ABSTRACT
Copper radical alcohol oxidases (CRO-AlcOx), which have been recently discovered among fungal phytopathogens, are attractive for the production of fragrant fatty aldehydes. With the initial objective to investigate the secretion of CRO-AlcOx by natural fungal strains, we undertook time course analyses of the secretomes of three Colletotrichum species (C. graminicola, C. tabacum, and C. destructivum) using proteomics. The addition of a copper-manganese-ethanol mixture in the absence of any plant-biomass mimicking compounds to Colletotrichum cultures unexpectedly induced the secretion of up to 400 proteins, 29 to 52% of which were carbohydrate-active enzymes (CAZymes), including a wide diversity of copper-containing oxidoreductases from the auxiliary activities (AA) class (AA1, AA3, AA5, AA7, AA9, AA11, AA12, AA13, and AA16). Under these specific conditions, while a CRO-glyoxal oxidase from the AA5_1 subfamily was among the most abundantly secreted proteins, the targeted AA5_2 CRO-AlcOx were secreted at lower levels, suggesting heterologous expression as a more promising strategy for CRO-AlcOx production and utilization. C. tabacum and C. destructivum CRO-AlcOx were thus expressed in Pichia pastoris, and their preference toward both aromatic and aliphatic primary alcohols was assessed. The CRO-AlcOx from C. destructivum was further investigated in applied settings, revealing a full conversion of C6 and C8 alcohols into their corresponding fragrant aldehydes.
IMPORTANCE In the context of the industrial shift toward greener processes, the biocatalytic production of aldehydes is of utmost interest owing to their importance for their use as flavor and fragrance ingredients. Copper radical alcohol oxidases (CRO-AlcOx) have the potential to become platform enzymes for the oxidation of alcohols to aldehydes. However, the secretion of CRO-AlcOx by natural fungal strains has never been explored, while the use of crude fungal secretomes is an appealing approach for industrial applications to alleviate various costs pertaining to biocatalyst production. While investigating this primary objective, the secretomics studies revealed unexpected results showing that under the oxidative stress conditions we probed, Colletotrichum species can secrete a broad diversity of copper-containing enzymes (laccases, sugar oxidoreductases, and lytic polysaccharide monooxygenases [LPMOs]) usually assigned to “plant cell wall degradation,” despite the absence of any plant-biomass mimicking compound. However, in these conditions, only small amounts of CRO-AlcOx were secreted, pointing out recombinant expression as the most promising path for their biocatalytic application.
KEYWORDS: copper radical oxidase, CAZymes, copper-containing enzymes, alcohol oxidase, filamentous fungi, phytopathogen, Colletotrichum, secretomics, flavors and fragrances
INTRODUCTION
The development of a new technological paradigm to support the shift from a fossil-based economy to a greener and more circular economy is at the heart of today’s bioeconomic challenges (1). Biotechnology is one of the main levers in this ongoing industrial transformation (2). The development of biotechnology largely relies on biocatalysis and thus require the discovery, control, and engineering of a variety of enzymes and microorganisms (3). The fungal kingdom represents a vast and untapped reservoir of biocatalysts (4), but to date, the enzymatic potential of filamentous fungi has mostly been studied in the frame of plant cell wall degradation (5–7). Fungal secretomics is a powerful approach to study the wide diversity of enzymes secreted and to formulate enzymatic cocktails with biotechnological relevance (8–10). Using suitable inducers, some fungal strains can secrete large amounts of oxidoreductases (11), including copper-containing enzymes. For instance, hypersecretory fungal strains from the Pycnoporus genus have been selected to produce laccases and use them in applied settings (12, 13).
Most of the copper-dependent enzymes are classified within the auxiliary activities (AA) class (14) in the CAZy database (www.cazy.org) (15). The founding member of copper radical oxidases (CROs), a galactose oxidase (FgrGalOx; EC 1.1.3.9), was discovered in 1959 from the secretome of Fusarium graminearum (16, 17). In 2013, fungal CROs have been classified within the AA5 family and further classified into subfamilies AA5_1 and AA5_2. AA5_1 contains glyoxal oxidases (GLOX; EC 1.2.3.15) that catalyze the oxidation of aldehydes to carboxylic acids, and AA5_2 gathers enzymes that oxidize the alcohol function of diverse substrates (e.g., galactose, galactose-containing polysaccharides and aromatic and aliphatic primary alcohols) to generate the corresponding aldehydes. More recently, the AA5_2 subfamily was shown to be more diverse than expected, encompassing notably various alcohol oxidases (CRO-AlcOx; EC 1.1.3.13) originating from the phytopathogen ascomycete fungi Colletotrichum and Magnaporthe spp. (18–20). CRO-AlcOx are monocopper and otherwise organic cofactor free enzymes that catalyze the oxidation of alcohols into aldehydes (with the concomitant reduction of O2 to H2O2). They offer appealing perspectives to produce aldehydes (21), which are indispensable intermediates in many chemical pathways (22, 23) and valuable ingredients for the flavor and fragrance industries (24, 25). In particular, saturated long-chain aldehydes (fatty aldehydes) are widely used for their citrus scent (26, 27) but are currently derived from fossil-based chemistry (28, 29) or extracted from natural plant materials, the supply of which is threatened by emerging diseases (30, 31). Therefore, alternative green and/or natural production pathways must be investigated (32).
To date, only pure recombinant forms of CRO-AlcOx have been harnessed in biocatalytic applications (18, 19, 21, 33), and no information on their secretion by natural fungal strains is available. Determining whether CRO-AlcOx–encoding fungal strains can secrete these enzymes under laboratory conditions is of strong interest for enzyme selection and technological applications. It is noteworthy that aside from the scope of pathogenesis (34–36), very few studies investigated the enzymatic biotechnological potential of Colletotrichum spp. (37–40). The use of natural fungal strains could be advantageous to identify and then use potential naturally secreted enzyme partners. In particular, both catalase and peroxidase are required to sustain in vitro the activity of isolated CRO-AlcOx and other AA5 (21, 33, 41, 42). Catalase acts as a H2O2 scavenger, while peroxidase is required for CRO-AlcOx activation (20, 33).
In this context, we investigated the ability of several fungal strains from the Colletotrichum genus to secrete CRO-AlcOx. While pursuing this primary goal, we observed the unexpected secretion of a large panel of plant-biomass-degrading oxidoreductases and chitin-related enzymes under oxidative stress conditions and in the absence of any plant cell wall inducers. We report here an integrated study, starting from fungal time course secretomics and including analysis of copper-containing oxidoreductase secretion profiles, with a final focus on the recombinant CRO-AlcOx production as well as characterization of these enzymes and their biocatalytic application to produce fatty aldehydes for the flavor and fragrance industries.
RESULTS
Selection of fungal strains and time course secretomic study.
To investigate the secretion of copper-containing enzymes, including AA5_2 CRO-AlcOx, by natural fungal strains and their potential use for bioconversion of fatty alcohols to fragrant aldehydes, we selected three fungal phytopathogens from the Colletotrichum genus for which genomic information is available. The first obvious choice was C. graminicola because the three AA5_2 it encodes have all been previously biochemically characterized (18, 43, 44), one of them being the first, most studied, and to date, most active CRO-AlcOx, namely, CgrAlcOx (18, 21, 33). By performing a BLAST analysis of the CgrAlcOx sequence against the genomes of some other Colletotrichum species we had access to, we identified two genes encoding AA5_2 in both C. tabacum and C. destructivum. For each strain, one gene encoded a putative AlcOx, and the other one encoded a putative aryl alcohol oxidase (AAO; based on the recent discovery of an AAO in the AA5_2 family) (44) (Fig. S1). These enzymes will hereafter be named CtaAlcOx/CdeAlcOx and CtaAAO/CdeAAO for C. tabacum and C. destructivum, respectively. Based on the few studies reporting the detection in some Ascomycota fungal secretomes of a range of copper-containing enzymes from the AA class, including the FgrGalOx and other AA5 (45–47), we identified copper, manganese, and ethanol as possible inducers of AA5. Copper is known to be crucial to activate the FgrGalOx and was used previously in combination with manganese to recover active FgrGalOx from fungal cultures (47). Ethanol is often used as an inducer of laccases (12) (other copper-containing AAs), and we noticed the presence of CROs in some secretomes induced with ethanol (45). We therefore elaborated two liquid growth media: (i) “YG” (for yeast extract-glucose) and (ii) “YG-CME” (for YG-copper-ethanol-manganese), with the same composition as YG but supplemented with CuSO4, manganese, and ethanol. A time course analysis was performed on each strain in both media at three time points (days 3, 7, and 14), and proteomic analyses were performed on the 16 secretomes generated (note that no samples were harvested at day 3 for C. graminicola in both YG and YG-CME due to insufficient growth). The quantification of total proteins revealed an increasing amount of proteins over time (except for C. destructivum) (Fig. S2), while capillary electrophoresis analysis revealed distinct profiles over time and between fungal strains (Fig. 1).
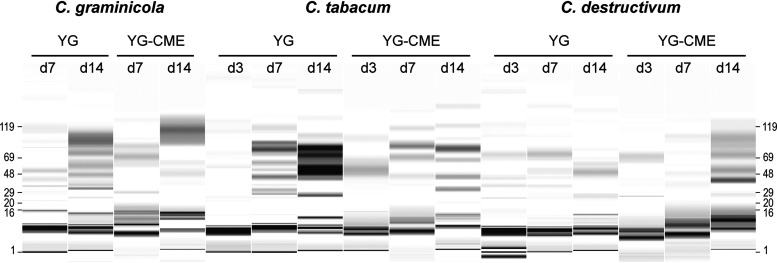
FIG 1.
