ABSTRACT
Introduction
Emerging novel infectious diseases and persistent pandemics with potential to destabilize normal life remain a public health concern for the whole world. The recent outbreak of pneumonia caused by Coronavirus infectious disease-2019 (COVID-19) resulted in high mortality due to a lack of effective drugs or vaccines. With a constantly increasing number of infections with mutated strains and deaths across the globe, rapid, affordable and specific detections with more accurate diagnosis and improved health treatments are needed to combat the spread of this novel pathogen COVID-19.
Areas covered
Researchers have started to utilize the recently invented clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (CRISPR/Cas)-based tools for the rapid detection of novel COVID-19. In this review, we summarize the potential of CRISPR/Cas system for the diagnosis and enablement of efficient control of COVID-19.
Expert opinion
Multiple groups have demonstrated the potential of utilizing CRISPR-based diagnosis tools for the detection of SARS-CoV-2. In coming months, we expect more novel and rapid CRISPR-based kits for mass detection of COVID-19-infected persons within a fraction of a second. Therefore, we believe science will conquer COVID-19 in the near future.
KEYWORDS: COVID-19, CRISPR/Cas, CRISPR/Cas9, CRISPR/Cas12, CRISPR/Cas13, SHERLOCK, DETECTR, diagnostics, infectious disease
1. Introduction
Coronavirus infectious disease-2019 (COVID-19) is caused by a severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). This COVID-19 first occurred in Wuhan, China, in late December 2019 [1,2]. Initially, the seriousness of COVID-19 was unknown. Later, this SARS-CoV-2 was identified to cause large-scale loss of life and severe human suffering in many countries. Since the pandemic of coronavirus (CoV) is increasing across the globe, scientists are focusing on developing the novel diagnostic tools, vaccines, and treatments. Some vaccines like Pfizer-BioTNech (BNT162b2), Moderna (mRNA-1273), Johnson & Johnson’s Janssen (JNJ-78436735), Astra Zeneca and Oxford’s Covishield, and Bharath Biotech’s Covaxin have been approved for emergency use for SARS-CoV-2. However, the spread of the virus has not yet been stopped. New mutant variants such as B.1.1.7, B.1.1.7.2, B.1.526, and P.1 of the CoVs are creating serious health concern especially in India.
CoVs are a family of positive-sense, single-stranded RNA (ssRNA) viruses, which infect nose, throat, and sinuses leading to neurological and respiratory diseases in humans [3,4]. Prior to SARS-CoV-2, there were six others major CoVs species including SARS-CoV and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) [4]. World Health Organization (WHO) declared that COVID-19 is a great infectious disease to the world [5]. Recent reports state that more than 161 countries are being affected severely due to COVID-19 [5,6]. Therefore, scientists are forced to develop efficient vaccines to stop the spread of this novel SARS-CoV-2. At present, the SARS-CoV-2 infected persons are being isolated for 7–28 days, and severely infected patients are kept under supportive care [7,8].
Scientists are collecting the blood samples from the pneumonia patients and isolating the genomic sequence of SARS-CoV-2 to identify the mutants [9]. Based on the genomic characters, biopharmaceutical companies employ diagnosis tests namely qRT-PCR and reverse transcription loop-mediated isothermal amplification (RT-LAMP) to confirm SARS-CoV-2 [10–12]. However, these detection techniques suffer by providing false negative results, which allow the infected patients to continue the spread of the virus. It also requires sophisticated instruments and skilled manpower. Hence, an easy, simple, and quick diagnostic method with accuracy is essential for rapid diagnose of SARS-CoV-2 infection.
Scientists started to explore modern molecular tools including genome-editing systems, especially the recently invented and noble prize winning clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated proteins (Cas) system for rapid diagnosis of SARS-CoV-2 [13,14]. CRISPR/Cas system has become a most popular genome-editing tool. This system uses a programmable protein that is attached to the targeted sequence with the help of guide RNA (gRNA) and creates double-stranded breaks (DSBs) [15]. There are many types of Cas proteins viz. Cas1, Cas2, Cas3, Cas6, Cas9, and Cas10 [16,17]. Among these, Cas9 protein is the efficient genome-editing system to target double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA). Following Cas9 protein, the Cas12, and Cas13 proteins receive more attention in diagnosing viral diseases. Cas12 protein cleaves ssDNA and dsDNA, whereas Cas13 protein cleaves ssRNA; therefore, these proteins play an important role in the diagnosis of CoVs. In this review, we summarize the potential of CRISPR/Cas systems including Cas9, Cas12, and Cas13 proteins utilized for the diagnosis of SARS-CoV-2 infection.
2. SARS-CoV-2
The SARS-CoV-2 has a linear, and positive-stranded RNA which transmits deadly infections to all eukaryotic organisms [18]. It belongs to the families Coronaviridae, Roniviridae, and Arteriviridae and to the order Nidovirales [19,20]. They are classified into four genera: 1) Alpha coronaviruses that include human coronavirus NL63 (hcOv-NL63), porcine transmissible gastroenteritis coronavirus (PTGEV); 2) Beta coronaviruses that include SARS-CoV, coronavirus HKU4, MERS-CoV, bovine coronavirus (BCoV), bat, mouse hepatitis coronavirus (MHV) and human coronavirus OC43; 3) Gamma that include avian infectious bronchitis coronavirus (IBV); and 4) Delta coronaviruses that include porcine delta coronavirus (pdCoV) [21–23]. Coronaviridae and Ronoviridae have large-sized RNA viral genomes ranging from 26 to 32 kbp. It is covered by a helical capsid formed by nucleocapsid protein (N) [24] and surrounded by an envelope protein (E). The structure of viral associated proteins includes the membrane protein (M) [25] and E protein [26] that aid in virus assembly, and the spike protein (S) that allows the virus to enter into the host. Among these, the S protein is large which appears like a crown. Due to the presence of crown-like structure (latin word corona meaning crown), this virus is termed as coronavirus [18,27]
3. Diagnosis of SARS-CoV-2 infection
The SARS-CoV-2 pandemic has broken out widely all over the world. WHO informed that COVID-19 disease is pandemic and spreading rapidly, therefore urged for an efficient diagnostic methods. Scientists first isolated the unknown pathogen from the pneumonia-affected person and did a metagenomic next-generation sequencing (mNGS) [28]. The novel CoV genome, which showed 79% similarity with the previously identified SARS-CoV [28,29]. Then CoV was diagnosed by the RT-PCR and chest computed tomography (CT) [30]. Readily available CT is used for detection in lung diseases, scientists used chest CT as an option to detect the CoVs [30]. Cheng et al. [31] tried to detect the novel CoV using chest CT [31]. They selected 8 CoV patients for CT imaging. They observed and reported that 63% patients had mild infections, 25% patients had moderate infections, and 13% had severe CoV symptoms. From these findings, they concluded that chest CT could provide early detection of CoV patients [31]. RT-PCR testing method was used as a highly specific messenger RNA detection method to detect the presence of CoV [31]. The RT-PCR test uses fluorescent dyes which enables the researchers to diagnose the suspects [31]. The Centers for Disease Control and Prevention from United States developed the primer which targets the viral nucleocapsid gene (N1 and N2), and WHO developed an assay that targets the virus RNA-dependent RNA polymerase gene and E gene [31]. However, RT-PCR requires excess reagents, skilled manpower and specialized equipment, which slows down the disease detection. Therefore, an advanced diagnostic method is needed to diagnose this emerging infection.
