Abstract
Host cellular receptors play key roles in the determination of virus tropism and pathogenesis. However, little is known about SARS-CoV-2 host receptors with the exception of ACE2. Furthermore, ACE2 alone cannot explain the multi-organ tropism of SARS-CoV-2 nor the clinical differences between SARS-CoV-2 and SARS-CoV, suggesting the involvement of other receptor(s). Here, we performed genomic receptor profiling to screen 5054 human membrane proteins individually for interaction with the SARS-CoV-2 capsid spike (S) protein. Twelve proteins, including ACE2, ASGR1, and KREMEN1, were identified with diverse S-binding affinities and patterns. ASGR1 or KREMEN1 is sufficient for the entry of SARS-CoV-2 but not SARS-CoV in vitro and in vivo. SARS-CoV-2 utilizes distinct ACE2/ASGR1/KREMEN1 (ASK) receptor combinations to enter different cell types, and the expression of ASK together displays a markedly stronger correlation with virus susceptibility than that of any individual receptor at both the cell and tissue levels. The cocktail of ASK-related neutralizing antibodies provides the most substantial blockage of SARS-CoV-2 infection in human lung organoids when compared to individual antibodies. Our study revealed an interacting host receptome of SARS-CoV-2, and identified ASGR1 and KREMEN1 as alternative functional receptors that play essential roles in ACE2-independent virus entry, providing insight into SARS-CoV-2 tropism and pathogenesis, as well as a community resource and potential therapeutic strategies for further COVID-19 investigations.
Subject terms: Proteomic analysis, Mechanisms of disease
Introduction
The global outbreak of COVID-19 caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a severe threat to human health.1,2 SARS-CoV-2, a member of the beta-coronavirus genus, is closely related to SARS-CoV, and both viruses use ACE2 as an entry receptor.3–5 SARS-CoV-2 is more than a respiratory virus, causing a complex array of symptoms and exhibiting multiorgan tropism.2,6,7 Host cellular receptors play key roles in determining viral tropism and pathogenesis. Some viruses can bind to multiple host receptors for viral attachment, cell entry, and diverse specific host responses, including inducing cytokine secretion, stimulation of the immune responses, and alteration of virus budding and release.8–11
However, apart from ACE2, little is known about SARS-CoV-2 receptors. Growing evidence suggest the involvement of other receptor(s) in SARS-CoV-2 host interactions. Although SARS-CoV-2 and SARS-CoV both share ACE2 as an entry receptor, the primary infection sites and clinical manifestations of these viruses exhibit significant differences.1,2,12–15 Additionally, SARS-CoV-2 has been detected in tissues with little ACE2 expression, including the liver, brain, and blood, and even the lung, where only a small subset of cells express ACE2.16,17 Moreover, several large-scale single-cell transcriptome analyses of patients with COVID-19 revealed many virus-positive cells without ACE2 expression,18,19 suggesting that SARS-CoV-2 might infect cells in an ACE2-independent manner. Therefore, a comprehensive investigation of SARS-CoV-2-interacting host receptome is needed.
The identification of receptors from virus-susceptible cells is limited to membrane proteins on specific cell types. We previously investigated ligand-receptor interactions using a cell-based assay, in which receptor-expressing cells were incubated with a tagged ligand and then an anti-tag for labeling and detection.20–22 This assay closely mimics ligand-receptor interactions that occur under physiological conditions and is regularly used to validate novel specific ligand-receptor interactions. Based on this method, we developed a high-throughput receptor profiling system that covers nearly all types of single- and multi-transmembrane human membrane proteins and used it to identify SARS-CoV-2 cellular receptors.
In this study, we identified ten previously uncharacterized receptor-like host factors that exhibit diverse binding properties to the spike (S) of SARS-CoV-2. Among them, ASGR1 or KREMEN1 is sufficient for the entry of SARS-CoV-2 but not SARS-CoV in vitro and in vivo. SARS-CoV-2 can infect cells via ACE2-dependent and ACE2-independent routes, in the latter ASGR1 and KREMEN1 play critical roles. The combined expression level of ASGR1, KREMEN1 and ACE2, correlates much better with SARS-CoV-2 susceptibility than that of any individual receptor at both the cell and tissue levels, suggesting that ASK receptors underlie SARS-CoV-2 tropism. Compared with the results obtained with individual antibodies, the blockage of ASK receptors simultaneously using a neutralizing antibody cocktail provides the most substantial inhibition of SARS-CoV-2 infection in human cell lines and lung organoids. Our study revealed an interacting host receptome of SARS-CoV-2 and demonstrated ASGR1 and KREMEN1 as alternative functional receptors, providing insight into SARS-CoV-2 tropism and pathogenesis as well as useful resources and potential drug targets for further COVID-19 investigations.
Results
Identification of SARS-CoV-2 S-binding membrane proteins
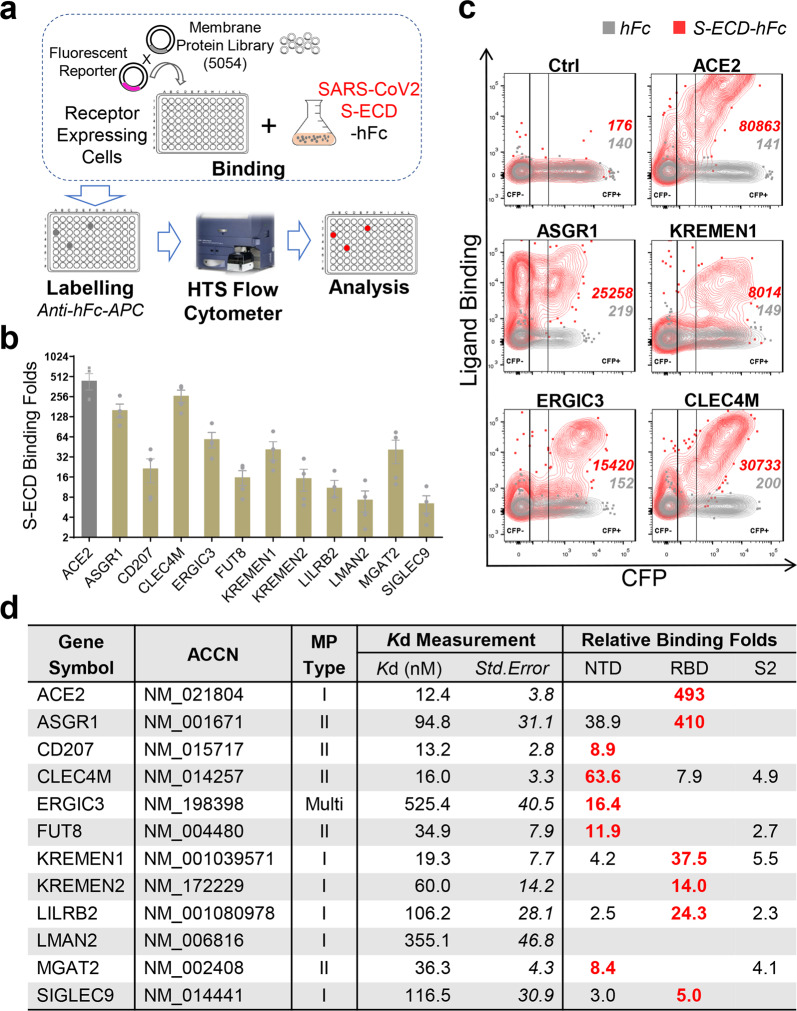
Given SARS-CoV-2 S protein is the major receptor-binding protein on the virion, we performed receptor profiling using S protein as the target. A total of 5054 human membrane protein-encoding genes (91.6% of the predicted human membrane proteins) were expressed individually on human HEK293E cells, and their binding with the extracellular domain of S protein (S-ECD) was measured (Fig. 1a; Supplementary information, Table. S1). Twelve membrane proteins were identified that specifically interact with the S-ECD (Fig. 1b, c; Supplementary information, Fig. S1a), including the previously reported ACE24,5 and SARS-CoV-specific CLEC4M (L-SIGN).23
Fig. 1. Genomic receptor profiling identifies twelve SARS-CoV-2 S-binding membrane proteins.
a Scheme of genomic receptor profiling. Plasmids encoding 5054 human membrane proteins were individually co-transfected with a CFP reporter into HEK293E cells. Cells were incubated with SARS-CoV-2 S-ECD-hFc protein, labeled using anti-hFc-APC antibody, then the binding was measured by flow cytometry. b Profiling identified S protein-binding receptor-like host factors. The relative binding of factors with S-ECD-hFc compared to that with hFc control in CFP+ cells were presented (means ± SEM, n = 4). c Representative flow dot plot showing the binding of S-ECD with top-ranked factors. d Characteristics of the identified interactions. ACCN, accession number; MP Type, membrane protein type. Binding Folds, relative binding of hFc-tagged NTD/RBD/S2 to hFc control. Major binding domain on S protein for each factor was labeled red.
The dissociation constants (Kd) of these interactions ranged from 12.4 to 525.4 nM (Fig. 1d; Supplementary information, Fig. S1b). ACE2 bound to S-ECD with a Kd of 12.4 nM, similar to the previously reported Kd of 15.3 nM,24 and ACE2, CD207, CLEC4M, and KREMEN1 all showed high-affinity with the S protein, with comparable Kd values. The binding domains on the S protein, including the receptor-binding domain (RBD), N-terminal domain (NTD) and S2 domain, were also examined. The RBD and NTD are the major binding sites for these membrane proteins. ACE2 only binds to the RBD, while CD207 and ERGIC3 bind exclusively with the NTD. The other proteins can bind to at least two domains, with CLEC4M, KREMEN1, and LILRB2 binding to all three domains, showing the highest binding with NTD, RBD, and RBD, respectively (Fig. 1d; Supplementary information, Fig. S2). Overall, these receptor-like host factors showed diverse S-binding patterns, and they have a diverse range of biological functions and signaling properties (Supplementary information, Fig. S3).
