Abstract
Respiratory syncytial viruses (RSVs) are an important cause of mortality worldwide and a major cause of respiratory tract infections in children, driving development of vaccine candidates. However, there are large gaps in our knowledge of the local evolutionary and transmission dynamics of RSVs, particularly in understudied regions such as the Middle East. To address this gap, we sequenced the complete genomes of 58 RSVA and 27 RSVB samples collected in a paediatric cohort in Amman, Jordan, between 2010 and 2013. RSVA and RSVB co-circulated during each winter epidemic of RSV in Amman, and each epidemic comprised multiple independent viral introductions of RSVA and RSVB. However, RSVA and RSVB alternated in dominance across years, potential evidence of immunological interactions. Children infected with RSVA tended to be older than RSVB-infected children [30 months versus 22.4 months, respectively (P value = 0.02)], and tended to developed bronchopneumonia less frequently than those with RSVB, although the difference was not statistically significant (P value = 0.06). Differences in spatial patterns were investigated, and RSVA lineages were often identified in multiple regions in Amman, whereas RSVB introductions did not spread beyond a single region of the city, although these findings were based on small sample sizes. Multiple RSVA genotypes were identified in Amman, including GA2 viruses as well as three viruses from the ON1 sub-genotype that emerged in 2009 and are now the dominant genotype circulating worldwide. As vaccine development advances, further sequencing of RSV is needed to understand viral ecology and transmission, particularly in under-studied locations.
Keywords: respiratory syncytial virus, paediatric cohort, next-generation sequencing, evolution, phylogenetic analysis, Bayesian analysis
Data Summary
All accession numbers of the genetic sequences used in this study are listed in the Supplementary material (available with the online version of this article).
Impact Statement.
With multiple respiratory syncytial virus (RSV) vaccine candidates in development, it is crucial to understand the evolutionary dynamics of the virus, including in regions where there is little data available, such as the Middle East. By investigating RSV in a paediatric cohort in Amman, Jordan, we were able to study the population dynamics of the virus over time, including the alternating dominance of the RSVA and RSVB subgroups, the association between viral genetics and symptom severity, and how different strains are introduced or persist over time in this region.
Introduction
Human respiratory syncytial virus (RSV) is a leading cause of severe lower respiratory tract infections in young children. RSV causes an estimated 3.2 million annual hospitalizations and 60 000 deaths globally, primarily in children younger than 1 year old [1]. In general, the infection in the first year of life is the most severe [2]. RSV also contributes to severe acute respiratory disease in the elderly, comparable to that of influenza [1, 3–5]. A vaccine to protect infants and the elderly from RSV is a global health priority, and multiple vaccine candidates and mAbs, protective against both antigenic subgroups, are in different stages of development [6, 7]. Palivizumab is currently the only preventive mAb that protects against severe RSV acute respiratory infection [8]. In preparation for future vaccine roll-outs, there have been expanded efforts to understand the evolution and genetic diversity of the virus, including whole-genome sequencing [9–11].
RSV is an orthopneumovirus in the newly defined family Pneumoviridae [12]. The viral genome consists of a negative ssRNA (~15 kb) strand that encodes 11 proteins. Two of the proteins are glycosylated, enabling fusion and attachment (F and G proteins, respectively) [7]. Two antigenic subgroups, RSVA and RSVB, are defined based on mAb reactivity to the glycoproteins [13]. Both antigenic subgroups co-circulate worldwide, causing seasonal epidemics, although RSVA is thought to be more prevalent and cause more severe illness than RSVB [11]. However, a higher prevalence of RSVB than of RSVA has been reported in several studies [14–16], and further understanding of the prevalence and immunological interactions between the antigenic subgroups is needed. Models of alternating epidemics have been proposed, driven by waning herd immunity [15, 17].
The molecular epidemiology of the virus has been characterized in multiple regions [10, 18–20], but there is a paucity of genetic sequence data for RSV, at least relative to influenza, and it remains unknown where new viral lineages originate and what drives their emergence. New RSV strains periodically emerge and become globally dominant, and 10 genotypes for RSVA and 13 RSVB genotypes have been identified to date [21, 22]. In the 1990s, a new RSVB genotype (BA) emerged with a duplication in the G gene and quickly disseminated globally [23, 24]. During the 2010–2011 winter season, a duplication in the G gene was detected among RSVA GA2 viruses, giving rise to the ON1 sub-genotype that also spread worldwide and became dominant in many countries [24–32]. The clinical impact of different genotypes has not been fully elucidated, as some studies report differences in disease severity [33–38], while others have seen similar clinical illness [36–38].
Intensive surveillance of RSV in infants in a paediatric cohort in Amman, Jordan, has revealed a high burden of RSV in hospitalized children [39–41]. To further understand the molecular evolution and transmission of RSV in this population, we obtained whole-genome sequences from 85 RSV samples collected during 2010–2013. We found that RSV epidemics in Jordan comprised multiple independent viral introductions of RSVA and RSVB, although one subgroup strongly dominated each year.
Methods
Sample and clinical data collection, and RSV diagnosis
All procedures were carried out in accordance with the ethical standards of the Jordanian Ministry of Health, University of Jordan and Vanderbilt University institutional review boards. After consent, trained local research staff obtained nasal and throat swabs. Standardized questionnaires were used to record demographic, clinical and socio-economic data. Parents were queried in Arabic, and bilingual research staff transcribed the information into an English-language case report form at the time of the interview. Demographic data, including age, sex, race and ethnicity, were recorded at the time of enrolment. Viral surveillance with clinical and demographic data in children<2 years old admitted with respiratory symptoms and/or fever was conducted at the Al-Bashir Government Hospital, Amman, Jordan from March 16 2010 to March 31 2013 [40, 41]. Nasal and throat swabs were collected and combined in transport medium (M4RT; Remel), aliquoted into MagMAX lysis/binding solution concentrate (Life Technologies), snap frozen, and stored at −80 °C. Original and lysis buffer aliquots were shipped on dry ice and were tested by reverse transcriptase (RT)-PCR for 11 respiratory viruses [RSV, human metapneumovirus (HMPV), human rhinovirus (HRV), influenza (flu) A, B and C, parainfluenza (PIV) virus 1, 2 and 3, adenovirus (ADV), and Middle East respiratory syndrome coronavirus (MERS-CoV)] [40].
From the 2010–2013 study period, a total of 3168 children's respiratory samples were processed for viral diagnostics, where>80 % tested positive for at least one virus, with RSV being the most common virus detected (44 %). Out of 1397 RSV-positive samples, 93 were randomly selected from all the 3 years with quantitative RT-PCR C t value <29 and were processed for whole-genome next-generation sequencing of RSV. Out of the 93 samples, 7 samples were taken from study subjects who had died due to RSV disease.
