Abstract
The present study is focused on the safety, technological characteristics, and probiotic evaluation of Enterococcus species from different artisanal raw milk dairy products, mainly cheeses with ripening. Apart from proteolytic and lipolytic activities, most enterococci show the ability to metabolize citrate and convert it to various aromatic compounds. Long-ripened cheeses therefore have a specific flavor that makes them different from cheeses produced from thermally treated milk with commercial starter cultures. In addition, enterococci are producers of bacteriocins effective against spoilage and pathogenic bacteria, so they can be used as food preservatives. However, the use of enterococci in the dairy industry should be approached with caution. Although originating from food, enterococci strains may carry various virulence factors and antibiotic-resistance genes and can have many adverse effects on human health. Still, despite their controversial status, the use of enterococci in the food industry is not strictly regulated since the existence of these so-called desirable and undesirable traits in enterococci is a strain-dependent characteristic. To be specific, the results of many studies showed that there are some enterococci strains that are safe for use as starter cultures or as probiotics since they do not carry virulence factors and antibiotic-resistance genes. These strains even exhibit strong health-promoting effects such as stimulation of the immune response, anti-inflammatory activity, hypocholesterolemic action, and usefulness in prevention/treatment of some diseases.
Keywords: Enterococcus spp., raw-milk cheeses, safety, technological characteristics, probiotic properties
1. Introduction
The genus Enterococcus is one of the main genera of the lactic acid bacteria (LAB) group, with over 50 species and subspecies [1]. Enterococci belong to the family Enterococcaceae. They are gram-positive, catalase- and oxidase-negative, non-spore-forming, facultative anaerobic cocci occurring either as single bacteria, in pairs, in short chains, or in groups [2,3]. They are found in a variety of habitats, such as the gastrointestinal tract of humans and animals and plants, soil, water, and foods of animal origin, especially cheeses and fermented sausages [4,5,6,7,8,9,10,11]. One of the reasons for their prevalence in diverse niches is their robust nature since the majority of them have the ability to grow at temperatures from 10 to 45 °C, in 6.5% NaCl, 40% bile, and pH from 4 to 9.6. Additionally, they can survive heating at 60 °C for 30 min [2,3].
In Southern European countries, such as Portugal, Spain, Italy, and Greece, enterococci are highly valued and used in cheese production as components of mixed starter cultures for development of taste and flavor during cheese ripening, probably through proteolysis, lipolysis, and citrate breakdown [3]. In addition, certain enterococci have the ability to produce bacteriocins active against relevant spoilage-causing and pathogenic microorganisms in foods (such as Listeria monocytogenes) and possess appropriate probiotic properties, which are strong arguments for their application in the production of fermented food [12,13,14,15]. The total viable count of enterococci in Mediterranean-type cheese curds is in the range between 104 and 106 CFU/g at the beginning of ripening and between 105 and 107 CFU/g at the end of cheese ripening, thereby contributing to their typical sensory properties [16].
On the other hand, Northern European countries have considered enterococci undesirable in the food industry because there is an opinion that enterococci, inasmuch as they are predominant in the gastrointestinal tract of humans and animals, are indicators of fecal pollution (enterococci were formerly known as the “fecal streptococci” or “Lancefield’s group D streptococci”), transmitting antibiotic-resistance genes and virulence factors [17,18]. However, the presence of enterococci in foods does not always have to be associated with fecal contamination [19], as they can enter food from other sources, such as water, animal feed, or the animal’s exterior [17]. The European Union (EU) established a maximum level for the presence of coliforms and Escherichia coli, both considered as indicators of hygiene, while no limit was set for enterococci [20]. Moreover, results obtained by Birollo et al. [21] showed that enterococci had little value as hygiene indicators in industrially produced foods. In accordance with these results, it is obvious that enterococci are ubiquitous in the environment and can be present in food without being of fecal origin [22].
Although enterococci exhibit useful benefits for the production of fermented food, they are also identified as opportunistic pathogens that can cause various human diseases, such as bacteremia, urinary tract infections, and endocarditis [23]. Two enterococci species are suggested as responsible for these infections—Enterococcus faecalis and Enterococcus faecium [24,25]. They show a high level of virulence and transmission of antibiotic resistance genes, particularly the vancomycin-resistant ones [26,27]. Despite recent knowledge that the pathogenesis of enterococci is a strain-dependent trait and is more common to clinical enterococci than to enterococci from food [28,29], Enterococcus species do not have a generally recognized as safe (GRAS) status [30], nor have they been included on the qualified presumption of safety (QPS) list [31,32]. In connection with the above, a previous study of ours [33] threw light on the diversity, antibiotic susceptibility, virulence traits, production of biogenic amines, and technological properties of 636 enterococci isolated from 55 artisanal dairy products. The results showed that only 0.8% of them satisfy safety criteria for use in the dairy industry and indicated that, although isolated from food, enterococci can be reservoirs of antibiotic-resistance and virulence genes as well as producers of biogenic amines, thereby stressing that detailed testing of each individual enterococcal strain is necessary before its potential use in the food industry [17].
Artisanal dairy products are mainly produced from raw milk, which is rich in a variety of autochthonous microorganisms that determine their typical aroma. Studies on the microbiota of traditional cheeses of Mediterranean and Western Balkan Countries produced mainly from raw milk of sheep, goats, or cows indicate that enterococci are a relevant component of the non-starter LAB. Findings obtained by LAB identification showed that about one-third of all isolated LAB are Enterococcus species [34,35,36,37,38,39,40]. Additionally, similar results were obtained by analyzing bacteria from traditional Brazilian cheeses [41]. Apart from dairy products, 20% of total bacteria isolated from raw cow’s milk belonged to Enterococcus species [42]. Enterococcus faecium, Enterococcus faecalis, and Enterococcus durans are found as the most prevalent species of enterococci in raw-milk cheeses [3,11,18,43,44,45,46,47,48,49,50,51,52,53,54]. In addition, Enterococcus italicus, Enterococcus galinarum, Enterococcus avium, Enterococcus casseliflavus, Enterococcus lactis, Enterococcus hermanniensis, and Enterococcus gilvus were isolated in a low percentage from sweet kajmaks, Turkish white cheese, artisanal Italian cheeses (the Toma Piemontese and Vastedda della valle Belìce cheeses), 60-day-old raw milk Zlatar cheese, artisanal Istrian raw-milk cheese, and Ezine nonstarter long-ripened white cheese, respectively [39,41,55,56,57,58,59,60]. Morandi et al. [61] described Enterococcus lactis sp. nov., a new species found during isolation of the autochthonous microflora of an Italian raw-milk cheese (Bitto).
Extensive study of enterococcal diversity in the Western Balkans region showed that enterococci are present in various types of autochthonous dairy products, such as white brined cheeses and salted kajmaks in Serbia; hard and soft fresh cheeses in Croatia; young cheeses, sweet creams, and sweet kajmaks in Bosnia and Herzegovina; and leafy, white and semi-hard cheeses and skorups in Montenegro (Figure 1) [11,33,47,48,52,53,62,63]. Most of them were identified as E. durans (44%), E. faecalis (35.9%), and E. faecium (17.9%), while less than 2% were E. italicus strains [33]. Similar data were reported by authors who determined the enterococcal population in artisanal dairy products manufactured in other regions [64,65].
Figure 1.
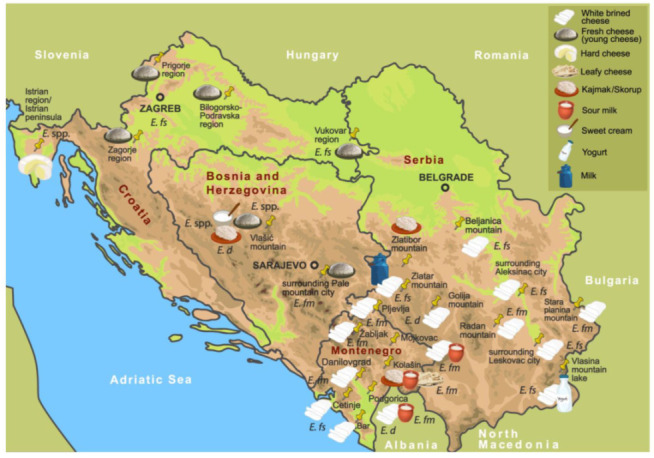
Artisanal raw milk dairy products manufactured in the region of Western Balkan Countries as a source of Enterococcus spp. Note: Most prevalent Enterococcus spp. in certain geographical localities: E. spp., Enterococcus species; E. fm, Enterococcus faecium; E. fs, Enterococcus faecalis; E. d, Enterococcus durans [11,54].
The aim of the present work is to point out advantages and disadvantages of enterococci isolated from raw-milk cheeses and summarize current knowledge about their safety, technological properties, and probiotic capacities on the one hand and stress their great potential for application in the food industry on the other, considering that some of these properties can lead to serious health problems in immunocompromised patients.
2. Safety of Using Enterococcus spp. in Dairy Food
According to the European Food Safety Authority (EFSA), members of the genus Enterococcus are not recommended for the QPS list [31] nor for GRAS status according to the Federal Food, Drug, and Cosmetic Act, USA [30], due to their controversial epidemiological status. In regard to their safety status, enterococci are assigned to risk group 2, together with other microorganisms carrying virulence factors [66]. The safety of dairy enterococci will be discussed here, together with recommended methods for its evaluation and the urgent need for new proposals and legislation to determine the safety of enterococci used in dairy and pharmaceutical industry.
On the one hand, enterococci have been associated with bacteremia and nosocomial infections mostly related to the presence of intrinsic or acquired antibiotic-resistance genes located on chromosomes, plasmids, or transposons [23,67] as well as to that of genes encoding virulence factors [68]. Moreover, a controversial correlation between E faecalis and pancreatic and colorectal cancers was recently suggested [69,70]. Besides the ability to cause diseases, the presence of resistance and virulence genes represents a huge threat to global health due to the possibility of horizontal transfer of these genes through the food chain to clinically relevant pathogens [71]. It is noteworthy in particular that vancomycin-resistant enterococci (VRE) represent a substantial problem in healthcare since vancomycin is often used as the last alternative in the treatment of nosocomial infections caused by multiple antibiotic-resistant enterococci [30]. However, while the horizontal transfer of resistance genes from E.faecalis to methicillin-resistant Staphylococcus aureus (MRSA) was reported [72,73], the results of Faron et al. [74] indicated that this horizontal transfer occurs at a very low frequency.