4. CRISPR/Cas system
CRISPR/Cas system is an RNA-guided endonuclease (RGEN)-based nucleic acid detecting tool that has changed the field of genomics, genome modification, gene therapy, and genome mapping [32,33]. The CRISPR and Cas proteins are adaptive immune systems against bacteria and archaea [34,35]. The CRISPR loci consist of a CRISPR array separated by a unique spacer sequence (30–40 bases). Adjacent to the CRISPR array, many Cas genes were discovered which encode the effector enzymes of the CRISPR system. The CRISPR-mediated immunity consists of three phases: (1) adaptation, (2) CRISPR RNA (crRNA) maturation, and (3) interference [36]. CRISPR/Cas system is divided into two classes based on the composition of the interference complex, namely Class 1 that includes types I, III, and IV, and Class 2 that includes types II, V, and VI [37,38]. Class 1 system utilizes crRNA with multi-effector complex to cleave target genome sequence, whereas the Class 2 system utilizes single multi-domain Cas protein with crRNA for interference (Table 1) [39]. In these classes, the type II CRISPR system has been widely adopted by the scientific community for more accurate and rapid diagnosis of infections [40–42]. Recently, CRISPR/Cas12 and Cas13 systems have been used in the diagnosis of human infections including bacteria and viruses [8].
Table 1.
Details on classification of CRISPR/Cas systems. The details such as class, type, protein name, target molecule, mechanism of spacer acquisition, processing of Pre-CRISPR, self vs non-self-discrimination, and host organism are included with their references
| Class | Type | Protein | Target | Spacer acquisition of CRISPR system | Interference of CRISPR system | Pre-CRISPR processing | Self vs non-self discrimination |
Host organism | References |
|---|---|---|---|---|---|---|---|---|---|
| Class 1 | I | Cas3 | ssDNA | Cas1/Cas2/ Cas4 | Cas7, Cas5, Cas8, and Cas3 | Cas6 | PAM | E. coli | [45] |
| III | Cas10 | ssDNA | Cas1/Cas2 | Cas7, Cas5, and Cas1 | Cas6 | CRISPR Repeat | S. epidermics | [43] | |
| IV | Csf1 | - | NA | Cas7, Cas5, and Csf1 | - | - | - | [34] | |
| Class 2 | II | Cas9 | dsDNA | Cas1/Cas2/ Cas4 | Cas9 | RNase III, and tracrRNA |
PAM |
S. thermophilus and S. pyogenes |
[15,44] |
| V | Cpf1 | ssDNA and dsDNA | Cas1/Cas2/ Cas4 | Cas12 | Cpf1 | PAM | F. novicida | [50] | |
| VI | C2c2 | ssRNA | Cas1/Cas2 | Cas13 | - | - | - | [57] |
PAM, protospacer adjacent motif; Cpf1, CRISPR-associated endonuclease in Prevotella and Francisella 1; tracrRNA, trans-activating CRISPR RNA, E. coli, Escherichia coli; S. epidermics, streptococcus epidermics; F. novicida, Francisella novicida
4.1. CRISPR/Cas9 system
The type II CRISPR/Cas system is a bacterial adaptive immune system, consisting of Cas9 nucleases and the guide-RNA (gRNA) [45–47]. The CRISPR/Cas9 system creates blunt dsDNA breaks that are repaired by non-homologous end joining (NHEJ) or homologous recombination (HDR) methods. The Cas9 nuclease isolated from Streptococcus pyogenes consists of two domains: (1) HNH endonuclease domain (single nuclease domain), and (2) RuvC endonuclease domain (three nuclease domains) [46,48]. The sgRNA which is derived from the fusion of crRNA and tracrRNA is complexed with the Cas9 nuclease targets the DNA through base pairing. This process targets the desired sequence with the help of protospacer adjacent motif (PAM)
4.2. CRISPR/Cas12 system
CRISPR/Cas12 is also known as CRISPR-associated endonuclease from Prevotella and Francisella 1 (cpf1); it belongs to the Class 2 type V associated nuclease [49,50]. CRISPR/Cas12 system is an efficient system that creates staggered cuts in both ssDNA and dsDNA. The Cas12 is derived from Acidaminococcus species Cas12a (AsCas12a) and Lachnospiraceae bacterium Cas12a (LbCas12a), which naturally evolves to fight against foreign genetic elements. The CRISPR/Cas12 system requires only the crRNA to create staggered cuts at the targeted sequence. The Cas12 nuclease contains two domains namely (1) nuclease lope (NUC) domain and (2) RuvC domain for cleavage activity [51,52]. Like the Cas9 system, the Cas12 system targets the region besides the PAM region. Once Cas12 starts to encounter, it initiates R-loop formation which forms base-pair hybrids between the crRNA and the target sequence [53]. During this step, it matches the <17 bp target sequence and establishes an R-loop formation. Once R-loop is formed, the Cas12 nuclease uses its active RuvC domain and creates staggered cuts at the targeted ssDNA or dsDNA sequence with the presence of PAM sequence [50,54].
4.3. CRISPR/Cas13 system
CRISPR/Cas13 system is referred to as C2c2 system which belongs to the Class 2 type VI [55,56]. CRISPR/Cas13 system is another important system in the CRISPR world because it targets only ssRNA but not ssDNA or dsDNA. CRISPR/Cas13 system requires only crRNA to recognize the target [57]. The Cas13 consists of alpha-helical recognition (REC) lobe with a helical-1 domain and an NUC lobe with two higher eukaryotic and prokaryotic nucleotide-binding protein (HEPN) domains [55,58]. These domains play a major role in RNA-targeted cleavage activity. Upon RNA binding, Cas13 protein gets activated by triggering the HEPN 1 domain to move the HEPN 2 domain, which will undergo a conformational change resulting in the cleavage of targeted ssRNA [59].
5. CRISPR/Cas9 system–mediated diagnosis of infectious diseases
CRISPR/Cas9 system has been adopted by the scientific community for the diagnosis of infectious diseases (Table 2). Pardee et al. [42] first demonstrated a simple novel method by combining CRISPR/Cas9 with isothermal amplification technique referred to as nucleic acid sequence-based amplification (NASBA) and detected Zika virus strain. The researchers utilized the dsDNA to intermediate the NASBA that served as a substrate for the Cas9 endonuclease. The sgRNA-Cas9 complex cleaves the resulting dsDNA, full-length strand (not truncated strands) activated the toehold switch sensors and led to color changes in order to identify different strains. From the result, investigators stated that this biomolecular platform resolved practical limitations and suggested that this could be used to develop molecular diagnostics for challenging the pandemic [42]. Guk et al. [41] developed a CRISPR/Cas9-mediated fluorescent in situ hybridization (DNA-FISH) method to detect methicillin-resistant Staphylococcus aureus (MRSA) [41]. In this method, the dCas9/sgRNA complex coupled with an SYBR green I fluorescent probe was used to target the mecA gene and recognize mecA gene associated with MRSA. The dCas9/sgRNA did not induce DNA cleavage but it recognizes the target DNA sequence, thus it is suitable for detection by FISH. The corresponding fluorescence was sufficient to efficiently detect the MRSA even at a lower concentration of 10 CFU/ml. From the result, researchers reported that dCas9/sgRNA-based detection is simple to detect various pathogens [41]. In another study, a combination of CRISPR/Cas9 with optical DNA mapping was employed to diagnose bacterial antibiotic-resistance genes [60]. In this demonstration, Cas9 and gRNA were fused that cleaved the targeted specific DNA sequences of resistance genes. Then fluorescent dye (YOYO-1) and netropsin bound to the resulting DNA based on AT-rich regions and detected each DNA segment of different resistant genes. The addition of multiple crRNA detected many different genes in each reaction. From these results, Muller et al. [60] concluded that CRISPR/Cas9 system in combination with optical DNA mapping could be applied for the detection of different pathogens (Figure 1).
Table 2.
