Abstract
This review summarizes the pathogenic mechanisms that underpin the monogenic epilepsies and discusses the potential of novel precision therapeutics to treat these disorders. Pathogenic mechanisms of epilepsy include recessive (null alleles), haploinsufficiency, imprinting, gain-of-function, and dominant negative effects. Understanding which pathogenic mechanism(s) that underlie each genetic epilepsy is pivotal to design precision therapies that are most likely to be beneficial for the patient. Novel therapeutics discussed include gene therapy, gene editing, antisense oligonucleotides, and protein replacement. Discussions are illustrated and reinforced with examples from the literature.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-021-01137-z.
Keywords: Epilepsy, Precision therapies, Genetics, Antisense oligonucleotides, Gene therapy
Introduction
Our understanding of the genetic underpinnings of early-onset pediatric epilepsies has completely transformed over the last decade, driven primarily by high-throughput DNA sequencing approaches. Initially, multigene and exome sequencing was applied in research cohorts of individuals with the developmental and epileptic encephalopathies (DEEs), leading to the discovery of multiple novel genes that harbor de novo variants [1–3]. In the intervening years, exome sequencing became routine in the research setting, leading to the identification of hundreds of genes that can cause many types of rare monogenic epilepsies that can be both inherited in a recessive manner, or arise de novo either in the germline or the soma. Nowadays, at least in high-income countries, multigene epilepsy gene panels are performed in a clinical setting in all new-onset unexplained epilepsy cases at large academic institutions [4, 5]. For those individuals without a positive diagnosis after initial panel testing, clinical exome sequencing is also performed. As a result, genetic testing can now lead to a diagnosis in 10–60% of individuals [4–8]. Diagnostic yields largely dependent on seizure type, onset, and response to therapies and the highest yields are in the DEEs and individuals with other neonatal or infantile-onset epilepsies.
We focus here on therapeutic approaches in these monogenic epilepsies. However, the more common epilepsies, including genetic generalized epilepsy (GGE) and non-lesional focal epilepsies, are enriched for rare variants in the same genes and pathways that have been previously implicated in monogenic conditions [9, 10]. Moreover, individual common variants and loci associated with GGE, focal epilepsies, and unclassified epilepsies have been putatively linked to known monogenic genes [11]. Collectively, this evidence suggests that rare and common variant epilepsy-risk factors will coalesce onto the same genes/pathways as monogenic epilepsies, though with less profound effects on individual protein function. Thus, precision therapies developed in these rarer epilepsies are likely to provide a viable avenue for treatment of a much broader spectrum of patients afflicted with epilepsy.
In this review, we discuss the recent progress towards precision therapeutics in the genetic epilepsies. We focus on three types of precision therapies: gene therapies, RNA therapeutics, or protein replacement, all of which aim to restore the protein levels of an epilepsy-associated gene to that of physiological levels that are not associated with disease. We define each of these approaches as:
Gene therapy, a DNA-based intervention, involves the replacement (by gene editing) or addition of the DNA sequence that codes for the faulty protein (i.e., the cDNA). This can involve delivery of the cDNA directly into cells, or editing of a patient’s own DNA to correct the disease-causing variant.
RNA therapeutics, an RNA-based intervention, most popular at the moment, are antisense oligonucleotides (ASOs) that can bind mRNA and both enhance or reduce the amount of protein that is ultimately generated from that mRNA transcript. There are other types of RNA therapeutics including RNA silencing (siRNA, RNAi), RNA modulation (miRNA, tRNA), and RNA agonists and antagonists (aptamers) that we do not discuss in depth here.
Protein replacement, a protein-based intervention, involves the production of a synthetic protein that replaces the mutant/absent protein in an individual.
Which of these therapies is most appropriate for a particular monogenic precision therapy will be based on a myriad of factors, but most importantly, the pathogenic mechanism that underpins that particular epilepsy. A pathogenic variant can alter protein in numerous ways, but in genetic terms, we consider null alleles (in recessive epilepsies), haploinsufficiency, gain-of-function, and dominant negative mechanisms. In this review, we describe the most promising precision therapeutic approaches in the genetic epilepsies using the modalities described above—gene replacement, editing, ASOs, and protein replacement. For each, we provide an overview of the basic premise, recent developments, and advantages and limitations of these approaches. Importantly, throughout the review, we highlight how the choice of precision therapy will rely on the pathogenic mechanism that underlies each genetic epilepsy and give specific examples from the rare monogenic epilepsies. Finally, given that this field is in its relevant infancy, we also give examples from other genetic conditions to highlight salient concepts.
Pathogenic Mechanisms in the Genetic Epilepsies
Accelerated gene discovery has enhanced our appreciation for the pathogenic mechanisms and myriad of biological pathways that can be perturbed in the epilepsies. Initially, epilepsy was hypothesized to be a channelopathy, supported by the identification of genetic variants in the sodium and potassium voltage-gated ion channels. However, unbiased gene discovery has now highlighted the disruption of the mTOR pathway, synaptic vesicle cycling, metabolism, chromatin remodeling, gene expression, and mRNA splicing in epilepsy [3]. In this review, we touch on some of these fundamental biological mechanisms when highlighting specific examples of therapies, but frame much of this review around the different pathogenic mechanisms, including null, haploinsufficiency, gain-of-function, and dominant negative mechanisms. We define each of these pathogenic mechanisms, with exemplars in epilepsy, but also, importantly, discuss why it is pivotal to understand which one (ideally) of these mechanisms underpin a particular monogenic epilepsy in order to develop safe and effective precision therapies.
Recessive Epilepsies Generally Cause Complete Loss of Function (Null Alleles)
In recessive epilepsies, pathogenic variants are required on both alleles (i.e., biallelic) for the condition to manifest; these variants can be the same (i.e., homozygous) or different (i.e., compound heterozygous). While originally thought to be rare, and more confined to consanguineous or genetically isolated populations, we now know that up to 38% of epilepsies can be due to recessive causes in the DEEs, though this number is likely to be less in the milder and more common epilepsy subtypes [12].
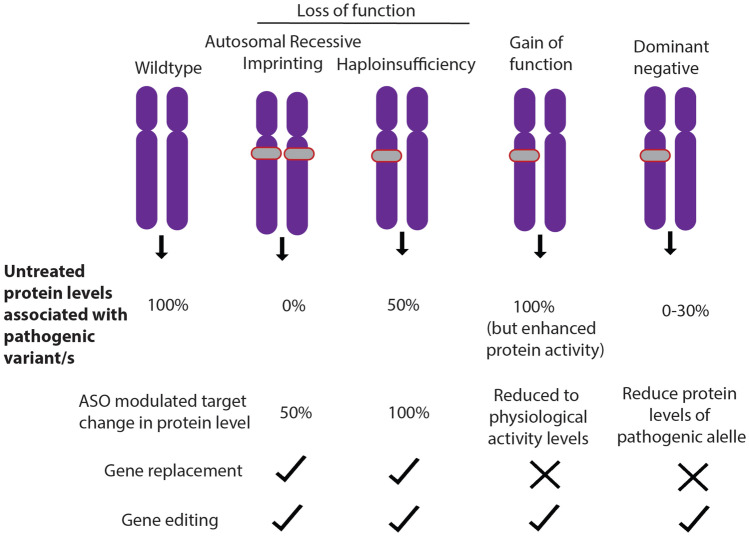
With rare exceptions, recessive pathogenic variants in epilepsy tend to be loss-of-function. These variants are often alterations to the amino acid sequence that introduce a premature truncating codon resulting in a degradation of the mRNA transcript in the cell through the process of nonsense-mediated decay (NMD). Occasionally, NMD may not be activated, and the transcript can code for a shortened protein that is generally non-functional. Truncating variants include nonsense, frameshifts, and variants at the donor/acceptor splice site that generally result in frameshifts. Missense variants can also result in loss-of-function if they affect protein function, often if they occur in a conserved domain, leading to an unstable protein or otherwise affects the tertiary structure of the protein. The net result of truncating or missense loss-of-function variants inherited in a recessive manner is that the proband essentially has no or very little functional protein, while parents with 50% residual protein levels are unaffected. Thus, any therapeutic approach that restores protein levels to ~50% will be an effective strategy (Fig. 1).
Fig. 1.
Pathogenic mechanisms and precision therapy approaches in the monogenic epilepsies. Autosomal recessive disorders result in no functional protein as pathogenic variants occur on both alleles (grey). Likewise, imprinting disorders result in no functional protein due to a pathogenic variant on one allele, while there is no protein from the imprinted allele. Gain-of-function variants can both increase protein function, or alter the protein function, often in a toxic manner. Dominant negative variants interfere with the functional protein made from the other allele resulting in less than 50% residual protein. The viable ASO approach is shown as well as whether this pathogenic mechanism can be targeted using gene replacement or editing
Haploinsufficiency
Loss of function alleles in autosomal dominant or de novo genetic models are generally associated with haploinsufficiency where a protein level of 50%, produced from the unaffected allele, is insufficient to prevent phenotypic manifestation, in this instance, epilepsy. There are a large number of genetic epilepsies that are known or hypothesized to cause epilepsy via haploinsufficient mechanisms, including a number of the most commonly implicated genes (Table 1). The goal of therapeutic approaches in haploinsufficient disorders is to restore protein levels as close as possible to 100%, i.e., protein levels similar to what is produced from two functional alleles (Fig. 1). For instance, pathogenic variants in SCN1A cause the DEE, Dravet syndrome, which results from haploinsufficiency of the sodium channel Nav1.1 [13]. Scn1a targeting ASOs were recently shown to effectively restore Nav1.1 in both cellular models and a mouse model to levels similar to wild-type cell/animals. The restoration of physiological levels of Nav1.1 also rescued the phenotypes of Scn1a knockdown in these mice, including premature death as well as reduced number and latency of spontaneous seizures [14].
