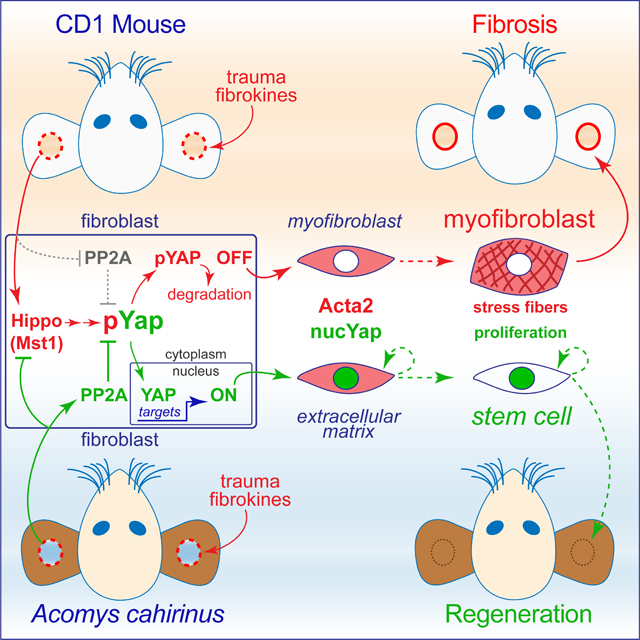
Summary
Spiny mice (Acomys cahirinus) are terrestrial mammals that evolved unique scar-free regenerative wound healing properties. Myofibroblasts (MFs) are the major scar-forming cell type in skin. We found that following traumatic injury to ear pinnae, MFs appeared rapidly in both Acomys and mouse yet persisted only in mouse. The timing of MF loss in Acomys correlated with wound closure, blastema differentiation, and nuclear localization of the Hippo pathway target protein Yap. Experiments in vitro revealed an accelerated PP2A-dependent dephosphorylation activity that maintained nuclear Yap in Acomys dermal fibroblasts (DFs) and was not detected in mouse or human DFs. Treatment of Acomys in vivo with the nuclear Yap-TEAD inhibitor verteporfin prolonged MF persistence and converted tissue regeneration to fibrosis. Forced Yap activity prevented and rescued TGFβ1-induced human MF formation in vitro. These results suggest Acomys evolved modifications of Yap activity and MF fate important for scar-free regenerative wound healing in vivo.
Keywords: regeneration, fibrosis, myofibroblast, Hippo-Yap, blastema, reprogramming, chrondrogenesis, adipocytes, hair follicle neogenesis, melanocytes
eTOC Blurb
Brewer et al. report that following ear pinnae injury myofibroblasts (MFs) appear rapidly in both Acomys (spiny mice) and Mus, but persist only in Mus. Acomys MFs in vitro resist TGFβ1-stimulated maturation via accelerated PP2A-dependent Yap phosphatase activity. Blocking Yap-TEAD interaction in vivo alters blastemal cell fate, inhibits regeneration, and promotes fibrosis.
Graphical Abstract
Introduction
Adult scar-free wound healing is a long sought clinical goal for improving chronic and traumatic injury outcomes. Most mammalian species exhibit scar-free wound repair during fetal or early post-natal development (Longaker et al., 1990; Lorenz and Adzick, 1993; Cass et al., 1997). However, this ability is lost shortly after birth transitioning to a scar-forming fibrotic response to tissue injuries (Longaker et al., 1994; Porrello et al., 2011). Indeed, progressive fibrosis underlies many degenerative diseases including cardiac failure, idiopathic pulmonary disease, chronic kidney disease, liver cirrhosis, and scleroderma (Raghu et al., 2006; Campanholle et al., 2013; Ho et al., 2014; Urban et al., 2015). Regrettably, few treatments exist for patients with these end stage fibrotic diseases other than organ transplantation (Friedman et al., 2013).
Species within the genus Acomys (spiny mouse) can completely shed their dorsal skin as an evolved adaptation to avoid predation (Seifert et al., 2012). Remarkably, healing of the shed skin results in full-thickness skin regeneration complete with hair follicles, sebaceous glands, cartilage, adipose tissue, nerves and blood vessels in the correct architectural organization without tissue overgrowth or scar formation (Seifert et al., 2012; Gawriluk et al., 2016; Matias Santos et al., 2016). Hence, while Acomys heal wounds in a manner functionally similar to lower order regenerative species, they are mammals with a physiology and a genome more closely related to other mammals including humans. Thus, spiny mice offer a unique opportunity for discovery that may find applications for novel anti-fibrotic treatments in humans.
Injury-induced MFs restore structural integrity of damaged tissue but limit regenerative potential in most mammalian models (Darby et al., 1990; Tomasek et al., 2002). The conventional view of MF function in wound healing is as a fibrotic matrix-producing cell that acquires contractile activity, a proinflammatory phenotype, and locally stiffens injured tissue (Hinz et al., 2012). However, recent studies have begun to redefine our understanding of MFs in wound healing. A surprising degree of MF plasticity in vivo together with a better understanding of the diversity of MF origins is emerging from genetic lineage tracking and single-cell studies (Driskell et al., 2013; Rinkevich et al., 2015; Fu et al., 2018; Guerrero-Juarez et al., 2019). MF to adipocyte transdifferentiation and interstitial MF to fibroblast reversion are a few recent examples (Plikus et al., 2017; Sanchez-Iranzo et al., 2018; Plikus et al., 2021). Moreover, transient MF induction and reversion is a necessary component for zebrafish scar-free cardiac regeneration (Sanchez-Iranzo et al., 2018). These data indicate that MFs are active participants in scar-free regenerative repair. Identifying the evolved adaptations in genus Acomys that produce MFs within a mammalian blastema that promote regeneration, rather than fibrosis, is an important first step to redirecting MF fate in human fibrotic diseases.
The molecular mechanisms Acomys utilize to promote repair independent of the canonical MF scarring response are currently unknown. One candidate pathway ideal for wound repair modification is Hippo-Yap (Dong et al., 2007). The Hippo-Yap pathway is a Ser/Thr protein kinase cascade that controls the nuclear localization and transcriptional coactivator functions of Yes-associated protein-1 (Yap1 or Yap) and transcriptional coactivator with PDZ binding motif (Taz) (Zhao et al., 2011). Genetic analyses of Hippo-Yap signaling in Drosophila and Mus demonstrate physiological roles in the development of proper organ size and architecture (Heallen et al., 2011; Zhao et al., 2011), and that Yap reactivation facilitates functional restoration of several organs with poor regenerative capacity including the adult heart (Leach et al., 2017; Moya and Halder, 2019). In this study, we sought to determine the role of MFs in Acomys scarless wound healing and investigate whether Yap nuclear localization and function contributes to tissue regeneration in spiny mice.
Results
Acomys exhibit transient myofibroblast formation followed by robust angiogenesis and chondrogenesis
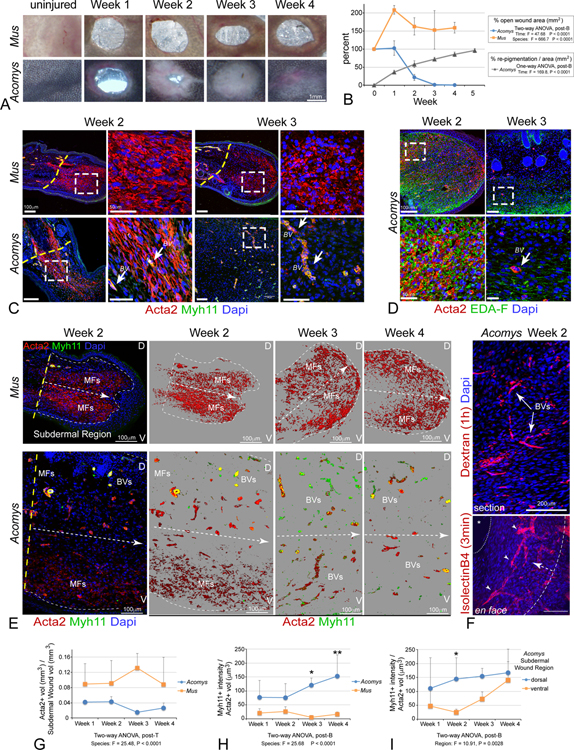
We hypothesized that evolved adaptations in the control of MF formation and regression underlie regenerative wound healing in Acomys. To test this hypothesis, we first used 2mm ear pinnae punch injuries in adult Mus musculus (outbred CD1 strain, referred to as Mus) compared to adult Acomys cahirinus (referred to as Acomys). The ear punch model produces a complete, confined, and highly reproducible injury to a complex tissue containing cartilage, adipocytes, muscle, nerves, blood vessels, melanocytes, epithelium, and hair follicles. As previously reported, Mus ear punch injuries rapidly formed scar tissue and remained open (Figure 1A), while Acomys injuries closed by 2–3 weeks post-injury with full restoration of pigmentation and no scar tissue by 5–6 weeks post injury (Figure 1A,B) (Seifert et al., 2012; Gawriluk et al., 2016; Matias Santos et al., 2016; Maden and Brant, 2019).
Figure 1: Acomys exhibit transient myofibroblast formation prior to functional revascularization and tissue regeneration.
(A, B) Time course (A) and quantification (B) of ear hole closure and repigmentation kinetics in Acomys and CD1 M.musculus (Mus); data are mean ± SD.
(C) Immunostaining for Acta2 (red) and Myh11 (green) at indicated time points. Arrows indicate Acta2+/Myh11+ double positive vascular smooth muscle containing blood vessels (BV). Note the transient appearance (weeks 1–2) and resolution of MFs (weeks 3–4) in Acomys compared to Mus. Dashed lines indicate amputation plane. Dashed boxes indicate area of magnification.
(D) EDA-Fibronectin (EDA,green) and Acta2 (red) immunostaining at weeks 2–3 in regenerating Acomys ear wounds.
(E) Temporal and spatial vasculature formation in Acomys versus fibrosis in Mus: Acta2 (red) and Myh11 (green). Dashed lines indicate tissue midline, with dorsal (D, top) and ventral (V, bottom) indicated.
(F) Functional vasculature revealed by intracardiac and/or retroorbital injection of Texas-red Dextran to fill vascular lumens (top, 1h, red) or Isolectin B4 (bottom, 3min, red) in regenerating Acomys ear wounds at 2-weeks post injury.
(G) Quantitation of Acta2+/Myh11− MF volume within the subdermal mesenchymal space reveals increased MF abundance in CD1 Mus vs Acomys.
(H) Quantitation of Acta2+/Myh11+ BVs temporal dynamics in Acomys vs CD1 Mus.
(I) Quantification of Acta2+/Myh11+ BVs spatial dynamics in Acomys reveals a dorsal-ventral temporal progression. All data are mean ± SD.
We performed additional histopathology to characterize tissue reorganization and composition more extensively during ear punch wound healing in Mus compared to Acomys (Figure S1). We found that Acomys ear pinnae contained lower levels of mature cross-linked collagen (Masson’s Trichrome) at all stages, by contrast to Mus, which accumulated and maintained dense deposits in the subdermal wound space covered by epidermis without hair follicle neogenesis (Figure S1A), similar to previous reports (Gawriluk et al., 2016). Alcian Blue and Safranin O counterstains revealed extensive cartilage regeneration in Acomys, including new outgrowth from the injured cartilage plate and de novo formation of new cartilage islands within regenerating tissue by 4 weeks that continued to mature and differentiate into organized structures by 5 weeks (Figure S1B,C). By contrast, the dense cross-linked collagen deposits in Mus injuries maintained disorganized structure indicative of fibrocartilage (Figure S1B). By 4 weeks in Acomys, we could also observe extensive de novo adipocyte formation, hair follicle neogenesis, and appearance of melanocytes migrating just under the dermis (Figure S1B,C). Furthermore, we found that even elastic fiber composition returned to Acomys newly regenerated adult ear tissue (Figure S1D), something that normally does not occur in human skin after birth (Le Page et al., 2019).
We further tested whether multiple 2mm ear punch injuries, given at the same time or in sequence, affected overall healing trajectory, and found that previous injuries in CD1 Mus did not grossly alter the time course of fibrosis nor the outcome of additional new injuries, and vice versa (Figure S2A). Similarly, previous injuries in Acomys did not alter regenerative healing of new injuries, which in turn did not alter the course of regenerative healing in earlier wounds (Figure S2A). Moreover, we observed that naturally occurring abdominal/flank skin injuries (fight wounds) also did not alter ear punch regeneration trajectory as they were also regeneratively healed (not shown). We also tested aged Acomys (>3ys old,) for their ability to regenerate 2mm ear punches, and found that although slightly delayed, even old animals from both sexes could fully regenerate multiple traumatic ear wounds (Figure S2A,B). These findings are suggestive of near complete local tissue architecture restoration in Acomys, even into old age.
A previous study suggested the lack of fibrosis in Acomys is due to lack of MF formation (Seifert et al., 2012). Although no single marker definitively labels MFs, upregulation of smooth muscle-α actin (Acta2) without co-upregulation of Myh11 is often used to assay for MF formation (Eyden, 2005, 2007; Hinz et al., 2007). Hence, we utilized multiple markers, double labeling, and timed injury strategies to more rigorously test for MF formation and differentiation in vivo between Mus and Acomys. To validate cross-species antigen recognition, we demonstrated anti-Acta2 and anti-Myh11 antibodies recognized single bands on western blots from primary ear fibroblasts growing in culture at the appropriate molecular weight and immunostained mature vascular smooth muscle cells (SMCs) localized to artery walls in uninjured ear tissue from both Mus and Acomys (Figure 1, S7A). Ear punch injury induced a robust Acta2+/Myh11− MF accumulation in both Mus and Acomys at early time points after injury (weeks 1 and 2, Figure 1C,G). Of interest, while Mus maintained robust numbers of MFs in fibrotic tissue that persisted though week 4 post injury, the Acomys MF population was only transient and completely resolved by 3 weeks after wounding (Figure 1C,G). Loss of Acomys MFs correlated with wound closure after which, Acta2 expression became limited to Acta2+/Myh11+ vascular SMCs in blood vessel walls (Figure 1C). Likewise, abundant expression of EDA-FN (Figure 1D), a splice variant of fibronectin (FN) required for MF activation and maturation (Serini et al., 1998) was found for Acomys MFs at 2 weeks after injury and not detected after ear hole closure one week later (Figure 1D). These findings indicate that the initial appearance of MFs after injury is common to both species, while resolution of MFs after 3 weeks is observed only in Acomys.
We developed a quantitative volumetric imaging approach to analyze MF appearance and distribution more extensively in both Mus and Acomys. Since individual Acta2+ MFs are difficult to quantify in sections, we took advantage of the overall bulk labeling of MFs in confocal 3D stacks to analyze MF subvolumes normalized to their respective tissue compartments. This approach highlighted the disappearance of Acta2+/Myh11− MFs and the expansion of Acta2+/Myh11+ vascular SMCs with respect to anatomic location, blood vessel structure, and vessel size in Acomys compared to Mus (Figure 1E). This analysis revealed the extensive number and distribution of vascular profiles in Acomys regenerating blastema at 2–4 weeks, compared to the relatively avascular scar tissue formed in Mus over the same time period. Intravascular injection of fluorescent dextran or isolectin B4 revealed that the nascent network of de novo blood vessels present at 2 weeks post-injury in Acomys were functional and continuous with the systemic circulation (Figure 1F). Quantification revealed both rodent species exhibited acute increases in MF numbers after injury, yet only Mus maintained a robust MF pool at the injury site after 2 weeks (Figure 1E,G). Interestingly, as Acomys MFs disappeared, blood vessels became more evident and were more abundant in dorsal regions initially, followed by more ventral regions after 2 weeks (Figure 1H,I). These data indicate that while Acomys and Mus both exhibit an evolutionarily conserved rapid induction of MF formation after tissue injury, Acomys evolved a transient MF phenotype that does not produce fibrosis, is associated with robust angiogenesis, chondrogenesis, hair follicle and adipocyte neogenesis, the return of melanocytes and elastin production, and may provide important cues for directing Acomys unique regenerative repair process.