Analysis by capillary electrophoresis of the Colletotrichum secretomes samples. Concentrated secretomes samples were diluted so that the equivalent of 87.5 μl (prior to concentration) of each secretome sample was injected, except for C. tabacum yeast extract and glucose with copper, ethanol, and manganese (YG-CME) day (d) 14 that was further diluted 3-fold to avoid a too-high protein concentration. The molecular weights (in kDa) of the protein ladder are displayed on the left- and right-hand sides of the image. A darker color corresponds to a higher concentration of protein.
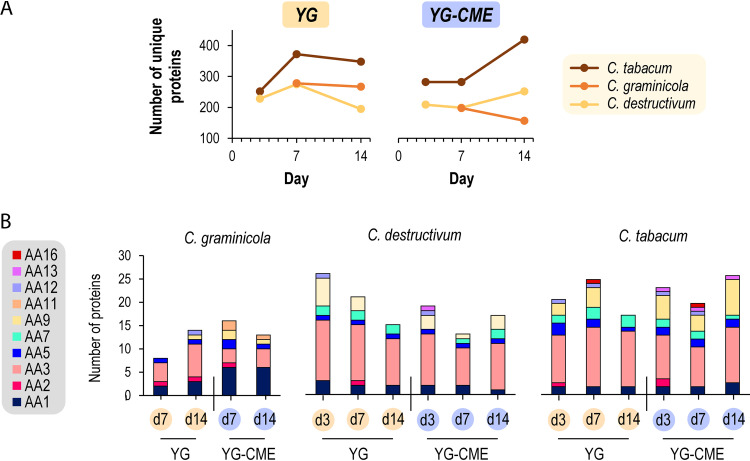
By performing proteomic analyses of the secretomes using LC-MS/MS and mass matching, we detected a high number of secreted proteins (between 150 and more than 400 different proteins, depending on the culture conditions; Supplementary Data Set; Fig. 2A) encompassing a large diversity of enzymes. Carbohydrate-active enzymes (CAZymes) accounted for ca. 29 to 52% of the total protein abundance in each secretome (Fig. S3), with a notable high diversity of enzyme from the AA class (Fig. 2B). Among them, a variety of laccases (AA1), peroxidases (AA2), flavo-oxidases (AA3) including cellobiose dehydrogenases (AA3_1-AA8), CROs (AA5), oligosaccharide oxidases (AA7), lytic polysaccharide monooxygenases (LPMOs; AA9, AA11, AA13, and AA16), and pyrroloquinoline quinone-dependent oxidoreductases (AA12) were detected. To determine whether this high number and diversity of proteins detected was the result of cell lysis (7, 48), we evaluated the proportion of proteins predicted as not secreted (i.e., without signal peptide) and checked more specifically the abundance of some known intracellular proteins (i.e., DNA-polymerase, proteasome endopeptidase complex, citrate synthase, malate dehydrogenase, transketolase, and ubiquitin-protein ligase) (Fig. S4). Despite the significant number of proteins predicted without signal peptide, we did not observe any clear trend that would account for an accumulation of intracellular proteins in the secretomes over time. Notably, some proteins without signal peptide are known to be secreted through unconventional secretion pathways in fungi (48, 49).
FIG 2.
Overview of the total number of proteins secreted over time in different media (A) and diversity of AA enzymes detected in the secretomes (B).
Effect of copper-manganese-ethanol supplementation on the secretome profiles.
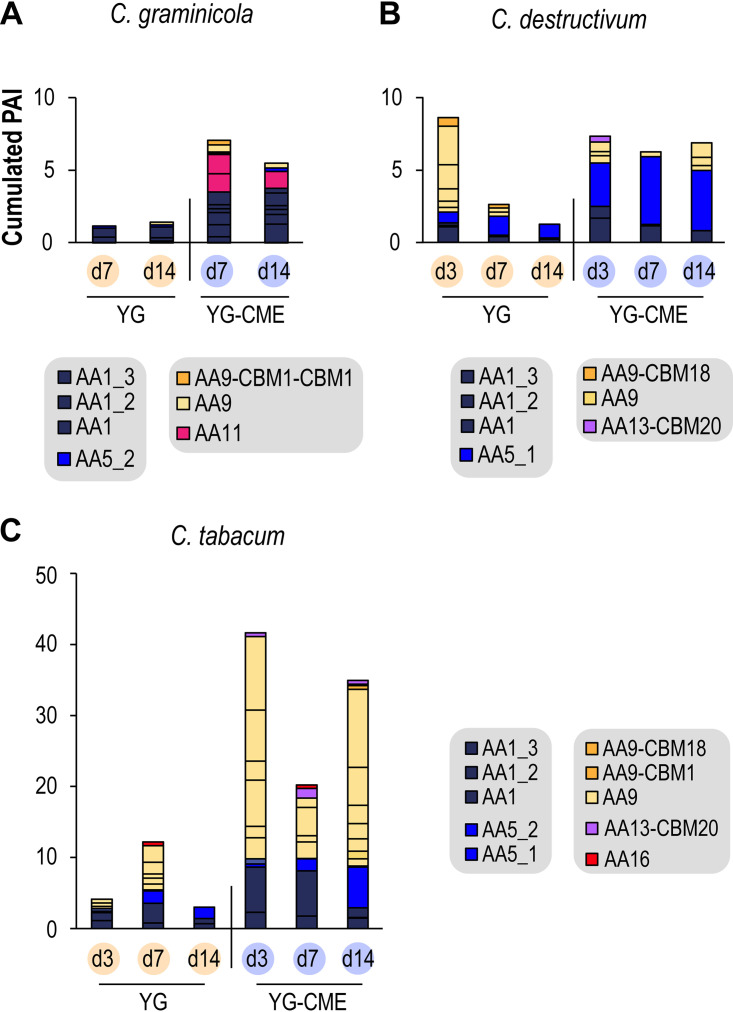
We then looked at the effect of supplementation with CME. At first glance, no clear effect was distinguishable on CAZyme diversity (Fig. 2 and Fig. S3), but when we focused on copper-containing enzymes (Fig. 3), a clear trend became apparent. Indeed, copper-containing enzymes were generally more abundant in the CME medium with a diversity of oxidases from the AA class, including AA1 laccases, AA5_1 GLOX, and AA9, AA11, AA13, and AA16 LPMOs. Furthermore, AA1_3, AA5_1, and AA9 were among the top five most abundant enzymes in YG-CME for the C. tabacum and C. destructivum secretomes, while no AA enzymes were identified in the top five ranking for the corresponding YG secretomes (Table S1). Remarkably, the presence of some AA9-CBM18 was detected in both the C. tabacum and C. destructivum secretomes (Fig. 3). This association of a chitin-binding domain from the CBM18 family (mainly found associated with chitin-active enzymes) to an AA9 domain is intriguing because AA9 LPMOs have not yet been described to act on chitin substrates. Strikingly AA9 LPMOs were particularly abundant at day 3 in YG medium for C. destructivum while drastically decreasing at day 7 and 14 (Fig. 3) as already observed in the secretomes of the basidiomycete fungus Laetisaria arvalis (50). Interestingly, C. tabacum AA9 LPMOs were also found in great abundancy at all time points in the YG-CME conditions.
FIG 3.
Abundance of copper-containing AAs in fungal secretomes from C. graminicola (A), C. destructivum (B), and C. tabacum (C). The y axis legend in panel A applies to all panels likewise (note the different scale for C. tabacum). For each enzyme family, the bar is subdivided according to the number of enzymes identified; for instance, C. tabacum at day 3 in YG-CME displayed six different auxiliary activities class (AA) 9. PAI, protein abundance index.
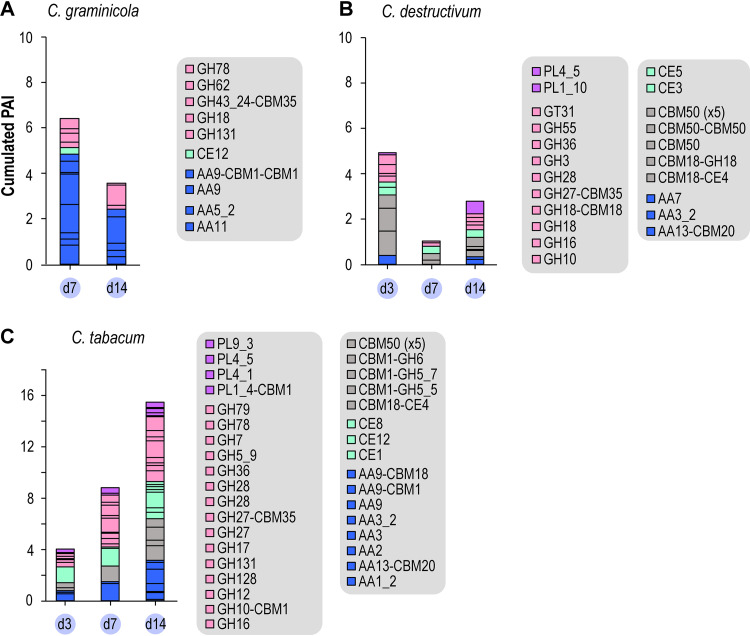
In addition, a diversity of glycoside hydrolases (GH), polysaccharide lyases (PL), carbohydrate esterase (CE), and AAs were detected exclusively in the CME medium (Fig. 4). Among these enzymes, we noticed the intriguing presence of hydrolases (GH18 and GH18-CBM18), oxidases (AA7 and AA11), and esterases (CE4-CBM18) potentially active on chitin or chito-oligosaccharides. Remarkably, a multimodular protein with five consecutive CBM50 chitin-binding domains (“CBM50 (×5)” in Fig. 4) but without any catalytic domain was detected in both the C. tabacum and C. destructivum secretomes. This protein is analogous to other chitin-binding proteins (i.e., LysM effector proteins) (see Discussion).
