Abstract
De novo variation in SCN2A can give rise to severe childhood disorders. Biophysical gain of function in SCN2A is seen in some patients with early seizure onset developmental and epileptic encephalopathy (DEE). In these cases, targeted reduction in SCN2A expression could substantially improve clinical outcomes. We tested this theory by central administration of a gapmer antisense oligonucleotide (ASO) targeting Scn2a mRNA in a mouse model of Scn2a early seizure onset DEE (Q/+ mice). Untreated Q/+ mice presented with spontaneous seizures at P1 and did not survive beyond P30. Administration of the ASO to Q/+ mice reduced spontaneous seizures and significantly extended life span. Across a range of behavioral tests, Scn2a ASO-treated Q/+ mice were largely indistinguishable from WT mice, suggesting treatment is well tolerated. A human SCN2A gapmer ASO could likewise impact the lives of patients with SCN2A gain-of-function DEE.
Keywords: Neuroscience
Keywords: Epilepsy
Introduction
Developmental and epileptic encephalopathies (DEEs) are devastating neurological disorders presenting during infancy and early childhood (1). The affected children have ongoing, refractory seizures in addition to profound global developmental delay, intellectual disability, and movement disorders (1). The prognosis of these patients is poor, marked by progressive disability and increased risk of early death (1). The limited treatment options, presence of comorbidities, and the need for long-term supported care represent a burden for affected patients, caregivers, and health services.
Only a decade ago little was known about DEE etiologies, but the employment of next-generation sequencing has identified single-gene de novo pathogenic variations as the main cause (2). The number of identified DEE genes is increasing, with those encoding ion channels, as the pivotal brain excitability regulators, being the most represented (2–4). Identification of the genetic cause and increased understanding of the mechanistic basis of DEEs have laid the foundation for the development of precision medicine therapeutic approaches. This is especially important as the current treatments primarily focus on the symptomatic control of seizures while the developmental aspects of the disease remain untended.
Several major technological advances have enabled the possibility of establishing effective targeted gene therapies for DEEs. After several decades of development, antisense oligonucleotides (ASOs) have recently been approved as a treatment for spinal muscular atrophy and Duchenne muscular dystrophy, which demonstrated that RNA-targeted therapies are a viable therapeutic strategy for these devastating neurogenetic disorders (5, 6). ASOs are single-stranded oligonucleotides that are designed to specifically target the RNA of interest through Watson-Crick base pairing, which results in RNase H1-mediated mRNA degradation, modulation of pre-mRNA splicing, or regulation of translational efficiency (7, 8).
SCN2A encodes the α-subunit of a voltage-gated sodium channel (Nav1.2) and is involved in the initiation and conduction of action potentials, particularly during early postnatal development when its expression is higher than that of other voltage-gated sodium channel isoforms (9, 10). In recent years, this gene was implicated in a range of neurodevelopmental disorders (11, 12). The phenotypic spectrum associated with SCN2A de novo variants includes schizophrenia, autism spectrum disorder, intellectual disability, and a myriad of epileptic syndromes including DEEs (11–15). Case history studies of patients with SCN2A DEE have identified 2 major clinical presentations. Based on the age of seizure onset, pharmacoresponsiveness, and disease severity, we recognize DEE with an early-onset seizures starting within the first 3 months of life and DEE with late-onset seizures commencing between 3 months and 4 years of age (13). Functional investigation of a number of SCN2A DEE pathogenic variations has shown that the early-onset seizure phenotype is caused by gain of function of SCN2A whereas the late-onset seizure DEE is linked to a loss-of-function patho-mechanism (11, 16). These studies correlate well with the observation that patients with early onset of seizures respond better to antiseizure drugs that nonselectively block sodium channel function, such as phenytoin (13). Moreover, this observation suggests that patients could be amenable to gene therapy designed to reduce SCN2A expression.
To corroborate this hypothesis, we engineered a mouse model of SCN2A DEE with early seizure onset based on the human equivalent of c.5645G>A variant, which results in the protein change p.R1882Q. This is one of the most recurrent pathogenic variants for this form of SCN2A DEE, with 6 cases reported to date (12). In some of these patients, phenytoin was reported to partially improve seizure outcomes (13). However, within the therapeutic window of phenytoin, seizures are not fully resolved and developmental impairment remains unaddressed. In vitro biophysical analysis confirmed the SCN2A p.R1882Q variant causes larger peak and persistent sodium currents, with dynamic clamp simulations predicting a higher action potential firing frequency (16).
Here, an antisense oligonucleotide (ASO) specifically designed to downregulate mouse Scn2a mRNA expression (Scn2a ASO) was delivered to the mouse model carrying the R1882Q corresponding variant (Q/+). The Q/+ mice develop spontaneous seizures at P1 and suffer premature death, thus recapitulating the severe symptoms observed in patients with early seizure onset DEE. We show that Scn2a ASO–mediated knockdown effectively prevents premature death and suppresses spontaneous seizures in the Q/+ mouse model. The Scn2a ASO–treated Q/+ mice behaved similarly to WT (+/+) mice in almost all other motor and psychosocial tests. Scn2a ASO is the first therapy to specifically address the disease phenotype of SCN2A early seizure onset DEE, positioning ASO therapy as a promising strategy for SCN2A gain-of-function diseases.
Results
The SCN2A gain-of-function DEE mouse model.
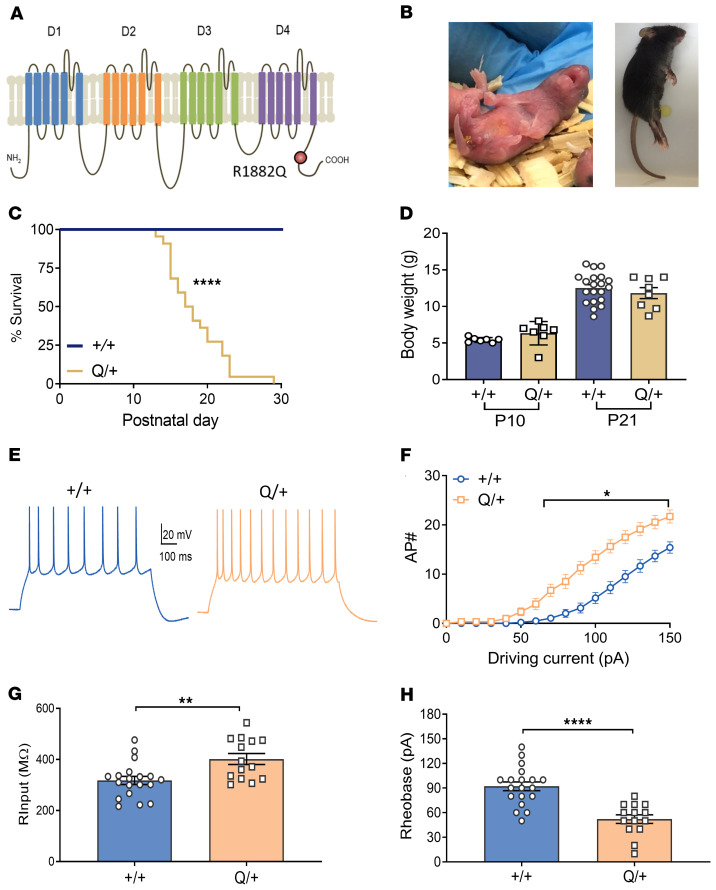
The SCN2A R1882Q variant is located at the C-terminal tail (Figure 1A). Mirroring the clinical presentation of SCN2A p.R1882Q DEE, Q/+ mice have a severe, early seizure onset phenotype. Spontaneous tonic seizures were observed as early as P1, which later developed into tonic-clonic seizures with hindlimb extension (Figure 1B). The earliest death occurred at P13 and the median survival was P18 (Figure 1C). No significant difference in body weight was found between Q/+ and +/+ mice when measured at P10 or P21 (Figure 1D). Because no Q/+ mice survived beyond P30, and they present with frequent seizures, there was a limited opportunity for behavioral characterization.
