Abstract
Entamoeba histolytica is believed to be devoid of peroxisomes, like most anaerobic protists. In this work, we provided the first evidence that peroxisomes are present in E. histolytica, although only seven proteins responsible for peroxisome biogenesis (peroxins) were identified (Pex1, Pex6, Pex5, Pex11, Pex14, Pex16, and Pex19). Targeting matrix proteins to peroxisomes is reduced to the PTS1-dependent pathway mediated via the soluble Pex5 receptor, while the PTS2 receptor Pex7 is absent. Immunofluorescence microscopy showed that peroxisomal markers (Pex5, Pex14, Pex16, Pex19) are present in vesicles distinct from mitosomes, the endoplasmic reticulum, and the endosome/phagosome system, except Pex11, which has dual localization in peroxisomes and mitosomes. Immunoelectron microscopy revealed that Pex14 localized to vesicles of approximately 90–100 nm in diameter. Proteomic analyses of affinity-purified peroxisomes and in silico PTS1 predictions provided datasets of 655 and 56 peroxisomal candidates, respectively; however, only six proteins were shared by both datasets, including myo-inositol dehydrogenase (myo-IDH). Peroxisomal NAD-dependent myo-IDH appeared to be a dimeric enzyme with high affinity to myo-inositol (Km 0.044 mM) and can utilize also scyllo-inositol, D-glucose and D-xylose as substrates. Phylogenetic analyses revealed that orthologs of myo-IDH with PTS1 are present in E. dispar, E. nutalli and E. moshkovskii but not in E. invadens, and form a monophyletic clade of mostly peroxisomal orthologs with free-living Mastigamoeba balamuthi and Pelomyxa schiedti. The presence of peroxisomes in E. histolytica and other archamoebae breaks the paradigm of peroxisome absence in anaerobes and provides a new potential target for the development of antiparasitic drugs.
Author summary
E. histolytica colonizes the human large intestine upon ingestion of amebic cysts. Trophozoites dwell in the hypoxic milieu of the intestinal lumen and can invade the epithelium to cause potentially lethal amebiasis. As an anaerobe, E. histolytica is dependent on glycolysis to generate ATP, whereas mitochondria are present in a greatly reduced form called mitosomes. Peroxisomes are believed to be absent in E. histolytica, as in other anaerobes, supported by the lack of typical peroxisomal pathways, including β-oxidation and catalase. However, we identified seven proteins involved in peroxisome biogenesis, the peroxins, and our investigations of their cellular localization indicated that E. histolytica possesses the anaerobic form of peroxisomes that we recently discovered in free-living anaerobic relatives. Proteomic analysis of anaerobic peroxisomes revealed the presence of highly active homodimeric myo-inositol dehydrogenase (myo-IDH), which catalyzes NAD-dependent oxidation of myo-inositol and other cyclitols. Since upstream and downstream pathways are absent in anaerobic peroxisomes, the role of myo-IDH is most likely to maintain intraperoxisomal redox potential. Discovery of anaerobic peroxisomes in E. histolytica and identification of myo-IDH that is absent in host cells provide a new avenue for the development of strategies against the parasite.
Introduction
Entamoeba histolytica is a causative agent of amoebiasis, one of the most prevalent parasitic diseases of humans. Over 65,000 lethal cases of amoebiasis per year have been reported worldwide [1,2], and E. histolytica prevalence has been estimated to reach 3.55% globally [3]. E. histolytica colonizes the human large intestine, and upon a not-well-understood trigger, the trophozoites invade the mucous barrier and cause dysentery and eventually extraintestinal amoebiasis [4]. Due to its adaptation to oxygen-poor environments and its parasitic lifestyle, the cellular organelles and metabolism of E. histolytica are highly modified [5]. Entamoeba does not possess classical mitochondria; instead, the cells harbor minimal versions, called mitosomes. The organelles do not contain DNA or exhibit classical energy metabolism, yet they are surrounded by a double membrane [6–8]. The only known mitosomal function is sulfate activation [9,10]. Energy metabolism is based on glycolysis in the cytosol, where glucose is converted to pyruvate, and extended glycolysis, that includes conversion of pyruvate to acetyl-CoA by pyruvate:ferredoxin oxidoreductase; acetyl-CoA is then utilized for ATP production by acetyl-CoA synthase [11]. The E. histolytica cytosol contains numerous vesicles, lysosomes, endosomes, and multivesicular bodies, whereas the endoplasmic reticulum (ER) and Golgi apparatus (GA) are difficult to recognize and, in the case of the Golgi, were once thought to be absent [12]. However, later studies showed that the ER forms a tubular structure similar to the ER of other eukaryotic cells [13,14], and a GA was visualized in the form of separated vesicles bearing some GA markers, such as Golgi-associated coatomer protein ɛ-COP [15,16]. Peroxisomes are single membrane-bound multifunctional organelles. A defining feature of classical peroxisome is the presence of peroxide-generating and detoxifying pathways with the marker enzyme catalase [17,18]. However, some specialized forms of the organelle do not contain these key pathways and play various different roles: glycosomes of trypanosomatids are known to contain the first six or seven glycolytic enzymes [19], and plant glyoxysomes engage in the glyoxalate cycle, whereas Woronin bodies of filamentous fungi serve as a physical barrier between two cells upon hyphal wounding [20].
Despite the wide range of metabolic diversity, all peroxisomes share a common de novo biogenesis pathway mediated by a set of specific proteins named peroxins (PEXs). In the model organism Saccharomyces cerevisiae, peroxisomal matrix proteins are synthesized in the cytosol and recognized by the receptor proteins Pex5 and Pex7 with C-terminal peroxisomal targeting signal (PTS)1 and N-terminal PTS2, respectively [21]. The receptor-cargo is then recruited by docking proteins (Pex13, Pex14 and Pex17) and a transient pore composed of Pex5-Pex14 is formed. The cargo is released into the lumen of peroxisomes while RING finger proteins (ubiquitin ligases Pex2, Pex10 and Pex12) are assembled onto the transition pore. The receptor is recycled from the membrane upon monoubiquinylation of a conserved cysteine or degraded upon polyubiquitinylation of a conserved lysine. The process of ubiquitinylation involves RING proteins, and ubiquitin-conjugated Pex4 is anchored to the membrane via Pex22. Release of the receptor from the membrane is facilitated by the Pex1/6 heterohexamer anchored to the membrane via Pex15, which is also responsible for deubiquitinylation [21]. Membrane proteins are incorporated into the peroxisomal membrane by the cycling chaperone Pex19, which recognizes the membrane PTS and recruits the protein to a membrane-anchored Pex3 [18]. The trafficking of glycosylated proteins from the ER is mediated by Pex16, which is absent in Saccharomyces species [18,22]. Similar to mitochondria, peroxisomes can undergo fission, exploiting dynamin-like proteins that are recruited by Pex11 [18].
Peroxisomes have been widely reported from aerobic organisms, and their evolution was suggested to be tightly connected with oxygen-dependent metabolic pathways, particularly β-oxidation of fatty acids, redox homeostasis, and bioenergetics [23,24]. Moreover, it has been shown in human mutant fibroblasts that peroxisomes could be formed by a fusion of vesicles derived from both the ER and mitochondria [25]. In this case, Pex3 and Pex14 were first inserted into the mitochondrial outer membrane, which later buds in the form of a vesicle; moreover, Pex16-containing vesicles were shown to originate from the ER. Mature peroxisomes were determined to be a union of the two distinct classes of pre-peroxisomal vesicles [25].
Peroxisomes are generally thought to be absent in anaerobic organisms, including E. histolytica [26]. Recently, however, anaerobic peroxisomes were described in the free-living anaerobic amoeba Mastigamoeba balamuthi, which bear an anaerobic form of mitochondria named hydrogenosomes [27]. M. balamuthi peroxisomes contain a major part of the pyrimidine biosynthesis pathway, enzymes required for coenzyme A synthesis, and carbohydrate metabolism, while β-oxidation and catalase are absent [27]. Moreover, the presence of peroxisomes was noted in another anaerobic member of the Archamoebae, Pelomyxa schiedti [28]. The discovery of anaerobic peroxisomes in free-living archamoebae prompted us to search for peroxisomes in evolutionarily related parasitic Entamoeba species. Indeed, we identified a minimal set of pex genes that provide E. histolytica with the capacity to form peroxisomes, and we partially characterized the peroxisomal proteome. The common feature of anaerobic peroxisomes in Archamoebae appears to be the metabolism of myo-inositol.
Results
Identification of putative PEX proteins in the genome of Entamoeba species
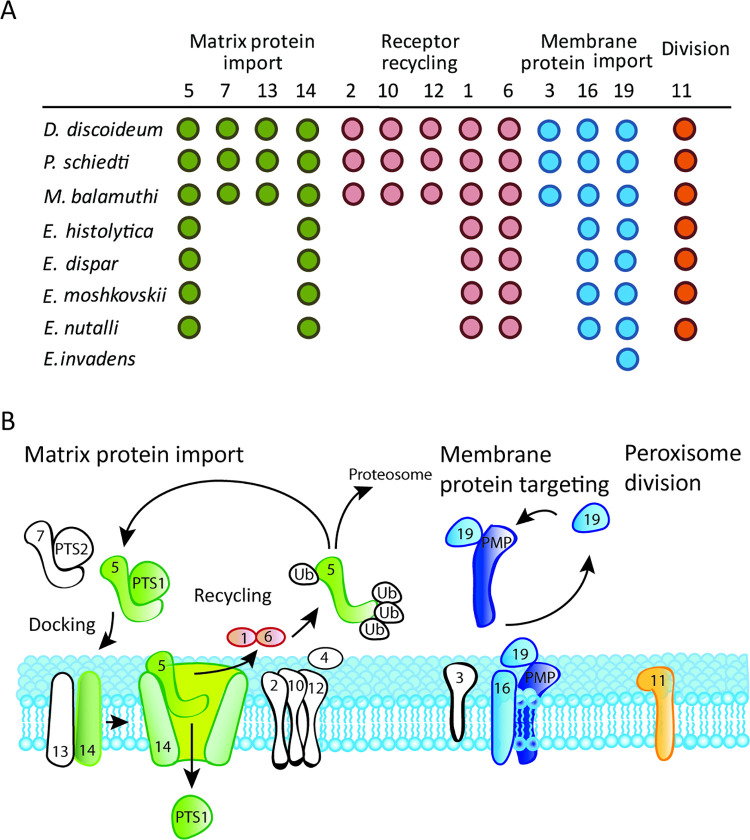
Initially, we searched for PEXs in the genomes of E. histolytica and four other Entamoeba species, the human parasites E. dispar and E. moshkovskii, a parasite of non-human primates E. nuttalli, and a reptile pathogen E. invadens. The searches, which were performed using PEXs of a related amoebozoan M. balamuthi and HMMs of other eukaryotic PEX proteins as queries, revealed the presence of a reduced set of seven PEXs in all primate species (Fig 1A and S1 Table). The identified PEXs include members of each functional category (Fig 1B). Components of matrix protein imports were represented by Pex5 and Pex14. Pex5 is a soluble receptor that recognizes the PTS1 of proteins to be imported by binding to a domain formed by tetratricopeptide repeats within its C-terminal half (Fig 2). A transient pore constituting of Pex14 and Pex5 is formed after the interaction of Pex5 with the docking complex by binding to characteristic motifs in the N-terminal half of Pex14 (Fig 2) [29]. Members of membrane protein import include Pex16, a protein involved in embedding of peroxisomal membrane proteins, and Pex19, a membrane protein receptor. Although E. histolytica Pex19 is considerably shorter than the human ortholog, its classification to the Pex19 family (PF04614) is strongly supported, although it lacks the N-terminal domain that is required for interaction with Pex3 [30,31] (Fig 2). We did not identify any member of the RING finger proteins of the receptor recycling machinery (Pex2, Pex10, Pex12); however, we detected putative Pex1 and Pex6, which are responsible for receptor deubiquitinylation (Fig 1B). Because Pex1 and Pex6 possess AAA domains that are present in other proteins and may lead to false-positive identification, we also performed a phylogenetic analysis of Pex1/Pex6 orthologs and related AAA proteins with different functions (Fig 3). This analysis supported the correct identification of putative Entamoeba Pex1 and Pex6. Finally, we identified Pex11, a multi-purpose protein, which is involved in peroxisomal proliferation by elongation and fission and metabolism via the formation of a membrane channel [32,33]. Surprisingly, we did not identify any PEXs in E. invadens except for Pex19 (Fig 1A). All identified proteins showed apparent divergence from canonical sequences of yeast and human orthologues, with protein sequence identity ranging from 8–23% (S2 Table). Taken together, our searches suggested that E. histolytica and three other Entamoeba species have a minimal set of seven PEXs that might be able to facilitate peroxisomal biogenesis, while E. invadens most likely lacks these organelles, with only Pex19 being present.
