Abstract
Pathogenic Legionella species grow optimally inside free-living amoebae to concentrations that increase risks to those who are exposed. The aim of this study was to screen a complete drinking water system and cooling towers for the occurrence of Acanthamoeba spp. and Naegleria fowleri and their cooccurrence with Legionella pneumophila, Legionella anisa, Legionella micdadei, Legionella bozemanii, and Legionella longbeachae. A total of 42 large-volume water samples, including 12 from the reservoir (water source), 24 from two buildings (influents to the buildings and exposure sites (taps)), and six cooling towers were collected and analyzed using droplet digital PCR (ddPCR). N. fowleri cooccurred with L. micdadei in 76 (32/42) of the water samples. In the building water system, the concentrations of N. fowleri and L. micdadei ranged from 1.5 to 1.6 Log10 gene copies (GC)/100 mL, but the concentrations of species increased in the cooling towers. The data obtained in this study illustrate the ecology of pathogenic Legionella species in taps and cooling towers. Investigating Legionella’s ecology in drinking and industrial waters will hopefully lead to better control of these pathogenic species in drinking water supply systems and cooling towers.
Keywords: Legionella, L. pneumophila, L. micdadei, L. anisa, L. bozemanii, L. longbeachae, N. fowleri, Acanthamoeba spp., water supply system, cooling towers
1. Introduction
Free-living amoebae are ubiquitous protozoa that have been found in various natural and engineered water systems, such as surface water, groundwater, drinking water supply systems, hot springs, and cooling towers [1,2,3,4,5,6,7]. Similar to free-living amoebae, Legionella spp. are also found in natural water bodies (surface water, groundwater, and hot springs) [8,9,10,11] and human-made systems (swimming pools, drinking water supply systems, and cooling towers) [12,13,14,15,16,17,18,19,20,21]. In their natural and engineered environments, free-living amoebae serve as a host, reservoir, and vehicle of pathogenic bacteria, including Legionella species [8,9,10,11,12,13,14,15,16,17,18,19,20,21].
Acanthamoeba and Naegleria are the two most common genera that are frequently isolated from aquatic environments [12,13,14,15,16] and serve as suitable hosts for Legionella species (17). Legionella species are associated with free-living amoebae in two developmental stages: the active stage, trophozoite (growth: Legionella spp. amplify to high concentrations [22]) and the resisting stage, cysts (survival: intracellular growth enables protection for Legionella spp. against chemical disinfectants in the water treatment system [23]). L. pneumophila, responsible for Legionnaires’ disease (LD) and Pontiac fever [24,25], has evolved a mechanism to invade the host cell by inducing phagocytosis and can avoid being digested by inhibiting the fusion of phagosomes with lysosomes [26,27]. Additionally, L. pneumophila growth within free-living amoebae has been shown to enhance the virulence factors of this species [28,29]. After intracellular amoebae replication of Legionella spp., the host cell is lysed, and once in the water column, Legionella has been shown to be less susceptible to disinfectants and can amplify to concentrations that are a risk to public health [30,31]. To that end, cooling towers and hot-water taps are key sources of human infections with Legionella species [32,33], ultimately posing a risk to human health via inhalation as these exposure sites are aerosol spreading units [34,35,36,37].
Specific species of Legionella, Naegleria, and Acanthamoeba have been known to cause disease in humans [38,39]. For example, L. pneumophila, L. anisa, L. micdadei, L. bozemanii, and L. longbeachae covers most of the incidence of LD [40]. Worldwide, L. pneumophila, L. longbeachae, and L. bozemanii account for 91.5%, 3.9%, and 2.4%, respectively, of LD [40]. Together, L. micadedi and L. anisa cover 1.8% of LD [40]. N. fowleri is the only species of Naegleria that causes primary amoebic meningoencephalitis (infection of the brain), a fatal disease of the central nervous system (CNS) [41]. Acanthamoeba spp. (9/20) causes cutaneous, ocular, and CNS infections, known as chronic granulomatous amebic encephalitis [39,42]. Because of such species relevance to human disease, the cooccurrence of amoebae and Legionella species are warranted further investigation in a complete water supply system.
There are limited studies examining their cooccurrence in a complete drinking water system and cooling towers, collectively. The cooccurrence of Legionella spp. and free-living amoebae has been examined solely in a drinking water supply system [43], a hospital water network [44], and cooling towers [45]. Each study focused mainly on the cooccurrence of L. pneumophila and various amoebae species by collecting a small volume (1 L or less) of the sample [43,44,45]. It is hypothesized that sampling a small volume of water may not represent the true distribution of pathogenic amoebae and coexisting Legionella spp. in the environment. Thus, the present paper is aimed at filling in the knowledge gaps in the distribution of pathogenic amoebae species and Legionella spp. by collecting a large volume of bulk water from a drinking water source, building water system, and cooling towers, all served by a groundwater source. Using ddPCR, this study addressed the following objectives (i) identify the cooccurrence of pathogenic Legionella spp. (L. pneumophila, L. anisa, L. longbeachae, L. bozemanii, and L. micdadei) and amoebae species (N. fowleri and Acanthamoeba spp.) in a complete water supply chain and cooling towers, (ii) examine the difference in occurrence and concentration of N. fowleri and Acanthamoeba spp. in a groundwater source, which supplied the building water system, and cooling towers, all located in the state of Michigan.
2. Results
N.fowleri occurrence rate was higher 40% (17/42) than detectable Acanthamoeba spp. 19% (8/42) (Table 1). In the drinking water system (RES_IN (influent pipe of the reservoir), RES_EF (effluent pipe of the reservoir), Fa (building one), and ERC (building two)), Acanthamoeba spp. only occurred in 8% (3/36) of the samples, while N. fowleri occurred in 33% (12/36). Acanthamoeba spp. only occurred in building Fa and the cooling towers, while. N. fowleri occurred in every sampling site except building Fa (Table 1). Water samples collected from the cooling towers were higher in percentage positives for Acanthamoeba spp. 83% (5/6) and N. fowleri 83% (5/6) than the drinking water system (Fa, ERC, and RES_IN, and RES_EF) (Table 1). The concentrations of Acanthamoeba spp. in building Fa ranged from 1.1 to 1.4 Log10 GC/100 mL. The concentrations of N. fowleri in the reservoir and building ERC ranged from 1.3 to 1.7 and 1.1 to 1.8 Log10 GC/100 mL, respectively (Table 1). The concentrations of Acanthamoeba spp. and N. fowleri in the cooling towers ranged from 2.0 to 2.5 and 1.3 to 2.4 Log10 GC/100 mL, respectively (Table 1).
Table 1.
Concentrations of amoebae and Legionella species increased in water samples collected from the cooling towers (CT) than in the drinking water system (RES_IN, RES_EF, Fa, and ERC). Influent, hot-, and cold-water samples were nested together for buildings Fa and ERC.
