Abstract
Scaffold molecules interact with multiple effectors to elicit specific signal transduction pathways. CIITA, a non-DNA-binding regulator of class II major histocompatibility complex (MHC) gene transcription, may serve as a transcriptional scaffold. Regulation of the class II MHC promoter by CIITA requires strict spatial-helical arrangements of the X and Y promoter elements. The X element binds RFX (RFX5/RFXANK-RFXB/RFXAP) and CREB, while Y binds NF-Y/CBF (NF-YA, NF-YB, and NF-YC). CIITA interacts with all three. In vivo analysis using both N-terminal and C-terminal deletion constructs identified critical domains of CIITA that are required for interaction with NF-YB, NF-YC, RFX5, RFXANK/RFXB, and CREB. We propose that binding of NF-Y/CBF, RFX, and CREB by CIITA results in a macromolecular complex which allows transcription factors to interact with the class II MHC promoter in a spatially and helically constrained fashion.
The major histocompatibility complex (MHC) class II proteins play a central role in the immune response. Extensive analysis has underscored that much of the fluctuation in class II MHC antigen expression can be attributed to changes at the transcriptional level (46, 47). In addition to the class II MHC molecules themselves, associative accessory molecules that are necessary for class II antigen MHC function appear to be controlled in a similar fashion. These associative molecules include the MHC class II-associated invariant chain (Ii) and the more recently described DM heterodimer. All class II MHC, Ii, and DM promoters share the unique presence of three DNA elements, called W, X, and Y, which are highly conserved and critical for promoter function (2, 15). The W-X-Y elements are not only important for constitutive gene expression in B cells but also critical for inducible gene expression. In addition to the conservation in sequence, the spacing between the X and Y elements is highly conserved at approximately two helical turns. Increasing the number of helical turns between these two elements preserves function, while disrupting this orientation destroys promoter activation. Our group previously hypothesized that this restrictive spacing may be required to align the X and Y elements on the same side of the DNA helix, thus allowing transcription factors which can bind these elements to directly interact or to participate in the assembly of a larger promoter complex (48, 49).
The Y box is a CCAAT motif, and it interacts with NF-Y/CBF (also known as YEBP/CP-1). NF-Y/CBF is composed of A, B, and C subunits (26, 27, 57), with the conserved core sequences of NF-YC (CBF-C) and NF-YB (CBF-A) forming a histone fold motif similar to the nucleosome subunits H2A and H2B (1). NF-Y/CBF plays a critical role in opening chromatin because mutation of the NF-Y/CBF-binding sites in both the DRA and Ii promoters results in the loss of protein binding across these promoters in intact cells (24, 54). NF-Y/CBF can preset chromatin for other transcriptional coactivators, such as the histone acetylase GCN5, p300, and pCAF (10, 19, 23). The X box is a bipartite sequence. X1 is bound by the trimeric transcription factor, RFX, formed by RFX5, RFXAP, and RFXANK/RFXB (12, 28, 45). The lack of RFX results in several subclasses of bare lymphocyte syndrome (BLS), a severe immunodeficiency attributed to the lack of class II MHC expression. RFX is required for both the constitutive and gamma interferon (IFN-γ) induction of class II MHC expression (5). The X2 element binds a protein complex, X2BP, which has been recently identified as the CREB protein (29).
Despite the extensive demonstration that X and Y box-binding proteins are important for class II MHC regulation, these proteins are constitutively expressed and cannot explain the cell-, tissue-, developmentally, and cytokine-inducible expression of class II MHC. A major puzzle of class II MHC gene control was solved by the seminal isolation of the class II transactivator, CIITA (43). CIITA was identified by complementation cloning of the DR− mutant B-cell line, RJ2.2.5. CIITA complemented not only the in vitro-generated RJ2.2.5 but also cell lines from type II (group A) BLS patients. CIITA is now shown to be required for IFN-γ, interleukin-4 (IL-4), IL-10, IL-1, transforming growth factor β, and lipopolysaccharide control of class II genes (4, 5, 18, 32, 44). CIITA is itself not a DNA-binding protein and has both conventional and unconventional features for a transcriptional regulator. It has acidic residues similar to those of other transcriptional activation domains (37, 58) followed by stretches rich in proline, serine, and threonine. CIITA also has a nuclear localization sequence as demonstrated by the analysis of a group A BLS cell line which lacks an exon-encoding sequence critical for nuclear translocation (9). However, unlike conventional transcription factors, CIITA does not contain homologies to known DNA-binding domains, and it does not appear to bind any of the class II MHC promoter elements. It also has three regions similar to GTP-binding consensus motifs that are important for function (6, 17, 55). These motifs can be replaced with their counterparts from Ras, a prototype GTPase and G protein. The presence of a functional GTP-binding motif in a transcription factor is first described for CIITA; thus, it may represent a novel class of transcriptional coactivator.
A crucial remaining question is the mode of action of CIITA. The most likely scenario is that CIITA interacts with RFX and NF-Y/CBF. Interaction between CIITA and RFX5 has been detected using an in vitro cell-free system, where CIITA and RFX5 were placed in a two-hybrid system to reveal functional interactions (39). This report shows that CIITA does not interact with NF-YA but does interact with the NF-YB and NF-YC subunits of NF-Y/CBF and the RFX5 and RFXANK/RFXB subunits of RFX, in addition to CREB. Detailed mapping analyses identified the distinct sequences within CIITA that are required for interaction with these first-tier transcription factors. RFXANK requires the N-terminal residues 1 to 335 of CIITA for interaction, while RFX5 requires the adjacent residues 336 to 612. Similarly, NF-YC requires residues 218 to 335, while NF-YB requires the adjacent residues 518 to 612. The C-terminal half of the molecule is not involved in these interactions. A model is suggested where RFX and NF-Y/CBF interact with CIITA in a highly specific fashion, to result in the stereospecific alignment of X and Y elements. This further suggests that CIITA may serve as a scaffold for the specific recruitment and binding of DNA-binding proteins, to cause the selective activation of class II MHC, Ii, and DM promoters.
MATERIALS AND METHODS
Plasmid constructs.
The construction of the wild-type DRA-CAT reporter gene and its mutant forms W(X+5)Y, W(X+10)Y, (W+5)XY, and (W+10)XY was described previously (49). DRA-CAT contains 141 bp of DRA promoter fused to the chloramphenicol acetyltransferase (CAT) gene. W(X+5)Y, W(X+10)Y, (W+5)XY, and (W+10)XY mutant forms of DRA-CAT contain either a 5-bp TGCAG or a 10-bp TGCAGGTCGC insertion. The constructs mutW, mutX, and mutY have been described previously (30).
The cDNA for cloning RFXANK/RFXB was generated using mouse mammary tumor virus reverse transcriptase (Bio-Rad) and random hexamers (Gibco-BRL) as primers from Raji total RNA isolated using the TRIzol reagent (Gibco-BRL). The cDNA was further amplified by PCR using oligonucleotide primers based on the published human RFXANK/RFXB sequence (28, 31). The DNA fragment generated was introduced into pcDNA3 mammalian cell expression vector (Invitrogen) by insertion into the EcoRV cloning site. The construct was characterized by automated DNA sequencing. With the RFXANK/RFXB as parent template, FlagRFXANK and mycRFXANK were constructed with the addition of Met-Flag and Met-myc 5′ to the original initiator start codon. FlagRFX5 and mycRFX5 were respectively produced from RFX5 (3) similar to the construction of epitope-tagged RFXANKs. The NotI-digested mycRFX5 PCR fragment was directly introduced into EcoRV- and NotI-digested pcDNA3, while FlagRFX5 was ligated into pcDNA3 via shuttling through PCRII (Invitrogen).