Fig 1. Prediction of PEX coding genes.
A. Predicted PEXs in E. histolytica and related Evosea species. B. Scheme of the peroxisomal machinery identified in Entamoeba species except for E. invadens. The white color indicates PEXs that were not identified. Ub, ubiquitin; PMP, peroxisomal membrane protein.
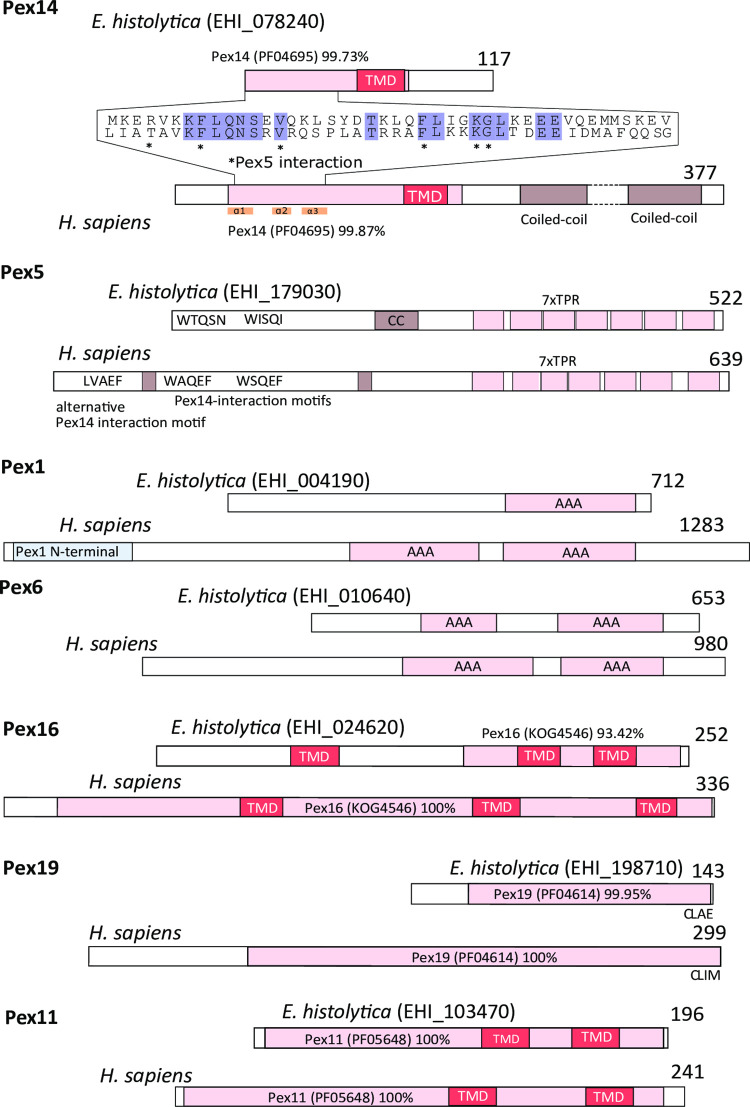
Fig 2. Domain structures of E. histolytica and Homo sapiens PEXs.
PFAM domain identifiers (in brackets) and probability percentages are indicated. TMD, transmembrane domain; TPR, tetratricopeptide domain; stars indicate Pex14 amino acid residues that are involved in Pex5 interactions [30].
Fig 3. Phylogeny of Pex1, Pex6 and other proteins with AAA domains.
The maximum likelihood tree was inferred with IQ-TREE using 51 protein sequences and 522 positions. Numbers at nodes of the tree indicate statistical support in the form of an ultrafast bootstrap of the IQ-Tree and posterior probability of the PhyloBayes analysis.
Identified PEXs are all expressed
Identification of the putative PEX proteins prompted us to test whether the proteins are expressed and to what extent. Total RNA was isolated using cells upon reaching a monolayer in the culture (approximately 48 hours). RT-qPCR showed that under these conditions, all PEXs were expressed, but the relative expression level was lower than that of the selected housekeeping enzymes pyruvate kinase, phosphofructokinase, and alcohol dehydrogenase (S3 Table). The low expression level of putative PEXs corresponded to previously published transcriptomic data [34]. The highest expression was observed for Pex11, while Pex19 was the least expressed. Because the regulation of genes involved in the same function might be linked, we decided to clone Pex5, 11, 14, 16, and 19 into a vector that allowed their overexpression with a polyHis-tag under the control of a strong lectin promotor [34,35]. The transfectants revealed a relative PEX overexpression from 31 (Pex11) to 580 (Pex5)-fold (S4 Table). However, overexpression of a particular PEX gene had a negligible effect on native expression of other PEXs, with the exception of Pex14, overexpression of which was accompanied by slightly increased expression of Pex16 (3-fold).
PEX proteins in E. histolytica localized to vesicles distinct from mitosomes, the ER and endosomal/lysosomal structures
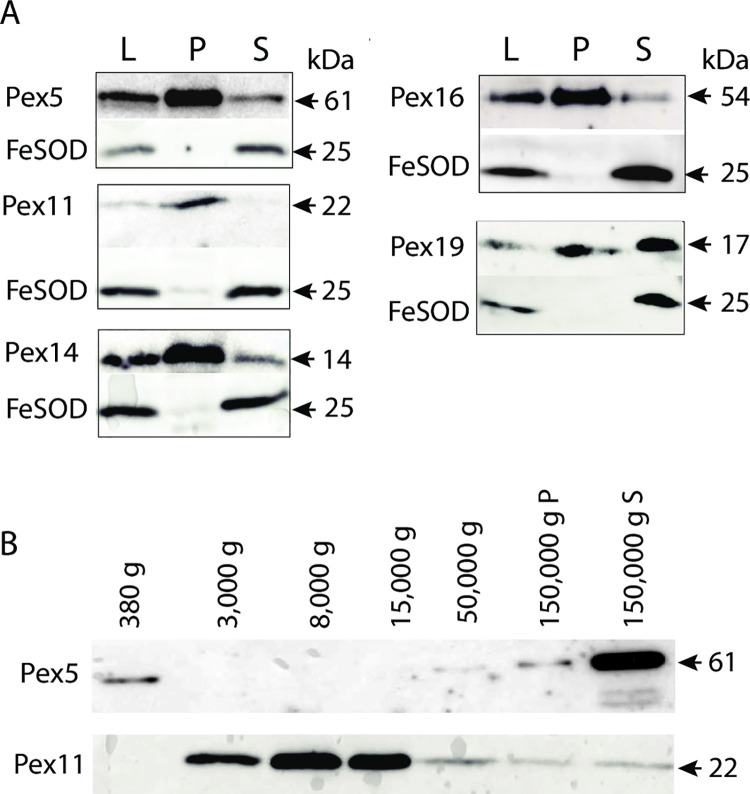
The subcellular localization of PEXs was studied in transfectants expressing Pex5, 11, 14, 16, and 19. First, we analyzed recombinant proteins using immunoblot analysis of the sedimentable (organelle) and soluble (cytosolic) fractions of transfectants (Figs 4 and S1). This analysis confirmed that all PEXs are expressed on the protein level, and more importantly, Pex5, 11, 14, and 16 were predominantly present in the sedimentable fraction, whereas the signal for Pex19 was comparably strong in the organellar fraction and the cytosol. Iron-containing superoxide dismutase (Fe-SOD) was used as a cytosolic marker (Fig 4A). For the soluble receptor Pex5, we expected to observe higher amounts of this protein in the cytosol. Thus, we performed a more detailed cell fractionation using differential centrifugation in five consecutive steps at 380–150,000 x g, and we used 0.05% Tween-20 in the buffer to limit protein aggregation. Under these conditions, a majority of Pex5 was in the soluble fraction, while the majority of the peroxisomal membrane Pex11 was present in sedimentable fractions (Fig 4B). This result indicates that Pex5 is transiently associated with organelles.
Fig 4. Expression of tagged PEXs in E. histolytica.
A. Western blot analysis of cell lysate (L), organelle (P) and cytosolic soluble (S) fractions. Fe-superoxide dismutase (FeSOD) was used as the cytosolic marker. B. Western blot analysis of Pex5 and Pex11 in seven fractions isolated by differential centrifugation using 0.05% Tween-20 to limit protein aggregation.
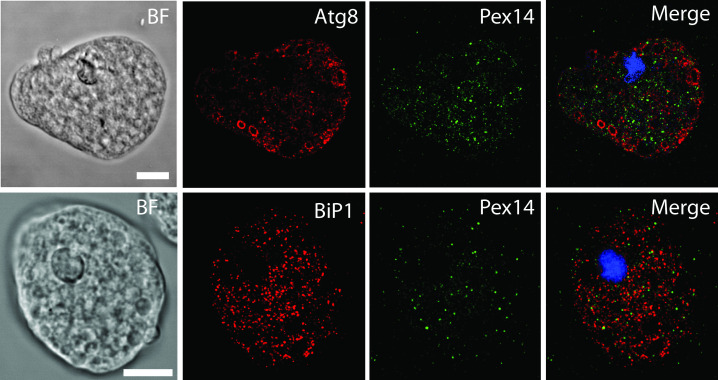
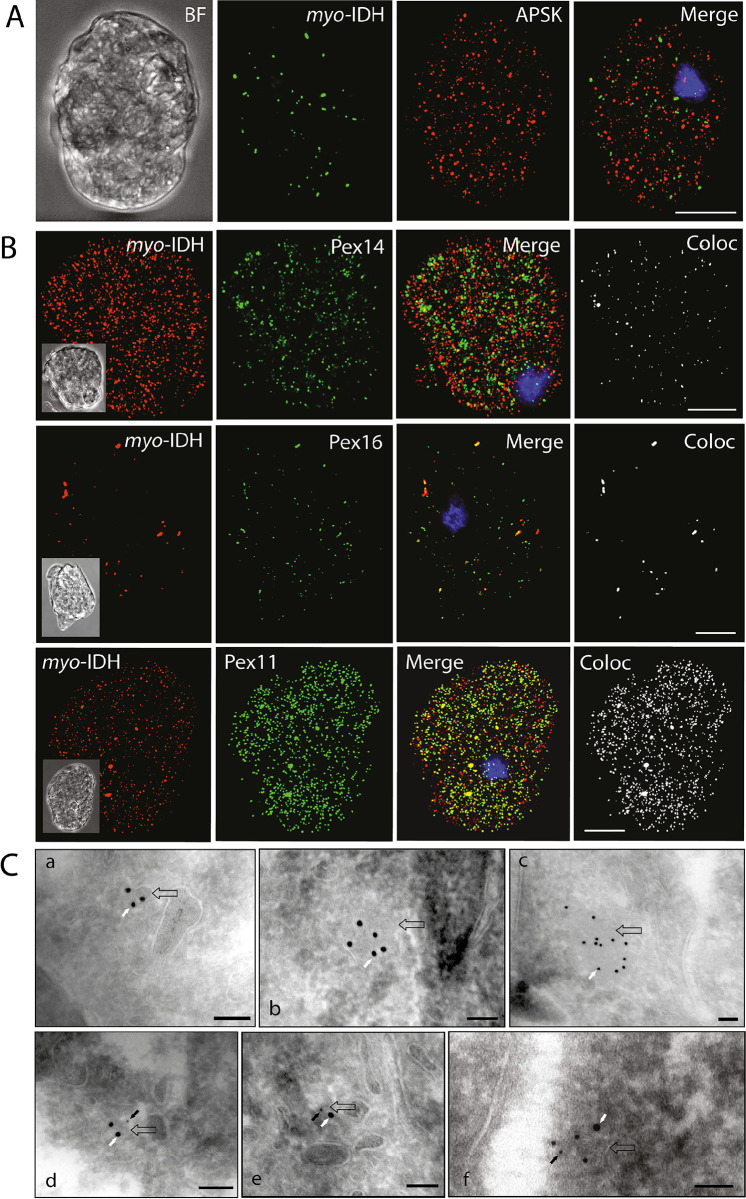
More detailed localization of PEXs was investigated using confocal microscopy. First, we attempted to distinguish putative peroxisomes from mitosomes (Fig 5A). We observed that both PEXs and APSK, a marker protein of mitosomes, labeled small round vesicles of similar sizes, but none of the PEXs colocalized with APSK. The Pearson’s correlation coefficient in colocalized volume (PCC) ranged between -0.072 to 0.023, which indicates no or negligible correlation between PEXs and APSK signals (S2 and S5 Figs and S5 Table). The only exception was Pex11, which partially localized to mitosomes with weak PCC r = 0.241. This observation was further supported by structured illumination microscopy (SIM)(Fig 5B). While Pex14 did not colocalize with the APSK signal, Pex11 was partially associated with mitosomes (Fig 5B). Pex14 also did not colocalize with other cellular vesicles and vacuoles labeled with Atg8(PCC r = 0.033) [36] and the ER marker BiP1(PCC r = 0.021) [37] (Figs 6 and S2 and S5 Tables). The number of putative peroxisomes counted for Pex14-labeled organelles was approximately 267±60 per 100 μm2 (mean ± S.D., n = 25). The specific vesicular localization of Pex14 and Pex16 was further supported by immunoelectron microscopy. Weak but highly specific signals for His-tagged Pex14 were associated with the membranes of round vesicles of approximately 90–100 nm in diameter (Fig 7A and 7B). Similarly, His-tagged Pex16 was observed at the vesicular membrane. We also observed Pex16-labeled vesicles that were partially surrounded by two more membranes, with Pex16 signals on the proximal and distal membranes. The character of these structures is unclear though they remind lamellar derivatives of the endoplasmic reticulum that deliver Pex16 and other membrane proteins for de novo formation of putative peroxisomes (Fig 7C and 7D) [38].