KREMEN1 and ASGR1 mediate SARS-CoV-2 entry
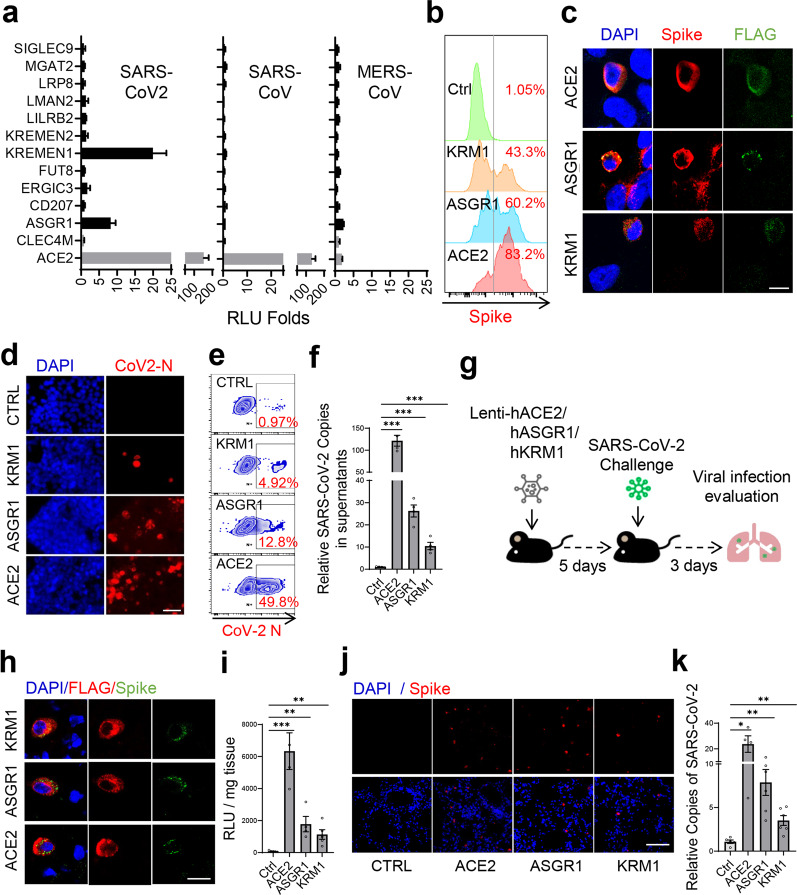
To determine whether these host factors can mediate virus entry independent of ACE2, ACE2 was further knocked out in HEK293T cells (Supplementary information, Fig. S4a, b), which exhibited markedly low sensitivity to SARS-CoV-2 and SARS-CoV.3,4,13 We ectopically expressed the factors in ACE2-KO HEK293T cells and infected cells separately with S-pseudotyped SARS-CoV-2, SARS-CoV or MERS-CoV. KREMEN1-expressing cells showed clear infection of SARS-CoV-2, as did ASGR1, although to a lower extent (Fig. 2a). ACE2 did not bind with ASGR1 or KREMEN1 (Supplementary information, Fig. S4c), further excluding the possibility that these two factors function through ACE2. Both KREMEN1 and ASGR1 are specific to SARS-CoV-2, whereas ACE2 mediates the entry of both SARS-CoV-2 and SARS-CoV (Fig. 2a). KREMEN1 and ASGR1 enabled efficient attachment of S-pseudotyped SARS-CoV-2 particles to the cell surface (Fig. 2b), and S protein was found to be co-localized with these two proteins after infection (Fig. 2c). ASGR1- and KREMEN1-dependent virus entry was confirmed with patient-derived authentic SARS-CoV-2, with ASGR1 promoting higher levels of viral infection and progeny release (Fig. 2d–f; Supplementary information, Fig. S4d). Thus, these results indicated that ASGR1 and KREMEN1 facilitate SARS-CoV-2 entry in vitro.
Fig. 2. KREMEN1 and ASGR1 directly mediate SARS-CoV-2 entry both in vitro and in vivo.
a Identified S-binding membrane proteins were individually ectopically expressed in ACE2-KO HEK293T cells, and the cells were then infected with S-pseudotyped SARS-CoV-2, SARS-CoV, and MERS-CoV, separately. Relative luciferase activities (RLU) to that of empty vector-transfected cells were measured 60 h post-infection (means ± SD, n = 3). b KREMEN1-, ASGR1-, or ACE2-transfected ACE2-KO HEK293T cells were incubated with S-pseudotyped SARS-CoV-2 on ice for 1 h, and viral attachment to cell surface was measured by spike staining and flow cytometry. c ACE2-KO HEK293T cells expressing FLAG-tagged KREMEN1, ASGR1, or ACE2 were infected with S-pseudotyped SARS-CoV-2 for 48 h, immunofluorescence was performed with antibodies against FLAG and S protein (scale bar, 50 μm). d–f KREMEN1-, ASGR1-, or ACE2-transfected ACE2-KO HEK293T cells were infected with authentic SARS-CoV-2, and immunofluorescence (d) and flow cytometry (e) were performed with an antibody against the N protein of SARS-CoV-2 72 h post-infection (scale bar, 50 μm); the viral titers in the cell supernatants (f) were measured by RT-qPCR of SARS-CoV-2 ORF1ab gene (means ± SEM, n = 4). g Experimental strategy to study the role of ASK receptors in mediating SARS-CoV-2 infection in mouse models. h, i Immunofluorescence staining of S protein (anti-S) and ASK receptors (anti-FLAG) (h) of the lungs of Lenti-ACE2/ASGR1/KREMEN1-transduced mice challenged with S-pseudotyped SARS-CoV-2 (scale bar, 200 μm); the viral loads in the lungs of each group (i) were measured by luciferase assay (means ± SEM, n = 4–6 biologically independent mice from each group). j, k S protein staining (j) of the lungs of Lenti-ACE2/ASGR1/KREMEN1-transduced mice challenged with SARS-CoV-2 (scale bar, 200 μm); the viral loads in the lungs of each group (k) were measured by RT-qPCR of the SARS-CoV-2 ORF1ab gene (means ± SEM, n = 4–6 biologically independent mice from each group). The statistical significance was evaluated by unpaired two-tailed Student’s t-tests, *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate whether ASGR1 and KREMEN1 can mediate SARS-CoV-2 entry in vivo, we prepared lentiviral particles encoding human ASGR1, KREMEN1 or ACE2, to transduce mice intranasally for 5 days, and then followed by challenge with S-pseudotyped SARS-CoV-2 for another 3 days (Fig. 2g). Compared to control mice, luciferase activity was significantly higher in the lungs of ASGR1-, KREMEN1- and ACE2-transduced mice, and S protein was observed to be co-localized with ASGR1, KREMEN1 and ACE2, separately (Fig. 2h, i). Given that mouse Ace2 exhibits low S-binding activity to support efficient SARS-CoV-2 entry,5,25,26 and similar findings were observed with mouse Kremen1 and Asgr1 (Supplementary information, Fig. S5), the above results indicated that human ASGR1 and KREMEN1 can mediate SARS-CoV-2 entry in vivo. For further confirmation, we challenged the transduced mice with authentic SARS-CoV-2. Although no significant difference in body weight was observed in hACE2-transgenic or Ad5-hACE2-transduced mice,27,28 which might be due to the short time of infection and limited number of cells being transduced (Supplementary information, Fig. S4e, f), immuno-fluorescence results showed that SARS-CoV-2 was clearly detected in the lungs of ACE2-expressing mice, and also in ASGR1- and KREMEN1-expressing mice, while low in control mice (Fig. 2j). Consistently, the virus titers in the lungs of ASGR1- and KREMEN1-expressing mice were significantly higher than in control mice (Fig. 2k). Taken together, these results demonstrated that ASGR1 and KREMEN1 are able to mediate SARS-CoV-2 entry both in vitro and in vivo.
Interaction of KREMEN1 and ASGR1 with SARS-CoV-2 S protein
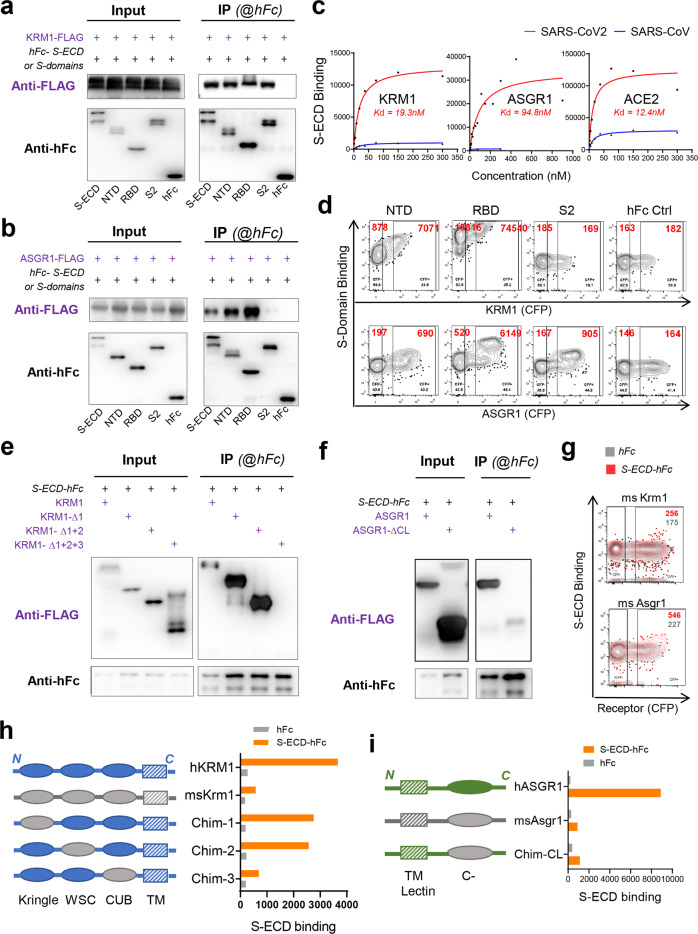
The direct interaction of SARS-CoV-2 S protein with KREMEN1 and ASGR1 was confirmed by co-immunoprecipitation (Co-IP) (Fig. 3a, b; Supplementary information, Fig. S6a, b). KREMEN1 and ASGR1 bound to S-ECD with Kds of 19.3 nM and 94.8 nM, respectively, the former Kd being comparable to that of ACE2 (12.4 nM). ACE2 exhibits the highest maximum binding capacity for S-ECD, being ~3- and ~10-fold that of ASGR1 and KREMEN1, respectively (Fig. 3c), consistent with the SARS-CoV-2 sensitivities of cells expressing these receptors (Fig. 2). In accordance with the finding that KREMEN1 and ASGR1 do not mediate SARS-CoV entry (Fig. 2a), both receptors showed low affinity to the SARS-CoV S protein (Fig. 3c; Supplementary information, Fig. S6c), confirming that KREMEN1 and ASGR1 are specific receptors of SARS-CoV-2 but not SARS-CoV.
