Abstract
Transcription of genes encoding molecular chaperones and folding enzymes in the endoplasmic reticulum (ER) is induced by accumulation of unfolded proteins in the ER. This intracellular signaling, known as the unfolded protein response (UPR), is mediated by the cis-acting ER stress response element (ERSE) in mammals. In addition to ER chaperones, the mammalian transcription factor CHOP (also called GADD153) is induced by ER stress. We report here that the transcription factor XBP-1 (also called TREB5) is also induced by ER stress and that induction of CHOP and XBP-1 is mediated by ERSE. The ERSE consensus sequence is CCAAT-N9-CCACG. As the general transcription factor NF-Y (also known as CBF) binds to CCAAT, CCACG is considered to provide specificity in the mammalian UPR. We recently found that the basic leucine zipper protein ATF6 isolated as a CCACG-binding protein is synthesized as a transmembrane protein in the ER, and ER stress-induced proteolysis produces a soluble form of ATF6 that translocates into the nucleus. We report here that overexpression of soluble ATF6 activates transcription of the CHOP and XBP-1 genes as well as of ER chaperone genes constitutively, whereas overexpression of a dominant negative mutant of ATF6 blocks the induction by ER stress. Furthermore, we demonstrated that soluble ATF6 binds directly to CCACG only when CCAAT exactly 9 bp upstream of CCACG is bound to NF-Y. Based on these and other findings, we concluded that specific and direct interactions between ATF6 and ERSE are critical for transcriptional induction not only of ER chaperones but also of CHOP and XBP-1.
Secretory and transmembrane proteins must fold properly in the endoplasmic reticulum (ER) prior to subsequent transport to subcellular compartments that reside in the secretory pathway (14, 19). This productive folding process, however, can be perturbed by a variety of physiological and environmental stress conditions that cause accumulation of unfolded proteins in the ER. Under such ER stress conditions, homeostasis of protein folding in the ER is maintained by interorganelle signaling from the ER to the nucleus, a process called the unfolded protein response (UPR) (20, 30). Thus, from yeast to humans, transcription of genes encoding molecular chaperones and folding enzymes in the ER is induced in the nucleus in response to unfolding in the ER. Mammalian ER stress-inducible proteins include molecular chaperones such as GRP78 (also known as BiP), GRP94, GRP170 (also known as ORP150), and calreticulin as well as folding enzymes such as peptidyl-prolyl-cis-trans-isomerase FKBP13, protein disulfide isomerase, and protein disulfide isomerase-like proteins ERp72, ERp61 (also known as ERp57 or GRP58), and ERp29 (references 13 and 20 and references therein), indicating that synthesis of the majority of proteins assisting or facilitating protein folding in the ER is coregulated. Thus, the cell can adjust the folding capacity in the ER quite effectively by simply controlling cellular UPR activity.
The mechanism of the UPR has been very well characterized for the budding yeast Saccharomyces cerevisiae. The transmembrane protein kinase Ire1p (also known as Ern1p) is considered to function as a sensor molecule of ER stress (8, 32), and the basic leucine zipper (bZIP) protein Hac1p is a yeast UPR-specific transcription factor (9, 31, 36) that activates transcription of target genes in the nucleus through binding to the cis-acting UPR element (34, 35). ER stress-induced splicing of HAC1 mRNA plays an essential role in connecting events in the ER and those in the nucleus (9, 21). The sequence-specific and nonsequential cleavage of the splice sites is carried out by Ire1p, and the splicing reaction is completed by the tRNA ligase Rlg1p (15, 22, 49, 50). This unique system allows yeast cells to synthesize the highly active transcription factor Hac1p only when they need to cope with unfolded proteins in the ER (6, 21, 33).
Despite the initial discovery of induction of GRP78 and GRP94 in mammalian cells more than 20 years ago (48), the mechanism of the mammalian UPR remained poorly understood until the recent discovery of several key components involved in signaling. Mammalian cells were shown to contain two proteins designated as IRE1α and IRE1β, which are homologous in both sequence and domain structure to yeast Ire1p (53, 56). Overexpression of IRE1α or IRE1β constitutively activated transcription of ER chaperone genes as in the case of yeast Ire1p, suggesting similarities in the stress sensing system between yeast and mammalian ER. In addition to regulation at the level of transcription, mammalian cells respond to ER stress by regulating translation (20); initiation of translation is blocked under ER stress conditions so that no more proteins can be translocated into the ER, where protein folding is prevented (40). It has recently been shown that the ER-resident transmembrane protein kinase PERK (also known as PEK) plays a major role in ER stress-induced translational attenuation (16, 17, 47). Interestingly, the luminal domain of PERK shows significant sequence homology to those of IRE1α and IRE1β, suggesting that these three proteins use similar mechanisms for sensing the presence of unfolded proteins in the ER. A very recent paper reported that the luminal domain of PERK can be functionally exchanged with that of IRE1 and that the chaperone protein GRP78 is directly involved in activating both IRE1 and PERK in response to ER stress (4).
Recently, a unique sequence consisting of 19 nucleotides (CCAAT-N9-CCACG) was shown independently in our laboratory and A. S. Lee's laboratory to be commonly present in the promoter regions of mammalian UPR target genes (43, 60). We demonstrated that this sequence, designated the ER stress response element (ERSE), is indeed the cis-acting element necessary and sufficient for induction by ER stress of at least three major ER chaperones, GRP78, GRP94, and calreticulin (60). We confirmed that the general transcription factor NF-Y (also known as CBF) (28) binds to the CCAAT part of ERSE as reported previously by Lee and coworkers (44). Thus, we proposed that the CCACG part of ERSE 9 bp downstream of the CCAAT part provides specificity in the mammalian UPR (60).
Using yeast one-hybrid screening, we isolated the bZIP protein ATF6 as a candidate for the mammalian UPR-specific transcription factor that binds to the CCACG part of ERSE (60). Subsequent analysis revealed an intriguing mechanism by which ATF6 is activated in response to ER stress (18). ATF6 is constitutively synthesized as a 90-kDa protein (p90ATF6), which is converted to a 50-kDa protein (p50ATF6) specifically in ER-stressed cells prior to induction of GRP78. The most important consequence of this processing was alteration of the subcellular localization of ATF6. p90ATF6 is a type II transmembrane glycoprotein embedded in the ER via the single transmembrane domain near the center of the molecule, suggesting that the bZIP-containing N-terminal region is located in the cytoplasmic side. In marked contrast, p50ATF6 is a soluble nuclear protein. C-terminal deletion analysis showed that ATF6 mutants representing the cytoplasmic region were accumulated in the nucleus and that their overexpression resulted in constitutive enhancement of the levels of both GRP78 mRNA and GRP78 protein. We thus proposed that upon ER stress, the N-terminal fragment facing the cytoplasm is released from the ER membrane by a proteolytic process, and the resulting p50ATF6 translocates into the nucleus and activates transcription of ER chaperone genes. However, as yet there is no evidence in support of a direct interaction between ATF6 and ERSE. In addition, it still remains to be determined how the proteolysis of ATF6 is regulated by putative sensor molecules of ER stress.
In mammals, the transcription factor CHOP (also known as GADD153) (11, 42) is known to be induced under the conditions of ER stress (58). We report here that the transcription factor XBP-1 (also called TREB5) (27, 61) is also induced by ER stress. It was previously reported that overexpression of murine IRE1β activated transcription of both the GRP78 and CHOP genes, whereas overexpression of mutant IRE1β lacking the cytoplasmic kinase domain showed dominant negative effects and thus mitigated induction by ER stress of both the GRP78 and CHOP genes (56), suggesting coregulation of GRP78 and CHOP.
In this report, we examined whether ERSE and ATF6 are involved in induction of CHOP and XBP-1 as well as in that of ER chaperones. We then asked whether ATF6 directly recognizes and binds to the CCACG part of ERSE.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, and antibiotics (100 U of penicillin/ml and 100 μg of streptomycin/ml). 293 cells were grown in Eagle's minimum essential medium supplemented with 10% horse serum, 2 mM glutamine, and antibiotics. Cells were maintained at 37°C in a humidified 5% CO2–95% air atmosphere. Transfection was carried out by the standard calcium phosphate method (45) essentially as described previously (60).
Construction of plasmids.
Recombinant DNA techniques were performed according to standard procedures (45). The mammalian expression plasmid pcDNA3.1(+) was obtained from Invitrogen (Carlsbad, Calif.). The full-length ATF6 expression plasmid pcDNA-ATF6 was constructed previously (60). Another full-length ATF6 expression plasmid, pCGN-ATF6 (670), as well as mutant ATF6 expression plasmids pCGN-ATF6 (373) and pCGN-ATF6 (366), has been described previously (18). pcDNA-ATF6 (373) and pcDNA-ATF6 (373) ΔAD were constructed by PCR-mediated amplification of the regions corresponding to amino acids 1 to 373 and 171 to 373 of ATF6, respectively, together with a stop codon followed by insertion of the resultant fragments into pcDNA3.1(+) after their sequences had been confirmed.
Based on the published sequence of the human XBP-1 gene (38), a 459-bp fragment of the XBP-1 promoter (the −330-to-+129 region; numbers indicate nucleotide positions relative to the transcription start site) was amplified by PCR from HeLa genomic DNA and cloned into the KpnI-BglI sites of the pGL3-Basic vector (Promega, Madison, Wis.), which contains the firefly luciferase coding sequence but lacks eukaryotic promoter or enhancer elements. Similarly, an 887-bp fragment of the human CHOP promoter (the −870-to-+17 region) (37) was amplified by PCR and cloned into the KpnI-HindIII sites of the pGL3-Basic vector. ERSE was eliminated from the XBP-1 or CHOP promoter by site-directed mutagenesis using an Exsite Site-Directed Mutagenesis Kit (Stratagene, La Jolla, Calif.). Various deletion mutants of the XBP-1 or CHOP promoter were prepared by PCR-mediated mutagenesis. Synthetic, double-stranded oligonucleotides encoding wild-type GRP78–ERSE1, XBP-1–ERSE1, CHOP–ERSE1, and CHOP-ERSE2, as well as those encoding point mutants of GRP78–ERSE1 and CHOP-ERSE1, were inserted into the XhoI-BglII sites of the pGL3-Promoter vector (Promega), which contains the simian virus 40 (SV40) minimal promoter upstream of the luciferase coding sequence.
