Significance
Pathogenic microbes inject virulence proteins, termed effectors, into the cells of their hosts where they sabotage the cellular machinery and promote the infection process. However, plant and animal cells have evolved the ability to sense the molecular activities of these pathogenic proteins and mount an immune response to protect against infection. Here we have identified a chemical that mimics the immune eliciting activity of a pathogenic effector protein and importantly, it also activates immune responses in plants. We showcase how the intricate molecular dialog of the host–pathogen interface can be used to uncover chemical immunogens that specifically activate the eukaryotic immune system to prevent disease.
Keywords: type III effector, immunity, chemical biology: zaractin, Arabidopsis, Pseudomonas syringae
Abstract
Pathogenic effector proteins use a variety of enzymatic activities to manipulate host cellular proteins and favor the infection process. However, these perturbations can be sensed by nucleotide-binding leucine-rich-repeat (NLR) proteins to activate effector-triggered immunity (ETI). Here we have identified a small molecule (Zaractin) that mimics the immune eliciting activity of the Pseudomonas syringae type III secreted effector (T3SE) HopF1r and show that both HopF1r and Zaractin activate the same NLR-mediated immune pathway in Arabidopsis. Our results demonstrate that the ETI-inducing action of pathogenic effectors can be harnessed to identify synthetic activators of the eukaryotic immune system.
Gram-negative bacterial plant pathogens such as Pseudomonas syringae use the type III secretion system (T3SS) to inject type III secreted effectors (T3SEs) into plant cells (1). A major function of T3SEs is to suppress plant immunity by targeting host proteins involved in disease resistance (2). However, plant nucleotide-binding leucine-rich-repeat (NLR) proteins can directly or indirectly recognize T3SEs and activate a response known as effector-triggered immunity (ETI), which can be accompanied by a hypersensitive cell death response (2, 3). The Arabidopsis thaliana NLR ZAR1 recognizes T3SE-induced complexes of ZED1-related kinases (ZRKs; RLCK XII-2 family) and PBS1-like (PBL) kinases (RLCK VII family) to activate ETI (4–7). T3SEs modify either ZRK and/or PBL kinases and promote their interaction, which activates ZAR1-mediated ETI (5, 6). For example, the Xanthomonas campestris AvrAC, uridylates PBL2 kinase, promoting an interaction with ZRK1 (also known as RKS1) that then acts as a nucleotide exchange factor to activate the ZAR1 resistosome (8, 9).
In addition to AvrAC, five P. syringae effector families have been shown to trigger ZAR1/ZRK-mediated immunity in Arabidopsis (4, 10–13). ZAR1-mediated immunity triggered by the acetyltransferase HopZ1a requires the ZRK kinase ZED1 and the functionally redundant PBL kinases SZE1 and SZE2 (4, 7). Interestingly, acetylation of ZED1 by HopZ1a can activate immunity, suggesting that modification of either ZRK or PBL kinases can activate the ZAR1 resistosome (4, 6). HopX1i-recognition also requires ZED1 and SZE1, but not SZE2 (13). HopO1c- and HopF1r-mediated immunity requires ZRK3, whereas HopBA1a recognition requires ZRK2, but no PBL kinase requirement has been identified (11, 13). Based on the genetic requirements of ZAR1-mediated ETI against P. syringae effectors and the model of AvrAC recognition, a general mechanism of ZAR1 activation likely involves T3SE perturbations of ZRK and/or PBL kinases that promote their interaction, which in turn activates the ZAR1 resistosome (8, 9).
We hypothesized that small molecules that mimic ETI-promoting effector perturbations would represent a powerful, targeted approach to activate plant immunity. Given the model of ZAR1 activation described above, we developed a chemical screen to identify small molecules that enhance ETI-inducing ZRK/PBL interactions. First, we show that the P. syringae T3SE HopF1r can enhance the interaction between ZRK3 and PBL27 and that PBL27 is a required component of ZAR1-mediated recognition of HopF1r. We then used our chemical screen to identify a small molecule (Zaractin) that can enhance the ZRK3/PBL27 interaction and activate ZAR1-dependent immunity. Overall, our results demonstrate that chemical mimicry of type III effector function can activate an NLR-mediated immune response, providing an approach to identify chemical immunomodulators.
Results
PBL27 Kinase Is Required for Recognition of HopF1r in Arabidopsis.