Fig. 3. Interaction of SARS-CoV-2 spike with KREMEN1 and ASGR1.
a, b Co-immunoprecipitation to detect the interaction of full-length KREMEN1 (a) or ASGR1 (b) with S-ECD, NTD, RBD or S2 domains respectively. c KREMEN1, ASGR1 or ACE2 expressing HEK293E cells were incubated with different concentrations of S-ECD-hFc of SARS-CoV2 or SARS-CoV, separately, and S-ECD binding was monitored by flow cytometry to determine Kd. d Binding of NTD, RBD, or S2 domains with HEK293E cells expressing the KREMEN1 or ASGR1. e, f Co-immunoprecipitation to detect the interaction of S-ECD with truncated forms of KREMEN1 (e) or ASGR1 (f). g Binding of S-ECD with HEK293E cells expressing mouse Kremen1 or Asgr1. h, i Chimeric KREMEN1 (h) and ASGR1(i) were constructed and transfected into HEK293E cells, and S-ECD binding were measured by flow cytometry.
ACE2 binds exclusively to the RBD, KREMEN1 binds to all three domains of S-ECD, with highest affinity for the RBD, and ASGR1 binds to both the NTD and the RBD, the latter also with higher affinity (Figs. 1d, 3a, b, d). Evidence indicates that the NTD is involved in the entry of coronaviruses including SARS-CoV-2,29–32 suggesting that ASGR1 and KREMEN1 may play a critical role in SARS-CoV-2 infection. We determined which domain(s) on KREMEN1 and ASGR1 are essential for S binding. KREMEN1 has three domains on its extracellular part, including Kringle, WSC and CUB domains, while there is one C-type lectin domain on that of ASGR1. Deletion of Kringle and WSC domains did not affect the S-binding activity of KREMEN1, which was abrogated by further deletion of the CUB domain, indicating that CUB domain is important for KREMEN1 to bind S protein (Fig. 3e). C-type lectin domain is important for ASGR1 to bind S protein, and deletion of C-type lectin domain abolished the interaction of ASGR1 with S-ECD (Fig. 3f). Because mouse Kremen1 and Asgr1 displayed low S-binding activities (Fig. 3g; Supplementary information, Fig. S5), chimeric human KREMEN1 or ASGR1 was then constructed. Consistently, substitution of CUB domain, but not Kringle or WSC domain, to that of mouse homolog substantially reduced the binding of human KREMEN1 to S-ECD, so did the C-type lectin domain for ASGR1 (Fig. 3h, i), further confirming that CUB domain on KREMEN1 and C-type lectin domain on ASGR1 are essential for these receptors to bind S protein.
KREMEN1 is a high-affinity DKK1 receptor that antagonizes canonical Wnt signaling33 and is also the entry receptor for a major group of enteroviruses.34 ASGR1 is an endocytic recycling receptor involved in serum glycoprotein homeostasis35 and has been reported to facilitate the entry of Hepatitis C virus.36 Because all three receptors directly mediate SARS-CoV-2 entry, we refer to ASGR1, KREMEN1, and ACE2 as the ASK (ACE2/ASGR1/KREMEN1) entry receptors.
KREMEN1 and ASGR1 play essential roles for ACE2-independent SARS-CoV-2 entry
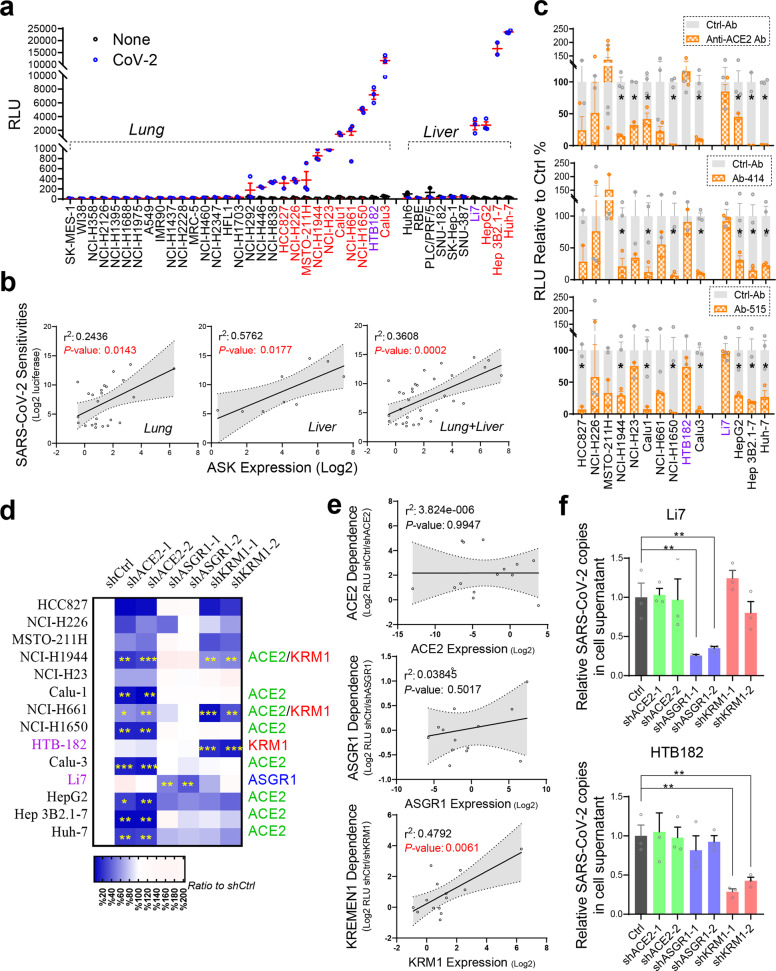
We then analyzed whether endogenous ASGR1 and KREMEN1 regulate SARS-CoV-2 entry and their relationship to ACE2. Given that lung and liver are the two major organs most frequently infected or damaged in patients with COVID-19,1,2,37,38 we screened 39 lung and liver cell lines with S-pseudotyped SARS-CoV-2. Thirteen lung cell lines and four liver cell lines were sensitive to SARS-CoV-2, showing significantly higher luciferase activities than mock-infected cells, including the commonly used lung cell lines Calu3 and Calu1 and the liver cell line Huh-7 (Fig. 4a). When considered individually, none of the ASK receptors showed a strong correlation with viral sensitivity in lung or liver cell lines (Supplementary information, Fig. S7a), but when the expression of the ASK receptors was combined, the correlation with viral sensitivity was highly significant in lung and/or liver cell lines (Fig. 4b), indicating that the ASK combination more closely predicts SARS-CoV-2 sensitivity.
Fig. 4. KREMEN1 and ASGR1 play essential roles in ACE2-independent SARS-CoV-2 entry.
a, b Twenty-nine lung cancer cell lines and ten liver cancer cell lines were infected with or without S-pseudotyped SARS-CoV-2. a Luciferase activities were measured 60 h post-infection (means ± SEM, n = 3). b Correlations of virus sensitivities with the combined ASK expression levels in the tested lung and/or liver cell lines (two-sided Pearson correlation analysis). c High SARS-CoV-2-sensitive cell lines (labeled red or purple in a) were infected with pseudotyped SARS-CoV-2 in the presence of different ACE2-S-blocking antibodies (at a final concentration of 2 µg/mL). Luciferase activity relative to control antibody treatment was measured 60 h post-infection (means ± SEM, n = 3). d, e KD of ACE2, ASGR1 or KREMEN1 in the indicated cell lines followed by infection with pseudotyped SARS-CoV-2. Luciferase activity relative to control was measured 60 h post-infection and shown as a heatmap (d) (means ± SEM, n = 3). The logarithm of the effect of receptor shRNA-mediated KD on virus entry was correlated with the expression level of corresponding receptors (e) (two-sided Pearson correlation analysis). f KD of ACE2, ASGR1 or KREMEN1 in HTB-182 and Li7 cells followed by infection with authentic SARS-CoV-2 at MOI of 1. Expression of the SARS-CoV-2 ORF1ab gene was examined in the cells 72 h post-infection by RT-qPCR (means ± SEM, n = 3). The statistical significance was evaluated by unpaired two-tailed Student’s t-tests, *P < 0.05, **P < 0.01, ***P < 0.001.
To determine whether cell-line viral entry requires ACE2, we selected 14 sensitive cell lines with luciferase activities higher than 500 and treated them with a commercial ACE2 neutralizing monoclonal antibody that blocks the ACE2–S interaction and SARS-CoV-2 entry (Supplementary information, Fig. S7b). In eight cell lines, SARS-CoV-2 S-mediated entry was significantly inhibited by ACE2 neutralizing antibody, whereas in the lung cell line HTB-182 and liver cell line Li7, which are highly sensitive to SARS-CoV-2, viral entry was unaffected (Fig. 4c). To verify this result, we treated the cells with two potent SARS-CoV-2 neutralizing monoclonal antibodies, Ab-414 and Ab-515, that we isolated from convalescent COVID-19 patients39 and that selectively interrupted the interaction of ACE2 with the S protein but had little effect on that of KREMEN1 or ASGR1 (Supplementary information, Fig. S7c). The blocking patterns of Ab-414 and Ab-515 on SARS-CoV-2 entry are highly consistent with that of the ACE2 neutralizing antibody (Fig. 4c). Viral entry into Calu3, NCI-H1650, Huh-7, and Hep3B cell lines was highly ACE2 dependent, with luciferase activity decreasing substantially by all three antibodies. However, it was unaffected by any antibody in HTB-182 and Li7 cells, confirming that SARS-CoV-2 entry into these two cell lines is ACE2 independent (Fig. 4c).
The contribution of ACE2, KREMEN1 or ASGR1 to viral entry in these sensitive cell lines was measured using knockdown (KD) experiments. ACE2 shRNA-mediated KD inhibited SARS-CoV-2 entry in most of the cell lines, correlated well with the effects of ACE2-related blocking antibodies (Fig. 4d; Supplementary information, Fig. S7d–f). ACE2 KD had no effects on viral entry into HTB-182 and Li7 cells, confirming that the entry process does not require ACE2. ASGR1 KD had little effect in lung cell lines but inhibited virus entry to different levels in liver cell lines, with the strongest effect being observed in Li7 cells (Fig. 4d; Supplementary information, Fig. S7e). KREMEN1 KD inhibited virus entry to varying degrees in six lung cell lines and three liver cell lines, among which three lung cell lines were significantly affected, including the ACE2-independent HTB-182 cell line (Fig. 4d; Supplementary information, Fig. S7e), indicating that KREMEN1 exerts a broader effect on viral sensitivity than ASGR1 in these cell lines. The results further demonstrated that the entry of SARS-CoV-2 in ACE2-independent Li7 and HTB-182 cells depends on ASGR1 and KREMEN1, respectively.
