Abstract
The field of sequencing is a topic of significant interest since its emergence and has become increasingly important over time. Impressive achievements have been obtained in this field, especially in relations to DNA and RNA sequencing. Since the first achievements by Sanger and colleagues in the 1950s, many sequencing techniques have been developed, while others have disappeared. DNA sequencing has undergone three generations of major evolution. Each generation has its own specifications that are mentioned briefly. Among these generations, nanopore sequencing has its own exciting characteristics that have been given more attention here. Among pioneer technologies being used by the third-generation techniques, nanopores, either biological or solid-state, have been experimentally or theoretically extensively studied. All sequencing technologies have their own advantages and disadvantages, so nanopores are not free from this general rule. It is also generally pointed out what research has been done to overcome the obstacles. In this review, biological and solid-state nanopores are elaborated on, and applications of them are also discussed briefly.
Keywords: DNA sequencing, Solid-state and biological nanopores, DNA sequencing generations
Introduction
DNA sequencing is significant in many fields such as forensic sciences, biology, genetics, molecular biology, archeology, and the likes. Nucleic acids are essential for continuing life since they constitute the genetic information of living matters. As the matter of fact, the ability to sequence the human genome has drastically outpaced our ability to interpret genetic variations. It has gained superabundant attention as the arrangement of nucleic acids in polynucleotide chains encompasses the information for the patrimonial and biochemical traits of living species. Since the discovery of the 3D structure of DNA by Watson and Crick in 1953 (Watson and Crick 1953; Zallen 2003), sequencing technology has experienced three generations of evolution which will be discussed in this review.
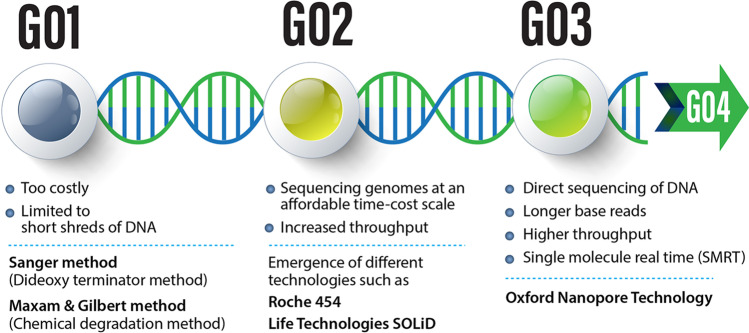
Nucleic acids sequencing is categorized into three generations (Fig. 1). During the first generation, short DNA shreds were sequenced. In the second generation, increasing throughput was achieved, alongside decreasing turnaround time and costs. Hence, at the end of the second generation, whole genome and transcriptome sequencing became more convenient. The third generation continues to surpass technological boundaries with capabilities in sequencing single molecules without prior amplification that was previously inconceivable. The second and the third generation are often referred to as “next-generation sequencing” (NGS).
Fig. 1.
A glance at DNA sequencing generations and some features of each generation
The growth of commercial sequencing platforms and optimization of experimental protocols have led to a huge growth in applications of DNA sequencing. There is special emphasis on integrating sequencing-related technologies like genomics, transcriptomics, proteomics, epigenomics, and metabolomics (Graw et al. 2021; Khella et al. 2021; Li et al. 2020; Reiter et al. 2021). The combination of these technologies with morphological and physiological techniques makes a general approach to unraveling biological systems possible (Philpott et al. 2020; Zhu et al. 2020).
First generation
The first protein sequence, of insulin, was determined in the early 1950s by Fred Sanger who devoted his scientific life to the determination of primary sequence (Heather and Chain 2016). During the first generation, efforts were focused on sequencing pure RNA species such as tRNA. At that time, researchers had borrowed the sequencing techniques from analytical chemistry that were just capable of measuring nucleotide composition, but not the order (Holley et al. 1961). In 1965, Robert Holley and colleagues invented a new method with ribonuclease treatment to generate RNA fragments and produced the first full nucleic acid sequence (Holley et al. 1965). The first sequencing procedure of alanine tRNA needed 3 years and five people working with 1 g of pure material detached from 140 g of yeast to specify 76 nucleotides (Holley et al. 1965). Molecular cloning protocols were so time-consuming that it had taken several years and was eventually replaced by in vitro amplification which was more efficiently, taking months instead of years (Lario et al. 1997). At the same time, Fred Sanger and colleagues fostered a method based on radiolabeled partial digestion of DNA fragments (Adams et al. 1969). This method labeled DNA strands with radioactive nucleotides to deduce its sequence (Padmanabhan et al. 1974; Wu 1970). However, this method again limited sequencing to tiny strands of DNA. Besides the challenges mentioned above, a remarkable methodological evolution of analytical chemistry and fractionation procedures was adapted to nucleic acid sequencing.
First-generation platforms include processes such as Maxam-Gilbert (chemical degradation) and Sanger (dideoxy terminator). As noted, they were capable of sequencing short slices of DNA. At the time of their development, first-generation techniques were monumental allowing researchers to begin to sequence DNA. More modern utilization of fluorescent labels in place of radioactive labels led to optimization of the currently recognized Sanger’s method (Lario et al. 1997).
The emergence of 2D fabrication methodology, which comprises electrophoresis and chromatography, had a significant influence on sequencing. This method provided researchers with significantly higher resolving power, originally employed by Coulson and Sanger in the “plus and minus” protocol which used Escherichia coli DNA polymerase I and DNA polymerase from bacteriophage T4 with different limiting nucleoside triphosphates. The products generated by polymerases were resolved by ionophoresis on acrylamide gels (França et al. 2002). Maxam and Gilbert also used it in their chemical cleavage technique (Maxam and Gilbert 1977; Sanger and Coulson 1975). The first DNA genome was sequenced with the aid of the plus and minus technique by Sanger and colleagues (Sanger et al. 1977). In contrast, the Maxam and Gilbert technique was quite different, and this method was widely adapted and could be considered the true arrival of “first-generation” DNA sequencing. The chief advantages of the Maxam-Gilbert technique compared with Sanger’s method are as follows: (1) sequencing could be done from the original DNA fragment, instead of from enzymic copies, (2) no PCR (polymerase chain reaction) is required, and (3) this method is less susceptible to mistakes with regard to sequencing of secondary structures or enzymic mistakes (França et al. 2002).
Sanger sequencing has provided the foundation for the growth of automatic DNA sequencing machines (Kambara et al. 1988; Luckey et al. 1990). These DNA sequencing machines were capable of reading no more than thousands of bases. Finally, newer sequencers like ABI PRISM that was outsourced from Leroy Hood research and manufactured by Applied Biosystems (Smith et al. 1986) were capable of simultaneously sequencing hundreds of samples (Ansorge 2009). This latter technology was employed in the now infamous Human Genome Project (HGP).