Non-conserved cell-autonomous responses to TGFβ1 in isolated Acomys dermal fibroblasts
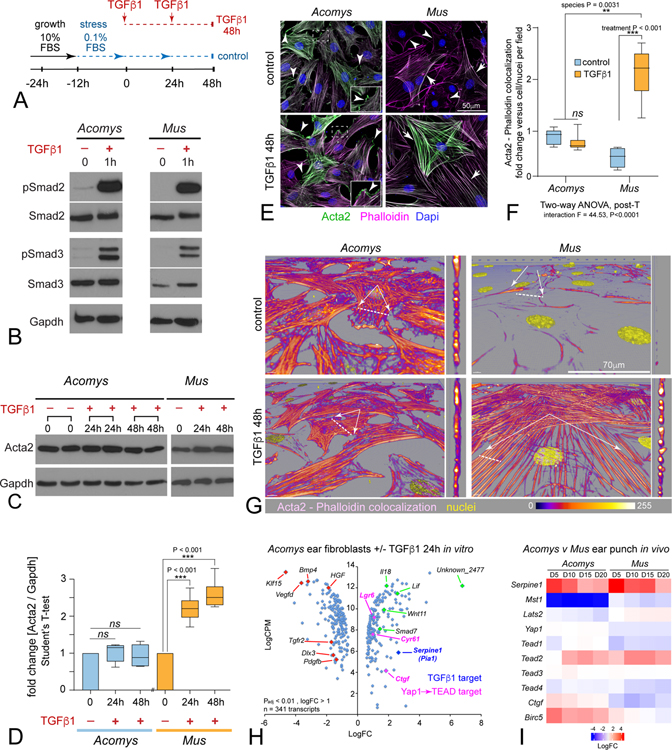
We generated primary dermal fibroblast (DF) cell lines from adult Mus and Acomys 2mm ear punches to further test the plasticity and cell signaling characteristics of this unique MF phenotype in Acomys. Mammalian fibroblasts growing in low-serum conditions differentiate to MFs via TGFβ1 signaling (Hinz et al., 2007). We grew Acomys and Mus primary ear fibroblasts to subconfluency, switched media to low serum, and treated cultures with TGFβ1 to promote MF differentiation (Figure 2A). TGFβ1-mediated MF induction can occur by both canonical SMAD and non-canonical Mitogen Activated Protein Kinase (MAPK) p38 mediated signaling (Derynck and Zhang, 2003). Western blots of Acomys and Mus DF cell lysates revealed that SMAD2, SMAD3 (Figure 2B) and p38 (not shown) were all phosphorylated in response to TGFβ1 (2ng/ml) demonstrating Acomys fibroblasts equally sense and respond to fibrokine signals when compared to Mus fibroblasts.
Figure 2: Acomys primary ear tissue fibroblasts respond to fibrokine signaling but resist terminal myofibroblast differentiation.
(A) Diagram for MF induction assay in response to TGFβ (2ng/ml).
(B) Western blot for Smad2 and Smad3 reveals phosphorylation in both Acomys and Mus primary ear fibroblasts after 1h TGFβ1.
(C) Western blot of Acta2 protein levels from Acomys and Mus in response to TGFβ1 at the indicated times: N ≥ 4 independent cultures, 2 replicates shown for Acomys.
(D) Quantification of Western blots reveals significant increase in Mus compared to no change in Acomys, two-way ANOVA, post-T as indicated; data are mean ± SD.
(E) Immunostaining Acta2 (green) in primary Acomys and Mus ear fibroblasts exposed to 48h TGFβ1 co-stained with Phalloidin (magenta), and Dapi (blue), even retaining Acta2+ lamellipodia with TGFβ1 exposure (insets), by contrast to Mus.
(F) Quantification of Acta2/Phalloidin colocalization levels after 48h TGFβ1 reveals similar significant increases in Mus compared to no change in Acomys, as indicated (see also Figure S3); data are mean ± SD.
(G) Confocal rotated higher power rendered views of individual fibroblasts reveal minimal structural or morphological differences in Acomys Acta2/Phalloidin colocalized short filaments by contrast to MFs with thick F-actin stress fibers spanning flattened cells in Mus.
(H) Scatter plot of RNAseq analysis (bullk) comparing Acomys DFs treated with TGFβ1+ versus stressed controls (24h): note lack of Acta2 transcriptional up- or down-regulation: Green, candidate up-regulated genes; Red, candidate down-regulated genes, Dark blue: internal positive control for TGFβ1 signaling (Serpine 1 / PAI-1); Magenta, Yap-TEAD target genes: see also Table S1, S2.
(I) Heatmap of available RNAseq data generated from in vivo bulk ear punch tissues comparing Acomys versus Mus wound healing series (Gawriluk et al., 2016) confirming conserved high fibrokine signaling (Serpine1) in both species, with non-conserved and coordinated Hippo (Mst1)-Yap network level transcriptional downregulation of negative upstream inhibitors (Mst1, Lats2) with concomitant upregulation of Yap-Tead targets (Birc5, Ctgf) in Acomys, by contrast to Mus.
Surprisingly, quantitative immunoblotting for Acta2 revealed Acomys DFs normally express Acta2 under control conditions, which remained constant upon exposure to TGFβ1 48h in vitro, by contrast to Mus (Figure 2C–D). Nevertheless, TGFβ1 induced EDA-F expression in Acomys DFs in vitro (Figure S2C), similar to injury-induced MFs in vivo (Figure 1D) by contrast to Mus, indicating functional yet non-conserved MF formation in Acomys. Immunolabeling confirmed Acomys DFs normally expressed diffuse Acta2, and after TGFβ1 exposure appeared not to assemble mature Acta2-containing stress fiber filaments while preserving homeostatic morphologies (Figure 2E). In Mus by contrast, TGFβ1 induced robust upregulation and assembly of mature Acta2+ stress fiber filaments and morphed into large flattened cells (Figure 2E). We considered the possibility that Acomys species may use Actg2 or Actb, instead of Acta2, to form stress fiber filaments in MFs, and employed pan F-actin-binding phalloidin-Texas Red to detect all F-actin types. Quantitation of Acta2/phalloidin colocalization confirmed Acomys maintained their normal levels, by contrast to Mus (Figure 2F, S3A). Under these conditions, it was evident Acomys DFs contained a meshwork of short F-actin filaments in control cells that largely maintained their normal length or diameter after exposure to TGFβ1, even retaining Acta2+ lamellepodial labeling (Figure 2E–G). By contrast, Mus DFs assembled parallel arrays of long thick F-actin filaments that traversed nearly the entire length of these large flattened cells (Figure 2G) (Goffin et al., 2006), a conserved structural and morphological response that was also observed in human SK5 epidermal fibroblasts treated with TGFβ1 (Figure S4A). We tested additional human fibroblast cell lines isolated from different axial levels and body origin, including abdominal skin (AB), aorta (AO) and coronary (CO) vessels for potential TGFβ1-mediated responses and found some variability in ACTA2 induction levels and structural organization (Figure S4B). We tested further by pre-labeling human cells with a lentiviral CMV-GFP (LV_CMV-GFP) tracker during MF induction, and confirmed a range of distinct single cellular morphologies coupled with variation in ACTA2 levels and structural organization (Figure S4C,D). Whether these data indicate true potential heterogenic differences between fibroblast axial origin versus fibrokine response remains to be determined, however recent studies indicate a strong likelihood for fibroblast contextual diversity in adult tissues (Lynch and Watt, 2018; Xie et al., 2018; Guerrero-Juarez et al., 2019; Buechler et al., 2021). Regardless of ACTA2, TGFβ1 induced the formation of large flattened cells with MF-like morphologies in all human lines (Figure S4D), an in vitro cellular response conserved in Mus but not in Acomys DF’s. Although Acomys DFs resisted the cellular phenotypes normally associated with MF differentiation in vitro, they still expressed ECM components (EDA-F) during initial in vitro and in vivo responses (Figure 1D, S2C). These data demonstrate Acomys ear fibroblasts evolved the ability to resist TGFβ1-mediated MF maturation in a cell-autonomous manner, consistent with their recently reported homeostatic response to increased reactive oxygen insult (Saxena et al., 2019).
We next used RNAseq analysis to more broadly analyze transcriptional responses initiated by TGFβ1 in Acomys DFs (Table S1,S2). We aligned transcripts from our stimulated fibroblasts against a recently published Acomys transcriptome (Mamrot et al., 2017) as a first step to identify differently expressed candidate genes. Increased expression of Serpine1 (plasminogen activator inhibitor-1, PAI-1), a well-known target of TGFβ1 signaling in mouse and human cells was the most significantly upregulated transcript accompanied by no change in Acta2 gene expression (Table S1, S2) confirming our biochemical and cellular data demonstrating activated TGFβ1 signaling occurs without traditional MF induction in isolated Acomys DFs in vitro, as well as in vivo (Figure 2H,I). Although Col8a2 and Col5a1 showed limited upregulation (~1.2 LogFC, Table S2) adding to Acomys transient MF ECM-structural role, they are two key collagens associated with limiting scar formation, promoting proliferation, and cause soft connective tissue, skin, tendon, ligament and corneal fibroblast disorders (Malfait et al., 2010; Aldave et al., 2013; Seet et al., 2017; Rong et al., 2017; Lv et al., 2018; Yokota et al., 2020). Lack of Col1a1 changes coupled with limited cross-linking enzymes Loxl2/4 changes (Table S2) (Piersma and Bank, 2019; Vallet and Ricard-Blum, 2019) support our histological studies (Figure S1) and further indicate functional yet altered MF trajectories in Acomys.
Further examination revealed a unique anti-fibrotic like profile with down regulation of known fibrotic signaling pathways (Pdgfb, Tgfr2, Vegfd, Bmp4, Hgf) and transcriptional regulators (Klf15, Dlx3) after exposure of Acomys DFs to TGFβ1 (Figure 2H). We also observed upregulation of genes and pathways considered more regenerative, angiogenic, and potentially anti-inflammatory, especially secreted factors (Lif, Cyr61, Wnt11, Il18, Ctgf) (Figure 2H). Indeed, Ctgf, one of the top 15 most significantly upregulated transcripts (Table S1), along with Lrg6, and Cyr61, all targets of Hippo-Yap signaling, strongly implicated a concerted transcriptional activation of this well-known developmental pathway in response to TGFβ1 signaling in Acomys DF cells (Figure 2H) (Shimomura et al., 2014; Mokalled et al., 2016). Re-examination of available in vivo bulk transcriptomic data generated from Acomys ear regeneration versus Mus fibrotic healing series (Gawriluk et al., 2016) confirms fibrokine activity in both species with increased Serpine1 levels (Figure 2I). Comparing expression of in vivo Hippo-Yap signaling components between Acomys and Mus confirmed a surprisingly non-conserved coordinated response in this critical homeostatic pathway. In Acomys, transcript levels for Mst1 and Lats2, the upstream negative regulators of Yap signaling, were downregulated (especially Mst1), while Yap1 was maintained (Figure 2I). By contrast, Lats2 expression appeared to increase while Yap1 levels decreased in Mus (Figure 2I). These differences predict a loss of Yap signaling during wound healing in Mus as opposed to a gain of Yap signaling in Acomys. In support of this notion, Ctgf and Birc5, well-known targets of Yap activity, were also upregulated in Acomys compared to Mus (Figure 2I). These data suggest evolved adaptations to tissue injury in Acomys include orchestrated gene expression network-level changes in key homeostatic signaling pathways underlying isolated cell autonomous responses to prevent fibrosis and promote regeneration as adult mammals, consistent with a more ancient adult vertebrate regeneration response program in Acomys that is conserved with some teleosts and other poikilotherms after injury (Wang et al., 2020).
Acomys dermal fibroblasts differentially regulate nuclear Yap activity in response to TGFβ1 signaling
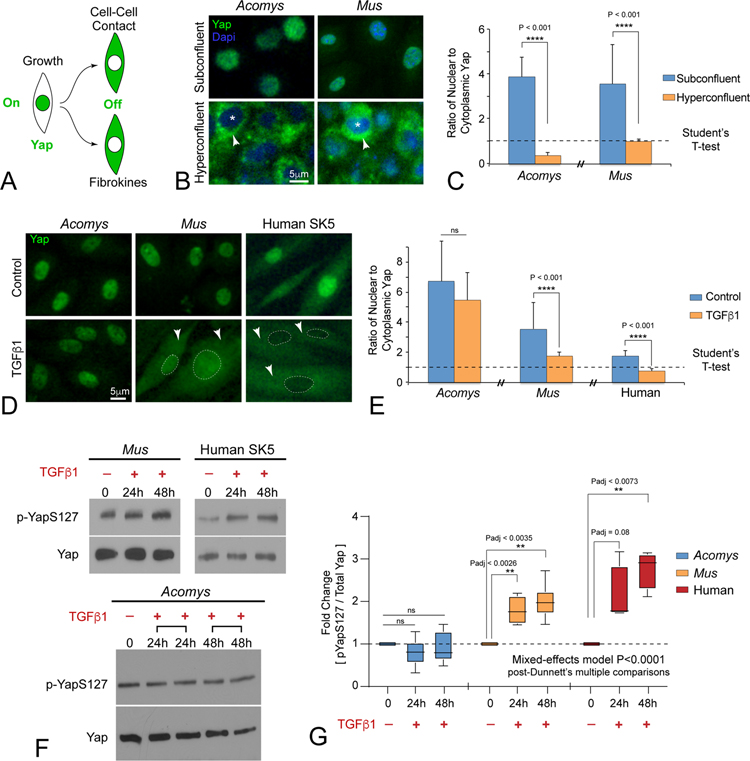
We hypothesized differential regulation of Hippo-Yap signaling contributes to Acomys regenerative wound healing properties. Although Hippo-Yap signaling activity is highly contextual, measuring Yap phosphorylation states governing nuclear versus cytoplasmic localization can be used as a proxy for pathway activity (Figure 3A) (Zhao et al., 2007). Unphosphorylated Yap is active and accumulates in the nucleus while Yap S127 phosphorylation converts Yap to an inactive nuclear protein that becomes sequestered in the cytoplasm and subject to further phosphorylation and degradation. Since Yap is a transcriptional coactivator, nuclear accumulation denotes activity at the level of gene expression (Zhao et al., 2010). We developed an image analysis assay to quantify nuclear versus cytoplasmic Yap in cultured DFs (Figure S3B). Quantification of Yap immunolocalization showed high nuclear/cytoplasmic (N/C) ratios at subconfluent density and low N/C ratios at hyperconfluent density in DFs from both species (Figure 3B,C). These data suggest that pathways controlled by cell-cell contact (Kim et al., 2011) regulate YAP phosphorylation and cytoplasmic localization in similar ways in both species. By contrast, upon exposure to TGFβ1 Acomys DFs maintained nuclear Yap while, Mus and human DFs under identical conditions exhibit cytoplasmic shuttling of Yap during MF induction (Figure 3D,E). Quantitative immunoblotting of Yap-S127 phosphorylation after TGFβ1 stimulation revealed that only a small portion of the total pool of Yap protein in Acomys cells is phosphorylated compared to a much larger fraction of pYap-S127/total Yap found in Mus and human cells (Figure 3F,G). TGFβ1 treatment also stimulated increases in pYap-S127 levels over time in Mus and human DFs while Acomys pYap-S127 levels remained stable (Figure 3F,G). These data reveal that in addition to transcriptional differences, further post-translational regulation of Yap phosphorylation in response to TGFβ1 is significantly different in Acomys DFs compared to Mus and human cells.
Figure 3: Acomys primary ear tissue fibroblasts evolved unique context-dependent regulation of Hippo pathway effector protein Yap.