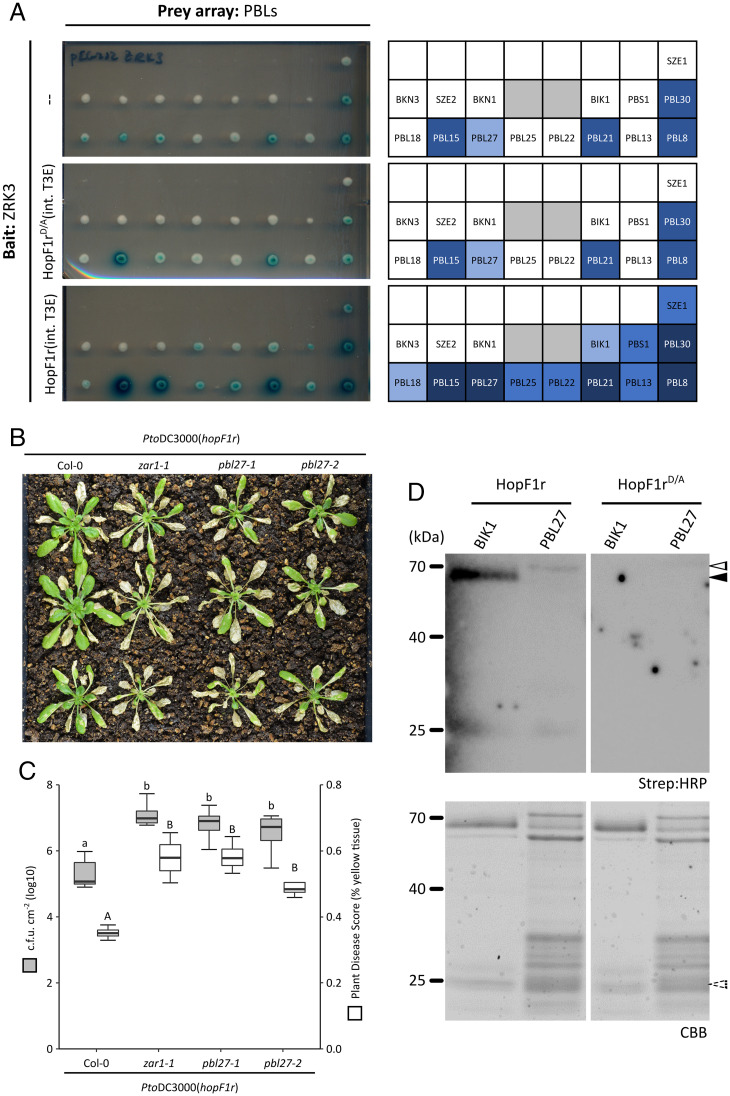
The Arabidopsis ZAR1-mediated ETI response against the P. syringae ADP ribosyltransferase T3SE HopF1r in Arabidopsis was previously shown to require ZRK3 kinase (11). However, HopF1r did not ADP ribosylate ZRK3, indicating that HopF1r may target PBL kinases rather than ZRK3 (11). We previously developed a yeast three-hybrid (Y3H) assay to screen for effector-enhanced ZRK/PBL interactions that can activate ZAR1 immunity (6, 13). Using this system, we demonstrated that type III effectors that activate ZAR1-mediated ETI can enhance several ZRK/PBL interactions, including those that activate ZAR1 such as ZRK1/PBL2 by AvrAC and ZED1/SZE1 by HopX1i (6, 13). We used this assay to determine whether HopF1r can induce ZRK/PBL complexes using ZRK3 as bait against PBL kinases (6). When wild-type HopF1r was expressed, we observed enhanced interactions between ZRK3 and several PBL kinases (PBL15, PBL27, PBL21, PBL30, PBL8, PBL25, PBL22, BIK1, PBS1, PBL13, and SZE1; SI Appendix, Table S1 and Fig. S1 and Fig. 1A), demonstrating that HopF1r can induce ZRK3/PBL interactions. These enhanced interactions were unchanged when the catalytic mutant of HopF1r (HopF1rD/A) (11) was coexpressed (Fig. 1A and SI Appendix, Fig. S1).
Fig. 1.
PBL27 is required for HopF1r-triggered immunity in Arabidopsis. (A) Y3H assays testing interactions between ZRK3 and a subset of PBL kinases in the absence of chromosomally integrated HopF1r (–, Top), or in the presence of HopF1rD/A (Middle) or HopF1r (Bottom). Interaction layouts are depicted on the Right. Gray boxes are not relevant to this study. Consolidated Y3H interaction data are presented in SI Appendix, Table S1. (B) Disease phenotypes of Col-0, zar1-1, pbl27-1, or pbl27-2 plants after spraying with PtoDC3000(hopF1r) at OD600 = 1.0. Symptoms pictured are 14 d postinfection. (C) Growth of PtoDC3000(hopF1r) (gray) or PIDIQ disease quantification (white) on Col-0, zar1-1, pbl27-1, and pbl27-2 Arabidopsis plants. Bacterial counts were taken at 3 d postinfection. Lowercase letters represent statistically significant differences (Tukey’s honest significant difference [HSD], P < 0.05). Experiments were replicated three times with similar results. Capital letters represent statistically significant differences for PIDIQ disease quantification (Tukey’s HSD, P < 0.05). pbl27-1 and pbl27-2 lines were genotyped for homozygosity. (D) ADP ribosylation assay using His::HopF1r or His::HopF1rD/A with GST::BIK1 or GST::PBL27. The black arrow indicates the size of GST-BIK1 (∼71 kDa); the white arrow indicates the size of GST::PBL27 (∼83 kDa). ADP ribosylated proteins were detected using Strep:HRP (Top) and protein loading was visualized with Coomassie Brilliant Blue (CBB; Bottom). Dashed arrow indicates the size of His::HopF1r (∼25 kDa; purified His::HopF1r and His::HopF1rD/A are depicted in SI Appendix, Fig. S6).
To determine whether one of the PBL kinases identified in our Y3H screen was required for HopF1r ETI, we tested PBL kinase T-DNA insertion lines for loss of HopF1r–triggered immunity (Fig. 1B and SI Appendix, Table S1 and Figs. S2 and S3). When the PBL T-DNA insertion lines corresponding to those with HopF1r-enhanced ZRK3 interactions were sprayed with PtoDC3000(hopF1r), we observed that only the pbl27-1 line was more susceptible compared to wild-type Col-0 plants (Fig. 1B and SI Appendix, Figs. S2 and S3). We observed a similar level of susceptibility to PtoDC3000(hopF1r) with a second pbl27 mutant line (pbl27-2) (Fig. 1B). The level of disease symptoms (i.e., yellow tissue) of pbl27 plants sprayed with PtoDC3000(hopF1r) was comparable to zar1-1 plants (Fig. 1B). These observations were verified by bacterial growth assays, which showed that PtoDC3000(hopF1r) grew to the same level on pbl27 plants as zar1-1 plants (Fig. 1C). The compromised HopF1r-ETI was not observed on mutant plants of six related PBL kinases, including BIK1, a well-characterized plant immunomodulator (SI Appendix, Figs. S3 and S4) (14). Furthermore, pbl27 plants did not show compromised HopO1c or HopX1i ETI, which are also ZAR1 dependent (SI Appendix, Fig. S5) (12).