A glance on second generation
While efforts were being made to develop large-scale sequencing, the next generation of DNA sequencers was gradually coming to the scene. A new technique appeared which was strikingly different from existing methods since it did not identify nucleotides with the aid of radio-labeling or fluorescently labeled deoxyribonucleotides (dNTPs). The new method consisted of a two-enzyme process in which adenosine triphosphate (ATP) sulfurylase was used to convert pyrophosphate into ATP which is then used as the substrate for luciferase, thus producing light proportional to the amount of pyrophosphate (Nyrén and Lundin 1985). Notwithstanding the distinctions, both Sanger’s method and this new technique (Pyrosequencing) are known as “sequence-by-synthesis” (SBS) techniques, whereas the application of DNA polymerase to crop the apparent output was still required. This breakthrough of the second-generation sequencing technology allowed genome sequencing at an affordable time-cost scale. Second-generation sequencers overcame first-generation sequencing limitations with the aid of the following approaches, including (1) emulsion polymerase chain reaction (PCR), (2) reversible terminator, (3) sequencing by oligonucleotide ligation and detection, and the likes (Dorado et al. 2021). In spite of being revolutionary with respect to the first generation, limitations remained such as the requirement to amplify DNA which would intrinsically introduce errors to the read sequence (Ozsolak et al. 2009).
The disadvantage of the improved Sanger sequencing equipment was the cost and time consumption, and the Human Genome Project is a prime example, costing 3 billion dollars and 13 years (Lander et al. 2001). In contrast, the latter technique possessed some specifications that were considered beneficial; natural nucleotides (instead of greatly modified dNTPs) could be observable in real time (Ronaghi et al. 1996). The major drawback of this method was that the noise in the signal-to-noise ratio yielded a non-linear readout above four or five similar nucleotides (Ronaghi 1998). Pyrosequencing was then licensed to 454 Life Sciences which developed into the first chief “next-generation sequencing” (NGS) technology. These sequencing devices boosted the read output by orders of magnitude and allowed researchers to sequence a single human’s genome thoroughly in 2 months at approximately one-hundredth of the cost of traditional capillary electrophoresis methods (Wheeler et al. 2008). The tremendous shift in sequencing appreciably enhanced the quantity of DNA which could be sequenced in a single run. In a typical run, over 25 million bases could be sequenced (Margulies et al. 2005).
In principle the concepts behind Sanger vs. NGS are similar where DNA polymerase adds fluorescent nucleotides one by one onto a growing DNA template strand. Each incorporated nucleotide is identified by its fluorescent tag. The critical difference between Sanger sequencing and NGS is sequencing volume. While the Sanger method only sequences a single DNA fragment at a time, NGS is massively parallel, sequencing millions of fragments simultaneously per run. This high-throughput process translates into sequencing hundreds to thousands of genes at one time. NGS also offers greater discovery power to detect novel or rare variants with deep sequencing.
After the success of NGS, some parallel sequencing techniques emerged. Among them, the Solexa method is the most recognized and is described in detail in the following references (Bentley et al. 2008; Fedurco et al. 2006). Throughout this second generation, technologies and techniques improved substantially, now capable of reading greater length, achieving more accuracy and even faster reads.
DNA sequencing abilities from 2004 until 2010 reduplicated every 5 months which was much faster than the pace of computing revolution growth embodied by Moore’s law that doubles every 2 years (Stein 2010). From 2007 until 2012, the overall expense of DNA sequencing per base plunged by four orders of magnitude (Wetterstrand 2017). Besides, some companies have appeared or disappeared which had their own influence. Some were capable of producing machines with faster read lengths, while the others produced machines with more accuracy or cheaper sequencing per base (Glenn 2011).
Third generation
Although there is no distinct boundary between various DNA sequencing generations, especially the margin between the second and third generations (Pareek et al. 2011), real-time sequencing, single-molecule sequencing (SMS), and uninvolved split from prior technologies could be considered as the prominent specifications of the third generation. The key feature of the third-generation technologies stems from the fact that it can accurately sequence long strands of nucleic acid without an intermediary and without previous retro transcription or amplification (Ozsolak et al. 2009). Several platforms recently became commercially available such as Helicos Bio Sensing, Pacific Biosciences, BGI Group Complete Genomics, and Oxford Nanopore Technology. Each platform has its own advantages and disadvantages (Blom 2021; Broseus et al. 2020), Thus, a multifold compound of them may be required for a deep analysis of gene phraseology (Ilgisonis et al. 2021). In addition, computational models like machine learning have been exerted to these analyses (Bobrovskikh et al. 2021; Liu et al. 2021). For example, Pacific Biosciences is capable of long reads in the order of 20 kb and is capable of retaining 300 kb (Hestand and Ameur 2019); nanopore sequencing is capable of reading 30 kb, extending to 2.3 Mb (Amarasinghe et al. 2020). To reach the full potential of the third generation, some disadvantages such as the demand for higher nucleic acid concentrations, in some platforms, should be addressed to remove the need for amplification (Amarasinghe et al. 2020; Bleidorn 2016; Feng et al. 2021; Jain et al. 2018; Wang et al. 2020). In 2015, the single-molecule real-time (SMRT) sequencing platform was perchance the mostly utilized technology of third generation (Van Dijk et al. 2014).
Nanopore sequencing might be the most favorable platform for the development of third-generation DNA sequencing. It is a branch of the immense field of using nanopores for the identification of biological and chemical molecules (Haque et al. 2013). As a matter of fact, the potential of nanopores for sequencing was established much earlier than the emergence of the second generation but was not well recognized in mainstream science until recently. Researchers showed that single-stranded DNA (ssDNA) or RNA could be steered across a lipid bilayer throughout α-hemolysin ion grooves by crossing channel barricades and temporarily blocking the flow of ion current (blockade current) commensurate to the protraction of the nucleic acid (Kasianowicz et al. 1996). The possibility of utilizing solid-state nanopores was more recently mentioned in the literature as a means to sequence double-stranded DNA (Dekker 2010). In the section below, the review will describe solid-state nanopores and their application in DNA sequencing.
The amalgamation of gene engineering and computer aided technology may form the foundations of the fourth generation of sequencing platforms. For example, Oxford Nanopore Technology (ONT) developed the nanopore technology to sequence distorted bases resulting from DNA passing through the nanopore (Mikheyev and Tin 2014). Nanopores and sequencing through them will be addressed later. A comparison between some features of different generation platforms is shown in Table 1.
Table 1.