(A) Diagram illustrating Yap localization as a readout for Hippo pathway activity.
(B, C) Immunofluorescent staining (B) and quantitation (C) for Yap (green) subcellular localization in Acomys and Mus primary ear tissue fibroblasts growing under homeostatic subconfluent and hyperconfluent cell densities in vitro (N ≥100 cells per treatment group). Asterisks indicate nucleus. Arrowheads indicate cytoplasm. Dapi serves as a nuclear localization marker. ns = not significant; data are mean ± SD; see also Figure S3B.
(D, E) Immunofluorescent staining (D) and quantitation (E) for subcellular localization of Yap after TGFβ1 (2ng/ml, 48h), arrowheads indicate cytoplasmic localization, dashed lines indicate nucleus: ns = not significant, N > 4 independent cultures; data are mean ± SD.
(F, G) Western blot (F) and quantitation (G) of pYap-S127 versus total Yap levels in during TGFβ1-mediated MF formation across species. Note that TGFβ1 stimulated Yap-S127 phosphorylation in Mus and human fibroblasts, but not in Acomys fibroblasts. ns = not significant. N ≥ 4 independent cultures; data are mean ± SD.
Nuclear YAP localization in Acomys dermal fibroblasts is maintained by a novel protein phosphatase activity
We next examined Yap protein sequence obtained by compiling all overlapping transcript reads from our Acomys RNAseq dataset and comparing those reads to the orthologous Mus Yap exon sequence data (Figure S5A). This approach generated 8–9 potential Yap isoforms present in Acomys DFs, the same number of isoforms present in multiple human and mouse tissues (Gaffney et al., 2012). Open reading frame analysis of the longest Acomys transcripts revealed a predicted peptide (490aa) that is 93% and 96% identical to human and mouse Yap1–2, respectively, with all key regulatory Ser/Thr/Tyr phosphorylation sites conserved (Figure S5). Therefore, regulation of Yap phosphorylation state, rather than sequence differences or isoform variants, contributes to retention of biologically active nuclear Yap in Acomys DFs versus Mus and human cells under identical conditions in vitro.
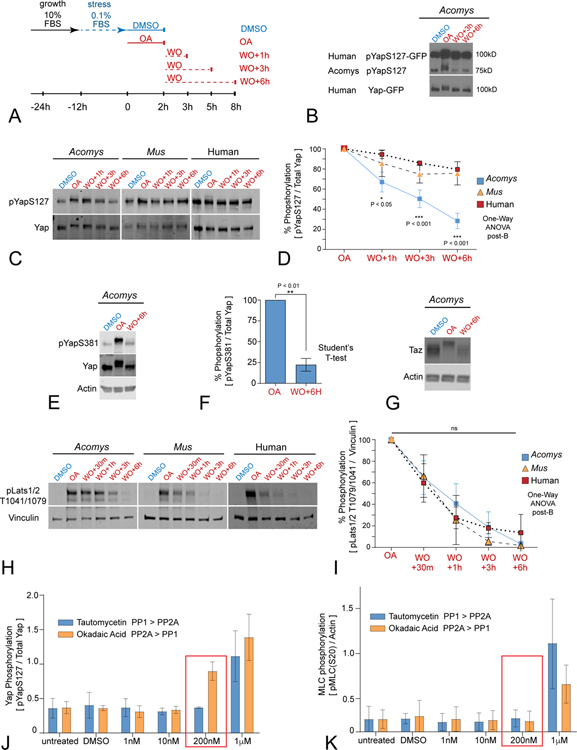
Yap dephosphorylation is largely mediated by PP1 and PP2A class serine/threonine (S/T) protein phosphatases (Schlegelmilch et al., 2011; Wang et al., 2011). We developed a pulse chase assay using okadaic acid (OA), a potent S/T phosphatase inhibitor, to determine the kinetics of Yap de-phosphorylation in Acomys vs Mus DFs (Figure 4A, S6A) (Swingle et al., 2007; Bialojan et al., 1988). OA treatment (200nM) allows phosphorylated Yap to accumulate, while OA washout allows PP1/PP2A-mediated Yap phosphatase kinetics to be determined. Changes in Yap phosphorylation on multiple serine and threonine residues (Figures 4B–D; S5C,D) are evident as a molecular weight gel shift observed on immunoblotting. We first tested for potential species differences in Yap dephosphorylation by establishing a stable Acomys DF cell line expressing human GFP-tagged YAP (hYap-GFP). We observed both hYap-GFP and endogenous Acomys Yap underwent a hypershift after OA pulsing followed by equally rapid dephosphorylation kinetics after washout (Figure 4B), confirming high Yap peptide conservation and regulation within identical cellular environments, and suggesting novel phosphatase activity contributes to Acomys unique capacity to maintain high levels of nuclear Yap activity.
Figure 4. PP2A-mediates accelerated Yap dephosphorylation kinetics and nuclear retention in Acomys primary ear tissue fibroblasts.
(A) Schematic diagram of dephosphorylation assay utilizing timed okadaic acid (OA) pulses, washouts (WO), followed by media replacement for indicated timepoints; see also Figure S6A.
(B) Western blot for pYap-S127 (Cell Signaling Technologies, CST) in Acomys fibroblasts expressing human Yap (hYap-GFP) after OA pulse and washout (N≥3). Note both species’ Yap share extensive hypershift and dephosphorylation kinetics over indicated time points.
(C, D) Western blot (C) and quantitation (D) of dephosphorylation kinetics for pYap-S127 (both CST and Abcam antibodies) in fibroblasts after OA pulse and washout. Note the rapid and extensive dephosphorylation of pYap-S127/total Yap in Acomys compared to the slow, limited dephosphorylation in pYap-S127/total Yap in Mus fibroblasts and human SK5 cells; statistics as noted; data are mean ± SD (N≥3); see also Figure S6C–D).
(E, F) Western blot (E) and quantitation (F) of dephosphorylation kinetics for pYap-S381 in Acomys fibroblasts after OA pulse and washout: note similar rapid dephosphorylation of pYap-S381/total Yap (E) as seen for pYap-S127/total Yap (C); data are mean ± SD (N≥3).
(G) Similar accelerated de-phosphorylation kinetics were observed for TAZ in Acomys fibroblasts after OA treatment and WO (N≥3); see also Figure S6D.
(H, I) Western blot (H) and quantification (I) of pLats1/2 dephosphorylation kinetics after OA pulse and washout. Vinculin is used for loading control. Note no significant statistical differences of Lats1/2 dephosphorylation kinetics between Acomys, Mus and human; one-way ANOVA post Brown-Forsythe, ns = not significant; data are mean ± SD (N≥3).
(J, K) Dose-dependent differential phosphatase inhibition reveals increased PP2A affinity for Yap dephosphorylation in Acomys. Quantitation of pYap-S127/total Yap (J) and pMyosin Light Chain-S20 (MLC) (K), red boxes indicate significant difference; data are mean ± SD (N≥3); see also Figure S6.
We designed a more specific phosphatase substrate-inhibitor strategy (Figure S6A) and first tested ERK dephosphorylation kinetics for potential differences between species. After OA-treatment, pERK levels were unaltered in both Acomys or Mus DFs (Figure S6B) in agreement with its role as a DUSP (dual specificity phosphatase) substrate (Sun et al., 1993). We directly compared the rate of endogenous Yap-S127 de-phosphorylation between Acomys, Mus, and human DFs after OA washout and found that Acomys DFs mediated Yap de-phosphorylation with significantly faster kinetics and to a much greater extent than Mus and human cells (Figure 4C,D). We also found that de-phosphorylation of the phospho-degron motif within the Yap TAD domain (S381-Mus/S397-Human) (Figure S5), which targets Yap to E3 dependent ubiquitination and proteasomal degradation (Zhao et al., 2010), likewise showed significantly rapid return to its normally low level in Acomys DFs (Figure 4E,F). We tested the specificity of Acomys PP1/PP2A de-phosphorylation activity by analyzing endogenous Taz and Lats1/2. We found that Acomys Taz, a Yap paralog, hypershifted in OA and appeared degraded after washout (Figure 4G) with lower levels overall in Acomys cells (Figure 4G), possibly reflecting an inverse relationship between Yap abundance and Taz stability (Finch-Edmondson et al., 2015). We found that rates of endogenous Lats1/2 de-phosphorylation were similar between Acomys, Mus, and human fibroblasts (Figure 4H, I), indicating conserved phosphatase regulation of the direct upstream regulator of Yap in the Hippo pathway.
To distinguish between PP1 and PP2A protein phosphatase activities in Acomys, we used selective inhibitors of each phosphatase subclass in a dose-response assay (Mitsuhashi et al., 2001; Fernandez et al., 2002): OA has greater affinity for PP2A, tautomycetin has greater affinity for PP1 (Figure S6A). We quantified changes in the levels of Acomys pYap-S127 versus Yap, compared to changes in the levels of pMyosin light chain (pMLC)-S20 versus actin (Figure S6C). We found increased pYap-S127/Yap at 200nM OA with no change in pYap-S127/total Yap at 200nM tautomycetin compared to vehicle-treated cells (Figure 4J–K, S6C). By contrast, pMLC-S20/actin levels did not change until higher concentrations (1uM) of the inhibitors were used (Figure 4K, S6C). Indeed, despite differences in levels, both Yap and Taz show similarly rapid PP2A-dependent dephosphorylation kinetics (Figure S6D). These data indicate that Yap/Taz de-phosphorylation is more sensitive to PP2A inhibition compared to pMLC-S20, a PP1 substrate (Figure 4K, S6C). We confirmed Acomys has high overall PP2A phosphatase activity by examining AKT kinetics, as pAKT(T308) is targeted by PP2A while pAKT(S473) is targeted by PHLPP phosphatases (Figure S6A) (Gao et al., 2005; Kuo et al., 2008). Although dephosphorylation at pAKT(Thr308) occurred much more rapidly than at pAKT(S473) indicating high PP2A activity (Figure S6E), pAKT(Thr308) levels actually increased after washout (1h) prior to PP2A engagement, by contrast to pYap-S127 (Figure 4C–D), indicating differential PP2A subunit regulation may contribute to more rapid Yap targeting. These findings suggest that Acomys DFs also evolved a novel cell-autonomous and high affinity PP2A-mediated surveillance function that rapidly and specifically dephosphorylates key residues controlling Yap nuclear localization (S127) and stability (S381) in the presence of fibrokine signals.
Spatial and temporal nuclear Yap activity correlates with ear tissue regeneration in Acomys versus fibrosis in Mus
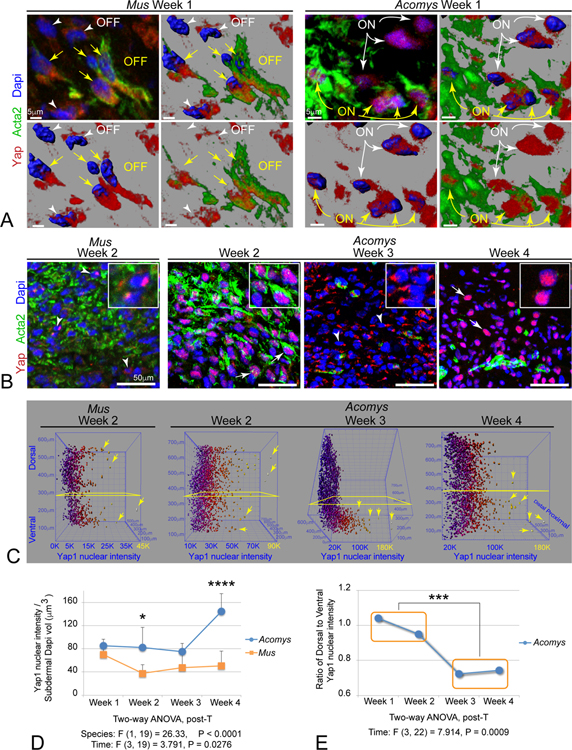
We validated whether predicted species differences in Yap signaling were apparent by immunolabeling tissue sections of Mus and Acomys ear punch injuries in vivo. Normally, Yap was restricted to the cytoplasmic compartments of epidermal and hair follicle populations and excluded from the mesenchyme (Figure S7A). By 1-week post-injury, Yap protein was present in Acta2+ MFs in both Mus and Acomys (Figure 5A–B, S7B). However, Yap protein was largely excluded from the nucleus in Mus, whereas Acomys MFs exhibited strong nuclear localization of Yap in both Acta2+ MFs and Acta2− cells within the regeneration blastema (Figure 5A–B, S7B, Movie S1). By 2 weeks, Acomys maintained strong nuclear Yap localization through week 2, just prior to wound closure, whereas Yap levels decreased in Mus (Figure 5B, S7C). Interestingly, Acomys Yap protein became largely relegated to the perinuclear cytoplasm in most mesenchymal cells at week 3, a time point where Acta2+ MFs disappeared (Figure 5B, S7D). However, by week 4, nuclear Yap localization was re-established in a majority of Acomys mesenchymal cells, especially those residing in the ventral region of the regenerating tissue during this period of intense cell patterning and differentiation (Figure 5B, S7E).
Figure 5. Biphasic nuclear Yap localization distinguishes Acomys regenerative vs CD1 Mus fibrotic wound healing in vivo.
(A) High powered magnifications and renderings of isolated immunolabeled Yap+ (red), Acta2+ (green), and rendered Dapi nuclei (blue) fibroblasts within Week 1 subdermal wound. Panels indicate progressive cellular and nuclear renderings to illustrate cytoplasmic distribution in Mus Acta2+ MFs (OFF) compared to predominately nuclear Yap in Acomys Acta2+ MFs (ON) (yellow arrows): note there are also some Acta2− cells in Mus that also exhibit cytoplasmic Yap localization (OFF, white arrowheads), and Acta2− cells in Acomys with strong nuclear Yap localization (ON, white arrows): see also Movie S1.
(B) Yap (red) and Acta2 (green) immunolabelling across injury sequence in vivo: note the lower overall levels of Yap in Mus at week 2, and the relocalization of nuclear Yap at week 1–2, to cytoplasmic at week 3, and back to predominant nuclear Yap by week 4 in Acomys.
(C) Examples of quantitative 3D heat map plots of individual nuclei from sections labeled in situ and rotated to view nuclear Yap levels (X axis, blue to yellow increasing intensity) in subdermal cells located along the dorsal-ventral axis (Y axis) and the proximal-distal axis (Z-axis) (see also Figure S3C,D).
(D) Quantitation of total subdermal cellular nuclear Yap levels over time reveals a significant biphasic distribution during temporal progression of Acomys ear trauma regeneration; data are mean ± SD.
(E) Quantitation of Acomys nuclear Yap localization reveals initial widespread subdermal distribution during early wound phase 1 (weeks 1–2) that becomes predominately ventrally restricted as wounds close and tissue repatterns during phase 2 (weeks 3–4); data are mean ± SD.
We developed a quantitative imaging approach to determine species and pattern differences in nuclear/cytoplasmic Yap localization in mesenchymal cells within the regenerating blastema in vivo (Figure S3C,D). Digitally segmented subdermal blastema nuclei were analyzed for nuclear Yap levels during each wound healing stage in both Mus and Acomys (Figure S3C,D). 3D plots of individual Acomys nuclei at the indicated timepoints clearly illustrate the different levels of nuclear Yap intensity within individual cells with respect to dorsal-ventral location across regeneration time (Figure 5C). Quantifying total nuclear Yap intensity within the subdermal wound compartment revealed statistically significant increases in nuclear Yap levels overall in Acomys cells, particularly just before and after wound closure, indicating divergent biphasic Yap activity compared to Mus (Figure 5B,D). Moreover, we observed quantitatively significant spatial and temporal differences in Acomys nuclear Yap localization during early proliferative phase (weeks 1–2) compared to later differentiating phase (weeks 3–4) (Figure 5E). Acomys unique spatial and temporal nuclear Yap patterning in vivo is consistent with our in vitro data showing that Acomys fibroblasts exhibit an increased capacity for maintaining nuclear localization of Yap. These data suggest that Acomys species naturally evolved a solution for mammalian tissue regeneration without fibrosis that includes alterations in Yap regulation across multiple levels: transcriptional downregulation of upstream negative regulators (Mst1,Lats2), combined with post-translational regulation of Yap phosphatase activity (PP2A-selective) that correlates with cell proliferation, cell fate, and restoration of dorsal-ventral patterning during tissue regeneration.
