Abstract
Phenotypic analysis by flow cytometry is one of the most utilized primary tools to study the hematopoietic system. Here, we present a complex panel designed for spectral flow cytometry that allows for the in-depth analysis of the mouse hematopoietic stem and progenitor compartments. The developed panel encompasses the hematopoietic stem cell (HSC) compartment, an array of multipotent progenitors with early marks of lineage specification and a series of progenitors associated with lymphoid, granulo-macrophagic, megakaryocytic and erythroid lineage commitment. It has a built-in redundancy for key markers known to decipher the fine architecture of the HSC compartment by segregating subsets with different functional potential. As a resource, we used this panel to provide a snapshot view of the evolution of these phenotypically defined hematopoietic compartments during the life of the animals. We show that by using a spectral cytometer, this panel is compatible with the analysis of GFP-expressing gene-reporter mice across the hematopoietic system. We leverage this tool to determine how previously described markers such as CD150, CD34, CD105, CD41, ECPR, and CD49b define specific HSC subsets and confirm that high expression of the transcription factor Gfi1 is a hallmark of the most primitive HSC compartment. Altogether, our results provide a convenient protocol to obtain in one analysis a more extensive view of the hematopoietic architecture in mouse models. Our results could also serve as a base for further development of high-end panels leveraging spectral flow cytometry beyond the 15-fluorochrome panel presented in this report.
Keywords: immunophenotyping, spectral flow cytometry, mouse hematopoietic hierarchy, hematopoietic stem cell compartments, hematopoietic progenitors, aging, gene-reporter mouse model
Hematopoiesis is a well-characterized step-wise process that develops from a small population of self-renewing multipotent hematopoietic stem cells (HSC) to an assembly of progenitors with diverse proliferation and differentiation potentials. Assessment of the diversity of the hematopoietic stem and progenitor cell (HSPC) compartments has become more and more refined and complex (1,2). Thus, the functional definitions of early HSPC populations have dramatically changed. Studies of HSCs self-renewal potential that were traditionally monitored through their ability to reconstitute the hematopoiesis in lethally irradiated recipients over a 4-month-period have been extended to secondary and even tertiary transplantation experiments to reveal HSC subsets with different differentiation and self-renewal potentials (3). HSPC compartments previously identified as multipotent have been established as heterogeneous and carrying early signs of lineage specification (4,5). Molecular descriptions of this heterogeneity through gene expression or epigenetic analyses at the single cell level have uncovered new paths of lineage specification and differentiation (6). Besides these sophisticated assays, phenotypic analysis by flow cytometry remains a key tool to characterize the hematopoietic hierarchy. Notably, flow cytometry is the primary experimental assay to analyze hematopoietic disruptions associated with broad physio-pathological conditions or to characterize the impact of targeted molecular disruption in genetically modified mice. Recent years have been associated with a steady increase of the number of cell surface markers able to identify different hematopoietic cell populations and characterize their fine composition (7). However, much remains to be done in the field to compare the different phenotypic characterizations that have developed over time and used by different groups. Here, we combine several previously described phenotyping strategies that characterize multiple critical nodes of the mouse hematopoietic hierarchy from HSC sub-groups to progenitors committed toward the lymphoid, granulo-macrophagic, megakaryocytic and erythroid lineages. We show that combined panel enables the direct comparison of previously described HSC subsets and provides a versatile tool for the routine analysis of the earliest mouse hematopoietic compartments.
Materials and Methods
Mice
Wild-type C57BL/6J (B6.SJL-Ptprca Pepcb/BoyJ) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) (#002014) and Gfi1-GFP reporter mice were generously provided by Pr. Grimes (CCHMC) (8). Mice were housed at the AAALAC-accredited animal facility of the Cincinnati Children’s Hospital Medical Center (CCHMC). All animal experiments were approved by the CCHMC Institutional Animal Care and Use Committee.
