Abstract
CDC42 encodes a highly conserved GTPase of the Rho family that is best known for its role in regulating cell polarity and actin organization. In addition, various studies of both yeast and mammalian cells have suggested that Cdc42p, through its interaction with p21-activated kinases (PAKs), plays a role in signaling pathways that regulate target gene transcription. However, recent studies of the yeast pheromone response pathway suggested that prior results with temperature-sensitive cdc42 mutants were misleading and that Cdc42p and the Cdc42p-PAK interaction are not involved in signaling. To clarify this issue, we have identified and characterized novel viable pheromone-resistant cdc42 alleles that retain the ability to perform polarity-related functions. Mutation of the Cdc42p residue Val36 or Tyr40 caused defects in pheromone signaling and in the localization of the Ste20p PAK in vivo and affected binding to the Ste20p Cdc42p-Rac interactive binding (CRIB) domain in vitro. Epistasis analysis suggested that they affect the signaling step at which Ste20p acts, and overproduction of Ste20p rescued the defect. These results suggest that Cdc42p is in fact required for pheromone response and that interaction with the PAK Ste20p is critical for that role. Furthermore, the ste20ΔCRIB allele, previously used to disrupt the Cdc42p-Ste20p interaction, behaved as an activated allele, largely bypassing the signaling defect of the cdc42 mutants. Additional observations lead us to suggest that Cdc42p collaborates with the SH3-domain protein Bem1p to facilitate signal transduction, possibly by providing a cell surface scaffold that aids in the local concentration of signaling kinases, thus promoting activation of a mitogen-activated protein kinase cascade by Ste20p.
In the budding yeast Saccharomyces cerevisiae, cells of the a and α mating types secrete pheromones that bind to G-protein-coupled receptors on the surfaces of cells of the opposite mating type, initiating a signaling cascade in which the βγ subunits of the G protein promote the activation of a mitogen-activated protein kinase (MAPK) cascade (4). This process appears to involve the binding of at least two proteins, the upstream kinase Ste20p and the scaffold protein Ste5p, to the liberated Gβγ subunits (11, 25, 40, 51). MAPK activation then promotes cell cycle arrest in G1 and stimulates the expression of several genes, including FUS1, leading ultimately to fusion of the mating partners (22).
The small GTPase Cdc42p and its guanine nucleotide exchange factor Cdc24p are essential for polarity establishment and subsequent bud formation (1, 38). In addition to their roles in cell polarity, these proteins have been proposed to play roles in signal transduction in response to mating pheromones (46, 47, 53). Temperature-sensitive cdc42 and cdc24 mutants have defects in α-factor-stimulated transcription of FUS1 and in maintaining G1 arrest at the restrictive temperature (46, 53). In addition, an interaction was detected by two-hybrid analysis between Gβ and Cdc24p (53), and Cdc42p-GTP was shown to bind to and activate Ste20p (46). These findings led to the hypothesis that Gβγ activated Cdc24p, causing GTP loading of Cdc42p and consequent activation of Ste20p, as an important part of the pheromone signaling pathway.
More recent experiments have cast doubt upon the existence of a Gβγ-Cdc24p-Cdc42p-Ste20p pathway. Mutations in CDC24 that abolished detectable interaction with Gβγ did not cause any defects in α-factor-stimulated FUS1 transcription or G1 arrest but rather were specifically defective in orientation of the mating projection towards the mating partner (33). Furthermore, mutations in STE20 that abolished detectable interaction with Cdc42p were also reported to be wild type with regard to α-factor-stimulated FUS1 transcription, G1 arrest, and mating (23, 37). Together, these studies indicated that neither the Gβγ-Cdc24p interaction nor the Cdc42p-Ste20p interaction was important for α-factor signaling. Furthermore, the polarity defect exhibited by temperature-sensitive cdc24 and cdc42 mutants triggers the morphogenesis checkpoint to delay the cells in G2 with abundant G1 cyclins (26), a state known to render cells unresponsive to α-factor (34). This raised the possibility that the signaling defect of these mutants might be an indirect consequence of their accumulation at a nonresponding stage of the cell cycle. Indeed, the transcriptional induction of FUS1 was found to be quite normal in cdc24 and cdc42 mutants that were first arrested in G1 by deprivation of G1 cyclins (35), raising the question of whether Cdc24p and Cdc42p play any role at all in α-factor signaling.
As illustrated by these studies, the interpretation of cdc42 and cdc24 phenotypes is complicated by the possibility of indirect effects stemming from the well-characterized polarity defects caused by these mutants at the restrictive temperature. To circumvent these problems, we performed a screen to identify α-factor-resistant alleles of CDC42 that could still perform polarization functions. In this paper we report the isolation and characterization of such mutants, supporting the notion that Cdc42p has some direct role in pheromone signaling. Our results further suggest that this signaling function operates through Cdc42p-dependent activation and localization of Ste20p.
MATERIALS AND METHODS
Yeast media and cell synchrony.
Yeast media (YEPD rich medium, synthetic complete medium lacking specific nutrients, and sporulation medium) have been described previously (13). YEPG and YEPS are the same as YEPD but with 2% galactose or sucrose instead of dextrose. Centrifugal elutriation to isolate early-G1-phase cells was performed as described previously (27).
Strains, plasmids, and PCR manipulations.
Standard media and methods were used for plasmid manipulations (2) and yeast genetic manipulations (13). The S. cerevisiae strains used in this study are listed in Table 1, and the plasmids used are listed in Table 2.
TABLE 1.
Strains used in this studya
| Strain name | Genotype |
|---|---|
| DLY1 | MATa bar1 |
| DLY3067 | MATa bar1 cdc42::LEU2::GAL1p-CDC42 |
| MOSY0023 | MATa bar1 cdc42::LEU2::GAL1p-CDC42 cla4::TRP1 |
| MOSY0090 | MATa bar1 cdc42::URA3 leu2::GAL1p-CDC42::LEU2 |
| MOSY0095 | MATα bar1 cdc42::LEU2::GAL1p-CDC42 |
| MOSY0106 | MATa bar1 cdc42::LEU2::GAL1p-CDC42 ste20::TRP1 |
| MOSY0178 | MATa bar1 cdc42-md1 |
| MOSY0252 | MATa bar1 ste20ΔCRIB |
| MOSY0268 | MATa bar1 cdc42::LEU2::GAL1p-CDC42 ste20ΔCRIB |
| MOSY0270 | MATa bar1 ste20ΔCRIB bem1::URA3 |
| JMY1128 | MATa bar1 bem1::URA3 |
| PPY911 | MATa bar1 cdc42-1 FUS1::FUS1-lacZ::HIS2 |
All strains are in the BF264-15Du (41a) background (ade1 his2 leu2-3,112 trp1-1 ura3Δns).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| pDLB377 | LEU2 GAL1p-CDC42 | See text |
| pDLB642 | pCR2.1 CDC42p-TDH3t | See text |
| pDLB643 | pCR2.1 CDC42p-CDC42-TDH3t | See text |
| pDLB644 | CEN URA3 CDC42p-TDH3t | See text |
| pDLB722 | 2μm URA3 CLA4 | Erfei Bi |
| pDLB1034 | pUNI-10 CDC42-Q61L | See text |
| pDLB1035 | pUNI-10 CDC42-D57Y | See text |
| pDLB1160 | pUNI-10 CDC42-V36A,Q61L | See text |
| pDLB1234 | pHB1-MYC3 CDC42-D57Y | See text |
| pDLB1235 | pHB1-MYC3 CDC42-Q61L | See text |
| pDLB1240 | pHB1-MYC3 CDC42-V36A,Q61L | See text |
| pDLB1242 | pHB1-MYC3 CDC42-Y40C,Q61L | See text |
| pDLB1277 | pUNI-10 CDC42-Y40C,Q61L | See text |
| pG11-4 | CEN URA3 GAL1p-STE11-4 | See text |
| pGEX-Ste20CRIB | GST-STE20-CRIB | See text |
| pGS11ΔN-T | CEN TRP1 GAL1p-GST-STE11ΔN | See text |
| pGFP-GS5ΔN-CTM | CEN TRP1 GAL1p-GFP-STE5ΔN-CTM | 40 |
| pGFP-GS5ΔN-Sec22 | CEN TRP1 GAL1p-GFP-STE5ΔN-Sec22 | 40 |
| pL19 | CEN URA3 GAL1p-STE4 | 50 |
| pL-GS5ΔN-CTM | CEN LEU2 GAL1p-STE5ΔN-CTM | 40 |
| pMOSB16 | pCR2.1 cdc42-V36A | See text |
| pMOSB29 | LEU2 GAL1p-CDC42-D57Y | See text |
| pMOSB36 | CEN URA3 cdc42-md1 | See text |
| pMOSB37 | CEN URA3 cdc42-md2 | See text |
| pMOSB38 | CEN URA3 cdc42-md12 | See text |
| pMOSB42 | CEN TRP1 cdc42-md1 | See text |
| pMOSB43 | CEN TRP1 cdc42-md2 | See text |
| pMOSB44 | CEN TRP1 cdc42-md12 | See text |
| pMOSB45 | CEN TRP1 CDC42 | See text |
| pMOSB47 | CEN TRP1 cdc42-V36A | See text |
| pMOSB53 | pCR2.1 cdc42-Y40C | See text |
| pMOSB55 | CEN URA3 CDC42 | See text |
| pMOSB134 | URA3 ste20ΔCRIB | See text |
| pMOSB175 | CEN TRP1 cdc42-Y40C | See text |
| pMOSB176 | CEN URA3 cdc42-Y40C | See text |
| pMOSB177 | CEN URA3 cdc42-V36A | See text |
| pNC252 | 2μm URA3 GAL1p-STE12 | 39 |
| pPB321 | 2μm URA3 BEM1 | 5 |
| pPP828 | CEN LEU2 GFP-STE20 | See text |
| pRL116 | CEN URA3 GFP-STE20 | 23 |
| pSB231 | CEN URA3 FUS1-lacZ | 48 |
| pVTU-Ste20 | 2μm URA3 ADH1p-STE20 | 21 |
| p3058 | CEN LEU2 FUS1p-lacZ | C. Boone |
To generate the GAL1p-CDC42 allele, the oligonucleotides DJL42-1 (5′-GC CGGAACTCAAAAGGGTAATTTCGTGAAAAACAATCATCGACTACGT CGTAAGGCCG-3′ and DJL42-2 (5′-TCAGTAGAAGGATATGACAAGG G-3′) were used to amplify a DNA fragment containing the LEU2 gene, the GAL1 promoter, and flanking CDC42 sequences using PCR with the plasmid pGAL-CDC42Sc (55) as a template (the underlined sequence in DJL42-1 is the genomic sequence 733 to 693 bp upstream of the CDC42 start codon, whereas in DJL42-2 it is the reverse complement of nucleotides 204 to 226 in the CDC42 open reading frame). The PCR fragment was transformed into DLY1, replacing the endogenous CDC42 promoter (1 to 693 bp upstream of the start codon) with LEU2 and the GAL1 promoter, creating DLY3067. Leu+ transformants were selected on galactose-containing plates, and promoter replacement was confirmed by the inability to grow on dextrose-containing plates (GAL1 promoter off) and by PCR.