To test whether HopF1r can ADP ribosylate PBL27, we conducted in vitro HopF1r ADP ribosyltransferase assays using PBL27 and BIK1 as substrates (11). The results showed that HopF1r, but not the catalytically inactive HopF1rD/A ADP ribosylated both PBL27 and BIK1 (Fig. 1D and SI Appendix, Fig. S6). Together, these results demonstrate that HopF1r can ADP ribosylate both PBL27 and BIK1, but only PBL27 is required for HopF1r-induced ETI.
The Small Molecule Zaractin Activates ZAR1-Dependent Immunity in Arabidopsis.
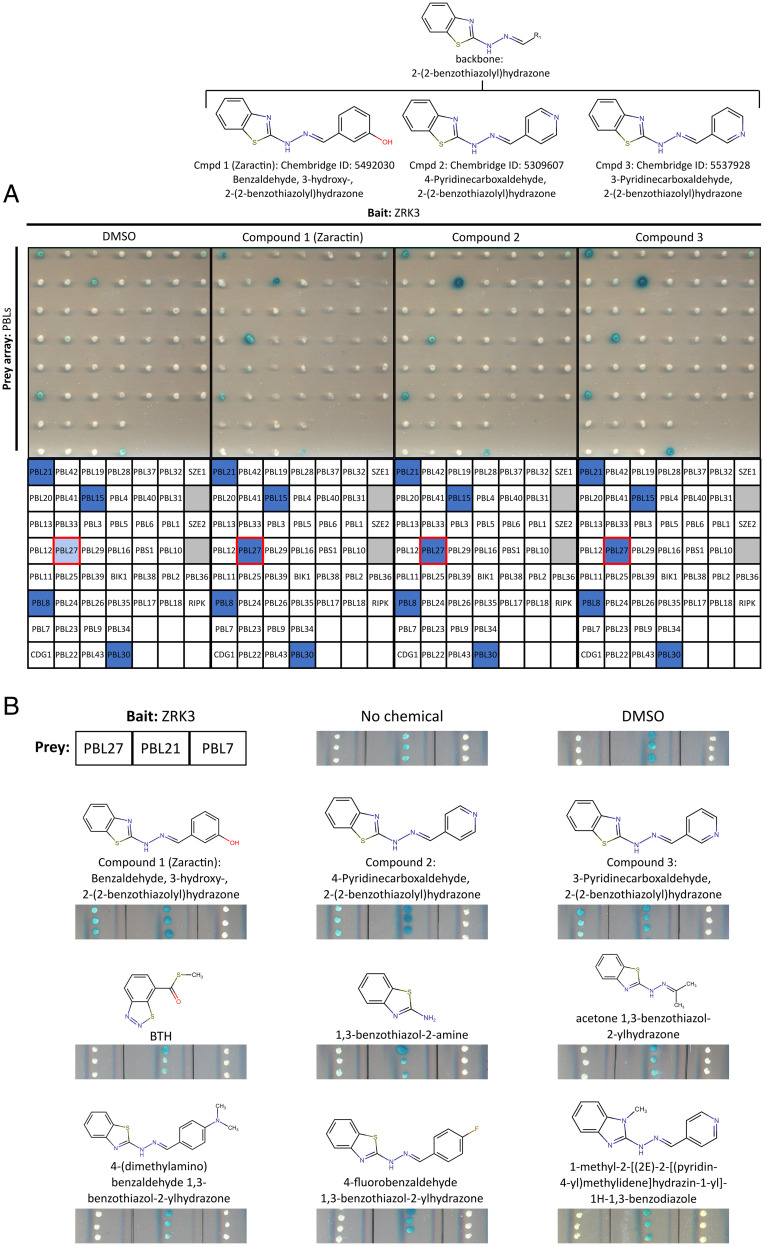
We developed a yeast two-hybrid (Y2H)-based chemical screen to identify small molecules that enhance the ZRK3/PBL27 or the ZRK1/PBL2 interaction, similar to the HopF1r- and AvrAC-enhanced interactions (Fig. 1A) (5, 6). We screened a collection of 4,182 compounds from a yeast-active chemical library, which has been shown to be enriched for bioactive compounds in organisms including Escherichia coli and Caenorhabditis elegans (15), and was used to identify small molecules that enhance Y2H interactions (16). Although no compounds were identified that induced the ZRK1/PBL2 interaction, our screen did identify several chemicals that enhanced the ZRK3/PBL27 interaction and among them, the top three strongest inducers on duplicate screening plates possessed the same benzothiazolyl hydrazone chemical backbone with a modified benzene ring R group (Fig. 2A and SI Appendix, Fig. S7). These compounds were benzaldehyde, 3-hydroxy-2-(2-benzothiazolyl)hydrazone (compound 1), 4-pyridinecarbozaldehyde, 2-(2-benzothiazolyl)hydrazone (compound 2), and 3-pyridinecarbozaldehyde, 2-(2-benzothiazolyl)hydrazone (compound 3) (Fig. 2A). We used an independent yeast two-hybrid assay to test whether the action of these three compounds was specific to enhance the ZRK3 and PBL27 interaction. Remarkably, the compounds did not enhance the interaction between ZRK3 and other PBL kinases or the interaction between PBL27 and ZED1 (Fig. 2A and SI Appendix, Fig. S8). Furthermore, we tested four additional compounds that possess the benzothiazolyl hydrazone chemical backbone, as well as the 1,3-benzothiazol-2-amine subgroup, and the immunity-inducing compound benzothiadiazole (BTH), structurally similar to our identified compounds (Fig. 2B) (17). Of all the chemicals tested, only the three identified in our screen (i.e., compounds 1 to 3) promoted the interaction between PBL27 and ZRK3 (Fig. 2B and SI Appendix, Table S2). Specific modifications of both the benzothiazolyl hydrazone backbone or the R group interfered with the ability to promote the ZRK3/PBL27 interaction, demonstrating that both of these chemical features are required for enhancing this interaction.
