Abstract
The recent advent of single-cell RNA-sequencing technology has provided new fundamental insights into the heterogeneity of the prostate epithelium. Several independent studies have described extensive heterogeneity of the luminal epithelial compartment, including a major division between a novel population of luminal cells located in the proximal region of the prostate ducts versus luminal cells located more distally. Proximal luminal cells as well as novel periurethral cells display increased progenitor potential in organoid culture and tissue reconstitution assays, but not in lineage-tracing analyses during prostate homeostasis, suggesting context-dependent plasticity of these populations. Here we describe and synthesize recent findings regarding the epithelial cell populations in the mouse prostate, draw comparisons to the human prostate, and address the relevance of these findings to prostate diseases and cancer.
Keywords: Stem cell, organoid, lineage-tracing, homeostasis, plasticity
The prostate gland has long been of biomedical interest due to its propensity to develop disease. Benign prostate hyperplasia and prostatitis are common diseases in older men with potentially significant effects on quality of life, and prostate cancer is the leading cancer diagnosis as well as the second most lethal cancer in American men [1]. To understand the initiation and progression of these diseases, it is necessary to understand their cellular origins within the benign prostate.
Recent findings using single-cell RNA-sequencing have provided important new insights into the heterogeneity and complex functionality of cell types in the mouse and human prostate epithelium, with considerable implications for understanding prostate disease. Here, we review these recent studies, place these findings in the context of previous work, and discuss future directions.
Anatomy and structure of the mouse and human prostate
Historically, the mouse and human prostate has been described as containing a pseudostratified epithelium that is composed of three primary epithelial cell types, corresponding to luminal cells, basal cells, and rare neuroendocrine cells [2]. However, despite their similar function in producing prostatic fluids, the mouse and human prostates display significant anatomical and histological differences. The mouse prostate is situated around the urethra, directly underneath the bladder, and is divided into four distinct pairs of lobes, corresponding to the anterior (AP), dorsal (DP), lateral (LP), and ventral (VP) prostate lobes (Fig. 1A); the dorsal and lateral lobes are frequently combined during dissection and designated as the dorsolateral prostate (DLP). These lobes contain branching ducts that produce and collect prostatic secretions, which drain through a proximal region into the urethra during ejaculation. Notably, each lobe has functional differences as they express different prostatic secretory proteins that are often lobe-specific [3–5].
Figure 1. Single-cell analysis of prostate epithelial heterogeneity.
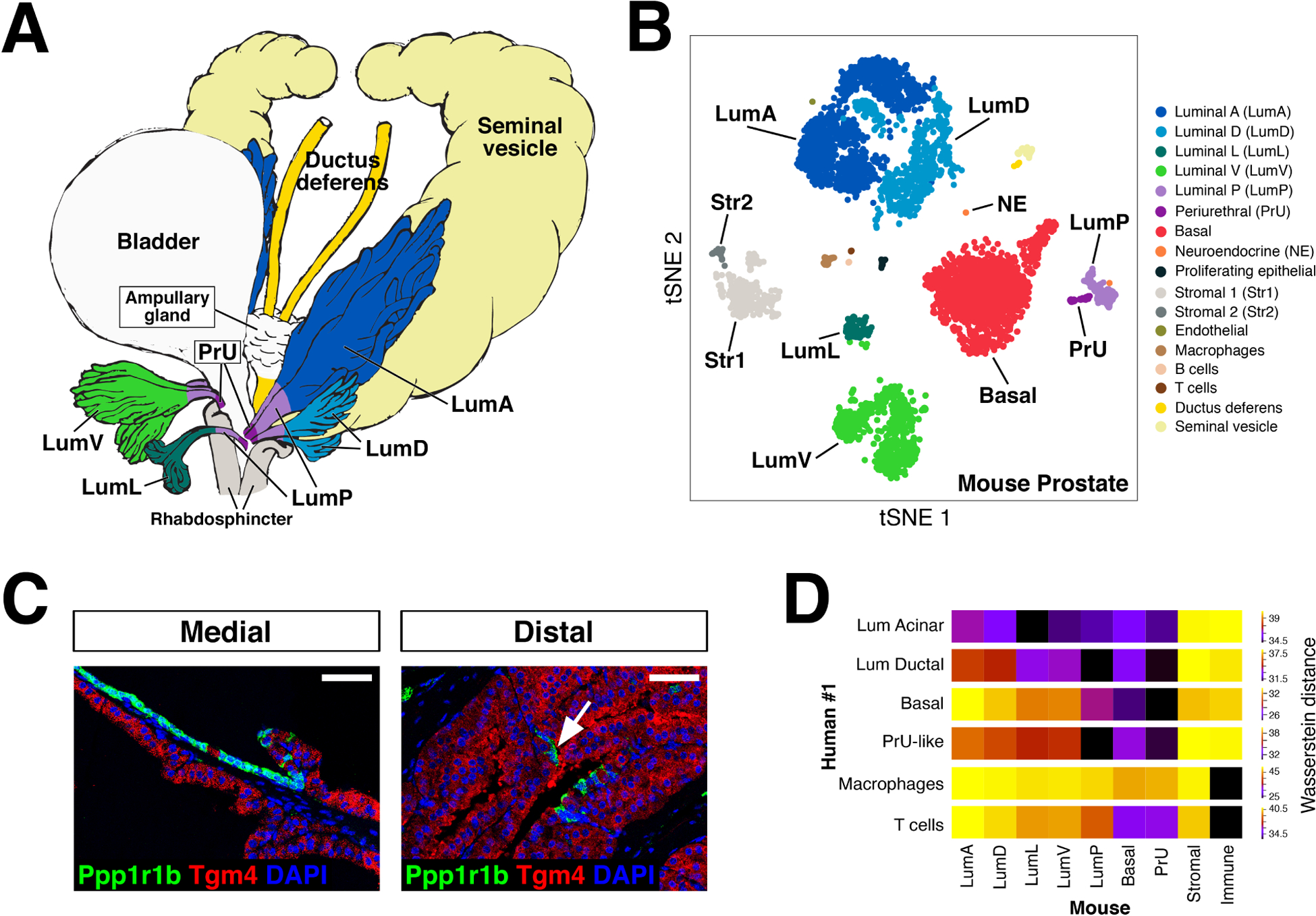
(A) Schematic model of prostate lobes, with the urethral rhabdosphincter partially removed to show the distribution of luminal epithelial populations as indicated. (B) t-distributed stochastic neighbor embedding (tSNE) plot of 5,288 cells from an aggregated dataset of two normal mouse prostates. (C) Immunofluorescence staining of a mouse dorsal prostate lobe, showing proximal luminal cells marked with Ppp1r1b and a medial border between proximal and distal luminal cells. (D) Heatmap visualization of transcriptional Wasserstein distances (also known as earth mover’s distances); this metric corresponds to transcriptional differences between populations, where less distance indicates transcriptional similarity, and greater distance indicates dissimilarity) between human and mouse prostate populations. Adapted from [8].
