ABSTRACT
The developmental programs that build and sustain animal forms also encode the capacity to sense and adapt to the microbial world within which they evolved. This is abundantly apparent in the development of the digestive tract, which typically harbors the densest microbial communities of the body. Here, we review studies in human, mouse, zebrafish and Drosophila that are revealing how the microbiota impacts the development of the gut and its communication with the nervous system, highlighting important implications for human and animal health.
KEY WORDS: Enteroendocrine cell, Gastroenterology, Gut-brain axis, Interoception, Microbiome, Stem cell
Introduction
The complex communities of microorganisms that colonize animals (microbiotas) have been identified as major environmental factors that can shape animal development. Indeed, research using a variety of models has continued to uncover remarkable ways in which microbiotas contribute to animal development and physiology, within individual life cycles and over evolutionary timescales (McFall-Ngai et al., 2013). Studies in extant representatives of early animal lineages, such as choanoflagellates (Woznica and King, 2018), echinoderms (Carrier and Reitzel, 2020) and cnidaria (Deines et al., 2017), indicate that host-microbe interactions have been abundant from the earliest stages of animal evolution. For example, even the primitive nervous system that develops in the cnidarian Hydra has been shown to perceive microbial stimuli (Murillo-Rincon et al., 2017) and exert control over resident microbes (Augustin et al., 2017). This ancient theme of host-microbial interactions has continued to shape the evolution, development and physiology of animal lineages. Furthermore, revelations from studies of host-microbial interactions have challenged traditional views of biological individuality and support a view of animal development and physiology as emergent outcomes of dynamic ecological interactions (Gilbert et al., 2012). These interactions are most salient in the digestive tract, which is the primary site of dietary nutrient absorption and often harbors the densest and most diverse microbial communities in the animal.
The purpose of this Review is to summarize our current understanding of how the microbiota contributes to the development of the digestive tract, with particular emphasis on the interactions between the intestinal epithelium and the nervous system. Although these interactions apply to all animals, the majority of this Review focuses on studies in humans, mice and zebrafish, with some comparison to the fruit fly, Drosophila. We focus on these animals for two main reasons. First, these animals together represent a relatively broad range of evolutionary distances, thereby facilitating the identification of mechanisms and principles that have been conserved since their respective last common ancestors (Benton and Donoghue, 2007). Second, historic research emphasis on these animal species has already afforded extensive insights into the mechanisms and principles underlying their host-microbe interactions and intestinal biology. Whereas emphasis on humans is motivated by human health interests, emphasis on model systems such as Drosophila, zebrafish and mice has been motivated by the abundance of experimental tools and genomic resources available in those systems. In particular, these model organisms permit the controlled experimental manipulation of host genotype as well as microbial, dietary and other environmental exposures (see Box 1).
Box 1. The experimental manipulation of microbiota.
Most animals used in developmental biology and physiology research are reared and maintained under laboratory conditions, in which they are colonized with microbial communities from their local environment. The study of host-microbe interactions requires control over microbial exposure and this can be achieved by multiple means. For example, broad spectrum antibiotics can be administered to reduce and modify microbiota and test their impact on a given host phenotype. However, this approach does not typically distinguish between the effects of reduced antibiotic-sensitive microbiota and residual antibiotic-resistant microbiota, or direct toxicological effects of the antibiotics on the host (Morgun et al., 2015). Other approaches include the use of animals reared under gnotobiotic conditions, for example those reared in the absence of microbiota (i.e. germ-free or axenic animals) or ex-germ-free animals colonized with defined microbial strains or communities. Germ-free animals display numerous developmental, physiological and immunological differences from conventionally-reared and otherwise colonized animals, which creates caveats for certain types of experiments (Kennedy et al., 2018). However, each of these differences underscore the large extent to which our concepts of ‘normal’ animal biology are implicitly determined by microbial inputs normally encountered throughout the life cycle. With all of these general approaches, a given microbial community, strain or product can be introduced to test its sufficiency to influence a given phenotype.
The choreography between microbiota and intestinal development
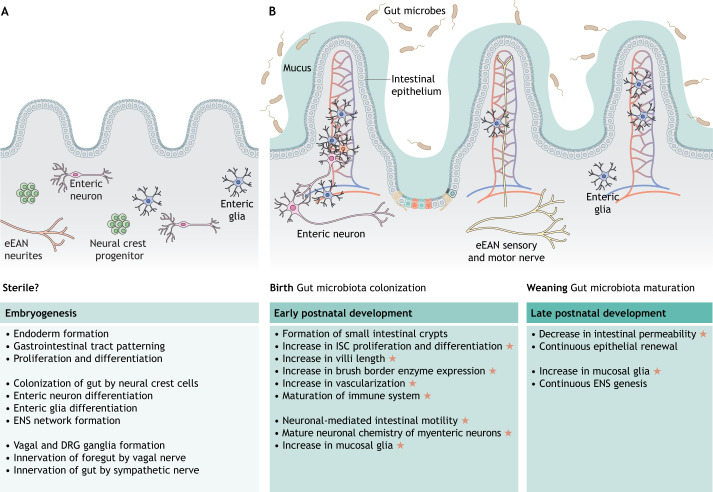
The orchestrated process of animal development is paralleled by the ways in which microbes in their surrounding environment begin to colonize and influence their developing bodies. It is notable that early embryonic development typically occurs in the protective confines of a chorion and other structures that provide a physical barrier excluding most, if not all, microbial cells from directly contacting the developing embryo. The process of birth or hatching then represents the first substantial contact of the animal with microbial cells, although a major exception is seen in the case of arthropods, which are able to engage in early endosymbiotic relationships with microbes (reviewed by Perlmutter and Bordenstein, 2020). Consequently, early development in healthy animals has traditionally been considered to occur in the absence of free-living microorganisms. This concept has been challenged in recent years based on the reported observations of microbial cells associated with mammalian embryos in utero (Chen and Gur, 2019) and the discovery that metabolites and products from the maternal microbiota in mice are able to enter the maternal-fetal circulation and thereby influence immune system development (Gomez de Agüero et al., 2016). The existence and prevalence of prenatal microbiota across animal species in natural or lab-reared habitats, or the extent to which they shape animal development, are active areas of research. However, the ability of researchers to routinely derive conventionally-reared animals into germ-free (GF) conditions suggests that the environment within the chorion is usually axenic, at least at the time of derivation. Regardless, the chorion and associated prenatal barriers effectively limit the extent of microbial contacts and exposures during early development, insulating those critical stages of the life cycle against microbial and other environmental influences (Fig. 1).
Fig. 1.
Gut microbiota regulate intestinal development. (A,B) Microbiota colonize the intestine immediately following birth and engage in co-evolved relationships with the host developmental program. Listed are the key steps and processes involved in pre- (A) and postnatal (B) development of the intestine. Orange stars indicate the developmental events that are known to be regulated by gut microbiota. Created using BioRender.com. DRG, dorsal root ganglion; eEAN, extrinsic enteric-associated neurons; ENS, enteric nervous system; ISC, intestinal stem cells.
At the time of birth or hatching, animals become exposed to new sources of microbes in their respective environments that can subsequently colonize their digestive tracts and influence their biology. In all vertebrates, the embryonic origin of the intestinal tract appears to be similar. Specifically, the endoderm gives rise to the epithelial layer that lines the gastrointestinal (GI) lumen, while mesenchymal cells within the lamina propria support the epithelium and, along with the epithelium, form the mucosa. Mesenchymal cells also form the outer strata including the submucosa, the muscularis and the serosa (Dauça, 1990). In addition to these structures, a plexus of branched interconnected vessels develops in the small intestine of mammals and zebrafish coincident with the period of microbiota assembly (Stappenbeck et al., 2002; Willms et al., 2021 preprint), suggesting that these two events might be connected. Indeed, colonization of GF mice with microbiota promotes increased vessel density in the small intestine via microbiota-induced extravascular protease-activated receptor (PAR) signaling (Reinhardt et al., 2012). More recently, research using gnotobiotic zebrafish has revealed that commensal gut microbiota promote transduction of vascular endothelial growth factor (VEGF)-dependent signals and facilitate intestinal vascularization (Willms et al., 2021 preprint) (Fig. 1).
In mammals, the maturity of the intestine at birth is generally dependent upon the length of the gestational period. In humans, organogenesis of the intestine is completed by 13 weeks of gestation (Montgomery et al., 1999) and the crypts and villi of the small intestine become organized and functional well before birth (Walthall et al., 2005). Postnatal maturation of the human GI tract includes maturation and proliferation of cells within the small and large intestinal epithelium (Walthall et al., 2005). Mice and rats have a relatively short gestational period and thus have a relatively immature intestine at birth (Henning, 1981). As a result, postnatal maturation in rodents involves replacing the immature epithelium with a mature epithelium (Carlile and Beck, 1983). The epithelial crypts of the rodent small intestine are formed after birth (Ariely, 1975) and the proliferation of crypt cells is low until weaning. In rats, a marked increase in cell division and migration occurs in intestinal crypts during weaning (postnatal day 15 to 30). This results in an increase in epithelial cell number and in the absorptive ability of the small intestine (Henning, 1981; Buts and De Meyer, 1981). This rapid and pronounced functional maturation of the gut happens during the suckling-weaning transition, a time when significant changes in gut microbiota occur (Derrien et al., 2019). As discussed in greater detail elsewhere (Torow and Hornef, 2017; Sommer and Bäckhed, 2013; Round and Mazmanian, 2009) and below, animals reared under germ-free conditions display attenuation or loss of features associated with normal intestinal and immune system development, including differentiation of the epithelial brush border, development of the gut-associated lymphoid tissues (GALT) and differentiation of distinct immune cell subsets. Identifying the specific microbes and microbial signals responsible for these aspects of intestinal maturation remains an important challenge in the field.
In humans, early infant microbiota composition is influenced by gestational age, birth mode, maternal microbiota, antibiotic exposure and early-life feeding practices. With the introduction of solid foods and discontinuation of breast milk feeding, a significant shift in gut microbial structure occurs. This results in the establishment of an adult-like microbiome, which is typically dominated by the bacterial phyla Bacteroidetes and Firmicutes by three years of age (Bäckhed et al., 2015; Dogra et al., 2015a,b; Yatsunenko et al., 2012; Sprockett et al., 2018). Mouse gut microbiota assembly is governed by similar processes (Pantoja-Feliciano et al., 2013; Carmody et al., 2015; Martínez, 2018; Morais et al., 2020). In addition to these temporal dynamics, the human and mouse gut microbiota also exhibit spatial dynamics along the anterior-posterior axis, with the highest densities occurring in the colon (Donaldson et al., 2016). Microbiota density also varies along the crypt-villus axis, with anaerobic Bacteroidetes and Firmicutes predominating in the luminal contents and crypt-associated communities that contain aerobic Proteobacteria taxa, similar to those found in the intestines of zebrafish and Drosophila (Pédron et al., 2012).
In oviparous animals such as zebrafish and Drosophila, the developing animal encounters environmental microbes immediately after hatching from the chorion. In zebrafish, larvae hatch from their protective chorion at a time point when the major steps of organogenesis of the digestive tract have finished. Interestingly, the zebrafish intestinal lumen becomes patent around the same time as the larva hatches, which permits the immediate colonization of the intestinal lumen with environmental microbes (Rawls et al., 2007). Zebrafish gut microbial community composition varies during development, dominated initially by aerobic Proteobacteria during larval stages, with anaerobic Fusobacteria and Firmicutes accumulating during adult stages (Rawls et al., 2004; Stephens et al., 2016). Functional genomic comparisons of intestinal epithelial gene expression between zebrafish and mice indicate conserved organization along the anterior-posterior axis, which in zebrafish consists of a small intestine, an ileum and a colon-like region (Wang et al., 2010; Lickwar et al., 2017). Although the mouse gut microbiota displays significant compositional differences along these different regions (van den Bogert et al., 2011), this has not been yet assessed in zebrafish.
The digestive tract in Drosophila has three distinct segments: the foregut, midgut and hindgut. The foregut and hindgut originate from ectoderm, whereas the midgut, which is where most food digestion occurs, is derived from the endoderm. During metamorphosis, the endodermal progenitors that are dispersed along the larvae midgut proliferate and differentiate to form a new adult midgut epithelium that eventually replaces the degenerated larval midgut. Unlike vertebrates, transovarial and parental microbial transmission (i.e. transmission inside the eggs) is common in Drosophila (Perlmutter and Bordenstein, 2020). Therefore, it is likely that some microbial colonization happens before hatching. Subsequently, gut microbial composition and density change with developmental stages and as flies age. Unlike mammals and zebrafish, Drosophila raised in the lab are associated with relatively simple bacterial communities, predominated by four core species from Firmicutes and Proteobacteria (Ludington and Ja, 2020).
The effects of gut microbiota on intestinal epithelial cell lineages
The intestine's apical surface is lined with a single layer of epithelial cells, collectively called the intestinal epithelium. The intestinal epithelium has a remarkable capacity for adaptation and self-renewal throughout life. Among the animal tissues that exhibit renewal capabilities, rates of renewal are fastest in the intestinal epithelium, made possible by intestinal stem cells (ISCs) located at the base of the epithelial crypts. Asymmetric division of ISCs yield cells that differentiate into secretory and absorptive cellular lineages. The latter primarily consists of enterocytes that function to absorb dietary nutrients. In mammals, the secretory cellular lineages include Paneth cells that specialize in production of antimicrobial proteins, goblet cells that specialize in regulated secretion of mucus, and enteroendocrine cells (EECs) that specialize in regulated secretion of hormones and neurotransmitters. In addition to these main lineages, the mammalian intestine contains additional rare epithelial lineages involved in immune responses, including tuft cells and M-cells (Fig. 2A).
Fig. 2.
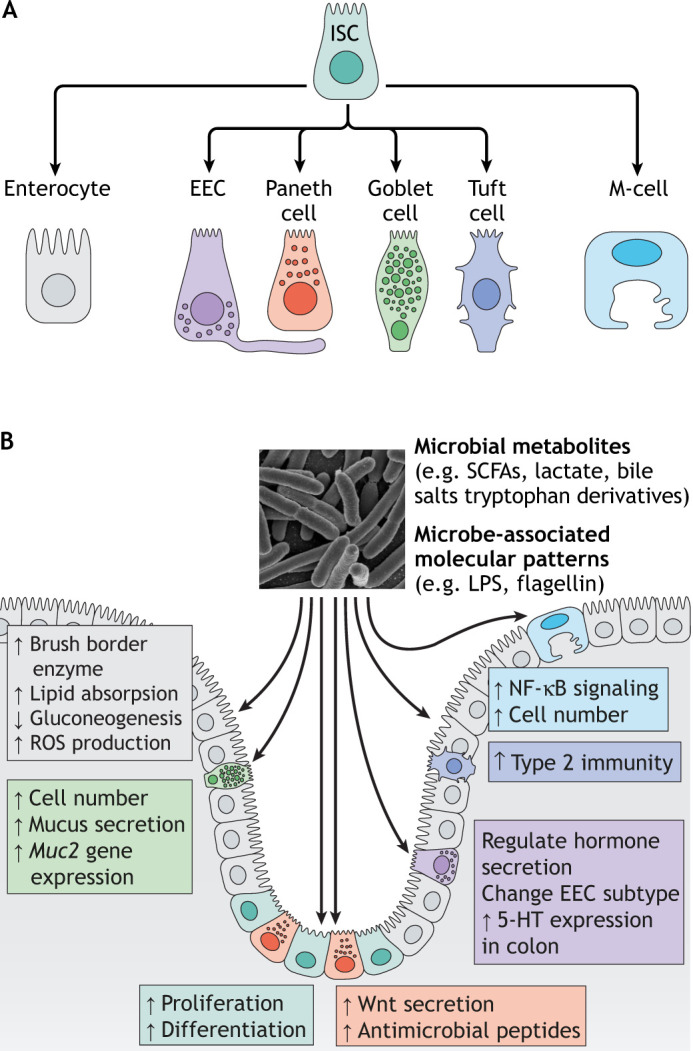
The gut microbiota regulates intestinal lineages. (A) In mammals, intestinal stem cells (ISCs) differentiate into absorptive cells (enterocytes), enteroendocrine cells (EECs), goblet cells, Paneth cells, tuft cells and M-cells. (B) Gut microbiota regulate the development and function of these intestinal cell types in various ways via microbial metabolites or microbial-associated molecular patterns. Created using BioRender.com. LPS, lipopolysaccharide; ROS, reactive oxygen species; SCFA, short chain fatty acid.
