Abstract
Stat4 is activated by the cytokines interleukin 12 and alpha interferon (IFN-α) and plays a significant role in directing development of naïve CD4+ T cells to the Th1 phenotype. Signal transducers and activators of transcription (STAT) proteins undergo phosphorylation on a conserved tyrosine residue, resulting in homo- and heterodimerization, nuclear translocation, and DNA binding. Stat4 can bind to single IFN-γ-activated sites (GASs) as a dimer or bind two tandem GASs as a pair of STAT dimers, or tetramer, stabilized through N-terminal domain (N domain) interactions between dimers. We uncovered an unexpected effect of the Stat4 N domain in controlling the proximal activation of Stat4 by tyrosine phosphorylation at activated receptor complexes. Mutation of the N domain at tryptophan residue W37, predicted to interrupt N domain dimer formation, unexpectedly prevented IFN-α-induced tyrosine phosphorylation of the Stat4 monomer, blocking dimer formation and nuclear translocation. Furthermore, N domains appear to exert private STAT functions, since interchanging the N domains between Stat1 and Stat4 prevented receptor-mediated tyrosine phosphorylation in one case and interrupted STAT-specific gene activation in another. Finally, replacement of the N domain of Stat1 with that of Stat4 abrogated the normal Stat2 dependence of Stat1 phosphorylation, again suggesting the domains are not equivalent. Thus, in addition to its role in STAT tetramerization, the conserved STAT N domain appears to participate in very proximal steps of receptor-mediated ligand-induced tyrosine phosphorylation.
The STAT (signal transducers and activators of transcription) family of transcription factors reside as latent cytoplasmic monomers which are phosphorylated on a conserved tyrosine residue in response to ligand-induced receptor activation (5, 15, 38). Following tyrosine phosphorylation, STAT proteins undergo homo- and heterodimerization via reciprocal interactions involving their conserved SH2 domains, followed by STAT dimer nuclear translocation and participation in transcriptional regulation of various cytokine responsive genes. STAT dimers bind the palindromic gamma interferon (IFN-γ)-activated sequence (GAS) TTCNmGAA, where m equals 3 for all STAT proteins except Stat6 (6, 7, 14, 19). STAT-specific binding site preferences have been identified involving both the central nucleotide core and sequences flanking the core palindrome (10, 25, 33, 34). Furthermore, higher-order interactions between STAT dimers, in which a tetrameric complex of two STAT dimers cooperatively binds two adjacent GAS elements, have been described for Stat1, Stat4, and Stat5 (42, 43, 47). It has been suggested that such tetramer formation is facilitated through interactions between the highly conserved N-terminal domain (N domain), facilitating STAT dimer binding to low-affinity, nonconsensus STAT binding sites (47). Other functions for the N domain have also been suggested, including roles in nuclear localization and binding to CREB-binding protein, or p300 (39, 48).
The specificity of STAT activation by cytokines is in part mediated by the selective interaction of their SH2 domains with distinct tyrosine-containing motifs located within the cytoplasmic domains of specific cytokine receptors. In addition, each STAT protein has private physiologic functions exerted presumably by selective activation of distinct target genes. One important STAT-dependent biologic function involves T helper differentiation. In particular, Stat4 activation plays a significant role in directing development of T helper type 1 (Th1) T cells from naive CD4+ precursors (16, 18, 41). Control of Th1 differentiation is exerted both by the tissue-restricted expression of Stat4 and by the limited activation of Stat4 by only certain cytokines. Stat4 is activated by interleukin 12 (IL-12) (1, 16), a cytokine which potently induces Th1 development (13, 22) through recruitment to a tyrosine-based motif in the IL-12 receptor β2 (IL-12Rβ2) subunit via the Stat4 SH2 domain (26). IL-12 activates Stat4 in all species examined, and the requirement for Stat4 in Th1 development has been confirmed by targeted deletion of Stat4 in mice (18, 41). IFN-α also activates Stat4 and induces Th1 development in human T cells (1, 32), but not mouse T cells (32, 46). Stat4 activation by IFN-α, however, does not involve direct binding to the cytoplasmic domain of the IFN-α receptor (IFN-αR), but instead occurs through an intermediate step (8). First, IFN-α signaling leads to phosphorylation of a conserved tyrosine in the receptor cytoplasmic domain that acts to recruit Stat2, which is subsequently phosphorylated on a conserved tyrosine 690. Stat2 serves as an adapter that binds the SH2 domains of both Stat1 and Stat4. Stat1 binds to tyrosine 690 of Stat2; however, Stat4 binds to a distinct region of Stat2, specifically to the most carboxy-terminal regions of Stat2. In summary, IL-12 and IFN-α each induce Th1 development and activate Stat4, but the Stat4 SH2 domain interaction differs between these receptor pathways.
How Stat4 promotes Th1 development is unclear. Stat4 could directly regulate activity of the IFN-γ gene. Recombinant Stat4 produces a footprint on specific sites within the IFN-γ gene promoter and first intron, sites which are low affinity and nonconsensus. The cooperative interaction of adjacent sites with Stat4 dimers binding as a tetramer via adjacent amino termini was suggested as a mechanism for augmenting IFN-γ gene expression (47), although the requirement for these sites in regulation of the native IFN-γ gene has not been established. Alternately, Stat4 may regulate expression of other signaling molecules or transcription factors that act in Th1 development. For example, Stat4 is required for the Th1-specific expression of the Ets transcription family member ERM (28), although a role for ERM in IFN-γ gene expression has not been demonstrated.
In addressing potential Stat4 tetramer interactions for IFN-γ regulation, we became interested in the role of the Stat4 N domain in mediating STAT dimer-dimer interactions. The structure of the STAT N domain was determined from the isolated N domain from Stat4, which was found to naturally pack as a dimer in the crystal (43). In the Stat4 N domain, composed of eight alpha helices which form a hook-like structure, a conserved tryptophan residue, W37, was shown to be engaged in critical internal polar interactions between interacting helices of reciprocal N domain subunits (43). For functional analysis, the role of this tryptophan was evaluated in Stat1 rather than Stat4; however, mutation of this tryptophan prevented tetramer formation of recombinant Stat1 protein and caused the loss of an IFN-γ augmentation of a synthetic promoter composed of multimerized GAS elements. Furthermore, mutation of this conserved tryptophan in Stat5 was recently shown to prevent the ability of Stat5 to undergo tetramer formation on the adjacent STAT sites present in the IL-2Rα chain promoter (17). So far, the role of this tryptophan-mediated N domain dimerization had not been evaluated for Stat4 nor evaluated in a system where native physiologic responses to Stat4 activation could be observed. To this end, we have carried out a mutational analysis of the Stat4 N domain for IFN-α- and IL-12-induced Stat4 activation using cell lines that lack Stat4 expression and primary T cells derived from Stat4-deficient mice. Surprisingly, our results point to additional roles of the Stat4 N domain beyond mediating tetramer formation on DNA. Our results indicate that the Stat4 N domain also can influence the ability of STAT proteins to undergo successful interactions with cytokine receptor complexes. Importantly, the W37A mutation within the N domain of Stat4 interferes with IFN-α-induced tyrosine phosphorylation of the Stat4 monomer, interrupting Stat4 activation before formation of the Stat4 dimer. This result precludes any conclusions regarding functional tetramer activity based on this mutation for Stat4. Finally, the data suggest that N-domains may be involved in targeting certain STATs to receptors, influencing their suitability as substrates for receptor-dependent kinases.
MATERIALS AND METHODS
Cytokines, antibodies, and reagents.
Recombinant human IFN-α A/D was a gift from U. Gubler (Hoffman-LaRoche, Nutley, N.J.). Recombinant human IFN-γ was a gift from R. Schreiber (Washington University School of Medicine). Polyclonal antisera specific for murine and human Stat1 and Stat4 were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). The peroxidase-conjugated antiphosphotyrosine antibody RC20 was purchased from Transduction Laboratories (Lexington, Ky.). The phycoerythrin (PE)-conjugated anti-human CD4 (anti-hCD4-PE) antibody used for cell sorting was purchased from Caltag (Burlingame, Calif.). DNA restriction and modifying enzymes used in molecular cloning were purchased from New England Biolabs (Beverly, Mass.). Oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, Iowa). The measurement of cytokines by enzyme-linked immunosorbent assay has been described previously (12).
Mutants and retroviral constructs.
We analyzed the interface between the two interacting subunits of the N domain dimer (43) by using LigPlot (45) to identify residues involved in dimer formation. This analysis indicated that residues Q36, W37, T40, and E66 made contacts at the interface, which were also noted in the first description of the N domain structure reported by Vinkemeier et al. (43).