Details of CRISPR/Cas systems utilized for detecting pathogenic viruses. The details such as name of the CRISPR/Cas system, used, name of the effector, amplification method, targeted viruses, type of nucleic acids, specificity and detection time are included with their references
| CRISPR/Cas systems |
System utilized | Effector | Amplification method | Target Virus |
Type of Nucleic acid | Specificity | Detection time | References |
|---|---|---|---|---|---|---|---|---|
| CRISPR/Cas12 | CRISPR/Cas12a | LbCas12a | RT-LAMP | SARS-CoV-2 | ssRNA | High | <40 minutes | [61] |
| CRISPR/Cas13 | SHERLOCK SHERLOCKv2 |
LwCas13a | RT-RPA | Dengue Virus | ssRNA | High | 5 hours | [78] |
| CRISPR/Cas13 | POC | LwCas13a | RT-PCR | Ebola Virus | ssRNA | High | <2 hours | [62] |
| CRISPR/Cas13 | SHERLOCK SHERLOCKv2 |
pspCas13a | RT-RPA | Influenza Virus | ssRNA | High | <2 hours | [63] |
| CRISPR/Cas12/13 | SHERLOCK DETECTR |
LwCas13 LbCas12a |
RT-RPA PCR |
White spot syndrome Virus | ssDNA and ssRNA | High | <1 hour | [64] |
| CRISPR/Cas13 | SHERLOCK | pspCas13 | RT-RPA | VSV | ssRNA | High | <1 hour | [63] |
| CRISPR/Cas13 | SHERLOCK | LbCas13 | RT-RPA | PRSSV | ssRNA | High | 3 hours | [65] |
| CRISPR/Cas13 | HOLMES | LbCas13 | RT-RPA | LCMV | ssRNA | High | 2 hours | [63] |
| CRISPR/Cas12 | HOLMES | LbCas12a | PCR | JEV | ssDNA | High | 1 hour | [66] |
| CRISPR/Cas13 | SHERLOCK | LwCas13 | RT-RPA | EBV | ssRNA | High | 1 hour | [67] |
| CRISPR/Cas13 | SHERLOCK | LwCas13 | RT-RPA | CPV-2 | ssRNA | High | 1 hour | [68] |
| CRISPR/Cas12 | DETECTR | LwCas13 | RT-RPA | ASFV | ssDNA | High | 1 hour | [69] |
| CRISPR/Cas12 | CRISPR-MTB-FISH | Cas12a | RPA | MRSA | ssDNA | High | 1 hour | [41] |
| CRISPR/Cas9 | Cas9 | SpdCas9 | PCR | M. Tuberculosis |
dsDNA | High | <1 hour | [70] |
| CRISPR/Cas9 | APC-Cas | SpdCas9 | PCR | Salmomella | dsDNA | High | 3 hours | [71] |
SHERLOCK, specific high-sensitivity enzymatic reporter unlocking; DETECTR, DNA endonuclease-targeted CRISPR trans-reporter; POC, point of care; HOLMES, HOur Low-cost multipurpose highly efficient system; FISH, fluorescent in situ hybridization; RT-LAMP, reverse transcription loop-mediated isothermal amplification; RT-RPA, reverse transcription-recombinase polymerase isothermal amplification; PCR-polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VSV, Vesicular stomatitis virus; PRSSV, Porcine reproductive and respiratory syndrome virus; LCMV, Lymphocytic choriomeningitis virus; JEV, Japanese encephalitis virus; H7N9, Avian influenza A virus; GRBV, Grapevine red blotch-associated virus; EBV, Epstein–Barr virus; CPV2, Canine parvovirus type 2 virus, ASFV, African swine fever virus; MRSA, Methicillin-resistant Staphylococcus aureus.
Figure 1.
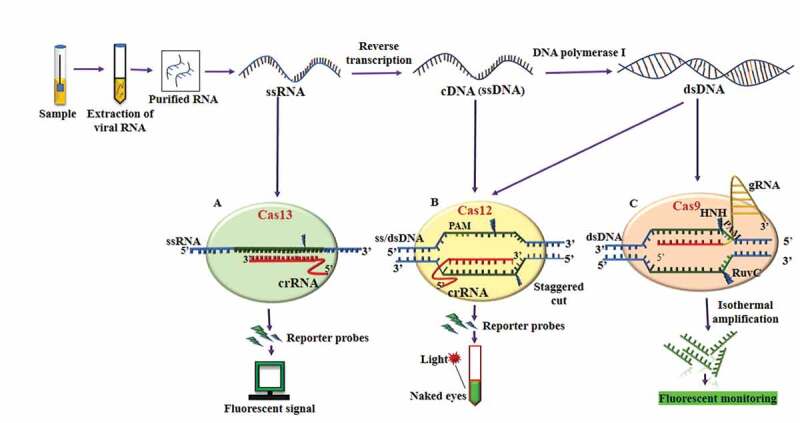
An overview of CRISPR/Cas system (Cas13, Cas12, and Cas9) based SARS-CoV-2 detection methods. Viral sample is collected from nasal, nasopharyngeal, and throat swabs of humans. The RNA is extracted from the virus and purified using specific RNA extraction kit. (A) The purified RNA is subjected to Cas13 system and using appropriate probe, the target is detected. (B) The purified RNA is subjected to reverse transcriptase (RT) and converted into complementary DNA (cDNA) and single-stranded DNA (ssDNA). The Cas12 system binds with the targeted sequence and discriminate its cleavage activity with the reporter probe and detects the virus. (C) The cDNA is converted into double-stranded DNA (dsDNA) using DNA polymerase I enzyme, which is recognized by the Cas9 system. It creates a double-stranded breaks (DSBs) and upon cleavage, isothermal amplification is used to detect the targeted virus
Researchers detected dual genes of SARS-CoV-2 in a single test using the CRISPR/Cas9 system [72]. They first designed a triple-line lateral flow strip (TL-LFA) and later designed two sgRNAs (sgRNA1 and sgRNA2) for targeting E and orf1ab genes with their scaffold sequences. The scaffold sequences are designed in such a way as having a binding site for employing AuNP-DNA probes. Then the sgRNAs were mixed with Cas9 and the complexes were run over the lateral flow strip, the AuNP-DNA probes bound to the scaffold sequence via DNA hybridization. Once AuNP accumulated, the colored bands are seen through the naked eyes that helps to detect the SARS-CoV-2. From these analyses, researchers confirmed that the CRISPR/Cas9 based TL-FLA strip assay would detect the SARS-CoV-2 accurately [72]. Recently, the ‘CRISPR/Cas9 eraser’ approach had been applied to detect the SARS-CoV-2 infection and swine flu virus [73]. CRISPR/Cas9 eraser was designed to accomplish a contamination-free RNA detection through RT-PCR. In addition, the researchers extended the CRISPR/Cas9 eraser to the PCR technique. They designed an artificial PAM site into the PCR products through primer designing and planned an approach named PAM-implanted PCR. They programmed conventional PCR primers by adding two extra cytosine (C) bases at the 3′ end that leads to the amplified products containing the PAM site (artificial PAM site). The sgRNA which is corresponding to the amplified PCR products (identical artificial PAM site) was used for Cas9/sgRNA recognition and cleavage. Through CRISPR eraser, the researchers spotted the SARS-Cov-2 and swine flu virus with the suitable strip test [73].
