Highlights
-
•
Antibiotics are used extensively in agriculture, livestock and in animal husbandry.
-
•
In agriculture, these are used to increase crop productivity; in animal husbandry and livestock to treat sick animals and as growth promoters in animal feed at controlled concentrations.
-
•
Bacteria are able to modify their genes under stress conditions by different mechanisms, and the resistance genes are cycled among agricultural soils and other different ecosystems through nutrient cycling.
-
•
A number of significant molecular strategies have been proposed to reduce the occurrence of antibiotic resistance in bacterial genomes .
Keywords: Agriculture, Antibiotic cycling, Antibiotic resistance, Livestock, Molecular strategies
Abstract
Antibiotic resistance is a massive problem rising constantly and spreading rapidly since the past decade. The major underlying mechanism responsible for this problem is an overuse or severe misuse of antibiotics. Regardless of this emerging global threat, antibiotics are still being widely used, not only for treatment of human infections, but also to a great extent in agriculture, livestock and animal husbandry. If the current scenario persists, we might enter into a post-antibiotic era where drugs might not be able to treat even the simplest of infections. This review discusses the current status of antibiotic utilization and molecular basis of antibiotic resistance mechanisms acquired by bacteria, along with the modes of transmittance of the resultant resistant genes into human pathogens through their cycling among different ecosystems. The main focus of the article is to provide an insight into the different molecular and other strategies currently being studied worldwide for their use as an alternate to antibiotics with an overall aim to overcome or minimize the global problem of antibiotic resistance.
1. Introduction
Antibiotics are substances given in a controlled amount, meant to kill or reduce the growth of microorganisms, particularly bacteria (Ghosh et al., 2007). These are usually naturally occurring substances and are mostly produced by microorganisms containing genes encoding resistance to different antibiotics they produce (Salyers et al., 1997). Besides their major applications in humans, these are used extensively in agriculture to increase crop productivity, in animal husbandry and livestock to treat sick animals, for prophylactic/metaphylactic purposes to prevent infections, and also as growth promoters in animal feed at controlled concentrations. When we look at the global consumption of various antibiotics, it has been reportedly found that approximately half of the antibiotics used in the entire animal husbandry are consumed in China, followed by US, Brazil, India and Germany (Laxminarayan et al., 2015). For livestock as well, it has been reported that in 2010, the largest antimicrobial consumer was China which was estimated to use up to 30% of the overall antimicrobial production globally; and at the current usage rate, India comes after China in the consumption of antimicrobials for livestock production and maintenance (Kleina et al., 2018). In 2015, among the developed countries, the leading consumers of antibiotics were USA, France, and Italy; and among the developing countries, India, China, and Pakistan topped the list where a 65% increase in consumption between 2000 and 2015 was reported globally (Ganguly et al., 2011). A study has also projected a 67% rise in antibiotic consumption by 2030 in various highly populated countries of the world including India (Van Boeckel et al., 2015). This extensive usage of antibiotics, although has resulted in the realization of an ever increasing demand of diverse agricultural and animal products; but their long-term application even at sub-therapeutic concentrations, directly on the fields or indirectly via animal manure, is adversely affecting the microbiota of agricultural soils.
Because of this overuse of antibiotics, the present century has also witnessed an extensive increase in the emergence of microbes which have been observed to modify their genes more rapidly and efficiently; helping them in developing resistance against different antibiotic groups, especially broad- spectrum antibiotics. Even the new generation antibiotics are reported to be inefficacious against such microbes, making this a major challenge for researchers to handle, and hence generating the need to find new methods to conquer this ever-rising issue. Development of new strategies to slowdown, or to provide an alternate solution to this emerging problem of multiple drug resistance is thus the need of the hour.
The present article is therefore aimed at shedding light on various problems generated globally due to an overuse of different types of antibiotics commonly used in agriculture and livestock; the rapid development of resistance by several bacteria against most of these commonly used antibiotics in agriculture; and some of the methods which are being researched upon to provide an alternate to antibiotics to overcome this major global issue. There is a dire need for the development of new alternatives to antibiotics, so that we are not only able to cure the existing diseases, but also because we need to become competent to prevent the initiation of new infections in the world.
2. Antibiotic consumption and development of antibiotic resistance in agriculture
Antibiotics are widely used in agriculture, livestock, poultry, fisheries and animal husbandry. In agriculture, antibiotics are most commonly used to prevent and cure various diseases in crops; whereas, in livestock and animal husbandry, these are most commonly used as growth promoting agents, and in preventing/ curing infections. There are at least 30 different antibiotics that are commonly used in agriculture and livestock, among which macrolides, penicillins and tetracyclines are the major ones (Laxminarayan et al., 2015) (Table 1). In animal husbandry alone, the average yearly consumption of antibiotics has been estimated as 172 mg/kg in pigs, 148 mg/kg in chicken, and 45 mg/kg in cattle worldwide (Van Boeckel et al., 2015).
Table 1.
Antibiotics used in agriculture and animal husbandry.
| Field/Area | Antibiotic used |
|---|---|
| Agriculture | Oxytetracycline, streptomycin, penicillin, oxolinic acid, gentamycin |
| Swine Production | Benzylpenicillins and tetracycline (most commonly used), sulfadimidine, sulfathiazole and trimethoprim, bacitracin, lincosamides, macrolides, floroquinolones, 3rd generation cephalosporins, colistin (Lekagul et al., 2018) |
| Chicken Production | Bacitracin, chlortetracycline, decoquinate, diclazuril, naracin, nicarbazin, monensin, penicillin, rebenedine hydrochloride, virginiamycin, colistin, tylosin, doxycycline, tiamulin, roxithromycin, amikacin |
| Cattle Production | Penicillin, tetracycline, ceftiofur, florfenicol, tilmicosin, enrofloxacin, and tulathromycin, phenicol, lincosamide, pleuromutilin, macrolide, polypeptide, streptogramin, carbadox, bambermycin |
Looking at the historical perspective of antibiotics usage in agriculture, streptomycin, an aminoglycoside, has been most commonly used in plant agriculture to treat diseases such as fire blight since early 1940’s. Till late 1940’s, due to a lack of effective bactericide alternatives for various plant diseases, there had been a decade-long dependence on streptomycin, thus resulting into an emergence of resistant strains against this antibiotic, and impeding the control of many diseases (Magnet et al., 2005, Mingoet et al., 1999). Various bacterial strains like Pseudomonas spp., and Xanthomonas campestris had been found to develop resistance against this antibiotic (Mac Manus et al., 1997). As a solution to this, amikacin, another aminoglycoside when introduced in late 1940s, was started being given in combination with other antibiotics. However, later on due to the resistance caused by aminoglycoside modification enzymes, other forms of aminoglycosides had to be proposed (Ramirez and Tomasky et al., 2017). Another class of antibiotics which came to be commonly used in 1950’s was tetracyclines. They were used for improvement in swine production and cattle production against both Gram-positive and Gram-negative bacteria, and for the control of other classes of micro-organisms such as eukaryotic protozoan parasites as well Roberts (2019). However, Shigella dysenteriae which causes bacterial dysentery first showed tetracycline resistance in the year 1953 Roberts (1996). Since then, mutations found in copious amount in various bacteria such as E.coli, Enterococcus, Staphylococcus, Streptococcus etc. were observed to cause resistance against tetracycline (Roberts, 1996, Cadena et al., 2018, Roberts, 2019). Methicillin, a β-lactam antibiotic acts by inhibiting penicillin-binding proteins (PBPs) that are involved in the synthesis of peptidoglycan layer surrounding the cell Stapleon and Taylor (2002). However, methicillin resistant Staphylococcus aureus (MRSA) also emerged soon, these were first isolated in the year 1961 in England and were initially found to be resistant against only β-lactam antibiotics (Brown and Reynolds, 1980). But with, the outbreak of MRSA, the prevalence of antibiotic resistance spread extensively during the 1980s, resulting into vancomycin becoming a more important drug as compared to penicillin, since it came into a wider use for the treatment of Gram-positive bacterial infections. Vancomycin was the antibiotic of choice until 2003 in treating MRSA infections, but since resistance to this agent also has rapidly developed recently, it has now become the drug of last resort for the treatment of MRSA (Swartz, 1994). Studies have also been conducted in which sulfonamide (sul) resistance in Psychrobacter, Enterococcus, and Bacillus sp. were reported for the first time in the year 2009. More recently, resistance has also been observed against fluoroquinolone, especially ciprofloxacin (CIP) which is used as a common treatment for Campylobacter caused gastroenteritis (Piddock, 1998). Further, a study proved the presence of erythromycin and other macrolide traces in livestock products such as liver, muscle, egg and milk (Petz et al., 1987).
The resistance was not only limited to animals and their products but was soon observed in agricultural soils. In one such study, Popowska et al. analyzed soils from agricultural fields and detected several genes (erm(C), erm(V), erm(X), msr(A), ole(B) and vga) responsible for erythromycin resistance in those soil samples (Popowska et al., 1987). Diverse, potentially mobile and abundant Antibiotic Resistance Genes (ARGs) of sulfonamides discovered in farm samples recommended that unchecked use of antibiotics was causing the emergence and release of ARGs in to the environment (Byrne et al., 2009). As a further confirmation of this fact, it has been observed that bacteria such as Citrobacter species, Enterobacter species, K. pneumonia, K. oxytoca, S. aureus, Proteus species and Y. enterocolitica have been found resistant to cephalosporins. Although, cephalosporin use is very restricted in food animals as compared to its use in humans, still resistance is being observed in various bacteria; this can only be explained by hypothesizing that the resistance is being transmitted from different environments to animals and then to humans (Wonhee et al., 2014). However, fifth generation cephalosporins are still in use.
In order to reduce the usage of antibiotics, and to generate a more effective method for disease resistance in crops, one of the strategies that humans have invented is the usage of genetically modified crops/transgenic crops. Transgenic crops are the ones in which insertion/deletion/silencing of the gene of interest is done in order to produce plants having desired qualities (Grifths et al. 2005). Insect resistant transgenic crops have also been introduced to save plants from insects and the pathogens that they carry on their body surface. Although, this helps minimize the economic burden on farmers by producing better crop yield, but recently, it has been observed that the resistance breakdown is increasing in target bacterial or insecticide population (Bawa and Anilakumar, 2013, Gilbert, 2013). Due to excessive cultivation of transgenic crops, high selection pressure is imparted on targeted insect population and weeds leading to evolution of new insect biotypes and emergence of superweeds posessing resistance against transgenic technology. Further, antibiotic resistance genes are also being transferred from the transgenic plants to the genome of non-target organisms such as non-transgenic crops and insects, for eg. Monarch butterfly feeding on milkweed leaves (Losey et al., 1999).