Fig 5. Cellular localization of PEXs in E. histolytica.
A. Immunofluorescence microscopy of His-tagged PEXs and the mitosomal marker APSK. B. Structured illumination microscopy of Pex11, Pex14, and APSK. BF, bright field. Scale bar: 10 μm.
Fig 6. Confocal microscopy of vesicles labeled with Pex14, ER marker BiP1 and lysosomal marker Atg8.
BF, bright field. Scale bar: 10 μm.
Fig 7. Immunoelectron microscopy detection of Pex14 and Pex16 in E. histolytica.
Pex14 (A,B) was detected using Ni-NTA conjugated with 5 nm gold particles (A) or anti-V5 antibody (B). Pex16 (C,D) was detected using rabbit (C) and mouse (B) anti-His antibodies and the corresponding antibody conjugated with 5 nm gold particles. Scale bar: 100 nm.
Proteomic analysis of the peroxisomal fraction and PTS1 predictions revealed the presence of inositol dehydrogenase in the peroxisomal matrix
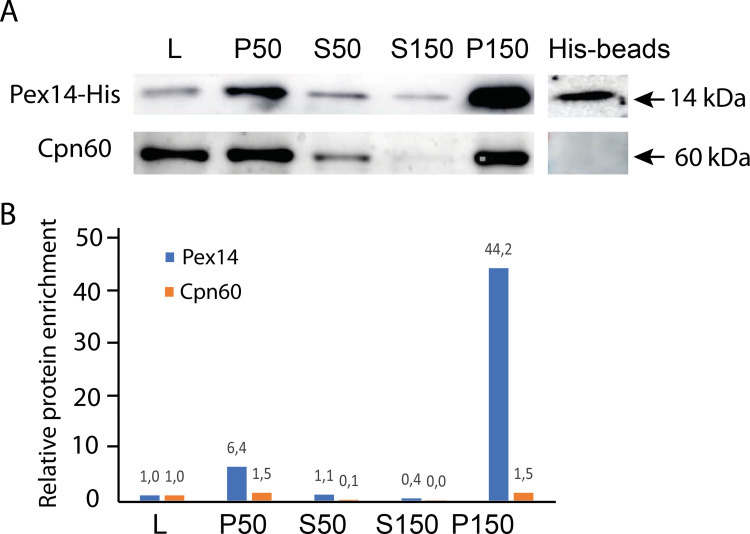
Initial experiments to isolate putative peroxisomes based on differential and gradient centrifugation did not allow effective separation of Pex14-localized organelles and mitosomes. Therefore, we decided to use affinity purification of the organelles. Pex14 was expressed with a C-terminal poly-His or V5 tag facing the cytosol [39], and putative peroxisomes were isolated from cell homogenates using magnetic beads conjugated with the corresponding antibody. In a pilot experiment, all steps were monitored by western blotting to assess enrichment of putative peroxisomes using an anti-His antibody and mitosome contamination using an anti-Cpn60 antibody (Figs 8 and S1B). Western blot analysis showed approximately 40-fold enrichment of the Pex14-His signal after differential centrifugation (150,000 x g pellet, Fig 8B), and the Pex14-His signal was separated from Cpn60 using anti-His antibody-conjugated beads (Fig 8A). A summary of LFQ mass spectrometry analyses of putative peroxisome-enriched fractions with His-tagged Pex14 (seven experiments) and V5-tagged Pex14 (six experiments) is given in S6 Table. In two experiments with V5-tagged Pex14, we omitted Tween 20 to estimate, which proteins are loosely associated with the organelle surface. Altogether, 655 proteins were identified in the peroxisome-enriched fraction, of which 24 were observed only in experiments without Tween 20 (S6 Table). In addition to tagged Pex14, the dataset contained two other PEXs, Pex5 and Pex11. Interestingly, although orthologs of RING complex E3 ubiquitin ligases Pex2, Pex10 and Pex12 and E2 ubiquitin-conjugating protein Pex4 are absent in E. histolytica, a single putative E3 ubiquitin ligase EHI_030770 and two ubiquitin-conjugating enzyme family proteins, EHI_083560 and EHI_048700, were identified in the proteome.
Fig 8. Affinity purification of peroxisomes.
A. Lysate of cells (L) expressing His-tagged Pex14 was used for differential centrifugation at 50 000 and 150 000 x g, and pellet (P) and soluble (S) fractions were analyzed by western blotting. In the last step, peroxisomes were purified using anti-His antibody-conjugated beads (His-beads) that were used for mass spectrometry. Antibodies against Cpn60 were used as mitosomal markers. B. Relative protein enrichment was calculated with densitometry of Pex14-His and Cpn60 signals.
Comparison with the proteome of M. balamuthi peroxisomes [27] revealed eight proteins in common, such as putative inositol dehydrogenase (IDH), long-chain fatty acid-CoA ligase, and malate dehydrogenase (S6 Table). We were also interested in the overlap between our dataset and the previously reported mitosomal proteome [9]. This comparison revealed 35 common proteins, of which four proteins detected in the mitosome-enriched fraction were between our top 100 peroxisomal candidates with the highest cumulative score (S6 Table). These proteins included putative IDH, which was previously localized in the cytosol [9], a hypothetical protein EHI_103470, which we recognized as Pex11, a hypothetical protein (EHI_170120) that was reported as the mitosomal membrane protein [40], and a microtubule-binding protein (S6 Table).
Next, we performed in silico prediction of PTS1 signals in proteins predicted in all available Entamoeba species using a machine learning algorithm (PTS1 ML) optimized according to the M. balamuthi peroxisomal proteome [27]. We rationalized that a high score for PTS1 ML prediction in E. invadens that seems to lack peroxisomes likely represents a false positive signal and that the absence of proteins in E. invadens in comparison to other species might be related to the absence of peroxisomes. Thus, the PTS1 ML score for E. invadens proteins was subtracted from the corresponding PTS1 ML score for E. histolytica (S7 Table). In addition, we used three other available tools for PTS1 prediction [41,42]. These analyses provided a set of 56 proteins with putative PTS1 predicted by at least two tools (S7 Table). The comparison of this dataset and proteomic data (S6 Table) revealed only six proteins present in the proteome of the putative peroxisome-enriched fraction with the predicted PTS1 signal using our strict criteria. These proteins include five hypothetical proteins of unknown function (EHI_045060, EHI_050510, EHI_183900, EHI_185440, and EHI_161040) and putative IDH (EHI_125740). The absence of genes for IDH and the hypothetical protein EHI_045060 in the E. invadens genome further support its possible associations with putative peroxisomes of E. histolytica (S7 Table).
Import of IDH to putative peroxisomes is PTS1-dependent
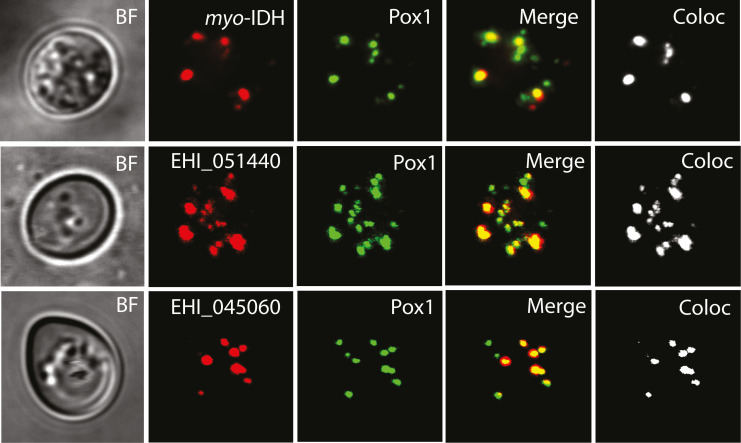
Predicted peroxisomal localization was evaluated for seventeen selected proteins (S7 Table). The proteins were expressed with an N-terminal mCherry tag in the yeast strain BY4742:POX1-EGFP expressing the integrated GFP-tagged peroxisomal marker protein acyl-CoA oxidase (Pox1). Three proteins, putative IDH (Fig 9), and hypothetical proteins EHI_051440 and EHI_045060 were observed in round vesicles in which they colocalized with Pox1, whereas other localizations were observed for the rest of the tested proteins (Fig 9 and S7 Table). The localization of IDH in E. histolytica was studied using a specific polyclonal antibody raised against the corresponding recombinant protein (S4 Fig). Confocal immunofluorescence microscopy revealed IDH-labeled punctual structures distinct from mitosomes visualized by an antibody against APSK (PCC r = -0.159)(Fig 10A). Furthermore, we investigated the colocalization of IDH in cells expressing His-tagged PEXs (Figs 10B and S2 and S5 Tables). IDH was detected in similar structures as above and partially colocalized with Pex14 (PCC r = 0.512, Fig 10B) and Pex16 (PCC r = 0.260). The strongest correlation (PCC r = 0.547) was observed between IDH and Pex11 (Fig 10B). The vesicular localization of IDH was confirmed by immunoelectron microscopy (Fig 10C). The double-labeling experiments revealed IDH signals with those of Pex16 within the vesicular structures(Fig 10C).
Fig 9. Localization of PTS1-containing candidate proteins in yeast peroxisomes.
Fluorescence microscopy of mCherry-tagged myo-IDH and two hypothetical proteins (EHI_051440 and EHI_045060) (red) expressed in yeast. Pox1 fused with GFP was used as a peroxisomal marker (green). Signal colocalization analysis (white) was performed by ImarisColoc software.
Fig 10. Localization of IDH in E. histolytica.
A. Detection of myo-IDH (green)and APSK (mitosomal marker protein, red) in wild- type cells. Bar = 10 μm. B. Detection of native myo-IDH (red) in cells expressing C-terminally His-tagged PEXs (green). Signal colocalization analysis (white) was performed by ImarisColoc software. Bar = 10 μm. C. Immunoelectron microscopy. Myo-IDH was detected using a rabbit polyclonal anti-myo-IDH antibody and anti-rabbit IgG conjugated to 15 nm gold particles (white arrows, a-f), Pex16 was detected using a mouse monoclonal anti-poly-His antibody, and anti-mouse IgG conjugated to 5 nm gold nanoparticles (black arrows, d-f). Empty arrows indicate membranes. Bars 100 μm.
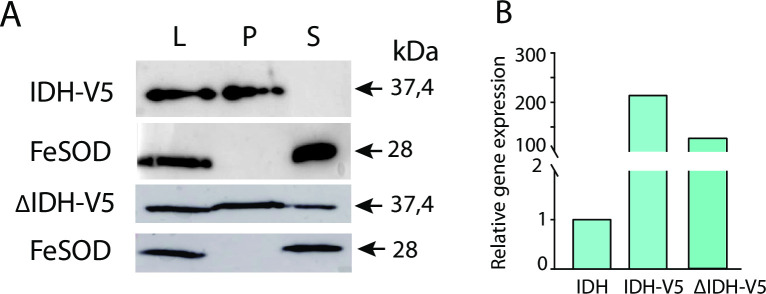
Finally, we expressed IDH in E. histolytica with a V5-tag at the N-terminus. Expression of the gene was confirmed by RT-qPCR and by immunoblotting analysis of cell fractions (Fig 11). IDH was present exclusively in the sedimentable organellar fraction, while the cytosolic marker Fe-SOD was present in the soluble fraction. Next, we prepared a truncated version of the protein lacking C-terminal PTS1 tripeptide. The truncated gene was transcribed at a comparable level as the complete V5-tagged gene (Fig 11B). The deletion of PTS1 tripeptide resulted in dual localization of IDH in the cytosol and the organellar fraction (Fig 11A).