Given the effects of ASK KD on SARS-CoV-2 entry, the sensitive cell lines can be divided into five dependency categories: ACE2, KREMEN1, ASGR1, multiple receptors, and undefined (Fig. 4d). Six cell lines (including Calu-3, Calu-1, and Huh-7) were included in the ACE2-dependent group. The KREMEN1 and ASGR1 groups included HTB-182 and Li-7, respectively. NCI-H1944 and NCI-H661 belonged to the multiple receptor-dependent group, in which both ACE2 and KREMEN1 contribute significantly. Four lung cell lines that showed low sensitivity to SARS-CoV-2 formed the undefined dependency group (Fig. 4a). Among the 14 sensitive cell lines tested, high ACE2 or ASGR1 expression did not correlate with either of the receptors being needed for SARS-CoV-2 entry, whereas KREMEN1 expression and KREMEN1 dependence for viral entry were highly correlated (Fig. 4e; Supplementary information, Fig. S7g). Overall, ACE2 plays the broadest role in both lung and liver cell lines, KREMEN1 functions in lung cell lines, and ASGR1 acts in liver cell lines.
To further demonstrate that viral entry into HTB-182 and Li-7 cells is dependent on KREMEN1 and ASGR1, respectively, we infected the cells with authentic SARS-CoV-2. Consistently, KD of KREMEN1 specifically reduced the viral infection in HTB-182 cells, as did ASGR1 KD in Li-7 cells (Fig. 4f; Supplementary information, Fig. S7h), confirming that KREMEN1 and ASGR1 are entry receptors in these two ACE2-independent cell lines. Taken together, our results revealed that SARS-CoV-2 utilizes distinct ASK receptors to gain entry into different cell types, and demonstrate ACE2-independent entry, in which KREMEN1 and ASGR1 play essential roles.
ASK receptors underlie SARS-CoV-2 tropism
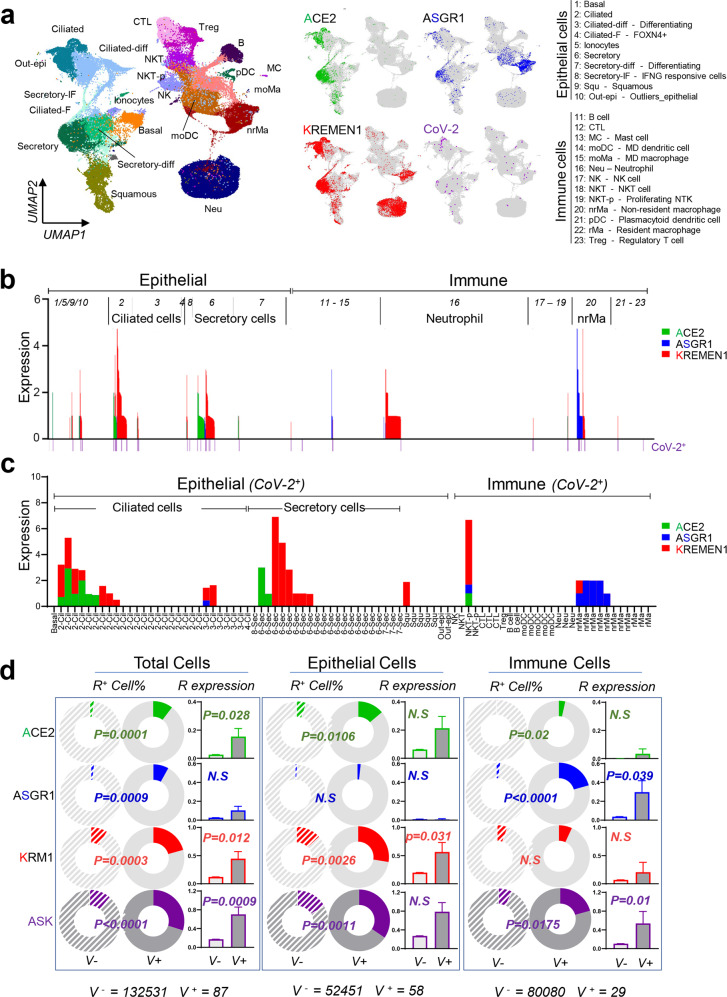
To investigate the clinical relevance of the ASK entry receptors for SARS-CoV-2 susceptibility, we analyzed a recently published single-cell sequencing (scRNA-seq) profile of the upper respiratory tract of 19 patients with COVID-19.18 The dataset was derived from nasopharyngeal/pharyngeal swabs and contains both the gene expression and virus infection status for individual cells, which are composed mainly of epithelial and immune populations. ACE2 is principally expressed in epithelial populations, as previously reported,18 whereas ASGR1 and KREMEN1 are enriched in both epithelial and immune populations (Fig. 5a). The majority of ASK+ cells express only one entry receptor (88.5%), and KREMEN1-expressing cells are the most abundant, being ~5-fold more numerous than either ACE2- or ASGR1-expressing cells (Fig. 5b; Supplementary information, Fig. S8a). SARS-CoV-2 is mainly observed in epithelial ciliated and secretory cells and immune non-resident macrophages (nrMa), which are also the major populations that express ASK receptors (Fig. 5b, c). Within SARS-CoV-2-positive cells (V+ cells), only 10.3% express ACE2 (Fig. 5c; Supplementary information, Fig. S8b), suggesting that other receptors will facilitate entry.
Fig. 5. ASK receptors are significantly correlated with SARS-CoV-2 susceptibility in the upper respiratory tract of COVID-19 patients.
a Distribution of ACE2, ASGR1, KREMEN1, and SARS-CoV-2 in different cell populations of the upper airway from COVID-19 patients. b Map of ASK expression levels and virus infection patterns in different cell populations. c ASK expression pattern in SARS-CoV-2-positive cells. d Correlations of virus susceptibility with ASK receptors individually or in combination based on the receptor-positive cell percentage and receptor expression level. P values were calculated by unpaired two-sided χ2 tests.
Correlating the presence of ACE2, ASGR1, and KREMEN1 entry receptors with SARS-CoV-2 susceptibility revealed that in all cells, the receptor-positive cell percentage was significantly higher in V+ cells than in V– cells for all three receptors (Fig. 5d). In epithelial populations, both ACE2 and KREMEN1 were substantially enriched in V+ cells, whereas in immune populations, particularly macrophages, only ASGR1 was correlated with virus susceptibility (Fig. 5d; Supplementary information, Fig. S8c). Epithelial ciliated and secretory cells are known target cells of SARS-CoV-2.17,18 ACE2 displayed a more significant correlation with the virus susceptibility of ciliated cells than KREMEN1, which was the only entry receptor that showed a high correlation with virus susceptibility in secretory cells (Supplementary information, Fig. S8c). Either in all cells or cell subpopulations, the ASK combination was usually more highly correlated with virus infection than individual receptors (Fig. 5d; Supplementary information, Fig. S8c). Similar result was obtained with another scRNA-seq dataset of experimentally SARS-CoV-2-infected human bronchial epithelial cells40 (Supplementary information, Fig. S9). These data further support our finding that the entry of SARS-CoV-2 in different cells relies on distinct entry receptors.
SARS-CoV-2 displays multi-organ tropism in COVID-19 patients.6,11,14,16,37 However, ACE2 alone cannot explain the multi-organ tropism of SARS-CoV-2. In virus-positive tissues, such as the brain, liver, peripheral blood (PB), and even lung, ACE2 expression is low or only detected in a small subset of cells15–17 (Supplementary information, Fig. S10a, b). To gain insights into the SARS-CoV-2 tissue tropism, we modeled a systemic host–SARS-CoV-2 interaction based on the tissue distribution of ASK entry receptors. ACE2 and ASGR1 are highly expressed in the gastrointestinal tract and liver, respectively, while KREMEN1 is broadly expressed throughout the body. In virus-positive tissues, we found that at least one of the entry receptors was expressed (Supplementary information, Fig. S10b). ASK receptor expression levels were correlated with virus infection rates in different tissues identified in a recent biopsy study,6 and the three receptors together showed a markedly stronger correlation with virus susceptibility than any individual receptor (Supplementary information, Fig. S10c), suggesting that the ASK receptors underlie SARS-CoV-2 tropism and can therefore better explain the multi-organ tropism of this virus.