Using mycRFX5 as a template, C-terminal truncation mutants mycRFX5 (1–570), mycRFX5(1–537), mycRFX5(1–501), mycRFX5(1–466), mycRFX5(1–427), mycRFX5(1–343), mycRFX5(1–307), mycRFX5(1–263), mycRFX5(1–225), mycRFX5(1–200), mycRFX5(1–169), and mycRFX5(1–135) were constructed by PCR using cloned Pfu DNA polymerase (Stratagene) with a common upper-strand primer complementary to pcDNA3 (5′-GCCCTCTAGATGCATGCTCG) and different lower-strand primers distributed along the human RFX5 sequence to introduce stop codons. The PCR-amplified fragments were subjected to HindIII restriction enzyme digestion and ligation into a HindIII-EcoRV-cleaved pcDNA3 fragment. Flag-tagged RFX5 N-terminal mutants FlagRFX5(46–617), FlagRFX5(79–617), FlagRFX5(121–617), FlagRFX5(155–617), and FlagRFX5(162–617) were generated in a way similar to the construction of C-terminal mycRFX5 deletions using upper-strand primers that complemented different regions of the RFX5 sequence, initiating at the position indicated in their names.
Full-length FlagNF-YA, FlagNF-YB, and mycNF-YB were generated using the previously described pGEM-GST-NF-YAq and pGEM-GST-NF-YB (25, 50, 54) as templates. Flag and myc sequences were introduced by PCR. The EcoRV-XhoI-digested PCR-amplified fragments were ligated into pcDNA3. The construction of full-length FlagNF-YC and mycNF-YC was similarly performed. PCR-amplified FlagNF-YC and mycNF-YC used CBF-C (41) as a template, and the products were introduced into pcDNA3 at the EcoRI site with the correct orientation verified by restriction enzyme cutting. The EcoRI-XhoI-digested PCR fragment of myc-tagged NF-YC was ligated into pcDNA3 cleaved with the same restriction enzymes.
The construction of FlagCIITA (6) and FlagCIITA(1–1017) (7) has been described previously. They were produced in the pcDNA3 expression vector (Invitrogen). Using FlagCIITA as a parent template, stop codons were introduced at designated residue positions to generate FlagCIITA(1–335), FlagCIITA(1–421), FlagCIITA(1–444), FlagCIITA(1–518), FlagCIITA(1–612), and FlagCIITA(1–793) using the QuickChange mutagenesis protocol (Stratagene). FlagCIITA (1–186) was constructed in a fashion identical to that of FlagRFX5 C-terminal series truncation mutants using FlagCIITA as template. pUC19-mycCIITA was generated by ligating overlapping oligonucleotides coding for the myc epitope tag, MEQKLISEEDL, to the N terminus of the CIITA cDNA and replacing the original initiator methionine. The EcoRI-SalI mycCIITA insert was transferred into EcoRI-XhoI-digested pcDNA3. mycCIITA(1–793) and mycCIITA(1– 1017) were constructed by introducing the SacII-XhoI restriction fragments of FlagCIITA(1–1017) and FlagCIITA(1–793) into mycCIITA SacII-XhoI sites, respectively. mycCIITA(1–518) and mycCIITA(1–612) were generated by changing residues 519 and 613 into a stop codon using the QuickChange mutagenesis kit (Stratagene). Flag epitope-tagged CIITA N-terminal deletions FlagCIITA(88–1130), FlagCIITA(148–1130), FlagCIITA(180–1130), FlagCIITA(218–1130), FlagCIITA(254–1130), FlagCIITA(300–1130), FlagCIITA(335–1130), FlagCIITA(518–1130), FlagCIITA(613–1130), and FlagCIITA(706–1130) were made in a way identical to the construction of Flag-tagged RFX5 N-terminal deletions.
pcDNA3myc is a generous gift from Yue Xiong, and Rous sarcoma virus (RSV)-CREB was kindly provided by Shannon Kenney, University of North Carolina (UNC) Lineberger Cancer Center. All plasmids were purified using a Qiagen column (Qiagen) prior to transfection. Detailed plasmid construction information is available upon request.
Antibodies.
With the GCG program (GCG, Madison, Wis.) as a search tool, peptide sequences QDVQKFSDNDKLC (at the N terminus of human RFX5) and HTEDNKRRTLQRNDC (at the C terminus of human NF-YC) were selected for the production of rabbit anti-RFX5 and rabbit anti-NF-YC antibodies, respectively. The synthetic peptides were conjugated to keyhole limpet hemocyanin (UNC Peptide Synthesis Core Facility) and were used to prepare antisera (Rockland Immunochemicals Inc.). Monoclonal anti-myc (9E10) was purified from 9E10 hybridoma culture medium using a protein A/G affinity column (Pierce). Monoclonal anti-Flag (M5), and monoclonal anti-Flag–agarose were purchased from Sigma. Rabbit anti-myc and rabbit anti-CREB were obtained from Santa Cruz. Goat anti-mouse immunoglobulin G (IgG) Dynabeads and sheep anti-rabbit IgG Dynabeads were from Dynal Corp. (Oslo, Norway).
Transient transfection and CAT assay.
Transient transfection and subsequent CAT assay were performed as described previously (54) using U373-MG cells, a glioblastoma multiform cell line. Briefly, 10 μg of DRA-CAT reporter plasmid was cotransfected with 10 μg of CIITA or its vector pcDNA3 into 4 × 106 U373-MG cells by electroporation. Transfected cells were incubated with or without 500 U of IFN-γ per ml. Cells were harvested 48 h later for assay of CAT levels as described previously (54). The quantitation was performed with a Molecular Dynamics PhosphorImager. Fold induction is calculated by dividing the percentage of acetylation in the presence of CIITA or IFN-γ by its corresponding controls.
In vitro transcription, translation, and binding assay.
Plasmids were in vitro transcribed and translated using TNT T7 transcription and translation coupled kit (Promega). For one reaction, 1 μg of each DNA as indicated in each figure legend was incubated with 40 μl of TNT T7 master mix in the presence of 2 μl of [35S]Met in a 50-μl reaction volume for 90 min at 30°C. Protein binding interactions were performed in a 1× TBST environment (1 M NaCl, 40 mM Tris [pH 7.5], 0.25% Tween 20) in a total volume of 55 μl. After 1 h of incubation, 1.8 μg of monoclonal anti-myc antibody (9E10) was added for 2 h before the addition of protein G agarose (Pierce) for another 3 h at 4°C. Samples were washed three times with 1× TBST, prepared according to the standard procedure, and half of the volume was subjected to sodium dodecyl sulfate (SDS)-PAGE electrophoresis (38). Gels were dried before exposure with X-OMAT film (Kodak).
Immunoprecipitation and Western blot analyses of in vivo protein-protein interaction.
COS7 cells were cultured in a 37°C incubator with 5% CO2 in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum. Cells were plated at 9 × 105 cells/100-mm-diameter dish and allowed to grow for 18 h. Cells were cotransfected with 3 μg of each plasmid as described in the figure or its legend for each experiment using Fugene 6 (Boehringer Mannheim) according to the manufacturer's instructions. After 30 to 40 h of culture, cells were washed twice with 1× phosphate-buffered saline and lysed with 1.5 ml of cold RIPA buffer (40) (0.1% SDS, 1% NP-40, 1% deoxycholate, 150 mM NaCl, 2 mM EDTA, 0.01 M sodium phosphate [pH 7.2], and 50 mM NaF) supplemented with a tablet of Complete protease inhibitor cocktail (Boehringer Mannheim) per 50 ml of solution. Immunoprecipitation and Western blotting were performed according to standard procedures (38, 40, 42). Blots were detected by enhanced chemiluminescence (Pierce) using Kodak X-OMAT film. Detailed information is available upon request.
RESULTS
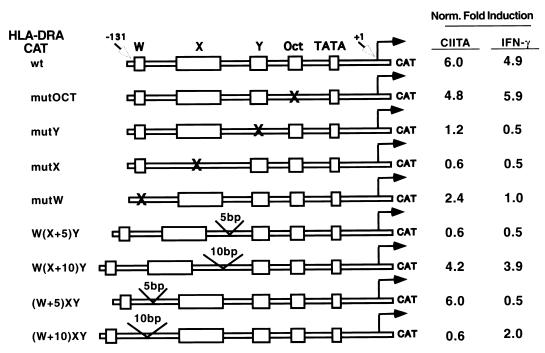
Regulation of the DRA promoter by CIITA requires the proper helical orientation of X and Y elements.
