Abstract
In prevalent cohort design, subjects who have experienced an initial event but not the failure event are preferentially enrolled and the observed failure times are often length-biased. Moreover, the prospective follow-up may not be continuously monitored and failure times are subject to interval censoring. We study the nonparametric maximum likelihood estimation for the proportional hazards model with length-biased interval-censored data. Direct maximization of likelihood function is intractable, thus we develop a computationally simple and stable expectation-maximization algorithm through introducing two layers of data augmentation. We establish the strong consistency, asymptotic normality and efficiency of the proposed estimator and provide an inferential procedure through profile likelihood. We assess the performance of the proposed methods through extensive simulations and apply the proposed methods to the Massachusetts Health Care Panel Study.
Keywords: left truncation, nonparametric maximum likelihood estimation, proportional hazards model, semiparametric efficiency
1 |. INTRODUCTION
Interval-censored data arise when a failure time is not recorded precisely but is rather known to lie within a time interval. Such data are encountered in prospective follow-up studies, where the ascertainment of the event of interest is made over a series of examination times. Regression analysis of unbiased interval-censored survival data has been extensively studied. In particular, nonparametric maximum likelihood estimation for the proportional hazards and transformation models have been studied by Huang (1996) and Zeng et al. (2016), respectively. Due to intractable likelihood, sieve estimation is also proposed for the proportional hazards model by Huang and Rossini (1997) and Cai and Betensky (2003), among others. A comprehensive review is given in Sun (2007).
Although sampling incident cases in a follow-up study is common, it may require a long follow-up period to observe enough failure events for meaningful analysis. Alternatively, a prevalent cohort design samples individuals who have experienced an initial event but not the failure event at enrollment, and is often considered as a more focused and economical design (Brookmeyer and Gail, 1987). However, subjects with a longer survival time are preferentially sampled in a prevalent cohort. When the incidence of the initial event is stationary over time, a prevalent cohort collects length-biased data (Wang, 1991; Shen et al., 2009), where the probability of observing the failure time is proportional to its value.
Prospective follow-up of a prevalent cohort can be subject to interval censoring. An example is the Massachusetts Health Care Panel Study (Chappell, 1991), where the time to loss of active life for elderly individuals were assessed approximately 1.25, 6, and 10 years after study recruitment. Since only functionally independent individuals were enrolled, subjects with a longer time to loss of active life were more likely to be sampled. Although a prevalent cohort is a biased sample that requires special methods for analysis, it may provide information that is otherwise unavailable in an incident cohort. For example, in an incident sampling design, the right tail of survival distribution may not be identified because of limited study duration. Using prevalent sampling design, the identifiable region for the survival distribution and marked variables that are observed only at the event occurrence could be enlarged (Chan and Wang, 2010). In the Massachusetts Health Care Panel Study data, even though the last monitoring time is 10 years after study recruitment, we can identify a survival distribution that ranges over 30 years (see Section 3.2), because individuals are event-free for a period before enrollment. An added advantage for interval-censored data is that, even when the monitoring time has a discrete distribution, we can identify the continuous survival distribution of the failure event because the event-free period before enrollment is typically continuous. For incident sampling with discrete monitoring time, in contrast, we can only identify a discrete survival distribution.
Statistical methodology for regression modeling of length-biased data are mostly proposed for uncensored and right-censored data. Wang (1996) and Chen (2010) considered uncensored data. For right-censored data, Qin and Shen (2010) proposed inverse weighted estimating equation and Huang and Qin (2012) proposed a composite likelihood approach for the proportional hazards model. Qin et al. (2011) considered the nonparametric maximum likelihood estimator and derived an expectation-maximization (EM) algorithm for computation, and showed that the estimator is efficient.
Even though there has been limited literature on length-biased interval-censored data, several methods were proposed for left-truncated interval-censored data without the length-biased assumption for the truncation time. In particular, Pan and Chappell (1998) considered the proportional hazards model and applied a gradient projection-based method for non-parametric maximum likelihood estimation, where the baseline survival function may be underestimated. Pan and Chappell (2002) considered the same model and suggested a marginal likelihood approach that avoids estimating the baseline hazards function and a monotone maximum likelihood approach assuming that the baseline distribution has a nondecreasing hazard function. Kim (2003) studied the special case of left-truncated current status data, where there is only one examination time, and established the asymptotic properties of the nonparametric maximum likelihood estimators. Recently, Wang et al. (2015) studied the additive hazard model with left-truncated interval-censored data and proposed a sieve estimation method.