To generate the cdc42::URA3 null allele, the oligonucleotides CDC42-5′ (5′-CTATTTTCCTGAGGAGATAGGTTAACAAACGAATTAGAGAAGGCGCGTTTCGGTGATGAC-3′) and CDC42-3′ (5′-GAGGCTCCAAGGCGGCCACGATAGCTTCATCGAATACATTCTTTCCTGATGCGGTATTTTC-3′) were used to amplify the URA3 gene using PCR with the plasmid pRS306 (45) as a template. The PCR product was transformed directly into a yeast strain containing an additional copy of CDC42 under the control of the GAL1 promoter (in this case, integrated at LEU2 using pDLB377, a YIp derivative of pGAL-CDC42Sc [55] generated by replacing the ClaI fragment containing pRS315 vector sequences with the corresponding fragment from pRS305 [45]). Ura+ transformants were selected on galactose-containing plates to replace the endogenous copy of CDC42 (from 45 bp upstream to 494 bp downstream of the start codon, removing all but the last 26 codons) with URA3, generating strain MOSY0090. Transformants were tested for the inability to grow on dextrose-containing plates (GAL1 promoter off), and gene replacement was confirmed by PCR.
cdc42-md1 was integrated into the endogenous CDC42 locus by one-step replacement of the cdc42::URA3 allele. A 1.2-kb EcoRI fragment containing cdc42-md1 was excised from pMOSB36 and transformed into MOSY0090 together with the TRP1-marked plasmid, pRS314 (45). Trp+ transformants were selected on galactose-containing plates and then selected for the ability to grow on dextrose-containing plates (GAL1 promoter off) containing 5-fluoroorotic acid (to kill URA3 cells [7]), and gene replacement was confirmed by PCR. cdc42-md1 was then back-crossed to separate it from the leu2::GAL1p-CDC42::LEU2 present in MOSY0090, generating MOSY0178.
To create pDLB643, the template for PCR mutagenesis of CDC42, the CDC42 promoter and coding sequences were fused to the transcription terminator sequences from TDH3. The oligonucleotides DJL42-3 (5′-CCACCGTCGATTCAAGGGTC-3′) and DJL42-4 (5′-AGATCTCTGAGCAAAGCG-3′) were first used to amplify CDC42 (from 366 bp upstream of the start codon to 30 bp downstream of the stop codon) using the plasmid YEp351-CDC42 (55) as a template, and this fragment was cloned into pCR2.1 (Invitrogen, Carlsbad, Calif.) to make pCR42-3/4. The TDH3 transcription terminator sequences (a 900-bp BamHI/BglII fragment from pAB23BX [43]) were then cloned into the BamHI site of pCR42-3/4 to make pDLB643. A 2-kb XhoI/BamHI fragment from pDLB643 was also cloned into the corresponding sites of pRS316 (45) to make pMOSB55.
To create pDLB644, the recipient plasmid for expressing the mutant alleles of CDC42, CDC42 promoter sequences were fused (via a unique BglII site) to the transcription terminator sequences from TDH3 and cloned into the vector pRS316 (45). The oligonucleotides DJL42-3 (see above) and DJL42-5 (5′-AGATCTGGAAGACCTAATACG-3′; the BglII site is underlined) were used to amplify a DNA fragment containing the CDC42 promoter (1 to 366 bp upstream of the start codon) using the plasmid YEp351-CDC42 (55) as a template, and this fragment was cloned into pCR2.1 (Invitrogen) to make pCR42-3/5. The TDH3 transcription terminator sequences were then cloned into the BamHI site of pCR42-3/5 to create pDLB642. The 1.2-kb XhoI/BamHI fragment from pDLB642 was then cloned into the corresponding sites of pRS316 (45) to make pDLB644. pDLB644 was linearized by digestion with BglII and transformed into yeast cells together with the mutagenized CDC42 PCR products which underwent gap repair to yield Ura+ colonies.
The cdc42-V36A, cdc42-Y40C, and cdc42-D57Y alleles were constructed using the ExSite PCR-based site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). To generate pMOSB16 (cdc42-V36A), PCR was performed using the oligonucleotides cdc-4 (5′-ACAGCGTTCGATAACTATGCGG-3′) and cdc-11 (5′-TGGAACATAGTCAGCTGGAAATTGATTCG-3′) with pDLB643 as a template. cdc-11 has a silent mutation which introduces a PvuII site (underlined). To generate pMOSB53 (cdc42-Y40C), PCR was performed using the oligonucleotides cdc-22 (5′-ACAGTGTTCGATAACTGTGCGGTGACTGTGATG-3′) and cdc-11 with pDLB643 as a template. To generate pMOSB29 (cdc42-D57Y), PCR was performed using the oligonucleotides cdc-18 (5′-TATACGGCCGGTCAAGAAG-3′) and cdc-19 (5′-AAACAAACCTAACGTATATGG-3′) with pDLB377 as a template. The mutants were sequenced to confirm the presence of the desired mutation and the absence of any other mutations.
To generate pMOSB176 and pMOSB177, PCR was performed using the oligonucleotides DJL42-3 (see above) and DJL42-6 (5′-CTACTACAGATATTACATGTGGCG-3′) to amplify cdc42-Y40C (pMOSB53 template) or cdc42-V36A (pMOSB16 template), respectively, and introduced into pDLB644 by gap repair. To generate pMOSB175 and pMOSB47, 2.1-kb BamHI/XhoI fragments containing cdc42 alleles were excised from pMOSB176 and pMOSB177, respectively, and cloned into the corresponding sites of pRS314 (45).
The cdc42-V36A,Q61L and cdc42-Y40C,Q61L alleles were generated by a two-step strategy in which we first removed residues 32 to 40 in CDC42-Q61L and then employed homologous recombination to repair that “gap” with sequences from cdc42-V36A or cdc42-Y40C. We first replaced amino acids 32 to 40 in CDC42-Q61L,C188S with a NotI site, using the ExSite PCR-based site-directed mutagenesis kit. PCR was performed with the oligonucleotides JMCDC42-1 (5′-CCGCGTCGGCTGGAAATTGATTCG-3′) and JMCDC42-2 (5′-CCGCGGTGACTGTGATGATTGG-3′) with pEG202-cdc42-Q61L,C188S (46) as a template. The resulting allele was then transferred to pOBD.CYH using a PCR and gap repair strategy involving sequential amplification of the allele with the oligonucleotides LC20-CDC42 (5′-AATTCCAGCTGACCACCATGCAAACGCTAAAGTGTG-3′) and RC20-CDC42 (5′-GATCCCCGGGAATTGCCATGCTACAAAATTGTAGATTTT-3′) followed by a second PCR using the oligonucleotides BD70F and BD70R (15) containing homology to the first primers and 70-nucleotide extensions with homology to pOBD.CYH. The resulting PCR product was transformed together with PvuII/NcoI-linearized pOBD.CYH into DLY1 to generate pOBD.CYH-cdc42-Q61L/C188S(Δ32–40) by gap repair. cdc42-V36A and cdc42-Y40C were then amplified by PCR using the oligonucleotides DJL42-3 and cdc-19 (see above) with pMOSB16 or pMOSB53 as a template and transformed into DLY1 together with NotI-cut pOBD.CYH-cdc42-Q61L/C188S(Δ32–40) to generate the gap repair products pOBD.CYH-cdc42-V36A/Q61L/C188S and pOBD.CYH-cdc42-Y40C/Q61L/C188S.
Plasmids for the expression of myc-tagged alleles of CDC42 in bacteria were made using the Univector system (28). CDC42 alleles were amplified by PCR using the oligonucleotides CDC42-UNI1 (5′-GGAATTCCATATGCAAACGCTAAAGTGTGTTGTTGTC-3′; the NdeI site is underlined, and the start codon is in boldface) and CDC42-UNI2 (5′-CCGAGCTCCTACAAAATTGTACATTTTTTACTTTTC-3′; the SacI site is underlined) with pGAL-CDC42(Q61L) (55), pMOSB29, pOBD.CYH-cdc42-V36A/Q61L/C188S, or pOBD.CYH-cdc42-Y40C/Q61L/C188S as a template. NdeI/SacI-digested PCR fragments were cloned into the respective sites of pUNI-10 (28), generating pDLB1034, -1035, -1160, and -1277. The pUNI-10-CDC42 plasmids were then recombined with pHB1-MYC3 using the Cre/lox system (28) to produce the bacterial expression plasmids pDLB1234, -1235, -1240, and -1242. All constructs were sequenced to confirm that no changes occurred as a result of PCR manipulations.
To construct pG11-4, STE11-4 was amplified by PCR and inserted downstream of the GAL1 promoter in pRD53* as a BamHI/XhoI fragment; pRD53* is a derivative of pRD53 (40) in which URA3 lacks a PstI site.
Plasmid pGS11ΔN-T was constructed by transferring the 3.5-kb SacI/XhoI fragment encoding GAL1p-GST-STE11ΔN from pRD-STE11-H3 (32) into the corresponding sites in pRS314 (45).
Plasmid pPP828 is a LEU2-marked derivative of pRL116 (23) created by homologous recombination in yeast using pUC4-ura3::LEU2 (31).
To express the Ste20p Cdc42p-Rac interactive binding (CRIB) domain as a recombinant glutathione S-transferase (GST) fusion protein, the oligonucleotides ste20-1 (5′-ATGTCATCTTCTATAACCACCGC-3′) and ste20-2 (5′-TGTTTGCAGGCGGTGTTG-3′) were used to amplify a DNA fragment encoding the Ste20p residues 328 to 428 by PCR using yeast genomic DNA as a template. The PCR fragment was cloned into pCR2.1 using the TA cloning kit (Invitrogen) and then excised using the flanking EcoRI sites and cloned into the EcoRI site of pGEX-KG, generating pGEX-Ste20CRIB. The construct was sequenced to confirm its orientation and that no PCR-induced mutations had occurred.
To generate the ste20ΔCRIB allele (23), the 3-kb SphI/KpnI fragment from pRS316-ste20(Δ334–369) (23) was cloned into the corresponding sites of YIplac211 (12), generating pMOSB134. pMOSB134 was digested at the unique XbaI site within STE20 (upstream of the deleted CRIB region) and transformed into DLY3067. Ura+ transformants containing tandem STE20-URA3-ste20ΔCRIB sequences were selected, and then “pop-out” events in which URA3-containing sequences between the STE20 and ste20ΔCRIB genes were excised by homologous recombination were selected by plating them on 5-fluoroorotic acid. The colonies were screened by PCR using the oligonucleotide ste20-2 (5′-GAGTTTGCAGGCGGTGTTG-3′) and ste20-9 (5′-AACCGTCCAAGCCTGAAG-3′) to determine whether the excision event left STE20 (558-bp PCR product) or ste20ΔCRIB (446-bp PCR product) as the sole remaining allele.