6. CRISPR/Cas12 system–mediated diagnosis
CRISPR/Cas12 is a smaller system; it does not require tracrRNA, and cleaves dsDNA or ssDNA via the RuvC domain. Unlike Cas9, the Cas12 recognizes a different PAM sequence (5′-TTN-3′), upstream of its target. Researchers developed a different method called DNA endonuclease-targeted CRISPR trans-reporter (DETECTR) based on Cas12 system (Table 2) [8]. The DETECTR technique utilizes type V enzyme to directly cleave ssDNA sequence in three-stage process: (1) gRNA directs the Cas12 enzyme to ssDNA with targeted viral genome, (2) While binding to its targeted viral genome, a ssDNA binds with the quencher molecule and reporter fluorophore to cleave randomly by the Cas12 enzyme, (3) This collateral cleavage is detected by the fluorescent signal released from quencher and fluorophore [8]. This system showed the advantage of the diagnosis method to detect even a single molecule of viral particle (Figure 2). Chen et al. [8] first reported the DETECTR [8]. DETECTR technique achieved the attomolar sensitivity for DNA detection. In this work, they combined the nonspecific ssDNA of Cas12a and reported the rapid detection of the virus. Utilizing DETECTR, they differentiated several human papillomaviruses (HPV16 and HPV18) from crude DNA extracts from clinical samples. The DETECTR technique identified HPV16 and HPV18 from clinical specimens and showed 25/25 and 23/25 results within 1 hour [8]. In addition, CRISPR/Cas12 was also used to diagnose the African Swine Fever Virus (ASFV) [74]. CRISPR/Cas12a was combined with fluorescent-based point of care (POC) system, which bound with and detected targeted ssDNA. Cas12/crRNA/ASFV DNA complex got activated upon binding and degraded the ssDNA. They detected the ASFV with a limitation of 1 pM within 2 hours. In addition, Cas12, crRNA and ASFV DNA complex continuously cleaved ssDNA complex, even after 24 hours, which displayed an improved detection limit of 100 femtomolar (fM). Cas12 system is very specific and can detect even up to a single nucleotide of targeted virus [74].
Figure 2.
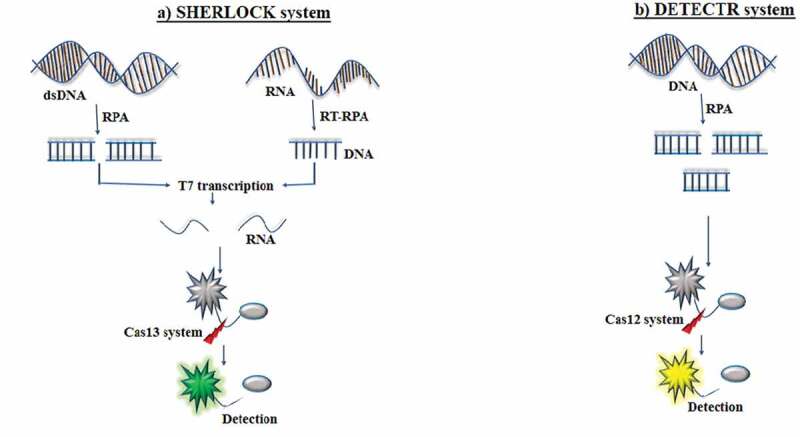
Mechanism of SHERLOCK and DETECTR system. (A) Targeted double-stranded DNA (dsDNA) or RNA is amplified with recombinase polymerase amplification (RPA) or reverse transcription (RT)-RPA. The RPA is coupled with T7 transcription to covert targeted RNA for detection by Cas13 system. This amplification steps in combination with the reporter probe enables specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) to detect the targeted sequence, (B) In DNA endonuclease-targeted CRISPR trans reporter (DETECTR), DNA is amplified with RPA. The Cas12 system pairs with the single-stranded DNA (ssDNA) of interest, the DNase activity of Cas12 system is initiated. This amplification step in combination with the reporter probe enables DETECR to detect the targeted sequence
Researchers started to utilize the DETECTR system for the SARS-Cov-2 detection. The Mammoth Biosciences, Inc., San Francisco, CA, first diagnosed the SARS-Cov-2 by targeting the N and E genes through CRISPR/Cas12 system [9]. They engineered Cas12-gRNAs to specifically target the SARS-CoV-2 and optimized the DETECTR assay for E and N genes. They run this enhanced DETECTR assay and visualized the detection of SARS-CoV-2 on a lateral flow strip. From the analysis, a faster detection was observed through DETECTR, which saved more than an hour. The sensitivity range of the DETECTR platform was 70–300 copies/µl input [9]. The protocol encompassed LAMP in combination with reverse transcription instead of recombinase polymerase amplification (RPA)-based amplification that resulted in detection of SARS-CoV-2 within 30 min [9]. Similarly, CRISPR/Cas12-based fast and portable SARS-CoV-2 detection was employed by Argentina and CASPR Biotech [75]. They collected saliva specimens from COVID-19 patients and reported that naturally occurring molecules present in saliva did not influence any inhibitory problems during the CRISPR/Cas12-based paper strip assay [75]. Additionally, a cross-institution from China checked an unambiguous detection of SARS-CoV-2 using the CRISPR/Cas12 system [76]. They designed crRNAs targeting orf1a, orf1b, N, and E genes of SARS and its related virus (Wuhan-Hu-1 strain; GenBank. No: MN908947), which detected the single nucleotide polymorphisms (SNPs). Based on the sensitivity and fluorescence intensity, E-crRNAmix was employed in screening SARS-CoV-2, whereas other target genes such as orf1a-crRNAmix, orf1b-crRNAmix, and N-crRNAmix also additionally confirmed the diagnosis of virus [76]. From the results, Feng and Li stated that CRISPR/Cas12-based detection could be utilized for the efficient and rapid diagnosis of COVID-19 (Figure 1) [76]. Recently, Jiang et al. [77] detected SARS-CoV-2 by developing a magnetic-pull-down-assisted colorimetric method associated with CRISPR/Cas12 system (M-CDC). For this method, they used gold nanoparticle (AuNP) probes to detect SARS-CoV-2. In addition, they screened 41 viral samples using M-CDC and observed performance of more than 95%, which is equal to the approved RT-PCR technique for detecting the virus. Thus M-CDC seems to be an efficient method for screening the SARS-CoV-2 without the need of sophisticated instruments [77].
7. CRISPR/Cas13 system–mediated diagnosis
The CRISPR/Cas13 mediated genome-editing is also utilized for a highly sensitive pathogen-detection (Table 3) [55,78]. The first reported platform of CRISPR/Cas13 based system is called as specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) (Figure 2) [79]. SHERLOCK technique combines isothermal and RPA or reverse transcription (RT)-RPA with nuclease activity of Cas13. The crRNA–Cas13 complex attached to the target nucleic acids cleaves the targeted nucleotides as well non-targeted RNA which are coupled with a fluorescent reporter that provides accurate fluorescent signal for real-time detection of pathogens. Gootenberg et al. [79] diagnosed E. coli and Pseudomonas aeruginosa with the DNA isolated from these strains using SHERLOCK system. They have also discriminated the isolates of Klebsiella pneumoniae with two types of resistance genes such as K. pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase 1. In addition, SHERLOCK allowed the discrimination of the targets that differ in only a single base-pair and distinguished successfully as African and American strains of ZIKA virus, serotype of dengue virus (DENV), cancer-related DNA target, and three ssRNA targets in a single reaction [79]. Gootenberg et al. [79] reported that this SHERLOCK could be used to detect targeted nucleic acids with higher sensitivity within 30 minutes including Zika virus and DENV. SHERLOCK has also been applied for the viral diagnosis [79]. Researchers developed a novel method by binding SHERLOCK with heating unextracted diagnostic samples to obliterate nucleases (HUDSON) for the detection of viral infection [80]e. This method detects the DENV in the saliva, blood, and serum of viral infected patient within 2 hours [80]. Such platforms paved the way to utilize this technique to battle against COVID-19 or any other infectious diseases. In 2020, SHERLOCK Biosciences, Inc. and Mammoth Biosciences, Inc. brought out a new combination for diagnostics of COVID-19 by utilizing SHERLOCK technique. They first amplified the viral RNA using RPA and performed in vitro transcription of amplified DNA into RNA. They designed sgRNA targeting S and Orf1ab genes and detected RNA using Cas13 nuclease. Then they used a commercially available paper stick for visual color readout and captures the cleaved reporter RNA. From the readout results, the researchers informed that this protocol would detect the SARS-CoV-2 target RNA sequence with a higher sensitivity of 10 to 100 copies/µl. The Mammoth Biosciences, Inc. stated that the SHERLOCK technique would detect the SARS-CoV-2 within an hour without any special instrumentation. Metsky et al. [81] evaluated the SARS-CoV-2 assay by utilizing CRISPR/Cas13 detection system [81]. The researchers created a website for CRISPR/Cas13 based assay designs to detect 67 viral species including the SARS-CoV-2. Using assay designs they screened the 4 SARS-CoV-2 designs with the help of CRISPR/Cas13 system. They incorporated synthetic RNA targets with fluorescent visualizers and detected 10 copies/μl of synthetic SARS-CoV-2-RNA [81].