Thus, although transgenic crops were introduced as a means to reduce disease-resistance in crops, which in turn should have decreased the use of antibiotics and hence the spread of antibiotic resistance; but, on the contrary, the excessive cultivation of transgenic crops has today resulted indirectly into an increase in the spread of antibiotic resistance. Therefore, as demonstrated in all the above quoted studies, it can be concluded that due to an overuse of antibiotics, the resistance among various micro-organisms against the commonly used antibiotics (both in agriculture and in livestock) is increasing at a rapid pace; and through nutrient cycling, the genes responsible for this resistance are spreading rapidly among different environments.
3. Mechanisms of antibiotic resistance adopted by different micro-organisms
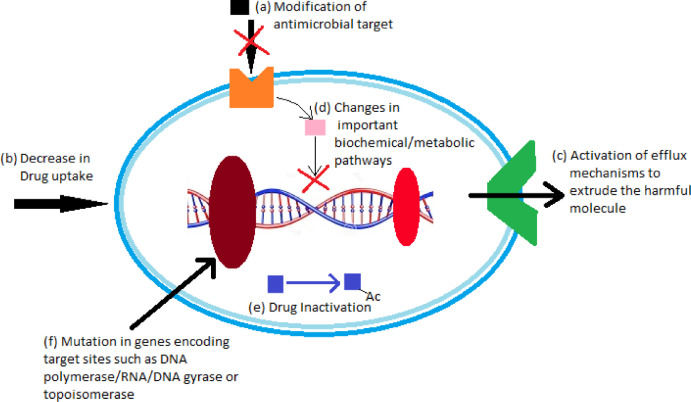
The antibiotic resistance mechanisms adopted by different micro-organisms can be broadly classified into: (i) mechanisms having a genetic basis of microbial resistance, and (ii) mechanisms with a mechanistic basis of antimicrobial resistance. The genetic resistance occurs due to mutations which result into a modification of antimicrobial targets, decrease in drug uptake, increase in efflux of molecules, and changes in metabolic pathways; and due to horizontal gene transfer via transformation, transduction and conjugation. On the other hand, mechanistic resistance occurs by modification of antimicrobial molecules, prevention of compounds from reaching antimicrobial targets, bypassing of target sites, and resistance due to global cell adaptive processes (Jose et al., 2017) (Fig. 1).
Fig. 1.
Mechanisms of antibiotic resistance adopted by bacteria
(a) Modification of antimicrobial targets- some bacterial membranes lack the target molecule for a particular antibiotic, and thus become impermeable to the passage of such antibiotics across their cell membranes; (b) decrease in drug uptake- a few bacteria modify their cell membranes in order to reduce or diminish the drug uptake; (c) activation of efflux mechanisms to extrude the harmful molecules-these mechanisms increase the efflux of antibiotics at a faster pace such that they are not able to produce much effect on the cell; (d) changes in important metabolic pathways-effect of antibiotics can be produced through many biochemical pathways, hence by incorporating changes in these pathways, bacteria rescue themselves from the effect of antibiotics; (e) drug inactivation- drugs are inactivated by processes such as acetylation or methylation once they enter in the cell, thus making them ineffective; (f) mutations in genes encoding target sites-drugs have targets such as DNA polymerase/ RNA/ DNA gyrase and topoisomerase etc., so the genes encoding such target molecules are mutated/ suspended by some bacteria to protect them from the effect of antibiotics (Fig. 1).
Besides these major mechanisms, more recently, a phenomenon called protein promiscuity has also been reported to be responsible for antibiotic resistance. According to classical biochemistry, proteins bind to their ligands/substrates in a specific manner via their pre-formed binding sites. However, several types of variations from this norm known as promiscuous behavior/protein promiscuity have been observed in the recent years (Gupta et al., 2020). A study on a drug, albicidin, has shown that even structural alteration in a drug may not help in defeating drug resistance (Rostock at al., 2018). Albicidin is an effective agent against both Gram negative and Gram positive bacteria that acts by inhibiting DNA gyrase (EC 5.99.1.3). However, in some cases, the bacteria incorporates resistance due to expression of drug binding proteins, as a result of which the compound is not available for binding to the gyrase. Rostock et al. in 2018, presented that the drug-binding domain of the protein AlbA, in the case of Klebsiella oxytoca, showed significant promiscuous behaviour by binding to different kinds of derivatives of albicidin (Rostock et al., 2018). These interactions make the antibiotics unavailable to their molecular target as they have dissociation constants in nanomolar range which leads to drug resistance. Another similar study represented the work on resistance development against the broad-spectrum antibiotic fosfomycin (Brown et al., 2009). The proteins FosA, FosB and FosX catalyze the reaction of the drug with glutathione, cysteine and water, respectively. The study shows that a protein, FosXmt may act as a progenitor of FosA and FosX, and this progenitor protein was observed to exhibit catalytically promiscuous activity, and found to have very low glutathione transferase and epoxide hydrolase activities. Compared with the mutants, they presented that only 10% difference in the sequence (from wild proteins) resulted in antibiotic resistance activity from its progenitor. On the other hand, in aminoglycosides (e.g. gentamycin, amikacin), antibiotic resistance is caused by enzymatic adenylation/phosphorylation/acetylation of the drugs. A kinase involved in phosphorylation, was found to exhibit high-level of promiscuity since it could bind to ten different aminoglycosides Fong and Berghuis (2002).
Thus, bacteria possessing many of the above listed antibiotic-resistance mechanisms against almost all antibiotics currently available in clinical practice have been reported to be present in several different environmental niches, thus making it very difficult to deal with rapidly spreading drug-resistant microbes.
4. Antibiotic and antibiotic resistance cycling through different ecosystems
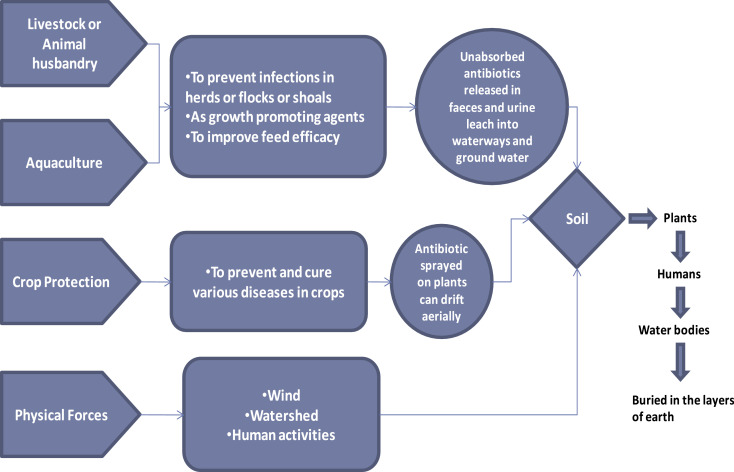
Since there is a strong connection between soil, plants and animals, the spread of antibiotics and antibiotic-resistance in our environment is not limited to any one ecosystem. Antibiotics travel among all these routes (Karesh et al., 2012), and create a network where ARGs also travel along with them (Fig. 2). Antibiotics are provided in animal feed, agriculture and aquaculture to improve growth, and in crop protection to prevent and cure several diseases. In case of animals, however, complete absorption of these antibiotics does not take place inside the gut, and they are excreted out of the body in the form of faeces and urine, which ultimately leads to the formation of manure. This manure which is a rich source of organic matter and nutrients, although helps in improving the fertility of soil, but additionally, it also becomes responsible for the transfer of traces of antibiotics from the animal kingdom to the soil ecosystem and ultimately to the plants (Chambers et al., 2009). Application of manure onto agricultural land results in proliferation of antibiotic resistance among soil bacteria. It has been reported that fresh and composted swine manure in agricultural soils leads to the development of tetracycline resistance after long term application (Walson et al., 2001). Use of manure near animal pens has also reported high level of chlortetracycline resistance, and the bacterial isolates obtained from such soils have been reported to encode genes for tetracycline resistance (Ghosh et al., 2007). A few studies also support that the application of manure incorporated with antibiotics to crops can also lead to the development of antibiotic resistance among different crops. Kumar et al. in 2005 reported development of tylosin resistance in corn, green onion and cabbage. In aquaculture also, antibiotics diffuse into waterways and pens surrounding the farm area. However, there are several other factors also, which are responsible for the movement of antibiotic resistance genes. These include physical forces like wind, watershed and human activities, which have resulted in an increase in resistance among common pathogenic bacteria. Research studies have shown high frequency of resistance in E.coli isolates associated with wild animals (Souza et al., 1999). Animals which are present at highest proximity to humans are more prone to incorporating antibiotic resistance in their gut bacteria as antibiotics are administered consciously to animals for rapid growth and high productivity to meet the daily requirements of the growing population.
Fig. 2.
Pathway of antibiotics and antibiotic resistant genes through agriculture and livestock.
From all these studies, it can be concluded that excessive application of animal manure leads to the persistence of antibiotic resistance genes in the soil bacteria, which ultimately gets transferred to plants, animals or humans through cycling. On the other hand, use of antibiotics as growth promoters also exerts a selective pressure in food animals; this appears to have created large reservoirs of exchangeable antibiotic resistance in different ecosystems (Witte et al., 2000).
5. Current strategies adopted by different governments to encounter the rising problem of antibiotic resistance
As discussed in previous sections, excessive and unrestricted use of antibiotics in agriculture to increase crop productivity, and as growth promoters in livestock rather than for controlling infections is increasing the emergence and spread of antibiotic resistance genes to a great extent. It has been predicted that by the year 2050, nearly 10 million deaths worldwide will be attributed to antimicrobial resistance (Aarestrup et al., 2010). At this time, when we understand the magnitude of this major problem, several programs have been acknowledged and initiated by the different Governments to reduce its abuse in farm animals and to preserve the miracle of antibiotics (Aerestrup et al., 2010). In America, this is monitored by US Department of Agriculture (USDA) to ensure that the antibiotic residues do not exceed the tolerance levels marked unsafe by FDA and USDA (Aarestrup et al., 2010). Various other countries such as Sweden, Denmark, India and China have also adopted regulatory measures to some extent (Culp et al., 2020, EMA and EFSA, 2017, Ranjalkar and Chandy 2019, Qu, 2019). The recommended options include, tracking down the antimicrobial use, setting up nation-wide targets for reduction of antibiotic usage, use of good health practices, having a track on veterinarians and health professionals to prescribe antimicrobials, increase in reliable diagnostics and better management practices to reduce disease risk O'Neil (2016).
The European Union has already applied a ban against antibiotics in a country-specific manner, which has abolished their misuse in farm animals (EMA and EFSA, 2017). Similarly, World Health Organization (WHO) and Institute of Medicine in USA have also imposed some restrictions. There are several other organizations such as Food and Drug Administration (FDA), Preservation of antibiotics for Medical Treatment Act (PAMTA), and Delivering Antibiotic Transparency in Animals (DATA) which are working on the same aspect by implying a ban on specific antibiotics. In addition to the measures initiated by the government, practicing genuine safety measures could also be a good method to bring down the transfer of resistant genes from farm animals to humans. To overcome this shift of resistant genes, strict withdrawal periods need to be followed before animals are processed for food.
