Abstract
Background:
Peanut and tree nut allergies are the most important causes of anaphylaxis. Co-reactivity to more than one nut is frequent, and co-sensitization in the absence of clinical data is often obtained. Confirmatory oral food challenges (OFCs) are inconsistently performed.
Objective:
To investigate the utility of the basophil activation test (BAT) in diagnosing peanut and tree nut allergy.
Methods:
The Markers Of Nut Allergy Study (MONAS) prospectively enrolled patients aged 0.5–17 years with confirmed peanut and/or tree nut (almond, cashew, hazelnut, pistachio, walnut) allergy or sensitization from Canadian (n=150) and Austrian (n=50) tertiary pediatric centers. BAT using %CD63+ basophils (SSClow/CCR3pos) as outcome was performed with whole blood samples stimulated with allergen extracts of each nut (0.001–1000 ng/mL protein). BAT results were assessed against confirmed allergic status in a blinded fashion to develop a generalizable statistical model for comparison to extract and marker allergen-specific IgE.
Results:
A mixed effect model integrating BAT results for 10 and 100 ng/mL of peanut and individual tree nut extracts was optimal. The area under the ROC curve (AUROC) was 0.98 for peanut, 0.97 for cashew, 0.92 for hazelnut, 0.95 for pistachio, and 0.97 for walnut. The BAT outperformed sIgE testing for peanut or hazelnut and was comparable for walnut (AUROC 0.95, 0.94, 0.92) in a sub-analysis in sensitized patients undergoing OFC.
Conclusions:
BAT can predict allergic clinical status to peanut and tree nuts in multi-nut sensitized children and may reduce the need for high-risk OFCs in patients.
Keywords: Allergy diagnosis, Basophil, Challenge tests, Food allergy, Pediatrics, Molecular allergology, Multiple nut allergy
Introduction
Peanut and tree nut allergy are among the most common food allergies in children across the globe.1–4 The prevalence of peanut and tree nut allergy ranges between 1–5% and 0.05–4.9%, respectively, depending on the population, definition of allergy used, and type of study.2,5,6 The rate of nut sensitization is also influenced by pollen sensitization, which has geographic variability. Peanut and tree nut allergies are not only common, but are the leading causes of life-threatening anaphylactic reactions.7,8 These allergies also negatively impact the health-related quality of life (HRQL) of affected patients and their families.
The diagnosis of peanut and tree nut allergy is a result of the combination of the clinical history of an immediate allergic reaction, skin prick testing (SPT) and specific IgE (sIgE) testing. The diagnosis can become challenging when there is no clear history of peanut or tree nut consumption, or if the SPT and sIgE results are equivocal.9 In addition, peanut allergic patients have increased rates of tree nut sensitization, and those presenting with a single tree nut sensitization are frequently sensitized to other tree nuts.2,10 However, rates of tree nut sensitization are higher than the rates of true allergy.2
Oral food challenges (OFC) are the gold-standard procedure in the diagnosis of food allergy, but they can be associated with the risk of serious allergic reactions and familial anxiety.11,12 OFCs also require medical resources that may not be readily available, especially in the context of multiple food allergies such as with individual tree nuts. OFCs are indicated when the clinical history, combined with results of SPT and sIgE testing, are equivocal, or in the absence of a clinical history. Although diagnostic decision values for SPT and sIgE have been reported in specific populations, there are patients who have results under these cut-offs who fail challenges, as well as those who can tolerate the foods despite having SPT or sIgE values above the cut-offs.13–15 The growing need for OFCs identifies the requirement for more accurate diagnostic tests in food allergy.
Molecular allergy diagnosis (MolAD) has been investigated in food allergy with the intention to separate cross-reactivity with other allergen families and pollen allergens.16 In particular, sensitization to the seed storage peanut protein Ara h 2 has been shown to be an important predictor of clinical reactivity.17,18 Similar heat-stable proteins such Ana o 3 (cashew and also pistachio), Cor a 9 and Cor a 14 (hazelnut), and Jug r 1 (walnut) have been identified for tree nuts.19–22 Studies investigating the relevance of these markers are encouraging but limited.
The basophil activation test (BAT), an in vitro assay using allergen stimulation in whole blood, has the potential to be used for diagnosing peanut and tree nut allergies.23 Flow cytometry is used to evaluate degranulation and/or activation of basophils after stimulation with the respective food allergens.24 It has been demonstrated to be useful due to its high specificity, and in particular when clinical history, SPT and sIgE results are equivocal.23,25
The purpose of The Markers of Nut Allergy Study (MONAS) was to determine, in a real-life setting in a large group of Canadian and Austrian children, the utility and accuracy of the BAT in the diagnosis of peanut and tree nut allergy.
Methods
Patient Population
Patients from 6-months to 17-years with peanut and/or tree nut (almond, cashew, hazelnut, pistachio, walnut) allergy or sensitization were prospectively recruited from the Hospital for Sick Children, Toronto, Canada (n=150) and the Medical University of Vienna, Austria (n=50) between May, 2017 and November, 2019.
Clinical allergy was defined as positive OFC, or a pre-test probability of >90% of allergic reaction (SPT ≥8mm and/or sIgE ≥15 kUA/L).10,14,26 Almond allergy was defined solely based on positive OFCs or recurrent strong clinical reaction upon proven exposure.23,27 Open OFCs, and not double-blind placebo-controlled food challenges (DBPCFCs), were performed as part of the clinical care provided at the allergy clinics at both sites. Non-allergic status was defined as tolerating an equivalent of a full serving of the food. Indeterminate allergic status was defined as sensitized individuals who were not known to tolerate the nut (no OFC, no history of tolerating nut).
Patients were eligible for enrollment to undergo further workup if they met the following inclusion criteria. 1) Peanut or tree nut allergy diagnosed based on an immediate allergic reaction to peanut or tree nut within the last 24 months, with corresponding positive sensitization, or a pre-test probability of >90% of allergic reaction (SPT≥8 mm and/or sIgE≥15 kUA/L). 2) Sensitized to peanut or tree nut, where sensitization was defined by SPT>3 mm and/or sIgE>0.35 kUA/L, who were scheduled to undergo OFCs to at least one of the nuts. 3) Peanut and tree nut sensitized SPT>3 mm and/or sIgE>0.35 kUA/L, but who regularly consume nuts. Exclusion criteria were as follows. 1) Age <6 months. 2) Unclear history of peanut or tree nut allergy with no OFC in allergy clinic. 3) Sensitization and <90% pre-test probability of allergic reaction.
All patients were clinically assessed and clinical data was collected using a standardized data collection form. SPT was performed using peanut and individual tree nut extracts (ALK-Abello, Horsholm, Denmark) using lancets. sIgE level (peanut, individual tree nut, MOLAD) was determined using ImmunoCAP (Thermo-Fisher, Uppsala, Sweden).
Oral food challenges
Open label OFCs were performed in accordance with internal standardized clinical protocols (see Online Repository).
Basophil activation tests
Heparinized whole blood was drawn and delivered to the research lab blinded. It was stored at room temperature and processed within <6 hours. Whole blood was stimulated with up to 7 serial 10-fold dilutions (0.001–1000 ng/ml) with each of peanut extract (ALK Abelló), hazelnut, pistachio, cashew, walnut, and almond allergen extract. Tree nut extracts were made in house and characterized (see Online Repository).
The BAT was performed in accordance with the manufacturer’s instructions (Bühlmann, Switzerland). Flow cytometric analysis was performed either with a CytoFLEX at the Canadian site (Beckman Coulter, United States) or a BD FACSCanto II (BD Bioscience, San Jose California) at the Austrian site. Basophils were gated as SSClow/CCR3+ cells. Basophil activation was determined as the percentage of activated basophils (%CD63+). Testing and analysis was performed in a blinded fashion.
In 9% of individuals who consented, either no blood was drawn (7.5%; n=15) or inclusion criteria were not met upon review (1.5%; n=3). 11.5% of BAT experiments were removed from analysis because of a baseline activation of >5% CD63 in the negative control (5%; n=10), because of a non-response to stimulation with either FMLP or FcƐRI cross-linking antibody (3.5%; n=7), or because of one with technical issues or five having too low basophil counts (3%; n=6).
Assessment of allergen specific sensitization patterns
In addition to the clinically performed allergen testing, sIgE was measured with the Allergy Explorer (ALEX) array (Macro Array Diagnostics, Austria). Patient serum was diluted 1:5 and incubated on the ALEX chip containing 156 extracts and 126 molecular allergen markers as described recently.28,29 The chips were imaged and quantified within the dynamic range of 0.3–40 kUA/L (Raptor, Macro Array Diagnostics). Positive sensitizations were defined as ≥0.30 kUA/L, as reported recently28,30.