A comparison between some features of different generations platforms (Solieri et al. 2013)
| Platform | Chemistry | PCR amplification | Starting DNA | Read length | Reads/run | TH/run | Run time | Disadvantages | Applications |
|---|---|---|---|---|---|---|---|---|---|
| Sanger sequencing | Asynchronous with base-specific terminator | Standard PCR | 0.5–1 mg | 700 | Few 1000 bp | 1 Mb | 2 h | PCR biases; low degree of parallelism; high cost of sequencing | Gene/genome sequencing |
| Roche 454 | Sequencing-by synthesis (pyrosequencing) | EmPCR | 1 μg for shotgun library and 5 μg for pair-end | > 400 | 1000,000 | 0.4–0.6Gb | 7–10 h | PCR biases; asynchronous synthesis; homopolymer run; base insertion and deletion errors; EmPCR is cumbersome and technically challenging | De novo genome sequencing, RNA-seq, resequencing/targeted re-sequencing |
| Illumina | Polymerase-based sequencing-by synthesis | Bridge amplification | <1 μg for single or pair-end | 75/2 × 100a | 40,000,000 | 3–6/200 | 3–4 days | PCR biases; low multiplexing capability of samples | De novo genome sequencing, RNA-seq, resequencing/ targeted re-sequencing, metagenomics, ChIP |
| SOLiD | Ligation-based sequencing | EmPCR | <2 μg for shotgun library and 5–20 μg for pair-end | 35–40 | 85,000,000 | 10–20Gb | 7 days | EmPCR is cumbersome and technically challenging PCR biases; long run time | Transcript counting, mutation detection, ChIP, RNA-seq, etc. |
| HeliScope | Polymerase (asynchronous extension | SM; no PCR | <2 μg, single end only | 25–50 | 1000,000,000 | 28Gb | 8 days | Asynchronous synthesis; homopolymer run; high instrument cost; short read lengths; high error rates compared with other reversible terminator chemistries | Resequencing, transcript counting, ChIP, RNA-seq |
| Polonator | Synchronous controlled synthesis | Em PCR | __ | 26 | 160,000,000 | 4.5Gb | 4 days | Low read length; emPCR is cumbersome and technically challenging | Bacterial genome, resequencing, SNPs and structural variants detection |
| PacBio | Phospholinked fluorescent nucleotides | SMRT | ∼1.5 μg (ideally 2–3 μg) | 1000–1200 | 100,000,000 | 100Gb/Hr | 8 h | High instrument cost; low number of sequence read per run; highest error rates compared with other NGS chemistries | De novo genome sequencing, RNA-seq, resequencing/targeted re-sequencing, metagenomics, SNPs and structural variants detection |
| CMOS non-optical sequencing | Template-directed DNA polymerase synthesis | __b | __ | __ | __ | __ | __ | __ | De novo genome sequencing |
Importance of sequencing generated data
A tremendous amount of data generated and collected, mostly during the second and the third generations of sequencing, requires new software and hardware to analyze. Thus, to address the big data generated, many fields such as mathematics, statistics, and bioinformatics are involved. Artificial intelligence, machine learning, and similar fields have been developed (Chachar et al. 2021; Jovčevska 2020). The importance of nucleic acid data is listed as several examples below: sequencing of nucleic acid technologies is the vaccine design to treat COVID-19 disease (Wang et al. 2021); the Human Genome Project (HGP) paved the way for whole-genome sequencing (Wang et al. 2021); nucleic acids could be utilized as well to put in store any sort of data in a dense and efficacious manner that could be recovered and decoded by sequencing (Wang et al. 2021); functional genomics used in diverse arenas such as medicine and agronomy; and could be inspection of disease resistance or abiotic and biotic stresses in animals and plants with impressive consequences in health programs (Jha et al. 2021). This is so vital in disease diagnostics and clinical treatments (Caspar et al. 2021).
In the last 2 decades, the quantity of total drug-resistant bacteria that are resistant to all familiar antibiotics, principally because of the misapply of antibiotics, have increased (Gaultney et al. 2020). This calls attention to the requirement of new abatement and action toward the set of tactics for pathogenic bacteria, discovering surrogates to antibiotics. The recently developed sequencing technologies are brought into play to attain this objective (Allue Guardia et al. 2021). In this schema, extremely conserved DNA methyl-transferases (MTases) are possible objectives to action infections for epigenetic inhibitors (Oliveira and Fang 2021). A simple comparison between various platforms, belonging to different generations, is provided in Table 2.
Table 2.
A comparison between different platforms from different generations (Lin et al. 2021)
| Reading length (kb) | Estimated cost per Gb (US $) | Throughput per flow cell (Gb) | Read accuracy (%) | |
|---|---|---|---|---|
| Sanger(1st) | <1 kb | 13,000 | / | >99.9 |
| Illumina(2nd) | 0.075–0.15 | 50–63 | 16–30 | >99.9 |
| PacBio(3rd) | 10–20 | 43–86 | 15–30 | >99 |
| ONT | 10–60 | 21–42 | 50–100 | 87–98 |
Nanopore sequencing
Nanopore sequencing is a new age of sequencing, rapidly grown to meet the gap in advancements to sustain the flair for larger read length, faster sequencing, and lower costs. In some published texts, nanopore sequencing is considered fourth generation of DNA sequencing technology (Lin et al. 2021). Nanopore technology is an encouraging platform that utilizes highly sensitive single-molecule detectors for DNA or RNA (Garalde et al. 2018; Kasianowicz et al. 1996). In addition, nanopore sensors are easily miniaturized and integrated into portable “lab-on-a-chip” devices (Roman et al. 2017). Despite the benefits of nanopore sequencing, complicated sample preparation and data processing algorithms remain challenges that need to be overcome (Bayley 2015; Kasianowicz et al. 1996; Deamer et al. 2016).
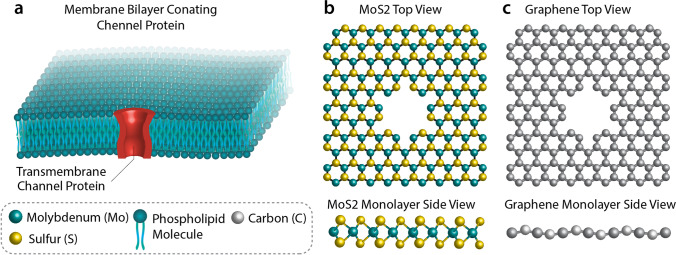
A nanopore is a perforation of nanometer size that can be constructed either by proteins or by artificial molecules. All nanopore types are utilized to sequence biological and chemical molecules at the nanoscale (Deamer and Branton 2002). Nanopore sequencing offers inexpensive and fast DNA sequencing without using labels (Rhee and Burns 2006). Some types of nanopores and its materials are shown in Fig. 2.
Fig. 2.