ISCs, enterocytes and EECs are ancient cellular programs present in all triploblastic animals, including Echinoderms and insects (García-Arrarás et al., 2019; Zeng and Hou, 2015). In vertebrates, the intestinal epithelium also evolved to produce goblet cells (Wallace et al., 2005; Ye et al., 2019). Paneth cells are found in mammals, and Paneth-like cells are found in some fish species such as salmon (Paulsen et al., 2001; Sveinbjornsson et al., 1996), but not in zebrafish. Neither tuft cells nor M-cells have been described in non-mammalian species, although recent single-cell RNA-seq studies of the zebrafish intestine have revealed cell clusters expressing markers of mammalian tuft cells and M-cells, suggesting similar cell lineages may exist in zebrafish (Willms et al., 2021 preprint; Wen et al., 2020 preprint). In Drosophila, the midgut intestinal epithelium is derived from endoderm and is composed of four different cell types: ISCs, enteroblast cells (EBs), EECs and enterocytes. The EBs are undifferentiated ISC daughters that can differentiate into EECs or enterocytes (Capo et al., 2019). The similarities and differences between intestinal epithelial composition and physiology in Drosophila, zebrafish and mice have been reviewed elsewhere (Ferguson and Foley, 2021).
Studies using GF mice, zebrafish and Drosophila have established that, although microbial colonization is not required for animal survival as long as nutritional requirements are met, it does exert diverse effects on GI tract development and physiology (Douglas, 2019; Kostic et al., 2013). Below, we describe the impacts of gut microbiota on each major intestinal epithelial cell type (Fig. 2B). The effects of microbiota on the emergent physiological function of the GI tract have been reviewed elsewhere (Sommer and Bäckhed, 2013; Zheng et al., 2020; Dabke et al., 2019).
Intestinal epithelial stem cells
The self-renewing and proliferative activity of ISCs, which maintain intestinal homeostasis under physiological and pathological conditions, is regulated by diverse developmental and environmental stimuli (Hou et al., 2017). ISCs in the mouse intestine reside at the crypt base and are interspersed by Paneth cells (Clevers, 2013). ISC proliferation requires Wnt signals that are produced by Paneth cells (Farin et al., 2016). Mice raised in GF conditions exhibit reduced crypt depth, and colonizing GF mice with commensal gut microbiota increases ISC proliferation and deepens crypt depth in the small and large intestine (Lesher et al., 1964; Peck et al., 2017). Supplementation of a Lactobacillus plantarum strain in conventionally-raised wild-type mice increases ISC proliferation in both the small and large intestine through the generation of lactate. Lactate stimulates the G-protein coupled receptor Gpr81 (also known as Hcar1) in Paneth and stromal cells causing them to secrete Wnt signals that act on ISCs to increase proliferation (Lee et al., 2018). Similarly, gnotobiotic studies in zebrafish suggest that gut microbiota increase ISC proliferation to maintain the healthy intestinal epithelium (Chen et al., 2010; Hooper et al., 2001; Cheesman et al., 2011). Studies in Drosophila also show that GF flies exhibit reduced numbers of mitotic stem cells and epithelial renewal rates compared with conventionally-raised flies (Buchon et al., 2009a,b; Amcheslavsky et al., 2009; Cronin et al., 2009).
Microbiota-induced signals from enterocytes and EECs also regulate ISC proliferation. For example, intestinal infection of adult fruit flies with Pseudomonas aeruginosa causes apoptosis of enterocytes; signaling from the damaged enterocytes in turn promotes dramatic ISC proliferation and oncogene expression in the intestinal epithelium (Apidianakis et al., 2009; Bonfini et al., 2016). In Drosophila, commensal Lactobacilli bacteria also increase ISC proliferation by stimulating enterocytes to produce reactive oxygen species (Wang et al., 2013; Jones et al., 2013; Buchon et al., 2009a,b). In addition to enterocytes, EECs regulate ISC proliferation, as EEC deletion in Drosophila significantly impairs diet-stimulated ISC proliferation in the gut (Amcheslavsky et al., 2014). Transcriptomic analysis of midguts from Drosophila infected with the pathogen Pseudomonas entomophila suggests that EECs may sense and respond to pathogenic bacteria (Dutta et al., 2015). However, whether and how EECs integrate commensal and pathogenic microbial signals to regulate ISC proliferation remains unknown.
Enterocytes
Enterocytes compose more than 80% of the intestinal epithelial cell population. They carry out essential GI tract functions by expressing digestive enzymes to facilitate nutrient digestion, absorbing and transporting nutrients and electrolytes, secreting fluids and antimicrobial peptides, and creating a physical barrier. Absorptive functions of enterocytes are mediated by the formation of brush border microvilli on their apical luminal surface. Studies suggest that gut microbiota strongly affect the development of this enterocyte brush border. For example, mice raised GF display reduced brush border differentiation and have reduced villus thickness (Sommer and Bäckhed, 2013). The intestinal brush border expresses numerous hydrolase enzymes, including glucosidases, alkaline phosphatase and γ-glutamyltransferase. The activities of some enzymes (i.e. sucrase, maltase, isomaltase, alkaline phosphatase) are not detected or are low at birth and begin or increase at weaning. Comparison of GF and conventionalized neonatal pigs suggests that enterocytes respond to microbial colonization by increasing their expression of the brush border enzymes lactase phlorizin hydrolase and aminopeptidase N (Willing and Van Kessel, 2009). Another study in gnotobiotic rats suggests that activity of sucrase displays a circadian pattern, with GF rats exhibiting much lower activity than conventionally-raised rats (Olsen and Korsmo, 1982). Similarly, colonization of GF zebrafish with normal microbiota increases intestinal proliferation and promotes the expression of intestinal alkaline phosphatase, which detoxifies bacterial lipopolysaccharide (LPS) (Bates et al., 2007).
Nutrient absorption and metabolism by small intestinal enterocytes are also regulated by microbiota. Studies in gnotobiotic zebrafish have revealed that microbiota colonization promotes intestinal absorption and extra-intestinal metabolism of dietary lipids (Semova et al., 2012; Sheng et al., 2018). This microbial influence is strain-specific (Semova et al., 2012) and genetic tools have been developed for one of the bioactive strains (Bae et al., 2016). In accord, a study has demonstrated that GF mice exhibit impaired lipid digestion and absorption compared with colonized controls, which likely contributes to their resistance to diet-induced obesity (Martinez-Guryn et al., 2018). In addition, the transplantation of small intestinal microbiota from mice fed a high-fat diet promotes lipid absorption in GF recipient mice, suggesting the small intestinal microbiota plays crucial roles in host adaptability to dietary lipids (Martinez-Guryn et al., 2018). The microbiota also induces the rhythmic expression of histone deacetylase 3 (HDAC3) in small intestinal epithelial cells, which drives oscillations in intestinal metabolic gene expression, especially for genes implicated in nutrient transport and lipid metabolism (Kuang et al., 2019). These results in zebrafish and mice suggest that the gut microbiota is a target for regulating dietary fat absorption. However, the microbial signals regulating intestinal lipid absorption remain poorly understood. A recent study using cultured intestinal epithelial cells indicates that fermentation products from gut commensals can affect enterocyte lipid metabolism (Araújo et al., 2020), although it remains unclear whether and how these microbial products contribute to the ability of the microbiota to promote dietary fat absorption in vivo (Semova et al., 2012; Martinez-Guryn et al., 2018). Recent studies have revealed that the ability of intestinal epithelial cells to adapt to dietary nutrients and microbes is heavily influenced by immune cells (Wang et al., 2017; Sullivan et al., 2021). A comprehensive understanding of the microbiota's influence on intestinal lipid metabolism will therefore need to consider microbes and nutrients in the physiochemical environment of the intestinal lumen along with epithelial and other non-epithelial cell types.
Gut microbiota also regulate enterocyte development in the small intestine. The major absorptive cell in the ileum of suckling-stage mammals is specialized for the uptake and digestion of milk macromolecules from the intestinal lumen to assist in immune system maturation (Gonnella and Neutra, 1984; Henning, 1985; Robberecht et al., 1971; Wilson et al., 1991; Weström et al., 2020). These specialized enterocytes exhibit an apical endocytic complex, are highly vacuolated and are enriched in large lysosomes (Gonnella and Neutra, 1984). In mammals, as the intestinal epithelium undergoes significant structural and biochemical changes during the suckling-to-weaning transition, these lysosome-rich enterocytes (LREs) are replaced with regular enterocytes (Muncan et al., 2011; Park et al., 2019). LREs are conserved in zebrafish (Park et al., 2019) and other fish (Rombout et al., 1985), persisting into adulthood (Lickwar et al., 2017; Wallace et al., 2005). Several studies in zebrafish have indicated that LRE function may be enhanced by microbiota colonization (Rawls et al., 2004; Bates et al., 2006). Whether gut microbiota affects the ileal transition from LRE to mature enterocyte in mammals, and whether LREs regulate the gut microbial ecology or immunology in the vertebrate gut, remain unknown.
Paneth cells
Paneth cells are specialized small intestinal epithelial cells that reside at the base of crypts and contribute to innate intestinal immunity by secreting a diverse repertoire of antimicrobial proteins (Ayabe et al., 2000). They also play a crucial role in maintaining gut microbial homeostasis and protect the intestine from the invasion of microbial pathogens (Bevins and Salzman, 2011). Paneth cell granule release can be directly triggered by gut microbial products, such as LPS and lipoteichoic acid from bacteria, but not fungal or protozoal products (Ayabe et al., 2000). LPS and other microbial-associated molecular patterns (MAMPs) stimulate Toll-like receptors (TLR) in Paneth cells, which trigger downstream antimicrobial signaling cascades through the TLR adaptor protein Myd88 (Ayabe et al., 2000). In response, Paneth cells increase their expression of multiple antimicrobial factors that constitute a chemical barrier, which reduces microbial penetration of the intestinal mucosa (Vaishnava et al., 2008; Kobayashi et al., 2005). Paneth cell numbers are also higher in the jejunum of conventional mice compared with GF controls (Schoenborn et al., 2019). In this context, commensal bacteria induce Paneth cells to secrete antimicrobial peptides that act to contain commensals within the intestinal lumen and to protect from potentially invasive pathogens.
Goblet cells
The mucus secreted by intestinal epithelial goblet cells covers the epithelial cell lining and functions as a lubricant that facilitates the transit of intestinal luminal contents and serves as a protective barrier against microbial invasion (Johansson et al., 2011). Studies using gnotobiotic rodents have revealed that microorganisms significantly affect mucus thickness and composition (Schroeder, 2019). Compared with conventionally raised rats, GF rats have fewer goblet cells and a thinner mucus layer (Sharma et al., 1995). A similar phenotype has been observed in gnotobiotic zebrafish (Troll et al., 2018). Bacterial products such as LPS and peptidoglycan are sufficient to establish conventional colon mucus properties in GF mice (Petersson et al., 2011). In addition to modulating goblet cell development and formation, gut microbiota directly stimulate goblet cell mucus secretion to modulate mucus layer properties. A recent study suggests that specialized goblet cells localized to the colonic crypt entrance directly respond to MAMP molecules to secrete Muc2 mucins (Birchenough et al., 2016). Goblet cells can also directly engage in transport of luminal antigens to underlying immune cells, thereby contributing to oral tolerance (Kulkarni et al., 2020). These dynamic properties coupled with our emerging understanding of goblet cell heterogeneity place goblet cells as key mediators of host-microbe homeostasis (Nyström et al., 2021).
Tuft cells and M-cells
Compared with other cell lineages in the intestinal epithelium, tuft cells and M-cells are relatively rare. Tuft cells are crucial for the gut to orchestrate parasite-targeted immune responses. In response to parasite infection, the tuft cell population greatly expands and secretes cytokines to facilitate parasite clearance (Howitt et al., 2016). It remains unclear how commensal gut microbiota regulate tuft cell formation and function. Succinate, a common by-product of microbiota, stimulates the succinate receptor (SUCNR1) expressed in the tuft cells to activate a type 2 immune response in the mouse small intestine, suggesting gut commensal may play a role in regulating tuft cells (Nadjsombati et al., 2018). M-cells reside in the epithelium adjacent to mucosa-associated lymphoid tissues, such as the Peyer's patches (PPs) of the small intestine. M-cells actively transport luminal antigens to underlying lymphoid follicles to initiate an immune response. Differentiation of the M-cell lineage is directly controlled by signaling through the NF-κB pathway; blocking receptor activator of NF-κB ligand (RANKL) depletes M-cells, which diminishes the maturation of PPs (Ramakrishnan et al., 2019). Gut microbiota directly modulate M-cell functions by providing antigens that elicit M-cell function. In addition, the presence of gut microbiota may increase M-cell differentiation and M-cell numbers by activating the NF-kB pathway, as suggested by previous studies in mice (Peng et al., 2020).
Enteroendocrine cells
EECs are hormone-secreting cells that are distributed along the entire GI tract. In mammals, EECs composed less than 1% of the intestinal epithelium population. However, collectively these cells form the largest endocrine organ. Traditionally, EECs have been classified by the hormones they secrete. However, recent single-cell RNA-seq studies suggest that EECs exhibit tremendous heterogeneity, with EECs located in different GI tract regions expressing diverse hormones. For example, ghrelin is primarily expressed by gastric EECs in mammals and is secreted during the fasting stage to promote hunger (Gribble and Reimann, 2019, 2016). EECs in the ileum and large intestine express high levels of peptide YY (PYY) and glucagon-like peptide 1 (GLP-1, derived from the glucagon gene), which potently regulate intestinal motility, satiation and insulin secretion (Gribble and Reimann, 2019).
There is considerable interest in understanding how microbiota affect EEC biology. GF or antibiotic-treated mice increase basal GLP-1 secretion and the expression of glucagon transcripts in the colon relative to colonized controls, which can be suppressed by administration of short chain fatty acids (SCFAs) or colonization of polysaccharide-fermenting bacteria (Wichmann et al., 2013). GF mice also exhibit reduced total EEC numbers in the ileum but increased EEC numbers in the colon (Duca et al., 2012). In addition to GLP-1, GF mice display elevated expression of the EEC hormone insulin-like peptide 5 (INSL5) in the colon (Lee et al., 2016). INSL5 is known to promote hepatic glucose production (Burnicka-Turek et al., 2012), therefore increased EEC INSL5 expression may contribute to the ‘fasting’-like metabolic phenotype of GF mice (Lee et al., 2016). In addition to secreting hormones associated with metabolism, there is a specific EEC cell lineage that secretes the neurotransmitter serotonin (5-HT). These 5-HT-secreting EECs are termed enterochromaffin cells (ECs). ECs have important roles in regulating intestinal motility and communicating irritant signals from the intestinal lumen to the enteric and central nervous systems (Bellono et al., 2017). GF mice have lower 5-HT levels (Wikoff et al., 2009) and display lower EC density in the colon, but not in the small intestine, compared with conventional mice (Yano et al., 2015). In addition, the colonization of GF mice with spore-forming bacteria promotes the formation of colonic ECs (Yano et al., 2015).
