Abstract
Srp1p (importin α) functions as the nuclear localization signal (NLS) receptor in Saccharomyces cerevisiae. The srp1-31 mutant is defective in this nuclear localization function, whereas an srp1-49 mutant exhibits defects that are unrelated to this localization function, as was confirmed by intragenic complementation between the two mutants. RPN11 and STS1 (DBF8) were identified as high-dosage suppressors of the srp1-49 mutation but not of the srp1-31 mutation. We found that Sts1p interacts directly with Srp1p in vitro and also in vivo, as judged by coimmunoprecipitation and two-hybrid analyses. Mutants of Sts1p that cannot interact with Srp1p are incapable of suppressing srp1-49 defects, strongly suggesting that Sts1p functions in a complex with Srp1p. STS1 also interacted with the second suppressor, RPN11, a subunit of the 26S proteasome, in the two-hybrid system. Further, degradation of Ub-Pro-β-galactosidase, a test substrate for the ubiquitin-proteasome system, was defective in srp1-49 but not in srp1-31. This defect in protein degradation was alleviated by overexpression of either RPN11 or STS1 in srp1-49. These results suggest a role for Srp1p in regulation of protein degradation separate from its well-established role as the NLS receptor.
Nucleocytoplasmic transport is a complex process mediated by interaction of transport cargoes with components of the nuclear pore complex along with other soluble factors (reviewed in references 9, 28, and 30). Import of most karyophilic proteins into the nucleus is energy dependent and mediated by short stretches of amino acids known as nuclear localization signals (NLSs). Proteins with classical NLSs, as exemplified by the simian virus 40 (SV40) large T antigen NLS and the nucleoplasmin NLS, are transported to the nucleus by a heterodimer composed of Srp1p (the Saccharomyces cerevisiae homolog of importin α [Kap60p]) and Kap95p (an S. cerevisiae homolog of importin β) (6, 10). Srp1p is the NLS receptor component that binds to the NLS itself, while Kap95p interacts with some components of the nuclear pore complex to facilitate movement of the Srp1p-Kap95p complex carrying a cargo protein from the cytoplasm to the nucleus (35, 36, 37).
There are several other receptor proteins, which have amino acid sequence similarity to importin β (Kap95p) and are grouped in the importin β family. These proteins directly bind cargo proteins such as ribosomal proteins and carry out a protein import function independently of Srp1p (reviewed in references 9, 28, and 30).
SRP1 was originally identified as a suppressor of certain temperature-sensitive (ts) mutations in RNA polymerase I (Pol I) in S. cerevisiae (54). Specific mutations in SRP1 were able to suppress ts mutations in the zinc-binding domains of RPA190 and RPA135, genes encoding the largest and second largest subunits of Pol I, respectively, but not other rpa190 and rpa135 ts mutations outside of this domain. These SRP1 suppressor mutations displayed no apparent phenotype on their own. Additional work has shown that different ts mutations in SRP1 cause different defects, which include protein import defects, nuclear segregation defects, altered microtubule morphology, and altered nucleolar morphology (24, 44, 55). Although the role of Srp1p in protein import is firmly established, it is not clear whether all of the diverse and allele-specific phenotypes displayed by these mutations are the consequences of defects in protein import. For example, while defects in NLS binding and NLS protein import were clearly demonstrated for the srp1-31 mutation (24, 44), no or only slight defects in NLS binding and protein import were observed for the srp1-49 mutation, which showed various other phenotypes (44, 55; unpublished data). Likewise, suppression of Pol I ts mutations by SRP1 mutations is difficult to explain on the basis of the Srp1p function as the NLS receptor (54, 55).
For these reasons, we considered and tested the possibility that Srp1p might have an additional function(s) distinct from the NLS receptor function in protein import. We now report our discovery that two mutations, srp1-49 and srp1-31, show intragenic complementation, indicating that the function affected by srp1-49 is clearly different from the function affected by srp1-31, i.e., the NLS receptor function. To obtain clues to the functions affected by srp1-49, high-dosage suppressors of srp1-49 were isolated. Two suppressors identified, STS1 and RPN11, suppressed only srp1-49 and not srp1-31. STS1 was originally identified (22) as a dosage-dependent suppressor of a mutation in SEC23, a gene required for endoplasmic reticulum (ER)-Golgi protein transport (56). Independently, DBF8, which is identical to STS1, was also identified as a gene essential for nuclear segregation and division (12). RPN11 was recently identified as a component of the regulatory particle of the 26S proteasome (8), suggesting a link between SRP1 and the ubiquitin-proteasome system. Further experiments designed to examine such a link have led to the conclusion that Srp1p, together with Sts1p, carries out a function in the regulation of protein degradation by the ubiquitin-proteasome system and this function is separate from its established function in NLS-dependent protein import into the nucleus. We report these experiments and also discuss previous observations which support this conclusion.
MATERIALS AND METHODS
Media, strains, and plasmids.
YEPD medium consists of 2% yeast extract, 1% Bacto-peptone (Difco Laboratories, Detroit, Mich.), and 2% glucose. Synthetic glucose (SGlu) medium (2% glucose, 0.67% yeast nitrogen base [Difco], and 0.5% Casamino acids [Difco]) was supplemented with required bases or tryptophan as described by Sherman et al. (42). Synthetic galactose (SGal) and synthetic raffinose (SRaff) media are the same as SGlu but with 2% galactose or 3% raffinose substituted for glucose, respectively. To make solid medium, 2% agar was added.
The yeast strains and plasmids used in this study are described in Table 1. Yeast plasmid transformations were performed as described by Gietz et al. (7). DNA sequencing was performed using an ABI373A DNA sequencer (PE Applied Biosystems). The SRP1 chromosomal locus was replaced with the srp1-31 or srp1-49 mutant allele (55) by plasmid integration and pop-out (24, 51).
TABLE 1.
Yeast strains and plasmids used
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| SFY526 | MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 canr gal4-542 gal80-538 URA3::GAL1-lacZ | Clontech |
| NOY388 | (W303-1a) MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 | |
| NOY397 | MATa/α ade2-1/ade2-1 ura3-1/ura3-1 his3-11/his3-11 trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 | |
| NOY514 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 Δsrp1::LEU2 pNOY166 (srp1-49 TRP1) | |
| NOY520 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 Δsrp1::LEU2 pNOY162 (SRP1 TRP1) | |
| JLY543 (=NOY612) | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 srp1-31 | 24 |
| JLY555 (=NOY613) | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 srp1-49 | 24 |
| NOY726 | MATa/α ade2-1/ade2-1 ura3-1/ura3-1 his3-11/his3-11 trp1-1/trp1-1 leu2-3,112/leu2-3,112 can1-100/can1-100 Δsts1::HIS3/STS1 | |
| NOY934 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 pNOY485 pUb-P-ek-βgal | |
| NOY937 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 srp1-49 pNOY485 pUb-P-ek-β-gal | |
| NOY938 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 srp1-49 pNOY486 pUb-P-ek-β-gal | |
| NOY939 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 srp1-49 pNOY487 pUb-P-ek-β-gal | |
| NOY940 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 srp1-31 pNOY485 pUb-P-ek-β-gal | |
| NOY943 | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 Δsts1::HIS3 pNOY471 | |
| NOY944 | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 Δsts1::HIS3 pNOY470 (STS1 TRP1) | |
| NOY945 | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 Δsts1::HIS3 pNOY479 | |
| NOY946 | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 Δsts1::HIS3 Δsrp1::LEU2 pNOY343 [sts1-11(E43G) URA3], pNOY166 (srp1-49 TRP1) | |
| NOY947 | MATα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 Δsts1::HIS3 pNOY488 (HA-STS1) | |
| NOY948 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-1,112 can1-100 pNOY478 (GAL-GFP STS1) | |
| NOY949 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-1,112 can1-100 pNOY482 (GAL-GFP-sts1ΔNLS1) | |
| NOY950 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-1,112 can1-100 pNOY484 [GAL-GFP sts1-11,12 (E43G, I162T)] | |
| NOY992 | Strain JLY543 containing plasmid pRS316-GAL | |
| NOY990 | Strain JLY543 containing plasmid pNOY258 | |