Viral RNA extraction, whole-genome sequencing, assembly and annotation
Extraction of the viral RNA was performed using 140 µl nasal wash sample in viral transport medium using the viral RNA mini kit (Qiagen). Four forward reverse transcription (RT) primers were designed and four sets of PCR primers were manually picked from primers designed across a consensus of complete RSV genome sequences as described before [11]. The four forward RT primers were diluted to 2 µM in water and pooled in equal volumes. cDNA was generated from 4 µl undiluted RNA, using the pooled forward primers and SuperScript III reverse transcriptase (Thermo Fisher Scientific). Four independent PCR reactions were performed on 2 µl cDNA template using either AccuPrime Taq DNA polymerase (Thermo Fisher Scientific) or Phusion high fidelity DNA polymerase (New England Biolabs) to generate four overlapping ~4 kb amplicons across the genome. Amplicons were verified with 1 % agarose gels, and excess primers and dNTPs were removed by treatment with exonuclease I (New England Biolabs) and shrimp alkaline phosphatase (Affymetrix) for 37 °C for 60 min, followed by incubation at 72 °C for 15 min. Amplicons were quantitated using a SYBR Green dsDNA detection assay (SYBR Green I nucleic acid gel stain; Thermo Fisher Scientific), and all four amplicons per genome were pooled in equal concentrations.
Samples were sequenced using both MiSeq (Illumina) and Ion Torrent PGM (Thermo Fisher Scientific) to overcome platform-specific errors. For the Illumina sequencing, libraries were prepared using the Nextera DNA sample preparation kit (Illumina) with half reaction volumes for MiSeq. Briefly, 25 ng pooled DNA amplicons were tagmented at 55 °C for 5 min. Tagmented DNA was cleaned with the ZR-96 DNA clean and concentrator kit (Zymo Research) and eluted in 25 µl resuspension buffer. Illumina sequencing adapters and barcodes were added to tagmented DNA via PCR amplification, where 20 µl tagmented DNA was combined with 7.5 µl Nextera PCR master mix, 2.5 µl Nextera PCR primer cocktail and 2.5 µl each index primer (Integrated DNA Technologies) for a total volume of 35 µl per reaction. Thermocycling was performed with 5 cycles of PCR, as per the Nextera DNA sample preparation kit protocol (3 min at 72 °C, denaturation for 10 s at 98 °C, annealing for 30 s at 63 °C and extension for 3 min at 72 °C) to create a dual-indexed library for each sample. After PCR amplification, 10 µl each library was pooled into a 1.5 ml tube, and the pool was cleaned twice with Ampure XP reagent (Beckman Coulter) to remove all leftover primers and small DNA fragments. The first cleaning used a 1.2× volume of the Ampure reagent, while the second cleaning used a 0.6× volume of the Ampure reagent. The cleaned pool was sequenced on the Illumina MiSeq v2 instrument (Illumina) with 300 bp paired-end reads.
In addition to Illumina sequencing, for Ion Torrent PGM (Thermo Fisher Scientific), 100 ng pooled DNA amplicons were sheared for 7 min and Ion-Torrent-compatible barcoded adapters were ligated to the sheared DNA using an Ion Xpress Plus fragment library kit (Thermo Fisher Scientific) to create 400 bp libraries. Libraries were pooled in equal volumes and cleaned with Ampure XP reagent (Beckman Coulter). Quantitative PCR was performed on the pooled, barcoded libraries to assess the quality of the pool and to determine the template dilution factor for emulsion PCR. The pool was diluted appropriately and amplified on Ion Sphere particles (ISPs) during emulsion PCR on an Ion One Touch 2 instrument (Thermo Fisher Scientific). The emulsion was broken, and the pool was cleaned and enriched for template-positive ISPs on an Ion One Touch ES instrument (Thermo Fisher Scientific). Sequencing was performed on the Ion Torrent PGM using 318v2 chips (Thermo Fisher Scientific).
RSV genome assembly and annotation
Sequence reads were sorted by barcode, trimmed and de novo assembled using CLC Bio’s clc assembler program, formerly known as clc novo assembly (http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/852/index.php?manual=De_novo_assembly.html), and the resulting contigs were searched against custom, full-length RSV nucleotide databases to find the closest reference sequence. All sequence reads were then mapped to the selected reference RSV sequence using CLC Bio’s clc_mapper_legacy, formerly called as clc_ref_assemble_long program (http://resources.qiagenbioinformatics.com/manuals/clcassemblycell/current/index.php?manual=Options_clc_mapper_legacy.html). At loci where both Ion Torrent and Illumina sequence data agreed on a variation (compared with the reference sequence), the reference sequence was updated to reflect the difference. A final mapping of all next-generation sequences to the updated reference sequences was performed with CLC Bio’s clc_mapper_legacy program. Curated assemblies were validated and annotated with the viral annotation software called Viral Genome ORF Reader, VIGOR 3.0 [42], before submission to GenBank. VIGOR was used to predict genes, perform alignments, ensure the fidelity of ORFs, correlate nucleotide polymorphisms with amino acid changes and detect any potential sequencing errors. The annotation was subjected to manual inspection and quality control before submission. All 85 sequences generated as part of this study were submitted to GenBank as part of the BioProject ID PRJNA262901 with accession number KX655615–KX655701 (KX655701, KX655688 and KX655625 are partial sequences).
Dataset assembly and phylogenetic analyses
In order to enrich the datasets of 58 RSVA and 27 RSVB Jordanian full-genome sequences, we retrieved all RSV whole-genome sequences from GenBank on July 21 2016, resulting in datasets of 771 and 331 sequences for RSVA and RSVB, respectively. The data were aligned using mafft v7.310 [43] and subsequently manually edited to accommodate the ORFs of all genes and the G gene duplication. To identify recombinant strains that would complicate the phylogenetic inferences, we analysed the datasets using the Recombination Detection Program Beta v4.94 [44]. This resulted in the exclusion of one recombinant RSVA sequence (JX015495) and four recombinant RSVB sequences (KJ939932, KJ672473, KJ627251, KJ939933) from locations other than Jordan. Maximum-likelihood trees were inferred using RAxML v7.2.6 [45] incorporating a general time reversible (GTR) model of nucleotide substitution with a gamma-distributed (Γ) rate variation among sites. To assess the robustness of each node, a 500 replicates bootstrap resampling process was performed. We also investigated the temporal signal of the datasets using TempEst [46]. A few sequences showing incongruent temporal patterns were excluded. Phylogenetic relationships were inferred for each of the data sets separately (669 RSVA sequences and 327 RSVB sequences) with a Bayesian phylogenetic approach using Markov Chain Monte Carlo available via the beast v1.8.4 package [47] and the high-performance computational capabilities of the Biowulf Linux cluster at the NIH, Bethesda, MD, USA (https://hpc.nih.gov/systems/). We used an uncorrelated relaxed molecular clock with branch rates drawn form a lognormal distribution to account for evolutionary rate variation among lineages, with a Skygrid demographic prior [48], and a GTR model of nucleotide substitution with gamma-distributed rate variation among sites. For viruses with only the year of viral collection available, the lack of tip date precision was accommodated by sampling uniformly across a 1 year window from January 1st to December 31st. The Markov chain Monte Carlo (MCMC) chain was run separately at least three times for each of the data sets and for at least 200 million iterations with sub-sampling every 20 000 iterations, using the beagle [49] library to improve computational performance. All parameters reached convergence, as assessed visually using Tracer v.1.6.0, with statistical uncertainty reflected in values of the 95 % highest posterior density (HPD). At least 10 % of the chain was removed as burn-in. Maximum clade credibility (MCC) trees were summarized using TreeAnnotator v1.8.4 and the trees were visualized in FigTree v1.4.3. The samples were collected from different neighbourhood zones in Amman, Jordan (11 and 9 zones for the antigenic subgroup A and B, respectively), which were aggregated into four spatial regions within Amman (North, Central, North-East and West regions) for our spatial analysis. To study the proportion of RSVA and RSVB circulating in the Amman population and globally, we merged the datasets of both RSV antigenic subgroups and estimated the proportion of RSVA and RSVB viruses, and their respective genotypes, circulating in 15 countries over time, using the R program [50].