Statistical analysis
Performance of BAT was examined against the allergic status to individual nuts using receiver-operating characteristic (ROC) curves via application of statistical models. All modeling and subsequent analyses were performed in R version 3.6.1 (R Core Team 2019). Figures were generated using the ggplot2 package in R.31 Bayesian logistic regression models were fit to predict the allergy status per nut per person using a variable slope, variable intercept mixed effect framework for each allergen concentration per allergen. Concentrations of 10 and 100 (ng/ml) were chosen for statistical modelling as these models resulted with the greatest area under the ROC (AUROC) while maintaining minimal model complexity. Details of original model and model coefficients can be found in the Online Repository. The final equation used in modelling was:
Where there is a variable slope for each concentration and variable intercept for each allergen.
An optimal threshold along the ROC curve was chosen using the Youden’s Index31, which corresponds to the maximum true positive rate (TPR) and minimum false positive rate (FPR) for each nut (this is identical to maximizing both the sensitivity and specificity). The same methodology was applied to the performance of BAT against OFC status to individual nuts.
External validation cohort
To determine the applicability of the established statistical model in an assay using a similar outcome measurement and whole blood stimulation (after ~24 hours at 4° C, with measurement of high CD63+ expression), with a different set of antibodies, and without IL-3 stimulation in vitro, external validation was performed in an independent cohort of peanut allergic patients with DBPCFC proven peanut allergy from the POISED trial (n=109 analyzed; n=9 non-responders; n=2 no data available). BAT results from the baseline visit were used. A detailed description of the patient population and methods was reported recently.32,33
Ethics approval
Research ethics board (REB) approval was obtained at both Austrian (EK Nr.: 2302/2016) and Canadian (1000053791) study sites.
Results
Study Population
Two-hundred patients were prospectively recruited for the study, 150 from Toronto and 50 from Vienna. Three were excluded because they had not undergone OFCs in the clinic by the end of the enrolment period (Figure 1). A total of 182 patients had blood samples available for MolAD, and 159 patients had BAT data for analysis. BAT analysis was conducted only with data that met the criteria defined above for clinical allergy or confirmed tolerance. The study population had a mean age of 8.5 years with male predominance. Fifty-two percent of patients had a diagnosis of asthma, 40% allergic rhino-conjunctivitis to environmental allergens, and 63% atopic dermatitis (Table 1).
Figure 1. Study design and analysis:
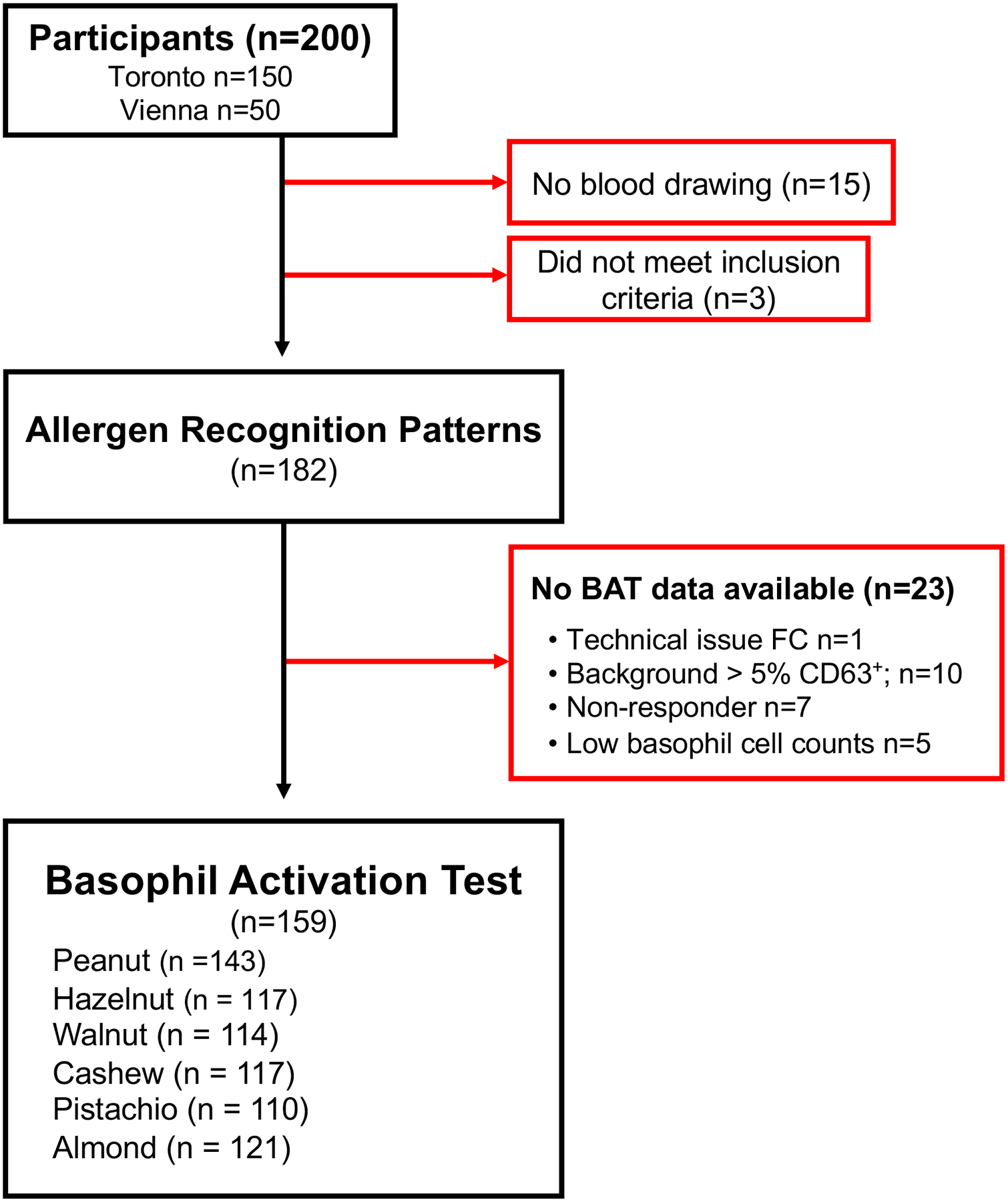
Among the 200 participants who were recruited for the MONAS study (150 Toronto, 50 Vienna), 3 did not meet inclusion criteria and 15 did not have their blood drawn. Of the 182 available samples, BAT data could not be obtained from 23 participants due to several factors including technical issues (n=1), high background activity (n=10), non-responder basophils (n=7) and low cell count (n=5). BATs were performed on a total of 159 samples for the tested nuts: peanut (n=143), hazelnut (n=117), walnut (n=114), cashew (n=117), pistachio (n=110), and almond (n=121). BAT measurements from these 159 samples were used for subsequent data analysis.
Table 1.
Demographics of patient population.
| Clinical characteristics | All patients (n=197) | Toronto (n=149) | Vienna (n=48) | P value |
|---|---|---|---|---|
| Mean age (IQR) | 8.53±4.70 (4.6–12.3) | 9.37 ± 4.67 (5.8–17.9) | 5.9 ± 3.77 (5.8–13.2) | <0.001 |
| Male sex (%) | 122 (62) | 90 (60) | 32 (67) | 0.44 |
| Asthma (%) | 102 (52) | 82 (55) | 20 (42) | 0.1 |
| Allergic rhinoconjunctivitis (%) | 80 (40) | 64 (43) | 16 (33) | 0.22 |
| Atopic Dermatitis (%) | 123 (63) | 100 (67) | 23 (48) | 0.01 |
| Food allergy other than peanut/tree nut (%) | 110 (56) | 92 (62) | 18 (38) | 0.003 |
| Previous anaphylaxis (%) | 87 (44%) | 61 (41) | 26 (54) | 0.13 |
Patients from the two sites were comparable with regards to sex, asthma, allergic rhino-conjunctivitis, concomitant egg allergy, and number of previous episodes of anaphylaxis (Table 1). Mean age of the children was higher in the Canadian group (9.4y vs. 5.9y) and Canadian individuals were more likely to have a history of atopic dermatitis (67% vs. 48%; p=0.01). Food allergies other than peanut, tree nut and sesame allergy, were more frequently diagnosed in the Canadian cohort (62% vs 38%, p=0.003 and 19% vs 2% p=0.004) respectively.
In total, 244 OFCs were performed in 107 patients. Results of 36 OFCs were positive. Mean SPT size and sIgE reported in Table E2 of the Online Repository. Individuals were most frequently sensitized to peanut (86%), hazelnut (76%) and pistachio (67%). No positive OFCs for pistachio or almond in either the Toronto or Vienna groups were reported. Astier scores based on challenge results indicated no difference regarding the severity of the clinical reaction between peanut, hazelnut, and walnut (2.6 ± 1.3; 2.0 ± 0.9 vs 2.4 ± 0.9; Table E2 the Online Repository).