Flow Cytometry
Bone marrow (BM) cells were flushed from mouse femurs and tibiae and treated with ACK (150 mM NH4Cl and 10 mM KHCO3) for 2 min at 4°C to lyse red blood cells (RBC). Short RBC lysis conditions were chosen as we observed a reduction of the erythoid progenitors (CFU-E and pre-CFU-E) in harsher condition. After treatment, cells were washed using Hank’s Buffered Salt Solution (HBSS, Gibco #14175-093) with 2% heat-inactivated fetal bovine serum (hereafter referred to as Staining Media, SM) and counted with a hemocytometer. For each analysis, two tubes of 107 unfractionated BM cells were simultaneously stained and combined for flow cytometry analysis. Reagents and optimized antibody dilutions are presented in Supplementary Table 1. BM cells were stained with unconjugated rat lineage-specific antibodies (Ter-119, Mac1, Gr-1, B220, CD5, CD3, CD4, CD8) in a volume of 500 μl of SM for 45 min at 4°C. Cells were washed with SM and stained with goat anti-rat PE-Cy5 secondary antibody in a volume of 400 μl of SM for 30 min at 4°C. After wash with SM, cells were stained with c-kit-APC-eFluor780, Sca1-PB, CD48-BV711, CD150-PE, Flk2-Biotin, FcγR-BV510, CD34-FITC, EPCR-PerCP-eFluor710, CD49b-PE/Dazzle594, CD41-BV605, CD105-APC, and CD127-BV785 antibodies in a volume of 200 μl for 30 min at 4°C in a 1:3 ratio of Brilliant Staining Buffer (BD #563-784)/SM. Secondary staining was performed with streptavidin-PE-Cy7 for 30 min at 4°C in a volume of 200 μl of SM. For dead cell exclusion, cells were washed with phosphate-buffered solution (PBS without serum, Corning #21-031-CV) and stained with Zombie NIR fixable viability kit for 15 min at room temperature. Cells were resuspended in SM and filtered through 70 μM nylon mesh before analysis. All single-stained controls, except Zombie NIR, were stained using UltraComp eBeads compensation beads used according the manufacturer’s instructions (1 drop, ~50 μl and stained with antibody concentrations listed in Supplementary Table 1. For the Zombie NIR single stained control, 106 healthy BM cells were mixed with 106 dead BM cells (obtained after 10 min treatment at 65°C) and resuspended in 100 μl Zombie NIR (1:200 dilution) for 15–30 min at room temperature. All antibody concentrations for single stained controls were optimized to ensure a brighter or equally bright signal as compared to the sample signal. Data were collected on a 4 Laser (16 Violet [405 nm] channels, 14 Blue [488 nm] channels, 10 Yellow-Green [561 nm] channels, 8 Red [640 nm] channels) Cytek Aurora spectral flow cytometer and analyzed with SpectroFlo software (Cytek Biosciences, Fremont, CA), which uses Ordinary Least Squares Linear Unmixing to deconvolute the different fluorescence spectra (9). Around 107 events (~10 min per sample) were recorded to ensure resolution of rare HSC populations. Comparison with “conventional” polychromatic cytometer were performed on a 5 Laser (UV [355 nm], Violet [405 nm], Blue [488 nm], Yellow-Green [561 nm], Red [635 nm]) BD LSRFortessa cell analyzer (BD Biosciences, San Jose, CA) (Supplementary Table 2). The same panel and controls were used with the exception of dead cell exclusion which was performed with Propidium Iodide (100 ng/ml) detected along with the lineage staining in the PE-Cy5 channel. Data was acquired, and compensation was performed using BD FACSDiva software (BD Biosciences).
Flow Cytometry Data Analysis
Data was analyzed using FlowJo v10.6.1 software (BD). Briefly, biexponential transformations were manually set for every fluorochrome to ensure accurate representation of event distributions and better data display. Offset histograms were presented in modal mode for better representation of small cell populations. An identical gating strategy was used for all presented samples. Time parameter was not used in the gating strategy, as no fluidic disturbances or signal anomalies were observed during sample acquisition. Gate positions were determined using fluorescence minus one (FMO) controls to delineate boundaries separating negative from positive staining. For HSC subsets defined within a continuum of marker expression, gates were adjusted based on population density to define low and high marker expressing populations. t-Distributed stochastic neighbor embedding (t-SNE) was used as an unsupervised nonlinear dimensionality reduction method to explore and visualize the multidimensional data generated with the panel (10). tSNE analyses of individual samples were performed on indicated populations based on all parameters (excluding forward scatter, side scatter, lineage, and dead cell parameters) with default FlowJo v10.6.1 software setting (Vantage point tree Algorithm; Iterations: 1000; perplexity: 30 Learning rate (eta): 128). Representative analyses are presented. Unmixed primary FCS files and FlowJo analysis Wsp files are available upon request.
Results
Development of a 14-Color Panel Describing Early Mouse Hematopoiesis
To facilitate a deeper characterization of the mouse hematopoietic tree, we combined several “classical” hematopoietic phenotyping strategies to encompass the entire early hematopoietic hierarchy from the HSC compartment to the diverse multipotent progenitor fractions and lineage-committed progenitors segregating the lymphoid, granulo-macrophagic, megakaryocytic, and erythroid cell fates (Fig. 1A). We included the original populations described by the Weissman laboratory as marking the commitment toward the lymphoid lineages (i.e., common lymphoid progenitor, CLP) or the myeloid lineages (i.e., common myeloid progenitor, CMP; granulo-macrophagic progenitor, GMP; megakaryocytic-erythroid progenitors, MEP) (11,12) (Fig. 1B). We further deciphered the hierarchy of the myelo-erythroid progenitors by combining the markers identified by the Bryder group to visualize the divergence of the megakaryocytic and erythroid potentials (13). Within the multipotent progenitor (MPP) compartment, we used Flk2 marker (14) and the signaling lymphocyte activation molecule (SLAM) family members CD48 and CD150 (15) to separate (i) a series of functionally distinct populations (classically denoted MPP2: LSK Flk2− CD48+ CD150+; MPP3: LSK Flk2− CD48+ CD150− and MPP4: LSK Flk2+ CD48+ CD150−) that carry the early marks of lineage specification (4) and (ii) a population of multipotent progenitors (denoted MPP5: LSK Flk2− CD48− CD150−) with limited reconstitution ability in transplantation experiments (16). We also used this set of markers to define a population denoted (HSC-SLAM: LSK Flk2− CD48− CD150+) that encompass all the long-term reconstitution potential detectable within the BM hematopoietic tissue (15). To fully reveal the diversity of this population, we combined several markers described in published reports to define HSC subsets with different self-renewal potential or different lineage output. This includes the CD34 marker that segregates the most immature MPP population (MPP1: LSK Flk2− CD48− CD150+ CD34+) with limited self-renewal potential (17). This also includes markers such as EPCR (also known as PROCR/CD201) (18,19), CD49b (Integrin α2) (20,21), CD41 (Integrin α2b) (3,22), or CD105 (Endoglin) (23,24). This particular combination of markers in one unique panel was designated to assess the phenotypic heterogeneity present in the HSC-SLAM compartment and determine how strategies used by different groups may overlap.