Previous results from this laboratory have shown that the regulation of class II MHC promoters is dependent on the highly specific arrangement of the W, X, and Y elements. The X and Y elements are evolutionarily conserved such that they are separated by a spacer that spans approximately two helical turns of DNA. The addition of complete helical turns (up to six) to this spacer does not alter promoter function. In dramatic contrast, addition or deletion of half-helical turns that would misalign the X and Y elements destroy the function of this promoter. Altering the distance between W and X also dramatically decreases promoter activity, although this does not depend on the helical orientation. These results lead to our hypothesis that X and Y binding proteins may have to bind to the same side of the DNA for proper direct interaction (48, 49). Furthermore, indirect interaction with a second-tier transcription factor may also require the stereospecific alignment of these DNA elements.
CIITA may represent a potential second-tier transcription factor. To address this hypothesis, experiments were performed to assess if the regulation of a class II MHC promoter by CIITA also requires stereospecific alignment of X and Y elements, and if the requirement parallels that of IFN-γ-induced promoter activation. The previous reports cited above showed that class II promoter activation in B-cell lines and by IFN-γ requires stereospecific alignment of these two promoter elements, while this experiment directly assesses if CIITA has a similar stereospecific requirement. As shown in Fig. 1, CIITA and IFN-γ induced similar levels of reporter gene expression under the control of a wild-type DRA promoter (row 1). A mutation in the W, X, or Y element greatly reduced both IFN-γ- and CIITA-induced promoter activation, as expected. In contrast, a mutation of the octamer element had little effect. The addition of half a helical turn (5 bp) to the spacer between X and Y [construct W(X+5)Y] destroyed DRA promoter activation by both CIITA and IFN-γ, while the addition of one helical turn (10 bp) [construct W(X+10)Y] restored most of the activity. This indicates that CIITA activates only class II promoters that maintain helical alignment of the X and Y elements. The addition of any distance between W and X, regardless of helical orientation, destroyed promoter activation by IFN-γ. However, only the addition of 10 bp, not 5 bp, destroyed promoter activation by CIITA. This suggests that overexpression of CIITA can overcome short-distance changes between W and X, although the mechanism is unclear. Together, these data suggest that the regulation of a class II MHC promoter by CIITA requires properly aligned X and Y and, to a lesser extent, W. It is then plausible that CIITA may serve as the second-tier protein that binds to RFX, CREB, and NF-Y/CBF, which in turn recognize X and Y elements.
FIG. 1.
Transactivation of the DRA promoter by CIITA requires stereospecifically aligned and/or properly spaced W-X-Y elements. The transactivation of different DRA promoter mutants by CIITA compared with that by IFN-γ is shown. Fold activation is calculated as CAT expression in the presence of CIITA or IFN-γ divided by CAT expression in its absence. Mutated W, X, Y, or octamer sequence is indicated by an “X.” W(X+5)Y and W(X+10)Y have insertions of 5 and 10 bp, respectively, between X and Y, while (W+5)XY and (W+10)XY have insertions of 5 and 10 bp, respectively, between W and X.
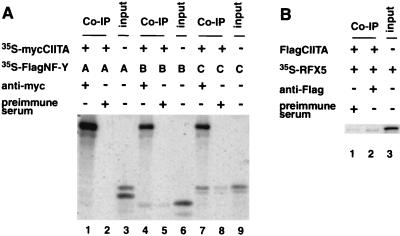
In vitro interaction of CIITA with NF-YB and -C and RFX5.
A cell-free, in vitro translation system was used to assess if CIITA interacts with subunits of NF-Y/CBF and RFX. myc-tagged CIITA (mycCIITA) was cotranslated with either NF-YA, -B, or -C subunits in the presence of [35S]methionine, followed by immunoprecipitation with an anti-myc antibody or preimmune serum (Fig. 2A). Anti-myc precipitated mycCIITA as expected. The mycCIITA protein did not coprecipitate NF-YA but did coprecipitate NF-YB and -C (lanes 1, 4, and 7). Coprecipitated proteins were easily detectable because they were radiolabeled. Samples treated with preimmune control serum also show bands corresponding to NF-YB and -C (lanes 2, 5, and 8); these are likely due to nonspecific binding of these proteins to serum and agarose beads. Based on the amount of NF-YA, -B, or -C (lanes 3, 6, and 9) that was added to each reaction, relatively more NF-YC was coprecipitated than NF-YB. A similar experiment was performed, except that RFX5 was used in place of NF-Y/CBF subunits (Fig. 2B). RFX5 was also coprecipitated with mycCIITA, in accordance with a previous study using yeast two-hybrid analysis (39), although a band was also detected in the control (lane 1). This is a typical problem that we have noticed with in vitro protein-protein interaction analysis; therefore, we performed most of the following analysis in cell extracts.
FIG. 2.
CIITA interacts with the B and C subunits of NF-Y/CBF and with RFX5 in a cell-free system. (A) CIITA interacts with NF-YC and NF-YB. 35S-labeled myc-tagged CIITA (mycCIITA) and 35S-labeled FlagNF-YA (lanes 1 and 2), FlagNF-YB (lanes 4 and 5), or FlagNF-YC (lanes 7 and 8) were produced by cotranslation of these products. The minus signs in the chart indicate the presence of either an appropriate empty control plasmid or an appropriate Ig control. The mixtures were immunoprecipitated with anti-myc (9E10) antibody (lanes 1, 4, and 7) or preimmune serum (lanes 2, 5, and 8) and anti-mouse IgG agarose. In vitro-translated 35S-labeled FlagNF-YA (lane 3), FlagNF-YB (lane 6), and FlagNF-YC (lane 9) were included to show the electrophoresis pattern of the expected translation products. The translated CIITA appears in the upper part of lanes 1, 4, and 7. Coprecipitated NF-YB (lane 4) and NF-YC (lane 7) are seen comigrating with the input product shown in lanes 6 and 9. (B) CIITA interacts with RFX5 in vitro. The experiment was performed similarly to that described for panel A, except that the in vitro-translated FlagCIITA was not radiolabeled, and only the RFX5 protein was radiolabeled. Lane 2 shows a weak band indicative of RFX5 which coprecipitated with the CIITA protein. Lane 3 shows the electrophoresis pattern of RFX5.
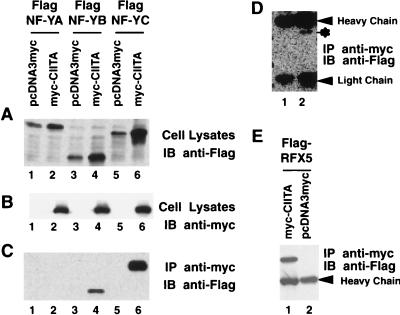
CIITA interacts with NF-YB and -C in cells.
It is crucial that any in vitro result is verified with in vivo findings. In tissues and cell lines tested to date, the detection of endogenous CIITA protein has been difficult due to its extremely low level of expression (5). To overcome this problem, cells were cotransfected with a CIITA expression vector and NF-YA, -B, or -C subunits. The CIITA gene was tagged with a myc epitope, while the NF-YA, -B, and -C subunits were tagged with a Flag epitope. As a control for loading, the NF-Y/CBF subunits from total lysate were detected by immunoblotting with anti-Flag (Fig. 3A), and CIITA was detected by immunoblotting with anti-myc (Fig. 3B). To determine if CIITA interacts with NF-Y/CBF subunits, cotransfected cells were lysed, and the cell lysate was incubated with the anti-myc antibody to precipitate the CIITA protein. The NF-Y/CBF subunits which coprecipitated with CIITA are shown in Fig. 3C. Immunoprecipitation of CIITA coprecipitated NF-YB (lane 4) and NF-YC (lane 6), but not NF-YA (lane 2). An empty vector control in place of the CIITA expression vector did not pull down NF-Y/CBF subunits (lanes 1, 3, and 5). A prolonged exposure of the portion of the gel that contained the NF-YA protein revealed a weak band corresponding to NF-YA. This indicates that the interaction of CIITA with NF-YA is very weak. These findings mirror that of the cell-free system shown in Fig. 2.