In addition to regulating colonic EECs, gut microbiota regulate ileal EEC gene expression and function. A transcriptomic analysis of GLP-1-secreting EEC cells (L-cells) from GF and conventional mice revealed that gut microbiota exert rapid and profound effects on ileal L-cells but not colonic L-cells (Arora et al., 2018). Previous studies have suggested that activation of G-protein-coupled bile acid receptor 1 (GPBAR1; also known as TGR5 or M-BAR) increases L-cell differentiation in the mouse ileum (Lund et al., 2020). It is well appreciated that the gut microbiota are a major regulator of bile salt signaling, raising the possibility that the development and function of the L-cell lineage are modulated by interactions among gut microbiota and bile salts.
Together, these studies indicate that gut microbiota exert specific influences on multiple EEC lineages dependent on their position in the intestinal region. It is well known that gastric and small intestinal EECs secrete many hormones that are important for nutrient processing and energy homeostasis (Gribble and Reimann, 2019). Whether and how gut microbiota regulate the development and function of gastric and small intestinal EECs is less clear. A recent study using zebrafish demonstrated that a high-fat diet acutely and temporarily impairs proximal intestinal EEC nutrient sensing ability, a phenotype called ‘EEC silencing’. Interestingly, the high-fat diet-induced EEC silencing was found to be strongly dependent on gut microbiota (Ye et al., 2019). Whether such diet- and gut microbiota-regulated EEC functional changes are conserved in the intestines of mammals or other animals remains unknown. Moreover, the mechanisms underlying gut microbiota-regulated EEC development remain unclear. A previous study using gnotobiotic zebrafish suggests that gut microbiota may inhibit Notch signaling and promote the differentiation of secretory cell lineages, including EECs and goblet cells (Troll et al., 2018). Interestingly, GF Drosophila intestines display an increased EEC cell ratio over total cells (Broderick et al., 2014). Nonetheless, how exactly gut microbiota regulate EEC lineage development and differentiation remains a crucial gap of knowledge in the field.
The effects of gut microbiota on the enteric nervous system and intestinal motility
The GI tract is wired by a complex enteric nervous system (ENS; see Glossary, Box 2). During embryonic development, neural crest cells that are derived from the hindbrain and sacral region migrate to the endoderm and give rise to ENS neurons and glia throughout the GI tract. The GI tract is also innervated by sensory and motor neurons (see Glossary, Box 2) from the sympathetic and parasympathetic nervous system, both of which belong to the autonomic nervous system (ANS; see Glossary, Box 2). Unlike those from the ENS, the neuron bodies of ANS neurons are not in the gut wall but reside in the brain stem or spinal cord. The function and homeostasis of the GI tract are orchestrated by the cooperative interaction of the ENS and the ANS.
Box 2. Glossary of terms associated with the ENS and eEANs.
Autonomic nervous system (ANS): The division of the peripheral nervous system that innervates visceral organs and is responsible for regulating involuntary body functions such as heartbeat and food digestion. It is divided into the parasympathetic nervous system and the sympathetic nervous system, which exhibit counteractive functions. The sympathetic nervous system governs the ‘fight or flight’ response, whereas the parasympathetic nervous system governs the ‘digest and rest’ response. Both systems innervate the digestive tract. The parasympathetic nervous system promotes digestion and intestinal motility, whereas the sympathetic nervous system predominantly exerts inhibitory effects on intestinal motility and mucosal secretion.
Enteric nervous system (ENS): The intrinsic nervous system in the gastrointestinal (GI) tract that governs that GI tract function. It is completely separate from the central nervous system (CNS) and can function independently of brain or spinal cord input.
Extrinsic enteric-associated neurons (eEANs): Neurons that have neurites innervating the digestive tract, but their neuronal bodies do not reside within the gut wall. In vertebrates, eEANs are composed of vagal sensory neurons, parasympathetic neurons, spinal sensory neurons and sympathetic neurons. In Drosophila, eEANs are primarily neurons in the ventral ganglion and the proventricular ganglion.
Motor/efferent neurons: Neurons that carry neural impulses away from the CNS to the target tissue to control its function.
Sensory/afferent neurons: Neurons that carry nerve impulses from sensory stimuli to the CNS.
Spinal sensory neurons: Neurons that carry peripheral sensory information to the spinal cord. Their neuron bodies reside in the dorsal root ganglia (DRG).
Sympathetic neurons: The sympathetic nerves run parallel to the spinal cord. The preganglion neurons are located within the spinal cord. Their axons project to the sympathetic neurons in the prevertebral ganglion, axons from which then innervate the target organs. Similar to vagal motor neurons, these neurons may directly regulate intestinal function or form synapses with the ENS to indirectly regulate intestinal function.
Vagal motor neurons: Neurons in the vagus system that carry neural impulses from the brain to peripheral target organs. Their neuronal bodies reside in the dorsal motor nucleus of the vagus in the hindbrain. They are the main components of the parasympathetic nervous system and may directly innervate the intestinal epithelium, blood vessels or intestinal smooth muscle. They also synapse with the ENS to indirectly regulate intestinal function.
Vagal sensory neurons: Neurons in the vagus system that carry peripheral sensory information to the brain. Their neuronal bodies reside in the nodose ganglia that are at the base of the skull and behind the carotid canal.
The ENS in different animal models
The ENS is a network of neurons and glia within the wall of the GI tract. In mammals, the ENS is organized into two layers of interconnected ganglia, the outer myenteric and the inner submucosal plexus, which control almost all aspects of GI physiology (Spencer and Hu, 2020). Enteric neurons are classified into distinct subtypes according to morphological characteristics, intrinsic electrophysiological properties or the combinatorial expression of neurotransmitters and neuropeptides (see Box 3) (Furness, 2000; Brookes, 2001; Qu et al., 2008). Both enteric neurons and glia are derived from neuroectodermal progenitors, which delaminate from the neural crest and colonize the gut during embryogenesis. Most of the adult ENS forms during embryogenesis; however, newly born enteric neurons and glia continue to be integrated into pre-existing functional neural circuits for several weeks after birth (Pham et al., 1991; Laranjeira et al., 2011; Hao and Young, 2009; Young et al., 2003). Postnatal neurogenesis and gliogenesis coincide with and contribute to maturation of the digestive tract intrinsic neural circuits and the acquisition of spontaneous and induced motility patterns (Roberts et al., 2007; Kabouridis and Pachnis, 2015).
Box 3. Functional and chemical coding within the ENS.
The ENS is chemically coded using more than 30 different neuron transmitters, which are usually co-localized with their function. Sensory neurons usually express choline acetyltransferase (Chat; the enzyme that synthesizes acetylcholine), calbindin (Calb), calcitonin gene-related peptide (Cgrp) and substance P (SP). Interneurons may express 5-HT, dynorphin (Dyn) and somatostatin (Sst). The excitatory muscle motor neurons that innervate and activate the longitudinal and circular muscles and the muscularis mucosae throughout the GI tract usually express Chat and SP. On the other hand, the inhibitor muscle motor neurons usually express inhibitory neurotransmitters such as Dyn, enkephalins and nitric oxide synthetase (NOS). In addition to the muscle motor neurons, the ENS contains secretory motor neurons that regulate the mucosal epithelium secretion and blood flow. These neurons control epithelial secretion through acetylcholine release, and vasoactive intestinal polypeptide (VIP) appears to be essential for them to control blood flow.
Other vertebrates, including zebrafish, also exhibit a conserved ENS. Because zebrafish lack the submucosa layer found in the guts of amniotes, the zebrafish ENS lacks submucosal ganglia, and myenteric neurons are typically arranged as single neurons or neuronal pairs (Shepherd and Eisen, 2011). The genetic programs and growth factors that control the differentiation, migration and intestinal colonization of neural crest cells that form intrinsic enteric neurons in zebrafish are similar to those seen in mammals (Ganz, 2018). The zebrafish ENS network is developed and functional by the larval stage (Ganz, 2018). However, in contrast to mammalian systems, enteric glia in zebrafish apparently do not develop until adulthood (McCallum et al., 2020). It remains unclear whether other intestinal lineages fulfill the function of enteric glia in zebrafish larvae. Moreover, although neuronal control of intestine function can be observed in almost all animal phyla, ENS neurons with bodies in the gut wall are not found outside of the vertebrates. For example, in Drosophila, intestinal function appears to be regulated by neurons within the central nervous system (CNS) and by peripheral ganglia (collectively termed ‘extrinsic enteric-associated neurons’; see discussion below).
Gut microbiota regulate the development of the ENS
Accumulating evidence suggests that gut microbiota may play essential roles in regulating ENS development and neurotransmitter coding. Gut microbiota colonization in 12-week-old GF mice promotes proliferation of nestin+ enteric neuronal precursors and induces neuronal 5-HT production in adult mouse intestines (De Vadder et al., 2018). GF mice display altered spontaneous circular muscle contractions and decreased nerve density in the jejunum and ileum compared to colonized controls (Collins et al., 2014). The microbiota's impact on enteric circuitry is also highlighted by the ability of probiotic microbes to affect neurotransmitter coding in enteric neurons (Kamm et al., 2004; Joshua, 1991). Translatome profiling of the duodenum-, ileum- and colon-associated ENS from GF and conventionally-raised mice suggests that gut microbiota regulate the ENS in a regionally-specific manner (Muller et al., 2020a,b).
Besides regulating enteric neurons, gut microbiota may also play a role in regulating the development and function of enteric glial cells. Enteric glia provide nourishment and mechanical support for neurons and they contribute to the regulation of synaptic transmission and communication between the nervous and immune systems (Grubišić and Gulbransen, 2017). Studies in gnotobiotic mice show that gut microbiota promote enteric glia migration from the intestinal submucosa to the mucosa, thereby increasing the abundance of enteric glia in the mucosal layer (Kabouridis et al., 2015). Further evidence is provided by the finding that piglets treated with the probiotic Pediococcus acidilactici display higher enteric glial cell numbers in the inner and outer submucosal plexuses (Di Giancamillo et al., 2010).
In addition to regulating enteric neuron development, gut microbiota may directly regulate enteric neuron activity. Gut microbiota are required for normal gut intrinsic primary afferent neuronal excitability (McVey Neufeld et al., 2013). GF mice show reduced intrinsic sensory neuron excitability, which can be corrected following conventionalization with normal microbiota (McVey Neufeld et al., 2013). A recent study using transcriptomic analysis suggests that gut microbiota upregulate the transcription factor aryl hydrocarbon receptor (AhR) in enteric neurons in the distal GI tract, enabling them to respond to the luminal environment, including microbially-derived tryptophan metabolites (Obata et al., 2020).
Gut microbiota regulate intestinal motility
The ENS regulates many intestinal functions and plays a central role in controlling intestinal motility. Human patients harboring genetic mutations that result in loss of enteric neurons develop life-threatening constipation (Amiel et al., 2008; Lake and Heuckeroth, 2013). Studies in gnotobiotic mice suggest that gut microbiota may play an essential role in regulating intestinal motility. GF mice exhibit significantly slower intestinal transit (Wichmann et al., 2013; Kleene et al., 1985), and GF or antibiotic-treated mice display abnormal colonic migrating motor complexes (MMC) (Vincent et al., 2018). Interestingly, in contrast with mice, GF zebrafish larvae display increased intestinal motility and transit time compared with conventionalized controls (Bates et al., 2006). Gut microbiota likely regulate intestinal motility in a microbial species-dependent manner. For example, monoassociation of GF mice with anaerobic bacteria, including Clostridium tabificum, Lactobacillus acidophilus and Bifidobacterium bifidum, stimulates small intestinal transit time. By contrast, aerobic strains such as Micrococcus luteus and Escherichia coli suppress or have no significant effects on small intestinal transit time (Husebye et al., 2001). Gut microbiota may regulate intestinal motility by modifying ENS neuropeptide expression and secretion. For example, colonizing GF mice with conventional gut microbiota reduces neuropeptide Y, which may result in reduced inhibitor control and therefore promote intestinal motility (Husebye et al., 2001).
The mechanisms by which microbiota impact ENS function and intestinal motility are less clear. Past research has focused on microbial fermentation products, especially the SCFAs produced by colonic bacteria from polysaccharide catabolism. The effects of SCFAs differ based on chain length. In vitro and in vivo rat studies have shown that butyrate, but not propionate or acetate, increases the proportion of ascending excitatory cholinergic neurons and increases colonic contractile activity (Soret et al., 2010). In a study performed on guinea pigs, butyrate was shown to increase propulsive motility in the proximal colon and increase the velocity of pellet propulsion through the distal colon. In contrast, both propionate and acetate decrease colonic propagations (Hurst et al., 2014). In addition to SCFAs, gasses including methane (CH4) and hydrogen sulfide (H2S) produced during anaerobic bacterial metabolism slow intestinal transit and inhibit spontaneous and agonist-induced intestinal rhythmic contractile activity, suggesting gaseous products of bacterial metabolism can play important roles in regulating intestinal motility (Pimentel et al., 2006; Medani et al., 2011). In addition to bacterial metabolites, the bacterium Vibrio cholerae was shown to accelerate intestinal transit time in zebrafish hosts through its type VI secretion system, suggesting the presence of specific microbial secreted effectors that regulate intestinal motility (Logan et al., 2018). Characterizing the microbial and host mechanisms underlying the microbial regulation of ENS development and intestinal function remains a major goal in the field.
The effects of gut microbiota on extrinsic enteric-associated neurons
Extrinsic enteric-associated neurons (eEANs; see Glossary, Box 2) – comprising sensory afferents and autonomic efferents – are equipped to sense multiple areas of the intestine simultaneously, to transmit information to other tissues, and to complement the intrinsic ENS in the control of gut function (Muller et al., 2020a,b) (Fig. 3). In vertebrates, extrinsic enteric-associated sensory neurons are clustered in ganglia outside of the brain or spinal cord. In contrast, the bodies of efferent neurons are located within the brainstem or spinal cord (Uesaka et al., 2016). The efferent branch of eEANs is also defined as the ANS. Based on physiological effects, the ANS can be divided into the sympathetic and parasympathetic nervous systems. The ANS is essential for maintaining physiological functions involving the cardiovascular, respiratory and GI systems, and it is intricately connected to higher brain systems involved in emotion and behavior (Mulkey and Plessis, 2018).
Fig. 3.
