Abstract
BACKGROUND
Immunotherapy has revolutionized the clinical outcomes of intractable cancer patients. Little is known about the intestinal nonpathogenic bacterial composition of hepatocellular carcinoma (HCC) patients treated by immunotherapy.
AIM
To determine whether there is a correlation between gut bacterial composition and prognosis in HCC patients.
METHODS
From September 2019 to March 2020, we prospectively collected fecal samples and examined the gut microbiome of 8 advanced HCC patients treated with nivolumab as a second- or third-line systemic treatment. Fecal samples were collected before the start of immunotherapy. Fecal samples of patients with progression during treatment were collected at the time of progression, and fecal samples of patients who showed good response to nivolumab were collected after 5-7 mo as follow-up. Metagenomic data from 16S ribosomal RNA sequencing were analyzed using CLC Genomics Workbench. Microbiome data were analyzed according to therapeutic response.
RESULTS
All 8 patients were male, of which 6 had underlying chronic hepatitis B. A higher Shannon index was found in the responders than in the non-responders after nivolumab therapy (P = 0.036). The unweighted beta diversity analysis also showed that the overall bacterial community structure and phylogenetic diversity were clearly distinguished according to therapeutic response. There was no significant difference in the diversity or composition of the patient gut microbiome according to the immunotherapy used. Several taxa specific to therapeutic response were designated as follows: Dialister pneumosintes, Escherichia coli, Lactobacillus reteri, Streptococcus mutans, Enterococcus faecium, Streptococcus gordonii, Veillonella atypica, Granulicatella sp., and Trchuris trichiura for the non-responders; Citrobacter freundii, Azospirillum sp. and Enterococcus durans for the responders. Of note, a skewed Firmicutes/Bacteroidetes ratio and a low Prevotella/Bacteroides ratio can serve as predictive markers of non-response, whereas the presence of Akkermansia species predicts a good response.
CONCLUSION
The current presumptive study suggests a potential role for the gut microbiome as a prognostic marker for the response to nivolumab in treatment of HCC patients.
Keywords: Microbiome, Nivolumab, Firmicutes/Bacteroidetes ratio, Prevotella/Bacteroides ratio, Hepatocellular carcinoma, Prognosis
Core Tip: Immune check point inhibitors are known to be an effective treatment option for advanced hepatocellular carcinoma (HCC), not only for second-line, but also as a first-line treatment. However, there are few predictive or prognostic markers for which patient group will have a good treatment response to immunotherapy or systemic therapy for HCC until now. Our study shows that non-responders to nivolumab in HCC patients have dysbiotic fecal composition, whereas a high Prevotella/Bacteroides ratio can predict a better response to nivolumab, highlighting a potential role for the gut microbiome as a prognostic marker for the response to nivolumab therapy.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the most frequent primary malignancy of the liver, and it is now the second leading cause of cancer death worldwide[1]. Curative HCC treatment is only feasible in the early stages and involves local ablative procedures, surgical resection, or liver transplantation. For patients not amenable to curative therapy and in those with metastatic disease, other systemic treatments, such as sorafenib, are available. Beyond the limits of standard therapies, immunotherapy has been introduced for the oncological treatment of various solid malignancies. Nivolumab, as the first programmed cell death protein-1 (PD-1) agent, has shown great promise in treatment of various cancers, including melanoma and squamous cell carcinoma of the head and neck[2,3]. Recently, it was approved as the second-line agent for advanced HCC patients who have experienced sorafenib failure[4,5]. In line with this growing clinical relevance, stratification of patients receiving nivolumab therapy is now required to predict the prognosis and tumor aggressiveness.