Blocking nuclear Yap signaling activity inhibits Acomys regenerative wound repair.
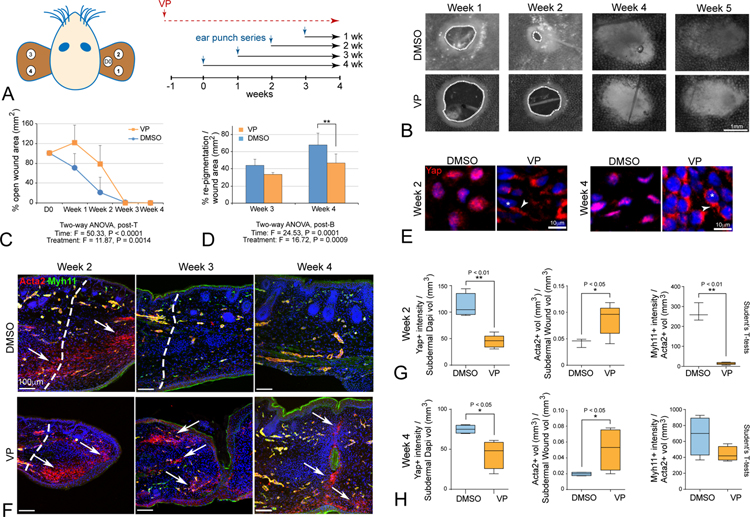
Since Acomys genetic tools are not available, we tested whether Yap signaling was necessary for Acomys ear tissue regeneration by using verteporfin (VP), a small molecule inhibitor of nuclear Yap-TEAD interactions that induces cytoplasmic sequestration of Yap (Liu-Chittenden et al., 2012). VP has previously been shown to disrupt Yap-TEAD activity with few demonstrated off target or cytotoxic effects (Chen et al., 2018; Konishi et al., 2018; Mohede et al., 2018). We pretreated adult Acomys (n=5–6 per group) with VP [100μg/g, DMSO or mineral oil] or equal volume of vehicle every other day for a week. Ear punches were then performed as depicted in Figure 6A, and VP or vehicle control injections were continued every 48h thereafter until 4 weeks post ear punch. Chronic VP treatment elicited no overt toxic effects and body weights of VP-treated versus control groups did not change over the full-time course of the experiment (Figure S7J). At the macroscopic level, VP-treatment clearly attenuated time to wound closure (Figure 6 B,C) and the rate of re-pigmentation (Figure 6B–D). At the microscopic level, immunolabeling revealed a clear reduction of nuclear Yap accompanied by increased cytoplasmic Yap in 2-week VP-treated animals (Figure 6E), accompanied by attenuated Ctgf immunolabeling in vivo (Figure S7F–I), a downstream Yap target gene expressed in dermal tissues (Kapoor et al., 2008; Morales et al., 2018). VP-treatment led to increased and persistent Acta2+/Myh11− MFs, especially in dorsal regions and along tissue closure points, which although appearing closed macroscopically, failed to fuse properly upon microscopic inspection (Figure 6F). After 2 weeks of VP treatment, quantitation revealed a ~50% reduction in Yap-positive nuclei in cells in the subdermal blastema accompanied by an increased content of MFs (total Acta2+ volume) and decreased angiogenesis (Acta2+/Myh11+ SMCs) (Figure 6G), which was maintained through week 4 (Figure 6H). These data reveal that inhibition of nuclear Yap-TEAD signaling severely attenuates ear tissue regeneration and prolongs the residence time of MFs, thus converting Acomys wound healing into a more Mus/human-like scar-forming response.
Figure 6: Yap activity is required for epimorphic ear regeneration and to prevent fibrosis in Acomys.
(A) Schematic diagram of chronic verteporfin (VP) treatment relative to ear punch timing in Acomys.
(B) Representative images of ear hole closure in verteporfin-treated (VP) and vehicle-treated (DMSO) Acomys. Dashed lines indicate border of open wound hole.
(C, D) Quantitation reveals significant attenuation of ear closure and decreased re-pigmentation in traumatic ear wounds due to VP (orange) relative to DMSO control (blue).
(E) VP treatment redistributes nuclear Yap to the cytoplasm in vivo at week 2 and maintained through week 4 (asterisks indicate nuclei, arrowheads indicate cytoplasmic Yap OFF), with attenuated in vivo Ctgf target labeling at week 1 and without body weight change (Figure S7F–J)
(F) Increased and persistent Acta2+/Myh11− MFs are present due to VP-treatment, and Acta2+/Myh11+ BV formation is delayed compared to control DMSO-treatment; note that microscopic views at week 3 indicate failure to close fully, with incomplete fusion and scab clearance (arrows, week 4).
(G, H) Quantitation for Acta2, Myh11 and Yap in VP-treated and DMSO-treated ear tissues at week 2 (G) and week 4 (H) traumatic ear repair: nuclear Yap is decreased, Acta2+/Myh11− MFs are increased, and Acta2+/Myh11+ vasculature formation is attenuated in VP-treated tissues: all data are mean ± SD.
Nuclear YAP signaling regulates early phase blastema formation and later differentiation stages during Acomys tissue regeneration
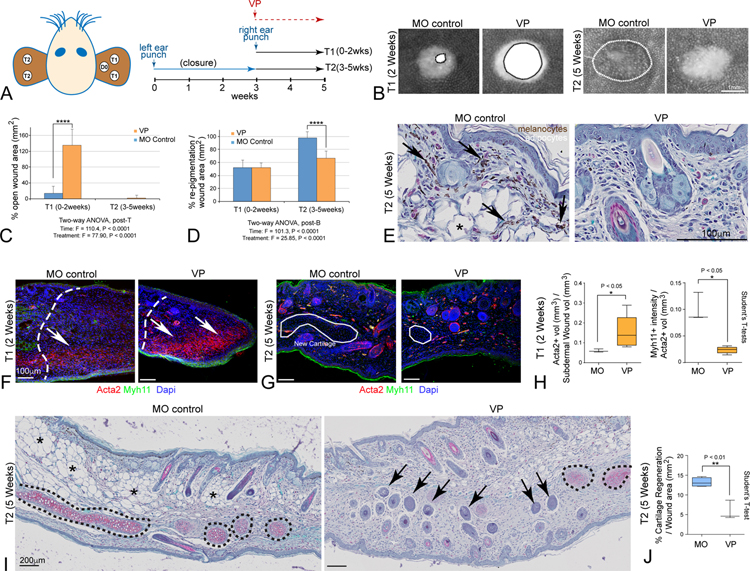
We modified our VP-treatment protocol to specifically test the role of nuclear Yap signaling during initial blastema formation (weeks 2–3) separately from later differentiation events occurring after wound closure (weeks 4–5) in the same animal (Figure 7A). We punched 2mm holes in one ear of adult Acomys (n = 4–6 per group) and allowed injuries to first undergo normal regenerative healing leading to closure by 3 weeks. Upon closure, additional 2mm punches were applied to the other ear and VP treatment, or vehicle control, was commenced for the final 2 weeks. This strategy allows separation of the early phase responses (T1) from the late phase responses (T2) with respect to VP treatment. Acute VP-treatment resulted in clear and statistically significant wound closure effects (T1) and inhibited re-pigmentation at later phases (T2) (Figure 7B–D). Histological examination confirmed pigment defects were due to lack of differentiated melanocytes near the dorsal regions at later phases (T2, Figure 7E). In the early blastema, VP-treatment increased Acta2+ MF labeling and inhibited angiogenesis (T1), while VP treatment in the late phase of regeneration (T2) resulted in disruption of tissue architecture (Figure 7F–H). Safranin O histology and quantification revealed VP treatment during the late phase of wound healing (T2-week 5) significantly reduced cartilage regeneration (Figure 7I–J, S3E). Strikingly, in addition to cartilage, both adipose and melanocyte progenitors were also lost or reprogrammed, potentially contributing to the large increases seen in ectopic hair follicle formation in ventral regions (Figure 7I, arrows). These data confirm an essential role for nuclear Yap signaling during early regenerative blastema formation and additional key roles in tissue patterning and cell fate potentials during later phases of Acomys tissue regeneration.
Figure 7. Yap regulates two distinct phases of mammalian epimorphic ear regeneration in Acomys.
(A) Schematic diagram depicts strategy for blocking Yap activity during initial Phase 1 trauma responses, and later Phase 2 patterning and differentiation in the same animal.
(B) Representative images of VP-treated versus mineral oil (MO)-treated animals at Time 1 (T1, week 2, left panels) and at Time 2 (T2, week 5, right panels).
(C, D) Quantitation of VP-mediated effects on closure rate (C) and re-pigmentation levels (D).
(E) High magnification histological view reveals loss of pigmentated melanocytes (arrows) and adipocytes (asterisks) after VP treatment.
(F, G) Increased Acta2+/Myh11− MFs are only observed in the initial blastema during T1 after VP-treatment (F, arrows), while Dapi labeled organized cartilage clusters were reduced during T2 VP-treatment (G, outlines).
(H) VP-treatment during T1 led to significantly increased Acta2+/Myh11− MFs and reduced Acat2+/Myh11+ BVs.
(I, J) Safranin O histology reveals significant loss of new de novo generated cartilage (black dashed lines; quantified in (I) and adipocytes (asterisks), accompanied by expansion of ectopically located hair follicles in ventral regions (arrows); all data are mean ± SD.
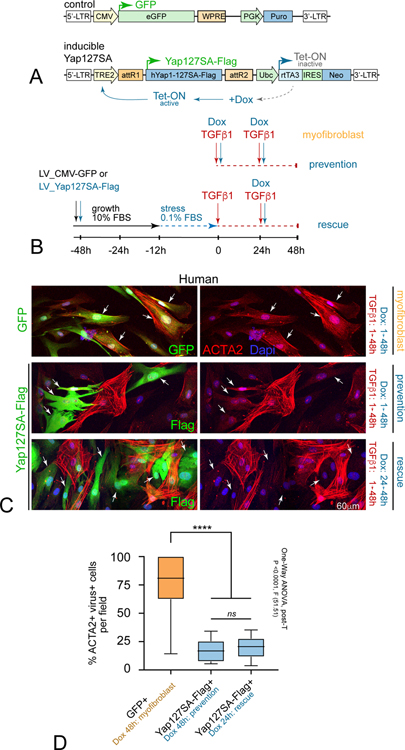
YAP activation prevents and rescues human myofibroblast formation in vitro
We next tested whether mimicking Acomys nuclear Yap might alter the MF differentiation response to TGFβ1 in human DFs. As a first step, we employed a doxycycline (Dox)-inducible Yap-S127A-Flag lentiviral expression construct (LV-Dox-Yap-127SA-Flag), which limits Mst/Lats-mediated phosphorylation and promotes Yap nuclear activity (Figure 8A) (Song et al., 2014). We transduced human SK5 fibroblasts with either LV-CMV-GFP or LV-Dox-Yap-127SA-Flag under growth conditions, and then analyzed TGFβ1-mediated MF differentiation at the time points indicated in transduced cells (Figure 8B). We found that human SK5 fibroblasts that received concurrent TGFβ1 and Dox for 2 days displayed statistically significant reduction in Acta2 levels in LV-Dox-Yap-127SA-Flag+ fibroblasts compared to neighboring uninfected cells or control GFP+ cells (Figure 8C,D). Moreover, transduced fibroblasts exposed to TGFβ1 for the first 24h to activate MF-induction pathways, and then treated with Dox + TGFβ1 during the second 24h also exhibited a statistically significant reduction in Acta2 levels and stress fiber formation in LV-Dox-Yap-127SA-Flag+ fibroblasts (Figure 8C,D). It remains to be determined whether this could be due to direct Yap inhibition of Acta2 gene expression, or possibly Yap-mediated cell reprogramming (Xie et al., 2012; Wang et al., 2012; Panciera et al., 2016; Yui et al., 2018). Nevertheless, these data provide proof-of-principle evidence that novel regenerative insights discovered in Acomys can be translated into treatments that could prevent or reverse fibrotic scarring in humans.
Figure 8. Translating Acomys novel Yap biology prevents and rescues human MF formation in vitro.
(A) Lentiviral strategy for conditionally increasing nuclear Yap activity: LV-GFP control (top) versus doxycycline (Dox) inducible constitutively active Yap (LV-YapS127A-Flag) (bottom).
(B) Human SK5 dermal fibroblasts were transduced with LV-GFP and LV-Yap-127SA viruses for 48h, serum-starved overnight, treated with TGFβ1 and/or Dox as indicated, and grown for another 48h to promote MF differentiation: note some wells were exposed to TGFβ1 for 24h prior to Dox to test rescue capacity).
(C) High-resolution confocal view show isolated cells co-immunolabeled for Acta2 (red) and LV-GFP control (top, green, arrows), by contrast to LV-Yap-127SA (bottom, green Flag tag, arrows) in human DFs after 48hrs indicated conditions: note concomitant TGFβ1 and Dox-treatment prevented Acta2+ stress fiber formation, and that even Dox addition after prior 24h TGFβ1-mediated MF initiation rescued this phenotype.
(D) Quantification of percent GFP/Flag+ cells per condition; data are mean ± SD.
Discussion
Mammalian tissue regeneration is extremely limited (Poss, 2010). Most adult mammals instead repair injury by mobilizing both inflammatory cells and ECM-secreting MFs that together form scar tissue. By contrast, three species of African spiny mice (genus Acomys) have now been shown to completely regenerate fully functional and anatomically correct skin without scarring after extensive loss of adult tissue (Seifert et al., 2012; Gawriluk et al., 2016; Matias Santos et al., 2016; Jiang et al., 2019; Maden and Brant, 2019). The regenerative mechanisms activated in adult Acomys wound repair have clear clinical implications for treating chronic fibrotic diseases and promoting tissue regeneration (Gurtner et al., 2008). The major findings we report here are: (1) MFs appear rapidly after injury in both Acomys and Mus ear tissue, but persist only in Mus; (2) isolated Acomys fibroblasts resist MF-like cytoskeletal remodeling when stimulated by TGFβ1, by contrast to Mus and human fibroblasts; (3) Acomys TGFβ1-stimulated fibroblasts maintain transcriptionally active nuclear Yap due to species-specific accelerated PP2A-dependent Yap dephosphorylation kinetics in vitro, (4) Acomys regenerating ear tissue in vivo exhibits transcriptional downregulation of Mst1 (Hippo pathway) with concomitant nuclear Yap localization and target gene upregulation, by contrast to cytosolic Yap and fibrotic scarring in Mus; (5) biphasic nuclear Yap-TEAD activity is necessary for preventing initial fibrotic scarring and promoting subsequent epimorphic regeneration in Acomys; (6) translating insights from Acomys novel Yap regulation to human fibroblasts in vitro both prevented and rescued TGFβ1-mediated MF differentiation.