Table 3.
Details on molecular diagnostic tools-based detection tests used for SARS-CoV-2. The details such as test name, type of test, source of sample, targeted genes, results, test time and name of the organization with their country are included
| Test Name | Type of test | Sample source | Targeted genes | Test time | Organization Name | Country |
|---|---|---|---|---|---|---|
|
iAMP COVID-19 detection Kit |
RT-RPA | Nasal, nasopharyngeal and throat swabs | ORF1 and N | <1.5 hours | Atila Biosystems Ltd. | United States |
| ID NOW COVID-19 | RT-RPA | Nasal, nasopharyngea, and throat swabs | RdRP | <5 minutes | Abbott Diagnostics Scarborough Ltd. |
United States |
| SARS-CoV-2 | RT-PCR | Nasal, nasopharyngeal, throat swabs, midturbinate nasal swabs and bronchoalveolar lavage fluid | N1 and RdRP | <1 hour | Diagnostic Molecular Laboratory |
United States |
| RT-PCR (COVID-19) | qRT-PCR | Nasal, nasopharyngeal, throat swabs, midturbinate nasal swabs, bronchoalveolar lavage fluid, aspirates, and sputum | N | 4 days | LabCorp Laboratory Corporation of America |
United States |
| ARIES SARS-CoV-2 assay | qRT-PCR | Nasal | ORF1 lab and N | 2 hours | Luminex Corporation |
United States |
| SARS-CoV-2 | RT-RPA | Respiratory samples | E and N | 30–40 minutes | Mammoth Biosciences |
United States |
| Accula SARS-CoV-2 | PCR | Throat and nasal swabs | N | 30 minutes | Mesa Biotech Inc. |
United States |
| QIAstat-Dx Respiratory SARS-CoV-2 | qRT-PCR | Nasopharyngeal | Viral RNA | 1 hour | Qiagen GmBH | United States |
| TaqPath COVID-19 combo Kit | qRT-PCR | Oropharyngeal, nasopharyngeal, and anterior nasal | ORF1, S, and N | - | Rutgers Clinical Genomics Laboratory |
United States |
| cobas SARS-CoV-2 | qRT-PCR | Nasal and nasopharyngeal | Viral RNA | 5 hours | Roche Molecular Systems. Ltd | United States |
| Viracor SARS-CoV-2 assay | qRT-PCR | Oropharyngeal, nasopharyngeal, and anterior nasal | N | 18 hours | Viracor Eurofins Clinical Diagnostics |
United States |
| BioFire COVID-19 | qRT-PCR | Nasopharyngeal | ORF1ab and ORF8 | 1. 5 hours | bioFire Defense LLC. |
United States |
| CDC-2019 | PCR | Oropharyngeal, Nasopharyngeal, and anterior nasal, nasal swabs, bronchoalveolar lavage fluid, aspirates and sputum | N | - | CDC-US | United States |
| Xepert Xpress SARS-CoV-2 | PCR | Nasal and nasopharyngeal | N2 and E | 45 Minutes | Cepheid | Australia, Canada, Singapore, and United States |
| Panther Fushion SARS-CoV-2 | PCR | Nasopharyngeal | ORF1 lab 1 and 2 | <3 hours | Hologic Inc. | Australia and United States |
|
MiRXES FORTITUDE KIT |
qRT-PCR | Nasopharyngeal | Viral RNA | 90 Minutes | mIREXES Pte Ltd. |
Singapore |
|
VitaPCR SARS-CoV-2 |
PCR | Nasal and nasopharyngeal | Viral RNA | 20 Minutes | Credo Diagnostics Biomedical Ptd. Ltd. |
Singapore |
|
Lyra SARS-CoV-2 |
qRT-PCR | Oropharyngeal and nasopharyngeal | Pp1ab | 75 Minutes | Diagnostics Hybrid Inc. |
Canada |
| Nucleic Acid Diagnostic Kit (COVID-19) | qRT-PCR | Oropharyngeal, nasopharyngeal, serum, blood, and fecus | ORF1 lab 1 and E | 30 Minutes | Sansure Biotech Inc. |
China |
|
Standard M nCOV RT Detection Kit |
qRT-PCR | Oropharyngeal swabs | E and RrRP1 | 50 Minutes | SD Biosensor |
South Korea |
RT-RPA, reverse transcription-recombinase-polymerase isothermal amplification; qRT-PCR, quantitative reverse transcriptase-polymerase chain reaction; N, nucleocapsid; ORF1, open reading frame; E, envelope; pp1ab, poly proteins.
Rauch et al. [82] applied Cas13-based-Ragged-Equitable-Scalable Testing (CREST) protocol to check the potency to detect SARS-CoV-2 [82]. This system involves the use of simple, effortless to operate, portable, and plastic-filter-based LED visualizers. In previously published works, researchers detected target RNAs using Cas13 nuclease with the help of lateral flow immunochromatography strips, but these strips demand a high cost. Therefore, they designed cost less P51 cardboard fluorescence visualizer (using 9 V battery) instead of immunochromatography strips and detected 10 copies of a target RNA molecule/ml. In addition, they used the smartphone camera for documenting results, and consequent uploading to the cloud to facilitate POC testing [82]. It advances the CRISPR based nucleic acid detection and shows its potency to shift on diagnostic and surveillance endeavors through direct screening even at a large group of populations [82]. Later, Tian et al. [83] group introduced the CRISPR/Cas13a assay for single-molecule RNA diagnosis which eliminated the method of nucleic acid extension and RT procedure. They reported that the CRISPR/Cas13a assay would be a powerful tool to detect the SARS-CoV-2 [83]. From this demonstration, they conclude that the CRISPR/Cas13 system could easily support both patients and public health practitioners for efficiently detecting the SARS-CoV-2 (Figure 1).
8. Economic feasibility of CRISPR/Cas system-based diagnosis
As an emergency situation for diagnosing the SARS-CoV-2, many industries started to discover economically viable kits by utilizing CRISPR/Cas system. As a result, they used DETECTR and SHERLOCK systems for rapid and accurate diagnosing of SARS-CoV-2 [84]. The Mammoth and Biosciences. Inc already started selling the DETECTR and their diagnostic kits in the market to detect viral and bacterial infections [85]. Therefore, the DETECTR and SHERLOCK methods play a major role in detecting SARS-CoV-2 within an hour or less with cost-effective detection. The other companies like Sigma-Aldrich also provide commercial CRISPR/Cas9 kits which detects bacterial and viral infections (Table 3). Recently, DIY scientists’ company offered CRISPR kits for individuals at a low cost of 200 USD. Hopefully, these CRISPR/Cas-based techniques will offer cheap and rapid diagnostics option for other novel pathogens in future.