However, in-spite of these restrictions and preventive measures, the current methods are not sufficient enough, as it has been observed that the microorganisms are still continuing to develop resistance at a very fast pace Spellberg and Gilbert (2014). Hence, due to this inability of the Government initiatives to completely control the emergence of antibacterial resistance, there is a need to develop other scientific strategies, which may be over and above the existing government initiatives.
6. Alternate scientific strategies to encounter antibiotic resistance
Molecular methods targeting DNA, RNA or proteins and other scientific methods targeting cell wall, cell membrane, an intracellular target, biosynthetic pathways, ribosomes etc. present several advantages in the field of medical biotechnology, including having a high potential to alleviate or reduce the phenomenon of antibiotic resistance. These methods are continuously spreading both in terms of technological advances and popularity. The present section therefore, focuses on evaluating the current state of science and the future aspects of different methodologies with respect to their possible application in the control of antibiotic resistance.
Table 2.
Different scientific scientific strategies for overcoming antibiotic resistance.
| S.No. | Scientific Strategy | Mode of action | References |
|---|---|---|---|
| Basic Scientific Strategies | |||
| iUse of Peptide antibiotics/antimicrobial peptides(AMPs) | Target cell membrane directly or an intracellular target. Can also trigger immune response to combat diseases. | Pasupuleti et al., 2012, Egorov et al., 2018 | |
| iiUse of Carbohydrate modified compounds | Target cell wall, biosynthetic pathway of peptidoglycan layer, small or large subunit of ribosome etc. | Ramirez et al., 2010, Jeong et al., 2017 | |
| iiiUse of Non-antibacterials | Enhance activity of conventional antibiotics | Pires et al., 2017 | |
| ivUse of Combinations of antibiotics andcompounds | Combinations of antibiotics are directed towards multiple targets | Silver et al., 2007, Deshayes et al., 2017 | |
| II | Advanced Molecular Strategies | ||
| iPhenotypic conversion of drug-resistant todrug-sensitive bacteria | Specific sequence insertion in plasmid DNA to produce specific proteins leading to conversion of drug-resistant to drug-sensitive bacteria | Guerrier et al., 1997, Toney et al., 1998, Alfonso et al., 2007, Jackson et al., 2016 | |
| iiApplication of DNA and mRNA vaccines | Specific sequence insertion in plasmid DNA or directly in mRNA to produce a humoral immune response | Pizza et al., 2000, Endmann et al., 2014, Jansen et al., 2018, Zhang et al., 2019 | |
| iiiBacteriophage therapy | Bacteriophages attach themselves on specific bacterial cells which result in lysing/killing of the host pathogenic bacteria, or phages insert their DNA into the bacterial genome and it gets replicated along with the host DNA machinery | Alisky et al., 1998, Malik et al., 2019, Malik et al., 2020 | |
| ivGene editing technology | Insertion, deletion or point mutation of specific genes | Schouten et al., 2006, Holme et al., 2012, Waltz, 2015, Kumar et al., 2020 | |
| vCRISPR/Cas gene editing system | Gene silencing or gene editing is done to inactivate gene causing resistance | Pak, 2014, Rodrigues et al., 2019 | |
6.1. Basic scientific strategies to overcome antibiotic resistance
The, various strategies designed to exploit recent advances in the field of biotechnology, including use of antimicrobial peptides, use of carbohydrate modified compounds, use of non-antibacterials and combination therapies for developing new antibacterial agents that may have the potential to overcome antibiotic resistance have been reviewed in the subsequent section.
6.1.1. Use Peptide antibiotics / antimicrobial peptides (AMPs) as an alternate to antibiotics
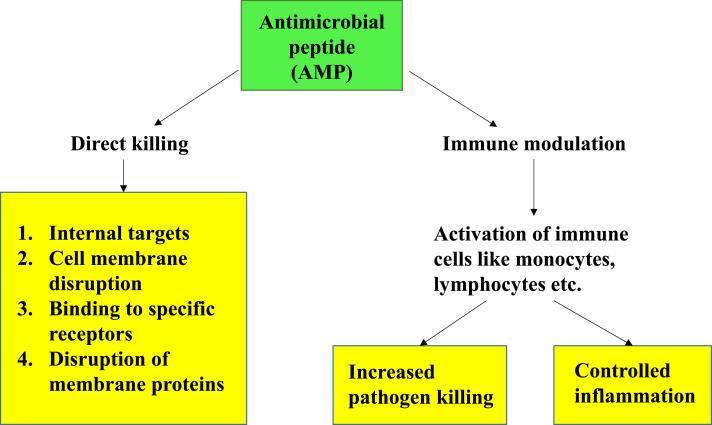
Peptide antibiotics/antimicrobial peptides, also known as host defense peptides, are short and generally positively charged peptides found in a wide variety of life forms like mammals, bacteria and fungi. The epithelial surfaces in mammals have a remarkable ability to provide physiologic functions in the face of frequent microbial invasions. Studies have shown that epithelial cells of the skin, respiratory, genitourinary and alimentary tract synthesize antimicrobial peptides. These molecules contribute to intrinsic mucosal immunity and save from simple microbial infections (Fig. 3). Their biological activity and level of expression can be altered, which can persuade the organisms to encounter microbial infections. On the other hand, by modifying cell targets or by single point mutations in the sequences of enzymes, the efficiency of antimicrobial drugs can be enhanced (Drlica et al., 2009, Nikolaidis et et al., 2014, Aldred et al., 2014, Hoopers and Jacob, 2016, Egorov et al., 2018).
Fig. 3.
Schematic representation of various mechanisms of action of AMPs.
AMPs are produced in nature either by ribosomal translation of mRNA (produced by all species of life) or by nonribosomal peptide synthesis (mainly produced by bacteria) (Sidhu and Nehra, 2021, Hancock and Chapple, 1999). Most AMPs commonly consist of 10–50 amino acids. They display an overall positive charge (ranging from +2 to +11), and contain up to 50% of hydrophobic residues in their structure (Yeaman and Yount, 2003, Hancock and Sahl, 2006, Pasupuleti et al., 2012). Based on their secondary structure, AMPs are commonly classified into α-helical, β-sheet, or peptides with extended/random-coil structure (Yeaman and Yount, 2003, Takahashi et al., 2010, Nguyen et al., 2011). AMPs act by targeting membrane directly or an intracellular target which must be reached by the means of translocation (Yeung et al., 2011, Nguyen et al., 2016, Malmsten, 2016). AMPs such as Pexiganan (Lamb and Wiseman, 1998, Lipsky et al., 2008), Omiganan, LTX-109 (Nilsson et al., 2015) have been developed for local application. Whereas, hLF1-11, novexatin, CZEN-002 are developed for direct administration against different types of fungal infections (Velden et al., 2009, Fjell et al., 2012, Fox, 2013). Several AMPs like LL-37, PXL01, hLF1-11 are also under different phases of drug development.
Apart from the direct administration or application of AMPs, there are several attempts going on to increase the production of AMPs in the body to boost immune responses to help combat infections. For eg. Vitamin D supplements are being evaluated for their applicability in treatment of bacterial infections as it has shown to directly modulate expression of AMPs (Yamshchikov et al., 2009, Wang et al., 2004, Weber et al., 2005).
This method is a wonderful approach towards reducing antibiotic resistance. However, it limits us to various points, mainly because of the fact that not a large number of peptide synthetases are currently well characterized, and also because AMPs are metabolically unstable. AMPs are also degraded by various factors and enzymes in the body. To achieve success in this method, a profuse amount of knowledge is required about structural activity and relationships of the secondary metabolites, which is currently not well known. It remains a tedious process to introduce an abundance of desired amino acid substitutions into bioactive peptides. Moreover, it also seems unlikely that random changes would lead to improved activities of these antibiotics. Therefore, it is required that the pathway of the attack of various antibiotics should be well known to achieve the discovery and synthesis of more refined drugs.
6.1.2. Use of Carbohydrate-modified compounds as an alternate to antibiotics
Carbohydrates play a crucial role in building the structural framework of all cells. They provide energy for metabolism and are a part of various intercellular processes (Ramirez et al., 2010). Various antibiotics today are carbohydrate-based, either they contain a glycan portion in their structure, or they target an enzyme or a receptor that is associated with carbohydrate metabolism. Modified carbohydrates serve as another kind of interesting molecules having the potential of reducing the problem of antibiotic-resistance (Thomas and Chi-Huey, 2001, Ge et al., 1999). However, for this kind of drug development, various targets such as biosynthesis pathway of bacterial cell wall, biosynthetic pathway of peptidoglycan layer, small and large subunit of ribosome and various other targets are necessary Thomas and Chi-Huey (2001). Although this technique holds a great potential in the development of carbohydrate-based therapeutics, it too comes with certain drawbacks. These compounds consist of hydroxyl groups having similar reactivity making their synthesis difficult. They bind to their target with relatively low affinity. Apart from this, medicinal chemists consist this as a non- interesting subject for development of newer drugs because carbohydrates are too complex for process development. Also, their bioavailability is restricted as they are excessively hydrophilic and they are not stable enough to allow oral administration. To overcome these problems, mimics of carbohydrates have been designed having improved properties such as specificity, stability, synthetic availability, affinity and hydrophobicity. These carbohydrate mimics target numerous natural processes in which glycans are involved, without any side effects (Chapleur, 1998, Sears and Wong, 1999, Jeong et al., 2017). Hence, carbohydrate based molecules have high potential in solving the problem of antibiotic resistance.
6.1.3. Use of non-antibacterials as an alternate to antibiotics
Non-antibacterials such as phenothiazines and other neuroleptics which enhance the activity of conventional antibiotics can also be used for the control of several infections (Pires et al., 2017). These substances induce changes in cell permeability and hence act as antimicrobials. They include compounds such as general anaesthetics, local anaesthetics, antihypertensive beta-adrenergic receptor antagonists, diuretics, anti-inflammatory drugs, proton pump inhibitors, calcium antagonists, psychotherapeutic compounds and antihistamines. This is a major but new research area, and a detailed information is required to work on various pathways involved in this field.
