sequence on the 3′ end and an 18-bp sequence that amplified LYS2 on the 5′ end. Transformants were selected on SC-Lys medium and screened by PCR analysis to confirm the disruption of UBP3.
The complete disruptions of stt4Δ, kex2Δ, snf2Δ, swi1Δ, and snf5Δ were made individually in a ppr2Δ/PPR2+ diploid strain (CMKy35) by replacing one copy of the entire target gene with the kanr gene (26). A snf2Δ mutation was also made in CMKy48 (PPR2/PPR2). Transformants were selected on G418 plates and replica plated to new G418 plates after 48 h. Correct integrants were identified by PCR analysis. PCR primers containing ∼45 bp of homology to the target gene on the 3′ end and 18 bp to the kanr gene on the 5′ end generated a fragment containing the kanr gene flanked by the upstream and downstream regions of the target gene.
The snf2Δ mutation in CMKy45 (ppr2Δ::URA3/ppr2Δ::hisG) was constructed by kanr cassette insertion and confirmed as described above. This heterozygous snf2Δ mutation resulted in a strain with severe morphological defects noted after several days. Thus, the transformants, once confirmed for the disruption, were immediately stored at −80°C in 15% glycerol–1 M sorbitol–YPD medium. The snf2Δ alleles were created in the heterozygous PPR2/ppr2Δ::URA3 diploid strains CMKy76 (YPH499) and CMKy78 (W303) by replacing the entire SNF2 gene with the kanr gene as in CH1305.
The snf2ΔHind mutation was made by replacing only the 500-bp HindIII fragment within SNF2 (1) with kanr in CMKy35 and was confirmed by PCR analysis.
The hta1/htb1Δ::LEU2 allele was made as previously described (33). The locus was disrupted in the diploid CMKy73, and upon sporulation, there was 2:2 survival. The viable spores were leucine auxotrophs. Several attempts to recover disruptions in the haploid synthetic lethal mutant 28A were not successful.
Allele rescue.
The synthetic mutant allele of SWI2-SNF2 was recovered by PCR (Vent polymerase [NEB]). The promoter region (480 bp) and the terminator region (360 bp) were also recovered. Due to the large size of SNF2 (5.1 kb), the mutant allele was recovered in three PCR fragments, each of which was subcloned into pRS314 and sequenced.
Linkage analysis for UBP3, KEX2, and STT4.
Mutant 70B [MATa ura3 leu2 lys2 ppr2Δ::his G (pJD4)] and a MATα ura3 leu2 lys2 ubp3Δ::LYS2 strain (derived from CMKy27) were mated, and Ura+ Lys+ diploids were selected. The diploid was sporulated, and 10 complete tetrads were examined. Growth on lysine was used to identify the ubp3Δ segregants. As the ppr2Δ allele did not contain an auxotrophic gene, the ppr2Δ allele was determined by PCR analysis on each segregant. Red segregants carried pJD4. As we have observed previously, pJD4 segregated very poorly, presumably due to the mutant CEN/ARS.
The mutant 44B [MATa ura3 leu2 lys2 ppr2Δ::hisG(pJD4)] was mated to a MATα ura3 leu2 lys2 kex2Δ::Kanr(pKEX2) strain (derived from CMKy39). Plasmid pKEX2 is a LEU2 library plasmid that contained KEX2. Ura+ Leu+ diploids were selected. The presence of both PPR2 and KEX2 on plasmids would complicate the analysis, so the diploid was grown in YPD for several generations and plated. A Ura− Leu− diploid was sporulated, and 10 complete tetrads were analyzed. The ppr2Δ allele was determined by PCR analysis. Kanr was used to identify the kex2Δ allele. The presence of pJD4 was indicated by a red color.
The mutant [56B MATa ura3 leu2 lys2 ppr2Δ::hisG(pJD4)] was mated to CMKy22 (MATα ura3 leu2 lys2 ppr2Δ::hisG trp1::hisG). Zygotes were selected, and diploids were confirmed by the inability to mate to tester strains. A diploid was transformed with a PCR-derived cassette designed to disrupt STT4 (same strategy presented above). PCR analysis was used to identify five diploids heterozygous for the stt4Δ mutation. The diploids were sporulated, and 10 complete tetrads were analyzed for each.
Recovery of swi-snf strains.
The initial work with the snf2Δ mutation in CH1305 showed poor growth and poor viability of spores. A modified method for strain recovery was used in all the tetrad analyses of swi-snf strains in order to unambiguously identify the genotype present at each spore position. Fresh, moist YPD plates were used for tetrad dissection. Plates were warmed to room temperature before use. After tetrads were dissected, each plate (plate A) was grown for 7 days and microscopically examined to determine the approximate number of cells at each spore position. Small colonies were streaked in patches on a fresh YPD plate (plate B). Plate A was then replica plated to both SC-Ura and SC-Trp media. Plate B was grown for 7 days. All viable patches were transferred to separate G418, SC-Ura, and SC-Trp plates.
Microscopic characterization of phenotypes.
Yeast cells were resuspended in H2O to a concentration of approximately 7 × 107 cells/ml, and cellular morphology was observed on a Zeiss photomicroscope equipped with Nomarski differential interference contrast optics (Carl Zeiss, Thornwood, N.Y.). To monitor nuclear morphology, yeast cells were harvested from an overnight culture, rinsed in cold methanol for 10 min to fix the cells, and resuspended in 1× phosphate-buffered saline. Cells were stained with 30 ng of 4′,6′-diamidine-2-phenylindole dihydrochloride (DAPI) per ml for 5 min. Stained cells (5 μl) were applied to a glass slide and examined using a Zeiss Axioskop fluorescence microscope and a 100× Neoflor objective (Carl Zeiss).
RESULTS
Rationale of screen.
S. cerevisiae contains a single copy of the gene encoding TFIIS (PPR2), and gene disruption strains have only modest phenotypes (76). Thus, to investigate the function of TFIIS in vivo, a synthetic lethal screen was initiated. Synthetic lethal interactions might uncover proteins whose function overlaps that of TFIIS or proteins that share regulatory or functional roles with TFIIS.
Results of synthetic lethal screen.
The synthetic lethal screen utilized the color phenotypes associated with the adenine biosynthetic pathway (43, 45). Approximately 75,000 mutagenized colonies were screened for a nonsectoring phenotype, and 187 mutants were identified that reproducibly produced nonsectoring colonies. Of these, 185 mutants were plasmid dependent. Eighty-nine mutants were recessive, and the survival of 55 of these depended upon the presence of PPR2. Of these, 49 mutants showed 2:2 segregation of the nonsectoring phenotype, indicating that each mutant resided in either one gene or a set of closely linked genes. Ten complementation groups were defined among these 49 mutants, and all were tested for growth on sucrose, raffinose, 6-azauracil, and medium lacking inositol and for growth at 37°C. The relevant phenotypes of the mutants are presented below.
A CEN/ARS genomic library was used to identify the genes that complemented the nonsectoring phenotype for each of the 10 complementation groups. Inserts from library plasmids that restored sectoring were end sequenced and identified from the genomic database. As the genomic library fragments were approximately 10 kb in length, subclones were used to define the gene that restored sectoring (Table 3). Four of the 10 complementation groups were analyzed in this way. For one complementation group, SNF2 was identified (Table 3), and allele rescue of the mutant locus allowed identification of the specific sequence change that led to the synthetic lethal phenotype. For the other three complementation groups (Table 4), phenotypic analysis suggested complementation rather than suppression of a mutation in a separate locus. To confirm complementation, each synthetic lethal mutant strain was crossed to a strain in which the suspected complementing gene had been disrupted (as described in Materials and Methods). Allelism was confirmed by the 2:2 segregation of synthetic lethal phenotypes following sporulation and tetrad dissection.
