Evolutionary analyses reveal conservation and divergence of epigenetic transition during early development across mammals.
Abstract
While mouse remains the most popular model, the conservation of parental-to-embryonic epigenetic transition across mammals is poorly defined. Through analysis of oocytes and early embryos in human, bovine, porcine, rat, and mouse, we revealed remarkable species-specific innovations as no single animal model fully recapitulates the human epigenetic transition. In rodent oocytes, transcription-dependent DNA methylation allows methylation of maternal imprints but not intergenic paternal imprints. Unexpectedly, prevalent DNA hypermethylation, paralleled by H3K36me2/3, also occurs in nontranscribed regions in porcine and bovine oocytes, except for megabase-long “CpG continents (CGCs)” where imprinting control regions preferentially reside. Broad H3K4me3 and H3K27me3 domains exist in nonhuman oocytes, yet only rodent H3K27me3 survives beyond genome activation. Coincidently, regulatory elements preferentially evade H3K27me3 in rodent oocytes, and failure to do so causes aberrant embryonic gene repression. Hence, the diverse mammalian innovations of parental-to-embryonic transition center on a delicate “to-methylate-or-not” balance in establishing imprints while protecting other regulatory regions.
INTRODUCTION
The states of chromatin encode epigenetic information that instructs gene expression without altering DNA sequence (1). Like genetic information, epigenetic marks can be robustly inherited following DNA replication and cell division, and some can be even transmitted from one generation to the next to exert inter- or transgenerational gene regulation (2). DNA methylation plays important roles in transcriptional regulation, genomic imprinting, X chromosome inactivation (XCI), etc. In mammals, DNA methylation is established by de novo DNA methyltransferase, including DNMT3A and DNMT3B, and is maintained during DNA replication by DNMT1 (3). On the other hand, histone modifications are prevalent epigenetic modifications regulating spatiotemporal gene expression and transcriptional memory (4). For example, H3K4me3 (histone H3 lysine 4 tri-methylation) deposited by the highly conserved COMPASS (complex proteins associated with Set1) complex generally correlates with gene activation and is a hallmark of promoter (5). By contrast, H3K27me3 is a repressive mark deposited by the PRC2 complex (Polycomb repressive complex 2) frequently at the promoters of developmental genes. H3K4me3 and H3K27me3 can also co-occupy the promoters of a set of developmental genes, forming bivalent marks (5, 6). Bivalency is proposed to keep genes in a silent state but poised for activation in response to stimuli. Deficiency of these epigenetic modifiers often leads to dysregulation of gene expression and developmental defects including embryonic lethality (5).
Genomic imprinting is a finely regulated epigenetic process that allows monoallelic gene expression, which is often governed by allele-specific DNA methylation at the imprinting control regions (ICRs) (7). The parent-of-origin-specific methylation is acquired during gametogenesis and is maintained after fertilization and throughout somatic development. In mouse oocytes, DNA methylation is established in a transcription-dependent manner, as SETD2 (SET domain containing 2) deposits H3K36me3 [and possibly H3K36me2 (8)], which further recruits DNMT3A to methylate transcribed regions (9). This allows the establishment of maternally methylated ICRs (maternal imprints) when oocyte-specific promoters upstream of these ICRs are exploited. In mouse sperm, Nuclear receptor binding SET domain protein 1 (NSD1), which deposits widespread H3K36me2, mediates de novo DNA methylation of the majority of intergenic regions, including paternal imprints, and a large fraction of genic regions (8). Apart from such canonical imprinting, germline-derived histone modifications can be also transmitted to embryos to regulate allele-specific gene expression. Specifically, H3K27me3 in mouse oocytes is briefly inherited after fertilization, leading to maternal allele-specific repression of several autosomal (e.g., Gab1, Sfmbt2, and Slc38a4) and X chromosome genes (Xist) (10). Such noncanonical genomic imprinting allows paternal-specific XCI in mouse early embryos. However, H3K27me3 controlled noncanonical imprinting is transient and is subsequently taken over by DNA methylation in extraembryonic tissues, where imprinting is maintained (11, 12). Intriguingly, imprinted XCI and H3K27me3-mediated imprinting appear to be missing in human early embryos (13, 14), suggesting species-specific regulatory mechanisms for genomic imprinting.
Besides genomic imprinting, extensive epigenetic reprogramming occurs genome wide during parental-to-embryonic transition in mammals. After fertilization, the genome undergoes extensive global DNA demethylation, a process that is largely conserved although with different demethylation kinetics and extents among different mammals (3, 15, 16). However, the distribution and reprogramming of histone modifications diverge substantially between mouse and human. For example, in most somatic and embryonic stem cells (ESCs), H3K4me3 and H3K27me3 are preferentially enriched at promoters (“canonical pattern”). In mouse oocytes, noncanonical H3K4me3 (ncH3K4me3) and H3K27me3 (ncH3K27me3) exist as broad distal domains present in nontranscribed, partially DNA methylated domains (PMDs) (17–20). ncH3K4me3 is briefly inherited to early embryos but resolves to a canonical pattern after zygotic genome activation (ZGA). While the function of ncH3K4me3 remains elusive, genetic evidence points to a role in transcriptional silencing of mouse oocytes and ZGA fidelity after fertilization (17, 19, 21). There is no ncH3K4me3 in human oocytes (13), where H3K4me3 exhibits a canonical pattern restricted at promoters. H3K27me3 in human oocytes also largely resembles that in somatic tissues and is globally depleted around ZGA, followed by restoration at developmental gene promoters as early as the morula stage (13, 22). The global depletion of H3K27me3 by ZGA suggests that oocyte-derived H3K27me3 unlikely acts as an imprinting mark in human (13). In addition, analysis of chromatin accessibility during the early development among bovine, human, and mouse suggested that the regulatory circuitry underlying bovine ZGA is more similar to human compared to mouse (23).
These conserved and species-specific epigenetic reprogramming events raise the key question of to what extent the knowledge we learned from early development in one species can be applied to other mammals. Are there potential logics or principles underlying diverse species-specific innovations in epigenetic reprogramming to achieve the same goal—parental-to-embryonic transition? Answers to these questions would be crucial to understand how life begins in mammals. In this study, we conducted a comprehensive analysis for epigenetic reprogramming in early development in five mammalian species. In addition to data from human (Homo sapiens) and mouse (Mus musculus) that we and others previously reported (13, 17, 18, 24, 25), we probed the transcriptomes, DNA methylomes, and several key histone marks (H3K4me3, H3K27me3, H3K36me2, and H3K36me3) in oocytes and selectively in early embryos across three additional placental mammalian species, including bovine (Bos taurus), porcine (Sus scrofa), and rat (Rattus norvegicus). These data reveal the notably diverse epigenetic landscapes during early embryogenesis among different mammals. Our integrative analyses uncovered potential principles underlying these seemingly complex reprogramming processes, as the establishment of genomic imprints and protection of nonimprints are delicately regulated and balanced in gametes to ensure the fidelity of embryonic transcription.
RESULTS
Mapping the transcriptomic and epigenomic landscapes in the oocytes and preimplantation embryos across placental mammals
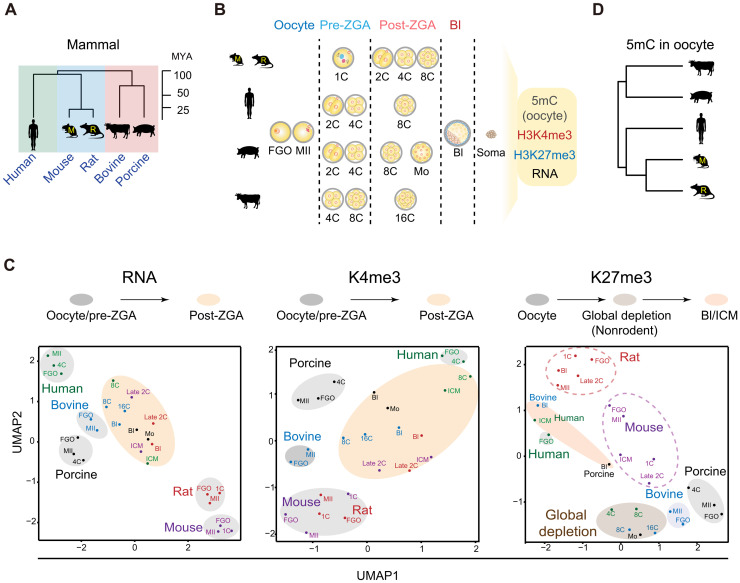
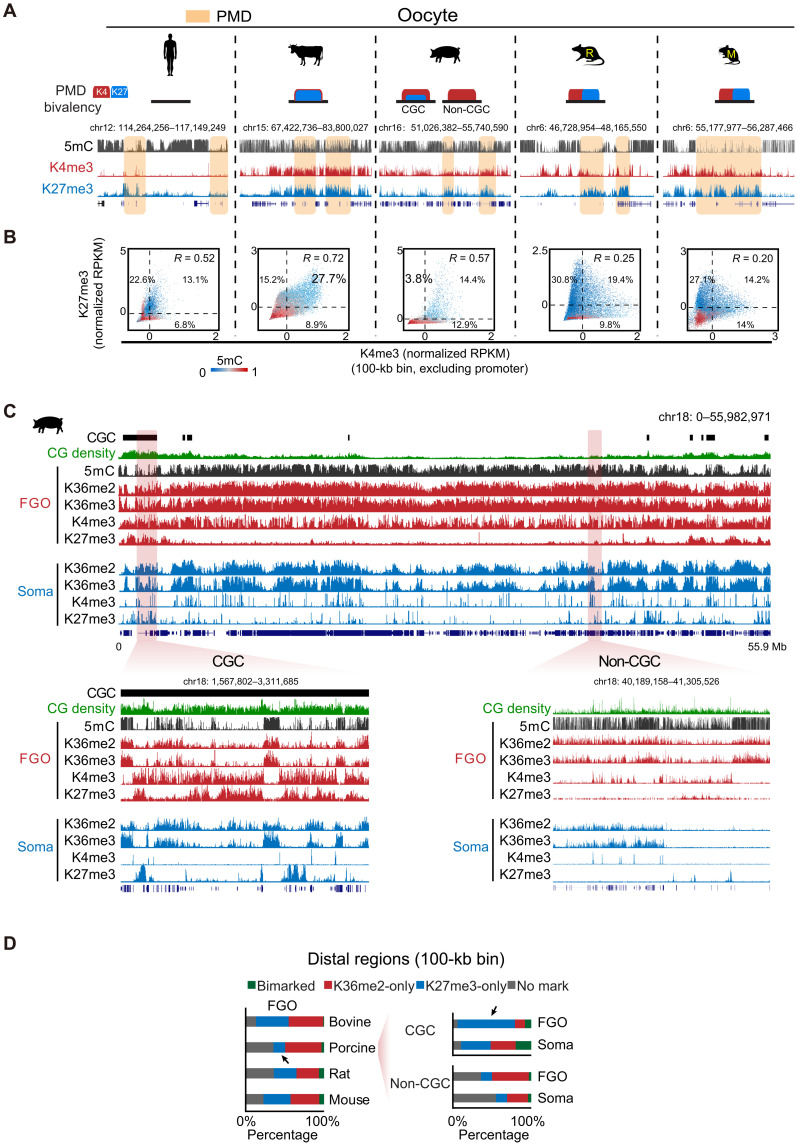
To investigate the conservation and divergence of epigenetic landscapes in mammalian early development, we conducted a systematic study for five placental mammalian species, human, bovine, porcine, rat, and mouse (Fig. 1A). In addition to previous epigenomic datasets we and others have reported in mammalian oocytes and early embryos (with the majority from human and mouse) (table S1) (13, 17, 18, 24–26), we further performed CUT&RUN (cleavage under targets & release using nuclease) (27) to probe H3K4me3 and H3K27me3 in the oocytes and preimplantation embryos of bovine, porcine, and rat (Fig. 1, A and B). We also conducted small-scale TELP-enabled methylome sequencing (STEM-seq), a low-input DNA methylome profiling method (28), to profile DNA methylomes in bovine and porcine oocytes. For each species, full-grown oocytes (FGOs), metaphase II (MII) oocytes, pre- and post-ZGA embryos, and blastocysts (Bl) were collected (Fig. 1B), with at least two replicates conducted (table S2). We also collected data from a panel of somatic cells as controls (see Fig. 1B legend). For histone marks in bovine, porcine, and rat, we performed H3K4me3 and H3K27me3 CUT&RUN in cumulus cells (CCs) as somatic controls. Immunostaining showed that H3K4me3 and H3K27me3 are strongly enriched in FGOs and MII oocytes in these three species, and the signals decrease substantially after fertilization (figs. S1 and S2). Semiquantitative analyses showed full or partial restoration of these marks by the blastocyst stage (figs. S1 and S2), except for H3K27me3 in rat, which remains relatively low even in blastocyst. These data are in line with reported global resetting after fertilization and subsequent restoration in bovine and porcine (29, 30). The CUT&RUN data are highly reproducible between replicates, except for H3K27me3 at 4- to 16-cell stage in bovine and two-cell to morula stage in porcine, when it is globally depleted and CUT&RUN cannot detect strong enrichment (discussed in detail later) (figs. S3 to S5). These data resemble a similar global loss of H3K27me3 in human early embryos (13). Note that for porcine early embryos, we used parthenogenetically activated (PG) instead of in vitro fertilized (IVF) embryos that are known to have polyspermy issue (31). Nevertheless, we also conducted CUT&RUN for H3K4me3 for IVF embryos at limited stages (eight-cell and blastocyst), which exhibits similar patterns with that from PG embryos (fig. S4, “IVF”). To investigate the relationship between these epigenetic marks and gene expression in each species, we also profiled transcriptomes for each stage in replicates (fig. S6, A to C, and table S2). As expected, the enrichment of H3K4me3 and H3K27me3 at gene promoters positively and negatively correlates with corresponding RNA levels, respectively, in each species in general (fig. S6, D and E). For example, Zar1, a well-known maternal gene, is enriched with H3K4me3 but not H3K27me3 in the oocytes and pre-ZGA embryos in all species. Slc34a2, a gene activated after major ZGA in each species, is decorated with strong canonical H3K4me3 peaks after ZGA but not H3K27me3. Note that while bovine and rat embryos showed expected ZGA timing (16-cell in bovine and late two-cell in rat) (fig. S6, A and C) (32), porcine embryos exhibit a delayed ZGA beyond the expected four- to eight-cell stage (32), perhaps due to in vitro culture. We therefore used the morula stage (fig. S6B) to represent the a post-ZGA stage in porcine for the subsequent analyses.
Fig. 1. Global view of the epigenomic landscapes in mammalian oocytes and preimplantation embryos.