Strains MOSY0023 and MOSY0106 were generated from a cross between MOSY0095 and BOY774 (cla4::TRP1) or BOY489 (ste20::TRP1), respectively. BOY489 and BOY774 were obtained from F. Cross (6).
The bem1::URA3 allele was generated by one-step gene replacement using an EcoRI-BamHI fragment from the plasmid pKO1 (9).
Strain PPY911 (40) contains a HIS2-marked FUS1-lacZ reporter integrated at the FUS1 locus that was introduced by transformation of the cdc42-1 strain DLY3032 with SphI-digested pFL-HIS2. pFL-HIS2 contains a 2-kb HIS2 HindIII fragment in place of the HinddIII/HindIII TRP1 fragment in pFL-TRPb, which itself contains a 0.9-kb TRP1 EcoRI/StuI fragment (along with pBluescript polylinker sequences from EcoRI-HincII) in place of the HindIII/StuI URA3 fragment of pSB286 (39).
Screen to identify α-factor-resistant cdc24 mutants.
The oligonucleotides DJL42-3 and DJL42-6 (see above) were used to amplify CDC42 (and TDH3 terminator sequences) by mutagenic PCR using pDLB643 as a template. Mutagenic PCRs were similar to standard PCR except for the addition of 0.2 mM MnCl2 and the use of an unbalanced deoxynucleoside triphosphate mix (0.5 mM dTTP, 0.5 mM dGTP, 0.1 mM dATP, and 0.1 mM dCTP [final concentrations]). The PCR products were transformed into DLY3067 together with BglII-digested (“gapped” plasmid) pDLB644, and Ura+ transformants were selected on dextrose-containing plates (to repress transcription of the genomic GAL1p-CDC42 allele) coated with 10 μg of α-factor (to select for α-factor-resistant cdc42 mutants). This amount of α-factor (custom synthesized by Research Genetics, Huntsville, Ala.) was more than 10 times the amount needed to completely inhibit growth of the bar1 parent strain. α-Factor-resistant colonies could in theory arise due to genomic mutations in other genes. To distinguish cdc42 mutants, colonies were tested for α-factor-resistant growth on galactose-containing plates, where the genomic (wild-type) CDC42 is expressed. Plasmids were isolated from colonies that were α-factor resistant when grown in dextrose-containing medium but not when grown in galactose-containing medium and were then retransformed into fresh DLY3067 to confirm that the plasmids were responsible for the α-factor-resistant growth phenotype. To generate the TRP1-containing plasmids pMOSB42 to -45, CDC42 alleles were transferred from URA3 plasmids (pMOSB36 to -38 and pMOSB55) as 2.1-kb BamHI/XhoI fragments to the corresponding sites of pRS314 (45).
Spot assays for growth arrest.
Cells harboring plasmid-borne CDC42 alleles were grown to stationary phase in dextrose-containing medium (to repress genomic GAL1p-CDC42) lacking nutrients appropriate to select for plasmid maintenance. The number of cells was determined using a hemocytometer, and diluted stocks were generated to spot 2 μl containing ∼1,250, ∼250, ∼50, and ∼10 cells onto plates containing the same selective dextrose medium either with or without 10 μg of α-factor per plate. Assuming a plate “volume” of 20 ml, this would translate to ∼0.3 μM α-factor.
Northern blot analysis.
RNA extraction, formaldehyde-agarose gel electrophoresis, capillary transfer, probe hybridization, and wash procedures were performed as described previously (10, 41, 42). The FUS1 probe was made from a 880-bp internal EcoRI/NheI fragment from the FUS1 gene, and the ACT1 probe was made from the entire ACT1 open reading frame, using [α-32P]dCTP (ICN Pharmaceuticals, Costa Mesa, Calif.) and the Prime-it II kit (Stratagene) according to the manufacturer's recommendations.
Flow cytometry.
Cells were processed for fluorescence-activated cell sorter (FACS) analysis as described (14), except that the DNA was stained with 1 μM Sytox (Molecular Probes, Eugene, Oreg.) in 50 mM Tris-HCl (pH 8.0) (instead of propidium iodide).
DIC microscopy.
Cells were viewed on an Axioskop apparatus (Zeiss, Thornwood, N.Y.) equipped with epifluorescence and differential interference contrast (DIC) optics. Images were captured by using a cooled-model charge-coupled device camera (Princeton Instruments, Princeton, N.J.).
Analysis of FUS1-lacZ expression.
β-Galactosidase assays were performed as described previously (40). Transformants in cdc42-1 strains were grown overnight at 28°C in selective synthetic medium containing 2% raffinose and 0.1% dextrose to an optical density at 660 nm of 0.3 to 0.6, preincubated for 2 h at 38.5°C, and then induced for 4 h with 2% galactose with or without 0.01 μM α-factor. Assays in CDC42 strains were performed at 30°C.
Production of recombinant proteins and binding assays.
Wild-type and mutant myc-tagged CDC42 alleles were expressed in Escherichia coli BL21(DE3) (Stratagene). Extracts were prepared in bacterial lysis buffer (750 mM sucrose, 100 mM NaCl, 100 mM Tris-HCl [pH 8.0], 5 mM EDTA) containing the protease inhibitors aprotinin (7.5 μg/ml; Sigma, St. Louis, Mo.), pepstatin (5 μg/ml; Sigma), leupeptin (10 μg/ml; Boehringer Mannheim, Indianapolis, Ind.), and phenylmethylsulfonyl fluoride (0.5 mM; Sigma). The cells were treated with 2 mg of lysozyme/ml for 20 min on ice. To remove genomic DNA, MgCl2 was added to 15 mM and DNase I was added to 50 μg/ml. The cells were lysed by 20 min of incubation at 4°C with 2 mg of deoxycholic acid/ml. Insoluble material was removed by centrifugation at 4°C for 10 min. GST-Ste20-CRIB was expressed in the protease-deficient E. coli BL21, extracts were prepared as described above, and the protein was purified using glutathione Sepharose 4B (Amersham Pharmacia Biotech, Piscataway, N.J.) as specified by the manufacturer.
Binding assays were performed by incubating the bacterial extracts containing Cdc42p-myc with either GST or GST-Ste20-CRIB immobilized on glutathione beads in 200 μl of binding buffer (10 mM Tris-HCl [pH 7.5], 85 mM NaCl, 6 mM MgCl2, 10% glycerol) at 4°C for 3 h. Binding reaction mixtures were washed three times at room temperature with wash buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 1 mM dithiothreitol, 0.1% Triton X-100). Bead-bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted to Immobilon-P nylon membranes (Millipore, Bedford, Mass.), and immunoblotted with monoclonal anti-myc antibodies (9E10; Santa Cruz Biotechnology, Santa Cruz, Calif.) using standard procedures (2). To confirm equal loading, the amount of GST-Ste20-CRIB in each lane was visualized by staining the membrane with India ink (42).
Localization of GFP-Ste20p.
Cells containing pRL116 or pPP828 were propagated for at least 24 h in selective liquid medium containing 2% dextrose and 0.008% adenine (to inhibit accumulation of red pigment in ade1 cells) at 30°C (GAL1p-CDC42 strains) or at 36°C (cdc42-1 strains). The cells were examined without fixation using a Nikon E600 epifluorescence microscope with 100× Plan Fluor and 50× Plan oil-immersion objectives. Images were collected using a cooled black and white charge-coupled device camera (DAGE-MTI).
RESULTS
Identification of recessive α-factor-resistant alleles of CDC42.
A hypothetical cdc42 mutant specifically defective in pheromone signal transduction should retain all functions necessary for cell polarity and proliferation but would fail to arrest in G1 upon exposure to α-factor. Thus, cells expressing such an allele as the sole source of Cdc42p should proliferate and form colonies on plates containing a growth-inhibitory dose of α-factor. We employed an error-prone PCR mutagenesis strategy to generate random mutations in CDC42 and recombined the resulting alleles into a low-copy-number plasmid in cells whose endogenous CDC42 was transcriptionally repressed (using the regulatable GAL1 promoter; see Materials and Methods for details). Transformants that promoted proliferation on α-factor-containing plates were selected. Plasmids containing putative CDC42 mutants were isolated from the rare α-factor-resistant colonies and retransformed into the starting strain to confirm that the α-factor resistance was due to the plasmid and not to a fortuitous genomic mutation. Three independently derived mutants (cdc42-md1, cdc42-md2, and cdc42-md12 [md for mating pathway defective]) were selected for further characterization (Fig. 1).
FIG. 1.
Isolation of α-factor-resistant cdc42 alleles. The results of growth arrest assays of strain DLY3067 (GAL1p-CDC42) containing plasmid pMOSB55 (CDC42), pMOSB36 (cdc42-md1), pMOSB37 (cdc42-md2), or pMOSB38 (cdc42-md12) are shown. The photographs show growth after 3 days at 30°C with (+) or without (−) α-factor.