Table 1.
Pathogenic mechanisms in the genetic epilepsies and therapeutic approaches
| Genetic mechanism | Examples of genes | Therapeutic approaches applied in preclinical models (reference) | NIH clinical trial study ID (approach) |
|---|---|---|---|
| Recessive (null) | CLN2 | ASO targeting a splice variant, upregulation of CLN2 [86] | |
| Haploinsufficiency | SCN1A, CHD2, SYNGAP1, DEPDC5 | SCN1A-ASO targeting an NMD-inducing exon (TANGO) [14] | NCT04442295 (TANGO ASO) |
| Gain-of-function | SCN8A, KCNT1 | RNAse H1–mediated knockdown of Scn8a [17] and Kcnt1 [18] | |
| Dominant negative | SMC1A, STXBP1 | ||
| Imprinted | UBE3A | ASO reducing the Ube3a-AS, increase paternal Ube3a expression [79] | NCT04259281 (UBE3A-AS targeting ASO) |
Gain-of-Function
Pathogenic variants that enhance the activity of a particular protein are known as gain-of-function; these types of protein alterations are generally associated with de novo/dominant disorders, though there are rare examples of putative recessive gain-of-function alleles in the epilepsies [15]. These gain-of-function alleles are almost always missense variants, for example, SCN8A variants in the voltage-gated sodium channel can lead to alterations in this sodium channel electrophysiological properties that shift the neuron to a more hyperexcitable state [16]. In theory, any therapeutic that can reduce the protein levels of this aberrant protein, e.g., Nav1.6, would be an effective strategy, ideally by targeting the mutant protein (Fig. 1). Some proof of principle studies have been performed with the DEE genes, KCNT1 and SCN8A (Table 1) [17, 18]. The Scn8a p.Arg1872Trp mouse model recapitulates many of the clinical features of individuals with DEE, including spontaneous seizures and premature lethality [19]. An Scn8a-targeting ASO that reduced both mutant and wild-type levels of the sodium channel Nav1.6 was recently shown to rescue the phenotypes of these mice; providing a proof of principle for this approach may be appropriate [17].
Dominant Negative
True examples of dominant negative mechanisms in epilepsy are rare, though genes such as STXBP1, KCNQ2, KCNQ3, and SMC1A [20–22] can harbor pathogenic missense variants that act in a dominant negative manner. This pathogenic mechanism occurs when some property of the mutant protein interferes with the function of the wild-type protein that is translated from the unaffected allele (Fig. 1). A common example is mutant proteins that can form aggregates in the cell that then sequester the wild-type protein, with the net result that protein levels are less than the 50% that one would expect from having a pathogenic variant on just one of two alleles. Dominant negative mechanisms are also common in proteins that form homodimers, i.e., the assembly of two or more protein monomers that make up a functional unit. Therapeutic approaches in dominant negative conditions are complex, in that an approach that simply increases the amount of protein will be ineffective, as the presence of the mutant protein will continue to interfere with function and, in the instance of aggregates, may even exacerbate the disease mechanism. However, there are some examples from animal models where this model may be effective. A GABRG2 truncating variant has been observed in individuals with febrile sensitive epilepsies, and the mouse model of this variant (p.Q390X) recapitulates many of the clinical features in humans [23]. Interestingly, this variant escapes nonsense-mediated decay and the truncated protein results in aggregates, affects trafficking of the GABA subunits, and disrupts formation of the GABA receptor, consistent with a gain-of-function phenotype. However, overexpression of the wild-type subunit was sufficient to rescue these phenotypes [23]. In this instance, an ASO that increased the levels of protein would theoretically be an effective strategy in individuals with similar GABRG2 pathogenic variants.
Thus, overall, the most effective strategy will likely need to be gene- and/or variant-specific; this could include targeting the mutant protein for degradation using allele-specific ASOs. However, if the cell is also sensitive to haploinsufficiency, this approach may need to be combined with another therapy that also increases the amount of wild-type protein. Finally, gene editing of the pathogenic allele with reversion to the wild type would be an effective putative therapy.
Dosage Sensitivity and Copy Number Variants
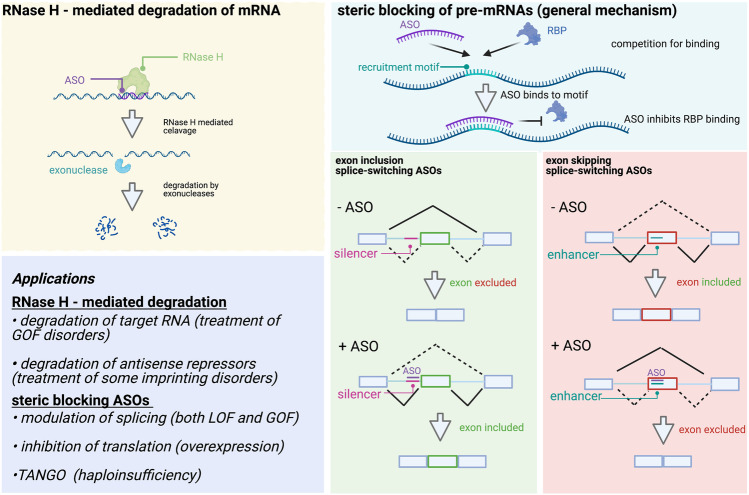
Copy number variants (CNVs) are regions of the genome > 1 kb that are either deleted or duplicated in the genome; they can be polymorphic in the population and associated with a variety of genetic conditions. Pathogenic CNVs account for an appreciable proportion of the genetic epilepsies, including up to 8% of DEEs [24], 10% of GGEs with intellectual disabilities [25, 26], and 1.5–3% of more common epilepsies [27]. These CNVs can either occur recurrently, where the size and position of the CNVs are the same (e.g., 15q13.3del, 16p13.11del) and non-recurrently [28, 29]. Precision therapies for CNVs are likely to be dependent on two primary factors: (1) whether the CNV is either a deletion or duplication, (2) the number of genes encompassed by the CNV and whether it is known which one or more of the genes primarily drive the clinical features. Whole or partial single-gene deletions generally cause haploinsufficiency and can be treated with steric blocking strategies as described in the ASO section (Fig. 2A), and in theory, reciprocal duplications, which lead to increased gene expression, may be treated with any RNase H approach that reduces the abundance of that protein (Fig. 2B). Gene dosage changes as a result of CNVs disrupting multiple genes will be more challenging and will require the identification of the “causative” genes driving the phenotype. In instances where a known epilepsy gene is encompassed, this gene is a logical target for modulations. However, for many CNVs, particularly recurrent microdeletions, one or more genes may be responsible for the phenotype, and will need to be targeted; there has been some recent progress in cellular and animal models to tease out the impact of gene dosage in multigene CNVs [30].
Fig. 2.
Mechanisms of gene regulation by ASOs. Left: RNase H–mediated regulation occurs by ASO binding and RNase H recruitment which cleaves the RNA component of DNA:RNA duplexes. The products of this cleavage are then further degraded by exonucleases, leading to reduced RNA and protein levels. Top right: General mechanism of steric blocking ASOs. ASOs compete for regulatory motifs that serve as RNA-binding protein (RBP) recruitment sites. ASO binding inhibits binding of regulatory RBPs by masking the recognition sequence. Bottom right: Examples of mechanisms by which ASOs can promote exon inclusion by masking silencer regions (left), or promote exon skipping by masking splicing enhancer regions (right). Bottom left: Applications of RNase H and steric blocking ASOs
Imprinting
Angelman syndrome, and more rarely Prader-Willi syndrome, is the only imprinting genetic conditions identified to date that are associated with seizures. There is a subset of genes in the genome that are subject to DNA methylation and thus gene expression silencing, depending on the parent from which that gene is inherited from; these genes are said to be “imprinted.” For instance, the UBE3A gene is expressed only from the maternally inherited allele, and the paternal allele is methylated in a complex process that involved expression of a long non-coding RNA (lncRNA) called UBE3A-AS (antisense) which ultimately prevents UBE3A gene expression. Thus, if UBE3A pathogenic variants, either deletions, truncations, or missense variants, are inherited from the mother, the individual has essentially no functional UBE3A and will manifest with Angelman syndrome, which can include a variety of seizure types [31]. Precision therapies are likely to target the process of gene repression from the paternal allele, leading to its expression of the paternal allele and thus physiological UBE3A expression.