| Free-Living Amoebae | Site Location | ||||
|---|---|---|---|---|---|
| Res_In | Res_EF | Fa | ERC | CT | |
|
Acanthamoeba spp. (%) |
0/6 (0%) |
0/6 (0%) |
3/15 (20%) |
0/9 (0%) |
5/6 (83%) |
|
Acanthamoeba spp. Min, and Max Geomean (Log10 GC/100 mL) |
ND | ND | 1.1, 1.4 |
ND | 2.0, 2.5 |
|
N.fowleri (%) |
5/6 (83%) |
1/6 (16%) |
0/15 (0%) |
6/9 (66%) |
5/6 (83%) |
|
N.fowleri Min, and Max Geomean (Log10 GC/100 mL) |
1.3, 1.7 |
1.5 a | ND | 1.1, 1.8 |
1.3, 2.4 |
| Legionella Species | |||||
|
Legionella spp. (23S rRNA) (%) |
(6/6) (100%) |
(6/6) (100%) |
15/15 (100%) |
9/9 (100%) |
6/6 (100%) |
|
Legionella spp. (23S rRNA) Geomean (Log10 GC/100 mL) |
3.1 | 2.7 | 2.3 | 4.3 | 4.5 |
|
L. pneumophila (%) |
5/6 (83%) |
5/6 (83%) |
6/15 (40%) |
3/9 (33%) |
5/6 (83%) |
|
L. pneumophila Geomean (Log10 GC/100 mL) |
1.6 | 1.8 | 1.6 | 1.4 | 2.8 |
|
L. micdadei (%) |
1/6 (17%) |
0/6 (0%) |
3/15 (20%) |
4/9 (44%) |
2/6 (33%) |
|
L. micdadei Geomean (Log10 GC/100 mL) |
1.5 | ND | 1.5 | 1.6 | 2.4 |
|
L. bozemanii (%) |
4/6 (67%) |
(6/6) (100%) |
2/15 (13%) |
4/9 (44%) |
6/6 (100%) |
|
L. bozemanii Geomean (Log10 GC/100 mL) |
1.5 | 1.7 | 1.4 | 1.5 | 2.9 |
|
L. longbeachae (%) |
0/6 (0%) |
0/6 (0%) |
8/15 (53%) |
4/9 (44%) |
3/6 (50%) |
|
L. longbeachae Geomean (Log10 GC/100 mL) |
ND | ND | 1.4 | 1.4 | 1.5 |
|
L. anisa (%) |
0/6 (0%) |
0/6 (0%) |
5/15 (33%) |
0/6 (0%) |
4/6 (67%) |
|
L. anisa Geomean (Log10 GC/100 mL) |
ND | ND | 1.4 | ND | 2.1 |
a Only value detected. ND: no data.
L. pneumophila was detected at every sample location (reservoir, buildings, and the cooling towers), and the concentration ranged from 1.4 to 2.8 log10 GC/100 mL (Table 1). The other four Legionella species were detected throughout the water supply system, and the concentrations ranged from 1.3 to 2.9 log10 GC/100 mL (Table 1). Four Legionella species (L. pneumophila, L. micdadei, L. bozemanii, and L. anisa) were higher in the cooling towers than in the drinking water system (Table 1). In contrast, L. longbeachae concentration remained relatively the same through the drinking water system and cooling towers (Table 1).
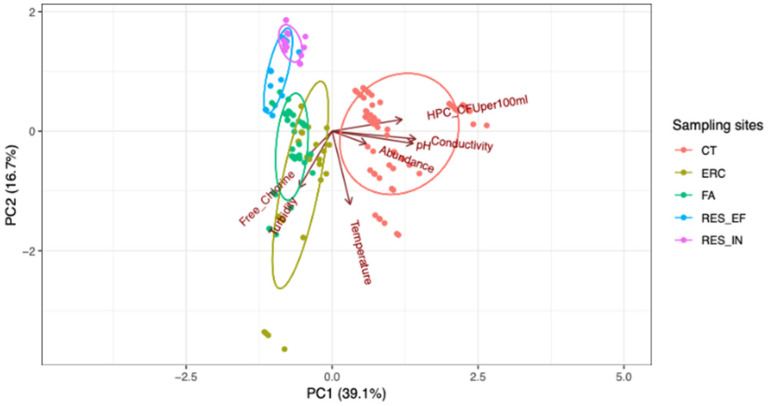
Figure 1 shows a clear separation between water samples collected from the cooling towers (CT) and those collected from the drinking water system (RES_IN, RES_EF, Fa, and ERC). The cooling towers’ chemical and microbial parameters are different from the drinking water system (Fa, ERC, and RES_IN, and RES_EF) (Table 2). For instance, the cooling towers are associated with higher bacterial abundance, pH, conductivity, HPC, and lower turbidity, and free chlorine (Table 2). Building Fa and ERC are characterized by free chlorine and turbidity, while reservoir clustering (RES_IN and RES_EF) are associated with lower temperature, abundance, pH, HPC, and conductivity (Table 2).
Figure 1.
Principal component analysis (PCA) biplot showed a clear separation between water samples collected from the cooling towers relative to those collected from the drinking water system (Fa, ERC, RES_IN, and RES_EF). Each data point represents each species from a particular sampling site (the observation). Samples from five sampling locations, color-coded based on sampling location. The absolute abundance of Legionella and amoebae species at each sampling site is depicted in the graph.
Table 2.
Water age impacted the microbial and chemical parameters in the building water system (Fa and ERC) and the cooling towers. Sample collection dates are as follows: reservoir (RES_IN and RES_EF), July 15th, 23rd, 29th and August 6th, 13th, and 20th; influent, hot-, and cold-water in building Fa, August 12th, and September 3rd, and 16th; influent, hot-, and cold-water in building ERC, August 19th, and September 9th, and 23rd; cooling towers, July 25th, 31st, and August 7th, 14th, and 21st. Sample replicates were collected on different days. Water age in the table is presented as hours, and it is defined as the time from the wells to the influent pipe, the influent pipe to the effluent pipe, the effluent pipe to the influent pipe of buildings, and the cooling towers. The dashes (–) indicate the same time as each respective building influent. Water quality parameters for building influent and potable water samples (hot-, and cold-water) are presented individually.
| Temperature (°C) | Total Chlorine (mg/L) | Free Chlorine (mg/L) | Turbidity NTU |
pH | Conductivity (mS) | HPC (CFU/100 mL) | Water Age (h) |
|---|---|---|---|---|---|---|---|
| RES_IN (N = 6) | |||||||
| 12.1 | 0 | 0 | 4.1 | 7.2 | 851 | 3.52 × 101 | 4.5 |
| RES_EF (N = 6) | |||||||
| 11.9 | 0.64 | 0.33 | 3.85 | 7.2 | 855 | 2.10 × 100 | 3.4 |
| Building F Influent (N = 3) | |||||||
| 26.8 | 0.41 | 0.35 | 8.4 | 7.3 | 897 | 8.57 × 104 | 9.2 |
|
Building Fa 1st Floor Cold; N = 3
(Hot Taps; N = 3) | |||||||
| 26.7 (28.6) |
0.16 (0.04) |
0.14 (0.02) |
3.06 (0.53) |
7.2 (7.1) |
867 (815) |
1.02 × 104 (7.3 × 103) |
– |
|
Building Fa 2nd Floor Cold; N = 3
(Hot Taps; N = 3) | |||||||
| 26.8 (28.8) |
0.05 (0.02) |
0.03 (0) |
3.37 (0.67) |
7.0 (6.9) |
856 (822) |
2.00 × 104 (3.15 × 103) |
– |
| Building ERC Influent (N = 3) | |||||||
| 31.5 | 0.31 | 0.20 | 12.5 | 7.4 | 883 | 4.32 × 105 | 20.8 |
|
Building ERC 1st Floor Cold (N = 3)
(Hot Taps; N = 3) | |||||||
| 23.5 (24.5) |
0.09 (0.04) |
0.03 (0) |
5.97 (6.27) |
7.6 (7.5) |
866 (847) |
4.38 × 105 (6.80 × 105) |
– |
| Cooling Towers (N = 6) | |||||||
| 25.3 | 0.49 | 0.08 | 1.94 | 8.2 | 2564 | 2.35 × 107 | 23.2 |
The chi-squared test of independence showed a significant association between two amoebae species and four pathogenic Legionella spp. in all 42 water samples: Acanthamoeba spp. and L. anisa (χ2 = 4.791, p = 0.0286), N. fowleri and L. micdadei (χ2 = 4.748, p = 0.0293), N. fowleri and L. pneumophila (χ2 = 4.356, p = 0.0369), N. fowleri and L. bozemanii (χ2 = 6.645, p = 0.0099). L. anisa-positive samples were observed in the presence or absence of Acanthamoeba spp. (Table 3). L. micdadei, L. bozemanii, and L. pneumophila-positive samples were also observed in the presence or absence of N. fowleri (Table 4, Table 5 and Table 6).
Table 3.
A total of 78% (33/42) of the water samples Acanthamoeba spp. and L. anisa co-exist or co-absent together.
| ap Value = 0.0286 * |
L. anisa Present |
L. anisa Absent |
Total |
|---|---|---|---|
|
Acanthamoeba spp. Present |
4 (9.52%) | 4 (9.52%) | 8 |
|
Acanthamoeba spp. Absent |
5 (11.90%) | 29 (69.05%) | 34 |
| Total | 9 (21.42%) | 33 (78.57%) | 42 |
a Strong p-value is ≤ 0.05. The asterisks (*) below represent the statistical significance between Acanthamoeba spp. and L. anisa; p = 0.0286; chi-square test.