Figure 1.
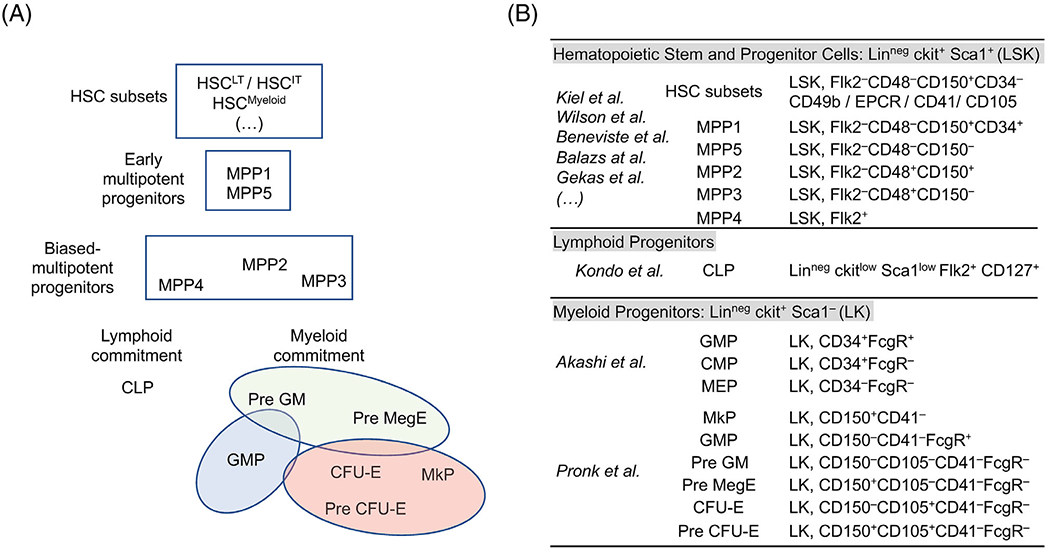
Definition of the hematopoietic populations of interest. (A) Cell populations of interest characterizing the murine hematopoietic hierarchy: Schematic shows (i) the heterogeneity of the earliest hematopoietic stem cells, including long term HSC subset (HSCLT) and its downstream intermediate-term progeny (HSCIT) as well as a HSC subset biased toward the myeloid lineage (HSCMyeloid); (ii) the most primitive MPP1 and MPP5 multipotent progenitors in addition to a series of lineage-biased multipotent progenitors, MPP2, MPP3, and MPP4 that are geared toward megakaryocytic, granulo-macrophagic and lymphoid differentiation, respectively; (iii) a compartment of lineage-committed progenitors containing the common lymphoid progenitors (CLP) that marks lymphoid-specification and a multitude of myeloid committed progenitors. This latter group includes two stages of granulocyte/macrophagic differentiation denoted PreGM and GMP as well as a hierarchy defining the megakaryocytic/erythroid (MegE) differentiation, composed of common MegE progenitors (Pre MegE) and specialized colony-forming erythroid (Pre CFU-E and CFU-E) and megakaryocytic (MkP) immature precursors. (B) Phenotypic characterization of various early hematopoietic stem and progenitor cell surface markers as defined by key seminal publications.
In our analysis, we used forward and side scatter to exclude cellular debris and doublets before investigating the cells for viability and removing mature hematopoietic cells that express one or more lineage markers (Ter119, Mac1, Gr1, B220, CD5, CD4, CD8, and CD3) (Fig. 2A). Within this lineage negative fraction, lineage-committed and multipotent progenitor fractions were defined based on the expression of c-kit and Sca1 (25,26). As previously described, CLPs were derived from the Linneg ckitlow Sca1low fraction (Fig. 2A, left panel), myeloid-committed progenitors from the Linneg ckit+ Sca1− (LK) population and the HSC/MPP populations from the Linneg ckit+ Sca1+ (LSK) compartment. Quality control for the spectral unmixing was performed by comparing the interaction of every parameter versus every other parameter within the live, lineage negative population (Supplementary Fig. 1). Gating for each population was determined with fluorescence minus one (FMO) controls with some adjustments based on population density or biological (different aged mice) comparisons (Supplementary Fig. 2). The results in wild-type adult mice, were consistent with previously published studies (11–13,15,27). Side-by-side comparisons between spectral and “conventional” polychromatic flow cytometers demonstrated consistent population distribution with minor differences in resolution (Supplementary Fig. 3). For the myeloid progenitors, high-dimensional analyses with tSNE (based on all parameters except lineage and live/dead) confirmed the complementarity of the panels described by Akashi et al. and Pronk et al. to study the granulo-macrophagic and erythro-megakaryocytic differentiation paths (Fig. 2A, central panel and Fig. 2B) (11,13). For the HSC compartment, we confirmed that CD49b and EPCR segregate the most immature CD34− HSC-SLAM fraction in two populations defined in the literature as long-term and intermediate-term HSCs (denoted HSCLT and HSCIT, respectively) (18–20) (Fig. 2A, right panel). The results demonstrates that CD49b and EPCR segregate identical populations based on percentage and cross-expression, with CD34lo CD49b− HSC-SLAM showing high EPCR expression and CD34lo EPCRhigh HSC-SLAM being CD49b negative. This was graphically illustrated by tSNE which (i) segregates the different MPP populations in LSK fraction and (ii) highlights the similarity of the HSC subsets defined based on the expression of CD49b and EPCR in the HSC-SLAM population (Fig. 2C,D). Altogether, these results establish a new panel to study early mouse hematopoietic hierarchy by spectral flow cytometry.