| NOY991 | Strain JLY543 containing plasmid pNPY285 | |
| Plasmids | ||
| pACT2 | GAL4 AD vector (LEU2 2μm Ampr) and carries the GAL4 transcriptional AD [GAL4(768–881), encoding Gal4p (768–881)], which is under control of the ADH promoter and followed by the region encoding an HA1-flu epitope tag and multicloning sites | 4 |
| pAS1 | GAL4 DBD vector (TRP1 CHY2 2μm Ampr) and carries the GAL4 DBD [GAL4(1–147), encoding Gal4p(1–147)], which is under control of the ADH promoter and followed by the region encoding an HA1-flu epitope tag and multicloning sites | 4 |
| pUC19 | 2,686-bp plasmid cloning vector | 53 |
| pRS314 | E. coli-yeast shuttle vector carrying TRP1, CEN6, and ARSH4 | 53 |
| pRS316 | E. coli-yeast shuttle vector carrying URA3, CEN6, and ARSH4 | 45 |
| pUb-P-ek-βgal | pGAL1::Ub-Pro-βgal 2μm Ampr | 45 |
| pRS316-GAL | E. coli-yeast shuttle vector carrying URA3, CEN6, and ARSH4 with the GAL1 promoter followed by multicloning sites | 23 |
| pNOY162 | Derivative of pRS314 carrying SRP1 | |
| pNOY163 | Derivative of pRS314 carrying srp1-31 | |
| pNOY166 | Derivative of pRS314 carrying srp1-49 | |
| pNOY258 | Derivative of pRS316-GAL with STS1 fused to the GAL1 promoter | |
| pNOY285 | Derivative of pRS316-GAL with RPN11 fused to the GAL1 promoter | |
| pNOY291 | Derivative of pGEX2T carrying GST-STS1 | |
| pNOY343 | Derivative of pNOY471 carrying sts1(E43G) instead of STS1 | |
| pNOY470 | Derivative of pRS314 carrying STS1 | |
| pNOY471 | Derivative of pRS316 carrying STS1 | |
| pNOY472 | Derivative of pAS1 carrying SRP1 | |
| pNOY473 | Derivative of pACT2 carrying STS1 | |
| pNOY474 | Derivative of pAS1 carrying STS1 | |
| pNOY475 | Derivative of pACT2 carrying RPN11 | |
| pNOY476 | Derivative of pRS314 carrying GFP-STS1 | |
| pNOY477 | Derivative of pNOY470 with a NotI site added after the ATG of STS1 | |
| pNOY478 | Derivative of pNOY258 carrying GFP-STS1 instead of STS1 | |
| pNOY479 | Derivative of pRS316 carrying GFP-STS1 | |
| pNOY480 | Derivative of pRS314 carrying sts1ΔNLS1 | |
| pNOY481 | Derivative of pNOY258 carrying sts1ΔNLS1 instead of STS1 | |
| pNOY482 | Derivative of pNOY478 carrying GFP-sts1ΔNLS1 instead of GFP-STS1 | |
| pNOY483 | Derivative of pNOY248 carrying sts1-11,12(E43G, I162T) instead of STS1 | |
| pNOY484 | Derivative of pNOY478 carrying GFP-sts1-11,12(E43G, I162T) instead of STS1 | |
| pNOY485 | Derivative of pRS314 containing GAL1 promoter | |
| pNOY486 | pNOY485 vector containing RPN11 gene fused to GAL1 promoter | |
| pNOY487 | pNOY485 vector containing STS1 gene fused to GAL1 promoter | |
| pNOY488 | Derivative of pRS314 carrying HA-STS1 | |
| pNOY3198 | Derivative of pGEX2T carrying GST-SRP1 | Pharmacia (2) |
| pNOY3234 | Derivative of pUC19 carrying STS1 | |
| pNOY3235 | Derivative of pNOY3234 with HIS3 replacing the PflMI-NsiI STS1 fragment | |
| pNOY3280 | Derivative of pGEX2T carrying GST-sts1ΔNLS1 |
AD, activation domain; DBD, DNA binding domain.
The STS1 gene was amplified by PCR using lambda genomic clone 70732 (American Type Culture Collection) as the template with the primers 5′ GGT ACA TAC TAG CGG CAG 3′ and 5′ CCG ACG ATG ACC ACT CAC 3′. The PCR product was cloned into the SmaI site of pUC19, yielding pNOY3234. A SalI digest was performed to remove STS1 and move it into the SalI site of pRS314 and pRS316 (45), creating pNOY470 and pNOY471, respectively. For STS1 genomic disruption, pNOY3234 was digested with PflMI and NsiI to remove most of the STS1 coding region, and this deletion was replaced with the HIS3 gene to make pNOY3235. pNOY3235 was digested with SalI, and the 1,934-bp fragment was gel isolated and digested with PvuII. The SalI-PvuII sts1::HIS3 fragment was then transformed into NOY397, and His+ transformants were selected (NOY726). NOY726 was sporulated, and tetrads were dissected and showed a 2:2 segregation of His− to His+, confirming that STS1 is an essential gene (12). NOY726 was transformed with pNOY470 or pNOY471, and Trp+ or Ura+ transformants were selected, respectively. Then the strains were sporulated and tetrads were dissected to create NOY943 and NOY944.
Two-hybrid system plasmids were prepared as derivatives of pAS1 and pACT2 (4). SFY526 (Clontech) was used as the reporter strain for all two-hybrid experiments. pNOY472 was constructed by inserting the NcoI-SacI and SacI-SalI SRP1 fragments from pNOY3198 into the NcoI and SalI sites of pAS1. STS1 was amplified using PCR with the template lambda clone 70732 and the primers 5′ CAT GCC ATG GGA TTT GAA TGG GGT TTT AAA CCC 3′ and 5′ CGG AAT TCT TAG TTA AAG GGC GAA TCA GTA G 3′. The PCR fragment was digested with NcoI and EcoRI and cloned into the corresponding sites of pACT2 (pNOY473). The NcoI-XhoI fragment from pNOY473 containing the STS1 gene was ligated into the NcoI and SalI sites of pAS1, creating pNOY474. An RPN11-containing fragment, obtained by digesting pNOY285 with SalI, was treated with T4 DNA polymerase (New England Biolabs) to fill in the overhang and then SacI digested. The blunt SacI fragment was ligated into the SmaI and SacI sites of pACT2, creating pNOY475.
pNOY476 was constructed by cloning a NotI fragment containing the open reading frame for green fluorescent protein (GFP) (a gift from Pamela Silver) into the NotI site of pNOY477. For constructing pNOY478, the GFP-STS1 fusion gene was derived from pNOY476 as a KpnI blunted EagI fragment. This fragment was ligated into pNOY258 previously digested with BamHI, blunt ended with T4 DNA polymerase, and then digested with EagI. pNOY479 was constructed by cloning the PflMI-BamHI fragment from pNOY476 into pNOY471. pNOY480 was constructed using site-directed mutagenesis and the oligonucleotide 5′ CTT GCT CCT CGT TGG CGT AGA CTC CAG CAG TTG GAA TC 3′ to remove NLS1 (21). pNOY481 was constructed by digesting pNOY258 with PflMI and SacII and replacing the STS1 fragment with the sts1ΔNLS1 PflMI-SacII fragment from pNOY480. pNOY482 is a derivative of pNOY478 made by exchanging the STS1 SacII fragment for the sts1ΔNLS1 SacII fragment from pNOY480. pNOY483 is a derivative of pNOY258 but with E43G and I162T. pNOY484 was constructed by moving the PflMI-SacI fragment from pNOY483 into pNOY478 digested with PflMI and SacI to remove STS1.
pNOY291 (glutathione-S-transferase [GST]-STS1) was constructed by PCR using 5′ TTT GGA TCC CAT ATG ATG GGC TTT GAA TGG GGT TTT AAA CCG AGC AGC AAA AT 3′ and 5′ CGC AAT TCT TAG TTA AAG GGC GAA TC 3′ and pNOY258 as the template. The fragment was digested with BamHI and EcoRI and cloned into the BamHI and EcoRI sites of pGEX2T (Pharmacia). pNOY3280 was produced similarly but with pNOY480 as the template. pNOY488 was constructed by ligating the 0.1-kb triple hemagglutinin epitope tag (HA-tag) NotI fragment derived from the GTEP plasmid (39) into the NotI site of pNOY478.
To construct the sts1(E43G) srp1-49 strain, NOY514 was mated with NOY945. The diploids were cured of the wild-type STS1 URA3 plasmid by streaking onto SGlu with 5-fluoroorotic acid and the resulting strain was transformed with pNOY343, which contains the sts1(E43G) mutation. The resulting diploid was sporulated, and tetrads were dissected. Spores that were Trp+, Leu+, His+, and Ura+ were selected (NOY946).
For the experiments to study the stability of the ubiquitin–Pro-β-galactosidase (Ub-P-βgal), strains were constructed by introducing plasmid pUb-P-ek-βgal (3) (obtained from A. Goldberg), carrying the gene for Ub-P-βgal fused to a GAL promoter, into the strains to be tested.
NLS peptide binding assay.
The NLS peptide binding assay was performed as described by Shulga et al. (44).
Protein pull-downs and immunoprecipitations.
GST fusion proteins were produced and purified as described previously (2). Srp1p was prepared from GST-Srp1p by thrombin cleavage, and free GST was removed using glutathione agarose. For GST pull-down reactions, 1 μg of input proteins was mixed together in 100 μl of buffer M (25 mM ammonium sulfate, 50 mM Tris-HCl [pH 8.0], 5% glycerol, 1 mM dithiothreitol, and 1 mM EDTA) and allowed to incubate on ice for 30 min. Then, 200 μl of buffer M plus 0.2% bovine serum albumin and 0.5% Tween 20 was added along with a 15-μl bed volume of glutathione-agarose beads washed in buffer M, and the reactions were incubated further for 45 min at 4°C with rotation. Beads were washed four times with 400 μl of buffer M. Sodium dodecyl sulfate (SDS) sample buffer was added to the beads, which were then boiled and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were blotted onto Immobilon (Millipore, Bedford, Mass.), and the blot was probed with sheep anti-Srp1 antibodies (54). SV40 large T antigen NLS peptide and reverse large T antigen peptide were added to reactions before incubations on ice and were described previously (2).