Fig. 2.
Specific enhancement of the ZRK3/PBL27 interaction by Zaractin and related compounds. (A) Y2H assay testing interactions between ZRK3 and a PBL array against chemicals that were identified as the strongest inducers of the ZRK3/PBL27 interaction (SI Appendix, Fig. S7). From Left to Right: DMSO, compound 1 (Zaractin), compound 2, and compound 3. All chemicals were tested at a concentration of 30 µM. Interaction layouts are depicted on the Right; the red box indicates the location of PBL27 on the array. Gray boxes are not relevant to this study. Chemical structures and their common backbone are displayed above their respective panels. (B) Y2H assay testing interaction between ZRK3 and PBL27, PBL21, or PBL7 with no chemical, DMSO, BTH, compound 1 (Zaractin), compound 2, compound 3, and structurally similar chemicals. All chemicals were tested at a concentration of 30 µM. PBL21 constitutively interacts with ZRK3 and PBL7 does not interact with ZRK3, nor is it induced by HopF1r or Zaractin. Chemical structures are displayed above their respective panels.
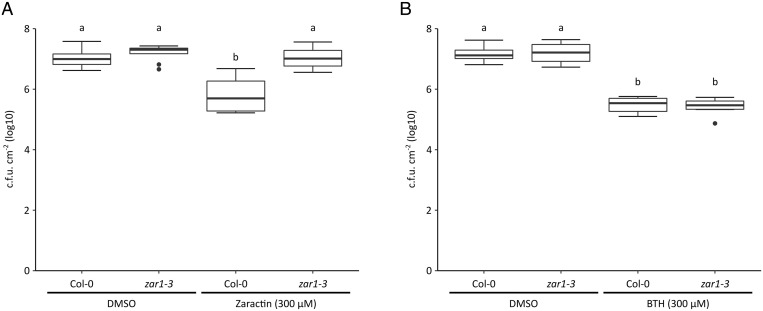
To test whether any of the three compounds identified in our chemical screen could activate ZAR1 immunity, Col-0 and zar1 Arabidopsis plants were initially treated with 150 μM of each of the chemicals 2 d before infection with PtoDC3000. Plants treated with compounds 1 and 3 showed significant but inconsistent reductions in bacterial growth that appeared to be ZAR1 dependent (SI Appendix, Fig. S9A). Compound 2 did not significantly reduce bacterial growth in any of our assays and was not pursued further (SI Appendix, Fig. S9A). A second test with compounds 1 and 3 at 300 μM, a concentration similar to that used for the immunity-inducing compound BTH (17), revealed that although Col-0 plants treated with both compounds showed less growth compared to the dimethylsulfoxide (DMSO) control, only compound 1–treated plants displayed consistently significant decreases in bacterial growth (Fig. 3A and SI Appendix, Fig. S9B). In contrast, compound 1–treated zar1 plants showed no significant difference in PtoDC3000 growth compared to the DMSO control (Fig. 3A and SI Appendix, Fig. S9B). We also confirmed this result with an independent insertion line of ZAR1: zar1-1 (SI Appendix, Fig. S10). BTH-treated zar1 plants showed significantly less growth compared to the DMSO control, similar to BTH-treated Col-0 plants demonstrating that, unlike compound 1, BTH-induced immunity is ZAR1 independent (Fig. 3B). In addition, a closely related compound (4-fluorobenzaldehyde 1,3-benzothiazol-2-ylhydrazone), which possesses a fluoride group on the benzene ring R group did not enhance the ZRK3/PBL27 interaction or promote immunity in Arabidopsis (Fig. 2B and SI Appendix, Fig. S11). Similar to the results obtained with HopF1r, plants infiltrated with a high dose of compound 1 did not trigger a hypersensitive cell death response (SI Appendix, Fig. S12) (11). We named compound 1 [benzaldehyde, 3-hydroxy-2-(2-benzothiazolyl)hydrazone] Zaractin for its activation of ZAR1-mediated immunity (Fig. 3A and SI Appendix, Fig. S10).
Fig. 3.
Zaractin triggers ZAR1-dependent immunity. Growth of PtoDC3000 on Col-0 or zar1-3 Arabidopsis plants treated with DMSO, (A) 300 µM compound 1 (Zaractin). (B) BTH (300 µM) 2 d before infection. Plants were sprayed with PtoDC3000 at OD600 = 1.0. Letters represent statistically significant differences (Tukey’s HSD, P < 0.05).
Zaractin-Induced Immunity Is Dependent on ZRK3 and PBL27.
