Significance
The liver is a source of excess lipid, atherogenic lipoproteins, and glucose in patients with type 2 diabetes. These factors predispose to micro- and macrovascular complications. The underlying pathophysiology is not well understood, and mechanistic insight into it may provide better tools to prevent, treat, and reverse the disease. Here, we propose an alternative explanation for this pathophysiologic conundrum by illustrating a transcriptional “logic” underlying the regulation of different classes of genes. These findings can be interpreted to provide an integrated stepwise model for the coexistence of lipid and glucose abnormalities in hepatic insulin resistance.
Keywords: diabetes, insulin sensitizers, drug failures, chromatin structure, animal models of human disease
Abstract
Abnormalities of lipid/lipoprotein and glucose metabolism are hallmarks of hepatic insulin resistance in type 2 diabetes. The former antedate the latter, but the latter become progressively refractory to treatment and contribute to therapeutic failures. It’s unclear whether the two processes share a common pathogenesis and what underlies their progressive nature. In this study, we investigated the hypothesis that genes in the lipid/lipoprotein pathway and those in the glucose metabolic pathway are governed by different transcriptional regulatory logics that affect their response to physiologic (fasting/refeeding) as well as pathophysiologic cues (insulin resistance and hyperglycemia). To this end, we obtained genomic and transcriptomic maps of the key insulin-regulated transcription factor, FoxO1, and integrated them with those of CREB, PPAR-α, and glucocorticoid receptor. We found that glucose metabolic genes are primarily regulated by promoter and intergenic enhancers in a fasting-dependent manner, while lipid genes are regulated through fasting-dependent intron enhancers and fasting-independent enhancerless introns. Glucose genes also showed a remarkable transcriptional resiliency (i.e., the ability to compensate following constitutive FoxO1 ablation through an enrichment of active marks at shared PPAR-α/FoxO1 regulatory elements). Unexpectedly, insulin resistance and hyperglycemia were associated with a “spreading” of FoxO1 binding to enhancers and the emergence of unique target sites. We surmise that this unusual pattern correlates with the progressively intractable nature of hepatic insulin resistance. This transcriptional logic provides an integrated model to interpret the combined lipid and glucose abnormalities of type 2 diabetes.
An impairment of the physiologic response to insulin, or insulin resistance, remains the central cause of type 2 diabetes and, together with declining insulin secretory capacity, its principal unmet treatment need (1). The pleiotropic nature of insulin resistance poses a therapeutic challenge by having different effects on different organs and different biological consequences within the same cell type, not to mention evidence of genetic heterogeneity (2, 3). Nowhere is this challenge more apparent than in the liver, a central organ in the pathogenesis of two key abnormalities in diabetes: increased production of atherogenic lipoproteins that increase the diabetic’s susceptibility to heart disease (1) and increased glucose production predisposing to microvascular complications (4). In addition, the progressive nature of the latter defect (5), together with declining β-cell function (6), likely underlies the therapeutic failure of antidiabetic drugs (7). Among drugs directly targeting hepatic glucose production, the diabetic pharmacopeia remains woefully limited to metformin (8).
Understanding whether the two central defects of hepatic insulin resistance harken back to a shared mechanism or arise independently has obvious implications for the discovery of new treatments (9). A useful conceptualization that has gained some consensus separates insulin signaling into FoxO1-dependent and Srebp1c-dependent branches. The former emanates from activation of Akt and allied kinases to regulate glucose metabolism, and the latter is relayed through mTOR to supervise lipid synthetic and turnover pathways (2). However, while the case for FoxO1 regulation of specific genes is strong, its genome-wide regulatory function in the broader context of the nutrient response has only been marginally addressed (10, 11). Therefore, the extent to which the lipid and glucose metabolic branches of insulin signaling share a common regulatory network remains unknown. Moreover, transcriptional networks are integrated, redundant units with overlapping functions. During fasting, as glucagon, catecholamine, and free fatty acids (FFA) levels rise, a host of factors are activated to modulate glucose and lipid mobilization. Besides FoxO, they include CREB, PPARs, CEBPs, and nuclear receptors (12). To address these questions, we undertook to generate a liver FoxO1 cistrome in different physiologic and pathophysiologic states and compare it with the CREB, PPAR-α, and glucocorticoid receptor cistromes. By leveraging a new mouse model developed for genome-wide interrogation of FoxO1 function (13), we discovered a FoxO1 transcriptional logic that provides insight into hepatic insulin action and resistance.
Results
In Vivo Features of Hepatic FoxO1 Translocation.
There is a dearth of primary data on the kinetics of hepatic FoxO1 localization in response to hormones and nutrients in the living organism. To optimize conditions for genome-wide chromatin immunoprecipitation sequencing (ChIP-seq), we performed immunohistochemistry in wild-type (WT) mice to determine the time and dose dependence of FoxO1 nucleocytoplasmic translocation in response to insulin. Insulin injection into the inferior vena cava (IVC) triggered rapid FoxO1 translocation that reached a plateau by 15 min (SI Appendix, Fig. S1A), with a median effective dose (ED50) of 0.02 U/kg (plasma level 0.4 ng/mL) (SI Appendix, Fig. S1B). In contrast, HNF4A remained nuclear throughout (SI Appendix, Fig. S1A). Thus, FoxO1 translocation is rapid and sensitive to physiological levels of insulin.
Next, we investigated translocation in response to fasting and refeeding. Following a physiologic 4-h fast, 1-h refeeding induced FoxO1 translocation (Fig. 1A). In contrast, a prolonged, 16-h fast resulted in decreased overall FoxO1 immunoreactivity. Subsequent refeeding for up to 4 h failed to translocate residual FoxO1 to the cytoplasm, while FoxO1 immunoreactivity increased and HNF4A immunoreactivity decreased after 2‐hr refeeding (SI Appendix, Fig. S1C). The reduced protein levels and delayed translocation are likely secondary to FoxO1 deacetylation (14–16). FoxO1 localization correlated with plasma glucose and insulin levels as well as liver Akt phosphorylation. Thus, rapid nuclear exclusion in the 4-h-fast/1-h-refeed design was associated with a modest rise of glucose and insulin levels (SI Appendix, Fig. S1D) and increased Akt phosphorylation (SI Appendix, Fig. S1E), whereas persistent nuclear localization in the 16-h fast/4-h refeed design was associated with hyperglycemia, hyperinsulinemia (SI Appendix, Fig. S1D), and reduced Akt phosphorylation (SI Appendix, Fig. S1E). Based on these findings, we selected the 4-h fast and 1-h refeed time points to assess the hepatic FoxO1 regulome.
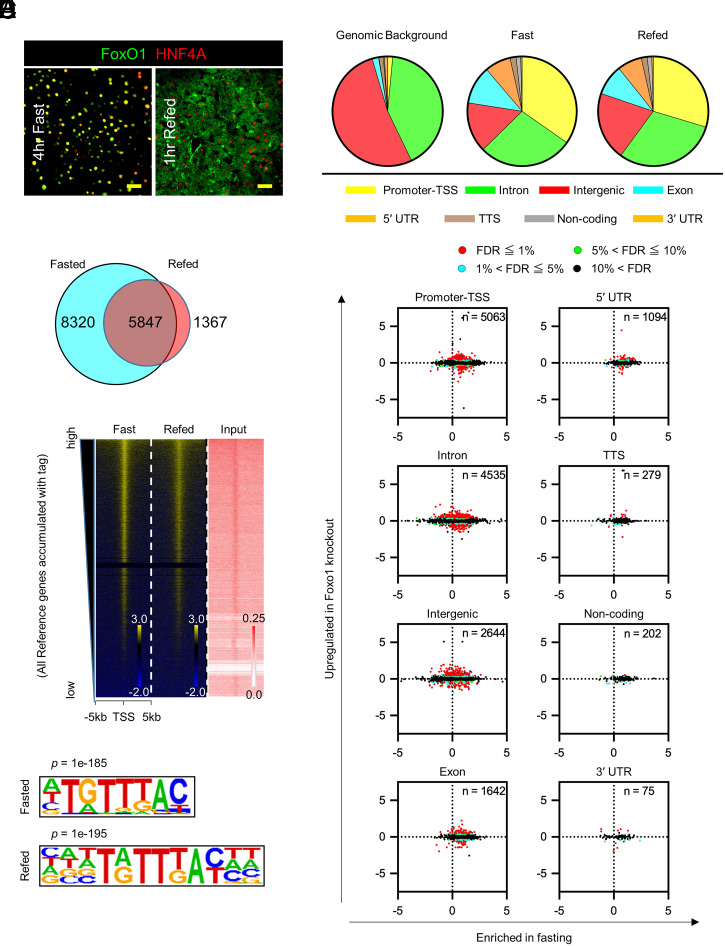
Fig. 1.
Distribution of genome-wide FoxO1 binding sites in the fast–refeed transition. (A) FoxO1 and HNF4-α immunohistochemistry in the liver. (Scale bar, 50 μm.) (B) Venn diagram of the number of FoxO1 peaks in fasted or refed conditions. (C) Distribution of FoxO1 peaks relative to annotated Reference Sequence (RefSeq) genes (color coded) compared with mouse genomic background. (D) Signal intensity plots of ChIP-seq data for FoxO1 compared to input chromatin. The highest level of binding occupancy of chromatin is at the top. (E) De novo motif analysis of the FoxO1 ChIP-seq. Logos of the recovered FoxO1 motif show position-specific probabilities for each nucleotide (P = 1×10−185 in fast, 1×10−195 in refeed). (F) Scatterplots of FoxO1 ChIP-seq peaks, expressed as log2 fold-change of FoxO1 tags between fast and refeed (horizontal axis) versus log2 fold-change of mRNA levels between WT and liver-specific FoxO1 knockout mice (vertical axis) for each genomic site. FoxO1 peaks detected in fasted or refed conditions were included in this analysis, and their number at each genomic annotation is shown inside each graph. Detailed information on peaks associated with genes whose FDR < 0.05 is in Dataset S1. Red = FDR < 1%; Blue = 1% ≤ FDR < 5%; Green = 5% ≤ FDR < 10%; Black = 10% ≤ FDR. See also SI Appendix, Figs. S1–S3.