Graphical representation of biological and 2D solid-state nanopores
Biological nanopores
Biological nanopores are also called transmembrane protein channels (Zeng et al. 2021). Biological nanopores are artificial or natural protein molecules produced by genetic engineering (Mohammad et al. 2012). Biological nanopores are generated by specified bacteria such as α-hemolysin pore protein (Bayley and Cremer 2001), MspA is from Mycobacterium smegmatis porin A (Zeng et al. 2021) (Derrington et al. 2010), and bacteriophage Phi-29 motor (Phi 29) is from Bacillus subtilis (Manrao et al. 2012). So α-hemolysin, MspA porin, and Phi 29 connector are some proteins that constitute pores. These biological nanopores are commonly utilized for smart drug delivery (Martinac et al. 2017; Martinac et al. 2020), disease diagnosis (Brown et al. 2021), protein sequencing (Hu et al. 2021), and gene sequencing (Quick et al. 2016). In laboratories, nanopores are inserted into a lipid bilayer film allowing manipulations and measurements to be undertaken (Briggs et al. 2018). Albeit there are abundant molecular channels, such as receptors and ligand-gated channels, that could be employed in sensing applications, but the main attention is paid to well-controlled pores that could be utilized as a single sensing element (Shen et al. 2020). Among these aforementioned ones, α-hemolysin is the first to be commonly used (Song et al. 1996).
Biological applications have inspired researchers to use technologies requiring synthetic and biological nanopores to detect gene sequences. These technologies have been extensively used in DNA sequencing (Heng et al. 2004; Manrao et al. 2012; Venkatesan and Bashir 2011; Wanunu 2012; Woodside et al. 2006) and even in RNA and protein sequencing as well (Depledge et al. 2019; Smith et al. 2019; Soneson et al. 2019; Xie et al. 1991). Besides, such technologies could be hired to determine the sequence of nucleic acids (Kono and Arakawa 2019; Lockhart and Winzeler 2000; Soneson et al. 2019; Xie et al. 1991).
Solid-state nanopores
Solid-state nanopores are principally produced in a thin film of materials such as graphene (single atom thickness sheet of carbon), silicon nitride (SiN), phosphorene, Al2O3 (Venkatesan et al. 2009) and HfO2 (Larkin et al. 2013). SiN, graphene, and phosphorene nanopores show superiorities over biological competitors like chemical and thermal stabilities, although this stability depends on the formation of the pore. There exist numerous techniques for producing solid-state nanopores such as “deploying and sculpting with ion beam” and “fabrication by electron beam” (Briggs et al. 2018). These pores can also be constructed using procedures such as electrochemical reactions, controlled breakdown, laser etching, and laser-assisted controlled breakdown (Feng et al. 2015b). However, the controlled chemical rectification of these nanopores is accessible though challenging (Brilmayer et al. 2020; Yusko et al. 2011). There are less restrictions with solid-state nanopores in contrast with biological ones; for example, solid-state nanopores can operate over wider temperature and voltage ranges. Besides, solid-state nanopores are more compatible and even more stable to solvent conditions, and they can be adjusted in diameter with sub-nanometer accuracy (Yuan et al. 2020). Si3N4 and SiO2 nanopores are among the most broadly employed nanopores, and their manufacturing is in accord with the complementary metal oxide semiconductor industrial integrated circuit processes. Ion etching in free-standing Si3N4 and SiO2 films using argon is the method by which these nanopores are produced (Tang et al. 2016).
Graphene holds unique chemical properties because of being electrically conductive and even much stronger than steel (Thompson and Milos 2011). Albeit graphene with its univalent layer character provides the optimal thickness (0.34 nm) for single-base resolution (Novoselov et al. 2016), MoS2 is the most frequently used two-dimensional (2D) material investigated for sequencing applications, because of the simple fabrication of MoS2 devices (Butler et al. 2013; Graf et al. 2019b; Tsutsui et al. 2011). It should be noted that the structure of single-layer plates and pores are not static, rather they are affected and distorted by electrostatic and hydrodynamic forces (Hernández-Ainsa et al. 2014; Plesa et al. 2014), even though it is recently shown that graphene nanopore is not a suitable candidate for sequencing DNA using ionic current. Since graphene and DNA nucleotides have strong hydrophobic interactions, DNA may stick to graphene which severely impacts translocation speed (Schneider et al. 2013). The major drawback of using graphene is its hydrophobic nature. Another point is the orientational fluctuations of nucleobases during DNA translocation through a graphene nanopore. From sequencing point of view, MoS2 can perform better than graphene. For example, signal to noise ratio and non-stickiness of DNA to MoS2 surface make it suitable (Graf et al. 2019a) (Henry et al. 2021). Instead, phosphorene nanopore and silicene (graphene like two-dimensional silicon) nanopore seem much more suitable (Henry et al. 2021). One of the main problems in detecting bases through solid-state nanopores is the fact that they have a low spatial resolution since dozens of bases can pass through them at any given moment (Yanagi et al. 2015).
In general, the thickness of a 2D single-layer material is approximately 3.0–11.0 Angstroms that is analogous to the gap between two successive nucleotides of an ssDNA which is almost 3.5–5.2 Angstroms (Liu et al. 2014).
As a theoretical example for nanopores’ applications, specifically graphene, an ssDNA is pulled through a nanopore whose diameter is comparable to single DNA bases. With the aid of molecular dynamics (MD) simulation, various parameters like pulling force or orientation of bases relative to the graphene plane or its normal axis are tracked during translocation of ssDNA can be resolved. In an unpublished work by the authors, the phosphorene atom of the DNA backbone is pulled through nanopore with constant velocity, and in addition to pulling force and base orientation, Vander Waals and electrostatic energies and forces are also tracked to see whether or not these parameters can yield an illustrious distinction between DNA bases (Fig. 3).
Fig. 3.
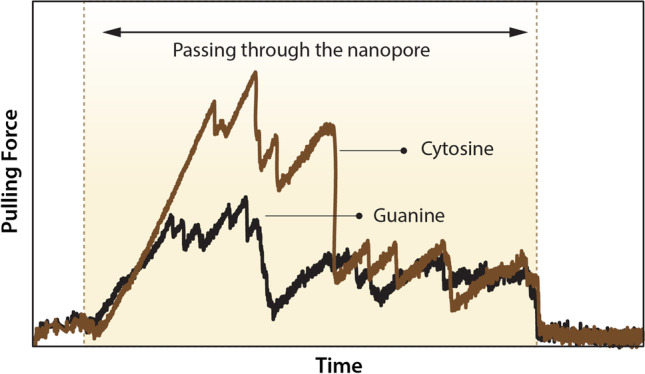
A schematic of SMD force vs. time curve (sample output of MD simulation) which is studied to investigate the distinction between bases
In addition, hexagonal boron nitride (hBN) is less hydrophobic than graphene. The thickness of hBN is comparable to the spacing between nucleotides (0.32–0.52 nm) in single-stranded DNA (ssDNA) (Zhao et al. 2014). It also shows other advantages over graphene in terms of its insulating property in high ionic strength solution and fewer defects made during the manufacturing process(Liu et al. 2013).
Several theoretical and experimental studies have proven MoS2 capabilities as a mono layer material in the form of the nanopore or nanoribbon (Feng et al. 2015a; Graf et al. 2019a; Liu et al. 2014). Moreover, graphene (Traversi et al. 2013), WS2 (Danda et al. 2017), and BN (Liu et al. 2013) have been demonstrated to detect DNA translocation. Up to now, none of solid-state nanopores have shown single-base resolution. Therefore, it is so crucial to proceed with studies to identify new materials, and two such prime candidate materials are phosphorene and silicene as mentioned earlier (Jose and Datta 2014; Zereshki et al. 2018). Both materials have properties that are ideal for base identification (Chen et al. 2017). Moreover, the biocompatibility and hydrophilicity of phosphorene makes it appropriate for biosensing applications (Cortés-Arriagada 2018; Kumawat et al. 2018).