In contrast, the human prostate is a walnut-sized organ that lacks a lobular structure. Instead, three contiguous but distinct zones can be distinguished histologically in the human prostate. The central zone is situated along the ejaculatory duct orifices through to the connection to the penile urethra, the transition zone is the smallest zone and encircles the upper prostatic urethra just anterior to the central zone, and the peripheral zone is the largest, encircling the lower prostatic urethra with lateral and posterior branches [6, 7]. Each zone consists of a series of compact, interconnected, tubular-acinar branching ducts that drain into the urethra through multiple different proximal regions. Whereas the central and transition zones contain a mix of unbranched proximal regions and branched glandular regions, the peripheral zone has a larger proportion of branched glandular ducts that contain secretory acinar cells. Notably, these zones have differential disease susceptibility, as the peripheral zone is the primary site of prostatitis and prostate adenocarcinoma, whereas the transition zone is the principal site of benign prostate hyperplasia [6].
Some features of luminal cells in the prostate are conserved between species. In mice and humans, the distal regions of the prostatic ducts (further from the urethra) have active secretory cells during homeostasis. Electron microscopy has shown that these distal luminal cells can secrete either through apocrine mechanisms, in which vesicles on the apical surface form a neck that is pinched closed during budding, or through merocrine mechanisms, in which the secretory content is exocytosed directly into the lumen [8, 9]. These distal luminal cells are larger and have a high ratio of cytoplasmic to nuclear volume, with the cytoplasm containing pronounced Golgi apparatus, rough endoplasmic reticulum, and free ribosomes, in addition to secretory vesicles toward the apical surface [8, 9]. In comparison, luminal cells in proximal ducts have a lower cytoplasmic/nuclear ratio and are more compact. In the mouse, the proximal luminal cells do not display notable secretory vesicles or organelles involved in protein production. However, these proximal luminal cells have a high density of mitochondria [8], which is sometimes described as a feature of progenitor cells, such as hematopoietic stem cells [10].
Earlier studies on heterogeneity of the prostate epithelium
Multiple lines of evidence have previously suggested the presence of heterogeneity within the basal and luminal compartments of the prostate epithelium. Notably, lineage-tracing in genetically-engineered mouse models has indicated the presence of distinct progenitor populations in specific contexts [11, 12]. In the hormonally-intact adult prostate, lineage-tracing studies have found that both the luminal and basal compartments are predominantly maintained by unipotent progenitors during homeostasis [13–15]. However, in response to perturbations such as tissue wounding or inflammation, basal epithelial cells can acquire bipotent properties and repair the epithelium by differentiating into luminal cells [16, 17]. In addition, basal bipotent progenitors can be detected in vivo during organogenesis by lineage-tracing in the pre-pubertal prostate for the first four weeks of post-natal development [18–20].
Luminal epithelial cells can also display bipotency in organoid culture and tissue recombination assays after isolation from dissociated prostate tissue [21, 22]. During organogenesis, bipotent luminal progenitors can also be detected, but are more short-lived than bipotent basal progenitors [23]. Luminal progenitors have also been described in the context of the regressed prostate after castration, in which castration-resistant Nkx3.1-expressing cells (CARNs) display bipotency in vivo, in organoid culture, and in tissue recombination assays [21, 24, 25]. Finally, a luminal progenitor that expresses the Sca-1 cell surface marker was shown to have bipotent features in organoid culture [26].
Epithelial heterogeneity has also been described along the proximal-distal axis in the mouse prostate. Analyses of prostate tissue following serial regression-regeneration cycles due to repeated androgen depletion and restoration in vivo identified BrdU label-retaining cells (LRCs) in the proximal prostate [27], which might correspond to slow-cycling progenitors. Another study using BrdU labeling during organogenesis found that LRCs were predominantly located in the proximal prostate epithelium versus the distal epithelium, and that the luminal epithelial compartment contained more LRCs than the basal compartment [28]. Similarly, a study using a histone-GFP fusion protein to mark LRCs during organogenesis identified a proximal luminal population with bipotent progenitor properties [29]. Consistent with these findings, a study of serial tissue recombination and renal grafting using proximal versus distal prostate tissue fragments found that only proximal regions could sustain serial grafting [30]. In addition, studies using flow-sorted cells from dissociated tissue have also suggested greater progenitor capacity in proximal prostate relative to distal regions [31–33]. Taken together, these studies have indicated regional differences within the prostate that correlate with epithelial heterogeneity and progenitor function.