There is mounting evidence that the gut microbiota can influence and modulate host immune responses; thus, microbiome profiling has been revealed as a predictor for response to immunotherapy among different groups and in different countries[6-8]. Moreover, some immunostimulatory bacterial species, including Akkermansia muciniphila[6], Bifidobacterium longum[7] and Bacteroidetes fragilis[8], were reported to elicit systemic immune responses and to reprogram the tumor microenvironment in mouse tumors treated with anti–cytotoxic T-lymphocyte-associated-4 and/or anti–PD-1 antibodies. The human gut microbiota has been shown to be associated with clinical responses to anti-PD-1/programmed death ligand 1 (PD-L1) immunotherapy in melanoma, non-small cell lung cancer, and renal cell carcinoma. However, the association between gut microbiota and response to nivolumab therapy is still not clear.
This study aimed to elucidate the impact of the gut microbiota on the prediction of prognosis in advanced HCC patients receiving nivolumab immunotherapy.
MATERIALS AND METHODS
Study design and patients
A total of 8 patients who received nivolumab (from September 2019 to August 2020) as second- or third-line treatment after sorafenib failure were included in the study. Tumor response was assessed using the Response Evaluation Criteria in Solid Tumors version 1.1[9]. The study was approved by the Chonnam National University Hwasun Hospital Institutional Review Board (CNUHH-2019-134). All patients signed a written informed consent form based on the principles of the Declaration of Helsinki. Patients were classified based on disease status (absence or progression at 12 mo after initiation of nivolumab therapy) and overall response (complete response, partial response, or stable disease for > 6 mo). Non-responders were those showing disease progression or stable disease for < 6 mo as well as those who died.
Collection of fecal samples and microbiome analysis
Fecal samples were collected prospectively, according to International Human Microbiome Standards guidelines (SOP_03_V1) at two time points — before the first nivolumab injection (< 1 mo, T0) and at the 3-mo follow-up (T1). The samples were immediately transferred on ice to our clinic and immediately stored at -80 °C until sample processing. Fecal genomic DNA was extracted as previously described[6-8]. DNA was extracted using the Cica Geneus® DNA Prep Kit (Kanto Chemical, Tokyo, Japan) following the manufacturer’s instructions. The fecal microbiome was assessed by sequencing various regions (V3-V4) of the 16S ribosomal RNA bacterial gene at months 0 and 2. Briefly, polymerase chain reaction amplification was performed using 16S universal primers targeting the V3-V4 region of the bacterial 16S ribosomal gene. The joint pair length was set to encompass 467 base pair amplicons using the 2 × 300 paired-end MiSeq kit (Illumina, San Diego, CA, United States). For each sample, a sequencing library was generated by adding sequencing adapters. Detection of the sequencing fragments was performed using MiSeq technology by Macrogen (Seoul, South Korea).
Metagenomic and network analysis
The targeted metagenomic sequences from the microbiota were analyzed using the bioinformatics pipeline established by Vaiomer (Labège, France) using the FROGS guidelines. Low-depth samples (less than 9000 sequences per sample) were removed from the analysis. Sequences were trimmed and merged and then clustered into operational taxonomic units (OTUs) using CLC Genomics Workbench v. 10.1.1 and CLC Microbial Genomics Module v. 2.5 (Qiagen, Hilden, Germany). Taxonomic assignment of these sequences was carried based on the National Center for Biotechnology Information taxonomy database, with an OTU cutoff of 3%. The most abundant sequences were considered representative of each cluster and assigned to a taxonomy level based CLC Microbial Genomics default values.
Alpha diversity metrics (richness and Shannon’s index) were calculated using the phyloseq R package based on rarefied OTU counts[10]. The beta diversity index was defined as the difference between the total number of species across the two groups and the number of species common to both[11]. Exploratory analysis of beta-diversity (between-sample diversity) was performed based on the Bray-Curtis measure of dissimilarity as a principal coordinate analysis.
For the hierarchical cluster analysis, Bray-Curtis metrics and complete linkage clustering were implemented. At deeper taxonomic levels (from the phylum to genus level), we performed linear discriminant analysis (LDA) effect size analysis based on the non-parametric factorial Kruskal-Wallis sum rank test, to detect bacterial taxa with significantly different abundance between responders and non-responders. Then, LDA effect size was used to estimate the size effect of each differentially abundant taxon based on the criteria of LDA ≥ 3.0 and P < 0.05. Volcano plots showed the estimated log 2-fold difference in OTU abundance between responders and non-responders.