Clinical data
We collected information on the patient age and bronchopneumonia status for the Amman paediatric cohort during 2010–2013. As we had collected this information, we investigated whether infection with a specific RSV antigenic subgroup was significantly associated with patient age or disease progression to bronchopneumonia. For this purpose, we used two statistical tests, namely the t-test (patient age as a continuous variable) and the chi-square test (bronchopneumonia as a binary outcome). Additionally, we also investigated the extent of RSV phylogenetic structure for bronchopneumonia (discrete trait) using the association index (AI). This metric quantifies the degree to which samples of patients with bronchopneumonia tend to cluster together relative to the expectation for randomized trait assignments. AI values close to 0 reflect strong phylogeny-trait correlation, whereas AI values close to 1 reflect the absence of phylogenetic structure for the trait [51]. For this purpose, we reconstructed the bronchopneumonia status at the ancestral nodes using the discrete diffusion models implemented in beast [52].
Results
Genetic diversity of RSV in a paediatric cohort in Amman, Jordan
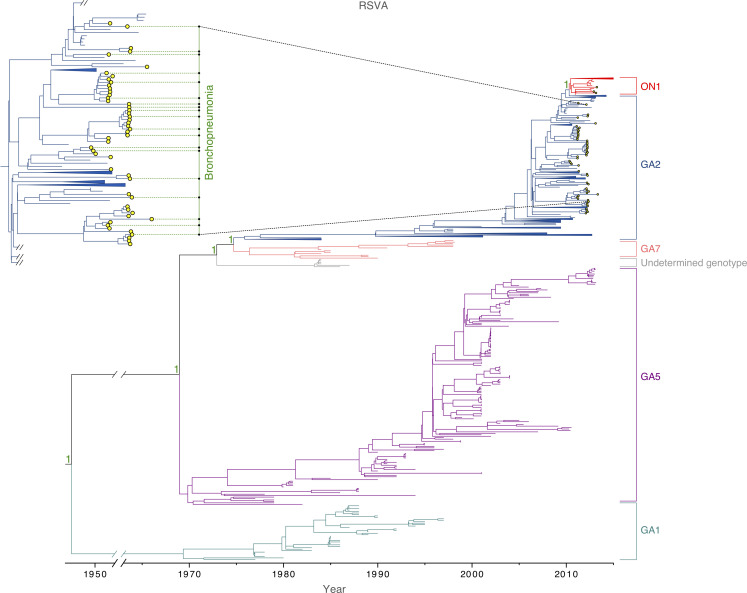
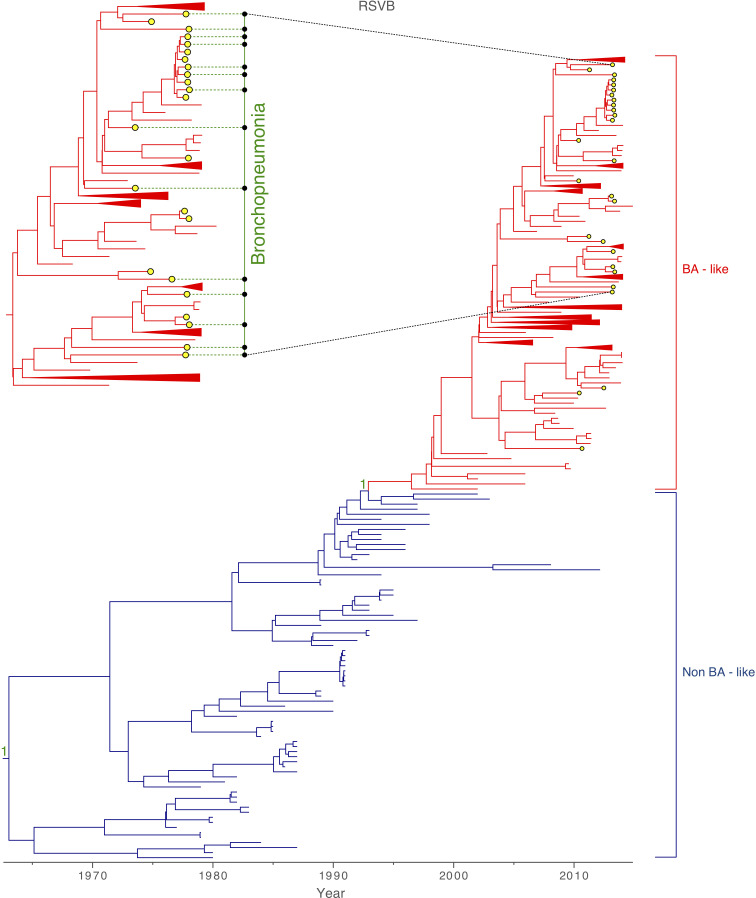
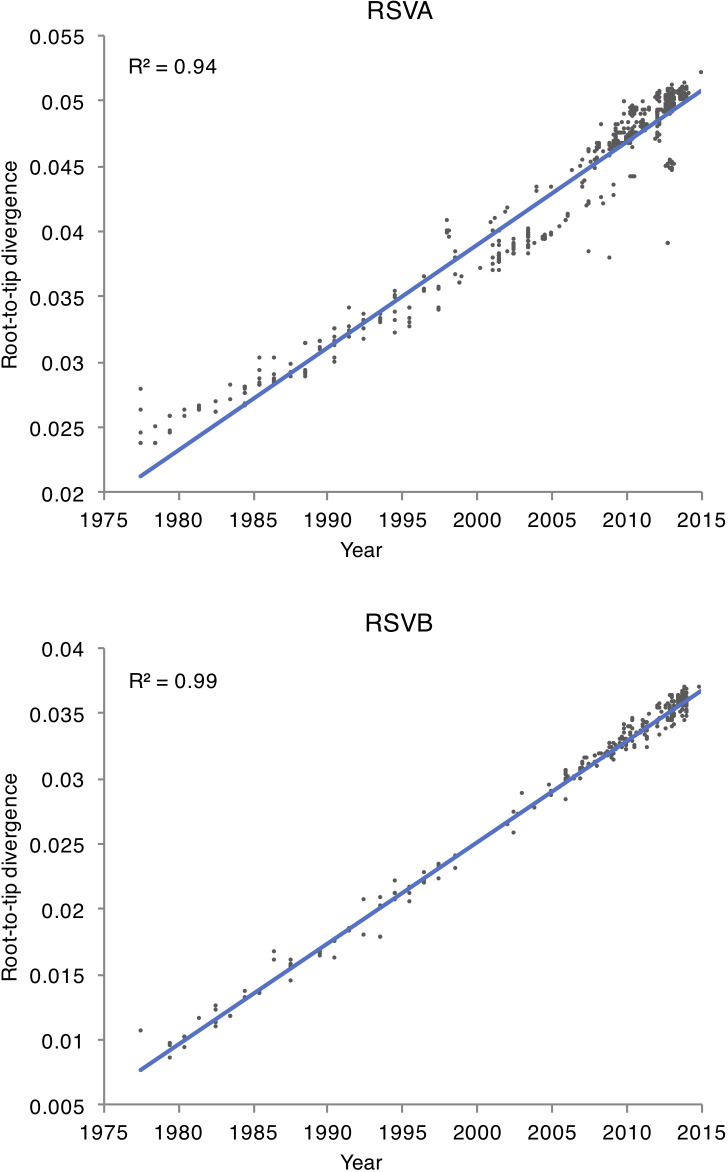
Among the 93 samples for which sequencing was attempted, 85 complete viral genomes were successfully sequenced from the Amman paediatric cohort during 2010–2013, 58 were identified as RSVA and 27 as RSVB. The mean age of patients with RSVA was 2.5 years (9.3 months – 5.4 years), and 1.8 years (8.3 months – 4.8 years) for RSVB. Most of the RSVA isolates collected in Amman belonged to the dominant GA2 genotype (55/58). Three samples, collected during October 2012 – February 2013, were positioned within the newly emerged ON1 sub-genotype, which is characterized by a 72 nucleotide duplication at the G gene [25] (Fig. 1). All RSVB samples were sub-genotyped as GB1 and positioned within the BA-like clade, which is defined by a duplication of 60 nucleotides in the G gene (Fig. 2) [23, 53]. A plot of genetic distance against time indicates that the evolution of RSVA and RSVB is strongly clock-like for both viruses, with correlation coefficients of 0.94 and 0.99, respectively (Fig. 3). RSVA and RSVB were found to be evolving at similar rates [RSVA 7.31×10−4 (95 % HPD 6.78×10−4 – 7.88×10−4) substitutions/site/year; RSVB 7.08×10−4 (95 % HPD 6.58×10−4 – 7.59×10−4) substitutions/site/year) (Table 1), estimates that are consistent with previous studies [11, 19]. However, the genetic diversity of RSVA traces back to a common ancestor that existed approximately 27 years earlier than RSVB (Table 1).
Fig. 1.
Global RSVA phylogeny. MCC tree inferred for 669 viruses sampled globally, coloured by genotype. The yellow-filled circles indicate 58 samples collected from a paediatric cohort in Amman, Jordan, which cluster within the GA2 genotype and ON1 sub-genotype. A few tips from countries other than Jordan are collapsed for visual clarity. Green dotted lines indicate samples from patients with bronchopneumonia. Posterior probabilities are indicated as support for nodes defining a genotype. A more detailed tree of RSVA samples collected in Amman is provided in Fig. 4.
