Abstract
R-spondin (RSPO) proteins constitute a family of four secreted glycoproteins (RSPO1–4) that have appeared as multipotent signaling ligands. The best-known molecular function of RSPOs lie within their capacity to agonize the Wnt/β-catenin signaling pathway. As RSPOs act upon cognate receptors LGR4/5/6 that are typically expressed by stem cells and progenitor cells, RSPO proteins importantly potentiate Wnt/β-catenin signaling especially within these proliferative stem cell compartments. Since multiple organs express LGR4/5/6 receptors and RSPO ligands within their stem cell niches, RSPOs can exert an influential role in stem cell regulation throughout the body. Inherently, over the last decade a multitude of reports implicated the deregulation of RSPOs in cancer development. First, RSPO2 and RSPO3 gene fusions with concomitant enhanced expression have been identified in colon cancer patients, and proposed as an alternative driver of Wnt/β-catenin hyperactivation that earmarks cancer in the colorectal tract. Moreover, the causal oncogenic capacity of RSPO3 overactivation has been demonstrated in the mouse intestine. As a paradigm organ in this field, most of current knowledge about RSPOs in cancer is derived from studies in the intestinal tract. However, RSPO gene fusions as well as enhanced RSPO expression have been reported in multiple additional cancer types, affecting different organs that involve divergent stem cell hierarchies. Importantly, the emerging oncogenic role of RSPO and its potential clinical utility as a therapeutic target have been recognized and investigated in preclinical and clinical settings. This review provides a survey of current knowledge on the role of RSPOs in cancer biology, addressing the different organs implicated, and of efforts made to explore intervention opportunities in cancer cases with RSPO overrepresentation, including the potential utilization of RSPO as novel therapeutic target itself.
Subject terms: Oncogenes, Mechanisms of disease, Cancer stem cells, Predictive markers
Introduction
The R-spondin (RSPO) family is represented by four genes RSPO1, 2, 3, and 4, encoding like-named secreted signaling proteins. Homologs of RSPOs are present amongst vertebrates and typically contain a thrombospondin type I repeat (TSR) domain, explaining their historical names as Human Protein With ThromboSpondin type I Repeat (hPWTSR) and Cysteine-rich single thrombospondin type I repeat containing protein (Cristin) [1, 2]. RSPO3 was the first member to be identified in a human fetal brain cDNA library in 2002, followed by the identification of mouse Rspo1 in 2004 (ref. [1, 3]). As Rspo1 expression was observed in the roof plate of the neural tube during mouse development, it was named Roof plate specific–Spondin (R-spondin). Subsequently, RSPO2 and RSPO4 were identified [4, 5]. Genetic mouse and human studies have revealed divergent and pivotal roles for the four RSPO members during development. Mutations in RSPO1 are linked with female-to-male XX sex reversal and Rspo1 knockout in mice revealed an important role in ovarian development [6, 7]. Rspo2 is involved in limb and respiratory tract development as well as craniofacial patterning and morphogenesis [8–11]. Rspo3 is essential for angiogenesis, vasculogenesis and placental development whereas genetic mutations in RSPO4 were detected in people with anonychia, characterized by the absence of finger and toe nails [12–17].
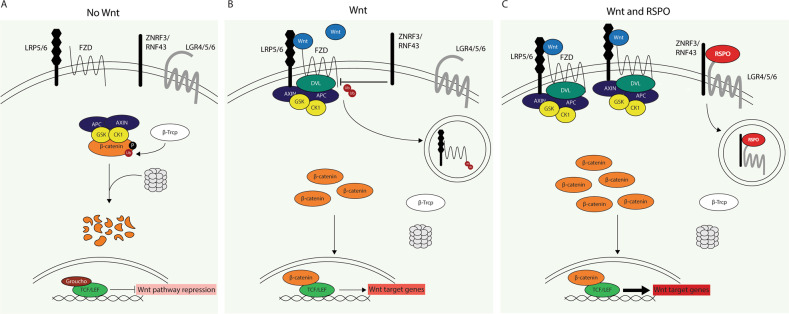
In 2004, Xenopus studies first described what is now the best-known molecular activity of R-spondin proteins: potentiation of the Wnt/β-catenin pathway, a crucial signaling pathway that regulates multiple fundamental processes including proliferation, stem cell control, tissue homeostasis and regeneration [5, 18]. Because of this fundamental role, the activity of the Wnt/β-catenin signaling pathway, in other words the downstream transcriptional activity of effector protein β-catenin, requires tight regulation which is executed at multiple levels. The central restraint of the pathway is provided by the intracellular APC containing destruction complex, which induces β-catenin degradation and as such inhibits the pathway (Fig. 1A). The pathway is activated upon binding of extracellular Wnt ligands to LRP5/6 and Frizzled (FZD) membrane receptors, leading to dissociation of β-catenin from the degradation complex, stabilization and nuclear translocation of β-catenin and subsequent transcriptional regulation of target genes in the nucleus (Fig. 1B). Another level of negative regulation is provided by ZNRF3 and RNF43, ubiquitin ligases that promote the degradation of LRP5/6 and FZD receptors, thereby reducing membranous Wnt receptor availability and subsequent downstream β-catenin signaling capacity [19, 20]. It is this latter ZNRF3/RNF43-mediated negative feedback loop that RSPO proteins interfere with, providing an additional level of canonical Wnt pathway regulation. All four RSPOs hold a conserved domain pattern composed of an N-terminal signal peptide, 2 cysteine rich furin like (FU1-FU2) domains, a thrombospondin (TSP) domain and a basic amino acid rich (BR) C-terminal domain. The FU1, FU2 and TSP domains enable RSPO proteins to bind ZNRF3/RNF43, Leucine-rich repeat-containing G-protein coupled receptors (LGR) 4–6 and heparin sulfate proteoglycans (HSPGs) respectively [2, 21–28]. Through interaction with ZNRF3/RNF43 and LGRs, RSPOs induce membrane clearance of ubiquitin ligases ZNRF3/RNF43, leading to enhanced Wnt receptor availability at the cell membrane and thereby potentiating Wnt ligand-mediated activation of the Wnt/β-catenin pathway (Fig. 1C)[19].
Fig. 1. The canonical Wnt pathway and the potentiating effect of RSPO.
A In the absence of canonical Wnt ligands the central destruction complex induces β-catenin degradation, restraining the transcription of Wnt target genes. B Canonical Wnt ligands induce dissociation of β-catenin from the degradation complex, leading to β-catenin accumulation, nuclear translocation and transcription of Wnt target genes. Ubiquitin ligases ZNRF3/RNF43 negatively regulate the Wnt pathway by internalizing and degrading membrane receptors LRP5/6 and FZD, thereby reducing Wnt receptor availability at the membrane. C RSPOs potentiate the canonical Wnt pathway by clearing negative regulators ZNRF3/RNF43 from the membrane, thereby increasing membranous Wnt receptor availability and potentiation of Wnt ligand-mediated pathway activation.
Despite the high homology among the four RSPOs, differences exist in their ability to bind LGRs and ZNRF3/RNF43 and to potentiate the Wnt/β-catenin pathway, where RSPO2 and RSPO3 show highest ZNRF3/RNF43 binding affinity and activity [21, 29]. Moreover, RSPO2 and RSPO3 can potentiate the Wnt/β-catenin pathway independently of LGR binding [27, 30]. This activity requires the binding of RSPOs to HSPGs with the TSP and BR domains in addition to binding to ZNRF3/RNF43 with the FU1 domain [27, 31]. Another study reported that RSPOs are also able to potentiate the Wnt/β-catenin pathway independent of ZNRF3/RNF43, where interaction of RSPO with LGR4 increases the affinity of scaffold protein IQGAP1 to bind DVL, resulting in LRP6 phosphorylation and potentiation of the Wnt/β-catenin pathway [32]. In addition to canonical Wnt signaling, RSPOs have also been implicated in non-canonical Wnt signaling in Xenopus embyros, where RSPO3 can modulate non-canonical Wnt/PCP signaling by binding to HSPG syndecan-4 and LGR4,5 to regulate gastrulation and head cartilage morphogenesis [28, 33]. Moreover, a recent Xenopus study described that RSPO3 exerts antagonistic effects on the BMP signaling route additionally, through binding of ZNRF3 and BMP receptor 1 A [34]. Thus, since their discovery multiple signaling activities have been attributed to RSPOs, especially in the regulation of canonical Wnt/β-catenin pathway but also beyond. In that perspective, more lessons will expectedly be learned considering the molecular activities of RSPOs.
The discovery that RSPOs represent ligands of the LGR4/5/6 receptors raised special interest, since these receptors are typically expressed by progenitor cells and stem cells, and as such the agonistic activity that RSPOs exert on Wnt/β-catenin signaling importantly influences the proliferative stem cell compartment [35–38]. As LGR5 was identified as a marker of stem cells in the intestine followed by the recognition of RSPOs representing LGR ligand, most knowledge on RSPO-LGR signaling currently exists in the field of the intestinal tract [33, 35, 39, 40]. However, RSPOs and LGR4/5/6 receptors are present in multiple organs and therefore RSPOs can influence stem cell regulation throughout the body. In accordance with this broad stem cell regulatory role, deregulated RSPO activity has increasingly been implicated in cancer development lately and RSPO alterations have been reported to occur in multiple cancer types as reviewed below (Table 1).
Table 1.