Cocktail of ASK antibodies substantially blocks SARS-CoV-2 entry
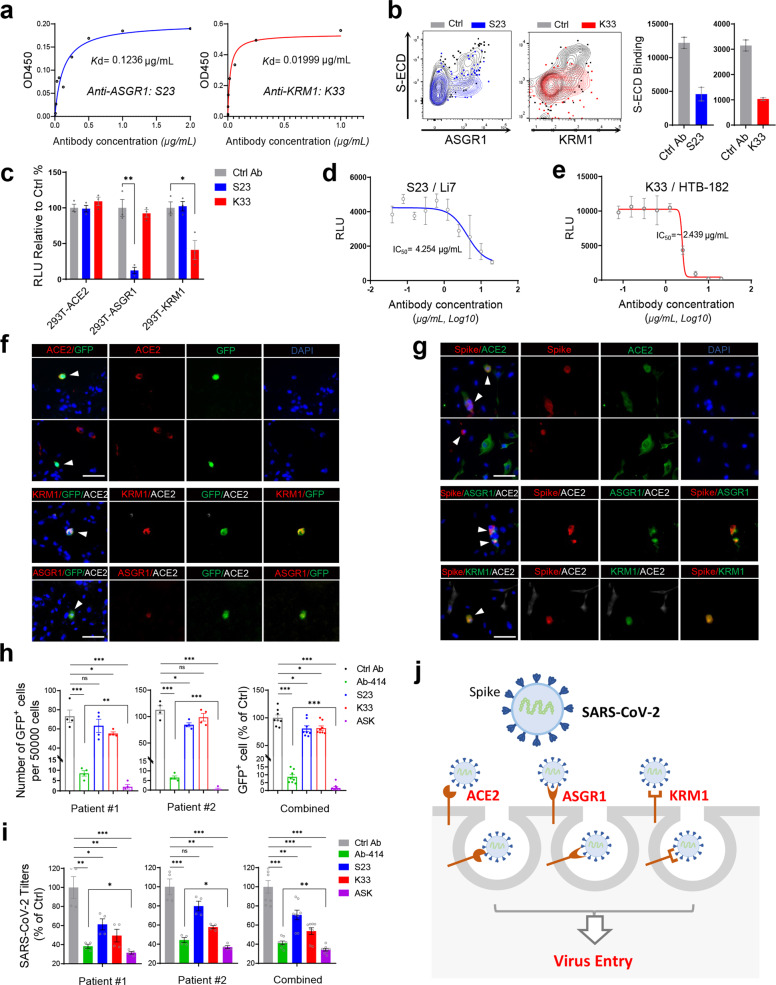
Our results suggest that simultaneously blocking the interaction between the S protein and ACE2/ASGR1/KREMEN1 should block SARS-CoV-2 entry more effectively than blocking any individual receptor, particularly in the complex tissues of the body. To this end, we developed monoclonal antibodies against ASGR1 and KREMEN1 and screened for their ability to block the interaction of the relevant receptor with the S protein. S23, an ASGR1 monoclonal antibody with a Kd of 0.12 µg/mL, and K33, a KREMEN1 monoclonal antibody with a Kd of 0.02 µg/mL, effectively blocked the binding of S-ECD to ASGR1 and KREMEN1, respectively (Fig. 6a, b; Supplementary information, Fig. S11a, b). The ability of these antibodies to block SARS-CoV-2 was then evaluated. S23 and K33 selectively blocked the SARS-CoV-2 S-mediated entry into ASGR1-293T and KREMEN1-293T cells, respectively, and had no effect on irrelevant ASK receptor-expressing cells (Fig. 6c). We also tested their efficacies in KREMEN1-dependent HTB-182 cells and ASGR1-dependent Li7 cells. S23 effectively blocked SARS-CoV-2 entry into Li-7 cells, with an IC50 of 4.25 µg/mL, K33 blocked virus entry into HTB-182 cells, with an IC50 of 2.43 µg/mL, and both antibodies had no effect on ACE2-dependent virus entry (Fig. 6d, e; Supplementary information, Fig. S11c). These results further confirm that the SARS-CoV-2 sensitivities of HTB-182 cells and Li-7 cells are dependent on KREMEN1 and ASGR1, respectively, and demonstrate that the neutralizing antibodies K33 and A23 can be used to selectively block KREMEN1- and ASGR1-mediated SARS-CoV-2 entry.
Fig. 6. Cocktail of ASK-related antibodies substantially blocks SARS-CoV-2 entry in human lung organoids.
a Kd values of the binding of the monoclonal antibodies S23 and K33 with ASGR1 and KREMEN1, respectively, measured by ELISA. b S23 and K33 antibodies (final concentration of 2 µg/mL) selectively block the interaction of SARS-CoV-2 S-ECD with ASGR1 and KREMEN1, respectively. S-ECD binding was measured by flow cytometry. A representative flow cytometry plot (left) and statistics (right) are shown. c S23 and K33 antibodies (final concentration of 2 µg/mL) selectively blocked SARS-CoV-2 S-mediated entry into HEK293T cells expressing ASGR1 and KREMEN1, respectively. Luciferase activity relative to control antibody treatment was measured 48 h post-infection (means ± SEM, n = 3). d S23 blocks SARS-CoV-2 S-mediated entry into Li7 cells with an IC50 of 4.25 µg/mL. Luciferase activity was measured 48 h post-infection (means ± SD, n = 2). e K33 blocks SARS-CoV-2 S-mediated entry into HTB-182 cells with an IC50 of 2.44 µg/mL. Luciferase activity was measured 48 h post-infection (means ± SD, n = 2). f, g Lung organoids were infected with rVSV-GFP/SARS-CoV-2 S chimeric virions (SARS-CoV-2/GFP) (f) or authentic SARS-CoV-2 (g) for 24 h or 48 h respectively, and immunostaining was performed with the indicated antibodies against ACE2, ASGR1 or KREMEN1 (scale bar, 50 μm). h, i Lung organoids from two patients were infected with SARS-CoV-2/GFP virus or authentic SARS-CoV-2 in the presence of the indicated antibodies (each at a final concentration of 4 µg/mL). The GFP+ cell numbers in each condition were counted under a fluorescence microscope 24 h post-infection (means ± SEM, n = 3 per organoid) (h), or the expression of SARS-CoV-2 ORF1ab gene in the cells was examined 48 h post infection by RT-qPCR (means ± SEM, n = 4 per organoid) (i). j Diagram of ASK receptors-mediated SARS-CoV-2 entry. The statistical significance was evaluated by unpaired two-tailed Student’s t-tests, *P < 0.05, **P < 0.01, ***P < 0.001.
To test whether an antibody cocktail would block virus entry more effectively than individual antibodies, we used a cocktail consisting of the K33 antibody and Ab-414, the SARS-CoV-2 neutralizing antibody that selectively blocks the ACE2-S interaction (Supplementary information, Fig. S7c), to treat NCI-H1944 and NCI-H661 cells, into which the entry of SARS-CoV-2 depends on both ACE2 and KREMEN1 (Fig. 4c, d; Supplementary information, Fig. S7e). Consistently, Ab-414 and K33 blocked the entry of SARS-CoV-2 (Supplementary information, Fig. S11d), confirming that the viral sensitivity of the two cell lines depends on both ACE2 and KREMEN1. The combination of Ab-414 and K33 showed a more potent inhibitory effect than the individual antibodies (Supplementary information, Fig. S11d), indicating that targeting of multiple entry receptors simultaneously blocks virus entry more effectively.
To test the antibodies in a more physiological condition, we conducted experiments using human lung organoids. The expression of ASK receptors was examined in human lung samples. ACE2, ASGR1, and KREMEN1 could clearly be detected, and little overlap in their expression was observed (Supplementary information, Fig. S11e), consistent with the scRNA-seq data (Fig. 5a, b; Supplementary information, Fig. S8a). To more clearly visualize virus infection, we infected human lung organoids with SARS-CoV-2 S-pseudotyped virus containing a GFP reporter. Overall, ~0.1–0.8% of cells were infected, as indicated by the GFP+ cells (Fig. 6g; Supplementary information, Fig. S11f), comparable to the percentage of ~0.1% observed in the upper respiratory tract of patients with COVID-19 (Fig. 5b). Immuno-fluorescence data revealed that both ACE2-expressing and non-expressing cells could be observed simultaneously among the infected GFP+ cells, and KREMEN1 or ASGR1 was detected in GFP+ACE2− cells (Fig. 6f). To verify these results, we infected lung organoids with authentic SARS-CoV-2, and observed both ACE2-expressing and non-expressing cells infected with SARS-CoV-2, and detected KREMEN1 or ASGR1 in SARS-CoV-2+ ACE2− cells (Fig. 6g). Thus, SARS-CoV-2 can infect lung organoids via both ACE2-dependent and ACE2-independent routes.
We treated lung organoids with ASK-targeting antibodies during infection with S-pseudotyped or authentic SARS-CoV-2. In addition to the ACE2-targeting Ab-414 antibody, both S23 and K33 antibodies significantly inhibited virus entry, and similar results were obtained with two human lung organoids (Fig. 6h, i), indicating that all receptors play critical roles during viral entry into lung organoids. Ab-414 was the most effective, followed by K33 and S23 (Fig. 6h, i), indicating that in human lung organoid, the ACE2 pathway is the major route for viral entry, and KREMEN1 and ASGR1 play important but more subsidiary roles. This finding is consistent with the results from our cell-line experiments and the scRNA-seq analysis of the airways of patients with COVID-19 (Figs. 4, 5). Compared with individual ASK antibodies, the ASK cocktail exerted a significantly higher inhibitory effect, providing the most substantial blockage of infection with either pseudotyped or authentic SARS-CoV-2, and the results were also observed in a second human lung organoid (Fig. 6h, i). Thus, KREMEN1 and ASGR1 function in parallel with ACE2 to mediate virus entry (Fig. 6j) and, therefore, simultaneously blocking the interaction between the viral S protein and the ASK receptors can more effectively block SARS-CoV-2 entry into cells.
Discussion
The affinity-based interactions between SARS-CoV-2 and cellular receptors are key determinants of virus tropism and pathogenesis. Identifying cells or tissues that express viral receptors should allow better characterization of the pathway for virus infection and improve the understanding of COVID-19 disease progression. SARS-CoV-2 can be detected in multiple organs, including the lungs, pharynx, heart, liver, brain, and kidneys,2,6,7 and is able to infect organoids from diverse tissues.41–45 However, the best characterized SARS-CoV-2 receptor, ACE2, is expressed in only a few cells in most tissues except the kidneys and digestive system.1,2,12–15 Our study identified a panel of SARS-CoV-2 receptors with diverse binding properties, functions, and tissue distributions. Integrating this panel of receptors with virological and clinical data should lead to the identification of infection and pathological mechanisms and targets.
Of the panel of identified SARS-CoV-2 S-interacting membrane proteins, two, ASGR1 and KREMEN1, function independently of ACE2, directly mediating virus entry both in vitro and in vivo. Among the 39 cell lines screened, the sensitivity of the HTB-182 and Li7 cell lines to SARS-CoV-2 are ACE2 independent, being unaffected by neither ACE2 KD, ACE2 neutralizing antibody nor ACE2-S-specific blocking antibodies. Furthermore, cells without apparent ACE2 expression can be infected by SARS-CoV-2 in lung organoids, and single-cell transcriptional analysis of COVID-19 patient airways revealed many virus-positive cells not expressing ACE2,18 a phenomenon that also observed in a large-scale single-cell transcriptome atlas of COVID-19 patients.19 Thus, the ACE2-independent SARS-CoV-2 entry occurs in multiple conditions. KREMEN1 mediates virus entry into HTB-182 cells and ASGR1 into Li7 cells. Blocking KREMEN1 and ASGR1 both significantly reduce virus entry into either cell line or into lung organoids. When KREMEN1 and ASGR blocking antibodies are combined with ACE2-targeting antibodies, the antibody cocktail displays a more potent blocking effect. Therefore, SARS-CoV-2 can infect cells via ACE2-dependent and independent routes, and ASGR1 and KREMEN1 play important roles in ACE2-independent SARS-CoV-2 entry.