Regarding the presence of virulence genes, previous results of ours indicated that the genes encoding gelatinase (gelE), cytolysin activator (cylA), hyaluronidase (hylEfm), aggregation substance (agg), collagen adhesin (ace), and enterococcal surface protein (esp) are sporadically detected in dairy isolates [68]. It should be emphasized that there are two groups of virulence factors: surface factors involved in the colonization of host cells and factors causing damage to the host’s tissues [75]. The virulence factors enabling host colonization, including aggregation substance (AS), cell wall adhesin (EfaA), collagen-binding protein (Ace), and enterococcal surface protein (Esp), provide for binding of enterococci cells to receptors on the host’s epithelium [16]. Hence, although colonization represents the first step in pathogenesis, adhesion factors alone are not necessarily involved in pathogenicity. The genes for hemolytic activity as well as for adhesions (esp and efaA) were found with high frequency in all tested E. faecalis strains [24]. Although the results of Popović et al. [68] indicated the agg and esp genes to be positively associated with probiotic features in dairy isolates of enterococci, this must unquestionably be taken with precaution since AS is part of the conjugation process responsible for exchange of genetic material between bacterial cells during the conjugative transfer of sex pheromone plasmids and could favor the horizontal transfer of antibiotic-resistance genes [76]. In addition, the esp gene encoding the Esp protein is located on the pathogenicity island (PAI), which also contains genes involved in antibiotic outflow [77]. Moreover, the Esp protein is involved in adhesion, colonization, and evasion of the immune system as well as in biofilm formation, an important feature in horizontal gene transfer and the occurrence of antibiotic resistance [78].
In order to use a particular strain as a food supplement, it is necessary to evaluate the presence/absence of transferable antibiotic resistance and/or virulence genes since the presence of genes encoding antibiotic resistance and virulence factors in enterococcal strains intended for starter culture in food production could have substantial effects on human and animal health due to potential horizontal gene transfer throughout the food chain or even cause the occurrence of disease [79].
The safety status of 636 dairy isolates belonging to the species E. durans, E. faecium, E. faecalis, E. italicus, and Enterococcus hirae was investigated previously using various microbiological and molecular methods [24,33,68]. Interestingly, while results obtained by the disk diffusion method recommended by the Clinical and Laboratory Standards Institute (CLSI) [37] revealed that 29.1% enterococci were antibiotic-susceptible, use of the microdilution method showed that among them even 57% were resistant to ciprofloxacin or gentamicin, indicating that the method used for antibiotic susceptibility testing should be carefully chosen [68]. The high frequency of antibiotic resistance in the tested dairy enterococcal isolates was in accordance with published data on the results of extensive use of antibiotics in livestock production [18,23,71,75]. The further spread of antibiotic-resistance genes in other food-associated bacteria could be the cause of disappearance of natural sources of LAB suitable for the dairy industry.
In the case of functional foods, the EFSA recommendations on strain safety should be followed [80]. According to those recommendations, enterococcal isolates from food must be considered individually, and the risks to health must be excluded for each particular strain. To be specific, strains that do not carry the IS16 marker, exclusively detected in clinical isolates of E.faecium [81] as well as the hylEfm and esp genes, can be regarded as safe [82]. Previously published data related to enterococci isolated from different environments indicated that virulence factors are more prevalent in E. faecalis isolates than in E. faecium strains [83,84].
Previous research designed to evaluate the safety status of dairy enterococci originating from artisanal dairy products from the Western Balkans showed that virulence genes were sporadically present in the analyzed isolates [68]. While the esp gene was previously detected in isolates originating from water, vegetables, and raw milk [85], the study of Popović et al. [68] for the first time revealed the presence of the esp gene in dairy isolates mostly associated with their adhesion properties, thereby pointing to its role in gut colonization rather than to virulence features. To be specific, the esp gene in dairy enterococci was associated to a greater extent with the agg, efaAfs, and efaAfm genes, which have a role in adhesion and colonization [68]. Although E. durans strains have been characterized as good candidates for use in the food and pharmaceutical industries, all of them had some of the tested virulence genes [68], suggesting that E. durans should be checked more rigorously. It is important to highlight that genes encoding degradative enzymes associated with tissue damage were not detected, with the exception of gelE [68]. To judge from published data, the gelE gene could be silent [83], a hoped-for possibility. Interestingly, only five isolates out of 636 tested enterococci strains were completely free of virulence factor genes [33]. In addition, 30 out of 75 analyzed strains were able to form a biofilm [68]. Published data suggest that biofilm formation is a common trait of commensal enterococci isolates from human feces, indicating that this trait is not necessarily involved in pathogenicity but could be associated with adhesion and colonization properties [86,87].
3. Technological Characteristics of Enterococci
Natural LAB isolates possess a system of catabolic enzymes for proteolysis, lipolysis, and citrate metabolism that is better adapted to the cheese environment than the one in commercial starter cultures [88]. Enterococci are highly important in traditional fermented foods, particularly in artisanal cheeses, due to their technological properties, viz., different activities (acidifying, proteolytic, and lipolytic), citrate utilization, and production of aromatic volatile compounds that provide the specific sensory characteristics of many cheese varieties [1,3]. Due to their desirable metabolic properties, it has been suggested that certain enterococci strains could be used as part of existing starter cultures in the production of various cheeses [3,30], such as Bitto [61], water-buffalo Mozzarella [89], feta [90], Venaco [91], Cebreiro [92], cheddar [93], Koopeh [94], Tulum [95], and Lighvan [96], known to consumers around the world.
3.1. Acidification Activity
The main characteristic of LAB is production of lactic acid by fermentation of lactose. In this way, various effects are achieved: (a) decrease of milk pH and coagulation of casein; (b) increase in acidity of the environment due to lowering of the milk’s pH value, which makes possible control of the growth of pathogenic and spoilage bacteria; (c) positive action of casein coagulation on the rheological properties of dairy products; and (d) determination of the final flavor quality of ripened cheeses as a result of acidification [11]. However, not all LAB have the ability to acidify milk. Some of them lower milk pH rapidly, some do so slowly, and some LAB do not acidify milk. Enterococci belong to the group of LAB, which in general exhibit low or medium milk-acidifying ability [22]. Numerous published data confirm the poor acidifying capacity of these bacteria in milk showing a pH below 5.0 after 24 h of incubation at 30–37 °C [97,98,99,100,101,102]. Examining 636 enterococcal strains isolated from various types of dairy products, investigators found only 27 isolates (4.2%) possessing the ability to form curd after 6 h of incubation at 37 °C [33]. Since starter cultures are defined as isolates, which produce sufficient acid to reduce the pH of milk to 5.3 in 6 h at 30–37 °C [103], enterococci with low acidifying ability would not be good candidates for starter cultures in cheese manufacture, but they could be useful as adjunct cultures in combination with high-capacity acidifiers due to exhibiting other technological characteristics that are desirable [100].
However, some authors reported good acidification ability of enterococci. Thus, for example, Ribeiro et al. [104] found six E. faecalis fast-acidifier strains that lowered the pH of UHT milk from a starting value of 6.48 to 5.13–4.87 over the course of 6 h at 30 °C. Enterococcus faecalis strain SLT13 reduced the milk´s pH to 4.29 after 18 h of growth at 37 °C [105], while two Enterococcus sp. isolates from Kashar cheese lowered it to 4.08 for 24 h [106]. According to results reported by de Paula et al. [107], better acidification values can be achieved by prolonging cultivation time. A few authors reported that E. faecalis strains reduce milk pH faster than E. faecium and E. durans strains [22,108]. In contrast, E. faecium strains from Turkish Tulum cheese showed faster fermentation activity than E. faecalis [109], while Jaouani et al. [110] did not find differences in the rate of acidification of milk between E. faecalis and E. faecium strains after 24 h.
In view of these contradictory findings, it is apparent that further research is needed in the future to clarify which Enterococcus species has the better acidifying ability or if this property is strain-specific.
3.2. Proteolytic Activity
The degradation of casein due to proteolytic and peptidolytic activities plays a major role in development of the texture and organoleptic properties of cheese [3,105]. The positive role of enterococci in cheese production is associated with their proteolytic activity [111]. Apart from the ability of enterococci to grow in an environment with a wide range of temperatures, high salt content, and low pH values, the predominance of enterococci in cheeses with a long ripening period [34,58,59] is made possible by their production of proteolytic enzymes, which provides them with the peptides and amino acids essential for their growth [111]. However, despite the fact that enterococci were found to be the predominant LAB group in raw milk dairy products [11], there are only a few studies treating the proteolytic system of enterococci in comparison with Lactobacillus and Lactococcus species [112,113]. The proteolytic enzymes of enterococci have been insufficiently examined, and this may be one of the reasons for the limited use of enterococci in the production of traditional cheeses at the industrial level.
Specific for the proteolytic system of enterococci is the fact that it is characterized by the presence of gelatinase, an extracellular zinc metalloprotease capable of hydrolyzing gelatin, elastin, collagen, and hemoglobin [1]. Gútiez et al. [114] showed that gelatinase present in E. faecalis isolates originating from food and the ambient environment was responsible for casein degradation and formation of bioactive peptides, a circumstance that can affect human health. Gelatinase is encoded by the gelE gene as a virulence factor, which plays a significant role in the pathogenicity of enterococcal strains [115]. However, gelE alone was not proven to be directly responsible for infection [116], nor was it established that the presence of functional gelatinase is associated with all virulence features since it is not required for enterococci to cause disease [117]. Based on these findings, it can be asserted that proteolytic enterococci from food containing gelatinase [115,117] does not have to be excluded from eventual commercial use in the food industry [118,119].
While some authors have reported high levels of proteolytic activity [98,109,120], others considered that enterococci have weak proteolytic activity [97,99]. The data in the literature are very diverse. For instance, only one of seven tested E. faecalis strains from an artisanal Pico cheese showed low proteolytic activity [104]. Twenty-one E. faecalis isolates from artisanal raw milk Zlatar cheese degraded casein poorly, but three E. faecalis strains completely degraded αs1- and κ-casein after 3 h of incubation and β-casein after 30 min of incubation, indicating that proteolytic activity is a strain-dependent property [35].
In line with the fact that gelatinase is more frequently detected in E. faecalis than in E. faecium [121,122], many studies reported that E. faecalis showed better proteolytic activity than other Enterococcus species [99,108,123]. Terzic-Vidojevic et al. [33] found that 111 (17.5%) out of 636 examined enterococci degraded β-casein and that the number of E. faecium and E. faecalis strains was approximately equal. On the other hand, Mrkonjic Fuka et al. [59], Dagdemir [124], and Cosentino et al. [125] obtained results indicating better caseinolytic activity in the species E. faecium. Interestingly, according to the observation of some authors [126,127], enterococci show higher proteolytic activities than other LAB, a fact that increases their significance for use in cheese production.
A deeper evaluation of the proteolytic activity of enterococci is needed in view of conflicting scientific knowledge about it, specific biochemical properties of the genus, and its potentially great importance for further use in the dairy industry as part of mixed starter cultures.
3.3. Lipolytic Activity
Enterococci are one of many bacterial groups that show lipolytic and esterolytic activity, producing both lipases and esterases, which hydrolyze triglycerides to free fatty acids, glycerol, and intermediates, such as mono- and diglycerides [128,129]. The lipolytic and esterolytic system of enterococci is therefore very useful in food fermentation, especially in the case of dairy and meat products that require ripening [10,22,128,130].