6.1.4. Combinations of existing antibiotics among themselves or with other compounds to enhance their activity
Analyses of antibiotic discovery and their execution as drugs has revealed that the agents which are directed against one particular protein target are less successful than those directed against various molecular targets, or, in-opposition to multi-subunit macromolecular machines or structures Silver (2007). Hence, as an advancement, the already discovered antibiotics are being investigated for their use in combinations, either among themselves, or with other biological molecules including antimicrobial peptides. In this direction, a few studies have already been conducted, whereas, others are under different stages of research. In one such study, Deshayes et al. (2017) have designed membrane permeable hybrid antibiotic peptide conjugates. According to their study, it is feasible to model and manufacture membrane active antibiotic peptide conjugates (MAAPCs) that combine multiple forms of antimicrobial activity, thus resulting in an unusually strong activity against sustained bacterial strains (Deshayes et al., 2017). Hence, antibiotics have been used in combinations to increase efficacy, where the combined effect is greater than the effect of a single antibiotic; to broaden the spectrum especially where the organism is unknown; to reduce the risk of antibiotic resistance and also to reduce host toxicity. A combination of antibiotics also leads to a reduction in the growth of unwanted bacteria/infection causing agents and an increase in the production of useful bacteria (Li et al., 2018). Acknowledging the fact that no antibiotic mixture is efficacious universally for every infection, one of the major drivers in amalgamating antibiotics was the need to produce better efficacy over discrete compounds. For example, the combination of penicillin with streptomycin was reported as early as 1950s (Jawetz et al., 1952), and sulfonamides with trimethoprim in 1968 (Bushby and Hichings, 1968); both combinations enhanced effectiveness and the spectrum of antibacterial activity. Similarly, using fixed dose combinations has proved vital for treatment success as the treatment periods for several drugs are long and this method improves compliance and reduces the development of resistance.
However, the use of antibiotic combinations comes with its possible downsides which mainly include: (a) A risk for adverse reactions- a combination of two or more drugs at times may lead to individual risks while causing reactions by interaction of drugs leading to different degrees of severity; (b) Antagonism- Antagonism is an unforeseen combination effect that cancels a part or all of the major effects of the drugs; (c) Unwanted rise in a whole lot of antibiotics used- instead of a proper diagnosed specific antibiotic treatment, if a broad-spectrum cocktail is used, it may result in a more severe impact on the patients microbiome with increasing selection of resistant bacteria as well as an increased risk of conditions such as diarrhoea.
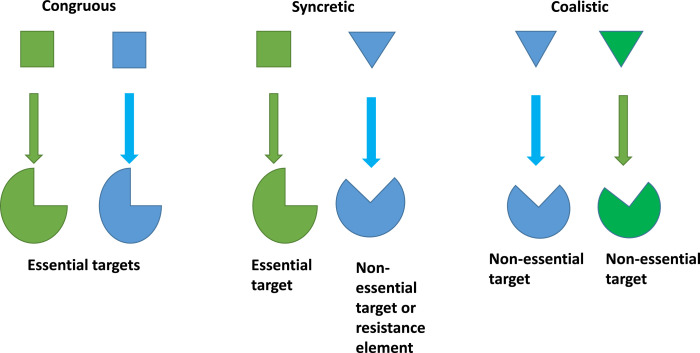
Considering the above mentioned drawbacks, researchers are now acknowledging the combinations of antibiotics with various compounds that enhance the activity of existing antibiotics. Some of these known adjuvants display synergy upon pairing with antibiotics. In a synergistic combination of antibiotics, the antibiotics have different targets. They can display either congruous (Moellering, 1983), syncretic combination (Kalan and Wright, 2011, Worthington and Melander, 2013, Wright, 2016, Gonzalez-Bello, 2017) or coalistic pairing (Tekin et al., 2016, Beppler et al., 2017, Tekin et al., 2018, Tyers and Wright, 2019) (Fig. 4).
Fig. 4.
Types of synergistic combinations
Congruous represents a combination in which two antibiotics target different important molecular processes, for eg. a combination of rifampin–isoniazid– pyrazinamide for the treatment of tuberculosis and a combination of penicillin with streptomycin to combat enterococcal infections. In a syncretic combination, one antibiotic targets an essential process, and another targets a non- antibiotic adjuvant, overall acting as resistance breakers. In a coalistic pair, the compounds do not have any antibiotic activity and they target non-essential but synthetically lethal gene functions.
This offers a very promising area for antibiotic discovery and development, but, this process requires extensive research on specific targets and a wide knowledge about the resistance pathways. Although yeast model is the most studied one in case of genetic interaction but, still there is no new development in targeting lethal gene functions in antibiotic drug discovery efforts. Despite these challenges, a renewed interest and effort is required in developing both congruous and syncretic drug combinations to address the antibiotic resistance crisis. Although the development of combination therapies is more complicated than for single agents, i.e. monotherapy; but finding new single-agent antibiotics has also proved near fruitless for over one-quarter of a century, and all existing antibiotics at some point will have to be compromised by increasing levels of resistance.
Efforts have been initiated in all the above mentioned directions by several scientific groups throughout the world, who have been carrying out research to devise different methodologies to encounter this prominent issue with different perspectives. As the problem of antibiotic resistance is the most threatening problem of the twenty-first century, the strategies explained in the above section hold some potential in overcoming this problem; but looking at the limitations of each of the above-mentioned strategies, new molecular and alternate strategies still need to be considered for effectively combating the issue of antibiotic resistance.
6.2. Advanced molecular strategies to overcome antibiotic resistance
Molecular methods have a general advantage of providing direct access to the total pool of DNA, RNA or protein in a sample, as it is estimated that more than 99% of the environmental bacteria are not readily cultured using standard methods (Allen et al., 2010). Using molecular methods, the targets from test samples can be extracted directly or visualized under the microscope. Another advantage is that the targets are also relatively unambiguous, such as, the DNA, RNA or proteins upon being sequenced using molecular methods, can be compared against publicly available databases, which proves to be a more specific method in contrast with the often laborious and ill-defied phenotypic assessment of pure cultures.
6.2.1. Phenotypic conversion of drug-resistant to drug-sensitive bacteria
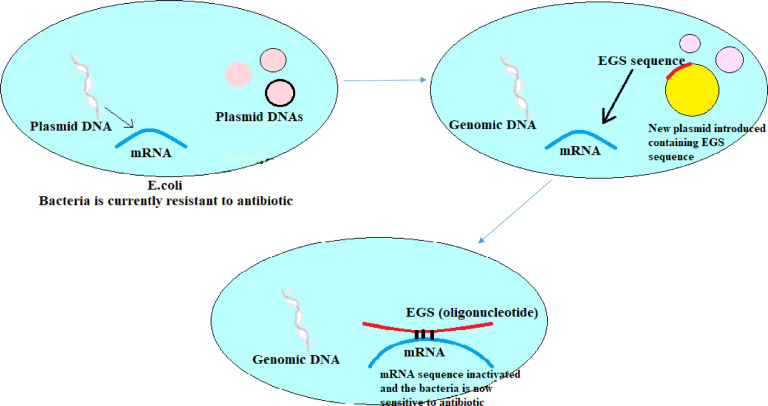
A promising strategy for the development of antibiotics is through antisense strategy involving RNase P-directed cleavage for inhibition of drug resistant activity in bacteria. Special genes are inserted in plasmids which code for small oligoribonucleotides (ORN) and are called External Guide Sequences (EGSs). These plasmids are then inserted in a bacteria like E.coli (Fig. 5). The property of these EGSs is to direct the RNase P to cleave the mRNAs which are transcribed to form proteins responsible for antibiotic resistance activity. Hence, they help in converting drug-resistant bacteria to drug-sensitive bacteria (Guerrier et al., 1997, Toney et al., 1998). EGS technology has been used to inhibit the expression of a broad range of genes, and this strategy has the potential for the development of novel treatments for many diseases. However, the major drawback of the use of EGS/ORN sequence is that these are rapidly degraded by nucleases.
Fig. 5.
Conversion of drug-resistant to drug-sensitive bacteria.
In this regard, Alfonso et al. (2007) conducted a study in which they demonstrated the use of EGS to reduce resistance towards amikacin by inhibition of expression of AAC(6)-Ib gene (gene which is responsible for resistance towards amikacin) (Alfonso et al., 2007). Their results proved that the use of external guide sequences is a viable strategy to conserve the potency of amikacin. They were also able to develop RNase P degrading ORN which were stable and viable in a nuclease environment known as locked nucleic acids (LNA) or (LNA)/DNA chimeric oligomers.
More recently, a new form of these sequences known as Bridge nucleic acids (BNAs) have also been developed, the BNA's have properties similar to LNA except that these are modified ribonucleotides that contain a bridge in their structures. For eg., a study was conducted on LNA and BNANC (second generation BNA), to compare their efficiency using the amikacin resistance aac(60)-Ib mRNA as target (Jackson et al., 2016). The researchers found that LNA/DNA gapmers were more efficient EGSs, whereas all BNANC/DNA gapmers showed very poor activity. Thus, although not much-researched, this strategy holds high potential in encountering the expanding problem of antibiotic-resistance. Furthermore, newer and exclusive BNA blends will be launched in the near future, providing great expectations from oligonucleotide-based fields of research and applications (Bistué et al., 2019).
6.2.2. DNA and mRNA vaccines
An effective way to prevent animals from becoming infected is vaccination. Better application of existing vaccines and development of new vaccines are crucial to tackle antibiotic resistance in order to reduce preventable disease and death. Also, vaccines serve as a great approach to limit the spread of antibiotic resistance, because it is due to the overuse of antibiotics only that the resistance is developing even against simplest of infections (Behr et al., 1999).
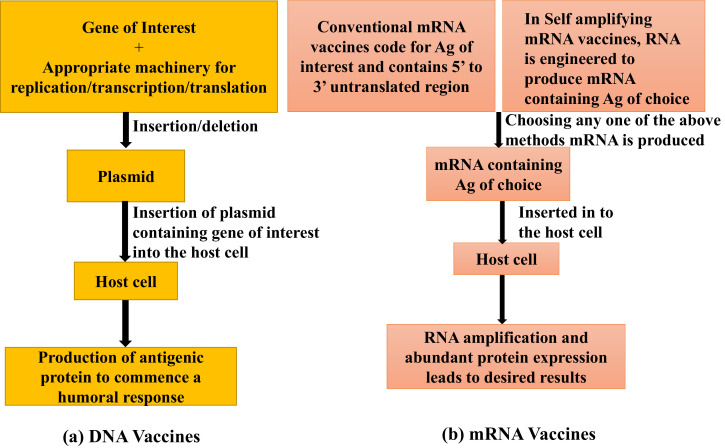
DNA vaccines serve as an important technology for vaccinating animals. These constitute insertion of the gene of interest into the plasmid along with appropriate machinery for replication, transcription and translation (Fig. 5). The plasmid is inserted into the host cell via suitable methods (direct injection, electroporation or gene gun etc.), where it results in the production of an antigenic protein that can commence cellular and humoral immune responses (Endmann et al., 2014). Both, conventional and self-amplifying mRNA (SAM) vaccines have been developed. Conventional mRNA-based vaccines code for the antigen of interest and contain 5′ and 3′ untranslated regions (UTRs), whereas in SAMs, the RNA is engineered in order to produce the RNA containing Ag of choice which enables RNA amplification and abundant protein expression (Pardi et al., 2018). The mRNA machinery does not interact with host DNA (Fig. 6). mRNA vaccines along with DNA vaccines have proved to be an attractive alternative to whole pathogen immunization. The mRNA vaccines were first tested in 1990’s, but not widely used due to factors such as omnipresent ribonucleases and their less stability. However, now there is a need for development of such vaccines which are specific and efficient (Zhang et al., 2019). These vaccines have various advantages like being non-infectious, non-integrating and egg-and-cell free; and also being capable of natural degradation, rapid production, and induction of B and T cell immune response.