Single-cell RNA sequencing analyses of the mouse prostate
Recent studies by multiple groups have employed single-cell RNA sequencing (scRNA-seq) technology to analyze the heterogeneity of the mouse prostate epithelium [8, 34–38]. While each study has discrete features, in combination they yield several major findings. First, there are distinct populations of luminal epithelial cells that differ in their transcriptional profiles (Fig. 1B). Our study of the entire prostate found that the distal luminal cells from each lobe correspond to distinct lobe-specific populations [8]. Consistent with this conclusion, three other studies of the entire prostate also observed multiple distal luminal populations [34, 35, 37]. Although one paper that examined a basal-enriched scRNA-seq dataset reported heterogeneity within the basal population [39], multiple other studies have described a single contiguous population of basal cells, whether a single lobe or the entire prostate was analyzed [8, 34–37] (Fig. 1B). Notably, the basis for the discrepancy between the single basal population described in these scRNA-seq analyses and the apparent multiple basal populations reported in previous literature [11, 12] remains unclear at present.
Second, there is a proximally-enriched luminal population within each lobe that represents a distinct population from the lobe-specific distal luminal populations (Fig. 1B,C). This proximal-distal distinction between luminal populations was observed within the anterior lobe in isolation [36], as well as in each lobe of the whole prostate [8, 34, 35, 37] (Fig. 1C). Unlike the distal luminal cells, which express common markers such as Nkx3.1 as well as secretory proteins that are more lobe-specific, the proximal luminal population is similar in each lobe, expressing a distinct cytokeratin profile as well as markers such as Sca-1 (Ly6a), Psca, and Trop2 (Tacstd2). This proximal luminal population can additionally be distinguished by its expression of Runx1 as well as Cytokeratin 4 (CK4), which can be used as the basis for lineage-tracing using corresponding Cre drivers (though it should be noted that many of these drivers also mark some basal cells) [34, 37]. Interestingly, rare single cells with features of the proximal luminal cells can be found in the distal regions of each lobe, where they are dispersed among the distal luminal cells [8, 34, 36] (Fig. 1C). At the bioinformatic level, gene ontology analyses revealed that the proximal luminal population is enriched for epithelial differentiation and tissue development pathways, and trajectory analysis indicated a trajectory from proximal to distal luminal populations [34], suggesting potential progenitor properties of proximal luminal cells.
Furthermore, several novel epithelial populations have been identified by individual scRNA-seq studies; these may be unique to specific studies due to differences in methodology or approach. In our study, we identified a unique periurethral cell population that is located near the connection of each lobe with the urethra, and is distinct from the proximal luminal population [8] (Fig. 1A,B). Cells from this periurethral population exhibit both luminal and basal features and share some transcriptional as well as histological features with both the urethral epithelium and the proximal luminal population. Importantly, the periurethral cells as well as the proximal luminal cells are prostatic in origin, since these populations are marked by lineage tracing with the Nkx3.1-Cre driver, which specifically marks the prostate epithelium and not the adjacent urethra [8, 40]; this finding argues against the conclusion that these populations are urethral in origin [35]. Notably, there is also evidence for this periurethral population in an independent study that analyzed the full proximal-distal extent of the prostate [35]. In addition, a rare Foxi1-positive luminal population that resembles ionocytes of the lung was identified in analyses of the anterior lobe [36].
Finally, these scRNA-seq studies have also identified different stromal and immune cell populations in the mouse prostate. At least two distinct mesenchymal populations with fibroblast-like characteristics have been identified in multiple studies [8, 36, 41] (Fig. 1B). One of these mesenchymal populations expresses signaling pathway molecules, whereas the second population expresses complement cascade, defense, and extracellular matrix molecules. In addition, a third stromal population of myofibroblasts has been described [41]. The first of these mesenchymal populations may correspond to stromal cells in the proximal region that express non-canonical Wnt ligands to inhibit proliferation of proximal epithelial cells [42]. In contrast, the second mesenchymal population inhibits the ability of basal cells to generate luminal cells in organoid culture [41].
It is important to note that there are several significant limitations that can lead to inconsistencies between these scRNA-seq studies of the prostate epithelium. For example, proximal populations can be missed or undetected if the corresponding tissue regions are not dissected, and tissue dissociation methods can differ between studies. Moreover, enzymatic digestion to dissociate tissues to single cells can introduce cellular stress and transcriptional changes that can also vary between populations, potentially leading to differences in population clustering. Furthermore, filtration methods to obtain single cells can lead to under-representation of larger and/or asymmetrically shaped cells, such as neuroendocrine cells and stromal cells. Finally, bioinformatic methods for analysis of scRNA-seq data can differ between studies. Since the majority of scRNA-seq data corresponds to noise [8, 43], methods to reduce noise and increase focus on true biological signals can lead to improved identification and clustering of cell populations.
Single-cell analyses of the human prostate
In spite of their anatomical and histological differences, single-cell RNA-sequencing has shown that the human prostate shares several major features with the mouse prostate. As in the mouse, human basal cells represent a single contiguous population, whereas there are multiple luminal populations [8, 34, 44]. Each zone of the human prostate contains a secretory acinar population, which resemble mouse distal luminal cells, as well as a ductal population with fewer secretory transcripts, which may resemble mouse proximal luminal cells [8, 34]. Notably, a human cell population with similarity to the mouse periurethral population was also identified, supporting the relevance of this novel population [8] (Fig. 1D).