RSPO alterations reported among cancer types.
| Organ | RSPO alteration | References |
|---|---|---|
| Intestine | Gene fusions | |
| EIF3E-RSPO2 | [49–51] | |
| PIEZO1-RSPO2 | [51] | |
| NRIPI-RSPO2 | [52] | |
| PRR15L-RSPO2 | [53] | |
| PTPRK-RSPO3 | [49–52] | |
| Overexpression | ||
| RSPO2 | [49, 50] | |
| RSPO3 | [43, 49, 50] | |
| Stomach | Gene fusions | |
| EMC2-RSPO2 | [67] | |
| HNF4G-RSPO2 | [67] | |
| Overexpression | ||
| RSPO3 | [64] | |
| Breast | Overexpression | |
| RSPO2 | [113] | |
| RSPO3 | [113, 115, 116] | |
| RSPO4 | [113, 116] | |
| Ovary | Overexpression | |
| RSPO1 | [116, 122] | |
| RSPO3 | [116] | |
| Prostate | Gene fusions | |
| GRHL2-RSPO2 | [131] | |
| Overexpression | ||
| RSPO3 | [130] | |
| Downregulation | ||
| RSPO3 | [132] | |
| Liver | Gene fusions | |
| SINE-RSPO2 | [137] | |
| Overexpression | ||
| RSPO2 | [136–138] | |
| Lungs | Gene fusions | |
| EIF3E-RSPO2 | [141] | |
| PTPRK-RSPO3 | [141] | |
| Overexpression | ||
| RSPO3 | [116, 140] | |
| Pancreas | Overexpression | |
| RSPO2 | [116, 142] | |
| Bladder | Overexpression | |
| RSPO3 | [143] |
Gastrointestinal tract
Intestine
The intestinal epithelium displays an exceptionally rapid turnover that is controlled by tightly balanced molecular signaling in conjunction with unique cellular build-up of the crypt-villus architecture. Canonical Wnt/β-catenin signaling plays a central role in fueling proliferation and self-renewal in the crypt region, from where most progeny cells migrate towards the villus region whilst differentiating. The intestinal stem cell niche housed in the crypt region holds a refined composition of cycling stem cells, being protected and instructed by neighboring Paneth cells, quiescent stem cells and transient amplifying cells. Importantly, the cycling stem cells that fuel the continuous epithelial renewal express the RSPO receptor LGR5, whilst LGR4 is expressed more broadly on cycling stem cells, transient amplifying cells and Paneth cells [35, 41]. RSPO3 ligand is produced by stromal cells that lie in close proximity to the crypt stem cells both in mouse intestine and human colon [42–44]. Within this crypt environment, paracrine regulation of Wnt/β-catenin signaling by RSPO and Wnt ligands provide instructive signals to the intestinal stem cell niche, albeit in distinct manners [45]. Whereas Wnt ligands agonize canonical Wnt/β-catenin signaling, they are incapable of inducing renewal of LGR5+ stem cells on their own [45]. Instead, Wnt ligands induce RSPO receptor expression, thereby optimizing conditions for RSPO ligands to exert their effects. RSPO ligands in their turn induce the self-renewal and expansion of stem cells, as such dictating the size of the intestinal stem cell pool [45]. In case of intestinal injury, stromal RSPO3 expression is elevated and is demonstrated to be indispensable for epithelial regeneration by inducing Wnt/β-catenin signaling in differentiated cells, probably through the LGR4 receptor [46]. Of note, LGR5+ stem cells are dispensable in these epithelial regeneration processes, indicating that RSPO3 ligand is essential and dominantly instructive in epithelial repair in the gut [46–48]. Taken together, in the non-transformed intestine, RSPO3 is produced in the pericryptal stroma and plays a fundamental role in controlling stem cell numbers and epithelial recovery through activation of the Wnt/β-catenin pathway.
In line with the central role of Wnt/β-catenin signaling in intestinal stem cell maintenance, hyperactivation of this pathway is the hallmark feature of colorectal cancer (CRC). In the majority of CRC patients, this hyperactivation is caused by either inactivating APC mutations or activating mutations in the β-catenin gene CTNNB1, both resulting in constitutive activation of the Wnt/β-catenin pathway, independent of Wnt ligand binding. Importantly, in 2012 it was found that 4–10% of CRC patients harbor gene fusions of the RSPO2 and RSPO3 genes with EIF3E and PTPRK respectively, co-occurring with enhanced expression of the considerate RSPO gene [49, 50]. These RSPO2 and RSPO3 gene rearrangements were found mutually exclusive with other Wnt pathway mutations, though co-occurring with either KRAS or BRAF mutations, suggestively serving as an alternative mechanism to achieve hyperactivation of the Wnt/β-catenin pathway and to hold oncogenic capacity [49, 50]. Following up on the initial discovery of RSPO gene fusions in CRC patients, other studies identified additional gene fusions of RSPO2 with PIEZO1, NRIPI and PRR15L and moreover, reported RSPO gene fusions to typically occur in traditional serrated adenoma (TSA) rather than conventional colon tumors [51–54]. In addition, another CRC patient subpopulation has been described that harbors high RSPO3 expression levels but seem to lack RSPO gene fusions or alternative Wnt pathway mutations [43]. Instead, in these tumors the elevated levels of RSPO3 are produced by the stromal compartment, and in line, most of these cases were of the CMS4 mesenchymal subtype [43]. These data suggest that enhanced RSPO3 expression by stromal cells can substitute for epithelial RSPO mutations in driving CRC. As RSPOs are secreted ligands, these findings support the plausibility that especially the cells that receive the RSPO signals, rather than the producing cells, determine the oncogenic response, therefore being most interesting in understanding the biology of RSPO-driven cancer. For CRC, the typical occurrence of RSPO gene fusions in TSA might be informative in this regard, and it has been proposed that this might point towards a different, TSA-like evolutionary trajectory for RSPO-mutant tumor development, distinct from conventional CRC [55]. However, details on the potential cell of origin and mutation selection along the tumorigenic cascade within RSPO-driven cancer remain to be unraveled.
Formal evidence for the causal oncogenic capacity of Rspo3 was provided by a mouse study where conditional Rspo3 overexpression consistently induced abundant intestinal tumor formation, demonstrating that augmentation of Rspo3 levels is causative in driving tumorigenesis [56]. RSPO3-driven tumors showed major expansion of crypt cells including LGR5+ stem cells, quiescent stem cells, Paneth cells and LGR4+ cells with modestly increased Wnt/β-catenin signaling [56]. Thus, enhanced Rspo3 levels induced a magnification of the proliferative, self-renewing crypt compartment. Adding up to the oncogenic capacity of Rspo3 overexpression, another mouse study showed that also the transgenic expression of either EIF3E-RSPO2 or PTPRK-RSPO3 gene fusion causally drives the formation of intestinal tumors, which comparably show expansion of proliferative cells and ectopic Paneth cells [57]. Inversely, targeted anti-RSPO3 treatment in a PTPRK-RSPO3 xenograft CRC model was shown to induce tumor differentiation whilst reducing growth, stem cell marker expression and canonical Wnt pathway activity [58]. Thus, these mouse studies demonstrated that RSPO gain of function, either through overexpression or genetic rearrangement, causally drives intestinal tumorigenesis, wherein deregulation of the proliferative stem cell compartment was shown to be involved. Notably, despite this and the occurrence of EIF3E-RSPO2 fusions and enhanced RSPO2 expression in CRC patients, some controversy exists considering the role of RSPO2 in CRC. Hence, RSPO2 has also been attributed tumor suppressive activities in CRC in some reports [59, 60].
In summary, during the last decade, studies in the intestinal tract have revealed that a subset of CRC patients harbors a gain in RSPO, which can act as oncogenic driver through fueling aberrant expansion of the crypt stem cell compartment. Currently, most of our knowledge on RSPOs in cancer is derived from studies in the intestine, and for this organ, aberrant RSPO activation is recognized as oncogenic driver.
Stomach
As in the intestine, Wnt signaling plays a crucial role in regulating epithelial turnover in the stomach and aberrant activation of the Wnt/β-catenin pathway is an established driver of gastric cancer [61–63]. In the homeostatic stomach, Wnt ligands and RSPO3 are expressed in the stroma neighboring the gland base that constitutes the gastric stem cell compartment [63, 64]. The stem cell compartment of mouse gastric antrum glands is composed of Lgr5+/Axin2+ cells at the base and more apical Lgr5-/Axin2+ cells [63]. Both these stem cell populations are capable of repopulating the gastric gland, giving rise to progenitor and differentiated cell types [37, 63–65]. The Lgr5-/Axin2+ cells appear to be the main driver of homeostatic epithelial turnover, repopulating the glands in 7 days, whereas Lgr5+/Axin2+ show relatively less proliferation and a gland turnover time of 10–14 days [63, 64]. In the stomach, RSPO3 is produced by myofibroblasts neighboring the stem cell compartment and plays a crucial role in regulating stem cell dynamics [64]. Interestingly, RSPO3 induces Lgr5+/Axin2+ stem cells to differentiate into secretory cells with antimicrobial activity, protecting the stem cell compartment against bacterial colonization [65]. In contrast, RSPO3 acts upon Lgr5-/Axin2+ cells by promoting their proliferation and expansion, probably through Lgr4 that is expressed on these cells [64]. Infection with Helicobacter pylori (H. pylori) enhances stromal Rspo3 expression and leads to expansion of proliferative Axin2+ stem cells and hyperplasia [64]. Importantly, H. pylori infection represents the main risk factor for the development of gastric cancer. Enhanced proliferation of gastric stem cells driven by RSPO3 upon H.pylori infection might contribute to this increased risk for cancer development [63, 66]. Despite interesting links have been revealed among stem cell (de)regulation, RSPO3 and H.pylori infection in the stomach, more research is needed to further assess their possible interplay in gastric carcinogenesis. With regard to genetic alterations that might underlie RSPO deregulation in gastric cancer patients, current knowledge is relatively limited. Two cases of RSPO2 gene fusions have been reported in gastric cancer patient-derived xenograft (PDX) material by one group [67].