Figure 1. Disease phenotype of the Q/+ mouse model.
(A) Schematic presentation of SCN2A channel depicting 4 domains (D1–D4), each comprised of 6 transmembrane regions, and the intracellular N and C-terminus of the channel. The p.R1882Q variant is predicted to affect the C-terminus of the channel. (B) Images of Q/+ mice undergoing spontaneous seizure at P1 (left) and P25 (right). (C) Survival curves of Q/+ and +/+ mice. (+/+ n = 19, Q/+ n = 22). ****P < 0.0001, log rank test. (D) Body weight measured on P10 and P21 (+/+ n = 7–20, Q/+ n = 7–8). (E) Representative voltage traces from a neuron injected with 100 pA current. Scale bar applies to all traces. (F) Input-output relationship generated for each injected current step (+/+ n = 3 mice, 20 cells, Q/+ n = 3 mice, 14 cells). *P < 0.05 (F (15, 512) = 6.301, 2-way ANOVA with Sidak’s multiple comparison. (G) Input resistance. **P < 0.005 (t = 3.22, df = 31). (H) Rheobase. **** P < 0.0001 (t = 5, df = 31), unpaired t test. Data are represented as mean ± SEM.
Dynamic clamp modeling predicted SCN2A p.R1882Q to increase action potential firing (16), but this has not been verified in vivo. Thus, whole-cell recording was performed in L2/3 cortical pyramidal neurons in brain slices of +/+ and Q/+ mice. In comparison to +/+ mice, pyramidal neurons from Q/+ mice showed overt gain-of-function phenotype, including higher action potential frequency (Figure 1, E and F). The maximal firing frequency was 15.4 ± 1.16 and 21.7 ± 1.32 in +/+ and Q/+ neurons, respectively. Furthermore, Q/+ neurons showed higher input resistance (Figure 1G) and lower rheobase (Figure 1H).
Scn2a ASO reduces Scn2a mRNA and protein levels.
An ASO targeting mouse Scn2a (GCTCATGTTACTCCTACCCT) was designed to test the hypothesis that reducing Scn2a expression would be therapeutic for Q/+ mice which express the SCN2A gain-of-function pathogenic variant. The Scn2a ASO was synthesized with five 2′-O-methoxyethyl (MOE) modified nucleotides at each end of the oligonucleotide, and 10 DNA nucleotides in the center. Because we aim to find an ASO therapy for SCN2A gain-of-function variants in general, the Scn2a ASO designed here is not allele specific.
Using a pan-ASO antibody (17), we confirmed broad ASO distribution throughout the brain of mice injected via the intracerebroventricular (i.c.v.) route at P1 (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI152079DS1). Quantification of overlapping ASO and neuronal marker in 10 different brain regions revealed an average of 97.6% ± 0.2% of neurons examined contained Scn2a ASO (Supplemental Table 1, n = 4 mice).
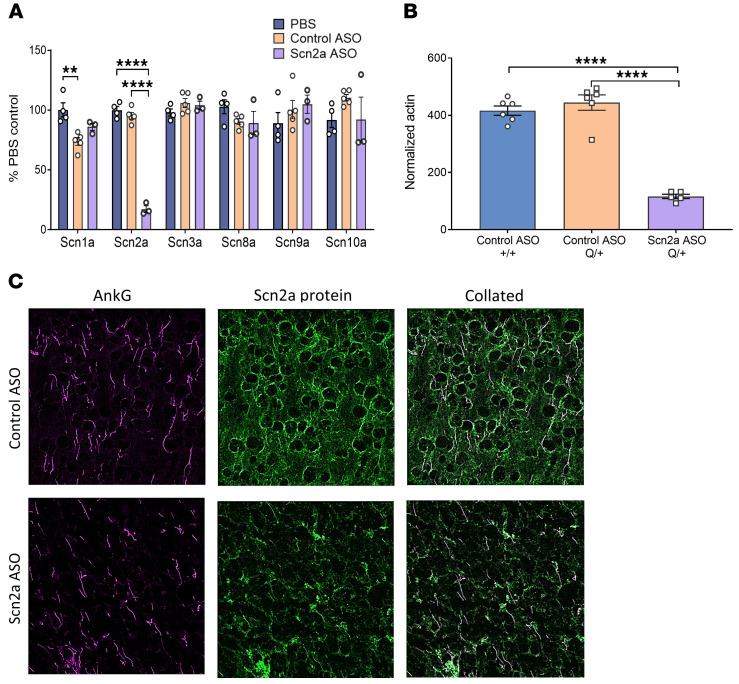
Scn2a ASO specifically reduced Scn2a mRNA and had no effect on the other voltage-gated sodium channels (Figure 2A). At the protein level, mass spectrometry determined a 73.9% reduction in Scn2a protein levels in the Scn2a ASO–treated mice when compared with the control nontargeting ASO group (Figure 2B). Using an anti-Scn2a protein antibody (Supplemental Figure 2), immunohistochemical analysis also provided qualitative data suggesting that Scn2a protein expression was reduced along the axonal initial segment and in neuropils (Figure 2C). To determine the effective dose for 50% (ED50) Scn2a mRNA reduction, mice were i.c.v. injected with Scn2a ASO at P1, P15, and P30 and Scn2a mRNA level measured 2 weeks after i.c.v. injection (Supplemental Figure 3). The Scn2a ASO ED50 determined for P1, P15, and P30 were 2 μg, 20 μg, and 33 μg, respectively.
Figure 2. Scn2a mRNA and protein expression were reduced in mice i.c.v. injected with Scn2a ASO ED80 at P1.
(A) Percentage of voltage-gated sodium channel isoform mRNA remaining in the cortex. n = 3–5 +/+ mice for each treatment group. **** P < 0.0001 (F (2, 9 = 212.6)), **P < 0.01 (F (2, 9 = 10.30)), 1-way ANOVA with Tukey’s multiple comparison. (B) Level of Scn2a protein normalized to actin as determined by mass spectrometry, n = 5 to 6 mice for each treatment group. **** P < 0.0001 (F(2, 14 = 79.80)), 1-way ANOVA with Tukey’s multiple comparison. (C) Z-stack images of cortical slices labelled by AnkG (magenta) and Scn2a protein (green). One independent experiment, n = 2–3 +/+ mice for each treatment group. Scale bar: 20 μm. Data are represented as mean ± SEM.
Scn2a ASO rescues premature death and seizure phenotype in Q/+ mice.
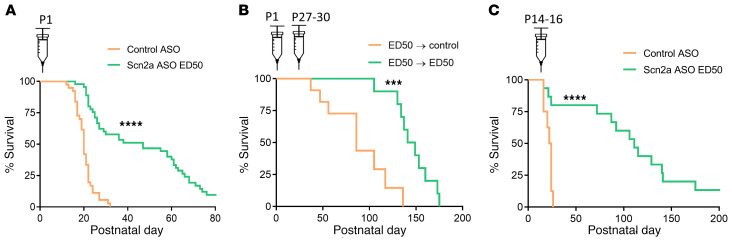
To test whether the Scn2a ASO can mitigate disease in the SCN2A early seizure onset DEE mouse model, we i.c.v. injected Q/+ mice with a single dose of Scn2a ASO ED50 or control ASO at P1. As expected, control ASO did not affect survival and median survival was P20 (n = 39 mice, Figure 3A). In sharp contrast, a single dose of Scn2a ASO ED50 at P1 more than doubled the median survival to P47 (n = 49 mice, Figure 3A). At experimental endpoint (P80), 9.63% of the Scn2a ASO ED50 group remained alive.
Figure 3. Scn2a ASO ED50 extends survival of Q/+ mice.
(A) Survival curves of Q/+ mice i.c.v. injected with Scn2a ASO ED50 or the negative control ASO at P1. Control ASO (50 μg, n = 39), Scn2a ASO ED50 (2 μg, n = 49). (B) Survival curves of Q/+ mice i.c.v. injected with Scn2a ASO ED50 (2 μg) at P1, then redosed with control ASO (170 μg, n = 11) or Scn2a ASO ED50 (33 μg, n = 15). (C) Survival curves of Q/+ mice i.c.v. injected with Scn2a ASO ED50 or control ASO at P14–P16. Control ASO (85 μg, n = 8), Scn2a ASO ED50 (20 μg, n = 15). Syringes indicate time of i.c.v. injection. ****P < 0.0001, ***P < 0.005, log rank test.