Contradictory data regarding the lipolytic activities of enterococci have been reported to date, the obtained results ranging from pronounced lipolytic activity [98], to relatively high lipolytic activity [99], to low lipolytic activity, or its absence [89,104,108,110]. Carrasco de Mendoza et al. [131] concluded that the lipolytic activity of enterococci in milk is strain dependent. Morandi et al. [132] found that E. faecium strains from dairy products in northwestern Italy were the most lipolytic, followed by the tested E. faecalis and E. durans strains. On the other hand, lipolytic ability was confirmed in one-third of enterococci isolated from an artisanal Istrian raw-milk cheese [59]. Enterococcus faecalis showed better lipolytic activity than E. faecium and E. durans [59,109]. Regardless of differences existing between Enterococcus species as well as between strains within the same species, strains with high lipolytic characteristics should be subjected to further examination as a potential commercial adjunct culture in production of fermented food.
3.4. Production of Aromatic Compounds
The bulk of aromatic compounds in cheese ripening is produced during citrate metabolism by the activity of LAB, the majority of which are often enterococci. Results obtained by Terzic-Vidojevic et al. [33] showed that 40.9% tested enterococci from various artisanal dairy products utilized citrate as the only source of carbon. During manufacturing and ripening of raw-milk cheeses, citrate can be degraded over different metabolic pathways, giving significant amounts of various aromatic compounds (mainly acetate, acetaldehyde, acetoin, and diacetyl) that are responsible for the specific and intense flavor of final raw-milk cheese compared to that of cheeses made with pasteurized milk [133]. The most significant contribution to the buttery and ‘‘buttermilk” aroma and flavor of dairy products was made by diacetyl, which is a volatile compound formed as an end product during the conversion of citrate to pyruvate [134]. The appearance of certain aromatic compounds in cheese is dependent on many factors that influence citrate metabolism, such as the type of LAB, cell density, culture condition, environment pH, and lactate concentration [135,136].
During the last several decades, the citrate metabolism of enterococci has been examined by a number of investigators. The obtained results showed that enterococci are better diacetyl-acetoin producers than other LAB [99,137]. It was established that enterococci produce numerous volatile compounds and contribute to the formation of the cheese aroma, especially during ripening [17,91,100,138]. In the study of Cárdenas et al. [139], a total of 41 volatile compounds were identified in experimental cheeses manufactured with E. faecium CECT 8849.
Not all LAB have the capacity to metabolize citrate [129]. Significant differences among Enterococcus species and strains were found with respect to diacetyl and acetoin production. The highest production of diacetyl was obtained with the strain E. faecium C1W5, followed by the strains E. faecalis N8W4 and N0W5 [140]. On the other hand, several authors reported that certain E. durans strains had better production of diacetyl compared to other Enterococcus species [33,101,125,141]. All 56 tested E. faecalis strains completely utilized both citrate and pyruvate after 16 h [99], and six out of seven E. faecalis strains from Pico cheese produced diacetyl, among which four strains showed medium and two strains low production of diacetyl [104]. In addition, it was shown that E. faecalis strains were better producers of acetoin than other enterococci strains, since 157 of 229 E. faecalis strains gave a positive reaction for acetoin production in contrast to 138 of 280 E. durans and 48 of 114 E. faecium strains [33].
To judge from previously published data, we can conclude that citrate utilization by enterococci is an important technological characteristic and that enterococci, as a predominant part of non-starter lactic acid bacteria of raw milk products, determine their specific flavor. However, various data can be found regarding the abilities of different enterococci species and strains to convert citrate and pyruvate to aromatic compounds, so it is obvious that citrate metabolism is a strain-specific property.
4. Probiotic Potential of Enterococcus spp.
Enterococci have been traditionally thought to be indicators of fecal contamination as well as a cause of nosocomial infections and food spoilage. For that reason, their safety status is still controversial, as was discussed above. However, many enterococcal dairy isolates have probiotic effects, thereby contributing positively to human and animal health. In spite of safety concerns and due to the lack of legislation, some of them have already been used in commercial probiotic products, such as Cylactins (Hoffmann-La Roche, Basel, Switzerland), Fargo 688s (Quest International, Naarden, Netherlands), ECOFLOR (Walthers Health Care, DenHaag, Netherlands), Symbioflor 1 (SymbioPharm, Herborn, Germany), and Cernivet® and FortiFlora® (containing E. faecium SF68®) (Cerbios-Pharma SA, Barbengo-Lugano, Switzerland) [142]. Notably, Symbioflor 1 (containing E. faecalis DSM 16431) has a history of long-term safe use, and its safety was proven by whole-genome sequencing (WGS) [142]. In recent years, WGS data have been increasingly used to identify potential probiotic strains as well as to characterize strains in terms of their potential functionality for health [80].
Probiotics have been defined as “live microorganisms that confer health benefits to the host when ingested in adequate amounts” [143,144]. Enterococci are generally widespread in nature due to their ability to survive harsh conditions, making them good probiotic candidates. One of the important characteristics of probiotics is the ability to survive the conditions of gastrointestinal tract [143,145]. Popović et al. [15] reported that 13 E. durans strains from traditional cheeses of the Western Balkan Countries showed good probiotic properties, such as surviving simulated gastric conditions and prolonged exposure to bile salts and pancreatic enzymes, pointing to their gut commensal origin.
Another important probiotic characteristic is antimicrobial activity. Enterococcal bacteriocins, so-called enterocins, mostly belong to Class-II bacteriocins [146]. Many enterococci simultaneously synthesize several bacteriocins active against a number of pathogens and could be good candidates for use as antibiotic replacements or food preservatives [32,147]. Numerous literature data report a strong antimicrobial action of certain E. durans, E. faecium, and E. faecalis strains against one or more pathogenic bacteria, indicating that natural dairy enterococcal isolates produce enterocins with a broad spectrum of activity [14,15,19,68,148,149,150,151].
One of the criteria for selection of probiotics could be adhesion ability to intestinal epithelial cells (IEC), a prerequisite for gut colonization and persistence as well as for competitive exclusion of pathogens [143,152]. However, the cell-surface proteins involved in colonization of enterococci are for the most part virulence factors, as noted above [87]. Adhesion to mucin and IEC of 13 E. durans dairy isolates originating from artisanal dairy products was reported previously [68]. Although strain-specific differences were noticed, all 13 isolates showed a high adhesion potential. Most of them harbor the efaAfs, efaAfm, agg, and esp genes associated with adhesion ability. Interestingly, the esp gene was detected in even as many as six out of 13 tested strains. Although according to EFSA [82], occurrence of the esp gene is an undesirable property in probiotic enterococcal strains, the study of Popović et al. [68] revealed that it was mostly correlated with genes having a role in gut colonization, viz., the agg, efaAfs, and efaAfm genes, while virulence factors important for tissue damage were not detected [68]. However, in two E. durans strains, detection of the esp gene was related to the ability to form a biofilm, a virulence factor important in enterococcal pathogenicity [68,153]. In addition, the ability to counteract the negative effects of pathogens by competitive exclusion is a highly desirable property that should be taken into account in the selection of probiotic bacteria. The study of Popović et al. [68] revealed that enterococcal dairy isolates were able to reduce adhesions of Escherichia coli ATCC 25,922 and Salmonella Enteritidis 654/7E to HT29-MTX. Similarly, Jin et al. [154] reported that adhesion of enterococci to IEC limits the excessive pathogen growth.
In view of all the above-mentioned controversial features of enterococci, it would appear that one of the safe options in seeking to exploit their health-promoting properties and avoid the risk of their potential virulence as well as the danger of horizontal transfer of genes encoding virulence factors and resistance to antibiotics is to use them as postbiotics, non-viable bacterial extracts, and metabolic by-products. The effectiveness of heat-killed E. faecium BGPAS1-3 as a potential postbiotic was reported by Popović et al. [15]. To be specific, the heat-killed BGPAS1-3 postbiotic exhibited the same strong anti-listerial effect inhibiting the adhesion of L. monocytogenes ATCC 19111 to differentiated Caco-2 IEC as live bacteria.
One of the most critical steps in pathogenesis is invasion by the pathogens and their passage through the selectively permeable intestinal epithelium barrier, a multi-protein complex between adjacent epithelial cells composed of what have been denoted as tight-junction proteins [155]. In particular, L. monocytogenes is one of the pathogens that can disrupt tight-junction transmembrane structures, thereby causing epithelial barrier dysfunction [156]. Interestingly, the heat-killed BGPAS1-3 postbiotic was able to prevent tight-junction disruption in the differentiated Caco-2 monolayer after infection by L. monocytogenes ATCC 19111 through stimulation of the expression of claudin and occludin, important tight-junction proteins in Caco-2 cells, suggesting that enterococci could be good regulators of the epithelial barrier´s function [15], particularly as a safe postbiotic and controllable therapeutic. Similarly, E. faecium NCIMB 10415 was shown to improve the intestinal barrier´s integrity [157].
Moreover, IEC were the place where the presence of pathogens was first recognized by pathogen-associated molecular patterns (PAMPs) with pathogen recognition receptors (PRRs), including toll-like receptors (TLRs), upon which IEC produce antimicrobial molecules and activate the innate immune response and stimulate the production of protective cytokines (such as IL-8) and that of transforming growth factor (TGF)-β [158,159,160,161,162]. IL-8 is secreted by IEC as well as by several other cell types and has an important role in the activation of leukocytes, initiating the acute inflammatory response in listeriosis [156]. On the other hand, TGF-β prevents inflammation-mediated epithelial damage, thereby protecting the epithelial barrier´s integrity [163]. Popović et al. [15] reported that the E. faecium BGPAS1-3 heat-killed postbiotic, besides having an antimicrobial anti-listerial effect, exhibits immunomodulatory activity through stimulation of the production of protective IL-8 and TGF-β in IEC as well as through modulation of MyD88-dependent TLR2 and TLR4 pathways. The findings are in accordance to other published data indicating that enterococci could be used as immunomodulators [164,165]. It is concluded that manipulation of TLR expression can be the way enterococci achieve immunomodulatory activity [166].
The immunomodulatory effect of enterococci can also be related to the production of short-chain fatty acids, particularly butyrate, such as in the case of E. durans M4-5 [167]. Moreover, enterococci exhibit an anticarcinogenic and hypocholesterolemic effect, e.g., E. faecium M74® and E. durans KLDS 6.0930 were shown to reduce the level of cholesterol in serum [168]. The importance of enterococcal probiotic strains has been confirmed not only in animals but also in humans, for example, the assessment of the effectiveness of E. faecium SF68® and E. faecalis Symbioflor 1 in humans for the treatment of antibiotics-associated diarrhea [169].