Considerations for X-Linked Disorders
Historically, disorders associated with genes on the X-chromosome have been referred to as dominant and recessive, based on whether the condition manifests in both sexes or males only, respectively. However, the usefulness of this categorization is disputed as pathogenic variants in X-linked genes are now known to be associated with a variety of clinical features in both sexes. Rather, here we highlight some of the more common X-linked epilepsy disorders and appropriate therapeutic approaches.
MECP2: Pathogenic loss-of-function variants in this gene cause Rett syndrome in females, a progressive neurodevelopmental disorder characterized by a period of normal development, followed by regression, seizures, and stereotypies. Males with loss-of-function MECP2 variants generally are less frequently observed, but are generally identified in individuals with severe DEE and early lethality [32]. Any therapeutic approach that increases MECP2 to physiological levels is likely to be effective. However, the amount of MECP2 protein that is produced will need to be tightly regulated as too much MECP2 is also pathogenic, evidenced by the severe ID and seizures that are evident in males with MECP2 duplication syndrome [33]. A similar sex-specific effect is observed for loss-of-function CDKL5 variants associated with DEE in females and a more severe phenotype in males [34].
PCDH19: Pathogenic variants in this gene exclusively affect females, while carrier males are unaffected. Females have an epilepsy characterized by focal seizures that tend to occur in clusters and are often precipitated by fever, as well as intellectual disability. This unusual inheritance pattern is owing to the random X-inactivation in females as a means of dosage compensation for genes on the X-chromosome. As a result, in females, some cells express wild-type PCDH19, while others either no PCDH19 (truncations) or mutant PCDH19 (missense); this is thought to cause cellular interference and ultimately seizures. In carrier males (truncations and pathogenic missense variants), all cells express the mutant allele, and thus, there is no cellular interference and males are unaffected [35]. Treatment options in females are likely to be complex but may theoretically involve silencing of the wild-type PCDH19 allele, or more challenging, reactivation of the silent PCDH19 on the inactive X in individuals with truncations.
Considerations for Gene-Level vs Variant-Level Therapeutic Approaches
While the field has made impressive progress of the last decade in understanding how pathogenic variants in epilepsy-associated genes affect protein function, we are also beginning to appreciate the complexities. For instance, genes can harbor pathogenic variants that can cause both gain- and loss-of-function, depending on the type and/or position of the variant in the protein, and both scenarios are associated with neurological conditions. The genes KCNQ2, SCN2A, and SCN8A, three of the genes most commonly implicated in genetic epilepsies, are good examples of this phenomenon.
KCNQ2: Loss-of-function pathogenic variants in this gene cause early-onset (first days of life) seizures that generally resolve by the first year of life. This condition is called benign familial neonatal epilepsy, though the use of the word “benign” is debated as 10–15% of individuals develop epilepsy later in life and can exhibit learning difficulties. Conversely, pathogenic gain-of-function missense variants are associated with seizure onset in the first weeks/months of life that do not resolve but rather evolve to a DEE [36]. However, as genetic testing has become more widespread, the clinical features associated with KCNQ2 pathogenic variants is evolving and can involve gain-of-function missense variants in individuals with neonatal encephalopathy without neonatal seizures, but can include non-epileptic myoclonus, hypomyelination, and infantile spasms [37, 38].
SCN2A: Pathogenic variants in this gene also exhibit a strong genotype–phenotype correlation. Gain-of-function missense variants are associated with infantile-onset seizures that can either evolve to DEE or resolve as in the case of benign infantile seizures. Meanwhile, pathogenic loss-of-function variants are one of the most common causes of autism spectrum disorder, and a small group of individuals also have childhood-onset epilepsy [39].
SCN8A: Gain-of-function pathogenic missense variants in this gene are associated with DEE, while loss-of-function variants are associated with intellectual disability and movement disorders [16].
It is highly likely that many epilepsy-associated genes will exhibit this scenario, and it is of the utmost importance that precision therapies are designed such that protein levels can be fine-tuned, as both too much and too little protein can be associated with poor clinical outcomes, including seizures. Similarly, we do not currently have a good grasp of just how much change to protein levels nor the number of neurons or cells that need to have physiological protein levels is required to rescue a phenotype in humans. These two considerations are likely to be one of the biggest challenges facing precision therapies in the future.
Therapeutic Approaches
Here we discuss each of the major therapeutic approaches that are being explored to correct the various pathogenic mechanisms underlying monogenic epilepsies including gene replacement and editing, ASOs, and protein replacement. For each therapeutic approach, a basic description of the technology and mechanisms, as well a discussion of advantages and disadvantages of the technology, is provided as well as an exploration of which pathogenic mechanism might be addressed by a particular therapeutic approach (Table 2).
Table 2.
Advantages and disadvantages of various therapeutic approaches
| Therapeutic approach | Gene replacement | Gene editing | ASOs | Protein replacement |
|---|---|---|---|---|
| Variant-specific | ✓ | ✓ | ||
| Gene-specific | ✓ | ✓ | ✓ | ✓ |
| Advantages | ||||
| One-time dose | ✓ | ✓ | ||
| Effective for truncated/deleted genes | ✓ | ✓ | ||
| Uses natural cell regulation | ✓ | ✓ | ||
| Disadvantages | ||||
| Recurrent dosing | ✓ | ✓ | ||
| Artificial regulation | ✓ | ✓ | ✓ | |
| Vector-based limitations | ✓ | ✓ | ||
| Off-target effects | ✓ | ✓ | ||
| High manufacturing cost | ✓ | ✓ | ✓ | |
Gene Replacement
Gene replacement is the process of inserting an extra copy of a defective gene into a cell through a viral vector to allow generation of a working protein [40]. Gene replacement has been successfully employed to significantly alter the course of numerous systemic and organ-specific monogenic conditions including the neurologic disorders, spinal muscular atrophy [41], and aromatic l-amino acid decarboxylase deficiency (AADC) [42]. There are four critical components of a successful gene replacement: the vector, the promotor, the gene, and the route of administration.
Viral vectors are required to transfer the gene of interest to the cells of interest. The viral vector, therefore, determines many critical properties of any gene transfer treatment. Most viral vectors being used clinically are non-integrating (DNA delivered to the cells are not integrated into the host genome), thus reducing the risk of adverse safety events such as cancer. The longevity of non-integrating gene replacement is suggested to be years based on many preclinical models [43], but exactly how long an effect will last in humans remains an open question [44] The vector also determines the cell-specific tropism (which cell types are targeted by the viral vector) and efficiency of gene transfer, issues that are particularly critical for neurologic diseases that may require transduction to specific neuronal or glial cell types. For some genes such as channelopathies, the number of cells that must be transfected could be relatively high [45]. Additionally, the vector should not be cytotoxic and in the case of neurologic diseases must be able to infect non-replicating cells. Finally, the viral vector will ideally have low immunogenicity to improve the overall safety of a potential gene replacement. Adeno-associated virus (AAV) is a commonly utilized class of viral vectors for neurologic diseases due to favorable qualities in each of these domains. The vector also determines the size of the genetic payload that can be delivered, generally less than 5 kb for AAV vectors [46]. In addition to viral vectors, non-viral, nanobiotechnology gene transfer technologies are being developed but have not yet been established as viable alternatives [47].
Promoters, attached to transduced gene, ensure that the gene is expressed in the cell of interest. In some cases, gene-specific promoters are used to allow for more natural endogenous regulation of the transduced gene; this can be very important in genes known to have tightly regulated expression such as MECP2 with both deficiency and overexpression syndromes [48]. In other cases, the approach is to utilize pan-neuronal promotors [49]. The promoter may also be important to ensure that the wrong cells do not express the transduced cell which may be toxic. The importance of this is influenced by the gene of interest, vector tropism, and route of administration [48]. Promotor choices are also influenced by the size of the gene of interest and vector choice as it influences the overall size of the payload [49, 50]. While promoters are required to get gene expression, they can be combined with inhibitors to more precisely regulate gene expression.
The gene of interest is perhaps the most straightforward part of development of a gene replacement; however, when the gene of interest is too large for most vectors, this becomes more complicated. Duchenne muscular dystrophy (DMD) gene replacement has focused extensively on this problem given the 2.6 Mb size of the dystrophin gene [51]. Most efforts have focused on symptom improvement with transduction of a portion of the gene, a micro-dystrophin gene replacement [52, 53]. While a micro-dystrophin replacement may not function as well as the full dystrophin gene, there could be significant amelioration of symptoms for some DMD patients [51]. Other large genes may not be as amenable to microgene development which would significantly limit the likelihood of successful gene replacement with current vector technology.