Fig 11. Effect of myo-IDH PTS1 tripeptide deletion.
A. Detection of N-terminally V5-tagged full-length myo-IDH (IDH-V5) and its C-terminal truncated version (ΔIDH-V5) in the cell lysate (L), organelle (P) and cytosolic (S) fractions using western blot analysis. FeSOD was used as the cytosolic marker. B. Relative expression of IDH-V5 and ΔIDH-V5 using RT-qPCR. The level of native IDH was determined in wild-type cells and used for standardization.
Collectively, the identification of IDH in the proteome of the putative peroxisome-enriched fraction, prediction of the PTS1 signal at the C-terminus with high statistical support, and localization studies in yeast and E. histolytica indicate that IDH is targeted to the matrix of E. histolytica organelle that contains PEXs, hereafter peroxisomes.
Peroxisomal IDH preferentially utilizes myo-inositol as a substrate
Identification of putative IDH in peroxisomes prompted us to investigate the substrate specificity of this enzyme. An interproscan search revealed the presence of the N-terminal NAD-binding Rossmann fold domain of the glucose–fructose oxidoreductase (GFO)_IDH_MocA protein family (PFAM: PF01408). These enzymes can utilize a broad spectrum of substrates, such as myo-inositol, scyllo-inositol, D-glucose, D-xylose, D-fructose, and 1,5-anhydro-D-fructose [43–47]. Protein sequence alignment of E. histolytica IDH to IDHs of other archamoebae and structurally characterized bacterial paralogs revealed the presence of conserved residues required for the binding of NAD+ (motifs I and II) and key residues of motifs III-VI that define the substrate-binding pocket (S3 Fig) [48]. However, diversity within motif IV did not allow reliable estimation of the enzyme substrate specificity (S3 Fig). Thus, recombinant E. histolytica IDH was expressed in E. coli, affinity purified (S4 Fig), and used to determine its kinetic parameters for various substrates (Table 1 and S5 Fig). The enzyme revealed dehydrogenase activity preferentially toward myo-inositol (hereafter myo-IDH), and this activity was dependent on NAD+, while negligible activity was detected with NADP+. The Michaelis constant Km for scyllo-inositol was approximately 10-fold higher than that for myo-inositol and those for D-glucose and D-xylose two orders of magnitude higher. The molecular mass of recombinant myo-IDH under reducing conditions using SDS-PAGE was 37.4 kDa, which corresponded well with the theoretical weight of 36.4 kDa, including the 6x-His tag. Molecular mass of the native enzyme was initially determined by size-exclusion chromatography. The peak of enzymatic activity was recovered at the elution volume corresponding to 41 kDa suggesting a momomeric structure (S6 Fig). However, because molecular mass determined by this approach might be affected by the protein shape, we also used multiangle light scattering (MALS). This analysis revealed that the molecular mass of the native recombinant enzyme was 77.4 (±4) kDa, suggesting a homodimeric structure with a minor contribution from the trimer (118.1± 22 kDa) (S7 Fig).
Table 1. Kinetic parameters of E. histolytica myo-IDH.
| Substrate | KmSubstrate (mean±SD) [mM] | Vmax (mean±SD) [μmol.min-1.mg-1] |
|---|---|---|
| myo-inositol | 0.044±0.004 | 5.451±0.456 |
| NAD+(myo-inositol)* | 0.038±0.004 | 1.002±0.017 |
| keto-2-inositol | 0.322±0.034 | 4.409±0.053 |
| NADH | 0.009±0.000 | 1.638±0.024 |
| scyllo-inositol | 0.229±0.020 | 6.204±0.477 |
| D-glucose | 8.576±1.234 | 4.782±0.174 |
| D-xylose | 13.223±1.900 | 4.582±0.095 |
| NADP+(myo-inositol)* | 1.201±0.532 | 0.826±0.113 |
*12.5 mM myo-inositol. Lineweaver-Burk plots are given in S5 Fig.
Peroxisomal IDH formed a monophyletic clade in phylogenetic reconstruction
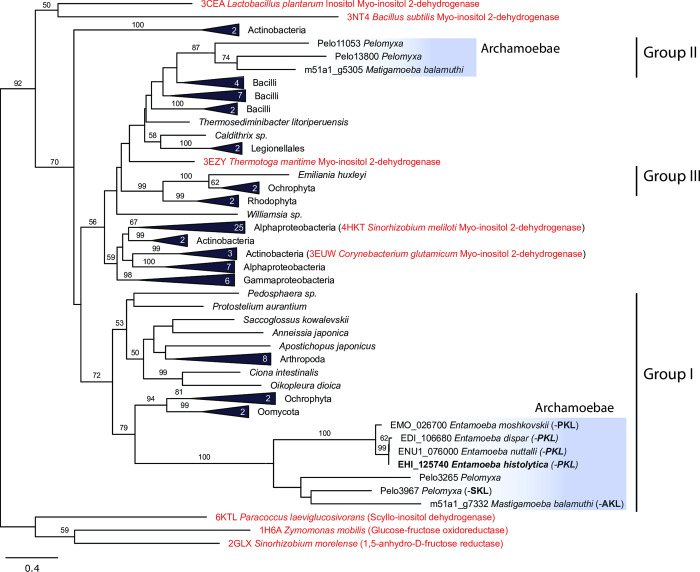
To investigate the evolution of E. histolytica myo-IDH, we performed phylogenetic analysis using eukaryotic (33) and prokaryotic (70) protein sequences, including those with known structures and functions (Figs 12 and S8). The eukaryotic sequences formed three distinct branches nested within bacterial orthologs. The IDH sequences identified in four Entamoeba species, M. balamuthi and P. schiedti, formed a monophyletic cluster of peroxisomal IDHs with high statistical support. All these sequences possessed PTS1 signals (-PKL, -SKL, -AKL), except for one of two P. schiedti IDH paralogs. The peroxisomal IDHs were placed within broader mostly eukaryotic branches (Group I). The closest orthologs appeared to be putative IDHs in ochrophytes and oomycotes, and we also identified IDH orthologs in the amoebozoan Planoprotostelium fungivorum and members of Hemochordata, Tunicata, Echinodermata, and Arthropoda. None of these orthologs possessed a PTS1 signal, although a rare PTS1 signal was noted for the IDH of P. fungivorum(-SKF). The cellular localization of most eukaryotic IDHs of Group I is likely cytosolic; however, IDHs of Stramenopiles that formed a sister group to peroxisomal IDHs were predicted to localize to mitochondria. In addition, IDHs of red algae with primary plastids and the haptophyte Emiliania huxleyi and two members of Ochrophyta with secondary plastids of red algal origin were predicted to target chloroplasts [49] (Group III) (S8 Table).
Fig 12. Phylogeny of myo-IDH.
The maximum likelihood tree was inferred with IQ-TREE using 103 protein sequences and 289 positions. Bootstrap support is given at nodes. Sequences with known crystal structures are highlighted in red. Numbers in triangles indicate the number of included sequences. PTS1 triplet is given in brackets.
Collectively, the evolutionary history of eukaryotic IDHs appeared to be rather complex. The observed topology suggests at least three independent lateral gene transfers of IDHs from bacteria to eukaryotes as well as possible transfer between eukaryotes via plastids of the red lineage. Peroxisomal myo-IDHs seem to have evolved from a common ancestor of Archamoebae independent of cytosolic IDH homologs. In E. invadens, myo-IDH was most likely secondarily lost.
Discussion
E. histolytica is believed to be devoid of peroxisomes, like most anaerobic protists [26,50]. In this work, we provided the first evidence that peroxisomes are present in this parasite, although E. histolytica peroxisomes seem to be remarkably reduced in comparison to their known peroxisome counterparts. E. histolytica contains only seven homologs of known PEXs (Pex1, Pex6, Pex5, Pex11, Pex14, Pex16, and Pex19) that may participate in organelle biogenesis. Targeting of matrix proteins to peroxisomes is reduced to the PTS1-dependent pathway mediated via the soluble Pex5 receptor, while the PTS2 receptor Pex7 is absent. Proteomic analyses of affinity-purified peroxisomes and in silico PTS1 predictions led to the identification of peroxisomal myo-IDH. Importantly, E. histolytica myo-IDH with PTS1 was shared with E. dispar, E. nutalli and E. moshkovskii and with evolutionarily related free-living M. balamuthi and P. schiedti, which all belong to the Archamoebae group of anaerobic protists, whereas E. invadens most likely lost peroxisomal myo-IDH together with peroxisomes.
The presence of peroxisomes in E. histolytica is supported by the localization of Pex5, Pex11, Pex14, Pex16, and Pex19 to numerous vesicles that are distinct from the structures of the ER, endosomal/lysosomal vesicles and mitosomes. Pex14 and Pex16 were detected in single membrane-bound vesicles of approximately 90–100 nm in diameter, which is within the range of anaerobic peroxisomes (80–440 nm) observed in M. balamuthi [27]. The matrix protein myo-IDH colocalized with Pox1 in yeast peroxisomes and within organelles labeled with Pex14, Pex16, and Pex11 in E. histolytica. Deletion of PTS1 tripeptide caused partial mistargeting of myo-IDH to the cytosol.
There are multiple metabolic interplays between mitochondria and peroxisomes, and in some cases, mitochondria may participate in peroxisome biogenesis [25]. However, the metabolism of E. histolytica mitochondria (mitosome) is reduced to a single pathway, and their protein import machinery contains only 2 out of 30 components known in model yeast mitochondria [9,51,52]. In this view, the observed reduction in peroxisomes is consistent with overall changes during the course of reductive evolution that formed Entamoeba species [5]. Most eukaryotes possess 13–17 PEXs, including humans and M. balamuthi, and four PEXs, Pex3, Pex10, Pex12, and Pex19, are considered peroxisomal markers absent only in organisms devoid of peroxisomes [53]. Three of these markers (Pex3, Pex10, and Pex12) were not identified in E. histolytica (Fig 1A). Pex3, together with Pex16 and Pex19, are involved in peroxisomal membrane protein (PMP) import. Pex3 facilitated the membrane docking of newly synthesized PMPs delivered via the shuttling receptor Pex19 directly from the cytosol (class I pathway for PMP import). Pex3 itself and other PMPs are recruited to the ER membrane by Pex16, which is cotranslationally inserted into the ER and subsequently delivered to peroxisomes through vesicular transport (class II pathway) [25,38]. We speculate that Pex16 might be sufficient for PMP import without a Pex3 contribution in E. histolytica. In support of this possibility, the formation of membrane structures with PMPs was observed in cells in which pex3 was deleted [54], and the budding of pre-peroxisomal vesicles was dependent on Pex19, a homolog of which we found in Entamoeba [55]. Moreover, the presence of Pex16 and Pex19 without Pex3 was noticed in Tetrahymena thermophila [50]. Nevertheless, we cannot rule out the possibility that E. histolytica possesses a highly divergent Pex3 that was not recognized by current bioinformatic tools. Indeed, the highly divergent trypanosomal Pex3 remained elusive for a long time, and it has been identified only recently [56,57]. However, the absence of the N-terminal domain of E. histolytica Pex19, which is essential for its interaction with Pex3 [30,31] is more consistent with the lack of Pex3 in this organism. Interestingly, Pex19 is the only peroxin, homolog of which we identified in E. invadens that likely lack peroxisomes. Thus, Pex19 may have a more general function in protein sorting, which is unrelated to peroxisomes. Pex10 and Pex12 (RING complex) belong to the Zn-RING finger E3 ubiquitin-protein ligase family that, together with the E2 conjugating protein Pex4, participate in Pex5 recycling.Pex5 with peroxisomal cargo associates with membrane-docking components to form a transient pore with Pex14. Upon the release of the cargo, Pex5 is dislocated from the membrane in an ATP-dependent manner by Pex1 and Pex6 (dislocase complex) [58], which we both identified in E. histolytica. Recognition of Pex5 by the dislocase complex is dependent on Pex5 ubiquitination via thiol ester bond at the conserved cysteine at the N-terminus by the RING complex [59]. The absence of Pex10, Pex12, and Pex4 in E. histolytica suggests that Pex5 might be recycled without a ubiquitination step. In support of this view, E. histolytica Pex5 lacks an N-terminal cysteine residue. Moreover, in Pichia pastoris, PTS receptors and Pex14 have been shown to form the minimal translocation machinery that can facilitate peroxisomal import independent of the RING complex [60]. Alternatively, other E3 and E2 family proteins may compensate for the absence of the RING complex and Pex4. E. histolytica possesses genes for at least six E3 ligases, of which the E3 ligase EHI_030770 was found in the proteome of the peroxisome-enriched fraction, although, unlike Pex10 and Pex12, EHI_030770 does not possess any predicted transmembrane domain (S7 Table). There are also two E2 ubiquitin conjugating (Ubc) proteins (EHI_083560, EHI_048700) in the proteome, of which EHI_083560 (named EhUbc5) was partially characterized and crystalized, however, its target proteins are not known [61]. In metazoans and fungi, Pex4 is absent and its function is compensated by Ubc-conjugating proteins [62,63]. However, replacement of Pex10 and Pex12 has not been observed thus far. In contrast, the absence of the PTS2 receptor Pex7 observed in E. histolytica is not unprecedented, and when Pex7 is present, PTS2 targets peroxisomes much less frequently than PTS1s [64]. For example, Pex7 was found in related M. balamuthi in which PTS2 was predicted in 15% of putative peroxisomal proteins [27]. The absence of Pex7 was noticed in nematodes, several arthropods, the rhodophyte Galdieria sulpharia, and the stramenopile Thalassiosira pseudonana [50,65].