(A) Phylogeny of the five placental mammals included in this study. MYA, million years ago; M, mouse; R, rat. (B) Schematic showing the oocytes and preimplantation embryos collected for each species and related datasets. The majority of datasets for human and mouse oocytes and early embryos were from published studies (13, 17, 18, 24–26). Control data from somatic tissues/cells (soma) were either generated in this study or previous studies. DNA methylation: human liver (73), bovine lung (74), porcine CCs (this study), rat left ventricle (75), and mouse cerebellum (76); H3K4me3 and H3K27me3: human liver (73), bovine CCs, porcine CCs, rat CCs (this study), and mouse cerebellum (77). (C) UMAP analysis of oocytes and early embryos among different species (color-coded) based on the RNA-seq (RNA), promoter H3K4me3 (K4me3) and promoter H3K27me3 (K27me3) data. Major developmental transitions are indicated above. (D) Hierarchical clustering of mammalian oocytes based on promoter DNA methylation (TSS ± 2.5 kb). 2C, late 2-cell embryo; 4C, 4-cell embryo; 8C, 8-cell embryo; 16C, 16-cell embryo; Mo, morula; Bl, blastocyst.
Global analysis reveals highly diverse epigenetic states of mammalian oocytes and early embryos across species
Using orthologous genes present in all five species, we first interrogate the global relationship of transcriptomes and epigenomes in the oocytes and early embryos across different mammals. Clustering analysis using RNA sequencing (RNA-seq) data showed that the oocytes and pre-ZGA embryos are clustered together in each species, consistent with the transcriptional silencing in pre-ZGA embryos and inheritance of maternal transcripts, but are separated among different species (Fig. 1C, RNA). However, after ZGA, transcriptomes deviate from oocyte and pre-ZGA embryos but instead tend to converge and cluster closer even from different species (Fig. 1C, RNA). These data agree with the major transcription transition around ZGA and support the hourglass model proposing developmental convergence of conservation toward the phylotypic period (33). Clustering analysis using promoter H3K4me3 data yields very similar results as RNA-seq, as a major transition occurs between oocyte/pre-ZGA embryos and post-ZGA embryos in most species (Fig. 1C, K4me3). The transition echoes the global resetting of H3K4me3 after ZGA from a broad, noncanonical form of H3K4me3 to a sharp, canonical form after ZGA (see fig. S6D for examples and discussed in detail later). Human, however, is an exception, as FGO also adopts a canonical form of H3K4me3 and is thus clustered closer to post-ZGA embryos (Fig. 1C, K4me3) (13). Unlike RNA-seq or H3K4me3, H3K27me3 shows much more diverse patterns among different species (Fig. 1C, K27me3). For mouse and rat, all stages are closely adjacent from oocytes to early embryos, echoing substantial inheritance of H3K27me3 throughout preimplantation development (discussed in detail later). For nonrodent mammals, two transitions were observed. Oocytes are separated away from postfertilization embryos, which further deviate from blastocysts or inner cell masses (ICMs) (Fig. 1C, K27me3). This likely reflects the global loss of H3K27me3 after fertilization (the first transition) and the subsequent restoration in ICM/blastocyst (the second transition). Human FGO again is clustered closer to blastocysts or ICMs presumably because its H3K27me3 adopts a canonical form. Last, hierarchical clustering analysis of DNA methylation [transcription start site (TSS) ± 2.5 kb] in mammalian oocytes shows that mouse and rat are adjacent to each other but are separated from bovine and porcine, with human falling in between but relatively closer to rodents (Fig. 1D). This result also recapitulates the phylogenetic relationship of these species (Fig. 1A). Thus, these data reveal highly diverse epigenetic reprogramming in early development among different species, and no single nonhuman species in this study seem to fully recapitulate the epigenetic states of human oocytes and early embryos. Given developmental timing varies among different species, we focused on the most representative stages in each species, including FGO, MII oocyte, pre-ZGA, post-ZGA, and ICM/blastocyst, for the subsequent analyses.
Unlike rodents, bovine and porcine oocytes show pervasive DNA methylation except for CGCs
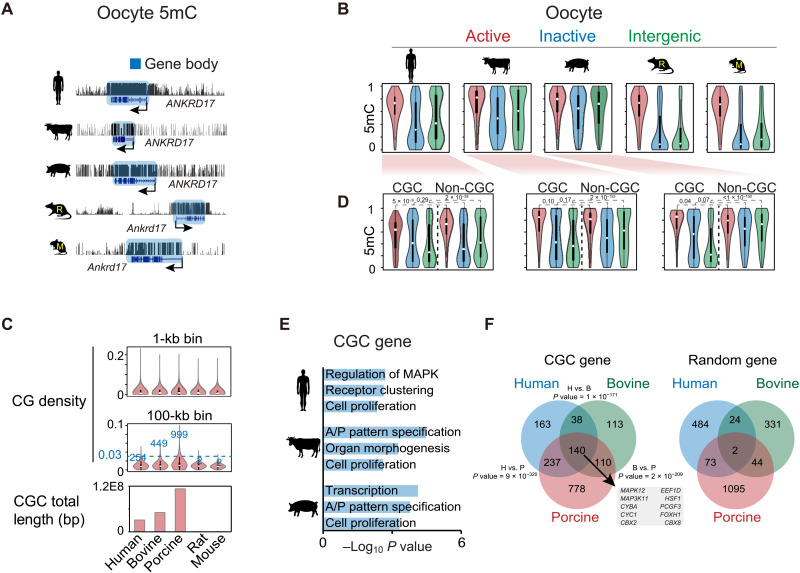
In mouse oocytes, DNA methylome is established in a transcription-dependent manner, leading to “highly methylated domains” (HMDs) in active gene bodies and PMDs in gene-poor and transcriptionally inactive regions (34). As expected, active gene bodies are highly methylated in the oocytes of all five species, supporting transcription-coupled DNA methylation (Fig. 2, A and B). Similar to findings in a recent study (16), the nonrodent mammals, including porcine, human, and bovine, show substantially higher global DNA methylation levels (53.1 to 61.1%) than that in rat (29.8%) or mouse (38.9%) (fig. S7A, top). This is echoed by the lower PMD coverages (20.7 to 29.6% in nonrodents compared to 43.4 to 57.4% in rodents) (fig. S7A, middle). Unexpectedly, a close examination revealed that, unlike that in rodents, nontranscribed regions tend to be highly methylated in the oocytes of porcine and bovine, followed by moderate methylation in human (Fig. 2B), as also confirmed by a reanalysis of published porcine oocyte methylome (fig. S7B, left) (16). This is also consistent with the hierarchical clustering result of oocyte DNA methylomes (Fig. 1D). The hypermethylation of nontranscribed regions does not appear to be due to the incomplete gene annotation in porcine and bovine, as annotated inactive genes also showed high methylation (Fig. 2B). When we restricted analyses to annotated genomes, the percentages of actively transcribed regions, based on RNA-seq, are comparable among all species (fig. S7A, bottom). These data indicate the existence of transcription-independent DNA methylation in porcine and bovine. It is worth mentioning that we cannot fully exclude the possibility that certain hypermethylated inactive gene bodies in porcine and bovine may reflect their previous transcription during early oogenesis.
Fig. 2. Oocyte DNA methylomes and CpG continents.
(A) The UCSC genome browser views showing DNA methylation at the ANKRD17 loci in mammalian oocytes. (B) Violin plots showing the DNA methylation levels in active gene bodies (red), inactive gene bodies (blue), and intergenic regions (green) in mammalian oocytes. (C) Violin plots showing the CG density in the genomes of different species. Top: CG density in 1-kb bins. Middle: CG density in 100-kb bins. The numbers of 100-kb bins with density more than 0.03 are shown. Bottom: Barplot showing the total CGC lengths in the genomes of the five species. (D) Violin plots showing the DNA methylation levels in active gene bodies (red), inactive gene bodies (blue), and intergenic regions (green) in CGC and non-CGC regions in human, bovine, and porcine oocytes. P values (Wilcoxon test) are also shown. (E) Barplot showing the enriched Gene Ontology (GO) terms for genes in CGCs in human, bovine, and porcine. A/P, anterior/posterior. (F) Venn diagram showing the overlap of orthologous genes in CGCs among human, bovine, and porcine. The P value (Fisher’s exact test) of shared genes for each species pair and overlapped genes among all three species are shown. The overlaps of random genes of equal numbers as CGC genes are also shown.
Intriguingly, a careful examination revealed that such hypermethylation of nontranscribed regions in porcine and bovine do have exceptions, where hypomethylation is evident in nontranscribed regions (fig. S7C). These regions, frequently residing near chromosome ends although far beyond the telomeric regions, show extraordinarily high levels of CGs that often extend over 100 kb and even megabase in distance (Fig. 2C, middle, and fig. S7, C and D). To distinguish them from the conventional CpG islands (CGIs), which are much smaller in size (around a few kilobase), we termed them CpG continents (CGCs). A search for long stretches of CG-rich domains that are over 100 kb (Materials and Methods) yielded 99.9-, 44.9-, and 25.4-Mb CGCs in porcine, bovine, and human, respectively, occupying 0.82 to 1.84% of the genomes (Fig. 2C, bottom). The sizes of individual CGCs range from 0.1 to 4.7 Mb in porcine, 2.9 Mb in bovine, and 2.8 Mb in human. By contrast, CGCs are nearly undetectable in rodents (Fig. 2C and fig. S7D). This is not due to globally elevated CG levels in nonrodent genomes as they show comparable CG levels with rodents in 1-kb bin analyses (Fig. 2C, top). In porcine oocytes, while active gene bodies show high levels of DNA methylation in both CGCs and non-CGCs, nontranscribed regions, particularly for intergenic regions, are highly methylated in non-CGCs but not in CGCs (Fig. 2D), as confirmed using a published methylome (16) (fig. S7B, left). A similar observation, although to a lesser extent, was made in bovine, followed by human (Fig. 2D). By contrast, both transcriptionally active and inactive regions are hypermethylated in somatic cells and sperm, regardless of CGCs or non-CGCs (fig. S7B, bottom middle and right). Thus, CGCs appear to avoid DNA methylation in nontranscribed regions specifically in oocytes, where additional mechanisms beyond their high CpG density must exist. Gene Ontology (GO) analysis showed that CGC regions tend to harbor developmental genes in bovine and porcine (Fig. 2E and table S3). In addition, 140 genes (based on orthologous genes) in CGCs are shared among human, bovine, and porcine (corresponding to 24.2% of human, 35.1% of bovine, and 11.1% of porcine CGC genes), which are significantly higher than random controls (Fig. 2F). These genes mainly function in cellular response to l-glutamine (P = 2 × 10−2) and JUN kinase activity (P = 3 × 10−2). These data indicate that CGCs exist in nonrodent mammalian genomes and harbor partially conserved genes.
CGCs harbor ICRs including key paternal ICRs
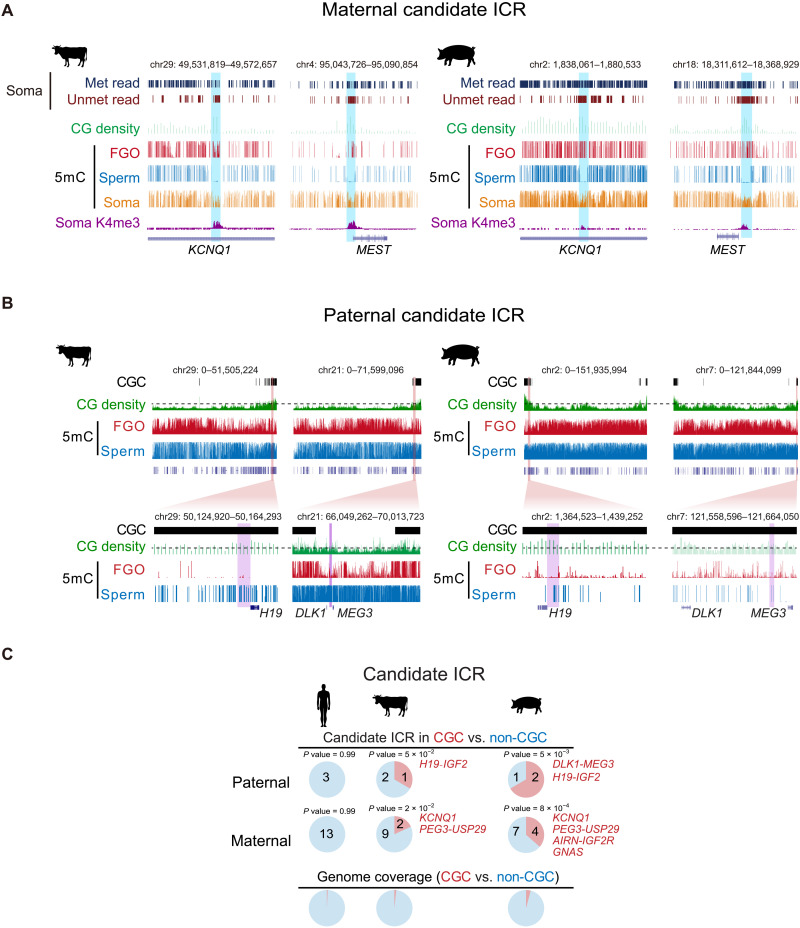
CGC regions also include a number of imprinted genes such as H19, IGF2, DLK1, and MEG3 (table S3). As imprinted genes are regulated by differentially methylated ICRs, we then asked whether ICRs are also enriched in CGCs. As ICRs are not well annotated in bovine, porcine, and rat and can be species specific, we focused on germline ICRs (gICRs) that are at least conserved between human and mouse (24, 35) and sought to identify their approximal locations with the following steps (Materials and Methods). (i) We assume that their methylation states (paternal or maternal) and proximity to the imprinted genes are largely conserved. (ii) Maternal ICRs are preferentially CGIs and marked by H3K4me3 in somatic cells. (iii) The putative ICRs are partially methylated (mCG/CG, 0.25 to 0.75) in somatic tissues and include both substantial percentages of reads that are fully methylated and fully unmethylated [at least 15% for each, with minimally 50 reads with at least three CGs, with parameters determined based on methylome data from annotated mouse ICRs (35)]. Overall, we manually identified putative positions of 16 imprints in bovine, porcine, and rat (Fig. 3A, fig. S8, and table S4). We focused on those in bovine and porcine as CGCs are largely absent from rat. We could not detect ICRs near GRB10 and HM13-MCTS2 loci in bovine and porcine, which were excluded from subsequent analysis. Of the three paternal ICRs, one (H19-IGF2, P = 5 × 10−2, Fisher’s exact test) and two (H19-IGF2 and DLK1-MEG3, P = 5 × 10−3) ICRs are present in CGCs of bovine and porcine, respectively (Fig. 3, B and C, table S4, and fig. S8, A and B). While the DLK1-MEG3 ICR in bovine missed the cutoff of CGC (0.024 versus 0.03, 100-kb bin), it is flanked by CGCs on both sides (~270 kb to the left and ~1 Mb to the right) (Fig. 3B). Both H19-IGF2 and DLK1-MEG3 ICRs are proximal to telomere in bovine and porcine (Fig. 3B). These two ICRs are critical regulators of bisexual reproduction as mutating these two regions allowed the efficient generation of bimaternal mice (36–38). These imprints are present in unmethylated intergenic regions in oocytes but are however hypermethylated in sperm (Fig. 3B). It should be noted that several maternal ICRs can also reside in oocyte CGCs (4 of 11 in porcine and 2 of 11 in bovine) (Fig. 3C and fig. S8B). They show high levels of DNA methylation presumably because they are in transcribed gene bodies. These data raise the possibility that CGCs may serve as safe harbors for key paternal ICRs against accidental DNA methylation in bovine and porcine. Intriguingly, human CGCs enrich neither developmental genes nor imprinted genes (Figs. 2E and 3C), suggesting that CGCs likely have additional functions beyond harboring imprints.
