Abstract
Nam9p is a protein of the mitochondrial ribosome. The respiration-deficient Saccharomyces cerevisiae strain MB43-nam9-1 expresses Nam9-1p containing the point mutation S82L. Respiratory deficiency correlates with a decrease in the steady level of some mitochondrially encoded proteins and the complete lack of mitochondrially encoded cytochrome oxidase subunit 2 (Cox2). De novo synthesis of Cox2 in MB43-nam9-1 is unaffected, indicating that newly synthesized Cox2 is rapidly degraded. Respiratory deficiency of MB43-nam9-1 is overcome by transient overexpression of HSP104, by deletion of HSP104, by transient exposure to guanidine hydrochloride, and by expression of the C-terminal portion of Sup35, indicating an involvement of the yeast prion [PSI+]. Respiratory deficiency of MB43-nam9-1 can be reinduced by transfer of cytosol from S. cerevisiae that harbors [PSI+]. We conclude that nam9-1 causes respiratory deficiency only in combination with the cytosolic prion [PSI+], presenting the first example of a synthetic effect between cytosolic [PSI+] and a mutant mitochondrial protein.
Most mitochondrial proteins are synthesized in the cytosol and are subsequently imported into the organelle. However, mitochondria contain a full transcriptional and translational machinery, made up of at least 127 imported proteins, which is responsible for the synthesis of eight major mitochondrially encoded proteins (32). Point mutations or small intragenic deletions in mitochondrial genes are termed [mit−] mutations. Yeast strains that carry this type of mutation are unable to grow on nonfermentable substrates but differ from the other respiration-deficient class of [rho−]/[rho0] mutants because [mit−] mutants retain the capacity for mitochondrial protein synthesis. nam9-1 was previously identified as a suppressor of the [mit−] ochre mutation cox2-V25 and was subsequently shown to suppress other ochre mutations in different mitochondrial genes. Wild-type Nam9p is a basic protein of 486 amino acids; in the mutant Nam9-1p, serine 82 is changed to leucine. The N-terminal domain of Nam9p reveals strong homology to the class of S4 ribosomal proteins from prokaryotes and eukaryotes (2–4). In Escherichia coli, S4 plays a key role in ribosome assembly and influences translational fidelity. Nam9p contains an additional C-terminal domain that shows no obvious homology with any known protein. Disruption of NAM9 perturbs mitochondrial DNA integrity and causes respiratory deficiency (4). Some mutations cause temperature-dependent loss of mitochondrial 15S rRNA (2). The combined properties indicate that Nam9p is a component of the mitochondrial ribosome and that Nam9-1p acts as a nonsense suppressor, suggesting that it is involved in ensuring translational fidelity (2, 4). (For a review about mitochondrial ribosomal proteins, see reference 30.) Here we present direct evidence that both Nam9p and Nam9-1p are stably bound to the small subunit of the mitochondrial ribosome.
The Saccharomyces cerevisiae strain MB43-nam9-1 expressing Nam9-1p fails to grow at 37°C and shows a delay of growth at 30°C on nonfermentable substrates. We show that the inability of the strain to respire at 30°C correlates with the lack of mitochondrially encoded cytochrome oxidase subunit 2 (Cox2) at steady-state level, whereas there is only moderate reduction of mitochondrially encoded Cox3, apocytochrome b (Cob), and Atp6, a subunit of the F0F1 ATPase. Although Nam9p is involved in mitochondrial translation, the mutant Nam9-1p does not interfere with the translation of Cox2 but rather with a subsequent step, most likely related to stability or assembly. These findings place NAM9 in a group of nuclear genes that exert a posttranscriptional effect on the accumulation of specific mitochondrially encoded subunits of the cytochrome c oxidase complex (5, 31, 50, 56; for a review, see references 24 and 32).
We reasoned that a screen for multicopy suppressors restoring normal respiration of the nam9-1 strain might reveal novel components of the mitochondrial translation machinery involved in either the fidelity of translation, the stability, and/or the assembly of Cox2. Unexpectedly we identified HSP104 as a strong multicopy suppressor of nam9-1. The cytosolic chaperone Hsp104 is not essential for the life of yeast but is induced during different conditions related to stress (45, 46, 59, 63). Under stress conditions, Hsp104 promotes the reactivation of denatured or aggregated proteins (34, 52). The effect of HSP104 on the phenotype of the nam9-1 strain is exceptional, because both overexpression and deletion allow growth on nonfermentable carbon sources. A similar behavior of Hsp104 has been observed only in the context of the replication of the yeast prion [PSI+]. The non-Mendelian cytoplasmic nonsense suppressor [PSI+] was identified in 1965 (13; for recent reviews see references 42, 65, and 80). How [PSI+] could be transmitted in a non-Mendelian fashion without being connected to any nucleic acid remained unknown for a long time. In 1994, Wickner proposed that [PSI+], like [URE3], is a yeast prion (78). [PSI+] is encoded by SUP35, and it is now well established that [PSI+] is the prion form of Sup35 (also called eRF3) (29, 55, 83). The C-terminal portion of Sup35 is essential for translational termination and cell viability. The N-terminal portion of Sup35 is dispensable for life but plays a key role in [PSI+] formation (54). Sup35 forms a complex with Sup45 (also called eRF1) that is involved in recognizing termination codons and releasing the translation product from the ribosome (68, 83). Recessive suppressor mutations of sup35 or sup45 relax the control of translational fidelity and lead to the readthrough of nonsense codons (70, 73).
Hsp104 was found to be essential for the propagation of [PSI+] in vivo (9). The ability of Hsp104 to actively disaggregate proteins is the key for understanding its effect on [PSI+]. There is evidence that the aggregates formed by [PSI+] are directly influenced by Hsp104 (29, 54, 64; for models of the mechanism, refer to references 39 and 44). The combined data of this study indicate that the genetic interaction of nam9-1 with HSP104 is connected to [PSI+]; the presence of [PSI+] is a prerequisite for the respiratory deficiency of the nam9-1 strain.
MATERIALS AND METHODS
Yeast strains and genetic procedures.
S. cerevisiae strains used in this study are listed in Table 1. Strain MB43-nam9-1, described by Dmochowska et al. (19), and its derivatives were used throughout this study.
TABLE 1.
Yeast strains
| Strain | Nuclear genotype | Mitochondrial genotype | Reference and comments |
|---|---|---|---|
| MB43-nam9-1 | MATa leu2-3,-112 ura3-1 his3 his4C nam9-1 | Wild type | 19 |
| MB43-NAM9 | MATa leu-2,-112 ura3-1 his3 his4C | Wild type | This study |
| MB43-nam9-1Δhsp104 | MATa leu2-3,-112 ura3-1 his3 his4C nam9-1 hsp104::LEU2 | Wild type | This study |
| MB43-nam9-1[HSP104↑] | MATa leu2-3,-112 ura3-1 his3 his4C nam9-1 [pA1/4] | Wild type | This study |
| MB43-nam9-1[mit−-V25] | MATa leu2-3,-112 ura3-1 his34 his4C nam9-1 | cox2-V25 | This study |
| CD112 | MATα kar1-1 ade2-1 his4-15 canR | cox2-V25 | 4 |
| JC25 | MATα kar1-1 ade1 his4-15 canR | Wild type | 12 |
| JC25/1 | MATα kar1-1 ade1 his4-15 canR | Wild type | This study; contains the cytoplasm of respiratory-deficient MB43-nam9-1 |
| V25 (Δcox2) | MATα ade1 op1 met3 | cox2-V25 | 37 |
| M3041 (Δcob) | MATα ade1 op1 | cob-M3041 | 27 |
| OT72 | MATα aro7-1 cyc1 his4-166 leu2-1 lys2-187 met8-1 trp5-48 ura3 [PSI+] | Wild type | Y. Chernoff, personal communication |
| OT74 | MATα aro7-1 cyc1 his4-166 leu2-1 lys2-187 met8-1 trp5-48 ura3 [psi−] | Wild type | Y. Chernoff, personal communication |
The wild-type NAM9 strain, isogenic to MB43-nam9-1, was generated using the two-step gene replacement method (36); first, NAM9 was cloned into the integrative URA3 plasmid pFL34, which was subsequently linearized in the coding region of NAM9; second, the linearized plasmid was transformed into MB43-nam9-1. Integration of the plasmid containing NAM9 and URA3 into the MB43-nam9-1 genome was confirmed by analysis of uracil prototrophy and respiratory competence. Homologous recombination forced by growth on 5-fluoroorotic acid resulted in the excision of the plasmid and loss of either NAM9 or nam9-1. A respiration-competent clone containing NAM9 is referred to as MB43-NAM9.