To express various subregions of ATF6 fused in frame to the DNA-binding domain of yeast Gal4p (amino acids 1 to 147; GAL4DB), the expression vector pBIND (Promega), which expresses GAL4DB under the control of the cytomegalovirus promoter, was used. Various subregions were amplified by PCR, and the resultant fragments were inserted into the XbaI site of pBIND after their sequences had been confirmed.
Luciferase assays.
HeLa cells in 96-well plates were transfected with 0.2 μg of a reporter plasmid carrying the firefly luciferase gene and 0.02 μg of the reference plasmid pRL-SV40 carrying the Renilla luciferase gene under the control of the SV40 enhancer and promoter (Promega) with or without 0.1 μg of an effector protein expression plasmid. After a 48-h incubation in fresh medium, cells were lysed in 20 μl of Passive Lysis Buffer (Promega). For induction of the UPR, cells were treated with 2 μg of tunicamycin/ml for 16 h prior to harvesting. Firefly luciferase and Renilla luciferase activities were measured with 5 μl of cell lysate using the Dual-Luciferase Reporter Assay System (Promega) and a Luminoskan luminometer (Labsystems, Helsinki, Finland). Relative activity was defined as the ratio of firefly luciferase activity to Renilla luciferase activity. Fold induction was defined as the ratio of induced to basal levels of activity.
To determine Gal4p-mediated transcriptional activity, HeLa cells in 24-well plates were transiently cotransfected with 0.1 μg of pBIND or a pBIND-derived plasmid and 0.5 μg of the reporter plasmid pG5luc (Promega). pG5luc contains five Gal4p binding sites in a minimal promoter upstream of the firefly luciferase gene. pBIND includes the Renilla luciferase gene to normalize transfection efficiency, the expression of which is controlled by the SV40 early promoter and growth hormone intron. Transfected cells were lysed in 100 μl of 1× Passive Lysis Buffer. Aliquots of 10 μl were used to determine firefly and Renilla luciferase activities.
Northern blot hybridization analysis.
Aliquots of 10 μg of total RNA extracted by the guanidine-phenol method were subjected to 1.2% agarose gel electrophoresis on a gel containing 2.2 M formaldehyde, blotted onto nylon membranes, and hybridized with 32P-labeled cDNA probes according to the standard procedure (45). Filters were exposed to X-ray film.
Electrophoretic mobility shift assay (EMSA).
Escherichia coli expression plasmids for human NF-YA, NF-YB, and NF-YC were kindly provided by Roberto Mantovani (University of Milan), and recombinant proteins were expressed separately in E. coli cells. The NF-Y trimer was reconstituted according to the procedure published previously (3); NF-YA, NF-YB, and NF-YC were solubilized with 8 M urea from inclusion bodies, mixed, and dialyzed against BC100 buffer (20 mM HEPES [pH 7.9], 100 mM KCl, and 10% glycerol). ATF6 (373) was translated in vitro using the TNT T7 quick coupled transcription-translation system (Promega). Nuclear extracts were prepared from HeLa cells as described previously (46). Anti-NF-YA antiserum was obtained from Rockland (Gilbertsville, Pa.). Anti-ATF6 antiserum was prepared and purified as described previously (18). Anti-CREB-RP antiserum was raised against the N-terminal region (amino acid 1 to 307) of CREB-RP fused with glutathione S-transferase, which had been expressed and purified from E. coli cells.
Double-stranded synthetic oligonucleotide probes were radiolabeled using the Klenow fragment of DNA polymerase I and [α-32P]dCTP (222 TBq/mmol) and purified by centrifugation through ProbeQuant G50 Micro Columns (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom). A 41-bp sequence present in the GRP78 promoter and containing GRP78–ERSE1 (GGAGGGCCTTCACCAATCGGCGGCCTCCACGACGGGGCTGG [underlined sequences perfectly match the consensus of ERSE]) was designated ERSE-CC. The CCAAT and/or CCACG part of ERSE-CC was mutated by multiple nucleotide replacements. Thus, the sequences of mutant ERSEs were as follows (with mutated sequences indicated by lowercase letters and ERSE consensus sequences underlined); ERSE-CM, GGAGGGCCTTCACCAATCGGCGGCCTgatgtACGGGGCTGG; ERSE-MC, GGAGGGCCTTCAgactaCGGCGGCCTCCACGACGGGGCTGG; and ERSE-MM, GGAGGGCCTTCAgactaCGGCGGCCTgatgtACGGGGCTGG. A 41-bp sequence present in the XBP-1 promoter and containing XBP-1–ERSE1 (GGGCGACGCTGGCCAATCGCGGAGGGCCACGACCGTAGAAA) was used as a probe. A 41-bp sequence present in the CHOP promoter and containing both CHOP–ERSE1 and CHOP–ERSE2 (GGCCAAAACCTACCAATCAGAAAGTGGCACGCCGGCATTGG (with underlined and boldface sequences denoting CHOP–ERSE1 and CHOP–ERSE2, respectively) was also used as a probe.
The binding buffer consisted of 20 mM HEPES (pH 7.9), 100 mM KCl, 10% glycerol, 1 mM MgCl2, 1 mM 2-mercaptoethanol, 0.1% Tween 20, and 20 μg of poly(dI-dC) · poly(dI-dC)/ml. Each of the oligonucleotide probes (0.1 pmol; ∼9,000 cpm) was incubated at 4°C for 1 h with 1 μl of in vitro-translated ATF6 (373) (∼800 fmol) in the presence or absence of recombinant NF-Y trimer (10 fmol) in a final volume of 10 μl. Samples were loaded onto nondenaturing gradient (5 to 20%) polyacrylamide gels and electrophoresed at 4°C at 20 mA for 100 min in native polyacrylamide gel electrophoresis (PAGE) buffer (25 mM Tris base, 192 mM glycine, 1 mM MgCl2, 1 mM 2-mercaptoethanol, 1% Tween 20, and 10% glycerol). Gels were then dried and exposed to X-ray film. Alternatively, a 32P-labeled probe (0.1 pmol; ∼6,000 cpm) was incubated at 4°C for 1 h with a HeLa cell nuclear extract (1 μg of proteins) in the presence or absence of 1 μl of in vitro-translated ATF6 (373) (∼800 fmol) in a final volume of 10 μl. Samples were loaded onto nondenaturing 4.5% polyacrylamide gels and electrophoresed at 4°C at 200 V for 150 min in 0.5× TBE buffer (45 mM Tris-borate and 1 mM EDTA). For supershift experiments, samples were treated with various antisera at 4°C for 1 h prior to incubation with a radiolabeled probe.
RESULTS
XBP-1 is induced by ER stress.
We previously obtained the mammalian bZIP type transcription factor XBP-1/TREB5 as a putative ERSE-binding protein in addition to ATF6 using yeast one-hybrid screening; both ATF6 and XBP-1 activated reporter gene transcription in an ERSE-dependent manner when expressed in yeast cells as fusion proteins with the activation domain of the yeast transcriptional activator Gal4p (60). Overexpression of full-length ATF6 alone in HeLa cells was sufficient to activate promoters of various mammalian ER chaperone genes even in the absence of ER stress (60). In marked contrast, overexpression of full-length XBP-1 alone showed little effect on the promoter activities of various ER chaperone genes (data not shown). Thus, the role of XBP-1 in the mammalian UPR is currently unclear.
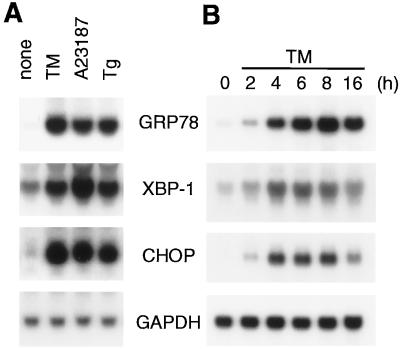
Interestingly, however, we found that XBP-1 mRNA was induced similarly to GRP78 mRNA and CHOP mRNA (Fig. 1A) by various chemicals that cause ER stress, i.e., tunicamycin (an inhibitor of protein N glycosylation [24]), A23187 (a calcium ionophore [59]), and thapsigargin (an inhibitor of the ER Ca2+ ATPase [25]). Furthermore, the time courses of induction of these three mRNAs were quite similar (Fig. 1B). These results prompted us to examine whether induction of XBP-1 or CHOP is mediated by ERSE as in the case of ER chaperones.
FIG. 1.
Induction of XBP-1 and CHOP transcripts by ER stress. (A) Effects of various ER stress-inducing reagents on the levels of XBP-1 and CHOP mRNA. Total RNA was extracted from HeLa cells in 60-mm dishes that had been treated for 8 h with either 2 μg of tunicamycin (TM)/ml, 3 μM A23187, or 100 nM thapsigargin (Tg) and analyzed by Northern blot hybridization. The filter was hybridized with one of the 32P-labeled DNA probes specific for GRP78, XBP-1, CHOP, or GAPDH mRNA and then stripped for hybridization with a different probe. (B) Time course of induction. HeLa cells were treated with 2 μg of tunicamycin/ml for the indicated periods before total RNA was extracted and analyzed by Northern blot hybridization as for panel A.