BAT discriminates between peanut and tree nut allergic and non-allergic patients
The main objective of this study was to determine whether the BAT could distinguish between allergic and non-allergic patients. The percentage of CD63 expression was used as the main outcome parameter. Activation via increasing concentrations of the individual nut extract (0.001–1000 ng/mL) in whole blood from allergic and non-allergic children were compared. Patients who were peanut and tree nut allergic displayed a statistically significant higher %CD63+ basophils (p<0.01) upon stimulation with 0.01–1000 ng/mL (cashew, hazelnut, peanut, walnut) or 0.1 ng-1000 ng (pistachio; Figure 2). Sensitivity and specificity values for the BAT were good for peanut, hazelnut, walnut, cashew, and pistachio (Table E3 in the Online Repository). Results for peanut were most consistent with high sensitivity and specificity (95.3% vs. 93.2%), good positive predictive values (PPV), and negative predictive values (NPV; 96% vs. 91%). Although the sensitivity for tree nuts was lower (85.4–91.7%), specificity was 100% for pistachio and walnut.
Figure 2. BAT shows increased proportion of activated CD63+ basophils in peanut and tree nut allergic patients compared to non-allergic patients.
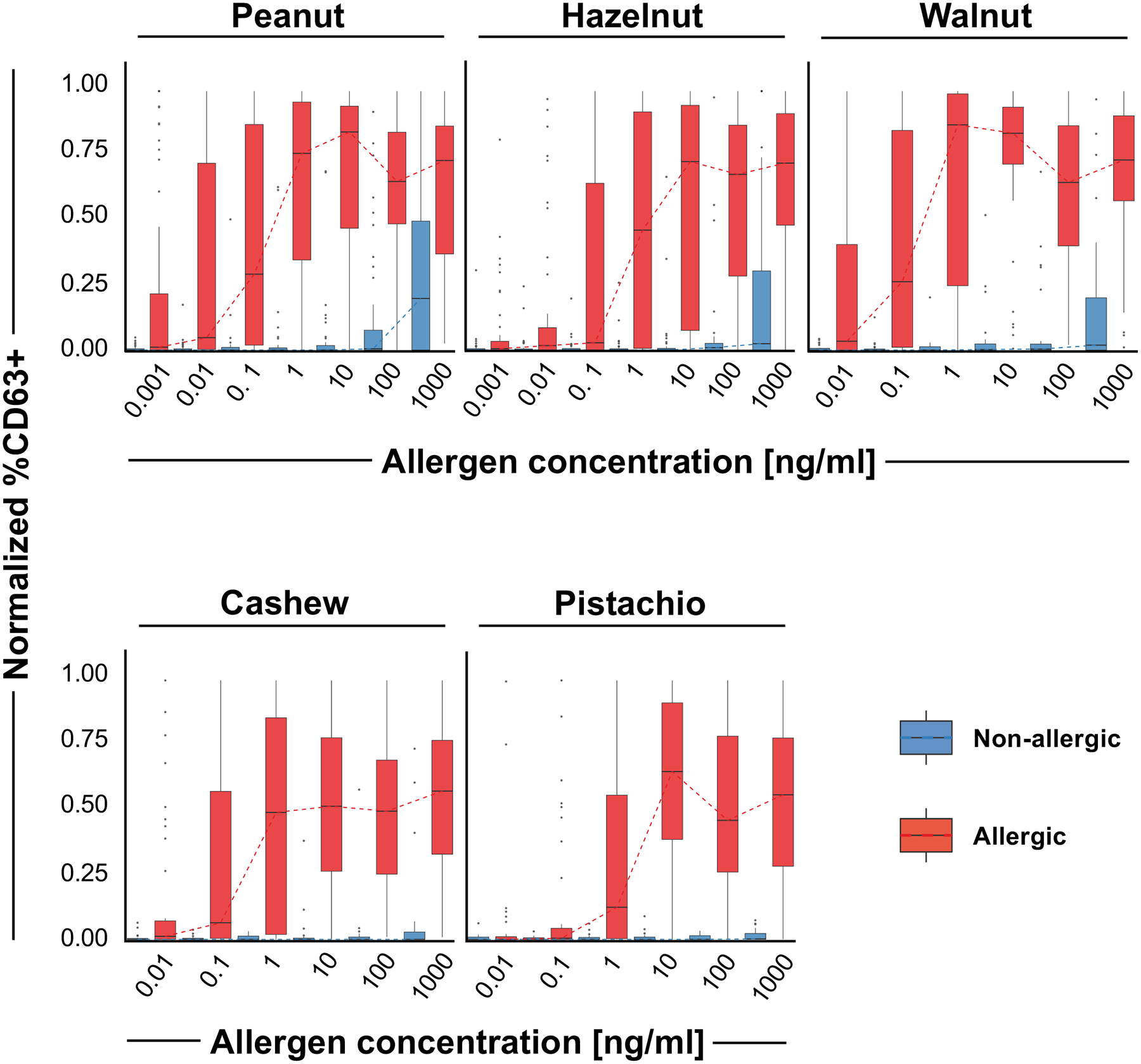
Boxplots depicting the distribution of normalized %CD63+ basophils for all samples of each nut and each concentration (0.001 to 1000 ng/mL). Allergic samples are depicted in red and non-allergic samples are depicted in blue (peanut allergic n=87, non-allergic n=46; hazelnut allergic n=48 non-allergic n=42, walnut allergic n=41, non-allergic n=35; cashew allergic n=50, non-allergic n=24; pistachio allergic n=45, non-allergic n=25). Dashed lines indicate the median values for each concentration. For each nut and concentration, the difference between allergic and non-allergic samples was statistically significant (p<0.05 Welch’s t-test), except for pistachio at 0.01 ng/mL concentration.
Mixed effect model integrating 10 and 100 ng/mL of peanut and individual tree nut stimulation is effective in diagnosing allergy
A mixed effect model integrating 10 and 100 ng/mL of peanut and individual tree nut extracts was selected to be optimal regarding specificity and sensitivity. A total of 429 observations over 147 participants were used in the model. Almond was excluded from statistical modeling because of the high rate of negative food challenges. The AUROC was 0.98 for peanut, 0.97 for cashew, 0.92 for hazelnut, 0.95 for pistachio, and 0.97 for walnut (Figure 3A). Individual data of normalized %CD63+ basophils of each nut for the concentrations 10 and 100 ng/mL are displayed in Figure S1 of the Online Repository. 39/48 patients who underwent almond OFCs had BAT performed. The negative predictive value of BAT for almond reactivity was 100% (using positive BAT criteria of >10% CD63+ basophils at concentrations of 10 and 100 ng/mL). Age did not influence the performance of the BAT in our patient cohort.
Figure 3.