Table 4.
A total of 76% (32/42) of the water samples N. fowleri and L. micdadei co-exist or co-absent together.
| ap Value = 0.0293 * |
L. micdadei Present |
L. micdadei Absent |
Total |
|---|---|---|---|
|
N.fowleri Present |
7 (16.67%) | 10 (23.81%) | 17 |
|
N.fowleri Absent |
3 (7.14%) | 22 (52.38%) | 25 |
| Total | 10 (23.8%) | 32 (76.19%) | 42 |
a Strong p-value is ≤ 0.05. The asterisks (*) below represent the statistical significance between N. fowleri and L. micdadei; p = 0.0293; chi-square test.
Table 5.
A total of 47% (20/42) of the water samples N. fowleri and L. bozemanii co-exist or co-absent together.
| ap Value = 0.0099 ** |
L. bozemanii Present |
L. bozemanii Absent |
Total |
|---|---|---|---|
|
N.fowleri Present |
13 (30.95%) | 4 (9.52%) | 17 |
|
N.fowleri Absent |
9 (21.43%) | 16 (38.10%) | 25 |
| Total | 22 (52.38%) | 20 (47.61%) | 42 |
a Strong p-value is ≤ 0.05. The asterisks (**) below represent the statistical significance between N. fowleri and L. bozemanii; p = 0.0099; chi-square test.
Table 6.
A total of 42% (18/42) of the water samples N. fowleri and L. pneumophila co-exist or co-absent together.
| ap Value = 0.0369 * |
L. pneumophila Present |
L. pneumophila Absent |
Total |
|---|---|---|---|
|
N.fowleri Present |
13 (30.95%) | 4 (9.52%) | 17 |
|
N.fowleri Absent |
11 (26.19%) | 14 (33.33%) | 25 |
| Total | 24 (57.14%) | 18 (42.85%) | 42 |
a Strong p-value is ≤ 0.05. The asterisks (*) below represent the statistical significance between N. fowleri and L. pneumophila; p = 0.0369; chi-square test.
The concordance percentage between the presence of Acanthamoeba spp. and L. anisa was 9.52%. The absence percentage of both species (Acanthamoeba spp. and L. anisa) was higher (69.05%) than the presence percentage (9.52%) (Table 3). A total of 78% of the time, either Acanthamoeba spp. and L. anisa co-exist or were co-absent together (Table 3). N. fowleri and three specific Legionella spp. (L. micdadei, L. bozemanii, and L. pneumophila) concordance positive percentage was 16.67%, 30.95%, and 30.95%, respectively (Table 4, Table 5 and Table 6). The absence percentage of N. fowleri and L. micdadei, L. bozemanii, and L. pneumophila were higher (52.38%, 38.10%, and 33.33%, respectively) than the positive percentage (Table 4, Table 5 and Table 6). A total of 76% of the time, either N. fowleri and L. micdadei, co-exist or were co-absent together (Table 4). In the absence of free-living amoebae, L. anisa, L. micdadei, L. bozemanii, and L. pneumophila occurred only 11%, 7%, 21%, and 26% of the time (it is hypothesized that amoebae hosts and/or the sloughing of the biofilm may have released Legionella species into the water supply system). Thus, the relationship that is observed between these two amoebae and Legionella species is driven by the absence percentage of both organisms (Table 3, Table 4, Table 5 and Table 6).
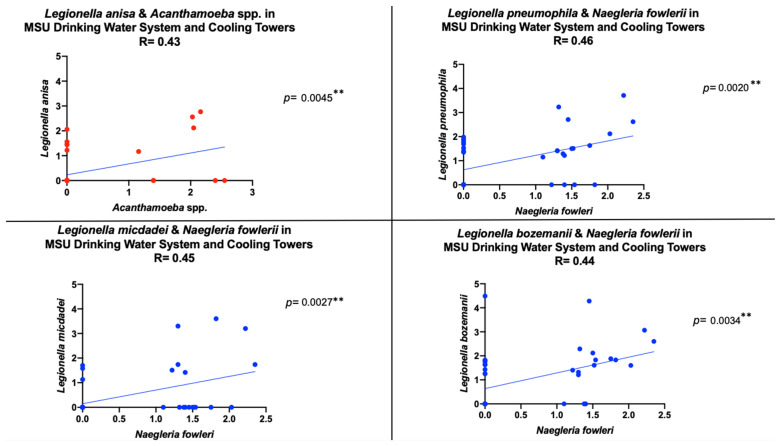
Figure 2 revealed a statistically significant positive correlation between Acanthamoeba spp. and L. anisa as well as between N. fowleri and L. micdadei, L. pneumophila, and L. bozemanii in all 42 water samples. Out of three influent water samples in the building Fa, Acanthamoeba spp. and L. anisa cooccurred in one sample (data not shown). Out of six cooling tower samples, Acanthamoeba spp. and L. anisa cooccurred in three water samples (data not shown). Thus, it is suggested that the relationship between Acanthamoeba spp. and L. anisa seen in Figure 2 was driven by the absence of both species in the drinking water system (except for building Fa) and the presence of both species in the cooling towers. Out of nine (influent and tap) water samples in building ERC, N. fowleri and L. micdadei cooccurred in three samples (data not shown). In the cooling tower samples, N. fowleri and L. micdadei cooccurred in 33% (1/6) of the water samples (data not shown). It is suggested that the relationship between N. fowleri and L. micdadei seen in Figure 2 was driven by the absence of these species.
Figure 2.
Statistically significant positive Pearson correlation between two amoeba species and four pathogenic Legionella spp. in 42 water samples. ** p < 0.01.
3. Discussion
This study revealed new quantitative information about the cooccurrence of five pathogenic Legionella species and two free-living amoebae species in a complete drinking water system. The chi-square test of independence showed a relationship between four Legionella species and two amoebae species. Interestingly, the relationship between Acanthamoeba spp. and L. anisa was driven by the absence of both species in the influent and effluent of the reservoir, as well as building ERC, and the presence of both species in the cooling towers (data not shown). The relationship between L. micdadei absence to N. fowleri absence, L. bozemanii absence to N. fowleri absence, and L. pneumophila absence to N. fowleri absence is driven by the building water quality and the cooling towers (data not shown). In this study, Acanthamoeba spp. and L. anisa, L. micdadei and N. fowleri, L. bozemanii and N. fowleri, and L. pneumophila and N. fowleri co-existence and absence occurred in 78%, 76%, 47%, and 42% of the water samples, respectively. The reason for this cooccurrence and collective absence is not completely clear; however, the water chemistry of the water supply system could be the contributing factor that is driving the microbial ecology. For example, the building water system (Fa and ERC) is characterized by free chlorine and turbidity, while the reservoir is associated with lower temperature, pH, HPC, and conductivity. The cooling towers are different from the drinking water system (reservoir and buildings) as they are associated with higher pH, conductivity, and lower free chlorine residual. Previous reports have also shown that Acanthamoeba spp. and N. fowleri were detected in a drinking water supply system [43] and cooling towers by qPCR [45].