In this paper, we study the nonparametric maximum likelihood estimation for the proportional hazards model with length-biased interval-censored data. Through introducing pseudo-truncated data and latent Poisson random variables, we develop a simple and computational stable EM algorithm. We establish the strong consistency and asymptotic normality of the proposed estimators and provide inference through a profile likelihood approach. We assess the performance of the proposed estimator and inferential procedures through extensive simulations and apply the proposed methods to the Massachusetts Health Care Panel Study data.
2 |. THE PROPOSED METHODOLOGY
2.1 |. Model and data
For individuals in the target population, let be the time to a failure event and be a p-vector of covariates. We assume that follows a proportional hazards model with a cumulative hazard function
where β is a p-vector of unknown regression parameters, and Λ(·) is an arbitrary increasing function with Λ(0) = 0.
For length-biased sampling, it is common to assume that the incidence rate of the initial event is constant over calendar time and , the truncation time, is uniformly distributed in [0, τ], where τ is the maximum support of (Wang, 1991; Qin et al., 2011). In a prevalent cohort study, a subject is included only if the failure time does not occur before the truncation time, that is, . We let T, A, and Z be the failure time, truncation time and covariates, respectively, in the prevalent cohort. Then, (T, A, Z) has the same joint distribution as conditional on . Suppose that the occurrence of the failure is not exactly observed but only determined at a sequence of examination times, denoted as . The failure time is then known to lie in the interval (L, R), where , , m = 1, …, M + 1}, U0 = A, and . In particular, if the failure occurs before the first examination time, then (L, R) = (A, U1); if the failure has not occurred at the last examination time, then . Let for m = 0, …, M, so that V0 = 0.
We assume the following non-informative sampling time condition, that M and are independent of (T, A) conditional on Z. For a length-biased sample of n subjects, the observed data are , where . The observed-data likelihood is then given by
| (1) |
The likelihood function involves τ, which is a constant related to study design but not a parameter of interest. In the E-step of the proposed EM algorithm, however, it is required to redistribute mass on [0, τ], and an approximation of τ is required. Let be the ordered sequence of all Li and . Following Qin et al. (2011), we approximate τ by tk, which converges to τ at a rate faster than n1/2, and therefore does not alter subsequent results.
2.2 |. Nonparametric maximum likelihood estimation
We adopt the nonparametric maximum likelihood estimation approach, where the estimator for Λ is a step function with nonnegative finite jumps at the ends of the intervals that bracket the failure times. Specifically, we let λ0, λ1,…, λk be the respective jump sizes at t0, t1,…,tk, where λ0 = 0. Write . We maximize the objective function
Direct maximization of is difficult due to a lack of analytical expressions. We introduce two layers of data augmentation and propose an EM algorithm to facilitate computation. First, to handle left truncation, we introduce pseudo-truncated data, which is also referred to as “ghost data” (Turnbull, 1976). In particular, let denote the pseudo-truncated samples corresponding to subject i, where are independent and identically distributed given Zi. Since the estimator for Λ only takes jump at , the failure time can only take values from . The number of truncated samples ni follows a negative binomial distribution with parameter
such that . Let . Given the total number follows a multinomial distribution with probabilities , where
| (2) |
By the missing information principle (Lai and Ying, 1994), the maximization of is equivalent to maximizing the conditional expectation of the log-likelihood function of the “complete-data” given the observed data. The “complete-data” log-likelihood function is given by
While we may propose an EM algorithm based on , its maximization step is still difficult to obtain, since β and λ cannot be separated in the complete-data log-likelihood function due to the interval censoring structure.
Therefore, we further introduce data augmentation based on independent Poisson random variables with means , where . The joint density function for is given by
Let and . Suppose that we observe Ni1 = 0 and Ni2 > 0. The observed-data likelihood for is equal to
Therefore, can be viewed as the observed log-likelihood function for with and as latent variables. In particular, the complete-data log-likelihood function based on is given by
Based on this formulation, we propose the following EM algorithm. In the E-step, we evaluate the conditional expectations of Wij and nij given the observed data. In particular, we have
and
where denote the conditional expectation with respect to the observed data . In the M-step, we maximize the expected complete-data log-likelihood function. We update λj by
and update β by solving
We iterate between the E-step and M-step until convergence. We denote the final estimators for β and λ as and .