TABLE 3.
Genes that rescue the synthetic mutant phenotype
| Synthetic lethal mutant | Gene(s) on recovered library plasmid | Plasmid | Rescue |
|---|---|---|---|
| 70B | SPT15 | pJD11 | − |
| PEA2 | pJD12 | − | |
| SPI1 | pJD7 | − | |
| UBP3 | pJD10 | + | |
| YER152 | pJD8 | − | |
| PET122 | pJD9 | − | |
| 44B | KEX2 | pLPKEX2 | + |
| 56B | STT4 | pLPSTT4a | + |
| UBC12 | pJD17 | − | |
| 28A | SNF2 | pLPSNF2a | + |
| YOR289 | pJD19 | − |
In these cases, the library plasmid contained both genes listed, but the alternate gene was unable to overcome the synthetic phenotype when tested alone.
TABLE 4.
Genes identified in synthetic lethal phenotype screen
| Mutation | Phenotype(s)
|
||
|---|---|---|---|
| UBP3 | KEX2 | STT4 | |
| Synthetic lethalitya | Slow growth | α-Specific sterile; higher permissive temperature than wild type | Temperature sensitive at 37°C, partially rescued by sorbitol; cannot grow on raffinose, sucrose, or galactose; 6-azauracil stimulates growth |
| Gene disruptiona | Slow growth (ubp3Δ) | α-Specific sterile; higher permissive temperature than wild type (kex2Δ) | Lethal (stt4Δ); conditional stt4 mutants are temperature sensitive and rescued by sorbitol |
| Double disruption | Very slow growth (ppr2Δ ubp3Δ) | Extremely sick (<18% viabilityb) in vegetatively growing cells (ppr2Δ kex2Δ) | |
Mutant characterized in a PPR2+ background.
Viability assessed by comparing cell count in liquid culture to the number of cells that could form colonies when plated.
The synthetic lethal mutant 70B was rescued by UBP3, encoding a ubiquitin protease. The ubp3Δ strain has a mild growth defect, and the ubp3Δ ppr2Δ strain, while viable, has a more severe growth defect (Table 4). The synthetic lethal phenotype of 70B segregated 2:2 with the severe growth defect of the ubp3Δ ppr2Δ strain. While confirming allelism or very tight linkage with UBP3, this result also indicated that the synthetic lethality of mutation 70B in UBP3 with ppr2Δ was allele specific.
The synthetic lethal mutant 44B was rescued by KEX2, encoding a prohormone protease located in the Golgi. Mutant 44B also exhibited phenotypes characteristic of kex2Δ strains: slow growth, α-specific sterility, and altered temperature tolerance (51). As both the synthetic lethal mutant 44B and the kex2Δ strain were α sterile, the MATa synthetic lethal mutant was mated to a MATα kex2Δ strain transformed with a KEX2-containing library plasmid. To simplify the analysis, a diploid which had lost both the library plasmid (KEX2 LEU2) and pJD4 (PPR2 ADE3 URA3) was used. Following sporulation and tetrad dissection, no segregants showed wild-type growth and all α segregants were sterile, as would be expected if the synthetic lethal mutation and kex2Δ were allelic. Moreover, both the kex2Δ (Kanr) and synthetic lethal allele (KanS) strains that were ppr2Δ were extremely sick on YPD medium, an observation consistent with the very low viability observed in liquid culture with the kex2Δ ppr2Δ strain. These results indicate an allelism or very tight linkage between the mutation in 44B and KEX2 and further suggest that mutant 44B contains a loss-of-function mutation in KEX2.
The synthetic lethal mutant 56B was rescued by STT4, encoding a phosphoinositol-4-kinase. A deletion of STT4 is lethal in the CH1305 strain background, as disruptions of stt4Δ could not be recovered in haploids under several conditions tested (data not shown). Thus, mutant 56B could not have suffered a total loss of function in STT4. However, mutant 56B does show phenotypes expected for conditional alleles of stt4, including temperature sensitivity and rescue by sorbitol (80) (Table 4). Synthetic lethal mutant 56B was mated to an STT4 ppr2Δ strain, and the resulting diploid was disrupted for STT4. Five diploids heterozygous for the stt4Δ mutation were identified. Each was sporulated, and tetrads were analyzed for each of the five. In this analysis, the STT4 disruption could occur in the diploid at either the wild-type or the mutant 56B locus (assuming allelism) on the complementary chromosomes. Any stt4Δ haploids themselves would be inviable. Of the five sporulation plates, three showed the 2:2 segregation for viability expected if the disruption had occurred in the STT4 locus and had replaced mutation 56B, leaving the wild-type copy of STT4. The other two stt4Δ heterozygotes had significantly reduced overall spore germination. In one case, only 4 of 40 spore positions were viable, and only 6 of 40 were viable in the second case. Each of these viable strains had a severe growth defect and appeared red (nonsectoring). The low viability is not surprising, as previous work with mutant 56B showed extremely poor spore viability associated with the synthetic lethal allele. These combined patterns of segregation suggest that mutant 56B represents a partial function allele of STT4.
Characterization of the interaction with SNF2.
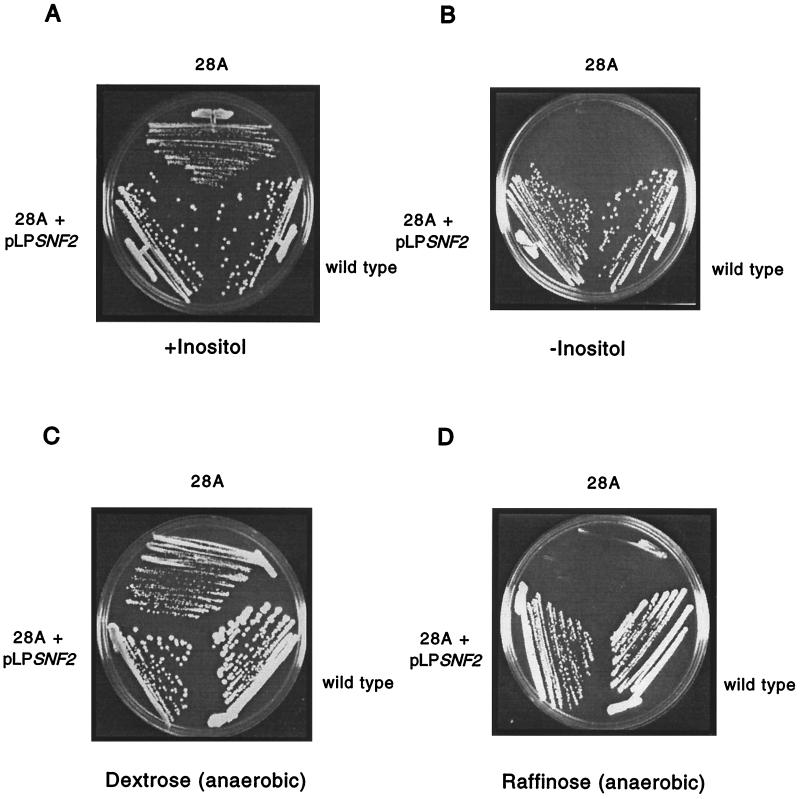
SWI2/SNF2 encodes a component of the Swi-Snf chromatin-remodeling complex and is the founding member of a gene family associated with chromosome structure changes (42). SWI2/SNF2 is not an essential gene (1), but the null mutant is characterized by a number of phenotypes, all shared by the mutant allele isolated in this screen (Fig. 1). For simplicity, SWI2/SNF2 will be referred to as SNF2. The snf2Δ strain cannot grow on medium containing raffinose, sucrose, and galactose under anaerobic conditions, and this mutant is also an inositol auxotroph. The raffinose sensitivity of the synthetic lethal mutant was complemented by cloned SNF2 (Fig. 1), strongly indicating that the mutation lay in SNF2.