The green fluorescent protein (GFP) retroviral vector GFP-RV has been described previously (31). The complete murine Stat4 cDNA was placed into GFP-RV by using the oligonucleotides (5′→3′) mST4 5′ (CAGCTGAGCCTGGACGGAGAGAGAG) and mST4 3′ (CGACAGTCGACTTCCTTCTTTAAAGTGTCC). Stat1 was cloned likewise by using mST1 5′ (CCGCTCGAGGATGTCACAGTGGTTCG) and mST1 3′ (CCGCTCGAGTGTCGCCAGAGAGAAATTC).
These oligonucleotides were used to generate a PCR product with Vent polymerase and the murine Stat1 and Stat4 cDNA as a template, digested with XhoI and SalI, and cloned into the unique XhoI site of GFP-RV.
For site-directed mutagenesis of Stat4, the QuickChange PCR-based strategy (Stratagene) was used with the following oligonucleotide sets (5′→3′): Q36A top (TCCGGCATCTGCTAGCTGCGTGGATTGAGACTCAAG) and Q36A bot (CTTGAGTCTCAATCCACGCAGCTAGCAGATGCCGGA), W37A top (CTGCTAGCTCAGGCGATTGAGACTCAAGAC) and W37A bot (GTCTTGAGTCTCAATCGCCTGAGCTAGCAG), T40A top (GCTCAGTGGATTGAGGCTCAAGACTGGGAAG) and T40A bot (CTTCCCAGTCTTGAGCCTCAATCCACTGAGC), and E66A top (CTAATACAATTGGATGCACAGTTGGGGCGGGTTT) and E66A bot (AAACCCGCCCCAACTGTGCATCCAATTGTATTAG), and K84A/R85A top (CACAATCTAGCGGCAATTAGAAAAGTTCTTCAGGGC) and K84A/R85A bot (CTAATTGCCGCTAGATTGTGAATCAATAGCAGA).
For mutation of murine Stat1, the following oligonucleotides were used (5′→3′): mSt1W37A top (TGGCCCAGGCGCTGGAAAAGCAAGACTGGGAG) and mSt1W37A bot (TTTTCCAGCGCCTGGGCCAGGTACTGTCTG).
To interchange the N domains of Stat1 and Stat4, we used site-directed mutagenesis by overlap extension. The Stat1 and Stat4 N domains were generated first as Vent polymerase PCR products by using the following oligonucleotides. For the Stat1 N domain, we used MST1 5′ (same as above) and S1S4 A8 bot (CTTCCCTTAAGCAGTTGTAGATGATCATGGACATC). For the Stat4 N domain, we used MST4 5′ (same as above) and S4S1 A8 bot (CTTCCTTCAGACAATTTGAAATTACCACAGCTAC).
The Stat1 and Stat4 carboxy domains were generated with Vent polymerase by using the following oligonucleotides. For the Stat1 carboxy domain, we used S4S1 A8 top (TAATTTCAAATTGTCTGAAGGAAGAAAGGAAG) and MST1 3′ (same as above). For the Stat4 carboxy domain, we used S1S4 A8 top (CATCTACAACTGCTTAAGGGAAGAGAGGAG) and MST4 3′ (same as above).
These PCR products were isolated and mixed in equimolar amounts and reamplified with MST1 5′ and MST4 3′ for the N1-C4 chimera and MST4 5′ and MST1 3′ for the N4-C1 chimera. These PCR products were digested with SalI and XhoI and cloned into the unique XhoI site of GFP-RV. All mutations were confirmed by direct sequencing.
T-cell culture and retroviral transduction.
Methods for activation and passage of DO11.10 T cells have been described previously (11–13). The Phoenix-Ampho or Phoenix-Eco packaging cell lines were transfected with the retroviral vectors described above by calcium phosphate precipitation (29). Twenty-four hours following transfection, the medium was replaced, and the retroviral supernatant was generated by culturing the cells at 32°C for 24 h. The Stat2-deficient U6A (20) and the Stat1-deficient U3A (23) cell lines were maintained in complete Iscove's minimal essential medium as previously described (23). The U3A and U6A cell lines were infected by spinning retroviral culture supernatants containing 4 μg of Polybrene (1,5-dimethyl-1,5-diazaundecamethylene polymethobromide; Sigma) per ml onto monolayers at 1,800 rpm for 30 min, followed by overnight culture. Infected cells were purified by fluorescence-activated cell sorting for GFP and/or hCD4 expression with anti-hCD4-PE secondary antibody. Sorted cells were expanded, were >90% pure, and stably expressed the retroviral marker by postsort analysis. T cells were infected as described previously (29).
Immunoprecipitation and immunoblotting.
Analysis of phosphotyrosine-containing STAT proteins was performed as described previously (16). Briefly, monolayers of fibroblasts were incubated with IFN-α A/D (1,000 U/ml) or IFN-γ (1,000 U/ml) for 20 min at 37°C. Whole-cell lysates were prepared, and STAT molecules were precipitated with specific polyclonal antibodies and protein G-Sepharose (Pharmacia, Piscataway, N.J.). Immunoprecipitates were resolved by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were transferred to nitrocellulose. Phosphotyrosine-containing proteins were detected by blotting with the peroxidase-conjugated RC20 antibody followed by enhanced chemiluminescence with the ECL system (Amersham, Arlington Heights, Ill.). The membranes were then stripped and reprobed with anti-Stat polyclonal antibodies followed by detection with peroxidase-conjugated goat anti-rabbit immunoglobulin (Jackson Immuno Research, West Grove, Pa.).
EMSA.
Electrophoretic mobility shift analysis (EMSA) was performed as follows. Nuclear extracts were prepared from cytokine-treated cells as previously described (16). Binding reactions consisted of 3 μg of nuclear extract, 1 μg of poly(dI-dC) (Pharmacia), 10 mM Tris-Cl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 5% (vol/vol) glycerol, and 2.5 × 104 cpm of Klenow-labeled probe in 20-μl reaction volumes. Reaction mixtures were incubated at room temperature for 30 min. DNA-binding complexes were resolved by nondenaturing 4.5% polyacrylamide gel electrophoresis for 2 h at 150 V followed by autoradiography. The following DNA probes were used in this study: M67SIE (44) (GTCGACATTTCCCGTAAATCGTCGA), Site 2+3 (47) (CGCGAAAATTTTAAGTGAATTTTTTGAGTTTCTTTTAAAATTTT), GAS c+n (24) (GTGCAGTTTCTTCTGAGAAGTACCAGACATTTCTGATAAGAGAG), Eα Y-box (40) (TCGACATTTTTCTGATTGGTTAAAAGTC), GAS c*+n (24) (GTGCAGTTTCTTCGTCTAAGTACCAGACATTTCTGATAAGAGAG [mutated bases are underlined]), and GAS c+n* (24) (GTGCAGTTTCTTCTGAGAAGTACCAGACATGGAGTCGCCGAGAG [mutated bases are underlined]).
RESULTS
Selective N domain mutations interfere with IFN-α-receptor-mediated Stat4 DNA binding.
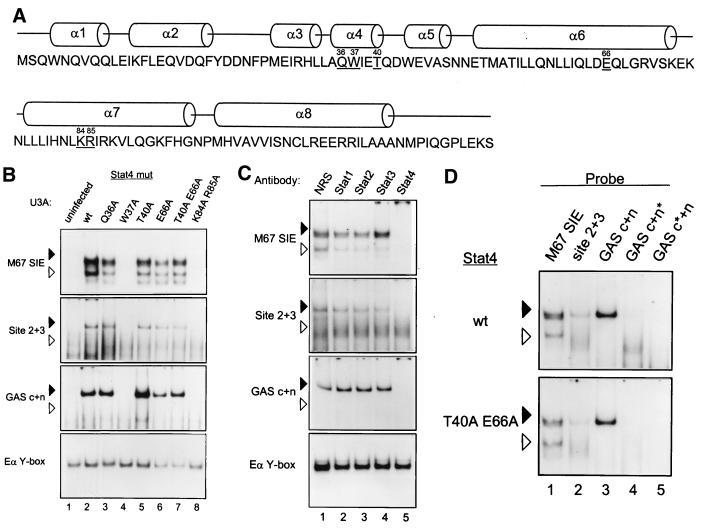
Functional data supporting formation of STAT tetramers was based on mutation of an invariant tryptophan residue, W37, which contacts the paired N domain residues E66 and Q63 (43). However, this mutation was made in Stat1 and Stat5 (17), but not Stat4, which was used for N domain structure analysis. To test functional activity in Stat4, we generated a larger series of mutations within the Stat4 N domain based on the predicted participation of residues in contacts at the interacting faces between the two N domain dimer subunits (Fig. 1A). In addition to the invariant tryptophan residue W37, residues Q36, T40, and E66 make contacts between the interacting N domain alpha helices surrounding W37. Thus, we made a series of single and double residue mutations of these residues in the context of full-length Stat4 and expressed these mutants by retroviral transduction to determine their functional effects on ligand-induced Stat4 activation and DNA binding (Fig. 1B).
FIG. 1.