Statistical analysis
Alpha diversity metrics were compared by Mann-Whitney tests; for comparisons involving more than two groups, Bonferroni's correction was applied. The LDA was performed, and the volcano plots generated, at http://huttenhower.sph.harvard.edu/galaxy/. Two-tailed student’s t test and the χ2 test were used to test for differences in the phenotypical characteristics of the microbiome using GraphPad Prism software (GraphPad Software Inc., San Diego, CA, United States).
RESULTS
Patients and sample
A single-site correlative study design was used to investigate the effects of gut microbiota on the efficacy of nivolumab in 8 adult HCC patients (Supplementary Table 1). All 8 patients received nivolumab as second- or third-line treatment after sorafenib failure. The median age was 62.5 (interquartile range: 58.0-66.25) years overall, and was not significantly different between the groups. All patients were male. Six patients (75%) were chronically infected with hepatitis B virus (HBV), and four patients (50%) had alcohol-related liver disease. All patients were Barcelona Clinic Liver Cancer stage C and stage IV by the modified Union for International Cancer Control stage system. While all 3 patients of the non-responder group had vascular invasion, only 1 patient (20%) had vascular invasion in the responder group. Presence of biliary invasion was 66.7% (2/3) in the non-responder group and 20% (1/5) in the responder group. The percentage of patients with an alpha-fetoprotein score ≥ 400 ng/mL at baseline was 66.7% (2/3) and 40% (2/5), each. Most patients (87.5%) were Child-Pugh class A, and only 1 patient in the good response group was Child-Pugh class B. All patients received three or more prior therapies before nivolumab.
Metagenome analysis
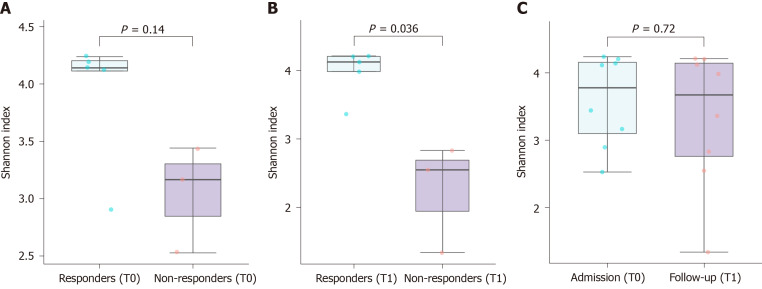
A total of 2027154 good-quality reads with a mean length of 301 base pairs were generated. A higher Shannon index was found in the responders when compared to the non-responders after nivolumab therapy (P = 0.036), reflecting a significantly higher species richness in the former group (Figure 1A and B). In contrast, there was no significant difference in alpha diversity within the same patients according to the nivolumab therapy (Figure 1C). The unweighted beta diversity analysis also showed that the overall bacterial community structure and phylogenetic diversity were similar between T0 and T1, but the responders and non-responders were clearly distinguished (Figure 2).
Figure 1.
Alpha diversity indices (Shannon index) of hepatocellular carcinoma patients based on treatment and prognosis. A: The Shannon index of the non-responders on admission (T0) was not significantly lower than that of the responders (P = 0.14); B: The Shannon index of the non-responders was significantly lower than that of the responders after nivolumab therapy (T1); C: Nivolumab therapy did not alter the Shannon index. HCC: Hepatocellular carcinoma.
Figure 2.
Composition of the gut microbiome in hepatocellular carcinoma patients is associated with the response to nivolumab. A: Heatmap showing the abundance of operational taxonomic units in responders (yellow) and non-responders (blue). The original comprehensive figure, including the names of bacterial taxa, is presented as Supplemental Figure 1; B: Unweighted beta diversity analysis showed that the overall bacterial community structure and phylogenetic diversity at T0 (light yellow and light blue) and T1 (yellow and blue) were similar; distinct clusters were not observed for responders (yellow) and non-responders (blue). Statistical values obtained by the PERMANOVA test are presented in Supplemental Table 2.