ERSE mediates induction of XBP-1 by ER stress.
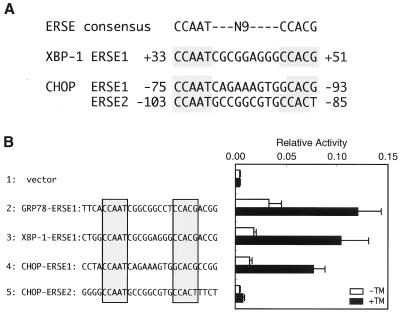
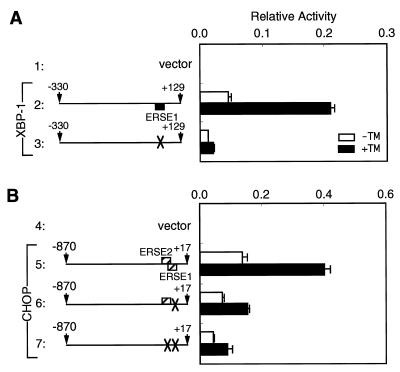
To analyze the cis-acting element(s) responsible for induction of XBP-1 by ER stress, the promoter region of XBP-1 (−330 to +129) was fused with the firefly luciferase gene and the resulting reporter plasmid was transiently introduced into HeLa cells (Fig. 2A). Treatment of transfected cells with tunicamycin for 16 h enhanced luciferase expression sixfold (Fig. 2A, line 2), comparable with the eightfold induction of XBP-1 mRNA by tunicamycin (see Fig. 1). Deletion analysis indicated that the region from nucleotide position −330 to −15 was dispensable for induction (Fig. 2A, lines 3 and 4) but that the region from position +32 to +65 was critical for induction (lines 5 to 7). Importantly, a sequence perfectly matching the consensus of ERSE, designated XBP-1–ERSE1, was present within the region from +32 to +65 (Fig. 3A), and XBP-1–ERSE1 conferred ER stress inducibility on a heterologous SV40 minimal promoter similarly to GRP78–ERSE1 (Fig. 3B). When XBP-1–ERSE1 was disrupted by multiple nucleotide replacements, the XBP-1 promoter lost the ability to respond to tunicamycin (Fig. 4A). Based on these results, we concluded that XBP-1–ERSE1 is the cis-acting element necessary and sufficient for induction of XBP-1 by ER stress.
FIG. 2.
Deletion analysis of the human XBP-1 (A) and CHOP (B) promoters. Various fragments derived from the human XBP-1 and CHOP promoter regions are shown schematically on the left. Numbers indicate nucleotide positions from the transcription start site. Solid and hatched boxes indicate the locations of ERSE motifs with perfect and considerable matches to the consensus, respectively (see Fig. 3A). Each of these fragments cloned immediately upstream of the firefly luciferase gene in the pGL3-Basic vector was transiently introduced into HeLa cells together with the pRL-SV40 reference plasmid as described in Materials and Methods. Relative luciferase activity in transfected cells incubated for 16 h with (solid boxes) or without (open boxes) 2 μg of tunicamycin (TM)/ml was determined, and averages from four independent experiments are presented with standard deviations (error bars). Fold induction was calculated by dividing the relative luciferase activity in TM-treated cells by that in untreated cells.
FIG. 3.
Presence of ERSE-like sequences in the human XBP-1 and CHOP promoters. (A) Comparison of ERSE-like sequences in the XBP-1 and CHOP promoters with the consensus. Nucleotides identical to the ERSE consensus are shaded. Numbers indicate the locations relative to the transcription start site. (B) Transcriptional activities of ERSE-like sequences in the XBP-1 and CHOP promoters. Oligonucleotides encoding each of the ERSE-like sequences (shaded) with indicated flanking nucleotides were inserted into the pGL3-Promoter vector, and the resulting plasmids were transiently introduced into HeLa cells with the pRL-SV40 reference plasmid. The relative luciferase activity in transfected cells incubated for 16 h with (solid boxes) or without (open boxes) 2 μg of tunicamycin (TM)/ml was determined, and averages from four independent experiments are presented with standard deviations (error bars).
FIG. 4.
Effects of disrupting ERSE on the activities of the human XBP-1 (A) and CHOP (B) promoters. Structures of the XBP-1 and CHOP promoters are shown schematically as described for Fig. 2. XBP-1–ERSE1, CHOP–ERSE1, and CHOP–ERSE2 were disrupted by mutating their sequences to agAtcN9CCACG, gatcTN9tacat, and gatccN9tgcga, respectively (mutated nucleotides are indicated by lowercase letters); disrupted ERSEs are marked by crosses. Each of the intact and mutant promoters was cloned immediately upstream of the firefly luciferase gene in the pGL3-Basic vector; their activities then were determined and are presented as described for Fig. 3.
ERSE mediates induction of CHOP by ER stress.
To identify the cis-acting element(s) responsible for induction of CHOP by ER stress, we analyzed the CHOP promoter (−870 to +17) fused to the firefly luciferase gene by transient transfection assays. The full-length promoter responded to tunicamycin treatment by enhancing luciferase expression approximately 3.5-fold (Fig. 2B, line 9). The extent of induction was much lower than that expected from the 18-fold induction of CHOP mRNA by tunicamycin (see Fig. 1). However, the high basal activity of the CHOP promoter, accounting for low inducibility, was also reported previously by others (37). Deletion of the region from −870 to −325 significantly affected neither basal nor induced expression (Fig. 2B, lines 10 to 14). On the other hand, deletion of the region from −325 to −105 markedly reduced basal expression, giving rise to increased inducibility up to approximately sixfold (Fig. 2B, lines 15 and 16). Further deletion of the region from −105 to −74 completely abolished the induction (line 17). We found that this indispensable region from −105 to −74 contained two ERSE-like sequences designated CHOP–ERSE1 and CHOP–ERSE2, which overlapped by 9 bp and were oriented in opposite directions (Fig. 3A).
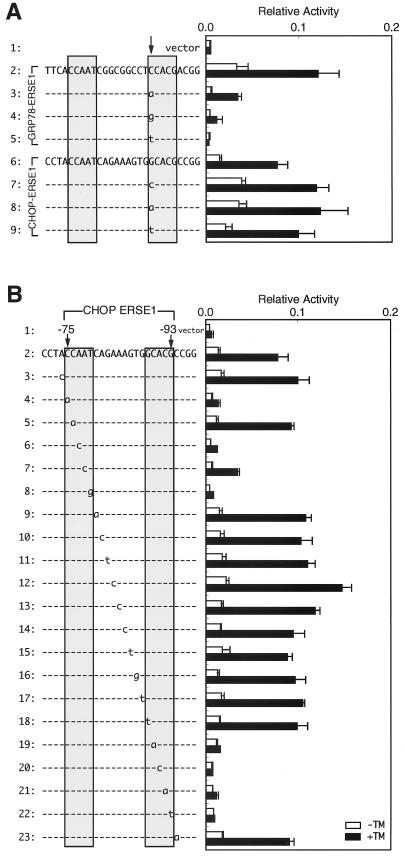
Interestingly, only CHOP–ERSE1 conferred ER stress inducibility on the SV40 minimal promoter (Fig. 3B). Transversion of G to T at the critical nucleotide −85 of CHOP–ERSE2 (the 3′-most nucleotide of the CCACG part) likely explains the inability of CHOP–ERSE2 to mediate transcriptional induction, because the same transversion inactivated GRP78–ERSE1 (60). In contrast, CHOP–ERSE1 was nearly as active as GRP78–ERSE1 despite the presence of a C-to-G alteration at the critical nucleotide −89 (the 5′-most nucleotide of the CCACG part). This activity was unexpected because the same alteration abolished the function of GRP78–ERSE1 (60). Therefore, we introduced all four nucleotides at the 5′-most side of the CCACG part of GRP78–ERSE1 or CHOP–ERSE1 and examined their effects (Fig. 5A). Compared with the wild-type GRP78–ERSE1 (Fig. 5A, line 2), mutant GRP78–ERSE1 carrying A at this position showed much lower basal activity but still retained the ability to respond to tunicamycin treatment (Fig. 5A, line 3). On the other hand, mutant GRP78–ERSE1 carrying G or T showed little or no activity (Fig. 5A, line 4 or 5, respectively). These results were consistent with our previous observation that C and A are preferred for the 5′-most nucleotide of the CCACG part of GRP78–ERSE1 (60). It should be noted that the pGL3-Promoter vector used in this study appeared to provide more-sensitive analysis of ERSE than the pGL2-Promoter vector used previously due to its lower basal activity. In marked contrast, the 5′-most nucleotide of the CCACG part of CHOP–ERSE1 could be replaced by any of the four nucleotides without significantly affecting ER stress inducibility (Fig. 5A, lines 6 to 9).
FIG. 5.
Identification of the nucleotides in CHOP–ERSE1 required to mediate transcriptional induction. (A) Effects of various nucleotides on the activities of GRP78–ERSE1 and CHOP–ERSE1. Nucleotide C at position −47 of GRP78–ERSE1 and nucleotide G at position −89 of CHOP–ERSE1 (marked by the arrow) were changed to other nucleotides as indicated by lowercase letters. The activities of wild-type and mutant ERSEs were determined and are presented as described for Fig. 3. (B) Effects of point mutations on the activity of CHOP–ERSE1. Each of the nucleotides (−74 to −94) in the human CHOP promoter was mutated by transversion as indicated by lowercase letters. The activities of wild-type and mutant ERSEs were determined and are presented as described for Fig. 3.