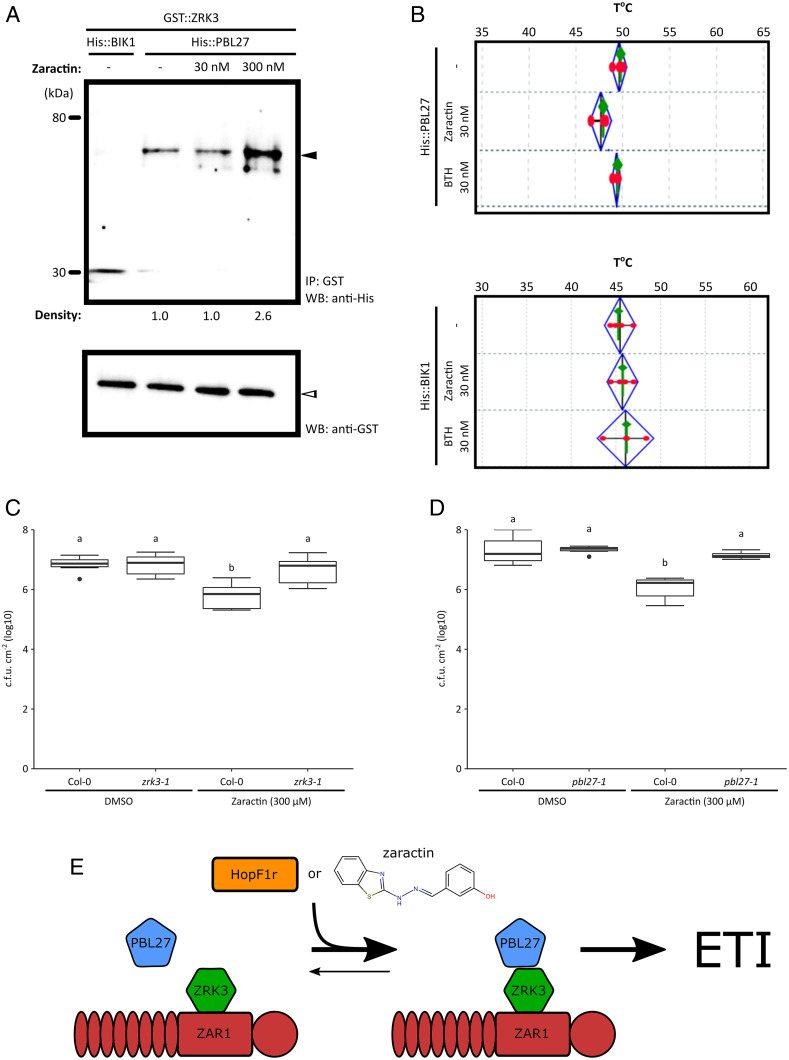
To validate our observations that Zaractin could enhance the interaction between PBL27 and ZRK3, we performed an in vitro glutathione S-transferase (GST) coimmunoprecipitation assay and found that the addition of 300 nM Zaractin increased the binding of PBL27 to ZRK3 (Fig. 4A and SI Appendix, Fig. S13). The interaction was specific as BIK1 did not bind to ZRK3 (Fig. 4A). Furthermore, we tested whether PBL27 could bind to Zaractin using a thermal shift assay (TSA). In the presence of BTH, we did not observe a shift in the melting temperature (Tm) for PBL27 or BIK1 (Fig. 4B and SI Appendix, Table S3). However, in the presence of 30 nM Zaractin, we observed a decrease in Tm for PBL27, which was not seen with BIK1, indicating that Zaractin, but not BTH, binds specifically to PBL27 (Fig. 4B and SI Appendix, Table S3).
Fig. 4.
Zaractin binds to PBL27, enhances the ZRK3-PBL27 interaction, and triggers ZRK3-, and PBL27-dependent immunity. (A) Purified His::PBL27 proteins were incubated with GST::ZRK3 proteins in the absence of Zaractin or the presence of Zaractin at 30 nM and 300 nM. GST::ZRK3 was immunoprecipitated using anti-GST magnetic microbeads (Bottom), and the interaction between His::PBL27 was demonstrated by His antibodies (Top). Density scan of the proteins showed ∼2.6-fold increase in binding of His::PBL27 to GST::ZRK3. As a negative control, purified His::BIK1 was incubated with GST::ZRK3-tagged proteins in the absence of Zaractin. This is representative of two independent experiments. Equal loading was demonstrated with GST antibodies (Bottom). IP, Immunoprecipitation; WB, Western blot. (B) Thermal shift assays showing the melting temperatures of His::PBL27 and His::BIK1 proteins in the absence or presence of Zaractin (30 nM) and BTH (30 nM). Results are representative of four independent biological replicates with four technical replicates in each experiment. Growth of PtoDC3000 on Col-0, (C) zrk3-1, or (D) pbl27-1 Arabidopsis plants treated with DMSO or 300 µM compound 1 (Zaractin) 2 d before infection. Plants were sprayed with PtoDC3000 at OD600 = 1.0. Letters represent statistically significant differences (Tukey’s HSD, P < 0.05). (E) A model of ZAR1-triggered immunity by HopF1r or Zaractin. HopF1r ADP ribosylates (red star) PBL27, leading to an induced interaction between PBL27 and ZRK3. The small molecule Zaractin mimics HopF1r function, which also promotes the PBL27/ZRK3 interaction and triggers ZAR1 immunity.
Since Zaractin enhanced the ZRK3/PBL27 interaction, we tested whether Zaractin-triggered immunity required ZRK3 and PBL27. Unlike Col-0 plants, Zaractin-treated zrk3 and pbl27 plants showed no significant difference in PtoDC3000 growth compared to the DMSO control (Fig. 4 C and D). We also confirmed these results with independent insertion lines of zrk3 and pbl27 (SI Appendix, Fig. S14). To confirm that Zaractin insensitivity was not a general property of T-DNA insertion lines, we treated zrk2 and pbl7 plants with Zaractin and observed significantly less PtoDC3000 growth compared to the DMSO control, like wild-type Col-0 plants (SI Appendix, Fig. S15).
Discussion
Zaractin is a chemical activator of ZAR1-dependent immunity in Arabidopsis. Zaractin binds to PBL27 and specifically enhances PBL27/ZRK3 interaction, resulting in immunity that is also PBL27 and ZRK3 dependent. We have also shown that PBL27 and ZRK3 are required for recognition of the P. syringae T3SE HopF1r by ZAR1. Since PBL27 has also been shown to contribute to immunity against P. syringae (18), it likely represents a virulence target of HopF1r that is guarded by ZAR1 (Fig. 1). We hypothesize that ADP ribosylation of PBL27 or its interaction with Zaractin promotes ZRK3 binding and activation of ZAR1 (Fig. 4E). The decreased Tm of PBL27 observed in the presence of Zaractin suggests that this compound may promote protein destabilization/unfolding (19) (Fig. 4B and SI Appendix, Table S3). Interestingly, we previously demonstrated that mutagenesis of PBL15 could enhance the interaction with the RLCK XII-2 kinase ZED1 (6). As such, the promotion of a destabilized protein conformation by effector modification, mutagenesis, or chemical binding could be a general mechanism that promotes PBL interactions with the RLCK XII-2 (ZRK) kinase family and activates ZAR1 immunity. The extent to which Zaractin mimics HopF1r activity will require further investigation of its mode of action, including the mapping of its respective binding/ADP ribosylation sites and the resulting consequences on the ZAR1 resistosome.