The expression of the ASK receptors showed little overlap in cells from human lung samples, as also seen in the scRNA-seq analysis of COVID-19 patient airways. In our functional screening of lung and liver cell lines, SARS-CoV-2 sensitivity was generally dependent on individual ASK entry receptors. In those cell lines that showed multiple receptor-dependent sensitivity, viral entry was found to principally require either ACE2 or KREMEN1. Together with virus susceptibility in COVID-19 airway epithelial and immune cells correlating with specific ASK receptors, it suggests that ASK entry receptors generally function independently of one another, and SARS-CoV-2 utilizes distinct ASK receptors to enter different cell types. The combined pattern of ASK expression displays a better correlation with viral sensitivity than any individual receptor in both cells and tissues. Indeed, the antibody cocktail blocking viral S protein interaction with ASK receptors blocked SARS-CoV-2 entry more effectively than individual antibodies in either cell lines or lung organoids, suggesting that the ASK receptors underlie SARS-CoV-2 multi-organ/cell tropism. Targeting ASK receptors simultaneously may offer a promising strategy to treat or prevent SARS-CoV-2 infection.
KREMEN1 is the entry receptor for a major group of enteroviruses,34 and ASGR1 facilitates the entry of Hepatitis C virus,36 indicating that different viruses can share the same receptors. ASGR1 and KREMEN1 do not mediate the entry of SARS-CoV, potentially explaining the difference of these two viruses in primary infection sites and clinical manifestations.1,2,12–15 Unlike ACE2, which only binds to the RBD of S protein, ASGR1 and KREMEN1 bind to both the RBD and NTD. The NTD has been implicated in coronavirus entry,31,32 and several neutralizing antibodies from convalescent COVID-19 patients recognize NTD,29,30 suggesting that the domain plays a role during SARS-CoV-2 infection, and that neutralizing antibodies against the NTD may act through ASGR1 or KREMEN1. Given the higher efficacy of the ASK antibody cocktail in blocking SARS-CoV-2 entry, identifying convalescent patient-derived antibodies that block S interactions with KREMEN1 and ASGR1 could be of clinical utility.
Several genome-wide CRISPR screens have been performed recently to identify host factors involved in SARS-CoV-2 infection.46–48 The cells used in the assays, Huh-7.5 and A549-ACE2, depend on ACE2 for viral entry. It is therefore not surprising that ASGR1 and KREMEN1 were not identified as candidates in these screens. However, the host factors identified in these screens do raise questions as to whether they are involved in ASGR1- and KREMEN1-mediated SARS-CoV-2 entry, and whether TMPRSS2, the protease essential for ACE2-dependent entry pathway,3 is also required. AXL, CD147 and NRP1 were recently identified as receptors or receptor-like host factors that bind specifically to S protein of SARS-CoV-2 to facilitate or promote viral entry.49–52 Although they showed low S-ECD binding activities in our screen, which might be due to the differences between cell-based and Co-IP-based methods in detecting ligand-receptor interactions, the proved functions of these proteins indeed reflect the complexity of SARS-CoV-2 interaction with the host receptome. Given the many entry-related host cell receptors involved, whether and how they cooperate to contribute to SARS-CoV-2 pathogenesis and the COVID19 pandemic need further study.
In addition to functioning in viral entry, virus–host receptor interactions could also induce cytokine secretion, apoptosis, and stimulation of the immune response, or alter virus budding and release.8–11 Our identified S-binding receptor-like host factors can be classified according to their functions in virus entry, immune regulation, the Wnt pathway, and protein trafficking (Supplementary information, Fig. S10a). The expression of the immune-related host factors are prominent in not only immune organs but also respiratory organs and the liver (Supplementary information, Fig. S10b), consistent with the respiratory manifestation and frequent liver injury seen in COVID-19 patients.1,2,37,38 Given that the immune group factors, CD207, CLEC4M, LILRB2, and SIGLEC9 are all mainly expressed in myeloid cells (Supplementary information, Fig. S12) and that COVID-19 is associated with hyperactivation of myeloid populations,11,53,54 it is possible that these factors may mediate monocyte and macrophage activation in COVID-19 and contribute to disease pathophysiology.
ERGIC3, LMAN2, and MGAT2, which are involved in protein trafficking, display approximately similar expression levels across most human tissues (Supplementary information, Fig. S10b). ERGIC3 and LMAN2 are the components of the endoplasmic reticulum-Golgi intermediate compartment (ERGIC), which is essential for coronavirus assembly and budding,55,56 while LMAN2 and ERGIC1 were recently found to interact specifically with the nonstructural proteins Nsp7 and Nsp10, respectively, of SARS-CoV-2.57 Whether and how they cooperate during the virus lifecycle will require further investigation. The expression of the factors belonging to the Wnt pathway is prominent in the salivary gland, tongue, esophagus, and brain (Supplementary information, Fig. S10b). Wnt/β-catenin signaling is critical in taste bud cell renewal and behavioral taste perception,58,59 and KREMEN1/2 plus FUT8 are all negative regulators of this pathway.33,60 Loss of smell and taste has frequently been observed in COVID-19 patients,61,62 suggesting that SARS-CoV-2 may act through these factors to affect Wnt/β-catenin signaling and, therefore, promote taste loss.
Taken together, alternative virus-binding receptors may exert context-dependent regulatory effects, leading to differential signaling outcomes, ultimately influencing infection patterns, immune responses, and clinical progression. Our study identified a panel of receptor-like host factors with diverse S-binding patterns, signaling properties, and tissue distributions, and demonstrated ASGR1 and KREMEN1 as alternative functional receptors that play essential roles in ACE2-independent virus entry, providing insight into the critical virus-host interactions of SARS-CoV-2, as well as a useful community resource and potential drug targets for further investigations of COVID-19.
Materials and methods
Authentic SARS-CoV-2 and ethics statement
The SARS-CoV-2 isolate was obtained from a clinical case in Shanghai, China (SARS-CoV-2/SH01/human/2020/CHN, GenBank ID: MT121215). SARS-CoV-2 was propagated in Vero E6 cells. Cells were collected 50 h post-infection and lysed using the freeze-thaw method. Virus-containing supernatants were collected by centrifugation at ~2500× g for 10 min, aliquoted and stored at –80 °C. All animal experiments and ethical approvals were approved by the Institutional Animal Care and Use Committee (IACUC) of Fudan University and the Center for Excellence in Molecular Cell Science (CEMCS). The human research studies were approved by the Institutional Review Board of CEMCS. Informed consent was obtained from all the participants in the present study. All procedures in this study regarding authentic SARS-CoV-2 virus were performed in a biosafety level 3 (BSL-3) facility at the Medical School of Fudan University.
Cell culture
All cell lines were incubated at 37 °C in the presence of 5% CO2 and normal levels of O2. Vero E6, HEK293T, Huh-7 and HTB-182 cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Thermo Fisher) supplemented with 10% FBS. Calu-3 cells were cultured in Minimum Essential Medium (MEM, Thermo Fisher) supplemented with 10% FBS, 1.5 g/L NaHCO3 and 0.11 g/L sodium pyruvate. Li-7 cells were cultured in RPMI-1640 (Thermo Fisher) supplemented with 10% FBS, 1.5 g/L NaHCO3, 2.5 g/L glucose, and 0.11 g/L sodium pyruvate. HEK293E cells were cultured in serum-free FreeStyle 293 medium (Thermo Fisher) at 37 °C with rotation at 120 rpm in the presence of 5% CO2 and a normal level of O2. The other cell lines mentioned in this study were obtained from the Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, and cultured in the suggested media and conditions. All media were supplemented with penicillin and streptomycin (Thermo Fisher, 100×). All cell lines were routinely tested using a mycoplasma contamination kit (R&D Systems).
Isolation of human lung cells and lung organoid culture
Nontumor lung tissue obtained from patients undergoing lung resection was dissected and minced with scissors, washed with 5 mL of DMEM (Gibco, C11995500BT) with Primocin (InvivoGen, ant-pm-2), and then digested with Collagenase II (Gibco, 17101015) for 1–2 h in a cell incubator at 37 °C with shaking. DMEM supplemented with 10% FBS (ExCell (Serum), FSP500) was added to terminate digestion. The suspension was strained through a 70-μm filter, and the cells were then collected by centrifugation at 500× g. If a visible red pellet was produced, erythrocytes were lysed in 8 mL of red blood cell lysis buffer (1 g/L KHCO3, 8.3 g/L NH4Cl, and 0.041 g/L EDTA-Na2·2H2O) for 4 min at room temperature before the addition of 24 mL of PBS and centrifugation at 500× g. The dissociated cells were washed, seeded in growth factor-reduced Matrigel (Corning) and cultured in Advance DMEM/F12 medium supplemented with 10 mM HEPES (Gibco), 2 mM GlutaMAX-1 (Gibco), 500× Primocin (InvivoGen), 1× B27 (Gibco), 1.56 mM N-acetylcysteine (Sigma), 10 mM nicotinamide (Gibco), 0.5 μM A83-01 (Tocris), 10 μM Y27632, 50 ng/mL EGF (Peprotech), 10 ng/mL FGF10 (Peprotech), 1 ng/mL FGF2 (Peprotech), 10% in-house-prepared R-Spondin1, 10% Noggin and 30% Wnt3a.
Transduction and infection of mice
Mice were anesthetized with Avertin (Sigma-Aldrich) and transduced intranasally with 2.5 × 107 FFUs of ACE2/ASGR1/KREMEN1-expressing lentivirus in 75 μL of DMEM (Gibco C11995500BT). The mice were infected intranasally with of 1 × 106 VPs of S-pseudotyped SARS-CoV-2 or 1 × 105 PFUs of authentic SARS-CoV-2 on day 5 after lentiviral transduction. Three days post infection, lungs were harvested for measurement of the virus titer.
Genomic receptor profiling
To prepare SARS-CoV-2 S-ECD-hFc- or control hFc-containing conditioned medium, pCMV-S-ECD-hFc or pCMV-secreted-hFc plasmid was transfected into HEK293E cells, and conditioned medium was collected 4 days post transfection and filtered through a 0.45-µm filter for screening. To prepare receptor-expressing cells, plasmids encoding 5054 human membrane proteins were individually cotransfected with CFP reporter vector (5:1) into HEK293E cells in 96 deep-well plates using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. A total of 2–5 × 104 membrane protein-expressing cells per well were collected 48 h after transfection, washed once with PBS/2% FBS and incubated with 1 mL of SARS-CoV-2 S-ECD or hFc control condition medium for 1 h on ice. The supernatant was discarded after centrifugation and washed once with PBS/2% FBS. The cells were then labeled with anti-hFc-APC (Jackson Lab, 109-605-003) antibody for 20 min and washed once with PBS/2% FBS. The binding of S-ECD to the cells was measured by HTS flow cytometry (BD Canto II). The flow data were analyzed with FlowJo software. The mean APC fluorescence intensity (APC-MFI) reflects the degree of ligand binding. The relative binding of receptors (CFP+ cells) to S-ECD-hFc compared with that to the hFc control was calculated.