To determine if survival could be further extended by repeated administration, Q/+ mice were first i.c.v. injected at P1 with Scn2a ASO ED50. At P27–P30, a second dose of Scn2a ASO ED50 or control ASO was administered. Q/+ mice that received 2 doses of Scn2a ASO ED50 had significantly longer life span than those redosed with the control ASO (median survival for control ASO: P86, n = 11; for Scn2a ASO ED50: P145, n = 15) (Figure 3B). This suggests that repeated dosing in later development maintained the therapeutic level of Scn2a ASO, effectively suppressing disease reemergence.
In practice, the ASO therapy would be delivered after genetic diagnosis and the DEE symptoms would have likely progressed before initiating ASO treatment. Therefore, it was critical to assess the efficacy of Scn2a ASO administration commencing at a later stage of disease development. Since Q/+ mice have spontaneous seizures at P1 and mortality was first observed at P13, Scn2a ASO ED50 was administered at the upper limit of P14–P16. We found that the life span of Q/+ mice was still significantly extended with the delayed therapeutic intervention. Moreover, in this treatment regime, Scn2a ASO ED50 extended median survival to P111 (n = 15, Figure 3C). The median survival for control ASO was P23 (n = 8, Figure 3C). These data strongly indicate that later therapeutic intervention with Scn2a ASO still has compelling disease reversing capability.
Once it was established that Scn2a ASO treatment significantly extended survival, we tested its seizure-modifying ability. After Scn2a ASO administration at P1, Q/+ mice were placed under 24-hour video monitoring at P21, and later at P30. Seizures above Racine score 4 were counted (18). During P21 video monitoring, the control ASO group had a total of 21 seizures (n = 8 mice), while only 1 seizure was observed in the Scn2a ASO ED50 group (n = 16 mice). In the P30 video monitoring, 1 seizure was observed in the Scn2a ASO ED50 group (n = 17 mice), while no mice survived in the control ASO group.
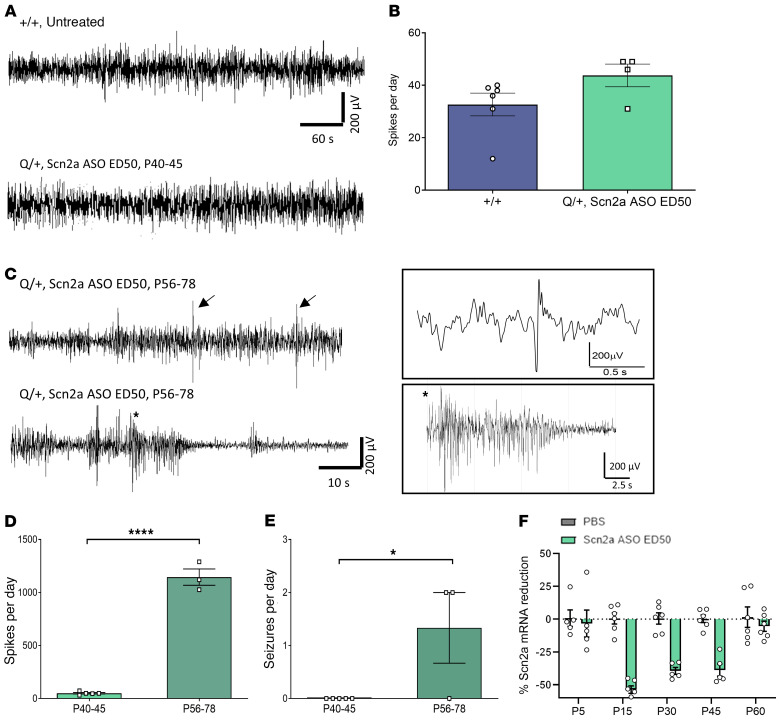
Since not all seizures have behavioral manifestation, 24-hour electrocorticography (ECoG) recordings were performed. No electrographic seizures were detected in Scn2a ASO ED50 Q/+ mice recorded at P40–P45. Furthermore, the number of spikes recorded on ECoG was similar to age-matched, untreated +/+ mice (Figure 4, A and B). When the ECoG was repeated at P56–P78, inter-ictal spikes were significantly increased compared with earlier P40–P45 recordings, and spontaneous seizures were detected (Figure 4, C–E).
Figure 4. Scn2a ASO ED50 suppresses seizure phenotype of Q/+ mice.
(A) Representative ECoG traces recorded at P40–P45. Scale bar applies to all traces. (B) Number of spikes during 24-hour ECoG recorded at P40–P45 (+/+ n = 6, Scn2a ASO ED50 Q/+ n = 4). (C) Representative ECoG traces recorded at P56–P78 in Q/+ mice treated with Scn2a ASO ED50 at P1. Top panel: arrows indicate inter-ictal spikes. Bottom panel: seizure followed by post-ictal depression recorded on ECoG. Trace in box showed time expanded view of spike and seizure. (D and E) Number of spikes (D) or seizures (E) determined at different time points in Q/+ mice i.c.v. injected with Scn2a ASO ED50 at P1 (P40–P45 n = 5, P56–P78 n = 3). ***P < 0.005 (t = 19.25, df = 6), *P < 0.05 (t = 2.739, df = 6), unpaired 2 tailed t test. (F) Time course of Scn2a mRNA reduction in mice i.c.v. injected with Scn2a ASO ED50 at P1, n = 5–6 per group per time point. Data are represented as mean ± SEM.
To determine the basis underlying seizure phenotype reemergence, we investigated the time course of Scn2a mRNA expression after Scn2a ASO ED50 administration at P1. Maximum Scn2a reduction (53.70% ± 2.82%) was observed at P15, which was then decreased to 39.45% ± 2.62% and 38.83% ± 4.65% at P30 and P45 respectively (Figure 4F). At P60 when seizure features reappeared in ECoG, Scn2a mRNA was only reduced by 5.46% ± 3.89% (Figure 4F). A simple linear regression was fitted from P15–P60 (equation y = 0.9849 × –70.86, R2 = 0.7361), and the time point at which Scn2a mRNA suppression was halved was P45. This strongly indicates that disease reemergence in Scn2a ASO ED50–treated Q/+ mice correlated with an increase in Scn2a expression from the initial knockdown levels.
Behavioral phenotype of Scn2a ASO ED50–treated Q/+ mice.
Voltage-gated sodium channels are critical for early rodent neurodevelopment, and nonselective sodium channel blockers such as phenytoin are known to cause neurotoxicity when administered to neonatal mice (19). Thus, a battery of behavioral tests was performed in Q/+ mice treated with Scn2a ASO (Figure 5A). As no untreated Q/+ mice survived after P30, comparisons in behavioral tests had to be made to age- and sex-matched untreated +/+ mice, instead of to the control ASO group.
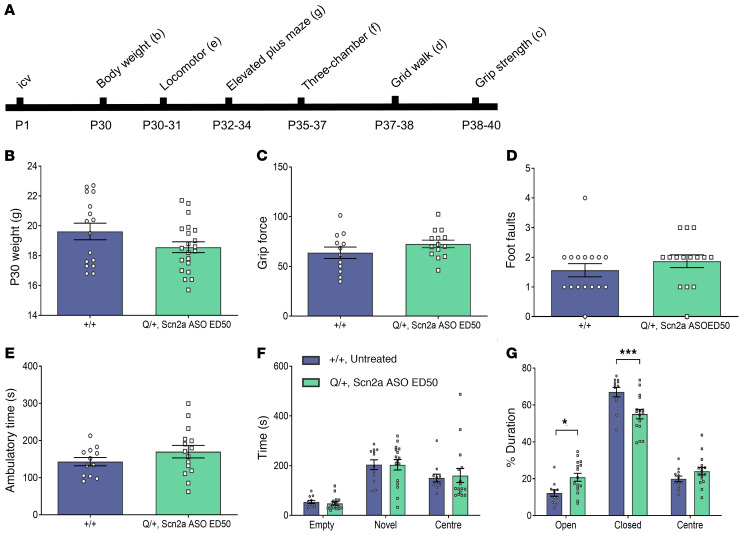
Figure 5. Behavioral comparison between Scn2a ASO ED50 treated Q/+ mice and untreated +/+ mice.