Selective loss of Stat4 DNA binding from mutations in the alpha 4 and alpha 6 N-domain helices. (A) Location of selected Stat4 N domain mutations. The schematic shows the sequence of the Stat4 N domain with the helical domains shown above the sequence. Underlined residues were selected for mutation to alanine. The numbers above the residue indicate its position relative to the initiation methionine. (B) EMSA activities of selected N domain mutations. U3A cells were left uninfected (uninfected) or infected with retrovirus expressing wild-type Stat4 (wt) or the indicated Stat4 mutants (Stat4 mut). Retrovirally infected cells were purified by cell sorting to >95%. Cells were treated with IFN-α, nuclear extracts were prepared, and EMSA was performed with the indicated probes. Arrowheads are positioned to indicate the relative migration positions between gels: solid arrowheads indicate lower mobility, and open arrowheads indicate higher mobility. Similar data were obtained in three independent experiments. (C) U3A cells infected with retrovirus expressing wild-type Stat4 were treated with IFN-α, nuclear extracts were prepared, and EMSA was performed with the indicated probes in the presence of 1 μg of normal rabbit serum (NRS) or the indicated anti-STAT antibody. (D) U3A cells infected with retrovirus expressing wild-type Stat4 or Stat4 containing a double mutation, T40A E66A, were treated with IFN-α, nuclear extracts were prepared, and EMSA was performed with the indicated wild-type probes (M67SIE, Site 2+3, and GAS c+n) or the GAS c+n probe mutated at either the consensus (GAS c*+n) or nonconsensus (GAS c+n*) site.
Wild-type and Stat4 mutants were expressed in U3A cells, a cell line that lacks expression of Stat1 due to a mutation (23) and which also lacks expression of endogenous Stat4. IFN-α-induced Stat4 activation was first analyzed by EMSA analysis with several STAT-specific DNA binding probes (Fig. 1B). The M67SIE probe contains a single high-affinity consensus STAT site (44) whose STAT binding should be unaffected by N domain-mediated tetramerization. Two additional probes were used to analyze potential tetramer interactions. The second probe, Site 2+3, is from the human IFN-γ first intron and contains two adjacent STAT sites, one of which is a STAT consensus and the second of which is a nonconsensus STAT site (47). The third probe, GAS c+n, from the murine IL-2Rα enhancer, was recently shown to contain two adjacent STAT complexes that could interact with Stat5 as a tetramer (17, 24). Probe EαY-box was used as a positive control for the nuclear extracts.
IFN-α treatment of U3A cells expressing Stat4 induced the formation of EMSA complexes using each of the STAT probes (Fig. 1B, lane 2). These complexes were not present in extracts of IFN-α-treated control U3A cells, which do not express Stat1 or Stat4 (Fig. 1B, lane 1), implying the complexes are specific to Stat4-expressing cells. Furthermore, supershift analysis revealed that the complexes were shifted only by Stat4 antibody (Fig. 1C, lane 5) and not by any other antibody to STATs potentially activated by IFN-α in these cells (Fig. 1C, lanes 1 to 4). Thus, these complexes are composed either exclusively of Stat4 or of Stat4 associated with other factors, but not factors entirely excluding Stat4. Stat4 binds the M67SIE probe as both a more rapidly migrating complex (Fig. 1D, top panel, lane 1), presumably the Stat4 dimer, and a more slowly migrating complex, consistent with Stat4 tetramers described earlier (47). Using the Site 2+3 and the GAS c+n probes (Fig. 1D, top panel, lanes 2 and 3), Stat4 binds only as the more slowly migrating complex, consistent with exclusive tetramer binding to these probes as described previously (17, 47). Mutation of either the nonconsensus (Fig. 1D, top panel, lane 4) or consensus (Fig. 1D, top panel, lane 5) site within the GAS c+n probe abrogated binding of Stat4. This confirms that both STAT sites are necessary for Stat4 binding and strongly suggests that the low-mobility complex represents a tetramer. Identical results were obtained with the Stat4 T40 E66A mutant (Fig. 1D, lower panel, lanes 1 to 5), suggesting that mutation of these residues at the N domain dimer interface is not sufficient to disrupt tetramer formation.
Surprisingly, mutation of the invariant tryptophan to alanine (W37A) caused the loss of all DNA binding (Fig. 1B, lane 4), inhibiting formation of complexes not only with the Site 2+3 and GAS c+n probes, but also with M67SIE, which contains only one high-affinity STAT binding site. In contrast, mutations of other residues at the dimer-dimer contact face (Q36A, T40A, E66A, and T40A E66A) did not block DNA binding, but appeared similar to wild-type Stat4 for each probe tested (Fig. 1B, lanes 3, 5, 6, and 7). Furthermore, mutation of the lysine and arginine residues K84A R85A present on the outer face of the alpha 8 helix of the N domain blocked IFN-α-induced EMSA complexes, similar to the W37A mutation (Fig. 1B, lane 8). Selective interruption of N domain-mediated tetramer formation would predict the loss of tetramer binding to the Site 2+3 and GAS c+n probes, but not necessarily that of dimers to the M67SIE probe. However, we surprisingly observed that mutations W37A and K84A R85A produced a more general inactivation of STAT DNA binding.
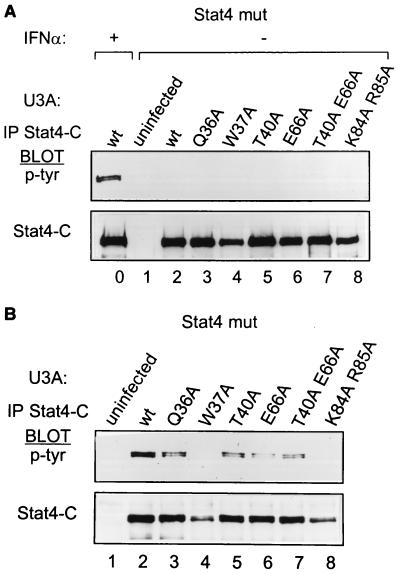
Selective N domain mutations interfere with IFN-α-receptor-mediated Stat4 tyrosine phosphorylation.
Conceivably, the general inhibition of DNA binding of the W37A Stat4 mutation resulted from a more general STAT inactivation, and not the selective loss of N domain-mediated tetramer formation. To test this hypothesis, we reexamined these mutants for early steps in STAT activation, beginning with tyrosine phosphorylation induced by the IFN-α receptor (Fig. 2). Wild-type U3A cells or U3A cells stably expressing wild-type or mutant Stat4 proteins were left untreated (Fig. 2A) or were treated with IFN-α for 20 min (Fig. 2B). Whole-cell extracts were prepared and immunoprecipitated with an antibody to the mouse Stat4 carboxy terminus, and Western blots were developed for phosphotyrosine. Wild-type Stat4 undergoes strong tyrosine phosphorylation in response to IFN-α (Fig. 2A, lane 0), as expected, migrating as a closely spaced doublet, possibly due to phosphoserine content. No Stat4 mutation tested here resulted in constitutive phosphorylation in unstimulated cells (Fig. 2A, lanes 2 to 8). This implies that these mutations did not interfere with potential phosphatase interactions, as was observed for the R31A mutation of Stat1 (35). In addition, Stat4 mutants Q36A, T40A, E66A, and T40A E66A (Fig. 2B, lanes 3, 5, 6, and 7) undergo tyrosine phosphorylation in response to IFN-α treatment, indicating successful receptor recruitment. Minor differences in the level of phosphorylation of these Stat4 mutants are likely due to slight differences in the level of purity of the sorted reconstituted cells, resulting in slight differences in Stat4 mutant input into the immunoprecipitation. Unexpectedly, the W37A Stat4 mutant completely lacked IFN-α-induced tyrosine phosphorylation (Fig. 2B, lane 4), indicating a block at this earliest point in STAT activation. Furthermore, the K84A R85A mutant, inactive in the DNA binding mentioned above, also lacked tyrosine phosphorylation (Fig. 2B, lane 8). These results indicate that the loss of DNA binding by the W37A and K84 R85 Stat4 mutants stems from their inability to undergo IFN-α-induced tyrosine phosphorylation, causing failure to even form STAT dimers.
FIG. 2.
Loss of IFN-α-induced Stat4 tyrosine phosphorylation from selective mutations in the Stat4 N domain. (A) U3A cells prepared with the retroviruses described in the legend to Fig. 1 were treated with IFN-α for 20 min (+) or left untreated (−), and whole-cell extracts were prepared and immunoprecipitated (IP) with 2 μg of antibody specific to the C terminus of murine Stat4. Phosphotyrosine (p-tyr) incorporation was determined with the antiphosphotyrosine antibody RC20, and blots were stripped and probed with an antimurine Stat4 antibody. (B) U3A cells prepared with the retroviruses described in the legend to Fig. 1 were treated with IFN-α for 20 min, and whole-cell extracts were prepared. Phosphotyrosine incorporation into Stat4 was determined as described for panel A.
N domains of Stat1 and Stat4 are not functionally interchangeable.