The route of administration is of particular importance for neurologic disorders. For most neurologic disorders, the goal is to transfer the replacement gene into as many cells in the central nervous system (CNS) as possible. In the case of epilepsy, the goal is likely to specifically include the majority of cortical cells (though some disorders may require cell-type-specific transduction). Direct CNS injection via lumbar puncture, intracerebroventricular, or intraparenchymal routes will often achieve higher rates of gene transfer by avoiding the blood–brain barrier. Additionally, higher doses of gene therapies seem to be better tolerated with injection directly into the CNS presumably by reducing the systemic exposure to both the viral vector and gene that may be toxic outside the CNS. Intraparenchymal injections can successfully transduce high percentages of cells in a relatively small area directly around the injection [54]. However, there is very limited diffusion outside that local area. This approach is likely to be most successful for disorders where the target cells are highly localized as seen in AADC deficiency [55, 56]. Clinical trials using intraparenchymal injections of the DDC gene into the bilateral putamina have been successful [42].
Advantages and Disadvantages of Gene Replacement
Gene replacement has several advantages over other therapeutic strategies. Gene replacement does not require an existing copy of the gene to be effective. Because gene replacement is providing a new copy of the gene of interest rather than manipulating the gene, the specific variant is less relevant and gene replacement could be effective for full gene deletions, gene truncating variants, or missense variants resulting in loss of function. In other words, as long as the pathogenic mechanism is null or haploinsufficiency, gene therapy could be effective. This makes gene replacement approaches potentially relevant to hundreds of monogenic causes of epilepsy and neurodevelopmental disorders. Additionally, as our understanding and technological advancements in vector and promotors improves, gene transfer will be increasingly applicable to a diverse set of genetic etiologies. Furthermore, gene therapy is attractive as it typically only requires a single treatment for long-lasting replacement of the gene of interest.
The disadvantages of gene therapy mostly stem from limitations of the vectors and promotors. Current vectors while promising still have limitations in terms of capacity, tropism, and transfer efficiency and immunogenicity. These limitations have reduced the impact of gene therapy in epilepsies compared to other conditions like SMA and AADC deficiency. Many genetic epilepsies have narrow gene dosing ranges and are associated with both loss- and gain-of-function phenotypes. For this reason, the goal of gene transfer in genetic epilepsies will often be widespread transfer of the gene of interest to most cortical neurons with a single new copy of the gene. Achieving sufficient cortical distribution safely with a single copy has proved challenging. Finally, gene replacement in many cases does not rely on the naturally occurring regulator mechanism of the gene being replaced. This is in part due to the space available to attach promotor and inhibitors to the gene being replaced. However, this can create challenges for dose-sensitive genes such as ion channel genes. On the other hand, this could be an advantage for many recessive conditions where a small dose increase may produce a large functional impact. Finally, it is worth noting that all vector-based therapies have a potential disadvantage of patients not being eligible for treatment if they have high baseline titers due to prior exposure to that family of viral vectors. Undoubtedly, many of these gene therapy limitations will be overcome in time with ongoing advancements in vector technology and improved understanding of the regulatory mechanisms of epilepsy-related genes.
Gene Editing
Gene or genome editing, also called as gene/genome engineering, is the process of adding, replacing, deleting, or otherwise altering the DNA sequence at a particular position in the human genome. In general, in this review, we concentrate on two different kinds of gene edits:
Correction of a single DNA nucleotide (the pathogenic variant) back to the reference allele (i.e., the wild-type or “normal” allele). This approach has been used for treatment of some disorders such as sickle cell disease, a form of beta thalassemia, and Leber congenital amaurosis, a form of retinal dystrophy [57, 58]. This is the more likely long-term approach that will be pursued in precision therapies as it is applicable to both loss- and gain-of-function mechanisms, as well as dominant negative.
Disruption: Creating a new mutation in the DNA such that the coding sequence of a particular gene is disrupted, effectively creating a truncating variant. In general, this approach is likely to only be used in cellular and animal models to study the effects of loss-of-function variants, and is not really being pursued as an effective therapeutic strategy as there are few instances where one would want to permanently delete a copy of a gene, except perhaps in the instance of dominant negative or gain-of-function alleles, but then specific targeting of the mutant allele is challenging
There are a number of different ways to perform gene editing; here we concentrate on the three most common methods:
Clustered regularly interspaced short palindromic repeats (CRISPR-Cas systems): The most popular of these systems is the CRISPR-Cas9 system that has revolutionized molecular biology and the biomedical sciences [59, 60]. CRISPR-Cas9 can induce double-stranded DNA breaks that are repaired naturally by the cell using non-homologous end-joining (NHEJ); this process often leads to insertions/deletions that result in a truncation and can be used to study gene disruption. Of greater relevance in therapeutics is repair of double-stranded breaks using homologous-directed repair (HDR) which leads to incorporation of a specific DNA sequence using a donor strand; this can be used for correction of a pathogenic variant. There are many different Cas systems that can recognize a variety of DNA sequences and induce DNA cleavage at those sites [61].
Prime editing does not rely on double-stranded breaks; rather, this technique makes use of a Cas protein that nicks the DNA on one strand, then uses an engineered reverse transcriptase and a guide RNA to introduce the desired edit. These edits can encompass both single nucleotide corrections and insertions/deletions less than 60 bp [62].
Base editing, likewise, does not rely on double-stranded breaks; rather, this system makes use of a Cas that does not cut DNA (dead-Cas) but targets the complex which includes a DNA-modifying enzyme (the base editor) to the target DNA sequence. Base editors can either modify cytosine or adenine residues and thus induce all DNA transitions, i.e., C > T, A > G, T > C, and G > A [63–65]. Other base editors are also under development to edit transversions (C > G, G > C).
The majority of gene editing systems are likely to be delivered using the same viral vectors described for gene replacement strategies. One of the unique challenges is the size of these gene editing systems, which, together with Cas-targeting, guide RNAs and repair/modify enzymes which are in the range of 6–8 kb, which are at or above most viral vector or other delivery systems. Thus, efforts are ongoing to slim down the size of these systems to carry only the most essential components and increase AAV efficiency. Moreover, a number of groups have used split approaches, where the different gene editing components are loaded into two separate AAVs for dual delivery, though the limitation of this approach is its reliance on dual delivery to the same cell [66].
There are very few examples of clinical use of gene editing in humans to date, though there are thousands of labs developing and implementing these systems in cellular and animal models. The first successful use of gene editing in humans was ex vivo in sickle cell anemia and in vivo for retinal dystrophy [57, 58]. In vivo CRISPR-based gene editing for hemophilia has also been achieved in mice [67] and pigs [68]. Base editing was also recently used to correct a dominant negative LMNA variant in mice; this pathogenic variant is causative of progeria, a monogenic disorder associated with accelerated aging and early death [69].
While more technically challenging and still in its infancy compared to ASOs, gene editing offers significant advantages, not least because this would be, in theory, a long-term and permanent therapeutic. Moreover, there would be no introduction of foreign DNA, and ideally, the only change to the individual’s DNA would be the correction of that specific pathogenic variant. However, currently, major limitations, not least concerns of off-target effects. Moreover, as with gene replacement approaches, delivery is still a challenge, as well as how many cells and of which type need to be targeted and “treated” to ameliorate symptoms. In the long term, it is likely that base and prime editors, and their next-generation iterations, are more likely to be used in therapeutics given the elimination of the concern of inducing double-stranded breaks with traditional CRISPR-Cas technologies. Finally, there are serious ethical considerations surrounding gene editing and the ability to permanently alter the DNA of an individual. As such, many oversight committees have been set up in the USA and across the world to develop guidelines on the use of these technologies, including a call for a moratorium on heritable (oocyte/sperm) genome editing [70].
ASOs
ASOs have recently been used in the treatment of genetic diseases such as spinal muscular atrophy [71, 72] and Duchenne muscular dystrophy [73]. ASOs are short synthetic oligonucleotides (usually 15–30 nucleotides) that intervene at the RNA level by base pairing with targets of interest. Since ASOs bind through complementarity with their target RNAs, they facilitate rapid, rational, personalized drug design, and have even been used to treat incredibly rare (n-of-1) diseases for which no suitable treatments exist.
ASOs function through two major mechanisms: (1) ASOs can downregulate target mRNA transcripts through recruitment of RNase H [74], which degrades the RNA target, and (2) ASOs can modulate mRNA processing by masking sequences that are recognized by RNA-binding proteins (RBPs) or RNA-RNA interactions [75]. Through these two modes of action, ASOs can either upregulate, downregulate, or alter the isoform of protein products in genes of interest. Here we consider the different mechanisms by which ASOs can regulate their targets, providing examples for how each strategy has been used in the development of therapies for patients with epilepsy (and other diseases).
Upregulation/Alteration of Gene Expression for Treatment of Loss of Function Disorders (Haploinsufficiency, Recessive, and Imprinting Disorders)
ASOs can upregulate target protein levels through a variety of mechanisms. ASOs have been shown to upregulate protein products of target transcripts by inhibiting translation of upstream open reading frames [76], by targeting translation inhibitory elements in 5′ UTRs [77], by increasing the levels of productive splicing [78], and by downregulation of silencing antisense long non-coding RNAs [79]. The latter two of these mechanisms have been used in developing ASOs for epileptic conditions and will be our primary focus.