The concentrations of the specific Legionella and amoebae species ranged from 1.3 to 2.9 and 1.1 to 2.5 log10 GC/100 mL, respectively, in the complete water supply system (from source to taps and cooling towers). The average concentration for both organisms (Legionella and amoebae species) was 1.6 log10 GC/100 mL. In this study, the cooling towers support high concentrations of Legionella and amoebae species relative to the building water system, and this fact is due to the water chemistry in the cooling towers (described above). Cooling towers have been the source of LD outbreaks [46]. This study further demonstrated that the cooling towers are a risk for LD outbreaks as this exposure site had increased concentrations of both Legionella species and its host, free-living amoebae. Currently, the commercial monitoring of cooling towers is solely focused on detecting L. pneumophila by culture-dependent methods and does not consider its host and vehicle of function, free-living amoebae [46,47,48,49]. Additionally, there are some limitations to the standard culture method [48,50]. For example, the culture method takes up to 10 days for results, it is incapable of enumerating viable but non-culturable cells (VBNC), and it is unable to identify Legionella species in a precise manner [50]. However, quantitative assessment using PCR can provide critical results on bacterial and amoebae concentrations to determine if there is amplification at a particular exposure site [51,52]. Digital droplet PCR was very useful and can be used to assess duplex assays for various species, as described in this study. Digital droplet PCR has been reported to show higher sensitivity and specificity for known concentrations of L. pneumophila in water samples than real-time quantitative PCR [53]. Additionally, ddPCR has been shown to quantify absolute, low DNA concentration and DNA in a host-pathogen interaction context [54,55]. Given such information, ddPCR was used to assess free-living amoebae as well as intra-amoebae Legionellae replication. While there are some limitations to assessing Legionella and amoebae host using ddPCR (it is incapable of distinguishing live and dead cells), there is an assumption that if all cells are culturable, the ratio of culture to the molecular method (PCR) is 7 CFU to 3 GC [56]. Thus, there is a need for a rapid, precise test to monitor the cooling towers. There is also a need for a greater comparison examining VBNC cells as well as live and dead cells to address the health risk by using and comparing molecular and culture methods to routinely quantify Legionella and amoebae species collected from various exposure sites (taps and cooling towers). A PCR method can serve as an early detection tool to warn the building owner of a potential outbreak, while the culture method can be used for the confirmation of the occurrence and concentration of these important species [51]. Overall, local and federal testing laboratories should incorporate a parallel method (culture and molecular method) to screen for both Legionella and host amoebae into their routine screening for taps and especially cooling towers.
The widespread of N. fowleri and Legionella species in the taps and cooling towers indicates an important health concern. As stated above, the cooling towers have different characteristics from the drinking water system; therefore, it is important to investigate free-living amoebae presence and concentration in a water supply system as they serve as vehicles for bacterial pathogens. In this study, N. fowleri was more often detected in the whole water supply system than Acanthamoeba spp. It is suggested that the microbial ecology and water chemistry affected the amplification of N. fowleri in the complete water supply system (water source, water age, and cooling towers). Consistent with this study, N. fowleri was frequently detected relative to Acanthamoeba spp. by PCR molecular technique in untreated and treated water [57], tap water [58], and from municipal drinking waters and recreational water sources [59]. This work presented herein demonstrated that L. micdadei and L. bozemanii were more often related to N. fowleri than Acanthamoeba spp., and the correlation between these species in the building water system and cooling towers observed separately (data not shown) could be due to the microbes being released into the water column from sloughing of the biofilms. Thus, it is important to identify the ecological factors that favor the presence of Legionella to improve its control in the built environment (taps, cooling towers). Several studies have focused on investigating and controlling Legionella [60,61,62] without considering the association with these host reservoirs, Naegleria and Acanthamoeba spp. It is hypothesized that if there is control of free-living amoeba, then one can subsequently control Legionella species in a water supply system.
A couple of studies have suggested that the concentrations of N. fowleri and L. pneumophila amplification in a distribution system may be seasonally influenced via water temperature [63,64]. In this study, the results showed that the detection of free-living amoeba and Legionella spp. was not due to ambient temperature. For example, pathogenic L. micdadei, L. bozemanii, and N. fowleri were detected more in a drinking water supply system that was supplied by a groundwater source (supplied by production wells), which is not affected by ambient temperatures such as surface waters. The occurrence of free-living amoeba and Legionella spp. may be affected by the built environment (i.e., water age). Water age has been observed to affect the detection of Legionella and amoebae species in a distribution system [59,64]. In this study, it is suggested that the increase in water age in the distribution system is associated with an increase in Legionella and amoebae species. Thus, water age may play a role in the occurrence of Legionella and amoebae species in a building with an increased water age, as seen in the ERC building (water age of 20.8 h) and the cooling towers (water age of 23.2 h).
Since free-living amoebae promote the growth of Legionella post-treatment in water systems [22], it is critical to gain an understanding of microbial-amoebae ecological relationships in large, complicated plumbing. The regression analysis (data not shown) revealed in this study showed that N. fowleri affects the occurrence of L. micdadei and L. bozemanii in building ERC, furthest away from the reservoir. The regression analysis emphasizes that it is a building issue, and the water quality in building ERC is driving the occurrence of N. fowlerii, L. micdadei, and L. bozemanii. However, the regression suggests that this observation will need to be further elucidated with a more extensive data set. An extensive data set such as increasing the total number of samples, sampling and analyzing the biofilm, and verifying the ddPCR results with DNA sequencing would overcome this study limitation. Overall, this study found a greater association between N. fowleri and two Legionella species.
4. Materials and Methods
4.1. Site Location and Sampling
Water samples were collected during July, August, and September of 2019 from the reservoir (influent: RES_IN and effluent: RES_EF), two research buildings (Fa and ERC), and six cooling towers on a large research institution that runs its own water system, all located in the state of Michigan. The reservoir contained two large storage tanks for the untreated (RES_IN) and treated water (RES_EF). The building characteristics (water usage, distance from the reservoir, and building age) are described below. The average monthly water usage (five-year average) and distance from the reservoir for Fa was 172,993 L/month and 4.7 km and for ERC was 738,533 L/month and 19.4 km. At the time of sample collection, the building age for Fa (construction year: 1948) and ERC (construction year 1986) were 71 and 33 years, respectively.
A total of 42 large-volume samples were collected from the RES_IN and RES_EF, two research buildings (Fa and ERC), and six cooling towers. The water supply system was supplied by a groundwater source. At the time of sampling, the groundwater source contained ~20 production wells; these wells drew the groundwater from an aquifer to the influent storage tank. The reservoir had two large water storage tanks: influent (untreated groundwater) and effluent (treated groundwater). Ten liters were collected from each site location (RES_IN and RES_EF), buildings (Fa and ERC), and cooling towers in a carboy containing 10% sodium thiosulfate to neutralize the residual chlorine. Ten liters of grab sample was collected from influent and effluent of the reservoir. For the building influent samples, 10 L grab sampling were collected from both building’s (Fa and ERC) influent sampling port. Building Fa influent pipe contained a sampling port, which was accessible for sample collection. Building ERC did not have a sampling port on the influent pipe; thus, it was decided to sample the nearest valve to the influent pipe, which was an eye-wash station located in a mechanical room, where the influent pipe entered the building. For the potable tap samples, 10 L composite sampling from the cold- and hot-water taps were collected as separate samples to evaluate and compare the water quality of the cold-water taps and the hot-water taps on the first and top floor, respectively. The volume of composite sampling was based on the number of sinks per floor. The points of use locations were sink faucets and showerheads located in bathrooms, locker rooms, and breakrooms. Building Fa had two floors, with two and three sinks on the first and second floors, respectively. Building ERC only had one floor, with 11 sinks and 2 showers. Because building Fa had two sinks on the first floor, 5 L of water was collected from each tap (cold and hot) and composited to make a total of two 10 L large samples for the first floor. This sampling regime was the same for the second floor of Fa and building ERC. Per sampling date, building Fa had a total of five samples, one from the influent sampling location and two composite samples per tap per floor (one cold-water composite sampling and one hot-water composite sampling/floor). Building ERC had a total of three samples: one for the influent sampling location and two composite samples (cold- and hot-water). There were several cooling towers on the research institution, but only six from the power plant on campus were sampled. There were sampling replicates per sampling site, and it is as follows: the reservoir and cooling towers were sampled six different times on different days, and each building was sampled three different times on different days. Thus, there were a total of 12 samples collected from the reservoir: influent (6 from RES_IN) and effluent (six from RES_EF), 6 from the cooling towers, and 24 from the buildings (15 from building Fa, nine from building ERC). Each water sample was processed immediately after collection.
4.2. Chemical and Physical Analyses
A 300 mL sample was collected separately from the large volume (mentioned above) for chemical and physical parameters. The water samples were analyzed for temperature and chlorine residuals (total and free) onsite using calibrated thermometers and the Test Kit Pocket Colorimeter II (HACH, Loveland, CO, USA) according to the manufacturer’s instructions. After sampling, conductivity, pH, and turbidity were measured offsite according to the manufacturers’ instructions using a Russell RL060C Portable Conductivity Meter (Thermo Scientific, Waltham, MA, USA), UltraBasic pH meter (Denver Instrument, Bohemia, NY, USA), and a Turbidity Meter code 1970-EPA (LaMotte Company, Chestertown, MD, USA).