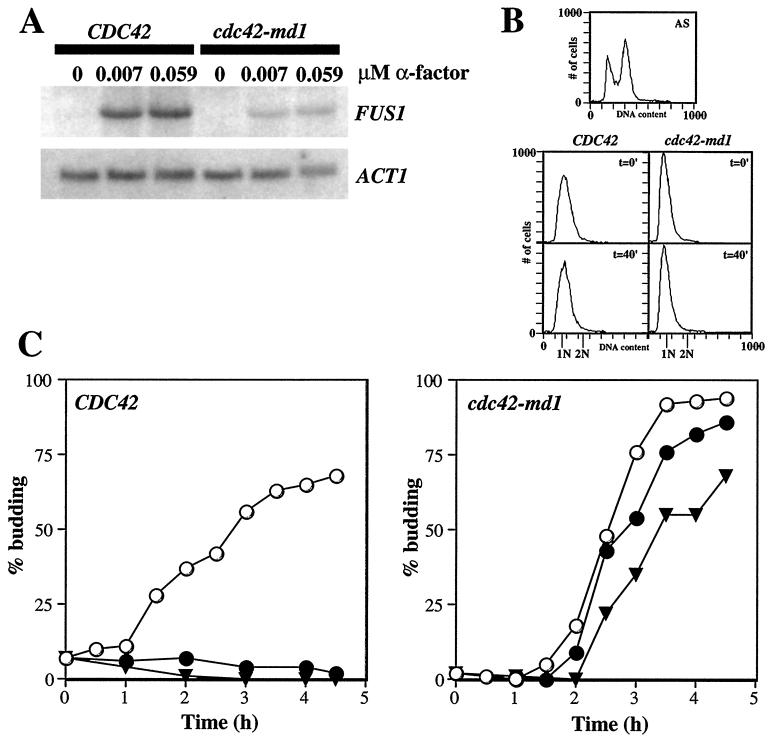
In addition to promoting growth on α-factor-containing plates (Fig. 1), strains bearing the cdc42-md alleles were significantly, though not completely, defective in α-factor-induced FUS1 transcription (see below) and in mating to a wild-type partner (data not shown). A possible explanation for the defect in FUS1 induction observed previously in temperature-sensitive cdc42 mutants is that the mutants accumulate at a nonresponding stage (post-G1) of the cell cycle (35). To address whether a similar situation might apply with cdc42-md mutants, we isolated early-G1-phase cells from cdc42-md1 and isogenic wild-type strains using centrifugal elutriation and monitored cell cycle progression and FUS1 mRNA accumulation upon treatment with different doses of α-factor. As shown in Fig. 2A, cdc42-md1 cells synchronized in early G1 phase displayed a defect in FUS1 induction compared to similarly treated wild-type cells, suggesting that these mutants are defective in responding to α-factor even when the cells are in a responsive stage of the cell cycle. Flow cytometric analysis confirmed that both mutant and wild-type cells remained in G1 for the duration of the 30-min α-factor treatment (Fig. 2B). In addition, cdc42-md1 mutants progressed into S phase (data not shown) and formed buds (Fig. 2C) with unaltered kinetics in the continuous presence of 0.015 μM α-factor, a concentration sufficient to completely arrest wild-type cells. However, 0.06 μM α-factor did cause cdc42-md1 cells to delay briefly in G1 (Fig. 2C), indicating that the signaling defect is not complete (consistent with the FUS1 induction data). In other experiments, we have found that cdc42-md mutants also show a FUS1 induction defect in cells arrested in G1 by depletion of the G1 cyclins Cln1p-Cln3p (data not shown). Together, these data strongly suggest that the signaling defect of cdc42-md mutants is not due to arrest at a nonresponding stage of the cell cycle.
FIG. 2.
FUS1 induction and cell cycle arrest in early-G1-phase cells. Cells containing two copies of wild-type CDC42 (DLY1 plus pMOSB55) or cdc42-md1 (MOSY0178 plus pMOSB36) were grown to exponential phase in sucrose-containing medium lacking uracil at 30°C (using two gene copies improved the morphology of cdc42-md1 cells, making the elutriation more effective). Small G1-phase cells were isolated by centrifugal elutriation and resuspended in YEPD medium at 30°C. Following a 10-min recovery period, α-factor was added to the indicated final concentrations and the incubation was continued with vigorous shaking. (A) Northern blot of FUS1 mRNA accumulation following 30 min of α-factor treatment. cdc42-md1 cells display severely reduced induction compared to wild-type cells. ACT1 mRNA on the same blots provides an internal control for loading and transfer. (B) FACS analysis demonstrating that both wild-type and cdc42-md1 cells were still in G1 phase after 40 min in YEPD medium without α-factor (the same time point used for the FUS1 mRNA analysis: a 10-min recovery period plus a 30-min α-factor treatment). The FACS profile of asynchronous wild-type cells (top) was used to determine the fluorescence signal equivalent to G1 (1 N) and G2 (2 N) DNA content. FACS profiles are shown for the indicated strains immediately following elutriation (t = 0′) or 40 min later in the absence of α-factor (t = 40′). (C) cdc42-md1 cells were not arrested in G1 phase in response to α-factor. Aliquots of cells were withdrawn at the indicated times, fixed, and examined by phase-contrast microscopy to determine the percentage of budded cells (n = 200). ○, no α-factor; ●, 0.015 μM α-factor; ▾, 0.059 μM α-factor.
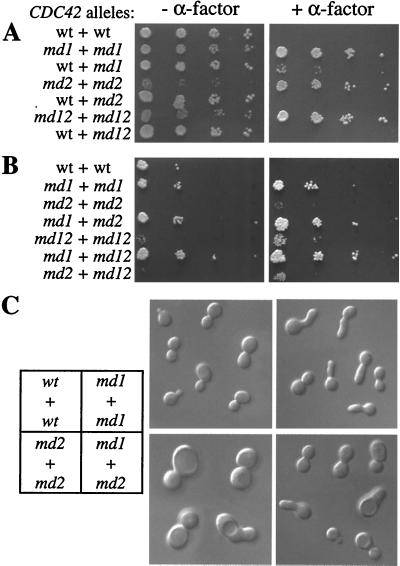
In principle, the cdc42-md mutants could be defective for a normal function of Cdc42p in α-factor signaling or they could encode forms of Cdc42p that can interfere with α-factor signaling even though wild-type Cdc42p might not play a role in signaling. To distinguish between these options, we performed a dominance test to determine whether cells containing both wild-type and mutant forms of CDC42 were α-factor resistant. As shown in Fig. 3A, the mutants were all recessive, in that cells containing plasmids expressing both wild-type and mutant Cdc42p were α-factor sensitive. Intriguingly, very tiny colonies were reproducibly observed in strains containing both cdc42-md1- and CDC42-expressing plasmids. This appears to be a dose-dependent effect such that cells with a high cdc42-md1/CDC42 plasmid ratio are selected for on α-factor plates: when this experiment was repeated in cells with an integrated copy of cdc42-md1 and a plasmid-borne copy of CDC42, no pheromone-resistant growth was observed (data not shown). The recessive behavior of these alleles suggests that Cdc42p wild-type function is required for an efficient pheromone response.
FIG. 3.
Intragenic-complementation analysis of the cdc42-md mutants. Plasmids containing the indicated pairs of CDC42 alleles were transformed into strain DLY3067. (A) Pheromone resistance of cdc42-md mutants is recessive. The photographs show growth after 3 days at 30°C with (+) and without (−) α-factor. The plasmid pairs were (in order) pMOSB55 plus pMOSB45, pMOSB36 plus pMOSB42, pMOSB55 plus pMOSB42, pMOSB37 plus pMOSB43, pMOSB55 plus pMOSB43, pMOSB38 plus pMOSB44, and pMOSB55 plus pMOSB44. wt, wild type. (B) cdc42-md mutants form a single complementation group with regard to α-factor resistance. Spot assays were performed as for panel A (except that fewer cells were spotted in this experiment). The plasmid pairs were (in order) pMOSB55 plus pMOSB45, pMOSB36 plus pMOSB42, pMOSB37 plus pMOSB43, pMOSB36 plus pMOSB43, pMOSB38 plus pMOSB44, pMOSB36 plus pMOSB44, and pMOSB37 plus pMOSB44. (C) cdc42-md mutants form two complementation groups with regard to cell morphology. Cells were grown to exponential phase and photographed using DIC optics. cdc42-md1 mutants exhibit broad necks and elongated buds (48% of cells), while cdc42-md2 mutants exhibit large round cells (>70% of cells); although some abnormal cells are present in the cdc42-md1 plus cdc42-md2 population (with generally less severe defects, perhaps due to plasmid loss), the majority (73%) exhibit normal morphology. The morphologies were similar in the presence and absence of α-factor. The plasmid pairs were pMOSB55 plus pMOSB45 (wt + wt), pMOSB36 plus pMOSB42 (md1 + md1), pMOSB37 plus pMOSB43 (md2 + md2), and pMOSB36 plus pMOSB43 (md1 + md2).
Intragenic complementation analysis of the cdc42-md mutants.
We took advantage of the recessive nature of these alleles to determine whether they had defects in the same or different functions required for the α-factor response. Cells containing all pairwise combinations of plasmids bearing different cdc42-md alleles were uniformly resistant to α-factor (Fig. 3B), indicating that no mutant could provide the signaling function(s) defective in another and suggesting that all three alleles were defective for the same function.
In addition to the α-factor-resistant phenotype, two of the mutants (cdc42-md2 and cdc42-md12) displayed a slow-growth phenotype (Fig. 1A and 3A) (interestingly, growth of these mutants was actually stimulated by α-factor), and all three mutants displayed aberrant cell morphologies when grown in the absence of α-factor (Fig. 3C) (a detailed analysis of the morphological phenotypes will be presented elsewhere). Like α-factor resistance, the slow growth and morphological phenotypes were recessive (Fig. 3A and data not shown). However, cells containing two plasmids expressing cdc42-md1 and cdc42-md2 or cdc42-md1 and cdc42-md12 but not cdc42-md2 and cdc42-md12 grew at wild-type rates and did not display morphological abnormalities (Fig. 3B and C and data not shown). This suggests that cdc42-md2 and cdc42-md12 share similar defects in vegetative growth while cdc42-md1 has a distinct defect: in cells containing both cdc42-md1 and cdc42-md2, or cdc42-md1 and cdc42-md12, all vegetative functions are provided by one of the alleles and cells grow normally. This finding of intragenic complementation for the vegetative-growth defects is in contrast to the lack of such complementation for the pheromone signaling defect (Fig. 3B), suggesting that the pheromone signaling defect is distinct from the vegetative-growth defects and cannot be explained as an indirect result of such defects.
Epistasis analysis of the cdc42-md mutants.
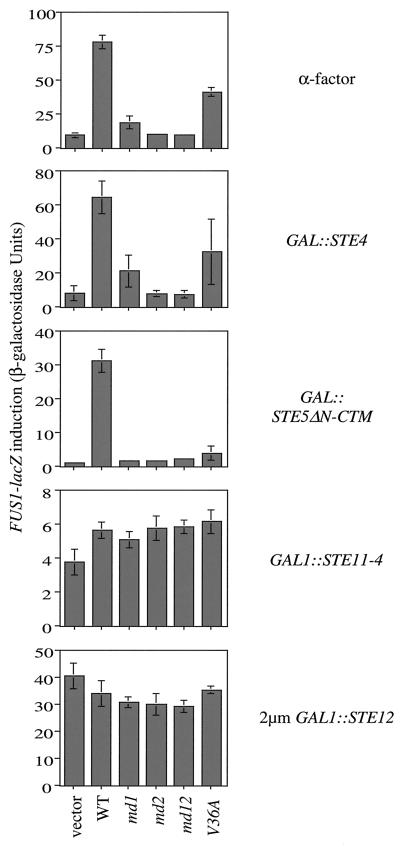
To determine where in the pheromone response pathway Cdc42p plays a role, we examined whether the cdc42-md mutants could block signaling that was initiated at various steps in the pathway. Induction of a FUS1-LacZ reporter construct was used as a readout of pathway activation. Like signaling induced by α-factor, the signals initiated by overexpression of the Gβ subunit Ste4p, or by a membrane-targeted Ste5p scaffold protein, were substantially blocked by cdc42-md mutants (Fig. 4). In contrast, signals initiated by the activated MEKK gene allele STE11-4 or by overexpression of the transcription factor Ste12p were unaffected by cdc42-md mutants (Fig. 4). This suggests that Cdc42p is required after Ste5p membrane recruitment for the activation of Ste11p. Previous studies have shown that this is precisely the step at which Ste20p is required (40).