(A) Timeline for behavioral studies. (B) Body weight measured at P30. (C) Grip force. (D) Number of foot faults in grid walk test. (E) Ambulatory time measured in locomotor chamber. (F) Time spent in the different compartments of 3-chamber social interaction test. (G) Duration spent in different arms of the elevated plus maze. ***P < 0.001, *P < 0.05 (F (2, 78) = 11.94), 2-way ANOVA with Sidak’s multiple comparison, n = 10–21 per genotype. Data are represented as mean ± SEM.
The Q/+ mice i.c.v. injected with Scn2a ASO ED50 at P1 had body weights similar to +/+ mice (Figure 5B). Looking at motor function, the Scn2a ASO ED50–treated Q/+ mice had limb muscle strength (Figure 5C) and number of foot faults similar to +/+ mice on grid walk test (Figure 5D). The Scn2a ASO ED50 treatment at P1 had no impact on the ambulatory time measured in locomotor assay (Figure 5E).
We further assessed the impact of Scn2a ASO on social function. In the 3-chamber social interaction test, Scn2a ASO ED50–treated Q/+ mice spent equivalent time interacting with a novel mouse as +/+ mice, indicating normal social approach (Figure 5F). Interestingly, on the elevated plus maze, Scn2a ASO ED50 Q/+ mice spent more time in the open arm and less time in the closed arm when compared with +/+ mice (Figure 5G).
Scn2a ASO ED80 shows higher efficacy but also changes multiple behaviors.
We next investigated whether increasing ASO dose to cause an 80% Scn2a reduction (Scn2a ASO ED80) would further improve survival and seizure outcomes. Scn2a ASO ED80 was determined to be 10 μg from dose-response curves for P1 administration (Supplemental Figure 3A). Q/+ mice were i.c.v. injected with Scn2a ASO ED80 at P1, and median survival was increased to P86 with this higher dose (Figure 6A). To monitor seizures, 24-hour video monitoring was performed. At P21 no seizures were observed (n = 10 mice) and at P30 only 1 seizure was observed (n = 13 mice). Consistently, ECoG found similar spikes between Scn2a ASO ED80 Q/+ mice and age-matched, untreated +/+ mice (Figure 6B). This indicates that the efficacy of Scn2a ASO is dose dependent.
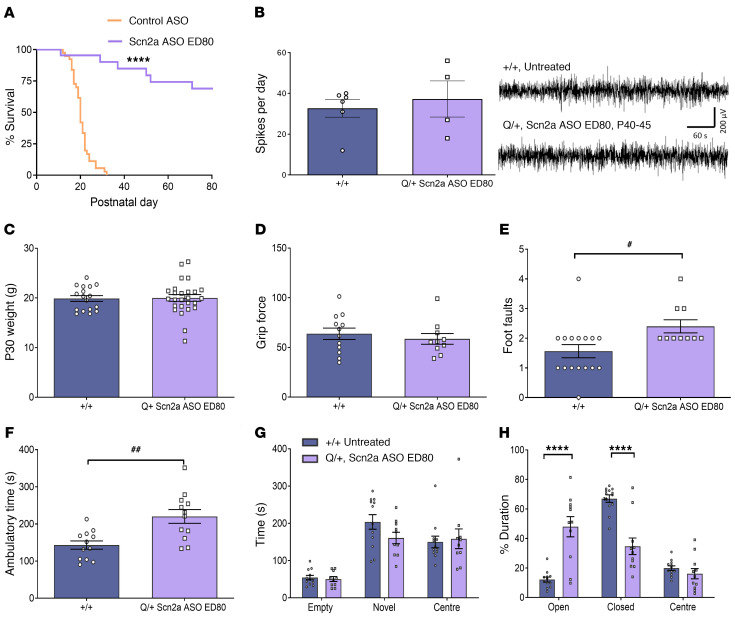
Figure 6. Effect of administering Scn2a ASO ED80 in Q/+ mice at P1.
(A) Survival curves of Q/+ mice i.c.v. injected with Scn2a ASO ED80 or the negative control ASO at P1. Control ASO (50 μg, n = 39), Scn2a ASO ED80 (10 μg, n = 22). (B) Left: number of spikes during 24-hour ECoG recorded at P40–P45 (+/+ n = 6, Scn2a ASO ED80 Q/+ n = 4). Right: representative ECoG traces. Scale bar applies to all traces. (C) Body weight measured at P30. (D) Grip force. (E) Number of foot faults in grid walk test. (F) Ambulatory time measured in locomotor chamber. (G) Duration spent in different arms of the elevated plus maze. (H) Time spent in the different compartments of 3-chamber social interaction test. ##P < 0.005 (t = 3.557, df = 22), #P < 0.05 (t = 2.518, df = 24), unpaired 2 tailed t test. ****P < 0.0001, (F (2, 63) = 36.72), 2-way ANOVA with Sidak’s multiple comparison, n = 10 to 26 per genotype. Data are represented as mean ± SEM.
The body weights of +/+ mice and Scn2a ASO ED80 treated Q/+ mice were similar as measured at P30 (Figure 6C). However, Scn2a ASO ED80 resulted in motor function changes that were not noted in the ED50 group. While the limb strength of Scn2a ASO ED80 Q/+ mice were similar to +/+ mice (Figure 6D) on the grid walk test, Scn2a ASO ED80 Q/+ mice had more foot faults than untreated +/+ mice (Figure 6E). They also showed higher ambulatory time in the locomotor assay (Figure 6F). The Scn2a ASO ED80 Q/+ mice also spent less time in the closed arm and more time in the open arm of the elevated plus maze (Figure 6G), and the magnitude of change was greater than in Scn2a ASO ED50 Q/+ mice. Taken together, these data indicate that while the ED80 dose resulted in better survival outcomes than the ED50 dose, it also caused more behavior changes.
Tolerability of Scn2a ASO ED50 in neonatal +/+ mice.
As part of the preclinical safety assessment, the tolerability of Scn2a ASO ED50 was examined in +/+ mice. It is critical to understand the safety of Scn2a ASO in the absence of gain-of-function variant, as Scn2a depletion in early rodent development is detrimental (20, 21). Consistently, administering Scn2a ASO ED80 into P1 +/+ mice resulted in early mortality and smaller body weight when compared with the control ASO group (Supplemental Figure 4). Early lethality and body weight reduction were not observed when the Scn2a ASO dose was lowered to ED50 (Figure 7A).
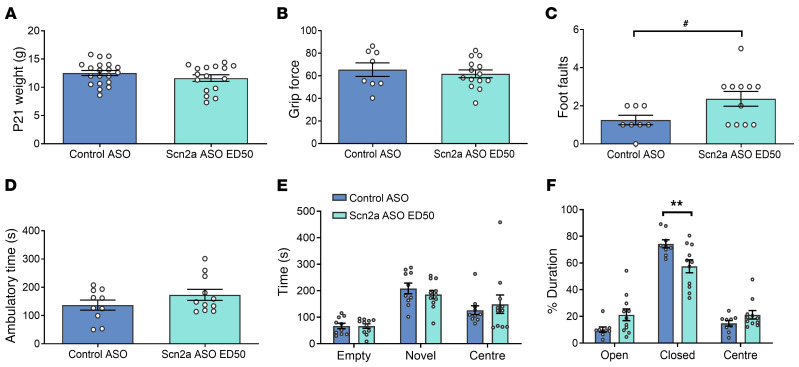
Figure 7. Effect of Scn2a ASO ED50 administration in P1 +/+ mice.