HSP104 was isolated from a genomic multicopy library based on pFL44 by complementation of the MB43-nam9-1 growth defect on 1% yeast extract–2% peptone–2% glycerol (YPGly). Besides isolating wild-type NAM9 three times, four different genes were found to enable MB43-nam9-1 to grow on nonfermentable substrates at 30°C but not at 37°C. The plasmid conferring the strongest suppression at 30°C was termed pA1/4. Sequencing of pA1/4 revealed that it contained only one complete open reading frame, bearing HSP104. A SaeI deletion, inactivating HSP104, abolished the effect of pA1/4 on the nam9-1 strain. The suppression was confirmed by isolation and retransformation of the plasmid into MB43-nam9-1. MB43-nam9-1 harboring pA1/4 is referred to as MB43-nam9-1[HSP104↑].
The disruption of HSP104 was generated in MB43-nam9-1 as previously described (9). The 1.2-kb ApaI-BglII fragment within the HSP104 coding region was replaced by LEU2. The disruption was confirmed by Southern analysis. The resulting strain is referred to as MB43-nam9-1Δhsp104.
Standard genetic manipulations were performed by published procedures (67). Diploids were selected by the complementation of auxotrophic markers or by micromanipulation. The [rho0] derivatives of the different strains were generated by ethidium bromide (EtBr) treatment (20). Cytoduction experiments were performed by standard procedures using the kar1-1 mutant strain CD112 and the [rho0] derivative of strain JC25. Cytoductants were selected by the presence of auxotrophic and mitochondrial markers (4, 12).
MB43-nam9-1[mit−-V25] was generated as follows. MB43-nam9-1[HSP104↑] was turned [rho0], and mitochondria from strain CD112 harboring the mutation V25 in COX2 (4) were introduced by cytoduction. Subsequently, MB43-nam9-1[mit−-V25][HSP104↑] was depleted from the HSP104-overproducing plasmid pA1/4 by growth on 5-fluoroorotic acid (36). The derivative of strain JC25 harboring cytoplasm from respiration-deficient MB43-nam9-1 is referred to as JC25/1.
OT72 and OT74, respectively, are isogenic [PSI+] and [psi−] derivatives of strain D1142-1A (77). Plasmids pEMBLyex4-SUP35 and pEMBLyex4-3ATG (72), encoding either full-length Sup35p or its C-terminal domain, were used for the transformation of MB43-nam9-1.
Media and growth conditions.
Rich media contained 1% yeast extract, 2% peptone, and either 2% glucose (YPD) or 2% glycerol. Minimal medium (2% glucose, 0.67% yeast nitrogen base without amino acids) was supplemented with amino acids, uracil, and adenine as required (67). All reagents were from Difco. When indicated, guanidine hydrochloride (GuHCl; Sigma) was added to a final concentration of 5 mM. For analysis of the respiratory phenotype, yeast strains were replica plated on YPGly and incubated for 3 days at 30°C.
In order to prevent glucose repression, the different MB43 strains were grown on minimal medium containing 0.5% glucose (low-glucose medium) at 30°C for biochemical characterization. As indicated in the figure legends, yeast grown on low-glucose medium was harvested either at an optical density at 600 nm (OD600) of 1.5 to 1.7, the point at which glucose was just consumed, or after an additional incubation of 12 h. Glucose exhaustion was determined with Glucostix (Bayer).
Import of in vitro-synthesized Nam9 and Nam9-1p into mitochondria.
The coding sequences of NAM9 and nam9-1, respectively, were amplified with Pfu polymerase (Stratagene) and cloned into pSP65 (Promega). In vitro transcription was done as previously described (57). In vitro translation of NAM9 and nam9-1 was performed in a yeast translation extract (25, 26). Import of Nam9p or Nam9-1p into isolated mitochondria was achieved basically as previously described (25). In brief, the translation reaction mixture containing either radiolabeled Nam9p or Nam9-1p was added to the prewarmed import reaction mixture containing import buffer (0.6 M sorbitol, 50 mM HEPES-KOH [pH 7.0], 50 mM KCl, 10 mM MgCl2, 2 mM KH2PO4, 5 mM methionine), 1 mg of fatty acid-free bovine serum albumin per ml, 2 mM NADH, 2 mM ATP, 20 mM creatine phosphate, 0.1 mg of creatine phosphate kinase per ml, and 1 mg of purified wild-type mitochondria per ml. The control reaction mixture without a membrane potential (−ΔΨ) contained 1 μg of valinomycin per ml. Import was carried out at 25°C, and aeration was ensured by recurrent agitation of the import reactions. Import was stopped by the addition of valinomycin (1-μg/ml final concentration) and chilling on ice. Precursor protein that had not crossed the mitochondrial membranes was removed by treatment of the intact mitochondria with 100 μg of proteinase K (Boehringer) per ml for 20 min at 4°C. Proteinase K was inhibited by addition of 1 mM phenylmethylsulfonyl fluoride. Mitochondria were reisolated, resuspended in import buffer containing 1 mM phenylmethylsulfonyl fluoride, and precipitated with 5% trichloroacetic acid (TCA).
Fractionation of yeast cells and purification of mitochondria.
Fractionation of yeast cells grown at 30°C on low-glucose medium was performed essentially as previously described (28), except that fractions corresponding to total yeast, cytosol, and crude mitochondria were collected. The Nycodenz purification step was omitted. For import experiments, yeast cells were grown and mitochondria were purified accurately as described (28).
Generation of a polyclonal antibody against Nam9p.
NAM9 was amplified by PCR using Pfu polymerase (Stratagene) and cloned into the E. coli expression vector pQE60 (Qiagen) in frame with the C-terminal hexahistidine tag. Expression was induced with isopropyl-β-d-thiogalactopyranoside according to the protocol of the manufacturer (Qiagen). Nam9p-His6 was purified under denaturing conditions by chromatography on P11 cellulose (Whatman) followed by affinity chromatography on Ni-nitrilotriacetic acid resin (Qiagen). A 53-kDa polypeptide corresponding to Nam9p was electroeluted by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), precipitated with acetone, and used for immunization of a rabbit according to the standard procedure (69). The antibody against Nam9p (α-Nam9) specifically reacts with a 53-kDa band in an extract of total yeast proteins and in mitochondrial extracts.
Isolation of mitochondrial ribosomes by density centrifugation and immunoprecipitation.
Mitochondria were solubilized by incubation in IP(+Mg) buffer (20 mM Tris-HCl [pH 7.5], 50 mM potassium acetate, 20 mM magnesium acetate, 2 mM dithiothreitol, 1% Triton X-100) for 5 min on ice. The lysate was clarified by centrifugation at 20,000 × g for 10 min and is referred to as mitochondrial extract. Aliquots containing 100 μg of total mitochondrial protein in a volume of 60 μl were either precipitated with 5% TCA (total) or loaded on top of a sucrose cushion [1.0 M sucrose in IP(+Mg) buffer] and centrifuged for 1 h at 250,000 × g. The pellet was separated from the supernatant after centrifugation. Total, supernatant, and pellet were separated by SDS-PAGE followed by immunoblotting with antibodies as indicated in the figure legends.
Immunoprecipitation reactions were performed with mitochondrial extract (100 μg of mitochondrial protein in a 600-μl reaction volume) in either IP(+Mg) buffer or IP(−Mg) buffer (20 mM Tris-HCl [pH 7.5], 50 mM potassium acetate, 2 mM magnesium acetate, 2 mM dithiothreitol, 1% Triton X-100). The reaction mixtures were incubated under gentle agitation in the presence of protein A-Sepharose beads (Pharmacia Biotech) preloaded with either anti-Nam9 or the corresponding preimmune serum. Incubation was carried out for 2 h to overnight at 4°C. Routinely, 10 μl of serum was incubated with 40 μl of preswollen protein A-Sepharose beads. Protein A-Sepharose beads were recovered by centrifugation and washed with IP(+Mg) buffer. The materials bound to protein A-Sepharose and the corresponding supernatants (unbound) were separated by SDS-PAGE and analyzed by immunoblotting.