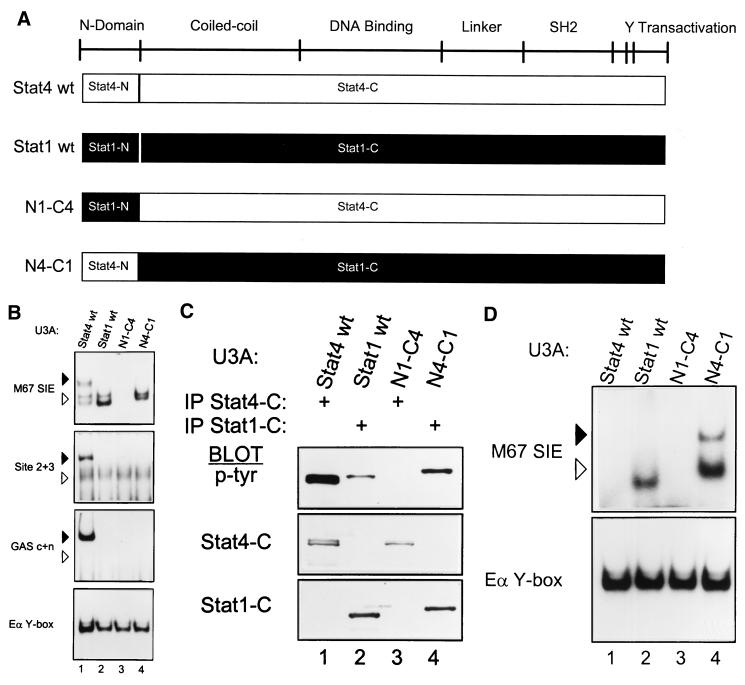
Reliable application of functional results of N domain mutations in Stat1 (43) or Stat5 (17) to Stat4 presumes some functional equivalence between the N domains of these proteins. To directly test this, we interchanged the N domains of Stat1 and Stat4, either placing the Stat4 N domain into Stat1 (Fig. 3A, N4-C1) or placing the Stat1 N domain into Stat4 (Fig. 3A, N1-C4). The chimeric splice was made in the alpha 8 helix of the N domain, since this region is highly conserved between Stat1 and Stat4 and is positioned on the opposite side of the structure from the dimer interaction face (43).
FIG. 3.
Functional noninterchangeability of Stat1 and Stat4 N domains. (A) Schematic of STAT structure showing the general positions of several domains (3). Below are diagrams of wild-type Stat4 (Stat4 wt) and Stat1 (Stat1 wt) and of two chimeras. In the N1-C4 chimera, the N domain of Stat1 (Stat1-N) is placed onto the Stat4 carboxy terminus (Stat4-C); in the N4-C1 chimera, the N domain of Stat4 (Stat4-N) is placed onto the Stat1 carboxy terminus (Stat1-C). cDNAs encoding these chimeras were placed into the retrovirus used for expression of the Stat4 mutants as described in Fig. 1. (B) U3A cells were infected with retroviruses expressing wild-type Stat4 (Stat4 wt) or Stat1 (Stat1 wt) and the N1-C4 and N4-C1 chimeras as indicated. Retrovirally infected cells were purified by cell sorting, treated with IFN-α, and nuclear extracts were prepared and analyzed by EMSA with the indicated probes as described in the legend to Fig. 1B. Arrowheads are positioned to indicate the relative migration positions between gels: solid arrowheads indicate lower mobility, and open arrowheads indicate higher mobility. Similar data were obtained in two other independent experiments. (C) The cells described for panel B were treated with IFN-α. Whole-cell extracts were prepared and immunoprecipitated with antibody to the Stat4 carboxy terminus (IP Stat4-C) or with antibody to the Stat1 carboxy terminus (IP Stat1-C) as indicated, and Western analysis was done for phosphotyrosine incorporation with the antiphosphotyrosine antibody RC20 (p-Tyr) as described above. Blots were stripped and reprobed with antimurine Stat4 antibody (Stat4-C) and antimurine Stat1 antibody (Stat1-C). (D) The cells described in the legend to panel B were treated with IFN-γ (1,000 U/ml) for 20 min. Nuclear extracts were prepared and analyzed by EMSA with the indicated probes.
Wild-type Stat1, Stat4, and both chimeric STAT proteins were stably expressed in U3A cells and activated by IFN-α, and their DNA binding was analyzed by EMSA with probes described in Fig. 1. Wild-type Stat1 and Stat4 bind with clearly distinguishable characteristics to these three probes. First, with the M67SIE probe, Stat4 forms complexes of both low and high mobility (Fig. 3B, upper panel, lane 1), whereas Stat1 forms only complexes of high mobility (lane 2). The high-mobility Stat4 complexes have been shown to represent dimers, and the low-mobility complexes represent tetramers (43). Furthermore, with the Site 2+3 and GAS c+n probes, Stat4 forms only low-mobility complexes, consistent with exclusive tetramer formation, whereas Stat1 does not form complexes at all with these probes (Fig. 3B). Notably, replacement of the Stat1 N domain with the Stat4 N domain (N4-C1) does not generate the lower-mobility complexes seen with wild-type Stat4 (Fig. 3B, lane 4), as would be predicted by strict interchangeability of the N domains. Rather, using the M67SIE probe, the N4-C1 chimera forms only high-mobility complexes migrating as a doublet, perhaps due to heterodimer formation (Fig. 3B, lane 4), and fails to form any complexes with the Site 2+3 or GAS c+n probes. Identical results were obtained when the chimeric junction was in the alpha 7 helix of the N domain (data not shown).
Further analysis of the N1-C4 chimera suggests a role for the Stat4 N domain in proximal receptor interactions regulating tyrosine phosphorylation of STATs, as suggested in Fig. 2. Placement of the Stat1 N domain into Stat4 (N1-C4) caused a complete loss of DNA binding with all of these probes (Fig. 3B, lane 3), potentially caused by loss of tyrosine phosphorylation similar to the W37A mutation. To test this, we examined both chimeras for IFN-α-induced tyrosine phosphorylation (Fig. 3C). As positive controls, Stat1 and Stat4 both undergo IFN-α-induced tyrosine phosphorylation as expected (Fig. 3C, lanes 1 and 2). In addition, N4-C1 undergoes tyrosine phosphorylation, consistent with its ability to bind the M67SIE probe (Fig. 3C, lane 4). In contrast, however, the N1-C4 chimera does not undergo IFN-α-induced tyrosine phosphorylation (Fig. 3C, lane 3). As further positive controls, we carried out direct Western analysis to ensure proper protein expression. The N1-C4 chimera is detected at the expected position by antisera to the C terminus of Stat4, and the N4-C1 chimera is detected by antisera to the C terminus of Stat1 (Fig. 3C, middle and lower panels). Thus, the N1-C4 chimera is properly expressed at the level of protein, but fails to become tyrosine phosphorylated, explaining its inability to bind all EMSA probes (Fig. 3B).
We next tested whether the Stat4 and Stat1 chimeras could be activated by IFN-γ (Fig. 3D) as measured by EMSA with probe M67. As expected, wild-type Stat4 was not activated by IFN-γ, whereas Stat1 was activated and bound to M67 as a high-mobility complex (Fig. 3D, lanes 1 and 2). Similar to IFN-α stimulation, the N1-C4 chimera was not activated by IFN-γ stimulation, likely due to the inability of the Stat4 SH2 domain to interact with the IFN-γ receptor. Surprisingly, the N4-C1 chimera produced both the low-mobility and high-mobility complexes on M67. This provides the first functional evidence that the Stat4 N domain can participate in tetramer formation in the context of N4-C1 homodimers induced by IFN-γ, but perhaps not in the context of N4-C1–Stat2–p48 complexes induced by IFN-α.
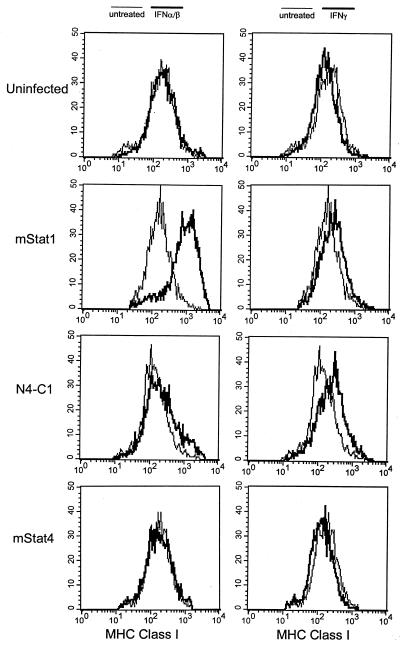
Transfer of the Stat4 N domain onto Stat1 (N4-C1) produced EMSA complexes with mobility similar to that of wild-type Stat1 following IFN-α stimulation (Fig. 3B, compare lanes 2 and 4), but produced additional complexes with low mobility similar to that of wild-type Stat4 following IFN-γ stimulation (Fig. 3D, compare lanes 2 and 4). To test for potential functional changes in the N4-C1 chimera, we analyzed IFN-α- and IFN-γ-induced expression of major histocompatibility complex (MHC) class I in U3A cells transfected to stably express Stat1, Stat4, or the N4-C1 chimera (Fig. 4). Since U3A cells lack Stat1, uninfected cells show no MHC class I induction by IFN-α or IFN-γ, as expected. Expression of Stat1, but not Stat4, restores IFN-α- and IFN-γ-induced expression of MHC class I, showing that this effect is specific to Stat1. Surprisingly, expression of the N4-C1 chimera in U3A cells failed to restore IFN-α-induced MHC class I expression (Fig. 4, left panel), despite the ability of this chimera to form EMSA complexes similar to those of wild-type Stat1. However, the N4C1 chimera was fully functional for IFN-γ-induced MHC class I induction (Fig. 4, right panel). This result suggests that the N domains of Stat1 and Stat4 are not fully functionally interchangeable for gene-specific transactivation events.