To determine whether the nucleotides required for the transcriptional activity of CHOP–ERSE1 conform to the consensus ERSE (CCAAT-N9-CCACG), we constructed a series of point mutants of CHOP–ERSE1 by transversion and analyzed their activities (Fig. 5B) as we reported previously for GRP78–ERSE1 (60). Mutations in four of the five nucleotides in the CCAAT part abolished or greatly reduced the activity (Fig. 5B, lines 4 to 8). Mutations in the CCACG part inactivated CHOP–ERSE1, except for the mutation of the 5′-most nucleotide (Fig. 5B, lines 18 to 22). On the other hand, neither point mutations in the N9 region (Fig. 5B, lines 9 to 17) nor point mutations in nucleotides outside of the ERSE consensus (Fig. 5B, lines 3 and 23) significantly affected the activity of CHOP–ERSE1, indicating that the nucleotides required for the activity of CHOP–ERSE1 are essentially identical to those required for the activity of GRP78–ERSE1. The preference for C as the 5′-most nucleotide in the CCACG part observed in GRP78–ERSE1 may be compromised in the case of CHOP–ERSE1 by nucleotides present in the N9 region or outside of the consensus sequence. As disruption of CHOP–ERSE1 and CHOP–ERSE2 by multiple nucleotide replacements greatly reduced the response of the CHOP promoter to tunicamycin treatment (Fig. 4B), we concluded that ERSE functions as the cis-acting element necessary and sufficient for induction of CHOP by ER stress. Nonetheless, because of the low inducibility of the reporter construct (see above), we cannot exclude the possibility that other mechanisms are also involved in the induction of CHOP by ER stress.
ER stress-induced proteolysis of ATF6 is involved in induction of XBP-1 and CHOP.
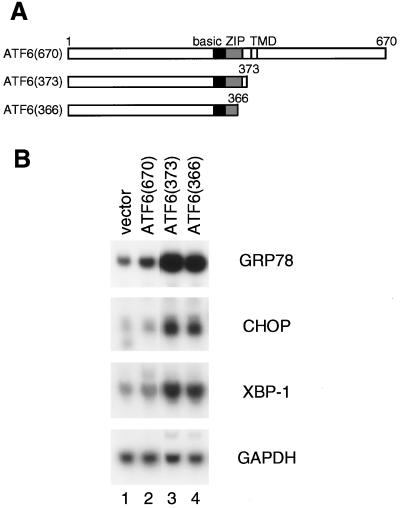
We then examined whether ATF6 is involved in transcriptional induction of XBP-1 and CHOP as in the case of ER chaperones. We reported previously (18) that transcription of the GRP78 gene was slightly activated by overexpression of full-length ATF6 (670), a membrane-bound form of ATF6, due to constitutively activated proteolysis of ATF6, whereas transcription of the GRP78 gene was markedly enhanced by overexpression of two ATF6 mutants, ATF6 (373) and ATF6 (366), representing p50ATF6, a soluble form, which can translocate into the nucleus (see the schematic structures in Fig. 6A). To examine the effects of ATF6 overproduction on the levels of endogenous mRNAs, we used 293 cells, which appear to exhibit higher transfection efficiencies than HeLa cells. The levels of all three mRNAs (GRP78 mRNA, CHOP mRNA, and XBP-1 mRNA) in 293 cells were constitutively elevated slightly by overexpression of p90ATF6 (Fig. 6B, lane 2) and markedly by two p50ATF6-like mutants (lanes 3 and 4) compared with the vector control (lane 1). These observations indicated that ER stress-induced proteolysis of ATF6 is a key regulatory step in transcriptional induction of CHOP and XBP-1 as well as ER chaperones.
FIG. 6.
Involvement of ER stress-induced proteolysis of ATF6 in transcriptional induction of the XBP-1 and CHOP genes. (A) Schematic structures of full-length ATF6, ATF6 (670), and its C-terminal deletion mutants ATF6 (373) and ATF6 (366). The locations of the basic region, leucine zipper (ZIP), and transmembrane domain (TMD) are indicated. Numbers indicate amino acid positions from the N terminus. (B) 293 cells cultured in 60-mm dishes were transiently transfected with 10 μg of pCGN alone (vector) or various pCGN-based ATF6 expression plasmids as indicated. At 48 h after transfection, total RNA was extracted and analyzed by Northern blot hybridization as for Fig. 1.
A dominant negative mutant form of ATF6 blocks induction of GRP78, XBP-1, and CHOP by ER stress.
To substantiate the importance of ATF6 in the mammalian UPR, we examined whether a mutant ATF6 would exhibit a dominant negative effect on the UPR. We sought to identify the transactivation domain of ATF6, which can be transplanted into other proteins, because a mutant transcription factor containing DNA-binding and dimerization domains but lacking an activation domain was expected to compete with endogenous wild-type protein for binding to its target element but to fail to activate transcription.
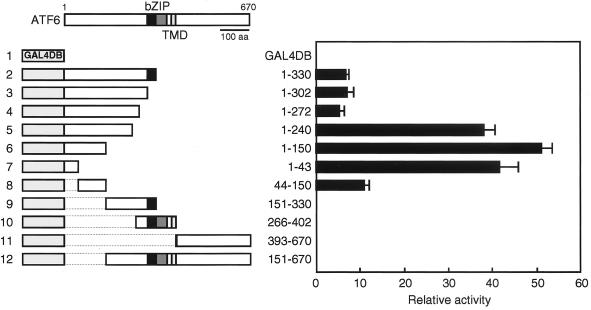
Various subregions of ATF6 were fused in frame to GAL4DB, and the transcriptional activities of these chimeric proteins were determined by measuring luciferase activities in lysates of HeLa cells cotransfected with the reporter plasmid pG5luc, which contains five Gal4p binding sites upstream of the firefly luciferase gene (Fig. 7). Transactivator activity was mapped to the N-terminal 150 amino acids (Fig. 7; compare line 6 with lines 9 to 12), and among these the N-terminal 43 amino acids appeared to make a major contribution (compare line 7 with line 8). Unfortunately, the expression level of each fusion protein could not be determined using a commercially available anti-GAL4DB antibody and therefore the reason for the increase in transactivation after deletion of amino acids 241 to 272 remains unclear (Fig. 7; compare line 4 with line 5); the short segment may have a negative effect on transcriptional activity or may affect the stability of the expressed protein. Our mapping results are consistent with a previous report (52) in which the N-terminal 273 amino acids of ATF6 were shown to activate transcription when fused with GAL4DB. The presence of a domain with potent transactivator activity in the N-terminal region ensures the ability of p50ATF6 to activate transcription in the nucleus.
FIG. 7.
Mapping of the transactivation domain of ATF6. (Left) Schematic structures of ATF6 as well as fusion proteins between GAL4DB and various ATF6 subregions. The dotted lines delineate the region deleted from the construct. The positions of the bZIP region and transmembrane domain (TMD) are indicated. (Right) Transcriptional activities of various fusion proteins. HeLa cells in 24-well plates were transiently transfected with each of the fusion plasmids together with the reporter plasmid pG5luc containing five Gal4p binding sites upstream of the firefly luciferase gene. Constitutively expressed luciferase activities were determined and normalized as described in Materials and Methods. Relative activities are presented as averages with standard deviations (error bars) from triplicate determinations of four independent transfections. The positive control supplied by the manufacturer (pBIND-Id and pACT-MyoD control vectors; Promega) showed a relative activity of 4.6 ± 0.3 in this assay.
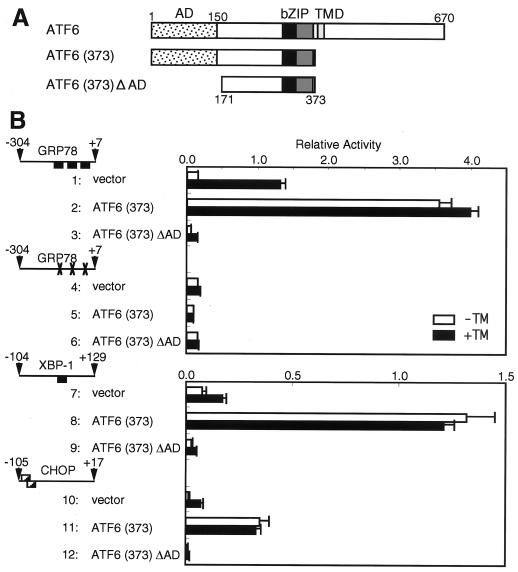
Based on the above findings, we constructed an ATF6 mutant, designated ATF6 (373) ΔAD, containing the bZIP region but lacking the activation domain (see the schematic structure in Fig. 8A). The effects of overexpression of ATF6 (373) ΔAD on the UPR were determined by measuring the activities of the GRP78, XBP-1, and CHOP promoters fused to firefly luciferase in transfected HeLa cells that were left untreated or treated with tunicamycin. We also examined the effects of overexpression of ATF6 (373) ΔAD on induction of endogenous GRP78, XBP-1, and CHOP mRNAs, but the results were not as clear as those expected from the promoter analysis, presumably because of low transfection efficiency even in 293 cells (data not shown).
FIG. 8.
Effects of overexpression of ATF6 mutants on the activities of the human GRP78, XBP-1, and CHOP promoters. (A) Schematic structures of full-length ATF6 and two mutants, ATF6 (373) and ATF6 (373) ΔAD. The locations of the activation domain (AD), bZIP region, and transmembrane domain (TMD) are marked. Numbers indicate amino acid positions relative to the N terminus. (B) Structures of intact and mutant promoters cloned immediately upstream of the firefly luciferase gene are shown schematically as in Fig. 4. A reporter plasmid and the pRL-SV40 reference plasmid were transfected into HeLa cells together with pcDNA3.1(+) (vector) or one of the mutant ATF6 expression plasmids pcDNA-ATF6 (373) and pcDNA-ATF6 (373) ΔAD as described in Materials and Methods. The relative luciferase activity in transfected cells incubated for 16 h with (solid boxes) or without (open boxes) 2 μg of tunicamycin (TM)/ml was determined, and averages from four independent experiments are presented with standard deviations (error bars).