In vivo labeling of mitochondrial translation products.
Yeast cells were grown on low-glucose medium to the point of glucose exhaustion as described above. The amount of cells corresponding to an OD600 of 0.5 was harvested and resuspended in 300 μl of buffer P (40 mM KPi [pH 7.0], 0.45% glucose, 0.0077% Complete Supplement Mixture–methionine [Bio 101]) and incubated on a shaker for 30 min at 30°C. Cycloheximide was added to a final concentration of 100 μg/ml. After 10 min at 30°C, the reaction mixtures were supplemented with 40 μCi of [35S]methionine (1,000 Ci/mM; ICN Biomedicals), and the incubation was continued for 30 min. The labeling reaction was stopped by rapid chilling of the cells on ice and simultaneous addition of unlabeled methionine to a final concentration of 20 mM. After centrifugation, cells were washed in CM buffer (40 mM KPi [pH 7.0]–100 μg of cycloheximide per ml–20 mM methionine), and subsequently total protein was extracted as previously described (81). Pulse-chase experiments were performed with cells grown on low-glucose medium to the point of glucose exhaustion as described above. Cell pellets corresponding to an OD600 of 2.5 were resuspended in 1.5 ml of buffer P. After addition of cycloheximide (final concentration, 100 μg/ml), labeling was started by addition of 200 μCi of [35S]methionine (1,000 Ci/mM). After gentle agitation of the reaction mixtures for 30 min at 30°C, unlabeled methionine was added to a concentration of 20 mM, and cells were reisolated, washed, and resuspended in chase buffer (40 mM KPi [pH 7.0], 20 mM methionine, 200 μg of acriflavine per ml, 100 μg of cycloheximide per ml). At the time points indicated below, aliquots of the cell suspension corresponding to an OD600 of 0.5 were withdrawn and analyzed as described below.
Miscellaneous.
Molecular biology techniques were performed by standard procedures (58). Yeast cells were transformed by the lithium acetate method (6). Protein concentration was estimated with bicinchoninic acid assay using bovine serum albumin as a standard (Pierce Chemical Co.). Samples containing radiolabeled Nam9p or Nam9-1p were analyzed on SDS–8 or 10% PAGE gels, followed by autoradiography.
For immunoblotting, proteins were separated by SDS-PAGE (40) or Tris-Tricine gels (62) and transferred electrophoretically to nitrocellulose. After incubation with antibody, visualization was with 125I-protein A (33) or anti-rabbit monoclonal antibody coupled to alkaline phosphatase (Promega). The polyclonal antibody against Hsp104 was from Stressgen, and the monoclonal antibodies against Cox2 and Cox3 were from Molecular Probes. All other antibodies were from the collection of G. Schatz, Biozentrum, Basel, Switzerland.
Mitochondrial translation products were analyzed by SDS–12 or 16% PAGE, followed by autoradiography (49). In order to assign the radioactive band corresponding to Cox2, strains V25 (37) and M3041 (27) containing stop mutations in the COX2 and COB genes, respectively, were used. Autoradiograms were quantified using ImageQuant, version 1.1, with local average background correction (Molecular Dynamics, Sunnyvale, Calif.). Kinetics of import were analyzed assuming a simple first-order process using Kaleidagraph 3.09 (Synergy Software).
RESULTS
Nam9p and Nam9-1p are bound to the mitochondrial ribosome.
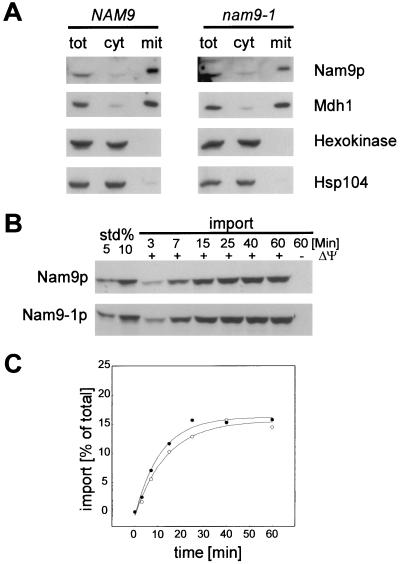
Strain MB43-nam9-1 shows a respiratory growth defect; however, the cells do not become [rho0] (19). In order to determine possible differences between Nam9p and Nam9-1p, we have compared their cellular localization and biochemical properties. Nam9p and Nam9-1p were expressed at similar levels, and both proteins were localized in the mitochondrial fraction after separation of the cytosol from the mitochondria (Fig. 1A). The minor levels of Nam9p and Nam9-1p observed in the cytosolic preparation were due to mitochondrial impurities, as shown by the similar distribution of Mdh1, a mitochondrial marker protein (Fig. 1A). The cytosolic marker proteins hexokinase and Hsp104 were absent from the mitochondrial fraction. Reflecting the steady-state distribution, in vitro-translated Nam9p and Nam9-1p were imported into isolated yeast mitochondria with similar efficiencies and in a potential-dependent manner (Fig. 1B). No shift in molecular mass was observed by SDS-PAGE after import, suggesting that Nam9p and Nam9-1p contain a noncleavable presequence (Fig. 1B). Comparison of the import kinetics of Nam9p and Nam9-1p revealed half times of 9.7 and 7.8 min, respectively, a difference that is within experimental error (Fig. 1C). The combined data confirm that Nam9p is a mitochondrial protein and show that the point mutation nam9-1 does not affect the expression level or intracellular localization.
FIG. 1.
Both Nam9p and Nam9-1p are localized in the mitochondria. (A) MB43-NAM9 and MB43-nam9-1 strains were grown on low-glucose medium, harvested at the point of glucose exhaustion, and fractionated as described in Materials and Methods. Two hundred micrograms of protein from total extract (tot), 200 μg of protein from postmitochondrial supernatant (cyt), and 20 μg of protein from crude mitochondria (mit) were analyzed by SDS-PAGE followed by immunodecoration with antibodies specific for Nam9p, Hsp104, mitochondrial malate dehydrogenase (Mdh1), and cytosolic hexokinase. (B) Nam9p and Nam9-1p were synthesized in a yeast translation system in the presence of [35S]methionine (for details, see Materials and Methods). In vitro import reactions into mitochondria were performed for the indicated times with either radiolabeled Nam9p or Nam9-1p in the presence (+ΔΨ) or absence (−ΔΨ) of a membrane potential. Precursor that had not crossed the mitochondrial membranes after the import reaction was removed by treatment with proteinase K. std 5% and std 10% correspond to 5 and 10% of the import reaction, respectively. (C) Kinetics of the import reactions shown in panel B. The amount of imported Nam9p or Nam9-1p is given as a percentage of the total amount of Nam9p or Nam9-1p added to the import reaction. ○, Nam9p; ●, Nam9-1p.
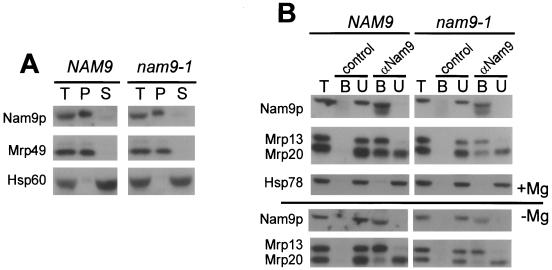
When mitochondrial ribosomes were isolated by sucrose density centrifugation, both Nam9p and Nam9-1p comigrated with a core ribosomal marker protein, suggesting that both proteins were bound to the ribosome (Fig. 2A). This result was confirmed by coimmunoprecipitation with an antibody specific for Nam9p. Incubation of mitochondrial extracts with Nam9p antibody not only leads to the quantitative depletion of Nam9p and Nam9-1p but also depletes Mrp13, a protein of the small ribosomal subunit, suggesting that Nam9p and Nam9-1p were tightly bound to the ribosome (Fig. 2B). However, a protein of the large ribosomal subunit (Mrp20) was only partially depleted from mitochondrial extracts by coimmunoprecipitation with Nam9p antibody. Conditions dissociating the two ribosomal subunits resulted in the coimmunoprecipitation of only Mrp13, whereas Mrp20 remained in the supernatant (Fig. 2B). The results indicate that Nam9p and Nam9-1p are quantitatively bound to the small subunit of the mitochondrial ribosome.
FIG. 2.