9. Potential of CRISPR/Cas system in SARS-CoV-2 diagnosis
CRISPR/Cas system has a great potential in various disciplines such as basic sciences, drug development, and developing molecular diagnostics against novel pathogens like SARS-CoV-2. The CRISPR/Cas systems could be a simpler tool to detect SARS-CoV-2 because these tools have successfully detected other pathogens with higher efficiency within 30–60 minutes. As already known, the recently invented CRISPR/Cas12/Cas13 systems cleave both ssDNA and ssRNA. Therefore, several researchers started to utilize the CRISPR/Cas systems for detecting pathogens and discovered fluorescent-labeled ssDNA/RNA probe for developing DNA-based diagnostic tool [86]. SHERLOCK technology uses the CRISPR/Cas13 system to identify the RNA and breaks collateral RNA along with targeted RNA. The non-targeted RNA is tagged to a fluorescent dye for identifying the targeted RNA. Zhang et al. [86] used CRISPR/Cas13 system-based SHERLOCK technique by utilizing two crRNAs which identify the signatures of SARS-CoV-2 [86]. Recently, US FDA approved SHERLOCK technology as an emergency use authorization (EUA) for the detection of SARS-Cov-2 [9]. Ding et al. [87] introduced All-In-One Dual CRISPR/Cas12 (AIOD-CRISPR) assay as an ultra-sensitive and faster assay to detect the target nucleic acid [87]. This AIOD-CRISPR assay mixtures are incubated at 37°C to prevent contaminations, which simplifies the detection procedure. Utilizing AIOD-CRISPR assay, they detected the RNA of HIV and SARS-Cov-2 within 60 minutes [87].
These overall studies proved the potential of the CRISPR/Cas system as an effective diagnostic tool for SARS-Cov-2 infections. Now we see only the beginning of the CRISPR based system for the diagnosis of SARS-CoV-2. Without a doubt, the potential of the CRISPR/Cas system can soon become the leading diagnostic tool for SARS-CoV-2 infection including the mutant strains.
10. Pros and cons of CRISPR/Cas-based diagnosis for infectious disease
In the modern era, many advanced diagnostic methods have been employed to diagnose and combat viral infections. The sudden emergency of novel COVID-19 virus has changed the life-style of humans. The major hurdles are the lack of proper equipments, time-consuming disease diagnosis methods, lack of low-cost techniques for COVID-19 diagnosis, and applying the diagnostic technique in remote areas (higher risk of disease transmission) especially in the developing countries. COVID-19 causes two types of infection: (1) symptomatic and (2) asymptomatic (causes a higher risk of getting unnoticed infectious disease transmission). Therefore, the availability of rapid, accurate diagnostic tools may help in screening the infected individuals and stop the chain of disease transmission by SARS-CoV-2 virus.
Recently invented CRISPR/Cas system plays a vital role in developing novel molecular diagnostic tools to detect SARS-CoV-2. The sensitive detection of COVID-19 infection by using CRISPR/Cas-based SHERLOCK, DETECTR, HOLMES and SHERLOCKv2 are good methods for rapid diagnosis and control of SARS-CoV-2 [35,88]. The CRISPR/Cas-based SHERLOCK and DETECTR techniques distinguish SNP, HPV, Zika, and DENV strains [88].
CRISPR-based diagnostic tools provide a great advantage than the currently used serologic tests, nucleic acid associated tests, and RT-PCR tests for the diagnosis of SARS-CoV-2. The major advantage of the CRISPR system is, it does not require any expensive experiments, highly sensitive, and large-scale screening of infected peoples can be detected within a few minutes that results in stopping the community spread of the SARS-CoV-2 and leads to reduce morbidity and mortality. Even though the standard diagnosis RT-PCR is effective and sensitive, it requires sophisticated instruments, expert manpower, and cost. Therefore, CRISPR-based detection fulfills all the requirements like being easy to use, fast, and applicable in all portable molecular labs. However, the demerits of the CRISPR/Cas system should also be validated before introducing in diagnosing the SARS-CoV-2. The disadvantage is an ‘off-target’ effect which results in false results even though multiple online software tools are available to reduce the off-target effects. Another major disadvantage is the Cas proteins utilized by the CRISPR system which have nucleotide mismatches tolerance that causes nonspecific binding to the targeted template sequences which results in false detection of the target. Also, apart from being a lab based and more speculative detection method, CRISPR-based tests may take more time to make it as a rapid and domestic strip-based test which are normally used for pregnancy tests for the diagnosis of SARS-CoV-2 virus.
Improvement in these techniques would detect the viral infection even faster. Recently, a CRISPR chip has been discovered that detects the virus within 15 min [89,90]. However, developing the new CRISPR/Cas-based diagnostic methods must be carefully validated and tested including off-target phenomenon before functioning, because it may lead to poor signaling and a mistake in the results.
11. Conclusions
The limitations in traditional diagnosis tools open novel ways to utilize recent molecular diagnostic tools for detecting pathogens very quickly and efficiently. CRISPR type II (Cas9), V (Cas12), and VI (Cas13) systems brought an advanced diagnostic method for rapid detection and control of the pathogens including SARS-CoV-2. In the near future, we can also expect a mobile-powered CRISPR/Cas-based alarm system that will provide an early warning if any viral infected patient is nearby. However, inventing the novel CRISPR-based diagnosis tools requires speed and accurate detection. In addition, the evaluation of risks correlated with the utilization of the CRISPR/Cas system for diagnosis of infectious disease will be an important risk factor for the future.
12. Expert opinion
Outbreak of the global pandemic COVID-19 due to SARS-CoV-2 has threatened the entire world. Rapid spread of further mutated SARS-CoV-2 thus caused severe health and economic burdens that require us to develop advanced and ideal strategies for the quick diagnosis and treatment of COVID-19 patients. Recently, SARS-CoV-2 infectious disease has moved to an alarming phase of the second wave in many countries especially India which witnessed a rapid increase of the infected cases and deaths. The spread and death rates are more severe during March–May 2021 in India compared to other countries. The major hidden threat of the COVID-19 disease is asymptomatic carriers. Increasing the frequency of mass-screening of the population is the only and primary strategy to identify and isolate the individuals from spreading the virus. It is really challenging especially in the countries like India. Utilizing specific diagnostic tools of SARS-CoV-2 with quick intervention is believed to check and control the spread of the infectious disease to others.
For an early and efficient detection of SARS-CoV-2, currently the RT-PCR test is being used. However, studies revealed that utilizing RT-PCR techniques to diagnose SARS-CoV-2 demands a longer time to get the results. Furthermore, RT-PCR results sometimes fails to recognize the infected patients as it depends on capturing and detecting the virus that does not deliver any additional evidence on other symptoms. Delaying the RT-PCR test of the collected upper and lower respiratory track samples decreased the viral dosage in the sample, which leads to the negative results of the samples collected. Therefore, combination of RT-PCR with X-ray, and antigen test results are required to stop the chain of COVID-19 transmission among individuals.
While WHO reported about the spread of SARS-CoV-2 infection all over the world, the CRISPR researchers without the second opinion, started to explore the CRISPR systems against the diagnosis of SARS-CoV-2 infection. Since the CRISPR system plays an effective role in defending against viral infection, scientists utilized it as a novel natural diagnostic tool against SARS-CoV-2 for human benefit. In this review, we have highlighted the potential of the Nobel Prize winning CRISPR/Cas system in detail for the effective diagnosis of SARS-CoV-2 infections. This review article covered all the studies on CRISPR/Cas system such as CRISPR/Cas9/12/13 systems till date with their methods for diagnosing the SARS-CoV-2 infection. Therefore, this article is timely and interesting that may help to develop more novel, rapid diagnostic tools based on CRISPR/Cas systems for the treatment of SARS-CoV-2 infection.