FIG 4.
Abundance of CAZymes identified exclusively in the YG-CME media from C. graminicola (A), C. destructivum (B), and C. tabacum (C). The y axis legend in panel A applies to all panels likewise (note the different scale for C. tabacum). For each enzyme family, the bar is subdivided according to the number of enzymes identified.
Presence of AA5 in the secretomes.
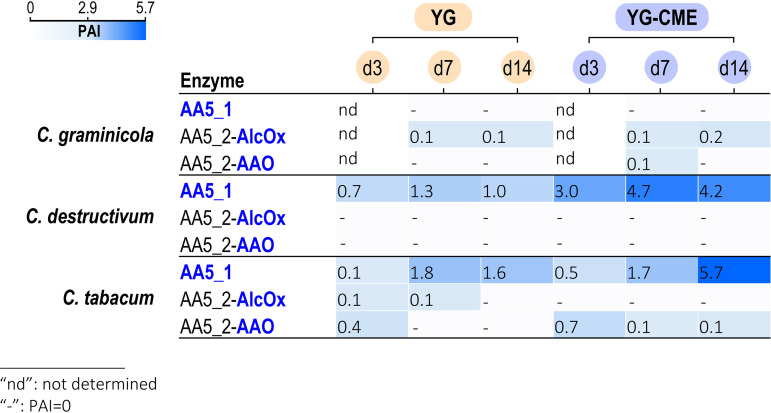
We next focused our analysis on AA5 CROs. Interestingly, a putative GLOX from the AA5_1 subfamily was detected in C. destructivum secretomes (Fig. 5), being the most abundant protein in the YG-CME secretome at day 7 (and the second most abundant at day 14; Table S1). The C. tabacum AA5_1 putative GLOX was also the third most abundant protein in the YG-CME secretome at day 14 (Table S1). We could also detect the occurrence of putative AlcOx and AAO from the AA5_2 subfamily in both C. graminicola and C. tabacum secretomes (Fig. 5). However, their relative abundance was quite low (between 0.05 and 0.13% of the total protein abundance index [PAI]), and they did not accumulate over time. No correlation with the presence of CME in the medium was seen except for the C. graminicola AAO. CROs are known to be activated by peroxidases (41, 51, 52). More specifically, the peroxidase partner responsible for the activation of CRO-AlcOx has been recently unveiled in fungal pathogens (20). Both genes occur as a tandem in the genomes of the Colletotrichum and Magnaporthe species harboring an AlcOx and are coexpressed in vivo prior to the plant-penetration stage (20). While some peroxidases from the AA2 family (Fig. 2) were found in some of the secretomes, the specific peroxidase partner of the AlcOx was not detected, reinforcing the authors’ claim that interaction with plants is needed for gene coexpression (20).
FIG 5.
Heat map of the abundance of AA5 detected in the fungal secretomes.
Together, these data provide unexpected information on the induction of secreted copper enzymes in Colletotrichum species. However, the low level (or even the absence for C. destructivum) of AA5_2 detected under these experimental conditions indicates that the direct implementation of such secretomes for the bioconversion of fatty alcohols is not a viable strategy. This was confirmed by preliminary conversion assays we carried out using the secretomes directly on a range of primary alcohols (data not shown). These tests were inconclusive, most likely because of the low level of AA5_2 secreted and were difficult to interpret given the complexity of the secretome samples (hundreds of enzymes, including various oxidases such as AA3 enzymes) (Fig. 2B). These results prompted us to produce the Colletotrichum CRO-AlcOx enzymes recombinantly in Pichia pastoris and to test their ability to convert alcohols of interest for the flavor and fragrance industries. The objective was to find CRO-AlcOx with potential interesting traits not predictable by sequence analysis such as (i) activity on unactivated fatty alcohols, (ii) minimized requirement in accessory enzymes, or (iii) reduced overoxidative activity.
Recombinant production and substrate specificity of CdeAlcOx and CtaAlcOx.
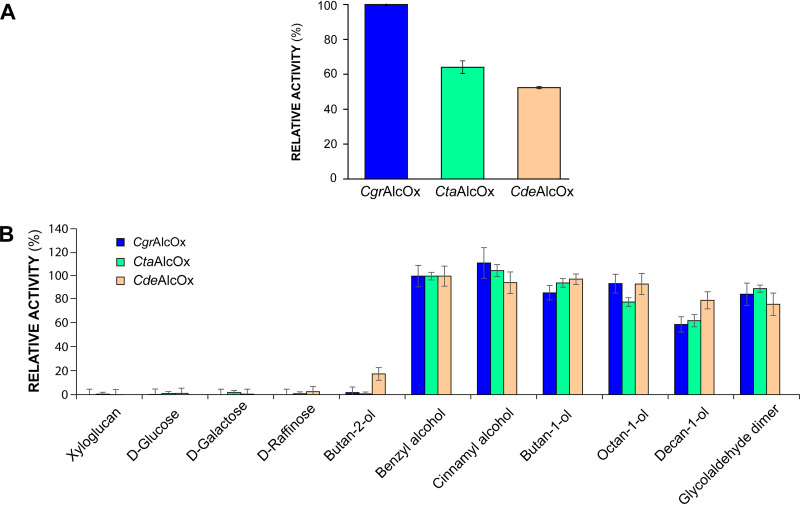
Detailed examination of the CdeAlcOx and CtaAlcOx sequences showed that the two AlcOx amino acid sequences share 90% identity with the CgrAlcOx and likewise comprise a single catalytic module, as with all other characterized CRO-AlcOx from Colletotrichum spp. (Fig. S1). The two AAOs share 85% identity with the recently characterized C. graminicola AAO (CgrAAO) (44) and possess an N-terminal PAN_1 domain, in addition to the catalytic module, similarly to the PAN_1 domain observed in the CgrAAO (Fig. S1). The function of the PAN_1 domain in AA5_2-AAOs is unclear, but such modules are known to mediate protein-–protein or protein–carbohydrate interactions (53). We successfully produced both CRO-AlcOx in P. pastoris (Fig. S5) with high titers for CtaAlcOx in flask culture (200 mg of purified enzyme per liter of culture) compared to CdeAlcOx (75 mg·liter−1) and CgrAlcOx (35 mg·liter−1). Activity screening using the 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS)/horseradish peroxidase (HRP) assay, with benzyl alcohol (BnOH; often used as benchmark with CRO-AlcOx) (18, 21, 44) used as reference, confirmed that both enzymes are indeed AlcOx and that they show similar substrate specificities compared to the canonical CgrAlcOx (Fig. 6). However, CdeAlcOx displayed a slightly higher specific activity on the secondary alcohol butan-2-ol and on the longer aliphatic substrate decan-1-ol. The latter observation could be related to the M173L substitution compared to CgrAlcOx, which could create a slightly more hydrophobic environment in the active site, favoring the accommodation of bulky alcohol substrates (Fig. S6). The pH-rate profiles determined for CdeAlcOx and CtaAlcOx (Fig. S7) revealed pH optima around 7.5 to 8 for both enzymes. For subsequent conversion assays and comparison with CgrAlcOx, only CdeAlcOx was retained because CtaAlcOx exhibited no noticeable differences in terms of substrate specificity. Nevertheless, CtaAlcOx remains attractive from a biotechnological point of view, being 5.7-fold more highly expressed than the CgrAlcOx in flask cultures.
FIG 6.
Comparative activity profiles of the three Colletotrichum AA5_2 alcohol oxidases (AlcOx). (A) Relative activity of the recombinant AlcOx measured by ABTS (2,2'-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)/horseradish peroxidase (HRP)–coupled assay on 3 mM BnOH. (B) Relative activity profiling of the recombinant AlcOx enzymes on a set of substrates, determined by ABTS/HRP-coupled assay, with activity on BnOH set as reference (activity on BnOH set as 100%). Substrate concentrations were as follows: 1% (wt/vol) xyloglucan; 50 mM for d-glucose, d-galactose, and d-raffinose; and 3 mM for butan-2-ol, butan-1-ol, benzyl alcohol, cinnamyl alcohol, octan-1-ol, decan-1-ol, and glycolaldehyde dimer. Reactions were performed with 1 nM CgrAlcOx and CtaAlcOx and 2 nM CdeAlcOx. Error bars show SD values (independent experiments, n = 3).
Biocatalytic production of fragrant aldehydes using CdeAlcOx.