Fig. 3. Enrichment of imprinted ICRs in CGCs in bovine and porcine.
(A) The UCSC genome browser views showing the locations of putative KCNQ1 and MEST ICRs in bovine and porcine. The sequenced reads with all CpGs (at least three) fully methylated (Met read) and fully unmethylated (Unmet read) in somatic tissues (Soma) [bovine, lung (74); porcine, muscle (78)], CG density (1-kb bin), DNA methylation (5mC) in oocyte, sperm [data are from (79) for bovine and (16) for porcine] and somatic tissues, and H3K4me3 (K4me3) in somatic tissues (CCs for both bovine and porcine) are shown. (B) The UCSC genome browser views (whole chromosome and zoomed-in) showing the chromosome end proximal localization of the putative paternal gICRs (purple shaded) H19-IGF2 and DLK1-MEG3 in CGC regions of bovine and porcine oocytes. CG density (1-kb bin) and DNA methylation in FGOs and sperm are also shown. The dashed lines for CG density indicate the cutoff of 0.03 (100-kb bin) used for calling CGCs. (C) Pie charts showing the percentages of paternal (top) and maternal (middle) gICRs located in CGC and non-CGC regions for human, bovine, and porcine. The genome coverages of CGC and non-CGC are shown as controls (bottom). P values (Fisher’s exact test) are also shown. Note the DLK1-MEG3 ICR is surrounded by CGCs in bovine.
Unlike rodents, H3K36me2 and H3K36me3 correlate with DNA methylation in both transcribed and nontranscribed regions in bovine and porcine oocytes
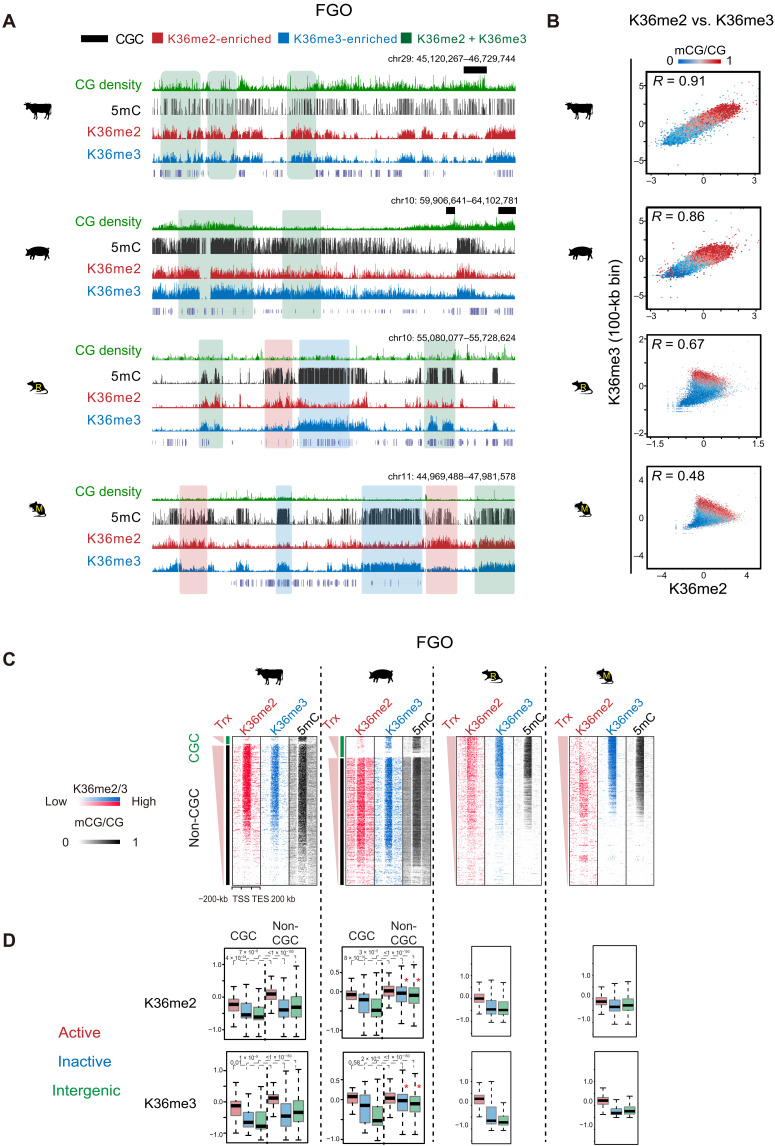
We next asked how DNA methylation is established in both transcribed and nontranscribed regions in porcine and bovine oocytes. De novo DNA methylation can be directed by both H3K36me2 and H3K36me3 in mouse germ line or ESCs (8, 9, 39). H3K36me3 is usually present in active gene bodies, while H3K36me2 can be found in both transcribed and nontranscribed regions. Therefore, we performed small-scale TELP-assisted rapid (STAR) chromatin immunoprecipitation sequencing (ChIP-seq) (17) for H3K36me2 and H3K36me3 in the oocytes of bovine, porcine, rat, and mouse, with CCs as controls. We chose STAR ChIP-seq as it worked well for H3K36me3 in mouse oocytes as we previously demonstrated (9). Both H3K36me2 (R = 0.73 to 0.92) and H3K36me3 (R = 0.72 to 0.94) data are reproducible between replicates (fig. S9A), and those in mouse FGOs well correlate with published data (fig. S9B, left) (8). The patterns of H3K36me2 and H3K36me3 in rat resemble those in mouse, as both marks correlate with DNA methylation, yet H3K36me2 is relatively weaker in regions with strong H3K36me3 (Fig. 4A). H3K36me2-enriched regions show relatively lower DNA methylation compared to H3K36me3-enriched regions (Fig. 4, A and B) as reported (8), suggesting that H3K36me2 may have a weaker activity in recruiting DNA methylation.
Fig. 4. The relationship of H3K36me2, H3K36me3, and DNA methylation in FGOs across mammals.
(A) The UCSC genome browser views showing DNA methylation, H3K36me2, and H3K36me3 enrichment in nonhuman FGOs. Regions covered by substantial H3K36me2 (K36me2-enriched), H3K36me3 (K36me3-enriched), and both marks (K36me2 + K36me3) are shaded in red, blue and green, respectively. CGCs are also shown in bovine and porcine. (B) Scatterplots comparing H3K36me2, H3K36me3, and DNA methylation in FGOs of all four species. DNA methylation levels in each bin are color-coded. The Pearson correlation coefficients are shown. (C) Heatmaps showing the enrichment of H3K36me2, H3K36me3, and DNA methylation in gene bodies and the flanking regions in FGOs of bovine, porcine, rat, and mouse. For bovine and porcine, CGC and non-CGC regions are separately shown. Genes are sorted by FPKM. Trx, transcription. (D) Boxplots showing the enrichment of H3K36me2 (top) and H3K36me3 (bottom) in active gene bodies (red), inactive gene bodies (blue), and intergenic regions (green) in the oocytes among the four species. CGC and non-CGC regions are separately shown for bovine and porcine. P values (Wilcoxon test) are also shown. Note that both H3K36me2 and H3K36me3 show high enrichment in inactive gene bodies and intergenic regions in non-CGCs in porcine FGOs (asterisk).
H3K36me2 and H3K36me3 appear to be more widespread in porcine, followed by bovine, and reassuringly highly correlate with DNA methylation (Fig. 4, A to C), supporting a conserved H3K36me2/3-mediated DNA methylation mechanism. Unexpectedly, H3K36me2 and H3K36me3 are much more similar to each other in porcine and bovine compared to rodents (Fig. 4, A to C, and fig. S9A). To further confirm the specificity of these antibodies, we also conducted H3K36me2 and H3K36me3 ChIP-seq in mouse ESCs (mESCs), where these two marks showed distinguishable patterns, with each data well recapitulating those reported previously (fig. S9B, right) (39). The concordance of these marks in bovine and porcine oocytes is not related to species in general, as they are well separated in bovine and porcine CCs from the same batch of experiments, with H3K36me2 enriched in both transcribed and nontranscribed regions, while H3K36me3 enriched in active gene bodies (fig. S9, A and C). Last, to rule out the possibility that the antibodies may cross-react when one of these marks is extremely low in abundance, we performed competitive and calibrated STAR ChIP-seq (cChIP-seq) by mixing samples of porcine FGOs with mESCs. The rationale is that, if H3K36me2 or H3K36me3 is extremely low, then providing competitive epitopes from mESCs would attract antibodies to the right epitopes. This experiment also provides a quantitative abundance of these marks in porcine FGOs. We confirmed that only less than 0.76% of reads from pure mESC samples can be mapped to porcine genome or vice versa, suggesting that the two genomes are highly divergent to allow separation of reads from mixed samples. Our cChIP-seq analysis of the FGO-mESC mixed samples again confirmed reproducible patterns of the two marks in porcine FGOs or mESCs (fig. S10A). Furthermore, H3K36me2 and H3K36me3 in porcine FGOs are more abundant than those in mESCs (fig. S10A and table S2), as further supported by immunofluorescent staining (fig. S10B). Therefore, these experimental results well validate the H3K36me2 and H3K36me3 data.
Despite their similarity in distributions, we attempted to identify regions with relatively more H3K36me2 than H3K36me3 (H3K36me2-enriched) and vice versa (H3K36me3-enriched) (fig. S11A). H3K36me3-enriched regions again show higher DNA methylation than H3K36me2-enriched regions, although the differences are much smaller than those in rodents (fig. S11B). Intriguingly, in porcine oocytes, both H3K36me2 and H3K36me3 are found in nontranscribed regions in non-CGCs (Fig. 4, C and D). By contrast, both marks are restricted to active gene bodies in CGCs (Fig. 4, C and D), consistent with transcription-correlated DNA methylation. SETD2-deposited H3K36me2/3 directs de novo DNA methylation mainly in transcribed regions in mouse oocytes (9), while NSD1-deposited H3K36me2 instructs the de novo DNA methylation in a large portion of both transcribed and nontranscribed regions in mouse male germ line (8). NSD3, an H3K36me2 methyltransferase, is highly expressed in porcine, followed by human and bovine, while Kdm2b, an H3K36me1/2 demethylase, is highly expressed in rodent oocytes but not in other species (fig. S11C). Last, it was shown that SETD2 is subjected to constant protein degradation, and stabilized SETD2 can deposit H3K36me3 in a polymerase II–independent manner (40). Future work is warranted to determine whether the widespread H3K36me2 in porcine and bovine oocytes may be caused by a combination of high NSD3 and low KDM2B. Once H3K36me2 is deposited, perhaps it can be converted to H3K36me3 through SETD2. In sum, both H3K36me2 and H3K36me3 correlate with DNA methylation in bovine and porcine oocytes, in both transcribed and nontranscribed regions, indicating different mechanisms of de novo DNA methylation establishment in mammalian oocytes.
H3K27me3 is depleted, while H3K36me2/3 pervades in non-CGCs in porcine oocytes
In mouse oocyte, a unique feature is the presence of broad domains of H3K4me3 and H3K27me3 in regions depleted of DNA methylation (PMDs) and H3K36me2/3 (9, 17, 19, 20). Mechanistically, H3K36me2/3 can antagonize PRC2 and, in turn, counteract H3K27me3 (41, 42). Nevertheless, such noncanonical H3K4me3 and H3K27me3 domains are absent in human oocytes (13) for unknown reasons. We sought to determine whether ncH3K4me3 and ncH3K27me3 may be present in other species and, if so, whether they may be reduced in porcine and bovine oocytes, given their pervasive H3K36me2/3 and DNA methylation. Both H3K4me3 and H3K27me3 are present in regions depleted of DNA methylation in nonhuman species (Fig. 5, A and B). Notably, H3K27me3 is particularly depleted in the porcine oocytes, especially in non-CGC regions (Fig. 5, C and D, and fig. S12, A to C) where H3K36me2/3 prevail. By contrast, H3K27me3 is strongly enriched in CGCs, where H3K36me2/3 is less abundant (Fig. 5D, right). As a result, H3K27me3 is also mainly enriched toward the chromosome ends (Fig. 5C and fig. S12B). Nevertheless, even in CGCs, H3K27me3 and H3K36me2/3-DNA methylation are again mutually exclusive, and few regions simultaneously bear the two antagonizing marks (Fig. 5D, right). We suspect that the overlapped regions are likely due to inaccurate calling of these broad domains. Notably, H3K27me3 is also more abundant in CGC regions compared to non-CGC regions in porcine CCs (Fig. 5, C and D, right), which is consistent with the notion that CG-rich regions recruit Polycomb group proteins (43), and CGCs are enriched for developmental genes (Fig. 2E). However, the depletion of H3K27me3 in non-CGC regions in oocytes is more notable, as even classic Polycomb targets also show minimal or no enrichment of H3K27me3 (fig. S12D, right). We noticed that the PRC2 key component genes EED and SUZ12 are strongly repressed in porcine and human oocytes but not the rest species (fig. S12E). The compromised PRC2 activity may contribute to the depletion of H3K27me3 in addition to the prevalent H3K36me2/3 in porcine oocytes. We postulate that CGC may serve as a strong “docking harbor” to recruit the residual PRC2 complex.
Fig. 5. Distribution of H3K4me3 and H3K27me3 and their relationship with H3K36me2/H3K36me3/DNA methylation in mammalian oocytes.
(A) The UCSC genome browser views showing the distribution of H3K4me3 and H3K27me3 and their relationship with DNA methylation in mammalian oocytes. The PMD regions are shaded. Schematic shows the states of bivalency (H3K4me3 and H3K27me3) in PMDs. (B) Scatterplots comparing distal H3K4me3 and H3K27me3 in each 100-kb bin (with Pearson correlation shown). DNA methylation levels in each bin are color-coded. The percentages of bins marked by both H3K4me3 and H3K27me3, H3K4me3-only, and H3K27me3-only are also shown in the plots. Note that bovine has high percentage of bivalent bins and porcine has very few H3K27me3-only bins. (C) The UCSC browser views showing H3K4me3, H3K27me3, and H3K36me2/3 in porcine oocytes. Histone marks in somatic CCs (soma) are shown as controls. Zoomed-in views of CGC and non-CGC are also shown. H3K27me3 is mainly enriched in CGCs. (D) Barcharts showing the percentages of the genome covered by H3K36me2 and H3K27me3 in the oocytes among different species (left). The percentages of genome covered by H3K36me2 and H3K27me3 in CGCs and non-CGCs are also shown for porcine FGOs (right). H3K27me3 shows limited overall coverage in the genome but shows high coverage in CGCs in porcine FGOs (arrows). The distribution in somatic CCs in porcine is calculated as controls.