Fig. 6.
Schematic representation of workflow of DNA and mRNA vaccines.
While it is doubtful that vaccines will be available for all kinds of infections, but, newer vaccines to prevent major infections are under development. Numerous comparable vaccines have already been developed, such as Bacille-Calmette-Guerin (BCG) vaccine to prevent tuberculosis (Pizza et al., 2000), haemophilus influenza type B (Hib) and pneumococcal conjugate vaccines (PCV7, PCV10, PCV13). Some vaccines are in the later stages of development against C. difficile (CDI) and Staphylococcus aureus infection (Jansen et al., 2018). The vaccine for Streptococcus agalactiae, also known as Group B Streptococcus (GBS) is at an early stage of development (Jansen et al., 2018). Many methods for vaccine approaches to prevent M.tuberculosis infections have been evaluated, however not many have been successful till date (Jansen et al., 2018). Hence, more research is mandatory with regard to the efficacy and safety of RNA and DNA vaccines prior to their wide use in domestic livestock and poultry.
Although, the immune systems have been developed to deal with infinite number of pathogens, there is no limit to how many vaccines can be given to an individual, be it a human or an animal. The most important consideration to stem the inappropriate use of antibiotics would always be an emphasis on reduction of their usage in agriculture and livestock. It is therefore vital to provide incentives and continuous investments to vaccine companies and for creating a market to develop and sell vaccines that are in public interest and also commercially viable. Considering the importance of vaccines for reducing AMR and global health, not following these trends could become a matter of public concern for many countries across the world.
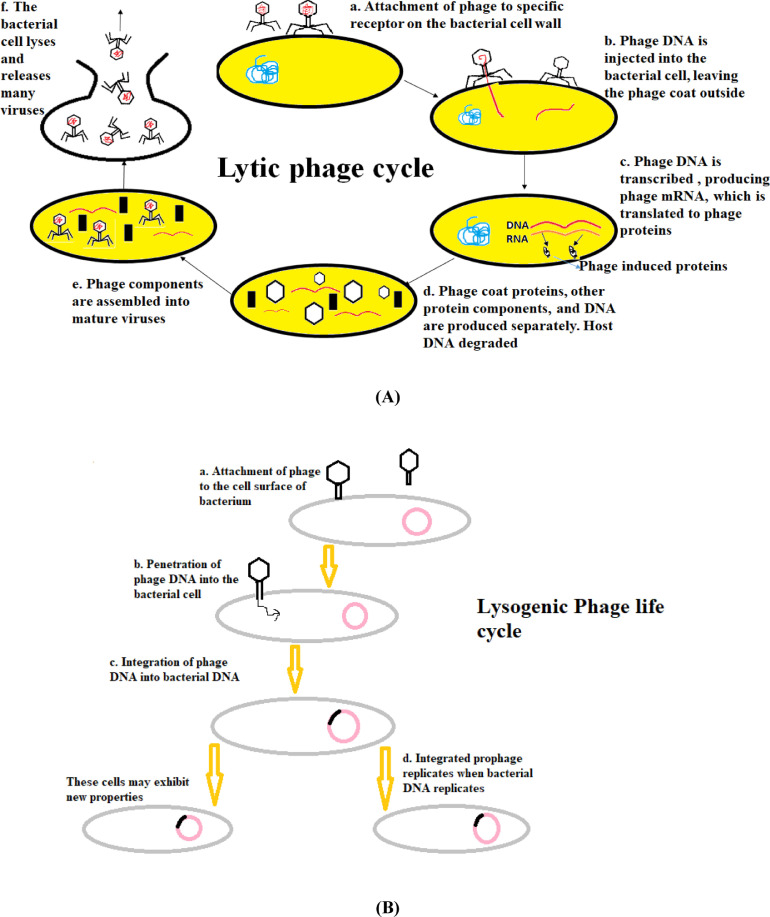
6.2.3. Bacteriophage therapy as an alternative to antibiotics
Bacteriophages or phages are viruses that infect bacteria and were discovered approximately a century ago. They have been used since then for therapeutic purposes by exploiting their lytic and lysogenic cycles (Fig. 7), by insertion of gene of interest to produce desired proteins. But, this was only practiced in a limited number of countries including Russia and adjoining regions. However, this mode of therapy had been highly neglected in the Western world. But, looking at its immense advantages, it is slowly picking up pace as a new mode of therapy to overcome antibiotic resistance. Bacteriophage therapy has proved to be an excellent alternative against many bacteria (Alisky et al., 1998). Studies have shown that bacteriophages could be used as a good scheme to treat systemic, mucosal and cutaneous infections, and they have also been suggested to be effective against multi-drug resistant bacteria. Phage therapy can be used against superbugs that are resistant to multiple antibiotics. Bacteriophages are very specific, therefore chances of developing secondary infections are circumvented. They are available where they are needed, as they replicate at the site of infection. One of the major advantages is that the phage-resistant bacteria remain susceptible to other phages having similar range of target organisms. The potential of phage therapy has been successfully determined against MDR uropathogenic E.coli (UPEC) for managing urinary tract infections (UTI's) as an alternative to chemotherapy (Malik et al., 2019, Malik et al., 2020).
Fig. 7.
(A) Lytic phage cycle and (B) Lysogenic phage cycle.
Phages can be exclusively used or can be used in combination with other antibacterial agents to improve their efficacy for biofilm related infections as well. Phages are natural predators of bacteria and have been considered favourable against bacterial biofilms. Lu and Collins (2007) genetically engineered the T7 phage and expressed a biofilm-degrading enzyme (dispersin B) which resulted in an improved efficacy of biofilm removal as compared to the wild-type phage (Lu et al., 2007, Esposito et al., 2017).
Various phage therapy studies have been conducted in animal models. Watanabe et al. (2007) researched a mouse model having gut deprived sepsis, and represented that phage delivery induced considerable defense against Pseudomonas aeruginosa (Watanabe et al., 2007). Fukuda et al. (2012) administered bacteriophage eye-drops using a model of P. keratitis and demonstrated high efficacy in P. aeruginosa elimination (Fukuda et al. 2012). Pabary et al. (2016) infected lungs of mice with P. aeruginosa and later delivered phage into it. They discovered a significant reduction in the bacterial load as well as a reduction in the spread of infection which was very well observed (Pabary et al., 2016).
However, bacteriophage therapy has a few shortcomings such as: bacteria may become resistant towards phages, and phages are specific and at times unstable at a low pH. But, studies are being conducted to overcome these problems. Further research developments may allow this field to pursue concerns regarding phage therapies and finally unlock their importance as antimicrobial agents.
6.2.4. Genetic modification technologies
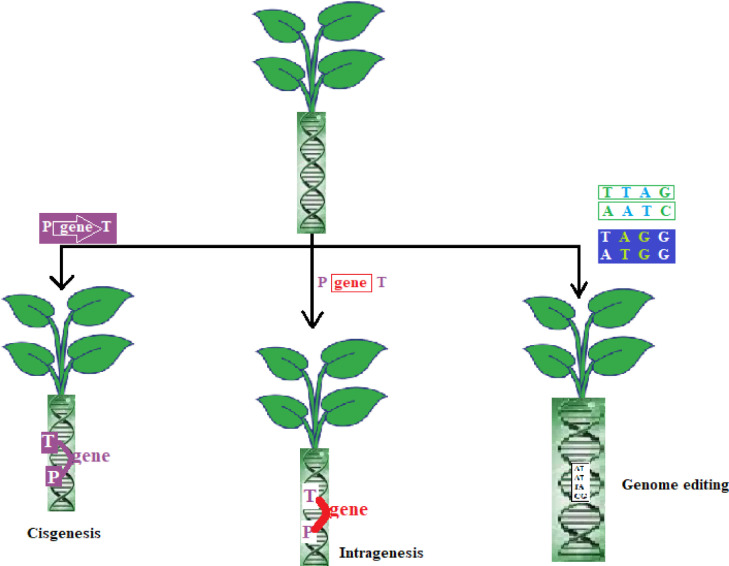
Genetic modifications in the field of agriculture mainly involve the production of transgenics. Transgenic crops, as discussed earlier have been used and developed by humans and possess various benefits such as better quality, better yield, resistance to pests and insects, and other desirable properties. But, they come with various disadvantages and are less accepted by consumers; so, in order to overcome these problems, alternate technologies such as cisgenesis, transgenesis, intragenesis and genome editing have been discovered to obtain improved plants/crops (Fig. 8).
Fig. 8.
Gene editing techniques.
6.2.4.1. Cisgenesis
In this technique, genetic modification is done by introduction of a copy of complete natural gene sequence consisting intron along with its own native promoter and terminator, in the sense orientation (Fig. 8). The desired cisgene can be taken from the crop species itself or from a sexually compatible plant species, just like conventional breeding (Schouten et al., 2006). However, in conventional plant breeding, undesired genes are also present along with the desired genes, whereas, this is not the case in cisgenic crops (they only contain desired genes). The deployment of cisgenes has been successfully carried out in plants to produce functions like resistance to scab in apple (Vanblaere et al., 2011), resistance to late blight in potato (Haverkort et. al., 2009), and increased phytase activity in barley (Holme et al., 2012).
6.2.4.2. Intragenesis
In this technology, for genetic modification, the desired gene is originated from the same crop species or sexually compatible plant species (in the sense or antisense orientation). However, it is not mandatory that the regulatory elements (promoter and terminator) are from the same gene (Fig. 8), thus giving novel genetic combinations (Rommens et al., 2007). Presence of regulatory elements from other genes might lead to a change in expression pattern of the desired gene. Various experiments have shown successful intragenesis such as, reduction of acrylamide (carcinogenic compounds) levels by tuber-specifc silencing of asparagine synthase-1 (StAst1) gene in potatoes. According to field trial results, the reduction in acrylamide forming potential of potatoes up to 70% was observed without affecting the tuber shape and yield (Chawla et al., 2012). Intragenic potato have been developed by J.R. Simplot Co., which exhibit multiple traits like resistance to bruising and discoloration, and a low acrylamide content (Waltz, 2015).
The potential risk associated with cisgenic plants is similar to traditionally bred plants, whereas, novel potential hazards are possible in case of intragenic and transgenic plants (EFSA 2012). The cisgenic and intragenic plants are considered as non-regulated (exempted from Biotechnology/GM organism regulation) by US Department of Agriculture (USDA), only if the introduced genetic elements are not derived from “plant pests”.