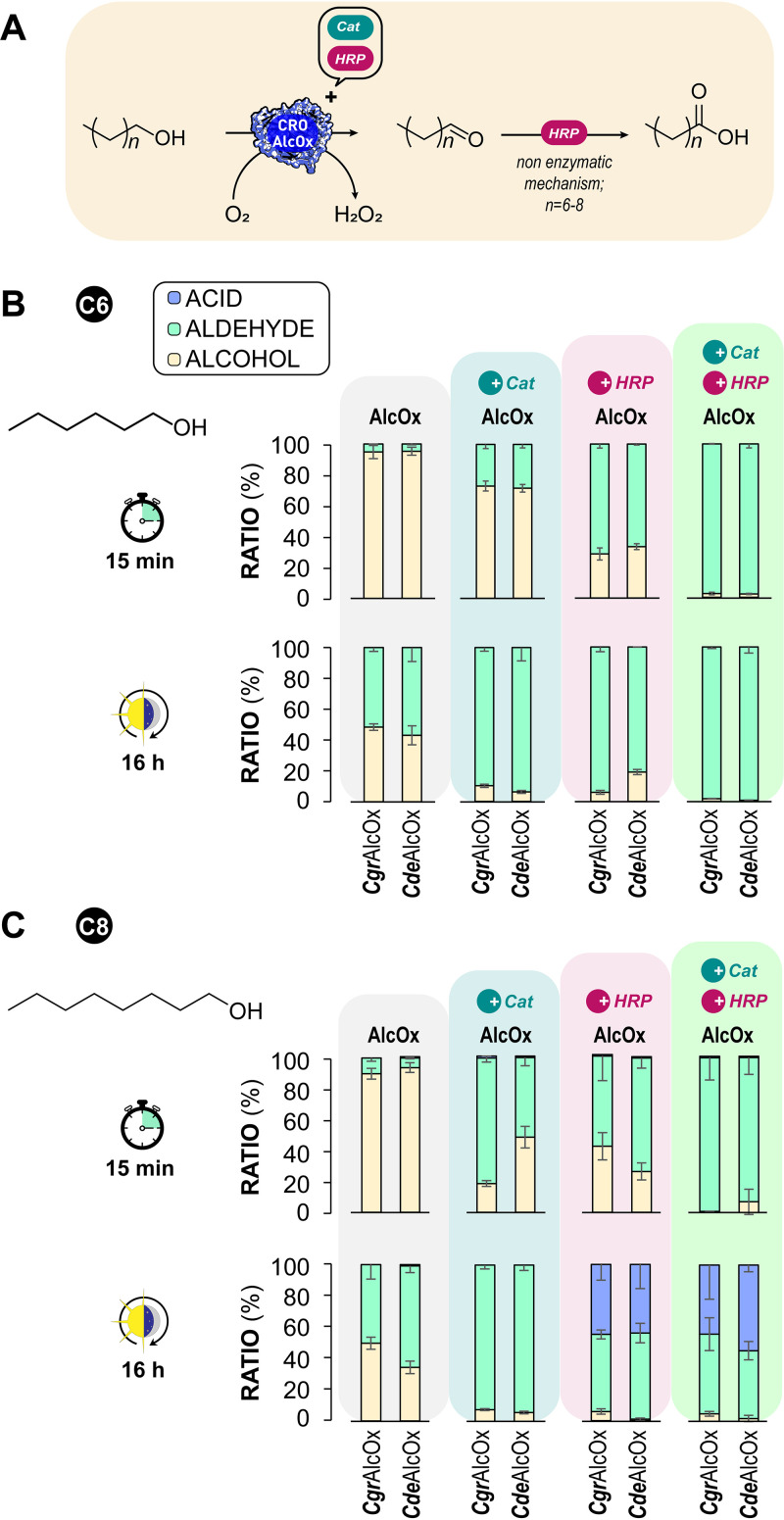
CdeAlcOx was subsequently probed for the conversion of hexan-1-ol and octan-1-ol (two precursors of fragrant aldehydes) using CgrAlcOx as a benchmark. For these experiments, we tested the impact of the reaction time (15 min versus 16 h conversion) and the addition of accessory enzymes (HRP and catalase) according to the parameters determined in a previous study (21). Conversion results showed that CdeAlcOx behave similarly to CgrAlcOx with all the tested substrates (Fig. 7). As previously observed with CgrAlcOx (21, 33), CdeAlcOx alone was unable to promote full alcohol conversion. However, when both HRP and catalase were added to the reaction mixture, full conversion was readily achieved in 15 min. Strikingly, the sole addition of catalase to the reaction mixture resulted in a significant improvement in substrate conversion that probably cannot be attributed only to its protective effect against H2O2. This may suggest an activating effect of catalase on CRO-AlcOx, similar to that of HRP.
FIG 7.
CgrAlcOx and CdeAlcOx mediated oxidation of some fatty alcohols. (A) Reaction scheme proposed (based on reference 21). (B) Conversion of hexan-1-ol. (C) Conversion of octan-1-ol. Reaction mixtures were incubated for either 15 min or 16 h. All reaction mixtures contained 3 mM substrate and 1 μM AlcOx in phosphate sodium buffer (50 mM, pH 8.0) at 23°C. Reactions varied as follows: no accessory enzyme added, addition of catalase (8 μM final), addition of HRP (12 μM final), addition of both catalase (0.5 μM) and HRP (12 μM final). The key in panel B applies for panel C likewise. Conversion products were analyzed by GC-FID. Error bars show SD values (independent experiments, n = 3).
We have previously demonstrated that upon long reaction times (16 h), partial overoxidation of >C6 fatty aldehydes was observed due to HRP, most likely through a nonenzymatic mechanism (Fig. 7A) (21). Carboxylic acids are undesired products in the frame of the F&F application targeted here but are valuable molecules in other fields (54). In agreement with our previous study (21), we observed here that carboxylic acid was not noticeable with hexan-1-ol as the substrate (Fig. 7B) but was observed after 16 h of conversion of octan-1-ol, only when HRP was present in the reaction (Fig. 7C).
We also observed previously (21) that in the case of benzaldehyde derivatives (i.e., 4-NO2-benzyl alcohol), some overoxidation was detected but mediated by the CgrAlcOx itself (Scheme S1) and dependent according to the propension of the aldehyde product to undergo hydration. To probe whether CdeAlcOx behaves in the same way as CgrAlcOx with 4-NO2-benzyl alcohol, we included this substrate in the panel of compounds assayed (Fig. S8). The conversion of 4-NO2-benzyl alcohol indeed led to the corresponding acid after 16 h of reaction even when HRP is omitted from the reaction (Fig. S8). Altogether, these results confirm that the behavior observed for the CgrAlcOx regarding accessory enzymes and overoxidation (21) applies to other CRO-AlcOx enzymes.
DISCUSSION
The time-resolved proteomic analyses performed here on Colletotrichum secretomes revealed interesting and unexpected results. We detected up to ca. 400 individual enzymes in the secretomes, with the maximum number found in the YG-CME medium, while former studies by our group usually yielded ca. 80 to 250 secreted proteins (8, 10, 55, 56). The high proportion of CAZymes in the secretomes is surprising considering the absence of any inducer to mimic plant cell walls. Strikingly, the addition of CME to the medium seemed to boost the secretion of AA enzymes. Moreover, our finding that some LPMOs (especially AA9) preferentially accumulated in the CME medium in the absence of any plant biomass raises questions about the molecular basis of the regulatory pathways driving their production and possibly their true biological function (57). In line with this observation, recent work suggests that some fungal LPMOs and LPMO-like proteins may be involved in biological functions distinct from plant biomass conversion; i.e., copper homeostasis and pathogenicity (58, 59), as well as fungal cell fusion (60). Other copper-containing AA enzymes exhibited higher accumulation in YG-CME cultures. Induction of the secretion of laccases could be either a result of the addition of ethanol to the medium (12) or a response to oxidative stress (61). Indeed, it is known that at high concentrations, copper contributes to generate reactive oxygen species by Fenton chemistry (62, 63).
The proteins secreted exclusively in YG-CME medium revealed a range of chitin-active enzymes, as well as nonenzymatic chitin-binding proteins (LysM proteins), some of which are released by fungal cells upon interaction with their plant hosts, to conceal their presence from the host immune system by preventing fungal chitin recognition by the plant or protecting fungal hyphae against plant chitinases (64–66). In the absence of the host or of any plant biomass, this observation suggests that the high concentration of CME in the medium could trigger some pathways related to pathogenesis. The presence of chitin active enzymes may also be indicative of a nutrient recycling in older senescing portions of the mycelium or of some fungal cell wall remodeling activity (67).
The relatively high abundance of GLOX from the AA5_1 subfamily in some YG-CME secretomes and the low secretion levels of AA5_2 AlcOx suggest distinct biological functions for these two types of CRO. AA5_1 GLOX are often described as enzymes acting on lignin in cooperation with some peroxidases in wood-decaying fungi (68). However, their high abundance here, under CME conditions, despite the absence of lignin, points toward a possible additional biological function of GLOX in Ascomycota related to filamentous growth or pathogenicity, as already demonstrated for the AA5_1 GLOX from the maize pathogen Ustilago maydis (69).
The primary objective of this study was to investigate the secretion of CRO-AlcOx by natural fungal strains with bioconversion potential. Under our experimental conditions, CRO-AlcOx are only sporadically secreted. Of note, a separate study very recently investigated the biological function of these enzymes and showed that they are cosecreted at low levels, over a short period of time during penetration of the plant host cuticle and spatially restricted to the puncturing site (20). Thus, with hindsight, the secretion of large amounts of CRO-AlcOx might be difficult to achieve without a proper signal to mimic the presence of plant host. Another option would be to identify and engineer the promoter region controlling coexpression of the genes encoding both the AlcOx and the associated peroxidase. However, for biocatalytic purposes, the recombinant expression of CRO-AlcOx remains the most promising alternative. We here confirmed that the knowledge gathered on sequence-structure-function of AA5_2 (18, 21, 44) can predict an AlcOx activity based on bioinformatic analysis and that various CRO-AlcOx can be easily produced heterologously using the yeast P. pastoris as host. CtaAlcOx was produced to high yield, which is promising for future upscaled production. The conversion assays performed with CdeAlcOx revealed that this enzyme can be used as a biocatalyst for the conversion of fatty alcohols. The behavior of CdeAlcOx in terms of overoxidation and accessory enzyme requirements suggests that the traits observed here and previously (20, 21, 33) are probably valid for all CRO-AlcOx.