Besides Zaractin, the only other small molecule known to activate NLR-mediated plant immunity is the insecticide fenthion, which activates the tomato NLR Prf in a Fen kinase–dependent manner (20). Although the mode of action of fenthion is uncharacterized, Fen kinase is involved in effector perception (21) and similar to Zaractin, fenthion may mimic an effector-induced perturbation of Fen kinase to activate Prf. Given the recent appreciation of the extensive cross-talk between the intracellular NLR and extracellular pattern-recognition receptor (PRR) immune pathways, we speculate that activation of ZAR1 by Zaractin may potentiate pattern-triggered immunity (PTI) by up-regulating PTI components (22).
Benzothiazolyl hydrazone derivatives have been shown to possess antimicrobial properties and antitumor activity (23, 24), but we report here their ability to promote plant immunity. BTH displays chemical similarity to Zaractin and is a well-established inducer of systemic acquired resistance in plants and is the active ingredient in the commercial pesticide Actigard (SI Appendix, Table S3) (17). However, its mode of action is distinct from Zaractin since it induces ZAR1-independent immunity (Fig. 3B), it does not promote PBL27/ZRK3 interaction (Fig. 2B), and it does not bind to PBL27 (Fig. 4B). Although we identified three benzothiazolyl hydrazone derivatives that enhanced PBL27/ZRK3 interaction, they displayed differences in their ability to induce immunity in Arabidopsis. Only Zaractin (compound 1) and compound 3 induced ZAR1-dependent immunity, with Zaractin being the most robust immunogen (SI Appendix, Fig. S9). We did not observe any induced resistance with compound 2 (SI Appendix, Fig. S9). Since all three compounds possess the same benzothiazolyl hydrazone backbone, the differences in bioactivity are likely due to the different modifications of the benzene ring R group, which may influence rates of cellular uptake/efflux, chemical stability, and/or potency. Further exploration of the Zaractin R group chemical space promises to yield derivatives with increased potency as immunogens and potentially expand their range of efficacy.
Overall, the ability to chemically activate NLR-mediated immunity represents a powerful approach to induce plant immunity. These results emphasize the importance of continued research on effector functions and how they activate ETI to harness the full spectrum of plant protection strategies.
Methods
Plant Material, Bacterial Strains, and Cloning.
Arabidopsis plants were grown in 12 h of light (130 to 150 μE m−2s−1) and 12 h of darkness at 21 to 22 °C in Sunshine Mix 1 soil supplemented with 20:20:20 fertilizer at 1 g/L. All P. syringae strains were grown at 28 °C in King’s B (KB) medium. KB medium was supplemented with the following antibiotics, where appropriate: 50 μg/mL rifampicin and 50 μg/mL kanamycin.
P. syringae expression constructs of HopF1r and HopF1rD/A (formerly HopF2a) were cloned into a Gateway-compatible multicopy broad-host range plasmid pBBR1 MCS-2 modified to create an in-frame C-terminal HA tag fusion as described previously (11).
Bacterial Spray Inoculation, In Planta Growth, and Hypersensitive Response Assays.
To screen for disease symptoms, P. syringae was resuspended in 10 mM MgCl2 and diluted to 8 × 108 CFU/mL (OD600 = 1.0), with 0.04% surfactant Silwet L-77 added. Five-week-old Arabidopsis plants were sprayed using Preval sprayers and the plants were covered to maintain high humidity. The covers were removed on day 3 and disease symptoms were monitored for up to 7 d. Images that best captured the resulting phenotypes were analyzed with plant immunity and disease image-based quantification (PIDIQ) to determine disease scores based on the proportion of yellow chlorotic tissue in each plant (25).
To quantify bacterial growth, Arabidopsis plants were sprayed as above and four disks (1 cm2) per plant (one disk per leaf) were harvested on day 3. Leaf disks from eight plants per treatment were ground in 1 mL sterile 10 mM MgCl2 and plated on KB with rifampicin (50 µg/mL) for colony quantification.
To examine the potential for chemical inhibition of bacterial growth, the Zaractin (Chembridge ID: 5492030) was diluted in ddH2O to a final concentration of 300 µM, with 0.02% surfactant Silwet L-77 added. Using a wooden popsicle stick, the diluted chemical was painted on the entire adaxial surface of 5-wk-old plant leaves (four leaves per plant) and covered for 2 d before infection. Spraying of the compound was not used to ration our limited chemical supplies. Following 2 d of chemical treatment, spray infection and quantification of PtoDC3000 were carried out as above.
For hypersensitive cell death response assays, P. syringae at 1.6 × 108 CFU/mL (OD600 = 0.2) or chemical (Zaractin or BTH) at 500 µM was syringe infiltrated into half leaves of 5-wk-old Arabidopsis plants. Hypersensitive response symptoms were monitored at 16 h postinfiltration.
ADP Ribosylation Assays.