Interestingly, E. histolytica Pex11 displayed dual localization in peroxisomes and mitosomes. Previously, mitochondrial localization of Pex11 was observed only in cells such as yeast and human fibroblasts that lack peroxisomes upon deletion of Pex3 and Pex19 [66,67]. It seems that Pex11 has some affinity to mitochondria, possibly via mitochondrial proteins, with which Pex11 interacts. Pex11 is involved in elongation and proliferation of peroxisomes, which share with mitochondria dynamin-related proteins (DRPs) for the peroxisomal division; Pex11 can interact with mitochondria via the ERMES complex [66] and the translocase of the outer mitochondrial membrane (TOM) complex receptor Tom22 [68]. Neither ERMES nor Tom22 are present in mitosomes of E. histolytica [69,70]; however, E. histolytica possesses two DRPs that are involved in the fission of mitosomes and may potentially interact with Pex11 [71]. Interestingly, Pex11 was previously identified in the mitosomal proteome, supporting its dual localization [9]. However, these proteomic data need to be considered with caution, as in our experiments, peroxisomes and mitosomes comigrated using standard density gradient separation and it is likely that peroxisomes contaminated the mitosomal proteome. We cannot rule out the possibility that the partial mitosomal localization of Pex11 was a result of its mislocalization due to protein overexpression. However, overexpression of Pex11 in E. histolytica estimated by RT-qPCR was the lowest compared to four other recombinant PEXs for which no association with mitosomes was observed.
The matrix of E. histolytica peroxisomes contains myo-IDH, which catalyzes reversible NAD+-dependent conversion of myo-inositol to keto-2-inositol. Myo-inositol is synthesized from glucose-6-phosphate by the activity of inositol 3-phosphate synthase and inositol-3-phosphatase, which are both present in E. histolytica. Myo-inositol is utilized for the synthesis of phosphatidylinositol and a spectrum of phosphoinositide derivatives that are produced by a set of phosphatidylinositol kinases and phosphatases [72,73]. These compounds have multiple roles as components of membrane lipids, they are used in cell signaling pathways, energy homeostasis, and as cytoprotective solutes [74,75]. In mammals, insects, yeast and Chlorophyta, the catabolism of myo-inositol is initiated by oxygen-dependent myo-inositol oxygenase that converts myo-inositol to glucuronic acid [76]. However, none of these eukaryotic biosynthetic or catabolic pathways required the activity of myo-IDH. Bacteria such as Klebsiella aerogenes can grow on myo-inositol and other cyclitols as sole carbon sources. Here, myo-IDH catalyzes the first step of the catabolic pathway, leading to the production of acetyl-CoA [77]. However, our searches did not identify any enzyme downstream from myo-IDH in this pathway in E. histolytica. The distribution of myo-IDH in eukaryotes is limited to a few lineages of algae [78] and invertebrates. The function of myo-IDH remains enigmatic; however, algae contain two isoenzymes that were predicted to localize in the cytosol and mitochondria. It could be hypothesized that predicted dual localization may allow shuttling of a reducing power between mitochondria and the cytosol via myo-inositol/keto-2-inositol transport across the mitochondrial membrane [78]. Interestingly, dual localization of myo-IDH in peroxisomes and the cytosol was predicted in M. balamuthi and P. schiedti, suggesting the possibility of NAD-linked redox shuttling between the cytosol and peroxisomes to maintain intraperoxisomal redox as in the case of NAD+-linked malate dehydrogenase and glycerol-3-phosphate dehydrogenase-dependent shuttle systems that maintain the redox balance in yeast peroxisomes [79]. In Entamoeba species, we found only peroxisomal myo-IDH, which makes this explanation less plausible, although dual localization of proteins with peroxisomal targeting under specific physiological conditions has been observed [80]. Although the biochemical context of E. histolytica myo-IDH needs further investigation, the enzyme displayed two unusual features. First, E. histolytica myo-IDH has a notably higher affinity for myo-inositol characterized by a Km value that is three and four orders of magnitude lower (Km 0.044 mM) than that of the characterized orthologs in Bacillus subtilis (Km 18 mM) and the red alga Galdieria sulphuraria (Km 430 mM). Similar to the bacterial enzyme, D-glucose and D-xylose can serve as substrates for E. histolytica myo-IDH with Km values increased to 8.5 mM and 13.2 mM, respectively. These values are still two orders of magnitude lower than those in the bacterial enzyme (167 mM and 190 mM) [43,81]. Second, E. histolytica myo-IDH appeared to be highly active as a homodimer. In B. subtilis and G. sulphuraria, the molecular weight of native myo-IDH was approximately 160 kDa, which indicated a homotetrameric structure, which has been confirmed by structural studies in B. subtilis [48] and Lactobacillus casei [82].
In conclusion, findings of peroxisomes in E. histolytica and previously in M. balamuthi erode the paradigm of peroxisome absence in anaerobes. It also suggested that a minimal set of only seven peroxins might be sufficient to build these organelles. Moreover, the specific parasite peroxisomes with a functional myo-IDH that is absent in host cells might be an interesting target for the development of antiparasitic drugs. However, this report represents only a starting point for further functional investigations of anaerobic peroxisomes in this important human parasite.
Materials and methods
Searches for peroxisomal proteins
Profile hidden Markov models (HMMs) of PEXs (Pex1, 2, 3, 4, 5, 6, 7, 10, 11, 12, 13, 14, 16, 19, 26) were collected from the EggNOG database of orthologous groups (version 5.0) [83]. Predicted proteins of E. histolytica were searched for homologs of PEXs using HMMER (version 3.3) [84]. Putative PEXs were analyzed by searching against the NCBI nr protein database using BLAST [85], EggNOG database using HMMER, and the Pfam and COG databases using HHpred [86]. PTS1 was predicted in the protein sequences of E. histolytica (https://amoebadb.org/amoeba/app) using a support vector machine with support vector classification (SVC)(scikit-learn.org). The model was trained using a dataset of 217 manually curated peroxisomal candidates selected from 11 amoebozoan proteomes, including experimentally verified peroxisomal proteins of M. balamuthi [27] with the omission of Entamoeba species (S1 Dataset). The predicted proteome of Entamoeba invadens, which lacks PTS1 receptors, served as the PTS1-negative set. The script and training data are available at https://github.com/vojtech-zarsky/PredictPTS1_ML. The SVC-based machine learning score (MLS) was calculated using ten C-terminal amino acid residues, and queries with a calculated MLS≥0.1 were predicted as PTS1. Three additional PTS1 prediction methods were used, including the PTS1 predictor at PeroxisomeDB (http://www.peroxisomedb.org/), PTS1 Predictor (https://mendel.imp.ac.at/pts1/) [42] and local searches for a strict consensus triplet motif (S/A/C)(K/R/H)L [87] and a relaxed motif (S/A/C/H/K/N/P/T)(K/R/H/N/Q/S)(L/I/M/F/A/V/Y) [88]. In a query sequence, the presence of amino acids of the strict motif was scored as 1, and any other amino acid residues in the relax motif were scored as 0.5. A sum of the triplet score TS ≥2.5 predicted PTS1.
Phylogenetic analysis
Representatives of the ATPases associated with various cellular activities (AAA) and protein families with various functions were selected from the Swiss-Prot protein database (https://www.uniprot.org/) and aligned with predicted Pex1 and Pex6 amino acid sequences using MAFFT (version 7) [89]. Representatives of GFO_IDH_MocA protein family were selected from the Swiss-Prot protein database and known crystal structures [47,48]. The alignments were trimmed using Block Mapping and Gathering with Entropy (BMGE) [90], and the phylogenetic tree was estimated using IQ-TREE (version 1.5) [91] with the LG+I+G4 model. Bootstrap support values were calculated using 500 bootstrap replicates. The Bayesian inference of the phylogenetic tree was estimated using PhyloBayes with the CAT mixture model [92]. The alignments are available in Mendeley Data, doi: 10.17632/dsszxzcc84.1.
Cultivation and transformation
E. histolytica cell line B2-5 derived from HM:IMSS was used for the study. The cell line was derived from Bernhard Nocht Institute of Tropical Medicine in Hamburg, Germany [93]. Cells were grown in TYI-S-33 medium [94] supplemented with 10% adult bovine serum (Merck/Sigma-Aldrich, St. Louis, MO, USA). E. histolytica transformations were performed using a lipofection protocol as described previously [95]. After transfection, Entamoeba cells were allowed to form a monolayer (approximately two days) before selection. Initially, 2 μg/ml G-418 or 1 μg/ml hygromycin was used, and after two days, the drug concentrations were increased to 20 μg/ml G-418 and 6 μg/ml hygromycin for at least one week of selection. The successful transformation was evaluated by real-time PCR (RT qPCR) and western blotting.
RNA isolation and RT qPCR
Total RNA was isolated using TRI Reagent (Merck/Sigma-Aldrich, St. Louis, MO, USA) and quantified using a Nanodrop 2000B (Thermo Fisher Scientific, Waltham, MA, USA). DNAse I treatment was performed for total RNA (2 μg) according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). cDNA amplification and RT-qPCR were performed in an RG-3000 cycler (Corbett Research/Qiagen, Hilden, Germany) using a one-step RT-qPCR kit (Merck/Sigma-Aldrich, St. Louis, MO, USA) as described [96]. Actin was used for normalization according to Meyer et al. [34]. The relative gene expression was measured using the 2-ΔΔCq method, and the statistical analysis was performed with a two-tailed Student’s t test using GraphPad Prism Version 7.0 [97]. All the primers are listed in S9 Table. Each experiment was performed in biological triplicates with technical duplicates.
Gene cloning and expression
Genes coding putative Pex5 (EHI_179030), Pex11 (EHI_103470), Pex14 (EHI_194840), Pex16 (EHI_024620), Pex19 (EHI_198710) and myo-IDH (EHI_125740) were amplified using specific primers (S9 Table) and cloned without stop codons into the pNeoCass vector [34] via KpnI and BamHI. A 6xHis tag or E. histolytica codon-optimized V5-tag sequence flanked by BglII and BamHI was added at the C-termini of PEXs. For myo-IDH, an E. histolytica codon-optimized V5-tag (S9 Table) was added at the N-terminus of the gene flanked by KpnI and BglII and cloned into pHygCass, a derivative of pNeoCass bearing hygromycin resistance cassette. A truncated version of the protein without three C-terminal amino acid residues (AAs) was prepared using PCR with the 3’ primer annealing to the920-939 bp position (S9 Table).
Expression and localization of proteins in S. cerevisiae
Peroxisomal candidates were amplified and subcloned into a modified pTVU100 vector [98] that allows protein expression in yeast with an N-terminal mCherry tag (pTVU100-N-mCherry). S. cerevisiae BY4742:POX1-EGFP (kindly provided by Zdena Palková, Charles University, Czech Republic) was transformed with the pTVU constructs and selected as described [27]. Transformed strains were incubated for 15–20 h in oleate medium prior to microscopy to stimulate peroxisome formation.
Cell fractionation
Cells (250 ml) were harvested [34], washed, and resuspended in MES isolation buffer (1 M sorbitol, 5 mM methanesulfonic acid, 1 mM KCl, 5 mM EDTA; pH 5.5). The pellet was resuspended in MES buffer containing inhibitors of proteases (0.5 mg/ml E-64, 10 μg/ml leupeptin, 50 μg/ml tosyl-L-lysyl-chloromethane hydrochloride [TLCK] and 1 tablet/50 ml Roche Complete Inhibitor Cocktail [Roche]) with or without 0.05% Tween-20. The mixture was homogenized using a Dounce homogenizer. The integrity of the cells was evaluated under microscopy, and homogenization proceeded until more than 95% of the cells were broken. Then, the homogenate was spun for 4 minutes at 380 x g to remove unbroken cells, and the supernatant was subjected either to organelle pull-down or differential centrifugation at 3,000, 9,000, 15,000, 50,000, and 150,000 x g steps (15 min per step). Alternatively, the supernatant was loaded on a 2 M sucrose-containing MES cushion and centrifuged at 50,000 x g for 20 min to separate soluble and sedimentable fractions.