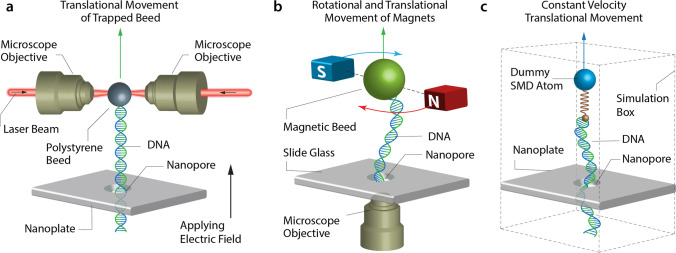
DNA translocates through solid-state nanopores very fast, up to 0.01–1 μs per base (Heerema et al. 2018). As a matter of choice, the DNA translocation velocity should be 1–100 base per microseconds in a nanopore to provide an acceptable signal from each nucleotide (Akahori et al. 2017). Thus, it is so essential to slow down DNA during translocation. Different methods have been examined to control translocation speed such temperature (Wanunu et al. 2008), electrolyte viscosity (Feng et al. 2015a), driving voltage (Liang et al. 2013), and ion concentration (Luan and Aksimentiev 2010). Alternative methods like two nanopores system (Langecker et al. 2011), optical tweezers (Keyser et al. 2006), optical trapping of a single DNA (Kim and Lee 2014), and magnetic tweezers (Peng and Ling 2009) have been used. A schematic of various nanopore sequencing approaches is depicted in Fig. 4.
Fig. 4.
Schematic of various approaches for sequencing; specifically exerting force on the strand to pull it. a Optical trapping, b magnetic trapping, and c molecular dynamic simulation
Conclusion
The first-generation methods, though revolutionary, suffered from disadvantages like being costly or being capable to sequence only small strands. The second-generation techniques presented modifications to genome sequencing at a reasonable time-cost scale and enhancing throughput while still required DNA amplification which would have made errors. The third generation could go several steps forward and attained the traits like direct sequencing, longer base reads, real-time sequencing, and single-molecule nature. One should be so optimistic to the future of the DNA sequencing grounded on new technologies, but there are still obstacles that should be overcome by researchers. What seems more progressive is quantum simulations that are more confidential but more cumbersome since they require much more computational costs.
Author contribution
M.M. Mohammadi: literature search and writing—original draft. O. Bavi: conceptualization, supervision, and writing—review and editing.
Funding
None
Declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams J, Jeppesen P, Sanger F, Barrell B. Nucleotide sequence from the coat protein cistron of R17 bacteriophage RNA. Nature. 1969;223:1009–1014. doi: 10.1038/2231009a0. [DOI] [PubMed] [Google Scholar]
- Akahori R, Yanagi I, Goto Y, Harada K, Yokoi T, Takeda K-I. Discrimination of three types of homopolymers in single-stranded DNA with solid-state nanopores through external control of the DNA motion. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-08290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allue Guardia A, Garcia JI, Torrelles JB. Evolution of drug-resistant mycobacterium tuberculosis strains and their adaptation to the human lung environment. Front Microbiol. 2021;12:137. doi: 10.3389/fmicb.2021.612675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe SL, Su S, Dong X, Zappia L, Ritchie ME, Gouil Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020;21:1–16. doi: 10.1186/s13059-020-1935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge WJ. Next-generation DNA sequencing techniques. New Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Bayley H. Nanopore sequencing: from imagination to reality. Clin Chem. 2015;61:25–31. doi: 10.1373/clinchem.2014.223016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley H, Cremer PS. Stochastic sensors inspired by biology. Nature. 2001;413:226–230. doi: 10.1038/35093038. [DOI] [PubMed] [Google Scholar]
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleidorn C. Third generation sequencing: technology and its potential impact on evolutionary biodiversity research. Syst Biodivers. 2016;14:1–8. doi: 10.1080/14772000.2015.1099575. [DOI] [Google Scholar]
- Blom MPK. Opportunities and challenges for high-quality biodiversity tissue archives in the age of long-read sequencing. Mol Ecol. 2021 doi: 10.1111/mec.15909. [DOI] [PubMed] [Google Scholar]
- Bobrovskikh AV, Doroshkov A, Mazzoleni S, Carteni F, Giannino F, Zubairova U. A sight on single-cell transcriptomics in plants through the prism of cell-based computational modeling approaches: benefits and challenges for data analysis. Front Genet. 2021;12:771. doi: 10.3389/fgene.2021.652974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs K, Madejski G, Magill M, Kastritis K, de Haan HW, McGrath JL, Tabard-Cossa V. DNA translocations through nanopores under nanoscale preconfinement. Nano Lett. 2018;18:660–668. doi: 10.1021/acs.nanolett.7b03987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilmayer R, Förster C, Zhao L, Andrieu-Brunsen A. Recent trends in nanopore polymer functionalization. Curr Opin Biotechnol. 2020;63:200–209. doi: 10.1016/j.copbio.2020.03.005. [DOI] [PubMed] [Google Scholar]
- Broseus L, Thomas A, Oldfield AJ, Severac D, Dubois E, Ritchie W. TALC: transcript-level aware long-read correction. Bioinformatics. 2020;36:5000–5006. doi: 10.1093/bioinformatics/btaa634. [DOI] [PubMed] [Google Scholar]
- Brown E, Freimanis G, Shaw AE, Horton DL, Gubbins S, King D (2021) Characterising foot-and-mouth disease virus in clinical samples using nanopore sequencing. Front Vet Sci 8:656256. 10.3389/fvets.2021.656256 [DOI] [PMC free article] [PubMed]
- Butler SZ, et al. Progress, challenges, and opportunities in two-dimensional materials beyond graphene. ACS Nano. 2013;7:2898–2926. doi: 10.1021/nn400280c. [DOI] [PubMed] [Google Scholar]
- Caspar SM, Schneider T, Stoll P, Meienberg J, Matyas G. Potential of whole-genome sequencing-based pharmacogenetic profiling. Pharmacogenomics. 2021;22:177–190. doi: 10.2217/pgs-2020-0155. [DOI] [PubMed] [Google Scholar]
- Chachar S, Liu J, Zhang P, Riaz A, Guan C, Liu S (2021) Harnessing current knowledge of DNA N6-methyladenosine from model plants for non-model crops. Front Genet 12:668317. 10.3389/fgene.2021.668317 [DOI] [PMC free article] [PubMed]
- Chen Y, Ren R, Pu H, Chang J, Mao S, Chen J. Field-effect transistor biosensors with two-dimensional black phosphorus nanosheets. Biosens Bioelectron. 2017;89:505–510. doi: 10.1016/j.bios.2016.03.059. [DOI] [PubMed] [Google Scholar]
- Cortés-Arriagada D. Phosphorene as a template material for physisorption of DNA/RNA nucleobases and resembling of base pairs: a cluster DFT study and comparisons with graphene. J Phys Chem C. 2018;122:4870–4880. doi: 10.1021/acs.jpcc.7b11268. [DOI] [Google Scholar]