Fig. 2.
Global RSVB phylogeny. MCC tree inferred for 327 viruses sampled globally, coloured by genotype. The yellow-filled circles indicate 27 samples collected from a paediatric cohort in Amman, Jordan, which cluster within the GB1 sub-genotype positioned within the BA-like clade. A few tips from countries other than Jordan are collapsed for visual clarity. Green dotted lines indicate samples from patients with bronchopneumonia. Posterior probabilities are indicated as support for nodes defining a genotype. A more detailed tree of RSVB samples collected in Amman is provided in Fig. 4.
Fig. 3.
Root-to-tip divergence. A plot of sampling time versus genetic distance, inferred from trees of RSVA and RSVB inferred using maximum-likelihood methods. Both RSVA and RSVB datasets display a strong temporal signal for a molecular clock, as shown by the high R 2.
Table 1.
Evolutionary rates and times of divergence for RSVA and RSVB
Values in parentheses represent 95 % HPD intervals.
|
RSV |
Evolutionary rate (substitutions/site/year) |
tMRCA (date in decimal format) |
|---|---|---|
|
RSVA |
7.31×10−4 (6.78×10−4 – 7.88×10−4) |
1937.54 (1917.58 – 1954.26) |
|
RSVB |
7.08×10−4 (6.58×10−4 – 7.59×10−4) |
1964.79 (1961.58 – 1967.84) |
Multiple introductions of RSV into the Amman cohort during each epidemic
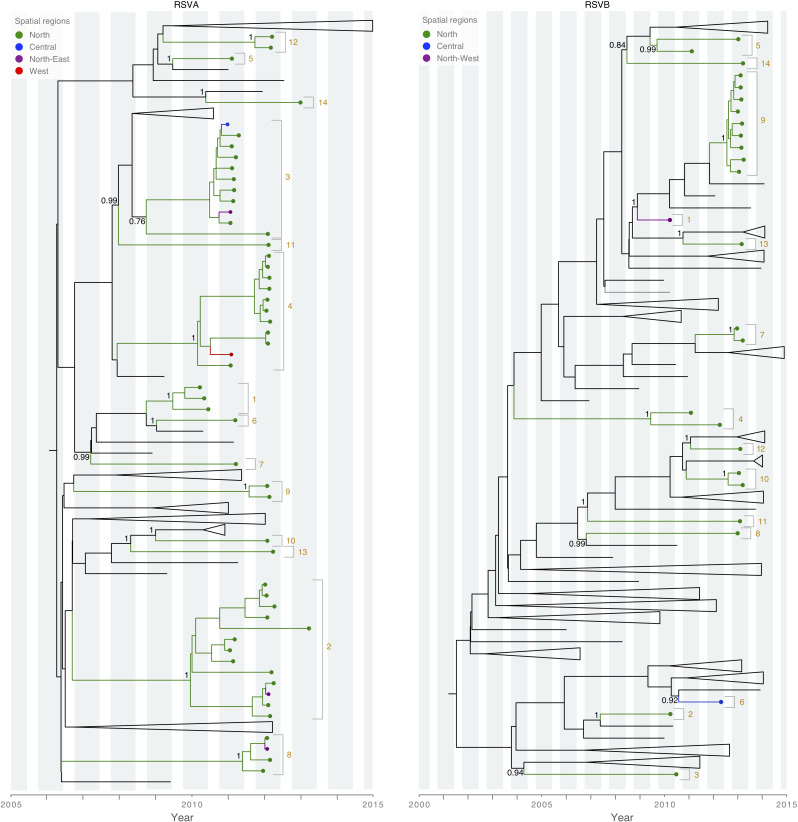
Our study period included four winter epidemic seasons, defined as September to June: 2009–2010, 2010–2011, 2011–2012 and 2012–2013. We found that each annual RSV epidemic in Amman was not a point-source outbreak, but instead seeded by multiple independent viral introductions, including both RSVA and RSVB lineages, into the Amman cohort population. The low availability of RSV sequence data from many countries makes it difficult to infer the precise number of viral introduction, but we estimated, conservatively, the number of introductions in each year (Fig. 4): five introductions during the 2009–2010 epidemic, eight during the 2010–2011 epidemic, seven during the 2011–2012 epidemic, and nine during the 2012–2013 epidemic. Despite the higher availability of RSVA sequences, the same number of total introductions of RSVA and RSVB were identified during the 4 years of study (n=14). Three RSVA (ON1) were identified during the 2012–2013 epidemic, representing at least one additional introduction since they belong to an entirely different genotype, but are not well supported on the tree. Viruses from multiple epidemic seasons sometimes clustered together, possibly representing persistence of RSV in Amman over multiple years. Similarly, the time to the most recent common ancestor (tMRCA) values were estimated to exceed 1 year for several viral introductions, based on the time-scaled MCC trees (Tables 2 and 3). However, the low availability of background RSV sequences from other locations means that we cannot exclude the possibility that genetically close viruses were re-imported each year from an unsampled location, rather than persisting locally.