Steroid hormone regulated organs
Breast
The mammary gland represents another organ where both RSPO and Wnt/β-catenin signaling have been implicated in stem cell regulation during homeostasis and carcinogenesis [68–74]. Although at first glance this involvement might seem comparable to the benchmark situation in the intestinal tract, it is important to realize that the mammary gland is a totally different, uniquely organized epithelial structure that is primarily instructed by steroid hormones estrogen and progesterone. The bilayered mammary epithelium consists of outer basal cells and inner luminal cells, latter being further segregated into luminal progenitor cells and mature luminal cells that express the estrogen receptor (ER) and progesterone receptor (PR). Steroid hormones regulate the exceptionally dynamic remodeling events that occur during puberty, menstrual cycles, pregnancy, lactation and involution. These processes require tightly controlled self-renewal, and the mammary epithelium constitutes a complex and unique, yet incompletely clarified hierarchy of co-existing progenitor and stem cell populations [38, 71, 74–83]. Mammary stem cells (MaSC) with repopulating capacity were firstly described to be part of the basal population [84, 85]. More recent studies report stem cells both in basal and luminal populations, and Wnt/β-catenin signaling has been implicated in the regulation of MaSCs [68, 71, 72, 74, 78–83]. In human breast, RSPO3 is expressed in ALDH+ cells, a cell population that has been proposed to represent (cancer) stem cells and luminal progenitor cells [86–88]. In the mouse mammary gland, RSPO1 has emerged as a key regulator of MaSCs, leading to defects in side-branching and alveologenesis upon its depletion [70, 73, 89, 90]. RSPO1 is produced by luminal progenitor cells, in proximity to mature luminal cells that produce Wnt4, which together cooperate in promoting the self-renewal of MaSCs [70, 73]. Moreover, RSPO1 and Wnt4 are synchronously upregulated upon steroid hormone signals during pregnancy, leading to Wnt/β-catenin signaling potentiation and fueling the expansion of basal cells and luminal progenitor cells [70, 73]. This collaborative RSPO1-Wnt4 action seems to represent the actual downstream executor of stem cell regulation, in response to upstream steroid hormone signals.
In line with the extensive stem cell hierarchy in the mammary gland, breast cancer is exceptionally heterogeneous, and uniquely classified based on the expression of the hormone receptors ER, PR and human epidermal growth factor receptor 2 (HER2). Triple negative breast cancer (TNBC) that lacks expression of these three receptors is the most aggressive subtype with poorest prognosis and most limited options for targeted treatment. Activation of the canonical Wnt pathway in breast cancer has been reported regularly, amongst multiple subtypes, though an association has been proposed especially with TNBC [91–100]. In striking contrast to CRC however, the majority of breast tumors lack mutations in APC or CTNNB1, obscuring the mutational cause of reported intracellular Wnt activation [68, 93]. A possible explanation for this might lie in the different tissue-specific dosages of canonical Wnt signaling activation that support tumor growth, where tumor growth in the mammary epithelium favors a relatively weaker level of Wnt/β-catenin activation compared to its intestinal counterpart [101–103]. Also, activation of the Wnt pathway might result from alterations in other pathway members [68, 104–107]. In this regard, RSPOs might represent additional candidates, supported by the self-renewal promoting effects that RSPO exerts in the normal mammary gland. The first indications that RSPOs might potentially represent mammary oncogenes come from studies in which Rspo1, Rspo2, and Rspo3 were identified as common integration sites of the mouse mammary tumor virus (MMTV) [108–111]. This was further supported by experiments where injection of cell lines overexpressing Rspo2 or Rspo3 in the mouse mammary gland resulted in mammary tumor formation, and distant metastases in case of Rspo2 (ref. [110, 112]). With regard to RSPOs in breast cancer patients, some reports have suggested a protumorigenic role for overexpressed RSPOs, mostly based upon associative studies and in vitro data [113–115]. Overexpression of RSPO2, RSPO3, and RSPO4 have been reported in breast tumors, with a particular occurrence in TNBC and being associated with reduced patient survival in case of RSPO2 upregulation [113, 115, 116]. Notably, EIF3E-RSPO2 fusion transcripts known to occur in CRC were not found in a group of 446 breast tumors tested [113]. This directed approach for these fusions specifically does however not exclude the possibility that other RSPO gene fusions might occur in breast cancer. Notably, the two cell lines HBcc-15 and BT549 that are derived from breast cancer patients do have EIF3E-RSPO2 gene fusions, and siRNA-mediated inhibition of RSPO2 in BT549 cells was shown to reduce the proliferation of this TNBC cell line [113]. Together these data point towards a protumorigenic role for RSPOs in breast cancer, though further research is needed to better establish this.
Ovary
In ovarian development RSPO1 has appeared as a crucial player, regulating female sex determination and ovarian differentiation in cooperation with Wnt4 (ref. [6, 7, 117–119]). RSPO1 and Wnt4 are expressed throughout ovarian development and influence cell proliferation and the entry of germ cells into meiosis by activating the Wnt/β-catenin signaling pathway [117, 119, 120]. In agreement with its essential role in ovarian development, the Wnt/β-catenin pathway has found to be frequently activated in ovarian cancer, being associated with epithelial-to-mesenchymal progression, chemotherapy resistance and poor prognosis [121]. Considering RSPOs in ovarian cancer, in silico analysis suggested relatively high RSPO1 mRNA expression in ovarian cancer, and another study reported high expression of RSPO1 and RSPO3 in ovarian tumor xenograft material [116, 122]. Also, SNPs in the RSPO1 locus have been identified as risk factors for ovarian cancers of serous histology [122, 123].
Moreover, a mouse study demonstrated that continuation of Rspo1 expression after birth, normally downregulated in the ovaries at this stage, resulted in impaired ovulation and Wnt/β-catenin-mediated formation of granulosa cell tumors at the onset of puberty, suggesting that aberrant RSPO1 holds oncogenic potential in the ovaries [124].
Prostate
Wnt/β-catenin signaling is crucial during prostate development and both Wnt and RSPO ligands are expressed within the developing urogenital tract [125–128]. In vitro studies have indicated that RSPOs promote the growth and luminal differentiation in prostate organoid cultures [127, 129]. In prostate cancer, aberrant regulation of RSPOs and Wnt/β-catenin pathway components have been described [130–133]. APC and CTNNB1 mutations are regularly found in prostate cancer [131, 133]. Moreover, RSPO2 gene fusions associated with elevated RSPO2 expression have been identified in prostate cancer patients, that were mutually exclusive with APC and CTNNB1 mutations [131]. Unlike in CRC, these prostate cancer cases harbored fusions of RSPO2 with GRHL2 instead of EIF3E (ref. [131]). Also, RSPO3 has been described as one of the genes being upregulated in prostate tumor stroma compared to healthy stroma [130]. In contrast, another group that studied gene expression data sets reported reduced levels of RSPO3 in prostate tumors compared to healthy tissue, with further expression loss in metastatic disease and RSPO3 loss correlating with an increased risk of relapse [132]. Thus, although RSPO fusions have been identified in prostate cancer patients and several reports have implicated RSPOs in prostate carcinogenesis, some controversy exists on the contribution of RSPOs to prostate cancer development.
Other organs
In addition to aforementioned cancers, RSPO activation has been implicated in tumorigenic processes in more tissues. In the liver, the RSPO-LGR pathway has been defined as a key regulator of zonation, size and regeneration [134, 135]. Several reports have described RSPO2 activation in liver cancer through distinct means [136–139]. Among these, RSPO2 gene fusions have been identified, co-occurring with increased RSPO2 expression levels, nuclear β-catenin localization and upregulation of Wnt target genes, resembling the situation of CRC cases with RSPO2 gene fusions [137]. Several other studies reported subsets of hepatocellular carcinoma that harbor RSPO2 copy number amplifications or enhanced RSPO2 mRNA expression associated with Wnt/β-catenin activation [136, 138, 139]. Also, it has been shown that overexpression of Rspo2 in a Trp53 loss background caused tumor formation in the mouse liver [138]. In lung cancer, a protumorigenic role for RSPOs has been proposed as enhanced expression of RSPO ligands was observed in a subset of lung cancer cases, and enhanced RSPO3 expression was associated with reduced patient survival [116, 140]. These studies reported no underlying RSPO gene fusions, and it was proposed that enhanced RSPO3 expression might have resulted from promoter demethylation and deficiency in tumor suppressor KEAP1 (ref. [116, 140]). Complementary in vitro and in vivo experiments suggested that RSPO3 promotes lung carcinogenesis through LGR4-IQGAP1 signaling [140]. Another group however did report EIF3E-RSPO2 and PTPRK-RSPO3 gene fusions in 1% and 2% of lung cancer patients respectively, being restricted to the squamous subtype of NSCLC [141]. Furthermore, enhanced RSPO expression and a tumor promoting role have also been described in pancreatic cancer and bladder cancer [116, 142, 143].