- Danda G, et al. Monolayer WS2 nanopores for DNA translocation with light-adjustable sizes. ACS Nano. 2017;11:1937–1945. doi: 10.1021/acsnano.6b08028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer DW, Branton D. Characterization of nucleic acids by nanopore analysis. Acc Chem Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- Deamer D, Akeson M, Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34:518–524. doi: 10.1038/nbt.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2(4):209–215. doi: 10.1142/9789814287005_0007. [DOI] [PubMed] [Google Scholar]
- Depledge DP, et al. Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nat Commun. 2019;10:1–13. doi: 10.1038/s41467-019-08734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrington IM, Butler TZ, Collins MD, Manrao E, Pavlenok M, Niederweis M, Gundlach JH. Nanopore DNA sequencing with MspA. Proc Natl Acad Sci. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorado G, Gálvez S, Rosales TE, Vásquez VF, Hernández P. Analyzing modern biomolecules: the revolution of nucleic-acid sequencing-review. Biomolecules. 2021;11:1111. doi: 10.3390/biom11081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedurco M, Romieu A, Williams S, Lawrence I, Turcatti G. BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res. 2006;34:e22–e22. doi: 10.1093/nar/gnj023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu K, Bulushev RD, Khlybov S, Dumcenco D, Kis A, Radenovic A. Identification of single nucleotides in MoS 2 nanopores. Nat Nanotechnol. 2015;10:1070–1076. doi: 10.1038/nnano.2015.219. [DOI] [PubMed] [Google Scholar]
- Feng Y, Zhang Y, Ying C, Wang D, Du C. Nanopore-based fourth-generation DNA sequencing technology. Genomics, Proteomics & Bioinformatics. 2015;13:4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Clemente JC, Wong B, Schadt EE. Detecting and phasing minor single-nucleotide variants from long-read sequencing data. Nat Commun. 2021;12:1–13. doi: 10.1038/s41467-021-23289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- França LT, Carrilho E, Kist TB. A review of DNA sequencing techniques. Q Rev Biophys. 2002;35:169–200. doi: 10.1017/S0033583502003797. [DOI] [PubMed] [Google Scholar]
- Garalde DR, et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods. 2018;15:201–206. doi: 10.1038/nmeth.4577. [DOI] [PubMed] [Google Scholar]
- Gaultney RA, et al. 4-Methylcytosine DNA modification is critical for global epigenetic regulation and virulence in the human pathogen Leptospira interrogans. Nucleic Acids Res. 2020;48:12102–12115. doi: 10.1093/nar/gkaa966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759–769. doi: 10.1111/j.1755-0998.2011.03024.x. [DOI] [PubMed] [Google Scholar]
- Graf M, Lihter M, Altus D, Marion S, Radenovic A. Transverse detection of DNA using a MoS2 nanopore. Nano Lett. 2019;19:9075–9083. doi: 10.1021/acs.nanolett.9b04180. [DOI] [PubMed] [Google Scholar]
- Graf M, et al. Fabrication and practical applications of molybdenum disulfide nanopores. Nat Protoc. 2019;14:1130–1168. doi: 10.1038/s41596-019-0131-0. [DOI] [PubMed] [Google Scholar]
- Graw S, Chappell K, Washam CL, Gies A, Bird J, Robeson MS, et al. Multi-omics data integration considerations and study design for biological systems and disease. Molecular Omics. 2021;17(2):170–185. doi: 10.1039/D0MO00041H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Li J, Wu H-C, Liang X-J, Guo P. Solid-state and biological nanopore for real-time sensing of single chemical and sequencing of DNA. Nano Today. 2013;8:56–74. doi: 10.1016/j.nantod.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heather JM, Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107:1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerema SJ, Vicarelli L, Pud S, Schouten RN, Zandbergen HW, Dekker C. Probing DNA translocations with inplane current signals in a graphene nanoribbon with a nanopore. ACS Nano. 2018;12:2623–2633. doi: 10.1021/acsnano.7b08635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JB, et al. Sizing DNA using a nanometer-diameter pore. Biophys J. 2004;87:2905–2911. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MB, Tumbapo M, Tayo BO. Identification of DNA bases using nanopores created in finite-size nanoribbons from graphene, phosphorene, and silicene. AIP Adv. 2021;11:035324. doi: 10.1063/5.0043000. [DOI] [Google Scholar]
- Hernández-Ainsa S, Misiunas K, Thacker VV, Hemmig EA, Keyser UF. Voltage-dependent properties of DNA origami nanopores. Nano Lett. 2014;14:1270–1274. doi: 10.1021/nl404183t. [DOI] [PubMed] [Google Scholar]
- Hestand MS, Ameur A (2019) The versatility of SMRT sequencing. Multidisciplinary Digital Publishing Institute. 10.3390/genes10010024 [DOI] [PMC free article] [PubMed]
- Holley RW, Apgar J, Merrill SH, Zubkoff PL. Nucleotide and oligonucleotide compositions of the alanine-, valine-, and tyrosine-acceptor “soluble” ribonucleic acids of yeast. J Am Chem Soc. 1961;83:4861–4862. doi: 10.1021/ja01484a040. [DOI] [Google Scholar]
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A (1965) Structure of a ribonucleic acid. Science 147(3664):1462–1465. https://www.jstor.org/stable/1715055#:~:text=https%3A//www.jstor.org/stable/1715055 [DOI] [PubMed]
- Hu ZL, Huo MZ, Ying YL, Long YT (2021) Biological nanopore approach for single-molecule protein sequencing. Angewandte Chemie 133(27):14862–14873. 10.1002/ange.202013462 [DOI] [PubMed]
- Ilgisonis E et al (2021) Genome of the single human chromosome 18 as a “gold standard” for its transcriptome. Front Genet 12:958. 10.3389/fgene.2021.674534 [DOI] [PMC free article] [PubMed]
- Jain M, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AB, Gali KK, Alam Z, Lachagari V, Warkentin TD. Potential application of genomic technologies in breeding for fungal and oomycete disease resistance in pea. Agronomy. 2021;11:1260. doi: 10.3390/agronomy11061260. [DOI] [Google Scholar]
- Jose D, Datta A. Structures and chemical properties of silicene: unlike graphene. Acc Chem Res. 2014;47:593–602. doi: 10.1021/ar400180e. [DOI] [PubMed] [Google Scholar]
- Jovčevska I. Next generation sequencing and machine learning technologies are painting the epigenetic portrait of glioblastoma. Front Oncol. 2020;10:798. doi: 10.3389/fonc.2020.00798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Nishikawa T, Katayama Y, Yamaguchi T. Optimization of parameters in a DNA sequenator using fluorescence detection. Bio/Technology. 1988;6:816–821. doi: 10.1038/nbt0788-816. [DOI] [Google Scholar]
- Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser UF, et al. Direct force measurements on DNA in a solid-state nanopore. Nat Phys. 2006;2:473–477. doi: 10.1038/nphys344. [DOI] [Google Scholar]
- Khella CA, Mehta GA, Mehta RN, Gatza ML. Recent advances in integrative multi-omics research in breast and ovarian cancer. Journal of Personalized Medicine. 2021;11:149. doi: 10.3390/jpm11020149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J-D, Lee Y-G. Trapping of a single DNA molecule using nanoplasmonic structures for biosensor applications. Biomedical Optics Express. 2014;5:2471–2480. doi: 10.1364/BOE.5.002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono N, Arakawa K. Nanopore sequencing: review of potential applications in functional genomics. Develop Growth Differ. 2019;61:316–326. doi: 10.1111/dgd.12608. [DOI] [PubMed] [Google Scholar]
- Kumawat RL, Garg P, Kumar S, Pathak B. Electronic transport through DNA nucleotides in atomically thin phosphorene electrodes for rapid DNA sequencing. ACS Appl Mater Interfaces. 2018;11:219–225. doi: 10.1021/acsami.8b17239. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Langecker M, Pedone D, Simmel FC, Rant U. Electrophoretic time-of-flight measurements of single DNA molecules with two stacked nanopores. Nano Lett. 2011;11:5002–5007. doi: 10.1021/nl2030079. [DOI] [PubMed] [Google Scholar]
- Lario A, González A, Dorado G. Automated laser-induced fluorescence DNA sequencing: equalizing signal-to-noise ratios significantly enhances overall performance. Anal Biochem. 1997;247:30–33. doi: 10.1006/abio.1996.9933. [DOI] [PubMed] [Google Scholar]
- Larkin J, Henley R, Bell DC, Cohen-Karni T, Rosenstein JK, Wanunu M (2013) Slow DNA transport through nanopores in hafnium oxide membranes. ACS Nano 7:10121–10128. 10.1021/nn404326f [DOI] [PMC free article] [PubMed]
- Li Y, Ma A, Mathé EA, Li L, Liu B, Ma Q. Elucidation of biological networks across complex diseases using single-cell omics. Trends Genet. 2020;36(12):951–966. doi: 10.1016/j.tig.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Cui P, Wang Q, Wu T, Ågren H, Tu Y. Theoretical study on key factors in DNA sequencing with graphene nanopores. RSC Adv. 2013;3:2445–2453. doi: 10.1039/C2RA22109H. [DOI] [Google Scholar]
- Lin B, Hui J, Mao H. Nanopore technology and its applications in gene sequencing. Biosensors. 2021;11:214. doi: 10.3390/bios11070214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, et al. Boron nitride nanopores: highly sensitive DNA single-molecule detectors. Adv Mater. 2013;25:4549–4554. doi: 10.1002/adma.201301336. [DOI] [PubMed] [Google Scholar]
- Liu K, Feng J, Kis A, Radenovic A. Atomically thin molybdenum disulfide nanopores with high sensitivity for DNA translocation. ACS Nano. 2014;8:2504–2511. doi: 10.1021/nn406102h. [DOI] [PubMed] [Google Scholar]
- Liu J, Fan Z, Zhao W, Zhou X. Machine intelligence in single-cell data analysis: advances and new challenges. Front Genet. 2021;12:807. doi: 10.3389/fgene.2021.655536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–836. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Luan B, Aksimentiev A. Electric and electrophoretic inversion of the DNA charge in multivalent electrolytes. Soft Matter. 2010;6:243–246. doi: 10.1039/B917973A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckey JA, Drossman H, Kostichka AJ, Mead DA, D'Cunha J, Norris TB, Smith LM. High speed DNA sequencing by capillary electrophoresis. Nucleic Acids Res. 1990;18:4417–4421. doi: 10.1093/nar/18.15.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac AD, Bavi N, Bavi O, Martinac B. Pulling MscL open via N-terminal and TM1 helices: a computational study towards engineering an MscL nanovalve. PLoS One. 2017;12:e0183822. doi: 10.1371/journal.pone.0183822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B, et al. Cell membrane mechanics and mechanosensory transduction. Curr Top Membr. 2020;86:83–141. doi: 10.1016/bs.ctm.2020.08.002. [DOI] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheyev AS, Tin MM. A first look at the Oxford Nanopore MinION sequencer. Mol Ecol Resour. 2014;14:1097–1102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- Mohammad MM, Iyer R, Howard KR, McPike MP, Borer PN, Movileanu L. Engineering a rigid protein tunnel for biomolecular detection. J Am Chem Soc. 2012;134:9521–9531. doi: 10.1021/ja3043646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoselov KS, Mishchenko A, Carvalho A, Castro Neto AH (2016) 2D materials and van der Waals heterostructures. Science 353(6298):aac9439–aac9439. 10.1126/science.aac9439 [DOI] [PubMed]
- Nyrén P, Lundin A. Enzymatic method for continuous monitoring of inorganic pyrophosphate synthesis. Anal Biochem. 1985;151:504–509. doi: 10.1016/0003-2697(85)90211-8. [DOI] [PubMed] [Google Scholar]
- Oliveira PH, Fang G. Conserved DNA methyltransferases: a window into fundamental mechanisms of epigenetic regulation in bacteria. Trends Microbiol. 2021;29:28–40. doi: 10.1016/j.tim.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R, Jay E, Wu R. Chemical synthesis of a primer and its use in the sequence analysis of the lysozyme gene of bacteriophage T4. Proc Natl Acad Sci. 1974;71:2510–2514. doi: 10.1073/pnas.71.6.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareek CS, Smoczynski R, Tretyn A. Sequencing technologies and genome sequencing. J Appl Genet. 2011;52:413–435. doi: 10.1007/s13353-011-0057-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Ling XS (2009) Reverse DNA translocation through a solid-state nanopore by magnetic tweezers. Nanotechnology 20(18):185101. 10.1088/0957-4484/20/18/185101 [DOI] [PMC free article] [PubMed]
- Philpott M, Cribbs AP, Brown T, Jr, Brown T, Sr, Oppermann U. Advances and challenges in epigenomic single-cell sequencing applications. Curr Opin Chem Biol. 2020;57:17–26. doi: 10.1016/j.cbpa.2020.01.013. [DOI] [PubMed] [Google Scholar]
- Plesa C, Ananth AN, Linko V, Gulcher C, Katan AJ, Dietz H, Dekker C. Ionic permeability and mechanical properties of DNA origami nanoplates on solid-state nanopores. ACS Nano. 2014;8:35–43. doi: 10.1021/nn405045x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530:228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter T, et al. Streamlining data-intensive biology with workflow systems. GigaScience. 2021;10:giaa140. doi: 10.1093/gigascience/giaa140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee M, Burns MA. Nanopore sequencing technology: research trends and applications. Trends Biotechnol. 2006;24:580–586. doi: 10.1016/j.tibtech.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Roman J, Jarroux N, Patriarche G, Français O, Pelta J, Le Pioufle B, Bacri L. Functionalized solid-state nanopore integrated in a reusable microfluidic device for a better stability and nanoparticle detection. ACS Appl Mater Interfaces. 2017;9:41634–41640. doi: 10.1021/acsami.7b14717. [DOI] [PubMed] [Google Scholar]