Nam9p and Nam9-1p localize to the small subunit of mitochondrial ribosome. (A) Extracts of crude mitochondria (100 μg) from MB43-NAM9 or MB43-nam9-1, respectively, were either precipitated with TCA (T) or separated into ribosomal pellet (P) and supernatant (S) by centrifugation through a sucrose cushion. After SDS-PAGE and Western blotting, samples were analyzed by immunodecoration with antibodies specific for Nam9p, Hsp60 (mitochondrial matrix), or Mrp49 (mitochondrial large ribosomal subunit). (B) Association of Nam9p with the mitochondrial ribosome was assessed by coimmunoprecipitation of the ribosomal components with antibodies specific for Nam9p (αNam9). Preimmune serum was used in the control reaction (control). Coimmunoprecipitation reactions were performed with 100 μg of total mitochondrial protein (T) under conditions that either stabilize (+Mg) or destabilize (−Mg) the interaction between the two ribosomal subunits. B, material bound to αNam9; U, material unbound to αNam9. T, B, and U were analyzed by Western blotting followed by immunodecoration with antibodies against Nam9p, Hsp78 (mitochondrial matrix), Mrp20 (mitochondrial large ribosomal subunit), and Mrp13 (mitochondrial small ribosomal subunit).
Translation of mitochondrially encoded proteins—steady-state levels and de novo synthesis.
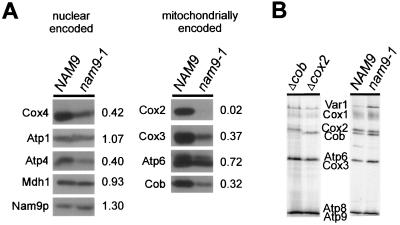
The steady-state levels of four mitochondrially encoded proteins (Cox2, Cox3, Cob, and Atp6) and of a selection of nucleus-encoded mitochondrial proteins were determined in strains MB43-NAM9 and MB43-nam9-1 grown under conditions that ensure expression of the respiratory chain components. In the nam9-1 strain, Cox3 and Cob levels were reduced to about 30%, and the Atp6 level was reduced to about 75%, compared to the wild-type control. However, Cox2 was undetectable in the nam9-1 strain (Fig. 3A). This result indicates that Nam9-1p differentially affects the steady-state levels of the mitochondrial translation products and suggests that the complete lack of Cox2—and possibly other respiratory chain components not tested here—accounts for the respiratory deficiency of the nam9-1 strain. Despite the reduction in steady-state levels, there was no significant effect of Nam9-1p on the de novo synthesis of any of these mitochondrially encoded polypeptides in vivo (Fig. 3B). Thus, the nam9-1 strain does not fail to synthesize Cox2. Rather, the lack of Cox2 at steady state must be due to an event downstream of translation. Cox2 is synthesized with a 15-amino-acid N-terminal presequence that is cleaved in the intermembrane space (35, 66). We were unable to detect the precursor form of de novo-synthesized Cox2 in the nam9-1 strain, suggesting that the N terminus of Cox2 had reached the intermembrane space before it was degraded (Fig. 4A). Processing of Cox2 prior to its degradation has been observed previously (60).
FIG. 3.
The absence of Cox2 in MB43-nam9-1 is due to an event downstream of translation. (A) The steady-state levels of mitochondrially encoded proteins were assessed in strains MB43-NAM9 and MB43-nam9-1, respectively. Crude mitochondria (50 μg of protein) were analyzed by immunoblotting with antibodies directed against Nam9p, Cox2, Cox3, Cox4, Atp1, Atp4, Atp6, and mitochondrial malate dehydrogenase (Mdh1). In order to determine the relative amounts of the various proteins in the nam9-1 and NAM9 strains, respectively, the amount of each protein present in the NAM9 strain was set to 1. Numbers to the right give the ratios between the nam9-1 and the NAM9 strains. (B) In vivo synthesis of mitochondrially encoded proteins in MB43-NAM9 and MB43-nam9-1, respectively. Mitochondrial translation was performed in the presence of [35S]methionine and cycloheximide as described in Materials and Methods and analyzed by SDS–12% PAGE. To ascertain the position of Cox2 and Cob in SDS-PAGE, the first two lanes show the mitochondrial translation products of the [mit−] strains M3041 (Δcob) and V25 (Δcox2), containing stop mutations in COB and COX2, respectively. As a result, the full-length translation products of the respective genes are absent. Var1, ribosomal protein. The Atp8 and Atp9 bands and the Cox3 and Atp6 bands are not separated from each other.
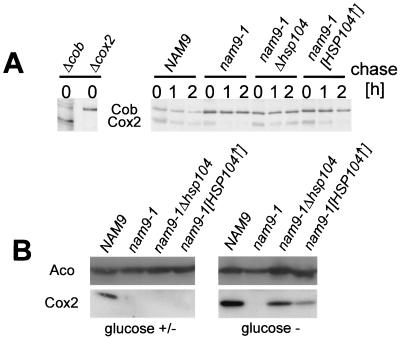
FIG. 4.
Deletion as well as overexpression of Hsp104 stabilizes Cox2 in the nam9-1 strain. (A) Stability of newly synthesized Cox2 in MB43-NAM9, MB43-nam9-1, MB43-nam9-1Δhsp104, and MB43-nam9-1[HSP104↑]. Cells were pulse-labeled with [35S]methionine in the presence of cycloheximide, and a chase reaction was performed for the times indicated (see Materials and Methods). The mitochondrial translation products from cells, corresponding to an OD600 of 0.5, were analyzed by SDS–16% PAGE. M3041 (Δcob) and V25 (Δcox2) were used as references for the migration of Cox2 and Cob, respectively. Note that Cox2 and Cob migrate in the reverse order in SDS–16% PAGE compared to SDS–12% PAGE (compare to Fig. 6). (B) Steady-state levels of Cox2 in strains MB43-NAM9, MB43-nam9-1, MB43-nam9-1 hsp104, and MB43-nam9-1[HSP104↑]. The respective strains were grown on low-glucose medium and harvested either at the point of glucose exhaustion (glucose +/−) or after an additional incubation of 12 h (glucose −) as described in Materials and Methods. Total protein was extracted from cells corresponding to an OD600 of 0.5. Western blots were decorated with antibodies directed against the mitochondrial matrix protein aconitase (Aco) and cytochrome c oxidase subunit 2 (Cox2).
A direct effect of Nam9-1p on proteins synthesized in the cytosol would not be expected, and indeed mitochondria isolated from the nam9-1 and the corresponding wild-type strain contained similar amounts of nucleus-encoded Nam9p and Nam9-1p, Mdh1, and Atp1 (Fig. 3A; compare also Fig. 1 and 2). However, the steady-state levels of the nucleus-encoded respiratory chain components Cox4 and Atp4 were reduced to about 40% of the wild-type control. The result is most likely explained by the imbalance of respiratory-chain subunits, which results in incomplete assembly and subsequent degradation of unassembled subunits, as has been previously observed by others (41, 50).
Both overexpression and inactivation of HSP104 suppress the effect of nam9-1 on mitochondrial function.