Figure 2.
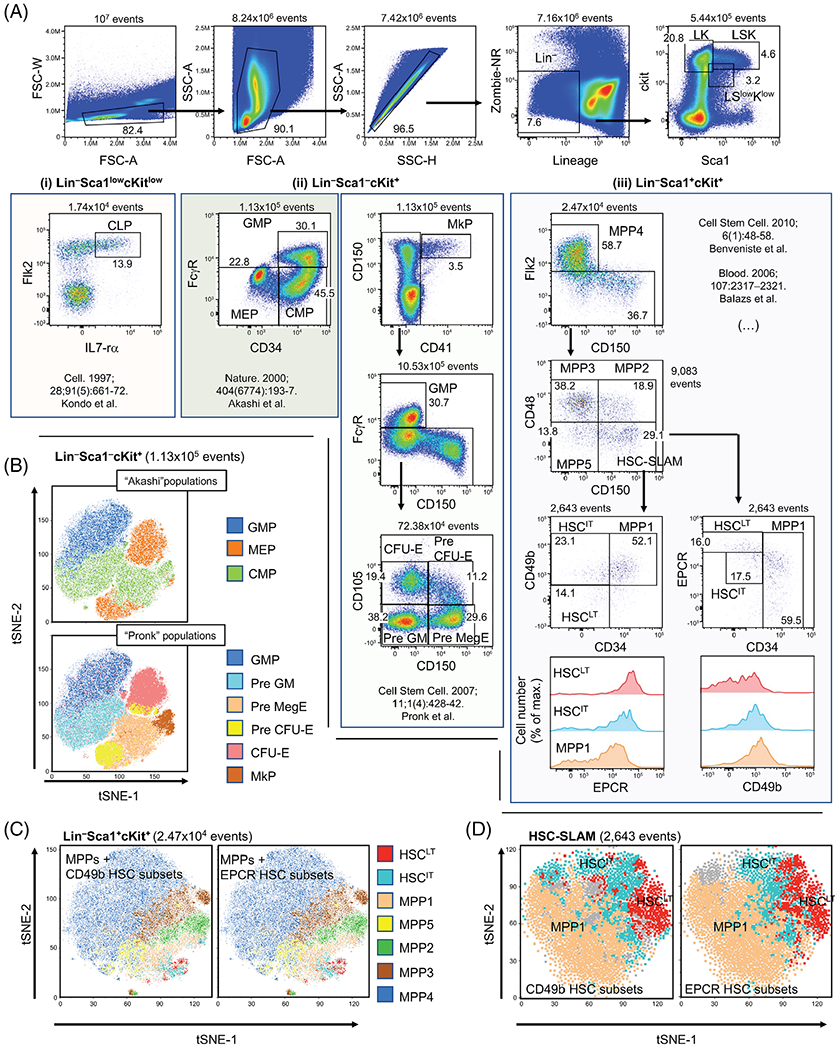
Combined analysis of the mouse hematopoietic stem and progenitor cell compartments. (A) Representative flow cytometry plots of the 14-parameter flow analysis on 8-week-old murine bone marrow (BM) cells: following erythrocyte lysis, BM cells were stained with fluorochrome-conjugated antibodies and a viability discriminator. BM cells were gated based on morphology to remove cellular debris and doublets. Lineage-negative (Lin−) live cells were analyzed for cKit and Sca1 expression: (i) Within the Lin− ckitlow sca1low population, the common lymphoid progenitor (CLP) was identified as described by Kondo et al. (12). (ii) Within the Lin−ckit+sca1−(LK) population, myeloid-committed progenitors (CMP, GMP, MEP, Pre GM, Pre MegE, Pre CFU-E, CFU-E and MkP) were identified as defined by Akashi et al. (11) or Pronk et al. (13). (iii) Within the Lin−ckit+sca1+ (LSK) population, early (MPP1 and MPP5) and lineage-biased (MPP2/MPP3/MPP4) multipotent progenitors were identified based on CD48, CD150 and CD34 expression (17). Comparative analysis of the hematopoietic stem cells (HSC) subsets was performed based on the expression of CD34, CD49b and EPCR (18,20). Figure is representative of seven independent experiments (see Supplementary Table 3). (B) High-dimensional analysis with t-distributed stochastic neighbor embedding (tSNE) (based on all parameters except lineage and live/dead) confirmed the complementarity of the results described by Akashi et al. (11) and Pronk et al. (13) to study the granulo-macrophagic and erythro-megakaryocytic differentiation paths. (C and D) tSNE analyses (based on all parameters except lineage and live/dead) segregates the different MPP populations in LSK fraction and indicates the similarity of the HSC subsets defined based on the expression of CD49b and EPCR in the LSK (C) and HSC-SLAM population (D).