(A) Body weight measured at P21. (B) Grip force. (C) Number of foot faults in grid walk test. #P < 0.05 (t = 2.207, df = 17), unpaired 2-tailed t test. (D) Ambulatory time measured in locomotor chamber. (E) Time spent in the different compartments of 3-chamber social interaction test. (F) Percentage duration spent in different arms of the elevated plus maze. **P < 0.01 (F (5, 54) = 8.512), 2-way ANOVA with Sidak’s multiple comparison, n = 8 to 20 per genotype. Data are represented as mean ± SEM.
The same behavioral studies performed on Q/+ mice were repeated in +/+ mice i.c.v. injected with Scn2a ASO ED50 or control ASO at P1. The Scn2a ASO ED50 treatment at P1 did not alter limb muscle strength (Figure 7B), ambulatory time in locomotor assay (Figure 7D), or social approach (Figure 7E) in +/+ mice. However, Scn2a ASO ED50 caused higher number of foot faults on the grid walk test in +/+ mice (Figure 7C). It should be noted that deficits in the grid walk assay were not observed for Q/+ mice administered ASOs at ED50, suggesting that a lower side effect profile may occur in the disease context. As observed previously in Scn2a ASO–treated Q/+ mice, Scn2a ASO ED50 +/+ mice also spent less time in the closed arm of the elevated plus maze (Figure 7F).
Discussion
Patients with the early seizure onset DEE caused by SCN2A gain-of-function variants have poor clinical outcomes, indicating an urgent need for novel treatments. The correlation between genetic variants, functional effect, and clinical phenotype is well defined (11, 12, 16), making SCN2A a credible candidate to trial ASO therapy to downregulate its expression. In this study, we demonstrated therapeutic benefits of ASO-mediated Scn2a mRNA and protein downregulation in a mouse model carrying one of the most recurrent SCN2A gain-of-function DEE variants. Critically, the Scn2a ASO ED50 treatment prolonged life span in the diseased mice and rescued the seizure phenotype when given at P1. With detailed testing of Scn2a ASO injection regimens, we showed that repeated administration provided better survival outcomes. In addition, ASO treatment at a later stage of disease development was also beneficial. These results are pertinent, as multiple dosing and therapy commencement after disease onset are the most likely scenarios expected in the clinical setting. Last, given the critical role of SCN2A in neurodevelopment and the fact that its loss of function has been linked to different disease phenotypes, we explored the therapeutic window of ASO-induced Scn2a downregulation. We identified that at the ED50 dose, efficacy was achieved with limited behavioral side effects in Q/+ mice. The ED80 dose achieved higher efficacy in terms of survival and seizure suppression but resulted in more behavioral changes in Q/+ mice.
Following 30 years of development, ASO technology has gained increasing recognition with the recent clinical success of ASO treatments for spinal muscular atrophy and Duchenne muscular dystrophy (5, 6). Recently, the ASO approach has demonstrated compelling efficacy in animal models of SCN1A and SCN8A epilepsies (22, 23). The ASO approach has several advantages over other gene therapy approaches. First, the introduced change does not affect genetic information carried in the DNA but targets the intermediate mRNA molecules and the resulting protein production, circumventing potential irreversible off-target changes to the patient genome. Second, the effect of ASO on RNA is transient and any potential adverse effects can be stopped simply by refraining from further ASO administration. In this regard, an alternative strategy currently in development is to administer a fully complementary ASO decoy to neutralize the targeting ASO (24). Further, the effect of ASO treatment is dose dependent and can be adjusted, enabling identification of a specific therapeutic window for each target and/or patient. Finally, ASOs show different modes of action, which can either affect the stability of mRNA or its splicing and interactions with other molecules (8).
To provide evidence that an ASO is a viable therapeutic strategy for SCN2A gain-of-function DEEs, we generated a knockin mouse model carrying a recurrent gain-of-function SCN2A variant linked to early seizure onset DEE. Compared with other existing SCN2A gain-of-function mouse models (25, 26), the Q/+ mouse model has a severe disease phenotype, presenting with spontaneous seizures and premature death, with all untreated mice dying before P30. Summation of all available data indicate our model recapitulates the severe and early seizure phenotype of SCN2A gain-of-function disease.
Despite the disease severity, a single bolus of Scn2a ASO ED50 administered at P1 was able to extend survival and reduce seizure frequency of Q/+ mice. This effect was correlated with ASO-mediated Scn2a downregulation at both the mRNA and protein levels. We demonstrated that the seizure phenotype reemerges when ASO is cleared from the brain over 60 days, which leads to the increase in Scn2a mRNA levels. A previous study showed that the in vivo half-life of ASO is several months (24). In the present study, the half-life of ASO is likely shorter, as it was administered at P1 and the rodent brain underwent rapid volume increase during development (27), thereby diluting the concentration of ASO in brain. Nevertheless, the seizure-suppressing effect of a single Scn2a ASO dose is still long-lasting, suggesting only infrequent dosing may be required in patients. Importantly, redosing at a later time point resulted in further protection from the severe seizure phenotype. It is noteworthy that reemergence of pathology that can be rescued by additional ASO treatment indicates that the acute impact of the pathogenic variant must underlie some proportion of the DEE phenotype. Importantly, we also showed that administration of Scn2a ASO 2 weeks after disease onset in mice was able to extend survival and reduce seizure frequency.
Testing of different dosing regimens has important implications for clinical development. Our data suggest a therapeutic regime may resemble the one currently employed for Nusinersen where upon initial “loading,” subsequent injections occur every 3 to 4 months (28–30). Last, as the ASO drug class cannot pass the blood-brain barrier, ASO therapies for neurological disorders such as DEE would likely be delivered clinically via intrathecal injections. Intrathecal injection has proven to be an effective route of ASO delivery and is a technique that can be safely performed in adults as well as children (31, 32).
Since SCN2A is critical for early postnatal development, the safety of ASO mediated Scn2a downregulation was explored in this study. The therapeutic window of Scn2a ASO depends on the disease state, that is, whether there is Scn2a gain-of-function. This was demonstrated by the contrasting effect of Scn2a ASO ED80 in Q/+ and in +/+ mice, where the same treatment prolonged survival in Q/+ mice but caused early mortality in +/+ mice. This suggests that a Scn2a gain-of-function variant would increase the tolerability to excessive Scn2a reduction.
Due to premature death and frequent spontaneous seizures, the behavioral profile of untreated Q/+ mouse model could not be ascertained. Despite these limitations, we were able to show that Q/+ mice administered with Scn2a ASO ED50 at P1 had a behavioral profile similar to untreated +/+ mice, with the exception in the elevated plus maze, where Scn2a ASO ED50 Q/+ mice spent more time in the open arm and less time in the closed arm of the elevated plus maze. Importantly, this altered behavior on the elevated plus maze was also observed in Scn2a ED50 +/+ mice, which do not express the gain-of-function allele. Therefore, altered elevated plus maze behavior is unlikely part of the SCN2A gain-of-function disease phenotype. Rather, it is likely attributed to exaggerated pharmacology caused by action against the intended target (33), in this case, Scn2a downregulation. Indeed, increased time in the open arm of the elevated plus maze was previously reported in mouse models of Scn2a haploinsufficiency (34, 35). Another aspect of exaggerated pharmacology was altered motor function, such as increased foot faults on grid walk test in Scn2a ED50 +/+ mice, which was not observed in Scn2a ED50 Q/+ mice. The behavioral data again highlights that Scn2a ASO exaggerated pharmacology is not only dose dependent, but also disease state dependent. Thus, a gain-of-function diagnosis is critical when considering Scn2a downregulatory ASO as a therapy.