- Ronaghi M. Real-time pyrophosphate detection for DNA sequencing. Science. 1998;281:363–364. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Sanger F, et al. Nucleotide sequence of bacteriophage φX174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Schneider GF, et al. Tailoring the hydrophobicity of graphene for its use as nanopores for DNA translocation. Nat Commun. 2013;4:1–7. doi: 10.1038/ncomms3619. [DOI] [PubMed] [Google Scholar]
- Shen B, Piskunen P, Nummelin S, Liu Q, Kostiainen MA, Linko V. Advanced DNA nanopore technologies. ACS Applied Bio Materials. 2020;3:5606–5619. doi: 10.1021/acsabm.0c00879. [DOI] [PubMed] [Google Scholar]
- Smith LM, et al. Fluorescence detection in automated DNA sequence analysis. Nature. 1986;321:674–679. doi: 10.1038/321674a0. [DOI] [PubMed] [Google Scholar]
- Smith AM, Jain M, Mulroney L, Garalde DR, Akeson M. Reading canonical and modified nucleobases in 16S ribosomal RNA using nanopore native RNA sequencing. PLoS One. 2019;14:e0216709. doi: 10.1371/journal.pone.0216709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solieri L, Dakal TC, Giudici P. Next-generation sequencing and its potential impact on food microbial genomics. Ann Microbiol. 2013;63:21–37. doi: 10.1007/s13213-012-0478-8. [DOI] [Google Scholar]
- Soneson C, Yao Y, Bratus-Neuenschwander A, Patrignani A, Robinson MD, Hussain S. A comprehensive examination of Nanopore native RNA sequencing for characterization of complex transcriptomes. Nat Commun. 2019;10:1–14. doi: 10.1038/s41467-019-11272-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1865. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- Stein LD. The case for cloud computing in genome informatics. Genome Biol. 2010;11:1–7. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Zhang D, Cui W, Zhang H, Pang W, Duan X. Fabrications, applications and challenges of solid-state nanopores: a mini review. Nanomaterials and Nanotechnology. 2016;6:35. doi: 10.5772/64015. [DOI] [Google Scholar]
- Thompson JF, Milos PM. The properties and applications of single-molecule DNA sequencing. Genome Biol. 2011;12:1–10. doi: 10.1186/gb-2011-12-2-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traversi F, et al. Detecting the translocation of DNA through a nanopore using graphene nanoribbons. Nat Nanotechnol. 2013;8:939–945. doi: 10.1038/nnano.2013.240. [DOI] [PubMed] [Google Scholar]
- Tsutsui M, Rahong S, Iizumi Y, Okazaki T, Taniguchi M, Kawai T. Single-molecule sensing electrode embedded in-plane nanopore. Sci Rep. 2011;1:1–6. doi: 10.1038/srep00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol. 2011;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- Venkatesan BM, Dorvel B, Yemenicioglu S, Watkins N, Petrov I, Bashir R. Highly sensitive, mechanically stable nanopore sensors for DNA analysis. Adv Mater. 2009;21:2771–2776. doi: 10.1002/adma.200803786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Qu L, Yang L, Wang Y, Zhu H. NanoReviser: an error-correction tool for nanopore sequencing based on a deep learning algorithm. Front Genet. 2020;11:900. doi: 10.3389/fgene.2020.00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, et al. Robust storage of Chinese language in a pool of small single-stranded DNA rings and its facile reading-out. Bull Chem Soc Jpn. 2021;94:53–59. doi: 10.1246/bcsj.20200201. [DOI] [Google Scholar]
- Wanunu M. Nanopores: a journey towards DNA sequencing. Phys Life Rev. 2012;9:125–158. doi: 10.1016/j.plrev.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanunu M, Sutin J, McNally B, Chow A, Meller A. DNA translocation governed by interactions with solid-state nanopores. Biophys J. 2008;95:4716–4725. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids: a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1097/BLO.0b013e31814b9304. [DOI] [PubMed] [Google Scholar]
- Wetterstrand K (2017) DNA sequencing costs: data from the NHGRI Genome Sequencing Program (GSP) http://www.genome.gov/sequencingcostsdata. Accessed 6 Nov 2021
- Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. Nucleotide sequence analysis of DNA: I. partial sequence of the cohesive ends of bacteriophage λ and 186 DNA. J Mol Biol. 1970;51:501–521. doi: 10.1016/0022-2836(70)90004-5. [DOI] [PubMed] [Google Scholar]
- Xie W, Chipman JG, Robertson DL, Erikson R, Simmons DL. Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing. Proc Natl Acad Sci. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi I, Ishida T, Fujisaki K, Takeda K-i. Fabrication of 3-nm-thick Si 3 N 4 membranes for solid-state nanopores using the poly-Si sacrificial layer process. Sci Rep. 2015;5:1–13. doi: 10.1038/srep14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Liu Y, Dai M, Yi X, Wang C. Controlling DNA translocation through solid-state nanopores. Nanoscale Res Lett. 2020;15:1–9. doi: 10.1186/s11671-020-03308-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko EC, et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nanotechnol. 2011;6:253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen DT. Despite Franklin's work, Wilkins earned his Nobel. Nature. 2003;425:15–15. doi: 10.1038/425015b. [DOI] [PubMed] [Google Scholar]
- Zeng X, et al. Nanopore technology for the application of protein detection. Nanomaterials. 2021;11:1942. doi: 10.3390/nano11081942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zereshki P, et al. Photocarrier dynamics in monolayer phosphorene and bulk black phosphorus. Nanoscale. 2018;10:11307–11313. doi: 10.1039/C8NR02540A. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xie Y, Liu Z, Wang X, Chai Y, Yan F. Two-dimensional material membranes: an emerging platform for controllable mass transport applications. Small. 2014;10:4521–4542. doi: 10.1002/smll.201401549. [DOI] [PubMed] [Google Scholar]
- Zhu C, Preissl S, Ren B. Single-cell multimodal omics: the power of many. Nat Methods. 2020;17:11–14. doi: 10.1038/s41592-019-0691-5. [DOI] [PubMed] [Google Scholar]