Affinity enrichment of peroxisomes
Peroxisomes were pulled down via C-terminally tagged Pex14 using a recombinant E. histolytica B2-5 strain expressing His-tagged or V5-tagged Pex14 and the untagged control. Cells were harvested and washed with NaPBS, and 10 mg of protein was used for pull-down experiments. The cells were resuspended in MES buffer supplemented with protease inhibitors with or without 0.05% Tween-20. The cells were homogenized on ice using a Dounce homogenizer, and the unbroken cells were removed. The cell lysate or the crude peroxisomal fraction (obtained from the homogenate ultracentrifugation at 150,000 x g for 15 min) was preincubated with magnetic beads containing non-specific mouse antibody (i. e. anti-His antibody for V5-tagged or anti-V5 antibody for His-tagged cells) to deplete the lysate of proteins binding nonspecifically; then, the precleared lysate was incubated with 50 μl of magnetic beads coupled with either anti-His or anti-V5 specific mouse antibody (MBL Life Science, Japan) for 90 min at 4°C on an overhead rotator. The beads were washed three times in 20 volumes of MES buffer. The proteins were identified by label-free quantitative-mass spectrometry.
Mass spectrometry and data processing
Label-free quantitative mass spectrometry (LFQ-MS) was performed according to standard procedures as described previously [70]. Briefly, the samples were digested with trypsin, and the peptides obtained were subjected to nanoliquid chromatography-MS. The MS/MS spectra were acquired using Thermo Orbitrap Fusion (Q-OT- qIT, Thermo Fisher Scientific), processed in Proteome Discoverer 2.1. (Thermo Fisher Scientific) and searched against the E. histolytica database (downloaded from AmoebaDB.org, containing 8,306 entries). The quantifications were performed with label-free algorithms using MaxQuant software (version 1.6.2.1) [99], and the data were analyzed using Perseus 1.6.1.3 software [100].
Dataset processing
Altogether 13 independent experiments were performed. The datasets were filtered using the following criteria: (i) the ratio of LFQ values for a given protein between cells expressing tagged Pex14 and wild-type cells was greater than 1, (ii) the protein was identified with at least 2 peptides, and (iii) the protein was identified in at least two independent experiments. The dataset from each experiment was sorted according to relative LFQ values, and each protein was scored: the protein with the highest relative LFQ was scored 100, and scores for other proteins were calculated successively [; x is the total number of proteins in a given dataset]. Annotations of the proteins were based on Release 51 (https://amoebadb.org/amoeba/app). The mass spectrometry proteomics data have been deposited to the ProteomeXchange consortium via the PRIDE partner repository [90] with identifier PXD026653 (http://www.ebi.ac.uk/pride/archive/projects/PXD026653).
Production of recombinant myo-IDH and polyclonal antibodies
Recombinant 6xHis tagged myo-IDH was produced in Escherichia coli BL21 Rosetta cells (Novagen, Merck/Sigma-Aldrich, St. Louis, MO, USA) using the pET42b expression vector (Novagen, Merck/Sigma-Aldrich, St. Louis, MO, USA). The protein was purified by affinity chromatography using Ni-nitrilotriacetic acid agarose under denaturing conditions according to the manufacturer’s protocol (Merck/Qiagen, Hilden, Germany) and used to raise polyclonal antibodies in rats and rabbits (Davids Biotechnologie GmbH, Regensburg, Germany). Enzymatically active myo-IDH was expressed as above and affinity purified under native conditions using imidazole according to the manufacturer’s protocol (Merck/Qiagen, Hilden, Germany).
Western blotting
Protein samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and probed using antibodies as indicated. Primary antibodies included mouse monoclonal anti-His [Merck/Qiagen, Hilden, Germany], mouse monoclonal anti-V5, 1:500 [Thermo Fisher Scientific, Waltham, MA, USA]; rabbit polyclonal anti-Cpn60, 1:1000 (a kind gift from T. Nozaki, Japan) antibodies; and rabbit polyclonal anti-myo-IDH, 1:500; and rabbit polyclonal anti-FeSOD antibodies, 1:1000 [101]. Anti-mouse or anti-rabbit horseradish peroxidase (HRP) conjugated secondary antibodies (Merck/Sigma-Aldrich, St. Louis, MO, USA) were used for detections. Visualization was performed with chemiluminescence (Immobilon Forte, Merck/Sigma-Aldrich, St. Louis, MO, USA) and images were obtained using ChemiDoc (BioRad, Hercules, USA).
Immunofluorescence microscopy
Slides for confocal microscopy were prepared as described [51]. Briefly, cells were fixed using 3% formaldehyde in PBS for 30 minutes and then detached by gentle sonication in a water bath. Cells were pelleted, washed and permeabilized with 0.05% saponin followed by treatment with 50 mM NH4Cl. Upon washing, the cells were blocked using 2% inactivated fetal bovine serum in PBS with 0.05% saponin (FNS) followed by overnight incubation with primary antibody in a refrigerator (mouse monoclonal anti-His [Merck/Qiagen, Hilden, Germany], mouse monoclonal anti-V5, 1:500 [Thermo Fisher Scientific, Waltham, MA, USA]; rabbit polyclonal anti-adenosine phosphosulfate kinase [APSK], 1:1000; anti-BiP1, 1:500; and anti-Atg8, 1:1000 [a kind gift from T. Nozaki, Japan]; and rabbit and rat polyclonal anti-myo-IDH, 1:500). Alexa Fluor-488 (1:1000) or Alexa Fluor-594 (1:1000) secondary antibodies were used for visualization (Thermo Fisher Scientific, Waltham, MA, USA). Slides were observed using a Leica SP8 FLIM confocal microscope (50 optical sections of 150 nm each). Images were deconvolved using Huygens 19.04 software (SVI) and further processed with Fiji software [102] and the Imaris 9.7.2 Package for Cell Biologists (Bitplane AG, Zurich, Switzerland).Voxel-based colocalization was performed using ImarisColoc. Costes’s automatic thresholding was applied to the images in the Z stack [103], and PCC in colocalized volume was calculated. The number of labeled organelles observed by confocal microscopy (20 optical slices of 1 μm each) in E. histolytica using His-tagged Pex14 as a marker was determined per 25 cells using Icy software [104]. The number of peroxisomes was expressed per 100 μm2.
Structured illumination microscopy (SIM) was performed on a 3D N-SIM microscope (Nikon Eclipse Ti-E, Nikon, Japan) equipped with a Nikon CFI SR Apo TIRF objective (100x oil, NA 1.49) as described [105].
Immunoelectron microscopy
Transmission electron microscopy was performed using E. histolytica cells overexpressing polyHis- or V5-tagged Pex14 and Pex16, and samples were processed as described previously [27]. The sections with V5-tagged Pex14 were blocked with 1% fish skin gelatin (FSG) and immunodecorated using mouse anti-V5 antibody (1:20, Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA) and goat anti-mouse antibody conjugated with 5 nm gold nanoparticles (BBI Solutions) for 1 h in 1% fish skin gelatin. Samples with polyHis-tagged Pex14 were blocked in buffer containing 1% bovine serum albumin/0.05% Tween 20 in 0.1 M HEPES for 1 h at room temperature. Labeling was performed using Ni-nitrilotriacetic acid (Ni-NTA) conjugated with 5 nm gold nanoparticles (Nanoprobes, Yaphank, USA). Sections with polyHis-tagged Pex16 were treated as described above or with mouse monoclonal anti-polyHis antibody (IgG2A, Merck/Sigma-Aldrich, St. Louis, MO, USA), and protein A conjugated with 5 nm gold nanoparticles (1:50)(CMC Utrecht) in 1% fish skin gelatin. Myo-IDH was detected using rabbit polyclonal anti-myo-IDH antibody and anti-rabbit IgG conjugated to 15 nm gold particles (Nanoprobes, Yaphank, USA).
Enzymatic assay
The activity of myo-IDH (EC 1.1.1.18) was measured spectrophotometrically at 340 nm and room temperature, as described [106]. The reaction mixture contained 1 mM NAD+ in 10 mM sodium pyrophosphate buffer, pH 9, 30 μg of the purified enzyme, and a substrate. The Km value was determined by measurement of the reaction velocity using myo-inositol (50 μM-25 mM), scyllo-inositol (370 μM-15 mM), D-glucose (5 mM-400 mM), D-xylose (5 mM-400 mM), and keto-2-inositol (12.5 μM—250 μM). For the Km of NAD+ and NADP+, 12.5 mM myo-inositol was used for both NAD+(50 μM—1 mM)and NADP+(500 μM—1.5 mM). Each measurement was performed in triplicates. The protein concentration was measured using the Lowry method. All chemicals were purchased from Sigma-Aldrich (Merck/Sigma-Aldrich, St. Louis, MO, USA).
Size exclusion chromatography and multiangle light scattering (MALS)
The native molecular mass of recombinant, 6xHis-tagged myo-IDH was determined using size-exclusion chromatography Superdex 200 Increase 10/300 GL column connected to BioLogic DuoFlow system (BioRad, Hercules, USA). The column was calibrated with BioRad gel filtration standard mixture (cat # 1511901), and the running buffer was 10 mM sodium pyrophosphate used for the enzymatic assay. Fractions of 0.5 ml were collected for monitoring the myo-IDH activity in the elution profile.
For MALS analysis, samples of enzymatically active myo-IDH (0,5–2 mg/ml) were injected onto an Agilent Biosec-3 column (4.6x300 mm) at a flow rate of 0.3 ml/min in 50 mM phosphate buffer (pH 7.0) and 300 mM NaCl at 15°C. The column was coupled with static light scattering (miniDAWN, Wyatt Technology), differential refractive index (Shodex RI-501) and Agilent 1260 Infinity II UV (Agilent Technologies) detectors. Data were analyzed using ASTRA software (Wyatt Technology).
Supporting information
(PDF)
The frequency plot is shown on a logarithmic scale (the high frequencies are shown in red-to-yellow, low frequencies are shown in blue-to-violet. The number indicates border bins. Pearson correlation coefficientsin volume are given in S5 Table.
(PDF)
(DOCX)
(PDF)
Each point was calculated from three measurements, error bars represent standard deviations.
(PDF)
Superdex 200 Increase 10/300 GL column connected to BioLogic DuoFlow system (BioRad) was used for the analysis.
(PDF)
Elution profile at three different concentrations, 2.0 mg/ml (red), 1.0 mg/ml (magenta) and 0.5 mg/ml (green), with the indicated molar mass.
(PDF)
(PDF)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Voxel-based colocalization with automatic thresholding was performed using ImarisColoc. Corresponding 2Dhistograms are given in S1 Fig.
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(TXT)
Acknowledgments
The authors wish to thank Michaela Marcinčiková for her excellent technical assistance, and Marie Olšinová for her help with fluorescence microscopy.
Data Availability
The mass spectrometry proteomics data are avilable from the ProteomeXchange consortium repository with identifier PXD026653. Alignments were provided to Mendeley data repository: DOI: 10.17632/dsszxzcc84.1. The script and training data are available at https://github.com/vojtech-zarsky/PredictPTS1_ML.