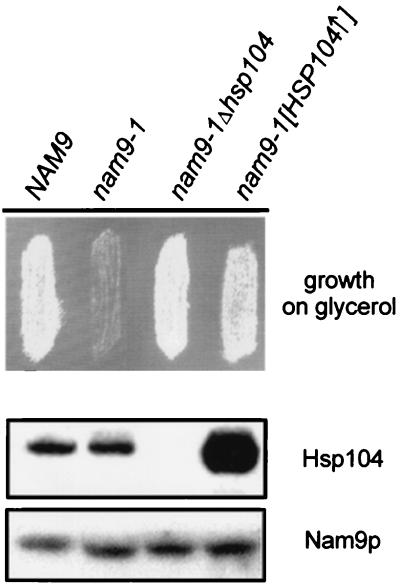
In order to identify components that cooperate with Nam9p during synthesis and assembly of the mitochondrially encoded subunits of the respiratory chain, we performed a multicopy suppressor screen. The plasmid conferring the strongest suppression contained a single open reading frame encoding the cytosolic chaperone Hsp104 (63). The effect of HSP104 was confirmed by transformation of the nam9-1 strain with a 2μm plasmid containing HSP104 (Fig. 5) and was also observed when HSP104 was expressed from a low-copy-number plasmid leading to only moderate overexpression (data not shown). Surprisingly, about 75% of the MB43-nam9-1 colonies retained the capability to respire when subsequently depleted of the plasmid expressing HSP104 (data not shown). This unusual behavior has been previously reported in the context of the yeast prion [PSI+], a structural variant of the cytosolic translation termination factor Sup35 that causes suppression of some nonsense mutations and behaves in a non-Mendelian fashion (13, 53, 54, 79). [PSI+] can be cured by transient overexpression as well as by deletion of HSP104 (9, 54) and also by treatment with low concentrations of GuHCl (22, 47, 75). Intrigued by the resemblance of the Hsp104 effect on nam9-1, we disrupted HSP104 in MB43-nam9-1. Indeed, in the absence of Hsp104, the nam9-1 mutant became respiration competent, and suppression was even stronger than by overproduction of Hsp104 (Fig. 5). The steady-state level of Nam9p and Nam9-1p was unaffected by varying the amount of Hsp104 and the ability of the cells to respire (Fig. 5). The effect of Hsp104 on the respiratory competence of the nam9-1 strain suggests that the posttranslational defect in Cox2 biogenesis should be cured by both deletion and overexpression of HSP104. To test this prediction, the stability of newly synthesized Cox2 was tested in the isogenic NAM9, nam9-1, nam9-1Δhsp104, and nam9-1[HSP104↑] strains. Newly synthesized Cox2 was significantly more stable in the NAM9 than in the nam9-1 strain. Deletion and overexpression of HSP104 stabilized newly synthesized Cox2 in the presence of Nam9-1p (Fig. 4A). We finally tested the effect of Hsp104 on the steady-state level of Cox2. The NAM9, nam9-1, nam9-1Δhsp104, and nam9-1[HSP104↑] strains were grown on low-glucose medium and were harvested either at the point of glucose exhaustion or after an additional incubation of 12 h. The four strains contained the mitochondrial marker protein aconitase in similar amounts (Fig. 4B). When the presence of glucose allowed growth by fermentation, Cox2 was detectable only in the NAM9 wild-type strain (Fig. 4B, glucose +/−). However, after glucose was consumed and respiration was required for growth, Cox2 was detectable in wild-type cells and in nam9-1Δhsp104 and nam9-1[HSP104↑] cells. Deletion of HSP104 led to a higher steady-state level of Cox2 than its overexpression (Fig. 4B, glucose −). This is in agreement with the finding that the nam9-1 phenotype was more strongly suppressed in the nam9-1Δhsp104 strain than it was in nam9-1[HSP104↑] strain (compare above). The combined data suggest that both deletion and overexpression of HSP104 enable the nam9-1 strain to grow on nonfermentable substrates, because Cox2 and possibly other mitochondrial proteins are stabilized.
FIG. 5.
Respiratory deficiency of MB43-nam9-1 is suppressed by the absence of or by overproduction of Hsp104. The respective derivatives of MB43 (compare to Table 1) were transferred to YPGly plates and incubated for 3 days at 30°C. In order to determine the content of Hsp104 and Nam9 or Nam9-1p, the strains were grown on low-glucose medium and harvested at the point of glucose exhaustion (see Materials and Methods). Total protein was extracted by alkaline lysis and analyzed by Western blotting using antibodies specific for Hsp104 and Nam9p.
The nam9-1 phenotype is dependent on the cytoplasmic prion [PSI+].
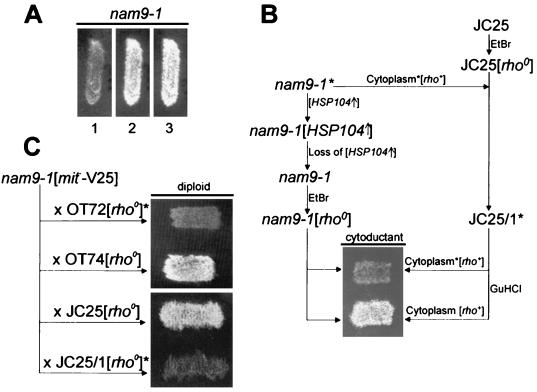
Growth on low concentrations of GuHCl cures yeast from [PSI+] and the prion [URE3] (22, 75, 78). When MB43-nam9-1 was grown on low concentrations of GuHCl, the respiratory growth defect was suppressed. Respiratory competence persisted in 85% of [rho+] cells after removal of GuHCl from the growth medium and was stable, as two independent clones showed no reversion to respiratory deficiency (Fig. 6A and data not shown; for the problem of detecting reversion to [PSI+] in nam9-1 strains, compare to Discussion below).
FIG. 6.
The nam9-1 mutation leads to respiratory deficiency only in the presence of a prion-like element. In order to test the respiratory competence of the MB43-nam9-1 derivatives (Table 1; see also Materials and Methods), the respective strains were transferred to YPGly plates and incubated for 3 days at 30°C. (A) MB43-nam9-1 was grown on YPGly (lane 1), YPGly containing 5 mM GuHCl (lane 2), and YPGly after transient growth on YPD containing 5 mM GuHCl (lane 3). (B) As outlined in the scheme, JC25 was turned [rho0] by treatment with EtBr and the resulting strain, JC25[rho0], was used as a recipient for the cytoplasm of respiration-deficient MB43-nam9-1. Strain JC25 containing the cytoplasm of respiration-deficient MB43-nam9-1 was termed JC25/1 and was used as the donor strain in a second cytoduction, either directly or after transient exposure to 5 mM GuHCl. The recipient strain for the second cytoduction was MB43-nam9-1[rho0], generated as follows: MB43-nam9-1 was transformed with a plasmid overexpressing Hsp104, the plasmid was cured, and finally the strain was treated with EtBr. For details compare Results and Materials and Methods. (C) Diploids generated by crosses of respiration-competent MB43-nam9-1[mit−-V25] with the [rho0] derivatives of the [PSI+] strain OT72, the [psi−] strain OT74, JC25, and JC25/1. The presence of a prion-like element in panels B and C is indicated by an asterisk.
The prion hypothesis predicts that curing of MB43-nam9-1 would not result in an irreversible genetic change but could be reversed by reintroduction of the prion. In S. cerevisiae, transfer of a prion from a donor to an acceptor strain can be achieved by cytoduction, a form of mating by which cytoplasm and mitochondria of a donor are transferred to a recipient strain without fusion of the nuclei. The kar1-1 strain JC25, defective in nuclear fusion, was turned [rho0] by treatment with EtBr and subsequently used for the cytoduction experiments (12). First, cytosol from the original, respiration-deficient MB43-nam9-1 was transferred to JC25[rho0], resulting in JC25/1. Second, JC25/1 served as donor strain for the recipient strain MB43-nam9-1[rho0], obtained by transient overexpression of HSP104 followed by treatment with EtBr. Transfer of cytoplasm from JC25/1 resulted in respiratory deficiency of MB43-nam9-1. When the same experiment was performed after JC25/1 was transiently grown on GuHCl, MB43-nam9-1 was respiration competent after cytoduction (Fig. 6B). We conclude that respiratory deficiency of MB43-nam9-1 depends on the presence of an element that can be reintroduced by cytoduction and is eliminated by treatment with GuHCl.
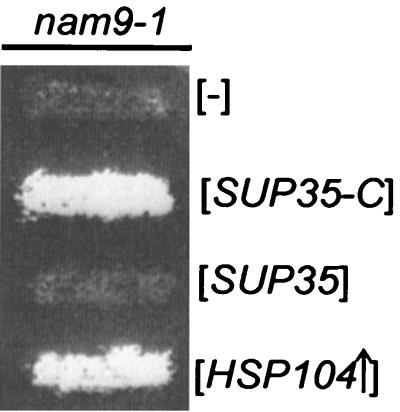
The possibility that [PSI+] causes respiratory deficiency when combined with nam9-1 was directly addressed with the following experiment. The C-terminal portion of Sup35 (Sup35-C) is sufficient for translation termination and does not become trapped in nam9-1 aggregates. Sup35-C exerts a dominant effect that eliminates [PSI+]-mediated nonsense suppression (54, 71, 72). When Sup35-C was expressed in MB43-nam9-1, the strain became respiration competent (Fig. 7). The expression of full-length Sup35 did not affect the respiration-deficient phenotype of nam9-1 (Fig. 7). The results identify the prion-like element contained in respiration-deficient MB43-nam9-1 as a structural variant of Sup35.
FIG. 7.