HopF1r and hopF1rD/A were cloned into the expression vector pET-28a to create an N-terminal His-tag fusion and transformed into E. coli BL21 codon+. For His-tag protein purification, bacterial cells induced with 1 mM isopropyl β- d-1-thiogalactopyranoside (IPTG) in 250 mL of LB medium were pelleted and lysed in His-binding buffer (20 mM sodium phosphate [pH 7.4], 0.5 M NaCl) with 5 mM phenylmethylsulfonyl fluoride (PMSF) and 0.5 mg/mL lysozyme followed by sonication. The clarified extracts were incubated for 30 min with 500 μL Ni Sepharose 6 Fast Flow (GE Healthcare). The columns were washed once with 10 mL His-binding buffer and washed twice with 10 mL His-binding buffer with 30 mM imidazole. Proteins were eluted with 10 mL His-binding buffer with 50 mM imidazole in 1-mL fractions. Purified proteins were verified by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis of elution fractions, and fractions containing purified protein were pooled and concentrated with Amicon Ultra-4 10,000 Da molecular weight cut-off (MWCO) (EMD Millipore). During concentration, purified proteins were exchanged with His-binding buffer and 20% (vol/vol) glycerol, snap frozen in liquid N2, and stored at −80 °C.
PBL27 and BIK1 were cloned into the Gateway-compatible expression vector pDEST15 to create an N-terminal GST tag fusion and transformed into E. coli BL21 codon+. For GST-tag protein purification, bacterial cells induced with 1 mM IPTG in 1 L of Luria-Bertani broth (LB) medium were pelleted and lysed in GST-binding buffer (50 mM Hepes-NaOH (pH 8.0), 500 mM NaCl, 1% [vol/vol] Triton X-100, 5% glycerol) with 5 mM PMSF and 0.5 mg/mL lysozyme followed by sonication. The clarified extracts were incubated for 1 h with 1 mL glutathione Sepharose 4B (GE Healthcare). The columns were washed once with 10 mL GST-binding buffer and followed by a 10-mL wash with GST-wash buffer 1 (50 mM Hepes-NaOH [pH 8.0], 100 mM NaCl, 1% Triton X-100, 5% glycerol) and 20 mL of GST-wash buffer 2 (50 mM Hepes-NaOH [pH 8.0], 100 mM NaCl, 5% glycerol). Purified proteins were eluted with GST-wash buffer 2 with 10 mM glutathione in 1-mL fractions. Purifications were verified by SDS-PAGE analysis of elution fractions, and fractions containing purified protein were pooled and concentrated with Amicon Ultra-4 10,000 Da MWCO (EMD Millipore). During concentration, purified proteins were exchanged with protein storage buffer (10 mM Hepes-NaOH [pH 7.5], 100 mM NaCl, 10% [vol/vol] glycerol), snap frozen in liquid N2, and stored at −80 °C.
For ADP ribosyltransferase assays, 230 ng of purified His::HopF2a or His::HopF2aD/A proteins were incubated with 0.5 μg GST::PBL27, GST::BIK1, in ADP ribosylation buffer containing 40 mM Hepes (pH 7.5), 5 mM MgCl2, 1 mM dithiothreitol (DTT), 60 μM adenosine triphosphate (ATP), 3 mM ADP ribose (Sigma-Aldrich), 10 μM β-nicotinamide adenine dinucleotide (β-NAD) (Sigma-Aldrich), and 20 μM 6-biotin-17-NAD (Trevigen) in 30 μL per reaction. Reactions were incubated at room temperature for 40 min and terminated by boiling in the SDS sample buffer for 5 min. The ADP ribosylation of proteins was detected by immunoblot using streptavidin–horseradish peroxidase conjugate (GE Healthcare).
Yeast Three-Hybrid Assays.
Using the LexA-based yeast two-hybrid system, pEG202 (HIS+) bait constructs expressing ZRK3 or ZRK1 were carried in the haploid yeast strain EGY48 (alpha mating type), while pJG4-5 (TRP+) prey constructs expressing PBL kinases were carried in the haploid yeast strain RFY206 (A mating type), which also carried the lacZ reporter plasmid pSH18-34 (URA+). To assess protein–protein interactions in the presence of a third protein, we used the yeast three-hybrid system described in Bastedo et al. (6), where a third protein of interest is integrated at the chromosomal HO locus of the haploid yeast strain EGY48.
The yeast strains EGY48 and RFY206 were mated by coincubation on nonselective YPD glucose agar for 2 d at 30 °C. The mated yeast strains were subjected to two rounds of selection (2 d of growth at 30 °C) on yeast nitrogen base (YNB) glucose lacking uracil, tryptophan, and histidine (−Ura −Trp −His). Following selection, diploid yeast strains were transferred to reporter plates with the above selection, supplemented with 1% raffinose, 2% galactose, 0.05 M sodium phosphate, 10 mg/mL X-gal and the appropriate selection, grown at 30 °C and monitored for up to 3 d.
Yeast Two-Hybrid Chemical Screen.
To prepare the reporter plates for the chemical Y2H assay, each well of a 96-well plate was aliquoted with 180 µL YNB agar medium lacking uracil, tryptophan, and histidine (−Ura −Trp −His), supplemented with 1% raffinose, 2% galactose, 0.05 M sodium phosphate, 10 mg/mL X-gal and with a different chemical from the yeast-active chemical library (15) (total of ∼4,182 chemicals) to a final concentration of 30 µM. After growing the diploid yeast strain expressing ZRK3 or ZRK1 (bait) and PBL27 or PBL2 (prey) on nonselective YPD glucose agar for 2 d at 30 °C, the strain was transferred to YNB glucose lacking uracil, tryptophan, and histidine (−Ura −Trp −His) and grown overnight at 30 °C. Following this diploid selection, the strain was transferred to the aforementioned 96-well chemical reporter plates and colonies were monitored for development of blue color for up to 3 d to assess interactions. A diploid yeast strain expressing ZED1 (bait), PBL15 (prey), and hopZ1a integrated at the HO locus was also present on the same plate in wells without chemical as a positive control as verified by Bastedo et al. (6).
Coimmunoprecipitation Assays.