Acomys have activated myofibroblasts during initial regeneration stages
We first tested whether lack of activated MFs was responsible for absence of fibrotic scarring in adult Acomys, as a previous report indicated MFs were not present during Acomys regenerative skin repair (Seifert et al., 2012). We found both Acomys and Mus exhibited an evolutionary conserved rapid induction of injury associated MFs. Formation of a MF-rich blastema in Acomys overlapped with an extensive expansion of a functioning vascular network not seen in MF-rich scar forming tissue in Mus. Upon wound closure in Acomys, a striking loss of MFs was observed without detectable increases in cell death. Delaying Acomys MF resolution with VP treatment inhibited angiogenic expansion, altered cell fates, and attenuated wound closure. These data point to evolved MF cellular adaptations in Acomys diverging from the classically defined MF phenotype, both preventing scar formation and promoting regenerative wound healing. Intriguingly, pro-regenerative MF repair responses were also noted in zebrafish cardiac regeneration (Sanchez-Iranzo et al., 2018). As Acomys MFs resolution is not due to apoptotic clearance as seen in other mammals (Desmouliere et al., 1995), it is conceivable transient MFs in Acomys serve as a precursor pool for other mesenchymal cell types required for regeneration (Plikus et al., 2017). Alternatively, MFs may revert back to a fibroblast phenotype as noted in zebrafish cardiac regeneration (Sanchez-Iranzo et al., 2018). Regardless, the similarities in MF behavior in both regenerative Acomys and zebrafish suggest MF adaptations in Acomys, unlike their scarring counterparts in Mus and humans, actively direct cell fate decisions favoring regeneration. Like macrophage diversity (Schulz et al., 2012), the possibility of MF subtypes promoting fibrosis or scar free wound healing (Driskell et al., 2013; Rinkevich et al., 2015; Jiang et al., 2018; Guerrero-Juarez et al., 2019; Buechler et al., 2021) underscores the importance of rigorous MF characterization in Acomys.
Isolated Acomys ear fibroblasts spontaneously acquire the traditional MF marker Acta2 but display behaviors unlike scar tissue-forming MFs. Despite robust TGFβ-pathway activation and target gene Serpine1 upregulation, our RNA seq analysis in vitro revealed Acomys MFs do not coordinately upregulate genes and proteins commonly associated with MFs such as Col1a1 or crosslinking enzymes Loxl2/4. MF activation is commonly assessed by assembly of actin subtypes into what is described as super-mature stress fiber formation (Tomasek et al., 2002). Immunofluorescence experiments revealed mechanically dissociated and isolated Acomys primary ear fibroblasts spontaneously upregulate Acta2, which embeds into meshworks of short F-actin filaments that resist overt changes in levels, lengths or diameters or overall cell size after TGFβ1-mediated MF induction, in contrast to both Mus and human MFs. These properties are reminiscent of Drosophila epithelial development where apical stress fibers sequester Wts and activate Yki (Lats1/2, Yap in vertebrates respectively) integrating cell proliferation responses with tissue forces (Fletcher et al., 2018; Lopez-Gay et al., 2020). Hence stress fiber formation and Yap activation exhibit an evolutionary similarity between mechanical regulatory landscapes in Drosophila embryos and regenerating Acomys fibroblasts. Nevertheless, Acomys transient MFs do upregulate Col8a2 and Col5a1 transcripts, although they may not be translated (Seet et al., 2017), and secrete ECM components (EDA-F) both in vitro (Figure S2C) and in vivo (Table S2; Figure 1D, S2C), contributing to MF ECM-functions prior their disappearance. We conclude Acomys evolved a solution to scar-forming wound repair that includes a cell autonomous block to MF maturation.
Evolutionary adaptations targeting Hippo-Yap-PP2A regulate Acomys regeneration
In experiments to characterize Acomys block to MF maturation, we discovered primary fibroblasts isolated from adult Acomys ear pinnae cell-autonomously upregulated several Hippo-Yap pathway-associated target genes in response to TGFβ1 in vitro, including Ctgf, Cyr61 and Birc5. Strikingly, these in vitro Hippo-Yap transcriptional responses to fibrokine signals mirrored transcriptional dyanmics during in vivo ear regeneration. Whereas Acomys regeneration program included downregulated Mst1 and increased Birc5 and Ctgf expression in vivo, confirmed by Ctgf immunolabeling in vivo, Mus by contrast exhibted the opposite transcriptional response during their fibrotic scarring program. At the cellular and protein level, Acomys DFs maintained nuclear Yap localization and activity in response to TGFβ1 stimulation in vitro, by contrast to scar forming Mus and human MFs. When Acomys DFs were maintained at post-confluent density, a surrogate for contact-mediated inhibition in vivo, YAP protein was found to be entirely cytoplasmic. Intriguingly, during the time Acomys newly regenerated tissues come into contact during wound closure, nuclear Yap also becomes sequestered into the cytoplasm. These findings indicate Acomys have not perturbed canonical Hippo signaling per se but rather have evolved unique adaptations in YAP signaling in response to tissue injury.
Additional biochemical and gene sequencing studies showed Acomys species-specific control of Yap nuclear localization is mediated by increased rates of PP2A catalytic subunit-dependent Yap phosphatase activity and not because of species-specific changes in Yap amino acid sequence. Moreover, Acomys PP2A phosphatase activity regulates both Yap spatial and temporal activity with S127 (cytoplasmic retention) and S381 (degradation) residues exhibiting similar increased rates of dephosphorylation. The targeted and rapid kinetics of PP2A-mediated pYap dephosphorylation in Acomys may be due to differential PP2A beta subunit usage, higher tier protein-protein complex formation (STRIPAK), or both (Couzens et al., 2013). Nevertheless, our studies reveal Acomys unique pro-regenerative Hippo-Yap signaling adaptations during adult wound-healing context evolved at both transcriptional and post-translational levels.
This is not the first time evolution has targeted the highly contextual Hippo-Yap-PP2A pathway to maintain or disrupt homeostasis (Liu et al., 2015; Liang et al., 2017; Dey et al., 2020). Context-dependent regulation of Yap localization has been noted in conjunction with normal progenitor proliferation and niche-mediated signals (Schlegelmilch et al., 2011; Hu et al., 2017). Likewise, evolutionary capture of pYap-PP2A interactions can both accelerate dephosphorylation kinetics leading to increased Yap activity and promote tumor formation (human polyomavirus), or prevent dephosphorylation to maintain low Yap activity for tumor cell killing (human adenovirus E4orf4) (Mui et al., 2015; Rouleau et al., 2016). Similarily, while Yap is normally upregulated in mouse and human intestinal cancer stem cells during disease progession, Yap activates a pro-regenerative reprogramming wound healing response to suppress proliferation and limit tumor formation (Cheung et al., 2020).
Nuclear Yap is required for Acomys epimorphic ear tissue regeneration
Blastema formation is considered a critical initiating event for epimorphic regenerative repair (Steen, 1970; Brockes et al., 2001; Tanaka, 2016). Our inhibitor studies with VP reveal nuclear Yap activity is required early in blastema formation. Interestingly, Acomys Yap nuclear localization was observed in multiple cell types within the blastema including MFs and stromal mesenchymal cells. This is consistent with the multiple biological effects of VP on the blastema, including failure of MF resolution upon wound closure and attenuated angiogenesis. Taken together, our data indicate that nuclear Yap activity is required early in the temporal sequence of regeneration (phase 1), and controls Acomys MF transitional behavior and plasticity, sustained angiogenic vascular network expansion, and inhibition of scar tissue formation.
Yap activity is also required during phase 2 of ear tissue regeneration in Acomys. Phase 2 is identified by post-wound closure tissue patterning and restoration of differentiated structures such as cartilage, muscle, adipose tissue, and hair follicles. Inhibition of nuclear Yap activity during phase 2 results in numerous tissue reconstitution defects suggesting Yap plays diverse roles across a broad range of cell types. Most striking is that VP treatment results in complete absence of melanocytes, adipocytes, and chondrocytes, without an overall loss of total cell number. Ventral tissue architectural organization is also disrupted with de-novo appearance of numerous ectopic immature hair follicles occupying the now absent cartilage plate. A possible role for YAP acting as a molecular switch to induce MF reprogramming to a mesenchymal stem cell-like phenotype capable of giving rise to chondrocytes, adipocytes, and melanocytes would be consistent with our data, and with scarless wound healing in general (Talele et al., 2015; Panciera et al., 2016; Plikus et al., 2017; Doeser et al., 2018; Cheung et al., 2020). Such a role for Yap signaling in Acomys ear pinnae tissue regeneration is reminiscent of models of sensory system regeneration in Xenopus retina (Hamon et al., 2019) and in chick hearing and balance organ hair cells (Kozlowski et al., 2020; Rudolf et al., 2020). Proliferative regeneration in these tissues is normally blocked as mice mature into adults, but overcome in both by forced expression of Yap5SA, a Hippo non-responsive form of Yap (Hamon et al., 2019; Rueda et al., 2019; Rudolf et al., 2020), or through mitogen and mechanical landscape alterations to inner ear sensory tissue leading to release of actin-sequestered endogenous Yap (Kozlowski et al., 2020). These data, together with the results reported here, provide support for the notion that studies of comparative regeneration across species may lead to translational outcomes (Bermingham-McDonogh and Reh, 2011). Since Acomys are mammals, their genomic adaptations promoting adult tissue regeneration may encounter fewer obstacles to translate into new therapeutics for human fibrotic diseases compared to non-mammalian models.
Limitations of the study
(1) Fibroblast diversity: DFs comprise subtypes with diverse embryonic origins, gene expression patterns, anatomical positions, and abilities for repair and regeneration (Driskell et al., 2013; Rinkevich et al., 2015; Philippeos et al., 2018; Guerrero-Juarez et al., 2019; Mascharak et al., 2021; Plikus et al., 2021). Future work will examine what DF subtypes are present in Acomys ear skin, which fibroblast subtype(s) participate in blastema formation, and whether MFs arise from one or multiple origins during wound healing. (2) Positional diversity: As the ear tissue is a unique appendage, whether DFs from other sites on the spiny mouse body exhibit the same MF fate plasticity, regulation of Yap activity, and lack of MF maturation that we describe here for ear DFs is yet to be determined. (3) Biomechanical signaling: Spiny mouse skin has reduced mechanical strength compared to murine skin presumably to allow for skin shedding (Seifert et al., 2012; Harn et al., 2021). It will be important to determine if lack of mechanical stiffening forces in Acomys plays a role in MF fate plasticity when compared to Mus. Individual MF fate tracking studies in Acomys are currently limited by lack of transgenic tools available for this species. (4) Yap signaling: Hippo-Yap signaling is highly context-dependent (Moya and Halder, 2019). The wound environment surrounding Acomys MFs differs from Mus in multiple ways. For example, spiny mice form a regeneration blastema at the wound site whereas mice do not (Gawriluk et al., 2016). Spiny mice exhibit sustained reactive oxygen species production, fewer pro-inflammatory macrophages, increased angiogenesis, and elevated Wnt signaling during wound healing compared to Mus (Seifert et al., 2012; Simkin et al., 2017; Maden and Brant, 2019; Gawriluk et al., 2020). Tensile strength and atomic force microscopy data show that the extracellular matrix stiffening that drives fibrotic mechanotransduction in Mus is greatly reduced or absent in Acomys (Seifert et al., 2012; Harn et al., 2021). Thus, the integrated cell surface signaling pathways that regulate Hippo-Yap pathway activity in Acomys DFs are different than Mus, suggesting that the nuclear context by which Yap activates fibrosis in mice (Liu et al., 2015; Liang et al., 2017; Mascharak et al., 2021) may not be present in Acomys. The absence of a nuclear context for fibrosis together with evolved differences in the two rodent genomes may allow Acomys Yap-TEAD coactivator complexes to occupy different gene targets and mediate different outcomes for wound healing in spiny mice compared to Mus, e.g., regeneration instead of scarring.
STAR METHODS
Lead Contact:
Further information and reasonable requests for resources and reagents should be directed to the Lead Contact, Mark W. Majesky, (mwm84@uw.edu).
Materials Availability:
Materials used in this study are available from the Lead Contact upon reasonable request. This study did not generate new unique reagents.
Data and Code Availability:
All data that support the findings of this study are available within the article and its supplemental information files and from the corresponding authors upon reasonable request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Animal husbandry and manipulations
The Seattle Children’s Research Institute’s (SCRI) Institutional Animal Care and Use Committee (IACUC) approved all animal procedures. Adult male and female Mus musculus (The Jackson Laboratory, CD1(ICR) and Acomys cahirinus (colony at SCRI) were maintained within the Seattle Children’s Research Institute’s onsite vivarium. CD1 mice were housed in a pathogen-free room maintained on 12:12 (Light:Dark) lighting schedule, Acomys were housed in a separate room maintained on 14:10 (Light:Dark) schedule, and all animals received food and water ad libitum. Acomys and Mus adult males and females were anaesthetized with 4% (v/v) vaporized isoflurane (Henry Schein Animal Health, Dublin, OH), and a 2mm thumb punch (Kent Scientific) was used to generate 2–3 punches across the medial ear pinna in the right and left ears, administered sequentially or concurrently as indicated: note, we did not detect overt sex differences at the current study level. For initial in vivo Yap inhibition experiments, adult Acomys received intraperitoneal injections of Verteporfin (VP, Tocris, 100mg/kg, 100mg/ml DMSO stock solution diluted 1:10 PBS prior to injection) every 3–4 days as described (Liu-Chittenden et al., 2012): controls received equivalent volume of 10% DMSO in PBS. For subsequent experiments, adult Acomys received intraperitoneal injections of VP (100mg/kg, 20mg/ml mineral oil stock solution) every 3–4 days: controls received equivalent volume of mineral oil. All animals received post procedure monitoring, including body weight measurements, all of which were stable over the experimental time course. Some animals received intravenous tail vein injection of fluorescent Dextran-Rhodamine 1h prior to sacrifice after 14 days, while others were lightly sedated with isoflurane and received 100 μg of isolectin B4 DyLight 594 (Vector Laboratories, DL-1207) by retroorbital injection and allowed to circulate for 2 minutes, then an intracardiac injection of isolectin B4 for 1 minute. Upon study completion, animals were terminally euthanized under anesthesia by cardiac perfusion with cold PBS, ears were photographed (Olympus SZX16 Stereomicroscope), tissue collected and analyzed as sections / whole mount processing to investigate vascularization.
METHOD DETAILS
Cell Lines:
Primary ear tissue fibroblasts were established from adult male Acomys cahirinus and CD1 Mus musculus (3–4-month-old) freshly biopsied ear tissue (2mm ear punches) using established procedures. The adult human fibroblast cell line SK5 was a generous gift from Dr. Elaine Raines at the University of Washington (Gronwald et al., 1988), source human male forearm skin (epidermis - fibroblasts in dermis) biopsy removed at the time of dialysis shunt placement. Other human cells include AB (HASFB or AbdSK = Human Abdominal skin fibroblasts from 36y male Caucasian abdomen, https://www.promocell.com/product/normal-human-dermal-fibroblasts-nhdf/); AO (HAFB = Human Aortic fibroblasts https://cellbiologics.com/index.php?route=product/product&keyword=H-6075&category_id=0&product_id=2425); and CO (HCFB =Human Coronary fibroblasts https://cellbiologics.com/index.php?route=product/product&product_id=2424%20#H-6049). All cell lines were grown on collagen coated tissue culture plates (5ug/cm2 BD Biosciences), maintained at the same culture conditions (DMEM 4.5g/L D-glucose, L-glutamine, 110mg/L sodium pyruvate supplemented with 10% FBS and 100 U/mL Pen-Strep, 37°C, 5.0% CO2) unless otherwise noted, and discarded after 15 passages in order to prevent gross genomic changes over long term in vitro culturing. For cell signaling experiments, ~80% confluent Acomys, Mus, and Human fibroblasts were serum starved for 12 hours (DMEM, 0.5% FBS and 100 U/ml Penicillin/Streptomycin; Serum Starvation Media/S.S.M) prior to addition of recombinant hTGFβ1 fibrokine (2ng/ml, unless stated otherwise, Cell Signaling Technologies) and/or other factors as indicated (Okadaic Acid [Abcam], Tautomycetin [Tocris]). Stable lines of Acomys ear fibroblasts expressing pEGFP-C3-hYAP2 (Addgene plasmid #17844) (Basu et al., 2003) were generated by transient transfection with FUGENE 6® (Promega). After 24 hours of plasmid uptake, stable cells were selected using G418 (Thermo Scientific) at a concentration of 500ug/ml for six days.