Co-IP and Kd measurement
Receptor-expressing cells were lysed with RIPA buffer (Sigma) and centrifuged for 15 min at 15,000 rpm and 4 °C, and the cell lysates were collected. Purified hFc-tagged S-ECD, RBD, NTD, or S2 domain proteins (final concentration of 10 µg/mL) were added to the cell lysate together with anti-FLAG beads (Sigma) or protein A beads (SMART Lifesciences), and the mixture was incubated at 4 °C overnight. The beads were washed three times with RIPA buffer, and the samples were prepared for western blot analysis with anti-hFc (ABclonal, AS002) or anti-FLAG (Smart Life Sciences, SLAB01) antibodies. For measurement of the Kd values, the receptor coding plasmid was cotransfected with the CFP reporter vector (5:1) into HEK293E cells. Cells were collected 48 h after transfection. Approximately 104 cells per well were used for binding with a series of diluted purified S-ECD-hFc proteins as described in the receptor profiling experiment. The flow data were analyzed with FlowJo software. The degree of ligand binding at each ligand concentration was calculated by normalizing the mean APC fluorescence intensity (APC-MFI) of receptor-positive cells with that at a ligand concentration of zero. The Kd and Bmax (maximum binding) values were calculated using Prism8 software.
Protein purification and western blot assay
For the purification of SARS-CoV-2 S-ECD-hFc, RBD-hFc, NTD-hFc, S2-hFc and SARS-CoV S-ECD, the plasmids were transfected into HEK293E cells, and conditioned medium was collected 4 days post transfection and filtered through a 0.45 µm filter. hFc-tagged proteins were purified using a Protein A affinity column and then desalted to PBS solution with an AKTA purifier system. The proteins were concentrated with a 10 kDa cutoff spin column (Amicon). For western blotting assay, the samples were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membrane was labeled with the primary antibody and then to the HRP-conjugated secondary antibody at the suggested concentration and detected with an ECL kit (Beyotime).
Constructs
For ACE2 knockout, ACE2 small guide RNA was constructed into pSLQ1651 (Addgene #51024) with a targeting sequence of CTTGGCCTGTTCCTCAATGGTGG. The ACE2 sgRNA plasmid or Cas9Bsd plasmid (Addgene #68343) was cotransfected with psPAX2 and pMD2G plasmids into HEK293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Lentiviruses were collected 72 h post transfection for the infection of HEK293T cells. The ACE2-KO stable HEK293T cell line was obtained by single-cell dilution. For the knockdown of ACE2, KREMEN1 or ASGR1, shRNAs were designed and constructed into the pLentilox3.7 vector using targeting sequences listed in supplementary oligo table. For lentiviral expression, ACE2, KREMEN1 or ASGR1 was constructed into pLVX-IRES-ZsGreen. pLentilox3.7-shRNA or pLVX constructs were cotransfected with the psPAX2 and pMD2G plasmids (at a ratio of 4:3:1) into HEK293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Lentiviruses were collected 72 h post transfection for the infection of target cells.
Quantitative reverse transcription PCR (RT-qPCR)
For the measurement of mRNA expression, total RNA was extracted using TRIzol (Invitrogen) and reverse transcribed into cDNA using SuperScript III Reverse Transcriptase (Invitrogen) according to the protocol provided. Real-time PCR was performed using Luna® Universal qPCR Master Mix (NEB). The mRNA levels were normalized to the level of GAPDH RNA transcripts present in the same sample, and the primers used in the assay are listed in the Supplementary information, Table S2.
Pseudotyped coronavirus production and infection
For HIV-based S-pseudotyped SARS-CoV-2, SARS-CoV and MERS-CoV, S protein-encoding pCDNA3.1 plasmids were mixed with pNL4-3.Luc.R vector separately at a ratio of 1:1 and transfected into HEK293T cells using Lipofectamine 2000. Virus-containing supernatant was collected 48–72 h posttransfection and filtered through a 0.45-µm PES membrane filter (Millipore). A replication-competent rVSV-GFP/SARS-CoV-2 chimeric virus was constructed by inserting the SARS-CoV-2 S-encoding sequence (lacking the C-terminal 18 codons) into the rVSVΔG backbone with a GFP reporter, which reportedly measures SARS-CoV-2-neutralizing antibody activity in patient plasma.63 The virus was passaged in Vero E6 cells. Cells were collected 50 h postinfection and lysed using the freeze-thaw method. Virus-containing supernatants were collected by centrifugation at ~2500× g for 10 min, aliquoted and stored at –80 °C. For infection, the cells were seeded into 96-well plates at ~2 × 104 cells per well, and 50 µL of virus-containing supernatant per well was added with or without antibodies at the indicated concentration. Luciferase activities were measured 48 h post infection using a Bright-Lumi™ Firefly Luciferase Reporter Gene Assay Kit (Beyotime, RG051M) and a multifunctional microplate reader (TECAN 200pro). GFP was detected by fluorescence microscope or flow cytometry (BD Canto II). For ASK antibody treatment, cells were treated with the indicated blocking antibodies (anti-ACE2-neutralizing mAb, Sino Biological, 10108-MM37; anti-ASGR1-blocking mAb (S23), homemade; anti-KREMEN1-blocking mAb (K33), homemade; SARS-CoV-2-neutralizing mAb 414 and mAb 515, homemade) at the indicated final concentration during infection.
Generation of ASGR1 and KREMEN1 mouse mAbs
Wild-type C57/BL6 mice were immunized subcutaneously with 0.1 mg of recombinantly expressed human ASGR1-ECD or KREMEN1-ECD proteins. Three weeks after the initial immunization, the animals were boosted two times at a 1-week interval. The serum titers were evaluated by indirect enzyme-linked immunosorbent assay (ELISA). B cell sorting and single-cell RT-PCR were performed according to a published protocol. Briefly, the mice were sacrificed, and spleen cells were labeled with antibodies (anti-mouse B220 AF700, Thermo Fisher, 56-0452-82; anti-mouse CD4 FITC, BioLegend, 100510; anti-mouse CD8 FITC, BioLegend, 100706; anti-mouse GR1 FITC, BioLegend, 108406; anti-mouse F4/80 FITC, BioLegend, 123108; anti-mouse IgG1 BV421, BD, 562580; anti-mouse IgM PE-Cy7, Thermo Fisher, 25-5790-82; and anti-mouse IgD APC-eF780, Thermo Fisher, 47-5993-82; each at 1:200) and PE/APC-labeled ASGR1-ECD or KREMEN1-ECD. Single antigen-specific memory B cells (B220+IgM−IgD−IgG+Antigen+) were sorted on a BD FACS Aria III into 96-well PCR plates containing 2.5 μL per well of lysis buffer (10 mM Dulbecco’s phosphate buffered saline (DPBS) and 4 U of mouse RNase inhibitor (NEB)). Reverse transcription and subsequent PCR amplification of heavy and light chain variable genes were performed using SuperScript III (Life Technologies) and the PCR mixture (SMART-Lifesciences). The PCR products were then purified using DNA FragSelect XP Magnetic Beads (SMART-Lifesciences) and cloned into vectors for the expression of mouse heavy and light chains. Heavy- and light-chain constructs were cotransfected into HEK293E cells using the transfection reagent PEI. After 4 days of expression, supernatants were harvested and analyzed by ELISA to obtain antigen-specific clones. For the antigen-recognizing clones, antibodies were purified using a Protein A affinity column and then desalted to PBS solution with an AKTA purifier system.
ELISA
Corning 96-well EIA/RIA plates were coated overnight at 4 °C with 0.5 μg/mL ASGR1-ECD-hFc, KREMEN1-ECD-hFc, or hFc alone. After washing with PBST, the plates were blocked using 3% nonfat milk in PBST for 1 h at 37 °C. Antibody supernatants or series dilutions of antibodies in PBST were added to each well and incubated at 37 °C for 1 h. Subsequently, the plates were washed with PBST and incubated for 30 min with anti-mouse IgG HRP-conjugated IgG (Thermo Fisher). After three washes with PBST, the immunoreactions were developed with tetramethylbenzidine (TMB) substrates and stopped by the addition of 2 mol/L sulfuric acid. The OD450 absorbance was detected using a multifunctional microplate reader (TECAN 200pro).
Authentic SARS-CoV-2 generation and infection
SARS-CoV-2/MT121215 was expanded in Vero E6 cells. Cells were collected 50 h postinfection and lysed using the freeze-thaw method. Virus-containing supernatants were collected by centrifugation at ~2500× g for 10 min, aliquoted and stored at –80 °C. For infection, targeted cells were incubated with fresh medium-diluted virus supernatant at MOI of 1 at 37 °C for 1 h, washed with 1× PBS three times and cultured in complete medium for the indicated time. The supernatant was collected for RT-qPCR with SARS-CoV-2 Orf1ab or N protein specific primers (Supplementary information, Table S2), or plaque assay using serial dilution with Vero E6 cells to measure virus titer. SARS-CoV-2 replication was examined by immuno-fluorescence and flow cytometry with anti-SARS-CoV-2 N or S protein antibodies (homemade by FUDAN BSL-3). For ASK antibody treatment, the cells were treated with the indicated antibodies as described in the pseudotyped virus infection section.
Immunofluorescence
For the immunofluorescence assay, cells were fixed, permeabilized at 4 °C for 30 min, incubated with the indicated antibodies (rabbit anti-ACE2 pAb, Proteintech, 21115-1-AP, 1:50; mouse anti-ASGPR1 mAb, Sino Biological, 10773-MM02, 1:50; rabbit anti-ASGPR1 mAb, Sino Biological, 10773-R011, 1:50; and mouse anti-KREMEN1 mAb, homemade, 1:100) at 4 °C overnight. The cells were washed twice with PBS, stained with labeled secondary antibodies (goat anti rabbit IgG/AF488, Thermo Fisher, A11034; goat anti mouse IgG/AF647, Thermo Fisher, A21236; goat anti human IgG/AF647, Thermo Fisher, A21455; goat anti mouse IgG/AF594, Thermo Fisher, A11005; each at 1:200) at 4 °C for 2 h, and subjected to 4′6-diamidino-2-phenylindole (DAPI, Sigma-Aldrich) staining for observation under a microscope imaging system (Olympus, Japan).