For coimmunoprecipitation of HA-Sts1p and Srp1p, NOY944 and NOY947 cells were grown to an A600 of ∼0.9 in 50 ml of synthetic glucose medium. Cells were harvested by centrifugation and washed with water. Extracts were prepared in buffer D (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 10% glycerol) by glass bead lysis. The protein concentrations were determined using Bradford reagent (Bio-Rad) and adjusted to be equal. Protein A-Sepharose and Srp1 antiserum were then added to extracts (containing approximately 1 mg of protein) and mixed for 4 h at 4°C. The beads were then washed with buffer A (50 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100), and samples were resolved on an SDS–10% polyacrylamide gel and analyzed by Western blotting using anti-HA antibody. The use of equivalent amounts of extracts in these experiments was always confirmed by Coomassie staining of gels after electrophoresis of samples.
Microscopy.
A Zeiss Axioplan 2 was used to visualize direct and indirect fluorescence. For direct fluorescence of strains containing galactose-inducible GFP constructs, strains were grown overnight in SRaff medium. Production of GFP fusion protein was induced by the addition of 3% galactose directly to the culture, and the cells were observed 3 to 4 h after induction.
Degradation of β-galactosidase test substrates.
Actively growing cells (A600, <1.0) were harvested by centrifugation and labeled in 200 μl of labeling buffer (50 mM sodium phosphate [pH 7.0], 2% galactose) with 0.25 mCi of EXPRES35S protein labeling mix (NEN) for 5 min at 30°C. Cells were then pelleted and resuspended in 400 μl of chase mix (2% yeast extract, 1% peptone, 2% glucose, 10 mM Met, 5 mM Cys, and 0.6 mg of cycloheximide per ml), and incubation was continued at 30°C. Aliquots were removed at specific time points and added to tubes containing 400 μl of buffer A with protease inhibitors (aprotinin, bestatin, pepstatin, leupeptin, and PMSF) and glass beads, and the tubes were rapidly frozen. The cells were lysed by vortexing, and radioactivity in trichloroacetic acid-precipitable material of the extracts was determined. Equal counts were used for immunoprecipitation reactions using anti-β-galactosidase antibodies (Promega) and protein A-Sepharose (Pharmacia). The protein A-Sepharose beads were washed with buffer A plus 0.1% SDS. Samples were analyzed by SDS-PAGE followed by autoradiography.
RESULTS
Intragenic complementation of srp1-31 and srp1-49.
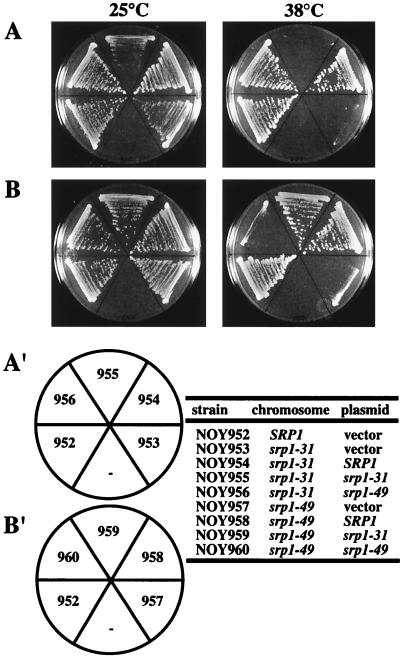
In previous work, ts defects in both in vitro NLS peptide binding and in vivo NLS protein import were observed for the srp1-31 mutation, whereas no or only very weak defects were observed for the srp1-49 mutation (24, 44). In order to test the possibility that these two mutations affected separate functions of Srp1p, we carried out a complementation test. Strains containing each of these two mutations on the chromosome were constructed and transformed with CEN plasmids containing SRP1, srp1-31, or srp1-49. The strains were then streaked on selective medium at 38°C to check for complementation of the ts growth phenotype by a different allele of srp1.
Ts growth of the srp1-31 strain could be complemented by a plasmid containing srp1-49, and ts growth of the srp1-49 strain could be complemented by a plasmid carrying srp1-31 (Fig. 1, sectors 956 and 959). Both mutant strains were complemented by the wild-type SRP1, confirming the recessive nature of the mutations, but not by plasmids containing the identical mutation or by the vector plasmid (Fig. 1). These results indicate that the functions affected by srp1-31 and srp1-49 are clearly different.
FIG. 1.
Complementation of srp1-31 and srp1-49 mutations. Strains carrying SRP1, srp1-31, or srp1-49 on the chromosome were transformed with plasmids containing SRP1, srp1-31, or srp1-49. The resulting strains, as listed in the figure, were streaked on synthetic glucose minus tryptophan plates and incubated at the indicated temperatures for 3 days. Vector represents pRS314.
Isolation of high-dosage suppressors of srp1-49.
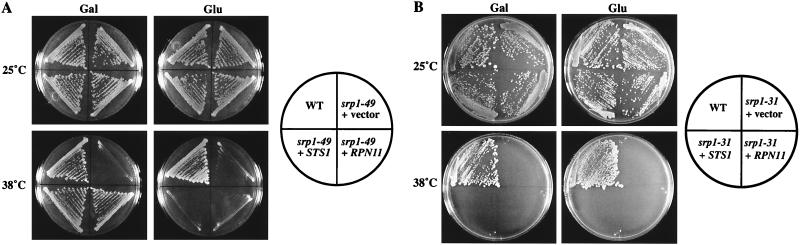
In order to understand the nature of the defect in the srp1-49 mutant, high-dosage suppressors of srp1-49 were isolated. The srp1-49 strains NOY514 and NOY613 were transformed with a galactose-inducible yeast cDNA library (23) and plated on glucose selective medium at 25°C for 2 days. Transformants were then replica plated onto galactose selective medium to induce expression of the cDNAs and allowed to grow at 38°C for an additional 3 to 5 days. Plasmid DNA was isolated from colonies that grew on galactose but not on glucose at 38°C, and DNA sequencing was performed to identify the dosage-dependent suppressors. Two suppressors of srp1-49 were identified in this way, STS1 and RPN11 (Fig. 2A). Plasmids carrying the STS1 or RPN11 suppressor were then transformed into strains carrying the srp1-31 mutation. No suppression was observed (Fig. 2B). Therefore, STS1 and RPN11 are allele-specific high-dosage suppressors of srp1-49.
FIG. 2.
Dosage-dependent suppression of srp1-49 but not srp1-31 by STS1 and RPN11. (A) The srp1-49 strains containing vector only (pRS316-GAL), a GAL-driven STS1 plasmid (pNOY258), and a GAL-driven RPN11 plasmid (pNOY285) are shown after growth on synthetic complete galactose (Gal) or glucose (Glu) plates lacking uracil as indicated for 3 days at 25 or 38°C. (B) The srp1-31 strains containing vector only, a GAL-driven STS1 plasmid, and a GAL-driven RPN11 plasmid were examined in the same way. WT, wild type.
Interaction of SRP1 with STS1.
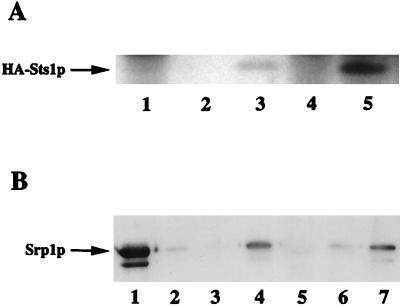
We first examined the possibility of whether the dosage-dependent suppression of srp1-49 by STS1 reflected a physical interaction of the two proteins encoded by these genes. An interaction of the two proteins was first indicated by the yeast two-hybrid system (Table 2). Second, we detected in crude cell extracts a complex containing Srp1p and Sts1p by coimmunoprecipitation. Extracts were prepared from a strain containing HA-tagged STS1 (NOY947) and a control strain without the HA-tag (NOY944). Srp1p was precipitated with Srp1p antiserum, and the precipitates were subjected to SDS-PAGE followed by Western blot using an anti-HA monoclonal antibody. As shown in Fig. 3A, HA-Sts1p was coimmunoprecipitated with Srp1p (lane 5; lane 3 is a negative control using extracts without Srp1p antiserum, which gave a very weak background signal). These two experiments strongly indicate that Srp1p and Sts1p interact in vivo.
TABLE 2.
Summary of two-hybrid assaysa
| Gene or mutation
|
β-Galactosidase activity | |
|---|---|---|
| AD | DBD | |
| STS1 | SRP1 | +++ |
| sts1(E43G I162T) | SRP1 | − |
| RPN11 | SRP1 | − |
| RPN11 | STS1 | ++ |
| RPN11 | None (pAS1) | − |
| STS1 | None (pAS1) | − |
| None (pACT2) | STS1 | − |
| None (pACT2) | None (pAS1) | − |
| p53 | T antigen | +++ |
Strain SFY526 was transformed with plasmids pACT2 (AD) and pAS1 (DBD) carrying the genes indicated. After growth on synthetic complete glucose medium lacking leucine and tryptophan for 4 to 5 days, β-galactosidase filter assays were performed as described by Steffan et al. (48). Color development was rated on a scale of + (blue color development after 1 h at 37°C) to +++ (blue color development in less than 20 min at 37°C). AD, activation domain; DBD, DNA binding domain.