FIG. 1.
Sensitive growth of synthetic lethal mutant 28A. (A) The synthetic lethal mutant 28A, a wild-type strain (CH1305), and mutant 28A transformed with the SNF2-containing library plasmid pLPSNF2 were streaked onto medium with inositol added. (B) The three strains were streaked onto medium without inositol. The plates were photographed after 4 days of growth at 30°C. (C) Wild-type (CH1305), mutant 28A, and mutant 28A transformed with the SNF2-containing library plasmid pLPSNF2 were streaked on dextrose medium. The plate was photographed after 4 days of anaerobic growth at 30°C. (D) The same strains were streaked onto raffinose medium and grown anaerobically for 4 days at 30°C prior to photography.
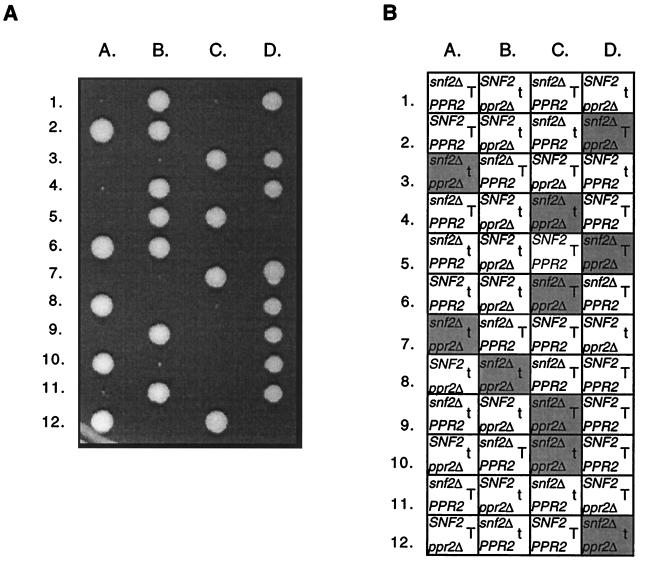
A disruption of SNF2 was made in a heterozygous ppr2Δ/PPR2 diploid, CMKy35. The resulting snf2Δ/SNF2 ppr2Δ/PPR2 diploid, CMKy38, was sporulated, and 12 tetrads were dissected. In each of the tetrads, two spores gave rise to healthy colonies while the other two spores were either inviable or gave rise to poorly growing colonies of variable size. As the viability of the snf2Δ strains was problematic in this strain background, plating modifications were necessary to improve germination and strain recovery following germination (see Materials and Methods). Subsequently, over 175 tetrads were analyzed from the CMKy38 diploid strain to permit a thorough analysis of the genotypes of every spore position and the number of cells resulting from each germination event. The results clearly demonstrate that an snf2Δ mutation is synthetically lethal in combination with a ppr2Δ mutation (Fig. 2).
FIG. 2.
A snf2Δ allele is synthetically lethal in combination with a ppr2Δ allele. (A) Tetrad analysis following sporulation of an snf2Δ::kanr/SNF2 ppr2Δ::URA3/PPR2 diploid (CMKy38). The tetrads were dissected on YPD medium, and the YPD plate was photographed after 7 days of growth at 30°C. (B) The snf2Δ::kanr spores were identified by their G418r phenotype, the ppr2Δ::URA3 spores were identified by their Ura+ phenotype, and TRP1 spores were identified by their Trp+ phenotype. The positions of inviable spores are shaded gray, the box containing the genotype deduced from the other segregants. The presence of the TRP1+ gene and the trp1 mutation indicated with a T and with a t, respectively.
Robust colonies occasionally arose in attempts to culture the double-disruption haploid. These strains were both Ura+ and G418r, indicating that the strain still contained the URA3 and kanr genes that had replaced PPR2 and SNF2, respectively. These double-deletion mutants that did grow were heterogeneous in size between the individual strains, but the large majority were very healthy, much more so than the snf2Δ strain itself. The low frequency with which they were identified (10−5) and the robust growth suggested that these were suppressor mutants. Interestingly, these putative suppressors obviate swi/snf mutant phenotypes, such as reduced growth on raffinose- and inositol-free medium, but do not appear to suppress the 6-azauracil sensitivity of ppr2Δ strains.
The entire 5.1-kb allele of SNF2 was rescued from the synthetic lethal mutant. Sequence analysis identified a nonsense mutation at nucleotide position 3385 that would be expected to produce a protein truncated in the ATPase domain. The genetic connection between PPR2 and SNF2 was examined in detail to explore the role of the Swi-Snf complex in transcript elongation and the possible role of TFIIS on a nucleosomal template. However, complex interactions with auxotrophic markers were examined first.
Early work with SWI/SNF had suggested that leucine auxotrophy would contribute to the phenotypic severity detected in swi/snf mutant strains (66). In that report, swi2/snf2 leu2 double mutant strains were found to require 300 μg of leucine per liter, 10 times the standard concentration (30 μg/ml). To allow comparisons of the interactions between swi2/snf2 and ppr2Δ directly, all characterizations were done in medium containing 300 μg of leucine per liter. Additionally, the observed effects were essentially identical in leu2 and LEU2+ backgrounds.
The TRP1+ genotype also contributed to the severity of phenotypes observed with snf2Δ strains. This effect is further described below, but the synthetic interactions between snf2 and trp1 initially complicated phenotypic interpretations with ppr2Δ. To avoid this, the interactions between ppr2Δ and snf2Δ were examined in TRP1+ strains. In CH1305, snf2Δ strains are much healthier in the presence of TRP1 on both rich medium (YPD) and minimal medium supplemented with tryptophan.
Indeed, suppressors of the double-deletion mutant only appeared when the strain contained TRP1+. In a trp1 background, the terminal cell count of the double-deletion mutant was between 20 and 100 cells after 1 week of incubation. In the presence of TRP1, the double-deletion mutant forms a small colony of approximately 100 to 1,000 cells after 1 week of incubation (Table 5). The explanation of this effect is not known, but it also extends to TRP5 (data not shown). As a result of these observations, the interactions between SNF2 and PPR2 were examined in a TRP+ background.
TABLE 5.
Comparison of the snf2Δ ppr2Δ interaction in three genetic backgrounds
| Strain | Growth of snf2Δ strainsa | Terminal cell count of snf2Δ ppr2Δ segregants |
|---|---|---|
| CH1305 | ||
| TRP1+ | + | 100–1,000 |
| trp1 | ± | 10–100 |
| W303 | + | 500–1,000 |
| YPH499 | ± | 2–5 |
+, 1-mm colonies or healthy growth in a lawn of cells after 7 days of incubation; ±, 0.1-mm colonies or light growth in a lawn of cells after 7 days of incubation.
Phenotypes of snf2Δ ppr2Δ cells.