FIG. 3.
CIITA interacts with NF-YB–NF-YC and RFX5 in cells. (A and B) Cells (9 × 105) were cotransfected with 3 μg of mycCIITA or its corresponding control vector pcDNA3myc and 3 μg of FlagNF-YA, FlagNF-YB, or FlagNF-YC as indicated. Cells were lysed with 1.5 ml of RIPA buffer 36 h after transfection, and 25 μl of the samples was subjected to SDS–10% polyacrylamide gel electrophoresis resolution. The expression of mycCIITA and FlagNF-YA, FlagNF-YB, or FlagNF-YC was analyzed by Western blotting using either anti-Flag (M5) (A) or anti-myc (9E10) (B) antibodies, which respectively detect the NF-Y subunits or CIITA. IB, immunoblotting. (C) Half of the cell lysate described for panel A was incubated with 3.7 μg of anti-myc (9E10) antibody for 1.5 h. The immunocomplexes were incubated at 4°C overnight with 15 μl of anti-mouse IgG Dynabeads. The association of CIITA with NF-YA (lane 2), NF-YB (lane 4), or NF-YC (lane 6) was studied by Western blotting using the anti-Flag (M5) antibody. IP, immunoprecipitation. (D) Prolonged exposure of the blot in panel C shows a weak interaction between CIITA and NF-YA (asterisk). H and L designate heavy and light chains of Ig, respectively. (E) CIITA interacts with RFX5. Similar to the experiments performed for panels A to D, FlagRFX5 was cotransfected with either mycCIITA or pcDNA3myc. Cell lysate was immunoprecipitated using the anti-myc (9E10) antibody. The associated FlagRFX5 was detected by anti-Flag (M5) antibody immunoblotting.
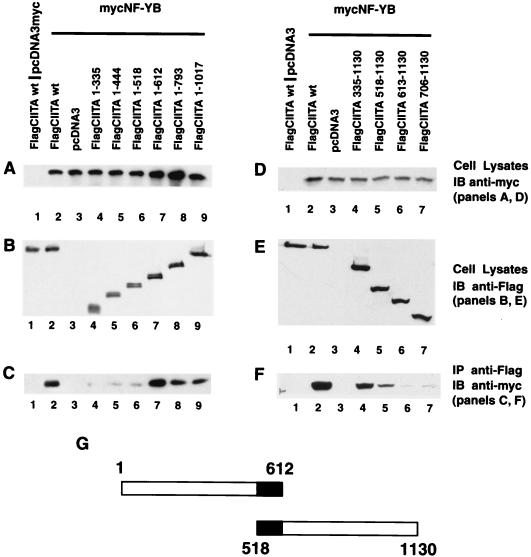
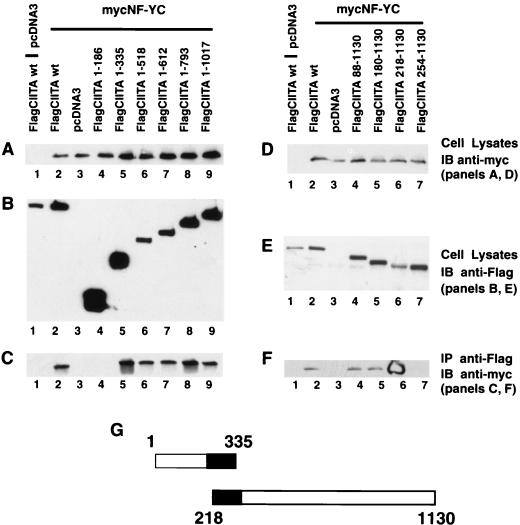
Mapping of the sequence within CIITA that is required for association with the NF-YB or -C subunit.
One working model is that distinct domains of CIITA function together to serve as a scaffold upon which DNA-binding transcription factors important in class II MHC gene transcription can be organized. To explore this possibility, it was necessary to determine the sequences within CIITA that are important for such interactions. This was achieved by constructing a series of CIITA deletion mutants and examining their association with the NF-Y/CBF subunits. As shown in Fig. 4A, six nested, C-terminal deletion mutants of CIITA were constructed. The amino acid sequence which remains in each construct is indicated; thus, CIITA(1–335) contains amino acids 1 through 335. Each FlagCIITA mutant was cotransfected with mycNF-YB or mycNF-YC. The expression of mycNF-YB (Fig. 4) or mycNF-YC (Fig. 5) was confirmed by immunoblotting of total cell lysate with an anti-myc antibody while the CIITA protein was detected using an anti-Flag antibody (Fig. 4B and E). NF-YB was coprecipitated with wild-type FlagCIITA and FlagCIITA(1–1017), -(1–793), and -(1–612) but not with FlagCIITA(1–335) (Fig. 4C). FlagCIITA(1–518) and -(1–444) consistently coprecipitated very small quantities of NF-YB. This indicates that CIITA(1–612) is required for optimal interactions with NF-YB. To further delineate the boundary of the N-terminal sequence of CIITA that is required for interaction with NF-YB, an additional four deletion mutants were made (Fig. 4D). Cell lysates were again analyzed for the expression level of recombinant molecules, and they were similar to wild-type controls (Fig. 4D and E). Deletion construct FlagCIITA(518–1130) retained interactive capacity, while FlagCIITA(613–1130) did not (Fig. 4F). This delineates the CIITA residues required for interaction with NF-YB as amino acids 518 to 612 (Fig. 4G). For the analysis of all deletion mutants, we caution that these interpretations are formed in the absence of structural information.
FIG. 4.
CIITA residues 518 to 612 interact with NF-YB in cells. To map the domain of CIITA that is required for interaction with NF-YB, a series of FlagCIITA C- and N-terminal deletion mutants were constructed. Each sample was cotransfected with 3 μg of FlagCIITA or its mutants and 3 μg of a myc-tagged NF-YB subunit. The subsequent immunoprecipitation, electrophoresis, and Western blotting procedures were performed as described in the Fig. 3 legend. (A) Expression of NF-YB in total cell lysate was detected by immunoblotting with the rabbit anti-myc antibody. (B) Expression of FlagCIITA and its mutants in total lysate was confirmed by immunoblotting with the anti-Flag (M5) antibody. (C) FlagCIITA and its C-terminal deletion mutants were used to coprecipitate mycNF-YB, which was detected by rabbit anti-myc antibody. (D to F) The same as panels A to C, respectively, except that N-terminal deletion mutants were used. (G) The two constructs which delineated the residues within CIITA that interacted with NF-YB. The black area marks the overlapping residues shared by these two constructs.
FIG. 5.
CIITA residues 218 to 335 interact with NF-YC in cells. To map the domain of CIITA that is required for interaction with NF-YC, a series of FlagCIITA C- and N-terminal deletion mutants were constructed. The experiment was performed similarly to that for Fig. 4. (A) Expression of NF-YC in total cell lysate was detected by immunoblotting with the rabbit anti-myc antibody. (B) Expression of FlagCIITA and its mutants in total lysate was confirmed by immunoblotting with the anti-Flag (M5) antibody. (C) FlagCIITA and its C-terminal mutants were used to coprecipitate mycNF-YC, which was detected by rabbit anti-myc antibody. (D to F) The same as panels A to C, respectively, except that N-terminal mutants were used. (G) The two constructs which delineated the residues within CIITA which interacted with NF-YC. See the Fig. 3 legend for more details.