Given the exclusive presence of H3K27me3 and H3K36me2/3-DNA methylation domains in all species, we sought to determine whether there is a causal relationship between them. Previously, we showed that, in mouse, loss of H3K36me3 causes ectopic invasion of H3K27me3 into former H3K36me3 territory (9). We then asked whether the converse is also true, by depleting Eed, the core component of PRC2, from mouse oocytes. Despite the markedly reduced H3K27me3, we found little changes of H3K36me3 in these mutant oocytes (fig. S12F). This agrees with the notion that, while H3K36me3 antagonizes PRC2 in vitro, H3K27me3 does not inhibit enzymatic activities of H3K36-specific methyltransferases (41).
Unlike H3K27me3, H3K4me3 is abundant in both CGC and non-CGC regions in porcine oocytes, and it also anticorrelates with H3K36me2/3 and DNA methylation (Fig. 5, A to C). In mouse oocytes, H3K4me3 and H3K27me3 tend to occupy distinct PMDs that are preferentially accessible and inaccessible, respectively (9, 17–19). The separation of H3K4me3 and H3K27me3 in PMDs is also similarly observed in rat (Fig. 5, A and B). Unexpectedly, nearly all strong H3K27me3 regions are also marked by H3K4me3 in porcine oocytes (Fig. 5, A to C), although the reverse is not true, as there exist many strong H3K4me3 domains without H3K27me3 in non-CGCs (Fig. 5C). In bovine oocytes, the colocalization is even more prevalent, occurring for most regions with strong H3K4me3 and H3K27me3 (Fig. 5, A and B). Collectively, these data reveal the notable depletion of H3K27me3 in non-CGCs in porcine oocytes. In addition, while both H3K4me3 and H3K27me3 are present in regions without H3K36me2/3-DNA methylation, their relationship differs among species (discussed in detail later).
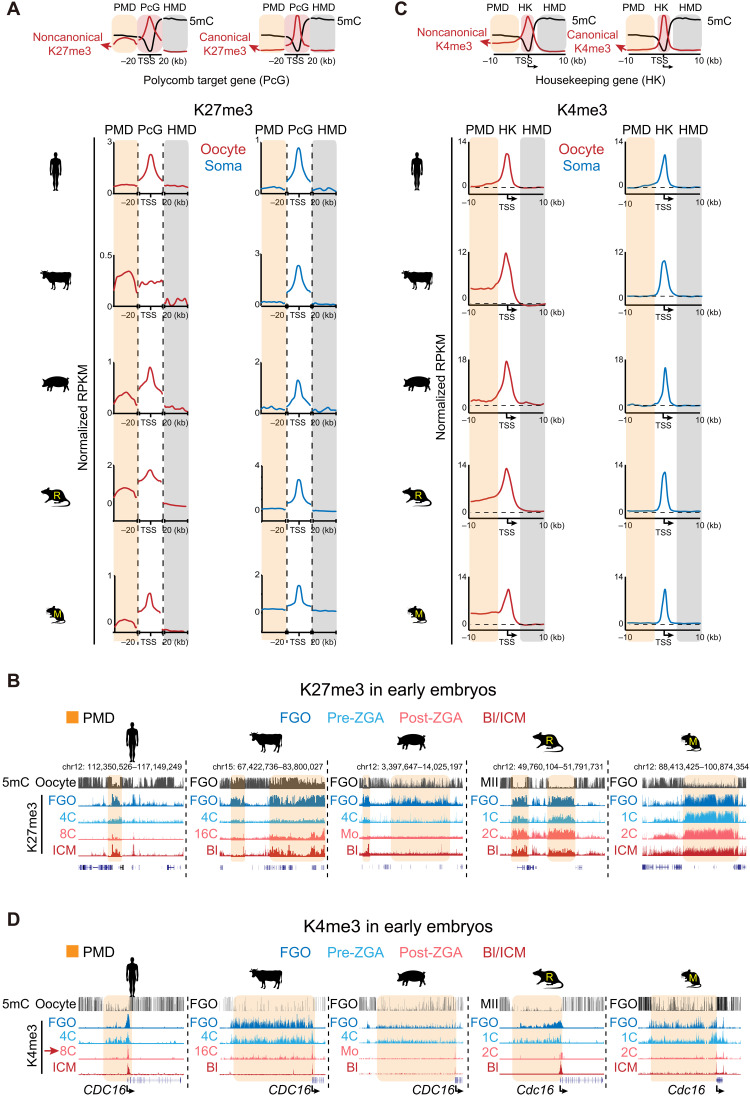
Oocyte H3K27me3-mediated noncanonical imprinting is likely restricted to rodents
In mice, promoter H3K27me3 from oocytes is erased shortly after fertilization in the one-cell embryos (18). By contrast, distal ncH3K27me3 domains from oocytes are briefly inherited until blastocyst, mediating noncanonical imprinting independent of DNA methylation (10, 18). However, such H3K27me3-mediated imprinting seems to be absent in human (13). To ask how prevalent such noncanonical imprinting is among mammals, we carefully examined H3K27me3 at both promoters and distal regions in oocytes and its fate after fertilization in all species. As H3K27me3 enrichment is also strongly related to CGCs (Fig. 5, C and D), we focused on the classic Polycomb group (PcG) targets (TSS ± 20 kb) and their adjacent PMDs (excluding the hypomethylated regions embedding PcG targets themselves) to ensure that their comparisons were done in regions with roughly comparable sequence and chromatin characteristics (Fig. 6A and Materials and Methods). The HMDs adjacent to PMDs are also included as negative controls. PcG targets show strong H3K27me3 in both FGOs and somatic cells (Fig. 6A and fig. S13A). Broad domains of H3K27me3 were observed in PMDs in nonhuman oocytes but not in somatic tissues (Fig. 6A and fig. S13A). As previously observed (Fig. 5, C and D, and fig. S12, A to C), in porcine oocytes, H3K27me3 is globally depleted in non-CGC regions but is enriched in CGCs (fig. S13B, bottom left). Bovine oocytes show strong H3K27me3 in both PMDs and PcG targets, where H3K27me3 promoter peaks appear to be masked by strong H3K27me3 in neighbor regions (Fig. 6A and fig. S13A). The strong H3K27me3 appears to correlate with the abundant presence of EED (embryonic ectoderm development) and SUZ12 (SUZ12 Polycomb repressive complex 2 subunit) and the repression of lysine demethylase 6A (KDM6A), the combination of which is unique to bovine (fig. S12E).
Fig. 6. H3K27me3 and H3K4me3 in mammalian oocytes and their reprogramming during preimplantation development.
(A) Top: Schematic model showing the distribution of H3K27me3 and DNA methylation in PMDs, promoters (TSS ± 20 kb) of Polycomb target genes (PcG) and highly methylated domains (HMDs). Bottom: Metaplots showing the enrichment of H3K27me3 in PMDs, promoters of PcG genes, and HMDs in mammalian FGOs (red). The same analysis was done in a somatic tissue (blue) as a control. Note that PMDs are adjacent to PcG target genes and do not include the hypomethylated regions harboring PcG target genes themselves. The HMDs are the closest ones to PMDs. (B) Snapshots showing the dynamics of H3K27me3 during preimplantation development of the five species. (C) Top: Schematic model showing the distribution of H3K4me3 and DNA methylation in PMDs and HMDs around housekeeping (HK) genes. Bottom: Metaplots showing the enrichment of H3K4me3 at PMDs, promoters, and HMDs around HK genes that have upstream PMDs in oocytes among the five species oocytes (red). The somatic tissue (blue) in each species is analyzed as a control. The dashed lines indicate H3K4me3 enrichment in HMDs serving as background levels. (D) The UCSC genome browser views showing the dynamics of H3K4me3 during preimplantation development of the five species. Four representative stages are shown (color-coded). The PMD regions are shaded.
We then asked whether oocyte H3K27me3 can be transmitted to early embryos after fertilization. Although we cannot separate the alleles in early embryos in all these species except for mouse, we reasoned that, as the paternal marks are often quickly erased after fertilization (44), the transmission of maternal mark would result in similar histone mark patterns between oocytes and early embryos. As in mouse, distal ncH3K27me3 in rat oocytes is transmitted to early embryos and persists until blastocyst (Fig. 6B and figs. S13C and S14A). Unexpectedly, unlike the complete loss of promoter H3K27me3 in one-cell mouse embryos, promoter H3K27me3 in rat is reduced but is partially inherited to blastocyst (figs. S13C and S14, B and C, compare mouse and rat). By contrast, in bovine and porcine, H3K27me3 was globally erased by the peri-ZGA stage, similar to that in human (Fig. 6B and fig. S13C), as evidenced by individual regions or all PcG targets (figs. S3, S4, and S14, A to C). Immunostaining confirmed the marked decrease of H3K27me3 in two- to four-cell and 4- to 16-cell stages in porcine and bovine, respectively (fig. S2), as reported (29, 30). Notably, promoter H3K27me3 is only partially restored in blastocyst in porcine (Fig. 6B and figs. S13C and S14, B and C), consistent with a previous immunostaining analysis (29), but is already strong in bovine blastocyst. This difference may reflect different epigenetic mechanisms, cellular composition, or developmental stage of blastocysts between the two species. We then asked what may account for the inheritance in rodents but extensive loss in other species for H3K27me3. Eed is not expressed at the pre-ZGA stages in human and porcine but is abundantly present in rodents (fig. S12E). In bovine, although the mRNA expression level of EED is high, it was reported that EED proteins were excluded from the nucleus in embryos before morula (30). Therefore, the absence of nuclear PRC2 key components in pre-ZGA embryos may partially account for the global loss of H3K27me3 in nonrodent mammals.
In mouse, oocyte-derived ncH3K27me3 mediates silencing of maternal Xist, leading to paternal-specific imprinted XCI in female embryos (10). In rat, broad ncH3K27me3 decorates the Xist locus from oocyte to four-cell stage and becomes weaker from the eight-cell stage (fig. S15). As Xist is activated as early as the two-cell stage, these data are consistent with the reported imprinted XCI in rat extraembryonic yolk sac (45). By contrast, H3K27me3 is absent from the Xist locus by ZGA in human, bovine, and porcine (fig. S15), agreeing with the absence of imprinted XCI in human preimplantation embryos, bovine trophectoderm lineage, and porcine placenta (14, 46, 47). Similar absence of H3K27me3 in human, bovine, and porcine was also observed near noncanonical imprinted autosomal genes identified in mouse, such as SFMBT2 (fig. S15). Collectively, our data suggest that oocyte ncH3K27me3 is present in nonhuman species, but its function in noncanonical imprinting appears to be restricted to rodents.
Noncanonical oocyte H3K4me3 is conserved among nonhuman mammals and is resolved to canonical H3K4me3 after ZGA
H3K4me3 in mouse oocyte is unique as, besides promoters, it is widely deposited in PMDs away from promoters, forming noncanonical H3K4me3 domains (17, 19, 20). Unexpectedly, noncanonical H3K4me3 is absent in human oocytes (13). Distal H3K4me3 occupies substantial percentages of the genomes in all species that we examined (14.4 to 24.7%) except in human (1.9%) (fig. S16A). Kdm5b, which encodes an H3K4me3 demethylase involved in removing ncH3K4me3 (17, 19, 20), appears to be mainly expressed in human oocytes (fig. S16B, red arrow). To better compare oocyte H3K4me3 in promoters and distal regions among different species, we focused on housekeeping (HK) genes [fragments per kilobase per million (FPKM) ≥ 5 for all stages for each species; n = 1992 to 3503; Materials and Methods]. Regions upstream of these promoters are usually nontranscribed intergenic regions and should be part of PMDs, while the downstream transcribed gene bodies should be HMDs (Fig. 6C). We confirmed the presence of PMDs near these genes in oocytes but not in somatic cells (fig. S16C, right). In all species except for human, H3K4me3 exhibits a similar noncanonical pattern with broad domains in PMDs in oocytes (figs. S14A and S16C, left, and Fig. 6C, with H3K4me3 in HMDs as background levels). The enrichment of ncH3K4me3 anticorrelates with DNA methylation levels in PMDs, with porcine showing the lowest H3K4me3 and highest DNA methylation (Fig. 6C and fig. S16C, right), consistent with its hypermethylated genome. Of note, although PMDs in porcine oocytes are less prevalent compared to those in rat and mouse (fig. S7A), the coverage of ncH3K4me3 is comparable (fig. S16A), as it occupies almost all PMDs in porcine but only a fraction of PMDs in rodent oocytes (Fig. 5A).
We then asked whether ncH3K4me3 can be inherited after fertilization. As in mouse (17, 19), ncH3K4me3 is briefly inherited from oocyte to the early embryos after fertilization in all nonhuman species (Fig. 6D and fig. S16D). After ZGA, ncH3K4me3 was resolved to a canonical pattern, with broad ncH3K4me3 in PMDs extensively lost, coinciding with the decrease of H3K4me3 signals in immunostaining (fig. S1) as reported for mouse (19). Notably, Kdm5b is activated after ZGA in all five species (fig. S16B). Knockdown of Kdm5b impairs both mouse and porcine preimplantation development (19, 20, 48), suggesting that this transition is likely important for embryonic development. Meanwhile, promoter H3K4me3 emerges at active genes (Fig. 6D). Promoter H3K4me3 at inactive genes such as developmental genes, however, appears decreased after ZGA (fig. S14, B and C), consistent with the notion that strong H3K4me3 only emerges at bivalent developmental gene promoters after implantation (18, 49). In sum, PMD-enriched ncH3K4me3 is conserved among nonhuman mammalian oocytes and transits from noncanonical to canonical patterns after ZGA.