Phenotypic Characterization of the Mouse Hematopoietic Tree at Different Ages
To further validate these results, we used the combined panel to illustrate the phenotypic changes of the hematopoietic tree occurring at different mouse stage of life (Fig. 3 and Supplementary Table 4). We included (i) 8-week-old adult mice as reference for steady-state hematopoiesis, (ii) 2-week-old mice, which correspond to a stage of quick development of the BM hematopoietic tissue characterized by the presence of cycling HSCs, (iii) 4-week-old mice when HSCs acquire their adult functional properties and finally 12-month-old mice as an example of aging hematopoiesis (28–31). We observed the expansion of the HSC-SLAM and MPP4 compartments during the transition between 2 and 4 weeks of age (Supplementary Fig. 4A and Supplementary Table 5). Adult HSC structure defined by number of HSCLT and HSCIT was established in 4-week-old mice (Supplementary Fig. 4B and Supplementary Table 5). As expected, we found an impairment of lymphoid specification pathway with age with progression reduction of the size of lymphoid-specified and lymphoid committed progenitors, MPP4 and CLP, respectively. 12-Month-old mice also showed the previously described expansion of the HSC-SLAM population with a dramatic reduction of the complexity of the compartment, which presents a phenotype similar to the most immature young HSCs (Fig. 3) (2). Notably, 12-month-old HSCs showed low CD34 expression and became homogeneously CD49b−, EPCRhigh and CD105+. Finally, the staining confirmed that old HSCs acquired CD41, a marker of myeloid-bias and aging (22,32). Altogether, the results further validate our gating strategy for the CD49b, CD105, and CD41 makers. They highlighted the progressive transition from an HSC heterogeneous immunophenotype in young adults that reflect their functional diversity to a homogeneous phenotype in aging mice where the proposed panels failed to reveal HSC diversity.
Figure 3.
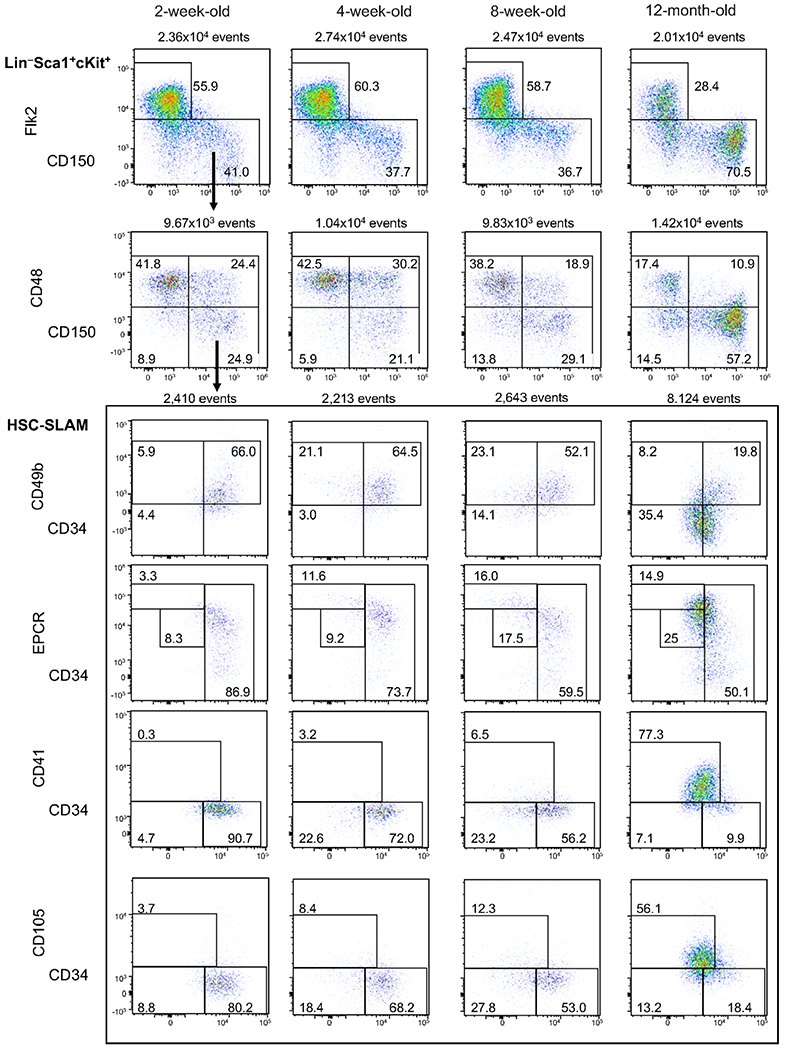
Complexity of the HSC compartment depending on age. Representative flow cytometry plots of the 14-parameter flow analysis on BM cells isolated from 2- and 4-week-old juvenile mice, 8-week-old adult mice and 12-month-old mice. Shown plots were gated on live LSK cells as indicated in Figure 2. Figure is representative of 3-7 independent experiments for each age group (see Supplementary Table 4).