FIG. 3.
Interaction of Srp1p and Sts1p as demonstrated by coimmunoprecipitation from cell extracts (A) and by the use of GST-Sts1p fusion protein (B). (A) Extracts were prepared from the strain carrying the HA-tagged STS1 gene (lanes 3 and 5) or the control strain without the HA-tag (lanes 2 and 4). The extracts were treated with anti-Srp1p antibodies (lanes 4 and 5) and protein A-Sepharose, and precipitated proteins were subjected to SDS-PAGE followed by Western analysis using a monoclonal anti-HA antibody to detect HA-Sts1p. Lane 1 is an antibody control without extracts. Lanes 2 and 3 are negative controls without antibody. (B) Srp1p was incubated with GST (lane 3), GST-Sts1p (lane 4), GST-Sts1ΔNLS1p (lane 5), GST-Sts1p plus 200 μM SV40 NLS peptide (lane 6), or GST-Sts1p plus 200 μM reverse NLS peptide (lane 7). Lane 2 did not receive any GST fusion protein. GST or GST fusion proteins were pulled down using glutathione-agarose beads and then subjected to SDS-PAGE followed by Western blot using anti-Srp1p antibodies to detect Srp1p. Lane 1 is 10% of the input Srp1p preparation.
In order to examine whether these two proteins interact directly in vitro, a GST-STS1 fusion gene was constructed. Purified Srp1p and GST-Sts1p, which were both produced in Escherichia coli as recombinant proteins, were mixed, and GST-Sts1p was pulled down using glutathione-agarose. Pulled-down proteins were separated by SDS-PAGE followed by Western blot analysis of Srp1p using anti-Srp1p antibodies. As shown in Fig. 3B (lane 4), a direct interaction between Srp1p and GST-Sts1p was indeed observed.
As described below, Sts1p is mostly localized to the nucleus. To determine if the interaction between Srp1p and Sts1p could be through an NLS in Sts1p, SV40 NLS peptide was added to the reaction mixture as a competitor in the GST pull-down experiments. NLS peptide was able to decrease the binding of GST-Sts1p to Srp1p (Fig. 3B, compare lane 6 to lane 4), while a peptide containing the same amino acids in reverse sequence was not (lane 7). Thus, the direct interaction of Srp1p with GST-Sts1p in vitro appears to take place through an interaction similar to that of Srp1p with classical NLS peptides.
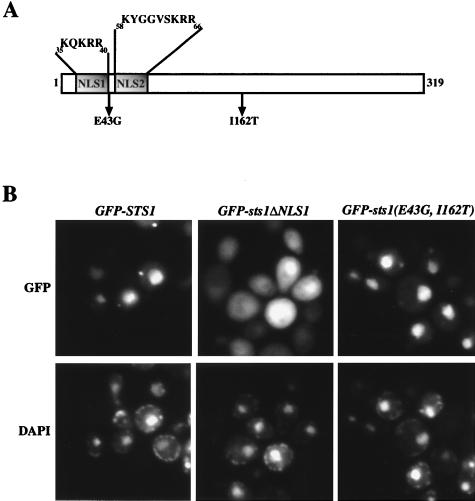
Examination of the Sts1p sequence revealed two basic regions in the amino-terminal end which could potentially serve an NLS function, NLS1 (35KQKRR39) and NLS2 (58KYGGVSKRR66) (Fig. 4A). Both regions were individually deleted using oligonucleotide-directed mutagenesis, and each deletion was tested for complementation of the sts1 genomic deletion. While the mutant carrying the deletion of NLS2 could still complement an sts1 deletion strain, one carrying the deletion of NLS1 did not (data not shown). The production of Sts1ΔNLS1 protein in yeast cells was confirmed by Western blot analysis using anti-HA antibody directed against an N-terminal HA epitope engineered into the deletion protein construct. Therefore, the failure of complementation was not due to lack of protein production. To further characterize NLS1, a fusion protein containing the NLS1 deletion, GST-Sts1ΔNLS1p, was isolated from E. coli as a recombinant protein and tested for its interaction with Srp1p in vitro. The mutant fusion protein was unable to interact with Srp1p (Fig. 3B, lane 5). These experimental results strongly suggest that Sts1p carries an NLS that resembles SV40 NLS and that the in vitro interaction of GST-Sts1p with Srp1p is through this NLS-like sequence of Sts1p. Thus, Sts1p may be a nuclear protein and its nuclear import from the cytoplasm may involve its binding to Srp1p. However, based on the results obtained by fractionation of cell extracts and by indirect immunofluorescence microscopy, it was previously reported that Sts1p was localized to the cytoplasm (1, 22). Therefore, the question of localization of Sts1p was reexamined.
FIG. 4.
Localization of Sts1p assayed by the use of GFP-Sts1 fusion protein. (A) Schematic representation of Sts1 protein, showing locations of NLS1 and NLS2 as well as two point mutations, E43G and I162T. (B) Strains carrying GFP-STS1, GFP-sts1ΔNLS1, and GFP-sts1-11,12(E43G, I162) were grown in SRaff at 30°C, and synthesis of the fusion proteins was induced by galactose. Localization of the protein was analyzed by direct fluorescence microscopy. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI).
Analysis of nuclear localization of Sts1p in the wild-type and srp1 mutant strains.
We constructed a strain containing a gene encoding a tagged GFP-Sts1 fusion protein under control of the galactose promoter (NOY948) and analyzed localization of the fusion protein after induction by galactose. As shown in Fig. 4B, GFP-Sts1 showed a clear nuclear localization in vivo by its fluorescence. This nuclear localization was also confirmed by indirect immunofluorescence using anti-GFP antibodies with a strain containing GFP-STS1 in an sts1 deletion background (data not shown). Effects of the NLS1 deletion on protein localization were then tested in vivo using a galactose-inducible GFP-sts1ΔNLS1 fusion gene. Compared to the nuclear localization of GFP-Sts1p, GFP-Sts1ΔNLS1p showed a more cytoplasmic distribution (Fig. 4B). The fact that GFP-Sts1ΔNLS1p was not excluded from the nucleus may suggest that there is another region in Sts1p with some NLS function. Taken together, these data strongly suggest that Sts1p is mainly localized to the nucleus and interacts with Srp1p via an NLS sequence resembling the SV40 NLS sequence to gain entry into the nucleus.
To prove that Sts1p indeed uses Srp1p for nuclear import, a plasmid containing galactose-inducible GFP-STS1 was transformed into an srp1-31 ts strain, which shows defects in NLS-dependent protein import at nonpermissive temperatures, as mentioned above. Localization of GFP-Sts1p was then followed after a temperature shift by direct fluorescence signal. GFP-Sts1p was found in both the nucleus and the cytoplasm after 4 h at 37°C (data not shown) (48a). Therefore, Sts1p uses Srp1p, at least in part, to gain entry into the nucleus. In contrast to the srp1-31 mutation, no defect in the localization of GFP-Sts1p in an srp1-49 strain was observed during up to 8 h of incubation at 37°C (data not shown) (48a), when many other drastic mutational defects such as an alteration in the nucleolar morphology had already taken place. Thus, suppression of srp1-49 by a high dosage of STS1 is not due to compensation of a (hypothetical) decrease in the nuclear transport of Sts1p in srp1-49 mutants.
Interaction between Srp1p and Sts1p required for suppression of srp1-49.
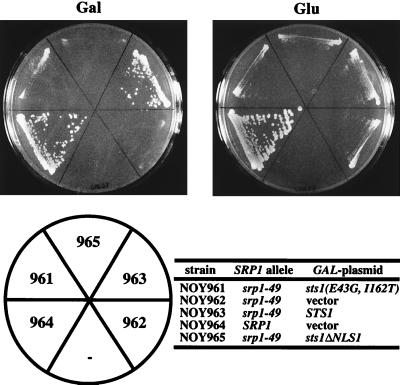
As described above, there is an interaction of Sts1p with Srp1p through the NLS1 sequence of Sts1p. We have found that this interaction may be prerequisite to suppression of srp1-49 by STS1 but is not sufficient for suppression. An additional interaction not involved in the protein import function must take place for suppression. The mutant gene sts1ΔNLS1 was put under control of the galactose promoter and transformed into the srp1-49 strain. Transformants were then tested for their ability to grow at the nonpermissive temperature in the presence of galactose. A high dosage of sts1ΔNLS1 was unable to suppress the growth defect of srp1-49 at 38°C on galactose (Fig. 5, strain 965 compared to 963). The results show that direct interaction with Srp1p through the NLS1 sequence may be required for a high dosage of STS1 to suppress srp1-49. Since Sts1ΔNLS1 protein is clearly present in the nucleus in addition to the cytoplasm, as mentioned above (see Fig. 4B), it appears that the direct interaction required for the suppression is probably not simply to achieve nuclear transport of Sts1p. This inference was further supported by the following experiments.
FIG. 5.