In summary, through introducing two layers of latent random variables, we proposed a stable computing algorithm to obtain the estimators that maximize the nonparametric likelihood function. The latent truncated “ghost data” were introduced to deal with the complications that arise from left truncation and the latent Poisson random variables were introduced to deal with the incomplete data caused by interval-censoring.
2.3 |. Asymptotic properties
In this section, we establish the strong consistency and asymptotic normality of the proposed estimators. We assume the following regularity conditions.
Condition 1. The true value of β, denoted by β0, belongs to the interior of a known compact set .
Condition 2. The true value Λ0(·) of Λ(·) is strictly increasing and continuously differentiable in [0, τ] with Λ0(0) = 0.
Condition 3. The covariate Z has bounded support and is not concentrated on any proper subspace of .
Condition 4. The examination times have finite support with the least upper bound τ. The number of potential examination times M is positive with E(M) < ∞. There exists a positive constant η such that . In addition, there exists a probability measure μ in such that the bivariate distribution function of (Um, Um+1) conditional on (M, Z) is dominated by μ × μ and its Radon-Nikodym derivative, denoted by , can be expanded to a positive and twice-continuously differentiable function in the set .
Conditions 1, 2, and 3 are standard conditions for failure time regression. Condition 4 pertains to the joint distribution of examination times. It requires that two adjacent examination times are separated by at least η; otherwise, the data may contain exact observations, which require a different theoretical treatment. The dominating measure μ is chosen as the Lebesgue measure if the examination times are continuous random variables and as the counting measure if the examinations occur only at a finite number of time points. The number of potential examination times M can be fixed or random, is possibly different among study subjects, and is allowed to depend on covariates.
We state the strong consistency of and the weak convergence of in two theorems.
Theorem 1.
Under Conditions 1–4, , and , where denotes the supremum norm on , and .
Theorem 2.
Under Conditions 1–4, converges weakly to a p-dimensional zero-mean normal random vector with a covariance matrix that attains the semiparametric efficiency bound.
The proofs of both theorems are provided in Appendix A.
2.4 |. Variance estimation
We estimate the covariance matrix of by a profile likelihood approach. Let , where is the set of bounded step functions with non-negative jumps at tl (l = 1,…, m). The maximizer can be obtained using the EM algorithm of Section 2.2 if we fix β and only update λ in the M-step. The profile log-likelihood function is defined as
Let be the ith subject’s contribution to . We estimate the covariance matrix of by the inverse of
where ej is the jth canonical vector in , and hn is a constant of order n−1/2. In the numerical studies, we used hn = 5n−1/2 as suggested by Zeng et al. (2016).
The above profile likelihood approach is different from that of Murphy and Vaart (2000). They estimate the covariance matrix of by the negative inverse of the Hessian matrix of pln(β) at , which is obtained by second order numerical differences. The estimated matrix may not be positive semidefinite, especially in small samples. Here, we estimate the covariance matrix by the inverse of the empirical covariance matrix of the gradient of using the first-order numerical differences, similar to Zeng et al. (2017). The calculation is quicker than the approach requiring second-order numerical differences, and the estimated covariance matrix is guaranteed to be positive semidefinite.
3 |. NUMERICAL STUDIES
3.1 |. Simulation
We conducted simulation studies to evaluate the performance of the proposed methods. We considered two covariates Z1 ∼ Bernoulli(0.5) and z2 ∼ Uniform(−0.5, 0.5). We set β = (0.5, 1)T, Λ(t) = 0.3t, and τ = 15. We generated the truncation time from Uniform(0, τ) and generated the sequence of potential examination times Um ∼ Um−1 + 0.1 + Uniform(0, 2) with U0 = . We set n = 100, 200, or 400 and examined 10000 replicates for each sample size. We compared the proposed nonparametric maximum likelihood method with the maximum conditional likelihood method of Pan and Chappell (1998), which is applicable to left-truncated interval-censored data. For coherent comparisons, we compute both estimators by EM algorithms, where the algorithm for the conditional likelihood estimator is an adaption of the proposed algorithm and is given in Appendix B. We set the initial value of β to 0 and the initial value of λl to 1/k.
Table 1 summarizes the simulation results on the estimation of β using the proposed and conditional likelihood approaches. The biases for the proposed estimators are small and decrease as sample size increases. The biases for the conditional likelihood estimators are larger than those for the proposed estimators for all sample sizes, but decrease as sample size increases. The variance estimators for using both approaches are accurate and the confidence intervals have proper coverage probabilities. As expected, the proposed estimator shows substantial efficiency gain compared to the conditional likelihood estimator. Web Figure S1 in the Supplementary Materials gives the estimated baseline survival functions. The nonparametric maximum likelihood estimation gives unbiased estimates, while the condition likelihood estimators tend to underestimate the true values, as indicated in Pan and Chappell (1998).