Therapeutic targeting of RSPO in cancer
Hyperactivation of the Wnt/β-catenin pathway has been linked to tumor development in multiple organs, and the underlying molecular alterations are divergent. In line, compelling efforts have been made to develop therapeutic agents that target the Wnt/β-catenin pathway at various levels, among which those intervening with FZD or LRP receptor activity or Wnt ligand maturation and secretion through porcupine inhibitors (PORCNi) [144]. Also the RSPO receptor LGR5 is subject of investigation as a candidate target for therapeutic intervention in cancer [145]. With regard to Wnt driven cancers, dichotomous distinction can be made between ligand-independent and ligand-dependent cases, including those with APC or CTNNB1 mutations versus those with RSPO or RNF43 mutations respectively [55].
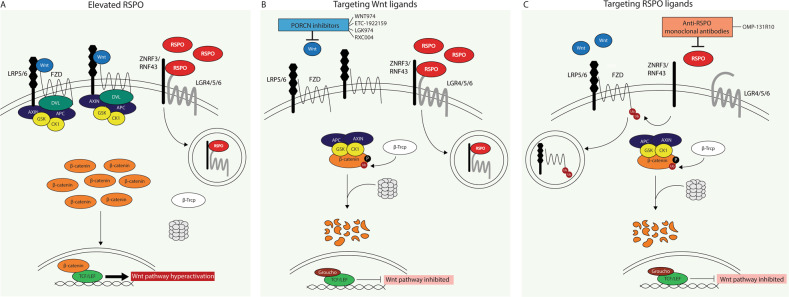
The ligand-dependent cases hold relatively more opportunities for targeted intervention. Specifically, with the growing indications for RSPO gene fusions/upregulation and a concomitant oncogenic role in several cancer types, RSPOs have emerged as promising candidate targets for therapeutic intervention and inherently as potential biomarkers predicting therapy responsiveness. Accordingly, some first studies have been published exploring the possibilities to inhibit tumor growth through targeting RSPO activity. These intervention strategies either directly targeted the RSPO ligands themselves or rather interfered more indirectly with Wnt ligand activity through PORCNi (Fig. 2). As the best described molecular activity of RSPOs lies within their capability. To amplify the signal of canonical Wnt ligands, aberrant RSPO expression would expectedly sensitize tumors to Wnt ligand blockade using PORCNi. Hence, PORCNi block the secretion of functionally active Wnt ligands and in their absence, RSPO ligands are impaired to exert any potentiating effects (Fig. 2B). Several preclinical studies have tested this using PORCNi in cancer cases with RSPO activation specifically [67, 146, 147]. Indeed, it was found consistently that PORCNi effectively reduced tumor growth whilst inducing tumor differentiation in PDX models of RSPO-fusion positive CRC and gastric cancers [67, 146, 147]. Currently, several clinical trials distinctively stratify cancer patients with genetic alterations in RSPO2/3 as inclusion criteria to investigate the efficacy of PORCNi, either or not combined with other drugs [148–151]. As these trials specifically take into consideration the RSPO status of the considerate cancer patients, the drug efficacy data to be obtained by these trials will expectedly provide useful information for further decision making towards targeted intervention strategies in cancer patients with RSPO overactivation.
Fig. 2. Schematic representation of therapeutic targeting opportunities in cancer cases with RSPO overactivation.
A Overexpression of RSPOs induces increased clearing of negative regulators ZNRF3/RNF43 from the membrane, thereby expanding membranous Wnt receptor availability and excessive activation of Wnt ligand-mediated pathway activation. B PORCN inhibitors block the availability of functional Wnt ligands, allowing the destruction complex to form and degrade β-catenin, resulting in inhibition of the Wnt pathway. Indicated PORCNi are tested in clinical trials for solid cancers considering the RSPO status. C Monoclonal anti-RSPO antibodies disable RSPOs to clear negative regulators ZNRF3/RNF43 from the membrane, causing ubiquitination and membrane clearance of Wnt receptors, as such inhibiting Wnt ligands to activate the pathway. Indicated antibody has been tested in a clinical trial for colorectal cancer.
Direct targeting of RSPO proteins themselves with anti-RSPO antibodies represents another possible intervention approach (Fig. 2C). Through this means, RSPO ligands are disabled to clear ZNRF3/RNF43 from the membrane, leading to Wnt receptor degradation and thereby to inhibited Wnt pathway activation. Of additive value, direct targeting of RSPOs might also interfere with potential oncogenic signaling activities beyond stimulating canonical Wnt signaling. Hence, RSPOs have been implicated in other signaling pathways, though possible oncogenic roles there are insufficiently clear yet. In cancer cases with RSPO overactivation, direct targeting of RSPOs themselves might be favorable, and several studies have addressed the efficacy of anti-RSPO antibodies [58, 116, 152]. Chartier et al. generated monoclonal antibodies against RSPO1–3, and showed that these inhibited tumor growth (both as single agent or in combination with chemotherapy) in multiple PDX cancer models with overexpression of the respective RSPO [116]. These included an ovarian tumor with RSPO1, pancreatic and colon tumors with RSPO2, and lung and CRC tumors with RSPO3 overexpression. Despite efficacy in most of the models, a minority of RSPO expressing tumors were not responsive [116]. Another study by Storm et al. used a CRC xenograft model specifically with a PTPRK-RSPO3 gene fusion, and showed that anti-RSPO3 effectively reduced tumor growth and induced differentiation [58]. Both studies demonstrated that differentiation induced by anti-RSPO3 treatment was accompanied by downregulation of Wnt target and stem cell related genes [58, 116]. In addition, another study by Fisher et al. tested anti-RSPO3 treatment on CRC PDX models harboring APC mutations. Although these were not sensitive to anti-RSPO3 treatment only, the combination of anti-RSPO3 with paclitaxel synergistically reduced tumor growth in most cases, being accompanied by reduced nuclear β-catenin, proliferation and CSC frequency against enhanced differentiation [152]. In addition to these results within solid tumor models, a recent study also showed beneficial effects of anti-RSPO3 treatment in certain acute myeloid leukemia PDX models, where anti-RSPO3 treatment effectively inhibited leukemia stem cells without harming healthy stem cells [153].
Recognizing the promising clinical potential of RSPOs as novel therapeutic targets, a clinical trial has been set-up that tested the safety and efficacy of the neutralizing monoclonal anti-RSPO3 antibody OMP131-R10 (Rosmantuzumab) in cancer patients with advanced solid tumors and metastatic CRC [154]. It was reported that OMP131-R10 was well-tolerated by patients, though serum bone markers appeared affected [155]. The trial was unfortunately halted in phase I as a consequence of insufficient evidence for clinical benefit [155]. However it seems that the inclusion criteria for this trial did not take into consideration the RSPO status. In that case, it is unknown whether any and how many patients were included in the trial that actually had a gain in RSPO3 specifically. Therefore, and given the multitude of indications for the relevant oncogenic role of RSPOs, it remains valuable to further investigate the clinical potential of anti-RSPO monoclonal antibodies specifically in cancer patients that harbor RSPO alterations.
Taken together, in line with the growing indications for the clinically relevant oncogenic role of RSPOs, some first avenues have been instigated to explore how we can potentially interfere with RSPO overactivation in cancer. Clinical trials addressing the efficacy of indirect and direct RSPO targeting strategies through PORCNi and anti-RSPO3 antibodies respectively will hopefully provide more insight beneficial to the development of novel treatment strategies against RSPO driven cancer.
Conclusions and perspectives
RSPO ligands are powerful regulators of stem cell maintenance and tissue homeostasis. In accordance with this influential role, aberrant RSPO activation has increasingly been implicated in cancer development over the last decade. RSPO alterations, mostly represented by gene fusions or upregulation, have been reported to occur in patients of multiple cancer types. In addition, several studies have demonstrated that RSPO overactivation causally drives tumorigenesis in the mouse intestine, and provided indications that abnormal expansion of the stem cell compartment seems part of the mechanism. Most of our current knowledge on the molecular activities of RSPOs have been obtained by studies in the intestinal tract. Although these provide solid indications and relevant insight, only the first part of the puzzle seems uncovered, leaving many questions still unanswered. Among these, it remains unclear how the pathologic RSPO alterations are mechanistically achieved. Though specific breakpoints in the RSPO genes as well as specific fusion partner genes are involved in reported RSPO rearrangements, it is currently unknown how and under which conditions the RSPO fusions arise and are selected for along the tumorigenic cascade. A possible cell of origin for RSPO-driven cancer has not been reported yet, and its identification might be complicated by the fact that RSPOs are secreted proteins. Also, it is insufficiently clear what the specific molecular activities are that RSPO proteins instigate on receiving cells and that underpin carcinogenesis. Notably, for all these questions, the answers likely differ per organ.