Suppression of srp1-49 by a high dosage of STS1 and sts1 mutants. Plasmids containing STS1, sts1-11,12(E43G, I162T), and sts1ΔNLS1 under control of the GAL promoter were transformed into the srp1-49 strain. Resulting strains and a control SRP1 strain were streaked onto synthetic complete galactose (Gal) or glucose (Glu) plates lacking uracil and incubated for 3 days at 38°C. Vector pRS314 was used as a plasmid control.
We isolated sts1 mutants that did not show an interaction with SRP1 in the two-hybrid assay. These mutants were then screened for loss of the ability to suppress srp1-49 under high-dosage expression. In this way, we found that an sts1 mutation containing a two-amino-acid substitution, sts1-11,12(E43G I162T), abolished both the ability to interact with SRP1 in the two-hybrid system (Table 2) and high-dosage suppression of srp1-49 (Fig. 5, strain 961; for the locations of the mutations, see Fig. 4A). We then examined whether this double mutation inhibited the nuclear localization of Sts1p. Like the wild-type GFP-Sts1p, the mutant GFP-Sts1(E43G, I162T)p was found to be localized to the nucleus (Fig. 4B). Thus, Sts1p localization to the nucleus is not sufficient for suppression of srp1-49. An additional interaction between Sts1p and Srp1p, which is detected by the two-hybrid system and is abolished by the E43G I162T double mutation, appears to be required for suppression of srp1-49. This additional interaction may or may not be a direct interaction and most likely takes place within the nucleus.
Combination of the sts1-11 and srp1-49 mutations.
The above sts1 double mutation, sts1-11,12, showed good complementation of growth defects caused by an sts1 deletion; no difference was observed in growth rate from the wild-type control (data not shown). Thus, the interaction disrupted by the double mutation and required for suppression of srp1-49 does not appear to be essential for normal growth in the context of the wild-type SRP1.
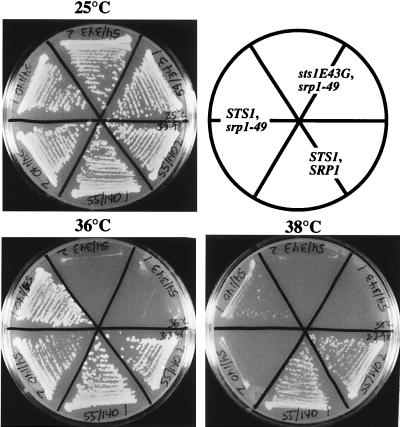
We also separated the two mutations, E43G and I162T, contained in the sts1 double mutant sts1-11,12 and examined them individually for the ability to interact with SRP1 in the two-hybrid system and for the ability to suppress srp1-49 by overexpression. Neither one of the mutations showed an interaction with SRP1 in the two-hybrid system, and each partially abolished the suppression of srp1-49 (data not shown). However, in the course of these experiments, we found that although the sts1-11(E43G) mutation itself did not show any growth defect phenotype, a strain which contained both srp1-49 and sts1-11 showed more severe temperature sensitivity than with srp1-49 only (Fig. 6). This result supports the above-described conclusion that the interaction of Sts1p and Srp1p is functionally important; a point mutation that abolishes this interaction in the two-hybrid system affects cell growth at high temperatures in the context of the srp1-49 mutation.
FIG. 6.
Temperature sensitivity of the sts1-11(E43G) srp1-49 double mutant. Strains containing combinations of wild-type STS1 or sts1(E43G) and wild-type SRP1 or srp1-49 were streaked onto synthetic complete glucose plates lacking tryptophan and uracil and grown for 3 days at 25, 36, or 38°C.
Interaction of STS1 and RPN11.
RPN11, the other high-dosage suppressor of srp1-49, did not show an interaction with SRP1 when tested by the two-hybrid system. However, it did show a positive interaction with STS1 in the two-hybrid system (Table 2). This result suggests that STS1 and RPN11 interact either directly or indirectly through other components and that this interaction might be related to RPN11 suppression of srp1-49.
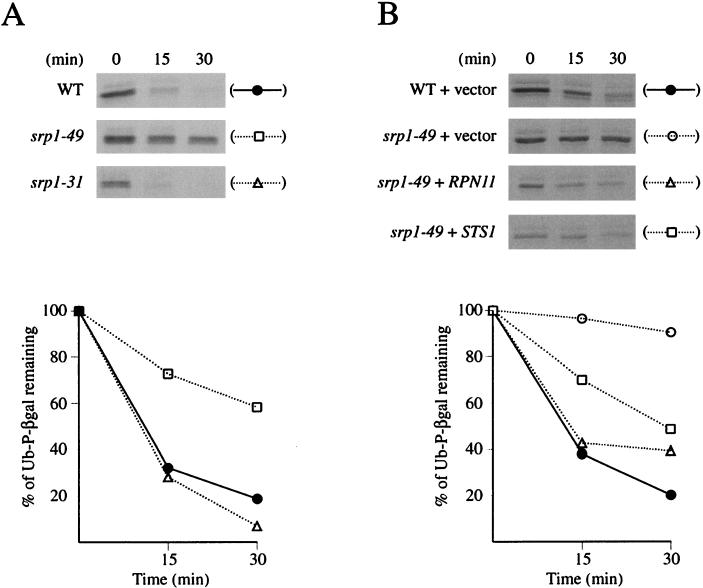
Degradation of Ub-P-βgal in srp1-49 and srp1-31.
The discovery that one of the high-dosage suppressors of srp1-49 was RPN11, a subunit of the 19S proteasome regulatory particle, led us to examine ubiquitin-dependent protein degradation in srp1-49 cells. For this purpose, we used Ub-P-βgal, a model substrate of the ubiquitin-proteasome pathway studied by previous investigators (3, 13). A plasmid carrying the gene for Ub-P-βgal fused to a GAL promoter was introduced into an srp1-49 strain, an srp1-31 strain, and a control wild-type SRP1 strain, as well as srp1-49 strains carrying an RPN11 or STS1 dosage suppressor plasmid. Degradation of Ub-P-βgal was then determined by pulse-chase and immunoprecipitation analysis (3, 13).
As shown in Fig. 7A and B, Ub-P-βgal was substantially more stable in srp1-49 than in the control strain, but no such stabilization was observed for srp1-31 (Fig. 7A). Significantly, both srp1-49 strains carrying the suppressor RPN11 and the suppressor STS1 showed a faster degradation rate approaching that shown by the control strains (Fig. 7B), although the amounts of radioactive Ub-P-βgal synthesized during the pulse-labeling period were reduced in these suppressed strains. We conclude that the srp1-49 mutation and its suppression by a high dosage of RPN11 or STS1 are correlated with a decrease in the rate of Ub-P-βgal degradation and its restoration, respectively. The results also demonstrate that the srp1-49 and srp1-31 mutations are clearly different in their effects on Ub-P-βgal degradation, confirming that the two mutations affect different functions of Srp1p. We note that the experiments shown in Fig. 7 were done at 30°C, which is a permissive temperature for these mutants. Similar results were also obtained when Ub-P-βgal degradation was assayed after a temperature shift from 30 to 38°C (data not shown). Thus, defects in the ability to degrade Ub-P-βgal observed in srp1-49 do not affect growth at 30°C but appear to cause growth defects at higher temperatures.
FIG. 7.
Degradation of Ub-P-βgal. Cells were grown overnight in SGal minimal medium at 30°C and labeled with 35S labeling mix for 5 min. The cells were resuspended in cycloheximide-containing chase mix, and aliquots were removed at the indicated times. The amount of labeled Ub-P-βgal in the extracts was analyzed by immunoprecipitation with anti-β-galactosidase antibodies followed by SDS-PAGE and quantification by phosphorimager analysis. (A) Degradation of Ub-P-βgal in wild-type (WT), srp1-49, and srp1-31 mutants (strains NOY934, NOY937, and NOY940, respectively). (B) Degradation of Ub-P-βgal in srp1-49 mutant in the presence of the suppressors RPN11 (NOY938) and STS1 (NOY939). Degradation of Ub-P-βgal was also assayed simultaneously in srp1-49 in the presence of vector (NOY937) and in wild-type cells (NOY934) as a control. The quantification of Ub-P-βgal is shown in the graphs below the autoradiograms.
Degradation of Ub-P-βgal involves the initial ubiquitination steps of the ubiquitin fusion degradation (UFD) pathway which are different from other ubiquitination pathways, e.g., the N-end rule pathway (13, 14, 16). Therefore, we tested degradation of another model substrate, Leu-βgal, which is degraded through the N-end rule pathway (3). We observed that Leu-βgal degradation was also significantly slower in srp1-49 than in the wild type (data not shown). Therefore, defects caused by the srp1-49 mutation appear not to be limited to the UFD system, but to reside in a more general step(s) in protein degradation shared by these two model substrates.
DISCUSSION
Genetic evidence for separable functions of SRP1.