Additional epithelial populations have been described in another single-cell study of the human prostate [44], which reported two novel populations resembling “hillock” and “club” cells that were first described in the lung epithelium. Hillock cells express basal markers, occur in multiple stratified layers, and are found closer to the urethra. Club cells are a subset of luminal cells that are not multi-layered, and are also found closer to the urethra. Both of these populations are predominantly found in the central and transition zones, whereas the conventional luminal cells are enriched in the peripheral zone [44].
Since the human prostate lacks a lobular structure, the relationship of the individual mouse prostate lobes to the human prostate has long remained unclear. Although there has not been a consensus regarding the relationship of mouse lobes to the human prostate [45–47], earlier gene expression profiling had suggested that mouse dorsolateral prostate was most similar to the human peripheral zone epithelium [3]. Interestingly, bioinformatic analyses of scRNA-seq data suggest that the human acinar luminal cells are most similar to the distal luminal cells from the mouse lateral lobe, but are relatively dissimilar to the distal luminal cells from the dorsal lobe [8] (Fig. 1D). Importantly, this finding implies that future studies of mouse prostate should distinguish between the dorsal and lateral lobes when feasible, particularly in the context of cancer models.
Functional and lineage-tracing analyses of mouse prostate epithelial populations
Based on these scRNA-seq data, the distinct epithelial populations of the mouse prostate can be isolated by tissue dissociation and flow cytometry, followed by analysis in functional assays. In general, these studies have shown consistent findings for basal epithelial cells, which efficiently generate organoids that contain both basal and luminal cells [8, 36] (Fig. 2A,B). Proximal luminal cells can also generate organoids efficiently, although less readily than basal cells, with the resulting organoids containing both basal and luminal cells [8, 34, 36, 37]. Periurethral cells also have elevated organoid formation efficiency, comparable to that of basal cells [8]. In contrast, distal luminal cells have a significantly reduced capacity to form organoids, even when a large number of cells are plated [8], indicating that basal, periurethral, and proximal luminal cells have higher progenitor potential in this organoid assay.
Figure 2. Functional analyses of mouse prostate epithelial populations in organoid and tissue reconstitution assays.
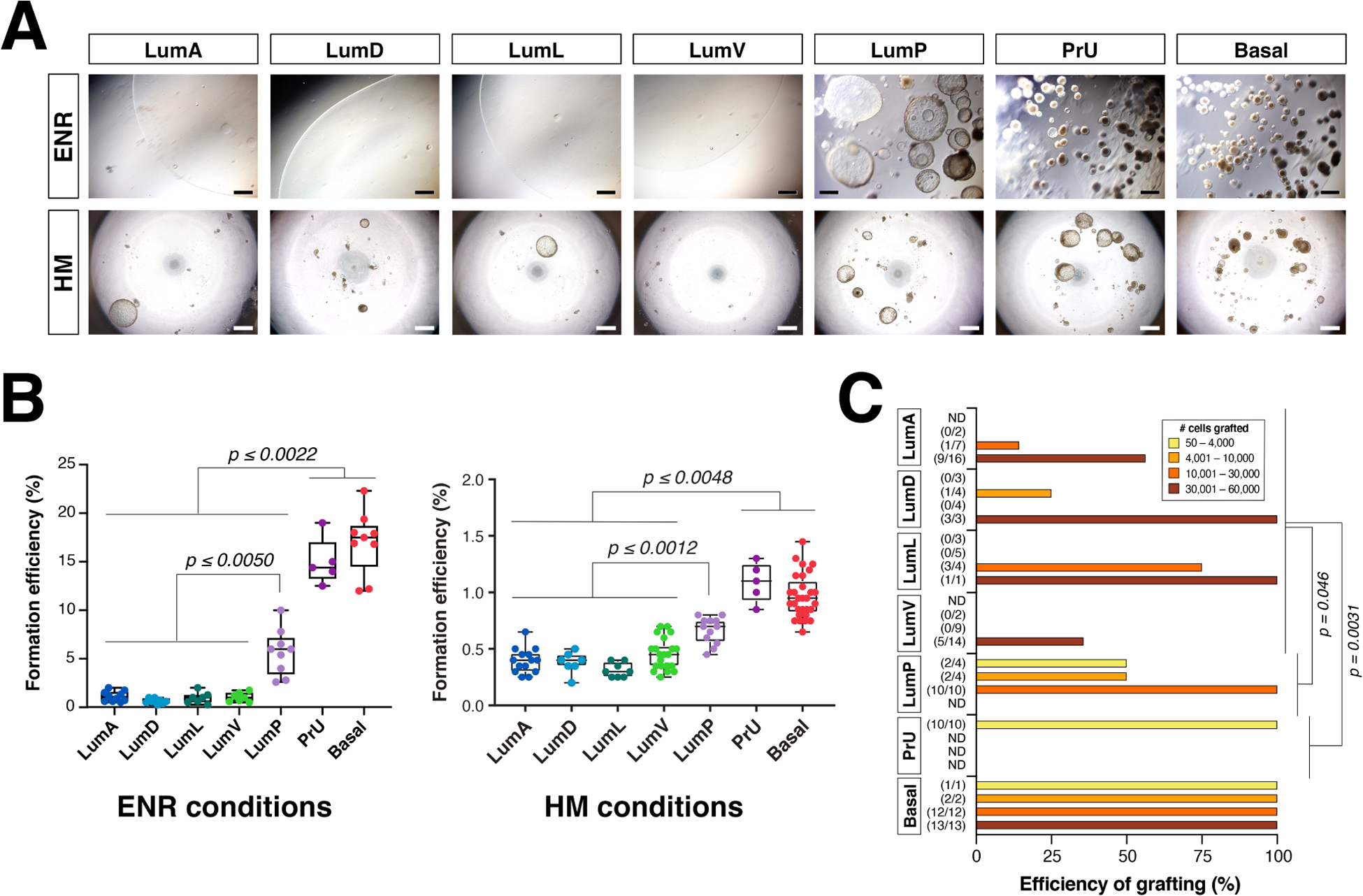
(A) Bright-field views of organoids grown from sorted epithelial cells of the indicated populations in two distinct culture conditions (ENR, Epidermal growth factor, Noggin, and R-spondin-1-containing media [54]; HM, hepatocyte media). (B) Organoid formation efficiency plots in the two culture conditions. (C) Grafting efficiency in tissue reconstitution assays. LumP, PrU, and Basal populations are significantly more efficient at generating grafts from smaller number of cells relative to distal luminal populations (LumA, LumD, LumL, and LumV). Adapted from [8].