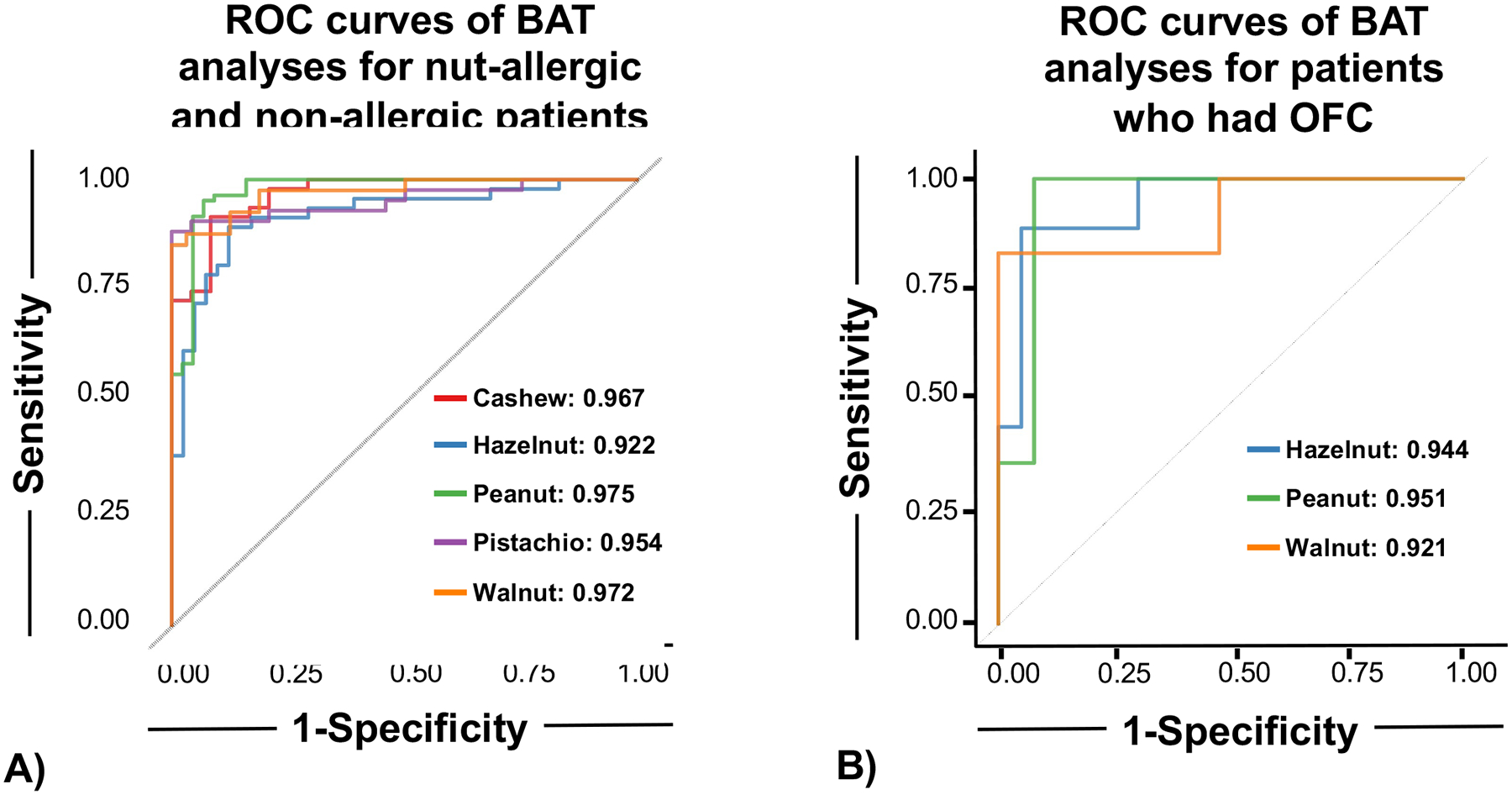
3A: Mixed effect model is effective in distinguishing between peanut and tree nut allergic and non-allergic patients. Models are fit using data from 10 ng/mL and 100 ng/mL concentrations. Allergic samples are in red and non-allergic samples are in blue. ROC curves for each individual nut. All nuts show similar model performance (AUROC for each nut is depicted in the plot legend). 3B: Basophil activation test distinguishes between OFC-positive and OFC-negative patients in those who underwent OFCs. The model was fitted using the same model equation using data from only OFC-confirmed samples at 10 ng/mL and 100 ng/mL concentrations. ROC curves for each individual nut with OFC-confirmed samples. All nuts show similar model performance (AUROC for each nut is depicted in the plot legend).
BAT separates allergic from non-allergic status in patients who underwent clinically indicated OFCs
To determine if the BAT could distinguish between allergic and non-allergic patients in children with equivocal/absent history and specific IgE <15kUA/L and/or SPT<8 mm, we investigated BAT results in patients who had positive OFCs versus those who had negative OFCs. For peanut, hazelnut, or walnut as a culprit allergen, those with positive OFCs had increased %CD63+ basophils at all nut concentrations compared to those who passed the OFC (Figure 4). Cashew, pistachio, and almond were excluded from modelling because of paucity of positive OFCs. The AUROC was 0.95 for peanut, 0.94 for hazelnut, and 0.92 for walnut (Figure 3B, Table E2 the Online Repository). Sensitivity was 100% for peanut with a specificity of 92.3%. Hazelnut and walnut BAT displayed high specificity (95% vs 100%) with a slightly reduced sensitivity.
Figure 4. Basophil activation test distinguishes between OFC-positive and OFC-negative patients in those who underwent OFCs:
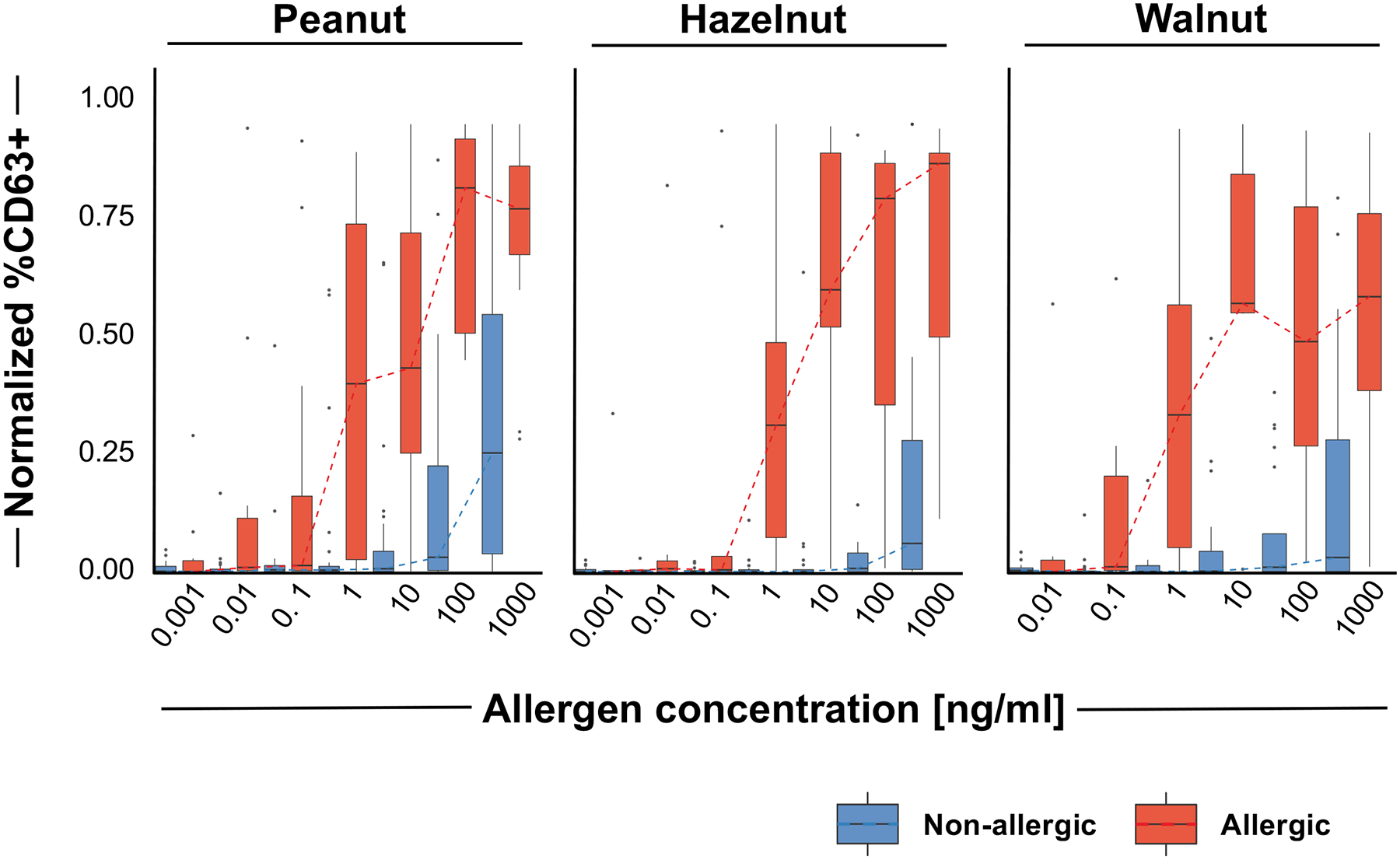
Boxplots depicting the distribution of normalized %CD63+ basophils for samples with OFC outcomes for peanut, hazelnut, or walnut at each concentration (0.001 to 1000 ng/mL). OFC positive samples are depicted in red and OFC negative samples are depicted in blue.
BAT outperforms extract-specific IgE testing for peanut and hazelnut and is equally predictive for walnut allergy
While high sIgE levels (≥15kUA/L) or SPT ≥8 mm to peanut and nut extracts are utilized in the diagnosis of food allergy, their high sensitivity combined with low specificity leads to overdiagnosis in case of equivocal history and lower sensitization values. In patients with clinical allergy, elevated levels of sIgE to extracts and molecular allergen markers were observed irrespective of the nature of the allergen (i.e. seed storage proteins, PR10, nsLTP; Tables E4–E9 in the Online Repository).
In patients with positive OFCs and equivocal histories, increased sIgE was demonstrated to the seed storage proteins Ara h 1, 2, 3, and 6 in peanut, Cor a 14 in hazelnut, and Jug r 1 and 2 in walnut, respectively (Tables E4b, E5b, E6b in the Online Repository). In contrast, allergens from the PR-10 and nsLTP families in peanut and hazelnut did not differ significantly between groups with the exception of Cor a 8, which was only detected at low levels of unclear significance (Table E5b in the Online Repository).