The role of importin α (karyopherin α) and its yeast homolog Srp1p in NLS-dependent nuclear protein import has been well established. Genetic experiments described in this article demonstrate that Srp1p carries out at least one additional important function, unrelated to its NLS receptor function. The first evidence for this conclusion is the intragenic complementation of the srp1-31 ts mutation and the srp1-49 ts mutation. It should be noted that intragenic complementation could be observed for a protein which functions as a homodimer consisting of two identical subunits; one mutational alteration at a site disrupting the functional conformation of the dimer could conceivably be suppressed by another “compensatory” amino acid alteration at a different site of the same protein molecule. However, the NLS receptor function of Srp1p is carried out as a monomer (or as a heterodimer complexed with Kap95). It was demonstrated that, in the absence of NLS peptides, purified Xenopus importin α forms aggregates at higher concentrations, but that importin α with a bound SV40-type NLS peptide remains a monomer (32). Crystal structure analysis of a 50-kDa fragment of Srp1p with and without an NLS peptide also supports this conclusion (5). Thus, intragenic complementation of srp1-31 and srp1-49 cannot be explained by the model of formation of a functional homodimer with NLS receptor function. Instead, this complementation demonstrates that the function affected by the srp1-49 mutation is separate from the NLS receptor function affected by the srp1-31 mutation. This intragenic complementation also shows that the mutational defect caused by srp1-31 is specific rather than a nonspecific heat inactivation of the protein; the mutant protein Srp1-31p is defective in the NLS protein transport function at nonpermissive temperatures but is able to carry out another function that is affected by the srp1-49 mutation at the same nonpermissive temperatures. We also note that both Srp1-31 and Srp1-49 mutant proteins are stable at 38°C for at least 9 h (unpublished experiments; 48a). This observation excludes the (unlikely) possibility that the observed intragenic complementation is a result of stabilization of a mutant protein(s) by some kind of interaction between the two proteins.
The second evidence is our finding that a high dosage of either STS1 or RPN11 suppresses srp1-49 but not srp1-31. This suppression in srp1-49 cells is correlated with suppression of decreased ubiquitin-dependent protein degradation, as assayed by the use of a model substrate, Ub-P-βgal. No such stabilization of Ub-P-βgal degradation was observed for srp1-31. It would be difficult to explain defects in Ub-P-βgal degradation observed in srp1-49 as a consequence of a defect in the NLS receptor-nuclear protein import function, as further discussed below.
The amino acid residue serine 116 affected by srp1-31 (S116F) (55) is an invariant residue among all published Srp1p homologs from a variety of organisms. The S116F mutation at this residue likely disrupts the structural integrity of the NLS binding site, as discussed by Conti and coworkers (5), based on the crystal structure of an Srp1p-NLS peptide complex, and this is indeed consistent with the experimental observations (44). The amino acid residue glutamic acid 145 altered by the srp1-49 mutation (E145K) (55) is located on the opposite, convex face of the Srp1p structure, away from the concave face where the known NLS binding sites are located. This is also consistent with our observations that mutation srp1-49 shows no defects in NLS binding or in the import of any protein substrate tested so far at nonpermissive temperatures (44; our unpublished experiments). As mentioned above, in vivo complementation of srp1-31 with srp1-49 implies the absence of any significant effect of this mutational alteration on the NLS receptor function of Srp1p.
One possible model to explain our observations is that the srp1-49 mutation is specifically defective in the transport of some proteasome components but is not defective in the transport of many other proteins, whereas the srp1-31 mutation is opposite, i.e., is defective in the transport of many NLS-containing proteins but not in the transport of the proteasome components. According to this model, Srp1p functions in protein transport using two entirely different mechanisms: one is the well-established mechanisms, i.e., by binding the classical SV40-nucleoplasmin-type NLS peptides at the concave face of the Srp1p protein structure; and the second is an ad hoc and undefined mechanism without using the classical NLS-Srp1p interaction. This second mechanism is not affected by the srp1-31 mutation but is somehow affected by the srp1-49 mutation; that is, this mechanism assumes binding of cargo proteins perhaps to the convex face of the protein rather than to the well-established NLS-interacting concave face. We originally considered the possibility that high-dosage suppression of srp1-49 by STS1 or RPN11 might reflect a possible failure of transport of Sts1p or Rpn11p or related proteins to the nucleus catalyzed by Srp1p. However, no defect in Sts1p transport to the nucleus was observed in srp1-49 mutant cells, as described in this paper, and no interaction of Srp1p with Rpn11p was detected in the two-hybrid system or in vitro. Thus, while we cannot exclude the above-mentioned model completely, we think that it is probably unlikely.
Ubiquitin-dependent protein degradation is affected by the srp1-49 mutation.
An important clue to the nature of the Srp1p function affected by the srp1-49 mutation came from the discovery of RPN11 as a high-dosage suppressor of the srp1-49 mutation. RPN11 is a component of the 19S regulatory particle of the proteasome and shows 65% identity to its human counterpart Poh1 (8, 47). Rpn11p contains a highly conserved sequence with similarity to the active-site Cys box seen in many deubiquitinating enzymes, and its possible function within the proteasome was speculated on in this connection (8). However, deubiquitination activity of isolated Rpn11p itself has not been demonstrated, and the significance of this similarity is not clear.
The Schizosaccharomyces pombe homolog of RPN11, pad1+, was originally isolated as a gene which confers resistance to staurosporine and other drugs when expressed in high dosage (43). The increased resistance to the drugs was explained by suggesting that overexpression of RPN11 caused stabilization of unstable transcription factors, such as Pap1, required for the expression of genes responsible for the drug resistance (31, 47). However, no experimental tests have been carried out to examine the validity of this explanation. Using the model substrate Ub-P-βgal for the ubiquitin-proteasome system, we have found that this model substrate is significantly more stable in the srp1-49 cells than in wild-type cells or the srp1-31 mutant cells. Overexpression of RPN11 in srp1-49 cells decreased the stability of this model substrate close to the wild-type level, although no such effects, i.e., further decrease in the stability, were observed in the control wild-type cells (Fig. 7 and unpublished experiments). It appears that the srp1-49 mutation causes some defects, directly or indirectly, in the activity of the ubiquitin-proteasome system and that overexpression of RPN11 somehow restores its activity. A similar mutational defect, increased stability of protein, was also observed for another model substrate, Leu-βgal. It is known that initial steps of degradation of Ub-P-βgal are through the UFD pathway, whereas those of Leu-βgal degradation take place through the N-end rule pathway (13, 14, 16). These two pathways do not share ubiquitination enzymes E2 or E3, but share later steps involving proteasome activity. Thus, the defect caused by the srp1-49 mutation might be in a step(s) involving the proteasome. However, mutations in proteasome subunits were often reported to cause accumulation of polyubiquitinated protein substrates. No increased accumulation of polyubiquitinated forms of Ub-Pro-βgal or Leu-βgal in the srp1-49 mutant was observed in our experiments. Therefore, it is possible that a ubiquitination-polyubiquitination step(s) of these model substrates is somehow affected by the mutation regardless of the kinds of E2-E3 ubiquitination enzyme systems.
Possible significance of an Srp1p-Sts1p complex in ubiquitin-dependent protein degradation.
The function of Srp1p affected by srp1-49, i.e., the function related to protein degradation, as discussed above, may be carried out by a complex containing Srp1p and Sts1p, and a direct interaction of these two proteins may be important for this function. First, Srp1p and Sts1p interact in vivo, as revealed in the two-hybrid system, and interact directly in vitro. Second, a protein complex containing these two proteins exists in cell extracts, as shown by coimmunoprecipitation experiments. Third, the ts phenotype of srp1-49 can be suppressed by a high dosage of STS1, and this suppression is abolished by an sts1 mutation that disrupts the Srp1p-Sts1p interaction detected by the two-hybrid system. Fourth, the same sts1 mutation, which does not cause growth defects by itself, causes a synthetic growth defect when combined with srp1-49, i.e., a more severe ts phenotype. In addition, RPN11 and STS1 interact in the two-hybrid system. Therefore, the main function of STS1 may also be related to protein degradation. In fact, we observed that overexpression of STS1 restores the decay rate of Ub-P-βgal in srp1-49 cells close to the wild-type level, and this may be the basis of suppression of the srp1-49 mutation by a high dosage of STS1. Several previous observations can be explained on the basis of the proposed new function (related to protein degradation) for both STS1 and SRP1, as discussed below.
First, SRP1 was originally identified as a suppressor of certain ts mutations in the A190 subunit of Pol I (54). Several point mutations of SRP1, such as SRP1-1 and SRP1-2, were isolated as suppressors. These mutations, when separated from the rpa190 mutations, did not show any phenotype by themselves, but the suppressor activity was dominant over the wild-type SRP1 (54, 55). The rpa190 ts mutations, suppressed by these SRP1 suppressors, such as rpa190-1 and rpa190-5, caused a large decrease in the amount of A190 at nonpermissive temperatures, apparently due to an instability of mutant A190 protein (26). One possibility that we considered based on the NLS receptor function of Srp1p was an increase in the efficiency of binding of A190 (or a complex containing A190, e.g., Pol I) to Srp1p, leading to an increased supply of A190 (or a Pol I precursor or Pol I) to the nucleus, counteracting the instability of the mutant protein. We tested this possibility by constructing a CEN plasmid carrying the srp1 gene (srp1-2,31) containing both the SRP1-2 suppressor mutation and the srp1-31 mutation. We found that the Srp1p carrying these two mutations was more severely ts in NLS binding in vitro than that carrying the srp1-31 mutation only, and yet the plasmid introduced into the rpa190-1 mutant suppressed the ts phenotype (our unpublished experiments). Thus, the suppressor activity was not abolished by inactivation of the NLS binding-NLS protein transport function. Therefore, we suggest that the suppressor mutations such as SRP1-2 affect another function of Srp1p which is related to protein degradation and counteract the degradation of the unstable mutant Pol I subunit protein.