ATF6 (373) was analyzed as a control. As expected from the results shown in Fig. 6, overexpression of ATF6 (373) resulted in constitutive and marked activation of the GRP78 promoter, which contained three functional ERSEs (Fig. 8B, lines 1 and 2). Importantly, elimination of three functional ERSEs from the GRP78 promoter abolished the enhancement (Fig. 8B; compare line 4 with line 5), confirming ERSE-dependent activation by ATF6. In marked contrast, overexpression of ATF6 (373) ΔAD blocked induction by tunicamycin of luciferase expressed from the GRP78 promoter with functional ERSEs (Fig. 8B; compare line 1 with line 3) but showed little effect on expression of luciferase from the GRP78 promoter without functional ERSEs (compare line 4 with line 6). These results indicated that ATF6 (373) ΔAD indeed exhibited a dominant negative effect on the UPR, as we expected. Essentially identical results were obtained with the XBP-1 and CHOP promoters (Fig. 8B, lines 7 to 12). We thus concluded that the function of ATF6 (or similar endogenous proteins) is critical for transcriptional induction of GRP78, XBP-1, and CHOP.
ATF6 directly binds to ERSE in vitro.
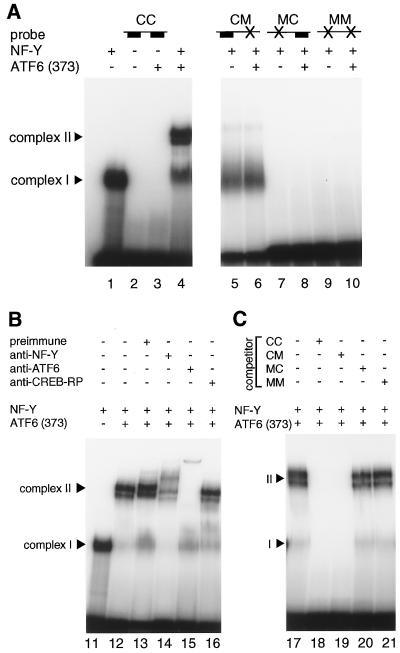
Previously, we failed to demonstrate direct interaction between ATF6 and ERSE when we analyzed full-length ATF6, a membrane-bound form, translated in vitro, despite the fact that ATF6 was isolated as an ERSE-binding protein in yeast one-hybrid screening (60). Therefore, we examined whether a soluble and active form of ATF6, ATF6 (373), could bind to ERSE directly. A 41-bp sequence containing GRP78–ERSE1 and its flanking nucleotides was used as a probe for EMSAs after labeling with 32P. To confirm the specificity of binding, two regions critical for the function of ERSE were mutated separately or simultaneously. For convenience, wild-type and mutant sequences are designated C (complete) and M (mutant), respectively. Thus, ERSE-CC indicates a perfect consensus sequence, CCAAT-N9-CCACG, whereas ERSE-CM, ERSE-MC, and ERSE-MM indicate mutated sequences CCAAT-N9-gatgt, gacta-N9-CCACG and gacta-N9-gatgt, respectively (with mutations indicated by lowercase letters) (see Materials and Methods).
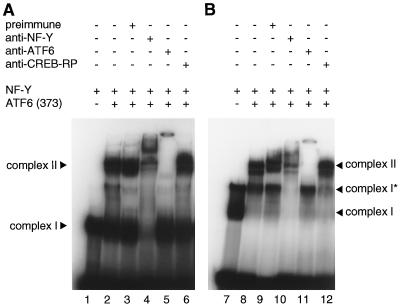
Recombinant NF-Y expressed and purified from E. coli cells bound to ERSE-CC, resulting information of a protein-DNA complex designated complex I (Fig. 9A, lane 1), as expected from the previous report (44). In contrast, ATF6 (373) translated in vitro alone did not exhibit any direct binding activity to ERSE-CC in EMSAs (Fig. 9A, lane 3). However, incubation of ATF6 (373) with ERSE-CC in the presence of NF-Y produced a novel protein-DNA complex designated complex II (Fig. 9A, lane 4). It was obvious that complex II consisted of NF-Y and ATF6 (373) bound to ERSE-CC because the mobility and/or formation of complex II was affected when mixtures of NF-Y and ATF6 (373) were treated with anti-NF-Y (Fig. 9B, lane 14) or anti-ATF6 (lane 15) antiserum prior to incubation with ERSE-CC, whereas pretreatment with preimmune (lane 13) or anti-CREB-RP (lane 16) antiserum showed no effects on complex II; CREB-RP is a bZIP protein closely related to ATF6 (60, 62), but anti-CREB-RP antiserum did not cross-react with ATF6 (data not shown).
FIG. 9.
Direct binding of ATF6 to ERSE in the presence of NF-Y. (A) 32P-labeled ERSE-CC containing CCAAT-N9-CCACG (lanes 1 to 4), ERSE-CM containing CCAAT-N9-gatgt (lanes 5 and 6), ERSE-MC containing gacta-N9-CCACG (lanes 7 and 8), or ERSE-MM containing gacta-N9-gatgt (lanes 9 and 10) was incubated with in vitro-translated ATF6 (373) in the presence (+) or absence (−) of recombinant NF-Y as indicated. Protein-DNA complexes formed were analyzed by EMSA as described in Materials and Methods. The positions of complexes I and II are indicated. (B) A mixture of in vitro-translated ATF6 (373) and recombinant NF-Y was treated with (+) or without (−) various antisera as indicated prior to incubation with 32P-labeled ERSE-CC. EMSA was carried out as for panel A. (C) The specific binding of in vitro-translated ATF6 (373) and recombinant NF-Y to 32P-labeled ERSE-CC was competed by unlabeled oligonucleotides in 100-fold molar excess as indicated. EMSA was carried out as for panel A.
Formation of complex II but not complex I was completely abolished by mutation in the CCACG part of ERSE (Fig. 9A, lanes 5 and 6). Neither complex I nor complex II was formed when the CCAAT part of ERSE was mutated (Fig. 9A, lanes 7 to 10). These results strongly indicated that ATF6 (373) indeed recognizes the CCACG part of ERSE and that it binds to ERSE only when the CCAAT part of ERSE is bound to NF-Y. We also performed competition experiments to further confirm the specificity of binding. The formation of complex II was completely blocked by a 100-fold molar excess of unlabeled ERSE-CC (Fig. 9C, lane 18) and ERSE-CM (lane 19), to which NF-Y can bind, but was not affected by ERSE-MC (lane 20) or ERSE-MM (lane 21), to which NF-Y cannot bind. Based on these results, we concluded that binding of NF-Y to the CCAAT part of ERSE is a prerequisite for ATF6 (373) to bind to the CCACG part of ERSE. This observation reflected the in vivo situation very well. As the CCAAT part of ERSE in the GRP78 promoter was shown to be occupied constitutively in HeLa cells (26), p50ATF6 liberated by ER stress-induced proteolysis can bind to the CCACG part of ERSE smoothly after its entrance into the nucleus.
ATF6 directly binds to XBP-1–ERSE and CHOP-ERSE.
We then examined whether ATF6 (373) binds to XBP-1–ERSE1 and CHOP-ERSE in a manner similar to its binding to GRP78–ERSE1. The results of EMSAs obtained with XBP-1–ERSE1 (Fig. 10A, lanes 1 to 6) were essentially the same as those with ERSE-CC (Fig. 9B, lanes 11 to 16). On the other hand, the results obtained with CHOP-ERSE were more complicated, because the probe used as CHOP-ERSE contained both CHOP-ERSE1 and CHOP-ERSE2 due to their overlap (see Materials and Methods), which would explain why two protein-DNA complexes designated complexes I and I*, were formed when CHOP-ERSE was incubated with NF-Y alone (Fig. 10B, lane 7). It was likely that complexes I and I* contained one and two NF-Y molecules bound to the probe, respectively, as both were supershifted by anti-NF-Y antiserum (Fig. 10B, lane 10; also data not shown). Importantly, incubation of CHOP-ERSE with NF-Y plus ATF6 (373) (Fig. 10B, lane 8) resulted in a decrease in the level of complex I and the appearance of complex II, as in the case of GRP78–ERSE1, whereas incubation of CHOP-ERSE with NF-Y plus control reticulocyte lysate did not give rise to complex II (data not shown). It should be noted that complex I* formation was not significantly affected under these conditions, suggesting that steric hindrance hampered the binding of ATF6 (373) to the probe to which two NF-Y molecules were bound; there was a space of only 5 bp between the GCACG part of CHOP–ERSE1 and the CCAAT part of CHOP–ERSE2 (see Materials and Methods). There might be certain mechanisms in vivo by which simultaneous occupation of two CCAAT sequences in CHOP-ERSE is prevented in order to allow binding of p50ATF6.
FIG. 10.
Direct binding of ATF6 to XBP-1–ERSE1 (A) and CHOP-ERSE (B). A mixture of in vitro-translated ATF6 (373) and recombinant NF-Y was treated with (+) or without (−) various antisera as indicated prior to incubation with 32P-labeled XBP-1–ERSE1 (lanes 1 to 6) or 32P-labeled CHOP-ERSE (lanes 7 to 12). The protein-DNA complexes formed were analyzed by EMSA as for Fig. 9. The positions of complexes I, I*, and II are indicated.
The mobility and/or formation of complex II were affected by pretreatment with anti-NF-Y (Fig. 10B, lane 10) or anti-ATF6 (lane 11) antiserum but were not affected by pretreatment with preimmune (lane 9) or anti-CREB-RP (lane 12) antiserum as expected. We confirmed that mutations in the CCACG part of XBP-1–ERSE1 or the GCACG part of CHOP-ERSE1 completely abolished the formation of complex II (data not shown). We thus concluded that ATF6 activated by ER stress-induced proteolysis stimulates transcription not only of ER chaperone genes but also of the genes encoding transcription factors XBP-1 and CHOP by direct binding to ERSE present in their promoter regions.