To identify the bacterial taxa associated with a good prognosis, different taxonomical levels were compared using LDA, and 36 and 4 species were found to be differentially abundant in the responders and non-responders, respectively (Figure 3). Of the bacterial species, Ruminococcus gnavus was abundant in the non-responders, while several bacterial taxa, including Clostridia, Prevotella 9, Rikenellaceae, Alistipes, the Christensenellaceae R-7 group, Dialister, Muribaculaceae, Desulfovibrionales, Deltaproteobacteria, the Eubacterium coprostanoligenes group, Acidaminococcaceae, the Lachnospiraceae NK4A136 group, Roseburia, Mitsuokella, Ruminiclostridium 9, Marinifilaceae, Lachnospiraceae, and Ruminococcaceae groups other than Ruminococcus gnavus were highly enriched in the responders. A volcano plot also showed several bacterial species specifically associated with the prognosis of nivolumab therapy in the HCC patients. Dialister pneumosintes, Escherichia coli, Lactobacillus reteri, Streptococcus mutans, Enterococcus faecium, Streptococcus gordonii, Veillonella atypica, Granulicatella sp., and Trchuris trichiura were specific to the non-responders, while Citrobacter freundii, Azospirillum sp., and Enterococcus durans were specific to the responders.
Figure 3.
Specific bacterial taxa are associated with the prognosis of nivolumab therapy in hepatocellular carcinoma patients according to linear discriminant analysis effect size analysis. A: Taxonomic cladogram based on linear discriminant analysis (LDA) using effect size (LEfSe) showing differences in fecal taxa. Dot size is proportional to the abundance of the taxon. Letters correspond to the following taxa: (a) Marinifilaceae, (b) Muribaculaceae, (c) Prevotella 9, (d) Alistipes, (e) Rikenellaceae, (f) the Lachnospiraceae NK4A136 group, (g) Roseburia, (h) the [Ruminococcus]gnavus group, (i) Ruminiclostridium 9, (j) the Ruminococcaceae NK4A214 group, (k) Ruminococcaceae UCG-002, (l) Ruminococcaceae UCG-003, (m) Ruminococcaceae UCG-005, (n) Ruminococcaceae UCG-010, (o) Ruminococcaceae UCG-014, (p) Ruminococcus 1, the (q) [Eubacterium]coprostanoligenes group, (r) Dialister, (s) Megamonas, (t) Mitsuokella, and (u) Salmonella; B: LDA scores for the differentially abundant taxa in the fecal microbiome of responders (yellow) and non-responders (blue). The length denotes the effect size for a taxon. P = 0.05 for the Kruskal-Wallis test; LDA score > 3; C: Volcano plot showing several bacterial taxa specifically related to the prognosis of nivolumab therapy in hepatocellular carcinoma patients. LDA: Linear discriminant analysis.