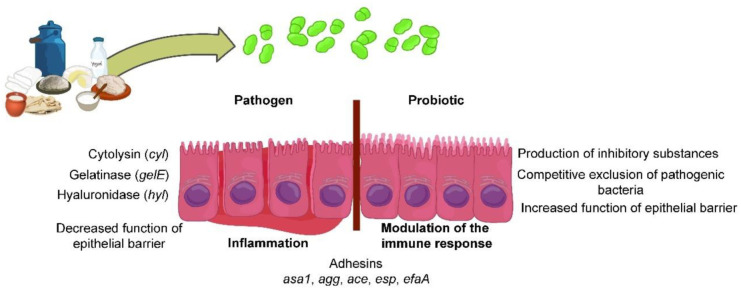
Safety and probiotic aspects of enterococci are summarized in Figure 2. Enterococci isolated from fermented dairy products show both a pathogenic and a probiotic potential. Their pathogenic potential is manifested in the synthesis of enzymes (cytolysin, gelatinase, hyaluronidase) that can degrade various proteins as well as whole cells, lower effectiveness of the epithelial barrier, and lead to inflammation. On the other hand, enterococci can have a probiotic effect that is realized through various mechanisms (synthesis of antimicrobial molecules, competitive exclusion of pathogens). Soluble and cell-bound molecules can enhance the epithelial barrier´s function and modulate the immune response. Various adhesins expressed on the cell surface (asa1, agg, ace, esp, efaA) play a role in cell binding to the host and colonization.
Figure 2.
Contrasting effects of enterococci-from emergence of pathogens to potential probiotic action.
5. Conclusions
As members of the LAB group, enterococci are well adapted for survival and persistence in various ecological niches. This review provides an update of information about Enterococcus species originating from raw-milk cheeses and their safety, technological characteristics, and probiotic capacity. It is known that enterococci are bacteria with “two faces” since they show desirable technological characteristics and probiotic properties but at the same time carry a number of virulence factors that make them undesirable for application in the food industry. The article presents numerous published data from which it can be concluded that there are no species of enterococci exclusively safe or exclusively unsafe for human health. All their properties, good or bad, are strain-specific.
Large numbers of in-vitro and in-vivo tests are needed to guarantee that a given Enterococcus strain is quite safe and suitable as a probiotic strain for potential application in the production of fermented foods. Modern techniques of molecular biology can help to obtain this knowledge and make it possible to develop improved legal standards and guidelines to ensure the faster introduction of enterococci for commercial purposes.
Acknowledgments
The authors would like to thank Dušan Radojević for figures preparation and Raymond Dooley, native English editor, for the proofreading of the manuscript.
Author Contributions
Conceptualization, A.T.-V., K.V., and N.P.; literature data collection, K.V., N.P., and M.T.; writing—original draft preparation, A.T.-V. and N.P.; writing—review and editing, A.T.-V. and N.G.; supervision, N.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Ministry of Education, Science, and Technological Development of the Republic of Serbia (Grant No. 173019), and agreement on the implementation and financing of research work in 2021, No: 451-03-9/2021-14/200042.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Graham K., Stack H., Rea R. Safety, beneficial and technological properties of enterococci for use in functional food application-a review. Crit. Rev. Food Sci. Nutr. 2020;60:3836–3861. doi: 10.1080/10408398.2019.1709800. [DOI] [PubMed] [Google Scholar]
- 2.Švec P., Franz C.M.A.P. The genus Enterococcus. In: Holzapfel W.H., Wood B.J.B., editors. Lactic Acid Bacteria: Biodiversity and Taxonomy. Wiley-Blackwell; Hoboken, NJ, USA: 2014. pp. 171–213. [Google Scholar]
- 3.Foulquie´ Moreno M.R., Sarantinopoulos P., Tsakalidou E., De Vuyst L. The role and application of enterococci in food and health. Int. J. Food Microbiol. 2006;106:1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Lebreton F., Willems R.J.L., Gilmore M.S. Enterococcus diversity, origins in nature, and gut colonization. In: Gilmore M.S., Clewell D.B., Ike Y., Shankar N., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection (Internet) Massachusetts Eye and Ear Infirmary; Boston, MA, USA: 2014. pp. 1–59. [PubMed] [Google Scholar]
- 5.Cho S., Jackson J.R., Frye J.G. The prevalence and antimicrobial resistance phenotypes of Salmonella, Escherichia coli and Enterococcus sp. in surface water. Lett. Appl. Microbiol. 2020;71:3–25. doi: 10.1111/lam.13301. [DOI] [PubMed] [Google Scholar]
- 6.Manson A.L., Van Tyne D., Straub T.J., Clock S., Crupain M., Rangan U., Gilmore M.S., Earl A.M. Chicken meat-associated enterococci: Influence of agricultural antibiotic use and connection to the clinic. Appl. Environ. Microbiol. 2019;85:e01559-19. doi: 10.1128/AEM.01559-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Švec P., Vandamme P., Bryndová H., Holochová P., Kosina M., Mašlaňová I., Sedláček I. Enterococcus plantarum sp. nov., isolated from plants. Int. J. Syst. Evol. Microbiol. 2012;62:1499–1505. doi: 10.1099/ijs.0.033357-0. [DOI] [PubMed] [Google Scholar]
- 8.Silva V., Peixoto F., Igrejas G., Parelho C., Garcia P., Carvalho I., Sousa M., Pereira J.E., Rodrigues A., Poeta P.A.C.Q.D. First report on vanA-Enterococcus faecalis recovered from soils subjected to long-term livestock agricultural practices in Azores archipelago. Int. J. Environ. Res. 2018;12:39–44. doi: 10.1007/s41742-018-0068-0. [DOI] [Google Scholar]
- 9.Said L.B., Klibi N., Dziri R., Borgo F., Boudabous A., Slamaa K.B., Torres C. Prevalence, antimicrobial resistance and genetic lineages of Enterococcus spp. from vegetable food, soil and irrigation water in farm environments in Tunisia. J. Sci. Food Agric. 2016;96:1627–1633. doi: 10.1002/jsfa.7264. [DOI] [PubMed] [Google Scholar]
- 10.Čolo J., Mihajlović S., Tolinački M., Popović D., Kojić M., Terzić-Vidojević A. Characterization of lactic acid bacteria isolated from Bosnian artisanal dry fermented sausage (Sudžuk) during fermentation. Genetika. 2015;47:819–832. doi: 10.2298/GENSR1503819C. [DOI] [Google Scholar]
- 11.Terzić-Vidojević A., Veljović K., Tolinački M., Živković M., Lukić J., Lozo J., Fira Đ., Jovčić B., Strahinić I., Begović J., et al. Diversity of non-starter lactic acid bacteria in autochthonous dairy products from Western Balkan Countries—Technological and probiotic properties. Food Res. Int. 2020;136:109494. doi: 10.1016/j.foodres.2020.109494. [DOI] [PubMed] [Google Scholar]
- 12.Qiao X., Du R., Wang Y., Han Y., Zhou Z. Isolation, characterisation and fermentation optimisation of bacteriocin-producing Enterococcus faecium. Waste Biomass Valorization. 2019;11:3173–3181. doi: 10.1007/s12649-019-00634-9. [DOI] [Google Scholar]
- 13.Elkenany R.M., Elsayed M.M., Eltaysh R.A., Zakaria A.I., El-Baz A.H. In vitro probiotic characteristics of Enterococcus species isolated from raw cow milk. Int. J. Probiotics Prebiotics. 2018;13:117–126. [Google Scholar]
- 14.Veljovic K., Fira D., Terzic-Vidojevic A., Abriouel H., Galvez A., Topisirovic L. Evaluation of antimicrobial and proteolytic activity of enterococci isolated from fermented products. Eur. Food Res. Technol. 2009;230:63–70. doi: 10.1007/s00217-009-1137-6. [DOI] [Google Scholar]
- 15.Popović N., Djokić J., Brdarić E., Dinić M., Terzić-Vidojević A., Golić N., Veljović K. The influence of heat-killed Enterococcus faecium BGPAS1-3 on the tight junction protein expression and immune function in differentiated Caco-2 cells infected with Listeria monocytogenes ATCC 19111. Front Microbiol. 2019;10:412. doi: 10.3389/fmicb.2019.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franz C.M.A.P., Stiles M.E., Schleifer K.H., Holzapfel W.H. Enterococci in foods–a conundrum for food safety. Int. J. Food Microbiol. 2003;88:105–122. doi: 10.1016/S0168-1605(03)00174-0. [DOI] [PubMed] [Google Scholar]
- 17.Giraffa G. Enterococci from foods. FEMS Microbiol. Rev. 2002;26:163–171. doi: 10.1111/j.1574-6976.2002.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 18.Câmara S.P.A., Dapkevicius A., Silva C.C.G., Malcata F.X., Dapkevicius M.L.E. Artisanal Pico cheese as reservoir of Enterococcus species possessing virulence and antibiotic resistance properties: Implications for food safety. Food Biotechnol. 2020;34:25–41. doi: 10.1080/08905436.2019.1710844. [DOI] [Google Scholar]
- 19.Popović N. Ph.D. Thesis. Faculty of Biology, University of Belgrade; Belgrade, Serbia: 2019. Examination of Probiotic and Immunomodulatory Characteristics of Natural Isolates of Enterococci on In Vitro and In Vivo Models. [Google Scholar]
- 20.Directive H. Council Directive 92/46/EEC of 16 June 1992 laying down the health rules for the production and placing on the market of raw milk, heat-treated milk and milk-based products. Off. J. Eur. Comm. 1992;268:1–32. [Google Scholar]
- 21.Birollo G.A., Reinheimer J.A., Vinderola C.G. Enterococci vs. nonlactic acid microflora as hygiene indicators for sweetened yoghurt. Food Microbiol. 2001;18:597–604. doi: 10.1006/fmic.2001.0435. [DOI] [Google Scholar]
- 22.Giraffa G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003;88:215–222. doi: 10.1016/S0168-1605(03)00183-1. [DOI] [PubMed] [Google Scholar]
- 23.Zaheer R., Cook S.R., Barbieri R., Goji N., Cameron A., Petkau A., Polo R.O., Tymensen L., Stamm C., Song J., et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020;10:3937. doi: 10.1038/s41598-020-61002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veljović K., Terzić-Vidojević A., Tolinački M., Mihajlović S., Vukotić G., Golić N., Kojić M. Molecular characterization of natural dairy isolates of Enterococcus faecalis and evaluation of their antimicrobial potential. In: Mack H.L., editor. Enterococcus Faecalis: Molecular Characteristics, Role in Nosocomial Infections and Antimicrobial Effects. (Bacteriology Research Developments) Nova Science Publishers Inc.; London, UK: 2014. pp. 123–135. [Google Scholar]
- 25.Jeeja V. Characterization of food and clinical isolates of enterococci. Acta Sci. Microbiol. 2019;2:56–61. [Google Scholar]