Binding of endogenous p50ATF6 to ERSE is hardly detected in nuclear extracts of ER-stressed cells due to its low abundance.
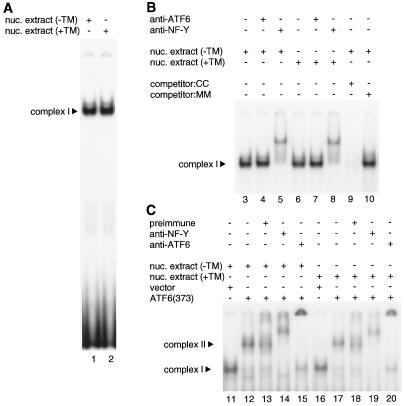
Using EMSA, we further examined whether we could detect binding of endogenous p50ATF6 to ERSE. Nuclear extracts of HeLa cells treated with or without tunicamycin for 4 h were incubated with 32P-labeled ERSE-CC (GRP78–ERSE1). As shown in Fig. 11A, a single binding activity was detected in unstressed cells (lane 1). This binding activity was specific to ERSE, because it was completely competed by a 100-fold molar excess of ERSE-CC (Fig. 11B, lane 9) but not by ERSE-MM (lane 10). This binding activity represented complex I, which we observed in Fig. 9, because it was supershifted by anti-NF-Y antiserum (Fig. 11B, lane 5) but not by anti-ATF6 antiserum (lane 4), consistent with constitutive occupation of the CCAAT part of ERSE in the GRP78 promoter in vivo (26).
FIG. 11.
ERSE-binding activities in nuclear extracts of HeLa cells. (A) 32P-labeled ERSE-CC was incubated with nuclear extracts prepared from HeLa cells that were left untreated (−TM) or treated (+TM) with 2 μg of tunicamycin/ml for 4 h. Protein-DNA complexes formed were analyzed by EMSA as described in Materials and Methods. The position of complex I is indicated. (B) Nuclear extracts of untreated HeLa cells (−TM) or those treated (+TM) with 2 μg of tunicamycin/ml for 4 h were incubated with (+) or without (−) various antisera as indicated prior to incubation with 32P-labeled ERSE-CC (lanes 3 to 8). Formation of complex I was competed by a 100-fold molar excess of unlabeled ERSE-CC (lane 9) or ERSE-MM (lane 10). EMSA was carried out as for panel A. Only specific binding is shown. The position of complex I is indicated. (C) Nuclear extracts of untreated HeLa cells (−TM) or those treated (+TM) with 2 μg of tunicamycin/ml for 4 h were mixed with in vitro-translated ATF6 (373) or control reticulocyte lysates (vector) and then incubated with (+) or without (−) various antisera as indicated prior to incubation with 32P-labeled ERSE-CC. EMSA was carried out as for panel A. Only specific binding is shown, and the positions of complexes I and II are indicated.
Contrary to our expectations, tunicamycin treatment affected neither the amount nor the mobility of complex I (Fig. 11A, lane 2, and Fig. 11B, lanes 6 to 8). We hypothesized that the inability to detect complex II in ER-stressed cells might have been due to a low abundance of p50ATF6 compared with the amount of NF-Y. We therefore added to nuclear extracts in vitro-translated ATF6 (373), the amount of which was estimated to be approximately 8,000-fold more than that of endogenous p50ATF6 present in the nuclear extract of tunicamycin-treated HeLa cells used for EMSA. As a result, complex II was formed regardless of tunicamycin treatment of HeLa cells (Fig. 11C, lanes 12 and 17), and complex II was supershifted by both anti-NF-Y (lanes 14 and 19) and anti-ATF6 (lanes 15 and 20) antisera but not by preimmune serum (lanes 13 and 18). We concluded that complex II formed in ER-stressed cells could not be detected by means of EMSA because of its low abundance.
The ATF6-binding activity of ERSE is correlated with its transcriptional activity.
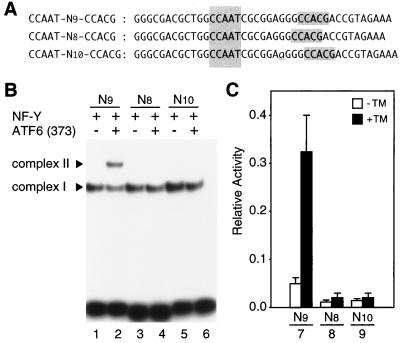
As the CCACG part of ERSE is always located 9 bp downstream of the CCAAT part in various promoter regions of ER chaperone genes (60), we examined how altering the spacing would affect both the ATF6-binding and transcription-inducing activities of ERSE. For this purpose, we analyzed the XBP-1 promoter because it contained a single ERSE sequence (XBP-1–ERSE1) necessary and sufficient for induction by ER stress (see above). Consistent with the results shown in Fig. 10, ATF6 (373) bound to XBP-1–ERSE1 (CCAAT-N9-CCACG) in the presence of NF-Y, resulting in the formation of complex II (Fig. 12B, lane 2). In marked contrast, NF-Y but not ATF6 was able to bind to mutant ERSE in which CCACG was separated from CCAAT by a spacer of 8 or 10 bp (Fig. 12B, lanes 3 to 6), indicating that spatial arrangement is critical for ERSE to accommodate simultaneous binding of NF-Y and ATF6. Most importantly, the ability of the XBP-1 promoter to respond to tunicamycin treatment (sixfold enhancement of the reporter luciferase expression [Fig. 12C, bars 7]) was abolished almost completely by altering the spacing in ERSE from 9 to 8 bp (bars 8) or 10 bp (bars 9), indicating that spatial arrangement is also critical for the in vivo function of ERSE. Thus, the ATF6-binding activities of ERSE-like sequences obtained in vitro with EMSA reflected very well their abilities to mediate induction by ER stress in vivo, determined by placing them in a natural promoter of a UPR target gene. These results further supported the notion that ATF6 plays a major role in transcriptional induction by ER stress of mammalian UPR target genes.
FIG. 12.
Effects of altering the spacing on the ATF6-binding and transcription-inducing activities of ERSE. (A) Nucleotide sequences of wild-type and mutant forms of XBP-1–ERSE1 analyzed. A 41-bp sequence containing XBP-1–ERSE1 and its surrounding nucleotides is shown and referred to as CCAAT-N9-CCACG. Sequences matching the consensus ERSE are shaded. One nucleotide, G, between the CCAAT and CCACG sequences was deleted to create CCAAT-N8-CCACG, while one nucleotide, A (indicated by the underlined lowercase letter), was inserted to create CCAAT-N10-CCACG. (B) Binding of ATF6 to wild-type or mutant XBP-1–ERSE1. The oligonucleotide probe CCAAT-N9-CCACG, CCAAT-N8-CCACG or CCAAT-N10-CCACG, the sequences of which are delineated in panel A, was incubated after labeling with 32P with (+) or without (−) in vitro-translated ATF6 (373) in the presence of NF-Y. Protein-DNA complexes formed were analyzed by EMSA. The positions of complexes I and II are indicated. C, Transcriptional response to ER stress of human XBP-1 promoter containing wild-type or mutant XBP-1-ERSE1. XBP-1-ERSE1 (CCAAT-N9-CCACG) was mutated to CCAAT-N8-CCACG or CCAAT-N10-CCACG as indicated in part A in the XBP-1 promoter which was then cloned immediately upstream of the firefly luciferase gene in the pGL3-Basic vector. Their transcriptional activities were determined and are presented as described in Fig. 3.
DISCUSSION
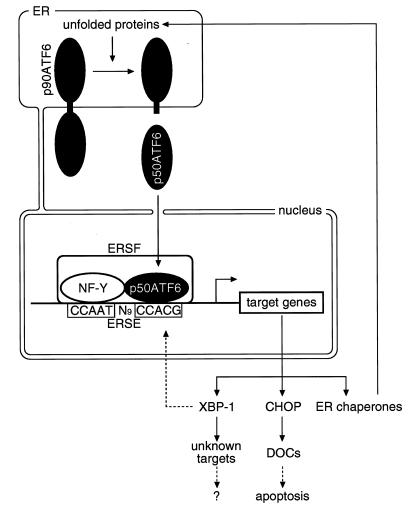
The cis-acting ERSE with a consensus of CCAAT-N9-CCACG is responsible for transcriptional induction of ER chaperones by unfolded proteins accumulated in the ER, and ATF6 was isolated as a putative ERSE-binding protein (60). We demonstrated here that ATF6 indeed recognizes the CCACG part of ERSE and binds directly to ERSE when the CCAAT part of ERSE is bound to the general transcription factor NF-Y (Fig. 9). In this regard, it was shown previously by in vivo genomic footprinting analysis that the CCAAT regions of the three functional ERSE sequences present in the GRP78 promoter are constitutively occupied in HeLa cells (26). The protein responsible for this constitutive occupation of the CCAAT part of ERSE is almost certainly NF-Y, because incubation of nuclear extracts of HeLa cells with ERSE gave rise only to complex I, which was supershifted by anti-NF-Y antiserum (Fig. 11). Importantly, addition of exogenous ATF6 to nuclear extracts of both unstressed and ER-stressed HeLa cells resulted in formation of complex II (Fig. 11), indicating that the properties of NF-Y are not specifically changed by ER stress. Thus, once p50ATF6 produced by ER stress-induced proteolysis is translocated into the nucleus, it can directly bind to ERSE and activate transcription of target genes in vivo as depicted in Fig. 13. We propose that the ER stress response factor (ERSF) binding to ERSE responsible for the mammalian UPR is a heterologous protein complex consisting of the constitutive component binding to the CCAAT part (NF-Y) and the inducible component binding to the CCACG part. The tightly regulated mechanism of activation (18), DNA-binding specificity (Fig. 9), and potent transcriptional activator activity present in the N-terminal region (Fig. 7) all indicate the importance of ATF6 as an inducible component of ERSF. Although it is possible that proteins other than p50ATF6, such as XBP-1, bind to the CCACG part of ERSE and modulate cellular UPR activity (see below), the importance of ATF6 (or similar proteins) in the mammalian UPR was further substantiated by the observation that an ATF6 mutant lacking the activation domain exhibited a dominant negative effect on induction of UPR target genes by ER stress (Fig. 8) and that the abilities of various ERSE-like sequences to bind to ATF6 were well correlated with their abilities to mediate induction by ER stress (Fig. 12).