Inhibition of Regulators
ASOs can achieve upregulation of genes of interest through RNAse H–mediated cleavage of repressive regulators of target genes. For example, this strategy has been used to develop therapeutics for Angelman syndrome. As mentioned above (see the “Imprinting” section), an ASO screen targeting UBE3A-AS was performed in mice and identified an ASO capable of downregulating the antisense silencer and restoring UBE3A levels [79]. Moreover, this ASO partially rescued cognitive defects associated with the disease in mouse models. Based on this success in preclinical models, several ASOs have moved to early phase clinical trials.
Splice-Switching ASOs
Splice-altering ASOs can be thought of as an upregulation mechanism since they lead to increased functional protein targets by promoting exon inclusion or exclusion. ASOs alter splicing of transcripts by targeting regions at or near splice sites on pre-mRNAs. Splicing is a complicated process regulated by the spliceosome, 5′ and 3′ splice sites, and cis-elements that can either enhance or limit exon inclusion [80]. Cis-elements are 4–8 nucleotide motifs that serve as binding sites for various RNA-binding proteins such as SR proteins and hnRNP proteins, and can either act as enhancers, which facilitate exon recognition, or silencers, which inhibit exon recognition by the spliceosome machinery. ASOs can directly mask splice sites or mask the enhancer and silencer regions by competing with the RBPs that normally bind these regions [75]. Thus, ASOs can both limit exon inclusion, by targeting enhancer regions, or promote inclusion, by targeting silencer regions.
Given the complexities associated with splicing, it is difficult to computationally predict the outcome of a single ASO event on splicing. Therefore, unbiased oligo walks have been utilized to simplify the design process by synthesizing and screening several ASOs that target many different sequences within the vicinity of a splice site [81].
Clinical Use
Eteplirsen and nusinersen are two splice-switching ASOs that highlight the power of ASOs in treatment of disease. These ASOs have been used in the treatment of DMD and SMA, respectively. Eteplirsen binds to exon 51 in dystrophin and promotes skipping of this exon, thereby correcting the reading frame in patients with a variety of frameshift (truncating) variants. Nusinersen, on the other hand, compensates for loss of SMN1 by promoting inclusion of exon 7 in SMN2, a homolog of SMN1. Exon 7 is normally skipped in mature SMN2 transcripts, and is unable to compensate for loss of SMN1. By promoting exon 7 inclusion, nusinersen can help SMN2 compensate for loss of SMN1.
TANGO
Targeted Augmentation of Nuclear Gene Output (TANGO) was recently developed to increase protein expression of target genes [78]. Aberrant RNAs that are the product of unproductive splicing events are rapidly degraded via nonsense-mediated decay (NMD). The prevalence of unproductive splicing events has been largely underappreciated due to low steady-state levels of these transcripts. Recent work has been able to uncover many of these “hidden” unproductive splicing events by stabilizing NMD targets by treating cells with cycloheximide, which inhibits translation and NMD. From this experimental approach, unproductive splicing events were found to occur in a substantial number of protein coding genes (> 7500 genes). Moreover, over 1200 of these genes were disease-associated.
In order to increase productive splicing, ASO walks were performed to identify ASOs that increased productive splicing events and functional protein levels. Indeed, using TANGO, researchers were able to increase protein levels by anywhere from 1.5- to 2.6-fold.
Clinical Use
Recently, researchers used TANGO to explore the efficacy of ASO therapeutics in Dravet syndrome caused by SCN1A haploinsufficiency [14]. An ASO screen identified ASOs that increase SCN1A levels by increasing productive splicing events in mice. Moreover, the ASO decreased the occurrence of seizures in these mice as well as sudden unexpected death in epilepsy (SUDEP). This study provides proof of principle in using ASOs to shift unproductive splicing events (the levels of which have been underappreciated), towards productive splicing events and correcting disease phenotypes. Given the prevalence of haploinsufficiency in the genetic epilepsies and the large number of genes that undergo unproductive splicing events, this strategy could pave the way for many new ASO drugs.
Downregulation of Gene Expression to Treat Gain-of-Function Disorders
Target downregulation is perhaps the most conceptually straightforward ASO mechanism. ASOs can downregulate target protein levels by selectively degrading target RNAs through RNase H–mediated cleavage or by functioning as a steric block to translation by binding near the start codon [82]. RNase H–mediated degradation is achieved through DNA-RNA duplex formation and selective cleavage of the phosphodiester backbone of the duplexed RNA [83]. RNase H shows both nuclear and cytoplasmic localization and can effectively target pre-mRNAs in the nucleus, as well as mature RNAs in the cytoplasm [84]. RNAse H is largely indiscriminate towards the exact sequence of DNA-RNA duplexes (so long as they are complementary) and specificity is conferred by the ASO, making this system modular and widely applicable to a number of diseases in which target downregulation is desired.
Clinical Use
RNase H–mediated cleavage is particularly well suited for targeting gain-of-function pathogenic variants in which target downregulation could rescue disease phenotypes. Indeed, recent work has shown that an ASO targeting SCN2A is capable of downregulating SCN2A and rescuing disease phenotype by reducing seizures and expanding life expectancy in a mouse model of SCN2A pathology [85]. Similarly, in the context of another DEE, an ASO targeting SCN8A reduced levels of Nav1.6 and ameliorated disease phenotype in a mouse model [17].
Advantages of ASO Therapies
Design
ASOs are attractive drug candidates due to their ease of design. Since ASOs rely on complementarity with the RNAs that they target, designing an ASO with high degrees of substrate specificity is much simpler than the rationale design of other small molecule drugs. The design and testing of ASOs has generally involved performing ASO “walks” in which several ASOs are designed to tile a region of interest.
Versatility
The general mechanisms of ASO treatment are versatile and can be applied to a wide range of diseases, as highlighted in the previous sections. GOF mutations can be treated through RNAse H–mediated cleavage. Haploinsufficiency can be addressed by promoting productive splicing. LOF mutations have also been treated by modulating splicing. The versatility of ASOs in treating many different types of diseases makes them attractive.
Cost
Due to their simple design, ASOs are cost-effective in comparison to vector-based approaches or enzyme replacement strategies. However, there remains a burden for patients with extremely rare genetic diseases who receive these treatments. While ASOs provide an ideal platform for treatment, the financial burden that these drugs place on families is a major challenge that requires creative solutions between funding agencies, researchers, companies, and families. Many biotech companies are understandably hesitant to develop n-of-1 therapeutics. However, the value gained in understanding better delivery and design approaches and how these translate from model systems to actual patients would likely provide value far greater than the money lost on developing n-of-1 therapeutics. If the insights gained from n-of-1 therapies can be translated into therapies that affect larger populations, mutually beneficial scenarios might be created in which the cost for the patient is reduced and, in return, drug developers gain valuable data and insights that would translate into better design principles for widely used drugs.
Rare Diseases
ASOs are ideal candidates for extremely rare genetic diseases due to their ease of design, low cost, and versatility. The most extreme example of this is milasen, an n-of-1 treatment used correct a pathologic splicing pattern in an individual with CLN7 Batten disease [86]. While there are still many barriers to widespread use of ASOs designed for a single patient, this case and others provide a path to individualized therapeutics (n-of-1).
Limitations of ASO Therapies
Off-Target Effects
Off-target effects can present challenges when designing ASOs. Counterintuitively, longer ASOs do not necessarily decrease off-target effects of RNAse H, in spite of their increased number of base pairing interactions due to RNAse H only needing 5 base pairs of complementarity to show enzymatic activity [87]. Moreover, other studies have shown that ASOs with imperfect complementarity to off-target transcripts can still downregulate these transcripts [88].
Splice-switching ASOs can be also prone to off-target effects. Recent work has shown a splice-switching ASO targeting PKM1 can produce mis-splicing events in 17 off-target transcripts [89]. Moreover, these mis-splicing events are difficult to predict and the choice of ASO chemistry can influence the extent of mis-splicing. The extent to which splice-switching ASOs modulate splicing of unintended target may present a hurdle in ASO design, and should be carefully considered and tested when designing ASOs.
A final concern is that off-target effects cannot be adequately tested in mouse models. While many mouse transcripts show large amounts of conservation to human transcripts, there are many differences across the transcriptome of mouse vs human. Similarly, since many transcripts show tissue-specific expression, choosing cell lines that accurately reflect the RNA composition of the tissue of interest (in this case brain) will improve the assessment of the off-target effects of ASOs. Therefore, special care should be taken in choosing the right model system when assessing on-target vs. off-target effects.
Delivery
Effective delivery of ASOs to the brain remains a key challenge in making effective ASO therapies. In general, intrathecal injections of ASOs seem to have fairly widespread distribution and uptake throughout the central nervous system in non-human primates and potentially humans [90–92]. What data is available suggest that ASOs may have better distribution than what is often achieved by current gene therapy vectors; however, it is not clear whether this is true across all neuronal and glial cell types and in all brain regions. It is also unclear to what degree this is impacted by the ASO sequence and backbone structure.