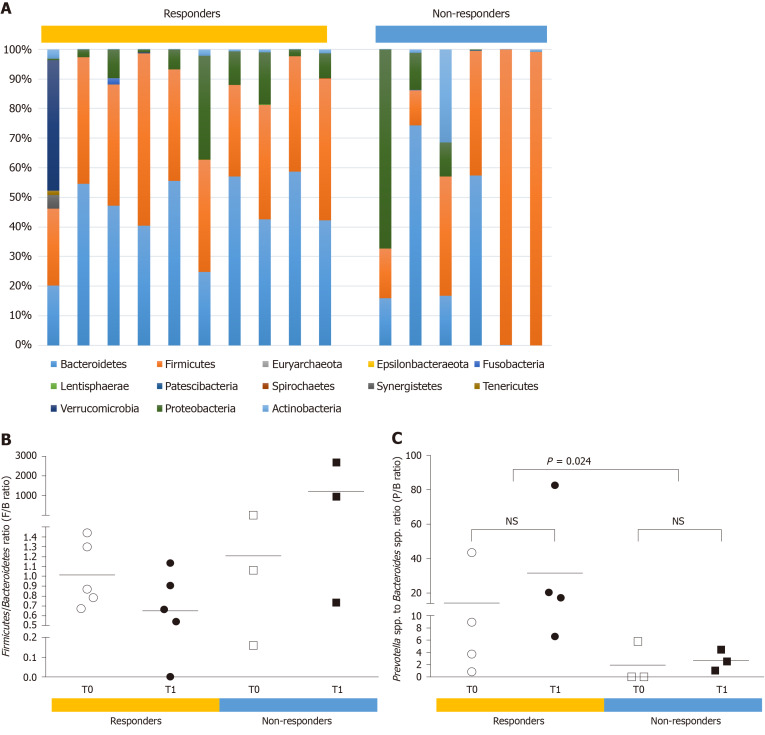
The gut microbiota composition in HCC patients was also described at the phylum and genus levels (Figure 4). At the phylum level, a skewed Firmicutes/Bacteroidetes ratio (< 0.5 or > 1.5) was more frequently found in the non-responders than in the responders (66.7% vs 10%, P < 0.05). The mean ratio of Prevotella species to Bacteroides species (P/B ratio) was significantly higher in the responders than in the non-responders (22.99 vs 2.312, P = 0.024). In addition, the presence of Akkermansia species was observed in two responders only, indicating that this could be a useful prognostic marker of the response to nivolumab therapy in advanced HCC patients.
Figure 4.
Potential prognostic markers of the response to nivolumab therapy in hepatocellular carcinoma patients. A and B: At the phylum level, a skewed Firmicutes/Bacteroidetes ratio (< 0.5 or > 1.5) was more frequent in the non-responders than the responders (66.7% vs 10.0%, P = 0.018); C: At the genus level, the ratio of Prevotella species to Bacteroides species (P/B ratio) was a prognostic marker of the response to nivolumab in hepatocellular carcinoma patients. The responders showed a significantly higher mean P/B ratio than the non-responders (22.99 vs 2.312, P = 0.024). NS: Statistically non-specific.
DISCUSSION
A number of clinical trials investigating the therapeutic potential of manipulation of the gut microbiota in non-HCC cancer patients has already begun[6-8,12]. Regarding HCC, however, only two Chinese studies have been reported to date[13,14]. Reduced alpha diversity was found in the non-responders in those studies, in line with our data. In addition, we found that several bacterial taxa, such as Dialister pneumosintes, Escherichia coli, Lactobacillus reteri, Streptococcus mutans, Enterococcus faecium, Streptococcus gordonii, Veillonella atypica, Granulicatella sp., and Trchuris trichiura, were specific to non-responders, while Citrobacter freundii, Azospirillum sp., and Enterococcus durans were specific to responders. The gut bacteria associated with the therapeutic response did not overlap with those in previous studies[13,14]. This might be partly due to differences in the techniques used to analyze samples, the reference databases, and geographical or racial/ethnic differences. While this study has not proven the clinical efficacy and benefit of our approach, it is feasible that it could reduce the therapeutic failure rate of nivolumab. Furthermore, it is not clear whether the aforementioned findings regarding the role of the gut microbiome in antitumor immune responses in animal models and patients with other tumor types also apply to patients with HCC.
Beyond dysbiosis, we suggested additional potential prognostic markers of the response to nivolumab therapy in HCC patients. We found that in all the responders, the Firmicutes/Bacteroidetes ratio ranged from 0.54 to 1.44 (mean of 0.88), whereas 66.7% of non-responders exhibited a highly skewed ratio (< 0.5 or > 1.5). Previous studies have shown elevated Firmicutes/Bacteroidetes ratio in the gut microbiota to be associated with obesity and older age[15], suggesting that it could be the result of dysbiosis arising from adaptation of individual microbial communities to long-term metabolic dysfunction[16]. Until recently, few studies have discussed the association between the Firmicutes/Bacteroidetes ratio and gastrointestinal cancer. Yu et al[17] reported that, in a rat model, precancerous lesions of gastric cancer had the highest Firmicutes/Bacteroidetes ratio. A reduced Firmicutes/Bacteroidetes ratio was also found to be related to the progression of liver diseases (liver cirrhosis and primary liver cancer)[18]. Notably, our data strongly support the notion that a highly skewed Firmicutes/ Bacteroidetes ratio can be used to predict a lack of response to nivolumab therapy. This could facilitate the selection of appropriate immunotherapies for advanced HCC patients.