Forthcoming, a better understanding on the molecular mechanisms of RSPOs with tissue-specific consideration is needed to provide well-founded directions for (pre)clinical studies. Current extensive indications for the oncogenic role of RSPOs however have already instigated the exploration of potential therapeutic opportunities and RSPOs have been recognized as promising therapeutic targets. Preclinical studies demonstrated that PORCNi and anti-RSPO antibodies efficiently inhibited tumor growth in PDX models of cancer with RSPO activation. Moreover, therapeutic targeting through both PORCNi and anti-RSPO3 antibodies are evaluated in clinical trials and will expectedly provide valuable information for further development of novel targeted intervention strategies.
Acknowledgements
This work was financially supported by the Netherlands Organization for Scientific Research (NWO/ZonMW VENI 016.186.138) and the Dutch Cancer Society (KWF Young Investigator Grant 10957).
Author contributions
Review conceptualization by ERMB. EJtS and EB co-wrote the manuscript. All authors have approved the final version.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen J-Z, Wang S, Tang R, Yang Q-Sh, Zhao E, CHao Y, et al. Cloning and identification of a cDNA that encodes a novel human protein with thrombospondin type I repeat domain, hPWTSR. Mol Biol Rep. 2002;29:287–92. doi: 10.1023/a:1020479301379. [DOI] [PubMed] [Google Scholar]
- 2.Nam JS, Turcotte TJ, Smith PF, Choi S, Jeong KY. Mouse cristin/R-spondin family proteins are novel ligands for the frizzled 8 and LRP6 receptors and activate β-catenin-dependent gene expression. J Biol Chem. 2006;281:13247–57. doi: 10.1074/jbc.M508324200. [DOI] [PubMed] [Google Scholar]
- 3.Kamata T, Katsube KI, Michikawa M, Yamada M, Takada S, Mizusawa H. R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim Biophys Acta - Gene Struct Expr. 2004;1676:51–62. doi: 10.1016/j.bbaexp.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Kim KA, Zhao J, Andarmani S, Kakitani M, Oshima T, Binnerts ME, et al. R-spondin proteins: A novel link to β-catenin activation. Cell Cycle. 2006;5:23–6. doi: 10.4161/cc.5.1.2305. [DOI] [PubMed] [Google Scholar]
- 5.Kazanskaya O, Glinka A, del Barco Barrantes I, Stannek P, Niehrs C, Wu W. R-Spondin2 is a secreted activator of Wnt/β-catenin signaling and is required for Xenopus myogenesis. Dev Cell. 2004;7:525–34. doi: 10.1016/j.devcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 6.Parma P, Radi O, Vidal V, Chaboissier MC, Dellambra E, Valentini S, et al. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat Genet. 2006;38:1304–9. doi: 10.1038/ng1907. [DOI] [PubMed] [Google Scholar]
- 7.Tomizuka K, Horikoshi K, Kitada R, Sugawara Y, Iba Y, Kojima A, et al. R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum Mol Genet. 2008;17:1278–91. doi: 10.1093/hmg/ddn036. [DOI] [PubMed] [Google Scholar]
- 8.Nam JS, Park E, Turcotte TJ, Palencia S, Zhan X, Lee J, et al. Mouse R-spondin2 is required for apical ectodermal ridge maintenance in the hindlimb. Dev Biol. 2007;311:124–35. doi: 10.1016/j.ydbio.2007.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki M, Kiyonari H, Nakamura H, Okamoto H. R-spondin2 expression in the apical ectodermal ridge is essential for outgrowth and patterning in mouse limb development. Dev Growth Differ. 2008;50:85–95. doi: 10.1111/j.1440-169X.2007.00978.x. [DOI] [PubMed] [Google Scholar]
- 10.Bell SM, Schreiner CM, Wert SE, Mucenski ML, Scott WJ, Whitsett JA. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development. 2008;135:1049–58. doi: 10.1242/dev.013359. [DOI] [PubMed] [Google Scholar]
- 11.Yamada W, Nagao K, Horikoshi K, Fujikura A, Ikeda E, Inagaki Y, et al. Craniofacial malformation in R-spondin2 knockout mice. Biochem Biophys Res Commun. 2009;381:453–8. doi: 10.1016/j.bbrc.2009.02.066. [DOI] [PubMed] [Google Scholar]
- 12.Aoki M, Mieda M, Ikeda T, Hamada Y, Nakamura H, Okamoto H. R-spondin3 is required for mouse placental development. Dev Biol. 2007;301:218–26. doi: 10.1016/j.ydbio.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 13.Kazanskaya O, Ohkawara B, Heroult M, Wu W, Maltry N, Augustin HG, et al. The Wnt signaling regulator R-spondin 3 promotes angioblast and vascular development. Development. 2008;135:3655–64. doi: 10.1242/dev.027284. [DOI] [PubMed] [Google Scholar]
- 14.Blaydon DC, Ishii Y, O’Toole EA, Unsworth HC, Teh MT, Rüschendorf F, et al. The gene encoding R-spondin 4 (RSPO4), a secreted protein implicated in Wnt signaling, is mutated in inherited anonychia. Nat Genet. 2006;38:1245–7. doi: 10.1038/ng1883. [DOI] [PubMed] [Google Scholar]
- 15.Ishii Y, Wajid M, Bazzi H, Fantauzzo KA, Barber AG, Blaydon DC, et al. Mutations in R-spondin 4 (RSPO4) underlie inherited anonychia. J Invest Dermatol. 2008;128:867–70. doi: 10.1038/sj.jid.5701078. [DOI] [PubMed] [Google Scholar]
- 16.Bergmann C, Senderek J, Anhuf D, Thiel CT, Ekici AB, Poblete-Gutiérrez P, et al. Mutations in the gene encoding the Wnt-signaling component R-spondin 4 (RSPO4) cause autosomal recessive anonychia. Am J Hum Genet. 2006;79:1105–9. doi: 10.1086/509789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brüchle NO, Frank J, Frank V, Senderek J, Akar A, Koc E, et al. RSPO4 is the major gene in autosomal-recessive anonychia and mutations cluster in the furin-like cysteine-rich domains of the Wnt signaling ligand R-spondin 4. J Invest Dermatol. 2008;128:791–6. doi: 10.1038/sj.jid.5701088. [DOI] [PubMed] [Google Scholar]
- 18.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Hao HX, Xie Y, Zhang Y, Zhang O, Oster E, Avello M, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–202. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 20.Koo BK, Spit M, Jordens I, Low TY, Stange DE, Van De Wetering M, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–9. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 21.Zebisch M, Xu Y, Krastev C, Macdonald BT, Chen M, Gilbert RJC, et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat Commun. 2013;4:2787. [DOI] [PMC free article] [PubMed]
- 22.Chen P, Chen X, Lin Z, Dev G, Chen P, Chen X, et al. The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev. 2013;1345–50. [DOI] [PMC free article] [PubMed]
- 23.Xie Y, Zamponi R, Charlat O, Ramones M, Swalley S, Jiang X, et al. Interaction with both ZNRF3 and LGR4 is required for the signalling activity of R-spondin. EMBO Rep. 2013;14:1120–6. doi: 10.1038/embor.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng WC, De Lau W, Madoori PK, Forneris F, Granneman JCM, Clevers H, et al. Structures of Wnt-antagonist ZNRF3 and its complex with R-spondin 1 and implications for signaling. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0083110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D, Huang B, Zhang S, Yu X, Wu W, Wang X. Structural basis for R-spondin receptors. Genes Dev. 2013;27:1339–44. doi: 10.1101/gad.219360.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu K, Xu Y, Rajashankar KR, Robev D, Nikolov DB. Crystal structures of Lgr4 and its complex with R-Spondin1. Structure. 2013;21:1683–9. doi: 10.1016/j.str.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lebensohn AM, Rohatgi R. R-spondins can potentiate WNT signaling without LGRs. Elife. 2018;7:1–18. doi: 10.7554/eLife.33126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohkawara B, Glinka A, Niehrs C. Rspo3 binds syndecan 4 and induces Wnt/PCP signaling via clathrin-mediated endocytosis to promote morphogenesis. Dev Cell. 2011;20:303–14. doi: 10.1016/j.devcel.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Park S, Cui J, Yu W, Wu L, Carmon KS, Liu QJ. Differential activities and mechanisms of the four r-spondins in potentiating wnt/-catenin signaling. J Biol Chem. 2018;293:9759–69. doi: 10.1074/jbc.RA118.002743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szenker-Ravi E, Altunoglu U, Leushacke M, Bosso-Lefèvre C, Khatoo M, Thi Tran H, et al. RSPO2 inhibition of RNF43 and ZNRF3 governs limb development independently of LGR4/5/6. Nature. 2018;557:564–9. doi: 10.1038/s41586-018-0118-y. [DOI] [PubMed] [Google Scholar]
- 31.Dubey R, Kerkhof PVan, Jordens I, Malinauskas T, Pusapati GV, Mckenna JK, et al. R-spondins engage heparan sulfate proteoglycans to potentiate WNT signaling. Elife. 2020;3:1–24. doi: 10.7554/eLife.54469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmon KS, Gong X, Yi J, Thomas A, Liu Q RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc Natl Acad Sci USA. 2014;111:E1221–9. [DOI] [PMC free article] [PubMed]