Repeat Doses and ASO Stability
Since ASO therapies modulate the expression of mutated genes, rather than correcting the mutation (as in CRISPR-based approaches), repeat treatments are necessary. In the case of nusinersen, 4 loading doses are administered, with the first three doses being administered in 14-day intervals and the fourth dose occurring 30 days after the third dose. Subsequent maintenance dosing is performed at 4-month intervals for the remainder of the patient’s life. Eteplirsen, on the other hand, is administered on a weekly basis.
In order to improve drug stability, numerous backbone modifications, such as phosphorothioate modifications, aim to improve the stability of ASOs, generating drugs that will maintain efficacy over time and limit the number follow-up treatments. A host of stabilizing modifications have been incorporated into ASO designs, making them resistant to cellular nucleases. Since RNAse H–mediated cleavage relies upon DNA-RNA duplex formation, stabilizing modifications cannot be incorporated throughout the entirety of the ASO sequence because they reduce RNAse H activity. Therefore, “gapmer” ASOs are widely used that contain an internal unmodified DNA region, flanked by regions that contain stabilizing modifications such as LNAs and phosphorothioate modifications. The internal unmodified DNA bases are sufficient for conferring enough DNA-RNA complementarity for RNAse H cleavage, while the flanking-modified bases make the gapmer resistant to cellular nucleases and add additional complementarity for target specificity.
dCas9-Based Alternatives to ASO Therapies
In this review, we have primarily focused on ASO therapeutics; however, the dCas9 and dCas13 proteins have also been leveraged for their regulatory abilities. These enzymes are catalytically inactive and therefore are not used for their gene editing capabilities, but rather as a means of targeting fusion proteins to targets of interest. For example, in the treatment of a mouse model, Dravet syndrome (SCN1A haploinsufficiency), a dCas9-VP160 (transcriptional activator) fusion, was targeted to the SCN1A promoter region via guide RNAs [93]. This experimental approach (1) robustly upregulated SCN1A expression, (2) rescued excitability in cortical interneurons, and (3) protected mice from hypothermia-induced seizures. Taken together, these results highlight the power of the dCas9 system, particularly in cases of haploinsufficiency, to upregulate genes of interest. Indeed, similar to ASOs, this system could be used in many different ways to modulate gene expression and/or RNA processing.
Protein Replacement
The goal of all the therapies discussed is to restore physiologic levels of protein in the relevant tissue and cell types. Protein replacement also known as enzyme replacement therapy (ERT) seeks to achieve this with direct injection of the protein. While there is a relatively strong track record of successful ERTs in lysosomal storage disorders (mucopolysaccharidosis types I, II, IV, and VI, as well as Fabry, Pompe, and Gaucher disease), one of the challenges has been getting the right amount of enzyme to the right tissues [94]. Historically, most ERTs have been administered via intravenous infusions with significant challenges getting the proteins past the blood–brain barrier. However, in 2017, the FDA approved the ERT cerliponase alfa for the treatment of neuronal ceroid lipofuscinosis type 2 (CLN2) which addresses this issue by direct intracerebroventricular administration of the recombinant enzyme tripeptidyl peptidase-1 (TPP1) [86].
Direct administration of the protein of interest bypasses some of the complicated processing and regulatory elements of our biology and may have advantages in some cases. There is also a strong historical knowledge of the manufacturing, delivery, and safety profile of ERTs that can be utilized. Additionally, similar to gene replacement, ERTs can be effective for recessive conditions even with full gene deletion. A challenge of ERTs is precision of dosing to particular tissues or cell types. While ERTs have been very successful for recessive degenerative conditions, there is often a reasonably large physiologic window in these conditions, and in some cases, a modest increase in protein levels may have a substantial impact on cell survival. While ERT has not been utilized for haploinsufficiency disorders to date, it is theoretically possible. However, many of these conditions have more tightly regulated physiologic expression and proper cell function would require more precise ERT dosing. Protein replacement has been explored in the preclinical space for conditions such as CDKL5 deficiency disorder, a disorder where some precision of dosing would likely be required. Future clinical trials of protein replacement in these types of disorders will be highly informative. ERT is unlikely to be a viable solution for dominant negative or gain-of-function mechanisms of disease, though it is possible that in some conditions an improvement could be derived from statistically overwhelming the proportion of protein that is dysfunctional.
Safety Considerations
There is still much that is not known about the safety profiles of these relatively new therapeutic classes. However, the evidence available to date does give us some patterns that are worth noting. As discussed throughout the manuscript, there are potential concerns regarding “off-target” effects of these therapies. Non-integrating gene replacement and enzyme replacement likely have the lowest concern in this regard, but dosing is the larger concern in this case. Off-target effects are a concern for ASOs and must be evaluated during the development phase but this concern is tempered by the non-permanent nature of the therapy. Gene edit has the highest standards in this regard as any off-target effects are likely permanent.
All vector-based technologies have safety considerations related to the vectors themselves and the generation of human immune response. Intravenous infusions of onasemnogene abeparvovec (gene replacement for SMA using AAV9) demonstrated a substantial risk for hepatotoxicity with 90% of patients demonstrating elevated liver enzymes and up to 34% classified as adverse events. Management generally involves prolonged courses of steroids which ranged from 1 to 8 months, resulting in significant morbidity. All patients recovered, and a minority were considered severe adverse reactions (> 20 × upper limit of normal). Steroid treatment is often prophylactic now for IV gene therapy protocols. There does seem to be a correlation between rising AAV titers and transaminitis [95]. It is not clear how variable this will be across different viral vectors and different routes of administration. Intraputaminal gene replacement for AADC deficiency also demonstrates the potential for serious adverse events including pyrexia requiring hospitalization, and transient dyskinesias but had less concern for hepatotoxicity [42]. Much remains to be learned regarding the ideal route of administration to maximize gene transfer to target tissues, minimizing safety concerns.
ASOs also likely have a different side effect profile depending on the route of administration, and we will focus on intrathecal injection as the most likely for epileptic disorders. Preclinical toxicity testing of ASOs via intrathecal administrations has demonstrated hindlimb weakness and loss of reflexes across a variety of animal models (rodent and non-human primate). The potential for this adverse effect does not seem to be ASO sequence-specific but more likely a potential adverse reaction to ASOs more generally with intrathecal administration [96]. Thus far, this has been reported less in human trials [41, 86], but this data remains limited, though there are several trials currently in progress that may be informative. This remains a potential adverse event that should be monitored closely. In general, ASOs seem to be well tolerated with intrathecal administration in humans but this remains a novel class of medications requiring vigilance to any potential side effects.