Tissue Preparation and Processing:
For paraffin embedding, excised tissues were placed in 4% PFA (w/v) and stored overnight at 4°C with agitation, washed three times in PBS, stored in 70% (v/v) ethanol, processed using a Tissue-Tek VIP automatic processor (Sakura), and cut at 8-m per section. Histological staining including Masson’s Trichrome, Alcian Blue, Saffron O and Verhoeff-Van Geisson’s stain were done in collaboration with Benaroya Research Institute or the University of Washington South Lake Union Imaging and Histology Core. For cryosectioning, tissue was embedded and frozen in O.C.T medium (TissueTech) using a dry-ice slurry/2-methybutanol mixture, and cut at 8–10μm per cryosection.
Immunofluorescence:
Tissue cryosections were thawed, washed with PBS, post fixed with 4% (w/v) PFA for 10 minutes. Post fixation, slides were washed three times for five minutes with PBS followed by permeabilization using 0.2% Triton-X100 in PBS (PBT) for 10 minutes. Slides were blocked (5% BSA, 2% normal goat serum in PBT) at room temperature for 1 hour, tissue sections were incubated in primary antibody overnight at 4°C in blocking solution (3% BSA, 0.2% Triton-X100 in PBS).Primary antibodies used included anti-Cleaved Caspase 3 (1/200, rabbit; Cell Signaling # 9661), anti-EDA-Fibronectin (1/200, mouse; Abcam# ab6328), anti-Flag (1/500,mouse; Sigma # F1804), anti-Smooth Muscle Myosin Heavy Chain (1/500, rabbit; Biomedical Technologies # BT562), anti-Acta2 (Smooth Muscle α-Actin) (1/2000, mouse; Sigma # A2547), anti-PCNA (1/100, mouse; Cell Signaling # 2586), anti-Vimentin (1/500, mouse; Sigma # V5255), anti-Vinculin (1/200, mouse; Sigma#V9131), anti-Yap (1/500,rabbit; Santa Cruz # sc-15407), anti-Ctgf (1/100, rabbit; Cell Signaling #86641T), anti-p44/42 MAPK (ERK1/2) (1/400, rabbit; Cell Signaling #4695), rabbit monoclonal anti-Phospho-AKT (Thr308) (244F9, Cell Signaling Technologies, 1/2000), rabbit Monoclonal anti-Phospho-AKT (S473) (D9E) XP (Cell Signaling Technologies Cat #4060, 1/2000), mouse monoclonal anti-AKT (40D4) (Cell Signaling Technologies, Cat#2920 1/1000). Slides were washed with PBS, and then incubated with ALEXA-Fluor 488- or 594-conjugated antibodies (Life Technologies) for 2 hours at room temperature in blocking solution. Cell nuclei were counterstained with Dapi (Molecular Probes) and mounted in 4% (w/v) propyl gallate anti-fade solution, or Fluoromount G (Fisher). For in immunolabeling adherent cell cultures, fibroblasts were plated into glass-bottom tissue culture multi-well plates (TissueTek) previously treated with acid, washed 3X in PBS, and then coated in poly-d-lysine (10mg/ml) and collagen as described. Cell culture media was removed, cells were washed with PBS, fixed with 4% PFA for 10min, washed again with PBS, and blocked for 1h prior to addition of primary antibody solutions. Cells were incubated overnight at 4°C, washed with PBS, ALEXA-conjugated secondary antibodies applied for 2h, counterstained with Dapi, and mounted as described.
Immunoblotting:
Cells were washed with ice-cold PBS (pH 7.4) and lysed at indicated time points using radioimmunoprecipitation assay buffer (50mM Tris, pH 7.4, 150mM NaCl, 1% TritonX-100, .5% Sodium deoxycholate, 0.1% SDS, 2mM EDTA, 2mM EGTA and 50mM NaF) supplemented with protease inhibitor cocktail (Sigma) and 10mM Na-Orthovanadate (Sigma). Normalization of protein concentrations was done using the Pierce™ BCA Protein Assay Kit (Thermo Scientific) and prepared in SDS sample buffer. Equal amounts of protein were resolved on SDS-Page and transferred to either a nitrocellulose (GE Healthcare) or PVDF (Thermo) depending on detection method. Membranes were incubated for 1 hour in LI-COR Biosciences block buffer. After washing, blots were incubated overnight at 4°C with antibodies including anti-β-tubulin (1/5000,mouse; Sigma Aldrich # BT7R), anti-GAPDH (1/5000, rabbit; Cell Signaling #5174), anti-pLats-T1079 (1/1000,rabbit; Cell Signaling # 9159), anti-Acta2 (Smooth Muscle α-Actin) (1/2000,mouse; Sigma # A2547), ant-Smad2(1/2000, rabbit; Cell Signaling # 5339), anti-pSmad2-S465/467 (1/1000, rabbit; Cell Signaling # 3108), anti-Smad3(1/2000, rabbit; Cell Signaling #9593), anti-pSmad3-S423/425 (1/1000, rabbit; Cell Signaling # 9520), anti-Taz (1/1000, rabbit #4883), anti-Yap (1/1000, mouse; Abcam # ab5670), anti-pYap-S127 (1/1000, rabbit; Cell Signaling # 4911), anti-pYapS381 (1/1000, rabbit Cell Signaling #13619), anti-Vinculin (1/5000, mouse; Sigma # V4505). After washing with TBST 0.1%, antigens were detected using either HRP-conjugated (Jackson Research Labs) or IR Dye-labeled (LI-COR Biosciences) secondary antibodies. HRP reaction was performed using ECL Prime (GE Healthcare) and exposed to x-ray film. Band intensity was quantified using Image J (National Institute of Health). IR dye secondaries were detected with infrared imaging system Odyssey (LI-COR Biosciences) and band intensity quantified using Image Studio Version 5.2 (LI-COR Bioscience). Immunoblots shown are presentative of experiments that were repeated a minimum of four independent times.
Transcriptomic Analysis:
Acomys primary ear fibroblasts were grown to ~90% confluency in 10% FBS DMEM, and then switched to 0.1%FBS DMEM (serum starve) overnight. Recombinant TGFβ1 (2ng/ml) or an equivalent volume of PBS was added to cells and cultured for 24h to induce a robust transcriptional response to TGFβ1 signaling (n=3 independent wells per treatment group). Afterwards, cells were washed with PBS and lysed in Trizol solution (Ambion). Total RNA was extracted, DNase treated, and purified (PureLink RNA Mini Kit, Thermo Fisher Scientific). RNA was processed with KAPA’s Stranded mRNA-Seq kit (Illumina) following the manufacturer’s protocol in duplicate for each sample. The resulting libraries were assessed for library quality using fragment length and number of cycles in real-time PCR. Passing samples were sequenced on a NextSeq 500 using a 300 cycle mid-output kit, with paired 150 basepair reads. Acomys annotated de novo transcriptome (Mamrot et al., 2017) was downloaded from the Zenodo data sharing resource (https://zenodo.org/record/808870#.WdfH-hNSzdc) and indexed with Kallisto version v0.43.1 (Bray et al., 2016). Expression of individual transcripts was performed using Kalisto’s quant command with ten bootstraps per sample. The resulting expression profiles were analyzed using Sleuth to determine differentially expressed genes between cells exposed to low serum with and without TGFβ1 (Pimentel et al., 2017): ~1/2 of the DE transcripts have annotated gene names from other species (mouse, rat, human) (Table S1).
Identification of Acomys Yap Peptide Sequence:
The exon sequences of mouse YAP1 were compared to our Acomys cahirinus genome (manuscript in preparation) using BLAT to identify Acomys YAP genomic loci. Our RNAseq data from Acomys ear fibroblasts identified 8–9 homologous Yap1 transcripts using CUFFLINKS, similar to the number of YAP isoforms identified in a variety of both mouse and human tissues (Gaffney et al., 2012). We compiled the transcript sequence information from those regions to identify their predicted Acomys YAP1 mRNA sequences. Open Reading Frame (ORF) analysis of the longest Acomys transcript revealed a potential start codon and contiguous ORF that generated a 490 amino acid peptide (ORFinder, NCBI) exhibiting high homology to human and mouse YAP1–2g isoform. Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/) was used for multiple peptide alignments, and NetPhos 3.1 (http://www.cbs.dtu.dk/services/NetPhos/) to predict high probability Serine/Theonine/Tyrosine phosphorylation sites (Figure S5).
Lentiviral Transduction:
Doxycycline-inducible human Flag-tagged Yap-S127A lentiviral plasmid (PIN20YAP1) was a generous gift of Dr. R. Johnson, and previously described (Song et al., 2014). Control lentiviral plasmid encoding CMV-based constitutive expression of GFP was purchased commercially (pLV[Exp]-Puro-CMV>EGFP, VectorBuilder). High-titer lentiviral particles for both constructs were generated (VectorBuilder), and primary CD1 Mus ear fibroblasts and human SK5 fibroblasts were transduced using polybrene as directed. Doxycycline (500 ug/ml) was added to media to induce Flag-Yap-S127A expression, which was confirmed by anti-Flag immunoblotting and Western blot (not shown) of cell lysates.
Quantitative Image Analysis:
Ear Wound Closure and Pigmentation:
Calibrated digital images of whole mount ears were captured prior to embedding (Olympus SZX16 Stereomicroscope), converted to greyscale in FIJI, and area of wound region interpolated using Surfaces function in Imaris 6.5 (Bitplane). Wound closure was calculated as the percent difference from starting area of a 2mm diameter punch to area size at time of sacrifice for timed injuries. Pigmentation levels were similarly determined by comparing percent difference from the average greyscale pixel intensity within the surrounding uninjured tissue, to the pixel intensity within the original 2mm diameter ear punch, using Surfaces function in Imaris. At least 4–6 animals were used per time point group. Snapshots were exported to Photoshop and panels assembled in Illustrator CS5–7 (Adobe).
Myofibroblast and New Blood Vessel Formation:
High-resolution image volumes from fluorescently labeled sections were collected using the same imaging conditions across all samples (Leica TCS SP5 confocal microscope) (n = 3–6 sections across ear punch injury). Representative mid-injury sections were normalized to Z-depth of 5.4–5.9mm within the image volume (Imaris, Bitplane) (n = 2 sections per animal per injury, n = 4–6 animals per injury and time point). The subdermal wound volume was manually traced from the internal subdermal boundary at the distal edge of the blastema to the proximal wound edge cartilage cleavage plane, and converted to 3D volume using Surfaces (Imaris, Bitplane). The volume of Acta2 immunolableling within this subdermal space was automatically rendered and segmented using Surfaces: channel = Acta2, smooth surface grain size = 0.01, background subtraction = false, enable split = 5.68, manual threshold A = 38.12, auto-threshold B = true, voxels > 30.0 (Imaris, Bitplane). The rendered Acta2 surface and subdermal wound volume were split into dorsal and ventral regions according to cartilage plane. Total Acta2 surface volume (mm3) per total subdermal wound volume (mm3) was determined using Statistics (Imaris, Bitplane). Dorsal and ventral Acta2 surface volumes (mm3) and subdermal wound volumes, respectively, as well as the ratio of dorsal to ventral Acta2 surface volumes (mm3) and subdermal wound volumes, were similarly determined. Myh11 immunolabeling was analyzed by further determining the intensity sum of Myh11 signal with the defined Acta2 surface volume (Surfaces, Imaris, Bitplane). All quantative data was exported to Excel. Snapshots of maximum intensity projection or blended volume views of representative sections and isolated cells were exported to Photoshop and panels assembled in Illustrator CS5–7 (Adobe).
Acta2 – Phalloidin colocalization levels:
Confocal stacks of equivalently stained and imaged Acomys and Mus cells were automatically thresholded in Imaris (Bitplane) using Pearson’s correlation, and resulting colocalization channel duplicated and quantified as the fold change per field (390×390μm) of view normalized to cell/nuclei number, n=3 wells, 3–10 fields per well). Snapshots of blended volume views of representative sections and isolated cells were exported to Photoshop and panels assembled in Illustrator CS5–7 (Adobe): human fibroblasts analyzed similarly. Additionally, some human fibroblasts transduced with LV-GFP were surface rendered based on soluble GFP-expression, digitally isolated and masked to compare Acta2 levels and organizational structure across heterogenic fibroblast morphologies using Imaris (Bitplane), with isolated cells exported to Photoshop and panels assembled in Illustrator CS5–7 (Adobe). Similarily, we quantified the number of Acta2+ LV_GFP+/ versus LV_Dox_Yap-127SA_Flag+ by automatically detecting all Dapi-labeled nuclei per field of view, thresholding nuclei using relaxed parameters to detect and count GFP+/Flag+ cells compared to their respective ACTA2 levels, calculated as percentage of ACTA2+/GFP+ versus ACTA2+/Flag+ cells per field of view 390X390 μm).
Nuclear Yap Localization:
For in vitro studies, Yap immunolabeling within the nuclear and cytoplasmic compartments were determined from epifluorescent images by placing paired 300nm diameter circles within the DAPI-labeled nucleus and adjacent cytoplasm within individual cells using Spots (Imaris, Bitplane), and intensity sum of Yap nuclear and cytoplasmic signal exported to Excel. The ratio of nuclear to cytoplasmic Yap signal was calculated per cell (n= >50 cells per group). Snapshots exported to Photoshop and panels assembled in Illustrator (Adobe). For in vivo studies, DAPI-labeled nuclei in the subdermal wound space were automatically rendered and segmented using Surfaces: channel = DAPI, smooth surface grain size = 0.4, background subtraction = 2.8, threshold = 21.85, enable split = 3.788, quality = 14.96, voxels > 10.0 (Statistics, Imaris). Total nuclear Yap signal was determined by calculating the intensity sum of Yap immunolabeling within the total chromatin nuclear surface compartment (n = 2 sections per timepoint per animal, n = 4–6 animals per timepoint). All data was exported to Excel. Single nuclei surfaces within representative subdermal 3D volumes were plotted using Vantage (Imaris) with respect to dorsal-ventral (Y-axis) / proximal-distal (X-axis) position, intensity sum of Yap nuclear signal (Z-axis), and color coded for Yap levels to exemplify single nuclei differences within the total pattern of Yap activity in the subdermal wound compartment. Snapshots of maximum intensity projection or blended volume views of representative sections and isolated cells / nuclei were exported to Photoshop and panels assembled in Illustrator CS5–7 (Adobe).
Cartilage Regeneration:
Saffron O-stained sections were scanned (Olympus VS120 Slide Scanner). The subdermal wound area and regions of new cartilage growth defined by Saffron O staining were traced, and the average area of regenerated cartilage per subdermal wound area was calculated (n = 6 sections per injury per time point; n = 4–6 animals per group; VS-Desktop Software). All data was exported to Excel. The difference between cartilage regeneration in control and Verteporfin treated animals was determined. Representative images were exported to Photoshop and panels assembled in Illustrator CS5–7 (Adobe); see Figure S3.
Statistical Analysis:
Unpaired Student’s T-test was used to compare two groups. One-way ANOVA with a Bonferroni, Brown-Forsythe, or Tukey post-hoc test was used for multiple comparisons of three or more groups over time. Mixed-effect models, Welch’s, and Two-way ANOVA with a Bonferroni, Tukey, or Dunnett’s post-hoc tests used for multiple comparisons of three or more groups and between species and over time. P < 0.05 was considered to be significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Statistical analyses and graphic presentation were all done using Prism 6 (GraphPad) and Excel and assembled using Illustrator CS5–7 (Adobe). Error bars on graphs ± standard deviation (SD) except when stated otherwise, and tests are noted in panels.