FoxO1 Regulome during Fasting and Refeeding.
To study the genome-wide regulation of FoxO1 with fasting and refeeding, we interrogated genome occupancy by FoxO1 using an anti-GFP antibody in FoxO1-Venus knockin mice (13) for ChIP to overcome the limitations of anti-FoxO1 antibodies. As reported (13), anti-FoxO1 antibodies detected the FoxO1-Venus fusion protein encoded by the modified Foxo1 locus (SI Appendix, Fig. S2 A and B). Comparison between the two antibodies at known FoxO1 target genes (Igfbp1, G6pc, and Pck1) confirmed the specificity and superior sensitivity of the GFP antibody (SI Appendix, Fig. S2C) (17). We next compared ChIP-qPCR and ChIP-seq using GFP antibody in FoxO1‐Venus mice in the same conditions (SI Appendix, Fig. S2 D–H). Both approaches demonstrated similar regulation of FoxO1 binding to Igfbp1, G6pc, and Pck1 but not to the unrelated Fkbp5. As the results were internally consistent, we performed further analysis with GFP antibody.
Genome-wide FoxO1 ChIP peak calling detected ∼1.5 × 104 peaks; ∼8 × 103 peaks unique to fasting, ∼103 to refeeding, and 5 × 103 found in both conditions but to different extents (Fig. 1B). Greater than 30% of FoxO1 sites localized to promoters/transcription start sites (TSS) (Fig. 1C). Signal intensity plots demonstrated that refeeding cleared FoxO1 binding to autosomes (Fig. 1D and SI Appendix, Fig. S3A) regardless of the distance from TSS (SI Appendix, Fig. S3B). Known (SI Appendix, Fig. S3C) and de novo motif analyses (Fig. 1E and SI Appendix, Fig. S4) retrieved the FoxO1 motif TGTTTAC (13). This motif was found in 17 and 29.3% of FoxO1 sites in fasted and refed conditions, respectively. The same motif was found in fasted and refed conditions (SI Appendix, Fig. S3D) and was evenly distributed between 1 and –5 Kb from TSS (SI Appendix, Fig. S3E) (18).
Next, we integrated ChIP-seq and RNA-sequencing (RNA-seq) data into a hepatic FoxO1 regulome. To identify FoxO1-regulated messenger RNAs (mRNAs), we induced somatic ablation of FoxO1 in liver by injecting Foxo1lox/lox mice with adeno-associated viruses (AAV)-Cre (A-FLKO) and documented its completeness and specificity by mRNA measurements and Western blotting of different tissues (SI Appendix, Fig. S3 F and G). After 3 wk, A-FLKO mice showed higher insulin sensitivity of the fast–refeed response compared to controls (SI Appendix, Fig S5 A and B). When subjected to high-fat diet (HFD) to induce insulin resistance, A-FLKO mice showed higher insulin sensitivity as early as day 3 after initiation of the diet (SI Appendix, Fig. S5 C and D). After 4 wk of HFD, we confirmed a significant improvement of glucose tolerance, higher insulin sensitivity, and lower gluconeogenesis in A-FLKO mice compared to controls (SI Appendix, Fig. S5 E–G). For RNA-seq analysis, we isolated livers from 4-h–fasted A-FLKO and control (A-WT) mice 3 wk after AAV injection. We plotted the log2 difference in DNA binding (FoxO1 ChIP-seq peak number in fasted versus refed animals) versus the log2 difference in gene expression between A-WT and A-FLKO mice (differentially expressed genes, DEGs). Thus, the effect of genotype lies along the vertical axis and that of fasting along the horizontal axis (Fig. 1F and Dataset S1).
Contingency analyses showed that genes regulated by FoxO1 in the fasted state had FoxO1 DNA binding sites at promoters/TSS (183 of 198, or 92.4%), followed by introns (260 of 344, 75.6%) and intergenic sites (181 of 281, 64.4%), respectively (P < 0.0001). These data provide initial, suggestive evidence of a FoxO1 transcriptional logic (i.e., genes regulated by FoxO1 in a fasting-dependent manner have a greater frequency of FoxO1 sites in their promoter/TSS).
FoxO1 Regulates Metabolic Genes through Active Enhancers.
In addition to metabolism, FoxO1 regulates cellular maintenance in a fasting-independent manner (19). We sought to understand the transcriptional logic of these diverging functions. We hypothesized that basic cellular functions are regulated through core promoters (generally within 1 kb from TSS and associated with housekeeping genes and developmental TFs) (20). Conversely, we surmised that metabolic genes are regulated through tissue-specific enhancers (12, 21). To test the hypothesis, we marked active enhancers by overlaying H3K27ac and H3K4me1 sites (22) in fasting and refeeding with FoxO1 sites (SI Appendix, Fig. S6A). We categorized three types of sites: “promoter enhancers” that mapped to promoter-TSS, “active enhancers” in regions other than promoters, and “enhancerless” regions.
Of 5,303 active enhancers colocalizing with FoxO1 sites genome wide, 2,975 were unique to fasting, 1,022 to refeeding, and 1,306 were found in both conditions (SI Appendix, Fig. S6B). The majority of FoxO1 sites mapped to active enhancers and a minority to promoter enhancers (Fig. 2A). The latter were more responsive to fasting/refeeding, with 81.5% (564/692) being cleared of FoxO1 in response to refeeding, while only ∼60% of active enhancers were (59.6% in intergenic regions, or 804/1348; and 67.9% in introns, or 1085/1597) (P < 0.0001).
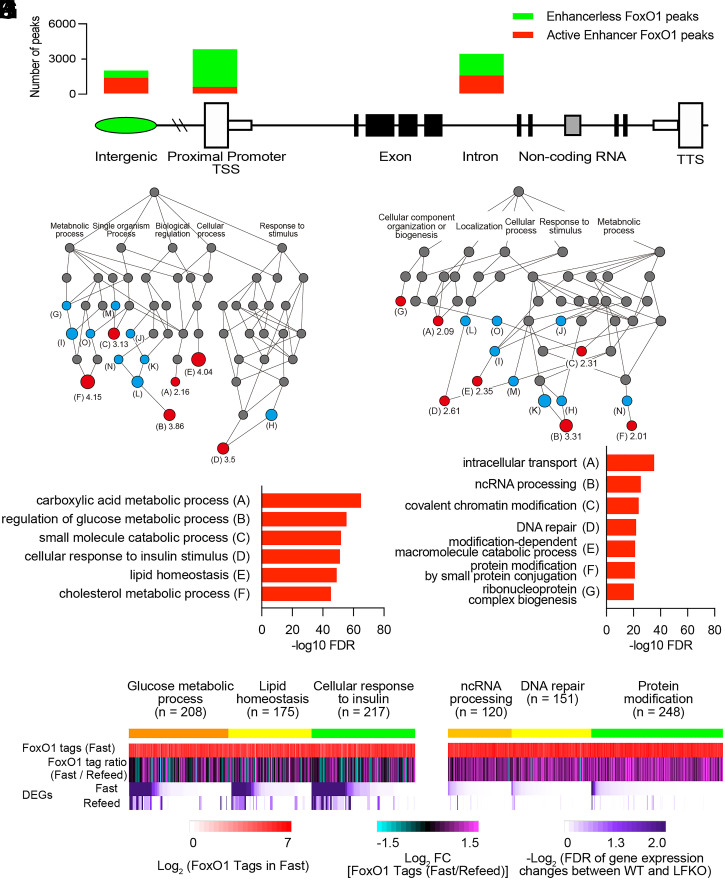
Fig. 2.
Comparison of the features of FoxO1 sites in active enhancers versus enhancerless promoter/TSS. (A) Bar diagram of FoxO1 in active enhancers (red) and in enhancerless (green) genomic location. The number of active enhancer/enhancerless at each genomic location is intergenic = 1795/849, 5′ UTR = 128/966, promoter/TSS = 760/4303, exon = 384/1258, intron = 2034/2501, noncoding = 44/158, TTS = 105/17, and 3′ UTR = 53/22. (B) DAG derived from GO of biological processes associated with 5,305 FoxO1 active enhancers by Genomic Regions Enrichment of Annotations Tool (GREAT) GO tools. Letters correspond to the groups shown in C and SI Appendix, Fig. S3C. Numbers indicate the term’s fold-enrichment. Red circles: fundamental ontologies in the hierarchy listed in C. Blue circles: additional enriched ontologies. Gray circles: parent ontologies. (C) List of GO in B and their –log10 FDR. (D–F) Heatmap alignments of ChIPseq FoxO1 binding in fast (D), fast/refeed ratio (E), and FDR of gene expression changes between WT and liver FoxO1 knockout mice (F) in GO related to glucose metabolic processes, lipid homeostasis, and cellular response to insulin genes as listed in B and C. (G and H) Same GO analysis as in B and C applied to 4,303 enhancerless FoxO1 sites in promoter/TSS. (I–K) Heatmap alignments, as in D–F, of GO related to ncRNA processing, DNA repair, and protein modification as listed in G and H. See also SI Appendix, Fig. S6.