Furthermore, we found that a high P/B ratio was a better prognostic marker of the response to nivolumab therapy outcome in HCC patients. Overall, we found that responders had a significantly higher P/B ratio than non-responders. This observation is partly supported by a recent study reporting an elevated P/B ratio in fecal samples obtained from advanced-stage gastrointestinal cancer patients receiving anti-PD-1/PD-L1 and an improved response to anti-PD-1/PD-L1 treatment[19]. In line with previous studies, we also found that the presence of Akkermansia species can serve as a good prognostic marker of the response to nivolumab therapy in advanced HCC patients[5,13]. Although we did not analyze the gut microbiome of non-HCC patients, but previous studies have shown that patients with HCC and underlying cirrhosis typically present with profound dysbiosis. Thus, it is tempting to speculate that underlying dysbiosis could contribute to immunotherapy failure in some patients and that gut microbiome modulation may have even more profound effects in HCC than other tumors.
In this study, all 5 responders and 1 of 3 non-responders were infected by HBV. However, this may reflect the relatively higher proportion of HBV-related HCC in the Korean population in general, rather than the positive impact of HBV infection on nivolumab outcome. The association of HBV in nivolumab treatment response is not clear yet. For further consideration, in the CheckMate 040 data, the efficacy and safety of nivolumab treatment in sorafenib-experienced patients with advanced HCC were comparable between the HBV-infected group and the non-infected group[5].
A major limitation of this study is the small size of its cohort, which did not provide sufficient statistical power. This preliminary data should be interpreted with caution and further studies enrolling larger numbers of subjects may, thus, reveal additional microbial patterns. In addition, translation of a prognosis-associated microbial signature may not be straightforward, and thus several inherent variabilities between individuals within each cohort should be also considered as potential confounding factors. Nevertheless, our data highlights the promising possibility that a feasible approach may be to combine several microbial features for prediction of nivolumab treatment.
CONCLUSION
The current study suggests that non-responders to nivolumab have dysbiotic fecal composition. Moreover, a high P/B ratio predicted a better response to nivolumab in HCC patients, albeit that cohort was relatively small. Further studies should help define the role of these bacteria and their potential as novel biomarkers. New insights into the pathophysiological relevance of intestinal dysbiosis in the prognosis of HCC may lead to innovative therapeutic solutions, such as supplementation with probiotics, to prevent primary resistance to therapy.
ARTICLE HIGHLIGHTS
Research background
Systemic chemotherapy for hepatocellular carcinoma (HCC) has an important role, and immunotherapy in HCC is challenging.
Research motivation
Predictive or prognostic markers for systemic treatment for HCC have not been elucidated.
Research objectives
To investigate the correlation between gut microbiome and treatment response in advanced HCC.
Research methods
Patients who were treated with nivolumab for HCC were identified from one tertiary hospital in South Korea from September 2019 to August 2020. Metagenomic data from 16S ribosomal RNA sequencing were analyzed according to therapeutic response.
Research results
A higher Shannon index was found in the responders than in the non-responders after nivolumab therapy (P = 0.036). The unweighted beta diversity analysis also showed that the overall bacterial community structure and phylogenetic diversity were clearly distinguished according to therapeutic response. A skewed Firmicutes/Bacteroidetes ratio and a low Prevotella/Bacteroides ratio can serve as predictive markers of non-response, whereas the presence of Akkermansia species predicts a good response.