- 33.Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep. 2011;12:1055–61. doi: 10.1038/embor.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee H, Seidl C, Sun R, Glinka A, Niehrs C. R-spondins are BMP receptor antagonists in Xenopus early embryonic development. Nat Commun. 2020;11:5570. doi: 10.1038/s41467-020-19373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barker N, Van EsJH, Kuipers J, Kujala P, Van Den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 36.Blaas L, Pucci F, Messal HA, Andersson AB, Ruiz EJ, Gerling M, et al. Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat Cell Biol. 2016;18:1346–56. doi: 10.1038/ncb3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 38.De Visser KE, Ciampricotti M, Michalak EM, Tan DWM, Speksnijder EN, Hau CS, et al. Developmental stage-specific contribution of LGR5+ cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol. 2012;228:300–9. doi: 10.1002/path.4096. [DOI] [PubMed] [Google Scholar]
- 39.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc Natl Acad Sci USA. 2011;108:11452–7. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Lau W, Barker N, Low TY, Koo BK, Li VSW, Teunissen H, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–7. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 41.Mustata RC, Van Loy T, Lefort A, Libert F, Strollo S, Vassart G, et al. Lgr4 is required for Paneth cell differentiation and maintenance of intestinal stem cells ex vivo. EMBO Rep. 2011;12:558–64. doi: 10.1038/embor.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greicius G, Kabiri Z, Sigmundsson K, Liang C, Bunte R, Singh MK, et al. PDGFRα+ pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci USA. 2018;115:E3173–81. doi: 10.1073/pnas.1713510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleeman SO, Koelzer VH, Jones HJ, Vazquez EG, Davis H, East JE, et al. Exploiting differential Wnt target gene expression to generate a molecular biomarker for colorectal cancer stratification. Gut. 2019;0:1–12. doi: 10.1136/gutjnl-2019-319126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang E, Yousefi M, Gruenheid S. R-spondins are expressed by the intestinal stroma and are differentially regulated during Citrobacter rodentium- and dssinduced colitis in mice. PLoS One. 2016;11:1–13. doi: 10.1371/journal.pone.0152859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5 + intestinal stem-cell self-renewal. Nature. 2017;545:238–42. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harnack C, Berger H, Antanaviciute A, Vidal R, Sauer S, Simmons A, et al. R-spondin 3 promotes stem cell recovery and epithelial regeneration in the colon. Nat Commun. 2019;10:4368. [DOI] [PMC free article] [PubMed]
- 47.Santos AJM, Lo YH, Mah AT, Kuo CJ. The intestinal stem cell niche: homeostasis and adaptations. Trends Cell Biol. 2018;28:1062–78. doi: 10.1016/j.tcb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castillo‐Azofeifa D, Fazio EN, Nattiv R, Good HJ, Wald T, Pest MA, et al. Atoh1 + secretory progenitors possess renewal capacity independent of Lgr5 + cells during colonic regeneration. EMBO J. 2019;38:1–16. doi: 10.15252/embj.201899984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinmura K, Kahyo T, Kato H, Igarashi H, Matsuura S, Nakamura S, et al. RSPO fusion transcripts in colorectal cancer in Japanese population. Mol Biol Rep. 2014;41:5375–84. doi: 10.1007/s11033-014-3409-x. [DOI] [PubMed] [Google Scholar]
- 50.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–4. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashimoto T, Ogawa R, Yoshida H, Taniguchi H, Kojima M, Saito Y, et al. EIF3E–RSPO2 and PIEZO1–RSPO2 fusions in colorectal traditional serrated adenoma. Histopathology. 2019;75:266–73. doi: 10.1111/his.13867. [DOI] [PubMed] [Google Scholar]
- 52.Sekine S, Ogawa R, Hashimoto T, Motohiro K, Yoshida H, Taniguchi H, et al. Comprehensive characterization of RSPO fusions in colorectal traditional serrated adenomas. Histopathology. 2017;71:601–9. doi: 10.1111/his.13265. [DOI] [PubMed] [Google Scholar]
- 53.Mizuguchi Y, Sakamoto T, Hashimoto T, Tsukamoto S, Iwasa S, Saito Y, et al. Identification of a novel PRR15L-RSPO2 fusion transcript in a sigmoid colon cancer derived from superficially serrated adenoma. Virchows Arch. 2019;475:659–63. doi: 10.1007/s00428-019-02604-x. [DOI] [PubMed] [Google Scholar]
- 54.Sekine S, Yamashita S, Tanabe T, Hashimoto T, Yoshida H, Taniguchi H, et al. Frequent PTPRK-RSPO3 fusions and RNF43 mutations in colorectal traditional serrated adenoma. J Pathol. 2016;239:133–8. doi: 10.1002/path.4709. [DOI] [PubMed] [Google Scholar]
- 55.Kleeman SO, Leedham SJ. Not all wnt activation is equal: Ligand-dependent versus ligand-independent wnt activation in colorectal cancer. Cancers. 2020;12:1–16. doi: 10.3390/cancers12113355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hilkens J, Timmer NC, Boer M, Ikink GJ, Schewe M, Sacchetti A, et al. RSPO3 expands intestinal stem cell and niche compartments and drives tumorigenesis. Gut. 2017;66:1095–105. doi: 10.1136/gutjnl-2016-311606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han T, Schatoff EM, Murphy C, Zafra MP, Wilkinson JE, Elemento O, et al. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun. 2017;8:1–12. doi: 10.1038/ncomms15945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Storm EE, Durinck S, De Sousa E, Melo F, Tremayne J, Kljavin N, Tan C, et al. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature. 2016;529:97–100. doi: 10.1038/nature16466. [DOI] [PubMed] [Google Scholar]
- 59.Wu C, Qiu S, Lu L, Zou J, Li WF, Wang O, et al. RSPO2-LGR5 signaling has tumour-suppressive activity in colorectal cancer. Nat Commun. 2014;5:149. [DOI] [PubMed]
- 60.Dong X, Liao W, Zhang L, Tu X, Hu J, Chen T, et al. RSPO2 suppresses colorectal cancer metastasis by counteracting the Wnt5a/Fzd7-driven noncanonical Wnt pathway. Cancer Lett. 2017;402:153–65. doi: 10.1016/j.canlet.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 61.Clements WM, Wang J, Sarnaik A, Kim OJ, MacDonald J, Fenoglio-Preiser C, et al. β-catenin mutation is a frequent cause of Wnt pathway activation in gastric cancer. Cancer Res. 2002;62:3503–6. [PubMed] [Google Scholar]
- 62.Koushyar S, Powell AG, Vincan E, Phesse TJ Targeting wnt signaling for the treatment of gastric cancer. Int J Mol Sci. 2020;21:3927. [DOI] [PMC free article] [PubMed]
- 63.Fischer AS, Sigal M. The role of Wnt and R-spondin in the stomach during health and disease. Biomedicines. 2019;7:1–15. doi: 10.3390/biomedicines7020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sigal M, Logan CY, Kapalczynska M, Mollenkopf HJ, Berger H, Wiedenmann B, et al. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature. 2017;548:451–5. doi: 10.1038/nature23642. [DOI] [PubMed] [Google Scholar]
- 65.Sigal M, Reinés M, del M, Müllerke S, Fischer C, Kapalczynska M, Berger H, et al. R-spondin-3 induces secretory, antimicrobial Lgr5+ cells in the stomach. Nat Cell Biol. 2019;21:812–23. doi: 10.1038/s41556-019-0339-9. [DOI] [PubMed] [Google Scholar]
- 66.Tomasetti C, Vogelstein B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li C, Cao J, Zhang N, Tu M, Xu F, Wei S, et al. Identification of RSPO2 Fusion Mutations and Target Therapy Using a Porcupine Inhibitor. Sci Rep. 2018;8:1–9. doi: 10.1038/s41598-018-32652-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Schie EH, van Amerongen R. Aberrant WNT/CTNNB1 signaling as a therapeutic target in human breast cancer: weighing the evidence. Front Cell Dev Biol. 2020;8:1–14. doi: 10.3389/fcell.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alexander CM. The Wnt signaling landscape of mammary stem cells and breast tumors. Prog Mol Biol Transl Sci. 2018;153:271–98. doi: 10.1016/bs.pmbts.2017.11.020. [DOI] [PubMed] [Google Scholar]
- 70.Cai C, Yu QC, Jiang W, Liu W, Song W, Yu H, et al. R-spondin1 is a novel hormone mediator for mammary stem cell self-renewal. Genes Dev. 2014;28:2205–18. doi: 10.1101/gad.245142.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/β-catenin- responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11:387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 72.Zeng YA, Nusse R. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell. 2010;6:568–77. doi: 10.1016/j.stem.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joshi PA, Waterhouse PD, Kannan N, Narala S, Fang H, Di Grappa MA, et al. RANK Signaling Amplifies WNT-Responsive Mammary Progenitors through R-SPONDIN1. Stem Cell Rep. 2015;5:31–44. doi: 10.1016/j.stemcr.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badders NM, Goel S, Clark RJ, Klos KS, Kim S, Bafico A, et al. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS One. 2009;4:e6594. [DOI] [PMC free article] [PubMed]