4.3. Microbiological Analysis
All samples were transported on ice to the laboratory and processed the same day. All samples collected for this study were tested for heterotrophic plate count (HPC) analyses using membrane filters (47 mm diameter, 0.45 μm pore size) (PALL Corporation, Port Washington, NY, USA) on m-HPC agar (Becton, Dickinson and Company, Difco™, Detroit, MI, USA). All plates were incubated for 48 ± 2 h at 35–37 °C, then enumerated for colony-forming units (CFU).
4.4. Water Sample Processing and DNA Extraction
The 10 L water samples were processed using a single-use Asahi REXEED-25S dialysis filter (Dial Medical Supply, Chester Springs, PA, USA). The Asahi REXEED filter was pretreated with 0.01% of sodium hexametaphosphate (used to trap microbial material onto each ultrafilter) and was used in a dead-end mode. After filtration, a high-pressure single-use elution fluid canister (Innovaprep LLC, Drexel, MO, USA) was used to concentrate the 10 L large volume to ∼50 mL. The 50 mL sample was aliquoted into 5–10 mL samples.
4.5. Molecular Analysis
For each sample, one 10 mL (remaining sample was stored at −80 °C) subsample was further filtered through a 47 mm, 0.45 μm polycarbonate filter (Whatman, Kent, U.K.) for DNA extraction. The polycarbonate filter was folded into a 1/8 shape with the contents of the filter folded to the inside. The filter was then transferred to a 2.0 mL polypropylene screw cap tube (VWR, Radnor, PA, USA), which contained 0.3 g of 212–300 μm acid-washed glass beads (Sigma, St. Louis, MO, USA) for a crude DNA extraction procedure, which is described below. DNA extraction was performed by adding 590 μL of AE buffer (Qiagen, Redwood City, CA, USA) to the samples to assist with lysing the cell membrane, and then bead milling using a FastPrep-24™ 5G Instrument MP Biomedicals (VWR, Radnor, PA, USA). Samples were bead-milled at 6000 rpm for one minute, then followed by centrifugation at 12,000× g for an additional minute (60 s). Approximately 400 μL of the supernatant was transferred to a new clean microcentrifuge tube and centrifuged at 12,000× g for an additional three minutes to pellet any remaining debris. Approximately 350 uL of extracted nucleic acid was eluted into a final clean microcentrifuge tube. Sixty microliters of the eluted volume were then aliquoted into several microcentrifuge tubes (∼five extraction replicates per sample) for storage at −80 °C to reduce the need for several freeze/thaw cycles. One aliquot per water sample was later used for PCR analysis (samples were held at −80 °C for up to 30 days before analysis).
4.6. Molecular Analysis of Acanthamoeba spp., N. fowleri, General Legionella spp., and Four Pathogenic Legionella Species
Droplet digital PCR (Bio-Rad Laboratories, Hercules, CA, USA) technology was performed according to the manufacturer’s instructions to analyze each sample for two amoebae species (Acanthamoeba spp. N. fowleri) and five pathogenic Legionella spp. (L. pneumophila, L. anisa, L. micdadei, L. bozemanii, and L. longbeachae). The primers and probes listed in this study are listed in Table 7. Duplex reactions were performed for four separate assays: the first assay consisted of Legionella spp. (data not shown) and L. pneumophila, the second assay comprised of L. micdadei and L. anisa, the third assay consisted of L. bozemanii and L. longbeachae, and the fourth assay contained Acanthamoeba spp. N. fowler (Table 7). All primers and probes were ordered from Eurofins Genomics Co., (Louisville, KY, USA).
Table 7.
Free-living amoebae primers and probes. 23S pan-Legionella is conserved by all species of Legionella, but the probes were uniquely designed to specifically identify each of these species: L. micdadei, L. anisa, L. bozemanii, and L. longbeachae. All primers and probes were designed and validated previously by the authors listed in the references below.
| Target Species | Primer/Probe Name | Primer/Probe Sequence | Amplicon Length (bp) | Reference |
|---|---|---|---|---|
| Acanthamoeba spp. | 18S rRNAF 18S rRNAR 18S rRNAP |
5′-CGACCAGCGATTAGGAGACG-3′ 5′-CCGACGCCAAGGACGAC-3′ 5′-FAM-TGAATACAAAACACCACCATCGGCGC-BHQ1-3′ |
63 | [65] |
| N. fowleri | ITSF ITSR ITSP |
5′-GTGAAAACCTTTTTTCCATTTACA-3′ 5′-AAATAAAAGATTGACCATTTGAAA-3′ 5′-HEX-GTGGCCCACGACAGCTTT-BHQ1-3′ |
69 | [66,67] |
| Legionella species | 23SF 23SR 23SP |
5′-CCCATGAAGCCCGTTGAA-3′ 5′-ACAATCAGCCAATTAGTACGAG TTAGC-3′ 5′-HEX-TCCACACCTCGCCTATCAACGTCGTAGT-BHQ1-3′ |
92 | [68] |
| L. pneumophila | mipF mipR mipP |
5′-AAAGGCATGCAAGACGCTATG-3′ 5′-GAAACTTGTTAAGAACGTCTTTCATTTG-3′ 5′-FAM-TGGCGCTCAATTGGCTTTAACCGA-BHQ1-3′ |
78 | [68] |
|
L. micdadei
L. anisa L. bozemanii L. longbeachae |
Pan-Legionella F Pan-Legionella R LmicdadeiP LanisaP Lbozemanii LlongbeachaeP |
5′-GTACTAATTGGCTGATTGTCTTG-3′ 5′-TTCACTTCTGAGTTCGAGATGG-3′ 5′-FAM-AGCTGATTGGTTAATAGCCCAATCGG-BHQ1-3′ 5′-HEX-CTCAACCTACGCAGAACTACTTGAGG-BHQ1-3′ 5′-FAM-TACGCCCATTCATCATGCAAACCAGnT-BHQ1-3′ 5′-HEX-CTGAGTATCATGCCAATAATGCGCGC-BHQ1-3′ |
Not available | [69] |
Positive controls using genomic DNA from amoebae, A. castellani strain Neff (ATCC No. 30010) and N. fowleri (ATCC No. 30174) and bacteria, L. pneumophila (ATCC No. No. 33152), L. anisa (ATCC No. 35292), L. micdadei (ATCC No. 33218), L. bozemanii (ATCC No. 33217), and L. longbeachae (ATCC No. 33462), were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). Each control (positive and negative) was run with each ddPCR plate. As part of the quality control, sample results were only considered for analysis when the reader accepted 10,000 or more droplets. Per assay, there was one ddPCR run with three biological (sample collection replicates) and three technical replicates (same extraction) tested. Biological and technical replicates were run for each sample to determine if there was a wide variation among sampling days and whether the assay results were reproducible, respectively.
Droplet digital PCR technology was performed according to the manufacturer’s instructions. In brief, the reaction mixture consisted of 2X supermix (no dUTP) (Bio-Rad Laboratories, Hercules, CA, USA), 900nM forward and reverse primers, and 250 nM for each organism (bacteria and amoebae) probe (Eurofins Genomics Co., Lousiville, KY, USA), and up to 330 ng of DNA template, to a final volume of 20 μL. Twenty microliters of the sample reaction mixtures were loaded into a DG8 cartridge (Bio-Rad Laboratories, Hercules, CA, USA), followed by 70 μL of droplet generator oil (Bio-Rad Laboratories, CA, USA). Droplets were generated using the QX200™ Droplet Generator Bio-Rad) and transfer of emulsified samples to a ddPCR 96-well plate (semi-skirted) and was performed according to manufacturer’s instructions (Instruction Manual, QX200™ Droplet Generator—Bio-Rad). The ddPCR plate was sealed with pierceable foil heat seals using a PX1TM PCR Plate Sealer (Bio-Rad, Laboratories, CA, USA). The plate was amplified using a Benchmark TC9639 thermal cycler (Benchmark Scientific Inc, Sayreville, NJ, USA). The cycling protocol was as follows: 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 sec and 57 °C for 1 min with a final 10 min cycle at 98 °C for 10 min. After endpoint amplification, droplets were read using a QX200 droplet reader (Bio-Rad QX200TM Droplet DigitalTM PCR System, Hecules, CA, USA). Two negative controls, a filtration blank (phosphate-buffered water) and a non-template control (molecular grade water), were run with each ddPCR plate. Negative and positive controls were used to determine contamination (if any) and the efficiency of the assay.