In conclusion, the work carried out in this study highlights that Colletotrichum species can secrete a range of copper-containing oxidoreductases from the AA class without any plant cell wall inducer. While CRO-AlcOx were detected in some of the secretomes, the direct utilization of fungal strains for bioconversion of fatty alcohols will require further efforts to achieve a high secretion yield of CRO-AlcOx and a deeper understanding of the factors controlling their secretion. The recombinant pathway therefore represents the most promising approach to develop their use in biocatalytic applications for the production of fragrant fatty aldehydes.
MATERIALS AND METHODS
General information.
All chemicals used for cultivation of fungi and microorganisms were reagent grade or higher. All chemicals used for biochemical assays were ≥95% purity. HRP type II and catalase from bovine liver were purchased from Sigma-Aldrich (Hamburg, Germany). Molar concentrations of HRP (molecular weight [MW], 33.89 kDa) and catalase (MW of monomer, 62.5 kDa) were estimated by Bradford assay using bovine serum albumin (BSA) as a standard. Precast 10% polyacrylamide gel Mini-PROTEAN TGX were purchased from Bio-Rad (Hercules, CA).
Natural and recombinant strains.
The following fungal strains were used: C. graminicola (M1.001, CBS 130836) (35), C. destructivum (LARS 709, CBS 520.97) (70), and C. tabacum (N150, CPC 18945) (70). The DNA sequences of CdeAlcOx and CtaAlcOx were obtained by blasting against the “raw” unannotated genomes of C. destructivum and C. tabacum, and further intron prediction was done using the Augustus web tool (http://bioinf.uni-greifswald.de/augustus/). DNA cloning and transformation of P. pastoris X33 with codon-optimized genes (for expression in P. pastoris) encoding CdeAlcOx and CtaAlcOx were carried out as described previously for other constructions (71). The recombinant P. pastoris X33 strains containing CgrAlcOx gene (GenBank EFQ30446.1) was recovered from previous work (18, 21). The selection of the most productive recombinant transformants was done through culture and expression in 24-well deep well plates with subsequent purification from supernatant by affinity chromatography on nickel–nitrilotriacetic acid (Ni-NTA) resin using an in-house automated procedure (71).
Fungal cultures and secretome harvesting.
Fungal strains were grown and allowed to sporulate on potato dextrose agar (PDA) medium plates in closed petri dishes at 23°C for 7 days under natural solar light without controlling the humidity and were stored at 4°C before utilization. Inocula were prepared by aseptically homogenizing the content of 90-mm petri dish cultures of Colletotrichum (containing fungal biomass [both mycelia and spores] and PDA medium) using an Ultra Turrax T25 homogenizer (IKA, Germany) for 1 min at 9,500 rpm in 80 ml of the culture medium (see below). 200 μl of this homogenate was used to inoculate 100 ml of liquid medium placed in a 500-ml baffled flask.
Liquid cultures were grown in two different media: YG and YG-CME. YG medium was composed of glucose (10 g·liter−1), ammonium tartrate (2 g·liter−1), yeast extract (1 g·liter−1), K2HPO4 (1 g·liter−1), MgSO4·7H2O (0.5 g·liter−1), KCl (0.5 g·liter−1), and 0.5 ml·liter−1 of trace elements stock solution composed of (NH4)6Mo7O24·4H2O (20 mg·ml−1), FeSO4·7H2O (100 mg·ml−1), ZnSO4·7H2O (140 mg·ml−1), B4O7Na2·10H2O (200 mg·ml−1). Glucose was autoclaved separately as a 10× stock solution. The trace elements stock solution was sterilized by filtration through a 0.22-μm polyether sulfone membrane (Merck-Millipore, Germany). Note that insoluble elements present in this solution were removed by the filtration step. YG-CME medium was prepared exactly as YG but was further supplemented with CuSO4·5H2O (500 μM), MnCl2 (500 μM), and 9 g·liter−1 ethanol; filtered; and sterilized (except for ethanol).
The inoculated cultures were placed in an Innova 42R incubator (New Brunswick, NJ) and allowed to grow at 25°C under natural light and shaking (at 150 rpm for C. destructivum and 105 rpm for C. tabacum and C. graminicola). The secretomes were harvested at days 3, 7, and 14 after inoculation. All samples were prepared in triplicate and pooled upon harvesting. Each harvested secretome was submitted to several filtration steps: (i) Miracloth (22- to 25-μm pore size) (Merck-Millipore); (ii) several glass microfiber filters used sequentially: 47-mm Whatman grade GF/D, A and F (GE Healthcare, USA); (iii) 0.45- and 0.22-μm polyether sulfone membranes (Merck-Millipore); and (iv) diafiltered onto a polyether sulfone membrane with a 10-kDa cutoff (Vivaspin, Sartorius, Göttingen, Germany) with 50 mM sodium phosphate buffer, pH 7.0, to a final volume of 1 to 7 ml. The secretomes were stored at 4°C until use. Their total protein concentration was evaluated by the Bradford method (protein assay, Bio-Rad, France) using a BSA standard protein.
Microfluidic capillary gel electrophoresis.
The concentrated secretomes samples were diluted so that 87.5 μl of the initially harvested secretomes (before concentration) were submitted to electrophoresis, except for the sample from C. tabacum YG-CME day 14, which was further diluted by 1/3 to avoid a too-high protein concentration. The samples were prepared in a 96-well plate using HT Protein Express Assay sample buffer (PerkinElmer, USA) supplemented with 36.4 mM (final concentration) of dithiothreitol. The samples were heated at 95°C for 10 min and subjected directly to LabChip GX II capillary gel electrophoresis (PerkinElmer) according to the manufacturer’s instructions. The electropherograms were analyzed using the LabChip GX II software (PerkinElmer).
LC–MS/MS protein identification.
For each secretome, 15 μg of proteins were loaded on a 10% Tris-glycine precast SDS-PAGE gel (Mini-PRO-TEAN TGX, Bio-Rad). After a short migration (0.5 cm) in the stacking gel, the gels were stained with Coomassie blue (Bio-Rad), and each electrophoresis track was cut into two 2-mm-wide strips. Proteomic identification was performed at the Plateforme d’Analyse Protéomique de Paris Sud-Ouest (PAPPSO, INRA, Jouy-en-Josas, France; http://pappso.inra.fr/), according to the protocol described by Amara et al. (72). Briefly, the digestion of the proteins contained in the gel strips was carried out according to a standard trypsinolysis process, using modified trypsin (Promega, France). Peptide analysis was performed by a NanoLC Ultra 2D system (Eksigent Technologies, CA) coupled to a Q-exactive mass spectrometer (Thermo Fisher Scientific, France) using electrospray ionization. Peptide attribution and protein annotation were performed by comparing mass spectrometry data with predicted proteins in the genomes of Colletotrichum graminicola M1.001 (https://mycocosm.jgi.doe.gov/Colgr1/Colgr1.home.html) and Colletotrichum higginsianum IMI 349063 (https://mycocosm.jgi.doe.gov/Colhig2/Colhig2.home.html) for both C. destructivum and C. tabacum strains. The internal contaminant database X!TandemPipeline software (version 0.2.30, PAPPSO, France) was also used. The protein annotation was completed manually using the CAZy database for the CAZymes annotation and using the BLASTP tool from the NCBI database (https://blast.ncbi.nlm.nih.gov) for other proteins.
Heterologous expression and purification.
Recombinant P. pastoris X33 strains were streaked on yeast extract-peptone-dextrose (YPD) agar plates containing Zeocin (100 μg·ml−1) and incubated for 3 days at 30°C. In 50-ml sterile conical tubes, 5 ml of YPD broth was inoculated with a P. pastoris transformant and incubated for 5 h (30°C, 160 rpm in an orbital shaker). The preculture was used to inoculate 0.2% (vol/vol) of 500 ml of buffered complex glycerol medium (BMGY) in a 2-liter flask and incubated (16 h) until the optical density at 600 nm (OD600 nm) reached 4–6. Then, the biomass was harvested by centrifugation (10 min, 16°C, 5,000 × g), resuspended in 100 ml of buffered complex methanol medium (BMMY) supplemented with CuSO4 (500 μM) and methanol (1% vol/vol) in a 500-ml flask, and shaken for 3 days in an orbital shaker (200 rpm, 16°C) with daily feeding of methanol (1% vol/vol). The culture was then centrifuged (10 min, 4°C, 5,000 × g), and the supernatant containing the secreted proteins was filtrated on a 0.45-μm cutoff membrane (Millipore, USA) followed by buffer exchange with Tris-HCl (50 mM, pH 8.5) performed by ultrafiltration through a 10-kDa cutoff polyether sulfone membrane (Vivacell 250, Sartorius Stedim Biotech GmbH, Germany). The buffered supernatant was loaded on a DEAE–20-ml HiPrep FF 16/10 anion-exchange chromatography column (GE Healthcare), equilibrated with Tris-HCl buffer (50 mM, pH 8.5), and connected to an Äkta Xpress system (GE Healthcare). Elution was performed by applying a linear gradient from 0 to 500 mM NaCl (in 50 mM Tris-HCl buffer, pH 8.5) with a flow rate set to 5 ml·min−1. The collected fractions were analyzed by SDS-PAGE (10% polyacrylamide precast gel; Bio-Rad) stained with Coomassie blue. Fractions containing the recombinant enzyme were pooled, concentrated, and exchanged to sodium phosphate buffer (50 mM, pH 7.0). The CgrAlcOx (ε280 = 101,215 M−1·cm−1), CdeAlcOx (ε280 = 94,225 M−1·cm−1), and CtaAlcOx (ε280 = 94,225 M−1·cm−1) concentrations were determined by UV absorption at 280 nm using a Nanodrop ND-200 spectrophotometer (Thermo Fisher Scientific). Final solutions (3 to 5.5 mg·ml−1) were flash-frozen using liquid nitrogen and stored at −80°C for long-term storage or stored at 4°C for immediate use.