FIG. 4.
Functional nonequivalence of the Stat4 and Stat1 N domains. (Left panels) Uninfected U3A cells (uninfected) or U3A cells expressing murine Stat1 (mStat1), the N4-C1 chimeric Stat protein (N4-C1), or wild-type Stat4 (mStat4) as described in the legend to Fig. 3 were either left untreated (light lines) or were treated with IFN-α (bold lines) for 48 h. (Right panels) Cells were left untreated (light lines) or treated with IFN-γ (bold lines) for 48 h. Results were analyzed by flow cytometry with a Becton Dickinson FACScan for surface expression of MHC class I by using a PE-conjugated anti-class I-specific antibody, clone Tu149 (Caltag).
Inhibition of tyrosine phosphatase does not rescue phosphorylation-defective mutants.
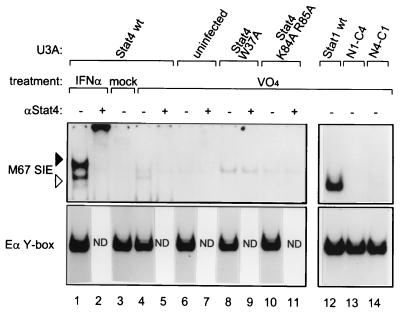
We wondered whether tyrosine phosphorylation of the W37A and K84A R85A Stat4 mutants and the N1-C4 chimera could be achieved independently of receptor signaling. Therefore, we treated U3A cells expressing wild-type or mutant STATs with sodium pervanadate to inhibit phosphatases (9). Nuclear extracts were prepared and analyzed by EMSA (Fig. 5). Pervanadate treatment of U3A cells containing wild-type Stat4 resulted in formation of a complex with mobility equal to that of the high-mobility complex generated by IFN-α treatment (Fig. 5, compare lanes 1 and 4), but did not generate the low-mobility tetramer complex. A complex of intermediate mobility was also observed, but it was not supershifted with an antibody to Stat4 (Fig. 5, lanes 4 and 5) and was present in uninfected U3A cells (Fig. 5, lanes 6 and 7), indicating that it does not contain Stat4. Neither the W37A nor the K84A R85A mutations of Stat4 produced a Stat4-containing complex after pervanadate treatment (Fig. 5, lanes 8 to 11). In addition, pervanadate treatment of U3A cells containing wild-type Stat1 produced a high-mobility complex (Fig. 5, lane 12), while treatment of cells containing the N1-C4 chimera produced no complexes (Fig. 5, lane 13), and treatment of cells containing the N4-C1 chimera produced a very weak high-mobility complex (Fig. 5, lane 14). Thus, the inability of various mutants to be activated by receptor ligation cannot be overcome by inhibition of phosphatase, suggesting a primary defect in their ability to become tyrosine phosphorylated at all, perhaps due to a lack of recognition by an appropriate kinase.
FIG. 5.
Inhibition of tyrosine phosphatase does not rescue phosphorylation-defective mutants. U3A cells either uninfected or expressing the indicated proteins were treated with IFN-α for 20 min (IFN-α), 500 μM H2O2 (mock), or 500 μM H2O2–500 μM NaVO4 (VO4) for 30 min as described previously (9). Nuclear extracts were prepared and analyzed by EMSA with the indicated probes in the absence (−) or presence (+) of antibody to the Stat4 carboxy terminus. wt, wild type; ND, not determined.
N domain mutations influence IL-12R-induced Stat4 tyrosine phosphorylation.
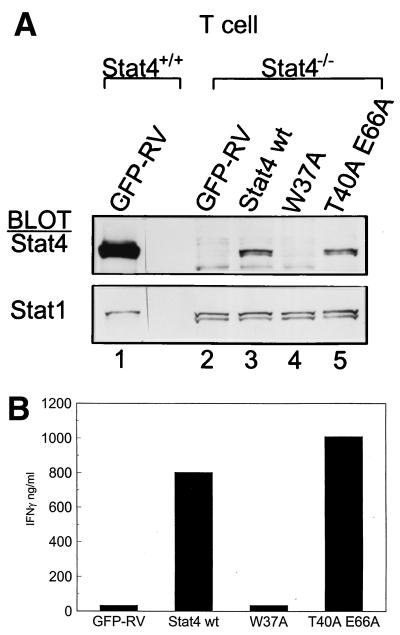
Since the W37A mutation of the Stat4 N-domain blocked tyrosine phosphorylation induced by IFN-α, we wondered whether this mutation would also block activation induced by IL-12. For this, we used Stat4-deficient T cells, which express IL-12Rs, but lack endogenous Stat4 (29, 41). Wild-type and Stat4-deficient DO11.10 T-cell receptor (TCR) transgenic cells were infected with control retrovirus or retrovirus expressing wild-type, W37A, or T40A E66A Stat4 proteins, purified by cell sorting, and analyzed for IL-12-induced Stat4 nuclear translocation (Fig. 6A). Since relatively small numbers of cells can be obtained in the infections of primary T cells, we used nuclear translocation as a surrogate assay for Stat4 tyrosine phosphorylation. As a further control, we examined nuclear translocation of Stat1, which is also activated by IL-12 (16). IL-12-treated wild-type T cells showed strong nuclear translocation of Stat4 (Fig. 6A, lane 1). Stat4-deficient T cells infected with control retrovirus (GFP-RV) showed only Stat1 nuclear location, but no Stat4 nuclear localization (Fig. 6A, lane 2), as expected. Stat4-deficient T cells infected with wild-type Stat4 showed IL-12-induced Stat4 nuclear translocation (Fig. 6A, lane 3), as expected, as did the T40A E66A mutant (lane 5). In contrast, the W37A Stat4 mutant showed no IL-12-induced nuclear translocation, implying an inability to undergo tyrosine phosphorylation by IL-12. Thus, these data suggest that the W37 mutation in the Stat4 N domain also disrupts activation induced by IL-12 signaling, as it did for IFN-α signaling.
FIG. 6.
Loss of IL-12-induced Stat4 nuclear translocation and Stat4-dependent IFN-γ production from mutations in the Stat4 N domain. (A) DO11.10 TCR transgenic T cells from wild-type (Stat4+/+) or Stat4-deficient (Stat4−/−) mice were activated as described previously (29) and infected with empty GFP-RV retrovirus (GFP-RV) or retrovirus expressing wild-type Stat4 (Stat4 wt) or W37A or T40A E66A N domain mutations as indicated. Cells were grown in the presence of 10 U of IL-12 per ml and expanded for 7 days, and the infected T cells were purified by positive cell sorting for expression of GFP and murine CD4. Purified T cells were expanded by restimulation with irradiated BALB/c splenocytes and ovalbumin and treated with IL-12, nuclear extracts were prepared, and direct Western analysis was done for Stat4 and Stat1. Similar results were obtained in two independent experiments. (B) T cells derived and purified as for panel A were analyzed for the production of IFN-γ in response to IL-12–IL-18 combined treatment. T cells were placed at 106 cells per well of a 48-well plate in 1 ml of medium and treated with IL-12 (10 U/ml) and IL-18 (100 U/ml) for 48 h, supernatants were harvested, and IFN-γ was measured by enzyme-linked immunosorbent assay.