Oocyte ncH3K27me3 is excluded from ncH3K4me3 domains in rodents, and ectopic spreading of H3K27me3 is linked to aberrant maternal silencing in early embryos
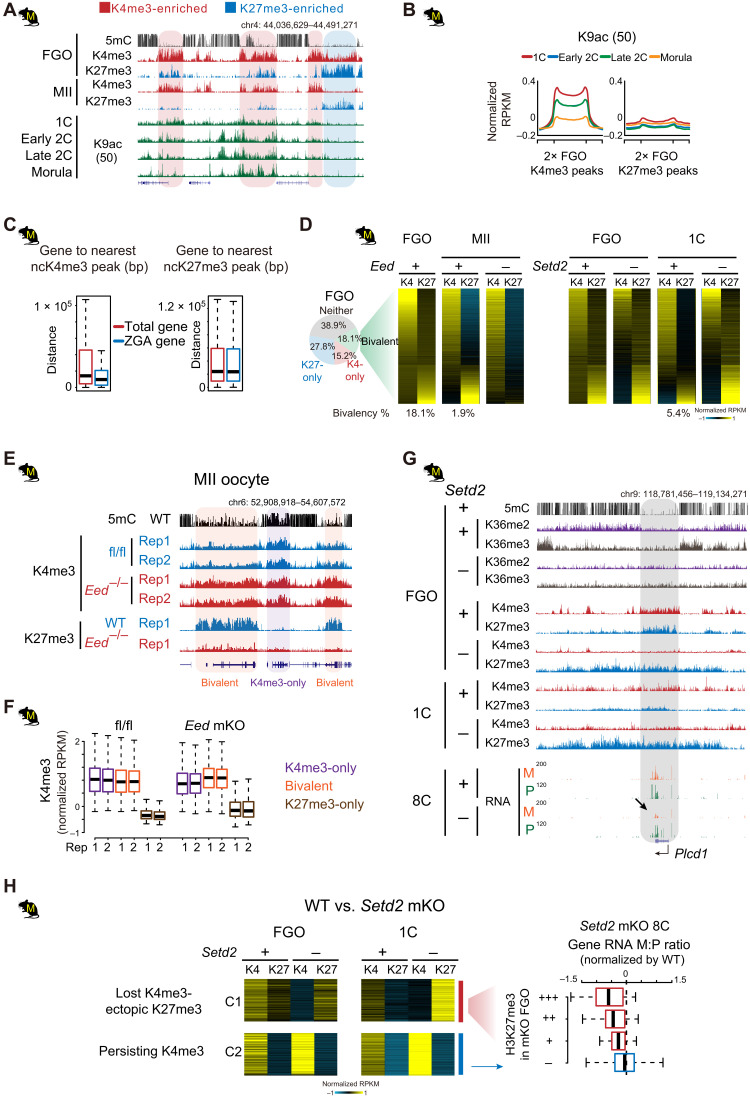
It is interesting that H3K27me3-dependent imprinting appears to be restricted to rodents. Mechanistically, this is likely related to the relatively faster ZGA in rodents (two-cell) compared to other mammals studied here (eight-cell and beyond) and the abundant PRC2 before ZGA (fig. S12E). While this allows increased opportunity for creating additional imprinted genes, it also potentially exposes embryos to risks of accidental silencing of embryonic genes. We hypothesized that crucial regulatory regions should be protected from invading H3K27me3 in rodent oocytes. Unlike those in porcine or bovine, oocyte H3K4me3 and H3K27me3 tend to be mutually exclusive in rodents (Fig. 5, A and B). These data raise the question that whether these oocyte ncH3K4me3 marked regions may be protected from H3K27me3 to avoid ectopic repression in early rodent embryos. By contrast, this is not necessary in nonrodent mammals, where H3K27me3 is globally erased by ZGA (Fig. 6B and fig. S2). ncH3K4me3 domains are preferentially enriched for histone acetylation in mouse embryos (Fig. 7, A and B) (50) and reside closer to ZGA genes (Fig. 7C), raising a possibility that they may harbor potential enhancers for mouse early embryos.
Fig. 7. Oocyte ncH3K27me3 is excluded from ncH3K4me3 domains in rodents.
(A and B) H3K9ac levels (50) at oocyte H3K4me3 or H3K27me3 domains in mouse embryos shown for an example locus (A) or meta-analysis (B). (C) Boxplots showing the distance of total genes and ZGA genes to the nearest ncH3K4me3 or ncH3K27me3 domains in mouse oocytes. (D) Left: Piechart showing the percentage of H3K27me3-only, H3K4me3-only, and bivalent bins in mouse FGOs. Middle right: Heatmaps showing the dynamics of H3K4me3 and H3K27me3 in bivalent domains (identified in FGOs) from FGO to one-cell in WT and Eed or Setd2 mKO mutants [data from (9)]. The percentages of bivalent bins at each stage in controls are also shown below. (E and F) Snapshots (E) and boxplots (F) showing the invasion of H3K4me3 to H3K27me3 marked regions in Eed mKO MII oocytes. H3K27me3 data in Eed mKO are from (80). (G) Aberrant spreading of H3K27me3 and the concomitant down-regulated maternal RNA reads at Plcd1 (arrow) in Setd2 mKO mutants. (H) Left: Heatmaps showing the spreading of H3K27me3 to H3K4me3-marked regions (5-kb bin) in Setd2 mKO FGOs and one-cell embryos [cluster 1 (C1)]. Right: Boxplot showing the maternal-to-paternal read count ratio (M:P) in Setd2 mKO eight-cell mutants (normalized by ratios in WT). Genes in C1 are classed into three groups according to the level of H3K27me3 (red) in Setd2 mKO FGOs. Genes in C2 that are marked by H3K4me3, but with no H3K27me3 spreading after Setd2 depletion, are similarly analyzed as control (blue). Setd2 mutant eight-cell embryos are derived from enucleated WT oocytes transferred with Setd2 mKO chromatin (9).
Despite the fact that H3K4me3 and H3K27me3 are largely nonoverlapping in mouse FGOs, a small subset of regions (18.1%) do carry both marks forming “bivalent domains” (Fig. 7, A and D) (18). These domains, however, resolve into either H3K4me3-only or H3K27me3-only domains in MII oocytes and one-cell embryos (maternal allele), with the ultimate results correlating to the initial H3K4me3 versus H3K27me3 relative enrichment in FGOs (Fig. 7D). The gradual resolution of bivalent domains may reflect the competition between repressive factors and active factors. We previously reported that H3K27me3 spreads across the genome in early-stage growing oocytes, before ncH3K4me3 appears gradually and invades many H3K27me3 domains during the transition to FGOs (18). Consistent with such “tug of war,” we found that the loss of H3K27me3 in mouse Eed maternal knockout (Eed mKO) MII oocytes causes increase of H3K4me3 in the former bivalent regions and H3K27me3 territories (Fig. 7, D to F). The loss of oocyte H3K27me3 has been shown to de-repress H3K27me3-controlled imprinted genes such as Xist, Gab1, Sfmbt2, and Slc38a4 (51, 52). Such derepression is caused by the loss of H3K27me3 rather than the invading ncH3K4me3, as it can be similarly achieved when removing H3K27me3 by injecting KDM6B in early embryos (51). We then asked what is the consequence of ectopic H3K27me3 spreading in oocytes. Notably, given H3K4me3 and H3K27me3 can coexist in nonrodent mammalian oocytes and most somatic cells at bivalent promoters (5, 6), it is unlikely that H3K4me3 itself antagonizes H3K27me3 in these regions. However, our previous study showed that H3K27me3 shows substantial spreading in Setd2 mKO FGOs (9). While many of such H3K27me3 spreading occurs in former H3K36me3 territories, consistent with their antagonism (41, 42), intriguingly, H3K27me3 also becomes dominant in former bivalent domains concomitant with the loss of H3K4me3 (Fig. 7D, right) (9). Although H3K36me2/3 is usually not present in bivalent PMDs, we speculate that, when H3K36me2/3 are lost in neighbor regions, H3K27me3 spreads and forms larger and strengthened domains, therefore tipping the balance toward H3K27me3 against H3K4me3 in the bivalent domains (as exemplified in Fig. 7G). To ask whether H3K27me3 spreading may have an impact on embryonic transcription, we specifically identified regions that show loss of H3K4me3 and increased/spreading H3K27me3 in Setd2 mutant FGOs. We confirmed that these regions are enriched for bivalent domains with H3K4me3 being relatively stronger (Fig. 7H). While they preferentially resolve into H3K4me3-only domains in wild-type (WT) one-cell embryos, they become H3K27me3-only domains in Setd2 mutant FGOs and persist in one-cell embryos (Fig. 7, G and H). While embryos derived from these oocytes are arrested at the one-cell stage, we were able to rescue them by transferring chromatin to enucleated WT oocytes, which can survive after fertilization before they die shortly after implantation (9). We then analyzed the RNA-seq data from eight-cell rescued embryos. Genes in these H3K4me3-to-H3K27me3 flipped sites preferentially show lower maternal-to-paternal ratios in expression (Fig. 7, G and H). Such effects are not found for genes in regions that retain H3K4me3 without acquiring ectopic H3K27me3 in Setd2 mutant oocytes (Fig. 7H and fig. S17A), suggesting that the increase or spreading of H3K27me3 to H3K4me3 domains in oocytes leads to aberrant gene repression in early embryos. In addition, we noticed that the spreading of H3K27me3 in Setd2 mutants also occurs in paternally methylated imprinted domains (fig. S17, B and C). Gpr1-Zbdf2, H19-Igf2, and Dlk1-Meg3 ICRs carry H3K27me3 in FGOs, which is then decreased (Gpr1-Zdbf2 and H19-Igf2) or lost (Dlk1-Meg3) in WT one-cell embryos but remains strong in Setd2 KO mutants (fig. S17, B and C). As these paternal imprint–controlled genes are inactive in WT embryos before implantation (fig. S17C), we cannot assess whether such persisting H3K27me3 affects their activation in early embryos. Together, these data indicate that the separation of H3K4me3 and H3K27me3 domains in rodent oocytes correlates with the presence of H3K27me3-mediated imprinting and may be involved in preventing accidental silencing of regulatory elements in early embryos.
DISCUSSION
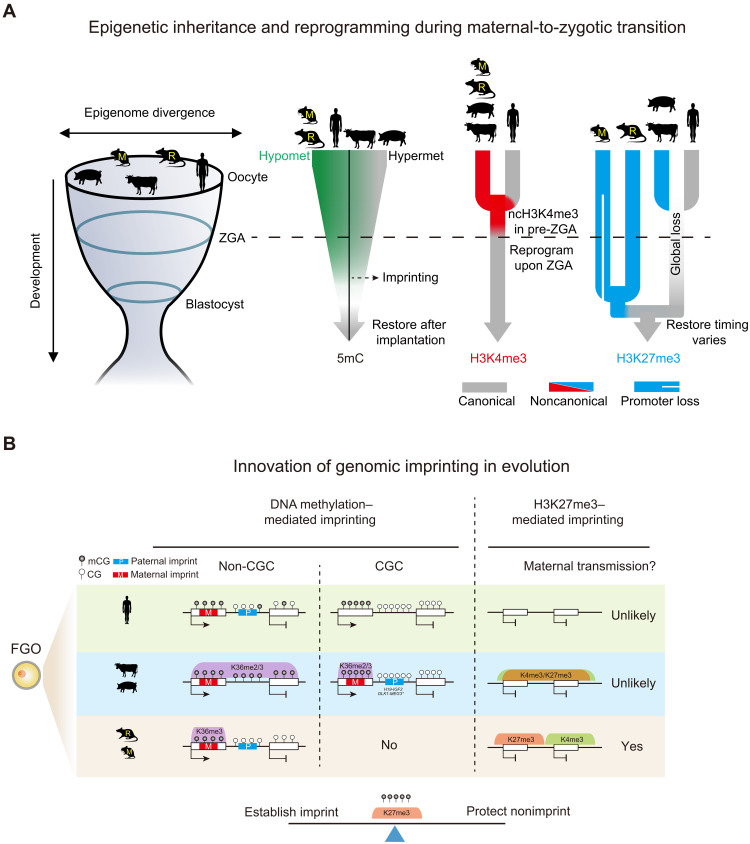
Gametic epigenomes are markedly reprogramed after fertilization in mammals. How epigenetic marks are established, inherited, and reset during mammalian early development are key questions to understand how life begins. It also remains enigmatic as to what extent these events are conserved among mammals. In this study, we investigated the landscapes of DNA methylome, H3K36me2, H3K36me3, H3K4me3, and H3K27me3 in the oocytes and preimplantation embryos of five mammalian species. Our study uncovered extensive species-specific innovations in epigenetic patterning of the parental epigenomes and their highly dynamic remodeling in mammalian early development (Fig. 8, A and B). These results also shed light on the possible “logic” of epigenetic inheritance and reprogramming underlying the complex patterning that may center on a fine balance in establishing genomic imprinting and protecting nonimprinting regions.
Fig. 8. Conservation and divergence of genomic imprinting and epigenetic reprogramming in mammalian early development.
(A) Schematic model showing the inheritance and reprogramming of DNA methylation, H3K4me3, and H3K27me3 during mammalian early development. Rodent oocytes show hypomethylated (Hypomet) DNA methylome while porcine oocytes show hypermethylated (Hypermet) DNA methylome. Human and bovine oocytes fall in between. After fertilization, DNA methylation is globally lost, except for genomic imprints, before being restored presumably after implantation. Oocyte ncH3K4me3 is conserved in nonhuman species. A “priming” state of H3K4me3 transiently appears in pre-ZGA human embryos (13). Both types of H3K4me3 are resolved to canonical patterns after ZGA. Broad ncH3K27me3 occurs in nonhuman oocytes. H3K27me3 is globally erased by the peri-ZGA stages in nonrodents. In rodents, promoter H3K27me3 is lost after fertilization in mouse but is partially retained in rat, while distal H3K27me3 is inherited until blastocysts. Embryo H3K27me3 is restored at variable stages (morula for human, blastocyst for bovine, and beyond blastocyst for porcine and rodents). DNA methylation, H3K4me3, and H3K27me3 show more similar patterns around implantation among species, supporting the “hourglass model” (left) (33). (B) Schematic model showing distinct mechanisms of genomic imprinting. DNA methylation–mediated imprinting is largely conserved among species. H3K36me2/3 correlated DNA methylation likely leads to the establishment of maternal imprints in the oocytes. Paternal imprints reside in nontranscribed regions. Some reside in CGCs (H19-IGF2 ICR for bovine, and H19-IGF2 and DLK1-MEG3 ICRs for porcine). Asterisk, DLK1-MEG3 ICR is not in CGC in bovine but is surrounded by CGCs. Oocyte-derived ncH3K27me3 can persist beyond ZGA and likely mediate noncanonical imprinting in rodents but unlikely in other species. Coincidently, ncH3K4me3 and ncH3K27me3 domains are segregated only in rodents. Oocyte finely balances the establishment of genomic imprints and the protection of nonimprints.
Resetting the parental epigenomes
Our study identified both conserved and divergent epigenetic reprogramming modes among mammals (Fig. 8, A and B). DNA methylation undergoes global resetting after fertilization, a process that is largely conserved among mammals (Fig. 8A) (2, 3). Our study showed that this is also true for histone marks (Fig. 8A). For example, most species we examined undergo marked transition for H3K4me3 (by ZGA) and H3K27me3 (by blastocyst). Even in mouse, maternal H3K27me3 is eventually taken over by embryonic version after implantation, suggesting that an epigenetic resetting is necessary to achieve efficient parental-to-zygotic transition.