- 26.Reyes K., Bardossy A.C., Zervos M. Vancomycin-resistant enterococci. Epidemiology, infection, prevention and control. Infect. Dis. Clin. N. Am. 2016;30:953–965. doi: 10.1016/j.idc.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Terkuran M., Turhan E.Ü., Erginkaya Z. The risk of vancomycin resistant enterococci infections from food industry. In: Malik A., Erginkaya Z., Erten H., editors. Health and Safety Aspects of Food Processing Technologies. Springer International Publishing; Cham, Switzerland: 2019. pp. 513–535. [Google Scholar]
- 28.Bonacina J., Suarez N., Hormigo R., Fadda S., Lechner M., Saavedra L. A genomic view of food-related and probiotic Enterococcus strains. DNA Res. 2017;24:11–24. doi: 10.1093/dnares/dsw043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montealegre M.C., Singh K.V., Murray B.E. Gastrointestinal tract colonization dynamics by different Enterococcus faecium clades. J. Infect. Dis. 2016;213:1914–1922. doi: 10.1093/infdis/jiv597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogier J.-C., Serror P. Safety assessment of dairy microorganisms: The Enterococcus genus. Int. J. Food Microbiol. 2008;126:291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 31.Ricci A., Allende A., Bolton D., Chemaly M., Davies R., Girones R., Herman L., Koutsoumanis K., Lindqvist R., Nørrung B., et al. Scientific opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to European Food Safety Authority (EFSA). Panel on Biological Hazards (BIOHAZ) EFSA J. 2017;15:e04664. doi: 10.2903/j.efsa.2017.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanchi H., Mottawea W., Sebei K., Hammami R. The genus Enterococcus: Between probiotic potential and safety concerns-an update. Front. Microbiol. 2018;9:1791. doi: 10.3389/fmicb.2018.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terzić-Vidojević A., Veljović K., Begović J., Filipić B., Popović D., Tolinački M., Miljković M., Kojić M., Golić N. Diversity and antibiotic susceptibility of autochthonous dairy enterococci isolates: Are they safe candidates for autochthonous starter cultures? Front. Microbiol. 2015;6:954. doi: 10.3389/fmicb.2015.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veljovic K., Terzic-Vidojevic A., Vukasinovic M., Strahinic I., Begovic J., Lozo J., Topisirovic L. Preliminary characterization of lactic acid bacteria isolated from Zlatar cheese. J. Appl. Microbiol. 2007;103:2142–2152. doi: 10.1111/j.1365-2672.2007.03450.x. [DOI] [PubMed] [Google Scholar]
- 35.Terzic-Vidojevic A., Veljovic K., Tolinacki M., Nikolic M., Ostojic M., Topisirovic L.J. Characterization of lactic acid bacteria isolated from artisanal Zlatar cheeses produced at two different geographical location. Genetika. 2009;41:117–136. doi: 10.2298/GENSR0901117T. [DOI] [Google Scholar]
- 36.Terzic-Vidojevic A., Lozo J., Topisirovic L.J. Dominant lactic acid bacteria in artisanal Pirot cheeses of different ripening period. Genetika. 2009;41:341–352. doi: 10.2298/GENSR0903341T. [DOI] [Google Scholar]
- 37.Terzic-Vidojevic A., Tolinacki M., Nikolic M., Veljovic K., Jovanovic S., Macej O., Topisirovic L. Artisanal Vlasina raw goat’s milk cheeses: Evaluation and selection of autochthonous lactic acid bacteria as starter cultures. Food Technol. Biotechnol. 2013;51:554–563. [Google Scholar]
- 38.Terzić-Vidojević A., Mihajlović S., Uzelac G., Golić N., Fira Ð., Kojić M., Topisirović L. Identification and characterization of lactic acid bacteria isolated from artisanal white brined Golija cows’ milk cheeses. Arch. Biol. Sci. 2014;66:179–192. doi: 10.2298/ABS1401179T. [DOI] [Google Scholar]
- 39.Terzic-Vidojevic A., Mihajlovic S., Uzelac G., Veljovic K., Tolinacki M., Nikolic M., Topisirovic L., Kojic M. Characterization of lactic acid bacteria isolated from artisanal Travnik young cheeses, sweet creams and sweet kajmaks over four seasons. Food Microbiol. 2014;39:27–38. doi: 10.1016/j.fm.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Golić N., Čadež N., Terzić-Vidojević A., Šuranská H., Beganović J., Lozo J., Kos B., Šušković J., Raspor P., Topisirović L. Evaluation of lactic acid bacteria and yeast diversity in traditional white pickled and fresh soft cheeses from the mountain regions of Serbia and lowland regions of Croatia. Int. J. Food Microbiol. 2013;166:294–300. doi: 10.1016/j.ijfoodmicro.2013.05.032. [DOI] [PubMed] [Google Scholar]
- 41.Margalho L.P., van Schalkwijk S., Bachmann H., Sant’Ana A.S. Enterococcus spp. in Brazilian artisanal cheeses: Occurrence and assessment of phenotypic and safety properties of a large set of strains through the use of high throughput tools combined with multivariate statistics. Food Control. 2020;118:107425. doi: 10.1016/j.foodcont.2020.107425. [DOI] [Google Scholar]
- 42.Karakas-Sen A., Karakas E. Isolation, identification and technological properties of lactic acid bacteria from raw cow milk. Biosci. J. Uberlândia. 2018;34:385–399. doi: 10.14393/BJ-v34n2a2018-34517. [DOI] [Google Scholar]
- 43.Slyvka I.M., Tsisaryk O.Y., Dronyk G.V., Musiy L.Y. Strains of lactic acid bacteria isolated from traditional Carpathian cheeses. Regul. Mech. Biosyst. 2018;9:62–68. doi: 10.15421/021808. [DOI] [Google Scholar]
- 44.Levkov V., Mojsova S., Nastova R., Srbinovska S., Gjorgovska N. Identification and phenotypic characteristics of lactic acid bacteria isolated from some traditional cheeses produced in the Republic of Macedonia. Maced. J. Anim. Sci. 2017;7:79–87. [Google Scholar]
- 45.Sofu A., Ekinci F.Y. Bacterial diversity dynamics of traditional Turkish Ezine cheese as evaluated by PCR-DGGE and SSCP analysis. Int. J. Dairy Technol. 2016;69:592–600. doi: 10.1111/1471-0307.12311. [DOI] [Google Scholar]
- 46.Pogačić T., D’Andrea M., Kagkli D.-M., Corich V., Giacomini A., Baldan E., Čanžek Majhenič A., Obermajer T., Rogelj I., Samaržija D. Biodiversity of microbial consortia isolated from traditional fresh sheep cheese Karakačanski skakutanac. Mljekarstvo. 2011;61:208–219. [Google Scholar]
- 47.Muruzović M., Mladenović K., Žugić-Petrović T., Čomić L. Characterization of lactic acid bacteria isolated from traditionally made Serbian cheese and evaluation of their antagonistic potential against Enterobacteriaceae. J. Food Process. Preserv. 2018;42:e13577. doi: 10.1111/jfpp.13577. [DOI] [Google Scholar]
- 48.Mrkonjić Fuka M., Engel M., Skelin A., Redžepović S., Schloter M. Bacterial communities associated with the production of artisanal Istrian cheese. Int. J. Food Microbiol. 2010;142:19–24. doi: 10.1016/j.ijfoodmicro.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Mrkonjić Fuka M., Wallisch S., Engel M., Welzl G., Havranek J., Schloter M. Dynamics of bacterial communities during the ripening process of different Croatian cheese types derived from raw ewe’s milk cheeses. PLoS ONE. 2013;8:e80734. doi: 10.1371/journal.pone.0080734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Başar Uymaz B., Akçelik N., Yüksel Z. Physicochemical and microbiological characterization of protected designation of origin Ezine cheese: Assessment of nonstarter lactic acid bacterial diversity with antimicrobial activity. Food Sci. Anim. Resour. 2019;39:804–819. doi: 10.5851/kosfa.2019.e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.González L., Fernández Cuadrillero A., Castro J.M., Bernardo A., Tornadijo M.E. Selection of lactic acid bacteria isolated from San Simón da Costa cheese (PDO) in order to develop an autochthonous starter culture. Adv. Microbiol. 2015;5:748–759. doi: 10.4236/aim.2015.511079. [DOI] [Google Scholar]
- 52.Kročko M., Čanigová M., Ducková V., Artimová A., Bezeková J., Poston J. Antibiotic resistance of Enterococcus species isolated from raw foods of animal origin in south west part of Slovakia. Czech J. Food Sci. 2011;29:654–659. doi: 10.17221/246/2010-CJFS. [DOI] [Google Scholar]
- 53.Jokovic N., Nikolic M., Begovic J., Jovcic B., Savic D., Topisirovic L. A survey of the lactic acid bacteria isolated from Serbian artisanal dairy product kajmak. Int. J. Food Microbiol. 2008;127:305–311. doi: 10.1016/j.ijfoodmicro.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 54.Bojanic Rasovic M., Mayrhofer S., Ochome A.A.M., Ajanovic E., Zunabovic M., Martinovic A., Domig K.J. Diversity of lactic acid bacteria isolated from traditional Montenegrin dairy products. Genetika. 2018;50:465–482. doi: 10.2298/GENSR1802465B. [DOI] [Google Scholar]
- 55.İspirli H., Demirbaş F., Dertli E. Characterization of functional properties of Enterococcus spp. isolated from Turkish white cheese. LWT-Food Sci. Technol. 2017;75:358–365. doi: 10.1016/j.lwt.2016.09.010. [DOI] [Google Scholar]
- 56.Fortina M.G., Ricci G., Acquati A., Zeppa G., Gandini A., Manachini P.L. Genetic characterization of some lactic acid bacteria occurring in an artisanal protected denomination origin (PDO) Italian cheese, the Toma piemontese. Food Microbiol. 2003;20:397–404. doi: 10.1016/S0740-0020(02)00149-1. [DOI] [Google Scholar]
- 57.Cruciata M., Gaglio R., Todaro M., Settanni L. Ecology of Vastedda della valle del Belìce cheeses: A review and recent findings to stabilize the traditional production. Food Rev. Int. 2019;35:90–103. doi: 10.1080/87559129.2018.1469142. [DOI] [Google Scholar]
- 58.Terzic-Vidojevic A., Vukasinovic M., Veljovic K., Ostojic M., Topisirovic L. Characterization of microflora in homemade semi-hard white Zlatar cheese. Int. J. Food Microbiol. 2007;114:36–42. doi: 10.1016/j.ijfoodmicro.2006.10.038. [DOI] [PubMed] [Google Scholar]
- 59.Mrkonjic Fuka M., Zgomba Maksimovic A., Tanuwidjaja I., Hulak N., Schloter M. Characterization of enterococcal community isolated from an artisan Istrian raw milk cheese: Biotechnological and safety aspects. Food Technol. Biotechnol. 2017;55:368–380. doi: 10.17113/ftb.55.03.17.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tezel B.U. Preliminary in-vitro evaluation of the probiotic potential of the bacteriocinogenic strain Enterococcus lactis PMD74 isolated from Ezine Cheese. J. Food Qual. 2019:4693513. [Google Scholar]