Gfi1 as a Marker of the Most Immature HSC Subset
Finally, we investigated the compatibility of this panel for the analysis of GFP-expressing gene-reporter mice. We focused on Gfi1, a well-described transcriptional factor regulating several key cell fate decisions across the hematopoietic system (33). We used our new panel on BM cells isolated from a knock-in Gfi1 reporter mouse that carries a GFP cassette inserted in the endogenous Gfi1 locus and allows for the monitoring of the activity of this locus at the single cell level. Despite the expected negative correlation in the negative populations due to high spectral similarity between CD34-FITC and GFP (Supplementary Fig. S5A and S5B), we were able to detect a specific GFP signal in all analyzed populations (Fig. 4A) (34). As expected for the myeloid progenitors, we found that Gfi1 expression increases during the granulo-macrophagic differentiation and conversely decreases during erythro-megakaryocytic differentiation (Fig. 4A, left panel). Similar trends were observed in the LSK fraction with the megakaryocytic-biased MPP2 showing lower Gfi1 expression than the lymphoid-biased MPP4 and myeloid-biased MPP3 (Fig. 4A, right panel). Consistent with a previous report, we confirmed that the highest Gfi1 expression occurs in the most immature HSC subset defined by CD49b and EPCR expression (35). To further illustrate this point, we used tSNE analysis to determine the most useful markers to identify the specific subsets in the CD34− HSC-SLAM compartment (Fig. 4B). This analysis confirmed that Gfi1-high expression coincides with the CD49b− EPCRhigh CD34lo HSC-SLAM set of cells. Lack of CD41 or high level of Sca1 expression seems to also mark the HSCLT fraction, although with reduced specificity. In contrast, high level of CD150 and CD105 expression appears not discriminant into the CD34− HSC-SLAM population. Altogether, our results validate the use of our panel in GFP-expressing gene-reporter mice and confirm that Gfi1 is highly expressed in the most immature HSC subset phenotypically identified so far.
Figure 4.
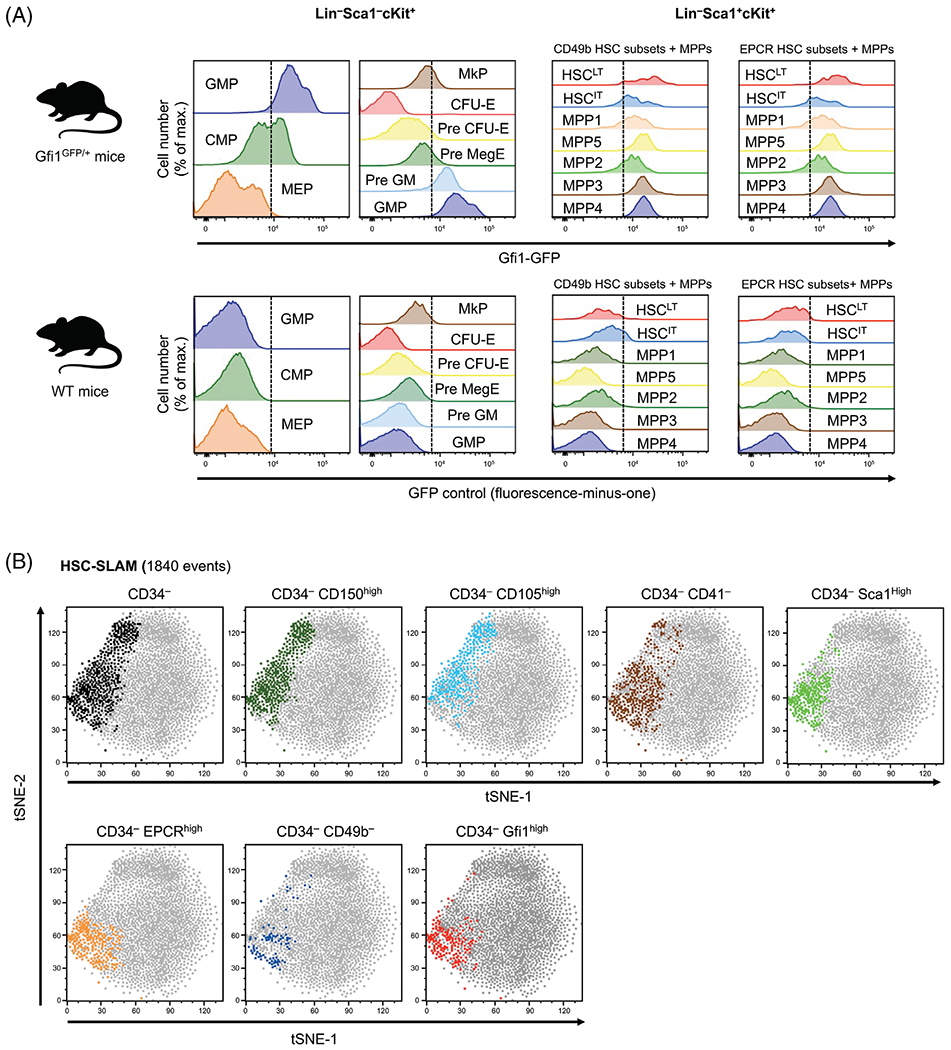
Gfi1 expression in hematopoietic stem and progenitor cells. (A) Analysis of Gfi1GFP/+ reporter mice: Heterogeneity of Gfi1 expression in (i) various myeloid-committed progenitors defined by Akashi et al. (11) and Pronk et al. (13) (left panels) and (ii) multipotent progenitors and EPCR-/CD49b-defined HSC subsets (right panels). Lower panel shows nonspecific fluorescence detection in the GFP channel for various hematopoietic compartments in WT mice. Dash line separates negative from positive signal based on the FMO. (B) tSNE analyses (based on all parameters except lineage and live/dead) on HSC-SLAM population (1840 events in the gate) demonstrating how expression of CD150, CD105, CD41, Sca1, EPCR, CD49b, or Gfi1 highlight the heterogeneity of the immature CD34− HSC-SLAM compartment. Figure is representative of two independent experiments.