TABLE 1.
Summary statistics for the simulation studies with length-biased assumption
| NPMLE |
CLE |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bias | SE | SEE | RMSE | CP | Bias | SE | SEE | RMSE | CP | ||
| n=100 | β 1 | 0.008 | 0.169 | 0.171 | 0.169 | 0.956 | 0.049 | 0.253 | 0.235 | 0.258 | 0.928 |
| β 2 | 0.015 | 0.295 | 0.317 | 0.295 | 0.965 | 0.089 | 0.444 | 0.436 | 0.453 | 0.942 | |
| n=200 | β 1 | 0.004 | 0.117 | 0.116 | 0.117 | 0.950 | 0.025 | 0.170 | 0.159 | 0.172 | 0.933 |
| β 2 | 0.005 | 0.206 | 0.212 | 0.206 | 0.957 | 0.044 | 0.296 | 0.290 | 0.299 | 0.943 | |
| n=400 | β 1 | 0.002 | 0.082 | 0.081 | 0.082 | 0.944 | 0.014 | 0.115 | 0.111 | 0.116 | 0.937 |
| β 2 | 0.002 | 0.144 | 0.146 | 0.144 | 0.951 | 0.024 | 0.200 | 0.199 | 0.202 | 0.949 | |
Note: NPMLE and CLE are the nonparametric maximum likelihood and conditional likelihood estimators. SE, SEE, RMSE, and CP are the empirical standard error, mean standard error estimator, root mean squared error, and empirical coverage probability of the 95% confidence interval, respectively.
We further assess the robustness of the nonparametric maximum likelihood estimator when the uniform assumption for the truncation time does not hold. In particular, we considered the same simulation setting but generated the truncation time from Exp(0.1) such that the stationary incidence assumption is violated. Table 2 shows the simulation results. The nonparametric maximum likelihood estimators are slightly biased, while the coverages of the 95% confidence intervals are acceptable. Even though the bias of the proposed estimator is larger than the conditional likelihood estimator when sample size is large (n = 400) and length-biased sampling is violated, the mean squared error of the proposed estimator is still smaller.
TABLE 2.
Summary statistics for the simulation studies without length-biased assumption
| NPMLE |
CLE |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bias | SE | SEE | RMSE | CP | Bias | SE | SEE | RMSE | CP | ||
| n=100 | β 1 | −0.039 | 0.160 | 0.167 | 0.165 | 0.953 | 0.045 | 0.244 | 0.231 | 0.248 | 0.933 |
| β 2 | −0.077 | 0.285 | 0.307 | 0.295 | 0.951 | 0.083 | 0.442 | 0.428 | 0.450 | 0.941 | |
| n=200 | β 1 | −0.044 | 0.111 | 0.113 | 0.119 | 0.933 | 0.023 | 0.164 | 0.157 | 0.165 | 0.935 |
| β 2 | −0.088 | 0.198 | 0.205 | 0.217 | 0.930 | 0.045 | 0.293 | 0.285 | 0.296 | 0.943 | |
| n=400 | β 1 | −0.047 | 0.078 | 0.078 | 0.091 | 0.899 | 0.012 | 0.113 | 0.109 | 0.113 | 0.938 |
| β 2 | −0.095 | 0.137 | 0.140 | 0.167 | 0.894 | 0.021 | 0.198 | 0.196 | 0.199 | 0.947 | |
Note: NPMLE and CLE are the nonparametric maximum likelihood and conditional likelihood estimators. SE, SEE, RMSE, and CP are the empirical standard error, mean standard error estimator, root mean squared error, and empirical coverage probability of the 95% confidence interval, respectively.
3.2 |. Massachusetts health care panel study
We apply the proposed methods to the Massachusetts Health Care Panel Study, which has been described and analyzed previously (Chappell, 1991; Pan and Chappell, 1998; Hudgens, 2005). The study aimed at assessing the risk at which elderly individuals lose active life, which is defined as a continued ability to perform various activities of daily living such as dressing and bathing. The study was first conducted in 1975 taking a baseline survey of Massachusetts residents over the age of 65. Since only subjects who were active at baseline were included, the time to loss of active life was subject to left truncation. Three follow-up waves were then taken at 1.25, 6, and 10 years after baseline to determine if subjects are still living actively, so the time to loss of active life was also interval-censored.