In general, the BAT outperformed both extract and molecular allergen sIgE testing when assessing OFC. In patients requiring OFC confirmation for peanut allergy, Ara h 2 sIgE was the best performing molecular allergen marker test with an AUROC of 0.92 (Figure 5), which was most comparable to the performance of the peanut BAT with an AUROC of 0.95 (Figure 4). For hazelnut allergy, Cor a 14 sIgE was the best MolAD marker with an AUROC of 0.82 (Figure 5), albeit secondary to the performance of the hazelnut BAT (0.94, Figure 4B). For walnut allergy, Jug r 1 sIgE had a comparable AUROC of 0.93 (Figure 4B, Figure 5).
Figure 5. Allergen-specific IgE to Ara h 2, Cor a 14, and Jug r 1 are the best molecular allergen diagnostic markers to distinguish between OFC-positive and OFC-negative patients:
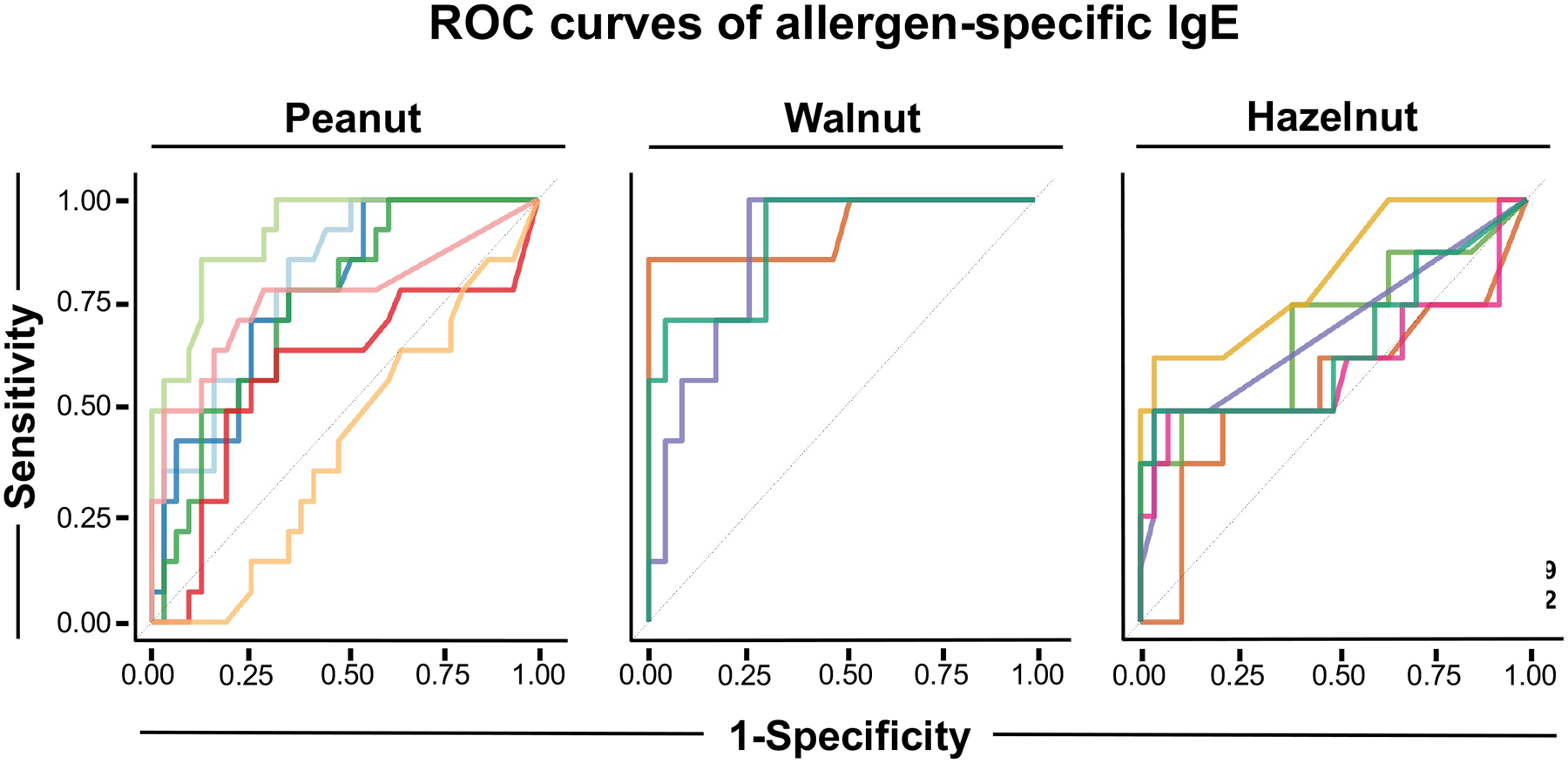
Each line depicts a ROC curve for each allergen from peanut, walnut and hazelnut, obtained by thresholding the raw allergen-specific IgE concentrations (kUA/L). The best performing sIgE markers for each nut were Ara h 2, Cor a 14, and Jug r 1 (AUROC for each allergen is depicted in the plot legend).
Validation of the mixed effect model in an external cohort of peanut allergic patients
To determine the applicability of the established statistical model, external validation was performed in an independent cohort of peanut allergic patients with DBPCFC proven peanut allergy and BAT data from the POISED trial (n=109 analyzed; n=9 non-responders; n=2 no data available).31 BAT values were compared with peanut allergic patients with positive OFC results from the MONAS cohort by utilizing the optimal cut-off determined in the MONAS cohort for the normalized fraction bound values of %CD63+ at 10 and 100 ng/mL of peanut extract. Utilizing this algorithm, the sensitivity in the POISED cohort was 0.88, as compared with sensitivity of 0.95 in the MONAS cohort (Figure S2 in the Online Repository). Specificity could not be calculated as data for negative peanut OFCs was not available from the POISED group.
Discussion
Peanut and tree nut allergies are the most frequent causes of food-associated anaphylaxis. Patients with peanut allergy or a single tree nut allergy often have co-sensitization to multiple tree nuts. It is likely that the test related cross-reactivity is higher than clinical co-reactivity, resulting in unnecessary avoidance. The main purpose of the MONAS study was to prospectively investigate the utility of the BAT in discriminating between peanut and tree nut allergic and peanut/tree nut sensitized individuals in a “real-life” setting in tertiary care allergy outpatient clinics. In addition, we compared BAT and allergen sIgE testing.
In our cohort of pediatric and adolescent patients from Canada and Austria, the BAT demonstrated the ability to predict the allergic clinical status in peanut and tree nut sensitized individuals.
Clinical allergy diagnosis is currently dominated by a combination of clinical history and extract-based SPT or sIgE testing.34 While DBPCFCs are the gold standard if reaction thresholds, delayed reactions, or more subjective symptoms are evaluated, open OFCs represent the standard in clinical settings to assess immediate reactions using objective symptoms. Objective criteria were utilized in the open OFCs in this study.15,35,36 The NUT CRACKER study, in line with our data, reported a higher rate of co-sensitization than co-allergy to tree nuts. With exception of highly cross-reactive tree nut pairs (pistachio/cashew and pecan/walnut), 30% of patients had a co-allergy.37 The Pronuts study assessed open OFC proven rate, or a high pre-test probability, of coexistent peanut, tree nut, and sesame seed allergy, and compared it to conventional food allergy diagnostic tests.38 We utilized a similar definition for the diagnosis of food allergy as the Pronuts study. They found that 60.7% of children had more than one nut or seed allergy. This is higher than previous studies which were mostly retrospective. This is comparable to our cohort with 52.6% of patients having an allergy to more than one nut. Using only BAT results, 51.2% had a concurrent positive BAT to at least one more nut. Both the Pronuts and the MONAS cohorts, are derived from patients referred to tertiary centers and multi-sensitized complex patients are more likely to be seen in these settings.
Our study suggests that the BAT is more accurate than conventional allergy diagnostic methods, including SPT and sIgE testing, in the diagnosis of peanut and tree nut allergy. SPT is associated with false-positive results and sIgE can produce false-negative results.39 For example, sIgE to peanut has a sensitivity of 75–100% and specificity of 17–63%.17,18,26,40–44 The performance of BAT in the diagnosis of food allergy has been investigated in multiple foods, including peanut25,44–47, hazelnut48–50, cow’s milk51,52, egg47,52, wheat53,54, shellfish55, sesame56, and in pollen-food syndromes57,58, with sensitivity and specificity ranging from 85–100%, and 75–100%, respectively.23 The performance of the BAT in tree nut allergy is not well characterized. BAT use in food allergy diagnosis has been suggested to be more accurate than sIgE in several studies.23,25,47,59,60 In one study of BAT use in peanut allergy, specificity and sensitivity was reported as between 94–96% and 95–100%, respectively.25 BAT reduced OFCs by two-thirds, superior to Ara h 2-sIgE and SPT.25 The BAT has also been studied to estimate the threshold of peanut allergic reactions.61 In our study, BAT specificity and sensitivity for peanut were 93% and 95%, respectively, comparable to what has been reported in the literature.