Discussion
Flow cytometry is widely used as an investigative tool to qualitatively and quantitatively assess the hematopoietic hierarchy (13,27,36). Here, we combined several previously published flow cytometry panels and used spectral flow cytometry to facilitate the analysis of the key hematopoietic nodes that reflect self-renewal and transplantation potentials as well as early specification and commitment toward the different hematopoietic lineages. This report also highlights the importance to account for the phenotypic heterogeneity of the HSC compartment to describe hematologic characteristics. Multiple phenotypic definitions of the HSC compartment can be found in the literature. Across laboratories, HSCs are defined in the LSK fraction based on the expression of (i) CD34 and Flk2, (ii) CD105, or (iii) the SLAM markers CD48 and CD150 (14,15,23,37). Further refinement of these heterogeneous compartments has been recently established based on the combination of these markers or the addition of new ones, such as EPCR, CD49b, and CD41 (18,20,22). The complex architecture of the HSC compartment and this heterogeneity of markers could be a source of misinterpretation for certain phenotypes and discrepancies between studies. By combining these markers, the described panel allows to assess the degree of phenotypic overlap between these HSC populations. We found a reassuring correlation between high expression of EPCR and the lack of expression of CD49b on CD34− HSC-SLAM, as both populations have been functionally shown to contain the most immature HSCLT subset with the strongest serial transplantation ability (20,38). Similarly, low EPCR expression and CD49b acquisition indicate a downstream intermediate HSCIT subset characterized functionally by time-restricted reconstitution ability (20,21). All other phenotypes based on high expression levels of CD150, Sca1, or CD105 failed to be fully discriminant in separating these two HSC subsets. We further leveraged spectral cytometry to detect GFP in the Gfi1-reporter mouse and showed that high expression of Gfi1 is a marker of the most immature HSCLT subset, highlighting its known contributions to HSC functions (33,35). Finally, our results emphasize the loss of HSC-SLAM phenotypic heterogeneity associated with age as 12-month-old mice showed by the acquisition of a homogeneous CD34−/lo EPCRhi CD49b− phenotype with increased expression of CD150, CD41, and CD105. Altogether, the protocol described here aims to serve as a base for deeper phenotypic characterization of the early mouse hematopoietic compartment. The report also provides useful reference points to reconcile the multiple phenotypes described in the literature for identification of these functionally diverse hematopoietic populations.
Supplementary Material
Acknowledgments
We acknowledge the assistance of the CCHMC Research Flow Cytometry core (supported by a National Institutes of Health grant S10OD025045). This work was supported by a National Institutes of Health grant (R01HL141418) to D.R. The authors would like to thank Jamie Fellers for proofreading the manuscript.
Grant sponsor: National Heart, Lung, and Blood Institute, Grant numberR01HL141418; Grant sponsor: National Institute of General Medical Sciences, Grant numberS10OD025045
Footnotes
Additional Supporting Information may be found in the online version of this article.
Conflict of Interest
M.D. is an employee and receives salaries of Cytek Biosciences. M.S. and D.R. declare no competing financial interests.