These functional distinctions between epithelial populations were also evident in tissue reconstitution assays. In this approach, dissociated epithelial cells are combined with embryonic urogenital mesenchyme cells, followed by renal grafting in an immunodeficient mouse host. Notably, basal as well as proximal luminal cells could form fully differentiated grafts from significantly fewer input cells than distal luminal cells (for example, 500 proximal luminal cells versus 20,000 distal luminal cells) [8] (Fig. 2C). Lobe-specific distal luminal populations could only generate basal cells as well as luminal cells of the same type and lobe-specificity in the resulting grafts, which typically were smaller and less differentiated. In contrast, basal, proximal luminal, and periurethral populations could generate well-differentiated grafts with multiple cell types, and proximal luminal cells were the most proficient at generating grafts containing multiple different distal luminal populations [8]. Thus, both organoid and tissue reconstitution assays indicate that basal, proximal luminal, and periurethral populations have greater proliferative and multipotent potential.
Lineage-tracing and explant culture assays have also been performed to assess the properties of these epithelial populations during development, homeostasis, and castration/regeneration. Two studies have used Cre-mediated lineage-tracing to examine the properties of proximal luminal cells. Lineage-marking of Runx1-positive cells in explanted urogenital sinus prior to prostate formation resulted in labeling of epithelial clones that respected the proximal-distal boundary, suggesting that proximal luminal cells are not progenitors of distal luminal cells [37]. Consistent with this finding, lineage-tracing of Ck4-positive and Psca-positive proximal cells through serial regression-regeneration cycles in vivo resulted in luminal clones that were restricted to the proximal region [34]. Interestingly, lineage-tracing of rare distal Ck4-positive or Psca-positive cells with proximal luminal features resulted in small distal clones that contained both proximal-like as well as distal-like luminal cells [34]. Taken together, these findings support the progenitor properties of proximal luminal cells in vivo, but also suggest that the function of these progenitors can vary depending on tissue context.
Furthermore, scRNA-seq and functional studies have provided new insights into interactions between the epithelial and stromal compartments in the prostate. Notably, analyses of expression of ligand-receptor pairs during regression after castration followed by androgen-mediated regeneration identified Nrg2 (neuregulin) and Fgf10 as stromal factors induced by androgens [36]. Addition of Neuregulin, and to a lesser extent Fgf10, could stimulate growth of both mouse and human prostate luminal cells in organoid culture, even when androgen receptor signaling was blocked. Interestingly, this stromal-epithelial interaction during androgen-mediated regeneration may parallel a role for Neuregulin expressed by the tumor microenvironment in promoting the emergence of castration-resistance in prostate cancer [48].
Lineage reconstruction analyses of the human prostate
Since human tissues are less experimentally accessible and cannot be manipulated in vivo using genetic engineering approaches, alternative methods have been used to investigate the clonal architecture of the human prostate epithelium. One important approach has used analyses of spatial patterns of mitochondrial DNA mutations in serial tissue sections to reconstruct lineage relationships. For example, earlier studies used this approach to analyze small regions of the prostate, and showed that basal and luminal epithelial cells could descend from a common progenitor [49, 50]. Subsequently, analyses of mutations in mitochondrial cytochrome C oxidase in serial sections of complete human prostate tissues revealed two distinct clonal patterns [51]. First, clones consisting of a small number of cells were found throughout the ducts without any apparent organization. Secondly, rare large clones (with greater than 100,000 cells) formed contiguous ribbons that stretched from near the urethra to the distal end of individual ducts. Since the cells at the proximal end of these large clones were predominantly basal, these findings suggested that multipotent basal cells near the urethral junction could generate the full extent of the prostate epithelium [51]. As proposed in this study, these urethra-adjacent progenitors generate progeny that migrate distally to form ribbons, such that the distal ducts tend to be monoclonal [51]. However, an important limitation of this study is that the lineage reconstruction methodology does not readily yield information about the temporal context in which these clones arose.
Recent work has addressed this issue by reconstructing clonal hierarchies from mutational phylogenies inferred from patterns of somatic mutations based on whole-genome sequencing [52]. This study concluded that the large clones extending from the urethra to the distal tips originate during fetal development, and large clones extending from the base of a duct to the distal tip arise during pubertal development. Small clones, whether located proximally or distally, were generated during adult tissue maintenance. Interestingly, mutational burden and telomere shortening increased in a proximal-distal direction, suggesting that distal cells have undergone greater numbers of cell divisions [52].
In combination, these studies raise the possibility that a periurethral progenitor population may be responsible for the generation of large clonal ribbons during human prostate organogenesis. In particular, these large clones mostly had basal cells at their proximal ends, but 12% had luminal cells, and additional clones contained luminal cells with 6–8 cells of their proximal tip [51], potentially consistent with the mixed basal/luminal features of the mouse periurethral population [8] (Fig. 1D). In contrast, the small clones generated during adult prostate homeostasis are consistent with the clones produced by proximal luminal cells in both proximal and distal locations in the mouse [34]. Thus, if this interpretation is correct, distinct epithelial progenitors would play different roles during development versus homeostasis of the human prostate.