Research conclusions
Gut microbiome has a potential role as a prognostic marker for the response to nivolumab in the treatment of HCC patients.
Research perspectives
Microbiome study before and/or follow up of treatment with immunotherapy for HCC patients could be a prognostic marker, and it can be a criterion for selecting which systemic treatment should be used for each patient as part of precision medicine.
ACKNOWLEDGEMENTS
We would like to thank all participants in this study.
Footnotes
Institutional review board statement: This study was carried out in accordance with all relevant institutional guidelines. The Ethics Committee of Chonnam National University Hwasun Hospital approved this study (CNUHH-2019-134) and written informed consent was obtained from all subjects.
Conflict-of-interest statement: All authors declare having no conflicts of interest.
Manuscript source: Unsolicited manuscript
Peer-review started: May 20, 2021
First decision: June 13, 2021
Article in press: November 3, 2021
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: South Korea
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tun HM S-Editor: Gong ZM L-Editor: A P-Editor: Gong ZM
Contributor Information
Min-Woo Chung, Department of Internal Medicine, Chonnam National University Hospital and College of Medicine, Hwasun 58128, Jeollanam-do, South Korea.
Moon-Ju Kim, Department of Parasitology and Tropical Medicine, Chonnam National University Medical School, Hwasun 58128, Jeollanam-do, South Korea.
Eun Jeong Won, Department of Parasitology and Tropical Medicine, Chonnam National University Medical School, Hwasun 58128, Jeollanam-do, South Korea; Department of Laboratory Medicine, Chonnam National University Hwasun Hospital, Hwasun 58128, Jeollanam-do, South Korea. parasite.woni@jnu.ac.kr.
Yu Jeong Lee, Department of Parasitology and Tropical Medicine, Chonnam National University Medical School, Hwasun 58128, Jeollanam-do, South Korea.
Yong-Woon Yun, Department of Preventive Medicine, Chonnam National University Medical School, Hwasun-eup, Hwasun 58128, Jeollanam-do, South Korea.
Sung Bum Cho, Department of Internal Medicine, Chonnam National University Hospital and College of Medicine, Hwasun 58128, Jeollanam-do, South Korea.
Young-Eun Joo, Department of Internal Medicine, Chonnam National University Hospital and College of Medicine, Hwasun 58128, Jeollanam-do, South Korea.
Jun-Eul Hwang, Department of Internal Medicine, Chonnam National University Hospital and College of Medicine, Hwasun 58128, Jeollanam-do, South Korea.
Woo Kyun Bae, Department of Internal Medicine, Chonnam National University Hospital and College of Medicine, Hwasun 58128, Jeollanam-do, South Korea.
Ik-Joo Chung, Department of Internal Medicine, Chonnam National University Hospital and College of Medicine, Hwasun 58128, Jeollanam-do, South Korea.
Myung Geun Shin, Department of Laboratory Medicine, Chonnam National University Hwasun Hospital, Hwasun 58128, Jeollanam-do, South Korea.
Jong Hee Shin, Department of Laboratory Medicine, Chonnam National University Hospital, Gwangju 61469, South Korea.
Data sharing statement
No additional data are available.