The data set includes 1286 subjects with enrollment age ranges from 65 to 97.3. Since the study population were defined to be over age 65, we consider the failure time as age at loss of active life minus 65. Since the subjects are active at the enrollment, the truncation time is the age at enrollment minus 65. We applied the proposed methods to study the association between loss of active life and gender.
Table 3 shows the estimation results for the regression parameter in the Massachusetts Health Care Panel Study. The point estimates from the proposed approach and the conditional likelihood approach are both positive, indicating that male subjects are associated with a higher risk of losing active life than females. The standard error estimate of the proposed nonparametric maximum likelihood estimator is smaller than that of the conditional likelihood estimator, so that at a 5% significance level, the null hypothesis of no association between gender and loss of active life is only rejected by the nonparametric maximum likelihood approach.
TABLE 3.
Estimation results for the regression parameter in the Massachusetts Health Care Panel Study
| NPMLE |
CLE |
|||||
|---|---|---|---|---|---|---|
| Covariate | Estimate | Std Err | p-value | Estimate | Std Err | p-value |
| Male | 0.144 | 0.059 | 0.014 | 0.133 | 0.076 | 0.081 |
Note: NPMLE and CLE are the nonparametric maximum likelihood and conditional likelihood estimators.
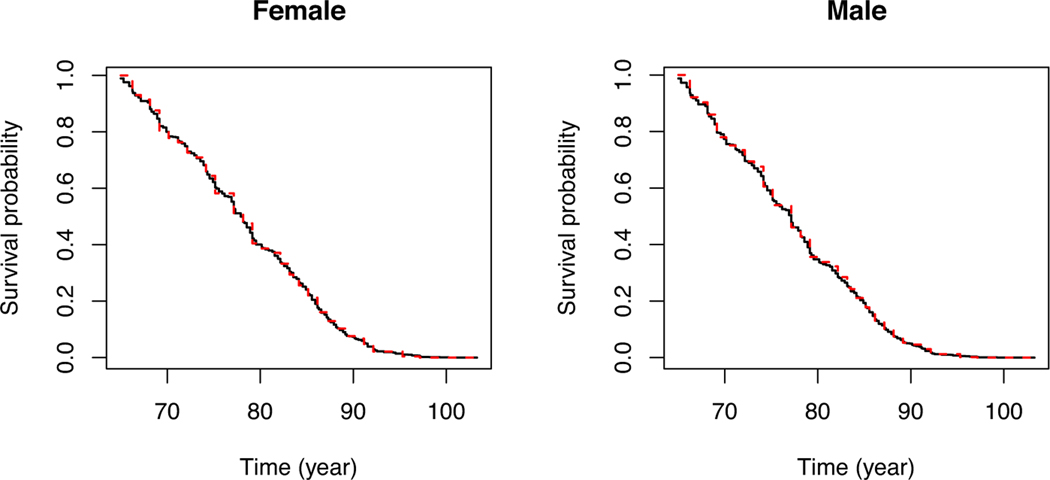
Figure 1 shows the estimated survival probabilities for male and female subjects using the two approaches. Even though they give similar estimates for the survival probabilities, the nonparametric maximum likelihood approach gives an estimate with finer jumps, resulting from the additional assumption on the truncation time. Using both approaches, the female subjects have a higher survival probability than the male subjects.
FIGURE 1.
Estimated survival probabilities for subgroups of subjects in the Massachusetts Health Care Panel Study. The solid and dashed curves pertain to the nonparametric maximum likelihood and conditional likelihood estimation approaches, respectively.
If the length-biased sampling assumption does not hold, the regression coefficients of the two methods would converge to different values. Therefore, we estimated the difference of the estimators and construct a 95% confidence interval by bootstrapping with 1000 replications, to see if the stationary assumption for the truncation time holds. The differences of the two estimators was 0.011 with a 95% confidence interval (−0.092, 0.107), indicating that the length-biased assumption is possibly valid.
4. | DISCUSSION
In this paper, we adopt the nonparametric maximum likelihood estimation where the estimator for Λ is a step function that is right-continuous which is usually considered in the literature. As mentioned by a reviewer, if Λ is only restricted to be nondecreasing, then the true maximizer of the likelihood should involve a left-continuous Λ, that is, Λ(t) = Λ(tj+1) on (tj, tj+1] for j = 0, …, m − 1. The two versions are asymptotically equivalent since any two adjacent step points get closer as sample size increases. In Web Appendix A of the Supplementary Materials, we implemented the version with left-continuous Λ and demonstrated that the numerical difference between the two versions is ignorable.