Next, we interrogated differences between genes with FoxO1 sites in active enhancers versus promoter/TSS and visualized them using directed acyclic graphs (DAG) (23). FoxO1 sites in active enhancers were overwhelmingly enriched in metabolic genes, with the top three ontologies being glucose metabolism, lipid homeostasis, and insulin response (false discovery rate [FDR] 10−40 to −70) (Fig. 2 B and C and SI Appendix, Fig. S6C). These gene ontologies showed a strong correlation with the fasting/refeeding ratio of FoxO1 DNA binding (Fig. 2 D and E and SI Appendix, Fig. S6D) (b = 0.09, P < 0.0001). In contrast, FoxO1 sites in promoter/TSS included genes related to intracellular transport, DNA repair, non-coding RNA (ncRNA) processing, and protein degradation (Fig. 2 G and H and SI Appendix, Fig. S6E) (FDR 10−20 to −40). These sites showed a lesser correlation with the fasting/refeeding ratio of FoxO1 binding (Fig. 2 I and J and SI Appendix, Fig. S6F) (b = 0.29, P < 0.0001). With regard to mRNA changes, metabolic genes were profoundly altered following FoxO1 ablation (Fig. 2F), while genes lacking active enhancers were largely unaffected by FoxO1 ablation (Fig. 2K). As a control, the active enhancer marker, H3K27ac, was unaffected by fasting and refeeding (b = 0.91, P < 0.0001) (SI Appendix, Fig. S6G). There were no differences in the FoxO1 motif associated with enhancer versus enhancerless sites (SI Appendix, Fig. S7).
These results indicate that the cell maintenance and metabolic functions of FoxO1 are ruled by distinct transcriptional logics: the former are governed by core promoters independently of fasting/refeeding, whereas the latter are governed by active enhancers and show a strong dependence on nutritional status (19).
FoxO1 Regulates Triglyceride and Cholesterol Genes through Introns.
The second most common genomic annotation of FoxO1 binding sites mapped to introns (Fig. 1C). The corresponding genes showed changes to their mRNAs following FoxO1 ablation, indicating that these sites are functionally relevant (Fig. 1F and Dataset S1). Statistical analyses of annotation distribution demonstrated that triglyceride metabolism genes were significantly enriched in introns (5 to 50 kb and –50 to –5 kb from TSS) (Fig. 3A), while glucose metabolism genes were enriched in intergenic and distal gene regions (50 to 500 kb from TSS and 5 to 50 kb regions) (Fig. 3 B and C) (P = 0.03). These differences were not the result of different length of glucose versus triglyceride genes (SI Appendix, Fig. S8A).
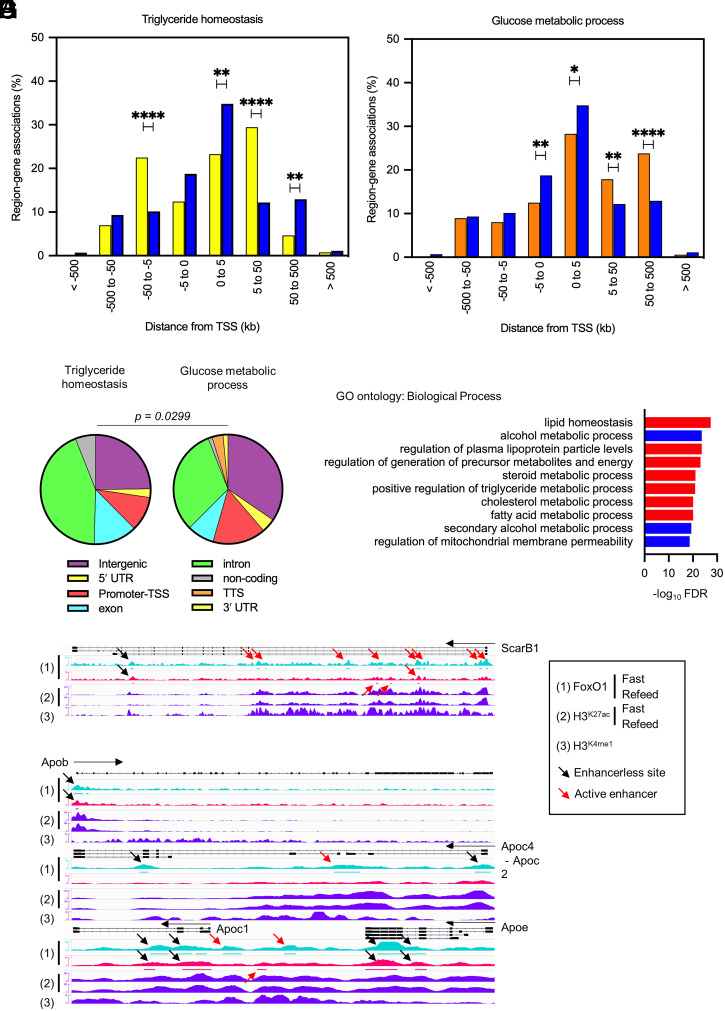
Fig. 3.
Different FoxO1 binding logic in triglyceride versus glucose metabolism genes. (A and B) Comparison between region–gene associations of triglyceride homeostasis (yellow bar) (A) or glucose metabolic process (orange bar), with set-wide FoxO1 binding sites (blue bar) as detected by FoxO1 ChIP-seq in fasted or refed conditions, binned by orientation and distance from TSS. *P < 0.05; **P < 0.01; ****P < 0.0001 by χ2 test. (C) Distribution of FoxO1 binding sites associated with triglyceride homeostasis or glucose metabolic process genes according to genomic annotation as in Fig. 1C. P < 0.03 by contingency analysis. (D) GO analysis of biological processes associated with 4,535 FoxO1 binding sites in introns using GREAT GO tools. (E–G) IGV Genome browser views of FoxO1 peaks and associated H3K27ac and H3K4me1 histone marks at selected apolipoprotein clusters (Apob and ApoC2/C4/C1/E Apob, Apoc4-c2, Apoc1, and Apoe) and ScarB1. Signals are normalized for the comparisons between fasted and refed conditions. FoxO1 signals are aligned with peak regions. Red arrows indicate active enhancers as detected by H3K27ac and H3K4me1 signals. FoxO1 peaks in introns are listed in Dataset S2. See also SI Appendix, Fig. S8.
Intron-enriched genes included nearly exclusively lipid, lipoproteins, and cholesterol genes (Fig. 3D). Nearly half of intron sites were associated with active enhancers (Fig. 2A). Next, we analyzed the functional regulation of these sites: are they modulated with fasting/refeeding, and do their mRNA change following FoxO1 ablation? Linear regression analyses demonstrated that FoxO1 binding to introns marked by active enhancers was more sensitive to fasting/refeeding than enhancerless introns (b = 0.19 versus 0.06) (SI Appendix, Fig. S8 B and C) and that the encoded mRNAs were more likely to be altered following FoxO1 ablation. For example, ScarB1 (24) (Fig. 3E), Angptl4, and Angptl8 (25) (Dataset S2) showed fasting-induced binding to introns with active enhancers as well as altered mRNA levels upon FoxO1 ablation. In contrast, the ApoB, ApoA1/C3/A4/A5, and C2/C4/C1/E clusters (26) showed fasting-independent FoxO1 binding to introns and preserved mRNA expression following FoxO1 ablation (Fig. 3 F and G and Dataset S2).
The transcriptional logic of the FoxO1 regulome emerging from these analyses suggests that a majority of glucose metabolism genes are governed through active enhancers and respond to fasting, whereas a majority of triglyceride, lipoprotein, and cholesterol genes are ruled by two distinct transcriptional logics: fasting-dependent active enhancers in introns and fasting-independent enhancerless introns.
We hypothesized that this differential regulation underlies hepatic insulin resistance. We tested the hypothesis using three models: 1) the ability of these genes to undergo compensatory changes in response to inactivation of FoxO1 function as a surrogate measure of insulin action (SI Appendix, Fig. S1), 2) comparative genomic analyses with other fasting-induced TFs to identify functional partners and redundancies, and 3) genome-wide FoxO1 ChIP-seq in insulin-resistant/hyperglycemic mice.
Transcriptional Resiliency of Glucose Metabolic Genes.
First, we sought to determine whether different modalities of FoxO1 regulation (intergenic and promoter/TSS versus intron) were associated with differential compensation by other TFs that may affect the pathophysiology of insulin resistance. To this end, we compared mRNA expression differences in a constitutive versus adult-onset somatic ablation of hepatic FoxO1 (27–29) and correlated these differences with ChIP-seq data.
We generated Alb-Cre:FoxO1fl/fl mice to induce constitutive hepatic FoxO1 ablation (C-FLKO) and compared gene expression differences between adult-onset (A-FLKO, described in Fig. 1) and constitutive (C-FLKO) knockouts according to nutritional state (fast versus refeed), genotype (WT versus FoxO1 ablation), and timing of ablation (A-FLKO versus C-FLKO) (SI Appendix, Fig. S9). t-distributed stochastic neighbor embedding (t-SNE) plots showed large differences in fasted versus refed gene expression patterns between A-FLKO and their matched controls (A-WT). In contrast, the differences between C-FLKO and C-WT were considerably blunted (Fig. 4A). We plotted fold-change and average gene expression in each WT/knockout pair as log-intensity ratios (M-values) versus averages (A-values). The number of differentially regulated genes in fasted C-FLKO mice decreased by 60% compared to A-FLKO (227 versus 585), whereas it was similar in refed conditions (301 versus 243) (Fig. 4 B and E and Dataset S3). Thus, a first conclusion is that chronic compensatory changes partially mask the effect of FoxO1 ablation on the fasting response.