6.2.4.3. Genome editing
Genome editing technology is the latest one, amongst all these genetic modification technologies. Genome editing technologies are used to mutate, knock-out, or replace a specific gene (Fig. 8). This technique makes the use of either a sequence-specific nuclease (SSN) to produce gene knock-out/knock-in, or synthetic oligonucleotides to incorporate specific point mutations in the specific DNA region (Songstad et al. 2017). Synthetic oligonucleotides have also been utilized recently for targeted editing in order to produce custom single nucleotide polymorphisms (SNPs). There are various other methods of genome editing discovered recently (Kumar et al., 2020). It is expected that genome-edited crops may be granted quicker regulatory consent which should lead to their worldwide acquisition in cultivation.
6.2.5. Use of the CRISPR/Cas gene editing system as an antimicrobial agent to decrease antibiotic resistance
Use of conventional broad spectrum antibiotics leads to the elimination of beneficial commensal bacteria and an increase in antibiotic resistance. CRISPR/Cas systems have been used to target specific virulence factors and antibiotic resistance genes in bacteria. Thus, they constitute as an appealing option for further development of configurable and sequence specific antimicrobials Bikard and Barrangou (2017). This technology has been considered as the most important discovery of this century in the field of biotechnology, and has paved the pathway to gene-editing for therapeutic purposes and establishment of engineered microbials. CRISPR is unique because antimicrobials developed by the use of this technology are able to kill bacteria based on genetic sequence or gene specificity. Since it is always desirable to eliminate only a selected group of bacteria within a species, this technology should prove beneficial in such cases. Also, it can be used to re-sensitize the bacteria to the antibiotics towards which it has developed resistance.
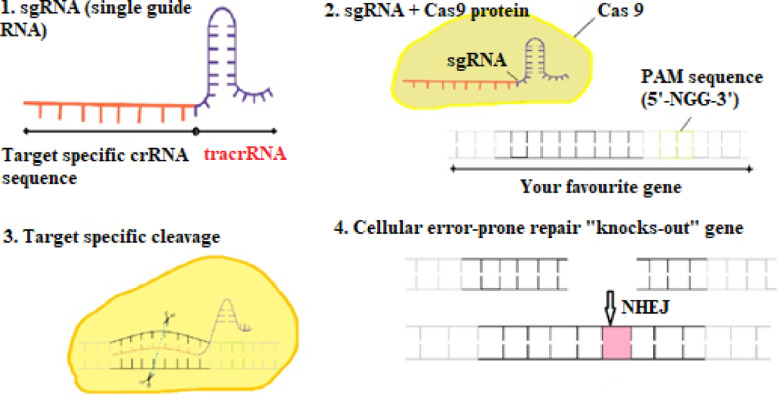
(Pak and SITN, 2014) In CRISPR/Cas technique, gene silencing (deletion of a segment of DNA) or gene editing (insertion of a segment of DNA) is done to inactivate the gene causing resistance (Pak, 2014). Rodrigues et al. (2019) engineered a plasmid to eliminate an ARG from Enterococcus faecalis, which is a part of benign intestinal flora (Rodrigues et al., 2019). However, when antibiotics kill beneficial bacteria in the intestine, E. faecalis becomes pathogenic and may acquire drug resistance. This has proved to be a major problem as resistant E. faecalis causes infections among patients in hospitals. However, gene editing has been successfully done using CRISPR-Cas9, and guide RNA to overcome this issue of drug resistance in E. faecalis. The guide RNA (homologous to the sequence in resistant DNA) directs the cell machinery to make cuts at specific places in the DNA for insertion of genes having a new sequence (Fig. 9), thus reversing antibiotic resistance (Bikard and Barangou, 2017, Gholizadeh et al., 2020).
Fig. 9.
Disruption caused by CRISPR-Cas9 system
(1) A single guide RNA (sgRNA), consists of crispr RNA sequence (crRNA, complementary to target DNA) + tracrRNA sequence (that binds to cas nuclease), (2) This sgRNA binds to recombinant Cas9 protein having activity similar to DNA endonuclease. (3) This complex then causes a target-specific ds-DNA cleavage. (4)The cleavage site is repaired by DNA repair pathway (non-homologous end joining, NHEJ). This leads to a process which is error-prone that may result in insertions or deletions (INDELs), which may disrupt gene function. Photospacer Adjacent Motif (PAM) is a short region (2-6 bp) which is present downstream of the DNA region which is being targeted for cleavage by CRISPR-Cas system.
CRISPR-Cas approach may therefore unlock novel ways for the development of modish antibiotics, which can wave-off MDR pathogens and differentiate between pathogenic and beneficial microorganisms. These systems can be utilized to selectively remove individual bacterial isolates/strains based on the sequence-specific manner, generating new opportunities for the treatment of MDR infections, the control of industrial fermentations and the study of microbial consortia.
7. Conclusion
In the present century, microbial infections have become a major clinical threat as there are rising concerns about the failure of treating diseases by using antibiotics due to resistance being developed by several bacteria for the commonly used antibiotics. A lot of evidence suggests that excessive utilization of antibiotics in agriculture and animal husbandry is contributing to the spread of this antibiotic resistance in different environments. Due to the application of manure from farm animals, spraying of antimicrobials in farmland, and practicing other activities to combat infections, transmittance of higher levels of antibiotic resistance genes have been observed at various levels of different ecosystems. In this review paper, we have described antibiotic resistance mechanisms that the bacteria have achieved over the years and also the most feasible techniques that are currently being used in this field to reduce the resistance caused by antibiotics. To control this problem of multi-drug resistance, a lot of strategies have been discovered and to our understanding, molecular methods have also been designed and applied but successful completion of control over this problem has not yet been achieved. Considering the fact that we can't completely cease the use of antibiotics because this is our primary, and in many cases the only method for treating infectious diseases; a more detailed study of environmental reservoirs and advanced remedial strategies to eradicate resistance to the lowest limit are crucial for our future ability to fight infections .
CRediT authorship contribution statement
Avantika Mann: Conceptualization, Methodology, Investigation, Resources, Data curation, Writing – original draft, Visualization. Kiran Nehra: Validation, Writing – review & editing, Supervision. J.S. Rana: Validation, Writing – review & editing, Supervision. Twinkle Dahiya: Conceptualization.
Declaration of Competing Interest
The authors want to state that they do not have any conflict of interest to declare.
Acknowledgement
The authors wish to acknowledge National Project Implementation Unit (NPIU), a unit of Ministry of Human Resource Development, Government of India, for the financial assistantship awarded to Ms. Avantika Mann through TEQIP-III project at Deenbandhu Chhotu Ram University of Science and Technology, Murthal, Sonipat, Haryana.
References
- Aarestrup F.M., Jensen V.F., Emborg H.D., Jacobsen E., Wegener H.C. Changes in the use of antimicrobials and the effects on productivity of swine farms in denmark. Am. J. Vet. Res. 2010;71:726–733. doi: 10.2460/ajvr.71.7.726. [DOI] [PubMed] [Google Scholar]
- Aldred K.J., Kerns R.J., Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfonso J.C., Soler Bistué, Ha H., Sarno R., Don M., Zorreguieta A., Tolmasky M.E. External guide sequences targeting the aac (6′)-Ib mRNA induce inhibition of amikacin resistance. Antimicrob. Agents and Chemother. 2007 doi: 10.1128/AAC.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisky J., et al. Bacteriophages show promise as antimicrobial agents. J. Infect. 1998;36:5–15. doi: 10.1016/s0163-4453(98)92874-2. [DOI] [PubMed] [Google Scholar]
- Allen H.K., Donato J., Wang H.H., Cloud-Hansen K.A., Davies J., Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- Bawa A.S., Anilakumar K.R. Genetically modifed foods: safety, risks and public concerns—a review. J. Food Sci. Technol. 2013;50:1035–1046. doi: 10.1007/s13197-012-0899-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr M., et al. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419. [DOI] [PubMed] [Google Scholar]
- Beppler, et al. When more is less: emergent suppressive interactions in three-drug combinations. BMC Microbiol. 2017;17:107. doi: 10.1186/s12866-017-1017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Barrangou R. Using CRISPR-cas systems as antimicrobials. Curr. Opin. Microbiol. 2017;37:155–160. doi: 10.1016/j.mib.2017.08.005. 37. [DOI] [PubMed] [Google Scholar]
- Bistué A.S., Zorreguieta A., Tolmasky M.E. Bridged nucleic acids reloaded. Molecules. 2019;24(12):2297. doi: 10.3390/molecules24122297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.F., Reynolds P.E. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 1980;122:275–278. doi: 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- Brown D.W., Schaab M.R., Birmingham W.R., Armstrong R.N. Evolution of the antibiotic resistance protein, FosA, is linked to a catalytically promiscuous progenitor. Biochemistry. 2009;48 doi: 10.1021/bi900078q. PMID: 19196010, PMCID: PMC2756217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushby S.R., Hitchings G.H. Trimethoprim, a sulphonamide potentiator. Br. J. Pharmacol. Chemother. 1968;33:72–90. doi: 10.1111/j.1476-5381.1968.tb00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne-Bailey K.G., Gaze W.H., Kay P., Boxall A.B.A., Hawkey P.M., Wellington E.M.H. Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob. Agents Chemother. 2009;53:696–702. doi: 10.1128/AAC.00652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena M., Durso L.M., Miller D.N., Waldrip H.M., Castleberry B.L., Drijber R.A., Wortman C. Tetracycline and sulfonamide antibiotic resistance genes in soils from nebraska organic farming operations. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.01283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H.F., Deleo F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapleur Y. Vol. 121. VCH, Weinheim; 1998. p. 6972. (Carbohydrate Mimics: Concepts and Methods). [DOI] [Google Scholar]
- Chawla R., Shakya R., Rommens C.M. Tuber-specifc silencing of asparagine synthetase-1 reduces the acrylamide-forming potential of potatoes grown in the feld without afecting tuber shape and yield. Plant Biotechnol. J. 2012;10:913–924. doi: 10.1111/j.1467-7652.2012.00720.x. [DOI] [PubMed] [Google Scholar]
- Culp E.J., Waglechner N., Wang W., Fiebig-Comyn A.A., Yen-Pang H., Koteva K., Sychantha D., Coombes B.K., Michael S., Van N., Brun Y.V., Wright G.D. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodeling. Nature. 2020;578:582–587. doi: 10.1038/s41586-020-1990-9. [DOI] [PubMed] [Google Scholar]
- Deshayes S., Xian W., Schmidt N.W., Kordbacheh S., Lieng J., Wang J., Zarmer S., et al. Designing hybrid antibiotic peptide conjugates to cross bacterial membranes. Bio Conjugate Chem. 2017;28:793–804. doi: 10.1021/acs.bioconjchem.6b00725. [DOI] [PubMed] [Google Scholar]