The iterative convex minorant (ICM) algorithm (Pan, 1999) is an alternative algorithm for the EM algorithm adopted in the paper to obtain the nonparametric maximum likelihood estimators for interval-censored data. Even though it is generally faster than the EM algorithm considered in the paper, it may become unstable for large datasets because it attempts to update a large number of parameters simultaneously using a quasi-Newton method (Zeng et al., 2016). Wang et al. (2016) also advocated the use of an EM algorithm by comparing it with the R package intcox (Henschel and Mansmann, 2013) that adopts the algorithm of Pan (1999). They found that ICM algorithm often exhibits larger biases, indicating that it may not converge to the true maximizer of the likelihood function.
In this paper, we studied the nonparametric maximum likelihood estimation of the proportional hazards model for length-biased interval-censored data. Although length-biasedness requires a stationary incidence distribution for the initial event, the proposed methods can be extended to situations when the incidence distribution follows a parametric model (Huang et al., 2015). In that case, the denominator of the individual components in Ln(β, Λ) need to be modified corresponding to the distribution of the truncation time, and the proposed EM algorithm can be adjusted accordingly.
The efficiency gain of the proposed nonparametric maximum likelihood estimators over the conditional likelihood estimators mainly comes from the information of the (uniform) distributional assumption on the truncation time. Relatively, the conditional likelihood estimators are more robust against the assumption. In practice, one need to carefully ascertain the assumption to apply the proposed approach. For the right-censored left-truncated data, graphical methods (Wang, 1991; Asgharian et al., 2006) and a goodness-of-fit test (Mandel and Betensky, 2007) have been proposed to test the length-biasedness assumption. In the numerical examples, we used a bootstrapped method to the difference of the nonparametric maximum likelihood and conditional likelihood approaches as an indirect test of length-biasedness. The diagnostic methods for right-censored data cannot be directly extended to the interval-censoring case, since the estimator for the survival function converges in a different, n1/3, rate. Formal tests for the length-biased assumption with interval-censored data will be developed in the future.
The individuals in the MHCPS data may also be subject to the risk of a competing cause, for example, death, such that the subjects who died before loss of active life were right-censored. In addition, there may be selection effect such that only alive subjects were included in the study. Therefore, the assumptions of conditional independent censoring and truncation times may be questionable. However, the information on the cause of right censoring is not available in MHCPS data, so we are not able to access the validity of the assumptions. The regression analysis of competing risks interval-censored data has been studied (Mao et al., 2017), however, no existing methods considered the scenario when the competing risks interval-censored data are also subject to left truncation. Methods incorporating such complications are important future research.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Prof Rick Chappell for sharing the data from Massachusetts Health Care Panel Study. We also thank the associate editor and two referees for constructive feedbacks that improve the paper. The authors were partially supported by the National Heart, Lung, and Blood Institute of the U.S. National Institutes of Health.
Appendix A
Proof of asymptotic results
We use to denote the empirical measure from n independent subjects and to denote the true probability measure. Write . Let L(β, Λ) be the observed-data likelihood for a single subject
where .
Write . Let be a step function that takes jumps only at t1, …, tk with for j = 1,…, k. Let
and
where DM = {Λ : Λ is increasing with Λ(0) = 0, Λ(τ) ≤ M}, and M < ∞. The proofs make use of two lemmas, whose proofs are given in Web Appendix B.
Lemma 1.
Under Conditions 1–4, the classes of functions is ℙ-Donsker.
Lemma 2.
Under Conditions 1–4,
Proof of Theorem 1
The jump points {t1,..., tk} depend on sample size n and for any ϵ > 0, ⋃jBϵ(tj) covers the support as n → ∞, where Br(t) is the open ball around tj with radius r. By the continuity of Λ0, converges uniformly to Λ0(t). It follows from Lemma 1 that the class is Donsker. By the concavity of the log function,
Since is bounded, for any subsequence of , we can find a further subsequence converging to . In addition, by Helly’s selection lemma, for any subsequence of , there exists a further subsequence that converges to some increasing function Λ*. We choose the converging subsequences of and such that we can obtain without loss of generality that and pointwise on any interior set of . Therefore,
such that the negative Kullback-Leibler information is positive. Therefore,
with probability 1. For any m ∈ {0,..., M}, we set Δm′ = 1 in the above equation m′ = m,..., M and take the sum of the resulting equations to obtain
Because m is arbitrary, we can replace Um in the above equation by any t ∈ . We take the logarithm and differentiate both sides with respect to t to find
such that β* = β0 and for t ∈ . Hence, Λ*(t) = Λ0(t) for t ∈ . We conclude that and for any t ∈ . Because Λ0 is continuous, converges uniformly to Λ0 on .