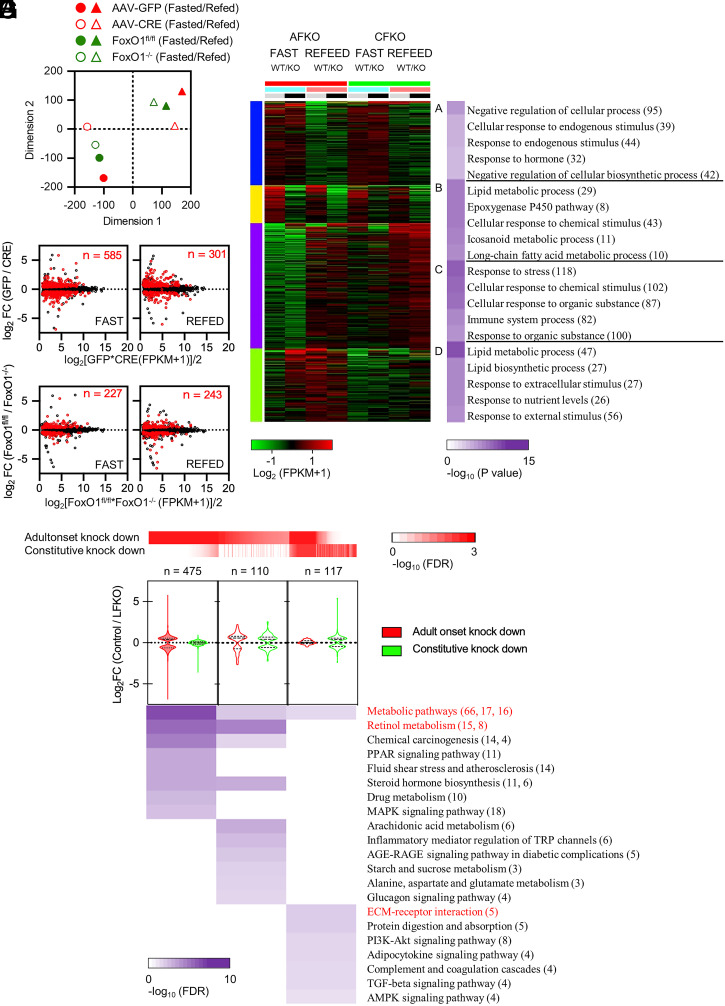
Fig. 4.
Resilience analysis of FoxO1-regulated genes. (A) t-SNE plot of RNA-seq data (n = 8). Circles indicate fasted animals and triangles refed animals. Filled red symbols: AAV-GFP–injected animals (A-WT in the text); empty symbols with red border: AAV-CRE–injected animals (A-FLKO in the text); green filled symbols: Foxo1loxp/loxp (C-WT in the text); empty symbols with green border: Alb-Cre/Foxo1flox/flox (C-FLKO in the text). (B–E) MA [log-intensity ratios (M-values) vs. averages (A-values)] scatterplots of average expression levels versus log2 fold-change induced by FoxO1 ablation in tag count within exons of ensemble gene bodies in fasted (B) or refed (C) A‐FLKO and fasted (D) or refed (E) C‐FLKO. Red dots represent DEGs (FDR ≤ 0.05). The number of DEGs is indicated in each box. (F) Enrichment analysis of k-Means clusters with molecular pathways underlying each category with top 1,000 variable genes among all samples used in A by iDEP tools. (G) GO analysis of DEGs in fasted conditions, shown in B and D, by Shiny GO tools. Red heatmap shows FDR of genes in A‐FLKO or C‐FLKO. Violin plots show log2 fold-change of gene expression between control and A-FLKO (red) or C-FLKO (green) for DEGs. Number of DEGs is indicated at the top. Purple heatmap shows FDR of each ontology described next to it. Red-colored ontologies indicate the top enriched term in each category. The number of genes in each ontology is shown in parenthesis in F and G. DEGs are listed in Dataset S3. See also SI Appendix, Fig. S9.
Next, we determined the ontologies of genes undergoing compensatory changes as a function of nutritional status (fast versus refeed), genotype (knockout versus WT), and timing of ablation (A-FLKO versus C-FLKO) (Fig. 4F). We identified four ontology groups (A through D). Immune, chemical, and stress response genes were either induced by fasting (Group A) or refeeding (Group C), but neither was affected by FoxO1 ablation. In contrast, lipid and fatty acid metabolism genes (Group B) decreased with fasting following FoxO1 knockout regardless of its timing (constitutive versus adult-onset). Interestingly, metabolic pathways, retinol and PPAR signaling, and steroid function genes (Group D) were impaired in fasted A-FLKO mice but not in C-FLKO mice (Fig. 4G). These genes undergo compensation following constitutive FoxO1 ablation; thus, we investigated them at a more granular level.
Group D included classical FoxO1 targets regulating insulin signaling (Irs2), gluconeogenesis (G6pc, Pck1, and Ppargc1a), glycolysis (Gck, Pfkfb1 and 3, and Ldhd), ketogenesis (Hmgcs1), and glucose/fatty acid partitioning (Pdk4) (Dataset S3). Other genes undergoing compensation included 17 members of the Cyp2 family and 6 members of the Cyp4 family of drug metabolizing enzymes and lipid metabolic and atherogenic genes (Angptl8 and lpl), Fgf21, Gdf15, Klf15, Slc13a5 (encoding INDY), Enho (encoding Adropin), Fmo3, and Asns.
Among genes involved in fatty acid synthesis or oxidation, apolipoproteins, and cholesterol trafficking, only Vldlr and Lpin1 showed >50% compensation. Thus, FoxO1-regulated glucose metabolism genes, as well as several metabolically important genes, undergo a compensatory response following constitutive FoxO1 ablation, whereas the majority of lipid metabolism genes don’t. We termed this finding transcriptional resiliency.
A FoxO1/PPAR-α Signature of Fasting-Inducible Enhancers.
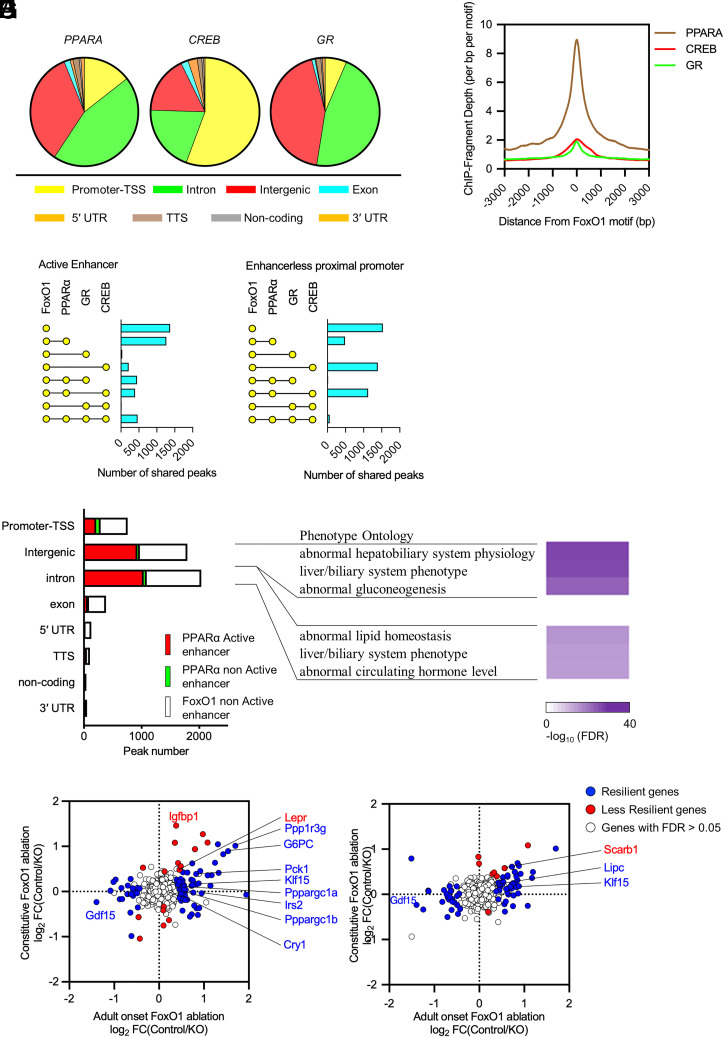
Transcriptional regulation of the fasting response involves several TFs, including CREB, GR, and PPAR-α (12). To understand the integration of these networks with FoxO1 and their potential role in the transcriptional resiliency observed after FoxO1 ablation, we compared the present dataset with published genome-wide ChIP-seq of these three factors (30, 31). Analyses of peak distribution demonstrated that CREB peaks are enriched at promoters, while GR and PPAR-α are enriched in introns and intergenic regions (Fig. 5A). When overlaid with FoxO1 sites, we found that colocalization of FoxO1/PPAR-α (Fig. 5B) prevailed at active intergenic and intron enhancers, in which approximately half of FoxO1 sites are shared with PPAR-α (Fig. 5 C and E). In contrast, trinomial combinations FoxO1/CREB/PPAR-α prevailed at enhancerless promoters (Fig. 5 D and E). At active enhancer sites, 11.2% of unique FoxO1 sites were associated with changes in gene expression following FoxO1 ablation, whereas only 5.4% of shared sites (FoxO1 and CREB or PPAR-α) were (P < 0.0001, SI Appendix, Table S1). This difference was not seen in enhancerless sites (6.09 versus 5.93%, respectively) (P = not significant [NS], SI Appendix, Table S1). Gene ontology analyses (GO) (Fig. 5F) showed that abnormal gluconeogenesis is the most significant annotation of FoxO1/PPAR-α shared intergenic peaks (FDR = 2.22 × 10−31), while lipid homeostasis is the most significant in introns (FDR = 2.01 × 10−21).