- Drlica K., Hiasa H., Kerns R., Malik M., Mustaev A., Zhao X. Quinolones: action and resistance updated. Curr. Top. Med. Chem. 2009;9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egorov A.M., Ulyashova M.M., Rubtsova M.Y. Bacterial enzymes and antibiotic resistance. Acta. Naturae. 2018;10:33–48. [PMC free article] [PubMed] [Google Scholar]
- EMA and EFSA Joint scientific opinion on measures to reduce the need to use antimicrobial agents in animal husbandry in the European Union, and the resulting impacts on food safety (RONAFA) EFSA J. 2017;15:1–332. doi: 10.2903/j.efsa.20174666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endmann A., Klünder K., Kapp K., Riede O., Oswald D., Talman E.G., Schroff M., Kleuss C., Ruiters M.H., Juhls C. Cationic lipid-formulated DNA vaccine against hepatitis B virus: immunogenicity of MIDGE-th1 vectors encoding small and large surface antigen in comparison to a licensed protein vaccine. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S., Simone G.D. Update on the main MDR pathogens: prevalence and treatment options. Infez Med. Riv. Period Eziologia Epidemiol Diagn Clin E TerDellePatolInfett. 2017;25:301–310. [PubMed] [Google Scholar]
- Fjell C.D., Hiss J.A., Hancock R.E., Schneider G. Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 2012;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- Fong D.H., Berghuis A.M. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 2002;21(15):2323–2331. doi: 10.1093/emboj/21.10.2323. EMBO J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.L. Antimicrobial peptides stage a comeback. Nat. Biotechnol. 2013;31:379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
- Fukuda K., Ishida W., Uchiyama J., Rashel M., Kato S., Morita T., et al. Pseudomonas aeruginosa keratitis in mice: effects of topical bacteriophage KPP12 administration. PloS One. 2012;7:e47742. doi: 10.1371/journal.pone.0047742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly N.K. Global antibiotic resistance partnership (GARP) Indian J. Med. Res. 2011;134:281–294. [PMC free article] [PubMed] [Google Scholar]
- Ge M., et al. Vancomycin derivatives that inhibit peptidoglycan biosynthesis without binding D-Ala-D-Ala. Science. 1999;284:507–511. doi: 10.1126/science.284.5413.507. [DOI] [PubMed] [Google Scholar]
- Gholizadeh P., Kose S., Dao S., Ganbarov K., Tanomand A., Dal T., Aghazadeh M., Ghotaslou R., Ahangarzadeh R.M., Yousefi B., Samadi K.H. Dovepress; 2020. How CRISPR-Cas System Could Be Used to Combat Antimicrobial Resistance; pp. 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., LaPara T.M. The effect of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007;1:191. doi: 10.1038/ismej.2007.31. -20. [DOI] [PubMed] [Google Scholar]
- Gilbert N. A hard look at GM crops. Nature. 2013;497:24–26. doi: 10.1038/497024a. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Bello C. Antibiotic adjuvants — a strategy to unlock bacterial resistance to antibiotics. Bioorg. Med. Chem. Lett. 2017;27:4221–4228. doi: 10.1016/j.bmcl.2017.08.027. [DOI] [PubMed] [Google Scholar]
- Grifths A.J.F., Wessler S.R., Lewontin R.C., Gelbart W.M., Suzuki D.T., Miller J.H. 8th (ed.) FreemanWH; New York: 2005. Introduction to Genetic Analysis.https://www.bio.bg.ac.rs/materijali_predmeta/med-eng-griffiths-an-introduction-to-genetic-analysis.pdf [Google Scholar]
- Guerrier C.T., Salavati R., Altman S. Phenotypic conversion of drug-resistant bacteria to drug sensitivity. Proc. Natl. Acad. Sci. USA. 1997;94:8468–8472. doi: 10.1073/pnas.94.16.8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.N., Alam A., Hasnain S.E. Protein promiscuity in drug discovery, drug-repurposing and antibiotic resistance. Biochimie. 2020;175:50–57. doi: 10.1016/j.biochi.2020.05.004. [DOI] [PubMed] [Google Scholar]
- Hancock R.E., Chapple D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999;43:1317–1323. doi: 10.1128/AAC.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R.E., Sahl H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- Haverkort J, Struik PC, Visser RGF, Jacobsen E. Applied biotechnology to combat late blight in potato caused by phytophthora infestans. Potato Res. 2009;52:249–264. doi: 10.1007/s11540-009-9136-3. [DOI] [Google Scholar]
- Holme I.B., Dionisio G., Brinch-Pedersen H., Wendt T., Madsen C.K., Vincze E., Holm P.B. Cisgenic barley with improved phytase activity. Plant. Biotechnol. J. 2012;10:237–247. doi: 10.1111/j.1467-7652.2011.00660.x. [DOI] [PubMed] [Google Scholar]
- Hooper D.C., Jacob G.A. Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. cold spring harb. Perspect. Med. 2016;6:1–21. doi: 10.1101/cshperspect.a025320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A., Jani S., Davies-Sala C., Alfonso J.C., Soler B., Zorreguieta A., Tolmasky M.E. Assessment of configurations and chemistries of bridged nucleic acids-containing oligomers as external guide sequences: a methodology for inhibition of expression of antibiotic resistance genes. Biol. Methods Protocols. 2016:1–8. doi: 10.1093/biomethods/bpw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen K.U., Anderson A.S. The role of vaccines in fighting antimicrobial resistance (AMR) Hum. Vaccines Immun. 2018;14:1–17. doi: 10.1080/21645515.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawetz E., Gunnison J.B., Bruff J.B., Coleman V.R. Studies on antibiotic synergism and antagonism: synergism among seven antibiotics against various bacteria in vitro. J. Bacteriol. 1952;64:29–39. doi: 10.1128/jb.64.1.29-39.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D., Joo S.W., Shinde V.V., Cho E., Jung S. Carbohydrate-based host-guest complexation of hydrophobic antibiotics for the enhancement of antibacterial activity. Molecules. 2017;22:1311. doi: 10.3390/molecules22081311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose M., Munita, Cesar A. Mechanisms of antibiotic resistance. Microbiol Spectr. 2017:4. doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalan L., Wright G.D. Antibiotic adjuvants: multicomponent anti-infective strategies. Expert Rev. Mol. Med. 2011;13 doi: 10.1017/S1462399410001766. [DOI] [PubMed] [Google Scholar]
- Karesh W.B., Dobson A., Lloyd-Smith J.O., Lubroth J., Dixon M.A., et al. Ecology of zoonoses: natural and unnatural histories. Lancet. 2012;380:1936–1945. doi: 10.1016/S0140-6736(12)61678-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleina E.Y., Van Boeckeld T.P., Martineza E.P., Panta S., Gandraa S., Levine S.A., Goossensh H., Laxminarayan R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. PNAS. 2018;15:3463–3470. doi: 10.1073/pnas.1717295115. www.pnas.org/cgi/doi/10.1073/pnas.1717295115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Gambhir G., Dass A., Tripathi A.K., Singh A., Jha A.K., Yadava P., Choudhary M., Rakshit S. Genetically modifed crops: current status and future prospects. Planta. 2020;251:91. doi: 10.1007/s00425-020-03372-8. [DOI] [PubMed] [Google Scholar]
- Lamb H.M., Wiseman L.R. Pexiganan acetate. Drugs. 1998;56:1047–1052. doi: 10.2165/00003495-199856060-00011. [DOI] [PubMed] [Google Scholar]
- Laxminarayan R., Van Boeckel T.P., Teillant A. The economic costs of withdrawing antimicrobial growth promoters from the livestock sector. OECD Food Agri. Fisheries Papers. 2015 doi: 10.1787/5js64kst5wvl-en. [DOI] [Google Scholar]
- Lekagul A., Tangcharoensathienb V., Yeung S. Patterns of antibiotic use in global pig production: a systematic review. Prevent. Vet. Med. 2018;160:85–98. doi: 10.1016/j.vas.2019.100058. [DOI] [PubMed] [Google Scholar]
- Li W., Neil M., O'Brien S., Holden J.A., Otvos L., Reynolds E.C., Separovic F., Hossain M.A., Wade J.D. Covalent conjugation of cationic antimicrobial peptides with a b-lactam antibiotic core. Peptide Sci. 2018;110:1–9. doi: 10.1002/pep2.24059. [DOI] [Google Scholar]
- Lipsky B.A., Holroyd K.J., Zasloff M. Topical versus systemic antimicrobial therapy for treating mildly infected diabetic foot ulcers: a randomized, controlled, double-blinded, multicenter trial of pexiganan cream. Clin. Infect. Dis. 2008;47:1537–1545. doi: 10.1086/593185. [DOI] [PubMed] [Google Scholar]
- Losey J.E., Rayor L.S., Carter M.E. Transgenic pollen harms monarch larvae. Nature. 1999;399:214. doi: 10.1038/20338. [DOI] [PubMed] [Google Scholar]
- Lu T.K., Collins J.J. Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. USA. 2007;104:11197–11202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacManus M.C. Mechanisms of bacterial resistance to antimicrobial agents. Am. J. Health Syst Pharm. 1997;54:1420–1433. doi: 10.1093/ajhp/54.12.1420. [DOI] [PubMed] [Google Scholar]
- Magnet S., Blanchard J.S. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 2005;105:477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- Malik S., Rana J.S., Nehra K. Escherichia phage PS6: a lytic phage for the bio-control of Escherichia coli causing urinary tract infections. Res J Biotechnol. 2020;15:1–9. doi: 10.1007/s12223-019-00750-y. [DOI] [Google Scholar]
- Malik S., Sidhu P.K., Rana J.S., Nehra K. Managing urinary tract infections through phage therapy: a novel approach. Folia Microbiologica. 2019 doi: 10.1007/s12223-019-00750-y. [DOI] [PubMed] [Google Scholar]
- Malmsten M. Interactions of antimicrobial peptides with bacterial membranes and membrane components. Curr. Top. Med. Chem. 2016;16:16–24. doi: 10.2174/1568026615666150703121518. [DOI] [PubMed] [Google Scholar]
- Mingeot-Leclercq M.P., Glupczynski Y., Tulkens P.M. Aminoglycosides: activity and resistance. Antimicrob. Agents Chemother. 1999;43:727–737. doi: 10.1128/aac.43.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering R.C. Jr. Rationale for use of antimicrobial combinations. Am. J. Med. 1983;75:4–8. doi: 10.1016/0002-9343(83)90088-8. [DOI] [PubMed] [Google Scholar]
- Nguyen L.T., Haney E.F., Vogel H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011;29:464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Nikolaidis I., Favini-Stabile S., Dessen A. Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci. 2014;23:243–259. doi: 10.1002/pro.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson A.C., Janson H., Wold H., Fugelli A., Andersson K., Håkangård C., et al. LTX-109 is a novel agent for nasal decolonization of methicillinresistant and -sensitive Staphylococcus aureus. Antimicrob. Agents Chemother. 2015;59:145–151. doi: 10.1128/aac.03513-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil J. Tackling drug-resistant infections globally: final report and recommendations. Rev. Antimicro. Resistance. 2016:84. https://amr-review.org/publications [Google Scholar]