A similar interpretation can be applied to the reported suppression of the rna15-2 mutation by a mutation in STS1 (1). Rna15p is an essential component of cleavage factor I that, together with Rna14p, is involved in cleavage and polyadenylation of mRNAs (27). For rna15-2, the observed defect in 3′-end mRNA processing was due to unstable Rna15-2 protein at high temperature. A mutation in STS1 partially corrects this protein instability and so partially rescues 3′-end mRNA processing (1). Since Srp1p and Sts1p appear to carry out a function as a complex, as shown in this paper, we suggest that this suppressor mutation in STS1 may involve stabilization of the unstable nuclear protein Rna15-2p. Increased amounts of the protein rescue the defect of rna15-2 at high temperature.
Liang et al. (22) originally discovered that a high dosage of STS1 suppresses the ts phenotype of sec23-11 but not the ts phenotype of sec23-1. These authors reported that a shift to the nonpermissive temperature leads to accumulation of the ER form of a secreted protein, invertase. Overexpression of STS1 alleviated this accumulation of invertase, which led them to suggest that STS1 corrects the secretion defects of this mutant. However, they have not followed the fate of the ER form of invertase, and so it is not clear if overexpression of STS1 leads to transport of the ER form of invertase to the cell surface. The ubiquitin-proteasome system is also involved in the degradation of mutant secretory proteins that are accumulated in the ER (reviewed in reference 34). It is therefore likely that a high dosage of STS1 stimulates the degradation of proteins accumulated in the ER, in the same way as it stimulates degradation of Ub-P-βgal in srp1-49 cells, leading to suppression of the ts phenotype of sec23-11. In fact, we found that like STS1, RPN11 can also suppress the ts defect of sec23-11 (our unpublished experiments) (48a), which supports our interpretation of the results published by Liang et al. (22).
Role of SRP1 and STS1 in mitosis.
Although the function of Srp1p as the NLS receptor in protein import has been well established, we have argued that Srp1p may also carry out an additional essential function(s) concerned with the regulation of protein degradation. It has been well established that regulated protein degradation through the ubiquitin-proteasome system is essential for progression of the cell cycle (reviewed in references 33 and 50). Mutations in proteasome components or in the anaphase-promoting complex (a ubiquitin protein ligase E3) are known to cause mitotic arrest. In fact, some mutations in RPN11, which encodes a proteasome regulatory subunit and is a high-dosage suppressor of srp1-49, have recently been shown to cause a mitotic arrest (38; see reference 31 for mutations in the S. pombe homolog pad1+). The basis of mitotic arrest observed in these instances were shown or interpreted to be due to mutational defects in the regulated proteolysis of certain key proteins, such as mitotic cyclins and Pds1p (Cut2), which were shown to be essential for progression of mitosis (33, 50, 52). Thus, the mitotic arrest phenotype evident in the srp1-49 mutant at nonpermissive temperatures (55) or after depletion of Srp1p (20) can now be explained on the basis of defects in protein degradation, as observed in the present work using model substrates for the ubiquitin-proteasome system. However, we have not determined the identity of the natural protein substrates whose stability is affected by the srp1-49 mutation. In this connection, we note that the srp1-31 mutation was reported to cause a clean mitotic arrest at nonpermissive temperatures and degradation of Clb2p is defective under these conditions (24). This observation was interpreted on the basis of defects in nuclear import of some cell cycle regulators (24). In our hands, however, mitotic arrests were clearly evident for srp1-49 but not for srp1-31 (our unpublished experiments). The reason for this discrepancy is not clear. As already discussed, we favor our interpretation that the mitotic arrest observed for srp1-49 is a consequence of defects in the function related to protein degradation and not a consequence of defects in nuclear import of hypothetical cell cycle regulatory proteins.
STS1, which was also called DBF8, was independently identified as a gene essential for nuclear division and segregation. Strains carrying dbf8 mutations show a mitotic arrest phenotype at nonpermissive temperatures similar to that seen for srp1-49 (12). This phenotype is consistent with our conclusion that Sts1p, like Rpn11p, plays a role in the regulation of protein degradation through the ubiquitin-proteasome system.
Another observation relevant to our conclusion on the new function of Srp1p separate from its NLS receptor function is a recent work by Matsusaka et al. (25) on the identification of S. pombe gene cut15+ as a homolog of SRP1. These investigators found that a ts mutation in cut15+ shows a mitotic arrest at nonpermissive temperatures, presumably due to defects in chromosome condensation. Surprisingly, the mutation caused a defect in NLS binding activity of the protein in vitro, and yet no defects in protein import were observed in vivo. They concluded that cut15+, a homolog of SRP1 is essential for mitotic chromosome condensation, but its role in nuclear protein import is dispensable (25). Their results support our conclusion that Srp1p has a function which is separable from the NLS receptor function. We note that chromosome condensation appears to be defective in srp1-49 mutants at nonpermissive temperatures (55) and in Srp1p-depleted cells (20, 55). It will be interesting to examine whether there is any defect in activities of the ubiquitin-proteasome system in the cut15 mutant analyzed.
We also note that a ts mutation (cse1-2) in CSE1, which encodes the yeast homolog of mammalian CAS, shows a mitotic arrest phenotype at nonpermissive temperatures which is very similar to srp1-49 (55), including a significant percentage of arrested cells with spindle orientation defects (41). Both yeast Cse1p and mammalian CAS have been demonstrated to function in recycling Srp1p back to the cytoplasm after completion of nuclear protein import (11, 18, 46). Consequently, the phenotypes of cse1 mutations and various genetic interactions between CSE1 and SRP1 have been explained solely on the basis of the functions of these two genes in nuclear protein transport. Schroeder et al. (41) reported suppression of srp1-49 but not srp1-31 by a high dosage of CSE1. Since srp1-49 does not show any defect in nuclear protein import, this observed suppression is not easy to explain on the basis of the Srp1p-recycling function of Cse1p. It is known that the CAS protein is associated with microtubules, including mitotic spindles (40), and its depletion by the use of antisense RNA leads to mitotic arrest in mammalian cells (29). The S. pombe Srp1p homolog, cut15 protein, was also shown to be clearly associated with the mitotic spindle, in addition to other intranuclear sites, during late mitosis (25). It is known that degradation of a microtubule-associated protein, Ase1p, is mediated by the anaphase-promoting complex-proteasome system in S. cerevisiae and that inappropriate expression of nondegradable Ase1p during G1 caused mitotic arrest (15). Thus, it is possible that Cse1p, like Srp1p, has functions important for mitosis that are independent of its role in nuclear transport and that suppression of defects in srp1-49 cells by a high dosage of CSE1 is related to the proposed function of regulation of protein degradation.
The proposed role of Srp1p in mitosis is also consistent with earlier observations that pendulin, one of the Drosophila homologs of Srp1p, rapidly translocates from the cytoplasm to the nucleus at the transition between G2 and M phase and is found associated with condensed metaphase chromosomes (19, 49). Such observations suggest that an important function of pendulin is related to mitosis. Since there are several distinct Srp1p homologs in higher eukaryotes (17 and references therein), there might be a functional differentiation among these homologs. In the yeast S. cerevisiae, only a single protein species, Srp1p, exists as an importin α homolog. Thus, one can speculate that the yeast Srp1p participates in a variety of nuclear functions through regulation of degradation of various regulatory proteins and that these regulatory functions might be divided among different Srp1p homologs in higher eukaryotes. One can also speculate that during an earlier time in evolution, Srp1p was a nuclear protein interacting with several other nuclear proteins, such as Sts1p, and carrying out a specific function(s) such as those discussed above, and that one of the importin β family members, that corresponding to the present-day Kap95p, was the import machinery for Srp1p. Therefore, the protein import function of Srp1p as a piggyback receptor might be a result of more recent evolution and be limited to only certain kinds of nuclear proteins. This speculative notion is supported by the fact that several importin β family members are engaged in nuclear import of a variety of proteins without using Srp1p (for review, see references 9, 28, and 30).
In conclusion, the work described in this paper strongly indicates that Srp1p carries out a function, almost certainly as a complex with Sts1p, separate from its function as the NLS receptor in nuclear protein import, and that this function is related to regulation of protein degradation through the ubiquitin-proteasome system. Elucidation of the molecular details of the proposed function is a subject for future studies.
ACKNOWLEDGMENTS
We thank M. Waterman for helpful discussions and critical advice on the manuscript. We thank J. Keener and K. Sutton in this laboratory for suggesting the complementation experiments and for excellent technical assistance, respectively. We thank J. Loeb for help in construction of mutant strains and are grateful to K. Madura, P. Silver, F. Kepes, and A. Goldberg for providing plasmids and strains. We also thank C. Carmen for help in preparation of the manuscript.
This work was supported by Public Health Service grant GM 35949.