Supplementary Material
Table S2: Full gene list of Acomys DFs stimulated with TGFβ1 for 24h in vitro, related to Figure 2.
Video S1: Nuclear Yap in Acomys versus cytoplasmic Yap in Mus MFs in vivo, related to Figure 5. This movie shows high-power views of isolated Acomys and Mus MFs in vivo within the blastema at week1 immunolabeled for Yap (red), Acta2 (green), and Dapi (blue rendered nuclei). Images are rotated around the y-axis to better show nuclear Yap localization in Acomys Acta2+ MFs compared to Mus MFs that exhibit cytoplasmic Yap localization.
Key Resources Table.
| SOURCE | IDENTIFIER | |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Actin (C4) | Millipore | Cat# MAB1501, RRID: AB_2223041 |
| Mouse monoclonal anti-Actin, alpha Smooth Muscle (clone 1A4) (Acta2) | Sigma-Aldrich | Cat# A2547, RRID: AB_476701 |
| Mouse monoclonal anti-EDA-Fibronectin (IST-9) | Abcam | Cat# ab6328, RRID: AB_305428 |
| Rabbit monoclonal anti-GAPDH (D16H11) | Cell Signaling Technology | Cat# 5174, RRID: AB_10622025 |
| Mouse monoclonal anti-GFP | Abcam | Cat# ab1218, RRID: AB_298911 |
| Mouse monoclonal anti-FLAG (M2) | Sigma-Aldrich | Cat# F1804, RRID: AB_262044 |
| Rabbit polyclonal anti-Myosin Light Chain (phospho S20) | Abcam | Cat# ab2480 RRID: AB_303094 |
| Rabbit polyclonal anti-Phospho-LATS1 (Thr1079) | Cell Signaling Technologies | Cat# 9159S, RRID: AB_10549072 |
| Rabbit monoclonal anti-Phospho-p38 MAPK (Thr180/Tyr182) (D3F9) | Cell Signaling Technologies | Cat#4511,RRID:AB_2139682 |
| Rabbit monoclonal anti-Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (D13.14.4E) | Cell Signaling Technologies | Cat# 4370, RRID: AB_2315112 |
| Rabbit monoclonal anti-p44/42 MAPK (Erk1/2) (137F5) | Cell Signaling Technologies | Cat# 4695 RRID:AB_390779 |
| Rabbit monoclonal anti-Phospho-Smad2 (Ser465/467) (138D4) | Cell Signaling Technologies | Cat# 3108, RRID:AB_490941 |
| Rabbit monoclonal anti-Phospho-Smad3 (Ser423/425) (C25A9) | Cell Signaling Technologies | Cat# 9520, RRID:AB_2193207 |
| Rabbit polyclonal anti-Phospho-YAP (Ser127) | Cell Signaling Technologies | Cat# 4911S, RRID:AB_2218913 |
| Rabbit monoclonal anti-Phospho-YAP (Ser397) (D1E7Y) | Cell Signaling Technologies | Cat# 13619, RRID:AB_2650554 |
| Rabbit monoclonal anti-Smad2 (D43B4) | Cell Signaling Technologies | Cat# 5339, RRID:AB_10626777 |
| Rabbit monoclonal anti- Smad3 (C67H9) | Cell Signaling Technologies | Cat# 9523, RRID:AB_2193182 |
| Rabbit polyclonal anti-Smooth Muscle Myosin Heavy Chain (Myh11) | Biomedical Technologies | Cat# BT-562, RRID:AB_10013421 |
| Rabbit polyclonal anti-TAZ (V386) | Cell Signaling Technologies | Cat# 4883S, RRID:AB_1904158 |
| Mouse monoclonal anti-Vinculin | Sigma-Aldrich | Cat# V9131, RRID:AB_477629 |
| Mouse monoclonal anti-YAP1 | Abcam | Cat# ab56701, RRID:AB_2219140 |
| Rabbit polyclonal anti-YAP (H-125) | Santa Cruz | Cat# sc-15407, RRID:AB_2273277 |
| Goat polyclonal anti-Mouse IgG (H+L), HRP | Jackson ImmunoResearch | Cat#115-035-166, RRID:AB_2338511 |
| Rabbit monoclonal anti-Ctgf (D8Z8U) | Cell Signaling Technologies | Cat# 86641T RRID:AB_2800085 |
| Rabbit monoclonal anti-Phospho-AKT (Thr308) (244F9) | Cell Signaling Technologies | Cat# 4056 RRID:AB_331163 |
| Rabbit Monoclonal anti-Phospho-AKT (S473) (D9E) XP | Cell Signaling Technologies | Cat# 4060 |
| Mouse monoclonal anti-AKT (40D4) | Cell Signaling Technologies | Cat# 2920 RRID:AB_10694382 |
| Goat polyclonal anti-Rabbit IgG (H+L), HRP | Jackson ImmunoResearch | Cat# 111-035-003, RRID:AB_2313567 |
| Goat polyclonal anti-Mouse IgG1 Secondary Antibody, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | Cat# A-21121, RRID:AB_2535764 |
| Goat polyclonal anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21131, RRID:AB_2535771 |
| Goat polyclonal anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-21135, RRID:AB_2535774 |
| Goat polyclonal anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11008, RRID:AB_143165 |
| Goat polyclonal anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | Cat# A-11012, RRID:AB_2534079 |
| IRDye® 680RD Goat anti-Mouse IgG (H+L) | LI-COR Biosciences | Cat# 925-68070, RRID:AB_2651128 |
| IRDye® 800CW Goat anti-Rabbit IgG (H+L) | LI-COR Biosciences | Cat# 925-32211, RRID:AB_2651127 |
| Bacterial and Virus Strains | ||
| pInducer20_Yap1S127A-Flag | Vector Builder (Song et al., 2014) | N/A |
| pLV[Exp]-Puro-CMV>EGFP | Vector Builder | VB900088-2243bzq |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dextran, Texas Red®, 70,000 MW | Thermo-Scientific | Cat#D1864 |
| Okadaic acid, Prorocentrum concavum | Sigma-Aldrich | Cat# O7885 CAS: 155751-72-7 |
| Phalloidin CF™594 | Biotium | Cat#00045 |
| Recombinant Human TGFβ1 | R&D Systems | Cat#240-B-002 Accession # P01137 |
| Tautomycetin | Tocris | Cat# 2305 CAS: 119757-73-2 |
| Verteporfin | United States Pharmacopeia | Cat# 1711461 USP CAS: 129497-78-5 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Acomys: A. C_E_M20 | This Study | N/A |
| Acomys: A. C_E_M20_ hYap-GFP | This Study | N/A |
| Acomys: A. C_E_N=6 | This Study | N/A |
| Mouse: M.M_E_CD1 | This Study | N/A |
| Human: SK5 Dermal | Gift of Elaine Raines (Gronwald et al., 1988) | RRID: CVCL_5H87 |
| Human: AB HASFB or AbdSK = Human Abdominal skin | Promocell | C-12352 |
| Human: AO HAFB = Human Aortic fibroblasts | CellBiologics | H-6075 |
| Human: CO HCFB =Human Coronary fibroblasts | CellBiologics | H-6049 |
| Experimental Models: Organisms/Strains | ||
| Acomys cahirinus: Egyptian Spiny Mouse | This study | N/A |
| Mus musculus: Crl:CD1(ICR) | Charles River | RRID:IMSR_CRL:22 |
| Recombinant DNA | ||
| pEGFP-C3-hYAP2 | Gift of Marius Sudol (Basu et al., 2003) | Addgene: #17844 |
| Software and Algorithms | ||
| Fiji (Image J) | http://fiji.sc/ | N/A |
| Imaris 7.4 | Bitplane | N/A |
| Image Studio Light V5.2 | https://www.licor.com/bio | N/A |
| VS-Desktop | Olympus | N/A |
| Prism7.03 | Graph Pad | N/A |
| Clustal O | www.ebi.ac.uk/Tools/msa/clustalo/ | |
| NetPhos 3.1 | (http://www.cbs.dtu.dk/services/NetPhos/) | |
| Other | ||
Highlights.
Ear injury-activated myofibroblasts (MFs) are transient in Acomys but persist in Mus
Acomys MFs in vitro resist maturation via rapid Yap dephosphorylation
Blocking Acomys Yap-TEAD interaction in vivo inhibits regeneration and promotes fibrosis
MF phenotypes in spiny mice may find application for novel anti-fibrotic therapies
Acknowledgments
We thank our colleagues Drs. David Beier and Barry Gumbiner at Seattle Children’s Research Institute for reagents and helpful discussions. We also thank Drs. Slobodan Beronja, Cyrus Ghajar, and Valeri Vasioukhin at the Fred Hutchison Cancer Research Center, Dr. Avril Somlyo at the University of Virginia, and Dr. Randy Johnson at the MD Anderson Cancer for advice and reagents. We acknowledge Dr. Robert Kao for technical expertise and helpful suggestions. The majority of work was supported by a grant from the W.M. Keck Foundation. Additional support was provided by NIH grants RO1-DK114149, RO1-HL123650, R21-OD023838, U24-DK115255, and T32-HL007312; from the Loie Power Robinson Stem Cell & Regenerative Medicine Fund, the Seattle Children’s Research Institute, and the Seattle Children’s Foundation.
Footnotes
Declaration of Interests
Authors declare no competing interests.
Supplemental Information
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2021.09.008
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldave AJ, Han J, and Frausto RF (2013). Genetics of the corneal endothelial dystrophies: an evidence-based review. Clin Genet 84, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, and Downward J (2003). Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14–3-3 and attenuation of p73-mediated apoptosis. Mol Cell 11, 11–23. [DOI] [PubMed] [Google Scholar]
- Bermingham-McDonogh O, and Reh TA (2011). Regulated reprogramming in the regeneration of sensory receptor cells. Neuron 71, 389–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialojan C, Ruegg JC, and Takai A (1988). Effects of okadaic acid on isometric tension and myosin phosphorylation of chemically skinned guinea-pig taenia coli. J Physiol 398, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nature biotechnology 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brockes JP, Kumar A, and Velloso CP (2001). Regeneration as an evolutionary variable. J Anat 199, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechler MB, Pradhan RN, Krishnamurty AT, Cox C, Calviello AK, Wang AW, Yang YA, Tam L, Caothien R, Roose-Girma M, et al. (2021). Cross-tissue organization of the fibroblast lineage. Nature 593, 575–579. [DOI] [PubMed] [Google Scholar]
- Campanholle G, Ligresti G, Gharib SA, and Duffield JS (2013). Cellular mechanisms of tissue fibrosis. 3. Novel mechanisms of kidney fibrosis. Am J Physiol Cell Physiol 304, C591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass DL, Bullard KM, Sylvester KG, Yang EY, Longaker MT, and Adzick NS (1997). Wound size and gestational age modulate scar formation in fetal wound repair. J Pediatr Surg 32, 411–415. [DOI] [PubMed] [Google Scholar]
- Chen J, You H, Li Y, Xu Y, He Q, and Harris RC (2018). EGF receptor-dependent YAP activation is important for renal recovery from AKI. J Am Soc Nephrol 29, 2372–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung P, Xiol J, Dill MT, Yuan WC, Panero R, Roper J, Osorio FG, Maglic D, Li Q, Gurung B, et al. (2020). Regenerative reprogramming of the intestinal stem cell state via hippo signaling suppresses metastatic colorectal cancer. Cell Stem Cell 27, 590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T, et al. (2013). Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci Signal 6, rs15. [DOI] [PubMed] [Google Scholar]
- Darby I, Skalli O, and Gabbiani G (1990). Alpha-smooth muscle actin is transiently expressed by myofibroblasts during experimental wound healing. Lab Invest 63, 21–29. [PubMed] [Google Scholar]
- Derynck R, and Zhang YE (2003). Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 425, 577–584. [DOI] [PubMed] [Google Scholar]
- Desmouliere A, Redard M, Darby I, and Gabbiani G (1995). Apoptosis mediates the decrease in cellularity during the transition between granulation tissue and scar. The American journal of pathology 146, 56–66. [PMC free article] [PubMed] [Google Scholar]
- Dey A, Varelas X, and Guan KL (2020). Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat Rev Drug Discov 19, 480–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doeser MC, Scholer HR, and Wu G (2018). Reduction of fibrosis and scar formation by partial reprogramming in vivo. Stem Cells 36, 1216–1225. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, and Pan D (2007). Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, et al. (2013). Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyden B (2005). The myofibroblast: a study of normal, reactive and neoplastic tissues, with an emphasis on ultrastructure. part 2 - tumours and tumour-like lesions. J Submicrosc Cytol Pathol 37, 231–296. [PubMed] [Google Scholar]
- Eyden B (2007). The myofibroblast: a study of normal, reactive and neoplastic tissues, with an emphasis on ultrastructure. J Submicrosc Cytol Pathol, 7–166. [PubMed] [Google Scholar]
- Fernandez JJ, Candenas ML, Souto ML, Trujillo MM, and Norte M (2002). Okadaic acid, useful tool for studying cellular processes. Curr Med Chem 9, 229–262. [DOI] [PubMed] [Google Scholar]
- Finch-Edmondson ML, Strauss RP, Passman AM, Sudol M, Yeoh GC, and Callus BA (2015). TAZ protein accumulation is negatively regulated by YAP abundance in mammalian cells. J Biol Chem 290, 27928–27938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher GC, Diaz-de-la-Loza MD, Borreguero-Munoz N, Holder M, Aguilar-Aragon M, and Thompson BJ (2018). Mechanical strain regulates the Hippo pathway in Drosophila. Development 145, dev159467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL, Sheppard D, Duffield JS, and Violette S (2013). Therapy for fibrotic diseases: nearing the starting line. Science translational medicine 5, 167sr161. [DOI] [PubMed] [Google Scholar]
- Fu X, Khalil H, Kanisicak O, Boyer JG, Vagnozzi RJ, Maliken BD, Sargent MA, Prasad V, Valiente-Alandi I, Blaxall BC, et al. (2018). Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J Clin Invest 128, 2127–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney CJ, Oka T, Mazack V, Hilman D, Gat U, Muramatsu T, Inazawa J, Golden A, Carey DJ, Farooq A, et al. (2012). Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene 509, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Furnari F, and Newton AC (2005). PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell 18, 13–24. [DOI] [PubMed] [Google Scholar]
- Gawriluk TR, Simkin J, Thompson KL, Biswas SK, Clare-Salzler Z, Kimani JM, Kiama SG, Smith JJ, Ezenwa VO, and Seifert AW (2016). Comparative analysis of ear-hole closure identifies epimorphic regeneration as a discrete trait in mammals. Nat Commun 7, 11164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawriluk TR, Simkin J, Hacker CK, Kimani JM, Kiama SG, Ezenwa VO, and Seifert AW (2020). Complex tissue regeneration in mammals is associated with reduced inflammatory cytokines and an influx of T cells. Front Immunol 11, 1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffin JM, Pittet P, Csucs G, Lussi JW, Meister JJ, and Hinz B (2006). Focal adhesion size controls tension-dependent recruitment of alpha-smooth muscle actin to stress fibers. J Cell Biol 172, 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwald RG, Grant FJ, Haldeman BA, Hart CE, O’Hara PJ, Hagen FS, Ross R, Bowen-Pope DF, and Murray MJ (1988). Cloning and expression of a cDNA coding for the human platelet-derived growth factor receptor: evidence for more than one receptor class. Proc Natl Acad Sci U S A 85, 3435–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat Commun 10, 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, and Longaker MT (2008). Wound repair and regeneration. Nature 453, 314–321. [DOI] [PubMed] [Google Scholar]
- Hamon A, Garcia-Garcia D, Ail D, Bitard J, Chesneau A, Dalkara D, Locker M, Roger JE, and Perron M (2019). Linking YAP to Muller Glia quiescence exit in the degenerative retina. Cell Rep 27, 1712–1725. [DOI] [PubMed] [Google Scholar]
- Harn HI, Wang SP, Lai YC, Van Handel B, Liang YC, Tsai S, Schiessl IM, Sarkar A, Xi H, Hughes M, et al. (2021). Symmetry breaking of tissue mechanics in wound induced hair follicle regeneration of laboratory and spiny mice. Nat Commun 12, 2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, and Martin JF (2011). Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, and Gabbiani G (2007). The myofibroblast: one function, multiple origins. Am J Pathol 170, 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B, Phan SH, Thannickal VJ, Prunotto M, Desmouliere A, Varga J, De Wever O, Mareel M, and Gabbiani G (2012). Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol 180, 1340–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YY, Lagares D, Tager AM, and Kapoor M (2014). Fibrosis--a lethal component of systemic sclerosis. Nat Rev Rheumatol 10, 390–402. [DOI] [PubMed] [Google Scholar]
- Hu JK, Du W, Shelton SJ, Oldham MC, DiPersio CM, and Klein OD (2017). An FAK-YAP-mTOR signaling axis regulates stem cell-based tissue renewal in mice. Cell Stem Cell 21, 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Correa-Gallegos D, Christ S, Stefanska A, Liu J, Ramesh P, Rajendran V, De Santis MM, Wagner DE, and Rinkevich Y (2018). Two succeeding fibroblastic lineages drive dermal development and the transition from regeneration to scarring. Nature cell biology 20, 422–431. [DOI] [PubMed] [Google Scholar]