FIG. 4.
Epistatis analysis of cdc42-md mutants. Strain PPY911 (cdc42-1) was transformed with pRS314 (vector), pMOSB45 (wild type [WT]), pMOSB42 (md1), pMOSB43 (md2), pMOSB44 (md12), or pMOSB47 (V36A), as indicated. Each of these strains was then transformed with the mating-pathway-activating plasmids pL19 (GAL1p-STE4 [GAL::STE4]), pL-GS5ΔN-CTM (GAL1p-STE5ΔN-CTM [GAL::STE5ΔN-CTM]), pG11-4 (GAL1p-STE11-4 [GAL1::STE11-4]), or pNC252 (2μm GAL1p-STE12 [2μm GAL1::STE12]) as indicated on the right. Transformants grown at restrictive temperature to inactivate cdc42-1 were induced with 2% galactose (or 2% galactose plus 0.01 μM α-factor; top). The bars indicate the means ± standard deviation for four to six independent transformants. Results similar to those shown with STE5ΔN-CTM were also seen with STE5-CTM, and results similar to those shown with GAL1p-STE11-4 were also seen with GAL1p-STE11ΔN (data not shown).
Sequence analysis of cdc42 mutants.
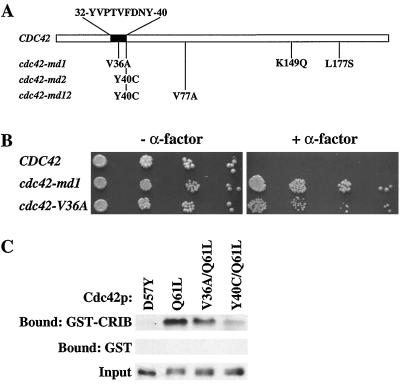
To determine the natures of the mutations that rendered CDC42 signaling defective, we recovered the mutant plasmids from yeast and sequenced the open reading frame of each mutant. As shown in Fig. 5A, all three mutants contained single-amino-acid changes in the “effector” domain of Cdc24p: a valine-to-alanine substitution at residue 36 in cdc42-md1 and a tyrosine-to-cysteine substitution at position 40 in both cdc42-md2 and cdc42-md12. In addition, cdc42-md1 contained substitutions at residues 149 and 177, and cdc42-md12 contained a valine-to-alanine substitution at residue 77 (Fig. 5A). Because the Y40C substitution was the only change in cdc42-md2, we assume that the α-factor resistance of cdc42-md12 also derives from this same change. Presumably the V77A mutation confers the somewhat-improved growth properties observed in cdc42-md12 compared to those of cdc42-md2. In a recent study, cdc42Y40C was reported to be nonviable (18). The discrepancy appears to be due to the fact that in our case the mutant is expressed from a low-copy-number (CEN ARS) plasmid while in that study it was integrated into the genome. To determine which changes were responsible for the α-factor resistance of the cdc42-md1 mutant, we constructed a mutant that contained only the V36A substitution. This mutant also conferred an α-factor-resistant phenotype (although not as strong as that conferred by cdc42-md1 [Fig. 4 and 5B]). We conclude that a mutation at either V36 (to A) or Y40 (to C) in the effector domain of Cdc42p renders the protein signaling defective.
FIG. 5.
Defective interaction of cdc42-md mutants with Ste20p. (A) Altered residues in cdc42-md mutants. (B) The V36A substitution confers α-factor resistance. Growth arrest assays of strain DLY3067 (GAL1p-CDC42) containing plasmid pMOSB55 (CDC42), pMOSB36 (cdc42-md1), or pMOSB177 (cdc42-V36A) were performed. The photographs show growth after 3 days at 30°C with (+) or without (−) α-factor. (C) Binding of cdc42 mutants to the Ste20p CRIB domain. The binding conditions were as described in Materials and Methods. The top row shows the amount of the indicated myc-tagged Cdc42p mutant that bound to immobilized GST-CRIB, the second row shows the amount of each Cdc42p mutant that bound to immobilized GST (to control for nonspecific adsorption), and the third row shows the amount of each Cdc42p mutant added to the binding reaction. India ink staining of the blots confirmed that equal amounts of GST or GST-CRIB were present in each lane. Similar results were obtained in four independent experiments. Recombinant Cdc42p was generated from pDLB1234 (D57Y, mimicking GDP-bound Cdc42p), pDLB1235 (Q61L, mimicking GTP-bound Cdc42p), pDLB1240 (V36A and Q61L), or pDLB1242 (Y40C and Q61L).
Defective interaction of Ste20p with cdc42-md mutants.
The Y40C mutation has been described in human CDC42 (which is 80% identical to yeast CDC42 and complements a cdc42 null mutation in yeast [44]), where it was found to render Cdc42p defective in binding to and activating p21-activated kinase (PAK), a mammalian Ste20p homologue (19). Combined with epistasis analysis (Fig. 4), this suggested that the cdc42-md mutants might be defective in binding to Ste20p. Binding of PAK family kinases to Cdc42p is mediated by the CRIB domain in these proteins (8). We produced the Ste20p CRIB domain as a GST fusion protein in E. coli and purified it using glutathione Sepharose beads. The wild type or V36A or Y40C mutants of Cdc42p were produced as myc-tagged proteins in E. coli and tested for binding to the GST-Ste20p-CRIB beads or to GST beads as a negative control. A Q61L substitution, which inhibits Cdc42p GTPase activity, was also engineered into the constructs to ensure that the proteins were GTP bound (a prerequisite for CRIB domain binding). As shown in Fig. 5C, the V36A substitution conferred a mild defect in Ste20p binding while the Y40C substitution conferred a severe defect.
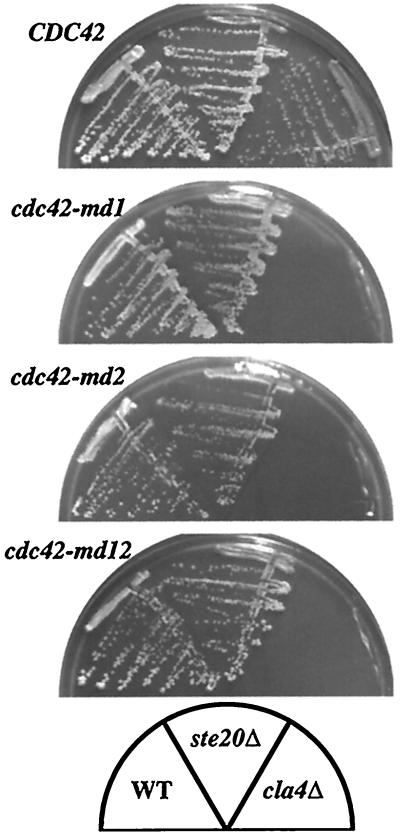
Ste20p shares a redundant essential function with the related PAK-family kinase Cla4p. The Cdc42p-Ste20p interaction has been reported to be critical for this function (23, 37). We found that cells containing cdc42-md mutants as the only source of Cdc42p were synthetically lethal with cla4Δ mutants (but not with ste20Δ mutants [Fig. 6]), suggesting that these mutants fail to interact with Ste20p in vivo as well as in vitro.
FIG. 6.
Synthetic lethality of cdc42-md mutants with cla4Δ but not ste20Δ mutants. Strains DLY3067 (GAL1p-CDC42; left), MOSY0106 (GAL1p-CDC42 ste20Δ; middle), and MOSY0023 (GAL1p-CDC42 cla4Δ; right) were transformed with pMOSB55 (CDC42), pMOSB36 (cdc42-md1), pMOSB37 (cdc42-md2), or pMOSB38 (cdc42-md12), as indicated. Cells were streaked out on dextrose-containing plates (to repress the genomic GAL1p-CDC42) lacking uracil (to select for plasmid maintenance) and incubated at 30°C for 3 days. WT, wild type.
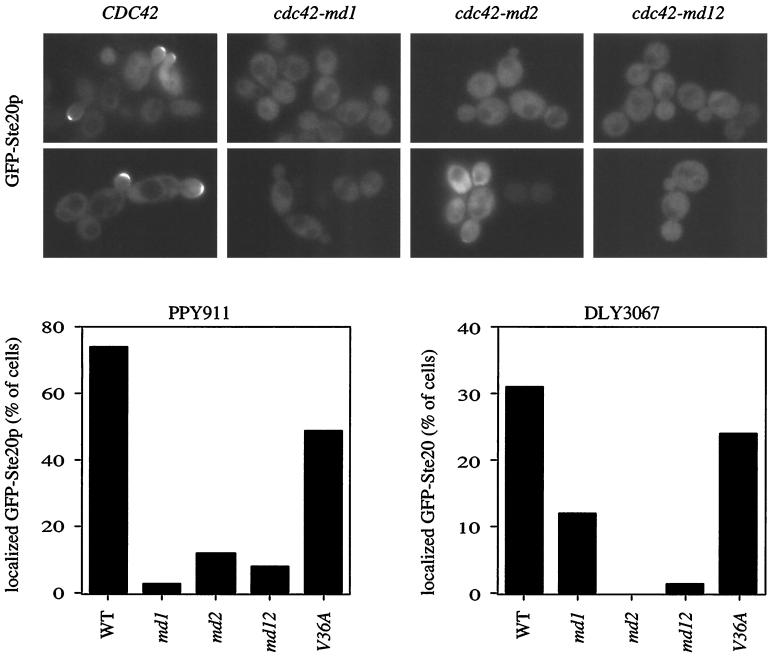
Defective localization of Ste20p in cdc42-md mutants.
Ste20p is concentrated at the bud tips of many cells during vegetative growth, and at the shmoo tips of cells responding to α-factor, in a manner that depends to a large extent on the CRIB domain (23, 37). GFP-Ste20p localization to bud tips was greatly reduced in all of the cdc42-md mutants (Fig. 7). Even cells containing both cdc42-md1 and cdc42-md2, which exhibited a wild-type growth rate and cell morphology, failed to localize GFP-Ste20p to the bud tip in most cells (see Fig. 11 below). We were unable to address the question of whether shmoo tip localization was affected because these cells were resistant to α-factor and did not make shmoos.
FIG. 7.
Ste20p localization in cdc42-md mutants. Strain PPY911 (cdc42-1) was transformed with pRL116 (GFP-STE20 [GFP-Ste20p]) and then with pMOSB45 (CDC45), pMOSB42 (cdc42-md1), pMOSB43 (cdc42-md2), pMOSB44 (cdc42-md12), or pMOSB47 (cdc42-V36A) as indicated. The cells were grown at 36°C to inactivate cdc42-1, and the localization of GFP-Ste20p was examined by fluorescence microscopy of living cells. Representative fields are shown (top), and the percentage of budded cells displaying GFP-Ste20p concentrated at the bud tips is quantitated (bottom left). The same set of plasmids was transformed into DLY3067 (GAL1p-CDC42), and the cells were grown on dextrose (to repress GAL1p-CDC42) and analyzed as above (bottom right). Several images were captured for each transformant, and all cells within those images were scored for whether GFP-Ste20 was detectably concentrated at the bud tip. n = 80 to 138 (PPY911 transformants) or n = 55 to 91 (DLY3067 transformants).