Conclusions
Over the last 20 years, the genomic revolution has uncovered thousands of single-gene neurogenetic disorders. Our increased understanding of the mechanisms of these diseases, as well as the fundamental principles and mechanics of the central dogma, has led to an explosion of novel strategies with the potential to cure or dramatically improve genetic epilepsies. Each of these strategies has advantages and disadvantages that may be better suited to specific pathogenic mechanisms. While many of these novel therapies remain in their infancy, they clearly have the potential to revolutionize the treatment of rare genetic epilepsies.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
GLC is sponsored by a Dravet Syndrome Foundation Research Grant. SD reports other from Upsher-Smith, other from Biomarin, other from Neurogene, other from Marinus, other from Ovid Therapeutics, grants from NIH, outside the submitted work.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Carvill GL, Heavin SB, Yendle SC, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nature genetics. 2013;45(7):825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epi4K Consortium. De novo mutations in epileptic encephalopathies. Nature, 501(7466), 217–221 (2013). [DOI] [PMC free article] [PubMed]
- 3.McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2015;15(3):304–316. doi: 10.1016/S1474-4422(15)00250-1. [DOI] [PubMed] [Google Scholar]
- 4.Truty R, Patil N, Sankar R, et al. Possible precision medicine implications from genetic testing using combined detection of sequence and intragenic copy number variants in a large cohort with childhood epilepsy. Epilepsia Open. 2019;4(3):397–408. doi: 10.1002/epi4.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helbig KL, Farwell Hagman KD, Shinde DN, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. 2016;18(9):898–905. doi: 10.1038/gim.2015.186. [DOI] [PubMed] [Google Scholar]
- 6.Perucca P, Scheffer IE, Harvey AS, et al. Real-world utility of whole exome sequencing with targeted gene analysis for focal epilepsy. Epilepsy Res. 2017;131:1–8. doi: 10.1016/j.eplepsyres.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Sánchez Fernández I, Loddenkemper T, Gaínza-Lein M, Sheidley BR, Poduri A. Diagnostic yield of genetic tests in epilepsy: A meta-analysis and cost-effectiveness study. Neurology. 2019;92(5):e418–428. doi: 10.1212/WNL.0000000000006850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Gotway G, Pascual JM, Park JY. Diagnostic yield of clinical next-generation sequencing panels for epilepsy. JAMA Neurol. 2014;71(5):650–651. doi: 10.1001/jamaneurol.2014.405. [DOI] [PubMed] [Google Scholar]
- 9.Consortium EK Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol. 2017;16(2):135–143. doi: 10.1016/S1474-4422(16)30359-3. [DOI] [PubMed] [Google Scholar]
- 10.Collaborative E. Ultra-Rare Genetic Variation in the Epilepsies: A Whole-Exome Sequencing Study of 17,606 Individuals. American journal of human genetics. 2019;105(2):267–282. doi: 10.1016/j.ajhg.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ILAE. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nature communications, 9(1), 5269 (2018). [DOI] [PMC free article] [PubMed]
- 12.Papuc SM, Abela L, Steindl K, et al. The role of recessive inheritance in early-onset epileptic encephalopathies: a combined whole-exome sequencing and copy number study. Eur J Hum Genet. 2019;27(3):408–421. doi: 10.1038/s41431-018-0299-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheffer IE, Nabbout R. SCN1A-related phenotypes: Epilepsy and beyond. Epilepsia. 2019;60(Suppl 3):S17–s24. doi: 10.1111/epi.16386. [DOI] [PubMed] [Google Scholar]
- 14.Han Z, Chen C, Christiansen A et al. Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome. Science translational medicine, 12(558) (2020). [DOI] [PubMed]
- 15.Lammertse HCA, van Berkel AA, Iacomino M et al. Homozygous STXBP1 variant causes encephalopathy and gain-of-function in synaptic transmission. Brain, (2019). [DOI] [PMC free article] [PubMed]
- 16.Meisler MH, Helman G, Hammer MF, et al. SCN8A encephalopathy: Research progress and prospects. Epilepsia. 2016;57(7):1027–1035. doi: 10.1111/epi.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenk GM, Jafar-Nejad P, Hill SF, et al. Scn8a Antisense Oligonucleotide Is Protective in Mouse Models of SCN8A Encephalopathy and Dravet Syndrome. Annals of neurology. 2020;87(3):339–346. doi: 10.1002/ana.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burbano LE, Li M, Jancovski N et al. Antisense oligonucleotide therapy for KCNT1 encephalopathy. bioRxiv, 2020.2011.2012.379164 (2020). [DOI] [PMC free article] [PubMed]
- 19.Bunton-Stasyshyn RKA, Wagnon JL, Wengert ER, et al. Prominent role of forebrain excitatory neurons inSCN8Aencephalopathy. Brain. 2019;142(2):362–375. doi: 10.1093/brain/awy324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu B, Mak JCH, Morris AP, et al. Functional analysis of epilepsy-associated variants in STXBP1/Munc18-1 using humanized Caenorhabditis elegans. Epilepsia. 2020;61(4):810–821. doi: 10.1111/epi.16464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu A, Xu X, Yang X, et al. The clinical spectrum of female epilepsy patients with PCDH19 mutations in a Chinese population. Clin Genet. 2016;91(1):54–62. doi: 10.1111/cge.12846. [DOI] [PubMed] [Google Scholar]
- 22.Orhan G, Bock M, Schepers D, et al. Dominant-negative effects of KCNQ2 mutations are associated with epileptic encephalopathy. Annals of neurology. 2014;75(3):382–394. doi: 10.1002/ana.24080. [DOI] [PubMed] [Google Scholar]
- 23.Huang X, Zhou C, Tian M, et al. Overexpressing wild-type γ2 subunits rescued the seizure phenotype in Gabrg2(+/Q390X) Dravet syndrome mice. Epilepsia. 2017;58(8):1451–1461. doi: 10.1111/epi.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mefford HC, Yendle SC, Hsu C, et al. Rare copy number variants are an important cause of epileptic encephalopathies. Annals of neurology. 2011;70(6):974–985. doi: 10.1002/ana.22645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullen SA, Carvill GL, Bellows S, et al. Copy number variants are frequent in genetic generalized epilepsy with intellectual disability. Neurology. 2013;81(17):1507–1514. doi: 10.1212/WNL.0b013e3182a95829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coppola A, Cellini E, Stamberger H, et al. Diagnostic implications of genetic copy number variation in epilepsy plus. Epilepsia. 2019;60(4):689–706. doi: 10.1111/epi.14683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niestroj LM, Perez-Palma E, Howrigan DP, et al. Epilepsy subtype-specific copy number burden observed in a genome-wide study of 17 458 subjects. Brain. 2020;143(7):2106–2118. doi: 10.1093/brain/awaa171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinzen EL, Radtke RA, Urban TJ, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. American journal of human genetics. 2010;86(5):707–718. doi: 10.1016/j.ajhg.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS genetics. 2010;6(5):e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu Y, Arbogast T, Lorenzo SM et al. Oligogenic Effects of 16p11.2 Copy-Number Variation on Craniofacial Development. Cell reports, 28(13), 3320–3328.e3324 (2019). [DOI] [PMC free article] [PubMed]
- 31.Tan WH, Bird LM. Angelman syndrome: Current and emerging therapies in 2016. Am J Med Genet C Semin Med Genet. 2016;172(4):384–401. doi: 10.1002/ajmg.c.31536. [DOI] [PubMed] [Google Scholar]
- 32.Kaur S, Christodoulou J. MECP2 Disorders. In: GeneReviews(®). Adam, MP, Ardinger, HH, Pagon, RA et al. (Eds.) (University of Washington, Seattle Copyright © 1993–2021, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved., Seattle (WA), 1993)
- 33.Van Esch H. MECP2 Duplication Syndrome. In: GeneReviews(®). Adam, MP, Ardinger, HH, Pagon, RA et al. (Eds.) (University of Washington, Seattle Copyright © 1993–2021, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved., Seattle (WA), 1993)
- 34.Olson HE, Demarest ST, Pestana-Knight EM, et al. Cyclin-Dependent Kinase-Like 5 Deficiency Disorder: Clinical Review. Pediatr Neurol. 2019;97:18–25. doi: 10.1016/j.pediatrneurol.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gecz J, Thomas PQ. Disentangling the paradox of the PCDH19 clustering epilepsy, a disorder of cellular mosaics. Curr Opin Genet Dev. 2020;65:169–175. doi: 10.1016/j.gde.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Miceli F, Soldovieri MV, Joshi N, Weckhuysen S, Cooper E, Taglialatela M. KCNQ2-Related Disorders. In: GeneReviews((R)). Adam, MP, Ardinger, HH, Pagon, RA et al. (Eds.) (University of Washington, SeattleUniversity of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved., Seattle (WA), 2018)
- 37.Mulkey SB, Ben-Zeev B, Nicolai J, et al. Neonatal nonepileptic myoclonus is a prominent clinical feature of KCNQ2 gain-of-function variants R201C and R201H. Epilepsia. 2017;58(3):436–445. doi: 10.1111/epi.13676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Millichap JJ, Miceli F, De Maria M, et al. Infantile spasms and encephalopathy without preceding neonatal seizures caused by KCNQ2 R198Q, a gain-of-function variant. Epilepsia. 2017;58(1):e10–e15. doi: 10.1111/epi.13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders SJ, Campbell AJ, Cottrell JR, et al. Progress in Understanding and Treating SCN2A-Mediated Disorders. Trends Neurosci. 2018;41(7):442–456. doi: 10.1016/j.tins.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kay MA, Glorioso JC, Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med. 2001;7(1):33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 41.Waldrop MA, Karingada C, Storey MA et al. Gene Therapy for Spinal Muscular Atrophy: Safety and Early Outcomes. Pediatrics, 146(3) (2020). [DOI] [PubMed]
- 42.Chien YH, Lee NC, Tseng SH, et al. Efficacy and safety of AAV2 gene therapy in children with aromatic L-amino acid decarboxylase deficiency: an open-label, phase 1/2 trial. Lancet Child Adolesc Health. 2017;1(4):265–273. doi: 10.1016/S2352-4642(17)30125-6. [DOI] [PubMed] [Google Scholar]