4.7. Statistical Analysis
Statistical analysis was performed in GraphPad Prism 8 software (GraphPad Software, San Diego, CA, USA). Contingency tables, chi-squared tests, and Pearson correlation were used to evaluate the associations between the occurrence of Legionella spp. and free-living amoebae. Additionally, the principal component analysis was also conducted to assess the associations between the cooccurrence of both organisms and different water quality parameters.
Sample concentrations were transformed from (GC)/100 mL into Log10 GC/100 mL. Biological data were expressed as geometric means, and chemical data were shown as arithmetic means with standard deviation [52]. Statistical results were interpreted at the level of significance p < 0.05.
5. Conclusions
Most of the public health agencies and guidance documents have water management programs that are focused on the risk of Legionella, without considering their host, Naegleria, and Acanthamoeba spp. Using ddPCR, N. fowleri was detected more often in a drinking water supply system and cooling towers. The chi-squared test showed that N. fowleri significantly cooccurred with three pathogenic Legionella spp. (L. pneumophila, L. micdadei, and L. bozemanii) in a drinking water supply system and cooling towers. In this study, L. micdadei and L. bozemanii were more often related to N. fowleri than Acanthamoeba species. Thus, by examining large volume (10 L), water ultrafiltrate concentrates from the groundwater source to exposure sites (taps and cooling towers) using ddPCR allowed for the detection of individual-specific Legionella and N. fowleri. Most importantly, the widespread of N. fowleri and Legionella species in the taps and cooling towers indicates an important health concern.
Acknowledgments
A.L.-J. thanks Matt Flood, Parker Kelly, and Rebecca Ives for their assistance in sampling.
Author Contributions
Conceptualization, A.L.-J. and J.B.R.; methodology, A.L.-J. analysis, A.L.-J.: investigation, A.L.-J.: resources, J.B.R.; writing—original draft preparation, A.L.-J.: writing—review and editing, A.L.-J. and J.B.R.; visualization, A.L.-J.: supervision, J.B.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the U.S. EPA Grant No. (FAIN): 83689001 and U.S. EPA agreement number R836890. The contents of this publication are solely the responsibility of the authors and do not represent the official views of the EPA.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Armand B., Motazedian M.H., Asgari Q. Isolation and identification of pathogenic free-living amoeba from surface and tap water of Shiraz City using morphological and molecular methods. Parasitol. Res. 2016;115:63–68. doi: 10.1007/s00436-015-4721-7. [DOI] [PubMed] [Google Scholar]
- 2.Lares-García C., Otero-Ruiz A., Gonzalez-Galaviz J.R., Ibarra-Gámez J.C., Lares-Villa F. Potentially pathogenic genera of free-living amoebae coexisting in a thermal spring. Exp. Parasitol. 2018;195:54–58. doi: 10.1016/j.exppara.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Baquero R.A., Reyes-Batlle M., Nicola G.G., Martín-Navarro C.M., López-Arencibia A., Guillermo Esteban J., Valladares B., Martínez-Carretero E., Piñero J.E., Lorenzo-Morales J. Presence of potentially pathogenic free-living amoebae strains from well water samples in Guinea-Bissau. Pathog. Glob. Health. 2014;108:206–211. doi: 10.1179/2047773214Y.0000000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delafont V., Brouke A., Bouchon D., Moulin L., Héchard Y. Microbiome of free-living amoebae isolated from drinking water. Water Res. 2013;47:6958–6965. doi: 10.1016/j.watres.2013.07.047. [DOI] [PubMed] [Google Scholar]
- 5.Retana-Moreira L., Abrahams-Sandí E., Cabello-Vílchez A.M., Reyes-Batlle M., Valladares B., Martínez-Carretero E., Piñero J.E., Lorenzo-Morales J. Isolation and molecular characterization of Acanthamoeba and Balamuthia mandrillaris from combination shower units in Costa Rica. Parasitol. Res. 2014;113:4117–4122. doi: 10.1007/s00436-014-4083-6. [DOI] [PubMed] [Google Scholar]
- 6.Canals O., Serrano-Suárez A., Salvadó H., Méndez J., Cervero-Aragó S., Ruiz de Porras V., Dellundé J., Araujo R. Effect of chlorine and temperature on free-living protozoa in operational man-made water systems (cooling towers and hot sanitary water systems) in Catalonia. Environ. Sci. Pollut. Res. Int. 2015;22:6610–6618. doi: 10.1007/s11356-014-3839-y. [DOI] [PubMed] [Google Scholar]
- 7.Ren K., Xue Y., Rønn R., Liu L., Chen H., Rensing C., Yang J. Dynamics and determinants of amoeba community, occurrence, and abundance in subtropical reservoirs and rivers. Water Res. 2018;146:177–186. doi: 10.1016/j.watres.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Shaheen M., Scott C., Ashbolt N.J. Long-term persistence of infectious Legionella with free-living amoebae in drinking water biofilms. Int. J. Hyg. Environ. Health. 2019;222:678–686. doi: 10.1016/j.ijheh.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Gomes T.S., Vaccaro L., Magnet A., Izquierdo F., Ollero D., Martínez-Fernández C., Mayo L., Moran M., Pozuelo M.J., Fenoy S., et al. Presence and interaction of free-living amoebae and amoeba-resisting bacteria in water from drinking water treatment plants. Sci. Total. Environ. 2020;719:137080. doi: 10.1016/j.scitotenv.2020.137080. [DOI] [PubMed] [Google Scholar]
- 10.Michel R., Hauröder-Philippczyk B., Müller K.-D., Weishaar I. Acanthamoeba from human nasal mucosa infected with an obligate intracellular parasite. Eur. J. Protistol. 1994;30:104–110. doi: 10.1016/S0932-4739(11)80203-8. [DOI] [Google Scholar]
- 11.Rowbotham T.J. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z., Lin Y.E., Stout J.E., Hwang C.C., Vidic R.D., Yu V.L. Effect of flow regimes on the presence of Legionella within the biofilm of a model plumbing system. J. Appl. Microbiol. 2006;101:437–442. doi: 10.1111/j.1365-2672.2006.02970.x. [DOI] [PubMed] [Google Scholar]
- 13.Thomas V., Herrera-Rimann K., Blanc D.S., Greub G. Biodiversity of amoebae and amoeba-resisting bacteria in a hospital water network. Appl. Environ. Microbiol. 2006;72:2428–2438. doi: 10.1128/AEM.72.4.2428-2438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas V., Loret J.-F., Jousset M., Greub G. Biodiversity of amoebae and amoebae-resisting bacteria in a drinking water treatment plant. Environ. Microbiol. 2008;10:2728–2745. doi: 10.1111/j.1462-2920.2008.01693.x. [DOI] [PubMed] [Google Scholar]
- 15.Rohr U., Weber S., Michel R., Selenka F., Wilhelm M. Comparison of Free-Living Amoebae in Hot Water Systems of Hospitals with Isolates from Moist Sanitary Areas by Identifying Genera and Determining Temperature Tolerance. Appl. Environ. Microbiol. 1998;64:1822–1824. doi: 10.1128/AEM.64.5.1822-1824.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buse H.Y., Lu J., Struewing I.T., Ashbolt N.J. Eukaryotic diversity in premise drinking water using 18S rDNA sequencing: Implications for health risks. Environ. Sci. Pollut. Res. 2013;20:6351–6366. doi: 10.1007/s11356-013-1646-5. [DOI] [PubMed] [Google Scholar]
- 17.Kilvington S., Price J. Survival of Legionella pneumophila within cysts of Acanthamoeba polyphaga following chlorine exposure. J. Appl. Bacteriol. 1990;68:519–525. doi: 10.1111/j.1365-2672.1990.tb02904.x. [DOI] [PubMed] [Google Scholar]