Spectrophotometric enzyme activity assays.
AlcOx initial rates were determined by monitoring H2O2 released during AlcOx oxidation of various substrates, using the coupled ABTS/HRP assay. Routine assays were performed in 96-well transparent microtiter plates (flat bottom, polystyrene; Greiner Bio-One, Austria) in a 200-μl final volume containing 0.25 mg/ml ABTS powder, 0.1 mg·ml−1 HRP powder (as provided by the supplier), 3 mM substrate (unless indicated otherwise), and 1 nM AlcOx in sodium phosphate buffer (50 mM, pH 8.0) at 23°C. The reactions were initiated by the addition of substrate to a premix containing all other reagents. Evolution of the absorbance at 414 nm (ABTS cation radical) was measured over time with a Tecan Infinite M200 (Tecan, Switzerland) plate reader. Oxidation of 1 mol of substrate by AlcOx consumes 1 equivalent of O2 and generates 1 equivalent of H2O2, which is in turn used by HRP as cosubstrate to oxidize 2 equivalents of ABTS. Insoluble substrates were prepared in acetone so that the final acetone concentration in the enzyme assay did not exceed 1% (vol/vol). Standard curve of know concentrations of H2O2 (1 to 40 μM) were made and used to quantify H2O2 production. One unit of AlcOx activity was defined as the amount of enzyme necessary to produce 1 μmol of H2O2/min.
Bioconversion assays.
Bioconversions were carried out in 4-ml clear borosilicate glass vials closed by screw caps with PTFE septum for a total reaction volume of 500 μl. Routine assay contained 3 mM substrate (prediluted in acetone), 1 μM AlcOx, various amounts of catalase (0 to 8 μM), and/or HRP (0 to 12 μM) in sodium phosphate buffer (50 mM, pH 8.0). The final reaction mixture contained 1% (vol/vol) acetone. The reaction mixtures were run at 23°C, under stirring at 190 rpm in an Innova 42R incubator (New Brunswick, NJ). The vials were placed lying down in the incubator. The reaction mixture was then acidified by addition of 10 μl of HCl (12 M) and products, and possible remaining substrates were extracted by adding 500 μl of cyclohexane/ethyl acetate mixture (1:1), followed by shaking and centrifugation for 5 min at 1,500 × g. The organic layer was transferred into a new vial by pipetting and analyzed with a GC-2014 apparatus (Shimadzu, Japan) equipped with a flame ionization detector (FID) and an Optima-σ-3 GC capillary column (30 m by 0.25 mm by 0.25 μm; Macherey-Nagel GmbH & Co., Germany). Nitrogen (200 kPa) was used as carrier gas. The injector and detector temperatures were set at 250°C, and temperature programs are described in Table S2. Heptanal or guaiacol were used as internal standards for either aliphatic or aromatic products analyses, respectively.
Data availability.
The protein sequences of the CdeAlcOx and CtaAlcOx were deposited in GenBank under the accession numbers MZ269520 and MZ269521, respectively.
ACKNOWLEDGMENTS
This study was supported by the French National Agency for Research (“Agence Nationale de la Recherche”) through the “Projet de Recherche Collaboratif International” ANR-NSERC (FUNTASTIC project, ANR-17-CE07-0047). We are grateful to Mane & Fils and the “Association Nationale Recherche Technologie” (ANRT) for funding the PhD fellowship of D.R. entitled “Discovery and structure–function study of new fungal copper radicals oxidases used as biocatalysts for the valorization of alcohols and plant biomass.” This Convention Industrielle de Formation par la Recherche grant 2017/1169 runs from 1 April 2018 to 1 April 2021. Fungal genome sequencing was supported by funding to R.J.O. (grant ANR-17-CAPS-0004-01).
We are grateful to Renaud Vincentelli (AFMB, CNRS, Marseille, France) for providing access to the microfluidic capillary gel electrophoresis apparatus.
D.R. carried out most of the experimental work and analyzed the data. S.S. was involved in the culture of fungi and production of the secretomes. L.O.C. and D.N. performed proteomics and analyzed data. E.D. and B.H. performed expert annotation of CAZymes. M.H. contributed to enzyme production in P. pastoris. S.G. performed the microfluidic capillary gel electrophoresis experiments. R.J.O. and N.L. provided the Colletotrichum strains and access to the associated genomic data. M.L. and J.-G.B. conceptualized the study. F.L., B.B., M.L., and J.-G.B. supervised the work. D.R. and J.-G.B. drafted the manuscript. All authors approved the final version of the manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Mickaël Lafond, Email: mickael.lafond@univ-amu.fr.
Jean-Guy Berrin, Email: jean-guy.berrin@inrae.fr.
Nicole R. Buan, University of Nebraska-Lincoln
REFERENCES
- 1.Wohlgemuth R, Twardowski T, Aguilar A. 2021. Bioeconomy moving forward step by step—a global journey. N Biotechnol 61:22–28. 10.1016/j.nbt.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Lokko Y, Heijde M, Schebesta K, Scholtès P, Van Montagu M, Giacca M. 2018. Biotechnology and the bioeconomy—towards inclusive and sustainable industrial development. N Biotechnol 40:5–10. 10.1016/j.nbt.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Pellis A, Cantone S, Ebert C, Gardossi L. 2018. Evolving biocatalysis to meet bioeconomy challenges and opportunities. N Biotechnol 40:154–169. 10.1016/j.nbt.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Meyer V, Basenko EY, Benz JP, Braus GH, Caddick MX, Csukai M, de Vries RP, Endy D, Frisvad JC, Gunde-Cimerman N, Haarmann T, Hadar Y, Hansen K, Johnson RI, Keller NP, Kraševec N, Mortensen UH, Perez R, Ram AFJ, Record E, Ross P, Shapaval V, Steiniger C, van den Brink H, van Munster J, Yarden O, Wösten HAB. 2020. Growing a circular economy with fungal biotechnology: a white paper. Fungal Biol Biotechnol 7:5. 10.1186/s40694-020-00095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hage H, Rosso M-N. 2021. Evolution of fungal carbohydrate-active enzyme portfolios and adaptation to plant cell-wall polymers. J Fungi 7:185. 10.3390/jof7030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glass NL, Schmoll M, Cate JHD, Coradetti S. 2013. Plant cell wall deconstruction by ascomycete fungi. Annu Rev Microbiol 67:477–498. 10.1146/annurev-micro-092611-150044. [DOI] [PubMed] [Google Scholar]
- 7.Arntzen MØ, Bengtsson O, Várnai A, Delogu F, Mathiesen G, Eijsink VGH. 2020. Quantitative comparison of the biomass-degrading enzyme repertoires of five filamentous fungi. Sci Rep 10:20267. 10.1038/s41598-020-75217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couturier M, Navarro D, Olivé C, Chevret D, Haon M, Favel A, Lesage-Meessen L, Henrissat B, Coutinho PM, Berrin J-G. 2012. Post-genomic analyses of fungal lignocellulosic biomass degradation reveal the unexpected potential of the plant pathogen Ustilago maydis. BMC Genomics 13:57. 10.1186/1471-2164-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandmontagne D, Navarro D, Neugnot-Roux V, Ladevèze S, Berrin J-G. 2021. The secretomes of Aspergillus japonicus and Aspergillus terreus supplement the Rovabio enzyme cocktail for the degradation of soybean meal for animal feed. J Fungi 7:278. 10.3390/jof7040278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filiatrault-Chastel C, Navarro D, Haon M, Grisel S, Herpoël-Gimbert I, Chevret D, Fanuel M, Henrissat B, Heiss-Blanquet S, Margeot A, Berrin J-G. 2019. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol Biofuels 12:55. 10.1186/s13068-019-1394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravalason H, Jan G, Mollé D, Pasco M, Coutinho PM, Lapierre C, Pollet B, Bertaud F, Petit-Conil M, Grisel S, Sigoillot J-C, Asther M, Herpoël-Gimbert I. 2008. Secretome analysis of Phanerochaete chrysosporium strain CIRM-BRFM41 grown on softwood. Appl Microbiol Biotechnol 80:719–733. 10.1007/s00253-008-1596-x. [DOI] [PubMed] [Google Scholar]
- 12.Lomascolo A, Record E, Herpoel-Gimbert I, Delattre M, Robert JL, Georis J, Dauvrin T, Sigoillot J-C, Asther M. 2003. Overproduction of laccase by a monokaryotic strain of Pycnoporus cinnabarinus using ethanol as inducer. J Appl Microbiol 94:618–624. 10.1046/j.1365-2672.2003.01879.x. [DOI] [PubMed] [Google Scholar]