FIG. 13.
Model for the mammalian UPR. ATF6 is synthesized as a precursor protein (p90ATF6) that anchors in the ER membrane under normal conditions. Upon accumulation of unfolded proteins in the ER, membrane-bound p90ATF6 is processed into a soluble and active form (p50ATF6). p50ATF6 translocates into the nucleus and directly binds to the CCACG part of ERSE, the CCAAT part of which is constitutively occupied by NF-Y. Thus, ERSF composed of NF-Y and p50ATF6 activates transcription of target genes. Target proteins include ER chaperones and two transcription factors, CHOP and XBP-1. Induced ER chaperones cope with unfolded proteins accumulated in the ER. Induced CHOP is likely to help the cells prepare for apoptosis by stimulating transcription of its target genes (referred to as DOCs). The roles of induced XBP-1 and its target genes are currently unknown. XBP-1 may function as a regulator of the UPR through ERSE.
Roy and Lee independently identified ERSE and reported that NF-Y and YY1 bind to the CCAAT part and CCACG part, respectively (43), although it is not clear how ubiquitous factor YY1 is specifically activated in response to ER stress. They also reported that the spacer (N9) region between the CCAAT and CCACG parts of ERSE is highly GC rich and showed that the sequence GGC in the N9 region is important for GRP78–ERSE1 activity. Furthermore, they identified binding activity to the GGC sequence in nuclear extracts of HeLa cells, which was found to be inducible by ER stress (43). However, we noted that the N9 region is not necessarily GC rich, as evidenced by the sequence of CHOP–ERSE1 (Fig. 3A). In addition, the sequence GGC is not present in the N9 regions of GRP94–ERSE1, GRP94–ERSE3, calreticulin-ERSE3, XBP-1–ERSE1, and CHOP-ERSE1, although all of these sequences were sufficient to mediate transcriptional induction when one copy of each was fused with a heterologous promoter in our highly sensitive assay system (60) (Fig. 3B). Thus, the role of the GGC-binding activity in the mammalian UPR remains unclear. At present we favor the idea that the N9 region is necessary to provide an optimal space that allows simultaneous binding of NF-Y and p50ATF6, because ATF6 (373) could not bind to ERSE even in the presence of NF-Y when CCACG was separated from CCAAT by a spacer of 8 or 10 bp (Fig. 12).
In addition to ER chaperones, several transcription factors are induced by ER stress in mammals, for example, CHOP and XBP-1 (Fig. 1). We showed here that both the CHOP and XBP-1 promoters contain functional ERSE sequences necessary and sufficient for induction (Fig. 2 to 5) and that ATF6 is involved in activating transcription of the CHOP and XBP-1 genes as well as ER chaperone genes (Fig. 6, 8, and 10). XBP-1 was originally identified as a protein binding to the cis-acting X box present in the promoters of human major histocompatibility complex class II genes (27), and also as a protein (TREB5) binding to the tax-responsive element present in the long terminal repeat of human T-cell leukemia virus type 1 (61). Recent studies on XBP-1 knockout mice revealed that the function of XBP-1 is essential for hepatocyte growth (41). Binding site selection experiments revealed that XBP-1 preferably binds to the sequence GATGACGTG(T/G)NNN(A/T)T (the underlined sequence is perfectly complementary to the sequence CCACG) (7); that would explain why XBP-1 was isolated as an ERSE-binding protein (60). Here, we identified XBP-1 as a target of the mammalian UPR, although the physiological significance of this finding remains unclear; overexpression of XBP-1 alone did not affect transcription of ER chaperone genes significantly in contrast to the case of ATF6 (data not shown). Since transactivation domains of XBP-1 have been identified and characterized previously (7, 29), it is still possible that XBP-1 may function as an inducible component of ERSF under certain as yet unknown conditions.
CHOP is a bZIP protein related to CCAAT/enhancer-binding protein and modulates transcription of target genes via heterodimerization with members of the CCAAT/enhancer-binding protein family (42, 55). Although CHOP was originally identified as GADD153, a protein inducible by growth arrest signals and DNA-damaging agents (11), subsequent analysis revealed that CHOP is induced by a variety of conditions that cause ER stress (5, 39, 58). We confirmed this observation (Fig. 1) and further demonstrated that the CHOP promoter contains functional ERSE sequences necessary and sufficient for induction by ER stress similarly to promoters of ER chaperone genes (Fig. 2 to 4). At present we do not know why the nucleotide requirement at the 5′-most side of the CCACG part of CHOP-ERSE1 was more tolerant than that in GRP78–ERSE1 (Fig. 5). Importantly, ATF6 was able to bind to CHOP-ERSE1 as well as to GRP78–ERSE1 in the presence of NF-Y (Fig. 10). Based on these and other results shown in Fig. 6 and 8, we concluded that CHOP is a target protein of the mammalian UPR and that its induction is mediated by ERSE and ERSF.
Expression of CHOP is known to be linked to programmed cell death, or apoptosis. Overexpression of CHOP leads to growth arrest (2) and promotes apoptosis (12), whereas mouse embryonic fibroblasts lacking CHOP exhibited significant resistance to ER stress-induced cell death (63). However, accumulating evidence suggests that induced CHOP appears not to be a primary determinant causing the cell to commit suicide when the function of the ER is severely impaired. Rather, induced CHOP may help the cell prepare for programmed cell death and activate the apoptotic process smoothly when there is no hope of overcoming the malfunction of the ER. The time course of induction of CHOP was similar to that of ER chaperones, indicating that cells are still coping with unfolded proteins accumulated in the ER when CHOP is induced. Although the apoptotic process was significantly delayed by the absence of CHOP in mouse embryonic fibroblasts, essentially all cells eventually died when treated with ER stress-inducing reagents (63). Target proteins of CHOP recently identified and designated as DOCs (downstream of CHOP) are mammalian orthologues of the Drosophila gene Tenm/Odz and novel homologues of the actin-binding proteins villin and gelsolin (57). Thus, none of these molecules has been shown to be directly involved in the process of programmed cell death. A novel form of carbonic anhydrase VI, identified as one of the DOCs, may promote apoptosis by increasing intracellular proton concentrations (51), because the membrane pore-forming activity of the proapoptotic regulator Bax is higher at lower pHs (1).
In conclusion, the present results provided a basis for the diversity of the mammalian UPR; a protein is induced by ER stress regardless of its subcellular localization only if its promoter contains a functional ERSE to which ERSF can bind. Primary targets of the UPR are, of course, ER chaperones, which cope with unfolded proteins in the ER. Non-ER proteins induced by ER stress may also have roles in ER-stressed cells, as discussed above for CHOP. In this connection, it is noteworthy that the other bZIP type transcription factors ATF-2 and ATF-4 were induced under conditions similar to ER stress (10, 23, 54). Further characterization of target proteins of the mammalian UPR will provide information useful for understanding how the cell balances survival with death under conditions of ER stress.
ACKNOWLEDGMENTS
We thank Roberto Mantovani for providing NF-Y expression vectors. We are grateful to Masako Nakayama, Seiji Takahara, and Tomoko Yoshifusa for technical assistance.