LITERATURE CITED
- 1.Haas S, Trumpp A, Milsom MD. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell 2018;22:627–638. [DOI] [PubMed] [Google Scholar]
- 2.Jurecic R Hematopoietic stem cell heterogeneity. Adv Exp Med Biol 2019;1169:195–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, Ema H, Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell 2013;154:1112–1126. [DOI] [PubMed] [Google Scholar]
- 4.Cabezas-Wallscheid N, Klimmeck D, Hansson J, Lipka DB, Reyes A, Wang Q, Weichenhan D, Lier A, von Paleske L, Renders S, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell 2014;15:507–522. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, Panero R, Patel SH, Jankovic M, Sun J, Calogero RA, Klein AM, Camargo FD. Clonal analysis of lineage fate in native haematopoiesis. Nature 2018;553:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 2016;537:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med 2010;207:1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yucel R, Kosan C, Heyd F, Moroy T. Gfi1:Green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J Biol Chem 2004;279:40906–40917. [DOI] [PubMed] [Google Scholar]
- 9.Futamura K, Sekino M, Hata A, Ikebuchi R, Nakanishi Y, Egawa G, Kabashima K, Watanabe T, Furuki M, Tomura M. Novel full-spectral flow cytometry with multiple spectrally-adjacent fluorescent proteins and fluorochromes and visualization of in vivo cellular movement. Cytometry Part A 2015;87A:830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res 2008;9:2579–2605. [Google Scholar]
- 11.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 2000;404:193–197. [DOI] [PubMed] [Google Scholar]
- 12.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 1997;91:661–672. [DOI] [PubMed] [Google Scholar]
- 13.Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell 2007;1:428–442. [DOI] [PubMed] [Google Scholar]
- 14.Adolfsson J, Borge OJ, Bryder D, Theilgaard-Monch K, Astrand-Grundstrom I, Sitnicka E, Sasaki Y, Jacobsen SE. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity 2001;15:659–669. [DOI] [PubMed] [Google Scholar]
- 15.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 2005;121:1109–1121. [DOI] [PubMed] [Google Scholar]
- 16.Pietras EM, Reynaud D, Kang YA, Carlin D, Calero-Nieto FJ, Leavitt AD, Stuart JM, Gottgens B, Passegue E. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 2015;17:35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 2008;135:1118–1129. [DOI] [PubMed] [Google Scholar]
- 18.Balazs AB, Fabian AJ, Esmon CT, Mulligan RC. Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood 2006;107:2317–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent DG, Copley MR, Benz C, Wohrer S, Dykstra BJ, Ma E, Cheyne J, Zhao Y, Bowie MB, Zhao Y, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood 2009;113:6342–6350. [DOI] [PubMed] [Google Scholar]
- 20.Benveniste P, Frelin C, Janmohamed S, Barbara M, Herrington R, Hyam D, Iscove NN. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell 2010;6:48–58. [DOI] [PubMed] [Google Scholar]
- 21.Qian P, He XC, Paulson A, Li Z, Tao F, Perry JM, Guo F, Zhao M, Zhi L, Venkatraman A, et al. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell 2016;18:214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gekas C, Graf T. CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood 2013;121:4463–4472. [DOI] [PubMed] [Google Scholar]
- 23.Chen CZ, Li M, de Graaf D, Monti S, Gottgens B, Sanchez MJ, Lander ES, Golub TR, Green AR, Lodish HF. Identification of endoglin as a functional marker that defines long-term repopulating hematopoietic stem cells. Proc Natl Acad Sci U S A 2002;99:15468–15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pronk CJH, Bryder D. Immunophenotypic identification of early myeloerythroid development. Methods Mol Biol 2018;1678:301–319. [DOI] [PubMed] [Google Scholar]
- 25.Okada S, Nakauchi H, Nagayoshi K, Nishikawa S, Miura Y, Suda T. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood 1992;80:3044–3050. [PubMed] [Google Scholar]
- 26.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science 1988;241:58–62. [DOI] [PubMed] [Google Scholar]
- 27.Eich M, Trumpp A, Schmitt S. OMIP-059: Identification of mouse hematopoietic stem and progenitor cells with simultaneous detection of CD45.1/2 and controllable green fluorescent protein expression by a single staining panel. Cytometry Part A 2019;95A:1049–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowie MB, Kent DG, Dykstra B, McKnight KD, McCaffrey L, Hoodless PA, Eaves CJ. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci U S A 2007;104:5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest 2006;116:2808–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Haan G, Lazare SS. Aging of hematopoietic stem cells. Blood 2018;131:479–487. [DOI] [PubMed] [Google Scholar]
- 31.Geiger H, Denkinger M, Schirmbeck R. Hematopoietic stem cell aging. Curr Opin Immunol 2014;29:86–92. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto R, Wilkinson AC, Ooehara J, Lan X, Lai CY, Nakauchi Y, Pritchard JK, Nakauchi H. Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell 2018;22:600–607.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phelan JD, Shroyer NF, Cook T, Gebelein B, Grimes HL. Gfi1-cells and circuits: Unraveling transcriptional networks of development and disease. Curr Opin Hematol 2010;17:300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roederer M Distributions of autofluorescence after compensation: Be panglossian, fret not. Cytometry Part A 2016;89A:398–402. [DOI] [PubMed] [Google Scholar]
- 35.Lee JM, Govindarajah V, Goddard B, Hinge A, Muench DE, Filippi MD, Aronow B, Cancelas JA, Salomonis N, Grimes HL, et al. Obesity alters the long-term fitness of the hematopoietic stem cell compartment through modulation of Gfi1 expression. J Exp Med 2018;215:627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson NK, Kent DG, Buettner F, Shehata M, Macaulay IC, Calero-Nieto FJ, Sanchez Castillo M, Oedekoven CA, Diamanti E, Schulte R, et al. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell Stem Cell 2015;16:712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science 1996;273:242–245. [DOI] [PubMed] [Google Scholar]
- 38.Gur-Cohen S, Itkin T, Chakrabarty S, Graf C, Kollet O, Ludin A, Golan K, Kalinkovich A, Ledergor G, Wong E, et al. PAR1 signaling regulates the retention and recruitment of EPCR-expressing bone marrow hematopoietic stem cells. Nat Med 2015;21:1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