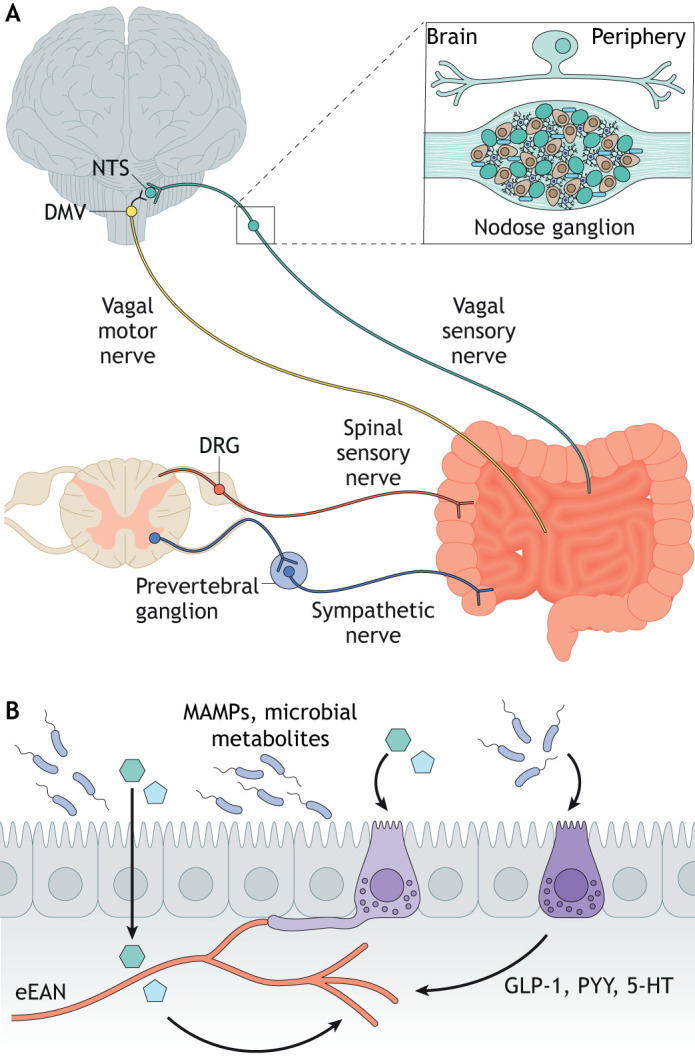
The gut microbiota regulates extrinsic enteric-associated neurons. (A) Vagal and spinal sensory and motor neurons innervate the mammalian digestive tract. Vagal and spinal sensory neuronal cell bodies reside in the ganglia adjacent to the brainstem or spine. A representative pseudounipolar vagal sensory neuron is shown above the nodose vagal ganglion. (B) Gut microbial signals may translocate across the intestinal epithelium and act on receptors on the nerve ends of extrinsic enteric-associated neurons (eEANs) to regulate eEAN function. Gut microbial stimulants also act on intestinal epithelial cells such as enteroendocrine cells (light and dark purple cells), which in turn may indirectly affect the development and function of eEANs. Created using BioRender.com. The nodose vagal ganglion drawing is based on Waise et al., 2018. DMV, dorsal motor nucleus of the vagus; DRG, dorsal root ganglion; MAMPs, microbe-associated molecular patterns; NTS, nucleus tractus solitarius.
Development of the vagal sensory and efferent systems in mammals
Vagal sensory and motor fibers (see Glossary, Box 2) are distributed throughout the GI tract, although colon vagal innervation is relatively sparse (Berthoud et al., 1991). The vagus nerve innervates the bowel from proximal to distal during embryogenesis (Ratcliffe et al., 2011). In mice, vagal nerve fibers first enter the esophagus at embryonic day (E)10 (Natarajan et al., 2002), the stomach at E11 (Baetge and Gershon, 1989), the duodenum at E14 (Murphy and Fox, 2007) and the distal small intestine by E16 (Ratcliffe et al., 2011). In rats, vagal sensory axons project to the gut before vagal motor axons arising from the nucleus ambiguous and the dorsal motor nucleus of the vagus (Rinaman and Levitt, 1993). These observations raise the possibility that vagal sensory neurons might provide signaling cues for efferent neurons to navigate the gut.
During embryonic and postnatal development, the synaptic connection between vagal sensory neurons and their target organs is established. A study in chicks suggests that more than half the precursor cells in vagal sensory ganglia (nodose ganglia) are lost during embryonic development, and the cell diameter of the surviving neurons increases (Harrison et al., 1994). The number of nodose ganglion neurons increases after hatching, and nodose ganglion neurons continue to develop, reaching their adult size by 2 weeks after hatching (Harrison et al., 1994). However, in the rat, from birth to 14 days postnatal development, the number of neurons in the nodose ganglion continues to decline while cell size continues to grow (Bakal and Wright, 1993). Distinct from brain neurons, vagal sensory neurons exhibit remarkable plasticity and regenerative capacity into adulthood. In adult rats, massive scale neurogenesis occurs in nodose ganglia following capsaicin-induced neuronal destruction (Gallaher et al., 2011; Czaja et al., 2008).
The mechanisms underlying vagal sensory neuron innervation, development and regeneration remain incompletely understood. In vitro culture studies suggest that the growth factor netrin 1 may play a role in attracting vagal sensory nerves into the GI tract. In culture, the neurites of nodose ganglion cells are attracted to foregut explants, but this can be blocked by netrin 1 antibodies (Ratcliffe et al., 2006). Other factors that are expressed in the developing gut and promote vagal innervation include brain-derived neurotrophic factor (BDNF) and neurotrophin 3 (Ernfors et al., 1994; Erickson et al., 1996). Besides attractive cues, vagal innervation is likely guided by repellent cues such as Slit. Indeed, neurites extending from nodose neurons are repelled by Slit2 in vitro, suggesting that Slit signaling may play a role in determining which cell types in the gut wall are innervated by vagal fibers (Goldberg et al., 2013).
The development of dorsal root ganglion sensory neurons and sympathetic efferent neurons in mammals
In developing mice, neural crest cells are located dorsally to the neural tube and migrate ventrally to the dorsal root ganglion (DRG) between E8.5 and E10 (Nascimento et al., 2018; Kasemeier-Kulesa et al., 2005). The peripheral projections of DRG sensory neurons can be detected at E12.5 (Taniguchi et al., 1997). In mammals, axons from DRG sensory neurons innervate both the proximal and distal intestine but are more prominent in the colon (Riera and Dillin, 2016). However, the exact timing of DRG neuron innervation of the GI tract is unclear. Sympathetic neurons (see Glossary, Box 2) that innervate the digestive tract arise from abdominal prevertebral ganglia comprising the coeliac, superior mesenteric and inferior mesenteric ganglia. Axons from sympathetic neurons grow towards their targets along arteries (Glebova and Ginty, 2005). In mice, sympathetic axons enter the gut wall around E15 (Hatch and Mukouyama, 2015). Many factors subsequently promote sympathetic axon innervation of the intestine. For example, the developing arteries express artemin (ARTN), endothelin (ET) and neurotrophin 3, which promote axon extension along arteries (Honma et al., 2002; Kuruvilla et al., 2004; Makita et al., 2008).
The development of eEANs in other model systems
The development of eEANs in other vertebrates has not been extensively studied. Like mammals, zebrafish larvae display the conserved structure of vagal sensory ganglia and a vagal motor nucleus in the brainstem (Olsson et al., 2008; Isabella et al., 2020; Barsh et al., 2017). The vagus nerve in zebrafish innervates the heart and, similar to mammals, the zebrafish vagal reflex is crucial in controlling heart rate and vasodilation (Stoyek et al., 2016). The vagus nerve also innervates pancreatic islets in zebrafish larvae, with sympathetic neuronal innervation of the pancreas being evident by juvenile stages (Yang et al., 2018). In addition, a recent study demonstrated that the vagus nerve in zebrafish larvae innervates the digestive tract and responds to enteric chemical and microbial stimuli (Olsson et al., 2008; Ye et al., 2021). These studies suggest that the afferent and efferent pathways in the parasympathetic nervous system are likely present and functional at the zebrafish larval stage. DRG neuronal differentiation in zebrafish begins in early larval stages but is not fully matured until the juvenile stage (An et al., 2002; McGraw et al., 2008). The formation and differentiation of the sympathetic ganglion is not initiated until the early juvenile stage (An et al., 2002). It remains unknown when during development the zebrafish sympathetic neuronal circuitry starts to be functional.
Unlike vertebrates, insects including Drosophila do not contain intrinsic enteric neurons with cell bodies in the intestinal wall. The neuronal bodies that elaborate axons to innervate the Drosophila intestine reside in the peripheral ganglia and brain (Lemaitre and Miguel-Aliaga, 2013; Schoofs et al., 2014; Cognigni et al., 2011). The neurites that extend from these neurons exhibit extensive sensory and efferent innervation of both intestinal muscles and epithelial cells, which are confined to discrete portions of the digestive tract (Cognigni et al., 2011). The eEANs in Drosophila are collectively referred to as the stomatogastric nervous system (SNS). In Drosophila, the neurites that innervate the proventriculus (a valve-like organ that gates the passage of food from the esophagus to the midgut), the anterior and posterior midgut, and the hindgut are derived from the ventral ganglion and the proventricular ganglion (Kuraishi et al., 2015). The neurons from these ganglia are derived from a small neurectodermal placode located in the foregut, that is, separate from the main neurectodermal domain of the segmented head and trunk that gives rise to the CNS (Hartenstein, 1997). The formation of the Drosophila SNS presents many parallels to the development of the vertebrate peripheral nervous system (PNS) (Hartenstein, 1997). Moreover, the neurons that innervate the Drosophila intestine appear to play a conserved role in relation to mammalian autonomic and intrinsic enteric neurons, in that they regulate appetite, intestinal secretion and intestinal transit (Cognigni et al., 2011).
Gut microbiota regulate eEAN function and development
The impact of gut microbiota on the development and function of eEANs is not well understood. A recent study that combined gnotobiotic mouse models with transcriptomics, circuit-tracing methods and functional manipulations demonstrated that the gut microbiota is crucial in modulating gut-extrinsic sympathetic neurons. Specifically, the absence of microbiota in GF mice led to increased expression of the neuronal transcription factor Fos. Colonization of GF mice with bacteria that produce SCFAs suppresses Fos expression in the gut sympathetic ganglia. This study further demonstrated that gut microbiota modulate sympathetic neuronal activity through a subset of distal intestine-projecting vagal sensory neurons. These results suggest that specific circuits exist to detect gut microbes and relay this information to areas of the CNS that, in turn, regulate gut physiology (Muller et al., 2020a,b).
Many other studies support the role of sensory eEANs in transducing enteric microbial information to the CNS. Cecal infection with the bacterial pathogen Campylobacter jejuni in mice leads to neuronal activation in the nucleus tractus solitarius, the first entrance of vagal afferents into the brain (Gaykema et al., 2004). Surgical disconnection of the vagal sensory nerve from the brain via vagotomy prevents the anxiety-reducing effect of beneficial Bifidobacterium longum in mice (Bercik et al., 2011). Similarly, the beneficial effects of Lactobacillus reuteri in a mouse autism model can be blocked following vagotomy (Sgritta et al., 2019). A recent study using zebrafish also suggests that specific enteric bacteria stimulate EECs to activate vagal sensory neurons through the production of tryptophan catabolites (Ye et al., 2021). Gut microbiota are also likely to modulate CNS function through spinal sensory nerve function. The branched-chain fatty acid isovalerate produced by gut bacteria directly activates EECs to stimulate the pelvic spinal sensory neurons (see Glossary, Box 2; Bellono et al., 2017). These studies highlight the crucial roles of eEANs in sensing and responding to diverse microbial stimuli.
Whether and how gut microbiota regulate development of the afferent and efferent autonomic nervous system remains almost entirely unknown, representing a major gap in our knowledge. In mammals, the differentiation, migration and innervation of vagal and sympathetic neurons occur during embryonic stages. However, neurogenesis, apoptosis and synaptic connections with target tissues and cells continue throughout postnatal development. As the gut microbiota typically colonizes the intestine during the postnatal stage, the development and function of the ANS might be altered. Gut microbiota also likely play crucial roles in controlling eEAN neurogenesis, apoptosis and synaptic connections with target intestinal cells. Gut microbiota can directly regulate eEANs through circulating metabolites (Yang and Chiu, 2017), and eEANs also receive signals from the intestinal epithelium, intestinal blood vessels and ENS (Critchley and Harrison, 2013). Therefore, it is possible that gut microbiota indirectly modulate eEAN development and function by initially acting on other intestinal cell types. In particular, several recent studies suggest that EECs form direct synaptic connections with eEANs to regulate eEAN activity (Kaelberer et al., 2018). Whether EECs regulate the development and innervation of eEANs is unknown. It also remains unclear when and how the synaptic connections between EECs and eEANs are formed and whether gut microbiota modulate EEC-EAN communication.
Specific metabolites produced by gut microbiota have been shown to directly or indirectly interact with eEANs (Fig. 3). For example, vagal sensory neurons express SCFA receptors (GPR41/43; also known as FFAR3/2) (Nøhr et al., 2015) and intraperitoneal SCFA injection activates vagal sensory neurons and inhibits food intake (Goswami et al., 2018). SCFAs are also shown to stimulate enteroendocrine cells in the intestinal epithelium, which in turn directly activate vagal sensory neurons through hormone or peptide secretion (Dalile et al., 2019) (Fig. 3). In addition to SCFAs, a recent study demonstrates that the microbial branched-chain fatty acid isovalerate can activate the olfactory receptor Olfr558 in ECs (Bellono et al., 2017). Serotonin released from ECs can then stimulate the mechanosensitive pelvic nerve (Bellono et al., 2017). In addition to SCFAs and branched-chain fatty acids, which are derived from bacterial carbohydrate metabolism, recent studies demonstrate that many other microbial amino acid metabolites might be crucial in mediating gut-eEAN communication (Liu et al., 2020). For example, gut microbiota can metabolize the essential aromatic amino acids phenylalanine, tyrosine and tryptophan into many bioactivate metabolites that can enter the circulation and interact with the PNS (Dodd et al., 2017). In addition, a recent study showed that the gut bacterial tryptophan metabolites indole and indole-3-aldehyde can stimulate the Trpa1 receptor in EECs to activate vagal sensory neurons as well as the ENS (Ye et al., 2021). Despite a wealth of data pointing to an association between gut microbial metabolites and CNS/PNS development and function, our understanding of the microbial sensory mechanisms used by animal hosts to recognize diverse microbial metabolites remains limited (Liu et al., 2020). Future studies using bioinformatics, synthetic biology and chemical screening to identify new receptors targeted by specific gut microbial metabolites should provide valuable insights that could further our understanding of how the gut microbiota interacts with the intestinal epithelium and how it communicates with and regulates eEAN development and function.
Conclusions and perspectives
The impact of gut microbiota on intestinal development directly affects many aspects of digestive tract physiology, including food digestion, nutrient absorption, intestinal motility, energy balance and immune function. In addition, gut microbiota significantly impact the development of the intestinal endocrine and neuronal systems, which directly impact how the intestine communicates chemical and physical stimuli from the gut to the rest of the body. With these abundant roles for gut microbiota under homeostatic conditions, it is perhaps not surprising that gut microbiota have also been implicated in diverse diseases and disorders. For example, causal roles for gut microbiota in the development of obesity and type 2 diabetes have been strongly supported by work in animal models as well as by human studies (Fan and Pedersen, 2021; Musso et al., 2010). Gut microbiota may directly impact metabolism by regulating intestinal cell development and nutrient absorption. In addition, gut microbiota may affect energy homeostasis by modulating eating behaviors and food choice. Gut EEC hormones and vagal signaling also play a crucial role in regulating appetite, satiation and feeding behavior. As such, abnormal gut hormonal and neuronal system development resulting from gut microbiome dysbiosis may directly contribute to the development of multiple metabolic and eating disorders (Tehrani et al., 2012; Fetissov and Hökfelt, 2019; Kim and de La Serre, 2018). Microbiota have also been implicated in other GI disorders including inflammatory bowel diseases (IBD) (Sartor and Wu, 2017) and colorectal cancer (Avril and DePaolo, 2021), as well as in bariatric surgery outcomes (Gutiérrez-Repiso et al., 2021) and the metabolism of diverse xenobiotics (Spanogiannopoulos et al., 2016).