Respiratory deficiency of MB43-nam9-1 can be overcome by expression of the C-terminal portion of Sup35. MB43-nam9-1 derivatives were transferred to YPGly plates and incubated for 3 days at 30°C. MB43-nam9-1 was transformed with empty control vector ([−]), pEMBLyex4-3ATG ([SUP35-C]), pEMBLyex4-SUP35 ([SUP35]), and pA1/4 ([HSP104↑]) (Table 1; see also Materials and Methods).
Because of its dominant character, nam9-1-mediated suppression of [mit−] mutations can be studied in heterozygous diploids (compare also to the introduction). In order to determine the effect of [PSI+] on yeast harboring nam9-1 and a [mit−] mitochondrial genome, we used the isogenic strains OT72 and OT74, which differ exclusively in their [PSI+] status: OT72 is [PSI+], and OT74 is [psi−] (Y. Chernoff, personal communication). The [rho+] mitochondrial genomes of OT72 and OT74 were eliminated by treatment with EtBr. Subsequently, MB43-nam9-1[mit−-V25], cured of the prion, was crossed with OT72[rho0] or OT74[rho0]. Only the [psi−] diploid was able to grow under respiratory conditions (Fig. 6C). JC25 and JC25/1 (compare to Fig. 6B) were used in an analogous experiment. A cross of MB43-nam9-1[mit−-V25] with JC25/1[rho0] resulted in a respiration-deficient diploid. In contrast, the diploid formed by the cross of JC25[rho0] with MB43-nam9-1[mit−-V25] was able to respire (Fig. 6C). The combined data show that the presence of [PSI+] negatively affects the function of mitochondria containing both mutant Nam9-1p and a [mit−-V25] mitochondrial genome. The effect of [PSI+] transferred from the well-characterized OT72 strain is indistinguishable from the effect of the factor present in the cytoplasm of respiration-deficient MB43-nam9-1.
DISCUSSION
Effect of Nam9-1p on mitochondrial translation products.
The mutant nam9-1 shows the genetic characteristics of an informational ribosomal suppressor. Using a cell-free in vitro system, it was shown that the respective suppressor mutants of the cytosolic S4 homologues generate an abnormally high level of errors during translation (48, 76). It is likely that Nam9-1p also increases the frequency of amino acid substitutions in mitochondrial translation products, leading to their decreased stability. A general decrease in stability of the mitochondrially synthesized proteins might well explain their reduced steady-state levels in the nam9-1 strain.
The fact that mitochondria lack Cox2 in the presence of Nam9-1p suggests an additional, more specific effect on a posttranslational event in the biogenesis of this subunit. Like seven of the eight mitochondrially synthesized proteins, Cox2 is embedded in the mitochondrial inner membrane (24, 32). The mitochondrial translation machinery thus can be regarded as a specialized apparatus, ensuring the production of integral membrane proteins and their assembly into complex multimeric structures. To this end, mitochondria contain membrane-bound translational activators interacting specifically with 5′ untranslated leaders of the mitochondrial mRNA (24). It has been suggested that translational activators and the 5′ untranslated region are required to tether mitochondrial translation complexes to the inner mitochondrial membrane, ensuring stability and correct assembly of mitochondrially encoded membrane proteins (60).
A number of mutations in nuclear genes affect posttranslational events in the biogenesis of Cox2 (32). We now report that a mutation in Nam9p also affects the stability of newly synthesized Cox2, while having only a moderate effect on its translation efficiency. This identifies Nam9p as the first example of a mitochondrial ribosomal protein involved in the stability of a mitochondrially encoded protein. The finding suggests that Nam9p, like translational activator proteins and the 5′ untranslated region of the mRNA, is involved in productively attaching the ribosome to the mitochondrial inner membrane. Nam9-1p might be defective in this process, leading to problems in the biogenesis of Cox2 and possibly other respiratory-chain components. It is interesting in this context that Nam9p contains a large C-terminal domain that is absent from the known bacterial and eukaryotic homologues.
Respiration-deficient MB43-nam9-1 harbors a prion variant of Sup35.
We present evidence that MB43-nam9-1 contains the prion [PSI+] or a related variant of Sup35. Respiratory deficiency of MB43-nam9-1 can be cured by overexpression and deletion of HSP104 or growth on low concentrations of GuHCl, exactly the same conditions that cure [PSI+] (9, 22, 46, 47, 54, 75). Consistent with a prion mechanism, transfer of [PSI+] cytoplasm to a [psi−] nam9-1 strain causes respiratory deficiency. Finally, expression of Sup35-C abolishes the respiratory deficiency of MB43-nam9-1, suggesting that depletion of functional Sup35 is responsible for the phenotype. It has been postulated as a genetic criterion for the presence of a yeast prion that overproduction of the protein leads to an increased de novo appearance of the respective prion (80). The frequency of spontaneous [PSI+] formation is on the order of 10−5 to 10−6, as determined by positive-selection systems (47). The frequency becomes at least 1 order of magnitude higher when Sup35 is overexpressed (8, 17). In contrast to the original studies on the reversion of [psi−] to [PSI+], the detection of de novo [PSI+] formation in MB43-nam9-1 must be based on loss of respiratory competence, a negative-selection system. Negative selection, however, is inapplicable for the detection of the expected low frequency of reversion. It has been recently discovered that [PSI+] variants cause phenotypes of different severity (16). [ETA+] is a weak, unstable variant of [PSI+] that is cured by overexpression and deletion of HSP104 (82). [PSI+] introduced to respiration-competent MB43-nam9-1 from strain OT72 reinduces respiratory deficiency. However, the original respiration-deficient MB43-nam9-1 isolate is not recognized by the [PSI+] tester strain BO512-4C (B. Ono, personal communication), which detects [PSI+] on the basis of SUQ5 ade2-1 suppression (data not shown). The genetic interaction between nam9-1 and [PSI+] is not restricted to a specific genetic background. We found that overexpression of HSP104 affects respiratory competence of a nam9-1[mit−-V25] strain derived from strain MHY500 (7), which is entirely unrelated to strain MB43 (data not shown).
How does cytosolic Sup35 affect mitochondrial function?
The cytosolic translation termination factor Sup35/[PSI+] affects mitochondrial protein synthesis in the presence of the mitochondrial nonsense suppressor nam9-1. The finding is unexpected because Sup35/[PSI+] and Nam9-1p are localized in different compartments of the cell. It is currently unknown what exactly connects the two proteins; however, one might envisage different scenarios. Possibly the physical presence of [PSI+] aggregates interferes with mitochondrial function. It has been reported that proteins can become trapped in [PSI+] aggregates (14). Depletion of a protein interacting with Nam9p might account for respiratory deficiency in the presence of nam9-1. It is also conceivable that [PSI+] aggregates might change the level of cytosolic chaperones available for the translocation of proteins to mitochondria, and as a consequence, the relative level of one or more components of the mitochondrial translation machinery might change. In this context, it is interesting that members of the Hsp70 chaperone family have been implicated in the maintenance of [PSI+] (10, 51). However, because expression of the C-terminal domain of Sup35 rescues respiratory deficiency of MB43-nam9-1 and for the reasons outlined below, we favor the hypothesis that a reduced level of functional Sup35 in combination with nam9-1 causes the phenotype.
A remarkable number of suppressors of [mit−] mutations in the mitochondrial DNA are part of the cytosolic translation machinery, suggesting that events in the cytosol exert great influence on the control of the translation process within mitochondria (1, 23, 43). Moreover, the majority of mutations in Sup35 and its partner protein Sup45 affecting termination of cytosolic translation cause respiratory deficiency (73, 74). Sup35 and Sup45 are distributed throughout the cytoplasmic-ribosome-enriched fractions (18) but are absent inside mitochondria (38). Interestingly, sup35 mutations are inherited in a Mendelian fashion, suggesting that the mitochondrial phenotype is not caused by a prion-like conformation adopted by the encoded mutant proteins. Recently it has been discovered that a mutant of Atp17 is able to suppress the respiratory deficiency of a Sup35 mutant (L. N. Mironova, personal communication). However, expression of the suppressor in the background of wild-type SUP35 leads to respiratory deficiency of a [psi−] but not of the corresponding [PSI+] strain. The finding presents another link between mitochondrial respiration and [PSI+], but the effect is opposite to nam9-1, which is manifested only in [PSI+] yeast.