- Jiang TX, Harn HI, Ou KL, Lei M, and Chuong CM (2019). Comparative regenerative biology of spiny (Acomys cahirinus) and laboratory (Mus musculus) mouse skin. Exp Dermatol 28, 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M, Liu S, Huh K, Parapuram S, Kennedy L, and Leask A (2008). Connective tissue growth factor promoter activity in normal and wounded skin. Fibrogenesis Tissue Repair 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NG, Koh E, Chen X, and Gumbiner BM (2011). E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci USA 108, 11930–11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Schuster RM, and Lentsch AB (2018). Proliferation of hepatic stellate cells, mediated by YAP and TAZ, contributes to liver repair and regeneration after liver ischemia-reperfusion injury. Am J Physiol Gastrointestinal and Liver Physiol 314, G471–G482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski MM, Rudolf MA, and Corwin JT (2020). EGF and a GSK3 inhibitor deplete junctional E-cadherin and stimulate proliferation in the mature mammalian ear. J Neurosci 40, 2618–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YC, Huang KY, Yang CH, Yang YS, Lee WY, and Chiang CW (2008). Regulation of phosphorylation of Thr-308 of Akt, cell proliferation, and survival by the B55alpha regulatory subunit targeting of the protein phosphatase 2A holoenzyme to Akt. J Biol Chem 283, 1882–1892. [DOI] [PubMed] [Google Scholar]
- Le Page A, Khalil A, Vermette P, Frost EH, Larbi A, Witkowski JM, and Fulop T (2019). The role of elastin-derived peptides in human physiology and diseases. Matrix biology : journal of the International Society for Matrix Biology 84, 81–96. [DOI] [PubMed] [Google Scholar]
- Leach JP, Heallen T, Zhang M, Rahmani M, Morikawa Y, Hill MC, Segura A, Willerson JT, and Martin JF (2017). Hippo pathway deficiency reverses systolic heart failure after infarction. Nature 550, 260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Yu M, Xia R, Song K, Wang J, Luo J, Chen G, and Cheng J (2017). Yap/Taz deletion in Gli(+) cell-derived myofibroblasts attenuates fibrosis. J Am Soc Nephrol 28, 3278–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, and Pan D (2012). Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 26, 1300–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Lagares D, Choi KM, Stopfer L, Marinkovic A, Vrbanac V, Probst CK, Hiemer SE, Sisson TH, Horowitz JC, et al. (2015). Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol 308, L344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longaker MT, Whitby DJ, Adzick NS, Crombleholme TM, Langer JC, Duncan BW, Bradley SM, Stern R, Ferguson MW, and Harrison MR (1990). Studies in fetal wound healing, VI. Second and early third trimester fetal wounds demonstrate rapid collagen deposition without scar formation. J Pediatr Surg 25, 63–68. [DOI] [PubMed] [Google Scholar]
- Longaker MT, Whitby DJ, Ferguson MW, Lorenz HP, Harrison MR, and Adzick NS (1994). Adult skin wounds in the fetal environment heal with scar formation. Ann Surg 219, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gay JM, Nunley H, Spencer M, di Pietro F, Guirao B, Bosveld F, Markova O, Gaugue I, Pelletier S, Lubensky DK, et al. (2020). Apical stress fibers enable a scaling between cell mechanical response and area in epithelial tissue. Science 370, eabb2169. [DOI] [PubMed] [Google Scholar]
- Lorenz HP, and Adzick NS (1993). Scarless skin wound repair in the fetus. West J Med 159, 350–355. [PMC free article] [PubMed] [Google Scholar]
- Lv ZT, Gao ST, Cheng P, Liang S, Yu SY, Yang Q, and Chen AM (2018). Association between polymorphism rs12722 in COL5A1 and musculoskeletal soft tissue injuries: a systematic review and meta-analysis. Oncotarget 9, 15365–15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MD, and Watt FM (2018). Fibroblast heterogeneity: implications for human disease. J Clin Invest 128, 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden M, and Brant JO (2019). Insights into the regeneration of skin from Acomys, the spiny mouse. Exp Dermatol 28, 436–441. [DOI] [PubMed] [Google Scholar]
- Malfait F, Wenstrup RJ, and De Paepe A (2010). Clinical and genetic aspects of Ehlers-Danlos syndrome, classic type. Genet Med 12, 597–605. [DOI] [PubMed] [Google Scholar]
- Mamrot J, Legaie R, Ellery SJ, Wilson T, Seemann T, Powell DR, Gardner DK, Walker DW, Temple-Smith P, Papenfuss AT, et al. (2017). De novo transcriptome assembly for the spiny mouse (Acomys cahirinus). Sci Rep 7, 8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascharak S, desJardins-Park HE, Davitt MF, Griffin M, Borrelli MR, Moore AL, Chen K, Duoto B, Chinta M, Foster DS, et al. (2021). Preventing Engrailed-1 activation in fibroblasts yields wound regeneration without scarring. Science 372, eaba2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias Santos D, Rita AM, Casanellas I, Brito Ova A, Araujo IM, Power D, and Tiscornia G (2016). Ear wound regeneration in the African spiny mouse Acomys cahirinus. Regeneration 3, 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Matsuura N, Ubukata M, Oikawa H, Shima H, and Kikuchi K (2001). Tautomycetin is a novel and specific inhibitor of serine/threonine protein phosphatase type 1, PP1. Biochem Biophys Res Commun 287, 328–331. [DOI] [PubMed] [Google Scholar]
- Mohede DCJ, de Jong IJ, Bank RA, and van Driel MF (2018). Verteporfin as a medical treatment in Peyronie’s disease. Sex Med 6, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokalled MH, Patra C, Dickson AL, Endo T, Stainier DY, and Poss KD (2016). Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354, 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales MG, Acuna MJ, Cabrera D, Goldschmeding R, and Brandan E (2018). The pro-fibrotic connective tissue growth factor (CTGF/CCN2) correlates with the number of necrotic-regenerative foci in dystrophic muscle. J Cell Commun Signal 12, 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya IM, and Halder G (2019). Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat Rev Mol Cell Biol 20, 211–226. [DOI] [PubMed] [Google Scholar]
- Mui MZ, Zhou Y, Blanchette P, Chughtai N, Knight JF, Gruosso T, Papadakis AI, Huang S, Park M, Gingras AC, et al. (2015). The human adenovirus type 5 E4orf4 protein targets two phosphatase regulators of the Hippo signaling pathway. J Virol 89, 8855–8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panciera T, Azzolin L, Fujimura A, Di Biagio D, Frasson C, Bresolin S, Soligo S, Basso G, Bicciato S, Rosato A, et al. (2016). Induction of expandable tissue-specific stem/progenitor cells through transient expression of YAP/TAZ. Cell Stem Cell 19, 725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippeos C, Telerman SB, Oules B, Pisco AO, Shaw TJ, Elgueta R, Lombardi G, Driskell RR, Soldin M, Lynch MD, et al. (2018). Spatial and single-cell transcriptional profiling identifies functionally distinct human dermal fibroblast subpopulations. J Invest Dermatol 138, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma B, and Bank RA (2019). Collagen cross-linking mediated by lysyl hydroxylase 2: an enzymatic battlefield to combat fibrosis. Essays Biochem 63, 377–387. [DOI] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, and Pachter L (2017). Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14, 687–690. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Guerrero-Juarez CF, Ito M, Li YR, Dedhia PH, Zheng Y, Shao M, Gay DL, Ramos R, Hsi TC, et al. (2017). Regeneration of fat cells from myofibroblasts during wound healing. Science 355, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Wang X, Sinha S, Forte E, Thompson SM, Herzog EL, Driskell RR, Rosenthal N, Biernaskie J, and Horsley V (2021). Fibroblasts: Origins, definitions, and functions in health and disease. Cell 184, 3852–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, and Sadek HA (2011). Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD (2010). Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet 11, 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu G, Weycker D, Edelsberg J, Bradford WZ, and Oster G (2006). Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 174, 810–816. [DOI] [PubMed] [Google Scholar]
- Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, Januszyk M, Krampitz GW, Gurtner GC, Lorenz HP, et al. (2015). Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 348, aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong SS, Ma STU, Yu XT, Ma L, Chu WK, Chan TCY, Wang YM, Young AL, Pang CP, Jhanji V, et al. (2017). Genetic associations for keratoconus: a systematic review and meta-analysis. Sci Rep 7, 4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau C, Pores Fernando AT, Hwang JH, Faure N, Jiang T, White EA, Roberts TM, and Schaffhausen BS (2016). Transformation by polyomavirus middle T antigen involves a unique bimodal interaction with the Hippo effector YAP. J Virol 90, 7032–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf MA, Andreeva A, Kozlowski MM, Kim CE, Moskowitz BA, Anaya-Rocha A, Kelley MW, and Corwin JT (2020). YAP mediates hair cell regeneration in balance organs of chickens, but LATS kinases suppress its activity in mice. J Neurosci 40, 3915–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda EM, Hall BM, Hill MC, Swinton PG, Tong X, Martin JF, and Poche RA (2019). The Hippo pathway blocks mammalian retinal Muller glial cell reprogramming. Cell Rep 27, 1637–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Iranzo H, Galardi-Castilla M, Sanz-Morejon A, Gonzalez-Rosa JM, Costa R, Ernst A, Sainz de Aja J, Langa X, and Mercader N (2018). Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc Natl Acad Sci USA 115, 4188–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Vekaria H, Sullivan PG, and Seifert AW (2019). Connective tissue fibroblasts from highly regenerative mammals are refractory to ROS-induced cellular senescence. Nat Commun 10, 4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. (2011). Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell 144, 782–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90. [DOI] [PubMed] [Google Scholar]
- Seet LF, Toh LZ, Chu SWL, Finger SN, Chua JLL, and Wong TT (2017). Upregulation of distinct collagen transcripts in post-surgery scar tissue: a study of conjunctival fibrosis. Dis Model Mech 10, 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, and Maden M (2012). Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 489, 561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serini G, Bochaton-Piallat ML, Ropraz P, Geinoz A, Borsi L, Zardi L, and Gabbiani G (1998). The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. J Cell Biol 142, 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura T, Miyamura N, Hata S, Miura R, Hirayama J, and Nishina H (2014). The PDZ-binding motif of Yes-associated protein is required for its co-activation of TEAD-mediated CTGF transcription and oncogenic cell transforming activity. Biochem Biophys Res Commun 443, 917–923. [DOI] [PubMed] [Google Scholar]
- Simkin J, Gawriluk TR, Gensel JC, and Seifert AW (2017). Macrophages are necessary for epimorphic regeneration in African spiny mice. Elife 6, e24623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Ajani JA, Honjo S, Maru DM, Chen Q, Scott AW, Heallen TR, Xiao L, Hofstetter WL, Weston B, et al. (2014). Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res 74, 4170–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen TP (1970). Origin and differentiative capacities of cells in the blastema of the regenerating salamander limb. Am Zool 10, 119–132. [DOI] [PubMed] [Google Scholar]
- Sun H, Charles CH, Lau LF, and Tonks NK (1993). MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell 75, 487–493. [DOI] [PubMed] [Google Scholar]
- Swingle M, Ni L, and Honkanen RE (2007). Small-molecule inhibitors of ser/thr protein phosphatases: specificity, use and common forms of abuse. Methods Mol Biol 365, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talele NP, Fradette J, Davies JE, Kapus A, and Hinz B (2015). Expression of alpha-smooth muscle actin determines the fate of mesenchymal stromal cells. Stem Cell Reports 4, 1016–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM (2016). The molecular and cellular choreography of appendage regeneration. Cell 165, 1598–1608. [DOI] [PubMed] [Google Scholar]
- Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, and Brown RA (2002). Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol 3, 349–363. [DOI] [PubMed] [Google Scholar]
- Urban ML, Manenti L, and Vaglio A (2015). Fibrosis--A common pathway to organ injury and failure. New Engl J Med 373, 95–96. [DOI] [PubMed] [Google Scholar]
- Vallet SD, and Ricard-Blum S (2019). Lysyl oxidases: from enzyme activity to extracellular matrix cross-links. Essays Biochem 63, 349–364. [DOI] [PubMed] [Google Scholar]
- Wang P, Bai Y, Song B, Wang Y, Liu D, Lai Y, Bi X, and Yuan Z (2011). PP1A-mediated dephosphorylation positively regulates YAP2 activity. PLoS One 6, e24288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu G, Gao X, Wang Y, Zhang W, Harmon EY, Zhi Z, Xu Z, Lennartz MR, Barroso M, Trebak M, Chen C, Zhou J (2012) The induction of yes-associated protein expression after arterial injury is crucial for smooth muscle phenotypic modulation and neointima formation. Arterioscler Thromb Vasc Biol 32, 2662–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hu CK, Zeng A, Alegre D, Hu D, Gotting K, Ortega Granillo A, Wang Y, Robb S, Schnittker R, et al. (2020). Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 369, eaaz3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Guo Y, Zhu T, Zhang J, Ma PX, and Chen YE (2012). Yap1 protein regulates vascular smooth muscle cell phenotypic switch by interaction with myocardin. J Biol Chem 287, 14598–14605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, Liu N, Kulur V, Yao C, Chen P, et al. (2018). Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep 22, 3625–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, McCourt J, Ma F, Ren S, Li S, Kim TH, Kurmangaliyev YZ, Nasiri R, Ahadian S, Nguyen T, et al. (2020). Type V collagen in scar tissue regulates the size of scar after heart injury. Cell 182, 545–562 e523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S, Azzolin L, Maimets M, Pedersen MT, Fordham RP, Hansen SL, Larsen HL, Guiu J, Alves MRP, Rundsten CF, et al. (2018). YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, and Guan KL (2010). A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev 24, 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, and Guan KL (2011). The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol 13, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. (2007). Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21, 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2: Full gene list of Acomys DFs stimulated with TGFβ1 for 24h in vitro, related to Figure 2.
Video S1: Nuclear Yap in Acomys versus cytoplasmic Yap in Mus MFs in vivo, related to Figure 5. This movie shows high-power views of isolated Acomys and Mus MFs in vivo within the blastema at week1 immunolabeled for Yap (red), Acta2 (green), and Dapi (blue rendered nuclei). Images are rotated around the y-axis to better show nuclear Yap localization in Acomys Acta2+ MFs compared to Mus MFs that exhibit cytoplasmic Yap localization.
Data Availability Statement
All data that support the findings of this study are available within the article and its supplemental information files and from the corresponding authors upon reasonable request.