A similar analysis was performed with NF-YC. NF-YC coprecipitated with all the CIITA C-terminal deletion mutants except FlagCIITA(1–186) (Fig. 5A to C). Additional analysis with the N-terminal deletion constructs shows stronger interactions up to FlagCIITA(218–1130) (Fig. 5D and E). A precipitous drop in interaction was observed with a further deletion (Fig. 5F, lane 7) despite a robust level of protein expression. It is notable that FlagCIITA(218–1130) has a lower level of expression than that of the other constructs (Fig. 5E) and yet interacted the most strongly (Fig. 5F, lane 6). The appearance of the signal in this sample is due to its intensity, which depleted the substrate. One interpretation is that the sequences at the N terminus preclude optimal interaction, and their removal in FlagCIITA(218–1130) caused enhanced interaction. Together, these results indicate that residues 218 to 335 of CIITA are required for optimal interaction with CIITA. They also map the NF-YC and NF-YB interaction sites to adjacent but different domains. All the biochemical association experiments in Fig. 5 and the following figures were repeated at least two more times.
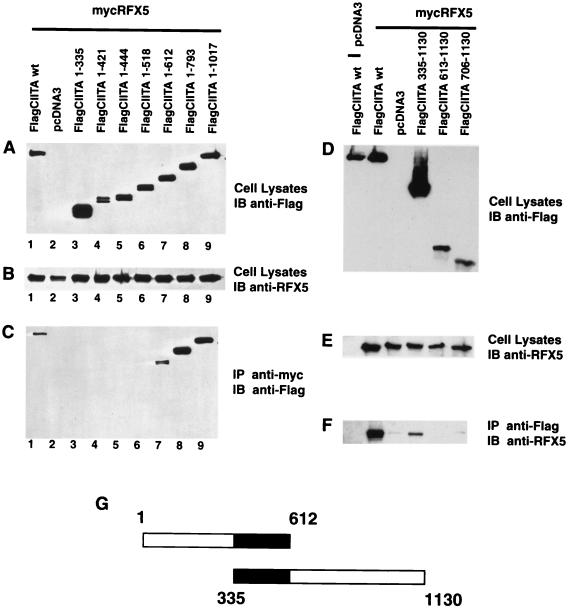
Mapping of sequences required for the association of CIITA and RFX5.
A previous study used CIITA as a bait and RFX5 as the test molecule in a yeast two-hybrid system to detect their interaction. Here we used an in vivo approach to address this issue and additionally mapped the interaction sites on both molecules. The approach is similar to that used in Fig. 3 to 5. The RFX5 protein was precipitated with an anti-myc antibody, and associated CIITA was detected by immunoblotting with the anti-Flag antibody. Figure 6A shows the expression of the nested CIITA deletion mutants in total cell lysate, and Fig. 6B shows the expression of RFX5 protein. CIITA(1–335) through CIITA(1–518) did not coprecipitate FlagRFX5, while mycCIITA(1–612) did. The inclusion of additional sequences in FlagCIITA(1–793) resulted in more coprecipitated proteins. This indicates that FlagCIITA(1–612) contains the minimal sequence required for interaction with RFX5, while additional sequences in CIITA(1–793) further enhanced this association. To further delineate the boundaries of interaction, a series of CIITA N-terminal mutations were also tested. As shown in Fig. 6D and E, FlagCIITA(335–1130) retained interaction with RFX5, while further deletion constructs did not. These data map the boundaries within CIITA that are required for interaction with RFX5 as residues 335 to 612 (Fig. 6G).
FIG. 6.
CIITA residues 335 to 612 interact with RFX5 in cells. To map the domain of CIITA that is required for interaction with RFX5, a series of FlagCIITA C- and N-terminal deletion mutants were constructed. The experiment was performed similarly to that for Fig. 4. (A) Expression of FlagCIITA and its mutants in total lysate was confirmed by immunoblotting with the anti-Flag (M5) antibody. (B) Expression of RFX5 in total cell lysate was detected by immunoblotting with the rabbit anti-RFX5 antibody. (C) FlagCIITA was coprecipitated with anti-myc and detected by immunoblotting with anti-Flag antibodies. (D to F) Similar to panels A to C, respectively, except that N-terminal mutants were used. (G) The two constructs which delineated the residues within CIITA which interacted with RFX5. See the Fig. 3 legend for more details.
To identify the sequences within RFX5 that are required for interaction with CIITA, a series of 17 RFX5 deletion constructs were produced: 11 are C-terminal mutants, and 6 are N-terminal mutants (Fig. 7). The expression of RFX5 in cell lysate was detected by immunoblotting with an anti-RFX5 antibody, and CIITA was detected using an anti-Flag antibody in Fig. 7A to C. Ten of these 11 RFX5 deletion mutants associated with CIITA; only mycRFX5(1–169) did not associate with CIITA. This indicates that mycRFX5(1–200) contains the minimum sequence required for association with CIITA.
FIG. 7.
RFX5 residues 121 to 200 interact with CIITA in cells. The sequence within RFX5 required for interaction with CIITA was mapped using a combination of either C-terminal RFX5 deletion constructs (A to C) or N-terminal deletion constructs. The experiment was done similarly to that described in the Fig. 4 legend. (A to C) FlagCIITA was cotransfected with mycRFX5 or its C-terminal mutants. (A and B) Expression of these proteins in cell lysates; (C) coprecipitated protein. The usage of 10 and 12% gels as indicated in the figure was necessary to optimize the differentiation of different mutant forms of RFX5. (D to F) mycCIITA was cotransfected with FlagRFX5 or its N-terminal mutants. These panels are similar to panels A to C, respectively. (G) The two constructs which delineated the residues within RFX5 which interacted with CIITA.
The N-terminal mutants were tested by immunoprecipitating mycCIITA with the appropriate antibody and then immunoblotting with anti-Flag antibody to detect the RFX5 protein (Fig. 7D to F). FlagRFX5(121–617) coprecipitated with CIITA (lane 6), while further deletions eliminated this interaction (lanes 7 to 9). This delineates the boundaries of the RFX5 domain that are required for interaction with CIITA as RFX5 residues 121 to 200.
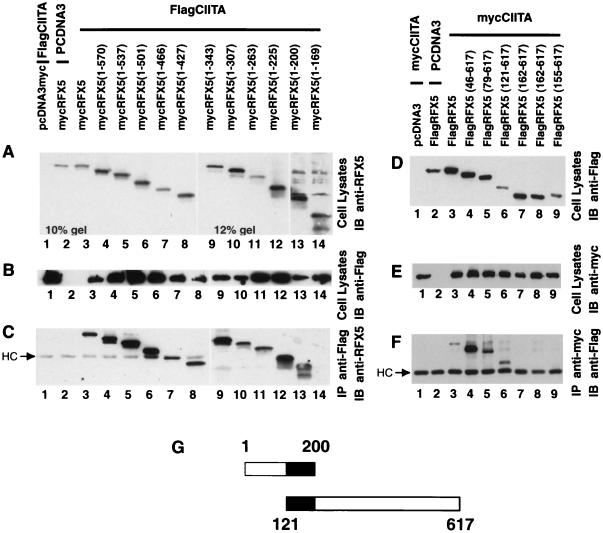
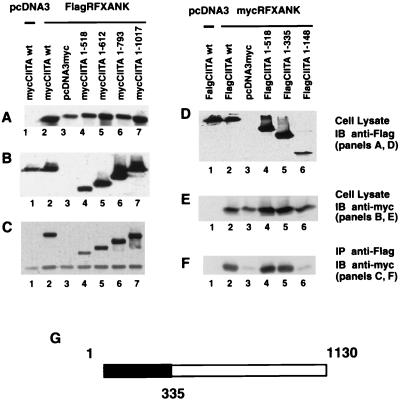
Mapping of sequences within CIITA that are required for association with RFXANK/RFXB and CREB.