Fig. 4.
Spatial patterns of RSV introductions in Amman, Jordan. Truncated MCC trees (RSVA, left; RSVB, right) with tip labels coloured by spatial region in Amman, Jordan. Tips from countries other than Jordan are collapsed for visual clarity. Discrete viral introductions are numbered, similarly to Tables 2 and 3, and shaded orange. Node support is indicated for posterior probabilities >0.75. Vertical grey bars indicate RSV epidemic seasons in Jordan, according to Halasa et al. [40].
Table 2.
Highly supported RSVA introductions in Amman, Jordan
|
Introduction |
No. of samples |
Posterior probability |
Spatial region |
tMRCA (date in decimal format)* |
Viral persistence |
Genotype |
|---|---|---|---|---|---|---|
|
1 |
3 |
1 |
North |
2009.56 (2009.18 – 2009.91) |
3 months |
GA2 |
|
2 |
13 |
1 |
North, North-East |
2009.56 (2009.02 – 2010.19) |
2 years and 2 months |
GA2 |
|
3 |
11 |
0.76 |
North, Central, North-East |
2008.65 (2008.07 – 2009.24) |
1 year and 2 months |
GA2 |
|
4 |
11 |
1 |
North, West |
2009.91 (2009.38 – 2010.39) |
1 year and 1 month |
GA2 |
|
5 |
1 |
1 |
North |
– |
– |
GA2 |
|
6 |
1 |
North |
– |
– |
GA2 |
|
|
7 |
1 |
0.99 |
North |
– |
– |
GA2 |
|
8 |
4 |
1 |
North, North-East |
2011.21 (2010.72 – 2011.63) |
2 months |
GA2 |
|
9 |
2 |
1 |
North |
2011.80 (2011.48 – 2012.04) |
1 month |
GA2 |
|
10 |
1 |
1 |
North |
– |
– |
GA2 |
|
11 |
1 |
0.99 |
North |
– |
– |
GA2 |
|
12 |
2 |
1 |
North |
2011.74 (2011.33 – 2012.07) |
13 days |
GA2 |
|
13 |
1 |
1 |
North |
– |
– |
GA2 |
|
14 |
1 |
1 |
North |
– |
– |
GA2 |
*Values in parentheses represent 95 % HPD intervals
Table 3.
Highly supported RSVB introductions in Amman, Jordan
|
Introduction |
No. of samples |
Posterior probability |
Spatial region |
tMRCA (date in decimal format)* |
Viral persistence |
Genotype |
|---|---|---|---|---|---|---|
|
1 |
1 |
1 |
North-West |
– |
– |
GB1 |
|
2 |
1 |
1 |
North |
– |
– |
GB1 |
|
3 |
1 |
0.94 |
North |
– |
– |
GB1 |
|
4 |
2 |
1 |
North |
2009.31 (2008.34 – 2010.22) |
1 year and 3 months |
GB1 |
|
5 |
2 |
0.99 |
North |
2009.86 (2009.26 – 2010.36) |
1 year and 11 months |
GB1 |
|
6 |
1 |
0.92 |
Central |
– |
– |
GB1 |
|
7 |
2 |
1 |
North |
2012.86 (2012.71 – 2012.96) |
3 months |
GB1 |
|
8 |
1 |
0.99 |
North |
– |
– |
GB1 |
|
9 |
9 |
1 |
North |
2012.46 (2012.22 – 2012.67) |
3 months |
GB1 |
|
10 |
2 |
1 |
North |
2012.63 (2012.33 – 2012.89) |
2 months |
GB1 |
|
11 |
1 |
1 |
North |
– |
– |
GB1 |
|
12 |
1 |
1 |
North |
– |
– |
GB1 |
|
13 |
1 |
1 |
North |
– |
– |
GB1 |
|
14 |
1 |
0.84 |
North |
– |
– |
GB1 |
*Values in parentheses represent 95 % HPD intervals.
Spatial patterns of RSVA and RSVB in Amman
To study the spatial dissemination of RSV within the Amman cohort population, four regions were defined among the city’s neighbourhoods from which viruses were successfully sequenced (Fig. 4). Four of the fourteen RSVA introductions were identified in multiple regions, consistent with spatial spread within the city. One RSVA introduction was identified in three of the four spatial regions defined within Amman. In contrast, although a large number of introductions of RSVB into Amman were identified (n=14), each introduction was confined to a single city region, and there was no evidence of dissemination of RSVB between regions (Tables 2 and 3).