- 43.Katz ML, Tecedor L, Chen Y, et al. AAV gene transfer delays disease onset in a TPP1-deficient canine model of the late infantile form of Batten disease. Science translational medicine. 2015;7(313):313ra180. doi: 10.1126/scitranslmed.aac6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Athanasopoulos T, Munye MM, Yáñez-Muñoz RJ. Nonintegrating Gene Therapy Vectors. Hematol Oncol Clin North Am. 2017;31(5):753–770. doi: 10.1016/j.hoc.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Yu W, Smolen CE, Hill SF, Meisler MH. Spontaneous seizures and elevated seizure susceptibility in response to somatic mutation of sodium channel Scn8a in the mouse. Hum Mol Genet, (2021). [DOI] [PMC free article] [PubMed]
- 46.Lynam L. Case management and critical pathways: friend or foe? Neonatal Netw, 13(8), 48–49, 51 (1994). [PubMed]
- 47.Liu L, Yang J, Men K, et al. Current Status of Nonviral Vectors for Gene Therapy in China. Hum Gene Ther. 2018;29(2):110–120. doi: 10.1089/hum.2017.226. [DOI] [PubMed] [Google Scholar]
- 48.Gadalla KKE, Vudhironarit T, Hector RD, et al. Development of a Novel AAV Gene Therapy Cassette with Improved Safety Features and Efficacy in a Mouse Model of Rett Syndrome. Mol Ther Methods Clin Dev. 2017;5:180–190. doi: 10.1016/j.omtm.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maturana CJ, Verpeut JL, Pisano TJ, et al. Small Alphaherpesvirus Latency-Associated Promoters Drive Efficient and Long-Term Transgene Expression in the CNS. Mol Ther Methods Clin Dev. 2020;17:843–857. doi: 10.1016/j.omtm.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray SJ, Foti SB, Schwartz JW, et al. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum Gene Ther. 2011;22(9):1143–1153. doi: 10.1089/hum.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duan D, Systemic AAV. Micro-dystrophin Gene Therapy for Duchenne Muscular Dystrophy. Mol Ther. 2018;26(10):2337–2356. doi: 10.1016/j.ymthe.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chamberlain JR, Chamberlain JS. Progress toward Gene Therapy for Duchenne Muscular Dystrophy. Mol Ther. 2017;25(5):1125–1131. doi: 10.1016/j.ymthe.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howard ZM, Dorn LE, Lowe J et al. Micro-dystrophin gene therapy prevents heart failure in an improved Duchenne muscular dystrophy cardiomyopathy mouse model. JCI Insight, (2021). [DOI] [PMC free article] [PubMed]
- 54.Richardson RM, Bankiewicz KS, Christine CW, et al. Data-driven evolution of neurosurgical gene therapy delivery in Parkinson’s disease. J Neurol Neurosurg Psychiatry. 2020;91(11):1210–1218. doi: 10.1136/jnnp-2020-322904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rosenberg JB, Chen A, De BP et al. Safety of Direct Intraparenchymal AAVrh.10-mediated CNS Gene Therapy for Metachromatic Leukodystrophy. Hum Gene Ther, (2020). [DOI] [PMC free article] [PubMed]
- 56.Sondhi D, Kaminsky SM, Hackett NR et al. Slowing late infantile Batten disease by direct brain parenchymal administration of a rh.10 adeno-associated virus expressing CLN2. Science translational medicine, 12(572) (2020). [DOI] [PMC free article] [PubMed]
- 57.Frangoul H, Altshuler D, Cappellini MD, et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N Engl J Med. 2021;384(3):252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 58.Maeder ML, Stefanidakis M, Wilson CJ, et al. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med. 2019;25(2):229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 59.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490–507. doi: 10.1038/s41580-019-0131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kurt IC, Zhou R, Iyer S, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nature biotechnology. 2021;39(1):41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakata RC, Ishiguro S, Mori H, et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nature biotechnology. 2020;38(7):865–869. doi: 10.1038/s41587-020-0509-0. [DOI] [PubMed] [Google Scholar]
- 65.Zhang X, Zhu B, Chen L, et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nature biotechnology. 2020;38(7):856–860. doi: 10.1038/s41587-020-0527-y. [DOI] [PubMed] [Google Scholar]
- 66.Ryu SM, Koo T, Kim K, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nature biotechnology. 2018;36(6):536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 67.Chen H, Shi M, Gilam A, et al. Hemophilia A ameliorated in mice by CRISPR-based in vivo genome editing of human Factor VIII. Scientific reports. 2019;9(1):16838. doi: 10.1038/s41598-019-53198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J, An B, Yu B, et al. CRISPR/Cas9-mediated knockin of human factor IX into swine factor IX locus effectively alleviates bleeding in hemophilia B pigs. Haematologica. 2021;106(3):829–837. doi: 10.3324/haematol.2019.224063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koblan LW, Erdos MR, Wilson C, et al. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature. 2021;589(7843):608–614. doi: 10.1038/s41586-020-03086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lander ES, Baylis F, Zhang F, et al. Adopt a moratorium on heritable genome editing. Nature. 2019;567(7747):165–168. doi: 10.1038/d41586-019-00726-5. [DOI] [PubMed] [Google Scholar]
- 71.Neil EE, Bisaccia EK. Nusinersen: A Novel Antisense Oligonucleotide for the Treatment of Spinal Muscular Atrophy. J Pediatr Pharmacol Ther. 2019;24(3):194–203. doi: 10.5863/1551-6776-24.3.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rigo F, Hua Y, Krainer AR, Bennett CF. Antisense-based therapy for the treatment of spinal muscular atrophy. J Cell Biol. 2012;199(1):21–25. doi: 10.1083/jcb.201207087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aartsma-Rus A, Krieg AM. FDA Approves Eteplirsen for Duchenne Muscular Dystrophy: The Next Chapter in the Eteplirsen Saga. Nucleic acid therapeutics. 2017;27(1):1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walder RY, Walder JA. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1988;85(14):5011–5015. doi: 10.1073/pnas.85.14.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Havens MA, Hastings ML. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic acids research. 2016;44(14):6549–6563. doi: 10.1093/nar/gkw533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang XH, Shen W, Sun H, Migawa MT, Vickers TA, Crooke ST. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nature biotechnology. 2016;34(8):875–880. doi: 10.1038/nbt.3589. [DOI] [PubMed] [Google Scholar]
- 77.Liang XH, Sun H, Shen W, et al. Antisense oligonucleotides targeting translation inhibitory elements in 5′ UTRs can selectively increase protein levels. Nucleic acids research. 2017;45(16):9528–9546. doi: 10.1093/nar/gkx632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lim KH, Han Z, Jeon HY, et al. Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nature communications. 2020;11(1):3501. doi: 10.1038/s41467-020-17093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518(7539):409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. Rna. 2008;14(5):802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5(4):e73. doi: 10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuespert S, Heydn R, Peters S et al. Antisense Oligonucleotide in LNA-Gapmer Design Targeting TGFBR2-A Key Single Gene Target for Safe and Effective Inhibition of TGFβ Signaling. Int J Mol Sci, 21(6) (2020). [DOI] [PMC free article] [PubMed]
- 83.Stein H, Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science. 1969;166(3903):393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- 84.Lai F, Damle SS, Ling KK, Rigo F. Directed RNase H Cleavage of Nascent Transcripts Causes Transcription Termination. Mol Cell. 2020;77(5):1032–1043.e1034. doi: 10.1016/j.molcel.2019.12.029. [DOI] [PubMed] [Google Scholar]
- 85.Li M, Jancovski N, Jafar-Nejad P et al. Antisense oligonucleotide therapy for SCN2A gain-of-function epilepsy. bioRxiv, 2020.2009.2009.289900 (2020).
- 86.Kim J, Hu C, Moufawad El Achkar C, et al. Patient-Customized Oligonucleotide Therapy for a Rare Genetic Disease. N Engl J Med. 2019;381(17):1644–1652. doi: 10.1056/NEJMoa1813279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu H, Lima WF, Crooke ST. Properties of cloned and expressed human RNase H1. The Journal of biological chemistry. 1999;274(40):28270–28278. doi: 10.1074/jbc.274.40.28270. [DOI] [PubMed] [Google Scholar]
- 88.Kamola PJ, Kitson JD, Turner G, et al. In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic acids research. 2015;43(18):8638–8650. doi: 10.1093/nar/gkv857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Scharner J, Ma WK, Zhang Q, et al. Hybridization-mediated off-target effects of splice-switching antisense oligonucleotides. Nucleic acids research. 2020;48(2):802–816. doi: 10.1093/nar/gkz1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramos DM, d’Ydewalle C, Gabbeta V, et al. Age-dependent SMN expression in disease-relevant tissue and implications for SMA treatment. J Clin Invest. 2019;129(11):4817–4831. doi: 10.1172/JCI124120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mazur C, Powers B, Zasadny K et al. Brain pharmacology of intrathecal antisense oligonucleotides revealed through multimodal imaging. JCI Insight, 4(20) (2019). [DOI] [PMC free article] [PubMed]
- 92.Sullivan JM, Mazur C, Wolf DA, et al. Convective forces increase rostral delivery of intrathecal radiotracers and antisense oligonucleotides in the cynomolgus monkey nervous system. J Transl Med. 2020;18(1):309. doi: 10.1186/s12967-020-02461-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Colasante G, Lignani G, Brusco S, et al. dCas9-Based Scn1a Gene Activation Restores Inhibitory Interneuron Excitability and Attenuates Seizures in Dravet Syndrome Mice. Mol Ther. 2020;28(1):235–253. doi: 10.1016/j.ymthe.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Safary A, Akbarzadeh Khiavi M, Mousavi R, Barar J, Rafi MA. Enzyme replacement therapies: what is the best option? Bioimpacts. 2018;8(3):153–157. doi: 10.15171/bi.2018.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chand D, Mohr F, McMillan H, et al. Hepatotoxicity following administration of onasemnogene abeparvovec (AVXS-101) for the treatment of spinal muscular atrophy. J Hepatol. 2021;74(3):560–566. doi: 10.1016/j.jhep.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 96.Korte S, Runge F, Wozniak MM, et al. Range of Neurological Signs in Cynomolgus Monkeys After Intrathecal Bolus Administration of Antisense Oligonucleotides. Int J Toxicol. 2020;39(6):505–509. doi: 10.1177/1091581820948454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.