Proof of Theorem 2.
Let
and . The score equations for β is given by
The score operator for Λ along the submodel dΛϵ,h = (1 + ϵh)dΛ for h ∈ L2(μ) is given by
Clearly,
and
We apply the Taylor series expansions at (β0, Λ0) to the right sides of the above two equations. In light of Lemma 2, the second-order terms are bounded by . Therefore,
and
dΛϵ,h, lΛβ(h) is the derivative of lΛ(h) with respect to β, and lΛΛ(h, − Λ0) is the derivative of lΛ(h) along the submodel dΛ0 + ϵd( − Λ0). All derivatives are evaluated at (β0, Λ0).
If the least favorable direction exists, we denote it as h* ∈ L2(μ)p. We first show the existence of h*, which is the solution of where is the adjoint operator of lΛ. We equip L2(μ) with an inner product defined as
On the same space, we define
It is easy to show that ∥·∥ is a seminorm on L2(μ). Furthermore, if ∥h∥ = 0, then . Thus, with probability 1, . By the arguments in the Lemma 2, h(t) = 0 for t ∈ . Clearly, ∥h∥ ≤ c < h, h >1/2 for some constant c by the Cauchy-Schwarz inequality. According to the bounded inverse theorem in Banach spaces, we have for another constant . By the Lax-Milgram theorem (Zeidler, 1995), there exists that satisfies
for t ∈ . Differentiation of the integral equations with respect to t yields
where q1(t) > 0, and qj (j = 1, 2, 3) and q4 are continuously differentiable functions. Thus, h* can be expanded to be a continuously differentiable function in with bounded total variations. It follows that
Using the arguments in the proof of Lemma 1, we can show that belongs to a Donsker class. Next, we show that is invertible. If the matrix is singular, then there exists an vector such that . It follows that, with probability 1, the score function along the submodel is zero. That is,
For any m ∈ {0,…, M}, we sum over all possible Δm′ with m′ = m,…, M to obtain
Because m is arbitrary, we can replace Um in the above equation by t ∈ . We differentiate both sides with respect to t to obtain
for any t ∈ . It then follows that υ = 0. Hence, the matrix is invertible.
Then, , and
The influence function for is the efficient influence function, such that converges weakly to a zero-mean normal random vector whose covariance matrix attains the semiparametric efficiency bound.
Appendix B
An EM algorithm for maximum conditional likelihood estimation
A nonparametric maximum conditional likelihood estimator is considered in Pan and Chappell (1998) for the proportional hazards model with left-truncated interval-censored data. A slight variation of the proposed EM algorithm can be used to compute their estimator, and is used in the numerical comparisons.
The observed-data conditional likelihood given the truncation time is given by
We estimate Λ nonparametrically such that the estimator for Λ is a step function that jumps only at t1,…, tk, which are the ordered sequence of all Li, Ri, and Ai. We maximize the objective function
We introduce a sequence of independent Poisson random variables with means . Let , and . The objective function can be viewed as the observed-data log-likelihood for {Ni1 = 0, Ni2 > 0 : i = 1,…, n} with as latent variables. We propose an EM algorithm. In the E-step, we evaluate
In the M step, we update λj by
and update β by solving
We iterate between the E-step and M-step until convergence.
Footnotes
SUPPORTING INFORMATION
The Web Appendices and Figure, referenced in Sections 3 and 4 and Appendix A, and the computation codes for the simulation studies are available with this paper at the Biometrics website on Wiley Online Library.
REFERENCES
- Asgharian M, Wolfson DB, and Zhang X. (2006). Checking stationarity of the incidence rate using prevalent cohort survival data. Stat Med 25, 1751–1767. [DOI] [PubMed] [Google Scholar]
- Brookmeyer R. and Gail MH (1987). Biases in prevalent cohorts. Biometrics 43, 739–749. [PubMed] [Google Scholar]
- Cai T. and Betensky RA (2003). Hazard regression for interval-censored data with penalized spline. Biometrics 59, 570–579. [DOI] [PubMed] [Google Scholar]
- Chan KCG And Wang M-C (2010). Backward estimation of stochastic processes with failure events as time origins. Ann Appl Stat 4, 1602–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell R. (1991). Sampling design of multiwave studies with an application to the Massachusetts health care panel study. Stat Med 10, 1945–1958. [DOI] [PubMed] [Google Scholar]
- Chen YQ (2010). Semiparametric regression in size-biased sampling. Biometrics 66, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschel V. and Mansmann U. (2013). intcox: Iterated convex minorant algorithm for interval-censored event data. R package version 0.9.3.