Some utility of the BAT has been demonstrated for the diagnosis of hazelnut allergy.48,49 In the recent NUT CRACKER study, for individual tree nuts (walnut, pecan, cashew, pistachio, hazelnut), BAT was superior to SPT with regards to specificity, but not sensitivity. However, comparability is limited, as only one concentration of allergen (1000 ng/ml) was used. We developed a mixed-effects model integrating 10 and 100 ng/mL of allergen. AUROC values were ≥0.95 for peanut and all tree nuts except hazelnut (0.92). In addition, the model was validated in an independent cohort of peanut allergic patients with positive OFCs. This is important for future approaches like meta-analysis of BAT data from clinical trials and comparable studies.
One of the strengths of the MONAS dataset is the direct comparison of conventional allergy testing (SPT, sIgE), MolAD, and BAT. MolAD data supporting the clinical value of peanut molecular allergen marker testing is strong.17,18,62 Less data exists on the clinical value of Cor a 9 and Cor a 14 for hazelnut allergy, Jug r 1 for walnut allergy, and Ana o 3 for cashew allergy.19–22,63 In our patients with equivocal history and sensitization, both Ara h 2 and peanut BAT were able to predict clinical reactivity (AUROC 0.92 vs 0.95). For the diagnosis of hazelnut allergy, the value of sIgE was limited (AUROC 0.65) whereas BAT was excellent (AUROC 0.94). Cor a 14 was the only relevant hazelnut allergen to predict allergy. BAT performed similarly to extract sIgE and Jug r 1 or Jug r 2 for walnut. Detailed MolAD co-sensitization matrix in our patients is displayed in Figure S3 in the Online Repository. Additional analyses combining MolAD with BAT data did not demonstrate significant superiority to the use of BAT alone (Table E10 in the Online Repository).
Our study differs from the Pronuts and the NUTCRACKER study as it was based on clinically relevant challenges. Challenges matched the clinical needs. The important message is that the majority of those patients who have an equivocal history or asymptomatic sensitization do not react during an OFC (14.8% MONAS, 26% Pronuts).38 Given the limited availability of OFCs, this study highlights the importance of a novel tool to aid in clinical decision making about the appropriate use of these scarce resources. The BAT could be a very important additional tool to SPT, sIgE tests, and MolAD testing to avoid unnecessary OFCs.
Another approach involves pooled OFCs based on BAT results. Applying our model to a group of participants with indeterminate allergic status, 66% of walnut, pistachio, cashew and hazelnut and 93.3% of peanut sensitized individuals in this group had negative BATs (Figure S4 in the Online Repository). Using the BAT-based prediction for the complete cohort, there would be a justification to do an OFC to almost 80% of the nuts they are currently avoiding. The anticipated result would be a negative challenge. Prospective studies are needed to confirm this finding.
Our results demonstrate that the BAT has the potential to decrease the number of OFCs needed, and allows for precision medicine in the diagnosis of peanut and tree nut allergy, in particular with multi-nut sensitizations. While there are technical limitations to performing the BAT in smaller clinical centres, patients who are being considered for an OFC could be referred for a BAT at specialized centres. Although indirect BAT tests have the potential to pool samples compared to direct BAT which uses fresh blood, they can require the same level of infrastructure (e.g. Need for cell cultures). In the case of a negative BAT, a low risk OFC can be conducted at the smaller clinical centre, instead of in a more resource-intensive hospital setting.
Other strengths of the MONAS study include that it was a prospective study, and was conducted at tertiary pediatric centres on two different continents with comparable birch pollen exposure. The prospective nature is important as it reflects the value of the respective tests in a setting where testing and challenging is limited by timing and availability.
Based on recent retrospective64 and prospective data37,65 almond SPT/sIgE shows low specificity and no sIgE cut offs with >90% predictive value are available. No patients had a positive almond OFC in our patients despite sIgE values of up to 53.4 kUA/L and SPT results up to 8 mm. Thus, in line with recent challenge proven data, routine testing to almond as part of panel testing should prompt OFCs even in the event of positive test results.
Disadvantages with the BAT include the need for rapid processing of the blood sample, although the POISED study used blood refrigerated for ~1 day before testing, and more resources. The non-responder rate was 4% in the MONAS cohort, comparable to what is reported in the literature.23 While we do not suggest that BAT should become first-line in the routine diagnosis of food allergy, it does have the potential to transform food allergy diagnosis in clinical scenarios with equivocal clinical history and multiple sensitizations. It can also be of important value for safe food introduction in high-risk populations.
In conclusion, the MONAS study demonstrates that the BAT is effective in diagnosing peanut and tree nut allergy in a real-world setting, and may be useful in reducing the number of resource-intensive OFCs needed. Additional prospective studies to investigate the ability of the BAT to reduce OFCs are required.
Supplementary Material
Acknowledgements
Lucy Duan has nothing to disclose. Alper Celik has nothing to disclose. Jennifer A. Hoang has nothing to disclose. Klara Schmidthaler has nothing to disclose. Delvin So has nothing to disclose. Xiaojun Yin has nothing to disclose. Christina M. Ditlof has nothing to disclose. Marta Ponce has nothing to disclose. Julia Upton reports grants and personal fees from ALK-Abelló A/S, personal fees from Bausch Health, personal fees from Kaleo, grants from DBV Technologies, grants from Regeneron, grants from Food Allergy Anaphylaxis Programme (SickKids), outside the submitted work; and Section Chair of Food Allergy and Anaphylaxis, Canadian Society of Allergy and Clinical Immunology; Healthcare Advisory Board, Food Allergy Canada. Jean-Soo Lee has nothing to disclose. Lisa Hung has nothing to disclose. Heimo Breiteneder has nothing to disclose. Chiara Palladino has nothing to disclose. Adelle R. Atkinson has nothing to disclose. Vy H.D. Kim has nothing to disclose. Alireza Berenjy has nothing to disclose. Maria Asper has nothing to disclose. David Hummel has nothing to disclose. Samantha Wong has nothing to disclose. Mara Alexanian-Farr has nothing to disclose. Ahuva Magder has nothing to disclose. Sharon Chinthrajah reports grants from NIAID, grants from AImmune, grants from DBV Technologies, grants from Astellas, grants from AnaptysBio, grants from Regeneron, grants from CoFAR, personal fees from Alladapt Therapeutics, personal fees from Genetech, personal fees from Novartis, personal fees from Before Brands, personal fees from Nurticia, during the conduct of the study; grants from NIAID, grants and other from AImmune, grants from DBV Technologies, grants from Astellas, grants from AnaptysBio, grants from Regeneron, grants from CoFAR, grants and other from All Adapt, grants and other from Genentech, grants and other from Novartis, grants and other from Before Brands, grants and other from Nutricia, other from Guidepoint Global, outside the submitted work. Kaori Mukai has nothing to disclose. Mindy Tsai reports grants from NIAID/NIH during the conduct of the study.
Kari Nadeau reports grants and other from NIAID, other from Novartis, personal fees and other from Regeneron, grants and other from FARE, grants from EAT, other from Sanofi, other from Astellas, other from Nestle, other from BeforeBrands, other from Alladapt, other from ForTra, other from Genentech, other from AImmune Therapeutics, other from DBV Technologies, personal fees from Astrazeneca, personal fees from ImmuneWorks, personal fees from Cour Pharmaceuticals, grants from Allergenis, grants from Ukko Pharma, other from AnaptysBio, other from Adare Pharmaceuticals, other from Stallergenes-Greer, other from NHLBI, other from NIEHS, other from EPA, other from WAO Center of Excellence, other from Iggenix, other from Probio, other from Vedanta, other from Centecor, other from Seed, from Immune Tolerance Network, from NIH, outside the submitted work; In addition, Dr. Nadeau has a patent Inhibition of Allergic Reaction to Peanut Allergen using an IL-33 Inhibitor pending, a patent Special Oral Formula for Decreasing Food Allergy Risk and Treatment for Food Allergy pending, a patent Basophil Activation Based Diagnostic Allergy Test pending, a patent Granulocyte-based methods for detecting and monitoring immune system disorders pending, a patent Methods and Assays for Detecting and Quantifying Pure Subpopulations of White Blood Cells in Immune System Disorders pending, a patent Mixed Allergen Compositions and Methods for Using the Same pending, and a patent Microfluidic Device and Diagnostic Methods for Allergy Testing Based on Detection of Basophil Activation pending. Stephen Galli reports grants from NIAID/NIH during the conduct of the study. Arun K. Ramani has nothing to disclose. Zsolt Szepfalusi has nothing to disclose. Thomas Eiwegger reports other from DBV, grants from Innovation fund Denmark, CIHR, other from Regeneron. He is the Co-I or scientific lead in three investigator initiated oral immunotherapy trials including the usage of biologicals supported by the Allergy and Anaphylaxis Program Sickkids and CIHR. He serves as associate editor for Allergy. He is on advisory boards for ALK.