Functional gastrointestinal disorders (FGIDs) are a group of prevalent GI diseases characterized by chronic or recurrent digestive symptoms, including abdominal pain or GI tract motility dysfunction (Drossman, 2006). Most FGID patients do not display identifiable structural or biochemical abnormalities in the GI tract (Drossman, 2006). Increasing evidence suggests that FGIDs may be attributed to abnormalities in the gut microbiome and dysfunctional gut-brain neuronal circuitry and communication (Collins, 2014; Mayer et al., 2014). FGIDs are common in both childhood and adults. Many studies suggest that early life events predispose individuals to childhood FGIDs and contribute to the development of FGIDs later in adulthood (Rasquin et al., 2006; Bonilla and Saps, 2013; Chitkara et al., 2008; Mendall and Kumar, 1998). These early life risk factors include enteric infection or antibiotic usage, both of which are associated with the disruption of normal gut microbiota (Bonilla and Saps, 2013; Mendall and Kumar, 1998). Perturbation of the gut microbiome during childhood development thus likely leads to the inappropriate formation and function of the ENS and eEANs, which would increase the risk of FGIDs.
In addition to metabolic and GI disorders, increasing studies have revealed that the gut microbiota is linked to many pediatric neurological diseases, including autism spectrum disorder (ASD) (Cryan et al., 2020). In addition to CNS-linked symptoms, many patients with ASD display complicated GI symptoms, including alteration of GI motility and increased visceral sensitivity. Furthermore, many ASD patients display dysregulated autonomic nervous system function and reduced vagal tone. Diverse genetic and environmental factors are associated with ASD symptoms including gut microbial dysbiosis (Li et al., 2017; Garcia-Gutierrez et al., 2020). However, it has been demonstrated that supplementation of probiotic microbes or fecal microbiota transplants may be beneficial for reversing ASD-associated social behavior deficits (Kang et al., 2019; Doenyas, 2018). Such gut microbiota may directly regulate central and peripheral neuronal programs through circulating metabolites, or perhaps by indirectly modulating CNS development and function through the enteroendocrine system and intrinsic or eEANs. Understanding the mechanisms by which gut microbiota regulate the development and function of intestine-associated hormonal and neuronal systems could thus provide new insights into the design of treatment strategies for disorders such as ASD and FGIDs.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by grants from the National Institutes of Health to L.Y. (K01-DK125527) and J.F.R. (P01-DK094779, R01-DK093399, R24-DK110492, R24-OD016761, R01-DK111857), and grants to J.F.R. from the Pew Charitable Trusts Innovation Fund, the Simons Foundation Autism Research Initiative, the Gordon and Betty Moore Foundation, and an Incubator Award from the Duke Institute for Brain Sciences, Duke University. Deposited in PMC for release after 12 months.
References
- Amcheslavsky, A., Jiang, J. and Ip, Y. T. (2009). Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4, 49-61. 10.1016/j.stem.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amcheslavsky, A., Song, W., Li, Q., Nie, Y., Bragatto, I., Ferrandon, D., Perrimon, N. and Ip, Y. T. (2014). Enteroendocrine cells support intestinal stem-cell-mediated homeostasis in Drosophila. Cell Rep. 9, 32-39. 10.1016/j.celrep.2014.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiel, J., Sproat-Emison, E., Garcia-Barcelo, M., Lantieri, F., Burzynski, G., Borrego, S., Pelet, A., Arnold, S., Miao, X., Griseri, P.et al. (2008). Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet. 45, 1-14. 10.1136/jmg.2007.053959 [DOI] [PubMed] [Google Scholar]
- An, M., Luo, R. and Henion, P. D. (2002). Differentiation and maturation of zebrafish dorsal root and sympathetic ganglion neurons. J. Comp. Neurol. 446, 267-275. 10.1002/cne.10214 [DOI] [PubMed] [Google Scholar]
- Apidianakis, Y., Pitsouli, C., Perrimon, N. and Rahme, L. (2009). Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc. Natl. Acad. Sci. USA 106, 20883-20888. 10.1073/pnas.0911797106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo, J. R., Tazi, A., Burlen-Defranoux, O., Vichier-Guerre, S., Nigro, G., Licandro, H., Demignot, S. and Sansonetti, P. J. (2020). Fermentation products of commensal bacteria alter enterocyte lipid metabolism. Cell Host Microbe 27, 358-375.e7. 10.1016/j.chom.2020.01.028 [DOI] [PubMed] [Google Scholar]
- Ariely, E. (1975). [Construction of total prostheses for patients with insufficient prosthesis support]. ZWR 84, 650-654. [PubMed] [Google Scholar]
- Arora, T., Akrami, R., Pais, R., Bergqvist, L., Johansson, B. R., Schwartz, T. W., Reimann, F., Gribble, F. M. and Bäckhed, F. (2018). Microbial regulation of the L cell transcriptome. Sci. Rep. 8, 1207. 10.1038/s41598-017-18079-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin, R., Schröder, K., Murillo Rincón, A. P., Fraune, S., Anton-Erxleben, F., Herbst, E.-M., Wittlieb, J., Schwentner, M., Grötzinger, J., Wassenaar, T. M.et al. (2017). A secreted antibacterial neuropeptide shapes the microbiome of Hydra. Nat. Commun. 8, 698. 10.1038/s41467-017-00625-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avril, M. and Depaolo, R. W. (2021). “Driver-passenger” bacteria and their metabolites in the pathogenesis of colorectal cancer. Gut Microbes 13, 1941710. 10.1080/19490976.2021.1941710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayabe, T., Satchell, D. P., Wilson, C. L., Parks, W. C., Selsted, M. E. and Ouellette, A. J. (2000). Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 1, 113-118. 10.1038/77783 [DOI] [PubMed] [Google Scholar]
- Bäckhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., Li, Y., Xia, Y., Xie, H., Zhong, H.et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 852. 10.1016/j.chom.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Bae, S., Mueller, O., Wong, S., Rawls, J. F. and Valdivia, R. H. (2016). Genomic sequencing-based mutational enrichment analysis identifies motility genes in a genetically intractable gut microbe. Proc. Natl. Acad. Sci. USA 113, 14127-14132. 10.1073/pnas.1612753113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetge, G. and Gershon, M. D. (1989). Transient catecholaminergic (TC) cells in the vagus nerves and bowel of fetal mice: relationship to the development of enteric neurons. Dev. Biol. 132, 189-211. 10.1016/0012-1606(89)90217-0 [DOI] [PubMed] [Google Scholar]
- Bakal, R. S. and Wright, L. L. (1993). Postnatal neuron death in the nodose ganglia of the rat. Dev. Neurosci. 15, 22-26. 10.1159/000111312 [DOI] [PubMed] [Google Scholar]
- Barsh, G. R., Isabella, A. J. and Moens, C. B. (2017). Vagus motor neuron topographic map determined by parallel mechanisms of hox5 expression and time of axon initiation. Curr. Biol. 27, 3812-3825.e3. 10.1016/j.cub.2017.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, J. M., Mittge, E., Kuhlman, J., Baden, K. N., Cheesman, S. E. and Guillemin, K. (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 297, 374-386. 10.1016/j.ydbio.2006.05.006 [DOI] [PubMed] [Google Scholar]
- Bates, J. M., Akerlund, J., Mittge, E. and Guillemin, K. (2007). Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371-382. 10.1016/j.chom.2007.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellono, N. W., Bayrer, J. R., Leitch, D. B., Castro, J., Zhang, C., O'Donnell, T. A., Brierley, S. M., Ingraham, H. A. and Julius, D. (2017). Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell 170, 185-198.e16. 10.1016/j.cell.2017.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton, M. J. and Donoghue, P. C. (2007). Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24, 26-53. 10.1093/molbev/msl150 [DOI] [PubMed] [Google Scholar]
- Bercik, P., Park, A. J., Sinclair, D., Khoshdel, A., Lu, J., Huang, X., Deng, Y., Blennerhassett, P. A., Fahnestock, M., Moine, D.et al. (2011). The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut-brain communication. Neurogastroenterol. Motil. 23, 1132-1139. 10.1111/j.1365-2982.2011.01796.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthoud, H. R., Carlson, N. R. and Powley, T. L. (1991). Topography of efferent vagal innervation of the rat gastrointestinal tract. Am. J. Physiol. Regul. Integr. Comp. Physiol. 260, R200-R207. 10.1152/ajpregu.1991.260.1.R200 [DOI] [PubMed] [Google Scholar]
- Bevins, C. L. and Salzman, N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356-368. 10.1038/nrmicro2546 [DOI] [PubMed] [Google Scholar]
- Birchenough, G. M. H., Nyström, E. E. L., Johansson, M. E. V. and Hansson, G. C. (2016). A sentinel goblet cell guards the colonic crypt by triggering Nlrp6-dependent Muc2 secretion. Science 352, 1535-1542. 10.1126/science.aaf7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfini, A., Liu, X. and Buchon, N. (2016). From pathogens to microbiota: how Drosophila intestinal stem cells react to gut microbes. Dev. Comp. Immunol. 64, 22-38. 10.1016/j.dci.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Bonilla, S. and Saps, M. (2013). Early life events predispose the onset of childhood functional gastrointestinal disorders. Rev. Gastroenterol. Mex. 78, 82-91. 10.1016/j.rgmx.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Broderick, N. A., Buchon, N. and Lemaitre, B. (2014). Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. mBio 5, e01117-14. 10.1128/mBio.01117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes, S. J. H. (2001). Classes of enteric nerve cells in the guinea-pig small intestine. Anat. Rec. 262, 58-70. [DOI] [PubMed] [Google Scholar]
- Buchon, N., Broderick, N. A., Chakrabarti, S. and Lemaitre, B. (2009a). Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23, 2333-2344. 10.1101/gad.1827009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon, N., Broderick, N. A., Poidevin, M., Pradervand, S. and Lemaitre, B. (2009b). Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5, 200-211. 10.1016/j.chom.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Burnicka-Turek, O., Mohamed, B. A., Shirneshan, K., Thanasupawat, T., Hombach-Klonisch, S., Klonisch, T. and Adham, I. M. (2012). INSL5-deficient mice display an alteration in glucose homeostasis and an impaired fertility. Endocrinology 153, 4655-4665. 10.1210/en.2012-1161 [DOI] [PubMed] [Google Scholar]
- Buts, J.-P. and de Meyer, R. (1981). Postnatal proximodistal development of the small bowel mucosal mass in growing rats. Biol. Neonate 40, 62-69. 10.1159/000241473 [DOI] [PubMed] [Google Scholar]
- Capo, F., Wilson, A. and Di Cara, F. (2019). The intestine of Drosophila melanogaster: an emerging versatile model system to study intestinal epithelial homeostasis and host-microbial interactions in humans. Microorganisms 7, 336. 10.3390/microorganisms7090336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile, A. E. and Beck, F. (1983). Maturation of the ileal epithelium in the young rat. J. Anat. 137, 357-369. [PMC free article] [PubMed] [Google Scholar]
- Carmody, R. N., Gerber, G. K., Luevano, J. M., Gatti, D. M., Somes, L., Svenson, K. L. and Turnbaugh, P. J. (2015). Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72-84. 10.1016/j.chom.2014.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier, T. J. and Reitzel, A. M. (2020). Symbiotic life of echinoderm larvae. Fron. Ecol. Evol. 7, 509. 10.3389/fevo.2019.00509 [DOI] [Google Scholar]
- Cheesman, S. E., Neal, J. T., Mittge, E., Seredick, B. M. and Guillemin, K. (2011). Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc. Natl. Acad. Sci. USA 108 Suppl. 1, 4570-4577. 10.1073/pnas.1000072107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. J. and Gur, T. L. (2019). Intrauterine microbiota: missing, or the missing link? Trends Neurosci. 42, 402-413. 10.1016/j.tins.2019.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. T., Malo, M. S., Moss, A. K., Zeller, S., Johnson, P., Ebrahimi, F., Mostafa, G., Alam, S. N., Ramasamy, S., Warren, H. S.et al. (2010). Identification of specific targets for the gut mucosal defense factor intestinal alkaline phosphatase. Am. J. Physiol. Gastrointest. Liver Physiol. 299, G467-G475. 10.1152/ajpgi.00364.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitkara, D. K., Van Tilburg, M. A. L., Blois-Martin, N. and Whitehead, W. E. (2008). Early life risk factors that contribute to irritable bowel syndrome in adults: a systematic review. Am. J. Gastroenterol. 103, 765-774. quiz 775 10.1111/j.1572-0241.2007.01722.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers, H. (2013). The intestinal crypt, a prototype stem cell compartment. Cell 154, 274-284. 10.1016/j.cell.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Cognigni, P., Bailey, A. P. and Miguel-Aliaga, I. (2011). Enteric neurons and systemic signals couple nutritional and reproductive status with intestinal homeostasis. Cell Metab. 13, 92-104. 10.1016/j.cmet.2010.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, S. M. (2014). A role for the gut microbiota in IBS. Nat. Rev. Gastroenterol. Hepatol. 11, 497-505. 10.1038/nrgastro.2014.40 [DOI] [PubMed] [Google Scholar]
- Collins, J., Borojevic, R., Verdu, E. F., Huizinga, J. D. and Ratcliffe, E. M. (2014). Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol. Motil. 26, 98-107. 10.1111/nmo.12236 [DOI] [PubMed] [Google Scholar]
- Critchley, H. D. and Harrison, N. A. (2013). Visceral influences on brain and behavior. Neuron 77, 624-638. 10.1016/j.neuron.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Cronin, S. J. F., Nehme, N. T., Limmer, S., Liegeois, S., Pospisilik, J. A., Schramek, D., Leibbrandt, A., Simoes, R. M., Gruber, S., Puc, U.et al. (2009). Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325, 340-343. 10.1126/science.1173164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan, J. F., O'riordan, K. J., Sandhu, K., Peterson, V. and Dinan, T. G. (2020). The gut microbiome in neurological disorders. Lancet Neurol. 19, 179-194. 10.1016/S1474-4422(19)30356-4 [DOI] [PubMed] [Google Scholar]
- Czaja, K., Burns, G. A. and Ritter, R. C. (2008). Capsaicin-induced neuronal death and proliferation of the primary sensory neurons located in the nodose ganglia of adult rats. Neuroscience 154, 621-630. 10.1016/j.neuroscience.2008.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabke, K., Hendrick, G. and Devkota, S. (2019). The gut microbiome and metabolic syndrome. J. Clin. Invest. 129, 4050-4057. 10.1172/JCI129194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalile, B., Van Oudenhove, L., Vervliet, B. and Verbeke, K. (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 16, 461-478. 10.1038/s41575-019-0157-3 [DOI] [PubMed] [Google Scholar]
- Dauça, M., Bouziges, F., Colin, S., Kedinger, M., Keller, M. K., Schilt, J., Simon-Assmann, P. and Haffen, K. (1990). Development of the vertebrate small intestine and mechanisms of cell differentiation. Int. J. Dev. Biol. 34, 205-218. [PubMed] [Google Scholar]