A similar strategy was used to map the domain within CIITA that is required for association with RFXANK/RFXB. The panel of Flag-tagged CIITAs was not used in this initial experiment as we encountered technical problems in the recognition of myc-tagged RFXANK/RFXB with anti-myc antibody. Instead, a new panel of Flag-tagged RFXANK and myc-tagged CIITA had to be constructed. As shown in Fig. 8A to C, FlagRFXANK and mutant forms of CIITA were detected in total cell lysate, and FlagRFXANK precipitated all mutant forms of mycCIITA. This indicates that residues 1 to 518 of CIITA interact with RFXANK. To better delineate the region that is required, additional C-terminal deletions were tested (Fig. 8D to F). In these latter experiments, we overcame the technical problems in the recognition of mycRFXANK by anti-myc antibodies. The experiment indicates that the N-terminal 335 residues of CIITA retained interaction with RFXANK but that a deletion mutant containing only the N-terminal residues 1 to 148 lost this interaction.
FIG. 8.
CIITA residues 1 to 335 interact with RFXANK/RFXB in cells. To map the domain of CIITA that is required for interaction with RFXANK/RFXB, a series of CIITA C-terminal deletion mutants were used. The experiment was performed similarly to that for Fig. 4. (A) DNAs used for cotransfection are indicated in the figure. The expression of FlagRFXANK in total lysate was validated by immunoblotting with the anti-Flag (M5) antibody. (B) The expression of mycCIITA and its mutant forms in total lysate was detected by immunoblotting with the anti-myc antibody. (C) Anti-Flag M2 antibody plus anti-mouse IgG Dynabeads was used to immunoprecipitate FlagRFXANK, and coprecipitated forms of mycCIITA were detected by immunoblotting with the anti-myc (9E10) antibody. (D to F) Similar to panels A through C, respectively, except that mycRFXANK and FlagCIITA deletion mutants were used. (G) The region of CIITA that is sufficient for interaction with RFXANK/RFXB.
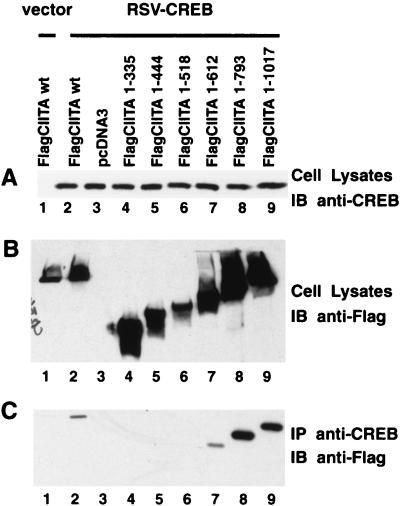
CREB is the protein that interacts with the 3′ half of the X box (also known as the X2 box). To determine if CREB associates with CIITA, cells were cotransfected with a plasmid containing the CREB gene driven by the RSV promoter and Flag CIITA or its various deletion mutants. The expression of CREB and CIITA is shown in Fig. 9A and B, while the coprecipitated CIITA is shown in Fig. 9C. FlagCIITA(1–793) strongly associated with CREB, while FlagCIITA(1–612) associated weakly. Further deletion mutants did not associate with CREB. This indicates that FlagCIITA(1–793) contains the sequences which exhibit optimal association with CREB, while CIITA(1–612) can still associate with CREB.
FIG. 9.
CIITA interacts with CREB. The figure is similar to Fig. 8, except that RSV-CREB was used in place of FlagRFXANK. Rabbit anti-CREB and anti-Flag (M5) immunoblottings were utilized to verify the expression of CREB (A) and that of FlagCIITA or its mutants (B) in total cell lysates. The interaction of CREB and CIITA was analyzed by immunoprecipitation with rabbit anti-CREB antibody plus anti-rabbit IgG Dynabeads, followed by immunoblotting with anti-Flag (M5) (C).
DISCUSSION
The past decade has witnessed an exponential increase in the number of proteins identified as important in signal transduction and in transcription; however, one of the greatest challenges that remain is understanding how these individual components can be assembled in different ways to produce distinct biological effects. The mitogen-activated protein kinase provides an excellent example of this dilemma. In yeast, these proteins have been linked to multiple signaling pathways including responses to mating pheromone, invasive growth, cell integrity, sporulation, and response to high osmolarity (16, 33, 52). Some of these kinase cascades involve the formation of a macromolecular complex, Ste5, which serves as a scaffold interacting with kinases of the pheromone mating pathway, thus promoting this pathway while diminishing the usage of these kinases in other pathways (22). In mammalian cells, scaffolding proteins such as the JNK-interacting protein have been found which preferentially promote the assembly of specific mitogen-activated protein kinases, to result in specific signal transduction pathways (11, 51, 56). Such a system has not been proposed for a protein involved in transcriptional activation, although it seems a logical and elegant possibility.
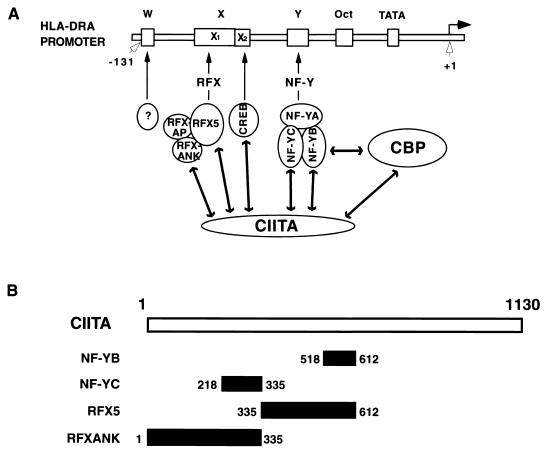
In this report, we show the tour de force analysis of CIITA's association with multiple transcription factors that bind the class II MHC promoter. Although such associations have been proposed, and the interaction with RFX5 has been analyzed, we believe that, for the field to advance, a more detailed analysis of all components is necessary. The results show that CIITA associates with NF-Y/CBF, RFX, and CREB. All three are ubiquitously expressed DNA-binding factors which recognize the prototype class II MHC promoter elements, but by themselves, these three factors are insufficient for the induction of class II MHC promoter. CIITA is additionally required. The data presented here suggest that assembly of the complex formed by the interaction of CIITA–NF-Y/CBF–RFX–CREB is an important step which preferentially brings together the DNA-binding proteins to increase their localized concentration at the site of a class II MHC promoter (Fig. 10A). Thus, by this definition, CIITA may serve as a transcriptional scaffold which enhances the assembly of a class II MHC-specific set of DNA-binding proteins.
FIG. 10.
A model depicting CIITA as a scaffold protein which interacts with DNA-binding transcription factors specific for class II MHC promoters. (A) A prototype class II MHC promoter containing the W-X-Y sequence is shown, along with DNA-binding proteins that recognize these sequences. Interactions of CIITA with NF-YB, NF-YC, RFX5, RFXANK/RFXB, and CREB is depicted in the model. Additionally, the interaction of CIITA with CBP is also shown. (B) A schematic summary of the distinct domains of CIITA which interact with NF-Y and RFX subunits.
The primary strategy is to utilize a nested series of CIITA deletion constructs to map amino acid sequences that are required for interaction with DNA-binding proteins. In one particular case, a nested series of RFX5 deletion constructs were also used. It is well appreciated that the results obtained with deletion constructs have to be interpreted with caution, because deletions may result in secondary or tertiary structural changes without removing the actual interaction sites. With this critical caveat and the necessity for future refined mutagenesis in mind, the sequences within CIITA that are required for interaction with NF-YB, NF-YC, RFX5, RFXANK/RFXB, and CREB were mapped. It is of interest to point out that CIITA interacts with NF-YB and -C but only minimally with NF-YA. In fact, the interaction with NF-YC appears to be the strongest, although quantitative measures and kinetics analyses are necessary to reach a definite conclusion. NF-YB and NF-YC both contain transactivator domains which can interact with basal transcription factors (8). Furthermore, the NF-YB subunit interacts with the coactivator for p300 (13). Together with the current finding, it appears that NF-YB and NF-YC interact with a number of factors.
The interaction of RFX5 with CIITA agrees with and extends previous findings using a yeast two-hybrid system (39). The site within RFX5 that is minimally required for interaction with CIITA lies between amino acids 121 and 200, thus defining a 79-amino-acid stretch that is important for interaction with CIITA. Conversely, the sequences within CIITA which are required to interact with RFX5 have not been previously studied, and it has been shown that they include residues 335 to 612. Likewise, the interaction of RFXANK/RFXB with CIITA has not been described previously, and they require CIITA residues 1 to 335.