As it is clear that the therapeutic window of Scn2a downregulatory ASO would be limited by exaggerated pharmacology, one limitation in this study is that only 2 Scn2a ASO doses were tested in Q/+ mice. While Scn2a ED80 caused greater efficacy than ED50 in terms of extending life span, it also resulted in more behavioral changes, akin to Scn2a haploinsufficiency. In contrast to ED80, the ED50 dose did not alter motor function, although it still increased the time Q/+ mice spent on the open arm of the elevated plus maze. This rodent behavior has been interpreted as reduced anxiety or increased risk-seeking activity (36), but it is unclear if this is a direct correlate with human anxiety behavior. Ideally, a Scn2a ASO dose should be efficacious and without any signs of exaggerated pharmacology. Future preclinical study could achieve this by administering Q/+ mice with a range of ASO doses below ED50, and assessing the survival, seizure, and behavioral outcomes at each lowered dose. The frequency of administration to maintain therapeutic level would also have to be explored. Our study already provides indirect evidence that ED40 is still of therapeutic benefit. In the experiment that tracked the time course of Scn2a ASO ED50 action from P1, Scn2a mRNA reduction was approximately 40% from P30–P45. We performed ECoG in Q/+ mice during that time period and detected no seizures or increases in inter-ictal spikes.
The clinical spectrum of SCN2A disorder continues to expand and many loss-of-function variants have been implicated in late seizure onset DEE and autism spectrum disorder. These patients would most likely not benefit from a SCN2A downregulatory ASO (11, 37). However, ASO technology can be used to upregulate gene and protein levels (38, 39), therefore also presenting an alternative therapeutic strategy for disorders caused by SCN2A loss-of-function variants.
In summary, this preclinical study demonstrated the therapeutic viability of ASO-mediated gene silencing for SCN2A gain-of-function disorders. The clinical development pathway for ASOs is well established and this study provides crucial efficacy and toxicology preclinical data for the SCN2A gain-of-function epilepsies that can be readily translated into clinical practice in the near future.
Methods
ASO preparation.
We screened a collection of ASOs designed to target various regions of the mouse Scn2a mRNA. After screening a few hundred ASOs for their ability to reduce Scn2a levels in cultured mouse primary cortical neurons, the lead ASOs were i.c.v. injected into P1 +/+ mice. The most optimal ASO candidate was selected based on criteria as previously described (40). Selection criteria include parameters such as in vivo activity and tolerability. An ASO targeting mouse Scn2a (GCTCATGTTACTCCTACCCT) and a nontargeting control ASO (CCTATAGGACTATCCAGGAA) were used for the studies. Both ASOs were developed and synthesized by Ionis Pharmaceuticals. ASOs were synthesized as described (38) and were 20 bp in length, with 5 MOE-modified nucleotides at each end of the oligonucleotide, and 10 DNA nucleotides in the center. The backbone of the ASOs consists of a mixture of phosphorothioate (PS) and phosphodiester (PO) linkages: 1-PS, 4-PO, 10-PS, 2-PO, and 2-PS (5′ to 3′). ASOs were reconstituted with sterile Ca2+ and Mg2+ free PBS (Gibco).
Animal model.
Male WT (+/+) mice or mice heterozygous for Scn2a p.1883Q, equivalent to human pathogenic variant p.R1882Q (Q/+), were commercially generated on the C57/Bl6N background using the TurboKnockout technology (Cyagen). The point variant encoding p.R1883Q (CGG to CAG) was introduced into exon 26 in the 5′ homology arm of the mouse Scn2a gene (NM_001099298.2). The targeting vector was generated by PCR using the BAC clone (Supplemental Figure 5). Within the targeting vector, the Neo cassette was flanked by LoxP sites, and diphtheria toxin A (DTA) was used for negative selection. The Neo cassette was engineered to be capable of self-deletion between the 2 LoxP sites without the need to breed to Flp deleter mice. This targeting vector was subsequently delivered into embryonic stem cells (ESCs) via electroporation. These ESCs were modified to generate 100% ESC-derived founder mice rather than chimeras, eliminating the need to screen for germline transmission. Targeting vector insertion was then confirmed by PCR screening, Southern blot, and sequencing. The targeted embryonic stem cell clones were then selected for blastocyst microinjection and implanted into surrogate dams. The F1 litters were used for experiments. The Scn2a knockout mouse model was generated on the C57/BL6J background (Australian National University) and contains a protein truncating variation, R851X. All animals were maintained in a temperature-controlled room, with a 12-hour light on/off cycle and free access to food and liquid.
Brain slice recording.
Mice (P12–P14) were deeply anesthetized with 4% isoflurane, followed by brain tissue isolation. The brain was immediately transferred into ice-cold cutting solution consisting of 125 mM Choline-Cl, 2.5 mM KCl, 0.4 mM CaCl2, 6 mM MgCl2, 1.25 mM NaH2PO4, 26 mM NaHCO3, and 20 mM d-glucose saturated with carbogen (95% oxygen and 5% carbon dioxide). Coronal sections (300 μm) were sliced on a vibratome (VT1200, Leica). The brain slices were incubated in artificial cerebral spinal fluid consisting of 125 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 1.25 mM NaH2PO4, 26 mM NaHCO3, and 10 mM d-glucose saturated with carbogen for at least 1 hour at room temperature before recording.
Individual slices were placed in a recording chamber on an upright microscope (Slicescope Pro 1000, Scientifica) and perfused with aCSF at a rate of 2 mL/minute at 32°C. Layer 2 or layer 3 excitatory neurons in the somatosensory cortex (S1) were identified with infrared-oblique illumination microscopy with a 40× objective lens (Olympus). Patch pipettes of 3 to 5 MΩ (Harvard Apparatus) were made using a puller (P-1000, Sutter Instruments) and were filled with internal solution consisting of 125 mM K-gluconate, 5 mM KCl, 2 mM MgCl2, 10 mM HEPES, 4 mM ATP-Mg, 0.3 mM GTP-Na, 10 mM phosphocretine, 10 mM EGTA and pH to 7.3 with an osmolarity of 280 mOsm. Whole-cell recording was made in current clamp mode using Axon Multiclamp 700B amplifiers (Molecular Devices). Data were acquired using pClamp v.10 software (Molecular Devices). Sampling frequency was 100 kHz and low-pass Bessel was filtered at 10 kHz (Digidata 1550, Molecular Devices). A holding current was injected to maintain membrane potential at approximately –70 mV. Neuronal excitability was determined by measuring voltage during a series of 800 ms steps from –60 to +300 pA in 10 pA increments every 2 seconds. Data were analyzed using the Axograph X software (Axograph).
Intracerebroventricular injection.
P1 mice were cryo-anesthetized, then a Hamilton syringe (32 G Huber point) was inserted midway between lambda and right eye. A total 2 μL injection containing PBS, control ASO, or Scn2a ASO was delivered at a depth of 2 mm below skin surface into the right ventricle. Pups were gently warmed until skin color returned to pink before being returned to home cage.
For mice over P14, i.c.v. injection was performed using stereotaxic apparatus (Kopf). Mice were anesthetized with 2% to 4% isoflurane and placed on a frame with the skull surface horizontal between lambda and bregma. A small piece of skull was removed by drilling, and an injection needle (30 G, PlasticsOne) was inserted at 0.25 mm posterior, 0.8 mm lateral to the bregma. The needle was lowered to a depth of 2.5 to 3 mm from brain surface. A total 5 μL injection containing PBS, control ASO, or Scn2a ASO was delivered into the right ventricle using the syringe pump (KD Scientific) at a rate of 0.5 μL/s. After 2–3 minutes, the needle was slowly withdrawn, and the incision was sutured.
Quantitative gene expression analysis (RT-qPCR).
Mice were i.c.v. injected with Scn2a ASO or control ASO (10 μg) at P1 for the voltage-gated sodium channel isoform qPCR (Figure 2A). Two weeks after i.c.v. injection, brains were harvested and snap-frozen in liquid nitrogen. Cortical tissue was homogenized and total RNA isolated using Trizol reagent according to the manufacturer’s instructions (Invitrogen). Contaminating genomic DNA was removed with DNAse I treatment (Ambion/Life Technologies). RNA was assayed for quality and quantity using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific). For RT-qPCR, 1μg DNase-treated purified RNA was analyzed using a Taqman RNA to Ct kit process (Applied Biosystems) with a Taqman probe to each sodium channel gene in duplex with the mouse endogenous gene Glucuronidase Beta (Gusb). RT-qPCR was performed on the ViiA 7 Real-Time PCR System (Applied Biosystems) using cycling conditions specified by kit protocols (Supplemental Table 2). All RT-qPCR reactions were performed in triplicate. Relative gene abundance values were calculated by normalization to Gusb and referenced to the control groups using the 2ΔΔCt method.