- 61.Morandi S., Cremonesi P., Povolo M., Brasca B. Enterococcus lactis sp. nov., from Italian raw milk cheeses. Int. J. Syst. Evol. Microbiol. 2012;62:1992–1996. doi: 10.1099/ijs.0.030825-0. [DOI] [PubMed] [Google Scholar]
- 62.Jokovic N., Vukasinovic M., Veljovic K., Tolinacki M., Topisirovic L. Characterization of non-starter lactic acid bacteria in traditionally produced homemade Radan cheese during ripening. Arch. Biol. Sci. 2011;63:1–10. doi: 10.2298/ABS1101001J. [DOI] [Google Scholar]
- 63.Pogačić T., Samaržija D., Corich V., D’Andrea M., Kagkli D.M., Giacomini A., Rogelj I. Microbiota of Karakačanski skakutanac, an artisanal fresh sheep cheese studied by culture-independent PCR-ARDRA and PCR-DGGE. Dairy Sci. Technol. 2010;90:641–648. doi: 10.1051/dst/2010004. [DOI] [Google Scholar]
- 64.Guo L., Li T., Tang Y., Yang L., Huo G. Probiotic properties of Enterococcus strains isolated from traditional naturally fermented cream in China. Microb. Biotechnol. 2016;9:737–745. doi: 10.1111/1751-7915.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Medina R., Katz M., González S., Oliver G. Characterization of the lactic acid bacteria in ewe milk and artisanal cheese from northwest Argentina. J. Food Prot. 2001;64:559–563. doi: 10.4315/0362-028X-64.4.559. [DOI] [PubMed] [Google Scholar]
- 66.European Parliament Directive 2000/54/EC of the European Parliament and of the Council of 18 September 2000 on the protection of workers from risks related to exposure to biological agents at work. Off. J. Eur. Comm. 2000;262:21–45. [Google Scholar]
- 67.Banwo K., Sanni A., Tan H. Technological properties and probiotic potential of Enterococcus faecium strains isolated from cow milk. J. Appl. Microbiol. 2013;114:229–241. doi: 10.1111/jam.12031. [DOI] [PubMed] [Google Scholar]
- 68.Popović N., Dinić M., Tolinački M., Mihajlović S., Terzić-Vidojević A., Bojić S., Djokić J., Golić N., Veljović K. New insight into biofilm formation ability, the presence of virulence genes and probiotic potential of Enterococcus sp. dairy isolates. Front. Microbiol. 2018;9:78. doi: 10.3389/fmicb.2018.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Almeida C.V., Taddei A., Amedei A. The controversial role of Enterococcus faecalis in colorectal cancer. Therap. Adv. Gastroenterol. 2018;11:1–11. doi: 10.1177/1756284818783606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maekawa T., Fukaya R., Takamatsu S., Itoyama S., Fukuoka T., Yamada M., Hata T., Nagaoka S., Kawamoto K., Eguchi H., et al. Possible involvement of Enterococcus infection in the pathogenesis of chronic pancreatitis and cancer. Biochem. Biophys. Res. Commun. 2018;506:962–969. doi: 10.1016/j.bbrc.2018.10.169. [DOI] [PubMed] [Google Scholar]
- 71.Jahan M., Zhanel G.G., Sparling R., Holley R.A. Horizontal transfer of antibiotic resistance from Enterococcus faecium of fermented meat origin to clinical isolates of E. faecium and Enterococcus faecalis. Int. J. Food Microbiol. 2015;199:78–85. doi: 10.1016/j.ijfoodmicro.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 72.Lemen S.W., Lewalter K. Antibiotic stewardship and horizontal infection control are more effective than screening, isolation and eradication. Infection. 2018;46:581–590. doi: 10.1007/s15010-018-1137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alonso V.P.P., Queiroz M.M., Gualberto M.L., Nascimento M.S. Klebsiella pneumonia carbapenemase (KPC), methicillin-resistant Staphylococcus aureus (MRSA), and vancomycin-resistant Enterococcus spp. (VRE) in the food production chain and biofilm formation on abiotic surfaces. Curr. Opin. Food Sci. 2019;26:79–86. doi: 10.1016/j.cofs.2019.04.002. [DOI] [Google Scholar]
- 74.Faron M.L., Ledeboer N.A., Buchan B.W. Resistance mechanisms, epidemiology, and approaches to screening for vancomycin-resistant Enterococcus in the health care setting. J. Clin. Microbiol. 2016;54:2436–2447. doi: 10.1128/JCM.00211-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fiore E., Van Tyne D., Gilmore M.S. Pathogenicity of enterococci. Microbiol. Spectr. 2019;7:GPP3-0053-2018. doi: 10.1128/microbiolspec.GPP3-0053-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sartingen S., Rozdzinski E., Muscholl-Silberhorn A., Marre R. Aggregation substance increases adherence and internalization, but not translocation, of Enterococcus faecalis through different intestinal epithelial cells in vitro. Infect. Immun. 2000;68:6044–6047. doi: 10.1128/IAI.68.10.6044-6047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leavis H., Top J., Shankar N., Borgen K., Bonten M., van Embden J., Willems R.J.L. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J. Bacteriol. 2004;186:672–682. doi: 10.1128/JB.186.3.672-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koch S., Hufnagel M., Theilacker C., Huebner J. Enterococcal infections: Host response, therapeutic, and prophylactic possibilities. Vaccine. 2004;22:822–830. doi: 10.1016/j.vaccine.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 79.Ramos S., Silva V., Dapkevicius M.L.E., Igrejas G., Poeta P. Enterococci, from harmless bacteria to a pathogen. Microorganisms. 2020;8:1118. doi: 10.3390/microorganisms8081118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.European Food Safety Authority (EFSA) Guidance on the characterisation of microorganisms used as feed additives or as production organisms. Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) EFSA J. 2018;16:e05206. doi: 10.2903/j.efsa.2018.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leavis H.L., Willems R.J., van Wamel W.J., Schuren F.H., Caspers M.P., Bonten M.J. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 2007;3:e7. doi: 10.1371/journal.ppat.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.European Food Safety Authority (EFSA) Guidance for assessing safety of Enterococcus faecium in animal feed. Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) EFSA J. 2012;10:2682. [Google Scholar]
- 83.Kiruthiga A., Padmavathy K., Shabana P., Naveenkumar V., Gnanadesikan S., Malaiyan J. Improved detection of esp, hyl, asa1, gelE, cylA virulence genes among clinical isolates of Enterococci. BMC Res. Notes. 2020;13:170. doi: 10.1186/s13104-020-05018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eaton T.J., Gasson M.J. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 2001;67:1628–1635. doi: 10.1128/AEM.67.4.1628-1635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anderson A.C., Jonas D., Huber I., Karygianni L., Wölber J., Hellwig E., Arweiler N., Vach K., Wittmer A., Al-Ahmad A. Enterococcus faecalis from food, clinical specimens, and oral sites: Prevalence of virulence factors in association with biofilm formation. Front Microbiol. 2016;6:1534. doi: 10.3389/fmicb.2015.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ch’ng J.-H., Chong K.K.L., Lam L.N., Wong J.J., Kline K.A. Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 2019;17:82–94. doi: 10.1038/s41579-018-0107-z. [DOI] [PubMed] [Google Scholar]
- 87.Elhadidy M., Zahran E. Biofilm mediates Enterococcus faecalis adhesion, invasion and survival into bovine mammary epithelial cells. Lett. Appl. Microbiol. 2014;58:248–254. doi: 10.1111/lam.12184. [DOI] [PubMed] [Google Scholar]
- 88.Gobbetti M., de Angelis M., di Cagno R., Mancini L., Fox P.F. Pros and cons for using non-starter lactic acid bacteria (NSLAB) as secondary/adjunct starters for cheese ripening. Trends Food Sci. Technol. 2015;45:167–178. doi: 10.1016/j.tifs.2015.07.016. [DOI] [Google Scholar]
- 89.Villani F., Coppola S. Selection of enterococcal strains for water-buffalo Mozzarella cheese manufacture. Ann. Microbiol. Enzim. 1994;44:97–105. [Google Scholar]
- 90.Sarantinopoulos P., Kalantzopoulos G., Tsakalidou E. Effect of Enterococcus faecium on microbiological, physicochemical and sensory characteristics of Greek Feta cheese. Int. J. Food Microbiol. 2002;76:93–105. doi: 10.1016/S0168-1605(02)00021-1. [DOI] [PubMed] [Google Scholar]
- 91.Casalta E., Zennaro R. Effect of specific starters on microbiological, biochemical and sensory characteristics of Venaco, a Corsican soft cheese. Sci. Alim. 1997;17:79–94. [Google Scholar]
- 92.Centeno J.A., Menendez S., Hermida M.A., Rodriguez-Otero J.L. Effects of the addition of Enterococcus faecalis in Cebreiro cheese manufacture. Int. J. Food Microbiol. 1999;48:97–111. doi: 10.1016/S0168-1605(99)00030-6. [DOI] [PubMed] [Google Scholar]
- 93.Gardiner G.E., Ross R.P., Wallace J.M., Scanlan F.P., Jagers P.P., Fitzgerald G.F., Collins J.K., Stanton C. Influence of a probiotic adjunct culture of Enterococcus faecium on the quality of Cheddar cheese. J. Agr. Food Chem. 1999;47:4907–4916. doi: 10.1021/jf990277m. [DOI] [PubMed] [Google Scholar]
- 94.Hassanzadazar H., Ehsani A., Mardan K. Antibacterial activity of Enterococcus faecium derived from Koopeh cheese against Listeria monocytogenes in probiotic ultra-filtrated cheese. Vet. Res. Forum. 2014;5:169–175. [PMC free article] [PubMed] [Google Scholar]
- 95.Yerlikaya O., Akbulut N. Potential use of probiotic Enterococcus faecium and Enterococcus durans strains in Izmir Tulum cheese as adjunct culture. J. Food Sci. Technol. 2019;56:2175–2185. doi: 10.1007/s13197-019-03699-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pirouzian H.R., Hesary J., Farajnia S., Moghaddam M., Ghiassifar S., Manafi M. Inclusion of Enterococcus faecalis and Enterococcus faecium to UF white cheese. Int. J. Nutr. Food Eng. 2010;4:405–408. [Google Scholar]
- 97.Andrighetto C., Knijff E., Lombardi A., Torriani S., Vancanneyt M., Kersters K., Swings J., Dellaglio F. Phenotypic and genetic diversity of enterococci isolated from Italian cheeses. J. Dairy. Res. 2001;68:303–316. doi: 10.1017/S0022029901004800. [DOI] [PubMed] [Google Scholar]
- 98.Durlu-Ozkaya F., Xanthopoulos V., Tunail N., Litopoulou-Tzanetaki E. Technologically important properties of lactic acid bacteria isolates from Beyaz cheese made from raw ewes’ milk. J. Appl. Microbiol. 2001;91:861–870. doi: 10.1046/j.1365-2672.2001.01448.x. [DOI] [PubMed] [Google Scholar]