IFN-γ production induced by IL-12 or IL-18 in CD4+ T cells is a Stat4-dependent activity (2). N domain interactions between Stat4 dimers bound to adjacent low-affinity, nonconsensus sites within the IFN-γ gene promoter and first intron were suggested as a mechanism for augmenting IFN-γ gene expression (47). Thus, to check for functional consequences of the W37A and T40A E66A mutations in IL-12-induced Stat4 activation, we analyzed these T cells for IFN-γ production (Fig. 6B). Stably infected T cells expressing the constructs shown in Fig. 5A were harvested and treated with IL-12 and IL-18 for 48 h, and supernatants were analyzed for IFN-γ production by ELISA (Fig. 6B). Stat4-deficient T cells infected with empty retrovirus failed to produce IFN-γ in response to IL-12 and IL-18 treatment (Fig. 6B), as expected, since Stat4 is required for Th1 development (18, 41). Reconstitution of Stat4-deficient T cells with wild-type Stat4 produced significantly higher IFN-γ at approximately 800 ng/ml following IL-12 and IL-18 treatment. In contrast, the W37A Stat4 mutant failed to restore IFN-γ production, whereas the T40 E66 Stat4 mutation reconstituted IFN-γ production to levels similar to those of wild-type Stat4 (Fig. 6B). Thus, the T40 E66 Stat4 mutation, although predicted by the crystal structure to be likely to disrupt N domain interactions, nonetheless functions normally for IFN-γ production. Thus, the W37A mutation in the Stat4 N domain generated a functional deficit in a Stat4-dependent response to IL-12 signaling, consistent with the observed loss of IL-12-induced nuclear translocation.
The Stat4 N domain allows Stat2-independent Stat1 activation.
The W37A and K84A R85A mutations in the Stat4 N domain, as well as a Stat4 N domain-Stat1 chimera, are unable to become phosphorylated in response to IFN-α (Fig. 2B and 3C). One possible explanation is that the Stat4 N domain is involved in the presentation of Stat4 to the receptor before ligand-mediated activation. In fact, a similar role for the Stat2 N domain has been suggested (21) based on the following observations. In U6A cells, phosphorylation of Stat1 is very weak, but can be restored by reintroducing Stat2. Furthermore, replacement of the N-terminal third of Stat1 with that of Stat2 allowed Stat1 to preassociate with the IFN-αR2 chain and become phosphorylated efficiently (21, 37). Therefore, we wondered if the Stat4 N domain could perform a similar function to the Stat2 N domain in allowing Stat1 to preassociate with the IFN-αR2 and become phosphorylated.
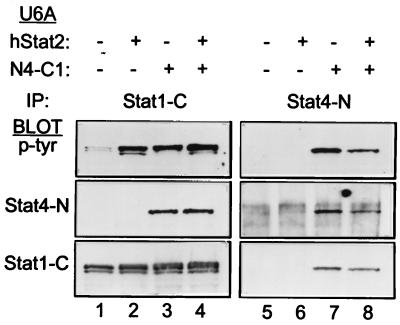
To test this, we expressed Stat1 and the N4-C1 chimera into U6A cells lacking Stat2 or into U6A cells reconstituted with Stat2 (Fig. 7). We then examined the requirement for Stat2 in Stat1 activation and in activation of the N4-C1 chimera. Cells were treated with IFN-α, and phosphotyrosine incorporation was assessed by immunoprecipitating cell lysates with antisera specific to the Stat1 carboxy terminus (Fig. 7, lanes 1 to 4) or the Stat4 amino terminus (lanes 5 to 8). IFN-α-induced Stat1 phosphorylation was very weak in U6A cells lacking Stat2 (Fig. 7, lane 1), as expected, but was significantly increased upon coexpression of Stat2 (Fig. 7, lane 2), in agreement with earlier reports (20, 30). In contrast, the N4-C1 chimera showed tyrosine phosphorylation that was independent of Stat2 expression (Fig. 7, lanes 7 and 8). As controls, direct Western blots were done to show the expected expression of the Stat4 N domain under only those conditions in which the N4-C1 chimera was expressed (Fig. 7, middle panels) and to show the expression of the Stat1 C terminus under the conditions expected (lower panels). Thus, the N domain of Stat4 conferred to Stat1 an ability to undergo IFN-α-induced tyrosine phosphorylation in a manner independent of Stat2. Since Stat1 and the N4-C1 chimera have identical SH2 domains, this suggests that the Stat4 N domain may provide Stat1 with the ability to preassociate with the receptor, a requirement normally provided by Stat2.
FIG. 7.
N domain involvement in Stat2-dependent Stat1 activation. U6A cells, which lack Stat2 expression, were infected (+) with bicistronic retrovirus expressing human Stat2 (hStat2) and human CD4 with truncation of the cytoplasmic domain, infected (+) with a bicistronic retrovirus expressing the N4-C1 STAT chimera (Fig. 3A) and GFP, infected with both viruses, or left uninfected (−) as indicated. Cells were purified to >95% by sorting of surface expression of the human CD4 marker for the hStat2 construct with an anti-hCD4-PE-conjugated antibody or by sorting for GFP expression for the N4-C1 STAT chimera, and doubly infected U6A cells were purified by sorting coexpression of both markers. Purified cells were treated with IFN-α for 20 min, and whole-cell extracts were prepared and immunoprecipitated with an antibody specific to the carboxy terminus of Stat1 (IP: Stat1-C) or the N terminus of Stat4 (IP: Stat4-N). Western analysis was done for phosphotyrosine with RC20 (p-tyr). Blots were stripped and reprobed with an antibody specific for the N terminus of Stat4 (Stat4-N) or carboxy terminus of Stat1 (Stat1-C).
W37A mutation of the Stat1 N domain interferes with IFN-α-mediated Stat1 DNA binding.
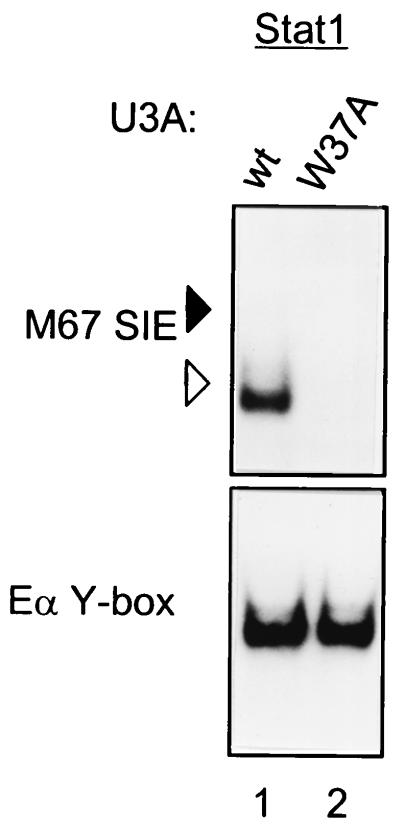
Previously, the importance of the invariant residue W37 in N domain dimers seen for Stat4 was tested by gel mobility shift assay with a W37A mutation of Stat1 purified from baculovirus-infected insect cells and in vitro phosphorylated by immunoprecipitated activated epidermal growth factor receptor kinase (43). To test the effect on DNA binding of this Stat1 mutation in a more physiologic system, we reconstituted U3A cells with the W37A Stat1 mutation and determined IFN-α-induced Stat1 activation by EMSA (Fig. 8). Wild-type Stat1 binds to probe M67SIE as a high-mobility complex, consistent with exclusive dimer binding to this single site probe (Fig. 8, lane 1), while the W37A mutant does not produce an EMSA complex (Fig. 8, lane 2). Since Stat1 appears to bind only as a dimer to this probe, the lack of binding of the mutant is not due to loss of the tetramer, but rather an earlier step in Stat1 activation. Thus, this role for the N domain may be conserved between various STATs.
FIG. 8.
W37A mutation of Stat1 causes loss of IFN-α-induced DNA binding. U3A cells were infected with wild-type Stat1 (wt) or Stat1 with a W37A mutation. Retrovirally infected cells were purified by cell sorting and treated with IFN-α for 20 min. Nuclear extracts were prepared, and EMSA was performed with the indicated probes. Arrowheads are positioned to indicate the relative migration positions between gels: solid arrowheads indicate lower mobility and open arrowheads indicate higher mobility.
DISCUSSION
To test whether N domain-mediated tetramerization is important for the authentic physiologic actions of Stat4, we mutated residues in the Stat4 N domain that were structurally implicated in dimer formation and tested their activities by expression in Stat4-deficient cells. We first generated a mutation in Stat4 corresponding to one previously analyzed for Stat1 (43) or Stat5 (17) and interpreted as disrupting Stat1 tetramer formation. In particular, mutation of the invariant tryptophan residue W37 in Stat1 was found to block IFN-γ-induced activity of a synthetic luciferase reporter containing tandem STAT-binding sites as an enhancer (43). The same mutation in Stat5a and Stat5b led to significant reduction in IL-2-induced activity of a luciferase reporter driven by the PRRIII enhancer of the IL-2Rα chain (17). In addition to mutating W37 of Stat4, we also mutated E66, which makes an important water-mediated contact with W37 in the N domain dimer interface and is spatially related to the K70A mutation reported by John et al. (17) for Stat5. We also mutated residue Q36, which makes reciprocal contacts with Q36 in the adjoining dimer, and T40 which contacts Q41, Q67, and R70 of the paired N domain dimer. Finally, we mutated the pair of basic residues K84 and R85 based on the slight similarity of this region to nuclear localization sequences (4), not expecting this mutation to influence tetramer formation. Surprisingly, the analysis of these mutations in Stat4 revealed that the Stat4 N domain likely plays an additional role beyond tetramer formation, acting in receptor-proximal interactions of Stat4 as a substrate for receptor-associated kinases.