Fig. 5.
Comparative analysis of fasting-inducible transcriptional factors. (A) Distribution of PPAR-α, CREB, and GR binding sites in fasted conditions. (B) Peak plot mapping the overlap of the FoxO1 (Fig. 1E) and PPAR-α, CREB, and GR peaks. (C and D) Intersection analyses of active enhancer (C) and enhancerless proximal promoter (D) in FoxO1 and PPAR-α, CREB, or GR enhancer peaks in fasting conditions. (E) Proportion of PAAR-α peaks with/without active enhancer marks in FoxO1 active enhancers in fasting conditions according to genomic annotation. (F) Heatmap with associated FDR of phenotype ontology terms of shared FoxO1/PPAR-α active enhancers (red bars in E) in intergenic regions and introns. (G and H) Resiliency plots of genes associated with shared FoxO1/PPAR-α active enhancers in intergenic regions (G) and introns (H). Plot show log2 fold-change induced by adult-onset versus constitutive liver FoxO1 ablation. Resilient genes (FDR ≤ 0.05 in AFKO or CFKO mice, showing lower fold-change and higher FDR value in CFKO mice than AFKO mice) are indicated by blue dots; nonresilient genes (FDR ≤ 0.05 in AFKO or CFKO mice) are marked by red dots, FDR > 0.05 in both mice by white dots. DEGs are listed in Dataset S4.
Next, we asked whether coregulation by FoxO1 and PPAR-α can explain the resiliency of gene expression. We plotted each FoxO1/PPAR-α shared peak with active enhancer marks versus changes to mRNA encoded by the respective genes in A-FLKO and C-FLKO (Fig. 5 G and H). In both intergenic (Fig. 5G) and intron (Fig. 5H) sites, >80% of FoxO1/PPAR-α coregulated genes showed a compensatory response to constitutive FoxO1 ablation (75 of 92 and 68 of 76, respectively). In intergenic sites, we found notable resilient glucose metabolism genes, such as Pck1, G6pc, Irs2, Ppargc1a and b, Ppp1r3g, Cry1, Gdf15 (32), and Klf15 (33) (Fig. 5G). In introns, we found lipid genes, such as Gdf15 and Lipc (Fig. 5H and Dataset S4). Thus, shared FoxO1/PPAR-α enhancers are more likely to undergo compensation when FoxO1 is inactive.
Enhancer Spreading of FoxO1 Binding in Insulin Resistance/Hyperglycemia.
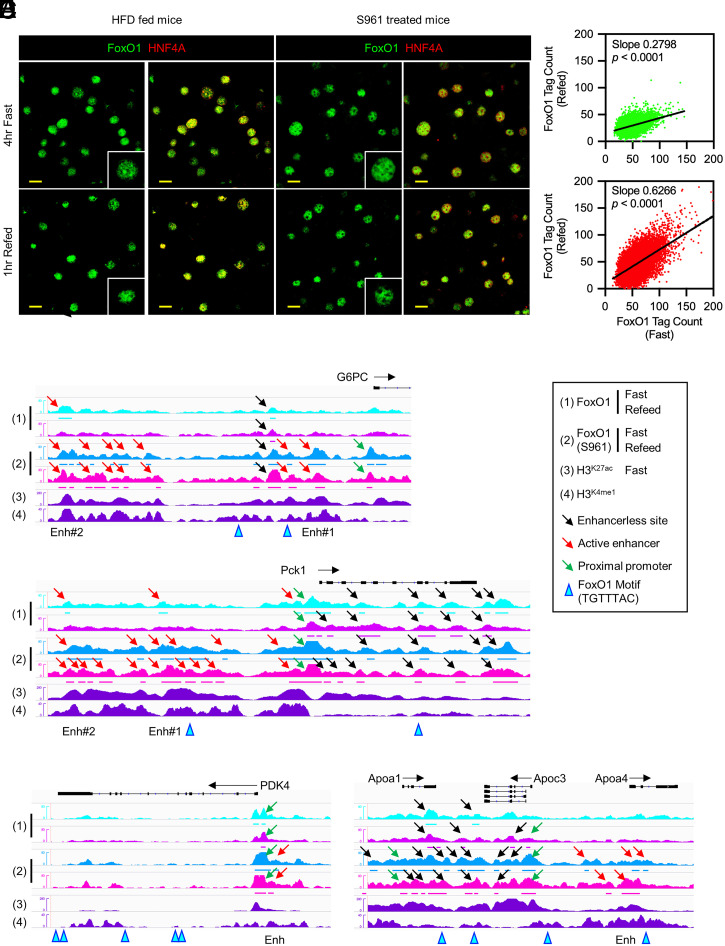
To evaluate the effects of insulin resistance and hyperglycemia on the FoxO1 regulome, we subjected FoxO1-Venus mice to HFD or treatment with the insulin receptor antagonist S961 (34). Both interventions impaired refeeding-induced FoxO1 translocation (Fig. 6A) and reduced refeeding-induced Akt phosphorylation, although with different time courses: 4 wk for HFD feeding and 1 d for S961 treatment (SI Appendix, Fig. S10A). In addition, phosphorylation of Akt after refeeding was blunted on day 7 after initiation of S961 treatment (SI Appendix, Fig. S10B). Plasma cholesterol levels were increased by HFD feeding but not by S961 treatment. Plasma triglycerides (TG) and non-esterified fatty acids levels were unaffected (SI Appendix, Fig. S10 C–E). Surgical implantation of osmotic pumps did not affect weight or food intake (SI Appendix, Fig. 10 F and G). S961 treatment induced marked hyperglycemia 1 d after initiation of treatment, peaked at day 3, and decreased slightly thereafter (SI Appendix, Fig. 10H). Plasma insulin levels peaked at day 1, followed by a decrease (SI Appendix, Fig.10I). These data indicate a rapid onset of insulin resistance within 24 h of initiation of treatment. To minimize the confounding effects of chronic S961 treatment, we selected day 1 for further analysis, when the effects on refeeding-induced glucose (SI Appendix, Fig. 10J) and insulin levels were already evident (SI Appendix, Fig. S10K).
Fig. 6.
Transition of FoxO1 binding sites in insulin resistance. (A) Immunohistochemistry of FoxO1 and HNF4-α after 4-h fasting or following 1-h refeeding in HFD-fed mice or insulin receptor antagonist (S961)-treated mice. (Scale bar, 20 μm.) (B) Scatterplots showing linear regression analysis of FoxO1 tag count in fasted versus refed conditions. Green: vehicle; red: S961-treated mice. (C–F) IGV Genome browser views of FoxO1 peaks with or without S961 treatment and associated H3K27ac and H3K4me1 marks of at G6pc, Pdk4, Angptl4/8, and ApoA1/C3/A4. “Enh” indicates the sites evaluated by ChIP-qPCR in the HFD feeding animal model. See also SI Appendix, Figs. S11 and S12.
We performed genome-wide ChIP-seq in livers of 4-h–fasted/1-h–refed mice treated with S961 versus vehicle for 1 d. Analysis of FoxO1 binding showed reduced clearance of sites in S961-treated mice than in vehicle controls after refeeding (b = 0.28 versus 0.62, Fig. 6B), consistent with impaired translocation (Fig. 6A). Importantly, we found new FoxO1 binding sites at active enhancers associated with glucose (intergenic/promoter/TSS) versus lipid genes (intron) (SI Appendix, Fig. S11A). Ontology analysis demonstrated significant enrichment in metabolic genes as well as PPAR-α signaling pathways (SI Appendix, Fig. S11B). We examined FoxO1 binding to representative genes of these two main transcription logics. Examples included intergenic/promoter enhancers of glucogenic (G6pc, Pck1, and Klf15) (Fig. 6 C and D and SI Appendix, Fig. S12A) and glucose–lipid metabolic partitioning genes (Pdk4) (Fig. 6E), as well as intron enhancers of lipid/cholesterol genes (ApoA1/C3/A4 and Scarb1) (Fig. 6F and SI Appendix, Fig. S12 B and C). These novel FoxO1 peaks were unaffected by fasting/refeeding and included sites with and without FoxO1 binding motifs. In contrast, novel FoxO1 peaks at enhancerless sites occurred less frequently. We confirmed these findings by targeted ChIP-qPCR in the less extreme model of HFD-fed mice (SI Appendix, Fig. S13). Thus, insulin resistance and hyperglycemia bring about an ectopic, dysregulated binding of FoxO1 at enhancer sites, which we term enhancer spreading.