Proportion of circulating RSV antigenic subgroups A and B
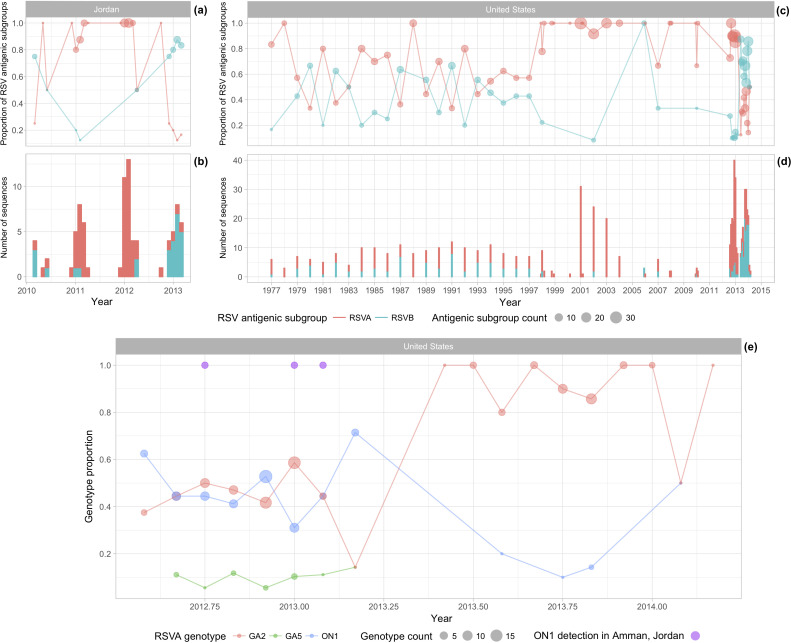
We investigated the frequency and distribution of RSV cases during the 2010–2013 period (Fig. S3) and observed limited variation across seasons. RSVA and RSVB co-circulated in Amman, but one antigenic subgroup dominated over the other during an epidemic. RSVA was dominant during the 2010/2011 and 2011/2012 winter epidemics, and RSVB was dominant during the 2009/2010 and 2012/2013 winter epidemics (Fig. 5a, b). Alternating dominance between RSVA and RSVB was also observed in the USA, the only country with a sufficiently long time series of genetic data to examine genotype interactions (Fig. 5c, d). The low availability of ON1 sequences in Jordan (n=3) prevented a similar examination of interactions between the RSVA ON1 and GA2 genotypes within Jordan. However, Fig. 5(e) shows that the cluster of ON1 viruses in Jordan during the 2012/2013 winter epidemic coincided with the emergence of ON1 in the USA.
Fig. 5.
Circulation of RSV in Jordan and the USA. The proportion of all available full-genome RSV sequences aggregated by antigenic subgroup for Jordan collected from 2010 to 2013 (a), for the USA from 1977 to 2014 (c) and aggregated by genotype for the USA from 2012 to 2014 (e). Circle sizes represent the number of sequences from each antigenic subgroup (a and c) or genotype (e) contributing to the proportion. Total number of RSV sequences from Jordan collected from 2010 to 2013 (b) and from the USA from 1977 to 2014 (d). Purple circles represent dates when genotype ON1 was identified in Amman, Jordan.
Association of viral genetics and clinical outcomes
RSVA-infected children tended to be older than RSVB-infected children (30 months versus 22.4 months, respectively) (two-tailed t-test P value=0.02) in this cohort. Approximately 32.8 % of RSVA-infected and 51.9 % of RSVB-infected patients developed bronchopneumonia (Figs 1 and 2), but this difference was not found to be significant (chi-square P value=0.06), given the small sample size. We observed little phylogenetic clustering by whether a child developed bronchopneumonia, with AI values very close to 1 for both antigenic subgroups: RSVA, AI=0.98 (95 % HPD interval 0.78–1.21); RSVB, AI=1.16 (95 % HPD interval 0.86–1.50).
Discussion
RSV imposes a high burden of respiratory disease on infants and children in Amman, Jordan [40]. By sequencing the complete genomes of 85 RSVA and RSVB samples from children in Amman, the largest whole-genome collection of RSVs from any country in the Middle East, this study provides insights into the genetic diversity of RSVA and RSVB in an under-studied region. We found that RSV epidemics in Amman comprised multiple co-circulating strains of both RSVA and RSVB, as observed in other regions [54], although one subgroup tends to strongly dominate (RSVA dominated seasons 2010/2011 and 2011/2012, and RSVB dominated seasons 2009/2010 and 2012/2013). The severity of clinical outcomes is difficult to predict from the genetic sequence of the virus alone, at least with the sample size used in this study, although the higher frequency of bronchopneumonia associated with RSVB (51.9 %) compared with RSVA (32.8 %) merits further study. The vast majority of RSV samples in Amman belonged to the most predominant RSVA and RSVB genotypes (GA2 and BA-like, respectively), with only three RSVA identified from the recently emerged ON1 sub-genotype. More intensive sequencing may be needed to detect lower frequency variants.
Our study benefitted from using whole-genome sequences, as the partial G gene may produce misleading results [53] and transmission patterns may be difficult to infer using shorter coding regions [19]. However, at this time there is far more background RSV sequence data available for the G gene for other countries, compared to whole-genome data, which limited our ability to differentiate independent viral introductions into Amman during an epidemic and infer their spatial origins. Compared to influenza A viruses, for which the global spatial ecology has been extensively characterized [55, 56], the paucity of RSV sequence data from many regions undermines efforts to understand how the virus moves spatially around the world between local epidemics. It is clear that RSV is continually being re-introduced into Amman, similar to the viral dynamics observed in other longitudinal studies of RSV, such as in Kenya [57]. However, it remains unclear whether global RSV evolution follows a metapopulation model or a source-sink model, and the extent to which viruses persist in a single location. Additionally, the limited number of RSV samples from Jordan prevented more sophisticated phylogeographical modelling that could unveil the dynamics within the city. In the future, it would be particularly interesting to compare the spatial dynamics of RSVA and RSVB. Our analysis revealed more instances of RSVA transmission across multiple regions of the city, but these inferences were based on very low sample sizes, particularly for RSVB, and require further study with larger data sets.
The ongoing development of candidate RSV vaccines underscores the need for further understanding of human immune responses, and the extent of cross-immunity between RSV strains of varying genetic distances. The potential interactions between RSVA and RSVB observed in Jordan and the USA, with sequential alternation in the transmission of antigenic subgroups and genotypes (Fig. S5), may result in subgroup- or genotype-specific immunity. These dynamics demonstrate that future vaccine compositions could benefit by including representative strains for both RSVA and RSVB, or even, representative strains for the different genotypes. Additionally, there is a need for further sequencing of RSV over sufficiently long time-series within discrete locations to characterize population dynamics and estimate competitive immunological interactions. The recent emergence of ON1 variants also provides an opportunity to examine competitive interactions between ON1 and previously dominant GA2 strains across multiple geographical locations, to better understand the strength and breadth of immune responses to different RSV strains. Going forward, long-term studies of RSV dynamics within defined populations will be essential to inform the design and potential impact of future RSV vaccines.
Data bibliography
Trovão N. S. Accession numbers of the genetic sequences used in this study are listed in the Supplementary material, available with the online version of this article (2019).
Supplementary Data
Funding information
Sample collection was supported by the UBS Optimus Foundation; and CTSA award UL1TR000445 from the National Center for Advancing Translational Sciences, NIH, USA. The sequencing and sequence data analysis was supported by the National Institute of Allergy and Infectious Diseases (NIAID)/NIH Genomic Centers for Infectious Diseases (GCID) program, grant U19-AI-110819 awarded to S. R. D. S. R. D. is also supported by the NIH-funded Tennessee Center for AIDS Research (grant P30 AI110527). N. S. T. is funded by the NIAID Center for Research on Influenza Pathogenesis (CRIP), contract HHSN272201400008C, which is part of the Centers of Excellence for Influenza Research and Surveillance (CEIRS). The contents of this article are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences nor the NIH.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All procedures were carried out in accordance with the ethical standards of the Jordanian Ministry of Health, University of Jordan and Vanderbilt University institutional review boards. Nasal and throat swabs were obtained by trained local research staff with consent. Standardized questionnaires were used to record demographic, clinical and socio-economic data. Parents were queried in Arabic, and bilingual research staff transcribed the information into an English-language case report form at the time of the interview.