- 13.Lomascolo A, Cayol J-L, Roche M, Guo L, Robert J-L, Record E, Lesage-Meessen L, Ollivier B, Sigoillot J-C, Asther M. 2002. Molecular clustering of Pycnoporus strains from various geographic origins and isolation of monokaryotic strains for laccase hyperproduction. Mycol Res 106:1193–1203. 10.1017/S0953756202006494. [DOI] [Google Scholar]
- 14.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. 2013. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels 6:41. 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper JAD, Smith W, Bacila M, Medina H. 1959. Galactose oxidase from Polyporus circinatus, Fr. J Biol Chem 234:445–448. 10.1016/S0021-9258(18)70223-8. [DOI] [PubMed] [Google Scholar]
- 17.Avigad G, Amaral D, Asensio C, Horecker BL. 1962. The d-galactose oxidase of Polyporus circinatus. J Biol Chem 237:2736–2743. 10.1016/S0021-9258(18)60220-0. [DOI] [PubMed] [Google Scholar]
- 18.Yin D, Urresti S, Lafond M, Johnston EM, Derikvand F, Ciano L, Berrin J-G, Henrissat B, Walton PH, Davies GJ, Brumer H. 2015. Structure–function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family. Nat Commun 6:10197. 10.1038/ncomms10197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oide S, Tanaka Y, Watanabe A, Inui M. 2019. Carbohydrate-binding property of a cell wall integrity and stress response component (WSC) domain of an alcohol oxidase from the rice blast pathogen Pyricularia oryzae. Enzyme Microb Technol 125:13–20. 10.1016/j.enzmictec.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Bissaro B, Kodama S, Hage H, Ribeaucourt D, Haon M, Grisel S, Simaan J, Beisson F, Forget S, Brumer H, Rosso M-N, O’Connell R, Lafond M, Kubo Y, Berrin J-G. 2021. Unravelling the role of alcohol copper radical oxidases in fungal plant pathogens. Res Square. 10.21203/rs.3.rs-493001/v1. [DOI] [Google Scholar]
- 21.Ribeaucourt D, Bissaro B, Guallar V, Yemloul M, Haon M, Grisel S, Alphand V, Brumer H, Lambert F, Berrin J-G, Lafond M. 2021. Comprehensive insights into the production of long chain aliphatic aldehydes using a copper-radical alcohol oxidase as biocatalyst. ACS Sustainable Chem Eng 9:4411–4421. 10.1021/acssuschemeng.0c07406. [DOI] [Google Scholar]
- 22.Liu J, Wu S, Li Z. 2018. Recent advances in enzymatic oxidation of alcohols. Curr Opin Chem Biol 43:77–86. 10.1016/j.cbpa.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Dong J, Fernández‐Fueyo E, Hollmann F, Paul CE, Pesic M, Schmidt S, Wang Y, Younes S, Zhang W. 2018. Biocatalytic oxidation reactions: a chemist’s perspective. Angew Chem Int Ed Engl 57:9238–9261. 10.1002/anie.201800343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surburg H, Panten J. 2016. Common fragrance and flavor materials: preparation, properties and uses, 6th ed, Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 25.Tsuzuki S. 2019. Higher straight-chain aliphatic aldehydes: importance as odor-active volatiles in human foods and issues for future research. J Agric Food Chem 67:4720–4725. 10.1021/acs.jafc.9b01131. [DOI] [PubMed] [Google Scholar]
- 26.Burdock GA. 2016. Fenaroli’s handbook of flavor ingredients. CRC Press LLC, Boca Raton, Florida. [Google Scholar]
- 27.Rouseff R, Perez-Cacho PR. 2007. Citrus Flavour, p 117–134. In Berger RG (ed), Flavours and fragrances: chemistry, bioprocessing and sustainability. Springer, Berlin, Germany. [Google Scholar]
- 28.Kohlpaintner C, Schulte M, Falbe J, Lappe P, Weber J, Frey GD. 2013. Aldehydes, aliphatic. Ullmann’s encyclopedia of industrial chemistry. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 29.Shah J, Arslan E, Cirucci J, O’Brien J, Moss D. 2016. Comparison of oleo- vs. petro-sourcing of fatty alcohols via cradle-to-gate life cycle assessment. J Surfactants Deterg 19:1333–1351. 10.1007/s11743-016-1867-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldwin E, Plotto A, Manthey J, McCollum G, Bai J, Irey M, Cameron R, Luzio G. 2010. Effect of liberibacter infection (Huanglongbing disease) of citrus on orange fruit physiology and fruit/fruit juice quality: chemical and physical analyses. J Agric Food Chem 58:1247–1262. 10.1021/jf9031958. [DOI] [PubMed] [Google Scholar]
- 31.Dala-Paula BM, Plotto A, Bai J, Manthey JA, Baldwin EA, Ferrarezi RS, Gloria MBA. 2018. Effect of Huanglongbing or greening disease on orange juice quality, a review. Front Plant Sci 9:1976. 10.3389/fpls.2018.01976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribeaucourt D, Bissaro B, Lambert F, Lafond M, Berrin J-G. 2021. Biocatalytic oxidation of fatty alcohols into aldehydes for the flavors and fragrances industry. Biotechnol Adv, in press. 10.1016/j.biotechadv.2021.107787. [DOI] [PubMed] [Google Scholar]
- 33.Forget S, Xia R, Hein JE, Brumer H. 2020. Determination of biocatalytic parameters of a copper radical oxidase using real-time reaction progress monitoring. Org Biomol Chem 18:2005–2184. 10.1039/c9ob02757b. [DOI] [PubMed] [Google Scholar]
- 34.Baroncelli R, Amby DB, Zapparata A, Sarrocco S, Vannacci G, Le Floch G, Harrison RJ, Holub E, Sukno SA, Sreenivasaprasad S, Thon MR. 2016. Gene family expansions and contractions are associated with host range in plant pathogens of the genus Colletotrichum. BMC Genomics 17:555. 10.1186/s12864-016-2917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, Altmüller J, Alvarado-Balderrama L, Bauser CA, Becker C, Birren BW, Chen Z, Choi J, Crouch JA, Duvick JP, Farman MA, Gan P, Heiman D, Henrissat B, Howard RJ, Kabbage M, Koch C, Kracher B, Kubo Y, Law AD, Lebrun M-H, Lee Y-H, Miyara I, Moore N, Neumann U, Nordström K, Panaccione DG, Panstruga R, Place M, Proctor RH, Prusky D, Rech G, Reinhardt R, Rollins JA, Rounsley S, Schardl CL, Schwartz DC, Shenoy N, Shirasu K, Sikhakolli UR, Stüber K, et al. 2012. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44:1060–1065. 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perfect SE, Hughes HB, O’Connell RJ, Green JR. 1999. Colletotrichum: a model genus for studies on pathology and fungal–plant interactions. Fungal Genet Biol 27:186–198. 10.1006/fgbi.1999.1143. [DOI] [PubMed] [Google Scholar]
- 37.Armesto C, Maia FGM, Monteiro FP, de Abreu MS. 2019. Exoenzymes as a pathogenicity factor for Colletotrichum gloeosporioides associated with coffee plants. Summa Phytopathol 45:368–373. 10.1590/0100-5405/191071. [DOI] [Google Scholar]
- 38.Onofre SB, Steilmann P, Bertolini J, Rotta D, Francini AS, Kagimura Y, Acirc S, Groff N, Mazzali L. 2011. Amylolytic enzymes produced by the fungus Colletotrichum gloeosporioides in rice semi-solid fermentation. J Yeast Fungal Res 2:28–32. [Google Scholar]
- 39.Tokuyasu K, Ohnishi-Kameyama M, Hayashi K. 1996. Purification and characterization of extracellular chitin deacetylase from Colletotrichum lindemuthianum. Biosci Biotechnol Biochem 60:1598–1603. 10.1271/bbb.60.1598. [DOI] [PubMed] [Google Scholar]
- 40.Rispoli FJ, Shah V. 2007. Mixture design as a first step for optimization of fermentation medium for cutinase production from Colletotrichum lindemuthianum. J Ind Microbiol Biotechnol 34:349–355. 10.1007/s10295-007-0203-y. [DOI] [PubMed] [Google Scholar]
- 41.Pedersen A, Birmingham WR, Rehn G, Charnock SJ, Turner NJ, Woodley JM. 2015. Process requirements of galactose oxidase catalyzed oxidation of alcohols. Org Process Res Dev 19:1580–1589. 10.1021/acs.oprd.5b00278. [DOI] [Google Scholar]
- 42.Kersten PJ. 1990. Glyoxal oxidase of Phanerochaete chrysosporium: its characterization and activation by lignin peroxidase. Proc Natl Acad Sci USA 87:2936–2940. 10.1073/pnas.87.8.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andberg M, Mollerup F, Parikka K, Koutaniemi S, Boer H, Juvonen M, Master E, Tenkanen M, Kruus K. 2017. A novel Colletotrichum graminicola raffinose oxidase in the AA5 family. Appl Environ Microbiol 83:e01383-17. 10.1128/AEM.01383-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mathieu Y, Offen WA, Forget SM, Ciano L, Viborg AH, Blagova E, Henrissat B, Walton PH, Davies GJ, Brumer H. 2020. Discovery of a fungal copper radical oxidase with high catalytic efficiency towards 5-hydroxymethylfurfural and benzyl alcohols for bioprocessing. ACS Catal 10:e3042–3058. 10.1021/acscatal.9b04727. [DOI] [Google Scholar]