Discussion
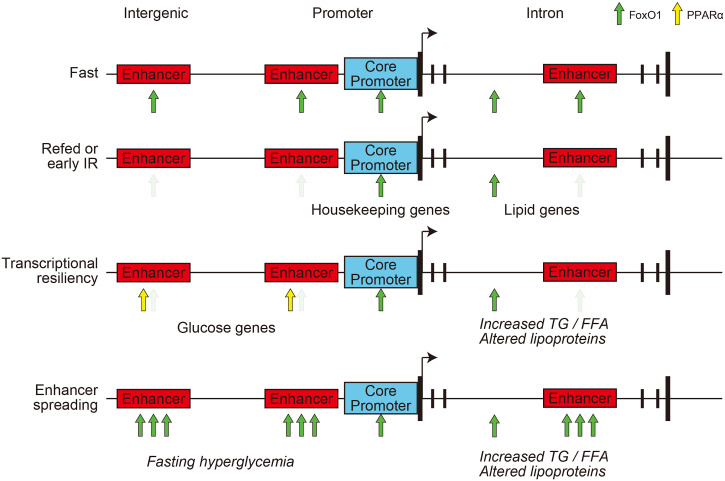
The present study provides insight into the differential regulation of glucose and lipid metabolism in response to nutrient changes and in insulin resistance, using a transcriptional logic model. There are obvious nontranscriptional components to these pathophysiologic states that are partly cell-nonautonomous (35), but the present study was designed to integrate the regulome of multiple TFs, including FoxO1, with the salient pathophysiologic features of hepatic insulin action and resistance. The main conclusions are the following: 1) The transcriptional logic of FoxO1 recasts the bifurcating model of insulin signaling to lipid versus glucose metabolism (36) by showing that glucose metabolic genes are governed by intergenic and promoter/TSS enhancers and lipid genes by a bipartite intron logic that includes fasting-dependent intron enhancers and fasting-independent enhancerless introns; 2) active enhancers of glucose metabolic genes show transcriptional resiliency, likely through shared PPAR-α/FoxO1 regulatory elements; and 3) insulin resistance and hyperglycemia result in the spreading of FoxO1 binding to enhancers, resulting in quantitative and qualitative abnormalities of FoxO1 marks (13). Based on these findings, we propose this model (Fig. 7): in the physiologic fasting/refeeding transition, FoxO1 is cleared more efficiently from enhancer-containing sites than from enhancerless sites. As the former are more tightly associated with glucose genes and the latter with lipid/lipoprotein genes, in the initial stages of insulin resistance, glucose genes can still be regulated, while regulation of lipid genes is impaired. This differential sensitivity can explain why lipid/lipoprotein abnormalities chronologically precede hyperglycemia in the progression of diabetes (37). As insulin resistance progresses, the gradual compensation of glucose versus lipid genes in response to chronic versus adult-onset FoxO1 ablation (transcriptional resiliency) can be interpreted to suggest that glucose genes can gradually become FoxO1-independent, allowing other transcription factors (likely PPAR-α) to induce their expression and hence glucose production to increase. In the clinically overt stage of the disease, as insulin resistance increases, FoxO1 binding to ectopic (or low-affinity) enhancers worsens fasting hyperglycemia and may possibly underlie therapeutic failures. The proposed model integrates in vivo pathophysiological and cell biological data with genome-wide assessments to explain a clinical conundrum that has important practical implications for treatment and drug development (1). This model also addresses two criticisms leveled at the FoxO-centric view of insulin action: 1) that FoxO1 sensitivity to insulin makes it an unlikely candidate as a mediator of insulin resistance (38) and 2) that transcription of candidate glucogenic genes alone does not fully explain increased hepatic glucose production (39). Indeed, the gamut of FoxO1 targets includes most genes involved in insulin action, and the failure to detect abnormalities in their expression following constitutive somatic ablation of FoxO1 can be explained by their resiliency.
Fig. 7.
Model of FoxO1 transcriptional logic in the pathogenesis of insulin resistance. In normal conditions, FoxO1 is cleared upon refeeding from resilient enhancers, enriched in glucose metabolism genes, but not in introns, and enriched in lipid metabolism genes. With the onset of insulin resistance, compensatory hyperinsulinemia can still clear FoxO1 from resilient enhancers, but not from introns, increasing serum lipoprotein and triglyceride levels. As insulin resistance progresses, compensation by PPAR-α and spreading of FoxO1 binding to additional sites increases expression of glucose metabolic genes, leading to fasting hyperglycemia with dyslipidemia.
To demonstrate a distinctive FoxO1 transcriptional logic, we decisively leveraged the ability to examine FoxO1 targets by genome-wide ChIP-seq (13). Previous studies have been limited by the sensitivity of available FoxO1 antibodies and have therefore detected fewer FoxO1 binding sites (10, 11, 40). There is a partial dissociation between the ChIP results, indicating that FoxO1 is still bound at several sites after refeeding, and the immunofluorescence that shows FoxO1 nuclear exclusion. However, ChIP is more sensitive than immunohistochemistry, being based on PCR amplification, and can detect lower levels of FoxO1 protein. The formation of different molecular complexes likely underlies the different modes of FoxO1 action. In this regard, we have previously shown that SIN3a is the FoxO1 corepressor at glucokinase, providing a mechanistic precedent for gene-specific targeting (9). The preferential regulation of FoxO1 by fasting/refeeding at active enhancers likely results from intrinsic and extrinsic factors such as higher DNA accessibility at active enhancers (41) and active enhancer–promoter interactions (42) that affect assembly of preinitiation complexes, initiation of transcription by RNA polymerase II, or transcription bursting (20).
Following FoxO1 ablation, expression of its targets can be compensated for by transcription factors acting synergistically, through its paralogue FoxO3, or reorganization of chromatin accessibility at sites where FoxO1 acts as a pioneer transcription factor (43), as shown with other FoxO isoforms (28). Interestingly, genes associated with glucose metabolism (G6pc, Pck1, Ppargc1a, Pdk4, and Klf15), but not those regulating general cellular responses, are selectively compensated for following FoxO1 ablation. FoxO1 peaks in these genes are cleared by refeeding but not in insulin-resistant conditions. These genes have been shown to play a role in diabetes in studies with insulin-resistant mice (27, 44–46). Among the genes involved in atherogenesis, Lpl and Angptl8 (47, 48) were affected only in A-FLKO mice. The distinct compensatory response between glucose and lipid genes can partly explain lipid abnormalities observed with changes in hepatic FoxO1 function, such as FoxO1 knockout or expression of constitutively active FoxO1 (29, 49).
There are parallels between our findings and recent evidence that immunocyte differentiation is controlled by an enhancer- or core promoter–driven logic (50). The former activity is cued by the overall activity of distal enhancers, while the latter is aligned with promoters. Although it remains unclear whether core promoters and enhancers represent different entities or synergistically regulate transcriptional bursting, enhancers are thought to be tissue-specific, consistent with our conclusion that they confer specificity on the metabolic functions of FoxO1 (21). Further work will be required to understand mechanistically the distinct binding affinities of FoxO1 to enhancer versus enhancerless regions in physiologic or insulin-resistant conditions. As there are no significant differences in the consensus FoxO1 binding motif of either region (SI Appendix, Fig. S7), we suggest that posttranscriptional modifications of the various components of the transcriptional complex or chromatin accessibility play a role in this process. With regard to the failure of Akt to induce FoxO1 translocation in insulin-resistant conditions (Fig. 6A and SI Appendix, Fig. S10A), this is consistent with prior studies of a FoxO1/Akt homeostatic loop, possibly mediated by Sestrins (51, 52). It is likely that phosphorylation by JNK and MST1 (53) and/or acetylation (15, 49) can account for the failure to exclude FoxO1 from the nucleus, as well as for the differential binding to enhancer versus enhancerless sites.
Our comparative analysis provides evidence of cooperative and noncooperative interactions with GR, CREB, and PPAR-α, the latter involving up to half of the FoxO1 sites in active enhancers. The extensive sharing of intergenic active enhancers of glucose genes by FoxO1 and PPAR-α dovetails with the different physiologic cues regulating these two TFs. During fasting, glycogenolysis precedes gluconeogenesis and the generation of FFA substrates that activate PPAR-α (54). Thus, we envision that FoxO1 and PPAR-α act in a physiologic relay to ensure continuity between the early and late fast. The significant overlap between FoxO1 and PPAR-α may also provide an explanation for the relatively mild phenotypes of liver-specific inactivation of FoxO1 (27) and PPAR-α (55). The interplay of FoxO1 and PPAR-α can also provide a mechanism to integrate direct effects of insulin on glucose production (mediated through FoxO1 nuclear exclusion) with indirect effect mediated by adipose-derived FFA that modulate PPAR-α. T.K. and D.A. have generated double liver-specific knockout mice to validate this hypothesis. Functional elucidation of their interactions, including validation of active enhancers for selected genes, will be important to determine key targets in glucose metabolism and their role in diabetes pathogenesis.
Experimental Model and Subject Details
Animals.
Mice were housed in a climate-controlled room on a 12-h light/dark cycle with lights on at 7:00 AM and off at 7:00 PM and were fed standard chow (PicoLab rodent diet 20, 5053; Purina Mills). Male mice of C57BL/6J background aged 8 to 12 wk were used. FoxO1-Venus mice have been described (13, 56). Briefly, to express GFP (Venus), we obtained the pCAG:myr-Venus plasmid. A 15-amino-acid linker sequence was placed between the carboxyl terminus of FoxO1 and N terminus of Venus to alleviate steric hindrance. We used bacterial artificial chromosome recombineering to generate FoxO1-Venus embryonic stem (ES) cells. To generate constitutive liver-specific FoxO1 knockouts, we crossed FoxO1lox/lox and Albumin-cre (57) transgenic mice. Adult-onset liver-specific FoxO1 knockout mice were generated by injection of 1 × 1011 purified viral particles (AAV8.TBG.eGFP or AAV8.TBG.Cre, Penn Vector Core) per mouse via tail vein. We performed metabolic analysis or killed animals on day 21 postinjection. To assess FoxO1 localization and other liver parameters, we took organs from 4-h–fasted (10:00 AM to 2:00 PM) or 4-h–fasted/1-h–refed mice. For prolonged fasting experiments, we removed food overnight (6:00 PM to 10:00 AM). Mice were killed 0, 1, 2, or 4 h after refeeding. For insulin treatment, we anesthetized 16-h–fasted mice with ketamine (100 mg/kg) and xylazine (10 mg/kg) intraperitoneal (i.p.), followed by injection of 1 U/kg insulin (NovoLog, Novo Nordisk) in the IVC. We collected blood and took the liver before and after insulin injection. Blood glucose was measured using CONTOUR NEXT ONE, Ascensia, and insulin with a mouse-specific enzyme-linked immunosorbent assay kit (Mercodia). All animal experiments were in accordance with NIH guidelines, approved and overseen by the Columbia University Institutional Animal Care and Use Committee.