Funding Statement
This study was funded by Czech Science Foundation (P305/11/1061), https://gacr.cz, European Regional Development Funds (CePaViP (CZ.02.1.01/0.0/0.0/16_019/0000759), https://www.msmt.cz/, University Research Centre of Charles University (UNCE, SCI/12), https://cuni.cz/UKEN-65.html, H2020 Spreading Excellence and Widening Participation MICOBION (H2020 No 810224) https://ec.europa.eu/programmes/horizon2020/en/h2020-section/spreading-excellence-and-widening-participation to JT, Jürgen Manchot Stiftung, https://www.im.nrw/juergen-manchot-stiftung to CK. Imaging Methods Core Facility at BIOCEV, and the Core Facility of the Institute of Parasitology and Biology Centre of the Czech Academy of Sciences were both supported by the MEYS CR (Large RI Project LM2018129 Czech-BioImaging) https://www.msmt.cz. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang H, Naghavi M, Allen C, Barber RM, Carter A, Casey DC, et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388: 1459–1544. doi: 10.1016/S0140-6736(16)31012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380: 2095–2128. doi: 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui Z, Li J, Chen Y, Zhang L. Molecular epidemiology, evolution, and phylogeny of Entamoeba spp. Infect Genet Evol. 2019;75: 104018. doi: 10.1016/j.meegid.2019.104018 [DOI] [PubMed] [Google Scholar]
- 4.Uddin MJ, Leslie JL, Petri WA. Host protective mechanisms to intestinal amebiasis. Trends Parasitol. 2021;37: 165–175. doi: 10.1016/j.pt.2020.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Žárský V, Klimeš V, Pačes J, Vlček Č, Hradilová M, Beneš V, et al. The Mastigamoeba balamuthi genome and the nature of the free-living ancestor of Entamoeba. Mol Biol Evol. 2021;38: 2240–2259. doi: 10.1093/molbev/msab020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makiuchi T, Nozaki T. Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie. 2014;100: 3–17. doi: 10.1016/j.biochi.2013.11.018 [DOI] [PubMed] [Google Scholar]
- 7.Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32: 1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x [DOI] [PubMed] [Google Scholar]
- 8.Ghosh S, Field J, Rogers R, Hickman M, Samuelson J. The Entamoeba histolyticamitochondrion-derived organelle (crypton) contains double-stranded DNA and appears to be bound by a double membrane. Infect Immun. 2000;68: 4319–4322. doi: 10.1128/IAI.68.7.4319-4322.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mi-ichi F, Yousuf MA, Nakada-Tsukui K, Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A. 2009;106: 21731–21736. doi: 10.1073/pnas.0907106106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mi-ichi F, Makiuchi T, Furukawa A, Sato D, Nozaki T. Sulfate activation in mitosomes plays an important role in the proliferation of Entamoeba histolytica. PLoS Negl Trop Dis. 2011;5: e1263. doi: 10.1371/journal.pntd.0001263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pineda E, Vázquez C, Encalada R, Nozaki T, Sato E, Hanadate Y, et al. Roles of acetyl-CoA synthetase (ADP-forming) and acetate kinase (PPi-forming) in ATP and PPi supply in Entamoeba histolytica. Biochim Biophys Acta—Gen Subj. 2016;1860: 1163–1172. doi: 10.1016/j.bbagen.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 12.Cavalier-Smith T. The origin of cells: a symbiosis between genes, catalysts, and membranes. Cold Spring Harb Symp Quant Biol. 1987;52: 805–824. doi: 10.1101/sqb.1987.052.01.089 [DOI] [PubMed] [Google Scholar]
- 13.Mazzuco A, Benchimol M, De Souza W. Endoplasmic reticulum and Golgi-like elements in Entamoeba. Micron. 1997;28: 241–247. doi: 10.1016/s0968-4328(97)00024-3 [DOI] [PubMed] [Google Scholar]
- 14.Teixeira JE, Huston CD. Evidence of a continuous endoplasmic reticulum in the protozoan parasite Entamoeba histolytica. Eukaryot Cell. 2008;7: 1222–1226. doi: 10.1128/EC.00007-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh SK, Field J, Frisardi M, Rosenthal B, Mai Z, Rogers R, et al. Chitinase secretion by encysting Entamoeba invadens and transfected Entamoeba histolytica trophozoites: localization of secretory vesicles, endoplasmic reticulum, and Golgi apparatus. Infect Immun. 1999;67: 3073–3081. doi: 10.1128/IAI.67.6.3073-3081.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barlow LD, Nývltová E, Aguilar M, Tachezy J, Dacks JB. A sophisticated, differentiated Golgi in the ancestor of eukaryotes. BMC Biol. 2018;16: 27. doi: 10.1186/s12915-018-0492-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabaldón T. Peroxisome diversity and evolution. Philos Trans R Soc B Biol Sci. 2010;365: 765–773. doi: 10.1098/rstb.2009.0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pieuchot L, Jedd G. Peroxisome assembly and functional diversity in eukaryotic microorganisms. Annu Rev Microbiol. 2012;66: 237–263. doi: 10.1146/annurev-micro-092611-150126 [DOI] [PubMed] [Google Scholar]
- 19.Allmann S, Bringaud F. Glycosomes: A comprehensive view of their metabolic roles in T. brucei. Int J Biochem Cell Biol. 2017;85: 85–90. doi: 10.1016/j.biocel.2017.01.015 [DOI] [PubMed] [Google Scholar]
- 20.van der Klei IJ, Veenhuis M. The Versatility of peroxisome function in filamentous fungi. Subcellular Biochemistry. Springer; New York; 2013. pp. 135–152. doi: 10.1007/978-94-007-6889-5_8 [DOI] [PubMed] [Google Scholar]
- 21.Walter T, Erdmann R. Current advances in protein import into peroxisomes. Protein J. 2019;38: 351–362. doi: 10.1007/s10930-019-09835-6 [DOI] [PubMed] [Google Scholar]
- 22.Mast FD, Rachubinski RA, Aitchison JD. Peroxisome prognostications: Exploring the birth, life, and death of an organelle. J Cell Biol. 2020;219: 1–13. doi: 10.1083/jcb.201912100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speijer D. Evolution of peroxisomes illustrates symbiogenesis. BioEssays. 2017;39: 1–8. doi: 10.1002/bies.201700050 [DOI] [PubMed] [Google Scholar]
- 24.Fransen M, Lismont C, Walton P. The peroxisome-mitochondria connection: How and why? Int J Mol Sci. 2017;18. doi: 10.3390/ijms18061126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugiura A, Mattie S, Prudent J, Mcbride HM. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature. 2017;542: 251–254. doi: 10.1038/nature21375 [DOI] [PubMed] [Google Scholar]
- 26.Gabaldón T, Ginger ML, Michels PAM. Peroxisomes in parasitic protists. Mol Biochem Parasitol. 2016;209: 35–45. doi: 10.1016/j.molbiopara.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 27.Le T, Žárský V, Nývltová E, Rada P, Harant K, Vancová M, et al. Anaerobic peroxisomes in Mastigamoeba balamuthi. Proc Natl Acad Sci U S A. 2020;117: 2065–2075. doi: 10.1073/pnas.1909755117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Záhonová K, Treitli SC, Le T, Škodová-Sveráková I, Hanousková P, Čepička I, et al. Anaerobic derivates of mitochondria and peroxisomes in the free-living amoeba Pelomyxa schiedti revealed by single-cell genomics. bioRxiv. 2021; 2021.05.20.444135. doi: 10.1101/2021.05.20.444135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neufeld C, Filipp F V., Simon B, Neuhaus A, Schüller, David, et al. Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19. EMBO J. 2009;28: 745–754. doi: 10.1038/emboj.2009.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt F, Treiber N, Zocher G, Bjelic S, Steinmetz MO, Kalbacher H, et al. Insights into peroxisome function from the structure of PEX3 in complex with a soluble fragment of PEX19. J Biol Chem. 2010;285: 25410–7. doi: 10.1074/jbc.M110.138503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato Y, Shibata H, Nakatsu T, Nakano H, Kashiwayama Y, Imanaka T, et al. Structural basis for docking of peroxisomal membrane protein carrier Pex19p onto its receptor Pex3p. EMBO J. 2010;29: 4083–93. doi: 10.1038/emboj.2010.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mindthoff S, Grunau S, Steinfort LL, Girzalsky W, Hiltunen JK, Erdmann R, et al. Peroxisomal Pex11 is a pore-forming protein homologous to TRPM channels. Biochim Biophys Acta—Mol Cell Res. 2016;1863: 271–283. doi: 10.1016/j.bbamcr.2015.11.013 [DOI] [PubMed] [Google Scholar]
- 33.Koch J, Pranjic K, Huber A, Ellinger A, Hartig A, Kragler F, et al. PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J Cell Sci. 2010;123: 3389–3400. doi: 10.1242/jcs.064907 [DOI] [PubMed] [Google Scholar]
- 34.Meyer M, Fehling H, Matthiesen J, Lorenzen S, Schuldt K, Bernin H, et al. Overexpression of differentially expressed genes identified in non-pathogenic and pathogenic Entamoeba histolytica clones allow identification of new pathogenicity factors involved in amoebic liver abscess formation. PLoS Pathog. 2016;12: e1005853. doi: 10.1371/journal.ppat.1005853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nickel R, Tannich E. Transfection and transient expression of chloramphenicol acetyltransferase gene in the protozoan parasite Entamoeba histolytica. Proc Natl Acad Sci U S A. 1994;91: 7095–7098. doi: 10.1073/pnas.91.15.7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Picazarri K, Nakada-Tsukui K, Tsuboi K, Miyamoto E, Watanabe N, Kawakami E, et al. Atg8 is involved in endosomal and phagosomal acidification in the parasitic protist Entamoeba histolytica. Cell Microbiol. 2015;17: 1510–1522. doi: 10.1111/cmi.12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanadate Y, Saito-Nakano Y, Nakada-Tsukui K, Nozaki T. Endoplasmic reticulum-resident Rab8A GTPase is involved in phagocytosis in the protozoan parasite Entamoeba histolytica. Cell Microbiol. 2016;18: 1358–73. doi: 10.1111/cmi.12570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173: 521–532. doi: 10.1083/jcb.200601036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barros-Barbosa A, Ferreira MJ, Rodrigues TA, Pedrosa AG, Grou CP, Pinto MP, et al. Membrane topologies of PEX13 and PEX14 provide new insights on the mechanism of protein import into peroxisomes. FEBS J. 2019;286: 205–222. doi: 10.1111/febs.14697 [DOI] [PubMed] [Google Scholar]
- 40.Santos HJ, Imai K, Hanadate Y, Fukasawa Y, Oda T, Mi-ichi F, et al. Screening and discovery of lineage-specific mitosomal membrane proteins in Entamoeba histolytica. Mol Biochem Parasitol. 2016;209: 10–17. doi: 10.1016/j.molbiopara.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 41.Schlüter A, Fourcade S, Domènech-Estévez E, Gabaldón T, Huerta-Cepas J, Berthommier G, et al. PeroxisomeDB: A database for the peroxisomal proteome, functional genomics and disease. Nucleic Acids Res. 2007;35: 815–822. doi: 10.1093/nar/gkl935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F. Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J Mol Biol. 2003;328: 567–579. doi: 10.1016/s0022-2836(03)00318-8 [DOI] [PubMed] [Google Scholar]
- 43.Ramaley R, Fujita Y, Freese E. Purification and properties of Bacillus subtilis inositol dehydrogenase. J Biol Chem. 1979;254: 7684–7690. doi: 10.1016/S0021-9258(18)36000-9 [DOI] [PubMed] [Google Scholar]
- 44.Dambe TR, Kühn AM, Brossette T, Giffhorn F, Scheidig AJ. Crystal Structure of NADP(H)-Dependent 1,5-Anhydro-D-fructose reductase from Sinorhizobium morelense at 2.2 Å resolution: Construction of a NADH-accepting mutant and its application in rare sugar synthesis. Biochemistry. 2006;45: 10030–10042. doi: 10.1021/bi052589q [DOI] [PubMed] [Google Scholar]
- 45.Kingston RL, Scopes RK, Baker EN. The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP. Structure. 1996;4: 1413–1428. doi: 10.1016/s0969-2126(96)00149-9 [DOI] [PubMed] [Google Scholar]
- 46.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27: 343–350. doi: 10.1093/bioinformatics/btq662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukano K, Ozawa K, Kokubu M, Shimizu T, Ito S, Sasaki Y, et al. Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification. PLoS One. 2018;13. doi: 10.1371/journal.pone.0198010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Straaten KE, Zheng H, Palmer DRJ, Sanders DAR. Structural investigation of myo-inositol dehydrogenase from Bacillus subtilis: implications for catalytic mechanism and inositol dehydrogenase subfamily classification. Biochem J. 2010;432: 237–247. doi: 10.1042/BJ20101079 [DOI] [PubMed] [Google Scholar]
- 49.Archibald JM. The puzzle of plastid evolution. Curr Biol. Cell Press; 2009. pp. R81–R88. doi: 10.1016/j.cub.2008.11.067 [DOI] [PubMed] [Google Scholar]