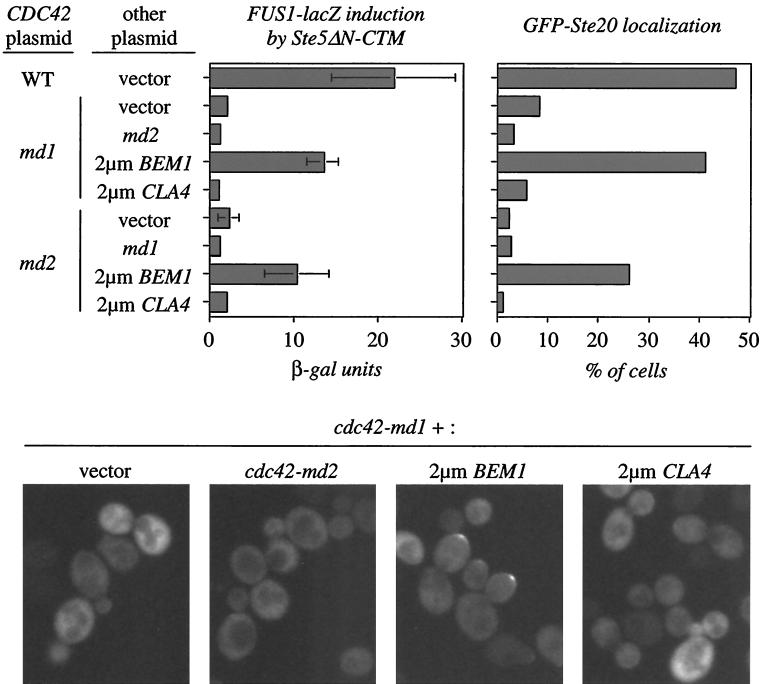
FIG. 11.
Overproduction of BEM1 restores Ste20p localization and signaling competence to cdc42-md strains. Strain PPY911 (cdc42-1) containing pL-GS5ΔN-CTM (for FUS1 induction) or pPP828 (for Ste20p localization) was transformed with the CDC42 plasmids pMOSB45 (wild type [WT]), pMOSB42 (md1), or pMOSB43 (md2) and then with YEp24 (vector), pMOSB36 (md1), pMOSB37 (md2), pPB321 (2μm BEM1), or pDLB722 (2μm CLA4) as indicated. FUS1-lacZ induction was measured as for Fig. 4; the bars indicate means ± SD of six independent transformants. GFP-Ste20p localization was quantitated as for Fig. 7 (200 cells counted), and representative cells are shown below.
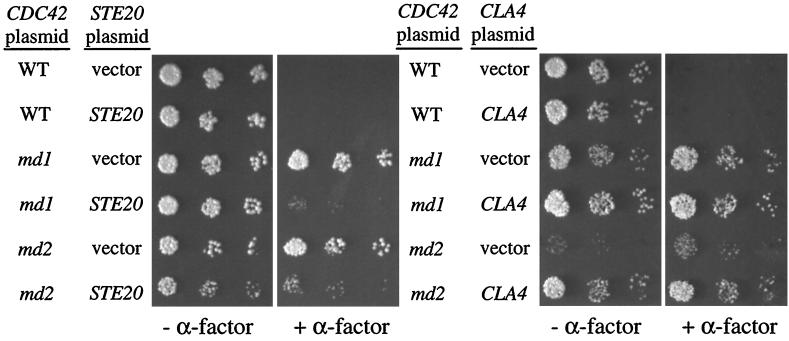
Suppression of the cdc42-md signaling defect by overexpression of Ste20p.
If the α-factor resistance phenotype of the cdc42-md mutants is due to defective interaction with Ste20p, then overexpression of Ste20p might suppress that phenotype. Indeed, overexpression of STE20 substantially suppressed the α-factor-resistant growth of cdc42-md mutants (Fig. 8). In contrast, overexpression of the related CLA4 did not suppress the cdc42-md signaling defect, although it did suppress the growth defect of cdc42-md2 mutants in the absence of α-factor (Fig. 8). These findings could be accommodated by at least two models: the observed suppression may be due to a bypass of the cdc42-md defects by the overexpressed kinases or to a restoration of a sufficiently effective interaction between the mutant Cdc42p and the kinases. In the latter model, the CRIB-binding defect of cdc42-md2 causes a vegetative growth defect due to impaired interaction with Cla4p and a signaling defect due to impaired interactions with Ste20p.
FIG. 8.
Overexpression of STE20 suppresses the cdc42-md signaling defect. Strain DLY3067 (GAL1p-CDC42) was transformed with pMOSB45 (wild type [WT]), pMOSB42 (md1), pMOSB43 (md2), or pMOSB44 (md12), and each strain was then transformed with pRS426 (vector), pVTU-Ste20 (STE20), or pDLB722 (CLA4). The photographs show growth after 3 days at 23°C with (+) or without (−) α-factor.
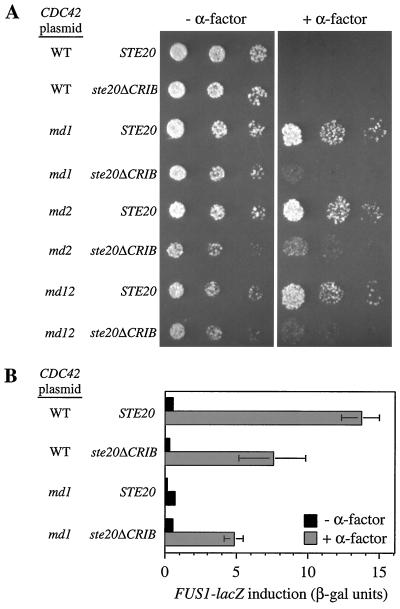
Restoration of pheromone signaling in cdc42-md mutants by the ste20ΔCRIB allele.
The findings presented above are consistent with the simple hypothesis that the Cdc42p-Ste20p interaction is a critical step in the α-factor signaling pathway: the cdc42-md mutants are defective for this interaction in vitro and in vivo, epistasis analysis suggests that Cdc42p acts at the same level of the pathway as Ste20p, and overexpression of Ste20p suppresses that the cdc42-md signaling defect. Nevertheless, the hypothesis that the Cdc42p-Ste20p interaction is important for signaling is contradicted by previous studies showing that a ste20 mutant lacking the CRIB domain (and therefore defective in Cdc42p interaction) did not affect signaling (23, 37). A possible resolution of this apparent paradox is suggested by recent evidence that the CRIB domain in PAK family kinases is part of an autoinhibitory domain, leading to a model in which Cdc42p (or Rac) proteins activate PAK family kinases by “relief of autoinhibition” (3, 49, 52, 54). For example, in the Pak1 kinase of Schizosaccharomyces pombe, a region overlapping the CRIB domain of Pak1 binds intramolecularly to the catalytic domain, yielding a “closed” conformation that can be “opened” either by binding to Cdc42p or by mutations in the CRIB domain (49). By analogy, the ste20ΔCRIB mutant may be competent for signal transduction because it no longer requires a normally critical interaction with Cdc42p for its activation. This hypothesis predicts that the ste20ΔCRIB mutant should bypass the requirement for Cdc42p in signaling, rendering cdc42-md strains sensitive to pheromone. Indeed, we found that ste20ΔCRIB restored significant α-factor signaling to the cdc42-md mutants, as assayed by growth inhibition (Fig. 9A) or FUS1 induction (Fig. 9B). This suggests that the ste20ΔCRIB allele encodes an activated form of Ste20p and supports the hypothesis that the Cdc42p-Ste20p interaction is normally important for pheromone signaling. We noticed, however, that ste20ΔCRIB did not completely restore α-factor sensitivity to cdc42-md mutants. This suggests that cdc42-md mutants have some defect in addition to the Ste20p activation defect.
FIG. 9.
ste20ΔCRIB largely restores α-factor sensitivity to cdc42-md strains. (A) Growth arrest. Strains DLY3067 (GAL1p-CDC42 STE20) and MOSY0268 (GAL1p-CDC42 ste20ΔCRIB) were transformed with pMOSB55 (wild type [WT]), pMOSB36 (md1), pMOSB37 (md2), or pMOSB38 (md12), as indicated. The photographs show growth after 3 days at 23°C with (+) or without (−) α-factor. (B) FUS1-lacZ induction. The same strains as in panel A harbored the FUS1-lacZ plasmid pSB231 plus either pMOSB45 (WT) or pMOSB42 (md1) and were assayed in glucose medium after 90 min with (+) or without (−) 0.01 μM α-factor. The bars indicate the means ± standard deviations of four transformants.
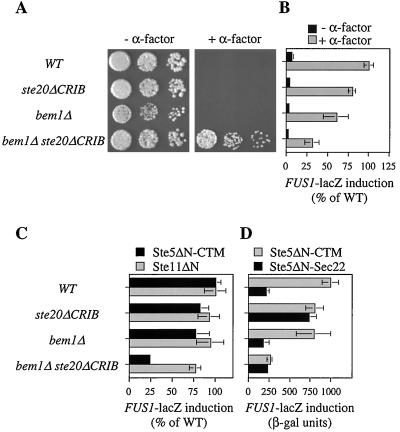
Contribution of Bem1p to Cdc42p- and Ste20-dependent signaling.
The BEM1 gene was discovered through its key role in polarity establishment (5, 9, 38). Subsequent studies showed that Bem1p can also modulate the pheromone response pathway, as Bem1p overexpression increases signaling while Bem1p removal decreases signaling (17, 29). Because the role of Bem1p in polarity establishment involves Cdc42p and Cdc24p (5, 38), we speculated that its role in pheromone-responsive signaling may also involve Cdc42p and therefore that the residual signaling defect observed in cdc42-md ste20ΔCRIB mutants might reflect reduced Bem1p function resulting from the cdc42-md mutations. Consistent with this hypothesis, we found that like cdc42-md ste20ΔCRIB mutants, bem1Δ ste20ΔCRIB mutants also displayed partial α-factor resistance (Fig. 10A) and reduced FUS1 induction (Fig. 10B) compared to ste20ΔCRIB single mutants. The signaling defect in bem1Δ ste20ΔCRIB mutants was also evident when the pathway was activated by membrane-targeted Ste5p but not when it was activated by the activated Ste11p derivative Ste11ΔN (Fig. 10C). This indicates that like Cdc42p, Bem1p acts downstream of Ste5p membrane recruitment to promote activation of Ste11p by Ste20p.
FIG. 10.