In Vivo Metabolic Studies.
We used male mice aged 8 to 12 wk. We performed insulin, glucose, and pyruvate tolerance tests after a 4-h (10:00 AM to 2:00 PM) or 16-h (6:00 PM to 10:00 AM) fast using intraperitoneal injection of 7.5 U/kg insulin (Humalog, Lilly), 2 g/kg glucose, or pyruvate. Blood glucose measurements were made from tail vein blood.
Primary Hepatocyte Isolation.
Primary hepatocyte isolation was performed as described (58). We anesthetized male mice with ketamine (100 mg/kg) and xylazine (10 mg/kg) i.p., cannulated the IVC with a 24-gauge catheter (Exel International), and infused 50 cc EGTA-based perfusion solution followed by 100 cc type I collagenase solution (Worthington Biochemicals). Following cell dissociation, we filtered cells with 100-mm mesh cell strainers and gradient centrifugation steps to purify cell suspension. Then, we suspended hepatocytes at 5 × 105 cells/mL in Medium 199 (Sigma), 10% fetal bovine serum (Life Technologies), and antibiotics (plating medium). After plating for 2 h on collagen-coated plates, we exchanged plating medium for 4 h.
Method Details
Chemicals and Antibodies.
Ketamine was from KetaSet and xylazine from AnaSed; medium 199, Hank's balanced salt solution, EGTA, Hepes, PenStrep, and Gentamycin from Life Technologies; collagen type 4 from Worthington; insulin (NovoLog) and S961 from Novo Nordisk A/S; sodium orthovanadate from New England Biolabs; and bovine serum albumin from Fisher Scientific. The 16% paraformaldehyde (PFA) was from Electron Microscopy Sciences and was diluted in sterile phosphate buffer solution to 4% final concentration. Anti-FoxO1 (for Western blot and immunohistochemistry, C29H4), anti-panAkt (for Western blot, 40D4) and phosphor-Akt (Ser473) (for Western blot, D9E), and normal Rabbit immunoglobulin G (IgG) (for ChIP, 2729) were from Cell Signaling. HNF4A (for immunohistochemistry, ab41898), and GFP (for ChIP, ab 290) and FoxO1 (for ChIP, ab39670) were from Abcam. H3K27ac (for ChIP, 39133) was from Active motif. Anti-GFP (immunohistochemistry, A-6455) was from Invitrogen.
Protein Analysis.
Livers were lysed in sonication buffer containing 20 mM Hepes pH 7.5, 150 mM NaCl, 25 mM ethylenediamine tetraacetic acid, 1% Nonidet P-40, 10% glycerol, 1 mM Na vanadate, 1 mM phenylmethylsulphonyl fluoride, and protease and phosphatase inhibitor mixture (Cell Signaling). We sonicated lysates for 100 s (5×, output 70%, 20 s/20 s) and centrifuged them for 15 min at 14,000 rpm. A total of 30 µg protein (Pierce BCA, Thermo Fisher Scientific) was subjected to sodium dodecyl sulphate–polyacrylamide gel electrophoresis. We used the following antibodies: Akt (1:2,000), phosphor-Akt (Ser473) (1:2,000), β-actin (1:1,000), FoxO1 (1:1,000) (all from Cell Signaling), and GFP (1:1,000) (Abcam, ab290). Densitometric analysis was performed using ImageJ software (NIH).
Immunohistochemistry.
We anesthetized 8- to 12-wk-old mice fasted or refed for various lengths of time and perfused them with 4% PFA through the IVC. Livers were collected, fixed in 4% PFA for 2-h, dehydrated in 30% sucrose overnight at 4 °C, embedded in optimal cutting temperature compound (Sakura), frozen to −80 °C, and cut into 7-µm sections. We used primary antibodies to FoxO1 (1:100; Cell Signaling Technology) and HNF4A (1:100; Abcam) and secondary anti-IgG antibodies conjugated with Alexa Fluor 488 and 555 for each of the species (1:1,000; Life Technologies). Immunofluorescence was visualized by the tyramide signal amplification fluorescence system (PerkinElmer).
RT-qPCR.
We lysed livers in 1 mL TRIzol, purified RNA using RNeasy Mini Kit (Qiagen), reverse-transcribed it with qScript cDNA Synthesis Kit (QuantaBio), and performed PCR with GoTaq qPCR Master Mix (Promega). qPCR primers used are listed. 18S forward: AAACGGCTACCACATCCAAG and reverse: AAACGGCTACCACATCCAAG. FoxO1 forward: TCCAGTTCCTTCATTCTGCACT and reverse: GCGTGCCCTACTTCAAGGATAA. FoxO3 forward: TGAACTCCTTGCGTCAGTCACC and reverse: CGGTGCTAGCCTGAGACATCAA. FoxO4 forward: TCAAGGACAAGGGTGACAGCAA and reverse: AGGGTTCAGCATCCACCAAGAG. Gene expression levels were normalized to 18S using the 2-DDCt method and are presented as relative transcript levels.
RNA-seq Library Constructions and Data Analysis.
We prepared the samples from three mice for each group and generated the libraries individually. Libraries for RNA-seq were prepared using the TruSeq Stranded mRNA Sample Prep Kit (Illumina), following the manufacturer’s protocol. Deep sequencing was carried out on the Illumina NextSEq. 500 platform using the NextSEq. 500/550 high-throughput kit version 2.5 (Illumina) in 75-base single-end mode according to the manufacturer’s protocol. Sequenced reads from the RNA-seq experiment were aligned to mouse genome mm10 using HISAT2. Cufflinks was used for transcript assembly. Gene expression levels were expressed as fragments per kilobase of exon per million mapped sequence reads, and Cuffdiff was used for statistical comparison.
ChIP Assays and ChIP-seq Library Construction.
The ChIP-IT High Sensitivity kit (Active Motif) was used for ChIP following the manufacturer’s protocol. We anesthetized 8- to 12-wk-old mice after 4-h fasting followed, or not, by 1-h refeeding and perfused them with 10 μM orthovanadate through the IVC. We harvested samples from the left lobe of liver tissues and pooled 100 mg samples from three individual replicates for further experiments. We obtained sheared chromatin from 300 mg liver extract using a S220 Focused-ultrasonicator (Covaris). Immunoprecipitation was performed using 4 µg anti-GFP antibody for 10 µg sheared chromatin. The specificity of the anti-GFP antibody was confirmed by Western blotting of liver extract. ChIP-seq libraries were constructed using KAPA Hyper Prep Kit (KAPA Biosystems) according to the manufacturer’s instructions. ChIP-seq libraries were quantified by Tapestation (Agilent) and sequenced on an Illumina NEXTseq (Illumina) with 75-base single-end mode.
ChIP-qPCR.
Real-time ChIP-qPCR was carried out with GoTaq® qPCR Master Mix (Promega, Madison, WI). The signal of binding events was normalized against input DNA for primer efficiency (Active Motif). qPCR primers used are listed. G6pc forward: GCCTCTAGCACTGTCAAGCAG and reverse: TGTGCCTTGCCCCTGTTTTATATG; Pck1 forward: TCCACCACACACCTAGTGAGG and reverse: AGGGCAGGCCTAGCCGAGACG; Igfbp1 forward: ATCTGGCTAGCAGCTTGCTGA and reverse: CCGTGTGCAGTGTTCAATGCT; Fkbp5 forward: TTTTGTTTTGAAGAGCACAGAA and reverse: TGTCAGCACATCGAGTTCAT. The other primers used for enhancer sites are followings. G6pc-Enh#1 forward: ATATACCGAGCACATGGCAG and reverse: CTGTGGTGATTCTAGGACTG; G6pc-Enh#2 forward: GTATGTTGTTAACCTGGACG and reverse: ACTTTGACGCTGGCTACTTC; Pck1-Enh#1 forward: ATAACCCTGGTAGGTCAGAG and reverse: TCTAGAAAACGTTGGGAAAA; Pck1-Enh#2 forward: CTGACAGTGCATTCGATTTA and reverse: CCCGTAACTAGGCACTGTTC; Apoa4-Enh forward: TTTGATCCTGGGTTCTGATC and reverse: CACCAACCTGAACTTTGTCT; PDK4-Enh forward: ATAGATCCCAGGTCGCTAGG and reverse: ATAGATCCCAGGTCGCTAGG.
ChIP-seq Data Analysis.
Reads were aligned to mouse genome mm10 using Bowtie2 software (59). The reads used in subsequent analysis passed Illumina’s purity filter, aligned with no more than two mismatches and mapped uniquely to the genome. Duplicate reads were removed with Picard tools. The tags were extended at their 3′-ends to 200 bp. Technical information of sequencing depth and aligned reads is summarized in SI Appendix, Table S2. Peak calling was performed by MACS 2.1.0 (60) with the P value cutoff of 10−7 for narrow peaks and with the q-value cutoff of 10−1 for broad peaks against the input DNA control sample. The TSS determined on mouse genome mm10 was used as measurement of the distance of each peak. HOMER software suite (61) was used to perform motif analysis, annotate peaks, such as promoter/TSS, introns, exons, intergenic, 5′ untranslated region (UTR), noncoding RNA, and 3′ UTR, merge files, and quantify data to compare peaks. For the detection of active enhancers, we used bedtools (62) by collecting the intersection of the peaks of TF and histone marks.
In Vivo Insulin-Resistance Model.