Since tetramer formation can be analyzed by EMSA (17, 47), we began by comparing the EMSA complexes generated by wild-type and mutant Stat4 proteins. U3A cells were used, since they lack expression of Stat1 by a mutation (23) and do not express endogenous Stat4 (8). Wild-type Stat4 generated complexes on a single high-affinity STAT site probe, M67SIE, whose mobilities were consistent with the formation of both dimers and tetramers, as previously described (17, 47). In contrast, with probes Site 2+3 and GAS c+n, each containing adjacent STAT sites, wild-type Stat4 generated only the complex whose mobility was consistent with the formation of a tetramer. Several mutations that were predicted to disrupt potential tetramer interactions based on the crystal structure of the Stat4 N domain dimer had no effect on binding to any of the three probes used in this study. For the W37A mutation, however, we observed loss of binding to the Site 2+3 and GAS c+n probes, consistent with the loss of tetramer formation. However, we also unexpectedly observed loss of both the lower- and higher-mobility complexes using the M67SIE probe, indicating loss of dimer formation as well.
Therefore, as a control, we examined the capacity for these Stat4 mutants to undergo IFN-α-induced tyrosine phosphorylation (Fig. 2). Surprisingly, the W37A Stat4 mutation had lost IFN-α-induced phosphotyrosine incorporation, suggesting a much earlier defect in activation caused by this mutant, but nonetheless explaining the loss of both tetramer and dimer formation. Mutations of the other residues within the dimer interface did not cause this loss of tyrosine phosphorylation. Thus, the W37A mutation of the Stat4 N domain prevented the STAT monomer from becoming a suitable substrate for the receptor-associated kinase during STAT activation.
Leonard and colleagues examined similar mutations in the N domain for Stat5 regulation of the IL-2Rα chain enhancer (17). In Stat5, the W37A and K70A mutations, as well as the W37A K70A double mutation, suggested a role for these residues in IL-2 induction via Stat5 activation of enhancer activity. The mutations were found to allow nuclear localization of Stat5 and thus were presumably phosphorylated, since phosphorylation is thought to be a prerequisite for STAT nuclear localization. However, that study did not explicitly examine phosphorylation of Stat5 mutants by Western blotting. In contrast, the W37A mutation in Stat4 blocked activation induced by both IL-12 and IFN-α.
We were surprised to find that mutation of Stat4 predicted by the crystal structure to disrupt N domain dimerization (i.e., T40A E66A) still bound DNA as a tetramer and was fully functional for Stat4-dependent IFN-γ production in developing T cells. This indicates that other residues may contribute to this apparently quite stable Stat4 tetramer. Interestingly, DNA binding site selection of dimeric and tetrameric Stat5 revealed that the most favorable inter-GAS spacing was 6 bp for Stat5a (36). This spacing positions the two Stat5 dimers on opposite sides of the DNA and allowed for additional stabilizing interactions between Stat5a core sequences according to a model based on the Stat1 structure. In our double site probes, the spacing of 11 bp between GAS elements would be predicted to position Stat4 dimers on the same side of the DNA and seemingly too far apart to allow interaction between core sequences. Optimal sites for Stat4 tetramer binding have not been determined, nor are examples of Stat4-dependent promoters currently known in which to carefully examine these issues. In essence, the role of the N domain for STAT-tetramer formation is still uncertain, and tetramer formation may involve interactions between other regions of the STAT molecule.
We noticed that in the alpha 7 helix of the Stat4 N domain, outside the dimer interaction face, a series of residues, KRIRKVL, exhibited some similarity to a monopartite nuclear localization signal (4). This region of the N domain is not well conserved between the STATs, and so may not be the location of elements that are generally involved in STAT nuclear translocation. However, we found that mutating two basic residues in this region, K84A and R85A, led to an unpredicted loss of tyrosine phosphorylation in response to IFN-α. A recent study (39) showed that deletion of the N domain of Stat1, as well as swapping the Stat2 N domain into Stat1 (CH2/1), resulted in defective nuclear translocation not attributed to lack of phosphorylation. Notably, in that study, the mutated STAT proteins were activated to undergo tyrosine phosphorylation by using an artificial receptor consisting of the extracellular domain of the human granulocyte colony-stimulating factor receptor fused to the transmembrane and intracellular domains of the IFN-αR α chain, fused to the Stat1 recruitment site (“y” motif). The use of this artificial receptor, which is very different in structure from the native IFN-αR used in our study, could potentially produce differences in requirements for receptor-induced tyrosine phosphorylation. Our chimeric protein, N4-C1, appears to be functional for nuclear translocation, since our EMSA studies were all performed with nuclear extracts. However, using the native IFN-αR, we see differences in tyrosine phosphorylation in the simple swap of N domains between Stat1 and Stat4, suggesting that there may be subtle interactions determining whether a molecule is a suitable substrate for the receptor-associated kinase.
We observed that in nuclear extracts from cytokine-activated cells, Stat4 binds to a high-affinity STAT site, M67SIE, as both low- and high-mobility complexes, whereas Stat1 binds only as a high-mobility complex. Furthermore, two probes containing adjacent STAT sites bind Stat4, but not Stat1. Since the DNA binding preference of Stat4 determined by random binding site selection is identical to that for Stat1 (47), we wondered whether the difference in probe binding could be explained by tetramer formation in the case of Stat4 and lack of tetramerization for Stat1. We found that a chimera containing the Stat4 N domain and Stat1 carboxy domain (N4-C1) still cannot bind to adjacent site probes, nor produces the low-mobility complex on M67SIE when activated by IFN-α treatment. However the same chimera does produce tetrameric complexes when activated by IFN-γ treatment. This suggests that tetramerization is a possible explanation for the difference in Stat4 and Stat1 binding to these probes at least for homodimer formation in the context of IFN-γ stimulation. Interestingly, we found that a chimera containing the Stat1 N domain and Stat4 carboxy domain (N1-C4) was unable to be phosphorylated after IFN-α or IFN-γ treatment, demonstrating that the N domains of STATs are not functionally interchangeable.
Since IL-12 as well as IFN-α activates Stat4, we asked whether N domain mutations would respond similarly to both stimuli. For the IL-12R complex, the Stat4 SH2 domain is directly recruited to a tyrosine motif Y800 (YLPSNID) in the IL-12Rβ2 subunit cytoplasmic domain (26). For the human IFN-αR complex, the Stat4 SH2 domain binds to activated Stat2 rather than to the receptor itself. We found that the W37A mutation of Stat4 blocked activation by both IL-12 and IFN-α, suggesting that although the SH2 domain interactions with either IFN-α and IL-12R complexes are different, there may be a preceding step in Stat4 activation common to both receptor pathways. Stark and colleagues have shown that Stat1 and Stat2 may be preassociated with the R2 subunit of the IFN-αR (IFNAR2) through the Stat2 amino terminus (21). No evidence for preassociation of Stat4 with either the IFN-α or IL-12R complex has yet been published. Our data showing that phosphorylation of the N4-C1 chimera is independent of Stat2 suggest that the Stat4 N domain may allow the preassociation of Stat1 with the IFNAR2, a role normally provided by Stat2. Interestingly, Ndubuisi et al. (27) found that the cytoplasmic pool of Stat3 resides in high-molecular-mass complexes termed “statosomes,” in which Stat3 may associate with the chaperone GRP58, rather than residing as a monomer. Conceivably, the Stat4 N domain interacts directly with cytokine receptors before activation, similar to Stat2, or alternately, adapter proteins may interact with the Stat4 N domain to regulate either preassociation with cytokine receptors or its participation in statosomes.
In summary, this study demonstrates that mutations in the Stat4 N domain can interrupt the proximal process of tyrosine phosphorylation by activated receptors. For Stat4, the W37A mutation prevents activation by interfering with the earliest step, before the formation of STAT dimers, preventing an in vivo analysis of tetramer formation. Studies of N domain interchange further indicate that N domains may not be freely interchangeable between different STATs for physiologic activation by cytokine receptors. Together, these results suggest that private activities mediated by some STAT N domains could influence association with the receptor or the ability to become a substrate for receptor-associated kinases.
ACKNOWLEDGMENTS
We thank George Stark and Stewart Leung for cell lines, Dominic Fenoglio for help with cell sorting, Kathy Frederick for animal husbandry, and Michael Martin Murphy for technical help.