[PSI+] wild-type strains exhibit enhanced tolerance to heat and chemical stress compared to the corresponding [psi−] strains (21), and it has been suggested that [PSI+] might be an evolutionarily beneficial state (61). Increased stress tolerance might be caused by [PSI+]-induced allosuppression generating one or more novel proteins bearing an extra C-terminal domain (21, 44). One or more nucleus-encoded proteins involved in the translation and stability of mitochondrially encoded proteins might be expressed in a C-terminally extended version(s) when low levels of functional Sup35 allow readthrough of nonsense codons. Either the presence of the extended translation product or the absence of the “normal” translation product might cause respiratory deficiency in combination with nam9-1. Candidates for cytosolic translation products affected by [PSI+] are components involved in mitochondrial translational fidelity (11, 15) as well as in export, turnover, or assembly of mitochondrially synthesized proteins (32). We have now started to determine specific differences in the mitochondrial protein content of nam9-1[PSI+], nam9-1[psi−], and the corresponding NAM9 wild-type strains. Our preliminary results indicate induction as well as repression of a defined set of nucleus-encoded proteins in the presence of nam9-1[PSI+]. The identification of these proteins should help us to understand the mechanism by which nam9-1 and [PSI+] interact and improve our understanding of the general interplay between mitochondrial and cytosolic translation.
ACKNOWLEDGMENTS
We are indebted to G. Schatz for giving A.C. the opportunity to work in his laboratory in the Biozentrum, for generous support, and for fruitful discussions throughout the project. We thank B. Szczesniak and R. Looser for excellent technical assistance. The pEMBLyex4-SUP35 and pEMBLyex4-3ATG plasmids and the OT72 and OT74 yeast strains were kind gifts of M. D. Ter-Avanesyan and Y. O. Chernoff. We thank U. Fünfschilling for help with the yeast translation extracts and B. Ono for strain BO512-4C. Critical reading of the manuscript by A. Paszewski, Y. Dubaquié, S. Merchant, C. Suzuki, and T. Lithgow is gratefully acknowledged.
This study was supported by State Committee for Scientific Research (KBN) grant 6PO4B02915 for A.C. and M.B., KBN grant 6PO4A03312 for A.C., Swiss National Science Foundation (SNSF) grant 7IP 051663 covering the costs of A.C.'s stay and research in the Biozentrum, University of Basel, and SNSF grant 3100-050954 to S.R.
REFERENCES
- 1.Altamura N, Dujardin G, Groudinsky O, Slonimski P P. Two adjacent nuclear genes, ISF1 and NAM7/UPF1, cooperatively participate in mitochondrial functions in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:49–56. doi: 10.1007/BF00277347. [DOI] [PubMed] [Google Scholar]
- 2.Biswas T K, Getz G S. The single amino acid changes in the yeast mitochondrial S4 ribosomal protein cause temperature-sensitive defect in the accumulation of mitochondrial 15S rRNA. Biochemistry. 1999;38:13042–13054. doi: 10.1021/bi990058u. [DOI] [PubMed] [Google Scholar]
- 3.Boguta M, Chacinska A, Murawski M, Szczesniak B. Expression of the yeast NAM9 gene coding for mitochondrial ribosomal protein. Acta Biochim Pol. 1997;44:251–258. [PubMed] [Google Scholar]
- 4.Boguta M, Dmochowksa A, Borsuk P, Wrobel K, Gargouri A, Lazowska J, Slonimski P P, Szczesniak B, Kruszewska A. NAM9 nuclear suppressor of mitochondrial ochre mutations in Saccharomyces cerevisiae codes for a protein homologous to S4 ribosomal proteins from chloroplasts, bacteria, and eucaryotes. Mol Cell Biol. 1992;12:402–412. doi: 10.1128/mcb.12.1.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnefoy N, Chalvet F, Hamel P, Slonimski P P, Dujardin G. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J Mol Biol. 1994;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- 6.Chen D C, Yang B C, Kuo T T. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 8.Chernoff Y O, Derkach I L, Inge-Vechtomov S G. Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr Genet. 1993;24:268–270. doi: 10.1007/BF00351802. [DOI] [PubMed] [Google Scholar]
- 9.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff Y O, Newnam G P, Kumar J, Allen K, Zink A D. Evidence for a protein mutator in yeast: role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colby G, Wu M, Tzagoloff A. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J Biol Chem. 1998;273:27945–27952. doi: 10.1074/jbc.273.43.27945. [DOI] [PubMed] [Google Scholar]
- 12.Conde J, Fink G R. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci USA. 1976;73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox B S, Tuite M F, McLaughlin C S. The psi factor of yeast: a problem in inheritance. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- 14.Czaplinski K, Ruiz-Echevarria M J, Paushkin S V, Han X, Weng Y, Perlick H A, Dietz H C, Ter-Avanesyan M D, Peltz S W. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decoster E, Vassal A, Faye G. MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J Mol Biol. 1993;232:79–88. doi: 10.1006/jmbi.1993.1371. [DOI] [PubMed] [Google Scholar]
- 16.Derkatch I L, Bradley M E, Zhou P, Chernoff Y O, Liebman S W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derkatch I L, Chernoff Y O, Kushnirov V V, Inge-Vechtomov S G, Liebman S W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Didichenko S A, Ter-Avanesyan M D, Smirnov V N. Ribosome-bound EF-1 alpha-like protein of yeast Saccharomyces cerevisiae. Eur J Biochem. 1991;198:705–711. doi: 10.1111/j.1432-1033.1991.tb16070.x. [DOI] [PubMed] [Google Scholar]
- 19.Dmochowska A, Konopinska A, Krzymowska M, Szczesniak B, Boguta M. The NAM9-1 suppressor mutation in a nuclear gene encoding ribosomal mitochondrial protein of Saccharomyces cerevisiae. Gene. 1995;162:81–85. doi: 10.1016/0378-1119(95)00311-s. [DOI] [PubMed] [Google Scholar]
- 20.Dujardin G, Pajot P, Groudinsky O, Slonimski P P. Long range control circuits within mitochondria and between nucleus and mitochondria. I. Methodology and phenomenology of suppressors. Mol Gen Genet. 1980;179:469–482. doi: 10.1007/BF00271736. [DOI] [PubMed] [Google Scholar]
- 21.Eaglestone S S, Cox B S, Tuite M F. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eaglestone S S, Ruddock L W, Cox B S, Tuite M F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folley L S, Fox T D. Reduced dosage of genes encoding ribosomal protein S18 suppresses a mitochondrial initiation codon mutation in Saccharomyces cerevisiae. Genetics. 1994;137:369–379. doi: 10.1093/genetics/137.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox T D. Translational control of endogenous and recoded nuclear genes in yeast mitochondria: regulation and membrane targeting. Experientia. 1996;52:1130–1135. doi: 10.1007/BF01952112. [DOI] [PubMed] [Google Scholar]
- 25.Fünfschilling U, Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol Biol Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia P D, Hansen W, Walter P. In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:675–682. doi: 10.1016/0076-6879(91)94049-i. [DOI] [PubMed] [Google Scholar]
- 27.Gargouri A F. Recherches sur les introns de l'ADN mitochondrial chez la levure Saccharomyces cerevisiae: mutations, suppressions et deletions genomiques d'introns. Gif-sur-Yvette, France: Institute of Molecular Genetics; 1989. [Google Scholar]
- 28.Glick B S, Pon L A. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- 29.Glover J R, Kowal A S, Schirmer E C, Patino M M, Liu J J, Lindquist S. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 30.Graack H R, Wittmann-Liebold B. Mitochondrial ribosomal proteins (MRPs) of yeast. Biochem J. 1998;329:433–438. doi: 10.1042/bj3290433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Green-Willms N S, Fox T D, Costanzo M C. Functional interactions between yeast mitochondrial ribosomes and mRNA 5′ untranslated leaders. Mol Cell Biol. 1998;18:1826–1834. doi: 10.1128/mcb.18.4.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grivell L A, Artal-Sanz M, Hakkaart G, de Jong L, Nijtmans L G, van Oosterum K, Siep M, van der Spek H. Mitochondrial assembly in yeast. FEBS Lett. 1999;452:57–60. doi: 10.1016/s0014-5793(99)00532-3. [DOI] [PubMed] [Google Scholar]