- De Vadder, F., Grasset, E., Mannerås Holm, L., Karsenty, G., Macpherson, A. J., Olofsson, L. E. and Bäckhed, F. (2018). Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc. Natl. Acad. Sci. USA 115, 6458-6463. 10.1073/pnas.1720017115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deines, P., Lachnit, T. and Bosch, T. C. G. (2017). Competing forces maintain the Hydra metaorganism. Immunol. Rev. 279, 123-136. 10.1111/imr.12564 [DOI] [PubMed] [Google Scholar]
- Derrien, M., Alvarez, A.-S. and De Vos, W. M. (2019). The gut microbiota in the first decade of life. Trends Microbiol. 27, 997-1010. 10.1016/j.tim.2019.08.001 [DOI] [PubMed] [Google Scholar]
- Di Giancamillo, A., Vitari, F., Bosi, G., Savoini, G. and Domeneghini, C. (2010). The chemical code of porcine enteric neurons and the number of enteric glial cells are altered by dietary probiotics. Neurogastroenterol. Motil. 22, e271-e278. 10.1111/j.1365-2982.2010.01529.x [DOI] [PubMed] [Google Scholar]
- Dodd, D., Spitzer, M. H., Van Treuren, W., Merrill, B. D., Hryckowian, A. J., Higginbottom, S. K., Le, A., Cowan, T. M., Nolan, G. P., Fischbach, M. A.et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648-652. 10.1038/nature24661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doenyas, C. (2018). Gut microbiota, inflammation, and probiotics on neural development in autism spectrum disorder. Neuroscience 374, 271-286. 10.1016/j.neuroscience.2018.01.060 [DOI] [PubMed] [Google Scholar]
- Dogra, S., Sakwinska, O., Soh, S.-E., Ngom-Bru, C., Brück, W. M., Berger, B., Brüssow, H., Lee, Y. S., Yap, F., Chong, Y.-S.et al. (2015a). Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio 6, e02419-14. 10.1128/mBio.02419-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra, S., Sakwinska, O., Soh, S.-E., Ngom-Bru, C., Brück, W. M., Berger, B., Brüssow, H., Karnani, N., Lee, Y. S., Yap, F.et al. (2015b). Rate of establishing the gut microbiota in infancy has consequences for future health. Gut Microbes 6, 321-325. 10.1080/19490976.2015.1078051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, G. P., Lee, S. M. and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20-32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas, A. E. (2019). Simple animal models for microbiome research. Nat. Rev. Microbiol. 17, 764-775. 10.1038/s41579-019-0242-1 [DOI] [PubMed] [Google Scholar]
- Drossman, D. A. (2006). The functional gastrointestinal disorders and the Rome III process. Gastroenterology 130, 1377-1390. 10.1053/j.gastro.2006.03.008 [DOI] [PubMed] [Google Scholar]
- Duca, F. A., Swartz, T. D., Sakar, Y. and Covasa, M. (2012). Increased oral detection, but decreased intestinal signaling for fats in mice lacking gut microbiota. PLoS ONE 7, e39748. 10.1371/journal.pone.0039748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, D., Dobson, A. J., Houtz, P. L., Gläßer, C., Revah, J., Korzelius, J., Patel, P. H., Edgar, B. A. and Buchon, N. (2015). Regional cell-specific transcriptome mapping reveals regulatory complexity in the adult Drosophila midgut. Cell Rep. 12, 346-358. 10.1016/j.celrep.2015.06.009 [DOI] [PubMed] [Google Scholar]
- Erickson, J. T., Conover, J. C., Borday, V., Champagnat, J., Barbacid, M., Yancopoulos, G. and Katz, D. M. (1996). Mice lacking brain-derived neurotrophic factor exhibit visceral sensory neuron losses distinct from mice lacking NT4 and display a severe developmental deficit in control of breathing. J. Neurosci. 16, 5361-5371. 10.1523/JNEUROSCI.16-17-05361.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors, P., Lee, K.-F., Kucera, J. and Jaenisch, R. (1994). Lack of neurotrophin-3 leads to deficiencies in the peripheral nervous system and loss of limb proprioceptive afferents. Cell 77, 503-512. 10.1016/0092-8674(94)90213-5 [DOI] [PubMed] [Google Scholar]
- Fan, Y. and Pedersen, O. (2021). Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 19, 55-71. 10.1038/s41579-020-0433-9 [DOI] [PubMed] [Google Scholar]
- Farin, H. F., Jordens, I., Mosa, M. H., Basak, O., Korving, J., Tauriello, D. V. F., De Punder, K., Angers, S., Peters, P. J., Maurice, M. M.et al. (2016). Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 530, 340-343. 10.1038/nature16937 [DOI] [PubMed] [Google Scholar]
- Ferguson, M. and Foley, E. (2021). Microbial recognition regulates intestinal epithelial growth in homeostasis and disease. FEBS J. [Epub ahead of print]. 10.1111/febs.15910 [DOI] [PubMed] [Google Scholar]
- Fetissov, S. O. and Hökfelt, T. (2019). On the origin of eating disorders: altered signaling between gut microbiota, adaptive immunity and the brain melanocortin system regulating feeding behavior. Curr. Opin. Pharmacol. 48, 82-91. 10.1016/j.coph.2019.07.004 [DOI] [PubMed] [Google Scholar]
- Furness, J. B. (2000). Types of neurons in the enteric nervous system. J. Auton. Nerv. Syst. 81, 87-96. 10.1016/S0165-1838(00)00127-2 [DOI] [PubMed] [Google Scholar]
- Gallaher, Z. R., Ryu, V., Larios, R. M., Sprunger, L. K. and Czaja, K. (2011). Neural proliferation and restoration of neurochemical phenotypes and compromised functions following capsaicin-induced neuronal damage in the nodose ganglion of the adult rat. Front. Neurosci. 5, 12. 10.3389/fnins.2011.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz, J. (2018). Gut feelings: studying enteric nervous system development, function, and disease in the zebrafish model system. Dev. Dyn. 247, 268-278. 10.1002/dvdy.24597 [DOI] [PubMed] [Google Scholar]
- García-Arrarás, J. E., Lefebre-Rivera, M. and Qi-Huang, S. (2019). Enteroendocrine cells in the Echinodermata. Cell Tissue Res. 377, 459-467. 10.1007/s00441-019-03053-3 [DOI] [PubMed] [Google Scholar]
- Garcia-Gutierrez, E., Narbad, A. and Rodríguez, J. M. (2020). Autism spectrum disorder associated with gut microbiota at immune, metabolomic, and neuroactive level. Front. Neurosci. 14, 578666. 10.3389/fnins.2020.578666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaykema, R. P. A., Goehler, L. E. and Lyte, M. (2004). Brain response to cecal infection with Campylobacter jejuni: analysis with Fos immunohistochemistry. Brain Behav. Immun. 18, 238-245. 10.1016/j.bbi.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Gilbert, S. F., Sapp, J. and Tauber, A. I. (2012). A symbiotic view of life: we have never been individuals. Q .Rev. Biol. 87, 325-341. 10.1086/668166 [DOI] [PubMed] [Google Scholar]
- Glebova, N. O. and Ginty, D. D. (2005). Growth and survival signals controlling sympathetic nervous system development. Annu. Rev. Neurosci. 28, 191-222. 10.1146/annurev.neuro.28.061604.135659 [DOI] [PubMed] [Google Scholar]
- Goldberg, D., Borojevic, R., Anderson, M., Chen, J. J., Gershon, M. D. and Ratcliffe, E. M. (2013). Slit/Robo-mediated chemorepulsion of vagal sensory axons in the fetal gut. Dev. Dyn. 242, 9-15. 10.1002/dvdy.23898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez de Agüero, M. and Ganal-Vonarburg, S. C., Fuhrer, T., Rupp, S., Uchimura, Y., Li, H., Steinert, A., Heikenwalder, M., Hapfelmeier, S. and Sauer, U.et al. (2016). The maternal microbiota drives early postnatal innate immune development. Science 351, 1296-1302. 10.1126/science.aad2571 [DOI] [PubMed] [Google Scholar]
- Gonnella, P. A. and Neutra, M. R. (1984). Membrane-bound and fluid-phase macromolecules enter separate prelysosomal compartments in absorptive cells of suckling rat ileum. J. Cell Biol. 99, 909-917. 10.1083/jcb.99.3.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami, C., Iwasaki, Y. and Yada, T. (2018). Short-chain fatty acids suppress food intake by activating vagal afferent neurons. J. Nutr. Biochem. 57, 130-135. 10.1016/j.jnutbio.2018.03.009 [DOI] [PubMed] [Google Scholar]
- Gribble, F. M. and Reimann, F. (2016). Enteroendocrine cells: chemosensors in the intestinal epithelium. Annu. Rev. Physiol. 78, 277-299. 10.1146/annurev-physiol-021115-105439 [DOI] [PubMed] [Google Scholar]
- Gribble, F. M. and Reimann, F. (2019). Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat. Rev. Endocrinol. 15, 226-237. 10.1038/s41574-019-0168-8 [DOI] [PubMed] [Google Scholar]
- Grubišić, V. and Gulbransen, B. D. (2017). Enteric glia: the most alimentary of all glia. J. Physiol. 595, 557-570. 10.1113/JP271021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Repiso, C., Moreno-Indias, I. and Tinahones, F. J. (2021). Shifts in gut microbiota and their metabolites induced by bariatric surgery. Impact of factors shaping gut microbiota on bariatric surgery outcomes. Rev. Endocr. Metab. Disord. [Epub ahead of print]. 10.1007/s11154-021-09676-8 [DOI] [PubMed] [Google Scholar]
- Hao, M. M. and Young, H. M. (2009). Development of enteric neuron diversity. J. Cell Mol. Med. 13, 1193-1210. 10.1111/j.1582-4934.2009.00813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, T. A., Stadt, H. A. and Kirby, M. L. (1994). Developmental characteristics of the chick nodose ganglion. Dev. Neurosci. 16, 67-73. 10.1159/000112090 [DOI] [PubMed] [Google Scholar]
- Hartenstein, V. (1997). Development of the insect stomatogastric nervous system. Trends Neurosci. 20, 421-427. 10.1016/S0166-2236(97)01066-7 [DOI] [PubMed] [Google Scholar]
- Hatch, J. and Mukouyama, Y. S. (2015). Spatiotemporal mapping of vascularization and innervation in the fetal murine intestine. Dev. Dyn. 244, 56-68. 10.1002/dvdy.24178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning, S. J. (1981). Postnatal development: coordination of feeding, digestion, and metabolism. Am. J. Physiol. 241, G199-G214. 10.1152/ajpgi.1981.241.3.G199 [DOI] [PubMed] [Google Scholar]
- Henning, S. J. (1985). Ontogeny of enzymes in the small intestine. Annu. Rev. Physiol. 47, 231-245. 10.1146/annurev.ph.47.030185.001311 [DOI] [PubMed] [Google Scholar]
- Honma, Y., Araki, T., Gianino, S., Bruce, A., Heuckeroth, R. O., Johnson, E. M. and Milbrandt, J. (2002). Artemin is a vascular-derived neurotropic factor for developing sympathetic neurons. Neuron 35, 267-282. 10.1016/S0896-6273(02)00774-2 [DOI] [PubMed] [Google Scholar]
- Hooper, L. V., Wong, M. H., Thelin, A., Hansson, L., Falk, P. G. and Gordon, J. I. (2001). Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881-884. 10.1126/science.291.5505.881 [DOI] [PubMed] [Google Scholar]
- Hou, Q., Ye, L., Huang, L. and Yu, Q. (2017). The research progress on intestinal stem cells and its relationship with intestinal microbiota. Front. Immunol. 8, 599. 10.3389/fimmu.2017.00599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt, M. R., Lavoie, S., Michaud, M., Blum, A. M., Tran, S. V., Weinstock, J. V., Gallini, C. A., Redding, K., Margolskee, R. F., Osborne, L. C.et al. (2016). Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329-1333. 10.1126/science.aaf1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst, N. R., Kendig, D. M., Murthy, K. S. and Grider, J. R. (2014). The short chain fatty acids, butyrate and propionate, have differential effects on the motility of the guinea pig colon. Neurogastroenterol. Motil. 26, 1586-1596. 10.1111/nmo.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye, E., Hellström, P. M., Sundler, F., Chen, J. and Midtvedt, T. (2001). Influence of microbial species on small intestinal myoelectric activity and transit in germ-free rats. Am. J. Physiol. Gastrointest. Liver Physiol. 280, G368-G380. 10.1152/ajpgi.2001.280.3.G368 [DOI] [PubMed] [Google Scholar]
- Isabella, A. J., Barsh, G. R., Stonick, J. A., Dubrulle, J. and Moens, C. B. (2020). Retinoic acid organizes the zebrafish vagus motor topographic map via spatiotemporal coordination of Hgf/Met signaling. Dev. Cell 53, 344-357.e5. 10.1016/j.devcel.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson, M. E. V., Larsson, J. M. H. and Hansson, G. C. (2011). The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 108 Suppl. 1, 4659-4665. 10.1073/pnas.1006451107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, R. M., Luo, L., Ardita, C. S., Richardson, A. N., Kwon, Y. M., Mercante, J. W., Alam, A., Gates, C. L., Wu, H., Swanson, P. A.et al. (2013). Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 32, 3017-3028. 10.1038/emboj.2013.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua, I. G. (1991). Neuropeptide Y-induced constriction in small resistance vessels of skeletal muscle. Peptides 12, 37-41. 10.1016/0196-9781(91)90163-J [DOI] [PubMed] [Google Scholar]
- Kabouridis, P. S. and Pachnis, V. (2015). Emerging roles of gut microbiota and the immune system in the development of the enteric nervous system. J. Clin. Invest. 125, 956-964. 10.1172/JCI76308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabouridis, P. S., Lasrado, R., Mccallum, S., Chng, S. H., Snippert, H. J., Clevers, H., Pettersson, S. and Pachnis, V. (2015). Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron 85, 289-295. 10.1016/j.neuron.2014.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelberer, M. M., Buchanan, K. L., Klein, M. E., Barth, B. B., Montoya, M. M., Shen, X. and Bohórquez, D. V. (2018). A gut-brain neural circuit for nutrient sensory transduction. Science 361, eaat5236. 10.1126/science.aat5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm, K., Hoppe, S., Breves, G., Schroder, B. and Schemann, M. (2004). Effects of the probiotic yeast Saccharomyces boulardii on the neurochemistry of myenteric neurones in pig jejunum. Neurogastroenterol. Motil. 16, 53-60. 10.1046/j.1365-2982.2003.00458.x [DOI] [PubMed] [Google Scholar]
- Kang, D.-W., Adams, J. B., Coleman, D. M., Pollard, E. L., Maldonado, J., Mcdonough-Means, S., Caporaso, J. G. and Krajmalnik-Brown, R. (2019). Long-term benefit of microbiota transfer therapy on autism symptoms and gut microbiota. Sci. Rep. 9, 5821. 10.1038/s41598-019-42183-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasemeier-Kulesa, J. C., Kulesa, P. M. and Lefcort, F. (2005). Imaging neural crest cell dynamics during formation of dorsal root ganglia and sympathetic ganglia. Development 132, 235-245. 10.1242/dev.01553 [DOI] [PubMed] [Google Scholar]
- Kennedy, E. A., King, K. Y. and Baldridge, M. T. (2018). Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 9, 1534. 10.3389/fphys.2018.01534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. S. and De La Serre, C. B. (2018). Diet, gut microbiota composition and feeding behavior. Physiol. Behav. 192, 177-181. 10.1016/j.physbeh.2018.03.026 [DOI] [PubMed] [Google Scholar]
- Kleene, K. C., Distel, R. J. and Hecht, N. B. (1985). Nucleotide sequence of a cDNA clone encoding mouse protamine 1. Biochemistry 24, 719-722. 10.1021/bi00324a027 [DOI] [PubMed] [Google Scholar]
- Kobayashi, K. S., Chamaillard, M., Ogura, Y., Henegariu, O., Inohara, N., Nuñez, G. and Flavell, R. A. (2005). Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731-734. 10.1126/science.1104911 [DOI] [PubMed] [Google Scholar]