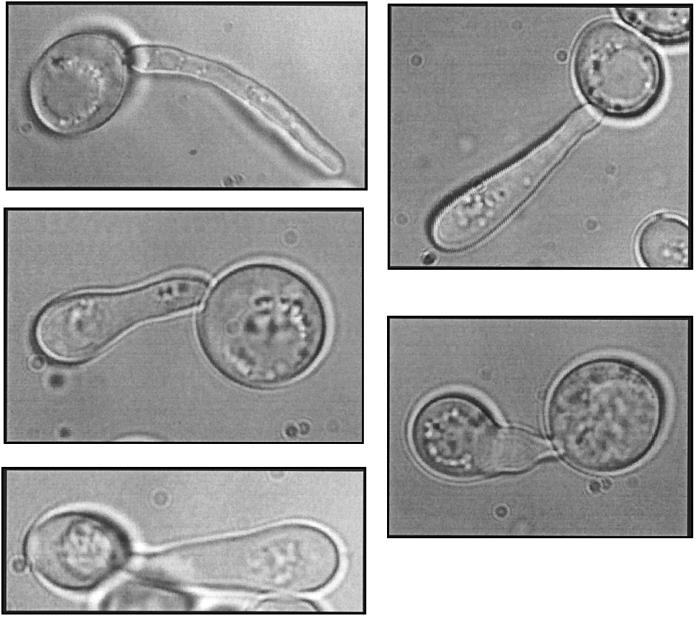
The haploid snf2Δ ppr2Δ double disruption was inviable, but the phenotypes of the SNF2/snf2Δ ppr2Δ/ppr2Δ heterozygous diploid CMKy46 were informative. This diploid strain had several unusual phenotypes, including a high frequency of lysis on rich (YPD) medium and complete lysis in medium of higher osmotic strength, such as sporulation medium. When the cells are examined microscopically, approximately 45% of the cells are characterized by highly elongated bud-like structures (Fig. 3). Intriguingly, highly elongated buds are also observed with some alleles of sth1, an essential SNF2-related component of the RSC chromatin remodeling complex (19). DAPI staining of the budded diploid CMKy46 demonstrated that the elongated bud structure contained a single nucleus. This nucleus was not located exclusively at the mother bud neck. A localization at the mother bud neck would have suggested a delay in nuclear division. The addition of 1 M sorbitol suppressed the hyperelongated bud phenotype and also reduced the doubling time of the diploid. Curiously, this diploid sporulates after 2 days of growth on YPD medium. Rich YPD medium contains both nitrogen and glucose, each of which normally represses the sporulation pathway. Thus, the SNF2/snf2Δ ppr2Δ/ppr2Δ diploid is overcoming both of these repressive sporulation signals, indicating a major deregulation within the cell.
FIG. 3.
Hyperelongated bud phenotype. The snf2Δ/SNF2 ppr2Δ/ppr2Δ diploid (CMKy46) under 100× magnification. Cells were visualized by differential interference contrast microscopy (see Materials and Methods). Approximately 45% of cells had this morphology.
Strain background affects the genetic interaction between PPR2 and SNF2.
The growth defect of the CH1305 snf2Δ strain was more severe than that reported for either an S288C snf2Δ::HIS3 strain (1) or an S288C snf2Δ::LEU2 strain (9). Furthermore, the ppr2Δ and snf2Δ alleles show no genetic interaction in S288C (G. Hartzog and F. Winston, personal communication). The disruptions of SNF2 that had been made in S288C differed in both the size and content of the disruption compared to those made in CH1305. The snf2Δ disruption in S288C had replaced 500 bp of SNF2 with HIS3 (1). The disruption in CH1305 replaced the entire open reading frame of SNF2 with the kanr gene. Thus, the difference between the PPR2 and SNF2 genetic interaction in S288C and CH1305 could have been due to the nature of the disruptions. To test this, the 500-bp region of SNF2 originally replaced in S288C by HIS3 was replaced with the kanr gene in CH1305. This CH1305-derived disruption strain (snf2ΔHind) had the same growth defects as the CH1305 strain with the complete kanr replacement of SNF2 (data not shown). This disruption was also synthetically lethal with ppr2Δ. These results suggest that the strain background, and not the size or location of the disruption, is the basis for the difference in both the growth defects and the genetic interaction between the ppr2 and snf2 alleles in S288C and CH1305.
Next, snf2Δ disruptions were constructed in three additional genetic backgrounds: Z321, a laboratory strain utilized by R. Young and N. Woychik (78); W303, a laboratory strain from R. Rothstein (70); and YPH499, a laboratory strain originally derived from S288C (64). The complete genotypes of the strains are listed in Table 1. In each case, a diploid strain was used to make the heterozygous SNF2/snf2Δ::kanr disruption. For Z321, the diploid was PPR2/PPR2. In both W303 and YPH499, a heterozygous PPR2/ppr2Δ::URA3 diploid was used. All three snf2Δ haploid strains derived from these diploids grew very poorly, although there was subtle variation among the three backgrounds in the severity of the growth defect. For YPH499, the strain originally derived from S288C, the snf2Δ strains were the least healthy of all strains tested. Incubation for 1 week was required to observe any visible growth. In Z321 and W303, the snf2Δ strain had a severe growth defect, but each was much more robust than the YPH499 snf2Δ strain.
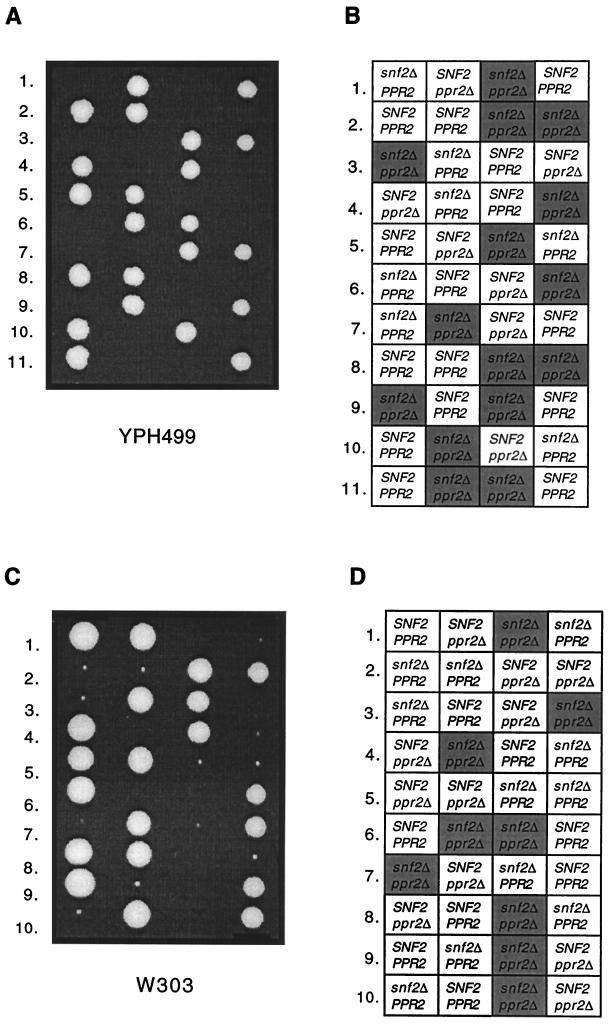
Strains W303 and YPH499/500 were used to examine the genetic interaction between ppr2Δ and snf2Δ across strain backgrounds. The tetrads derived from both W303 and YPH499/500 SNF2/snf2Δ PPR2/ppr2Δ heterozygous diploids showed a clear synthetic lethal interaction between ppr2Δ and snf2Δ (Fig. 4; results summarized in Table 5). The severity of the growth defect caused by an snf2Δ mutation alone affected the terminal cell count of either double deletion mutant. That is, the growth defect of a snf2Δ strain was similar in W303 and CH1305, and the terminal cell counts of the snf2Δ ppr2Δ cells in each background were similar. The double-deletion mutants of W303 grew to approximately 500 to 1,000 cells after 1 week, and suppressors could be recovered, as was observed with CH1305. In contrast, the YPH499/500 background gave snf2Δ strains that grew extremely slowly, and the terminal cell count of the snf2Δ ppr2Δ segregants was only two to five cells.