Perspectives and conclusions
It is increasingly evident that cell behaviors can be different in normal physiology versus abnormal or perturbed states. In this regard, the progenitor characteristics of the proximal luminal population might be highly context-dependent. Notably, proximal luminal cells and periurethral cells display enhanced progenitor characteristics in organoid and tissue reconstitution assays [8, 34, 36]. In contrast, proximal luminal cells do not show enhanced progenitor features relative to distal luminal cells in tissue explants at developmental stages [37]. In adult prostate homeostasis, proximal luminal cells also do not show increased ability to generate ribbons of progeny, and their clonal descendants do not cross the proximal/distal boundary [34, 36]. Together, these data suggest that proximal luminal cells do not exhibit significant progenitor function in normal development and homeostasis. However, in altered contexts such as tissue repair during wound healing, it is conceivable that they might acquire multipotent progenitor properties. Notably, basal epithelial cells play a similar role in tissue repair processes in the prostate and other tissues [15, 17, 53]. Thus, proximal luminal cells may represent a plastic reserve progenitor population that can respond to injury.
Such epithelial plasticity may also be relevant for understanding the origins of prostatic diseases. If distinct cell populations respond differentially to oncogenic insults, understanding the cell of origin may provide insights into appropriate disease therapy. For example, distinct epithelial populations display different responses to androgen-deprivation, which represents the mainstay of prostate cancer treatment. In particular, basal cells, proximal luminal cells, and periurethral cells are largely unaffected by castration, whereas distal luminal populations shift their transcriptomic profiles to acquire proximal-like features [36, 37], suggesting that the proximal luminal population could be relevant for understanding the response to androgen deprivation. Thus, a deeper understanding of the molecular and functional properties of these populations may provide key insights into prostate cancer initiation, progression, and therapy-response.
Table 1.
Summary of mouse prostate cell populations reported by single-cell RNA-sequencing studies.a
| Author | Crowley 2020 | Karthaus 2020b | Guo 2020 | Joseph 2020 | Mevel 2020 | Kwon 2019c |
|---|---|---|---|---|---|---|
| Reference | [8] | [36] | [34] | [35] | [37] | [41] |
| Population | ||||||
| Basal | Basal | Basal | Basal | Basal | Basal | |
| Distal luminal | ||||||
| Anterior | LumA | Lum1 | LumB | LumAP | LumB | |
| Dorsal | LumD | LumB | LumDLP | LumB | ||
| Lateral | LumL | LumB | LumDLP | LumC | ||
| Ventral | LumV | LumA | LumVP | LumA | ||
| Proximal luminal | LumP | Lum2 | LumC | Urethral | LumD | |
| Periurethral | PrU | Urethral | ||||
| Ionocytes | Lum3 | |||||
| Stromal 1 | Stromal 1 | Mes 1 | Stromald | R1 | ||
| Stromal 2 | Stromal 2 | Mes 2 | Stromald | R2 | ||
| Stromal 3 | R3 |
Blank cells indicate that the population was not described or detected. Shaded cells indicate that the corresponding tissue or cell types were not analyzed. Immune populations are not shown in this table.
This study only analyzed the anterior prostate lobe.
This study only analyzed stromal populations.
This study identified two distinct stromal populations, but their relationship to the stromal populations in other studies is unclear.
Highlights.
Single-cell RNA-seq has shown extensive heterogeneity in the prostate epithelium
Luminal epithelial cells are divided between proximal and distal compartments
Proximal luminal cells display greater progenitor potential in ex vivo assays
Prostate epithelial heterogeneity is conserved between mouse and human
Acknowledgements
We apologize to many colleagues whose work has not been cited due to space constraints. This work was supported by grants from the National Institutes of Health R01CA238005 and R01CA251527 (M.M.S.), the Prostate Cancer Foundation (M.M.S.), and National Science Foundation DGE-16-44869 (L.C.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer Statistics, 2021, CA Cancer J Clin, 71 (2021) 7–33. [DOI] [PubMed] [Google Scholar]
- [2].Toivanen R, Shen MM, Prostate organogenesis: tissue induction, hormonal regulation and cell type specification, Development, 144 (2017) 1382–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Berquin IM, Min Y, Wu R, Wu H, Chen YQ, Expression signature of the mouse prostate, J Biol Chem, 280 (2005) 36442–36451. [DOI] [PubMed] [Google Scholar]
- [4].Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y, The endocrinology and developmental biology of the prostate, Endocr Rev, 8 (1987) 338–362. [DOI] [PubMed] [Google Scholar]
- [5].Kinbara H, Cunha GR, Boutin E, Hayashi N, Kawamura J, Evidence of stem cells in the adult prostatic epithelium based upon responsiveness to mesenchymal inductors, Prostate, 29 (1996) 107–116. [DOI] [PubMed] [Google Scholar]
- [6].McNeal JE, The zonal anatomy of the prostate, Prostate, 2 (1981) 35–49. [DOI] [PubMed] [Google Scholar]
- [7].McNeal JE, Anatomy of the prostate and morphogenesis of BPH, Prog Clin Biol Res, 145 (1984) 27–53. [PubMed] [Google Scholar]