To methylate or not: How oocytes establish maternal DNA methylation imprints and avoid methylation of paternal imprints
Despite the global resetting, certain epigenetic memories, such as those carried by DNA methylation imprints, are transmitted to embryos (Fig. 8, A and B) (7). Nevertheless, the mechanisms underlying the establishment of imprints and gametic DNA methylome are only recently elucidated. Mouse oocyte is almost exclusively methylated in transcribed regions through SETD2 and H3K36me3 to allow the establishment of maternal imprints when oocyte-specific promoters upstream of ICRs are exploited (Fig. 8, A and B) (9, 34). Another important function of such gene body–specific methylation, which is often overlooked, is that it also avoids methylation of intergenic regions where paternal imprints also reside. Therefore, the gene body–specific methylation invention in oocytes addresses both needs. Such necessity is however less so for sperm, where paternal imprints, which are predominantly in intergenic regions, are naturally methylated, while maternal imprints, which are predominantly in CGI-rich promoters, are naturally unmethylated. However, an unexpected finding from this study is that substantial fractions of nontranscribed regions are also methylated in bovine and porcine oocytes (Fig. 2B), suggesting that transcription-dependent methylation is not an absolute prerequisite for mammalian oocyte methylome. Such pervasive methylation appears to raise risks of inappropriate methylation of key regulatory elements. In particular, paternal imprints, once aberrantly methylated in oocytes, presumably can survive post fertilization demethylation. An innovation in porcine and bovine oocytes is to restrict paternal imprints to CGCs, where DNA methylation is limited to transcribed regions, thus potentially providing a “safe harbor” for paternal imprints (Fig. 8B). High CG regions are known to be refractory to DNA methylation (53). Nevertheless, CGCs are not always immune to DNA methylation as these regions are highly methylated in sperm and somatic cells. Therefore, additional antagonizing factors are required to constantly guard against DNA methylation. CG-rich regions can also attract H3K4me3 and H3K27me3 (53), as observed for CGCs in porcine oocytes. Both H3K4me3 and H3K27me3 have been shown to antagonize DNA methylation (54, 55). The strong enrichment of H3K27me3 in CGC in porcine oocytes is particularly notable given a weakly expressed PRC2 complex and the global depletion of H3K27me3 in non-CGC regions. Perhaps the presence of CGCs, coupled by the pervasive presence of antagonizing H3K36me2/3 in non-CGC regions, greatly facilitates the recruitment of the limited PRC2. Last, the presence of CGCs in nonrodent species itself is highly interesting, given CG-rich regions are prone to be mutated and lost once methylated. How they can be maintained during evolution and whether they have functions beyond harboring paternal imprints are exciting future questions.
How widespread methylation of nontranscribed regions occurs in porcine and bovine oocytes remains unknown. Mechanistically, DNA methylomes of porcine and bovine oocyte are still highly correlated with H3K36me2 and H3K36me3, suggesting that H3K36 methylation dependency is likely still conserved. However, a surprise comes from the finding that H3K36me2 and H3K36me3 tend to occupy similar regions in porcine and bovine oocyte genomes. While H3K36me3 is usually restricted to transcribed regions, H3K36me2 is often deposited in intergenic regions and inactive gene bodies (8, 39). Intriguingly, neither mark seems to strictly follow this rule in porcine oocytes, as they both tend to occupy active and inactive regions in non-CGC regions but reside mainly in active regions in CGCs. Our FGO-mESC competitive and calibrated ChIP-seq experiments (fig. S10A) argue against the possibility that these antibodies cross-react when one mark is extremely low. How such unique regulation of H3K36 methylation is achieved is a fascinating question that awaits future investigations, and the answers may greatly further our fundamental understanding of these marks.
To methylate or not: Species-specific innovation of H3K27me3-mediated imprinting
It is interesting that maternal H3K27me3 can survive past ZGA and potentially regulate genes only in rodents but unlikely in nonrodent mammals (Fig. 8B). Why this strategy was adopted specifically by rodents remains unclear, but we propose that the faster ZGA in rodents just one cell cycle after fertilization renders the embryos more susceptible to acquire new imprints during evolution, which may provide extra controls of “parental conflicts” (56). Similar to DNA methylation, the establishment of H3K27me3 imprints in oocyte also comes with the needs to protect nonimprinting regions from accidental H3K27me3 deposition. H3K4me3 domains, which are enriched for regulatory elements, are often mutually exclusive from H3K27me3 domains specifically in rodent oocytes. We showed that ectopic H3K27me3 spreading into H3K4me3 domains can cause aberrant embryonic gene expression in mouse when inherited (Fig. 7, G and H). What excludes H3K27me3 from H3K4me3 marked regions in rodents remain unclear. KDM6A can interact with the mixed lineage leukemia protein (MLL)-containing H3K4 methyltransferase complex, resulting in the removal of H3K27me3 concomitant with H3K4me3 deposition (57). Kdm6a is not expressed in bovine oocytes, moderately expressed in porcine, but is highly expressed in mouse (fig. S12E). Whether KDM6A or other factors are involved in excluding H3K27me3 from regulatory elements remains to be determined in future studies.
The unique epigenome of human oocytes and early embryos
Last, one of the most notable findings from our study is the uniqueness of human oocytes and early embryos. Unlike all other species, human oocytes lack ncH3K4me3 and ncH3K27me3 (13). Although the underlying mechanisms remain unknown, this appears to be associated with several factors. Kdm5b, encoding an H3K4me3 demethylase, is only expressed in human oocytes (fig. S16B). Similar to porcine, EED and SUZ12 are only lowly expressed in human oocytes (fig. S12E). Nevertheless, why human oocytes can manage without broad H3K4me3 and H3K27me3 remains unclear. Maternal deficiency of Eed leads to sublethality in mouse (52), although this is, in part, attributed to H3K27me3-mediated imprinting, which is perhaps not required for human. ncH3K4me3 was proved to be involved in transcriptional silencing in mouse oocyte, and the deficiency of its methyltransferase KMT2B leads to severe oocyte defects in mouse and embryonic lethality (17, 21). Other mechanisms may compensate these functions in human. These data call for cautions when extrapolating knowledge learned from animal models to human. The selection of relevant species will be crucial when studying a particular biological event in human. For example, human appears to share more similarity with bovine and porcine in the global loss of H3K27me3 after fertilization, the possible transcription factors regulating early embryos (23), and germline imprinting regulators zinc finger protein 445 (ZNF445)] (16). Nonhuman primate–related research and the emerging human organoids provide exciting opportunities to decipher molecular circuitry governing human early development in the future. Together, we envision that this study will open new avenues for both understanding fundamental biological questions on how a totipotent embryo arises and improving related applications including IVF and animal cloning.
MATERIALS AND METHODS
Animal
C57BL/6N mice and Sprague Dawley rats were purchased from Beijing Vital River. Mice and rats were maintained under specific pathogen–free condition, and all animal experiments were performed in compliance with the guidelines of Tsinghua University (mice) and Institute of Zoology, Chinese Academy of Sciences (rats), respectively.
Collection of mammalian oocytes, preimplantation embryos, and CCs
For the collection of rat oocytes and preimplantation embryos, females were superovulated by injection of pregnant mare serum gonadotropin (PMSG) (150 IU/kg) intraperitoneally, followed by injection of human chorionic gonadotropin (hCG) (300 IU/kg) 51 to 52 hours later. All oocytes and embryos were collected in M2 medium (Sigma-Aldrich, M7167) supplemented with 5 μM MG-132 (Sigma-Aldrich, SML1135). FGOs were collected 48 hours after PMSG injection without hCG administration. MII oocytes were collected 14 hours after hCG treatment. Different stages of preimplantation embryos were collected at the following time points after hCG treatment and mating with male rats: one-cell, 18 to 20 hours; late two-cell, 58 to 60 hours; four-cell, 70 hours; eight-cell, 86 hours; blastocyst, 120 hours.
For the collection of porcine oocytes and preimplantation embryos, FGOs were collected from slaughterhouse-derived ovaries using Hepes buffer [113 mM NaCl, 3.2 mM KCl, 2.0 mM NaHCO3, 0.42 mM NaH2PO4, 2.1 mM CaCl2·2H2O, 0.45 mM MgCl2·6H2O, 10 mM Hepes, 1.9% (v/v) sodium DL-lactate solution (Sigma-Aldrich, L-4263), penicillin-streptomycin (10 IU/ml; Gibco, 15140-122), and bovine serum albumin (BSA) (3 mg/ml; Sigma-Aldrich, A-9647)]. In vitro matured MII oocytes were obtained from FGOs that were cultured in Medium 199 (Sigma-Aldrich, M-4530) supplemented with 10% (v/v) follicular fluid, penicillin-streptomycin (10 IU/ml), PMSG (1 IU/ml), hCG (1 IU/ml), epidermal growth factor (EGF) (10 ng/ml; Sigma-Aldrich, E-4127), cysteine (0.1 mg/ml; Sigma-Aldrich, C-7352), and 10% (v/v) fetal bovine serum (FBS) (Gibco, 10099-141) under the condition of 5% CO2 at 38.5°C for 42 to 44 hours. The MII oocytes were then subjected to PG. After removing the granulosa cells, MII oocytes were treated with porcine zygote medium-3 (PZM-3) [108 mM NaCl, 10 mM KCl, 25 mM NaHCO3, 0.35 mM KH2PO4, 5 mM hypotaurine (Sigma-Aldrich, H-1384), 2 mM calcium l-lactate hydrate, 0.4 mM MgSO4·7H2O, 0.2% (v/v) phenol red solution, 0.2 mM sodium pyruvate (Sigma-Aldrich, P-2256), 1 mM l-glutamine (Sigma-Aldrich, G-3126), 1% (v/v) minimum essential medium nonessential amino acid (NEAA) (Sigma-Aldrich, M-7145), 2% (v/v) BME amino acids (EAA) (Sigma-Aldrich, B-6766), and BSA (3 mg/ml) containing 5 μM ionomycin calcium salt (Sigma-Aldrich, I-0634) for 5 min, followed by 4-hour treatment in dark in PZM-3 medium containing 2 mM 6-(dimethylamino) purine (Sigma-Aldrich, D-2629)]. The activated oocytes were then transferred to PZM-3 culture medium (day 0) under the culture condition of 5% CO2 at 38.5°C. Medium was changed every other day. Two-cell, four-cell, eight-cell, morula embryos, and blastocyst were collected at 24, 48, 48, 96, and 168 hours after fertilization, respectively.
For the collection of bovine oocytes and preimplantation embryos, FGOs were collected from slaughterhouse-derived ovaries using Hepes buffer. MII oocytes were obtained from FGOs that were cultured in M199 medium supplemented with penicillin-streptomycin (10 IU/ml), EGF (25 ng/ml), follicle stimulating hormone (50 ng/ml; Sigma-Aldrich, F-2293), LH (1 μg/ml; Sigma-Aldrich, L-9773), β-estradiol (1 μg/ml; Sigma-Aldrich, E-8875), heparin (25 μg/ml; Sigma-Aldrich, H-0777), insulin-like growth factor (40 ng/ml; Sigma-Aldrich, I-3769), and 10% (v/v) FBS under the condition of 5% CO2 at 38.5°C for 22 to 24 hours. MII oocytes were then subjected to IVF using frozen/thawed semen in Bracket and Oliphant’s medium under the culture condition of 5% CO2 at 38.5°C. Sixteen to 18 hours later (day 0), the fertilized zygotes were transferred to Charles Rosenkrans + amino acids medium (CR1aa) containing 108 mM NaCl, 3 mM KCl, 25 mM NaHCO3, 0.5 mM calcium lactate, 1 mM l-glutamine, 1% (v/v) NEAA, 2% (v/v) EAA, and BSA (6 mg/ml). At day 2, half of the medium was change to modified Charles Rosenkrans 2 amino acid medium (mCR2aa) containing 94 mM NaCl, 2.7 mM KCl, 22.5 mM NaHCO3, 0.7 mM MgCl2·6H2O, 1 mM KH2PO4, 0.34 mM sodium pyruvate, 1.3 mM glucose, 13 mM Hepes, 0.9 mM calcium galactose, BSA (6 mg/ml), 10% (v/v) FBS, 1 mM l-glutamine, 1% (v/v) NEAA, and 2% (v/v) EAA. At days 4 and 6, medium was changed to fresh mCR2aa medium. Four-cell, eight-cell, 16-cell embryos, and blastocyst were collected at 48, 48, 84, and 168 hours after fertilization, respectively.
The zona pellucida of all oocytes and preimplantation embryos in this study were removed using Tyrode’s solution (Sigma-Aldrich, T1788) (for rats) or 0.5% (m/v) Protease from Streptomyces griseus (Sigma-Aldrich, P8811) (for bovine and porcine). The polar bodies in MII oocytes or one-cell embryos were also carefully removed using a glass pipette. CCs surrounding the mammalian FGOs were collected as somatic controls.
Culture of mouse and rat ESCs
R1 mESCs were cultured on 0.1% gelatin-coated dishes in Dulbecco’s modified Eagle’s medium (Gibco, catalog no. 11995065) containing 15% FBS (Hyclone, SH30396.03), leukemia inhibitory factor (LIF) (1000 IU/ml; Merck Millipore, ESG1107), penicillin (100 IU/ml) and streptomycin (100 μg/ml) (Merck Millipore, TMS-AB2-C), 2 mM GlutaMAX (Gibco, 35050061), 1% EmbryoMax nucleosides (Merck Millipore, ES-008-D), 0.1 mM nonessential amino acids (Gibco, 25-025-CIR), and 0.1 mM 2-mercaptoethanol (Gibco, 21985023). Rat ESCs were cultured on 0.1% gelatin-coated dishes in 2i/LIF medium [N2B27 medium supplemented with 1 μm of PD0325901 (Tocris, 41921), 3 μm of CHIR99021 (Tocris, 4423), and LIF (1000 IU/ml)].
Immunofluorescent analysis
Mammalian oocytes or embryos were fixed with 4% paraformaldehyde for 30 min at room temperature and permeabilized with 0.5% Triton X-100 for 20 min at room temperature. Samples were then blocked with 1% BSA for 1 hour at room temperature or overnight at 4°C, followed by incubation with primary antibody overnight at 4°C (H3K4me3, 1:200, in-house; H3K27me3, 1:1000, Cell Signaling Technology, 9733s). The next day, after washing three times with 0.1% bovine serum albumin (BSA) in PBS, samples were incubated with the secondary antibody and Hoechst33342 for 1 hour at room temperature. Images were taken with the LSM780 confocal microscope system (Zeiss). For immunofluorescence intensity analysis, images were analyzed with Fiji software. The level of certain histone modification is indicated as log2-transformed nuclear/cytoplasmic fluorescence intensity.
CUT&RUN library preparation and sequencing
CUT&RUN was performed as previously reported (27) with modifications. Briefly, after removing the zona pellucida with Tyrode’s solution (Sigma-Aldrich, T1788) or 0.5% (m/v) Protease from S. griseus (Sigma-Aldrich, P8811), mammalian oocytes and early embryos were incubated with Concanavalin-coated magnetic beads (Polyscience, 86057) for 10 min at room temperature on a thermomixer at 400 rpm. Samples were then incubated with primary antibody (H3K4me3, in-house; H3K27me3, Cell Signaling Technology, 9733s) at a ratio of 1:100 at 4°C overnight on a thermomixer at 400 rpm. The next day, after washing for one time, beads were incubated with protein A-MNase (pA-MNase) (to a final concentration of 400 to 700 ng/ml) (a gift from S. Henikoff’s laboratory) at 4°C for 3 hours on a thermomixer at 400 rpm. After washing two times, targeted digestion was performed by adding 2 μl of 100 mM CaCl2 for 30 min on ice, followed by termination by adding an equal volume of 2× stop buffer. Samples were then incubated at 37°C for 20 min for fragment releasing. The total samples or supernatants were digested with proteinase K [New England Biolabs (NEB), P8107S] and purified using phenol:chloroform:isoamyl alcohol (25:24:1, v/v) followed by ethanol purification at −80°C overnight. The next day, DNA was purified and subjected to TruSeq library preparation using the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, E7645S). Sequencing was done using the HiSeq X Ten system (Illumina) according to the manufacturer’s protocol.