Data analysis and statistics
Gene Ontology enrichment analysis was performed using R Bioconductor. For construction of the host–virus interaction map, the receptor expression in each tissue was obtained from the Human Protein Atlas (https://www.proteinatlas.org/). The mRNA expression level was normalized by dividing the expression level by the Kd of each receptor. The viral infection rates of tissues were obtained from the study published by Puelles et al. Cluster analysis was performed using the R package. For scRNA-seq profiling of the upper airway tract of patients with COVID-19, the count, viral load and metadata were obtained from Magellan COVID-19 Data Explorer at https://digital.bihealth.org. χ2 tests and Student’s t-tests were performed to compare the receptor percentages and receptor expression values among different cell populations, respectively. The correlation analysis was performed with GraphPad Prism software. All tests were two-sided, and a P value < 0.05 was designated to indicate significance.
Supplementary information
Acknowledgements
We thank Cheng Cheng Zhang of UT Southwestern Medical Center and Guy Riddihough of Life Science Editors for the discussions on and revision of the paper and the staff at the BSL3 Laboratory of Fudan University for helping with the experiments. We thank Aihua Zheng of Institute of Zoology, Chinese Academy of Science, for providing replication-competent rVSV-GFP/SARS-CoV-2 S chimeric virions. This study was funded by the National Natural Science Foundation of China (81873438, 81873922, 81971921, 81830054 and 81772723, 32125013), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16020905), the Basic Frontier Science Research Program of Chinese Academy of Sciences (No. ZDBS-LY-SM015), the National Key R&D Program of China (2020YFA0509002 and 2017YFA0505500), and the National Key Project for Infectious Diseases of China (2018ZX10301208 and 2018ZX10302207-004-002).
Author contributions
Z.L., M.L., Y.X., Yun Z. and D.G. conceived the project. Y.G., XY.Z. and J.C., with help from J.Z., X.J., J.W., J.Y., X.Z., and Yun Z., performed the receptor profiling studies and the characterization of receptor-ligand interactions. M.L., H.G., J.X., X.J. and J.W. assessed the virus sensitivity of cell lines. J.C., J.H. and D.G. performed the organoid experiments. J.C., H.G. and Y.W., with help from G.S., X.J., F.L., G.H., Yuanfei Z., S.D., Yunkai Z., R.Z. and D.Q., performed the virus-related experiments. Z.L., M.L., Y.G., J.C., H.G. and Y.W. performed the bioinformatics analysis and analyzed the data. Z.L., M.L., Y.X., G.X., Yun Z. and D.G. wrote the paper.
Data availability
The receptor expression levels in each tissue were obtained from the Human Protein Atlas (https://www.proteinatlas.org/). The receptor expression levels in PBMC populations were obtained from the Human Protein Atlas (https://www.proteinatlas.org/) and Human Cell Atlas (http://immunecellatlas.net/). The receptor expression levels in stable cell lines were obtained from Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle/). ScRNA-seq profiles of the upper airway tract of patients with COVID-19 and metadata were obtained from the Magellan COVID-19 Data Explorer at https://digital.bihealth.org. ScRNA-seq profiles of SARS-CoV-2 infected human bronchial epithelial cells (HBECs) in vitro were from NCBI GEO dataset GSE166766. All data supporting the findings of this study are available within the paper or in the supplementary information.
Competing interests
M.L., Z.L., Yun Z., H.G. and Y.X. are listed as inventors on a pending patent application for the newly identified S receptors described in this paper. The other authors declare no competing interests.
Footnotes
These authors contributed equally: Yunqing Gu, Jun Cao, Xinyu Zhang, Hai Gao, Yuyan Wang, Jia Wang, Juan He, Xiaoyi Jiang.
Change history
4/1/2022
A Correction to this paper has been published: 10.1038/s41422-022-00654-6
Contributor Information
Yun Zhao, Email: yunzhao@sibcb.ac.cn.
Dong Gao, Email: dong.gao@sibcb.ac.cn.
Youhua Xie, Email: yhxie@fudan.edu.cn.
Min Luo, Email: luo_min@fudan.edu.cn.
Zhigang Lu, Email: zhiganglu@fudan.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-021-00595-6.
References
- 1.Zhu N, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China (vol 395, pg 497, 2020) Lancet. 2020;395:496–496. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li WH, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–27. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puelles VG, et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- 8.Schneider-Schaulies J. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 2000;81:1413–1429. doi: 10.1099/0022-1317-81-6-1413. [DOI] [PubMed] [Google Scholar]
- 9.Milone MC, Fitzgerald-Bocarsly P. The mannose receptor mediates induction of IFN-alpha in peripheral blood dendritic cells by enveloped RNA and DNA viruses. J. Immunol. 1998;161:2391–2399. [PubMed] [Google Scholar]
- 10.Tseng CT, Perrone LA, Zhu H, Makino S, Peters CJ. Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J. Immunol. 2005;174:7977–7985. doi: 10.4049/jimmunol.174.12.7977. [DOI] [PubMed] [Google Scholar]
- 11.Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukassen S, et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu H, et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1:e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zou L, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou YJ, et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell. 2020;182:429–446 e414. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hikmet F, et al. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16:e9610. doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sungnak W, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chua RL, et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020;38:970–979. doi: 10.1038/s41587-020-0602-4. [DOI] [PubMed] [Google Scholar]
- 19.Ren X, et al. COVID-19 immune features revealed by a large-scale single cell transcriptome atlas. Cell. 2021;184:1895–1913. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng M, et al. A motif in LILRB2 critical for Angptl2 binding and activation. Blood. 2014;124:924–935. doi: 10.1182/blood-2014-01-549162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng M, et al. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature. 2018;562:605–609. doi: 10.1038/s41586-018-0615-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng J, et al. Inhibitory receptors bind ANGPTLs and support blood stem cells and leukaemia development. Nature. 2012;485:656–660. doi: 10.1038/nature11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeffers SA, et al. CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA. 2004;101:15748–15753. doi: 10.1073/pnas.0403812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, et al. Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2. Cell Discov. 2020;6:68. doi: 10.1038/s41421-020-00210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bao L, et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 28.Sun J, et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell. 2020;182:734–743 e735. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 30.Chi X, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERSCoV, and beyond. Trends Microbiol. 2015;23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang N, et al. Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD. Cell Rep. 2019;28:3395–3405 e3396. doi: 10.1016/j.celrep.2019.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao B, et al. Kremen proteins are Dickkopf receptors that regulate Wnt/beta-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 34.Staring J, et al. KREMEN1 Is a Host Entry Receptor for a Major Group of Enteroviruses. Cell Host Microbe. 2018;23:636–643 e635. doi: 10.1016/j.chom.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Seidah NG, Chretien M, Mbikay M. The ever-expanding saga of the proprotein convertases and their roles in body homeostasis: emphasis on novel proprotein convertase subtilisin kexin number 9 functions and regulation. Curr. Opin. Lipidol. 2018;29:144–150. doi: 10.1097/MOL.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 36.Saunier B, et al. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J. Virol. 2003;77:546–559. doi: 10.1128/JVI.77.1.546-559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao F, et al. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833 e1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phipps MM, et al. Acute Liver Injury in COVID-19: Prevalence and Association with Clinical Outcomes in a Large US Cohort. Hepatology. 2020;72:807–817. doi: 10.1002/hep.31404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan J, et al. Human-IgG-Neutralizing Monoclonal Antibodies Block the SARS-CoV-2 Infection. Cell Rep. 2020;32:107918. doi: 10.1016/j.celrep.2020.107918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravindra NG, et al. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol. 2021;19:e3001143. doi: 10.1371/journal.pbio.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han Y, et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacob F, et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell. 2020;27:937–950 e939. doi: 10.1016/j.stem.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salahudeen AA, et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588:670–675. doi: 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L, et al. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell. 2020;27:125–136 e127. doi: 10.1016/j.stem.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou J, et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 46.Daniloski Z, et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184:92–105 e116. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider WM, et al. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell. 2021;184:120–132 e114. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann HH, et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280 e265. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cantuti-Castelvetri L, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daly JL, et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang K, et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S, et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 54.Zhou YG, et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl. Sci. Rev. 2020;7:998–1002. doi: 10.1093/nsr/nwaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lontok E, Corse E, Machamer CE. Intracellular targeting signals contribute to localization of coronavirus spike proteins near the virus assembly site. J. Virol. 2004;78:5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gordon DE, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaillard D, et al. beta-catenin is required for taste bud cell renewal and behavioral taste perception in adult mice. PLoS Genet. 2017;13:e1006990. doi: 10.1371/journal.pgen.1006990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu F, et al. Wnt-beta-catenin signaling initiates taste papilla development. Nat. Genet. 2007;39:106–112. doi: 10.1038/ng1932. [DOI] [PubMed] [Google Scholar]
- 60.Kurimoto A, et al. The absence of core fucose up-regulates GnT-III and Wnt target genes: a possible mechanism for an adaptive response in terms of glycan function. J. Biol. Chem. 2014;289:11704–11714. doi: 10.1074/jbc.M113.502542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menni C, et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020;26:1037–1040. doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xydakis MS, et al. Smell and taste dysfunction in patients with COVID-19. Lancet Infect. Dis. 2020;20:1015–1016. doi: 10.1016/S1473-3099(20)30293-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The receptor expression levels in each tissue were obtained from the Human Protein Atlas (https://www.proteinatlas.org/). The receptor expression levels in PBMC populations were obtained from the Human Protein Atlas (https://www.proteinatlas.org/) and Human Cell Atlas (http://immunecellatlas.net/). The receptor expression levels in stable cell lines were obtained from Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle/). ScRNA-seq profiles of the upper airway tract of patients with COVID-19 and metadata were obtained from the Magellan COVID-19 Data Explorer at https://digital.bihealth.org. ScRNA-seq profiles of SARS-CoV-2 infected human bronchial epithelial cells (HBECs) in vitro were from NCBI GEO dataset GSE166766. All data supporting the findings of this study are available within the paper or in the supplementary information.