This work was supported by AI34580 and AI/DK39676 and a grant from the Juvenile Diabetes Foundation. K.M.M. is an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bacon C M, Petricoin E F, Ortaldo J R, Rees R C, Larner A C, Johnston J A, O'Shea J J. Interleukin 12 induces tyrosine phosphorylation and activation of STAT4 in human lymphocytes. Proc Natl Acad Sci USA. 1995;92:7307–7311. doi: 10.1073/pnas.92.16.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter L L, Murphy K M. Lineage-specific requirement for signal transducer and activator of transcription (Stat)4 in interferon gamma production from CD4(+) versus CD8(+) T cells. J Exp Med. 1999;189:1355–1360. doi: 10.1084/jem.189.8.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 4.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 5.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 6.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 7.Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 8.Farrar J D, Smith J D, Murphy T L, Murphy K M. Recruitment of Stat4 to the human interferon-alpha/beta receptor requires activated Stat2. J Biol Chem. 2000;275:2693–2697. doi: 10.1074/jbc.275.4.2693. [DOI] [PubMed] [Google Scholar]
- 9.Haque S J, Flati V, Deb A, Williams B R. Roles of protein-tyrosine phosphatases in Stat1 alpha-mediated cell signaling. J Biol Chem. 1995;270:25709–25714. doi: 10.1074/jbc.270.43.25709. [DOI] [PubMed] [Google Scholar]
- 10.Horvath C M, Wen Z, Darnell J E., Jr A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh C S, Heimberger A B, Gold J S, O'Garra A, Murphy K M. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an alpha beta T-cell-receptor transgenic system. Proc Natl Acad Sci USA. 1992;89:6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsieh C S, Macatonia S E, O'Garra A, Murphy K M. Pathogen-induced Th1 phenotype development in CD4+ alpha beta-TCR transgenic T cells is macrophage dependent. Int Immunol. 1993;5:371–382. doi: 10.1093/intimm/5.4.371. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh C S, Macatonia S E, Tripp C S, Wolf S F, O'Garra A, Murphy K M. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 14.Ihle J N. STATs: signal transducers and activators of transcription. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 15.Ihle J N, Kerr I M. Jaks and Stats in signaling by the cytokine receptor superfamily. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 16.Jacobson N G, Szabo S J, Weber-Nordt R M, Zhong Z, Schreiber R D, Darnell J E, Jr, Murphy K M. Interleukin 12 signaling in T helper type 1 (Th1) cells involves tyrosine phosphorylation of signal transducer and activator of transcription (Stat)3 and Stat4. J Exp Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John S, Vinkemeier U, Soldaini E, Darnell J E, Jr, Leonard W J. The significance of tetramerization in promoter recruitment by Stat5. Mol Cell Biol. 1999;19:1910–1918. doi: 10.1128/mcb.19.3.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan M H, Sun Y L, Hoey T, Grusby M J. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 19.Leonard W J, O'Shea J J. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 20.Leung S, Qureshi S A, Kerr I M, Darnell J E, Jr, Stark G R. Role of STAT2 in the alpha interferon signaling pathway. Mol Cell Biol. 1995;15:1312–1317. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Leung S, Kerr I M, Stark G R. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol Cell Biol. 1997;17:2048–2056. doi: 10.1128/mcb.17.4.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manetti R, Parronchi P, Giudizi M G, Piccinni M P, Maggi E, Trinchieri G, Romagnani S. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J Exp Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer W K, Reichenbach P, Schindler U, Soldaini E, Nabholz M. Interaction of STAT5 dimers on two low affinity binding sites mediates interleukin 2 (IL-2) stimulation of IL-2 receptor alpha gene transcription. J Biol Chem. 1997;272:31821–31828. doi: 10.1074/jbc.272.50.31821. [DOI] [PubMed] [Google Scholar]
- 25.Mikita T, Campbell D, Wu P, Williamson K, Schindler U. Requirements for interleukin-4-induced gene expression and functional characterization of Stat6. Mol Cell Biol. 1996;16:5811–5820. doi: 10.1128/mcb.16.10.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naeger L K, McKinney J, Salvekar A, Hoey T. Identification of a STAT4 binding site in the interleukin-12 receptor required for signaling. J Biol Chem. 1999;274:1875–1878. doi: 10.1074/jbc.274.4.1875. [DOI] [PubMed] [Google Scholar]
- 27.Ndubuisi M I, Guo G G, Fried V A, Etlinger J D, Sehgal P B. Cellular physiology of STAT3: where's the cytoplasmic monomer? J Biol Chem. 1999;274:25499–25509. doi: 10.1074/jbc.274.36.25499. [DOI] [PubMed] [Google Scholar]
- 28.Ouyang W, Jacobson N G, Bhattacharya D, Gorham J D, Fenoglio D, Sha W C, Murphy T L, Murphy K M. The Ets transcription factor ERM is Th1-specific and induced by IL-12 through a Stat4-dependent pathway. Proc Natl Acad Sci USA. 1999;96:3888–3893. doi: 10.1073/pnas.96.7.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ouyang W, Ranganath S H, Weindel K, Bhattacharya D, Murphy T L, Sha W C, Murphy K M. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 30.Qureshi S A, Leung S, Kerr I M, Stark G R, Darnell J E., Jr Function of Stat2 protein in transcriptional activation by alpha interferon. Mol Cell Biol. 1996;16:288–293. doi: 10.1128/mcb.16.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranganath S, Ouyang W, Bhattarcharya D, Sha W C, Grupe A, Peltz G, Murphy K M. GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol. 1998;161:3822–3826. [PubMed] [Google Scholar]
- 32.Rogge L, D'Ambrosio D, Biffi M, Penna G, Minetti L J, Presky D H, Adorini L, Sinigaglia F. The role of Stat4 in species-specific regulation of Th cell development by type I IFNs. J Immunol. 1998;161:6567–6574. [PubMed] [Google Scholar]
- 33.Schindler U, Wu P, Rothe M, Brasseur M, McKnight S L. Components of a Stat recognition code: evidence for two layers of molecular selectivity. Immunity. 1995;2:689–697. doi: 10.1016/1074-7613(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 34.Seidel H M, Milocco L H, Lamb P, Darnell J E J, Stein R B, Rosen J. Spacing of palindromic half sites as a determinant of selective STAT (signal transducers and activators of transcription) DNA binding and transcriptional activity. Proc Natl Acad Sci USA. 1995;92:3041–3045. doi: 10.1073/pnas.92.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shuai K, Liao J, Song M M. Enhancement of antiproliferative activity of gamma interferon by the specific inhibition of tyrosine dephosphorylation of Stat1. Mol Cell Biol. 1996;16:4932–4941. doi: 10.1128/mcb.16.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soldaini E, John S, Moro S, Bollenbacher J, Schindler U, Leonard W J. DNA binding site selection of dimeric and tetrameric Stat5 proteins reveals a large repertoire of divergent tetrameric Stat5a binding sites. Mol Cell Biol. 2000;20:389–401. doi: 10.1128/mcb.20.1.389-401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stancato L F, David M, Carter-Su C, Larner A C, Pratt W B. Preassociation of STAT1 with STAT2 and STAT3 in separate signalling complexes prior to cytokine stimulation. J Biol Chem. 1996;271:4134–4137. doi: 10.1074/jbc.271.8.4134. [DOI] [PubMed] [Google Scholar]
- 38.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 39.Strehlow I, Schindler C. Amino-terminal signal transducer and activator of transcription (STAT) domains regulate nuclear translocation and STAT deactivation. J Biol Chem. 1998;273:28049–28056. doi: 10.1074/jbc.273.43.28049. [DOI] [PubMed] [Google Scholar]
- 40.Szabo S J, Gold J S, Murphy T L, Murphy K M. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc. Mol Cell Biol. 1993;13:4793–4805. doi: 10.1128/mcb.13.8.4793. . (Erratum, 13:5928.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thierfelder W E, van Deursen J M, Yamamoto K, Tripp R A, Sarawar S R, Carson R T, Sangster M Y, Vignali D A, Doherty P C, Grosveld G C, Ihle J N. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 42.Vinkemeier U, Cohen S L, Moarefi I, Chait B T, Kuriyan J, Darnell J E., Jr DNA binding of in vitro activated Stat1 alpha, Stat1 beta and truncated Stat1: interaction between NH2-terminal domains stabilizes binding of two dimers to tandem DNA sites. EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 43.Vinkemeier U, Moarefi I, Darnell J E, Jr, Kuriyan J. Structure of the amino-terminal protein interaction domain of STAT-4. Science. 1998;279:1048–1052. doi: 10.1126/science.279.5353.1048. [DOI] [PubMed] [Google Scholar]
- 44.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding element confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace A C, Laskowski R A, Thornton J M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 46.Wenner C A, Guler M L, Macatonia S E, O'Garra A, Murphy K M. Roles of IFN-gamma and IFN-alpha in IL-12-induced T helper cell-1 development. J Immunol. 1996;156:1442–1447. [PubMed] [Google Scholar]
- 47.Xu X, Sun Y L, Hoey T. Cooperative DNA binding and sequence-selective recognition conferred by the STAT amino-terminal domain. Science. 1996;273:794–797. doi: 10.1126/science.273.5276.794. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]