- [8].Crowley L, Cambuli F, Aparicio L, Shibata M, Robinson BD, Xuan S, Li W, Hibshoosh H, Loda M, Rabadan R, Shen MM, A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors, eLife, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fullwood NJ, Lawlor AJ, Martin-Hirsch PL, Matanhelia SS, Martin FL, An analysis of benign human prostate offers insights into the mechanism of apocrine secretion and the origin of prostasomes, Sci Rep, 9 (2019) 4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].de Almeida MJ, Luchsinger LL, Corrigan DJ, Williams LJ, Snoeck HW, Dye-independent methods reveal elevated mitochondrial mass in hematopoietic stem cells, Cell Stem Cell, 21 (2017) 725–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li JJ, Shen MM, Prostate stem cells and cancer stem cells, Cold Spring Harb Perspect Med, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Xin L, Cells of origin for prostate cancer, Adv Exp Med Biol, 1210 (2019) 67–86. [DOI] [PubMed] [Google Scholar]
- [13].Choi N, Zhang B, Zhang L, Ittmann M, Xin L, Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation, Cancer Cell, 21 (2012) 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu J, Pascal LE, Isharwal S, Metzger D, Ramos Garcia R, Pilch J, Kasper S, Williams K, Basse PH, Nelson JB, Chambon P, Wang Z, Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate, Mol Endocrinol, 25 (2011) 1849–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang ZA, Mitrofanova A, Bergren SK, Abate-Shen C, Cardiff RD, Califano A, Shen MM, Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity, Nat Cell Biol, 15 (2013) 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kwon OJ, Zhang L, Ittmann MM, Xin L, Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin, Proc Natl Acad Sci USA, 111 (2014) E592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Toivanen R, Mohan A, Shen MM, Basal progenitors contribute to repair of the prostate epithelium following induced luminal anoikis, Stem cell reports, 6 (2016) 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ousset M, Van Keymeulen A, Bouvencourt G, Sharma N, Achouri Y, Simons BD, Blanpain C, Multipotent and unipotent progenitors contribute to prostate postnatal development, Nat Cell Biol, 14 (2012) 1131–1138. [DOI] [PubMed] [Google Scholar]
- [19].Tika E, Ousset M, Dannau A, Blanpain C, Spatiotemporal regulation of multipotency during prostate development, Development, 146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wuidart A, Ousset M, Rulands S, Simons BD, Van Keymeulen A, Blanpain C, Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells, Genes Dev, 30 (2016) 1261–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, Badani KK, McKiernan JM, Benson MC, Hibshoosh H, Shen MM, Single luminal epithelial progenitors can generate prostate organoids in culture, Nat Cell Biol, 16 (2014) 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, Vries RG, Cuppen E, Chen Y, Sawyers CL, Clevers HC, Identification of multipotent luminal progenitor cells in human prostate organoid cultures, Cell, 159 (2014) 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shibata M, Epsi NJ, Xuan S, Mitrofanova A, Shen MM, Bipotent progenitors do not require androgen receptor for luminal specification during prostate organogenesis, Stem cell reports, 15 (2020) 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chua CW, Epsi NJ, Leung EY, Xuan S, Lei M, Li BI, Bergren SK, Hibshoosh H, Mitrofanova A, Shen MM, Differential requirements of androgen receptor in luminal progenitors during prostate regeneration and tumor initiation, eLife, 7 (2018) e28768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu HL, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM, A luminal epithelial stem cell that is a cell of origin for prostate cancer, Nature, 461 (2009) 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kwon OJ, Zhang L, Xin L, Stem Cell Antigen-1 identifies a distinct androgen-independent murine prostatic luminal cell lineage with bipotent potential, Stem Cells, 34 (2016) 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsujimura A, Koikawa Y, Salm S, Takao T, Coetzee S, Moscatelli D, Shapiro E, Lepor H, Sun TT, Wilson EL, Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis, J Cell Biol, 157 (2002) 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ceder JA, Aalders TW, Schalken JA, Label retention and stem cell marker expression in the developing and adult prostate identifies basal and luminal epithelial stem cell subpopulations, Stem Cell Res Ther, 8 (2017) 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang D, Jeter C, Gong S, Tracz A, Lu Y, Shen J, Tang DG, Histone 2B-GFP label-retaining prostate luminal cells possess progenitor cell properties and are intrinsically resistant to castration, Stem cell reports, 10 (2018) 228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Goto K, Salm SN, Coetzee S, Xiong X, Burger PE, Shapiro E, Lepor H, Moscatelli D, Wilson EL, Proximal prostatic stem cells are programmed to regenerate a proximal-distal ductal axis, Stem Cells, 24 (2006) 1859–1868. [DOI] [PubMed] [Google Scholar]
- [31].Burger PE, Xiong X, Coetzee S, Salm SN, Moscatelli D, Goto K, Wilson EL, Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue, Proc Natl Acad Sci USA, 102 (2005) 7180–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xin L, Lawson DA, Witte ON, The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis, Proc Natl Acad Sci USA, 102 (2005) 6942–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON, Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics, Proc Natl Acad Sci USA, 105 (2008) 20882–20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Guo W, Li L, He J, Liu Z, Han M, Li F, Xia X, Zhang X, Zhu Y, Wei Y, Li Y, Aji R, Dai H, Wei H, Li C, Chen Y, Chen L, Gao D, Single-cell transcriptomics identifies a distinct luminal progenitor cell type in distal prostate invagination tips, Nat Genet, 52 (2020) 908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Joseph DB, Henry GH, Malewska A, Iqbal NS, Ruetten HM, Turco AE, Abler LL, Sandhu SK, Cadena MT, Malladi VS, Reese JC, Mauck RJ, Gahan