BIK1 and PBL27 were cloned into the expression vector pET-28a with His tag at the N terminus. The ZRK3 was cloned into the expression vector pGEX-4T3 with GST tag at the N terminus. The proteins were expressed in ArcticExpress (DE3) E. coli strain according to the manufacturer’s instructions (Agilent Technologies). The His-tagged proteins were purified using the Capturem His-tagged purification kit (Takara) and GST-tagged proteins were purified with glutathione Sepharose 4B (GE Health Care). All the proteins were desalted with Amicon Ultra-0.5 centrifugal filter devices (Millipore). The purified proteins were confirmed by running on a SDS-PAGE and stained with Coomassie blue stain and Western blot analysis. The His-tagged proteins were incubated with 1/5,000 dilution of the anti-His (C-term horseradish peroxidase [HRP]) antibodies (Invitrogen) and the GST-tagged proteins were incubated with 1/10,000 dilution of the GST-HRP antibodies (Santa Cruz Biotechnologies), followed by detection by SuperSignal chemiluminescent substrate (Thermo Fisher). The images were processed in the iBright CL750 imaging system (Invitrogen).
For the coimmunoprecipitation experiments, 1 µg of GST::ZRK3 protein was incubated with either 1 µg BIK1::His or PBL27::His proteins with or without Zaractin in 50 mM Tris·HCl (pH 7.5), 50 mM NaCl for 1.5 h at room temperature. Subsequently, 50 µL anti-GST magnetic microbeads (Miltenyi Biotec) were used to pull down the protein complex. The beads were washed two times with 50 mM Tris·HCl (pH 7.5), 50 mM NaCl, and processed to detect the tagged proteins as before.
Thermal Shift Assay.
TSAs were performed with purified His::BIK1 and His::PBL27 proteins. The TSA was performed in Quant Studio 3 (Applied Biosystems) with the Protein Thermal Shift dye kit (Applied Biosystems) in a 20-µL reaction volume containing 0.1 mg/mL of each protein with or without Zaractin and BTH (Sigma) in 0.1% DMSO. The PCR conditions were used according to manufacturer’s instructions. Tm data were generated using the Boltzmann method. ΔTm was calculated by comparing the Tm values for each protein without the inhibitors to those with the inhibitors. Data were collected at 1 °C intervals from 25 °C through 99 °C and analyzed with Protein Thermal Shift software version 1.4 (Thermo Fisher Scientific).
Supplementary Material
Acknowledgments
We thank members of the D.D. and D.G. laboratories for valuable input on the project. This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) postgraduate awards (D.S. and A.M.), NSERC Discovery grants (D.G. and D.D.), and the Centre for the Analysis of Genome Evolution and Function (D.D. and D.G.). T-DNA lines were provided by the Arabidopsis Biological Resource Center unless otherwise noted. We thank Daphne Goring at the University of Toronto for providing the bkn1 and sze2 mutant lines. We thank Jacqueline Monaghan at Queen’s University for providing the bik1 mutant line.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116570118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Büttner D., He S. Y., Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 150, 1656–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan M., Seto D., Subramaniam R., Desveaux D., Oh, the places they’ll go! A survey of phytopathogen effectors and their host targets. Plant J. 93, 651–663 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Jones J. D. G., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Lewis J. D., et al. , The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc. Natl. Acad. Sci. U.S.A. 110, 18722–18727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang G., et al. , The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18, 285–295 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Bastedo D. P., et al. , Perturbations of the ZED1 pseudokinase activate plant immunity. PLoS Pathog. 15, e1007900 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu C., et al. , Two arabidopsis receptor-like cytoplasmic kinases SZE1 and SZE2 associate with the ZAR1-ZED1 complex and are required for effector-triggered immunity. Mol. Plant 12, 967–983 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Wang J., et al. , Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364, eaav5870 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Wang J., et al. , Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364, eaav5868 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Lewis J. D., Wu R., Guttman D. S., Desveaux D., Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 6, e1000894 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seto D., et al. , Expanded type III effector recognition by the ZAR1 NLR protein using ZED1-related kinases. Nat. Plants 3, 17027 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Laflamme B., et al. The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 367, 763–768 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Martel A., et al. , Immunodiversity of the Arabidopsis ZAR1 NLR Is conveyed by receptor-like cytoplasmic kinase sensors. Front. Plant Sci. 11, 1290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang X., Zhou J.-M., Receptor-like cytoplasmic kinases: Central players in plant receptor kinase-mediated signaling. Annu. Rev. Plant Biol. 69, 267–299 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Wallace I. M., et al. , Compound prioritization methods increase rates of chemical probe discovery in model organisms. Chem. Biol. 18, 1273–1283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toh S., Holbrook-Smith D., Stokes M. E., Tsuchiya Y., McCourt P., Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chem. Biol. 21, 988–998 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Lawton K. A., et al. , Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 10, 71–82 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Shinya T., et al. , Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 79, 56–66 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Cimmperman P., et al. , A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys. J. 95, 3222–3231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin G. B., et al. , A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell 6, 1543–1552 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosebrock T. R., et al. , A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan M., Ngou B. P. M., Ding P., Xin X. F., PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Easmon J., Pürstinger G., Thies K. S., Heinisch G., Hofmann J., Synthesis, structure-activity relationships, and antitumor studies of 2-benzoxazolyl hydrazones derived from alpha-(N)-acyl heteroaromatics. J. Med. Chem. 49, 6343–6350 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Swamy D. K., Pachling S. P., Bhagat T. M., Synthesis, characerization, antibacterial and antifungal studies on metal complexes with benzothiazolyl hydrazone. Rasayan J. Chem. 5, 208–213 (2012). [Google Scholar]
- 25.Laflamme B., Middleton M., Lo T., Desveaux D., Guttman D. S., Image-based quantification of plant immunity and disease. Mol. Plant Microbe Interact. 29, 919–924 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.