Recently, CRISPR/Cas-based SHERLOCK diagnostic tool has been successfully approved by the US-FDA as an efficient diagnostic tool to detect SARS-CoV-2 infection. This approval has encouraged CRISPR researchers to find many novel diagnosis kits based on the CRISPR system. After SHERLOCK, the CRISPR/Cas-based DETECTR system joins the list in diagnosing the SARS-COV-2. DETECTR system takes less than 15 minutes to detect SARS-CoV-2. Multiple groups are actively involved in developing diagnostic kits utilizing CRISPR-based tools. However, all these techniques should come into the marker for diagnosing SARS-CoV-2 infections at a low cost before the outburst of a deadly third wave throughout the world.
It will not be surprising if we expect more CRISPR-based kits in the coming months for novel and rapid tests for mass detection of SARS-CoV-2 infected persons within a fraction of seconds. This may help to detect the infected persons more efficiently with portable detection kits in rural parts of developing countries like India where testing rate is limited due to lack of skilled manpower and RT-PCR instruments. Science has overcome many pandemics and diseases like polio, smallpox, chicken pox, hepatitis, malaria, etc. by prudent application of diagnostic tools and treatments. The best example is the eradication of polio by several countries. Similarly, science will conquer COVID-19 in the near future and CRISPR/Cas system may play a key role in the efficient diagnosis and treatment of this mass-spreading disease.
Funding Statement
The research works in our lab are funded by the Department of Biotechnology, Ministry of Science and Technology, Government of India under grant No: [BT/PR21321/GET/119/76/2016].
Article highlights
We have highlighted the potential of the Nobel Prize-winning CRISPR/Cas system for the effective diagnosis of COVID-19 infections.
SARS-CoV-2 and its newly evolving mutant variants such as B.1.1.7, B.1.526, P.1 of the CoVs are creating serious health concerns worldwide.
With a constantly increasing number of infections and deaths across the globe, rapid, affordable, and specific detections with more accurate diagnosis and improved health treatments are needed to combat the spread of COVID-19.
Presently RT-PCR testing method has been widely used as a highly specific detection method to detect the presence of SARS-CoV-2, but it has its own limitations.
The CRISPR/ type II (Cas9), V (Cas12), and VI (Cas13)-based SHERLOCK, DETECTOR tools are projected for the rapid detection of novel COVID-19 are compared.
We discuss the potential of the CRISPR/Cas system for the rapid diagnosis to enable efficient control of the COVID-19 infections.
Declaration of interest
The author(s) have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Cascella M, Rajnik M, Cuomo A, et al. Features, evaluation and treatment coronavirus (COVID-19). Statpearls [internet]. StatPearls Publishing; 2020. [PubMed] [Google Scholar]
- 2.Li H, Liu S-M, Yu X-H, et al. Coronavirus disease 2019 (COVID-19): current status and future perspective. Int J Antimicrob Agents. 2020;55(5):105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battegay M, Kuehl R, Tschudin-Sutter S, et al. 2019-novel Coronavirus (2019-nCoV): estimating the case fatality rate–a word of caution. Swiss Med Wkly. 2020;150:w20203. [DOI] [PubMed] [Google Scholar]
- 4.Corman VM, Eckerle I, Bleicker T, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance 2012;17(39):20285. [DOI] [PubMed] [Google Scholar]
- 5.Organization WH . World Health Organization coronavirus disease 2019 (COVID-19) situation report. 2020.
- 6.Organization WH. Novel coronavirus (2019-nCoV). Situat Rep. 2020;28 [Google Scholar]
- 7.Alhazzani W, Møller MH, Arabi YM, et al. Surviving Sepsis Campaign: guidelines on the management of critically ill adults with Coronavirus Disease 2019 (COVID-19). Intensive Care Med. 2020;46(5):854–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JS, Ma E, Harrington LB, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387): 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∙∙ Introduced a new method termed DNA endonuclease-targeted CRISPR trans-reporter (DETECTR) based on Cas12 system.
- 9.Broughton JP, Deng X, Yu G, et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∙∙ First detected the SARS-CoV-2 infection using CRISPR/Cas12 system.
- 10.Bhadra S, Jiang YS, Kumar MR, et al. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV). PLoS One. 2015;10(4):e0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang N, Luo C, Liu H, et al. Characterization of a new member of alphacoronavirus with unique genomic features in rhinolophus bats. Viruses 2019;11(4):379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaki AM, Van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. [DOI] [PubMed] [Google Scholar]
- 13.Garneau JE, Dupuis M-È, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010;468(7320):67–71. [DOI] [PubMed] [Google Scholar]
- 14.Gasiunas G, Barrangou R, Horvath P, et al. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci. 2012;109:2579–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mota DS, Marques JM, Guimarães JM, et al. CRISPR/Cas Class 2 systems and their applications in biotechnological processes. Genet Mol Res. 2020;20:1–10. [Google Scholar]
- 17.Makarova KS, Wolf YI, Koonin EV. Classification and nomenclature of CRISPR-Cas systems: where from here? Cris J. 2018;1(5):325–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fung TS, Liu DX. Human coronavirus: host-pathogen interaction. Annu Rev Microbiol. 2019;73(1):529–557. [DOI] [PubMed] [Google Scholar]
- 20.Gorbalenya A, Baker S, Baric R, et al. Coronaviridae study group of the international committee on taxonomy of viruses. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;2020:3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121(1):190–193. [DOI] [PubMed] [Google Scholar]
- 22.Kahn JS, McIntosh K. History and recent advances in coronavirus discovery. Pediatr Infect Dis J. 2005;24(11):S223–S227. [DOI] [PubMed] [Google Scholar]
- 23.Tyrrell DAJ, Bynoe ML. Cultivation of viruses from a high proportion of patients with colds. Lancet 1966;287(7428):76–77. [DOI] [PubMed] [Google Scholar]
- 24.Lan J, Ge J, Yu J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020;581(7807):215–220. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann H, Pöhlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12(10):466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vennema H, Godeke GJ, Rossen JW, et al. Nucleocapsid‐independent assembly of coronavirus‐like particles by co‐expression of viral envelope protein genes. Embo J. 1996;15(8):2020–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mehta P, McAuley DF, Brown M, et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou Y, Liu-Jia-Zi Shao F-S. Perioperative anaphylaxis: a potential hazard to the safety of surgical patients. Chin Med J (Engl). 2020;133(5):609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lei J, Li J, Li X, et al. CT imaging of the 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology 2020;295(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng Z, Lu Y, Cao Q, et al. Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a single-center study in Shanghai, China. Am J Roentgenol. 2020;215(1):1–6. [DOI] [PubMed] [Google Scholar]; ∙∙ First study was performed using chest CT for detecting SARS-CoV-2.
- 32.Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009;155(3):733–740. [DOI] [PubMed] [Google Scholar]
- 33.Van Der Oost J, Westra ER, Jackson RN, et al. Unravelling the structural and mechanistic basis of CRISPR–Cas systems. Nat Rev Microbiol. 2014;12(7):479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koonin EV, Makarova KS. Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol Evol. 2017;9(10):2812–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Li S, Wang J, et al. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37(7):730–743. [DOI] [PubMed] [Google Scholar]
- 36.Sternberg SH, Richter H, Charpentier E, et al. Adaptation in CRISPR-Cas systems. Mol Cell. 2016;61(6):797–808. [DOI] [PubMed] [Google Scholar]
- 37.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connell MR. Molecular mechanisms of RNA targeting by Cas13-containing Type VI CRISPR–Cas systems. J Mol Biol. 2019;431(1):66–87. [DOI] [PubMed] [Google Scholar]
- 39.Makarova KS, Wolf YI, Alkhnbashi OS, et al. An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guk K, Keem JO, Hwang SG, et al. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens Bioelectron. 2017;95:67–71. [DOI] [PubMed] [Google Scholar]
- 42.Pardee K, Green AA, Takahashi MK, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016;165(5):1255–1266. [DOI] [PubMed] [Google Scholar]; ∙∙ First published the rapid, low cost detection of viral strain using CRISPR/Cas9 system.