JC, Hutchinson RC, Roehrborn CG, Baker LA, Vezina CM, Strand DW, Urethral luminal epithelia are castration-insensitive cells of the proximal prostate, Prostate, 80 (2020) 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Karthaus WR, Hofree M, Choi D, Linton EL, Turkekul M, Bejnood A, Carver B, Gopalan A, Abida W, Laudone V, Biton M, Chaudhary O, Xu T, Masilionis I, Manova K, Mazutis L, Pe’er D, Regev A, Sawyers CL, Regenerative potential of prostate luminal cells revealed by single-cell analysis, Science, 368 (2020) 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mevel R, Steiner I, Mason S, Galbraith LC, Patel R, Fadlullah MZ, Ahmad I, Leung HY, Oliveira P, Blyth K, Baena E, Lacaud G, RUNX1 marks a luminal castration-resistant lineage established at the onset of prostate development, eLife, 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Barros-Silva JD, Linn DE, Steiner I, Guo G, Ali A, Pakula H, Ashton G, Peset I, Brown M, Clarke NW, Bronson RT, Yuan GC, Orkin SH, Li Z, Baena E, Single-cell analysis identifies LY6D as a marker linking castration-resistant prostate luminal cells to prostate progenitors and cancer, Cell reports, 25 (2018) 3504–3518 e3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang X, Xu H, Cheng C, Ji Z, Zhao H, Sheng Y, Li X, Wang J, Shu Y, He Y, Fan L, Dong B, Xue W, Wai Chua C, Wu D, Gao WQ, He Zhu H, Identification of a Zeb1 expressing basal stem cell subpopulation in the prostate, Nature communications, 11 (2020) 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kruithof-de Julio M, Shibata M, Desai N, Reynon M, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM, Canonical Wnt signaling regulates Nkx3.1 expression and luminal epithelial differentiation during prostate organogenesis, Dev Dyn, 242 (2013) 1160–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kwon OJ, Zhang Y, Li Y, Wei X, Zhang L, Chen R, Creighton CJ, Xin L, Functional Heterogeneity of Mouse Prostate Stromal Cells Revealed by Single-Cell RNA-Seq, iScience, 13 (2019) 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wei X, Zhang L, Zhou Z, Kwon OJ, Zhang Y, Nguyen H, Dumpit R, True L, Nelson P, Dong B, Xue W, Birchmeier W, Taketo MM, Xu F, Creighton CJ, Ittmann MM, Xin L, Spatially restricted stromal Wnt signaling restrains prostate epithelial progenitor growth through direct and Iindirect mechanisms, Cell Stem Cell, 24 (2019) 753–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Aparicio L, Bordyuh M, Blumberg AJ, Rabadan R, A random matrix theory approach to denoise single-cell data, Patterns, 1 (2020) 100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Henry GH, Malewska A, Joseph DB, Malladi VS, Lee J, Torrealba J, Mauck RJ, Gahan JC, Raj GV, Roehrborn CG, Hon GC, MacConmara MP, Reese JC, Hutchinson RC, Vezina CM, Strand DW, A cellular anatomy of the normal adult human prostate and prostatic urethra, Cell reports, 25 (2018) 3530–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ittmann M, Anatomy and histology of the human and murine prostate, Cold Spring Harb Perspect Med, 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, Simons BW, Ward JM, Robinson BD, Chu GC, Loda M, Thomas G, Borowsky A, Cardiff RD, Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee, Cancer Res, 73 (2013) 2718–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, Humphrey PA, Sundberg JP, Rozengurt N, Barrios R, Ward JM, Cardiff RD, Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee, Cancer Res, 64 (2004) 2270–2305. [DOI] [PubMed] [Google Scholar]
- [48].Zhang Z, Karthaus WR, Lee YS, Gao VR, Wu C, Russo JW, Liu M, Mota JM, Abida W, Linton E, Lee E, Barnes SD, Chen HA, Mao N, Wongvipat J, Choi D, Chen X, Zhao H, Manova-Todorova K, de Stanchina E, Taplin ME, Balk SP, Rathkopf DE, Gopalan A, Carver BS, Mu P, Jiang X, Watson PA, Sawyers CL, Tumor microenvironment-derived NRG1 promotes antiandrogen resistance in prostate cancer, Cancer Cell, 38 (2020) 279–296 e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Blackwood JK, Williamson SC, Greaves LC, Wilson L, Rigas AC, Sandher R, Pickard RS, Robson CN, Turnbull DM, Taylor RW, Heer R, In situ lineage tracking of human prostatic epithelial stem cell fate reveals a common clonal origin for basal and luminal cells, J Pathol, 225 (2011) 181–188. [DOI] [PubMed] [Google Scholar]
- [50].Gaisa NT, Graham TA, McDonald SA, Poulsom R, Heidenreich A, Jakse G, Knuechel R, Wright NA, Clonal architecture of human prostatic epithelium in benign and malignant conditions, J Pathol, 225 (2011) 172–180. [DOI] [PubMed] [Google Scholar]
- [51].Moad M, Hannezo E, Buczacki SJ, Wilson L, El-Sherif A, Sims D, Pickard R, Wright NA, Williamson SC, Turnbull DM, Taylor RW, Greaves L, Robson CN, Simons BD, Heer R, Multipotent basal stem cells, maintained in localized proximal niches, support directed long-ranging epithelial flows in human prostates, Cell reports, 20 (2017) 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Grossmann S, Hooks Y, Wilson L, Moore L, O’Neill L, Martincorena I, Voet T, Stratton MR, Heer R, Campbell PJ, Development, maturation, and maintenance of human prostate inferred from somatic mutations, Cell Stem Cell, 28 (2021) 1262–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Centonze A, Lin S, Tika E, Sifrim A, Fioramonti M, Malfait M, Song Y, Wuidart A, Van Herck J, Dannau A, Bouvencourt G, Dubois C, Dedoncker N, Sahay A, de Maertelaer V, Siebel CW, Van Keymeulen A, Voet T, Blanpain C, Heterotypic cell-cell communication regulates glandular stem cell multipotency, Nature, 584 (2020) 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H, Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts, Nature, 469 (2011) 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]