- Huang J. (1996). Efficient estimation for the proportional hazards model with interval censoring. Ann Stat 24, 540–568. [Google Scholar]
- Huang C-Y and Qin J. (2012). Composite partial likelihood estimation under length-biased sampling, with application to a prevalent cohort study of dementia. J Am Stat Assoc 107, 946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. and Rossini AJ (1997). Sieve estimation for the proportional-odds failure-time regression model with interval censoring. J Am Stat Assoc 92, 960–967. [Google Scholar]
- Huang C-Y, Ning J, and Qin J. (2015). Semiparametric likelihood inference for left-truncated and right-censored data. Biostatistics 16, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudgens MG (2005). On nonparametric maximum likelihood estimation with interval censoring and left truncation. J R Stat Soc B 67, 573–587. [Google Scholar]
- Kim JS (2003). Efficient estimation for the proportional hazards model with left-truncated and “Case 1” interval-censored data. Stat Sin 13, 519–537. [Google Scholar]
- Lai TL and Ying Z. (1994). A missing information principle and M-estimators in regression analysis with censored and truncated data. Ann Stat 22, 1222–1255. [Google Scholar]
- Mandel M. and Betensky RA (2007). Testing goodness of fit of a uniform truncation model. Biometrics 63, 405–412. [DOI] [PubMed] [Google Scholar]
- Mao L, Lin D-Y, and Zeng D. (2017). Semiparametric regression analysis of interval-censored competing risks data. Biometrics 73, 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SA and Vaart AW (2000). On profile likelihood. J Am Stat Assoc 95, 449–465. [Google Scholar]
- Pan W. (1999). Extending the iterative convex minorant algorithm to the Cox model for interval-censored data. J Comput Graph Stat 8, 109–120. [Google Scholar]
- Pan W. and Chappell R. (1998). Computation of the NPMLE of distribution functions for interval censored and truncated data with applications to the Cox model. Comput Stat Data Anal 28, 33–50. [Google Scholar]
- Pan W. and Chappell R. (2002). Estimation in the Cox proportional hazards model with left-truncated and interval-censored data. Biometrics 58, 64–70. [DOI] [PubMed] [Google Scholar]
- Qin J. and Shen Y. (2010). Statistical methods for analyzing right-censored length-biased data under Cox model. Biometrics 66, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Ning J, Liu H, and Shen Y. (2011). Maximum likelihood estimations and EM algorithms with length-biased data. J Am Stat Assoc 106, 1434–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Ning J, and Qin J. (2009). Analyzing length-biased data with semiparametric transformation and accelerated failure time models. J Am Stat Assoc 104, 1192–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J. (2007). The Statistical Analysis of Interval-Censored Failure Time Data. New York: Springer. [Google Scholar]
- Turnbull BW (1976). The empirical distribution function with arbitrarily grouped, censored and truncated data. J R Stat Soc B 38, 290–295. [Google Scholar]
- Wang M-C (1991). Nonparametric estimation from cross-sectional survival data J Am Stat Assoc 86, 130–143. [Google Scholar]
- Wang M-C (1996). Hazards regression analysis for length-biased data. Biometrika 83, 343–354. [Google Scholar]
- Wang P, Tong X, Zhao S, and Sun J. (2015). Regression analysis of left-truncated and case i interval-censored data with the additive hazards model. Commun Stat Theory Methods 44, 1537–1551. [Google Scholar]
- Wang L, McMahan CS, Hudgens MG, and Qureshi Z. (2016). A flexible, computationally efficient method for fitting the proportional hazards model to interval-censored data. Biometrics 72, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler E. (1995). Applied Functional Analysis—Applications to Mathematical Physics. New York: Springer. [Google Scholar]
- Zeng D, Mao L, and Lin DY (2016). Maximum likelihood estimation for semiparametric transformation models with interval-scensored data. Biometrika 103, 253–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng D, Gao F, and Lin DY (2017). Maximum likelihood estimation for semiparametric regression models with multivariate interval-censored data. Biometrika 104, 505–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.