- 75.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes Dev. 2014;28:1143–58. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tharmapalan P, Mahendralingam M, Berman HK, Khokha R. Mammary stem cells and progenitors: targeting the roots of breast cancer for prevention. EMBO J. 2019;38:1–19. doi: 10.15252/embj.2018100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, et al. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 2013;3:70–8. doi: 10.1016/j.celrep.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature. 2014;506:322–7. doi: 10.1038/nature12948. [DOI] [PubMed] [Google Scholar]
- 79.Wang D, Cai C, Dong X, Yu QC, Zhang XO, Yang L, et al. Identification of multipotent mammary stemcells by protein C receptor expression. Nature. 2015;517:81–4. doi: 10.1038/nature13851. [DOI] [PubMed] [Google Scholar]
- 80.Davis FM, Lloyd-Lewis B, Harris OB, Kozar S, Winton DJ, Muresan L, et al. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat Commun. 2016;7:13053. [DOI] [PMC free article] [PubMed]
- 81.Wuidart A, Ousset M, Rulands S, Simons BD, Van Keymeulen A, Blanpain C. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 2016;30:1261–77. doi: 10.1101/gad.280057.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–93. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 83.Scheele CLGJ, Hannezo E, Muraro MJ, Zomer A, Langedijk NSM, Van Oudenaarden A, et al. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature. 2017;542:313–7. doi: 10.1038/nature21046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 85.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 86.Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. [DOI] [PMC free article] [PubMed]
- 87.Colacino JA, Azizi E, Brooks MD, Harouaka R, Fouladdel S, McDermott SP, et al. Heterogeneity of human breast stem and progenitor cells as revealed by transcriptional profiling. Stem Cell Rep. 2018;10:1596–609. doi: 10.1016/j.stemcr.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chadi S, Buscara L, Pechoux C, Costa J, Laubier J, Chaboissier MC, et al. R-spondin1 is required for normal epithelial morphogenesis during mammary gland development. Biochem Biophys Res Commun. 2009;390:1040–3. doi: 10.1016/j.bbrc.2009.10.104. [DOI] [PubMed] [Google Scholar]
- 90.Chadi S, Polyte J, Lefevre L, Castille J, Ehanno A, Laubier J, et al. Phenotypic and molecular alterations in the mammary tissue of R-spondin1 knock-out mice during pregnancy. PLoS One. 2016;11:1–15. doi: 10.1371/journal.pone.0162566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH. Wnt/β-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol. 2010;176:2911–20. doi: 10.2353/ajpath.2010.091125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, et al. β-catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci USA. 2000;97:4262–6. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, et al. Β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol. 2011;24:209–31. doi: 10.1038/modpathol.2010.205. [DOI] [PubMed] [Google Scholar]
- 94.He Y, Liu Z, Qiao C, Xu M, Yu J, Li G. Expression and significance of Wnt signaling components and their target genes in breast carcinoma. Mol Med Rep. 2014;9:137–43. doi: 10.3892/mmr.2013.1774. [DOI] [PubMed] [Google Scholar]
- 95.Jonsson M, Borg Å, Nilbert M, Andersson T. Involvement of adenomatous polyposis coli (APC)/β-catenin signalling in human breast cancer. Eur J Cancer. 2000;36:242–8. doi: 10.1016/s0959-8049(99)00276-2. [DOI] [PubMed] [Google Scholar]
- 96.López-Knowles E, Zardawi SJ, McNeil CM, Millar EKA, Crea P, Musgrove EA, et al. Cytoplasmic localization of â-catenin is a marker of poor outcome in breast cancer patients. Cancer Epidemiol Biomark Prev. 2010;19:301–9. doi: 10.1158/1055-9965.EPI-09-0741. [DOI] [PubMed] [Google Scholar]
- 97.Prasad CP, Gupta SD, Rath G, Ralhan R. Wnt signaling pathway in invasive ductal carcinoma of the breast: Relationship between β-catenin, disheveled and cyclin D1 expression. Oncology. 2008;73:112–7. doi: 10.1159/000120999. [DOI] [PubMed] [Google Scholar]
- 98.Ozaki S, Ikeda S, Ishizaki Y, Kurihara T, Tokumoto N, Iseki M, et al. Alterations and correlations of the components in the Wnt signaling pathway and its target genes in breast cancer. Oncol Rep. 2005;14:1437–43. doi: 10.3892/or.14.6.1437. [DOI] [PubMed] [Google Scholar]
- 99.Karayiannakis AJ, Nakopoulou L, Gakiopoulou H, Keramopoulos A, Davaris PS, Pignatelli M. Expression patterns of β-catenin in in situ and invasive breast cancer. Eur J Surg Oncol. 2001;27:31–6. doi: 10.1053/ejso.1999.1017. [DOI] [PubMed] [Google Scholar]
- 100.Li S, Li S, Sun Y, Li L. The expression of β-catenin in different subtypes of breast cancer and its clinical significance. Tumor Biol. 2014;35:7693–8. doi: 10.1007/s13277-014-1975-0. [DOI] [PubMed] [Google Scholar]
- 101.Gaspar C, Fodde R. APC dosage effects in tumorigenesis and stem cell differentiation. Int J Dev Biol. 2004;48:377–86. doi: 10.1387/ijdb.041807cg. [DOI] [PubMed] [Google Scholar]
- 102.Bakker ERM, Hoekstra E, Franken PF, Helvensteijn W, Van Deurzen CHM, Van Veelen W, et al. β-Catenin signaling dosage dictates tissue-specific tumor predisposition in Apc-driven cancer. Oncogene. 2013;32:4579–85. doi: 10.1038/onc.2012.449. [DOI] [PubMed] [Google Scholar]
- 103.Gaspar C, Franken P, Molenaar L, Breukel C, Van Der Valk M, Smits R, et al. A targeted constitutive mutation in the Apc tumor suppressor gene underlies mammary but not intestinal tumorigenesis. PLoS Genet. 2009;5:e1000547. [DOI] [PMC free article] [PubMed]
- 104.Suzuki H, Toyota M, Caraway H, Gabrielson E, Ohmura T, Fujikane T, et al. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer. 2008;98:1147–56. doi: 10.1038/sj.bjc.6604259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Veeck J, Wild PJ, Fuchs T, Schüffler PJ, Hartmann A, Knüchel R, et al. Prognostic relevance of Wnt-inhibitory factor-1 (WIF1) and Dickkopf-3 (DKK3) promoter methylation in human breast cancer. BMC Cancer. 2009;9:1–13. doi: 10.1186/1471-2407-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Veeck J, Geisler C, Noetzel E, Alkaya S, Hartmann A, Knüchel R, et al. Epigenetic inactivation of the secreted frizzled-related protein-5 (SFRP5) gene in human breast cancer is associated with unfavorable prognosis. Carcinogenesis. 2008;29:991–8. doi: 10.1093/carcin/bgn076. [DOI] [PubMed] [Google Scholar]
- 107.Ai L, Tao Q, Zhong S, Fields CR, Kim WJ, Lee MW, et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27:1341–8. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 108.Callahan R, Mudunuri U, Bargo S, Raafat A, McCurdy D, Boulanger C, et al. Genes affected by mouse mammary tumor virus (MMTV) proviral insertions in mouse mammary tumors are deregulated or mutated in primary human mammary tumors. Oncotarget. 2012;3:1320–34. doi: 10.18632/oncotarget.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Klijn C, Koudijs MJ, Kool J, ten Hoeve J, Boer M, de Moes J, et al. Analysis of tumor heterogeneity and cancer gene networks using deep sequencing of MMTV-induced mouse mammary tumors. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0062113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Theodorou V, Kimm MA, Boer M, Wessels L, Theelen W, Jonkers J, et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet. 2007;39:759–69. doi: 10.1038/ng2034. [DOI] [PubMed] [Google Scholar]
- 111.Gattelli A, Zimberlin MN, Meiss RP, Castilla LH, Kordon EC. Selection of early-occurring mutations dictates hormone-independent progression in mouse mammary tumor lines. J Virol. 2006;80:11409–15. doi: 10.1128/JVI.00234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Klauzinska M, Baljinnyam B, Raafat A, Rodriguez-Canales J, Strizzi L, Endo Greer Y, et al. Rspo2/Int7 regulates invasiveness and tumorigenic properties of mammary epithelial cells. J Cell Physiol. 2012;227:1960–71. doi: 10.1002/jcp.22924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Coussy F, Lallemand F, Vacher S, Schnitzler A, Chemlali W, Caly M, et al. Clinical value of R-spondins in triple-negative and metaplastic breast cancers. Br J Cancer. 2017;116:1595–603. doi: 10.1038/bjc.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tocci JM, Felcher CM, García Solá ME, Kordon EC, R-spondin-mediated WNT. signaling potentiation in mammary and breast cancer development. IUBMB Life. 2020;72:1546–59. doi: 10.1002/iub.2278. [DOI] [PubMed] [Google Scholar]
- 115.Tocci JM, Felcher CM, García Sola ME, Goddio MV, Zimberlin MN, Rubinstein N, et al. R-Spondin3 is associated with basal-progenitor behavior in normal and tumor mammary cells. Cancer Res. 2018;78:4497–511. doi: 10.1158/0008-5472.CAN-17-2676. [DOI] [PubMed] [Google Scholar]
- 116.Chartier C, Raval J, Axelrod F, Bond C, Cain J, Dee-Hoskins C, et al. Therapeutic targeting of tumor-derived r-spondin attenuates b-catenin signaling and tumorigenesis in multiple cancer types. Cancer Res. 2016;76:713–23. doi: 10.1158/0008-5472.CAN-15-0561. [DOI] [PubMed] [Google Scholar]