- Kostic, A. D., Howitt, M. R. and Garrett, W. S. (2013). Exploring host-microbiota interactions in animal models and humans. Genes Dev. 27, 701-718. 10.1101/gad.212522.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang, Z., Wang, Y., Li, Y., Ye, C., Ruhn, K. A., Behrendt, C. L., Olson, E. N. and Hooper, L. V. (2019). The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science 365, 1428-1434. 10.1126/science.aaw3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, D. H., Gustafsson, J. K., Knoop, K. A., Mcdonald, K. G., Bidani, S. S., Davis, J. E., Floyd, A. N., Hogan, S. P., Hsieh, C.-S. and Newberry, R. D. (2020). Goblet cell associated antigen passages support the induction and maintenance of oral tolerance. Mucosal Immunol. 13, 271-282. 10.1038/s41385-019-0240-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraishi, T., Kenmoku, H. and Kurata, S. (2015). From mouth to anus: functional and structural relevance of enteric neurons in the Drosophila melanogaster gut. Insect Biochem. Mol. Biol. 67, 21-26. 10.1016/j.ibmb.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Kuruvilla, R., Zweifel, L. S., Glebova, N. O., Lonze, B. E., Valdez, G., Ye, H. and Ginty, D. D. (2004). A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell 118, 243-255. 10.1016/j.cell.2004.06.021 [DOI] [PubMed] [Google Scholar]
- Lake, J. I. and Heuckeroth, R. O. (2013). Enteric nervous system development: migration, differentiation, and disease. Am. J. Physiol. Gastrointest Liver Physiol. 305, G1-G24. 10.1152/ajpgi.00452.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laranjeira, C., Sandgren, K., Kessaris, N., Richardson, W., Potocnik, A., Vanden Berghe, P. and Pachnis, V. (2011). Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J. Clin. Invest. 121, 3412-3424. 10.1172/JCI58200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. S., De Vadder, F., Tremaroli, V., Wichmann, A., Mithieux, G. and Bäckhed, F. (2016). Insulin-like peptide 5 is a microbially regulated peptide that promotes hepatic glucose production. Mol. Metab. 5, 263-270. 10.1016/j.molmet.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.-S., Kim, T.-Y., Kim, Y., Lee, S.-H., Kim, S., Kang, S. W., Yang, J.-Y., Baek, I.-J., Sung, Y. H., Park, Y.-Y.et al. (2018). Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe 24, 833-846.e6. 10.1016/j.chom.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Lemaitre, B. and Miguel-Aliaga, I. (2013). The digestive tract of Drosophila melanogaster. Annu. Rev. Genet. 47, 377-404. 10.1146/annurev-genet-111212-133343 [DOI] [PubMed] [Google Scholar]
- Lesher, S., Walburg, H. E., Jr. and Sacher, G. A.Jr. (1964). Generation cycle in the duodenal crypt cells of germ-free and conventional mice. Nature 202, 884-886. 10.1038/202884a0 [DOI] [PubMed] [Google Scholar]
- Li, Q., Han, Y., Dy, A. B. C., Hagerman, R. J. (2017). The gut microbiota and autism spectrum disorders. Front. Cell Neurosci. 11, 120. 10.3389/fncel.2017.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar, C. R., Camp, J. G., Weiser, M., Cocchiaro, J. L., Kingsley, D. M., Furey, T. S., Sheikh, S. Z. and Rawls, J. F. (2017). Genomic dissection of conserved transcriptional regulation in intestinal epithelial cells. PLoS Biol. 15, e2002054. 10.1371/journal.pbio.2002054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Hou, Y., Wang, G., Zheng, X. and Hao, H. (2020). Gut microbial metabolites of aromatic amino acids as signals in host-microbe interplay. Trends Endocrinol. Metab. 31, 818-834. 10.1016/j.tem.2020.02.012 [DOI] [PubMed] [Google Scholar]
- Logan, S. L., Thomas, J., Yan, J., Baker, R. P., Shields, D. S., Xavier, J. B., Hammer, B. K. and Parthasarathy, R. (2018). The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc. Natl. Acad. Sci. USA 115, E3779-E3787. 10.1073/pnas.1720133115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludington, W. B. and Ja, W. W. (2020). Drosophila as a model for the gut microbiome. PLoS Pathog. 16, e1008398. 10.1371/journal.ppat.1008398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, M. L., Sorrentino, G., Egerod, K. L., Kroone, C., Mortensen, B., Knop, F. K., Reimann, F., Gribble, F. M., Drucker, D. J., De Koning, E. J. P.et al. (2020). L-cell differentiation is induced by bile acids through GPBAR1 and paracrine GLP-1 and serotonin signaling. Diabetes 69, 614-623. 10.2337/db19-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita, T., Sucov, H. M., Gariepy, C. E., Yanagisawa, M. and Ginty, D. D. (2008). Endothelins are vascular-derived axonal guidance cues for developing sympathetic neurons. Nature 452, 759-763. 10.1038/nature06859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez, I., Maldonado-Gomez, M. X., Gomes-Neto, J. C., Kittana, H., Ding, H., Schmaltz, R., Joglekar, P., Cardona, R. J., Marsteller, N. L., Kembel, S. W.et al. (2018). Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. eLife 7, e36521. 10.7554/eLife.36521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Guryn, K., Hubert, N., Frazier, K., Urlass, S., Musch, M. W., Ojeda, P., Pierre, J. F., Miyoshi, J., Sontag, T. J., Cham, C. M.et al. (2018). Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe 23, 458-469.e5. 10.1016/j.chom.2018.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, E. A., Savidge, T. and Shulman, R. J. (2014). Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology 146, 1500-1512. 10.1053/j.gastro.2014.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccallum, S., Obata, Y., Fourli, E., Boeing, S., Peddie, C. J., Xu, Q., Horswell, S., Kelsh, R. N., Collinson, L., Wilkinson, D.et al. (2020). Enteric glia as a source of neural progenitors in adult zebrafish. eLife 9, e56086. 10.7554/eLife.56086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcfall-Ngai, M., Hadfield, M. G., Bosch, T. C. G., Carey, H. V., Domazet-Lošo, T., Douglas, A. E., Dubilier, N., Eberl, G., Fukami, T., Gilbert, S. F.et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 110, 3229-3236. 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgraw, H. F., Nechiporuk, A. and Raible, D. W. (2008). Zebrafish dorsal root ganglia neural precursor cells adopt a glial fate in the absence of neurogenin1. J. Neurosci. 28, 12558-12569. 10.1523/JNEUROSCI.2079-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey Neufeld, K. A., Mao, Y. K., Bienenstock, J., Foster, J. A. and Kunze, W. A. (2013). The microbiome is essential for normal gut intrinsic primary afferent neuron excitability in the mouse. Neurogastroenterol. Motil. 25, e183-e88. 10.1111/nmo.12049 [DOI] [PubMed] [Google Scholar]
- Medani, M., Collins, D., Docherty, N. G., Baird, A. W., O'Connell, P. R. and Winter, D. C. (2011). Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm. Bowel Dis. 17, 1620-1625. 10.1002/ibd.21528 [DOI] [PubMed] [Google Scholar]
- Mendall, M. A. and Kumar, D. (1998). Antibiotic use, childhood affluence and irritable bowel syndrome (IBS). Eur. J. Gastroenterol. Hepatol. 10, 59-62. 10.1097/00042737-199801000-00011 [DOI] [PubMed] [Google Scholar]
- Montgomery, R. K., Mulberg, A. E. and Grand, R. J. (1999). Development of the human gastrointestinal tract: twenty years of progress. Gastroenterology 116, 702-731. 10.1016/S0016-5085(99)70193-9 [DOI] [PubMed] [Google Scholar]
- Morais, L. H., Golubeva, A. V., Moloney, G. M., Moya-Pérez, A., Ventura-Silva, A. P., Arboleya, S., Bastiaanssen, T. F. S., O'Sullivan, O., Rea, K., Borre, Y.et al. (2020). Enduring behavioral effects induced by birth by caesarean section in the mouse. Curr. Biol. 30, 3761-3774.e6. 10.1016/j.cub.2020.07.044 [DOI] [PubMed] [Google Scholar]
- Morgun, A., Dzutsev, A., Dong, X., Greer, R. L., Sexton, D. J., Ravel, J., Schuster, M., Hsiao, W., Matzinger, P. and Shulzhenko, N. (2015). Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut 64, 1732-1743. 10.1136/gutjnl-2014-308820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkey, S. B. and Plessis, A. D. (2018). The critical role of the central autonomic nervous system in fetal-neonatal transition. Semin. Pediatr. Neurol. 28, 29-37. 10.1016/j.spen.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, P. A., Matheis, F., Schneeberger, M., Kerner, Z., Jové, V. and Mucida, D. (2020a). Microbiota-modulated CART+ enteric neurons autonomously regulate blood glucose. Science 370, 314-321. 10.1126/science.abd6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, P. A., Schneeberger, M., Matheis, F., Wang, P., Kerner, Z., Ilanges, A., Pellegrino, K., del Mármol, J., Castro, T. B. R., Furuichi, M.et al. (2020b). Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature 583, 441-446. 10.1038/s41586-020-2474-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muncan, V., Heijmans, J., Krasinski, S. D., Büller, N. V., Wildenberg, M. E., Meisner, S., Radonjic, M., Stapleton, K. A., Lamers, W. H., Biemond, I.et al. (2011). Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat. Commun. 2, 452. 10.1038/ncomms1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo-Rincon, A. P., Klimovich, A., Pemöller, E., Taubenheim, J., Mortzfeld, B., Augustin, R. and Bosch, T. C. G. (2017). Spontaneous body contractions are modulated by the microbiome of Hydra. Sci. Rep. 7, 15937. 10.1038/s41598-017-16191-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, M. C. and Fox, E. A. (2007). Anterograde tracing method using DiI to label vagal innervation of the embryonic and early postnatal mouse gastrointestinal tract. J. Neurosci. Methods 163, 213-225. 10.1016/j.jneumeth.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso, G., Gambino, R. and Cassader, M. (2010). Obesity, diabetes, and gut microbiota: the hygiene hypothesis expanded? Diabetes Care 33, 2277-2284. 10.2337/dc10-0556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadjsombati, M. S., Mcginty, J. W., Lyons-Cohen, M. R., Jaffe, J. B., Dipeso, L., Schneider, C., Miller, C. N., Pollack, J. L., Nagana Gowda, G. A., Fontana, M. F.et al. (2018). Detection of succinate by intestinal tuft cells triggers a type 2 innate immune circuit. Immunity 49, 33-41.e7. 10.1016/j.immuni.2018.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento, A. I., Mar, F. M. and Sousa, M. M. (2018). The intriguing nature of dorsal root ganglion neurons: linking structure with polarity and function. Prog. Neurobiol. 168, 86-103. 10.1016/j.pneurobio.2018.05.002 [DOI] [PubMed] [Google Scholar]
- Natarajan, D., Marcos-Gutierrez, C., Pachnis, V. and De Graaff, E. (2002). Requirement of signalling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development 129, 5151-5160. 10.1242/dev.129.22.5151 [DOI] [PubMed] [Google Scholar]
- Nøhr, M. K., Egerod, K. L., Christiansen, S. H., Gille, A., Offermanns, S., Schwartz, T. W. and Møller, M. (2015). Expression of the short chain fatty acid receptor GPR41/FFAR3 in autonomic and somatic sensory ganglia. Neuroscience 290, 126-137. 10.1016/j.neuroscience.2015.01.040 [DOI] [PubMed] [Google Scholar]
- Nyström, E. E. L., Martinez-Abad, B., Arike, L., Birchenough, G. M. H., Nonnecke, E. B., Castillo, P. A., Svensson, F., Bevins, C. L., Hansson, G. C. and Johansson, M. E. V. (2021). An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 372, eabb1590. 10.1126/science.abb1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata, Y., Castaño, Á., Boeing, S., Bon-Frauches, A. C., Fung, C., Fallesen, T., de Agüero, M. G., Yilmaz, B., Lopes, R., Huseynova, A.et al. (2020). Neuronal programming by microbiota regulates intestinal physiology. Nature 578, 284-289. 10.1038/s41586-020-1975-8 [DOI] [PubMed] [Google Scholar]
- Olsen, W. A. and Korsmo, H. A. (1982). Sucrase metabolism in germfree rats. Am. J. Physiol. 242, G650-G653. 10.1152/ajpgi.1982.242.6.G650 [DOI] [PubMed] [Google Scholar]
- Olsson, C., Holmberg, A. and Holmgren, S. (2008). Development of enteric and vagal innervation of the zebrafish (Danio rerio) gut. J. Comp. Neurol. 508, 756-770. 10.1002/cne.21705 [DOI] [PubMed] [Google Scholar]
- Pantoja-Feliciano, I. G., Clemente, J. C., Costello, E. K., Perez, M. E., Blaser, M. J., Knight, R. and Dominguez-Bello, M. G. (2013). Biphasic assembly of the murine intestinal microbiota during early development. ISME J. 7, 1112-1115. 10.1038/ismej.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J., Levic, D. S., Sumigray, K. D., Bagwell, J., Eroglu, O., Block, C. L., Eroglu, C., Barry, R., Lickwar, C. R., Rawls, J. F.et al. (2019). Lysosome-rich enterocytes mediate protein absorption in the vertebrate gut. Dev. Cell 51, 7-20.e6. 10.1016/j.devcel.2019.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, S. M., Sveinbjørnsson, B. and Robertsen, B. (2001). Selective staining and disintegration of intestinal eosinophilic granule cells in Atlantic salmon after intraperitoneal injection of the zinc chelator dithizone. J. Fish Biol. 58, 768-775. 10.1111/j.1095-8649.2001.tb00529.x [DOI] [Google Scholar]
- Peck, B. C. E., Mah, A. T., Pitman, W. A., Ding, S., Lund, P. K. and Sethupathy, P. (2017). Functional transcriptomics in diverse intestinal epithelial cell types reveals robust microRNA sensitivity in intestinal stem cells to microbial status. J. Biol. Chem. 292, 2586-2600. 10.1074/jbc.M116.770099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pédron, T., Mulet, C., Dauga, C., Frangeul, L., Chervaux, C., Grompone, G. and Sansonetti, P. J. (2012). A crypt-specific core microbiota resides in the mouse colon. mBio 3, e00116-12. 10.1128/mBio.00116-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, C., Ouyang, Y., Lu, N. and Li, N. (2020). The NF-κB signaling pathway, the microbiota, and gastrointestinal tumorigenesis: recent advances. Front. Immunol. 11, 1387. 10.3389/fimmu.2020.01387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter, J. I. and Bordenstein, S. R. (2020). Microorganisms in the reproductive tissues of arthropods. Nat. Rev. Microbiol. 18, 97-111. 10.1038/s41579-019-0309-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson, J., Schreiber, O., Hansson, G. C., Gendler, S. J., Velcich, A., Lundberg, J. O., Roos, S., Holm, L. and Phillipson, M. (2011). Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 300, G327-G333. 10.1152/ajpgi.00422.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, T. D., Gershon, M. D. and Rothman, T. P. (1991). Time of origin of neurons in the murine enteric nervous system: sequence in relation to phenotype. J. Comp. Neurol. 314, 789-798. 10.1002/cne.903140411 [DOI] [PubMed] [Google Scholar]
- Pimentel, M., Lin, H. C., Enayati, P., Van Den Burg, B., Lee, H.-R., Chen, J. H., Park, S., Kong, Y. and Conklin, J. (2006). Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G1089-G1095. 10.1152/ajpgi.00574.2004 [DOI] [PubMed] [Google Scholar]
- Qu, Z.-D., Thacker, M., Castelucci, P., Bagyánszki, M., Epstein, M. L. and Furness, J. B. (2008). Immunohistochemical analysis of neuron types in the mouse small intestine. Cell Tissue Res. 334, 147-161. 10.1007/s00441-008-0684-7 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, S. K., Zhang, H., Ma, X., Jung, I., Schwartz, A. J., Triner, D., Devenport, S. N., Das, N. K., Xue, X., Zeng, M. Y.et al. (2019). Intestinal non-canonical NFκB signaling shapes the local and systemic immune response. Nat. Commun. 10, 660. 10.1038/s41467-019-08581-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasquin, A., Di Lorenzo, C., Forbes, D., Guiraldes, E., Hyams, J. S., Staiano, A. and Walker, L. S. (2006). Childhood functional gastrointestinal disorders: child/adolescent. Gastroenterology 130, 1527-1537. 10.1053/j.gastro.2005.08.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, E. M., Setru, S. U., Chen, J. J., Li, Z. S., D'Autréaux, F. and Gershon, M. D. (2006). Netrin/DCC-mediated attraction of vagal sensory axons to the fetal mouse gut. J. Comp. Neurol. 498, 567-580. 10.1002/cne.21027 [DOI] [PubMed] [Google Scholar]