BEM1 is important for signaling in ste20ΔCRIB mutant strains. (A) Growth arrest. The photographs show growth after 2 days at 30°C with (+) and without (−) α-factor. (B) FUS1p-lacZ induction by α-factor. Strains harboring a FUS1p-lacZ reporter plasmid (p3058) were treated with and without 0.1 μM α-factor in duplicate for 1 and 2 h with similar results; the data were expressed as percent mean induced β-galactosidase activity in the wild-type (WT) strain (427 U at 1 h; 835 U at 2 h) and combined, with each bar representing the mean ± standard deviation (SD) of four measurements. (C) FUS1p-lacZ induction by Ste5ΔN-CTM or Ste11ΔN was assayed in strains harboring p3058 and either pGFP-GS5ΔN-CTM or pGS11ΔN-T after 4 h of induction with galactose; the bars indicate the means ± SD of four to six transformants, expressed as percent mean induced β-galactosidase activity in the WT strain (1,024 U for Ste5ΔN-CTM; 644 U for Ste11ΔN). (D) FUS1p-lacZ induction in strains harboring p3058 and either pGFP-GS5ΔN-CTM (Ste5ΔN-CTM) or pGFP-GS5ΔN-Sec22 (Ste5ΔN-Sec22) was assayed after 4 h of induction with galactose; the bars indicate the means ± SD of eight transformants. The strains in all panels were DLY1 (WT), MOSY0252 (ste20ΔCRIB), JMY1128 (bem1Δ), and MOSY0270 (bem1Δ ste20ΔCRIB).
We also compared the effects of bem1Δ and ste20ΔCRIB mutations on signaling by Ste5p that was targeted to different subcellular locations (Fig. 10D). Remarkably, while Ste20pΔCRIB was mildly defective at activating Ste5p that was targeted to the plasma membrane (Ste5ΔN-CTM), it showed increased ability to activate Ste5p that was targeted primarily to internal membranes (Ste5ΔN-Sec22 [40]). Consequently, in ste20ΔCRIB cells, the four- to fivefold signaling advantage of plasma membrane-targeted Ste5p over internal-membrane-targeted Ste5p was eliminated. These results support the notion that Ste20pΔCRIB is an active but delocalized kinase. Loss of Bem1p did not mimic the effect of ste20ΔCRIB; instead, bem1Δ reduced the ability of Ste20pΔCRIB to support signaling by Ste5ΔN-Sec22 and Ste5ΔN-CTM to similar extents, indicating that Bem1p can affect Ste20p- and Ste5p-dependent signaling even when both components are mislocalized.
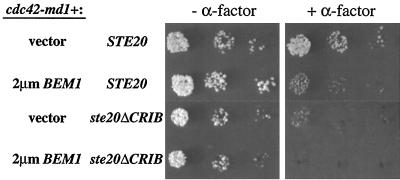
If a part of the signaling defect of cdc42-md mutants is due to defective Bem1p function, as argued above, then Bem1p overexpression might be expected to improve signaling in cdc42-md cells. Indeed, we found that a high-copy-number BEM1 plasmid substantially improved signaling in response to membrane-targeted Ste5p in cdc42-md1 and cdc42-md2 mutant strains (Fig. 11). Bem1p overexpression also suppressed the growth and morphology defects of cdc42-md mutants (data not shown). However, this cannot account for the improved signaling because a high-copy-number CLA4 plasmid (which also suppressed the growth and morphology defects) or introduction of both cdc42-md1 and cdc42-md2 (leading to complementation of the growth and morphology defects) failed to suppress the signaling defect (Fig. 11). Strikingly, Bem1p overexpression (but not Cla4p overexpression or cdc42-md combinations) also suppressed the Ste20p localization defect in cdc42-md mutants (Fig. 11). This result is consistent with previous studies indicating that Bem1p interacts with Ste20p (24) and strongly suggests that Bem1p is acting to promote signaling at the level of Ste20p, consistent with our epistasis results above. When combined, the introduction of Ste20pΔCRIB (to bypass the Ste20p activation defect) and excess Bem1p (to bypass a possible Bem1p defect) into the cdc42-md1 mutant completely suppressed its α-factor resistance phenotype (Fig. 12). Together, these data suggest that the signaling defect of cdc42-md mutants can be accounted for by their simultaneous failures to activate Ste20p and to promote Bem1p function.
FIG. 12.
Bem1p and Ste20pΔCRIB cooperate to restore α-factor sensitivity to a cdc42-md1 strain. Strains DLY3067 (GAL1p-CDC42 STE20) and MOSY0268 (GAL1p-CDC42 ste20ΔCRIB) were transformed with pMOSB42 (cdc42-md1) and either pRS426 (vector) or pPB321 (2μm BEM1), as indicated. The photographs show growth after 3 days at 30°C with (+) and without (−) α-factor.
DISCUSSION
A role for Cdc42p in signal transduction in response to α-factor.
We have isolated cdc42 mutants that display recessive defects in α-factor signaling while retaining the ability to proliferate. All of these cdc42-md mutants had morphogenesis defects in addition to their signaling defect. However, the signaling defect is unlikely to be an indirect result of the morphogenesis defects for the following reasons. First, overexpression of CLA4 suppressed the morphogenesis defects of cdc42-md mutants but not the signaling defect. Second, two pairwise combinations of cdc42-md alleles displayed intragenic complementation for their morphogenesis defects but not for their signaling defects. Third, a synchronous early-G1-phase population of cdc42-md1 mutant cells isolated by centrifugal elutriation displayed a signaling defect even during G1 phase, a time when cells are uniformly sensitive to α-factor and Cdc42p-dependent polarity has yet to be established. It is difficult to see how a signaling defect in this synchronous culture could arise due to any indirect cell cycle or morphogenesis problems. In summary, these results strongly suggest that Cdc42p plays a role in α-factor-induced signaling that is separate from its roles in morphogenesis, confirming the suggestions from earlier studies with temperature-sensitive cdc42 mutants (46, 47, 53).
The conclusion that Cdc42p is important for α-factor signaling is at odds with a study that found no signaling defect in G1-arrested temperature-sensitive cdc42 or cdc24 mutants (35). In that study, cells arrested in G1 by deprivation of G1 cyclins were subsequently shifted to the restrictive temperature for the cdc42 or cdc24 mutants and then exposed to relatively high doses of α-factor. Under these conditions, induction of FUS1 transcription was unaffected by the cdc42 or cdc24 mutations, suggesting that Cdc42p is not required for α-factor signaling and affects signaling indirectly as a result of cell cycle perturbation (35). However, an earlier study found Cdc24p and Cdc42p to be required for the maintenance of G1 arrest in cells that had been prearrested by α-factor (46), which is not explainable by a model in which the signaling defects of cdc24 and cdc42 mutants are due simply to accumulation of cells at a nonresponsible stage of the cell cycle.
While our observations support a role for Cdc42p in pheromone-responsive signal transduction, they do not suggest that Cdc42p is a pathway intermediate that gets activated in response to pheromone. Instead, our work is consistent with the view (40) that Cdc42p plays a permissive role in maintaining cellular competence for signaling. For instance, Cdc42p may be constitutively required for the establishment of relatively long-lived pre-signaling complexes containing active Ste20p (see below). Transmission of the pheromone signal could then utilize this preactivated pool of Ste20p without any further immediate involvement of Cdc42p. In this hypothesis, the cdc42-md mutants have a signaling defect because they fail to establish this competent pool of Ste20p, while the temperature-sensitive cdc42-1 inactivation regimen used by Oehlen and Cross (35) did not result in a signaling defect because sufficient Ste20p activation was established prior to the temperature shift to allow subsequent signaling. Notable in this regard is evidence from human PAK family members that GTPase-dependent establishment of the open conformation allows for PAK autophosphorylation, which subsequently interferes with regeneration of the closed conformation and thus makes the kinase temporarily GTPase independent (30, 52).
Ste20p is a key Cdc42p target involved in pheromone signaling.
By several criteria, the cdc42-md mutants are defective in interacting with Ste20p: they are defective (to varying extents) in binding to the Ste20p CRIB domain in vitro, they fail to localize Ste20p to bud tips in vivo, and they display synthetic lethality with cla4 mutants. Furthermore, epistasis analysis suggests that cdc42-md mutants are defective in signal transduction at the same step in the pathway as ste20 mutants (i.e, the activation of Ste11p following membrane targeting of Ste5p). Finally, the signaling defect of cdc42-md mutants is largely suppressed by overproduction of Ste20p. These data strongly support the hypothesis that the Cdc42p-Ste20p interaction is important for efficient signaling in the pheromone response pathway, as originally proposed (46, 53). However, more recent studies have argued that the Cdc42p-Ste20p interaction is dispensable for pheromone response, based on the signaling competence of a version of Ste20p (Ste20pΔCRIB) that cannot bind to Cdc42p. We found that Ste20pΔCRIB largely restored signaling competence to strains containing the cdc42-md mutants. This observation suggests that Ste20pΔCRIB is an activated version of Ste20p that no longer requires interaction with Cdc42p for its activation, consistent with current models for PAK activation which involve a relief of autoinhibition mechanism (3, 49, 54). We therefore conclude that interaction of Cdc42p with Ste20p is normally required for Ste20p to participate in the pheromone response pathway. Importantly, however, we do not find it necessary to propose that pheromone regulates either GTP loading of Cdc42p, the Cdc42p-Ste20p interaction, or Ste20p kinase activity. Instead, the simplest model consistent with available data is that access of Ste20p to its substrates is the pheromone-regulated step, with the effect of Cdc42p on Ste20p kinase activity being a preexisting condition that is independent of pheromone exposure (see reference 40 for further discussion).
Cdc42p may play a second role together with Bem1p in pheromone signaling.
Although Ste20p appears to be the major Cdc42p target for signal transduction, our results suggest the existence of a second role for Cdc42p in this process, because cdc42-md mutants displayed a residual signaling defect even in the presence of the activated ste20ΔCRIB allele. Given the strong links between Cdc42p and the SH3-domain-containing scaffold protein Bem1p that have been established through studies of cell polarity in yeast (5, 9, 38), we suspected that this second role might involve Bem1p (which has also been shown to modulate the strength of α-factor signaling [17, 29]). Consistent with this hypothesis, Bem1p overexpression partially suppressed the signaling defect of cdc42-md mutants and Bem1p overexpression together with ste20ΔCRIB could fully suppress the α-factor-resistant growth of cdc42-md mutants.