- 33.Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- 34.Hänninen A L, Simola M, Saris N, Makarow M. The cytoplasmic chaperone hsp104 is required for conformational repair of heat-denatured proteins in the yeast endoplasmic reticulum. Mol Biol Cell. 1999;10:3623–3632. doi: 10.1091/mbc.10.11.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He S, Fox T D. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaiser C, Michaelis S, Mitchel A. Methods in yeast genetics. A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 37.Kruszewska A, Szczesniak B, Claisse M. Recombinational analysis of OXI1 mutants and preliminary analysis of their translation products in S. cerevisiae. Curr Genet. 1980;2:45–51. doi: 10.1007/BF00445693. [DOI] [PubMed] [Google Scholar]
- 38.Kushnirov V V. Cardiology Research Center, Moskow, USSR. 1990. Ph.D. thesis. Structure and functional organisation of the SUP2 (SUP35) gene controlling translational fidelity in yeast. [Google Scholar]
- 39.Kushnirov V V, Ter-Avanesyan M D. Structure and replication of yeast prions. Cell. 1998;94:13–16. doi: 10.1016/s0092-8674(00)81216-7. [DOI] [PubMed] [Google Scholar]
- 40.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 41.Lemaire C, Robineau S, Netter P. Molecular and biochemical analysis of Saccharomyces cerevisiae cox1 mutants. Curr Genet. 1998;34:138–145. doi: 10.1007/s002940050378. [DOI] [PubMed] [Google Scholar]
- 42.Liebman S W, Derkatch I L. The yeast [PSI+] prion: making sense of nonsense. J Biol Chem. 1999;274:1181–1184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- 43.Linder P, Slonimski P P. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc Natl Acad Sci USA. 1989;86:2286–2290. doi: 10.1073/pnas.86.7.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindquist S. Mad cows meet psi-chotic yeast: the expansion of the prion hypothesis. Cell. 1997;89:495–498. doi: 10.1016/s0092-8674(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 45.Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci USA. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindquist S, Patino M M, Chernoff Y O, Kowal A S, Singer M A, Liebman S W, Lee K H, Blake T. The role of Hsp104 in stress tolerance and [PSI+] propagation in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1995;60:451–460. doi: 10.1101/sqb.1995.060.01.050. [DOI] [PubMed] [Google Scholar]
- 47.Lund P M, Cox B S. Reversion analysis of [psi−] mutations in Saccharomyces cerevisiae. Genet Res. 1981;37:173–182. doi: 10.1017/s0016672300020140. [DOI] [PubMed] [Google Scholar]
- 48.Masurekar M, Palmer E, Ono B I, Wilhelm J M, Sherman F. Misreading of the ribosomal suppressor SUP46 due to an altered 40 S subunit in yeast. J Mol Biol. 1981;147:381–390. doi: 10.1016/0022-2836(81)90490-3. [DOI] [PubMed] [Google Scholar]
- 49.Mulero J J, Fox T D. Reduced but accurate translation from a mutant AUA initiation codon in the mitochondrial COX2 mRNA of Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:383–390. doi: 10.1007/BF00281787. [DOI] [PubMed] [Google Scholar]
- 50.Nakai T, Yasuhara T, Fujiki Y, Ohashi A. Multiple genes, including a member of the AAA family, are essential for degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol Cell Biol. 1995;15:4441–4452. doi: 10.1128/mcb.15.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newnam G P, Wegrzyn R D, Lindquist S L, Chernoff Y O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol Cell Biol. 1999;19:1325–1333. doi: 10.1128/mcb.19.2.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parsell D A, Kowal A S, Singer M A, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 53.Patino M M, Liu J J, Glover J R, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- 54.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 55.Paushkin S V, Kushnirov V V, Smirnov V N, Ter-Avanesyan M D. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 56.Poutre C G, Fox T D. PET111, a Saccharomyces cerevisiae nuclear gene required for translation of the mitochondrial mRNA encoding cytochrome c oxidase subunit II. Genetics. 1987;115:637–647. doi: 10.1093/genetics/115.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rospert S, Schatz G. Protein translocation into mitochondria. In: Celis J E, editor. Cell biology, a laboratory handbook. Vol. 2. San Diego, Calif: Academic Press; 1998. pp. 277–285. [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Sanchez Y, Taulien J, Borkovich K A, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchirico M E, Fox T D, Mason T L. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J. 1998;17:5796–5804. doi: 10.1093/emboj/17.19.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santoso A, Chien P, Osherovich L Z, Weisman J S. Molecular basis of a yeast prion species barrier. Cell. 2000;100:277–288. doi: 10.1016/s0092-8674(00)81565-2. [DOI] [PubMed] [Google Scholar]
- 62.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 63.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 64.Schirmer E C, Lindquist S. Interactions of the chaperone Hsp104 with yeast Sup35 and mammalian PrP. Proc Natl Acad Sci USA. 1997;94:13932–13937. doi: 10.1073/pnas.94.25.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Serio T R, Lindquist S L. [PSI+]: an epigenetic modulator of translation termination efficiency. Annu Rev Cell Dev Biol. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- 66.Sevarino K A, Poyton R O. Mitochondrial membrane biogenesis: identification of a precursor to yeast cytochrome c oxidase subunit II, an integral polypeptide. Proc Natl Acad Sci USA. 1980;77:142–146. doi: 10.1073/pnas.77.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 68.Stansfield I, Jones K M, Kushnirov V V, Dagkesamanskaya A R, Poznyakovski A I, Paushkin S V, Nierras C R, Cox B S, Ter-Avanesyan M D, Tuite M F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Suissa M, Reid G A. Preparation and use of antibodies against insoluble membrane proteins. Methods Enzymol. 1983;97:305–311. doi: 10.1016/0076-6879(83)97142-2. [DOI] [PubMed] [Google Scholar]
- 70.Surguchov A P, Beretetskaya Y V, Fominykch E S, Pospelova E M, Smirnov V N, Ter-Avanesyan M D, Inge-Vechtomov S G. Recessive suppression in yeast Saccharomyces cerevisiae is mediated by a ribosomal mutation. FEBS Lett. 1980;111:175–178. doi: 10.1016/0014-5793(80)80786-1. [DOI] [PubMed] [Google Scholar]
- 71.Ter-Avanesyan M D, Dagkesamanskaya A R, Kushnirov V V, Smirnov V N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ter-Avanesyan M D, Kushnirov V V, Dagkesamanskaya A R, Didichenko S A, Chernoff Y O, Inge-Vechtomov S G, Smirnov V N. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 73.Ter-Avanesyan M D, Zimmermann J, Inge-Vechtomov S G, Sudarikov A B, Smirnov V N, Surguchov A P. Ribosomal recessive suppressors cause a respiratory deficiency in yeast Saccharomyces cerevisiae. Mol Gen Genet. 1982;185:319–323. doi: 10.1007/BF00330805. [DOI] [PubMed] [Google Scholar]
- 74.Tikhomirova V L, Inge-Vechtomov S G. Sensitivity of sup35 and sup45 suppressor mutants in Saccharomyces cerevisiae to the anti-microtubule drug benomyl. Curr Genet. 1996;30:44–49. doi: 10.1007/s002940050098. [DOI] [PubMed] [Google Scholar]
- 75.Tuite M F, Mundy C R, Cox B S. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vincent A, Liebman S W. The yeast omnipotent suppressor SUP46 encodes a ribosomal protein which is a functional and structural homolog of the Escherichia coli S4 ram protein. Genetics. 1992;132:375–386. doi: 10.1093/genetics/132.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wakem L P, Sherman F. Isolation and characterization of omnipotent suppressors in the yeast Saccharomyces cerevisiae. Genetics. 1990;124:515–522. doi: 10.1093/genetics/124.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wickner R B. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 79.Wickner R B, Masison D C, Edskes H K. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 80.Wickner R B, Taylor K L, Edskes H K, Maddelein M L, Moriyama H, Roberts B T. Prions in Saccharomyces and Podospora spp.: protein-based inheritance. Microbiol Mol Biol Rev. 1999;63:844–861. doi: 10.1128/mmbr.63.4.844-861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yaffe M P, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou P, Derkatch I L, Uptain S M, Patino M M, Lindquist S, Liebman S W. The yeast non-Mendelian factor [ETA+] is a variant of [PSI+], a prion-like form of release factor eRF3. EMBO J. 1999;18:1182–1191. doi: 10.1093/emboj/18.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]