Statement of Funding:
This work was supported by The Hospital for Sick Children and the Food Allergy and Anaphylaxis Program at the Hospital for Sick Children, the Austrian Science Fund (FWF) SFB F4615; FWF Doctoral Program W 1248-B13 MCCA; Integrated Genomic and Functional Studies of Immunotherapy for Multi-Food Allergy 5U19 AI104209-07; T cell Reagents for Allergy 5U01 AI140498-03; Effects of IgE Blockade on T Cells in Food Allergy R01AI140134-01; Sean N. Parker Center for Allergy and Asthma Research at Stanford University; Crown Family Foundation; Bunning Sunshine Foundation.
References
- 1.Ben-Shoshan M, Harrington DW, Soller L, et al. A population-based study on peanut, tree nut, fish, shellfish, and sesame allergy prevalence in Canada. Journal of Allergy and Clinical Immunology. 2010;125(6):1327–1335. [DOI] [PubMed] [Google Scholar]
- 2.McWilliam V, Koplin J, Lodge C, Tang M, Dharmage S, Allen K. The Prevalence of Tree Nut Allergy: A Systematic Review. Current Allergy and Asthma Reports. 2015;15(9):54. [DOI] [PubMed] [Google Scholar]
- 3.Sicherer SH, Muñoz-Furlong A, Godbold JH, Sampson HA. US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. Journal of Allergy and Clinical Immunology. 2010;125(6):1322–1326. [DOI] [PubMed] [Google Scholar]
- 4.Venter C, Hasan Arshad S, Grundy J, et al. Time trends in the prevalence of peanut allergy: three cohorts of children from the same geographical location in the UK: Time trends in peanut allergy. Allergy. 2010;65(1):103–108. [DOI] [PubMed] [Google Scholar]
- 5.Bunyavanich S, Rifas-Shiman SL, Platts-Mills TAE, et al. Peanut allergy prevalence among school-age children in a US cohort not selected for any disease. Journal of Allergy and Clinical Immunology. 2014;134(3):753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke AE, Elliott SJ, St Pierre Y, Soller L, La Vieille S, Ben-Shoshan M. Comparing food allergy prevalence in vulnerable and nonvulnerable Canadians. J Allergy Clin Immunol Pract. 2020. [DOI] [PubMed] [Google Scholar]
- 7.Bohlke K, Davis RL, DeStefano F, Marcy SM, Braun MM, Thompson RS. Epidemiology of anaphylaxis among children and adolescents enrolled in a health maintenance organization. Journal of Allergy and Clinical Immunology. 2004;113(3):536–542. [DOI] [PubMed] [Google Scholar]
- 8.Perry TT, Matsui EC, Conover-Walker MK, Wood RA. Risk of oral food challenges. Journal of Allergy and Clinical Immunology. 2004;114(5):1164–1168. [DOI] [PubMed] [Google Scholar]
- 9.Sicherer SH, Wood RA. Advances in Diagnosing Peanut Allergy. The Journal of Allergy and Clinical Immunology: In Practice. 2013;1(1):1–13. [DOI] [PubMed] [Google Scholar]
- 10.Maloney JM, Rudengren M, Ahlstedt S, Bock SA, Sampson HA. The use of serum-specific IgE measurements for the diagnosis of peanut, tree nut, and seed allergy. J Allergy Clin Immunol. 2008;122(1):145–151. [DOI] [PubMed] [Google Scholar]
- 11.Upton JEM, Bird JA. Oral food challenges: Special considerations. Ann Allergy Asthma Immunol. 2020;124(5):451–458. [DOI] [PubMed] [Google Scholar]
- 12.Bird JA, Leonard S, Groetch M, et al. Conducting an Oral Food Challenge: An Update to the 2009 Adverse Reactions to Foods Committee Work Group Report. J Allergy Clin Immunol Pract. 2020;8(1):75–90 e17. [DOI] [PubMed] [Google Scholar]
- 13.van Nieuwaal NHG, Lasfar W, Meijer Y, et al. Utility of peanut-specific IgE levels in predicting the outcome of double-blind, placebo-controlled food challenges. Journal of Allergy and Clinical Immunology. 2010;125(6):1391–1392. [DOI] [PubMed] [Google Scholar]
- 14.Sampson HA. Utility of food-specific IgE concentrations in predicting symptomatic food allergy. Journal of Allergy and Clinical Immunology. 2001;107(5):891–896. [DOI] [PubMed] [Google Scholar]
- 15.Sampson HA, Gerth van Wijk R, Bindslev-Jensen C, et al. Standardizing double-blind, placebo-controlled oral food challenges: American Academy of Allergy, Asthma & Immunology–European Academy of Allergy and Clinical Immunology PRACTALL consensus report. Journal of Allergy and Clinical Immunology. 2012;130(6):1260–1274. [DOI] [PubMed] [Google Scholar]
- 16.Kattan JD, Wang J. Allergen Component Testing for Food Allergy: Ready for Prime Time? Current Allergy and Asthma Reports. 2013;13(1):58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebisawa M, Moverare R, Sato S, Maruyama N, Borres MP, Komata T. Measurement of Ara h 1-, 2-, and 3-specific IgE antibodies is useful in diagnosis of peanut allergy in Japanese children. Pediatr Allergy Immunol. 2012;23(6):573–581. [DOI] [PubMed] [Google Scholar]
- 18.Klemans RJ, van Os-Medendorp H, Blankestijn M, Bruijnzeel-Koomen CA, Knol EF, Knulst AC. Diagnostic accuracy of specific IgE to components in diagnosing peanut allergy: a systematic review. Clin Exp Allergy. 2015;45(4):720–730. [DOI] [PubMed] [Google Scholar]
- 19.Buyuktiryaki B, Cavkaytar O, Sahiner UM, et al. Cor a 14, Hazelnut-Specific IgE, and SPT as a Reliable Tool in Hazelnut Allergy Diagnosis in Eastern Mediterranean Children. J Allergy Clin Immunol Pract. 2016;4(2):265–272 e263. [DOI] [PubMed] [Google Scholar]
- 20.Flores Kim J, McCleary N, Nwaru BI, Stoddart A, Sheikh A. Diagnostic accuracy, risk assessment, and cost-effectiveness of component-resolved diagnostics for food allergy: A systematic review. Allergy. 2018;73(8):1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange L, Lasota L, Finger A, et al. Ana o 3-specific IgE is a good predictor for clinically relevant cashew allergy in children. Allergy. 2017;72(4):598–603. [DOI] [PubMed] [Google Scholar]
- 22.Masthoff LJ, Mattsson L, Zuidmeer-Jongejan L, et al. Sensitization to Cor a 9 and Cor a 14 is highly specific for a hazelnut allergy with objective symptoms in Dutch children and adults. J Allergy Clin Immunol. 2013;132(2):393–399. [DOI] [PubMed] [Google Scholar]
- 23.Santos AF, Lack G. Basophil activation test: food challenge in a test tube or specialist research tool? Clinical and Translational Allergy. 2016;6(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebo DG, Bridts CH, Hagendorens MM, Aerts NE, De Clerck LS, Stevens WJ. Basophil activation test by flow cytometry: Present and future applications in allergology. Cytometry Part B: Clinical Cytometry. 2008;74B(4):201–210. [DOI] [PubMed] [Google Scholar]
- 25.Santos AF, Douiri A, Bécares N, et al. Basophil activation test discriminates between allergy and tolerance in peanut-sensitized children. Journal of Allergy and Clinical Immunology. 2014;134(3):645–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts G, Lack G. Diagnosing peanut allergy with skin prick and specific IgE testing. J Allergy Clin Immunol. 2005;115(6):1291–1296. [DOI] [PubMed] [Google Scholar]
- 27.Baker MG, Kattan JD. Review of 400 consecutive oral food challenges to almond. Annals of Allergy, Asthma & Immunology. 2019;122(2):189–192. [DOI] [PubMed] [Google Scholar]
- 28.Hoang JA, Mashouri P, Dai R, et al. Extract and component-specific sensitization patterns in Canadian moderate-to-severe preschool asthmatics. Allergy. 2019;74(12):2519–2521. [DOI] [PubMed] [Google Scholar]
- 29.Hoang JA, Celik A, Lupinek C, et al. Modelling the conversion between specific IgE test platforms for nut allergens in children and adolescents. Allergy. 2020. [DOI] [PubMed] [Google Scholar]
- 30.Duan L, Hoang JA, Kothari A, Eiwegger T, Vadas P. Shellfish allergy is a risk factor for cricket anaphylaxis. J Allergy Clin Immunol Pract. 2020. [DOI] [PubMed] [Google Scholar]
- 31.Wickham H. ggplot2: Elegant Graphics for Data Analysis.: Springer-Verlag; New York; 2016. [Google Scholar]