- 18.De Giglio O., Napoli C., Apollonio F., Brigida S., Marzella A., Diella G., Calia C., Scrascia M., Pacifico C., Pazzani C., et al. Occurrence of Legionella in groundwater used for sprinkler irrigation in Southern Italy. Environ. Res. 2019;170:215–221. doi: 10.1016/j.envres.2018.12.041. [DOI] [PubMed] [Google Scholar]
- 19.Shen S.-M., Chou M.-Y., Hsu B.-M., Ji W.-T., Hsu T.-K., Tsai H.-F., Huang Y.-L., Chiu Y.-C., Kao E.-S., Kao P.-M., et al. Assessment of Legionella pneumophila in recreational spring water with quantitative PCR (Taqman) assay. Pathog. Glob. Health. 2015;109:236–241. doi: 10.1179/2047773215Y.0000000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishizaki N., Sogawa K., Inoue H., Agata K., Edagawa A., Miyamoto H., Fukuyama M., Furuhata K. Legionella thermalis sp. Nov., isolated from hot spring water in Tokyo, Japan. Microbiol. Immunol. 2016;60:203–208. doi: 10.1111/1348-0421.12366. [DOI] [PubMed] [Google Scholar]
- 21.Leoni E., Catalani F., Marini S., Dallolio L. Legionellosis Associated with Recreational Waters: A Systematic Review of Cases and Outbreaks in Swimming Pools, Spa Pools, and Similar Environments. Int. J. Environ. Res. Public Health. 2018;15:1612. doi: 10.3390/ijerph15081612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dey R., Bodennec J., Mameri M.O., Pernin P. Free-living freshwater amoebae differ in their susceptibility to the pathogenic bacterium Legionella pneumophila. FEMS Microbiol. Lett. 2009;290:10–17. doi: 10.1111/j.1574-6968.2008.01387.x. [DOI] [PubMed] [Google Scholar]
- 23.Greub G., Raoult D. Microorganisms Resistant to Free-Living Amoebae. Clin. Microbiol. Rev. 2004;17:413–433. doi: 10.1128/CMR.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDade J.E., Shepard C.C., Fraser D.W., Tsai T.R., Redus M.A., Dowdle W.R. Legionnaires’ disease: Isolation of a bacterium and demonstration of its role in other respiratory disease. N. Engl. J. Med. 1977;297:1197–1203. doi: 10.1056/NEJM197712012972202. [DOI] [PubMed] [Google Scholar]
- 25.Kaufmann A.F., McDade J.E., Patton C.M., Bennett J.V., Skaliy P., Feeley J.C., Anderson D.C., Potter M.E., Newhouse V.F., Gregg M.B., et al. Pontiac fever: Isolation of the etiologic agent (Legionella pneumophilia) and demonstration of its mode of transmission. Am. J. Epidemiol. 1981;114:337–347. doi: 10.1093/oxfordjournals.aje.a113200. [DOI] [PubMed] [Google Scholar]
- 26.Abu Kwaik Y., Gao L.-Y., Stone B.J., Venkataraman C., Harb O.S. Invasion of Protozoa by Legionella pneumophila and Its Role in Bacterial Ecology and Pathogenesis. Appl. Environ. Microbiol. 1998;64:3127–3133. doi: 10.1128/AEM.64.9.3127-3133.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albert-Weissenberger C., Cazalet C., Buchrieser C. Legionella pneumophila—A human pathogen that co-evolved with fresh water protozoa. Cell. Mol. Life Sci. 2006;64:432. doi: 10.1007/s00018-006-6391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cirillo J.D., Falkow S., Tompkins L.S. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 1994;62:3254–3261. doi: 10.1128/iai.62.8.3254-3261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nora T., Lomma M., Gomez-Valero L., Buchrieser C. Molecular mimicry: An important virulence strategy employed by Legionella pneumophila to subvert host functions. Future Microbiol. 2009;4:691–701. doi: 10.2217/fmb.09.47. [DOI] [PubMed] [Google Scholar]
- 30.Dupuy M., Mazoua S., Berne F., Bodet C., Garrec N., Herbelin P., Ménard-Szczebara F., Oberti S., Rodier M.-H., Soreau S., et al. Efficiency of water disinfectants against Legionella pneumophila and Acanthamoeba. Water Res. 2011;45:1087–1094. doi: 10.1016/j.watres.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 31.Hwang M.G., Katayama H., Ohgaki S. Effect of intracellular resuscitation of Legionella pneumophila in Acanthamoeba polyphage cells on the antimicrobial properties of silver and copper. Environ. Sci. Technol. 2006;40:7434–7439. doi: 10.1021/es060412t. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen T.M.N., Ilef D., Jarraud S., Rouil L., Campese C., Che D., Haeghebaert S., Ganiayre F., Marcel F., Etienne J., et al. A community-wide outbreak of legionnaires disease linked to industrial cooling towers—How far can contaminated aerosols spread? J. Infect. Dis. 2006;193:102–111. doi: 10.1086/498575. [DOI] [PubMed] [Google Scholar]
- 33.Buse H.Y., Schoen M.E., Ashbolt N.J. Legionellae in engineered systems and use of quantitative microbial risk assessment to predict exposure. Water Res. 2012;46:921–933. doi: 10.1016/j.watres.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 34.Ferré M.R.S., Arias C., Oliva J.M., Pedrol A., García M., Pellicer T., Roura P., Domínguez A. A community outbreak of Legionnaires’ disease associated with a cooling tower in Vic and Gurb, Catalonia (Spain) in 2005. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2009;28:153–159. doi: 10.1007/s10096-008-0603-6. [DOI] [PubMed] [Google Scholar]
- 35.Türetgen I., Sungur E.I., Cotuk A. Enumeration of Legionella pneumophila in cooling tower water systems. Environ. Monit. Assess. 2005;100:53. doi: 10.1007/s10661-005-7058-3. [DOI] [PubMed] [Google Scholar]
- 36.Bollin G.E., Plouffe J.F., Para M.F., Hackman B. Aerosols containing Legionella pneumophila generated by shower heads and hot-water faucets. Appl. Environ. Microbiol. 1985;50:1128–1131. doi: 10.1128/aem.50.5.1128-1131.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borella P., Montagna M.T., Romano-Spica V., Stampi S., Stancanelli G., Triassi M., Neglia R., Marchesi I., Fantuzzi G., Tatò D., et al. Legionella Infection Risk from Domestic Hot Water. Emerg. Infect. Dis. 2004;10:457–464. doi: 10.3201/eid1003.020707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraser D.W., Tsai T.R., Orenstein W., Parkin W.E., Beecham H.J., Sharrar R.G., Harris J., Mallison G.F., Martin S.M., McDade J.E., et al. Legionnaires’ disease: Description of an epidemic of pneumonia. N. Engl. J. Med. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 39.Visvesvara G.S., Moura H., Schuster F.L. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 40.Yu V.L., Plouffe J.F., Pastoris M.C., Stout J.E., Schousboe M., Widmer A., Summersgill J., File T., Heath C.M., Paterson D.L., et al. Distribution of Legionella Species and Serogroups Isolated by Culture in Patients with Sporadic Community-Acquired Legionellosis: An International Collaborative Survey. J. Infect. Dis. 2002;186:127–128. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 41.Visvesvara G. Free-Living Amebae as Opportunistic Agents of Human Disease. J. Neuroparasitol. 2010;1:1689–1693. doi: 10.4303/jnp/N100802. [DOI] [Google Scholar]
- 42.Booton G.C., Visvesvara G.S., Byers T.J., Kelly D.J., Fuerst P.A. Identification and Distribution of Acanthamoeba Species Genotypes Associated with Nonkeratitis Infections. J. Clin. Microbiol. 2005;43:1689–1693. doi: 10.1128/JCM.43.4.1689-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valciņa O., Pūle D., Mališevs A., Trofimova J., Makarova S., Konvisers G., Bērziņš A., Krūmiņa A. Cooccurrence of Free-Living Amoeba and Legionella in Drinking Water Supply Systems. Medicina. 2019;55:492. doi: 10.3390/medicina55080492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muchesa P., Leifels M., Jurzik L., Barnard T.G., Bartie C. Detection of amoeba-associated Legionella pneumophila in hospital water networks of Johannesburg. S. Afr. J. Infect. Dis. 2018;33:72–75. doi: 10.4102/sajid.v33i3.8. [DOI] [Google Scholar]