References
- 1.Wallace MC, Preen D, Jeffrey GP, Adams LA. The evolving epidemiology of hepatocellular carcinoma: a global perspective. Expert Rev Gastroenterol Hepatol. 2015;9:765–779. doi: 10.1586/17474124.2015.1028363. [DOI] [PubMed] [Google Scholar]
- 2.Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M, Rutkowski P, Ferrucci PF, Hill A, Wagstaff J, Carlino MS, Haanen JB, Maio M, Marquez-Rodas I, McArthur GA, Ascierto PA, Long GV, Callahan MK, Postow MA, Grossmann K, Sznol M, Dreno B, Bastholt L, Yang A, Rollin LM, Horak C, Hodi FS, Wolchok JD. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrington KJ, Ferris RL, Blumenschein G Jr, Colevas AD, Fayette J, Licitra L, Kasper S, Even C, Vokes EE, Worden F, Saba NF, Kiyota N, Haddad R, Tahara M, Grünwald V, Shaw JW, Monga M, Lynch M, Taylor F, DeRosa M, Morrissey L, Cocks K, Gillison ML, Guigay J. Nivolumab versus standard, single-agent therapy of investigator's choice in recurrent or metastatic squamous cell carcinoma of the head and neck (CheckMate 141): health-related quality-of-life results from a randomised, phase 3 trial. Lancet Oncol. 2017;18:1104–1115. doi: 10.1016/S1470-2045(17)30421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, Kim TY, Choo SP, Trojan J, Welling TH Rd, Meyer T, Kang YK, Yeo W, Chopra A, Anderson J, Dela Cruz C, Lang L, Neely J, Tang H, Dastani HB, Melero I. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yau T, Hsu C, Kim TY, Choo SP, Kang YK, Hou MM, Numata K, Yeo W, Chopra A, Ikeda M, Kuromatsu R, Moriguchi M, Chao Y, Zhao H, Anderson J, Cruz CD, Kudo M. Nivolumab in advanced hepatocellular carcinoma: Sorafenib-experienced Asian cohort analysis. J Hepatol. 2019;71:543–552. doi: 10.1016/j.jhep.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 6.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 7.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, Chang EB, Gajewski TF. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, Poirier-Colame V, Roux A, Becharef S, Formenti S, Golden E, Cording S, Eberl G, Schlitzer A, Ginhoux F, Mani S, Yamazaki T, Jacquelot N, Enot DP, Bérard M, Nigou J, Opolon P, Eggermont A, Woerther PL, Chachaty E, Chaput N, Robert C, Mateus C, Kroemer G, Raoult D, Boneca IG, Carbonnel F, Chamaillard M, Zitvogel L. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Magurran AE. An index of diversity. In: Measuring Biological Diversity. Afr J Aquat Sci . 2004;29:285–286. [Google Scholar]
- 11.Veech JA, Summerville KS, Crist TO, Gering JC. The additive partitioning of species diversity: recent revival of an old idea. Oikos. 2002;99:93–99. [Google Scholar]
- 12.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, Jiang W, Cai S, Zhao P, Song R, Li P, Qin N, Fang W. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer. 2019;7:193. doi: 10.1186/s40425-019-0650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Ye J. Characterization of gut microbiota in patients with primary hepatocellular carcinoma received immune checkpoint inhibitors: A Chinese population-based study. Medicine (Baltimore) 2020;99:e21788. doi: 10.1097/MD.0000000000021788. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Grigor'eva IN. Gallstone Disease, Obesity and the Firmicutes/Bacteroidetes Ratio as a Possible Biomarker of Gut Dysbiosis. J Pers Med. 2020;11 doi: 10.3390/jpm11010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payne AN, Chassard C, Zimmermann M, Müller P, Stinca S, Lacroix C. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr Diabetes. 2011;1:e12. doi: 10.1038/nutd.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu C, Su Z, Li Y, Liu K, Chu F, Liu T, Chen R, Ding X. Dysbiosis of gut microbiota is associated with gastric carcinogenesis in rats. Biomed Pharmacother. 2020;126:110036. doi: 10.1016/j.biopha.2020.110036. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Wu YN, Chen T, Ren CH, Li X, Liu GX. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat Dis Int. 2019;18:149–157. doi: 10.1016/j.hbpd.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Peng Z, Cheng S, Kou Y, Wang Z, Jin R, Hu H, Zhang X, Gong JF, Li J, Lu M, Wang X, Zhou J, Lu Z, Zhang Q, Tzeng DTW, Bi D, Tan Y, Shen L. The Gut Microbiome Is Associated with Clinical Response to Anti-PD-1/PD-L1 Immunotherapy in Gastrointestinal Cancer. Cancer Immunol Res. 2020;8:1251–1261. doi: 10.1158/2326-6066.CIR-19-1014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.