REFERENCES
- 1.Amrani N, Dufour M-E, Bonneaud N, Lacroute F. Mutations in STS1 suppress the defect in 3′ mRNA processing caused by the rna15-2 mutation in Saccharomyces cerevisiae. Mol Gen Genet. 1996;252:552–562. doi: 10.1007/BF02172401. [DOI] [PubMed] [Google Scholar]
- 2.Azuma Y, Tabb M M, Vu L, Nomura M. Isolation of a yeast protein kinase that is activated by the protein encoded by SRP1 (Srp1p) and phosphorylates Srp1p complexed with nuclear localization signal peptides. Proc Natl Acad Sci USA. 1995;92:5159–5163. doi: 10.1073/pnas.92.11.5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 4.Bai C, Elledge E J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 5.Conti E, Yu M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 6.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 7.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glickman M H, Rubin D M, Fried V A, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 11.Hood J K, Silver P A. Cse1p is required for export of Srp1p/importin-α from the nucleus in Saccharomyces cerevisiae. J Biol Chem. 1999;273:35142–35146. doi: 10.1074/jbc.273.52.35142. [DOI] [PubMed] [Google Scholar]
- 12.Houman F, Holm C. DBF8, an essential gene required for efficient chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6350–6360. doi: 10.1128/mcb.14.9.6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson E S, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson E S, Philip C M, Ota I M, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 15.Juang Y L, Huang J, Peters J M, McLaughlin M E, Tai C Y, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- 16.Koegl M, Hoppe T, Schlenker S, Ulrich H D, Mayer T U, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 17.Kohler M, Speck C, Christiansen M, Bischoff F R, Prehn S, Haller H, Gorlich D, Hartmann E. Evidence for distinct substrate specificities of Importin α family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunzler M, Hurt E C. Cse1p functions as the nuclear export receptor for importin α in yeast. FEBS Lett. 1998;433:185–190. doi: 10.1016/s0014-5793(98)00892-8. [DOI] [PubMed] [Google Scholar]
- 19.Kussel P, Frasch M. Pendulin, a Drosophila protein with cell cycle-dependent nuclear localization, is required for normal cell proliferation. J Cell Biol. 1995;129:1491–1507. doi: 10.1083/jcb.129.6.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kussel P, Frasch M. Yeast Srp1p, a nuclear protein related to Drosophila and mouse pendulin, is required for normal migration, division, and integrity of nuclei during mitosis. Mol Gen Genet. 1995;248:351–363. doi: 10.1007/BF02191602. [DOI] [PubMed] [Google Scholar]
- 21.Lehming N, McGuire S, Brickman J M, Ptashne M. Interactions of a Rel protein with its inhibitor. Proc Natl Acad Sci USA. 1995;92:10242–10246. doi: 10.1073/pnas.92.22.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang S, Lacroute F, Kepes F. Multicopy STS1 restores both protein transport and ribosomal RNA stability in a new yeast sec23 mutant allele. Eur J Cell Biol. 1993;62:270–281. [PubMed] [Google Scholar]
- 23.Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loeb J D J, Schlenstedt G, Pellman D, Kornitzer D, Silver P A, Fink G R. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsusaka T, Imamoto N, Yoneda Y, Yanagida M. Mutations in fission yeast Cut15, an importin α homolog, lead to mitotic progression without chromosome segregation. Curr Biol. 1998;8:1031–1034. doi: 10.1016/s0960-9822(07)00425-3. [DOI] [PubMed] [Google Scholar]
- 26.McCusker J H, Yamagishi M, Kolb J M, Nomura M. Suppressor analysis of temperature-sensitive RNA polymerase I mutations in Saccharomyces cerevisiae: suppression of mutations in a zinc-binding motif by transposed mutant genes. Mol Cell Biol. 1991;11:746–753. doi: 10.1128/mcb.11.2.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate: sequence analysis reveals an RNA-binding domain in the RNA 15 protein. Mol Cell Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- 29.Ogryzko V V, Brinkmann E, Howard B H, Pastan I, Brinkmann U. Antisense inhibition of CAS, the human homologue of the yeast chromosome segregation gene CSE1, interferes with mitosis in HeLa cells. Biochemistry. 1997;36:9493–9500. doi: 10.1021/bi970236o. [DOI] [PubMed] [Google Scholar]
- 30.Pemberton L F, Blobel G, Rosenblum J S. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 31.Penney M, Wilkinson C, Wallace M, Javerzat J P, Ferrel K, Seeger M, Dubiel W, McKay S, Allshire R, Gordon C. The pad1+ gene encodes a subunit of the 26 S proteasome in fission yeast. J Biol Chem. 1998;273:23938–23945. doi: 10.1074/jbc.273.37.23938. [DOI] [PubMed] [Google Scholar]
- 32.Percipalle P, Jonathan P, Butler G, Finch J T, Jans D A, Rhodes D. Nuclear localization signal recognition causes release of importin-α from aggregates in the cytosol. J Mol Biol. 1999;292:263–273. doi: 10.1006/jmbi.1999.3077. [DOI] [PubMed] [Google Scholar]
- 33.Peters J-M, King R W, Deshaies R J. Cell cycle control by ubiquitin-dependent proteolysis. In: Peters J-M, et al., editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum Press; 1998. pp. 345–387. [Google Scholar]
- 34.Plemper R K, Wolf D H. Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem Sci. 1999;24:266–270. doi: 10.1016/s0968-0004(99)01420-6. [DOI] [PubMed] [Google Scholar]
- 35.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radu A, Moore M S, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 37.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 38.Rinaldi T, Ricci C, Porro D, Bolotin-Fukuhara M, Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol Biol Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roof D M, Meluh P B, Rose M D. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherf U, Pastan I, Willingham M C, Brinkmann U. The human CAS protein which is homologous to the CSE1 yeast chromosome segregation gene product is associated with microtubules and mitotic spindle. Proc Natl Acad Sci USA. 1996;93:2670–2674. doi: 10.1073/pnas.93.7.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroeder A J, Chen X H, Xiao Z, Fitzgerald-Hayes M. Genetic evidence for interactions between yeast importin α (Srp1p) and its nuclear export receptor, Cse1p. Mol Gen Genet. 1999;261:788–795. doi: 10.1007/s004380050022. [DOI] [PubMed] [Google Scholar]
- 42.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 43.Shimanuki M, Saka Y, Yanagida M, Toda T. A novel essential fission yeast gene pad1+ positively regulates pap1+-dependent transcription and is implicated in the maintenance of chromosome structures. J Cell Sci. 1995;108:569–579. doi: 10.1242/jcs.108.2.569. [DOI] [PubMed] [Google Scholar]
- 44.Shulga N, Roberts P, Gu Z, Spitz L, Tabb M M, Nomura M, Goldfarb D S. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sikorski R A, Heiter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solsbacher J, Maurer P, Bischoff E R, Schlenstedt G. Cse1p is involved in export of yeast importin α from the nucleus. Mol Cell Biol. 1998;18:6805–6815. doi: 10.1128/mcb.18.11.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spatoro V, Toda T, Craig R, Seeger M, Dubiel W, Harris A L, Norbury C. Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26S proteasome subunit. J Biol Chem. 1997;272:30470–30475. doi: 10.1074/jbc.272.48.30470. [DOI] [PubMed] [Google Scholar]
- 48.Steffan J S, Keys D A, Vu L, Nomura M. Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol Cell Biol. 1998;18:3752–3761. doi: 10.1128/mcb.18.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Tabb M M. Genetic and functional analysis of SRP1, the yeast nuclear localization signal receptor. Ph.D. thesis. University of California; 1998. , Irvine. [Google Scholar]
- 49.Torok I, Strand D, Schmitt R, Tick G, Torok T, Kiss I, Mechler B M. The overgrown hematopoietic organs-31 tumor suppressor gene of Drosophila encodes an importin-like protein accumulating in the nucleus at the onset of mitosis. J Cell Biol. 1995;129:1473–1489. doi: 10.1083/jcb.129.6.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Townsley F M, Ruderman J V. Proteolytic ratches that control progression through mitosis. Trends Cell Biol. 1998;8:238–244. doi: 10.1016/s0962-8924(98)01268-9. [DOI] [PubMed] [Google Scholar]
- 51.Winston F, Chumley F, Fink G R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:221–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]
- 52.Yanagida M. Fission yeast cut mutations revisited: control of anaphase. Trends Cell Biol. 1998;8:144–149. doi: 10.1016/s0962-8924(98)01236-7. [DOI] [PubMed] [Google Scholar]
- 53.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 54.Yano R, Oakes M, Yamagishi M, Dodd J A, Nomura M. Cloning and characterization of SRP1, a suppressor of temperature-sensitive RNA polymerase I mutations, in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5640–5651. doi: 10.1128/mcb.12.12.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yano R, Oakes M, Tabb M M, Nomura M. Yeast Srp1p has homology to armadillo/plakoglobin/β-catenin and participates in apparently multiple nuclear functions including the maintenance of nucleolar structure. Proc Natl Acad Sci USA. 1994;91:6880–6884. doi: 10.1073/pnas.91.15.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshihisa T, Barlowe C, Scheckman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1469–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]