STAR ChIP-seq library preparation and sequencing
STAR ChIP-seq was performed as previously described (17). Briefly, samples were lysed followed by fragmented by micrococcal nuclease (MNase, Sigma-Aldrich, N3755-200UN) at 37°C for 5 min. After being terminated, the supernatant containing chromatin was incubated with 1 μg primary antibody (H3K36me2, Active motif, 61019; H3K36me3, Active motif, 61021) at 4°C overnight with rotation. The next day, 100 μg of Protein A/G dynabeads (mixed at 1:1, Thermo Fisher Scientific) was added to each sample and incubated at 4°C for 2 to 3 hours with rotation. The beads were then washed five times with 150 μl of radioimmunoprecipitation assay buffer and once with 150 μl of LiCl buffer. For each sample, beads were resuspended with 28 μl of H2O, 1 μl of 10× Ex Taq buffer (Takara, RR006B) and 1 μl of proteinase K (NEB, P8107S) and incubated at 55°C for 90 min, followed by incubation at 72°C for 40 min to inactivate the proteinase K. Samples were then subjected to TruSeq library preparation using NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, E7645S). Libraries were sequenced using the HiSeq X Ten system (Illumina) according to the manufacturer’s protocol.
RNA library preparation and sequencing
The RNA libraries for mammalian oocytes and preimplantation embryos were prepared using Smart-seq2 method as previously reported (58).
STEM-seq library preparation
The whole genome bisulfite libraries for bovine and porcine FGOs were constructed with STEM-seq as previously described (28). Briefly, the oocytes were first lysed with 10 μl of lysis buffer and 1 μl of protease K at 55°C for 3 hours. Protease K was inactivated at 72°C for 40 min. After lysis, the spike-in λ-DNA (Promega, D150A) was added with a mass ratio of 1:200. The mixture was treated with EpiTect Fast Bisulfite Conversion Kit (Qiagen, 59824) following the manufacture’s instruction, with a modified protocol: denature for 8 min at 95°C, incubate at 60°C for 25 min, and repeat the procedure once. Last, the purified converted DNA was subjected to the TELP (tailing-extension-ligation-PCR) library construction process as previously reported (59).
CUT&RUN and STAR ChIP-seq data processing
All reads were first processed by TrimGalore (v.0.6.4) with default parameters to remove the reads of low quality and also the adapters. The filtered reads were then mapped to different genomes (rat, rn6; bovine, bosTau8; and porcine, susScr11) by Bowtie2 (v2.2.5) (60) with the default parameters. All multiple mapped reads and the duplicates were removed with MarkDuplicates.jar. All unique mapped reads were then used for the downstream RPKM (reads per kilobase per million of sequenced reads) calculation. The Pearson correlation was calculated for each sample with two biological replicates (10-kb bin). Replicates with good reproducibility were pooled for downstream analysis.
Competitive and calibrated ChIP-seq
We designed competitive and calibrated ChIP-seq to validate the specificity of H3K36me3 (Active motif, 61021) and H3K36me2 (Active motif, 61019) and quantify the relative abundance of these two marks in porcine FGOs and mESCs. One hundred porcine FGOs and 250 mESCs were mixed together and subjected to STAR ChIP-seq as described above. Sequencing reads were mapped separately to mouse and porcine genomes, respectively. The relative abundance was calculated as the ratio for uniquely mapped reads per (cell number × ploidy) between porcine FGOs (ploidy=4) and mESCs (ploidy=2) (set as 1).
RNA-seq data processing
All RNA-seq datasets were mapped to the different genomes (rat, rn6; bovine, bosTau8; porcine, susScr11) using Tophat v2.1.1 (61). Gene expression was then calculated by cufflinks 2.2.1 (62).
STEM-seq data processing
All STEM-seq datasets were first treated with cutadapt v1.11 (63), and the low-quality reads and adaptors were removed before mapping. All filtered reads were then aligned to different genomes (rat, rn6; bovine, bosTau8; porcine, susScr11) using Bismark (v0.7.0; bowtie2 2.1.0) (64) with default parameters. Multimapped reads and polymerase chain reaction duplicates were removed also with Bismark. The uniquely mapped reads were used for CpG methylation calling with bismark_methylation_extractor. Only the CpG sites that were covered at least three times were used for further analysis. The spiked-in λ-DNA (1:200) was used for conversion rate calculation.
Validation of DNA methylation, histone modification, and RNA datasets
To evaluate the reproducibility of STEM-seq/CUT&RUN/STAR ChIP-seq/RNA-seq datasets, at least two biological replicates were performed for each sample. For the STEM-seq samples, the average methylation values were calculated for 10-kb bins across the entire genome. Bins detected in both samples were used for the Pearson correlation calculation. For CUT&RUN and STAR ChIP-seq samples, the RPKM value for each 100–base pair (bp) bin was first calculated for all samples/replicates. Then, the average RPKM values were calculated for 10-kb bins across the entire genome. Bins detected in both samples were used for the Pearson correlation calculation. For RNA-seq samples in each species, the FPKM values (FPKM of sequenced fragments) for all genes were generated, and the Spearman correlations across all stages within species were calculated.
UMAP and hierarchical analysis for transcriptomes and epigenomes across species
Genes that have orthologs in all five species in this study were used for UMAP (uniform manifold approximation and projection) analysis (65). Ensembl BioMart (release 96) (66) was used to find one-to-one and one-to-many orthologs for all five species as previously reported (67). For RNA-seq, dynamically expressed genes were identified by removing genes that are silenced or expressed in all samples among the five species. Only genes that are expressed (FPKM ≥ 5) in more than one sample, but fewer than half of all samples (n = 25) were further selected for downstream UMAP analysis. For H3K4me3 and H3K27me3, only genes that are covered (containing at least one sequencing read at their promoters) at all stages across the five species were selected for UMAP analysis. For DNA methylation, the average methylation values at TSS±2.5 kb regions were calculated for all stages across species. R package “umap” was used for the UMAP analysis with parameter n_neighbors = 5 for RNA and H3K27me3 and n = 8 for H3K4me3. Hierarchical clustering was conducted with Cluster 3.0 with the Pearson correlation coefficients. Java TreeView was used to visualize the hierarchical clustering result.
Quantification of DNA methylation
The methylation level was calculated as the total methylated counts divided by the total counts across all reads covering each single CG or 1-kb bin. For single CG site, only CGs covered more than three times were selected for downstream analysis.
Identification of PMDs
All PMDs in mouse, rat, and human were identified as previously described (68). In general, the average methylation level for each 10-kb bin was first calculated, and only bins with at least 20 CpGs were included. Those hypomethylated bins (mCG/CG ≤ 0.4) were identified and merged into PMDs. For porcine and bovine oocytes, because of the different global methylation levels, sequence depths, and CG densities, different cutoffs for PMD identification (bins with at least five CGs) were used. Specifically, hypomethylated 1-kb bins (mCG/CG ≤ 0.5 for bovine and mCG/CG ≤ 0.6 for porcine) were identified and further merged into PMDs. For H3K4me3 analysis, PMD-HMD in promoter nearby regions were chosen to minimize the background differences from different regions in the genome (such as gene deserts versus gene dense regions). PMDs located upstream of the HK genes were identified. Similarly, for H3K27me3 analysis, PMDs nearest to the Polycomb target gene-containing PMDs (but excluding the PMDs where PcG target genes themselves reside) were identified; HMDs most adjacent to the identified PMDs were identified as controls.
CGC identification
The CG density within 1- and 100-kb bin was first calculated with fastaFromBed in each species. Bins with CG density higher than 0.03 in 100-kb bins were identified and merged to generate CGCs.
Identification of putative maternal and paternal ICRs in bovine, porcine, and rat oocytes
The putative maternal and paternal ICRs in bovine, porcine, and rat oocytes were identified through the following steps. (i) Only gICRs which are differentially methylated between oocytes and sperm were considered. (ii) Given that gICRs may not be present in all species, those that are at least conserved between human and mouse were chosen for identification, so that they are more likely to be present in bovine and porcine. (iii) The proximity to the imprinted genes and methylation states (paternal or maternal) of gICRs were assumed to be conserved among mammals. (iv) Maternal ICRs are preferentially CGIs and marked by H3K4me3 in somatic cells. (v) The putative ICRs are partially methylated (mCG/CG, 0.25 to 0.75) in somatic tissues and include both substantial percentages of both fully methylated and fully unmethylated reads [at least 15% for each, with minimally 50 reads with at least three CGs, with parameters determined based on methylome data from annotated mouse ICRs (35)].
Identification of bivalent/Polycomb group protein (PcG) target genes, HK genes, and inactive genes for each species
H3K4me3 and H3K27me3 datasets in ESCs for human (13), bovine (69), rat, mouse (18), porcine (induced pluripotent stem cells) (70) were used to identify the bivalent genes or PcG target genes. Briefly, genes marked with both H3K4me3 and H3K27me3 in pluripotent stem cells were identified as bivalent genes. Genes that are expressed in all cell types in oocytes and early embryos (FPKM > 5) were identified as HK genes. Genes that are silenced in all cell types in oocytes and early embryos (FPKM < 1) were identified as inactive genes.
Identification of promoter and distal peaks for histone modifications
H3K36me2, H3K36me3, H3K4me3, and H3K27me3 peaks were called using MACS2 with the parameters –broad –nomodel –nolambda. Peaks with very weak signals (summed RPKM < 3.5) were removed for each sample in further analyses. Peaks that are at least 2.5 kb away from annotated promoters were identified as distal peaks.
Promoter and bin-based histone modification enrichment analysis
Gene annotations (human, hg19; bovine, bosTau8; porcine, susScr11; rat, rn6; mouse, mm9) downloaded from UCSC were used for the promoter (TSS ± 2.5 kb) or 5-kb/100-kb bin enrichment analysis. The whole genome z score normalized RPKM was used for calculation. Z score was calculated with the following formula: for a given promoter or bin i: zi = (xi − μ)/σ, where zi is the normalized RPKM value; xi is the RPKM value before normalization; μ and σ are the mean and SD of all promoter or bins RPKM values for each single sample.
Analysis for H3K36me2, H3K36me3, and DNA methylation
For genome-wide analysis of H3K36me2-H3K36me3-DNA methylation relationship in FGO, the level of DNA methylation for each 100-kb bin was color-coded in the H3K36me2-H3K36me3 scatterplot. For gene bodies and the flanking regions analysis, the RPKM values of histone marks for each 100-bp bin for all samples were generated. Bins with no reads were assigned to zero. The whole genome z score normalized RPKM was used for calculation. The gene body and the flanking regions were split into 20 bins, respectively. The average RPKM value was calculated for each bin and shown in the heatmap.
GO analysis
GO analysis was performed according to the DAVID web tools (version 6.8) (71). All terms listed in the figures show P value less than 0.05.
Allele assignment of RNA-seq reads for Setd2 mutant early embryos
The Setd2 mutant early embryo data were from our previous study (9) and are based on a hybrid F1 mouse embryo [PWK/PhJ (paternal) × C57BL/6N (maternal)]. All RNA-seq reads were aligned to the genomes of the PWK and C57BL strains (mm9) separately with Tophat v2.0.11 and normalized as previously described (9, 72). Briefly, for each gene, only reads located in exons were counted, and the normalized allelic value for each allele was the total count × 1000/total exon length. Only genes with combined read count (maternal + paternal) more than 50 in the Setd2 mutant embryos were selected, and the maternal versus paternal RNA read ratio (M:P) was calculated for both WT and mutant embryos. The final M:P ratio for mutants was further normalized by that from WT for the same gene.
Acknowledgments
We thank S. Henikoff (Fred Hutchinson Cancer Research Center) for sharing pA-MNase for CUT&RUN. We thank M. Lorincz and J. Brind’Amour (University of British Columbia) for their help in data analysis and discussion. We are grateful to members of the W. Xie’s laboratory for discussion and comments during the preparation of the manuscript and the Animal Center and Biocomputing Facility at Tsinghua University for their support. X.L and Y.Z. are supported by the postdoctoral fellowships from the Tsinghua-Peking Joint Center for Life Sciences. W. Xie is a recipient of an HHMI International Research Scholar award.
Funding: This work was supported by National Natural Science Foundation of China, 31725018, 31988101, and 31830047 (to W. Xie); National Key R&D Program of China, 2019YFA0508900 (to W. Xie); Tsinghua-Peking Center for Life Sciences (to W. Xie); Beijing Municipal Science and Technology Commission grant, Z181100001318006 (to W. Xie); National Fund for Natural Sciences of China, 31772697 (to Y.G.); National Key Research and Development Program, 2019YFA0110800 (to W.L.); Strategic Priority Research Program of the Chinese Academy of Sciences, XDA16030403 (to W.L.); and National Natural Science Foundation of China, 31621004 (to W.L.).
Author contributions: X.L., Y.Z., and W. Xie conceived and designed the project. X.L. performed most of the experiments including the CUT&RUN, Smart-seq2, and STAR ChIP-seq. Y.Z. performed the STEM-seq experiment, analyzed most of the data, and prepared the figures in consultation with X.L. and W. Xie. X.L., Lijuan Wang, and H.W. collected the oocytes and embryos of bovine and porcine with the help of C.C., Z.L., X.W., and H.N. Leyun Wang collected the oocytes and embryos of rat with the help of S.M. Q.X. and Y.X. helped with the Eed mKO mouse data. X.L., Lijuan Wang, and F.K. collected mouse oocytes with the help of L.L. X.L., Y.Z., and W. Xia generated the model figure. X.L., Y.Z., and W. Xie interpreted the data and wrote the manuscript. W.L., Y.G., and W. Xie supervised this project. All authors discussed the results and contributed to the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All data have been deposited to GEO with the accession number GSE163620.