Our observations suggest that the cdc42-md mutants are defective in activation of the Ste11p-Ste7p-Fus3p MAPK cascade. This step involves the interaction of Gβγ with the Ste5p scaffold protein (associated with Ste11p, Ste7p, and Fus3p), resulting in recruitment of Ste5p to the plasma membrane (40) and also the interaction of Gβγ with Ste20p (25). These interactions likely serve to raise the local concentrations of Ste20p and its substrate Ste11p (bound to Ste5p), thereby initiating MAPK cascade activity. The previously described effects of Bem1p on pheromone signaling (20, 29) are compatible with its acting anywhere in the pathway from receptor to the MAPK Fus3p. Our experiments indicate that Bem1p affects steps following Ste5p membrane recruitment, since both loss and overproduction of Bem1p affects signaling by membrane-targeted Ste5p, even in cells with the activated ste20ΔCRIB allele. In contrast, signaling by the activated Ste11p derivative Ste11ΔN was comparatively insensitive to the loss of Bem1p. We also observed that Bem1p can affect the localization of Ste20p, and previous studies showed that Bem1p could form complexes with both Ste20p and Ste5p (24, 29). Together, these results suggest that Bem1p influences activation of the Ste5p-associated kinase cascade by Ste20p, perhaps by helping to bring these proteins together. It should be noted that earlier work observed effects of Bem1p overproduction on the residual pheromone response of ste20Δ cells (20, 29); the mechanism for this residual response has not been determined, but it may rely on inefficient substitution for Ste20p by another PAK family kinase (e.g., Cla4p), which would be compatible with our suggestion that Bem1p affects the step in which Ste11p is activated by the Ste20p (or substitute) kinase.
The dramatic effect of Bem1p overexpression on Ste20p localization in cdc42-md mutants provides a suggestion as to how Bem1p may function. During vegetative growth, recruitment of Ste20p to the bud tip by Cdc42p may be assisted by Bem1p-mediated local retention of Ste20p. Similarly, during pheromone response, recruitment of Ste5p and Ste20p to a region of the plasma membrane by Gβγ may be assisted by local retention of these proteins promoted by Bem1p. Our results suggest some collaboration between Cdc42p and Bem1p in pheromone response. Thus, Cdc42p and Bem1p may each help to provide a cell surface scaffold that facilitates the association and/or local retention of Ste5p (with its associated kinases) and Ste20p, allowing the local concentrations of these signaling components to be raised above the threshold required for efficient signal transduction.
Conclusions.
In aggregate, the analysis of the pheromone-resistant cdc42 alleles presented here combined with previous studies suggest a novel paradigm for the “signaling” role of Cdc42p that is quite distinct from the paradigm established for the ras GTPase. Cdc42p is required for Ste20p activation, but there is little evidence to suggest that pheromone signals stimulate this activation; instead, pheromone signaling may make use of a preexisting constitutive Ste20p activity. In addition, Cdc42p (acting with Bem1p) helps increase the local concentration of Ste20p together with other signaling components, promoting signal transduction by a proximity effect (36). Providing a surface conducive to the local concentration of signaling reactants could be a common contributor to eukaryotic signaling pathways, especially those that rely on regulatable protein-protein interactions rather than diffusible second messengers (16). It remains to be seen to what extent these types of signaling roles will be generally applicable for other Rho family GTPases and other cells.
ACKNOWLEDGMENTS
We thank members of the Pringle and Lew laboratories for many valuable discussions. We also thank E. Bi, C. Boone, F. Cross, E. Leberer, and M. Peter for providing plasmids or strains. Thanks to Lynn Martinek and Mike Cook from the Duke Cancer Center Flow Cytometry Shared Resource for help with the flow cytometry.
J.J.M. was supported by American Cancer Society postdoctoral fellowship PF-98-008. This work was supported by grants from the Worcester Foundation and the Millipore Foundation, by NIH grant GM57769 to P.M.P., and by NIH grant GM53050 and American Cancer Society grant RPG-98-046-CCG to D.J.L.
REFERENCES
- 1.Adams A E M, Johnson D I, Longnecker R M, Sloat B F, Pringle J R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1995. [Google Scholar]
- 3.Bagrodia S, Cerione R A. Pak to the future. Trends Cell Biol. 1999;9:350–355. doi: 10.1016/s0962-8924(99)01618-9. [DOI] [PubMed] [Google Scholar]
- 4.Bardwell L, Cook J G, Inouye C J, Thorner J. Signal propagation and regulation in the mating pheromone response pathway of the yeast Saccharomyces cerevisiae. Dev Biol. 1994;166:363–379. doi: 10.1006/dbio.1994.1323. [DOI] [PubMed] [Google Scholar]
- 5.Bender A, Pringle J R. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benton B K, Tinkelenberg A, Gonzalez I, Cross F R. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–173. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 8.Burbelo P D, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 9.Chenevert J, Corrado K, Bender A, Pringle J, Herskowitz I. A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature. 1992;356:77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- 10.Elder R T, Loh E Y, Davis R W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc Natl Acad Sci USA. 1983;80:2432–2436. doi: 10.1073/pnas.80.9.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y, Song L Y, Kincaid E, Mahanty S K, Elion E A. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [PubMed] [Google Scholar]
- 14.Haase S B, Lew D J. Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol. 1997;283:322–332. doi: 10.1016/s0076-6879(97)83026-1. [DOI] [PubMed] [Google Scholar]
- 15.Hudson J R, Jr, Dawson E P, Rushing K L, Jackson C H, Lockshon D, Conover D, Lanciault C, Harris J R, Simmons S J, Rothstein R, Fields S. The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form. Genome Res. 1997;7:1169–1173. doi: 10.1101/gr.7.12.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter T. Signaling—2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 17.Kao L R, Peterson J, Ji R, Bender L, Bender A. Interactions between the ankyrin repeat-containing protein Akr1p and the pheromone response pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:168–178. doi: 10.1128/mcb.16.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozminski K G, Chen A J, Rodal A A, Drubin D G. Functions and functional domains of the GTPase Cdc42p. Mol Biol Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamarche N, Tapon N, Stowers L, Burbelo P D, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- 20.Leberer E, Chenevert J, Leeuw T, Harcus D, Herskowitz I, Thomas D Y. Genetic interactions indicate a role for Mdg1p and the SH3 domain protein Bem1p in linking the G-protein mediated yeast pheromone signalling pathway to regulators of cell polarity. Mol Gen Genet. 1996;252:608–621. doi: 10.1007/BF02172407. [DOI] [PubMed] [Google Scholar]
- 21.Leberer E, Dignard D, Harcus D, Thomas D Y, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signaling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 23.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall J E, Thomas D Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas D Y, Leberer E. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- 25.Leeuw T, Wu C, Schrag J D, Whiteway M, Thomas D Y, Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- 26.Lew D J, Reed S I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lew D J, Reed S I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993;120:1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Li M Z, Leibham D, Cortez D, Elledge S J. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- 29.Lyons D M, Mahanty S K, Choi K Y, Manandhar M, Elion E A. The SH3-domain protein Bem1 coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4095–4106. doi: 10.1128/mcb.16.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin G A, Bollag G, McCormick F, Abo A. A novel serine kinase activated by rac1/CDC42Hs-dependent autophosphorylation is related to PAK65 and STE20. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. . (Erratum, 14:4385.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosch H U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neiman A M, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc Natl Acad Sci USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nern A, Arkowitz R A. A GTP-exchange factor required for cell orientation. Nature. 1998;391:195–198. doi: 10.1038/34458. [DOI] [PubMed] [Google Scholar]
- 34.Oehlen L J W M, Cross F R. G1 cyclins CLN1 and CLN2 repress the mating factor response pathway at Start in the yeast cell cycle. Genes Dev. 1994;8:1058–1070. doi: 10.1101/gad.8.9.1058. [DOI] [PubMed] [Google Scholar]
- 35.Oehlen L J W M, Cross F R. The role of Cdc42 in signal transduction and mating of the budding yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:8556–8559. doi: 10.1074/jbc.273.15.8556. [DOI] [PubMed] [Google Scholar]
- 36.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 37.Peter M, Neiman A M, Park H-O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 38.Pringle J R, Bi E, Harkins H A, Zahner J E, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- 39.Pryciak P M, Hartwell L H. AKR1 encodes a candidate effector of the βγ complex in the Saccharomyces cerevisiae pheromone response pathway and contributes to control of both cell shape and signal transduction. Mol Cell Biol. 1996;16:2614–2626. doi: 10.1128/mcb.16.6.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pryciak P M, Huntress F A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed S I, Ferguson J, Groppe J. Preliminary characterization of the transcriptional and translational products of the Saccharomyces cerevisiae cell division cycle gene CDC28. Mol Cell Biol. 1982;2:415–425. doi: 10.1128/mcb.2.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Richardson H E, Wittenberg C, Cross F, Reed S I. An essential G1 function for cyclin-like proteins in yeast. Cell. 1989;59:1127–1133. doi: 10.1016/0092-8674(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Schild D, Brake A J, Kiefer M C, Young D, Barr P J. Cloning of three multifunctional de novo purine biosynthetic genes by functional complementation of yeast mutants. Proc Natl Acad Sci USA. 1990;87:2916–2920. doi: 10.1073/pnas.87.8.2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinjo K, Koland K G, Hart M J, Narasimhan V, Johnson D I, Evans T, Cerione R A. Molecular cloning of the gene for the human placental GTP-binding protein Gp (G25K): identification of this GTP-binding protein as the human homolog of the yeast cell-division cycle protein CDC42. Proc Natl Acad Sci USA. 1990;87:9853–9857. doi: 10.1073/pnas.87.24.9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon M N, De Virgilio C, Souza B, Pringle J R, Abo A, Reed S I. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 47.Stevenson B J, Ferguson B, De Virgilio C, Bi E, Pringle J R, Ammerer G, Sprague G F., Jr Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 1995;9:2949–2963. doi: 10.1101/gad.9.23.2949. [DOI] [PubMed] [Google Scholar]
- 48.Trueheart J, Boeke J, Fink G R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tu H, Wigler M. Genetic evidence for Pak1 autoinhibition and its release by Cdc42. Mol Cell Biol. 1999;19:602–611. doi: 10.1128/mcb.19.1.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whiteway M, Hougan L, Thomas D Y. Overexpression of the STE4 gene leads to mating response in haploid Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:217–222. doi: 10.1128/mcb.10.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whiteway M S, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas D Y, Leberer E. Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science. 1995;269:1572–1575. doi: 10.1126/science.7667635. [DOI] [PubMed] [Google Scholar]
- 52.Zenke F T, King C C, Bohl B P, Bokoch G M. Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem. 1999;274:32565–32573. doi: 10.1074/jbc.274.46.32565. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Z S, Leung T, Manser E, Lim L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42p and its activator Cdc24p. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Z S, Manser E, Chen X Q, Chong C, Leung T, Lim L. A conserved negative regulatory region in alphaPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol Cell Biol. 1998;18:2153–2163. doi: 10.1128/mcb.18.4.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziman M, O'Brien J M, Ouellette L A, Church W R, Johnson D I. Mutational analysis of CDC42Sc, a Saccharomyces cerevisiae gene that encodes a putative GTP-binding protein involved in the control of cell polarity. Mol Cell Biol. 1991;11:3537–3544. doi: 10.1128/mcb.11.7.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]