- 50.Jansen RLM, Santana Molina C, Van Den Noort M, Devos DP, Van Der Klei IJ. Comparative genomics of peroxisome biogenesis proteins: making sense of the PEX mess. bioRxiv. 2020; 2020.12.16.423121. doi: 10.1101/2020.12.16.423121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolezal P, Dagley MJ, Kono M, Wolynec P, Likić VA, Foo JH, et al. The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica. PLoS Pathog. 2010;6: e1000812. doi: 10.1371/journal.ppat.1000812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiedemann N, Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem. 2017;86: 685–714. doi: 10.1146/annurev-biochem-060815-014352 [DOI] [PubMed] [Google Scholar]
- 53.Schlüter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A. The evolutionary origin of peroxisomes: An ER-peroxisome connection. Mol Biol Evol. 2006;23: 838–845. doi: 10.1093/molbev/msj103 [DOI] [PubMed] [Google Scholar]
- 54.Knoops K, Manivannan S, Cepińska MN, Krikken AM, Kram AM, Veenhuis M, et al. Preperoxisomal vesicles can form in the absence of Pex3. J Cell Biol. 2014;204: 659–668. doi: 10.1083/jcb.201310148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jansen RLM, Klei IJ. The peroxisome biogenesis factors Pex3 and Pex19: multitasking proteins with disputed functions. FEBS Lett. 2019;593: 457–474. doi: 10.1002/1873-3468.13340 [DOI] [PubMed] [Google Scholar]
- 56.Banerjee H, Knoblach B, Rachubinski RA. The early-acting glycosome biogenic protein Pex3 is essential for trypanosome viability. Life Sci Alliance. 2019;2: e201900421. doi: 10.26508/lsa.201900421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalel VC, Li M, Gaussmann S, Delhommel F, Schäfer A-B, Tippler B, et al. Evolutionary divergent PEX3 is essential for glycosome biogenesis and survival of trypanosomatid parasites. Biochim Biophys Acta—Mol Cell Res. 2019;1866: 118520. doi: 10.1016/j.bbamcr.2019.07.015 [DOI] [PubMed] [Google Scholar]
- 58.Miyata N, Fujiki Y. Shuttling echanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol Cell Biol. 2005;25: 10822–10832. doi: 10.1128/MCB.25.24.10822-10832.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Williams C, Van Den Berg M, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem. 2007;282: 22534–22543. doi: 10.1074/jbc.M702038200 [DOI] [PubMed] [Google Scholar]
- 60.Ma C, Schumann U, Rayapuram N, Subramani S. The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol Biol Cell. 2009;20: 3680–3689. doi: 10.1091/mbc.e09-01-0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bosch DE, Siderovski DP. Structural determinants of ubiquitin conjugation in Entamoeba histolytica. J Biol Chem. 2013;288: 2290–2302. doi: 10.1074/jbc.M112.417337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grou CP, Carvalho AF, Pinto MP, Wiese S, Piechura H, Meyer HE, et al. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J Biol Chem. 2008;283: 14190–14197. doi: 10.1074/jbc.M800402200 [DOI] [PubMed] [Google Scholar]
- 63.Platta HW, Hagen S, Reidick C, Erdmann R. The peroxisomal receptor dislocation pathway: To the exportomer and beyond. Biochimie; 2014;98: 16–28. doi: 10.1016/j.biochi.2013.12.009 [DOI] [PubMed] [Google Scholar]
- 64.Lazarow PB. Chapter 3.1.7. The import receptor Pex7p and the PTS2 targeting sequence. Biochim Biophys Acta—Mol Cell Res. 2006;1763: 1599–1604. doi: 10.1016/j.bbamcr.2006.08.011 [DOI] [PubMed] [Google Scholar]
- 65.Žárský V, Tachezy J. Evolutionary loss of peroxisomes–not limited to parasites. Biol Direct. 2015;10: 74. doi: 10.1186/s13062-015-0101-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mattiazzi Ušaj M, Brložnik M, Kaferle P, Žitnik M, Wolinski H, Leitner F, et al. Genome-wide localization study of yeast pex11 identifies peroxisome-mitochondria interactions through the ERMES complex. J Mol Biol. 2015;427: 2072–2087. doi: 10.1016/j.jmb.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148: 931–944. doi: 10.1083/jcb.148.5.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eckert JH, Johnsson N. Pex10p links the ubiquitin conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J Cell Sci. 2003;116: 3623–3634. doi: 10.1242/jcs.00678 [DOI] [PubMed] [Google Scholar]
- 69.Wideman JG, Muñoz-Gómez SA. The evolution of ERMIONE in mitochondrial biogenesis and lipid homeostasis: An evolutionary view from comparative cell biology. Biochim Biophys Acta. 2016;1861: 900–912. doi: 10.1016/j.bbalip.2016.01.015 [DOI] [PubMed] [Google Scholar]
- 70.Makki A, Rada P, Žárský V, Kereïche S, Kováčik L, Novotný M, et al. Triplet-pore structure of a highly divergent TOM complex of hydrogenosomes in Trichomonas vaginalis. PLoS Biol. 2019;17: e3000098. doi: 10.1371/journal.pbio.3000098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Makiuchi T, Santos HJ, Tachibana H, Nozaki T. Hetero-oligomer of dynamin-related proteins participates in the fission of highly divergent mitochondria from Entamoeba histolytica. Sci Rep. 2017;7: 13439. doi: 10.1038/s41598-017-13721-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma S, Bhattacharya S, Bhattacharya A. PtdIns(4,5)P 2 is generated by a novel phosphatidylinositol 4-phosphate 5-kinase in the protist parasite Entamoeba histolytica. FEBS J. 2019;286: 2216–2234. doi: 10.1111/febs.14804 [DOI] [PubMed] [Google Scholar]
- 73.Nakada-Tsukui K, Watanabe N, Maehama T, Nozaki T. Phosphatidylinositol kinases and phosphatases in Entamoeba histolytica. Front Cell Infect Microbiol. 2019;9. doi: 10.3389/fcimb.2019.00150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Michell RH. Inositol derivatives: Evolution and functions. Nat Rev Mol Cell Biol. 2008;9: 151–161. doi: 10.1038/nrm2334 [DOI] [PubMed] [Google Scholar]
- 75.Chatree S, Thongmaen N, Tantivejkul K, Sitticharoon C, Vucenik I. Role of inositols and inositol phosphates in energy metabolism. Molecules. 2020;25: 5079. doi: 10.3390/molecules25215079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xing G, Hoffart LM, Diao Y, Prabhu KS, Arner RJ, Reddy CC, et al. A coupled dinuclear iron cluster that is perturbed by substrate binding in myo-inositol oxygenase. Biochemistry. 2006;45: 5393–5401. doi: 10.1021/bi0519607 [DOI] [PubMed] [Google Scholar]
- 77.Yoshida K, Yamaguchi M, Morinaga T, Kinehara M, Ikeuchi M, Ashida H, et al. myo-Inositol catabolism in Bacillus subtilis. J Biol Chem. 2008;283: 10415–10424. doi: 10.1074/jbc.M708043200 [DOI] [PubMed] [Google Scholar]
- 78.Gross W, Meyer A. Distribution of myo-inositol dehydrogenase in algae. Eur J Phycol. 2003;38: 191–194. doi: 10.1080/1364253031000121705 [DOI] [Google Scholar]
- 79.Al-Saryi NA, Al-Hejjaj MY, van Roermund CWT, Hulmes GE, Ekal L, Payton C, et al. Two NAD-linked redox shuttles maintain the peroxisomal redox balance in Saccharomyces cerevisiae. Sci Rep. 2017;7: 11868. doi: 10.1038/s41598-017-11942-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ast J, Stiebler AC, Freitag J, Bölker M. Dual targeting of peroxisomal proteins. Front Physiol. 2013;4: 297. doi: 10.3389/fphys.2013.00297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stein R, Schnarrenberger C, Gross W. Myo-inositol dehydrogenase from the acido- and thermophilic red alga Galdieria sulphuraria. Phytochemistry. 1997;46: 17–20. doi: 10.1016/S0031-9422(96)00830-8 [DOI] [Google Scholar]
- 82.Bertwistle D, Vogt L, Aamudalapalli HB, Palmer DRJ, Sanders DAR. Purification, crystallization and room-temperature X-ray diffraction of inositol dehydrogenase LcIDH2 from Lactobacillus casei BL23. Acta Crystallogr Sect F Struct Biol Commun. 2014;70: 979–983. doi: 10.1107/S2053230X14011595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47: D309–D314. doi: 10.1093/nar/gky1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46: W200–W204. doi: 10.1093/nar/gky448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altschul S. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25: 3389–3402. doi: 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33: W244–W248. doi: 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Subramani S, Koller A, Snyder WB. Import of peroxisomal matrix and membrane proteins. Annu Rev Biochem. 2000;69: 399–418. doi: 10.1146/annurev.biochem.69.1.399 [DOI] [PubMed] [Google Scholar]
- 88.Emanuelsson O, Elofsson A, Von Heijne G, Cristóbal S. In silico prediction of the peroxisomal proteome in fungi, plants and animals. J Mol Biol. 2003;330: 443–456. doi: 10.1016/s0022-2836(03)00553-9 [DOI] [PubMed] [Google Scholar]
- 89.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30: 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10. doi: 10.1186/1471-2148-10-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32: 268–274. doi: 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21: 1095–1109. doi: 10.1093/molbev/msh112 [DOI] [PubMed] [Google Scholar]
- 93.Biller L, Schmidt H, Krause E, Gelhaus C, Matthiesen J, Handal G, et al. Comparison of two genetically related Entamoeba histolytica cell lines derived from the same isolate with different pathogenic properties. Proteomics. 2009;9: 4107–4120. doi: 10.1002/pmic.200900022 [DOI] [PubMed] [Google Scholar]
- 94.Clark CG, Diamond LS. Methods for cultivation of luminal parasitic protists of clinical importance. Clin Microbiol Rev. 2002;15: 329–341. doi: 10.1128/CMR.15.3.329-341.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baxt LA, Rastew E, Bracha R, Mirelman D, Singh U. Downregulation of an Entamoeba histolytica rhomboid protease reveals roles in regulating parasite adhesion and phagocytosis. Eukaryot Cell. 2010;9: 1283–1293. doi: 10.1128/EC.00015-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Narayanasamy RK, Castañón-Sanchez CA, Luna-Arias JP, García-Rivera G, Avendaño-Borromeo B, Labra-Barrios ML, et al. The Entamoeba histolytica TBP and TRF1 transcription factors are GAAC-box binding proteins, which display differential gene expression under different stress stimuli and during the interaction with mammalian cells. Parasit Vectors. 2018;11: 153. doi: 10.1186/s13071-018-2698-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 98.Malínská K, Malínský J, Opekarová M, Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol Biol Cell. 2003;14: 4427–4436. doi: 10.1091/mbc.e03-04-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13: 2513–2526. doi: 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13: 731–740. doi: 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 101.Bruchhaus I, Brattig NW, Tannich E. Recombinant expression, purification and biochemical characterization of a superoxide dismutase from Entamoeba histolytica. Arch Med Res. 1992;23: 27–29. Available: https://pubmed.ncbi.nlm.nih.gov/1340312/ [PubMed] [Google Scholar]
- 102.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9: 676–682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Costes S V., Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86: 3993–4003. doi: 10.1529/biophysj.103.038422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Chaumont F, Dallongeville S, Chenouard N, Hervé N, Pop S, Provoost T, et al. Icy: an open bioimage informatics platform for extended reproducible research. Nat Methods. 2012;9: 690–696. doi: 10.1038/nmeth.2075 [DOI] [PubMed] [Google Scholar]
- 105.Štáfková J, Rada P, Meloni D, Žárský V, Smutná T, Zimmann N, et al. Dynamic secretome of Trichomonas vaginalis: Case study of β-amylases. Mol Cell Proteomics. 2018;17: 304–320. doi: 10.1074/mcp.RA117.000434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Berman T, Magasanik B. The pathway of myo-inositol degradation in Aerobacter aerogenes. Dehydrogenation and dehydration. J Biol Chem. 1966;241: 800–806. doi: 10.1016/S0021-9258(18)96836-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
The frequency plot is shown on a logarithmic scale (the high frequencies are shown in red-to-yellow, low frequencies are shown in blue-to-violet. The number indicates border bins. Pearson correlation coefficientsin volume are given in S5 Table.
(PDF)
(DOCX)
(PDF)
Each point was calculated from three measurements, error bars represent standard deviations.
(PDF)
Superdex 200 Increase 10/300 GL column connected to BioLogic DuoFlow system (BioRad) was used for the analysis.
(PDF)
Elution profile at three different concentrations, 2.0 mg/ml (red), 1.0 mg/ml (magenta) and 0.5 mg/ml (green), with the indicated molar mass.
(PDF)
(PDF)
(XLSX)
(XLSX)
(DOCX)
(DOCX)
Voxel-based colocalization with automatic thresholding was performed using ImarisColoc. Corresponding 2Dhistograms are given in S1 Fig.
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(TXT)
Data Availability Statement
The mass spectrometry proteomics data are avilable from the ProteomeXchange consortium repository with identifier PXD026653. Alignments were provided to Mendeley data repository: DOI: 10.17632/dsszxzcc84.1. The script and training data are available at https://github.com/vojtech-zarsky/PredictPTS1_ML.