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Legends for tables S1 to S4
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4
REFERENCES AND NOTES
- 1.Allis C. D., Jenuwein T., The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 17, 487–500 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Eckersley-Maslin M. A., Alda-Catalinas C., Reik W., Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Greenberg M. V. C., Bourc’his D., The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 20, 590–607 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Suganuma T., Workman J. L., Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80, 473–499 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Jambhekar A., Dhall A., Shi Y., Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 20, 625–641 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S., A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Li Y., Sasaki H., Genomic imprinting in mammals: Its life cycle, molecular mechanisms and reprogramming. Cell Res. 21, 466–473 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shirane K., Miura F., Ito T., Lorincz M. C., NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat. Genet. 52, 1088–1098 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Xu Q., Xiang Y., Wang Q., Wang L., Brind’Amour J., Bogutz A. B., Zhang Y., Zhang B., Yu G., Xia W., Du Z., Huang C., Ma J., Zheng H., Li Y., Liu C., Walker C. L., Jonasch E., Lefebvre L., Wu M., Lorincz M. C., Li W., Li L., Xie W., SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat. Genet. 51, 844–856 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Chen Z., Zhang Y., Maternal H3K27me3-dependent autosomal and X chromosome imprinting. Nat. Rev. Genet. 21, 555–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z., Yin Q., Inoue A., Zhang C., Zhang Y., Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci. Adv. 5, eaay7246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna C. W., Perez-Palacios R., Gahurova L., Schubert M., Krueger F., Biggins L., Andrews S., Colome-Tatche M., Bourc’his D., Dean W., Kelsey G., Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol. 20, 225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia W., Xu J., Yu G., Yao G., Xu K., Ma X., Zhang N., Liu B., Li T., Lin Z., Chen X., Li L., Wang Q., Shi D., Shi S., Zhang Y., Song W., Jin H., Hu L., Bu Z., Wang Y., Na J., Xie W., Sun Y. P., Resetting histone modifications during human parental-to-zygotic transition. Science 365, 353–360 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Petropoulos S., Edsgard D., Reinius B., Deng Q., Panula S. P., Codeluppi S., Reyes A. P., Linnarsson S., Sandberg R., Lanner F., Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 167, 285 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu P., Guo H., Ren Y., Hou Y., Dong J., Li R., Lian Y., Fan X., Hu B., Gao Y., Wang X., Wei Y., Liu P., Yan J., Ren X., Yuan P., Yuan Y., Yan Z., Wen L., Yan L., Qiao J., Tang F., Single-cell DNA methylome sequencing of human preimplantation embryos. Nat. Genet. 50, 12–19 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Ivanova E., Canovas S., Garcia-Martinez S., Romar R., Lopes J. S., Rizos D., Sanchez-Calabuig M. J., Krueger F., Andrews S., Perez-Sanz F., Kelsey G., Coy P., DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin. Epigenetics 12, 64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B., Zheng H., Huang B., Li W., Xiang Y., Peng X., Ming J., Wu X., Zhang Y., Xu Q., Liu W., Kou X., Zhao Y., He W., Li C., Chen B., Li Y., Wang Q., Ma J., Yin Q., Kee K., Meng A., Gao S., Xu F., Na J., Xie W., Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 537, 553–557 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Zheng H., Huang B., Zhang B., Xiang Y., Du Z., Xu Q., Li Y., Wang Q., Ma J., Peng X., Xu F., Xie W., Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 63, 1066–1079 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Dahl J. A., Jung I., Aanes H., Greggains G. D., Manaf A., Lerdrup M., Li G., Kuan S., Li B., Lee A. Y., Preissl S., Jermstad I., Haugen M. H., Suganthan R., Bjoras M., Hansen K., Dalen K. T., Fedorcsak P., Ren B., Klungland A., Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 537, 548–552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H., Zhang Y., Gao Y., Gao S., Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Andreu-Vieyra C. V., Chen R. H., Agno J. E., Glaser S., Anastassiadis K., Stewart A. F., Matzuk M. M., MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLOS Biol. 8, e1000453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W., Chen Z., Yin Q., Zhang D., Racowsky C., Zhang Y., Maternal-biased H3K27me3 correlates with paternal-specific gene expression in the human morula. Genes Dev. 33, 382–387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halstead M. M., Ma X., Zhou C., Schultz R. M., Ross P. J., Chromatin remodeling in bovine embryos indicates species-specific regulation of genome activation. Nat. Commun. 11, 4654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okae H., Chiba H., Hiura H., Hamada H., Sato A., Utsunomiya T., Kikuchi H., Yoshida H., Tanaka A., Suyama M., Arima T., Genome-wide analysis of DNA methylation dynamics during early human development. PLOS Genet. 10, e1004868 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smallwood S. A., Lee H. J., Angermueller C., Krueger F., Saadeh H., Peet J., Andrews S. R., Stegle O., Reik W., Kelsey G., Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat. Methods 11, 817–820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brind’Amour J., Kobayashi H., Richard Albert J., Shirane K., Sakashita A., Kamio A., Bogutz A., Koike T., Karimi M. M., Lefebvre L., Kono T., Lorincz M. C., LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat. Commun. 9, 3331 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skene P. J., Henikoff J. G., Henikoff S., Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 13, 1006–1019 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y., Xiang Y., Yin Q., Du Z., Peng X., Wang Q., Fidalgo M., Xia W., Li Y., Zhao Z. A., Zhang W., Ma J., Xu F., Wang J., Li L., Xie W., Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 50, 96–105 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Cao Z., Li Y., Chen Z., Wang H., Zhang M., Zhou N., Wu R., Ling Y., Fang F., Li N., Zhang Y., Genome-Wide Dynamic Profiling of Histone Methylation during Nuclear Transfer-Mediated Porcine Somatic Cell Reprogramming. PLOS ONE 10, e0144897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ross P. J., Ragina N. P., Rodriguez R. M., Iager A. E., Siripattarapravat K., Lopez-Corrales N., Cibelli J. B., Polycomb gene expression and histone H3 lysine 27 trimethylation changes during bovine preimplantation development. Reproduction 136, 777–785 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Dang-Nguyen T. Q., Somfai T., Haraguchi S., Kikuchi K., Tajima A., Kanai Y., Nagai T., In vitro production of porcine embryos: Current status, future perspectives and alternative applications. Anim. Sci. J. 82, 374–382 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Piliszek A., Madeja Z. E., Pre-implantation Development of Domestic Animals. Curr. Top. Dev. Biol. 128, 267–294 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Irie N., Kuratani S., The developmental hourglass model: A predictor of the basic body plan? Development 141, 4649–4655 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Sendzikaite G., Kelsey G., The role and mechanisms of DNA methylation in the oocyte. Essays Biochem. 63, 691–705 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie W., Barr C. L., Kim A., Yue F., Lee A. Y., Eubanks J., Dempster E. L., Ren B., Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawahara M., Wu Q., Takahashi N., Morita S., Yamada K., Ito M., Ferguson-Smith A. C., Kono T., High-frequency generation of viable mice from engineered bi-maternal embryos. Nat. Biotechnol. 25, 1045–1050 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Li Z., Wan H., Feng G., Wang L., He Z., Wang Y., Wang X. J., Li W., Zhou Q., Hu B., Birth of fertile bimaternal offspring following intracytoplasmic injection of parthenogenetic haploid embryonic stem cells. Cell Res. 26, 135–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong C., Xie Z., Yin Q., Dong R., Yang S., Wu Y., Yang L., Li J., Parthenogenetic haploid embryonic stem cells efficiently support mouse generation by oocyte injection. Cell Res. 26, 131–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinberg D. N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K. N., Horth C., McGuire J. T., Xu X., Nikbakht H., Lemiesz A. E., Marchione D. M., Marunde M. R., Meiners M. J., Cheek M. A., Keogh M. C., Bareke E., Djedid A., Harutyunyan A. S., Jabado N., Garcia B. A., Li H., Allis C. D., Majewski J., Lu C., The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature 573, 281–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharya S., Workman J. L., Regulation of SETD2 stability is important for the fidelity of H3K36me3 deposition. Epigenetics Chromatin 13, 40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan W., Xu M., Huang C., Liu N., Chen S., Zhu B., H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 286, 7983–7989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitges F. W., Prusty A. B., Faty M., Stützer A., Lingaraju G. M., Aiwazian J., Sack R., Hess D., Li L., Zhou S. L., Bunker R. D., Wirth U., Bouwmeester T., Bauer A., Ly-Hartig N., Zhao K. H., Chan H. M., Gu J., Gut H., Fischle W., Müller J., Thomä N. H., Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Blackledge N. P., Rose N. R., Klose R. J., Targeting Polycomb systems to regulate gene expression: Modifications to a complex story. Nat. Rev. Mol. Cell Biol. 16, 643–649 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia W., Xie W., Rebooting the epigenomes during mammalian early embryogenesis. Stem Cell Reports 15, 1158–1175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wake N., Takagi N., Sasaki M., Non-random inactivation of X chromosome in the rat yolk sac. Nature 262, 580–581 (1976). [DOI] [PubMed] [Google Scholar]
- 46.Min B., Park J. S., Jeon K., Kang Y.-K., Characterization of X-chromosome gene expression in bovine blastocysts derived by in vitro fertilization and somatic cell nuclear transfer. Front. Genet. 8, 42 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu D., Du X., Wang J., Chen L., Wang Y., Xu H., Zhao Y., Zhao S., Pang Y., Liu Y., Hao H., Zhao X., Du W., Dai Y., Li N., Wu S., Zhu H., No imprinted XIST expression in pigs: Biallelic XIST expression in early embryos and random X inactivation in placentas. Cell. Mol. Life Sci. 76, 4525–4538 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J., Zhang H., Wang X., Dobbs K. B., Yao J., Qin G., Whitworth K., Walters E. M., Prather R. S., J. Zhao, Impairment of preimplantation porcine embryo development by histone demethylase KDM5B knockdown through disturbance of bivalent H3K4me3-H3K27me3 modifications. Biol. Reprod. 92, 72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiang Y., Zhang Y., Xu Q., Zhou C., Liu B., Du Z., Zhang K., Zhang B., Wang X., Gayen S., Liu L., Wang Y., Li Y., Wang Q., Kalantry S., Li L., Xie W., Epigenomic analysis of gastrulation identifies a unique chromatin state for primed pluripotency. Nat. Genet. 52, 95–105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang G., Zhang L., Liu W., Qiao Z., Shen S., Zhu Q., Gao R., Wang M., Wang M., Li C., Liu M., Sun J., Wang L., Liu W., Cui X., Zhao K., Zang R., Chen M., Liang Z., Wang L., Kou X., Zhao Y., Wang H., Wang Y., Gao S., Chen J., Jiang C., Dux-mediated corrections of aberrant H3K9ac during 2-cell genome activation optimize efficiency of somatic cell nuclear transfer. Cell Stem Cell 28, 150–163.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Inoue A., Jiang L., Lu F., Suzuki T., Zhang Y., Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue A., Chen Z., Yin Q., Zhang Y., Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev. 32, 1525–1536 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deaton A. M., Bird A., CpG islands and the regulation of transcription. Genes Dev. 25, 1010–1022 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ooi S. K. T., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.-P., Allis C. D., Cheng X., Bestor T. H., DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brinkman A. B., Gu H., Bartels S. J., Zhang Y., Matarese F., Simmer F., Marks H., Bock C., Gnirke A., Meissner A., Stunnenberg H. G., Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 22, 1128–1138 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore T., Haig D., Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 7, 45–49 (1991). [DOI] [PubMed] [Google Scholar]
- 57.Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K., PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282, 20395–20406 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Picelli S., Faridani O. R., Bjorklund A. K., Winberg G., Sagasser S., Sandberg R., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Peng X., Wu J., Brunmeir R., Kim S. Y., Zhang Q., Ding C., Han W., Xie W., Xu F., TELP, a sensitive and versatile library construction method for next-generation sequencing. Nucleic Acids Res. 43, e35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L., Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10 (2011). [Google Scholar]
- 64.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Becht E., McInnes L., Healy J., Dutertre C. A., Kwok I. W. H., Ng L. G., Ginhoux F., Newell E. W., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W., BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Lesch B. J., Silber S. J., McCarrey J. R., Page D. C., Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat. Genet. 48, 888–894 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Lister R., Pelizzola M., Dowen R. H., Hawkins R. D., Hon G., Tonti-Filippini J., Nery J. R., Lee L., Ye Z., Ngo Q. M., Edsall L., Antosiewicz-Bourget J., Stewart R., Ruotti V., Millar A. H., Thomson J. A., Ren B., Ecker J. R., Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bogliotti Y. S., Wu J., Vilarino M., Okamura D., Soto D. A., Zhong C., Sakurai M., Sampaio R. V., Suzuki K., Izpisua Belmonte J. C., Ross P. J., Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc. Natl. Acad. Sci. U.S.A. 115, 2090–2095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao S., Xie D., Cao X., Yu P., Xing X., Chen C. C., Musselman M., Xie M., West F. D., Lewin H. A., Wang T., Zhong S., Comparative epigenomic annotation of regulatory DNA. Cell 149, 1381–1392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dennis G. Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A., DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 (2003). [PubMed] [Google Scholar]
- 72.Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W., Li W., Li Y., Ma J., Peng X., Zheng H., Ming J., Zhang W., Zhang J., Tian G., Xu F., Chang Z., Na J., Yang X., Xie W., The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016). [DOI] [PubMed] [Google Scholar]
- 73.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhou Y., Liu S. L., Hu Y., Fang L. Z., Gao Y. H., Xia H., Schroeder S. G., Rosen B. D., Connor E. E., Li C. J., Baldwin R. L., Cole J. B., Van Tassell C. P., Yang L. G., Ma L., Liu G. E., Comparative whole genome DNA methylation profiling across cattle tissues reveals global and tissue-specific methylation patterns. BMC Biol. 18, 85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johnson M. D., Mueller M., Adamowicz-Brice M., Collins M. J., Gellert P., Maratou K., Srivastava P. K., Rotival M., Butt S., Game L., Atanur S. S., Silver N., Norsworthy P. J., Langley S. R., Petretto E., Pravenec M., Aitman T. J., Genetic analysis of the cardiac methylome at single nucleotide resolution in a model of human cardiovascular disease. PLOS Genet. 10, e1004813 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hon G. C., Rajagopal N., Shen Y., McCleary D. F., Yue F., Dang M. D., Ren B., Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat. Genet. 45, 1198–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V., Ren B., A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang Y., Fan X., Yan J., Chen M., Zhu M., Tang Y., Liu S., Tang Z., A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 49, 1313–1329 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duan J. E., Jiang Z. C., Alqahtani F., Mandoiu I., Dong H., Zheng X., Marjani S. L., Chen J., Tian X. C., Methylome dynamics of bovine gametes and in vivo early embryos. Front. Genet. 10, 512 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Du Z., Zheng H., Kawamura Y. K., Zhang K., Gassler J., Powell S., Xu Q., Lin Z., Xu K., Zhou Q., Ozonov E. A., Véron N., Huang B., Li L., Yu G., Liu L., Yeung W. K. A., Wang P., Chang L., Wang Q., He A., Sun Y., Na J., Sun Q., Sasaki H., Tachibana K., Peters A. H. F. M., Xie W., Polycomb group proteins regulate chromatin architecture in mouse oocytes and early embryos. Mol. Cell 77, 825–839.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 81.Wang L., Zhang J., Duan J., Gao X., Zhu W., Lu X., Yang L., Zhang J., Li G., Ci W., Li W., Zhou Q., Aluru N., Tang F., He C., Huang X., Liu J., Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Legends for tables S1 to S4
References
Tables S1 to S4