For HFD-induced insulin resistance, animals were fed either standard or high-fat chow (Rodent Diet with 60 kcal% fat, D12492i; Research Diets Inc.) beginning at 8 wk of age for 4 wk. For S961 treatment, vehicle (normal saline) or 10 nmol S961 was loaded into Alzet osmotic pumps 2001 and implanted subcutaneously in the back of the mice. Mice were killed up to 7 d after implantation.
Quantification and Statistical Analyses
Values are presented as means ± SEM and analyzed using Prism 8.2.1 (GraphPad Software, Inc.). We used unpaired Student’s t test for normally distributed variables for comparisons between two groups, one-way ANOVA followed by Bonferroni post hoc test for multiple comparisons, and Pearson’s correlation coefficient to investigate the relationship between two variables. χ2 tests are applied for contingency analysis. We used a threshold of P < 0.05 to declare statistical significance.
Acknowledgments
We thank members of the D.A. and A.K. laboratories, Dr. Utpal B. Pajvani, Dr. Rebecca A. Haeusler, Dr. Michael J. Kraakman, and Dr. Daisuke Komura for insightful discussions of the data, Dr. Katia Basso for help with ChIP experiments, and Mr. Thomas Kolar and Ms. Ana Maria Flete (Columbia University) for exceptional technical support. This work was supported by NIH Grants DK57539 and DK63608 (Columbia Diabetes Research Center).
Footnotes
Competing interest statement: D.A. is founder, director, and chair of the advisory board of Forkhead Biotherapeutics, Corp.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2102222118/-/DCSupplemental.
Data Availability
NGS data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GSE151546) (63). Previously published data were used for this work: GSE31039 (64), GSE35262 (31), and GSE72084(30).
References
- 1.Ferrannini E., DeFronzo R. A., Impact of glucose-lowering drugs on cardiovascular disease in type 2 diabetes. Eur. Heart J. 36, 2288–2296 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Haeusler R. A., McGraw T. E., Accili D., Biochemical and cellular properties of insulin receptor signalling. Nat. Rev. Mol. Cell Biol. 19, 31–44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen M. C., Shulman G. I., Mechanisms of insulin action and insulin resistance. Physiol. Rev. 98, 2133–2223 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergman R. N., Iyer M. S., Indirect regulation of endogenous glucose production by insulin: The single gateway hypothesis revisited. Diabetes 66, 1742–1747 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Rizza R. A., Pathogenesis of fasting and postprandial hyperglycemia in type 2 diabetes: Implications for therapy. Diabetes 59, 2697–2707 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Accili D., Insulin action research and the future of diabetes treatment: The 2017 banting medal for scientific achievement lecture. Diabetes 67, 1701–1709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monnier L., Colette C., Dunseath G. J., Owens D. R., The loss of postprandial glycemic control precedes stepwise deterioration of fasting with worsening diabetes. Diabetes Care 30, 263–269 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Evans M., Morgan A. R., Yousef Z., What next after metformin? Thinking beyond glycaemia: Are SGLT2 inhibitors the answer? Diabetes Ther. 10, 1719–1731 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langlet F., et al. , Selective inhibition of FOXO1 activator/repressor balance modulates hepatic glucose handling. Cell 171, 824–835.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan W., et al. , FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 29, 4223–4236 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin D. J., et al. , Genome-wide analysis of FoxO1 binding in hepatic chromatin: Potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic Acids Res. 40, 11499–11509 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldstein I., Hager G. L., Transcriptional and chromatin regulation during fasting - The genomic era. Trends Endocrinol. Metab. 26, 699–710 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo T., et al. , Identification of C2CD4A as a human diabetes susceptibility gene with a role in β cell insulin secretion. Proc. Natl. Acad. Sci. U.S.A. 116, 20033–20042 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiang L., Banks A. S., Accili D., Uncoupling of acetylation from phosphorylation regulates FoxO1 function independent of its subcellular localization. J. Biol. Chem. 285, 27396–27401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banks A. S., et al. , Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab. 14, 587–597 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim-Muller J. Y., et al. , FoxO1 deacetylation decreases fatty acid oxidation in β-cells and sustains insulin secretion in diabetes. J. Biol. Chem. 291, 10162–10172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nitzsche A., et al. , RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One 6, e19470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heintzman N. D., et al. , Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Accili D., Arden K. C., FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117, 421–426 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Haberle V., Stark A., Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 19, 621–637 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calo E., Wysocka J., Modification of enhancer chromatin: What, how, and why? Mol. Cell 49, 825–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raisner R., et al. , Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 24, 1722–1729 (2018). [DOI] [PubMed] [Google Scholar]
- 23.McLean C. Y., et al. , GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S. X., et al. , FoxO transcription factors are required for hepatic HDL cholesterol clearance. J. Clin. Invest. 128, 1615–1626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehrlich K. C., Lacey M., Ehrlich M., Tissue-specific epigenetics of atherosclerosis-related ANGPT and ANGPTL genes. Epigenomics 11, 169–186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allan C. M., Taylor S., Taylor J. M., Two hepatic enhancers, HCR.1 and HCR.2, coordinate the liver expression of the entire human apolipoprotein E/C-I/C-IV/C-II gene cluster. J. Biol. Chem. 272, 29113–29119 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto M., Pocai A., Rossetti L., Depinho R. A., Accili D., Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 6, 208–216 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Haeusler R. A., Kaestner K. H., Accili D., FoxOs function synergistically to promote glucose production. J. Biol. Chem. 285, 35245–35248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haeusler R. A., et al. , Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat. Commun. 5, 5190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein I., et al. , Transcription factor assisted loading and enhancer dynamics dictate the hepatic fasting response. Genome Res. 27, 427–439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boergesen M., et al. , Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol. Cell. Biol. 32, 852–867 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel S., et al. , GDF15 provides an endocrine signal of nutritional stress in mice and humans. Cell Metab. 29, 707–718.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi Y., et al. , KLF15 enables rapid switching between lipogenesis and gluconeogenesis during fasting. Cell Rep. 16, 2373–2386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vikram A., Jena G., S961, an insulin receptor antagonist causes hyperinsulinemia, insulin-resistance and depletion of energy stores in rats. Biochem. Biophys. Res. Commun. 398, 260–265 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Lin H. V., Accili D., Hormonal regulation of hepatic glucose production in health and disease. Cell Metab. 14, 9–19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown M. S., Goldstein J. L., Selective versus total insulin resistance: A pathogenic paradox. Cell Metab. 7, 95–96 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Baldi S., et al. ; RISC Investigators, Influence of apolipoproteins on the association between lipids and insulin sensitivity: A cross-sectional analysis of the RISC Study. Diabetes Care 36, 4125–4131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Titchenell P. M., Chu Q., Monks B. R., Birnbaum M. J., Hepatic insulin signalling is dispensable for suppression of glucose output by insulin in vivo. Nat. Commun. 6, 7078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samuel V. T., et al. , Fasting hyperglycemia is not associated with increased expression of PEPCK or G6Pc in patients with Type 2 Diabetes. Proc. Natl. Acad. Sci. U.S.A. 106, 12121–12126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalvisa A., et al. , Insulin signaling and reduced glucocorticoid receptor activity attenuate postprandial gene expression in liver. PLoS Biol. 16, e2006249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson R., Sandelin A., Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71–87 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Zabidi M. A., et al. , Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 518, 556–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brent M. M., Anand R., Marmorstein R., Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 16, 1407–1416 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong X. C., et al. , Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 8, 65–76 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubota N., et al. , Dynamic functional relay between insulin receptor substrate 1 and 2 in hepatic insulin signaling during fasting and feeding. Cell Metab. 8, 49–64 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Lu M., et al. , Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat. Med. 18, 388–395 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimada M., et al. , Suppression of diet-induced atherosclerosis in low density lipoprotein receptor knockout mice overexpressing lipoprotein lipase. Proc. Natl. Acad. Sci. U.S.A. 93, 7242–7246 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiao X., et al. , Angiopoietin-like protein 8 accelerates atherosclerosis in ApoE-/- mice. Atherosclerosis 307, 63–71 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Qiang L., et al. , Increased atherosclerosis and endothelial dysfunction in mice bearing constitutively deacetylated alleles of Foxo1 gene. J. Biol. Chem. 287, 13944–13951 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida H., et al. , Immunological Genome Project, The cis-regulatory atlas of the mouse immune system. Cell 176, 897–912.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto M., Han S., Kitamura T., Accili D., Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J. Clin. Invest. 116, 2464–2472 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hay N., Interplay between FOXO, TOR, and Akt. Biochim. Biophys. Acta 1813, 1965–1970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Greer E. L., Brunet A., FOXO transcription factors in ageing and cancer. Acta Physiol. (Oxf.) 192, 19–28 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Pawlak M., Lefebvre P., Staels B., Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 62, 720–733 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Montagner A., et al. , Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut 65, 1202–1214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuo T., et al. , Induction of α cell-restricted Gc in dedifferentiating β cells contributes to stress-induced β-cell dysfunction. JCI Insight 5, e128351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Postic C., et al. , Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Cook J. R., Langlet F., Kido Y., Accili D., Pathogenesis of selective insulin resistance in isolated hepatocytes. J. Biol. Chem. 290, 13972–13980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng J., Liu T., Qin B., Zhang Y., Liu X. S., Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan A. R., BEDTools: The Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics 47, 11.12.1-34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.T. Kitamoto, A. Okabe, A. Kaneda, D. Accili, Characterization of chromatin and gene expression changes during fasting and refeeding in mouse liver. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE151546. Deposited 1 June 2020. [Google Scholar]
- 64.ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
NGS data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GSE151546) (63). Previously published data were used for this work: GSE31039 (64), GSE35262 (31), and GSE72084(30).