- Ratcliffe, E. M., Farrar, N. R. and Fox, E. A. (2011). Development of the vagal innervation of the gut: steering the wandering nerve. Neurogastroenterol. Motil. 23, 898-911. 10.1111/j.1365-2982.2011.01764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls, J. F., Samuel, B. S. and Gordon, J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA 101, 4596-4601. 10.1073/pnas.0400706101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls, J. F., Mahowald, M. A., Goodman, A. L., Trent, C. M. and Gordon, J. I. (2007). In vivo imaging and genetic analysis link bacterial motility and symbiosis in the zebrafish gut. Proc. Natl. Acad. Sci. USA 104, 7622-7627. 10.1073/pnas.0702386104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, C., Bergentall, M., Greiner, T. U., Schaffner, F., Östergren-Lundén, G., Petersen, L. C., Ruf, W. and Bäckhed, F. (2012). Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature 483, 627-631. 10.1038/nature10893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera, C. E. and Dillin, A. (2016). Emerging role of sensory perception in aging and metabolism. Trends Endocrinol. Metab. 27, 294-303. 10.1016/j.tem.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Rinaman, L. and Levitt, P. (1993). Establishment of vagal sensorimotor circuits during fetal development in rats. J. Neurobiol. 24, 641-659. 10.1002/neu.480240509 [DOI] [PubMed] [Google Scholar]
- Robberecht, P., Deschodt-Lanckman, M., Camus, J., Bruylands, J. and Christophe, J. (1971). Rat pancreatic hydrolases from birth to weaning and dietary adaptation after weaning. Am. J. Physiol. 221, 376-381. 10.1152/ajplegacy.1971.221.1.376 [DOI] [PubMed] [Google Scholar]
- Roberts, R. R., Murphy, J. F., Young, H. M. and Bornstein, J. C. (2007). Development of colonic motility in the neonatal mouse-studies using spatiotemporal maps. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G930-G938. 10.1152/ajpgi.00444.2006 [DOI] [PubMed] [Google Scholar]
- Rombout, J. H. W. M., Lamers, C. H. J., Helfrich, M. H., Dekker, A. and Taverne-Thiele, J. J. (1985). Uptake and transport of intact macromolecules in the intestinal epithelium of carp (Cyprinus carpio L.) and the possible immunological implications. Cell Tissue Res. 239, 519-530. 10.1007/BF00219230 [DOI] [PubMed] [Google Scholar]
- Round, J. L. and Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313-323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor, R. B. and Wu, G. D. (2017). Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 152, 327-339.e4. 10.1053/j.gastro.2016.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn, A. A., Von Furstenberg, R. J., Valsaraj, S., Hussain, F. S., Stein, M., Shanahan, M. T., Henning, S. J. and Gulati, A. S. (2019). The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut Microbes 10, 45-58. 10.1080/19490976.2018.1474321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs, A., Hückesfeld, S., Surendran, S. and Pankratz, M. J. (2014). Serotonergic pathways in the Drosophila larval enteric nervous system. J. Insect Physiol. 69, 118-125. 10.1016/j.jinsphys.2014.05.022 [DOI] [PubMed] [Google Scholar]
- Schroeder, B. O. (2019). Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep. 7, 3-12. 10.1093/gastro/goy052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semova, I., Carten, J. D., Stombaugh, J., Mackey, L. C., Knight, R., Farber, S. A. and Rawls, J. F. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277-288. 10.1016/j.chom.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgritta, M., Dooling, S. W., Buffington, S. A., Momin, E. N., Francis, M. B., Britton, R. A. and Costa-Mattioli, M. (2019). Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 101, 246-259.e6. 10.1016/j.neuron.2018.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, R., Schumacher, U., Ronaasen, V. and Coates, M. (1995). Rat intestinal mucosal responses to a microbial flora and different diets. Gut 36, 209-214. 10.1136/gut.36.2.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng, Y., Ren, H., Limbu, S. M., Sun, Y., Qiao, F., Zhai, W., Du, Z.-Y. and Zhang, M. (2018). The presence or absence of intestinal microbiota affects lipid deposition and related genes expression in zebrafish (Danio rerio). Front. Microbiol. 9, 1124. 10.3389/fmicb.2018.01124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, I. and Eisen, J. (2011). Development of the zebrafish enteric nervous system. Methods Cell Biol. 101, 143-160. 10.1016/B978-0-12-387036-0.00006-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, F. and Bäckhed, F. (2013). The gut microbiota — masters of host development and physiology. Nat. Rev. Microbiol. 11, 227-238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- Soret, R., Chevalier, J., De Coppet, P., Poupeau, G., Derkinderen, P., Segain, J. P. and Neunlist, M. (2010). Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 138, 1772-1782. 10.1053/j.gastro.2010.01.053 [DOI] [PubMed] [Google Scholar]
- Spanogiannopoulos, P., Bess, E. N., Carmody, R. N. and Turnbaugh, P. J. (2016). The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273-287. 10.1038/nrmicro.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, N. J. and Hu, H. (2020). Enteric nervous system: sensory transduction, neural circuits and gastrointestinal motility. Nat. Rev. Gastroenterol. Hepatol. 17, 338-351. 10.1038/s41575-020-0271-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprockett, D., Fukami, T. and Relman, D. A. (2018). Role of priority effects in the early-life assembly of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 15, 197-205. 10.1038/nrgastro.2017.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappenbeck, T. S., Hooper, L. V. and Gordon, J. I. (2002). Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 99, 15451-15455. 10.1073/pnas.202604299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, W. Z., Burns, A. R., Stagaman, K., Wong, S., Rawls, J. F., Guillemin, K. and Bohannan, B. J. M. (2016). The composition of the zebrafish intestinal microbial community varies across development. ISME J. 10, 644-654. 10.1038/ismej.2015.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyek, M. R., Quinn, T. A., Croll, R. P. and Smith, F. M. (2016). Zebrafish heart as a model to study the integrative autonomic control of pacemaker function. Am. J. Physiol. Heart Circ. Physiol. 311, H676-H688. 10.1152/ajpheart.00330.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, Z. A., Khoury-Hanold, W., Lim, J., Smillie, C., Biton, M., Reis, B. S., Zwick, R. K., Pope, S. D., Israni-Winger, K., Parsa, R.et al. (2021)., γδ T cells regulate the intestinal response to nutrient sensing. Science 371, eaba8310. 10.1126/science.aba8310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveinbjornsson, B., Olsen, R. and Paulsen, S. (1996). Immunocytochemical localization of lysozyme in intestinal eosinophilic granule cells (EGCs) of Atlantic salmon, Salmo salar L. J. Fish Dis. 19, 349-355. 10.1111/j.1365-2761.1996.tb00373.x [DOI] [Google Scholar]
- Taniguchi, M., Yuasa, S., Fujisawa, H., Naruse, I., Saga, S., Mishina, M. and Yagi, T. (1997). Disruption of semaphorin III/D gene causes severe abnormality in peripheral nerve projection. Neuron 19, 519-530. 10.1016/S0896-6273(00)80368-2 [DOI] [PubMed] [Google Scholar]
- Tehrani, A. B., Nezami, B. G., Gewirtz, A. and Srinivasan, S. (2012). Obesity and its associated disease: a role for microbiota? Neurogastroenterol. Motil. 24, 305-311. 10.1111/j.1365-2982.2012.01895.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torow, N. and Hornef, M. W. (2017). The neonatal window of opportunity: setting the stage for life-long host-microbial interaction and immune homeostasis. J. Immunol. 198, 557-563. 10.4049/jimmunol.1601253 [DOI] [PubMed] [Google Scholar]
- Troll, J. V., Hamilton, M. K., Abel, M. L., Ganz, J., Bates, J. M., Stephens, W. Z., Melancon, E., Van Der Vaart, M., Meijer, A. H., Distel, M.et al. (2018). Microbiota promote secretory cell determination in the intestinal epithelium by modulating host Notch signaling. Development 145, dev155317. 10.1242/dev.155317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uesaka, T., Young, H. M., Pachnis, V. and Enomoto, H. (2016). Development of the intrinsic and extrinsic innervation of the gut. Dev. Biol. 417, 158-167. 10.1016/j.ydbio.2016.04.016 [DOI] [PubMed] [Google Scholar]
- Vaishnava, S., Behrendt, C. L., Ismail, A. S., Eckmann, L. and Hooper, L. V. (2008). Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA 105, 20858-20863. 10.1073/pnas.0808723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bogert, B., Leimena, M. M., de Vos, W. M., Zoetendal, E. G. and Kleerebezem, M. (2011). Functional intestinal metagenomics. In Handbook of Molecular Microbial Ecology II (ed. F.J. de Bruijn), pp. 175-190. 10.1002/9781118010549.ch18 [DOI] [Google Scholar]
- Vincent, A. D., Wang, X.-Y., Parsons, S. P., Khan, W. I. and Huizinga, J. D. (2018). Abnormal absorptive colonic motor activity in germ-free mice is rectified by butyrate, an effect possibly mediated by mucosal serotonin. Am. J. Physiol. Gastrointest. Liver Physiol. 315, G896-G907. 10.1152/ajpgi.00237.2017 [DOI] [PubMed] [Google Scholar]
- Waise, T. M. Z., Dranse, H. J. and Lam, T. K. T. (2018). The metabolic role of vagal afferent innervation. Nat. Rev. Gastroenterol. Hepatol. 15, 625-636. 10.1038/s41575-018-0062-1 [DOI] [PubMed] [Google Scholar]
- Wallace, K. N., Akhter, S., Smith, E. M., Lorent, K. and Pack, M. (2005). Intestinal growth and differentiation in zebrafish. Mech. Dev. 122, 157-173. 10.1016/j.mod.2004.10.009 [DOI] [PubMed] [Google Scholar]
- Walthall, K., Cappon, G. D., Hurtt, M. E. and Zoetis, T. (2005). Postnatal development of the gastrointestinal system: a species comparison. Birth Defects Res. B Dev. Reprod. Toxicol. 74, 132-156. 10.1002/bdrb.20040 [DOI] [PubMed] [Google Scholar]
- Wang, Z., Du, J., Lam, S. H., Mathavan, S., Matsudaira, P. and Gong, Z. (2010). Morphological and molecular evidence for functional organization along the rostrocaudal axis of the adult zebrafish intestine. BMC Genomics 11, 392. 10.1186/1471-2164-11-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K., Zhang, T., Dong, Q., Nice, E. C., Huang, C. and Wei, Y. (2013). Redox homeostasis: the linchpin in stem cell self-renewal and differentiation. Cell Death Dis. 4, e537. 10.1038/cddis.2013.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Kuang, Z., Yu, X., Ruhn, K. A., Kubo, M. and Hooper, L. V. (2017). The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 357, 912-916. 10.1126/science.aan0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, J., Padilla Mercado, G., Volland, A., Doden, H. L., Lickwar, C. R., Crooks, T., Kakiyama, G., Kelly, C., Cocchiaro, J. L., Ridlon, J. M.et al. (2021). Fxr signaling and microbial metabolism of bile salts in the zebrafish intestine. Sci. Adv. 7, eabg1371. 10.1126/sciadv.abg1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weström, B., Arévalo Sureda, E., Pierzynowska, K., Pierzynowski, S. G. and Pérez-Cano, F.-J. (2020). The Immature gut barrier and its importance in establishing immunity in newborn mammals. Front. Immunol. 11, 1153. 10.3389/fimmu.2020.01153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann, A., Allahyar, A., Greiner, T. U., Plovier, H., Lundén, G. Ö., Larsson, T., Drucker, D. J., Delzenne, N. M., Cani, P. D. and Bäckhed, F. (2013). Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe 14, 582-590. 10.1016/j.chom.2013.09.012 [DOI] [PubMed] [Google Scholar]
- Wikoff, W. R., Anfora, A. T., Liu, J., Schultz, P. G., Lesley, S. A., Peters, E. C. and Siuzdak, G. (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 106, 3698-3703. 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing, B. P. and Van Kessel, A. G. (2009). Intestinal microbiota differentially affect brush border enzyme activity and gene expression in the neonatal gnotobiotic pig. J. Anim. Physiol. Anim. Nutr. 93, 586-595. 10.1111/j.1439-0396.2008.00841.x [DOI] [PubMed] [Google Scholar]
- Willms, R. J., Jones, L. O., Hocking, J. C. and Foley, E. (2021). A cell atlas of microbe responsive processes in the zebrafish intestine. bioRxiv. 10.1101/2020.11.06.371609v2 [DOI] [PubMed] [Google Scholar]
- Wilson, J. M., Whitney, J. A. and Neutra, M. R. (1991). Biogenesis of the apical endosome-lysosome complex during differentiation of absorptive epithelial cells in rat ileum. J. Cell Sci. 100, 133-143. 10.1242/jcs.100.1.133 [DOI] [PubMed] [Google Scholar]
- Woznica, A. and King, N. (2018). Lessons from simple marine models on the bacterial regulation of eukaryotic development. Curr. Opin. Microbiol. 43, 108-116. 10.1016/j.mib.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, N. J. and Chiu, I. M. (2017). Bacterial signaling to the nervous system through toxins and metabolites. J. Mol. Biol. 429, 587-605. 10.1016/j.jmb.2016.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. H. C., Kawakami, K. and Stainier, D. Y. R. (2018). A new mode of pancreatic islet innervation revealed by live imaging in zebrafish. eLife 7, e34519. 10.7554/eLife.34519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, J. M., Yu, K., Donaldson, G. P., Shastri, G. G., Ann, P., Ma, L., Nagler, C. R., Ismagilov, R. F., Mazmanian, S. K. and Hsiao, E. Y. (2015). Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161, 264-276. 10.1016/j.cell.2015.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., Magris, M., Hidalgo, G., Baldassano, R. N., Anokhin, A. P.et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222-227. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, L., Mueller, O., Bagwell, J., Bagnat, M., Liddle, R. A. and Rawls, J. F. (2019). High fat diet induces microbiota-dependent silencing of enteroendocrine cells. eLife 8, e48479. 10.7554/eLife.48479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, L., Bae, M., Cassilly, C. D., Jabba, S. V., Thorpe, D. W., Martin, A. M., Lu, H.-Y., Wang, J., Thompson, J. D., Lickwar, C. R.et al. (2021). Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell Host Microbe 29, 179-196.e9. 10.1016/j.chom.2020.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, H. M., Bergner, A. J. and Müller, T. (2003). Acquisition of neuronal and glial markers by neural crest-derived cells in the mouse intestine. J. Comp. Neurol. 456, 1-11. 10.1002/cne.10448 [DOI] [PubMed] [Google Scholar]
- Zeng, X. and Hou, S. X. (2015). Enteroendocrine cells are generated from stem cells through a distinct progenitor in the adult Drosophila posterior midgut. Development 142, 644-653. 10.1242/dev.113357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, D., Liwinski, T. and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492-506. 10.1038/s41422-020-0332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]