FIG. 4.
A snf2Δ allele is synthetically lethal with a ppr2Δ allele in two unrelated genetic backgrounds. (A) Tetrad analysis following sporulation of a snf2Δ::kanr/SNF2 ppr2Δ::URA3/PPR2 diploid derived from YPH499/500 (CMKy77). The tetrads were dissected on YPD medium, and the YPD plate was photographed after 7 days of growth at 30°C. (B) The snf2Δ::kanr spores were identified by their G418r phenotype, and ppr2Δ::URA3 spores were identified by their Ura+ phenotype. Inviable spore positions are marked with a shaded box containing the genotype deduced from the other segregants. (C) Tetrad analysis following sporulation of an snf2Δ::kanr/SNF2 ppr2Δ::URA3/PPR2 diploid derived from W303 (CMKy79). The tetrads were dissected on YPD medium, and the YPD plate was photographed after 7 days of growth at 30°C. (D) The snf2Δ::kanr spores were identified by their G418r phenotype, and ppr2Δ::URA3 spores were identified by their Ura+ phenotype. Inviable spore positions are marked with a shaded box containing the genotype deduced from the other segregants.
In S288C, snf2Δ phenotypes can be suppressed by deletion of one of the two gene pairs encoding H2A and H2B (HTA1 and HTB1) (33). Thus, it was of interest to test whether a similar disruption could suppress the synthetic lethality between snf2Δ and ppr2Δ in CH1305. However, in contrast to S288C, HTA1-HTB1 is apparently essential in segregants derived from either a heterozygous snf2Δ/SNF2 ppr2Δ/PPR2 HTA1-HTB1/(hta1-htb1)Δ diploid (CMKy73) or the synthetic lethal snf2 mutant itself, both in the CH1305 background. The essential nature of the HTA1-HTB1 allele has also been observed in the W303 genetic background (P. Kaufman, personal communication).
To determine if the difference between the S288C snf2Δ strain and the CH1305 snf2Δ strain was caused by one gene, the two snf2Δ strains were mated. However, the homozygous snf2Δ/snf2Δ diploid was unable to sporulate. This inability of homozygous snf2Δ diploids to sporulate has been observed previously (66). This strain difference is being further pursued to understand if the differences can be revealing about transcriptional control affected by chromosome structural changes.
Synthetic lethality with other components of the SWI/SNF complex.
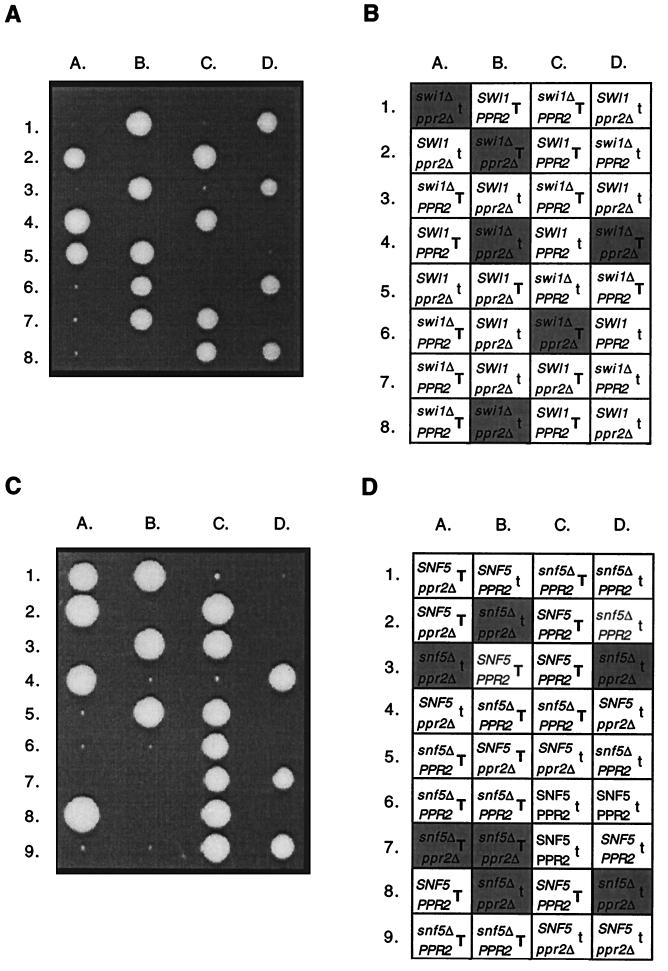
To determine if the synthetic lethal effects could be extended to additional SWI/SNF genes, SWI1 and SNF5 were tested for genetic interactions with PPR2. These genes were selected as two well-characterized components of the Swi-Snf complex, each required for its activity. A total of 68 tetrads were examined following sporulation of a heterozygous swi1Δ/SWI1 ppr2Δ/PPR2 diploid, CMKy60, and synthetic lethality was clearly observed (Fig. 5). The results were very similar to those obtained with an snf2Δ mutation. The TRP1 locus affected both the growth of swi1Δ strains and the terminal cell count of the swi1Δ ppr2Δ strains. However, the terminal cell count of swi1Δ ppr2Δ strains was higher than observed for the snf2Δ ppr2Δ strains. The swi1Δ ppr2Δ strains formed a small colony of approximately 500 to 1,000 cells. Apparent suppressors of the synthetic lethality again accumulated with a frequency estimated to be 10−5.
FIG. 5.
Both swi1Δ and snf5Δ alleles are synthetically lethal in combination with a ppr2Δ allele. (A) Tetrad analysis following sporulation of an swi1Δ::kanr/SWI1 ppr2Δ::URA3/PPR2 diploid (CMKy60). The tetrads were dissected on YPD medium, and the YPD plate was photographed after 7 days of growth at 30°C. (B) The swi1Δ::kanr spores were identified by their G418r phenotype, the ppr2Δ::URA3 spores were identified by their Ura+ phenotype, and TRP1 spores were identified by their Trp+ phenotype. Inviable spore positions are marked with a shaded box containing the genotype deduced from the other segregants. The presence of the TRP1+ gene is indicated with a T, and that of the trp1 mutation is indicated with a t. (C) Tetrad analysis following sporulation of an snf5Δ::kanr/SNF5 ppr2Δ::URA3/PPR2 diploid (CMKy64). The tetrads were dissected on YPD medium, and the YPD plate was photographed after 8 days of growth at 30°C. (D) The snf5Δ::kanr spores were identified by their G418r phenotype, the ppr2Δ::URA3 spores were identified by their Ura+ phenotype, and TRP1 spores were identified by their Trp+ phenotype. Inviable spore positions are marked with a shaded box containing the genotype deduced from the other segregants. The presence of the TRP1+ gene is indicated with a T, and that of the trp1 allele is indicated with a t.
Seventy-four tetrads were examined following sporulation of an snf5Δ/SNF5 ppr2Δ/PPR2 diploid, CMKy64. Synthetic lethality was also observed between null alleles of SNF5 and PPR2 (Fig. 5). Again, TRP1 affected the growth of the snf5Δ strain and the terminal cell count of the snf5Δ ppr2Δ strain. With TRP1, the snf5Δ ppr2Δ strain grew to 100 to 1,000 cells. Suppressors appeared at approximately the same frequency as for the snf2Δ ppr2Δ and swi1Δ ppr2Δ strains.