- 99.Sarantinopoulos P., Andrighetto C., Georgalaki M.D., Rea M.C., Lombardi A., Cogan T.M., Kalantzopoulos G., Tsakalidou E. Biochemical properties of enterococci relevant to their technological performance. Int. Dairy J. 2001;11:621–647. doi: 10.1016/S0958-6946(01)00087-5. [DOI] [Google Scholar]
- 100.Abeijón M.C., Medina R.B., Katz M.B., González S.N. Technological properties of Enterococcus faecium isolated from ewe’s milk and cheese with importance for flavour development. Can. J. Microbiol. 2006;52:237–245. doi: 10.1139/w05-136. [DOI] [PubMed] [Google Scholar]
- 101.Jamaly N., Benjouad A., Comunian R., Daga E., Bouksaim M. Characterization of Enterococci isolated from Moroccan dairy products. Afr. J. Microbiol. Res. 2010;4:1768–1774. [Google Scholar]
- 102.Aspri M., Bozoudi D., Tsaltas D., Hill C., Papademas P. Raw donkey milk as a source of Enterococcus diversity: Assessment of their technological properties and safety characteristics. Food Control. 2017;73:81–90. doi: 10.1016/j.foodcont.2016.05.022. [DOI] [Google Scholar]
- 103.Beresford T.P., Fitzsimons N.A., Brennan N.L., Cogan T.M. Recent advances in cheese microbiology. Int. Dairy J. 2001;11:259–274. doi: 10.1016/S0958-6946(01)00056-5. [DOI] [Google Scholar]
- 104.Ribeiro S.C., Coelho M.C., Todorov S.D., Franco B.D.G.M., Dapkevicius M.L.E., Silva C.C.G. Technological properties of bacteriocin-producing lactic acid bacteria isolated from Pico cheese an artisanal cow’s milk cheese. J. Appl. Microbiol. 2014;116:573–585. doi: 10.1111/jam.12388. [DOI] [PubMed] [Google Scholar]
- 105.Ziadi M., M’hir S., Dubois-Dauphin R., Chambellon E., Yvon M., Thonart P., Hamdi M. Analysis of volatile compounds, amino acid catabolism and some technological properties of Enterococcus faecalis strain SLT13 isolated from artisanal Tunisian fermented milk. Br. Microbiol. Res. J. 2016;14:17309. doi: 10.9734/BMRJ/2016/17309. [DOI] [Google Scholar]
- 106.Turhan İ., Öner Z. Determination of starter culture properties of lactic acid bacteria isolated from cheese. GIDA. 2014;39:9–15. [Google Scholar]
- 107.de Paula P.L.M., de Moraes M.L., Schueler J., de Souza N.A.A., Furlaneto M.C., Maia L.F., Katsuda M.S. Enterococcus faecium in artisanal ripening cheese: Technological and safety aspects. Res. Soc. Dev. 2020;9:e299119452. doi: 10.33448/rsd-v9i11.9452. [DOI] [Google Scholar]
- 108.Suzzi G., Caruso M., Gardini F., Lombardi A., Vannini L., Guerzoni M.E., Andrighetto C., Lanorte M.T. A survey of the enterococci isolated from an artisanal Italian goat’s cheese (semicotto caprino) J. Appl. Microbiol. 2000;89:267–274. doi: 10.1046/j.1365-2672.2000.01120.x. [DOI] [PubMed] [Google Scholar]
- 109.Tuncer Y. Some technological properties of phenotypically identified enterococci strains isolated from Turkish Tulum cheese. Afr. J. Biotechnol. 2009;8:7008–7016. [Google Scholar]
- 110.Jaouani I., Abbassi M.S., Ribeiro S.C., Khemiri M., Mansouri R., Messadi L., Silva C.C.G. Safety and technological properties of bacteriocinogenic enterococci isolates from Tunisia. J. Appl. Microbiol. 2015;119:1089–1100. doi: 10.1111/jam.12916. [DOI] [PubMed] [Google Scholar]
- 111.Wilkinson M.G., Guinee T.P., O’Callaghan D.M., Fox P.F. Autolysis and proteolysis in different strains of starter bacteria during Cheddar cheese ripening. J. Dairy Res. 1994;61:249–262. doi: 10.1017/S0022029900028260. [DOI] [Google Scholar]
- 112.González L., Sacristán N., Arenas R., Fresno J.M., Tornadijo M.E. Enzymatic activity of lactic acid bacteria (with antimicrobial properties) isolated from a traditional Spanish cheese. Food Microbiol. 2010;27:592–597. doi: 10.1016/j.fm.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 113.Pessione A., Lamberti C., Cocolin L., Campolongo S., Grunau A., Giubergia S., Eberl L., Riedel K., Pessione E. Different protein expression profiles in cheese and clinical isolates of Enterococcus faecalis revealed by proteomic analysis. Proteomics. 2012;12:431–447. doi: 10.1002/pmic.201100468. [DOI] [PubMed] [Google Scholar]
- 114.Gútiez L., Gómez-Sala B., Recio I., del Campo R., Cintas L.M., Herranz C., Hernández P.E. Enterococcus faecalis strains from food, environmental, and clinical origin produce ACE-inhibitory peptides and other bioactive peptides during growth in bovine skim milk. Int. J. Food Microbiol. 2013;166:93–101. doi: 10.1016/j.ijfoodmicro.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 115.Chajęcka-Wierzchowska W., Zadernowska A., Łaniewska-Trokenheim L. Virulence factors of Enterococcus spp. presented in food. LWT-Food Sci. Technol. 2017;75:670–676. doi: 10.1016/j.lwt.2016.10.026. [DOI] [Google Scholar]
- 116.Galloway-Peña J.R., Bourgogne A., Qin X., Murray B.E. Diversity of the fsr-gelE region of the Enterococcus faecalis genome but conservation in strains with partial deletions of the fsr operon. Appl. Environ. Microbiol. 2011;77:442–451. doi: 10.1128/AEM.00756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Medeiros A.W., Pereira R.I., Oliveira D.V., Martins P.D., D’azevedo P.A., Van der Sand S., Frazzon J., Frazzon A.P.G. Molecular detection of virulence factors among food and clinical Enterococcus faecalis strains in South Brazil. Braz. J. Microbiol. 2014;45:327–332. doi: 10.1590/S1517-83822014005000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Domann E., Hain T., Ghai R., Billion A., Kuenne C., Zimmermann K., Chakraborty T. Comparative genomic analysis for the presence of potential enterococcal virulence factors in the probiotic Enterococcus faecalis strain Symbioflor 1. Int. J. Med. Microbiol. 2007;297:533–539. doi: 10.1016/j.ijmm.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 119.Worsztynowicz P., Olejnik-Schmidt A., Bialas W., Grajek W. Identification and partial characterization of proteolytic activity of Enterococcus faecalis relevant to their application in the dairy industry. Acta Biochim. Pol. 2019;66:61–69. doi: 10.18388/abp.2018_2714. [DOI] [PubMed] [Google Scholar]
- 120.Tulini F.L., Hymery N., Haertlé T., Le Blay G., De Martinis E.C.P. Screening for antimicrobial and proteolytic activities of lactic acid bacteria isolated from cow, buffalo and goat milk and cheeses marketed in the southeast region of Brazil. J. Dairy Res. 2016;83:115–124. doi: 10.1017/S0022029915000606. [DOI] [PubMed] [Google Scholar]
- 121.Franz C.M., Muscholl-Silberhorn A.B., Yousif N.M., Vancanneyt M., Swings J., Holzapfel W.H. Incidence of virulence factors and antibiotic resistance among enterococci isolated from food. Appl. Environ. Microbiol. 2001;67:4385–4389. doi: 10.1128/AEM.67.9.4385-4389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cariolato D., Andrighetto C., Lombardi A. Occurrence of virulence factors and antibiotic resistances in Enterococcus faecalis and Enterococcus faecium collected from dairy and human samples in North Italy. Food Control. 2008;19:886–892. doi: 10.1016/j.foodcont.2007.08.019. [DOI] [Google Scholar]
- 123.Perin L.M., Belvisio S., dal Bello B., Nero L.A., Cocolin L. Technological properties and biogenic amines production by bacteriocinogenic lactococci and enterococci strains isolated from raw goat’s milk. J. Food Prot. 2017;80:151–157. doi: 10.4315/0362-028X.JFP-16-267. [DOI] [PubMed] [Google Scholar]
- 124.Dağdemir E. Ph.D. Thesis. Graduate School of Natural and Applied Science, Atatürk University Science and Technology; Erzurum, Turkey: 2006. Identification of Lactic Acid Bacteria Isolated from Pickled White Cheeses and the Possibilities of Using Some Selected Isolates as Culture; p. 190. [Google Scholar]
- 125.Cosentino S., Pisano M.B., Corda A., Fadda M.E., Piras C. Genotypic and technological characterization of enterococci isolated from artisanal Fiore Sardo cheese. J. Dairy Res. 2004;71:444–450. doi: 10.1017/S002202990400041X. [DOI] [PubMed] [Google Scholar]
- 126.Savitri M., Kumar V., Kumari A., Angmo K., Bhalla T.C. Isolation and characterization of lactic acid bacteria from traditional pickles of Himachal Pradesh, India. J. Food Sci. Technol. 2017;54:1945–1952. doi: 10.1007/s13197-017-2629-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tulini F.L., Biscola V., Choiset Y., Hymery N., Le Blay G., De Martinis E.C.P., Chobert J.-M., Haertlé T. Evaluation of the proteolytic activity of Enterococcus faecalis FT132 and Lactobacillus paracasei FT700, isolated from dairy products in Brazil, using milk proteins as substrates. Eur. Food Res. Technol. 2015;241:385–392. doi: 10.1007/s00217-015-2470-6. [DOI] [Google Scholar]
- 128.García-Cano I., Rocha-Mendoza D., Ortega-Anaya J., Wang K., Kosmerl E., Jiménez-Flores R. Lactic acid bacteria isolated from dairy products as potential producers of lipolytic, proteolytic and antibacterial proteins. Appl. Microbiol. Biotechnol. 2019;103:5243–5257. doi: 10.1007/s00253-019-09844-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Esteban-Torres M., Mancheño J.M., de Las Rivas B., Muñoz R. Production and characterization of a tributyrin esterase from Lactobacillus plantarum suitable for cheese lipolysis. J. Dairy Sci. 2014;97:6737–6744. doi: 10.3168/jds.2014-8234. [DOI] [PubMed] [Google Scholar]
- 130.Dinçer E., Kıvanç M. Lipolytic activity of lactic acid bacteria isolated from Turkish pastırma. Anadolu Univ. J. Sci. Technol. C-Life Sci. Biotechnol. 2018;7:12–19. doi: 10.18036/aubtdc.306292. [DOI] [Google Scholar]
- 131.Carrasco de Mendoza M.S., Scarinci M.S., Huerto-Garat H.E., Simonetta A.C. Technological properties of enterococci in lactic starters: Acidifying and lipolytic activities. Microbiol. Aliment. Nutr. 1992;10:289–293. [Google Scholar]
- 132.Morandi S., Brasca M., Andrighetto C., Lombardi A., Lodi R. Technological and molecular characterisation of enterococci isolated from north–west Italian dairy products. Int. Dairy J. 2006;16:867–875. doi: 10.1016/j.idairyj.2005.09.005. [DOI] [Google Scholar]