For the dose-response curves (Supplemental Figure 3), mice were i.c.v. injected with a range of Scn2a ASO at P1, P15, or P30. Two weeks after i.c.v. injection, brains were harvested and snap-frozen in liquid nitrogen. Total RNA was isolated using the Trizol reagent according to the manufacturer’s protocols (Thermo Fisher Scientific). Contaminating genomic DNA was removed with DNAse treatment (DNA-free Reagents, Ambion/Life Technologies). For RT-qPCR, oligo-dT primed cDNA was synthesized from 500 ng total RNA using Murine Moloney Leukemia Virus Reverse Transcriptase (Promega). RT-qPCR was performed on the ViiA 7 Real-Time PCR System, using SYBR green technology and GoTaq qPCR master mix (Promega) according to the manufacturer’s protocols. The primers used for Scn2a detection were forward TGCTGTGCGGAAATCTGCC and reverse CGGATGCTCAAGAGAGACTGG. Relative gene expression values were obtained by normalization to the endogenous reference gene RPL32 (forward GAGGTGCTGCTGATGTGC, reverse GGCGTTGGGATTGGTGACT) and referenced to the control group using the 2ΔΔCt method.
Immunohistochemistry.
To confirm the specificity of the anti-Scn2a protein antibody, brain sections were collected from P0 Scn2a homozygous knockout (R851X) mice as negative controls (Supplemental Figure 2). As shown in Figure 2C, mice were i.c.v. injected with control ASO or Scn2a ASO (10 μg) at P1, and brain tissue was isolated between P12–P15 and immediately frozen with liquid nitrogen. For Scn2a protein visualization, coronal sections (10 μm) were cut on a cryostat (CM1850, Leica) and mounted on Superfrost plus microscope slides (VWR). After air-drying at room temperature for 30 minutes, sections were fixed in precooled acetone for 8 minutes. Sections were blocked for 1 hour in room temperature with Tris buffered saline supplemented with 3% goat serum and 0.3% Triton. Sections were then incubated with the following antibodies: polyclonal rabbit anti-AnkyrinG antibody (clone H-215, catalog sc28-561, Santa Cruz) at 1:1000, and monoclonal mouse anti-Scn2a protein (anti-Nav1.2 Na channel antibody, clone K69/3, catalog 75-024, Neuromab) at 1:500 overnight at 4°C. After 5 washes, sections were incubated with fluorescently labelled secondary antibodies: donkey anti-rabbit-Alexa 647 at 1:800 (catalog A-31573, Invitrogen) and goat anti-mouse-Alexa 488 at 1:500 (catalog A11-001, Invitrogen) for 1 hour at room temperature. To identify the nuclei, sections were then stained with DAPI (4′,6-Diamidine-2′-phenylindole dihydrochloride, catalog D9542, Sigma-Aldrich). Slides were covered with Prolong Gold Antifade (Invitrogen) and stored at –30°C. The L2/3 layer of somatosensory cortex was examined, and confocal images captured using a Zeiss Axio 780 upright microscope, using a PL-APO 40x/NA 1.4 oil objective and according to Nyquist criterion.
For ASO visualization (Supplemental Figure 1), mice were i.c.v. injected with ASO 10 μg at P1 were perfused with 4% paraformaldehyde at P14. The brains were isolated and sequentially equilibrated in 10% to 30% sucrose (in 0.1 M PB) at 4°C. The brains were then embedded in OCT, frozen in chilled isopentane, and stored at –80°C until use. Sagittal sections (30 μm) were cut using a cryostat (CM1850, Leica) and mounted on positively charged slides. Sections were processed for immunohistochemistry to label neurons using NeuN (1:500; anti-NeuN antibody, clone A60, catalog ABN90, Merck Millipore), Calbindin (1:1000; mouse monoclonal, clone L109/57, catalog 75-448, UC Davis), and ASO backbone (1:7500; Ionis; ref. 17) with guinea pig and rabbit polyclonal primary antibodies. Tissue was incubated with primaries diluted in 5% normal goat serum and 0.15% Triton-X in 0.1 M PB overnight and washed several times before incubating with secondary antibodies, donkey anti–guinea pig Alexa594 (catalog 20170, Biotium) and donkey anti-rabbit Alexa 647 (catalog A-31573, Invitrogen) for 2 hours at room temperature. Sections were then washed in 0.1 M PB, DAPI-stained (catalog D9542, Sigma-Aldrich) and mounted using ProLong Diamond antifade reagent (Thermo Fisher Scientific) and cover slipped. Confocal images were acquired using a Zeiss Axio 780 upright microscope. Mosaic and z-step images were taken with a PL-APO 20x/NA0.8 air objective and high-resolution images using the PL-APO 40x/NA 1.4 oil objective and according to Nyquist criterion. All images were then deconvolved using Huygens Essential (V15.10; Scientific Volume Imaging).
Quantification of ASO-positive neurons.
For the quantification of ASO-positive neurons (Supplemental Table 1), the percentage of NeuN/ASO-positive cells was measured using ImageJ software (V1.53g). Images were acquired as outlined in immunohistochemistry (see above) and antibodies included ASO backbone, DAPI, NeuN for the neocortex, and calbindin-positive cerebellum Purkinje neurons, which are NeuN negative; Alexa Fluor 647, 594, and 488 dyes were used respectively as secondaries (Invitrogen).
Regions of interest (ROIs) were quantified across the whole brain at approximately 600 μm intervals using serial-sectioned coronal brain slices. ROIs are summarized and shown in Supplemental Figure 1 and were determined using DAPI and NeuN labelling and based on regions outlined in the mouse brain atlas (41). At least 3 areas were sampled per ROI, per animal; area per image was 910 μm2. For each image data set a maximum intensity projection was created from an optical z-stack of 20 μm with a 2 μm step size (0.3 μm/pixel xy) for ASO and neuron markers. Preprocessing included background subtraction and conversion to 8-bit binary image. NeuN positive cell count was obtained using analyze particles (size filter 35–300 μm2) with an average of 8000 NeuN-positive cells analyzed across each mouse brain to confirm spatial overlap with the ASO fluorescence signal. A no primary control (NPC) image was also acquired to determine background signal for the ASO channel, and threshold was determined using the signal intensity profile. NPC images were acquired using the same confocal image settings, including laser power, gain, and digital offset, as those used for all experimental data (n = 4 mice).
Mass spectrometry.
Mice were i.c.v. injected with ASO (10 μg) at P1 and brains were collected and snap frozen at P12–P15. Approximately 100 mg cortical tissue was removed and added in a 1:5 ratio to extraction buffer, which consisted of 1× protease inhibitor, 1% sodium deoxycholate, and 100 mM TEAB (triethylammonium bicarbonate). Samples were vortexed and sonicated using the Bioruptor Plus (Diagenode) until completely homogenized. Protein concentration was determined using the bicinchoninic acid assay (BCA assay) (Thermo Fisher Scientific). Protein reduction was performed with DL-Dithiothreitol (DTT) to a final concentration of 5 mM, for 30 minutes at 60°C. Samples were then alkylated in the dark with iodoacetamide (IAA) to a final concentration of 10 mM for 30 minutes at 37°C. Samples were quenched with additional DTT to a final concentration of 10 mM and incubated at 37°C for 30 minutes. The pH was checked for optimal trypsin digestion. A quantity of 100 μg protein was added to Axygen Maxymum Recovery low-bind microcentrifuge tubes, followed by the addition of trypsin in a 1:40 ratio, and incubated at 37°C for 16 hours (overnight).