The synthetic lethality for all three tested components of the Swi-Snf complex was complemented by expression of PPR2 in the double-disruption strain. The carboxy-terminal half of the TFIIS protein was sufficient for functional complementation; this portion of the protein is sufficient to complement both the 6-azauracil sensitivity of ppr2Δ strains and the in vitro activities of TFIIS for cleavage and readthrough by RNA polymerase II (12, 13, 53; N. Shimasaki and C. M. Kane, submitted for publication).
Lack of common phenotypes.
To further investigate the synthetic lethal interaction between null alleles of PPR2 and SWI/SNF, ppr2Δ and swi/snfΔ strains were tested for common phenotypes that might indicate a gene or set of genes regulated by both TFIIS and the Swi-Snf complex. As mentioned, swi/snf mutants have several distinct phenotypes, including poor anaerobic growth on raffinose, sucrose, and galactose and inositol auxotrophy (30, 46, 47, 58, 59). In contrast, the ppr2Δ strain grew identically to wild-type cells on raffinose, sucrose, and galactose in anaerobic conditions and was not an inositol auxotroph. Cells disrupted for PPR2 are sensitive to both 6-azauracil and mycophenolic acid (20, 35), and this sensitivity was detectably greater for ppr2Δ strains than for swi1Δ, snf5Δ, or snf2Δ strains (data not shown).
DISCUSSION
A major implication from these results is that the Swi-Snf complex may be needed to promote efficient transcription elongation. Previous work has focused primarily on the effects of the Swi-Snf complex on preinitiation events, although a connection with DNA replication (21) and possibly transcript elongation (3, 6, 67) has also been suggested. The synthetic lethal approach was undertaken to help understand the function of TFIIS in vivo, and the genes identified have suggested potential functional interactions between TFIIS and chromatin-remodeling machinery. Synthetic lethality was observed between a ppr2Δ mutation and swi1Δ, snf2Δ, and snf5Δ mutations. These results strongly suggest that the combined loss of the function of the Swi-Snf complex and TFIIS forms the basis of the synthetic lethality. The basic unit of chromatin consists of DNA wrapped around nucleosomes, which is then compacted into higher-order structures. Protein complexes which are capable of altering the structure of chromatin have been identified in S. cerevisiae, Drosophila melanogaster, and metazoans (reviewed in references 7 and 42). The different chromatin-remodeling complexes have distinct biochemical activities but share a few basic features. Each complex contains a subunit with homology to DNA-dependent ATPases (the SNF2 family), and all tested complexes show ATP-dependent chromatin remodeling of nucleosomal templates. The two identified chromatin-remodeling complexes in yeast cells are the Swi-Snf complex (reviewed in references 60 and 77) and the RSC complex (10). The Swi-Snf complex also associates with at least one RNA polymerase II holoenzyme complex (75).
It is easy to rationalize overlapping functions between TFIIS and the Swi-Snf complex. Certainly, the genetic interaction between swi/snfΔ and ppr2Δ alleles demonstrates only that the Swi-Snf complex and TFIIS share some essential function, whether through completely independent transcriptional control mechanisms on an essential gene (or genes) or through overlapping transcription functions on at least some genes. Recently, the need for a functional Swi-Snf complex throughout transcription has been reported (3), and it is possible that this requirement occurs during the elongation process. The involvement of the Swi-Snf complex in elongation also was suggested previously from the results of in vitro experiments (6). Indeed, the NPH-I protein of vaccinia virus, shown to affect elongation, is also an SNF2 homolog (16). In support of a link between elongation and chromatin remodeling, swi/snf strains are modestly sensitive to 6-azauracil and mycophenolic acid, both of which reduce cellular nucleotide pools and presumably slow RNA polymerases (62). Indeed, the overall transcription levels in the cell are lower in the presence of 6-azauracil (48). Under reduced nucleotide concentrations, the polymerase might stall and arrest more often, and in the absence of the Swi-Snf complex and TFIIS, it might not efficiently recover from arrest.
The terminal phenotypes of swi/snfΔ ppr2Δ cells include loss of osmoregulation and possibly cell cycle control. These phenotypes suggest a major loss of gene regulation in the cell as opposed to the specific disruption of a pathway independently regulated by the Swi-Snf complex and TFIIS. Transcription of chromatin templates by RNA polymerase II likely relies on a combination of chromosome-remodeling and elongation-stimulatory factors for efficient movement along the template. Eliminating TFIIS or the Swi-Snf complex individually might impede the polymerase, but neither is essential. The phenotypes of swi/snf mutants are more severe than those of ppr2 mutants, and this result emphasizes a role for the Swi-Snf complex beyond that carried out by TFIIS. Thus, transcript elongation is likely to be the overlapping function, and additional roles for the Swi-Snf complex in transcription (7) and replication (21) have been documented. In addition, the interaction with PPR2 could result from a combination of transcription effects, during initiation for the Swi-Snf complex and elongation for TFIIS.
Subtle differences were observed in the genetic interactions between ppr2Δ and each of swi1Δ, snf2Δ, and snf5Δ alleles. The combination of snf5Δ and ppr2Δ mutations was the most deleterious to the cell. The double-deletion mutants arrested at an average of 50 cells. The interaction with an snf2Δ mutation was slightly less severe, with the snf2Δ ppr2Δ strains arresting at an average of 200 cells. The combination of swi1Δ and ppr2Δ was the least deleterious, as double mutant strains continued to grow to an average of 500 cells. One simple explanation for the differences may be that different null SWI/SNF mutations disrupt the Swi-Snf complex to different degrees (C. Peterson, personal communication). There may be residual Swi-Snf complex activity in swi1Δ strains, not present in snf5Δ strains, that allows the swi1Δ ppr2Δ cells to survive longer. As ppr2Δ strains share none of the swi/snf mutant phenotypes, it does not appear that PPR2 has a selective role in the regulation of the identified genes whose expression is impaired in swi/snf mutants. In addition, preliminary results using microarray analysis do not highlight genes known to be affected in swi/snf mutants (C. Seidel and C. M. Kane, unpublished results).
The unusual phenotypes of an SNF2+/snf2Δ ppr2Δ/ppr2Δ diploid appear very relevant to the reduced viability of snf2Δ ppr2Δ cells. The results suggest that genes important in osmoregulation and cell wall maintenance are especially sensitive to the interaction between SWI/SNF and PPR2. In this regard, it is noteworthy that swi/snfΔ ppr2Δ double-disruption spores can germinate and divide for several generations, but when these cells are observed under higher magnification, the morphology of the cells is extremely aberrant, including highly elongated buds. A high degree of lysis is also evident. These phenotypes were not observed for swi1Δ, snf2Δ, or snf5Δ strains in a PPR2 background. Likewise, the ppr2Δ strain itself does not exhibit these phenotypes. Taken together, the results from both the diploid and haploid double-disruption mutants indicate that defects in osmoregulation and cell wall maintenance may be the primary cause of the lethality in swi/snfΔ ppr2Δ cells.