- 133.Kilcawley K.N. Cheese flavour. In: Fox P.F., Guinee T.P., Cogan T.M., Mc Sweeney P.L.H., editors. Fundamentals of Cheese Science. 2nd ed. Springer; Boston, MA, USA: 2017. pp. 443–474. [Google Scholar]
- 134.Smit G., Smit B.A., Engels W.J.M. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 2005;29:591–610. doi: 10.1016/j.fmrre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 135.Blaya J., Barzideh Z., LaPointe G. Symposium review: Interaction of starter cultures and nonstarter lactic acid bacteria in the cheese environment. J. Dairy Sci. 2018;101:3611–3629. doi: 10.3168/jds.2017-13345. [DOI] [PubMed] [Google Scholar]
- 136.Pretorius N., Engelbrecht L., Du Toit M. Influence of sugars and pH on the citrate metabolism of different lactic acid bacteria strains in a synthetic wine matrix. J. Appl. Microbiol. 2019;127:1490–1500. doi: 10.1111/jam.14401. [DOI] [PubMed] [Google Scholar]
- 137.Garabal J.I., Rodríguez-Alonso P., Centeno J.A. Characterization of lactic acid bacteria isolated from raw cows’ milk cheeses currently produced in Galicia (NW Spain) LWT-Food Sci. Technol. 2008;41:1452–1458. doi: 10.1016/j.lwt.2007.09.004. [DOI] [Google Scholar]
- 138.Sarantinopoulos P., Kalantzopoulos G., Tsakalidou E. Citrate metabolism by Enterococcus faecalis FAIR-E 229. Appl. Environ. Microbiol. 2001;67:5482–5487. doi: 10.1128/AEM.67.12.5482-5487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cárdenas N., Arroyo R., Calzada J., Peirotén A., Medina M., Rodríguez J.M., Fernández L. Evaluation of technological properties of Enterococcus faecium CECT 8849, a strain isolated from human milk, for the dairy industry. Appl. Microbiol. Biotechnol. 2016;100:7665–7677. doi: 10.1007/s00253-016-7616-3. [DOI] [PubMed] [Google Scholar]
- 140.Nieto-Arribas P., Seseña S., Poveda J.M., Chicón R., Cabezas L., Palop L. Enterococcus populations in artisanal Manchego cheese: Biodiversity, technological and safety aspects. Food Microbiol. 2011;28:891–899. doi: 10.1016/j.fm.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 141.Serio A., Chaves-López C., Paparella A., Suzzi G. Evaluation of metabolic activities of enterococci isolated from Pecorino Abruzzese cheese. Int. Dairy J. 2010;20:459–464. doi: 10.1016/j.idairyj.2010.02.005. [DOI] [Google Scholar]
- 142.Baccouri O., Boukerb A.M., Farhat L.B., Zébré A., Zimmermann K., Domann E., Cambronel M., Barreau M., Maillot O., Rincé I., et al. Probiotic potential and safety evaluation of Enterococcus faecalis OB14 and OB15, isolated from traditional Tunisian Testouri cheese and Rigouta, using physiological and genomic analysis. Front. Microbiol. 2019;10:881. doi: 10.3389/fmicb.2019.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Food and Agriculture Organization of the United Nations. World Health Organization . Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation. Food and Agriculture Organization of the United Nations; Rome, Italy: World Health Organization; Rome, Italy: 2006. pp. 2–3. FAO food and nutrition paper. [Google Scholar]
- 144.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 145.Nishiyama K., Sugiyama M., Mukai T. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms. 2016;4:34. doi: 10.3390/microorganisms4030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ness I.F., Diep D.B., Ike Y. Enterococcal bacteriocins and antimicrobial proteins that contribute to niche control. In: Gilmore M.S., Clewell D.B., Ike Y., Shankar N., editors. Enterococci: From Commensals to Leading Causes of Drug Resistant Infection (Internet) Massachusetts Eye and Ear Infirmary; Boston, MA, USA: 2014. pp. 1–33. [PubMed] [Google Scholar]
- 147.Ramiah K., Ten Doeschate K., Smith R., Dicks L.M.T. Safety assessment of Lactobacillus plantarum 423 and Enterococcus mundtii ST4SA determined in trials with Wistar rats. Probiotics Antimicrob. Proteins. 2009;1:15–23. doi: 10.1007/s12602-009-9010-2. [DOI] [PubMed] [Google Scholar]
- 148.Tosoni N.F., Perini H.F., Terra M.R., Katsuda M.S., Furlaneto M.C., Maia L.F. Antimicrobial activity of enterocin obtained from Enterococcus durans on Shiga-like toxin-producing Escherichia coli. Ciência Rural. 2019;49:e20190297. doi: 10.1590/0103-8478cr20190297. [DOI] [Google Scholar]
- 149.Pieniz S., Andreazza R., Anghinoni T., Camargo F., Brandelli A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control. 2015;37:251–256. doi: 10.1016/j.foodcont.2013.09.055. [DOI] [Google Scholar]
- 150.Vimont A., Fernandez B., Hammami R., Ababsa A., Daba H., Fliss I. Bacteriocin-producing LCW 44: A high potential probiotic candidate from raw Camel milk. Front. Microbiol. 2017;8:865. doi: 10.3389/fmicb.2017.00865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Furlaneto-Maia L., Ramalho R., Rocha K.R., Furlaneto M.C. Antimicrobial activity of enterocins against Listeria sp. and other food spoilage bacteria. Biotechnol. Lett. 2020;42:797–806. doi: 10.1007/s10529-020-02810-7. [DOI] [PubMed] [Google Scholar]
- 152.Živković M., Miljković M.S., Ruas-Madiedo P., Markelić M.B., Veljović K., Tolinački M., Soković S., Korać A., Golić N. EPS-SJ exopolisaccharide produced by the strain Lactobacillus paracasei subsp. paracasei BGSJ2-8 is involved in adhesion to epithelial intestinal cells and decrease on E. coli association to Caco-2 cells. Front. Microbiol. 2016;7:286. doi: 10.3389/fmicb.2016.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hashem Y.A., Amin H.M., Essam T.M., Yassin A.S., Aziz R.K. Biofilm formation in enterococci: Genotype-phenotype correlations and inhibition by vancomycin. Sci. Rep. 2017;7:5733. doi: 10.1038/s41598-017-05901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jin L., Marquardt R., Zhao X. A strain of Enterococcus faecium (18C23) inhibits adhesion of enterotoxigenic Escherichia coli K88 to porcine small intestine mucus. Appl. Environ. Microbiol. 2000;66:4200–4204. doi: 10.1128/AEM.66.10.4200-4204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luissint A.-C., Parkos C.A., Nusrat A. Inflammation and the intestinal barrier: Leukocyte–epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology. 2016;151:616–632. doi: 10.1053/j.gastro.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Drolia R., Tenguria S., Durkes A.C., Turner J.R., Bhunia A.K. Listeria adhesion protein induces intestinal epithelial barrier dysfunction for bacterial translocation. Cell Host Microbe. 2018;23:470–484. doi: 10.1016/j.chom.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bednorz C., Guenther S., Oelgeschläger K., Kinnemann B., Pieper R., Hartmann S., Tedin K., Semmler T., Neumann K., Schierack P., et al. Feeding the probiotic Enterococcus faecium strain NCIMB 10415 to piglets specifically reduces the number of Escherichia coli pathotypes that adhere to the gut mucosa. Appl. Environ. Microbiol. 2013;79:7896–7904. doi: 10.1128/AEM.03138-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vijay K. Toll-like receptors in immunity and inflammatory diseases: Past, present, and future. Int. Immunopharmacol. 2018;62:338. doi: 10.1016/j.intimp.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.D’Orazio S. Innate and adaptive immune responses during Listeria monocytogenes infection. Microbiol. Spectr. 2019;7:GPP3-0065-2019. doi: 10.1128/microbiolspec.GPP3-0065-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Onyiah J.C., Colgan S.P. Cytokine responses and epithelial function in the intestinal mucosa. Cell Mol. Life Sci. 2016;73:4203–4212. doi: 10.1007/s00018-016-2289-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rochman Y., Spolski R., Leonard W.J. New insights into the regulation of T cells by gamma(c) family cytokines. Nat. Rev. Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ihara S., Hirata Y., Koike K. TGF-β in inflammatory bowel disease: A key regulator of immune cells, epithelium, and the intestinal microbiota. J. Gastroenterol. 2017;52:777–787. doi: 10.1007/s00535-017-1350-1. [DOI] [PubMed] [Google Scholar]
- 163.Rachakonda G., Vu T., Jin L., Samanta D., Datta P.K. Role of TGF-β-induced Claudin-4 expression through c-Jun signaling in non-small cell lung cancer. Cell. Signal. 2016;28:1537–1544. doi: 10.1016/j.cellsig.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 164.Sparo M., Delpech G., Batisttelli S., Basualdo J.A. Immunomodulatory properties of cell wall extract from Enterococcus faecalis CECT7121. Braz. J. Infect. Dis. 2014;18:551–555. doi: 10.1016/j.bjid.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Carasi P., Racedo S.M., Jacquot C., Elie A.M., Serradell M.L., Urdaci M.C. Enterococcus durans EP1 a promising anti-inflammatory probiotic able to stimulate sIgA and to increase Faecalibacterium prausnitzii abundance. Front. Immunol. 2017;8:88. doi: 10.3389/fimmu.2017.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Torres D., Barrier M., Bihl F., Quesniaux V.J., Maillet I., Akira S., Ryffel B., Erard F. Toll-like receptor 2 is required for optimal control of Listeria monocytogenes infection. Infect. Immun. 2004;72:2131–2139. doi: 10.1128/IAI.72.4.2131-2139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Avram-Hananel L., Stock J., Parlesak A., Bode C., Schwartz B. E durans strain M4-5 isolated from human colonic flora attenuates intestinal inflammation. Dis. Colon Rectum. 2010;53:1676–1686. doi: 10.1007/DCR.0b013e3181f4b148. [DOI] [PubMed] [Google Scholar]
- 168.Hlivak P., Odraska J., Ferencik M., Ebringer L., Jahnova E., Mikes Z. One-year application of probiotic strain Enterococcus faecium M-74 decreases serum cholesterol levels. Bratisl Lek Listy. 2005;106:67–72. [PubMed] [Google Scholar]
- 169.Yamaguchi T., Miura Y., Matsumoto T. Antimicrobial susceptibility of Enterococcus strains used in clinical practice as probiotics. J. Infect. Chemother. 2013;19:1109–1115. doi: 10.1007/s10156-013-0633-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.