- 45.Copete LS, Chanagá X, Barriuso J, López-Lucendo MF, Martínez MJ, Camarero S. 2015. Identification and characterization of laccase-type multicopper oxidases involved in dye-decolorization by the fungus Leptosphaerulina sp. BMC Biotechnol 15:74. 10.1186/s12896-015-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeiner CA, Purvine SO, Zink EM, Paša-Tolić L, Chaput DL, Haridas S, Wu S, LaButti K, Grigoriev IV, Henrissat B, Santelli CM, Hansel CM. 2016. Comparative analysis of secretome profiles of manganese(II)-oxidizing ascomycete fungi. PLoS One 11:e0157844. 10.1371/journal.pone.0157844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Markus Z, Miller G, Avigad G. 1965. Effect of culture conditions on the production of d-galactose oxidase by Dactylium dendroides. Appl Microbiol 13:686–693. 10.1128/am.13.5.686-693.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura N, Ueda M. 2018. Evaluation of unconventional protein secretion by Saccharomyces cerevisiae and other fungi. Cells 7:128. 10.3390/cells7090128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vivek-Ananth RP, Mohanraj K, Vandanashree M, Jhingran A, Craig JP, Samal A. 2018. Comparative systems analysis of the secretome of the opportunistic pathogen Aspergillus fumigatus and other Aspergillus species. Sci Rep 8:6617. 10.1038/s41598-018-25016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Navarro D, Rosso M-N, Haon M, Olivé C, Bonnin E, Lesage-Meessen L, Chevret D, Coutinho PM, Henrissat B, Berrin J-G. 2014. Fast solubilization of recalcitrant cellulosic biomass by the basidiomycete fungus Laetisaria arvalis involves successive secretion of oxidative and hydrolytic enzymes. Biotechnol Biofuels 7:143. 10.1186/s13068-014-0143-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cleveland L, Coffman RE, Coon P, Davis L. 1975. Role of the copper in galactose oxidase. Biochemistry 14:1108–1115. 10.1021/bi00677a003. [DOI] [PubMed] [Google Scholar]
- 52.Wohlschlager L, Kracher D, Scheiblbrandner S, Csarman F, Ludwig R. 2021. Spectroelectrochemical investigation of the glyoxal oxidase activation mechanism. Bioelectrochemistry 141:107845. 10.1016/j.bioelechem.2021.107845. [DOI] [PubMed] [Google Scholar]
- 53.Tordai H, Bányai L, Patthy L. 1999. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett 461:63–67. 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- 54.Balaraman E, Khaskin E, Leitus G, Milstein D. 2013. Catalytic transformation of alcohols to carboxylic acid salts and H2 using water as the oxygen atom source. Nat Chem 5:122–125. 10.1038/nchem.1536. [DOI] [PubMed] [Google Scholar]
- 55.Poidevin L, Berrin J-G, Bennati-Granier C, Levasseur A, Herpoël-Gimbert I, Chevret D, Coutinho PM, Henrissat B, Heiss-Blanquet S, Record E. 2014. Comparative analyses of Podospora anserina secretomes reveal a large array of lignocellulose-active enzymes. Appl Microbiol Biotechnol 98:7457–7469. 10.1007/s00253-014-5698-3. [DOI] [PubMed] [Google Scholar]
- 56.Ravalason H, Grisel S, Chevret D, Favel A, Berrin J-G, Sigoillot J-C, Herpoël-Gimbert I. 2012. Fusarium verticillioides secretome as a source of auxiliary enzymes to enhance saccharification of wheat straw. Bioresour Technol 114:589–596. 10.1016/j.biortech.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 57.Agger JW, Isaksen T, Várnai A, Vidal-Melgosa S, Willats WGT, Ludwig R, Horn SJ, Eijsink VGH, Westereng B. 2014. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proc Natl Acad Sci USA 111:6287–6292. 10.1073/pnas.1323629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Labourel A, Frandsen KEH, Zhang F, Brouilly N, Grisel S, Haon M, Ciano L, Ropartz D, Fanuel M, Martin F, Navarro D, Rosso M-N, Tandrup T, Bissaro B, Johansen KS, Zerva A, Walton PH, Henrissat B, Leggio LL, Berrin J-G. 2020. A fungal family of lytic polysaccharide monooxygenase-like copper proteins. Nat Chem Biol 16:345–350. 10.1038/s41589-019-0438-8. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Santamarina S, Probst C, Festa RA, Ding C, Smith AD, Conklin SE, Brander S, Kinch LN, Grishin NV, Franz KJ, Riggs-Gelasco P, Lo Leggio L, Johansen KS, Thiele DJ. 2020. A lytic polysaccharide monooxygenase-like protein functions in fungal copper import and meningitis. Nat Chem Biol 16:337–344. 10.1038/s41589-019-0437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonçalves AP, Heller J, Span EA, Rosenfield G, Do HP, Palma-Guerrero J, Requena N, Marletta MA, Glass NL. 2019. Allorecognition upon fungal cell–cell contact determines social cooperation and impacts the acquisition of multicellularity. Curr Biol 29:3006–3017.e3. 10.1016/j.cub.2019.07.060. [DOI] [PubMed] [Google Scholar]
- 61.Kaur K, Sharma A, Capalash N, Sharma P. 2019. Multicopper oxidases: biocatalysts in microbial pathogenesis and stress management. Microbiol Res 222:1–13. 10.1016/j.micres.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 62.Gaetke LM, Chow CK. 2003. Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 189:147–163. 10.1016/s0300-483x(03)00159-8. [DOI] [PubMed] [Google Scholar]
- 63.Kohen R, Nyska A. 2002. Oxidation of biological systems: oxidative stress phenomena, antioxidants, redox reactions, and methods for their quantification. Toxicol Pathol 30:620–650. 10.1080/01926230290166724. [DOI] [PubMed] [Google Scholar]
- 64.Sánchez-Vallet A, Tian H, Rodriguez-Moreno L, Valkenburg D-J, Saleem-Batcha R, Wawra S, Kombrink A, Verhage L, de Jonge R, van Esse HP, Zuccaro A, Croll D, Mesters JR, Thomma BPHJ. 2020. A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PLoS Pathog 16:e1008652. 10.1371/journal.ppat.1008652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akcapinar GB, Kappel L, Sezerman OU, Seidl-Seiboth V. 2015. Molecular diversity of LysM carbohydrate-binding motifs in fungi. Curr Genet 61:103–113. 10.1007/s00294-014-0471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahara H, Hacquard S, Kombrink A, Hughes HB, Halder V, Robin GP, Hiruma K, Neumann U, Shinya T, Kombrink E, Shibuya N, Thomma BPHJ, O’Connell RJ. 2016. Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol 211:1323–1337. 10.1111/nph.13994. [DOI] [PubMed] [Google Scholar]
- 67.Emri T, Molnár Z, Szilágyi M, Pócsi I. 2008. Regulation of autolysis in Aspergillus nidulans. Appl Biochem Biotechnol 151:211–220. 10.1007/s12010-008-8174-7. [DOI] [PubMed] [Google Scholar]
- 68.Daou M, Faulds CB. 2017. Glyoxal oxidases: their nature and properties. World J Microbiol Biotechnol 33:87. 10.1007/s11274-017-2254-1. [DOI] [PubMed] [Google Scholar]
- 69.Leuthner B, Aichinger C, Oehmen E, Koopmann E, Müller O, Müller P, Kahmann R, Bölker M, Schreier PH. 2005. A H2O2-producing glyoxal oxidase is required for filamentous growth and pathogenicity in Ustilago maydis. Mol Genet Genomics 272:639–650. 10.1007/s00438-004-1085-6. [DOI] [PubMed] [Google Scholar]
- 70.Damm U, O’Connell RJ, Groenewald JZ, Crous PW. 2014. The Colletotrichum destructivum species complex—hemibiotrophic pathogens of forage and field crops. Stud Mycol 79:49–84. 10.1016/j.simyco.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haon M, Grisel S, Navarro D, Gruet A, Berrin J-G, Bignon C. 2015. Recombinant protein production facility for fungal biomass-degrading enzymes using the yeast Pichia pastoris. Front Microbiol 6:1002. 10.3389/fmicb.2015.01002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amara S, Perrot T, Navarro D, Deroy A, Benkhelfallah A, Chalak A, Daou M, Chevret D, Faulds CB, Berrin J-G, Morel-Rouhier M, Gelhaye E, Record E. 2018. Enzyme activities of two recombinant heme-containing peroxidases, Tv DyP1 and Tv VP2, identified from the secretome of Trametes versicolor. Appl Environ Microbiol 84:e02826-17. 10.1128/AEM.02826-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S8, Scheme S1, Tables S1 and S2. Download aem.01526-21-s0001.pdf, PDF file, 0.8 MB (834.1KB, pdf)
Supplemental Data Set. Download aem.01526-21-s0002.xlsx, XLSX file, 0.1 MB (132KB, xlsx)
Data Availability Statement
The protein sequences of the CdeAlcOx and CtaAlcOx were deposited in GenBank under the accession numbers MZ269520 and MZ269521, respectively.