Curiously, mutations in STT4 also cause osmosensitivity and cell wall defects that can be rescued by osmotic stabilizers. This gene can complement one of the other recessive mutations that is synthetically lethal with ppr2Δ (Table 4). Some conditional mutants in the essential RSC chromatin-remodeling complex also show highly elongated buds (11, 19). The mechanism that results in such buds is not known in either the rsc or the swi/snf ppr2 mutants. Additionally, temperature-sensitive alleles of genes encoding members of the RSC complex arrest at the G2/M boundary, another phenotype shared with conditional alleles of stt4 (11, 80). Mutations in the CLN genes, essential regulators of the cell cycle, also led to elongated buds (49). Thus, the phenotypes of the snf2Δ/SNF2 ppr2Δ/ppr2Δ diploid might result from either cell cycle defects or the loss of osmoregulation and cell wall integrity, but these two hypotheses need not invoke alternate mechanisms. The cell cycle phenotypes might also be related to the sporulation observed on rich medium of the SNF2/snf2Δ ppr2Δ/ppr2Δ diploid. Mutations in many genes lead to derepressed sporulation, and these include cell cycle mutants. Since TFIIS is believed to be a general regulator of transcription elongation, it is certainly reasonable that the sporulation phenotype of the snf2Δ/SNF2 ppr2Δ/ppr2Δ diploid might result from a loss of regulation of several genes important in the cell cycle.
The synthetic lethality between ppr2Δ and stt4 is worth further comment. A synthetic lethal mutant which contains a mutant allele of STT4 shares many swi/snf mutant phenotypes, but it also grows better in the presence of 6-azauracil than without the drug. Perhaps a reduction in transcription of TFIIS-regulated genes coupled with mutations in this phosphoinositol 4-kinase cripple the cell, although a more direct connection to chromatin remodeling is possible (81). In S. cerevisiae, Stt4 is a likely candidate to create the precursor to phosphatidylinositol 4,5-biphosphate (17, 34) and in human cells, phosphatidylinositol 4,5-biphosphate appears to target the human BAF chromatin remodeling complex to chromatin in vitro (81). The BAF complex contains BAF53, a human homolog of Arp7 and Arp9, the two actin-related subunits of the Swi-Snf complex in S. cerevisiae (8, 61). Mutations in either of these genes in yeast cells also result in swi/snf mutant phenotypes.
As a final note from this analysis, the identification of KEX2 in this screen (Table 4) marks the second time it has shown genetic interactions with the RNA polymerase II transcription machinery (51). The genetic interaction between the kex2Δ and ppr2Δ alleles resulted in severe sickness in the double-deletion mutant, characterized by a dramatic reduction in viability. Kex2 is a prohormone protease located in the Golgi. Null mutations in KEX2 also suppress mutations in the largest subunit of RNA polymerase II, RPB1 (51). While the nature of this suppression is unknown, Kex2 may process a protein that directly or indirectly modifies the biochemical properties of RNA polymerase II or the levels of its nucleotide substrates. The absence of this protein caused by the loss of Kex2 may create an RNA polymerase II dependent on the activity of TFIIS. All these possible connections are provocative, but it is essential to note that although the results might suggest a mechanistic linkage, mutations in many types of genes confer overlapping phenotypes (28). Further study is needed to test the interactions among the genes identified in this screen.
Several complementation groups from this synthetic lethal screen remain to be characterized, but as yet there is no evidence for a functional homolog that shares a mechanistically overlapping essential function with TFIIS. An additional component of the Swi-Snf complex, Tfg3, also has strong genetic interactions with TFIIS, and its genetic relationship to TFIIS is very different from that between TFIIS and other members of the Swi-Snf complex (J. Davie and C. Kane, unpublished data). The results presented here also predict that loss of TFIIS function might be compensated for by activities in addition to the Swi-Snf complex that promote efficient elongation. With a growing number of described proteins that impact elongation by RNA polymerase II, additional interactions with chromatin-remodeling components are likely to be found.
ACKNOWLEDGMENTS
We thank Danesh Moazed, Alexander Johnson, Grant Hartzog, Fred Winston, and Marian Carlson for kind gifts of reagents. We thank Danesh Moazed, Grant Hartzog, Fred Winston, Craig Peterson, Jeremy Thorner, Chau Huynh, Jasper Rine, and Paul Kaufman for helpful discussions and sharing unpublished observations.
This work was supported by a grant from the National Institutes of Health awarded to C.M.K. (GM54012). J.K.D. was supported by a Howard Hughes Medical Institute predoctoral fellowship.
REFERENCES
- 1.Abrams E, Neigeborn L, Carlson M. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3643–3651. doi: 10.1128/mcb.6.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggar S R, Crabtree G R. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 5.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 6.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 7.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 8.Cairns B R, Erdjument-Bromage H, Tempst P, Winston F, Kornberg R D. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 9.Cairns B R, Levinson R S, Yamamoto K R, Kornberg R D. Essential role of Swp73p in the function of yeast Swi/Snf complex. Genes Dev. 1996;10:2131–2144. doi: 10.1101/gad.10.17.2131. [DOI] [PubMed] [Google Scholar]
- 10.Cairns B R, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg R D. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 11.Cao Y, Cairns B R, Kornberg R D, Laurent B C. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol Cell Biol. 1997;17:3323–3334. doi: 10.1128/mcb.17.6.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie K R. Ph.D. thesis. Berkeley: University of California; 1995. [Google Scholar]
- 13.Christie K R, Awrey D E, Edwards A M, Kane C M. Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, P37. J Biol Chem. 1994;269:936–943. [PubMed] [Google Scholar]
- 14.Clark A B, Dykstra C C, Sugino A. Isolation, DNA sequence, and regulation of a Saccharomyces cerevisiae gene that encodes DNA strand transfer protein alpha. Mol Cell Biol. 1991;11:2576–2582. doi: 10.1128/mcb.11.5.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connelly S, Manley J L. RNA polymerase II transcription termination is mediated specifically by protein binding to a CCAAT box sequence. Mol Cell Biol. 1989;9:5254–5259. doi: 10.1128/mcb.9.11.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng L, Shuman S. Vaccinia NPH-I, a DExH-box ATPase, is the energy coupling factor for mRNA transcription termination. Genes Dev. 1998;12:538–546. doi: 10.1101/gad.12.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desrivières S, Cooke F T, Parker P J, Hall M N. MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J Biol Chem. 1998;273:15787–15793. doi: 10.1074/jbc.273.25.15787. [DOI] [PubMed] [Google Scholar]
- 18.Ding H F, Rimsky S, Batson S C, Bustin M, Hansen U. Stimulation of RNA polymerase II elongation by chromosomal protein HMG-14. Science. 1994;265:796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- 19.Du J, Nasir I, Benton B K, Kladde M P, Laurent B C. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics. 1998;150:987–1005. doi: 10.1093/genetics/150.3.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 21.Flanagan J F, Peterson C L. A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res. 1999;27:2022–2028. doi: 10.1093/nar/27.9.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerring S L, Spencer F, Hieter P. The CHL 1 (CTF 1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 1990;9:4347–4358. doi: 10.1002/j.1460-2075.1990.tb07884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 24.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- 25.Grant S G, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press; 1991. [PubMed] [Google Scholar]