Footnotes
Abbreviations: AI, association index; HPD, highest posterior density; MCC, maximum clade credibility; RSV, respiratory syncytial virus; tMRCA, time to the most recent common ancestor.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. The Lancet. 2017;390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogra PL. Respiratory syncytial virus: the virus, the disease and the immune response. Paediatr Respir Rev. 2004;5:S119–S126. doi: 10.1016/S1526-0542(04)90023-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falsey AR, Walsh EE. Respiratory syncytial virus infection in elderly adults. Drugs Aging. 2005;22:577–587. doi: 10.2165/00002512-200522070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee N, Lui GCY, Wong KT, Li TCM, Tse ECM, et al. High morbidity and mortality in adults hospitalized for respiratory syncytial virus infections. Clin Infect Dis. 2013;57:1069–1077. doi: 10.1093/cid/cit471. [DOI] [PubMed] [Google Scholar]
- 5.Falsey AR, McElhaney JE, Beran J, van Essen GA, Duval X, et al. Respiratory syncytial virus and other respiratory viral infections in older adults with moderate to severe influenza-like illness. J Infect Dis. 2014;209:1873–1881. doi: 10.1093/infdis/jit839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins D, Trujillo C, Keech C. Advances in RSV vaccine research and development – a global agenda. Vaccine. 2016;34:2870–2875. doi: 10.1016/j.vaccine.2016.03.109. [DOI] [PubMed] [Google Scholar]
- 7.Borchers AT, Chang C, Gershwin ME, Gershwin LJ. Respiratory syncytial virus – a comprehensive review. Clin Rev Allergy Immunol. 2013;45:331–379. doi: 10.1007/s12016-013-8368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andabaka T, Nickerson JW, Rojas-Reyes MX, Rueda JD, Bacic Vrca V, et al. Monoclonal antibody for reducing the risk of respiratory syncytial virus infection in children. Cochrane Database Syst Rev. 2013;4:CD006602. doi: 10.1002/14651858.CD006602.pub4. [DOI] [PubMed] [Google Scholar]
- 9.Rebuffo-Scheer C, Bose M, He J, Khaja S, Ulatowski M, et al. Whole genome sequencing and evolutionary analysis of human respiratory syncytial virus A and B from Milwaukee, WI 1998-2010. PLoS One. 2011;6:e25468. doi: 10.1371/journal.pone.0025468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agoti CN, Otieno JR, Munywoki PK, Mwihuri AG, Cane PA, et al. Local evolutionary patterns of human respiratory syncytial virus derived from whole-genome sequencing. J Virol. 2015;89:3444–3454. doi: 10.1128/JVI.03391-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schobel SA, Stucker KM, Moore ML, Anderson LJ, Larkin EK, et al. Respiratory syncytial virus whole-genome sequencing identifies convergent evolution of sequence duplication in the C-terminus of the G gene. Sci Rep. 2016;6:26311. doi: 10.1038/srep26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Afonso CL, Amarasinghe GK, Bányai K, Bào Y, Basler CF, et al. Taxonomy of the order Mononegavirales: update 2016. Arch Virol. 2016;161:2351–2360. doi: 10.1007/s00705-016-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grint P. Respiratory syncytial virus (RSV) in man and animals. J R Soc Med. 1989;82:56–57. doi: 10.1177/014107688908200127. [DOI] [Google Scholar]
- 14.Hall CB, Walsh EE, Schnabel KC, Long CE, McConnochie KM, et al. Occurrence of groups A and B of respiratory syncytial virus over 15 years: associated epidemiologic and clinical characteristics in hospitalized and ambulatory children. J Infect Dis. 1990;162:1283–1290. doi: 10.1093/infdis/162.6.1283. [DOI] [PubMed] [Google Scholar]
- 15.Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. Circulation patterns of genetically distinct group A and B strains of human respiratory syncytial virus in a community. J Gen Virol. 1998;79:2221–2229. doi: 10.1099/0022-1317-79-9-2221. [DOI] [PubMed] [Google Scholar]
- 16.Esposito S, Piralla A, Zampiero A, Bianchini S, Di Pietro G, et al. Characteristics and their clinical relevance of respiratory syncytial virus types and genotypes circulating in northern Italy in five consecutive winter seasons. PLoS One. 2015;10:e0129369. doi: 10.1371/journal.pone.0129369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White LJ, Waris M, Cane PA, Nokes DJ, Medley GF. The transmission dynamics of groups A and B human respiratory syncytial virus (hRSV) in England & Wales and Finland: seasonality and cross-protection. Epidemiol Infect. 2005;133:279–289. doi: 10.1017/S0950268804003450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katzov-Eckert H, Botosso VF, Neto EA, Zanotto PM. Phylodynamics and dispersal of hRSV entails its permanence in the general population in between yearly outbreaks in children. PLoS One. 2012;7:e41953. doi: 10.1371/journal.pone.0041953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan L, Coenjaerts FEJ, Houspie L, Viveen MC, van Bleek GM, et al. The comparative genomics of human respiratory syncytial virus subgroups A and B: genetic variability and molecular evolutionary dynamics. J Virol. 2013;87:8213–8226. doi: 10.1128/JVI.03278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinelli M, Frati ER, Zappa A, Ebranati E, Bianchi S, et al. Phylogeny and population dynamics of respiratory syncytial virus (RSV) A and B. Virus Res. 2014;189:293–302. doi: 10.1016/j.virusres.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Trento A, Casas I, Calderón A, Garcia-Garcia ML, Calvo C, et al. Ten years of global evolution of the human respiratory syncytial virus BA genotype with a 60-nucleotide duplication in the G protein gene. J Virol. 2010;84:7500–7512. doi: 10.1128/JVI.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duvvuri VR, Granados A, Rosenfeld P, Bahl J, Eshaghi A, et al. Genetic diversity and evolutionary insights of respiratory syncytial virus a ON1 genotype: global and local transmission dynamics. Sci Rep. 2015;5:14268. doi: 10.1038/srep14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trento A, Galiano M, Videla C, Carballal G, García-Barreno B, et al. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol. 2003;84:3115–3120. doi: 10.1099/vir.0.19357-0. [DOI] [PubMed] [Google Scholar]
- 24.Hotard AL, Laikhter E, Brooks K, Hartert TV, Moore ML. Functional analysis of the 60-nucleotide duplication in the respiratory syncytial virus Buenos Aires strain attachment glycoprotein. J Virol. 2015;89:8258–8266. doi: 10.1128/JVI.01045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eshaghi A, Duvvuri VR, Lai R, Nadarajah JT, Li A, et al. Genetic variability of human respiratory syncytial virus A strains circulating in Ontario: a novel genotype with a 72 nucleotide G gene duplication. PLoS One. 2012;7:e32807. doi: 10.1371/journal.pone.0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comas-García A, Noyola DE, Cadena-Mota S, Rico-Hernández M, Bernal-Silva S. Respiratory syncytial virus-A ON1 genotype emergence in central Mexico in 2009 and evidence of multiple duplication events. J Infect Dis. 2018;217:1089–1098. doi: 10.1093/infdis/jiy025. [DOI] [PubMed] [Google Scholar]