- 43.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008;322(5909):1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sapranauskas R, Gasiunas G, Fremaux C, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39(21):9275–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brouns SJJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008;321:960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lintner NG, Kerou M, Brumfield SK, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J Biol Chem. 2011;286(24):21643–21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mojica FJM, Díez‐Villaseñor C, Soria E, et al. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36(1):244–246. [DOI] [PubMed] [Google Scholar]
- 48.Ceasar SA, Rajan V, Prykhozhij SV, et al. Insert, remove or replace: a highly advanced genome editing system using CRISPR/Cas9. Biochim Biophys Acta Mol Cell Res. 2016;9:2334–2344. [DOI] [PubMed] [Google Scholar]
- 49.Lewis KM, Ke A. Building the class 2 CRISPR-Cas arsenal. Mol Cell. 2017;65(3):377–379. [DOI] [PubMed] [Google Scholar]
- 50.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015;163(3):759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao P, Yang H, Rajashankar KR, et al. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016;26(8):901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamano T, Nishimasu H, Zetsche B, et al. Crystal structure of Cpf1 in complex with guide RNA and target DNA. Cell 2016;165(4):949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anders C, Niewoehner O, Duerst A, et al. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014;513(7519):569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fonfara I, Le Rhun A, Chylinski K, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42(4):2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.East-Seletsky A, O’Connell MR, Knight SC, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016;538(7624):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smargon AA, Cox DBT, Pyzocha NK, et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016;353(6299):aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu L, Li X, Ma J, et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell 2017;170(4):714–726. [DOI] [PubMed] [Google Scholar]
- 59.Liu L, Li X, Wang J, et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell 2017;168(1–2):121–134. [DOI] [PubMed] [Google Scholar]
- 60.Muller V, Rajer F, Frykholm K, et al. Direct identification of antibiotic resistance genes on single plasmid molecules using CRISPR/Cas9 in combination with optical DNA mapping. Sci Rep. 2016;6(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.James PB, Deng XD, Yu GX, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qin P, Park M, Alfson KJ, et al. Rapid and fully microfluidic Ebola virus detection with CRISPR-Cas13a. ACS Sens. 2019;4(4):1048–1054. [DOI] [PubMed] [Google Scholar]
- 63.Freije CA, Myhrvold C, Boehm CK, et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell. 2019;76(5):826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan TJ, Dhar AK, Cruz-Flores R, et al. Rapid, CRISPR-based, field-deployable detection of white spot syndrome virus in shrimp. Sci Rep. 2019;9(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang Y, Deng Y, Li T, et al. Visual detection of porcine reproductive and respiratory syndrome virus using CRISPR‐Cas13a. Transbound Emerg Dis. 2020;67(2):564–571. [DOI] [PubMed] [Google Scholar]
- 66.Li Y, Mansour H, Wang T, et al. Naked-eye detection of grapevine red-blotch viral infection using a plasmonic CRISPR Cas12a assay. Anal Chem. 2019;91(18):11510–11513. [DOI] [PubMed] [Google Scholar]
- 67.Wu Y, Liu S-X, Wang F, et al. Room temperature detection of plasma Epstein–Barr virus DNA with CRISPR–Cas13. Clin Chem. 2019;65(4):591–592. [DOI] [PubMed] [Google Scholar]
- 68.Khan H, Khan A, Liu Y, et al. CRISPR-Cas13a mediated nanosystem for attomolar detection of canine parvovirus type 2. Chin Chem Lett. 2019;30(12):2201–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bai J, Lin H, Li H, et al. Cas12a-based on-site and rapid nucleic acid detection of African swine fever. Front Microbiol. 2019;10:2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Qian L, Wei W, et al. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth Biol. 2017;6(2):211–216. [DOI] [PubMed] [Google Scholar]
- 71.Shen J, Zhou X, Shan Y, et al. Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction. Nat Commun. 2020;11(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xiong E, Jiang L, Tian T, et al. Simultaneous dual‐gene diagnosis of SARS‐CoV‐2 based on CRISPR/Cas9‐mediated lateral flow assay. Angew Chemie. 2021;133(10):5367–5375. [DOI] [PubMed] [Google Scholar]
- 73.Lin W, Tian T, Jiang Y, et al. A CRISPR/Cas9 eraser strategy for contamination‐free PCR end‐point detection. Biotechnol Bioeng. 2021;118(5):2053–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∙ New approach of CRISPR/Cas9 eraser was demostrated to detect the SARS-CoV-2 and swine flu virus.
- 74.He Q, Yu D, Bao M, et al. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens Bioelectron. 2020;154:112068. [DOI] [PubMed] [Google Scholar]
- 75.Satyanarayana M. A COVID-19 diagnostic that uses CRISPR gets a nod from the FDA chemical & engineering news. 2020.
- 76.Feng M, Li X. Land cover mapping toward finer scales. Sci Bull. 2020;65(19):1604–1606. [DOI] [PubMed] [Google Scholar]
- 77.Jiang Y, Hu M, Liu AA, et al. Detection of SARS-CoV-2 by CRISPR/Cas12a-enhanced colorimetry. ACS Sens. 2021;6(3):1086–1093. [DOI] [PubMed] [Google Scholar]
- 78.Abudayyeh OO, Gootenberg JS, Essletzbichler P, et al. RNA targeting with CRISPR–Cas13. Nature 2017;550(7675):280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336): 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∙ Demonstrated the SHERLOCK system for viral diagnosis.
- 80.Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018;360(6387):444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Metsky HC, Freije CA, Kosoko-Thoroddsen T-SF, et al. CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design. BioRxiv 2020:1–11. [Google Scholar]
- 82.Rauch S, Roth N, Schwendt K, et al. mRNA based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus neutralizing antibodies and mediates protection in rodents. BioRxiv 2020:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]; ∙ Describes the Cas13 based-Ragged-Equitable-Scalable Testing (CREST) protocol to check the potency of detecting the SARS-CoV–2.
- 83.Tian T, Shu B, Jiang Y, et al. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano. 2020;15:1167–1178. [DOI] [PubMed] [Google Scholar]
- 84.Bhattacharyya RP, Thakku SG, Hung DT. Harnessing CRISPR effectors for infectious disease diagnostics. ACS Infect Dis. 2018;4(9):1278–1282. [DOI] [PubMed] [Google Scholar]
- 85.Katalani C, Booneh HA, Hajizade A, et al. CRISPR-based diagnosis of infectious and noninfectious diseases. Biol Proced Online. 2020;22:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang F, Abudayyeh OO, Gootenberg JS. A protocol for detection of COVID-19 using CRISPR diagnostics. A Protoc Detect COVID-19 using Cris diagnostics. 2020;8. [Google Scholar]
- 87.Ding X, Yin K, Li Z, et al. All-in-one dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. Preprint. Posted online March 21, 2020. BioRxiv. [Google Scholar]
- 88.Batista AC, Pacheco LGC. Detecting pathogens with Zinc-Finger, TALE and CRISPR-based programmable nucleic acid binding proteins. J Microbiol Methods. 2018;152:98–104. [DOI] [PubMed] [Google Scholar]
- 89.Gootenberg JS, Abudayyeh OO, Kellner MJ, et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018;360(6387):439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hajian R, Balderston S, Tran T, et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat Biomed Eng. 2019;3:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]