- 28.Hampsey M. A review of phenotypes in Saccharomyces cerevisiae. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D, Jessee J, Bloom F R. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 30.Happel A M, Swanson M S, Winston F. The SNF2, SNF5 and SNF6 genes are required for Ty transcription in Saccharomyces cerevisiae. Genetics. 1991;128:69–77. doi: 10.1093/genetics/128.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartzog G A, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirashima S, Hirai H, Nakanishi Y, Natori S. Molecular cloning and characterization of cDNA for eukaryotic transcription factor S-II. J Biol Chem. 1988;263:3858–3863. [PubMed] [Google Scholar]
- 33.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 34.Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y. Phosphatidylinositol-4-phosphate-5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem. 1998;273:15779–15786. doi: 10.1074/jbc.273.25.15779. [DOI] [PubMed] [Google Scholar]
- 35.Hubert J C, Guyonvarch A, Kammerer B, Exinger F, Liljelund P, Lacroute F. Complete sequence of a eukaryotic regulatory gene. EMBO J. 1983;2:2071–2073. doi: 10.1002/j.1460-2075.1983.tb01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huxley C, Green E D, Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- 37.Ito T, Xu Q, Takeuchi H, Kubo T, Natori S. Spermatocyte-specific expression of the gene for mouse testis-specific transcription elongation factor S-II. FEBS Lett. 1996;385:21–24. doi: 10.1016/0014-5793(96)00340-7. [DOI] [PubMed] [Google Scholar]
- 38.Izban M G, Luse D S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 39.Kanai A, Kuzuhara T, Sekimizu K, Natori S. Heterogeneity and tissue-specific expression of eukaryotic transcription factor S-II-related protein mRNA. J Biochem. 1991;109:674–677. doi: 10.1093/oxfordjournals.jbchem.a123439. [DOI] [PubMed] [Google Scholar]
- 40.Kipling D, Kearsey S E. Function of the S. cerevisiae DST1/PPR2 gene in transcription elongation. Cell. 1993;72:12. doi: 10.1016/0092-8674(93)90044-q. [DOI] [PubMed] [Google Scholar]
- 41.Kipling D, Kearsey S E. TFIIS and strand-transfer proteins. Nature. 1991;353:509. doi: 10.1038/353509a0. [DOI] [PubMed] [Google Scholar]
- 42.Kornberg R D, Lorch Y. Chromatin-modifying and -remodeling complexes. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 43.Koshland D, Kent J C, Hartwell L H. Genetic analysis of the mitotic transmission of minichromosomes. Cell. 1985;40:393–403. doi: 10.1016/0092-8674(85)90153-9. [DOI] [PubMed] [Google Scholar]
- 44.Koulich D, Orlova M, Malhotra A, Sali A, Darst S A, Borukhov S. Domain organization of Escherichia coli transcript cleavage factors GreA and GreB. J Biol Chem. 1997;272:7201–7210. doi: 10.1074/jbc.272.11.7201. [DOI] [PubMed] [Google Scholar]
- 45.Kranz J E, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laurent B C, Treitel M A, Carlson M. Functional interdependence of the yeast SNF2, SNF5, and SNF6 proteins in transcriptional activation. Proc Natl Acad Sci USA. 1991;88:2687–2691. doi: 10.1073/pnas.88.7.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laurent B C, Treitel M A, Carlson M. The SNF5 protein of Saccharomyces cerevisiae is a glutamine- and proline-rich transcriptional activator that affects expression of a broad spectrum of genes. Mol Cell Biol. 1990;10:5616–5625. doi: 10.1128/mcb.10.11.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lennon J C, 3rd, Wind M, Saunders L, Hock M B, Reines D. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:5771–5779. doi: 10.1128/mcb.18.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malone E A, Fassler J S, Winston F. Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol Gen Genet. 1993;237:449–459. doi: 10.1007/BF00279450. [DOI] [PubMed] [Google Scholar]
- 51.Martin C, Young R A. KEX2 mutations suppress RNA polymerase II mutants and alter the temperature range of yeast cell growth. Mol Cell Biol. 1989;9:2341–2349. doi: 10.1128/mcb.9.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mote J, Jr, Ghanouni P, Reines D. A DNA minor groove-binding ligand both potentiates and arrests transcription by RNA polymerase II: elongation factor SII enables readthrough at arrest sites. J Mol Biol. 1994;236:725–737. doi: 10.1006/jmbi.1994.1185. [DOI] [PubMed] [Google Scholar]
- 53.Nakanishi T, Shimoaraiso M, Kubo T, Natori S. Structure-function relationship of yeast S-II in terms of stimulation of RNA polymerase II, arrest relief, and suppression of 6-azauracil sensitivity. J Biol Chem. 1995;270:8991–8995. doi: 10.1074/jbc.270.15.8991. [DOI] [PubMed] [Google Scholar]
- 54.Olmsted V K, Awrey D E, Koth C, Shan X, Morin P E, Kazanis S, Edwards A M, Arrowsmith C H. Yeast transcript elongation factor (TFIIS), structure and function. I. NMR structural analysis of the minimal transcriptionally active region. J Biol Chem. 1998;273:22589–22594. doi: 10.1074/jbc.273.35.22589. [DOI] [PubMed] [Google Scholar]
- 55.Orphanides G, LeRoy G, Chang C H, Luse D S, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 56.Orphanides G, Wu W H, Lane W S, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 57.Otero G, Fellows J, Li Y, de Bizemont T, Dirac A M, Gustafsson C M, Erdjument-Bromage H, Tempst P, Svejstrup J Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 58.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 59.Peterson C L, Kruger W, Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991;64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 60.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 61.Peterson C L, Zhao Y, Chait B T. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem. 1998;273:23641–23644. doi: 10.1074/jbc.273.37.23641. [DOI] [PubMed] [Google Scholar]
- 62.Reines D, Conaway R C, Conaway J W. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr Opin Cell Biol. 1999;11:342–346. doi: 10.1016/S0955-0674(99)80047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 64.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stebbins C E, Borukhov S, Orlova M, Polyakov A, Goldfarb A, Darst S A. Crystal structure of the GreA transcript cleavage factor from Escherichia coli. Nature. 1995;373:636–640. doi: 10.1038/373636a0. [DOI] [PubMed] [Google Scholar]
- 66.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 67.Sudarsanam P, Cao Y, Wu L, Laurent B C, Winston F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 1999;18:3101–3106. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swanson M S, Malone E A, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. . (Erratum, 11:4286.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swanson M S, Winston F. SPT4, SPT4, and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics. 1992;132:325–336. doi: 10.1093/genetics/132.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 71.Umehara T, Kida S, Hasegawa S, Fujimoto H, Horikoshi M. Restricted expression of a member of the transcription elongation factor S-II family in testicular germ cells during and after meiosis. J Biochem. 1997;121:598–603. doi: 10.1093/oxfordjournals.jbchem.a021627. [DOI] [PubMed] [Google Scholar]
- 72.Umehara T, Kida S, Yamamoto T, Horikoshi M. Isolation and characterization of a cDNA encoding a new type of human transcription elongation factor S-II. Gene. 1995;167:297–302. doi: 10.1016/0378-1119(95)00634-6. [DOI] [PubMed] [Google Scholar]
- 73.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weaver Z A, Kane C M. Genomic characterization of a testis-specific TFIIS (TCEA2) gene. Genomics. 1997;46:516–519. doi: 10.1006/geno.1997.5073. [DOI] [PubMed] [Google Scholar]
- 75.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 76.Wind M, Reines D. Transcription elongation factor II. Bioessays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 78.Woychik N A, Lane W S, Young R A. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J Biol Chem. 1991;266:19053–19055. [PubMed] [Google Scholar]
- 79.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 80.Yoshida S, Ohya Y, Goebl M, Nakano A, Anraku Y. A novel gene, STT4, encodes a phosphatidylinositol 4-kinase in the PKC1 protein kinase pathway of Saccharomyces cerevisiae. J Biol Chem. 1994;269:1166–1172. [PubMed] [Google Scholar]
- 81.Zhao K, Wang W, Rando O J, Xue Y, Swiderek K, Kuo A, Crabtree G R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]