- Pabary R., Singh C., Morales S., Bush A., Alshafi K., Bilton D., et al. Antipseudomonal bacteriophage reduces infective burden and inflammatory response in murine lung. Antimicrob Agents Chemother. 2016;60:744–751. doi: 10.1128/AAC.01426-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak E., SITN . Harvard University; 2014. The Graduate School of Arts and Sciences.http://sitn.hms.harvard.edu/flsh/2014/crispr-a-game-changing-genetic-engineering-technique [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasupuleti M., Schmidtchen A., Malmsten M. Antimicrobial peptides: key components of the innate immune system. Crit. Rev. Biotechnol. 2012;32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- Petz M., Solly R., Lymburn M., Clear M.H. Thin-layer chromatographic determination of erythromycin and other macrolide antibiotics in livestock products. J. Ass. Official Anal. Chem. 1987;70:691–697. PMID: 3624179. [PubMed] [Google Scholar]
- Piddock L.J.V. Overuse of fluoroquinolones in human and veterinary medicine can breed resistance. BMJ. 1998;317:1029–1030. doi: 10.1136/bmj.317.7165.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D.P., Melo L.D.R, Boas D.V., Sillankorva S., Azeredo J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 2017;39:48–56. doi: 10.1016/j.mib.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Pizza M., et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459. [DOI] [PubMed] [Google Scholar]
- Popowska M., Rzeczycka M., Miernik A., Balska A.K., Walsh F., Duffy B. Influence of soil use on prevalence of tetracycline, streptomycin, and erythromycin resistance and associated resistance genes. Anti. Microb Agents and Chemother. 1987 doi: 10.1128/AAC.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Huang Y., Lv X. Crisis of antimicrobial resistance in china: now and the future. Front. Microbiol. 2019 doi: 10.3389/fmicb.2019.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M.S., Tolmasky M.E. Amikacin: uses, Resistance, and Prospects for Inhibition. Molecules. 2017;22:2267. doi: 10.3390/molecules22122267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M.S., Tolmasky M.E. Aminoglycoside modifying enzymes. Drug Resist. Update. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjalkar J., Chandy S.J. India's National Action Plan for antimicrobial resistance – an overview of the context, status, and way ahead. J. Family Med. Prim. Care. 2019;8(6):1828–1834. doi: 10.4103/jfmpc.jfmpc_275_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, M.C., 2019. Tetracycline Therapy: Update. CID 36, 462–7. doi: 10.1086/367622. [DOI] [PubMed]
- Roberts M.C. Tetracycline resistance determinants: mechanisms of action, regulation of expression, genetic mobility, and distribution. FEMS Microbiol. Rev. 1996;19:1–24. doi: 10.1111/j.1574-6976.1996.tb00251.x. [DOI] [PubMed] [Google Scholar]
- Rodrigues M., McBride S.W., Hullahalli K., Palmer K.L., Duerkop B.A. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant enterococci. Antimicrobial. Agents Chemother. 2019 doi: 10.1128/AAC.01454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommens C.M., Haring M.A., Swords K., Davies H.V. The intragenic approach as a new extension to traditional plant breeding. Trends Plant Sci. 2007;12:397–403. doi: 10.1016/j.tplants.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Rostock L., Driller R., Gratz S., Kerwat D., Eckardstein L.V., Petras D., Kunert M., Alings C., Schmitt F.J., Friedrich T., Wahl M.C., Loll B., Mainz A., Sussmuth R.D. Molecular insights into antibiotic resistance - how a binding protein traps albicidin. Nat. Commun. 2018;9:3095. doi: 10.1038/s41467-018-05551-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers A.A., Amabile-Cuevas C.F. Why are antibiotic resistance genes so resistant to elimination. Antimicrob agents chemother. 1997;11:2321–2325. doi: 10.1128/AAC.41.11.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten H.J., Krens F.A., Jacobsen E. Cisgenic plants are similar to traditionally bred plants. EMBO Rep. 2006;7:750–753. doi: 10.1038/sj.embor.7400769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears P., Wong C.H. A highly active catalyst for the room-temperature amination and suzuki coupling of aryl chlorides. Angew. Chem. 1999;(16):2413–2416. doi: 10.1002/(sici)1521-3773(19990816)38:16<2413::aid-anie2413>3.0.co;2-h. 10.1002/(sici)1521-3773(19990816)38:16<2413::aid-anie2413>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Sidhu P.K., Nehra K. Purification and characterization of bacteriocin Bac23 extracted from Lactobacillus plantarum PKLP5 and its interaction with silver nanoparticles for enhanced antimicrobial spectrum against food-borne pathogens. LWT. 2021;139 doi: 10.1016/j.lwt.2020.110546. [DOI] [Google Scholar]
- Silver L.L. Multi-targeting by monotherapeutic antibacterials. Nat. Rev. Drug Discov. 2007;6:41–55. doi: 10.1038/nrd2022. [DOI] [PubMed] [Google Scholar]
- Songstad DD, Petolino JF, Voytas DF, Reichert NA. Genome Editing of Plants. Crit Rev Plant Sci. 2017;36:1–23. doi: 10.1080/07352689.2017.1281663. [DOI] [Google Scholar]
- Souza, Rocha V.M., Valera A., Eguiarte L.E. Genetic structure of natural populations of Escherichia coli in wild hosts on different continents. Appl. Environ. Microbiol. 1999;65:3373–3385. doi: 10.1128/AEM.65.8.3373-3385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B., Gilbert D.N. The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis. 2014;59:71–75. doi: 10.1093/cid/ciu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton P., Taylor P.W. Methicillin resistance in Staphylococcus aureus mechanisms and modulation. Sci Prog. 2002;85:57–72. doi: 10.3184/003685002783238870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz M.N. Hospital acquired infections: diseases with increasingly limited therapies. Proc. Natl. Acad. Sci. 1994;91:2420–2427. doi: 10.1073/pnas.91.7.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi D., Shukla S.K., Prakash O., Zhang G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie. 2010;92:1236–1241. doi: 10.1016/j.biochi.2010.02.023. [DOI] [PubMed] [Google Scholar]
- Tekin E., et al. Enhanced identification of synergistic and antagonistic emergent interactions among three or more drugs. J. R. Soc. 2016;13 doi: 10.1098/rsif.2016.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekin E., et al. Prevalence and patterns of higher-order drug interactions in Escherichia coli. NPJ Syst. Biol. Appl. 2018;4:31. doi: 10.1038/s41540-018-0069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R.K., Chi-Huey W. Carbohydrate-based antibiotics: a new approach to tackling the problem of resistance. Angew. Chem. Int. Ed. 2001;40:3508–3533. doi: 10.1002/1521-3773(20011001)40:19<3508::AID-ANIE3508>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Toney J.H., et al. Antibiotic sensitisation using biphenyl tetrazolesas potent inhibitors of Bacteroidesfragilismetallo-b-lactamase. Chem. Biol. 1998;5:185–196. doi: 10.1016/s1074-5521(98)90632-9. [DOI] [PubMed] [Google Scholar]
- Tyers M., Wright G.D. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019;17:141–155. doi: 10.1038/s41579-018-0141-x. [DOI] [PubMed] [Google Scholar]
- Van Boeckel T.P., Brower C., Gilbert M., Grenfell B.T., Levin S.A., Robinson T.P., Teillant A., Laxminarayan R. Global trends in antimicrobial use in food animals. PNAS. 2015;112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanblaere T., Szankowski I., Schaart J., Schouten H., Flachowsky H., Broggini G.A., Gessler C. The development of a cisgenic apple plant. J. Biotechnol. 2011;154:304–311. doi: 10.1016/j.jbiotec.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Velden W.J., Van Iersel T.M., Blijlevens N.M., Donnelly J.P. Safety and tolerability of the antimicrobial peptide human lactoferrin 1-11 (hLF1-11) BMC Med. 2009;7:44. doi: 10.1186/1741-7015-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walson J.L., Marshall B., Pokhrel B.M., Kafle K.K., Levy S.B. Carriage of antibiotic-resistant fecal bacteria in Nepal reflects proximity to Kathmandu. J. Infect. Dis. 2001;184:1163–1169. doi: 10.1086/323647. [DOI] [PubMed] [Google Scholar]
- Waltz E. USDA approves next-generation GM potato. Nat Biotechnol. 2015;33:12–13. doi: 10.1038/nbt0115-12. [DOI] [PubMed] [Google Scholar]
- Wang T.T., Nestel F.P., Bourdeau V., Nagai Y., Wang Q., Liao J., et al. Cutting edge: 1, 25-dihydroxyvitamin D3 is a direct inducer of antimicrobial peptide gene expression. J. Immunol. 2004;173:2909–2912. doi: 10.4049/jimmunol.173.5.2909. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Matsumoto T., Sano G., Ishii T., Tateda K., Sumiyama Y., et al. Efficacy of bacteriophage therapy against gut-derived sepsis caused by Pseudomonas aeruginosa in mice. Antimicrob. Agents Chemother. 2007;51:446–452. doi: 10.1128/AAC.00635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G., Heilborn J.D., Chamorro Jimenez C.I., Hammarsjo A., Törmä H., Stahle M. Vitamin D induces the antimicrobial protein hCAP18 in human skin. J. Invest. Dermatol. 2005;124:1080–1082. doi: 10.1111/j.0022-202X.2005.23687.x. [DOI] [PubMed] [Google Scholar]
- Witte W. Selective pressure by antibiotic use in livestock. Int. J. Antimicrob. Agents. 2000;16:19–24. doi: 10.1016/S0924-8579(00)00301-0. [DOI] [PubMed] [Google Scholar]
- Wonhee S., Jared L.C., Zhanel G.G., Nicolau D.P. Comparison of in vivo and in vitro pharmacodynamics of a humanized regimen of 600 milligrams of ceftaroline fosamil every 12 hours against staphylococcus aureus at initial inocula of 106 and 108 CFU per milliliter. Am. Soc. Microbiol. 2014;58:6931–6933. doi: 10.1128/AAC.03652-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington R.J., Melander C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 2013;31:177–184. doi: 10.1016/j.tibtech.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G.D. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol. 2016;24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- Yamshchikov A.V., Desai N.S., Blumberg H.M., Ziegler T.R., Tangpricha V. Vitamin D for treatment and prevention of infectious diseases: a systematic review of randomized controlled trials. Endocr. Pract. 2009;15:438–449. doi: 10.4158/ep09101.orr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman M.R., Yount N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- Yeung A.T., Gellatly S.L., Hancock R.E. Multifunctional cationic host defence peptides and their clinical applications. Cell. Mol. Life Sci. 2011;68:2161–2176. doi: 10.1007/s00018-011-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Maruggi G., Shan H., Li J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019;10:594. doi: 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]