- 117.Chassot A, Ranc F, Gregoire EP, Roepers- HL, Taketo MM, Camerino G, et al. Activation of b -catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet. 2008;17:1264–77. [DOI] [PubMed]
- 118.Maatouk DM, Mork L, Chassot A, Chaboissier M, Capel B. Disruption of mitotic arrest precedes precocious differentiation and transdifferentiation of pregranulosa cells in the perinatal Wnt4 mutant ovary. Dev Biol. 2013;383:295–306. doi: 10.1016/j.ydbio.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chassot AA, Gregoire EP, Lavery R, Taketo MM, de Rooij DG, Adams IR, et al. RSPO1/β-Catenin signaling pathway regulates oogonia differentiation and entry into meiosis in the mouse fetal ovary. PLoS One. 2011;6:e25641. [DOI] [PMC free article] [PubMed]
- 120.Chassot AA, Bradford ST, Auguste A, Gregoire EP, Pailhoux E, de Rooij DG, et al. WNT4 and RSPO1 together are required for cell proliferation in the early mouse gonad. Dev. 2012;139:4461–72. doi: 10.1242/dev.078972. [DOI] [PubMed] [Google Scholar]
- 121.Teeuwssen Fodde. Wnt signaling in ovarian cancer stemness, EMT, and therapy resistance. J Clin Med. 2019;8:1658. doi: 10.3390/jcm8101658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015;47:164–71. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schindler AJ, Watanabe A, Howell SB. LGR5 and LGR6 in stem cell biology and ovarian cancer. Oncotarget. 2018;9:1346–55. doi: 10.18632/oncotarget.20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.De Cian MC, Pauper E, Bandiera R, Vidal VPI, Sacco S, Gregoire EP, et al. Amplification of R-spondin1 signaling induces granulosa cell fate defects and cancers in mouse adult ovary. Oncogene. 2017;36:208–18. doi: 10.1038/onc.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Francis JC, Thomsen MK, Taketo MM, Swain A β-catenin is required for prostate development and cooperates with Pten loss to drive invasive carcinoma. PLoS Genet. 2013;9:e1003180. [DOI] [PMC free article] [PubMed]
- 126.Simons BW, Hurley PJ, Huang Z, Ross AE, Miller R, Marchionni L, et al. Wnt signaling though beta-catenin is required for prostate lineage specification. Dev Biol. 2012;371:246–55. doi: 10.1016/j.ydbio.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luo W, Rodriguez M, Valdez JM, Zhu X, Tan K, Li D, et al. Lgr4 is a key regulator of prostate development and prostate stem cell differentiation. Stem Cells. 2013;31:2492–505. doi: 10.1002/stem.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mehta V, Abler LL, Keil KP, Schmitz CT, Joshi PS, Vezina CM. Atlas of Wnt and R-spondin gene expression in the developing male mouse lower urogenital tract. Dev Dyn. 2011;240:2548–60. doi: 10.1002/dvdy.22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, Van Boxtel R, Wongvipat J, et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell. 2014;159:163–75. doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Planche A, Bacac M, Provero P, Fusco C, Delorenzi M, Stehle JC, et al. Identification of prognostic molecular features in the reactive stroma of human breast and prostate cancer. PLoS One. 2011;6:e18640. [DOI] [PMC free article] [PubMed]
- 131.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mesci A, Lucien F, Huang X, Wang EH, Shin D, Meringer M, et al. RSPO3 is a prognostic biomarker and mediator of invasiveness in prostate cancer. J Transl Med. 2019;17:1–11.. doi: 10.1186/s12967-019-1878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Murillo-Garzón V, Kypta R. WNT signalling in prostate cancer. Nat Rev Urol. 2017;14:683–96. doi: 10.1038/nrurol.2017.144. [DOI] [PubMed] [Google Scholar]
- 134.Planas-Paz L, Orsini V, Boulter L, Calabrese D, Pikiolek M, Nigsch F, et al. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat Cell Biol. 2016;18:467–79. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- 135.Rocha AS, Vidal V, Mertz M, Kendall TJ, Charlet A, Okamoto H, et al. The angiocrine factor rspondin3 is a key determinant of liver zonation. Cell Rep. 2015;13:1757–64. doi: 10.1016/j.celrep.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 136.Kan Z, Zheng H, Liu X, Li S, Barber TD, Gong Z, et al. Whole-genome sequencing identifies recurrent mutations in hepatocellular carcinoma. Genome Res. 2013;23:1422–33. doi: 10.1101/gr.154492.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Longerich T, Endris V, Neumann O, Rempel E, Kirchner M, Abadi Z, et al. RSPO2 gene rearrangement: A powerful driver of β-catenin activation in liver tumours. Gut. 2019;68:1287–96. doi: 10.1136/gutjnl-2018-317632. [DOI] [PubMed] [Google Scholar]
- 138.Conboy CB, Vélez‐Reyes GL, Tschida BR, Hu H, Kaufmann G, Koes N, et al. R‐spondin 2 drives liver tumor development in a yes‐associated protein‐dependent manner. Hepatol Commun. 2019;3:1496–509. doi: 10.1002/hep4.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, et al. Genomic portrait of resectable hepatocellular carcinomas: Implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–82. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- 140.Gong X, Yi J, Carmon KS, Crumbley CA, Xiong W, Thomas A, et al. Aberrant RSPO3-LGR4 signaling in Keap1-deficient lung adenocarcinomas promotes tumor aggressiveness. Oncogene. 2015;34:4692–701. doi: 10.1038/onc.2014.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Karkera J, Martinez G, Bell K, Portale J, Gaffney D, Lorenzi MV, et al. Identification of R-spondin fusions in NSCLC. [abstract]. Proc AACR-IASLC Jt Conf Mol Origigins Lung cancer.
- 142.Ilmer M, Boiles AR, Regel I, Yokoi K, Michalski CW, Wistuba II, et al. RSPO2 enhances canonical wnt signaling to confer stemness-associated traits to susceptible pancreatic cancer cells. Cancer Res. 2015;75:1883–96. doi: 10.1158/0008-5472.CAN-14-1327. [DOI] [PubMed] [Google Scholar]
- 143.Chen Z, Zhou L, Chen L, Xiong M, Kazobinka G, Pang Z, et al. RSPO3 promotes the aggressiveness of bladder cancer via Wnt/β-catenin and Hedgehog signaling pathways. Carcinogenesis. 2019;40:360–9. doi: 10.1093/carcin/bgy140. [DOI] [PubMed] [Google Scholar]
- 144.Blagodatski A, Poteryaev D, Katanaev VL. Targeting the Wnt pathways for therapies. Mol Cell Ther. 2014;2:28. doi: 10.1186/2052-8426-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morgan RG, Mortensson E, Williams AC. Targeting LGR5 in Colorectal Cancer: Therapeutic gold or too plastic? Br J Cancer. 2018;118:1410–8. doi: 10.1038/s41416-018-0118-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Madan B, Ke Z, Harmston N, Ho SY, Frois AO, Alam J, et al. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene. 2016;35:2197–207. doi: 10.1038/onc.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Picco G, Petti C, Centonze A, Torchiaro E, Crisafulli G, Novara L, et al. Loss of AXIN1 drives acquired resistance to WNT pathway blockade in colorectal cancer cells carrying RSPO 3 fusions. EMBO Mol Med. 2017;9:293–303. doi: 10.15252/emmm.201606773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Study of WNT974 in Combination With LGX818 and Cetuximab in Patients With BRAF-mutant Metastatic Colorectal Cancer (mCRC) and Wnt Pathway Mutations. Available from: https://clinicaltrials.gov/ct2/show/NCT02278133.
- 149.A Study to Evaluate the Safety and Tolerability of ETC-1922159 as a Single Agent and in Combination With Pembrolizumab in Advanced Solid Tumours. Available from: https://clinicaltrials.gov/ct2/show/NCT02521844.
- 150.A study of LGK974 in Patients With Malignancies Dependent on Wnt Ligands. Available from: https://clinicaltrials.gov/ct2/show/NCT01351103.
- 151.A study to Assess Efficacy of RXC004 +/- Nivolumab in Ring Finger Protein 43 (RNF43) or R-spondin (RSPO) Aberrated, Metastatic, Microsatellite Stable, Colorectal Cancer After Progression on Standard of Care (SOC). Available from: https://clinicaltrials.gov/ct2/show/NCT04907539.
- 152.Fischer MM, Yeung VP, Cattaruzza F, Hussein R, Yen WC, Murriel C, et al. RSPO3 antagonism inhibits growth and tumorigenicity in colorectal tumors harboring common Wnt pathway mutations. Sci Rep. 2017;7:1–9. doi: 10.1038/s41598-017-15704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Salik B, Yi H, Hassan N, Santiappillai N, Vick B, Connerty P, et al. Targeting RSPO3-LGR4 signaling for leukemia stem cell eradication in acute myeloid leukemia. Cancer Cell. 2020;38:263–278. doi: 10.1016/j.ccell.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 154.A Phase 1a/b Dose Escalation Study of the Safety, Pharmacokinetics, and Pharmacodynamics of OMP-131R10. Available from: https://clinicaltrials.gov/ct2/show/NCT02482441.
- 155.Bendell J, Eckhardt SG, Hochster HS, Morris VK, Strickler J, Kapoun AM, et al. Initial results from a phase 1a / b study of OMP-131R10, a first-in-class anti-RSPO3 antibody, in advanced solid tumors and previously treated metastatic colorectal cancer (CRC) Eur J Cancer. 2016;69:S29–30. [Google Scholar]