REFERENCES
- 1.Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, Bernasconi L, Bernard A, Mermod J J, Mazzei G, Maundrell K, Gambale F, Sadoul R, Martinou J C. Inhibition of Bax channel-forming activity by Bcl-2. Science. 1997;277:370–372. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- 2.Barone M V, Crozat A, Tabaee A, Philipson L, Ron D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 1994;8:453–464. doi: 10.1101/gad.8.4.453. [DOI] [PubMed] [Google Scholar]
- 3.Bellorini M, Zemzoumi K, Farina A, Berthelsen J, Piaggio G, Mantovani R. Cloning and expression of human NF-YC. Gene. 1997;193:119–125. doi: 10.1016/s0378-1119(97)00109-1. [DOI] [PubMed] [Google Scholar]
- 4.Bertolotti A, Zhang Y, Hendershot L M, Harding H P, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 5.Carlson S G, Fawcett T W, Bartlett J D, Bernier M, Holbrook N J. Regulation of the C/EBP-related gene gadd153 by glucose deprivation. Mol Cell Biol. 1993;13:4736–4744. doi: 10.1128/mcb.13.8.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman R E, Walter P. Translational attenuation mediated by an mRNA intron. Curr Biol. 1997;7:850–859. doi: 10.1016/s0960-9822(06)00373-3. [DOI] [PubMed] [Google Scholar]
- 7.Clauss I M, Chu M, Zhao J L, Glimcher L H. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 1996;24:1855–1864. doi: 10.1093/nar/24.10.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J S, Shamu C E, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 9.Cox J S, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 10.Estes S D, Stoler D L, Anderson G R. Normal fibroblasts induce the C/EBP beta and ATF-4 bZIP transcription factors in response to anoxia. Exp Cell Res. 1995;220:47–54. doi: 10.1006/excr.1995.1290. [DOI] [PubMed] [Google Scholar]
- 11.Fornace A J, Jr, Nebert D W, Hollander M C, Luethy J D, Papathanasiou M, Fargnoli J, Holbrook N J. Mammalian genes coordinately regulated by growth arrest signals and DNA-damaging agents. Mol Cell Biol. 1989;9:4196–4203. doi: 10.1128/mcb.9.10.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedman A D. GADD153/CHOP, a DNA damage-inducible protein, reduced CAAT/enhancer binding protein activities and increased apoptosis in 32D c13 myeloid cells. Cancer Res. 1996;56:3250–3256. [PubMed] [Google Scholar]
- 13.Gething M J. Guidebook to molecular chaperones and protein-folding catalysts. Oxford, United Kingdom: Oxford University Press; 1997. [Google Scholar]
- 14.Gething M J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez T N, Sidrauski C, Dorfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding H P, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 17.Harding H P, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 18.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helenius A, Marquardt T, Braakman I. The endoplasmic reticulum as a protein folding compartment. Trends Cell Biol. 1992;2:227–231. doi: 10.1016/0962-8924(92)90309-b. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman R J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 21.Kawahara T, Yanagi H, Yura T, Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawahara T, Yanagi H, Yura T, Mori K. Unconventional splicing of HAC1/ERN4 mRNA required for the unfolded protein response: sequence-specific and non-sequential cleavage of the splice sites. J Biol Chem. 1998;273:1802–1807. doi: 10.1074/jbc.273.3.1802. [DOI] [PubMed] [Google Scholar]
- 23.Kokame K, Kato H, Miyata T. Homocysteine-respondent genes in vascular endothelial cells identified by differential display analysis. GRP78/BiP and novel genes. J Biol Chem. 1996;271:29659–29665. doi: 10.1074/jbc.271.47.29659. [DOI] [PubMed] [Google Scholar]
- 24.Kozutsumi Y, Segal M, Normington K, Gething M J, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 25.Li W W, Alexandre S, Cao X, Lee A S. Transactivation of the grp78 promoter by Ca2+ depletion. A comparative analysis with A23187 and the endoplasmic reticulum Ca2+-ATPase inhibitor thapsigargin. J Biol Chem. 1993;268:12003–12009. [PubMed] [Google Scholar]
- 26.Li W W, Sistonen L, Morimoto R I, Lee A S. Stress induction of the mammalian GRP78/BiP protein gene: in vivo genomic footprinting and identification of p70CORE from human nuclear extract as a DNA-binding component specific to the stress regulatory element. Mol Cell Biol. 1994;14:5533–5546. doi: 10.1128/mcb.14.8.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liou H C, Boothby M R, Finn P W, Davidon R, Nabavi N, Zeleznik-Le N J, Ting J P, Glimcher L H. A new member of the leucine zipper class of proteins that binds to the HLA DRα promoter. Science. 1990;247:1581–1584. doi: 10.1126/science.2321018. [DOI] [PubMed] [Google Scholar]
- 28.Maity S N, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 29.Matsuzaki Y, Fujisawa J, Yoshida M. Identification of transcriptional activation domain of TREB5, a CREB/ATF family protein that binds to HTLV-1 enhancer. J Biochem (Tokyo) 1995;117:303–308. doi: 10.1093/jb/117.2.303. [DOI] [PubMed] [Google Scholar]
- 30.Mori K. Tripartite management of unfolded proteins in the endoplasmic reticulum. Cell. 2000;101:451–454. doi: 10.1016/s0092-8674(00)80855-7. [DOI] [PubMed] [Google Scholar]
- 31.Mori K, Kawahara T, Yoshida H, Yanagi H, Yura T. Signalling from endoplasmic reticulum to nucleus: transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 32.Mori K, Ma W, Gething M J, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 33.Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci USA. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein-response element in Saccharomyces cerevisiae. J Biol Chem. 1998;273:9912–9920. doi: 10.1074/jbc.273.16.9912. [DOI] [PubMed] [Google Scholar]
- 35.Mori K, Sant A, Kohno K, Normington K, Gething M J, Sambrook J F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikawa J, Akiyoshi M, Hirata S, Fukuda T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic Acids Res. 1996;24:4222–4226. doi: 10.1093/nar/24.21.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park J S, Luethy J D, Wang M G, Fargnoli J, Fornace A J, Jr, McBride O W, Holbrook N J. Isolation, characterization and chromosomal localization of the human GADD153 gene. Gene. 1992;116:259–267. doi: 10.1016/0378-1119(92)90523-r. [DOI] [PubMed] [Google Scholar]
- 38.Ponath P D, Fass D, Liou H C, Glimcher L H, Strominger J L. The regulatory gene, hXBP-1, and its target, HLA-DRA, utilize both common and distinct regulatory elements and protein complexes. J Biol Chem. 1993;268:17074–17082. [PubMed] [Google Scholar]
- 39.Price B D, Calderwood S K. Gadd45 and Gadd153 messenger RNA levels are increased during hypoxia and after exposure of cells to agents which elevate the levels of the glucose-regulated proteins. Cancer Res. 1992;52:3814–3817. [PubMed] [Google Scholar]
- 40.Prostko C R, Brostrom M A, Malara E M, Brostrom C O. Phosphorylation of eukaryotic initiation factor (eIF) 2 alpha and inhibition of eIF-2B in GH3 pituitary cells by perturbants of early protein processing that induce GRP78. J Biol Chem. 1992;267:16751–16754. [PubMed] [Google Scholar]
- 41.Reimold A M, Etkin A, Clauss I, Perkins A, Friend D S, Zhang J, Horton H F, Scott A, Orkin S H, Byrne M C, Grusby M J, Glimcher L H. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 42.Ron D, Habener J F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 43.Roy B, Lee A S. The mammalian endoplasmic reticulum stress response element consists of an evolutionarily conserved tripartite structure and interacts with a novel stress-inducible complex. Nucleic Acids Res. 1999;27:1437–1443. doi: 10.1093/nar/27.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy B, Li W W, Lee A S. Calcium-sensitive transcriptional activation of the proximal CCAAT regulatory element of the grp78/BiP promoter by the human nuclear factor CBF/NF-Y. J Biol Chem. 1996;271:28995–29002. doi: 10.1074/jbc.271.46.28995. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Shapiro D J, Sharp P A, Wahli W W, Keller M J. A high-efficiency HeLa cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y, Vattem K M, Sood R, An J, Liang J, Stramm L, Wek R C. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shiu R P, Pouyssegur J, Pastan I. Glucose depletion accounts for the induction of two transformation-sensitive membrane proteins in Rous sarcoma virus-transformed chick embryo fibroblasts. Proc Natl Acad Sci USA. 1977;74:3840–3844. doi: 10.1073/pnas.74.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sidrauski C, Cox J S, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell. 1996;87:405–413. doi: 10.1016/s0092-8674(00)81361-6. [DOI] [PubMed] [Google Scholar]
- 50.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 51.Sok J, Wang X Z, Batchvarova N, Kuroda M, Harding H, Ron D. CHOP-dependent stress-inducible expression of a novel form of carbonic anhydrase VI. Mol Cell Biol. 1999;19:495–504. doi: 10.1128/mcb.19.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thuerauf D J, Arnold N D, Zechner D, Hanford D S, DeMartin K M, McDonough P M, Prywes R, Glembotski C C. p38 mitogen-activated protein kinase mediates the transcriptional induction of the atrial natriuretic factor gene through a serum response element; a potential role for the transcription factor ATF6. J Biol Chem. 1998;273:20636–20643. doi: 10.1074/jbc.273.32.20636. [DOI] [PubMed] [Google Scholar]
- 53.Tirasophon W, Welihinda A A, Kaufman R J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsujimoto A, Nyunoya H, Morita T, Sato T, Shimotohno K. Isolation of cDNAs for DNA-binding proteins which specifically bind to a tax-responsive enhancer element in the long terminal repeat of human T-cell leukemia virus type I. J Virol. 1991;65:1420–1426. doi: 10.1128/jvi.65.3.1420-1426.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ubeda M, Wang X Z, Zinszner H, Wu I, Habener J F, Ron D. Stress-induced binding of the transcription factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X-Z, Harding H P, Zhang Y, Jolicoeur E M, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X Z, Kuroda M, Sok J, Batchvarova N, Kimmel R, Chung P, Zinszner H, Ron D. Identification of novel stress-induced genes downstream of chop. EMBO J. 1998;17:3619–3630. doi: 10.1093/emboj/17.13.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X Z, Lawson B, Brewer J W, Zinszner H, Sanjay A, Mi L J, Boorstein R, Kreibich G, Hendershot L M, Ron D. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Mol Cell Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wooden S K, Li L J, Navarro D, Qadri I, Pereira L, Lee A S. Transactivation of the grp78 promoter by malfolded proteins, glycosylation block, and calcium ionophore is mediated through a proximal region containing a CCAAT motif which interacts with CTF/NF-I. Mol Cell Biol. 1991;11:5612–5623. doi: 10.1128/mcb.11.11.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida H, Haze K, Yanagi H, Yura T, Mori K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins; involvement of basic-leucine zipper transcription factors. J Biol Chem. 1998;273:33741–33749. doi: 10.1074/jbc.273.50.33741. [DOI] [PubMed] [Google Scholar]
- 61.Yoshimura T, Fujisawa J, Yoshida M. Multiple cDNA clones encoding nuclear proteins that bind to the tax-dependent enhancer of HTLV-1: all contain a leucine zipper structure and basic amino acid domain. EMBO J. 1990;9:2537–2542. doi: 10.1002/j.1460-2075.1990.tb07434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu C, Johansen F E, Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–4966. doi: 10.1128/mcb.17.9.4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot R T, Remotti H, Stevens J L, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]