- 45.Scheikl U., Sommer R., Kirschner A., Rameder A., Schrammel B., Zweimüller I., Wesner W., Hinker M., Walochnik J. Free-living amoebae (FLA) co-occurring with Legionellae in industrial waters. Eur. J. Protistol. 2014;50:422–429. doi: 10.1016/j.ejop.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheikl U., Tsao H.-F., Horn M., Indra A., Walochnik J. Free-living amoebae and their associated bacteria in Austrian cooling towers: A 1-year routine screening. Parasitol. Res. 2016;115:3365–3374. doi: 10.1007/s00436-016-5097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edagawa A., Kimura A., Doi H., Tanaka H., Tomioka K., Sakabe K., Nakajima C., Suzuki Y. Detection of culturable and nonculturable Legionella species from hot water systems of public buildings in Japan. J. Appl. Microbiol. 2008;105:2104–2114. doi: 10.1111/j.1365-2672.2008.03932.x. [DOI] [PubMed] [Google Scholar]
- 48.Barrette I. Comparison of Legiolert and a Conventional Culture Method for Detection of Legionella pneumophila from Cooling Towers in Québec. J. AOAC Int. 2019;102:1235–1240. doi: 10.5740/jaoacint.18-0245. [DOI] [PubMed] [Google Scholar]
- 49.Steinert M., Emödy L., Amann R., Hacker J. Resuscitation of viable but nonculturable Legionella pneumophila Philadelphia JR32 by Acanthamoeba castellanii. Appl. Environ. Microbiol. 1997;63:2047–2053. doi: 10.1128/aem.63.5.2047-2053.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whiley H., Taylor M. Legionella detection by culture and qPCR: Comparing apples and oranges. Crit. Rev. Microbiol. 2016;42:65–74. doi: 10.3109/1040841X.2014.885930. [DOI] [PubMed] [Google Scholar]
- 51.Young C., Smith D., Wafer T., Crook B. Rapid Testing and Interventions to Control Legionella Proliferation Following a Legionnaires’ Disease Outbreak Associated with Cooling Towers. Microorganisms. 2021;9:615. doi: 10.3390/microorganisms9030615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Logan-Jackson A.R., Flood M., Rose J.B. Enumeration and characterization of five pathogenic Legionella species from large research and educational buildings. Environ. Sci. Water Res. Technol. 2020 doi: 10.1039/D0EW00893A. [DOI] [Google Scholar]
- 53.Falzone L., Gattuso G., Lombardo C., Lupo G., Grillo C.M., Spandidos D.A., Libra M., Salmeri M. Droplet digital PCR for the detection and monitoring of Legionella pneumophila. Int. J. Mol. Med. 2020;46:1777–1782. doi: 10.3892/ijmm.2020.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneiders S., Hechard T., Edgren T., Avican K., Fällman M., Fahlgren A., Wang H. Spatiotemporal Variations in Growth Rate and Virulence Plasmid Copy Number during Yersinia Pseudotuberculosis Infection. Infect. Immun. 2021;89:e00710-20. doi: 10.1128/IAI.00710-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Bai R., Zhao Z., Tao L., Ma M., Ji Z., Jian M., Ding Z., Dai X., Bao F., et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci. Rep. 2018;38:BSR20181170. doi: 10.1042/BSR20181170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang G., Benson R., Pelish T., Brown E., Winchell J.M., Fields B. Dual detection of Legionella pneumophila and Legionella species by real-time PCR targeting the 23S-5S rRNA gene spacer region. Clin. Microbiol. Infect. 2010;16:255–261. doi: 10.1111/j.1469-0691.2009.02766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abdul Majid M.A., Mahboob T., Mong B.G., Jaturas N., Richard R.L., Tian-Chve T., Phimphila A., Mahaphonh P., Aye K.N., Aung W.L., et al. Pathogenic waterborne free-living amoebae: An update from selected Southeast Asian countries. PLoS ONE. 2017;12:e0169448. doi: 10.1371/journal.pone.0169448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gabriel S., Khan N.A., Siddiqui R. Occurrence of free-living amoebae (Acanthamoeba, Balamuthia, Naegleria) in water samples in Peninsular Malaysia. J. Water Health. 2019;17:160–171. doi: 10.2166/wh.2018.164. [DOI] [PubMed] [Google Scholar]
- 59.Ambrose M., Kralovic S.M., Roselle G.A., Kowalskyj O., Rizzo V.J., Wainwright D.L., Gamage S.D. Implementation of Legionella Prevention Policy in Health Care Facilities: The United States Veterans Health Administration Experience. J. Public Health Manag. Pract. 2020;26:E1. doi: 10.1097/PHH.0000000000000986. [DOI] [PubMed] [Google Scholar]
- 60.Donohue M.J., King D., Pfaller S., Mistry J.H. The sporadic nature of Legionella pneumophila, Legionella pneumophila Sg1 and Mycobacterium avium occurrence within residences and office buildings across 36 states in the United States. J. Appl. Microbiol. 2019;126:1568–1579. doi: 10.1111/jam.14196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozak N.A., Lucas C.E., Winchell J.M. Identification of Legionella in the environment. Methods Mol. Biol. 2013;954:3–25. doi: 10.1007/978-1-62703-161-5_1. [DOI] [PubMed] [Google Scholar]
- 62.Breiman R.F. Impact of Technology on the Emergence of Infectious Diseases. Epidemiol. Rev. 1996;18:4–9. doi: 10.1093/oxfordjournals.epirev.a017915. [DOI] [PubMed] [Google Scholar]
- 63.Marciano-Cabral F., Jamerson M., Kaneshiro E.S. Free-living amoebae, Legionella and Mycobacterium in tap water supplied by a municipal drinking water utility in the USA. J. Water Health. 2010;8:71–82. doi: 10.2166/wh.2009.129. [DOI] [PubMed] [Google Scholar]
- 64.Wang H., Masters S., Hong Y., Stallings J., Falkinham J.O., Edwards M.A., Pruden A. Effect of disinfectant, water age, and pipe material on occurrence and persistence of Legionella, mycobacteria, Pseudomonas aeruginosa, and two amoebas. Environ. Sci. Technol. 2012;46:11566–11574. doi: 10.1021/es303212a. [DOI] [PubMed] [Google Scholar]
- 65.Rivière D., Szczebara F.M., Berjeaud J.-M., Frère J., Héchard Y. Development of a real-time PCR assay for quantification of Acanthamoeba trophozoites and cysts. J. Microbiol. Methods. 2006;64:78–83. doi: 10.1016/j.mimet.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 66.Pélandakis M., Serre S., Pernin P. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J. Eukaryot. Microbiol. 2000;47:116–121. doi: 10.1111/j.1550-7408.2000.tb00020.x. [DOI] [PubMed] [Google Scholar]
- 67.Grimm D., Ludwig W., Brandt B., Michel R., Schleifer K.-H., Hacker J., Steinert M. Development of 18S rRNA-Targeted Oligonucleotide Probes for Specific Detection of Hartmannella and Naegleria in Legionella—Positive Environmental Samples. Syst. Appl. Microbiol. 2001;24:76–82. doi: 10.1078/0723-2020-00017. [DOI] [PubMed] [Google Scholar]
- 68.Nazarian E.J., Bopp D.J., Saylors A., Limberger R.J., Musser K.A. Design and implementation of a protocol for the detection of Legionella in clinical and environmental samples. Diagn. Microbiol. Infect. Dis. 2008;62:125–132. doi: 10.1016/j.diagmicrobio.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Cross K.E., Mercante J.W., Benitez A.J., Brown E.W., Diaz M.H., Winchell J.M. Simultaneous detection of Legionella species and L. anisa, L. bozemanii, L. longbeachae and L. micdadei using conserved primers and multiple probes in a multiplex real-time PCR assay. Diagn. Microbiol. Infect. Dis. 2016;85:295–301. doi: 10.1016/j.diagmicrobio.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.