- 32.Chinthrajah RS, Purington N, Andorf S, et al. Sustained outcomes in oral immunotherapy for peanut allergy (POISED study): a large, randomised, double-blind, placebo-controlled, phase 2 study. The Lancet. 2019;394(10207):1437–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai M, Mukai K, Chinthrajah RS, Nadeau KC, Galli SJ. Sustained successful peanut oral immunotherapy associated with low basophil activation and peanut-specific IgE. Journal of Allergy and Clinical Immunology. 2020;145(3):885–896.e886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eiwegger T, Hung L, San Diego KE, O’Mahony L, Upton J. Recent developments and highlights in food allergy. Allergy. 2019;74(12):2355–2367. [DOI] [PubMed] [Google Scholar]
- 35.Boyce JA, Assa’ad A, Burks AW, et al. Guidelines for the diagnosis and management of food allergy in the United States: report of the NIAID-sponsored expert panel. J Allergy Clin Immunol. 2010;126(6 Suppl):S1–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venter C, Pereira B, Voigt K, et al. Prevalence and cumulative incidence of food hypersensitivity in the first 3 years of life. Allergy. 2008;63(3):354–359. [DOI] [PubMed] [Google Scholar]
- 37.Elizur A, Appel MY, Nachshon L, et al. NUT Co Reactivity - ACquiring Knowledge for Elimination Recommendations (NUT CRACKER) study. Allergy. 2018;73(3):593–601. [DOI] [PubMed] [Google Scholar]
- 38.Brough HA, Caubet J-C, Mazon A, et al. Defining challenge-proven coexistent nut and sesame seed allergy: A prospective multicenter European study. Journal of Allergy and Clinical Immunology. 2020;145(4):1231–1239. [DOI] [PubMed] [Google Scholar]
- 39.Andorf S, Borres MP, Block W, et al. Association of Clinical Reactivity with Sensitization to Allergen Components in Multifood-Allergic Children. The Journal of Allergy and Clinical Immunology: In Practice. 2017;5(5):1325–1334.e1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernard H, Paty E, Mondoulet L, et al. Serological characteristics of peanut allergy in children. Allergy. 2003;58:1285–1292. [DOI] [PubMed] [Google Scholar]
- 41.Wainstein BK, Studdert J, Ziegler M, Ziegler JB. Prediction of anaphylaxis during peanut food challenge: usefulness of the peanut skin prick test (SPT) and specific IgE level. Pediatr Allergy Immunol. 2010;21(4 Pt 1):603–611. [DOI] [PubMed] [Google Scholar]
- 42.DunnGalvin A, Daly D, Cullinane C, et al. Highly accurate prediction of food challenge outcome using routinely available clinical data. J Allergy Clin Immunol. 2011;127(3):633–639 e631–633. [DOI] [PubMed] [Google Scholar]
- 43.Johannsen H, Nolan R, Pascoe EM, et al. Skin prick testing and peanut-specific IgE can predict peanut challenge outcomes in preschoolchildren with peanut sensitization. Clin Exp Allergy. 2011;41(7):994–1000. [DOI] [PubMed] [Google Scholar]
- 44.Glaumann S, Nopp A, Johansson SG, Rudengren M, Borres MP, Nilsson C. Basophil allergen threshold sensitivity, CD-sens, IgE-sensitization and DBPCFC in peanut-sensitized children. Allergy. 2012;67(2):242–247. [DOI] [PubMed] [Google Scholar]
- 45.Glaumann S, Nopp A, Johansson SG, Borres MP, Nilsson C. Oral peanut challenge identifies an allergy but the peanut allergen threshold sensitivity is not reproducible. PLoS One. 2013;8(1):e53465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Javaloyes G, M.J. G, Garcia Nunez G, et al. Performance of Different in Vitro Techniques in the Molecular Diagnosis of Peanut Allergy. J Investig Allergol Clin Immunol. 2012;22(7):508–513. [PubMed] [Google Scholar]
- 47.Ocmant A, Mulier S, Hanssens L, et al. Basophil activation tests for the diagnosis of food allergy in children. Clinical & Experimental Allergy. 2009;39(8):1234–1245. [DOI] [PubMed] [Google Scholar]
- 48.Brandstrom J, Nopp A, Johansson SG, et al. Basophil allergen threshold sensitivity and component-resolved diagnostics improve hazelnut allergy diagnosis. Clin Exp Allergy. 2015;45(9):1412–1418. [DOI] [PubMed] [Google Scholar]
- 49.Lotzsch B, Dolle S, Vieths S, Worm M. Exploratory analysis of CD63 and CD203c expression in basophils from hazelnut sensitized and allergic individuals. Clin Transl Allergy. 2016;6:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Worm M, Hompes S, Fiedler EM, Illner AK, Zuberbier T, Vieths S. Impact of native, heat-processed and encapsulated hazelnuts on the allergic response in hazelnut-allergic patients. Clin Exp Allergy. 2009;39(1):159–166. [DOI] [PubMed] [Google Scholar]
- 51.Ciepiela O, Zwiazek J, Zawadzka-Krajewska A, Kotula I, Kulus M, Demkow U. Basophil activation test based on the expression of CD203c in the diagnostics of cow milk allergy in children. Eur J Med Res. 2010;15:21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato S, Tachimoto H, Shukuya A, et al. Basophil activation marker CD203c is useful in the diagnosis of hen’s egg and cow’s milk allergies in children. Int Arch Allergy Immunol. 2010;152 Suppl 1:54–61. [DOI] [PubMed] [Google Scholar]
- 53.Carroccio A, Brusca I, Mansueto P, et al. A comparison between two different in vitro basophil activation tests for gluten- and cow’s milk protein sensitivity in irritable bowel syndrome (IBS)-like patients. Clin Chem Lab Med. 2013;51(6):1257–1263. [DOI] [PubMed] [Google Scholar]
- 54.Tokuda R, Nagao M, Hiraguchi Y, et al. Antigen-induced expression of CD203c on basophils predicts IgE-mediated wheat allergy. Allergol Int. 2009;58(2):193–199. [DOI] [PubMed] [Google Scholar]
- 55.Ebo DG, Bridts CH, Hagendorens MM, De Clerck LS, Stevens WJ. Scampi Allergy: From Fancy Name-Giving to Correct Diagnosis. J Allergy Clin Immunol. 2008;18(3):223–230. [PubMed] [Google Scholar]
- 56.Raap U, Wieczorek D, Schenck F, Kapp A, Wedi B. The basophil activation test is a helpful diagnostic tool in anaphylaxis to sesame with false-negative specific IgE and negative skin test. Allergy. 2011;66(11):1496–1497. [DOI] [PubMed] [Google Scholar]
- 57.Ebo DG, Hagendorens MM, Bridts CH, Schuerwegh AJ, De Clerck LS, Stevens WJ. Flow cytometric analysis of in vitro activated basophils, specific IgE and skin tests in the diagnosis of pollen-associated food allergy. Cytometry B Clin Cytom. 2005;64(1):28–33. [DOI] [PubMed] [Google Scholar]
- 58.Erdmann SM, Sachs B, Schmidt A, et al. In vitro analysis of birch-pollen-associated food allergy by use of recombinant allergens in the basophil activation test. Int Arch Allergy Immunol. 2005;136(3):230–238. [DOI] [PubMed] [Google Scholar]
- 59.Rubio A, Vivinus-Nebot M, Bourrier T, Saggio B, Albertini M, Bernard A. Benefit of the basophil activation test in deciding when to reintroduce cow’s milk in allergic children. Allergy. 2011;66(1):92–100. [DOI] [PubMed] [Google Scholar]
- 60.Santos AF, Du Toit G, Douiri A, et al. Distinct parameters of the basophil activation test reflect the severity and threshold of allergic reactions to peanut. J Allergy Clin Immunol. 2015;135(1):179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chinthrajah RS, Purington N, Andorf S, et al. Development of a tool predicting severity of allergic reaction during peanut challenge. Ann Allergy Asthma Immunol. 2018;121(1):69–76 e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jappe U, Breiteneder H. Peanut allergy-Individual molecules as a key to precision medicine. Allergy. 2019;74(2):216–219. [DOI] [PubMed] [Google Scholar]
- 63.Sato S, Moverare R, Ohya Y, et al. Ana o 3-specific IgE is a predictive marker for cashew oral food challenge failure. J Allergy Clin Immunol Pract. 2019;7(8):2909–2911 e2904. [DOI] [PubMed] [Google Scholar]
- 64.Baker MG, Kattan JD. Review of 400 consecutive oral food challenges to almond. Ann Allergy Asthma Immunol. 2019;122(2):189–192. [DOI] [PubMed] [Google Scholar]
- 65.Brough HA, Caubet JC, Mazon A, et al. Defining challenge-proven coexistent nut and sesame seed allergy: A prospective multicenter European study. J Allergy Clin Immunol. 2020;145(4):1231–1239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