It is most interesting that RFX5 and RFXANK/RFXB interact with adjacent but not overlapping regions of CIITA and that this pattern is also observed for NF-YB and NF-YC (Fig. 10B). It is likely that this binding of NF-Y and RFX subunits to adjacent N-terminal domains of CIITA results in a high local concentration of these DNA-binding proteins. This high concentration enhances the interaction of NF-Y and RFX, which has been shown to be an important component of class II promoter activation.
We appreciate the caveat that if two proteins interact with the same residues within CIITA, then the possibility exists that one of the two may be binding directly while the other binds indirectly through the first. The construct CIITA(1–612) yielded a much weaker association with RFX5. Interestingly, this pattern of greater association with CIITA(1–793) and weaker association with CIITA(1–612) is identical for RFX5 and CREB. More-specific mutagenesis is necessary to determine if the CREB and RFX interaction sites are identical. If they are distinct, then it is more likely that these two molecules are directly interacting with CIITA. If they are identical, then it is possible that RFX5 and CREB are associated in vivo and that they are interacting with CIITA as a unit. In other words, only one of these two molecules is interacting directly with CIITA, while the other is indirectly interacting with CIITA through association with its partner, either RFX5 or CREB.
The interaction of CIITA with the DNA-binding transcription factors which specifically bind class II MHC promoters explains much, but not all, of the earlier observations in the field. Earlier work revealed that the class II MHC promoter elements X and Y are highly conserved in evolution and correlatively in function (2, 48). The hypothesis is that proteins binding to these elements have to interact directly or indirectly with a second-tier protein in a highly restrained fashion to cause promoter activation. The interaction of CIITA with NF-Y/CBF, RFX, and CREB now explains these data and additionally identifies CIITA as a second-tier protein (see model in Fig. 10A). As depicted in the model, CIITA interacts with NF-YB, NF-YC, RFX5, RFXANK/RFXB, and CREB proteins, likely resulting in a higher local concentration of essential factors at the DRA promoter. These concerted interactions may also stabilize the macromolecular complex containing all these transcription factors. In addition to interactions among CIITA and its partner proteins, interaction between the X and Y binding proteins also has been reported (34, 54). Specifically, our group reported that an anti-NF-Y antibody can coprecipitate X1 as well as X2 binding activities. Another group showed that X and Y binding proteins cooperate in the binding of their target sites (35). What is yet unknown is whether this constitutes direct or indirect interactions through CIITA. Performance of such experiments in cells with or without CIITA expression would be of interest.
The interaction of CIITA with NF-Y/CBF, RFX, and CREB also explains the role of these factors in causing promoter opening in a sequence-specific manner. These previous experiments were all based on genomic footprint analyses which permitted detection of protein-DNA interactions in intact cells. The results observed with IFN-γ-inducible cell lines show that the occupancy of class II, Ii, and DM promoters by proteins in vivo is dependent on CIITA, NF-Y/CBF, and RFX (53). Direct analysis of cell lines which lack either RFX or CIITA shows a requirement for these two molecules in opening the class II promoters (5, 36, 55). The evidence linking NF-Y/CBF is less direct, because NF-Y/CBF is a critical factor for cell growth and proliferation, and mutants lacking this factor have yet to be identified. An approach was taken to study the role of NF-Y/CBF by in vivo footprint analysis of a panel of stable transfectants harboring either wild-type or mutant promoters (24, 54). The results showed that the ablation of protein binding to a mutated Y element abolished protein binding throughout the promoter. Mutating X has a lesser but similar effect. Combined, it appears that CIITA, NF-Y/CBF, and RFX all play a prominent role in opening the class II MHC promoter.
One likely explanation of the involvement of CIITA, NF-Y/CBF, and RFX in promoter opening is provided by the recent evidence that CIITA and NF-Y/CBF can interact with the histone acetylases CREB-binding protein (CBP) and p300 (14, 21). Recruitment of the histone acetylases causes histone acetylation and likely results in the opening of promoters. It is tempting to propose that CIITA brings together DNA-binding factors to target class II MHC promoters, while tethered CBP causes histone acetylation, promoter opening, and gene transcription (Fig. 10A). However, one unsatisfactory aspect of this model, as it applies to most studies of histone acetylase interaction with transcription factors, is that the promoter sites are presumably not accessible prior to histone acetylation, and therefore it is unclear how a specific DNA-binding factor would target the class II MHC promoter elements.
Another caveat regarding this model is that in BLS patient-derived cell lines which lack class II MHC expression, the lack of RFX is tightly correlated with the lack of in vivo footprints, while the lack of CIITA is not (20), and yet the class II promoter is inactive in both cases. It is possible that, in B-cell lines, another factor can substitute for the function of CIITA or alternatively that the DNA-binding proteins may be expressed at a higher level to compensate for the lack of CIITA. Nonetheless, CIITA serves an essential function in the transcriptional induction of class II MHC in B cells.
In conclusion, this work provides strong evidence for the in vivo interaction of distinct domains of CIITA with the DNA-binding proteins that are involved in class II MHC promoter activation. Functionally, this explains the observation that CIITA can activate class II promoters only when X and Y elements are stereospecifically aligned. Structurally, CIITA may represent a scaffold protein important in the transcriptional activation of a class of genes, all coding for proteins important for class II MHC-mediated antigen processing. At present, it is not entirely clear if the presence of class II MHC promoter enhances these interactions, although we have not noted any difference in these associations upon the addition of template DNA in vitro. More detailed and refined analysis will be necessary to address this point.
ACKNOWLEDGMENTS
We appreciate the helpful discussions of Beverly Errede (UNC Department of Biochemistry and Biophysics) and Brian K. Martin and the secretarial assistance of Gina Horton.
This study was supported by grants from the National Institutes of Health (AI45580, AI41751, and AI29564 to J.P.-Y.T.) and the National Multiple Sclerosis Society (7815 to J.P.-Y.T.).