- 27.Song J, Zhang Y, Wang H, Shi J, Sun L, et al. Emergence of ON1 genotype of human respiratory syncytial virus subgroup A in China between 2011 and 2015. Sci Rep. 2017;7:5501. doi: 10.1038/s41598-017-04824-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otieno JR, Agoti CN, Gitahi CW, Bett A, Ngama M, et al. Molecular evolutionary dynamics of respiratory syncytial virus group A in recurrent epidemics in coastal Kenya. J Virol. 2016;90:4990–5002. doi: 10.1128/JVI.03105-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thongpan I, Mauleekoonphairoj J, Vichiwattana P, Korkong S, Wasitthankasem R, et al. Respiratory syncytial virus genotypes NA1, ON1, and BA9 are prevalent in Thailand, 2012-2015. PeerJ. 2017;5:e3970. doi: 10.7717/peerj.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabatabai J, Prifert C, Pfeil J, Grulich-Henn J, Schnitzler P. Novel respiratory syncytial virus (RSV) genotype ON1 predominates in Germany during winter season 2012-13. PLoS One. 2014;9:e109191. doi: 10.1371/journal.pone.0109191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierangeli A, Trotta D, Scagnolari C, Ferreri ML, Nicolai A, et al. Rapid spread of the novel respiratory syncytial virus a ON1 genotype, central Italy, 2011 to 2013. Euro Surveill. 2014;19:20843. doi: 10.2807/1560-7917.ES2014.19.26.20843. [DOI] [PubMed] [Google Scholar]
- 32.Panayiotou C, Richter J, Koliou M, Kalogirou N, Georgiou E, et al. Epidemiology of respiratory syncytial virus in children in Cyprus during three consecutive winter seasons (2010-2013): age distribution, seasonality and association between prevalent genotypes and disease severity. Epidemiol Infect. 2014;142:2406–2411. doi: 10.1017/S0950268814000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laham FR, Mansbach JM, Piedra PA, Hasegawa K, Sullivan AF, et al. Clinical profiles of respiratory syncytial virus subtypes A and B among children hospitalized with bronchiolitis. Pediatr Infect Dis J. 2017;36:808–810. doi: 10.1097/INF.0000000000001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshihara K, Le MN, Okamoto M, Wadagni ACA, Nguyen HA, et al. Association of RSV-A ON1 genotype with increased pediatric acute lower respiratory tract infection in Vietnam. Sci Rep. 2016;6:27856. doi: 10.1038/srep27856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinello RA, Chen MD, Weibel C, Kahn JS. Correlation between respiratory syncytial virus genotype and severity of illness. J Infect Dis. 2002;186:839–842. doi: 10.1086/342414. [DOI] [PubMed] [Google Scholar]
- 36.Otieno JR, Kamau EM, Agoti CN, Lewa C, Otieno G, et al. Spread and evolution of respiratory syncytial virus a genotype ON1, coastal Kenya, 2010-2015. Emerg Infect Dis. 2017;23:264–271. doi: 10.3201/eid2302.161149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fodha I, Vabret A, Ghedira L, Seboui H, Chouchane S, et al. Respiratory syncytial virus infections in hospitalized infants: association between viral load, virus subgroup, and disease severity. J Med Virol. 2007;79:1951–1958. doi: 10.1002/jmv.21026. [DOI] [PubMed] [Google Scholar]
- 38.McIntosh ED, De Silva LM, Oates RK. Clinical severity of respiratory syncytial virus group A and B infection in Sydney, Australia. Pediatr Infect Dis J. 1993;12:815–819. doi: 10.1097/00006454-199310000-00004. [DOI] [PubMed] [Google Scholar]
- 39.Haynes AK, Manangan AP, Iwane MK, Sturm-Ramirez K, Homaira N, et al. Respiratory syncytial virus circulation in seven countries with global disease detection regional centers. J Infect Dis. 2013;208 (Suppl. 3):S246–S254. doi: 10.1093/infdis/jit515. [DOI] [PubMed] [Google Scholar]
- 40.Halasa N, Williams J, Faouri S, Shehabi A, Vermund SH, et al. Natural history and epidemiology of respiratory syncytial virus infection in the Middle East: hospital surveillance for children under age two in Jordan. Vaccine. 2015;33:6479–6487. doi: 10.1016/j.vaccine.2015.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khuri-Bulos N, Lawrence L, Piya B, Wang L, Fonnesbeck C, et al. Severe outcomes associated with respiratory viruses in newborns and infants: a prospective viral surveillance study in Jordan. BMJ Open. 2018;8:e021898. doi: 10.1136/bmjopen-2018-021898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Sundaram JP, Stockwell TB. VIGOR extended to annotate genomes for additional 12 different viruses. Nucleic Acids Res. 2012;40:W186–W192. doi: 10.1093/nar/gks528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 44.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 46.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the beast 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, et al. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol. 2013;30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370–1376. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team Vienna: R Foundation for Statistical Computing; 2014. https://www.R-project.org [Google Scholar]
- 51.Trovão NS, Suchard MA, Baele G, Gilbert M, Lemey P. Bayesian inference reveals host-specific contributions to the epidemic expansion of influenza A H5N1. Mol Biol Evol. 2015;32:3264–3275. doi: 10.1093/molbev/msv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bose ME, He J, Shrivastava S, Nelson MI, Bera J, et al. Sequencing and analysis of globally obtained human respiratory syncytial virus A and B genomes. PLoS One. 2015;10:e0120098. doi: 10.1371/journal.pone.0120098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan L, Lemey P, Houspie L, Viveen MC, Jansen NJG, et al. Genetic variability among complete human respiratory syncytial virus subgroup A genomes: bridging molecular evolutionary dynamics and epidemiology. PLoS One. 2012;7:e51439. doi: 10.1371/journal.pone.0051439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, et al. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemey P, Rambaut A, Bedford T, Faria N, Bielejec F, et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 2014;10:e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otieno JR, Kamau EM, Oketch JW, Ngoi JM, Gichuki AM, et al. Whole genome analysis of local Kenyan and global sequences unravels the epidemiological and molecular evolutionary dynamics of RSV genotype ON1 strains. Virus Evol. 2018;4:vey027. doi: 10.1093/ve/vey027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.