REFERENCES
- 1.Baxevanis A D, Arents G, Moudrianakis E N, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benoist C, Mathis D. Regulation of major histocompatibility complex class-II genes: X, Y, and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- 3.Brickey W J, Wright K L, Zhu X, Ting J P. Analysis of the defect in IFN-γ induction of MHC class II genes in G1B cells: identification of a novel and functionally critical leucine-rich motif (62-LYLYLQL-68) in the regulatory factor X5 transcription factor. J Immunol. 1999;163:6622–6630. [PubMed] [Google Scholar]
- 4.Chang C H, Fontes J D, Peterlin M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin K-C, Mao C, Skinner C, Riley J L, Wright K L, Moreno C S, Stark G R, Boss J M, Ting J P Y. Molecular analysis of G1B and G3A IFN gamma mutants reveals that defects in CIITA or RFX result in defective class II MHC and Ii gene induction. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 6.Chin K-C, Li G X, Ting J P Y. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: generation of transdominant-negative mutants. Proc Natl Acad Sci USA. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin K-C, Li G X, Ting J P Y. Activation and transdominant suppression of MHC class II and HLA-DMB promoters by a series of C-terminal CIITA deletion mutants. J Immunol. 1997;159:2789–2794. [PubMed] [Google Scholar]
- 8.Coustry F, Maity S N, Sinha S, de Crombrugghe B. The transcriptional activity of the CCAAT-binding factor CBF is mediated by two distinct activation domains, one in the CBF-B subunit and the other in the CBF-C subunit. J Biol Chem. 1996;271:14485–14491. doi: 10.1074/jbc.271.24.14485. [DOI] [PubMed] [Google Scholar]
- 9.Cressman D E, Chin K C, Taxman D J, Ting J P. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–171. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- 10.Currie R A. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 11.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 12.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faniello M C, Bevilacqua M A, Condorelli G, de Crombrugghe B, Maity S N, Avvedimento V E, Cimino F, Costanzo F. The B subunit of the CAAT-binding factor NFY binds the central segment of the co-activator p300. J Biol Chem. 1999;274:7623–7626. doi: 10.1074/jbc.274.12.7623. [DOI] [PubMed] [Google Scholar]
- 14.Fontes J D, Kanazawa S, Jean D, Peterlin B M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaur L K, Heise E R, Ting J P. Conservation of the promoter region of DRA-like genes from nonhuman primates. Immunogenetics. 1992;35:136–139. doi: 10.1007/BF00189524. [DOI] [PubMed] [Google Scholar]
- 16.Gustin M C, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harton J A, Cressman D E, Chin K C, Der C J, Ting J P. GTP binding by class II transactivator: role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 18.Itoh-Lindstrom Y, Piskurich J F, Felix N J, Wang Y, Brickey W J, Platt J L, Koller B H, Ting J P. Reduced IL-4-, lipopolysaccharide-, and IFN-gamma-induced MHC class II expression in mice lacking class II transactivator due to targeted deletion of the GTP-binding domain. J Immunol. 1999;163:2425–2431. [PubMed] [Google Scholar]
- 19.Jin S, Scotto K W. Transcriptional regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kara C J, Glimcher L H. In vivo footprinting of MHC class II genes: bare promoter in the bare lymphocyte syndrome. Science. 1991;252:709–712. doi: 10.1126/science.1902592. [DOI] [PubMed] [Google Scholar]
- 21.Kretsovali A. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levin D E, Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linhoff M W, Wright K L, Ting J P Y. CCAAT-binding factor NF-Y and RFX are required for in vivo assembly of a nucleoprotein complex that spans 250 base pairs: the invariant chain promoter as a model. Mol Cell Biol. 1997;17:4589–4596. doi: 10.1128/mcb.17.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maity S N, Vuorio T, de Crombrugghe B. The B subunit of a rat heteromeric CCAAT-binding transcription factor shows a striking sequence identity with the yeast Hap2 transcription factor. Proc Natl Acad Sci USA. 1990;87:5378–5382. doi: 10.1073/pnas.87.14.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maity S N, Sinha S, Ruteshouser E C, de Crombrugghe B. Three different polypeptides are necessary for DNA binding of mammalian heteromeric CCAAT binding factor. J Biol Chem. 1992;267:16574–16580. [PubMed] [Google Scholar]
- 27.Mantovani R, Pessara U, Tronche F, Li X Y, Knapp A M, Pasquali J L, Benoist C, Mathis D. Monoclonal antibodies to NF-Y define its function in MHC class II and albumin gene transcription. EMBO J. 1992;11:3315–3322. doi: 10.1002/j.1460-2075.1992.tb05410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 29.Moreno C S, Beresford G W, Louis-Plence P, Morris A C, Boss J M. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 30.Moses H, Panek R B, Benveniste E N, Ting J P. Usage of primary cells to delineate IFN-gamma-responsive DNA elements in the HLA-DRA promoter and to identify a novel IFN-gamma-enhanced nuclear factor. J Immunol. 1992;148:3643–3651. [PubMed] [Google Scholar]
- 31.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. . (Erratum, 10:399. [DOI] [PubMed] [Google Scholar]
- 32.O'Keefe G M, Nguyen V T, Benveniste E N. Class II transactivator and class II MHC gene expression in microglia: modulation by the cytokines TGF-beta, IL-4, IL-13 and IL-10. Eur J Immunol. 1999;29:1275–1285. doi: 10.1002/(SICI)1521-4141(199904)29:04<1275::AID-IMMU1275>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Posas F, Takekawa M, Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr Opin Microbiol. 1998;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- 34.Reith W, Siegrist C A, Durand B, Barras E, Mach B. Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc Natl Acad Sci USA. 1994;91:554–558. doi: 10.1073/pnas.91.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reith W, Kobr M, Emery P, Durand B, Siegrist C A, Mach B. Cooperative binding between factors RFX and X2bp to the X and X2 boxes of MHC class II promoters. J Biol Chem. 1994;269:20020–20025. [PubMed] [Google Scholar]
- 36.Rigaud G, De Lerma Barbaro A, Nicolis M, Cestari T, Ramarli D, Riviera A P, Accolla R S. Induction of CIITA and modification of in vivo HLA-DR promoter occupancy in normal thymic epithelial cells treated with IFN-gamma: similarities and distinctions with respect to HLA-DR-constitutive B cells. J Immunol. 1996;156:4254–4258. [PubMed] [Google Scholar]
- 37.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Activation of class II MHC genes requires both the X box region and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 18.1–18.74J. [Google Scholar]
- 39.Scholl T, Mahanta S K, Strominger J L. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sefton B M. Analysis of protein phosphorylation. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 18.2.1–18.2.8. [Google Scholar]
- 41.Sinha S, Maity S N, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Springer T A. Analysis of protein. In: Ausubel M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 10.16.1–10.16.11. [Google Scholar]
- 43.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 44.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 45.Steimle V, Durand B, Barras E, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 46.Steimle V, Reith W, Mach B. Major histocompatibility complex class II deficiency: a disease of gene regulation. Adv Immunol. 1996;61:327–340. doi: 10.1016/s0065-2776(08)60870-6. [DOI] [PubMed] [Google Scholar]
- 47.Ting J P, Zhu X. Class II MHC genes: a model gene regulatory system with great biologic consequences. Microbes Infect. 1999;1:855–861. [PubMed] [Google Scholar]
- 48.Vilen B J, Cogswell J P, Ting J P. Stereospecific alignment of the X and Y elements is required for major histocompatibility complex class II DRA promoter function. Mol Cell Biol. 1991;11:2406–2415. doi: 10.1128/mcb.11.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vilen B J, Penta J F, Ting J P Y. Structural constraints within a trimeric transcriptional regulatory region: constitutive and interferon-gamma inducible expression of the HLA-DRA gene. J Biol Chem. 1992;267:23728–23734. [PubMed] [Google Scholar]
- 50.Vuorio T, Maity S N, de Crombrugghe B. Purification and molecular cloning of the “A” chain of a rat heteromeric CCAAT-binding protein. Sequence identity with the yeast HAP3 transcription factor. J Biol Chem. 1990;265:22480–22486. [PubMed] [Google Scholar]
- 51.Whitmarsh A J, Davis R J. Structural organization of MAP-kinase signaling modules by scaffold. Trends Biochem Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 52.Widmann C, Gibson S, Jarpe M B, Johnson G L. Mitogen-activated protein kinase: conservation of a three-kinase module. Physiol Rev. 1999;79:143–180. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 53.Wright K L, Ting J P. In vivo footprint analysis of the HLA-DRA gene promoter: cell-specific interaction at the octamer site and up-regulation of X box binding by interferon gamma. Proc Natl Acad Sci USA. 1992;89:7601–7605. doi: 10.1073/pnas.89.16.7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright K L, Vilen B J, Itoh-Lindstrom Y, Moore T L, Li G, Criscitiello M, Cogswell P, Clarke J B, Ting J P Y. CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA binding transcription factors. EMBO J. 1994;13:4042–4053. doi: 10.1002/j.1460-2075.1994.tb06721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright K L, Chin K-C, Linhoff M, Skinner C, Brown J A, Boss J M, Stark G R, Ting J P Y. CIITA stimulation of transcription factor binding to MHC class II and associated promoters in vivo. Proc Natl Acad Sci USA. 1998;95:6267–6272. doi: 10.1073/pnas.95.11.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yasuda J, Whitmarsh A J, Cavanagh J, Sharma M, Davis R J. The JIP group of mitogen-activated protein kinase scaffold proteins. Mol Cell Biol. 1999;19:7245–7254. doi: 10.1128/mcb.19.10.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeleznik-Le N, Azizkhan J C, Ting J P Y. Affinity-purified CCAAT-box-binding protein (YEBP) functionally regulates expression of a human class II major histocompatibility complex gene and the herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci USA. 1991;88:1873–1877. doi: 10.1073/pnas.88.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H, Glimcher L H. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]