Deoxycholate was precipitated using 10% formic acid (FA). Samples were spiked with heavy peptide standard mix (JPT Peptide Technologies) and centrifuged at 12,000g for 10 minutes to pellet the sodium deoxycholate. The supernatant was transferred to new low-bind tubes. Solid phase extraction clean up occurred using the AssayMAP Bravo Platform (Agilent) on C18 plates using the standard protocol from the manufacturer. Samples were dried down and stored in –80°C before being resuspended in 2% acetonitrile, 0.05% trifluoroacetic acid to prepare for mass spectrometry (MS).
Agilent Technologies 6495 triple quadrupole LC-MS/MS was used for the relative quantification and detection of Scn2a protein and other target peptides. The method was optimized using Skyline software (MacCoss Lab, University of Washington) and Agilent Mass Hunter software. The transitions and method settings can be found in the supplementary material.
Spontaneous seizure monitoring.
Q/+ mice were placed under 24-hour video monitoring at P21 and P30 in their home cage using a digital camera (EVO2, Pacific Communications). Videos were reviewed by 2 experimenters and seizures over Racine score 4 were noted (18).
Electrocorticography (ECoG).
Mice (P30–P35) were anesthetized with 2% to 4% isoflurane and placed in a stereotaxic apparatus (Kopf) with the skull surface horizontal between lambda and bregma. Epidural electrodes, 1 reference and 2 recording (Pinnacle Technology), were placed on the brain surface. A ball of silver wire placed on top of the skull surface was used as a ground electrode. All electrodes were connected to a headmount (Pinnacle Technology) and secured to the skull with dental cement. Mice were allowed to recover for a minimum of 5 days before ECoG recording. Brain cortical activity was sampled at 250 Hz for 24 hours (Pinnacle Technology), with low pass and high pass filters of 40 Hz and 0.5 Hz, respectively. During recording, mice could move freely in the cage with food and water ad libitum. ECoG data were analyzed using ClampFit 10.7 (Molecular Devices), and a spike was identified when the amplitude was 2.5 times baseline and duration was shorter than 80 ms.
Behavioral tests.
All behavioral experiments were performed during the light phase between the 10:00 and 17:00 hours, with a minimum of overnight recovery between tests. Mice were acclimatized to the testing rooms for at least 30 minutes before experimentation. All equipment was thoroughly cleaned and disinfected with 70%–80% ethanol between trials or testing of each mouse.
Locomotor test.
Spontaneous locomotor activity was measured automatically with a laser-beam system monitoring distance and movements according to prescribed parameters (MedAssociates). Each mouse was placed in the center of the activity cage (27 × 27 cm) for 30 minutes under dimmed lighting.
Grid walk test.
Each mouse was placed on an elevated grid apparatus (30 cm) and allowed to freely walk along a 1 m grid of stainless steel bars (3 mm diameter) normally spaced 1 cm apart. The test was performed after 3 successful training runs. For the test, the grid bars in the last 50 cm were replaced with one of several set patterns that contain missing bars at a gap on 1 or 2 bars. Behaviors on the last 50 cm grid were recorded with a digital camera (EVO2, Pacific Communications). Videos were reviewed and the number of foot faults (total miss or deep slip) were noted (42).
Grip strength.
A grip strength meter (Bioseb) was used to assess strength and control of the fore and hind paws. Mice were rested on the angled stainless-steel mesh assembly, grasping the mesh with all 4 limbs. Mice were gently pulled by the tail away from the mesh, and the maximum force prior to release of the paws was recorded. Four trials were performed by 2 experimenters on each mouse.
Elevated plus maze.
The base of the maze was custom made of Perspex containing 2 open arms (7 × 31 cm) and 2 enclosed arms (7 × 31 × 15 cm) extending from a central platform (5 × 5 cm) and was elevated 40 cm above the floor. At the beginning of the experiment, each mouse was placed in the center of the maze facing the enclosed arm. Trial duration was 10 minutes. The duration on the open and closed arms of the elevated plus maze was tracked by the TopScan software (Clever Sys Inc).
Three-chamber social interaction test.
Mice were subjected to 3-chamber social test to assess social interaction behaviors. For mice i.c.v. injected at P1, the 3-chamber apparatus consisted of a Plexiglas rectangular box (40 × 38 cm) divided into 3 compartments with 2 transparent walls. The 2 openings on the walls allowed free access into each of the 3 chambers. A wired cage of equal size (16 × 10 cm) was placed in each of the 2 outside compartments (16 × 38 cm). The interaction zone was defined as 2.5 cm in front of the wired cage. Movement and duration were measured by TopScan software (Clever Sys Inc.). During habituation (5–10 minutes), the mouse was placed in the middle chamber and could freely explore all 3 chambers. An unfamiliar, age-, strain-, and sex-matched novel mouse was placed in the wired cage on the nonpreferred side, as determined from habituation. Activity of the test subject was then monitored for a further 10 minutes.
Data analysis.
All data are presented as mean ± SEM. Dose-response curves were fitted with data points using Motulsky regression. All statistical analyses were performed by GraphPad Prism 7, and significance was determined when P was less than 0.05.
Study approval.
All studies involving animals were carried out in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by the Florey Institute Animal Ethics Committee.
Author contributions
ML, NJ, PJN, and LEB designed and carried out experiments, performed data analyses, and drafted the manuscript. B Rollo, S Pachernegg, A Sedo, and JH performed qPCR experiments and analyses. KR and A Sedo performed immunohistochemistry experiments and analyses. LD and LJ performed behavioral experiments and analyses. TB and B Roberts performed mass spectrometry experiments and analyses. A Soriano, AN, KD, SM, CAR, FR, and S Petrou designed and coordinated the study. All authors read and contributed to the revision of manuscript.
Supplementary Material
Acknowledgments
We would like to thank Lynley Cordeiro for logistical and technical support for this project. This study was supported by National Health and Medical Research Council (NHMRC) program grant 10915693, NHMRC fellowship GNT1005050 (to S Petrou), RogCon Biosciences, and Praxis Precision Medicines.
Version 1. 12/01/2021
Electronic publication
Footnotes
B Roberts’s present address is: Monash University, Clayton, Victoria, Australia.
Conflict of interest: S Petrou, AN, and KD received payments from RogCon and Praxis Precision Medicines. PJN, FR, and A Soriano are employees of Ionis Pharmaceuticals.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(23):e152079.https://doi.org/10.1172/JCI152079.
See the related Commentary at All our knowledge begins with the antisenses.
Contributor Information
Melody Li, Email: melody.li@florey.edu.au.
Nikola Jancovski, Email: jancovski.n@florey.edu.au.
Lisseth E. Burbano, Email: lburbanoportilla@gmail.com.
Kay Richards, Email: kay.richards@florey.edu.au.
Lisa Drew, Email: lisajcdrew@gmail.com.
Alicia Sedo, Email: alicia.sedo@florey.edu.au.
Jacqueline Heighway, Email: j.heighway@florey.edu.au.
Svenja Pachernegg, Email: svenja.pachernegg@mcri.edu.au.
Armand Soriano, Email: asoriano@ionisph.com.
Linghan Jia, Email: Linghan.jia.mail@gmail.com.
Todd Blackburn, Email: t.blackburn@unimelb.edu.au.
Blaine Roberts, Email: blaine.roberts@emory.edu.
Alex Nemiroff, Email: alex@rogconbio.com.
Kelley Dalby, Email: kdalby@praxismedicines.com.
Snezana Maljevic, Email: snezana.maljevic@florey.edu.au.
Christopher A. Reid, Email: careid@unimelb.edu.au.
Frank Rigo, Email: FRigo@ionisph.com.
Steven Petrou, Email: steven.petrou@florey.edu.au.
References
- 1.Scheffer IE, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58(4):512–521. doi: 10.1111/epi.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maljevic S, et al. Models for discovery of targeted therapy in genetic epileptic encephalopathies. J Neurochem. 2017;143(1):30–48. doi: 10.1111/jnc.14134. [DOI] [PubMed] [Google Scholar]
- 3.Mulley JC, et al. Channelopathies as a genetic cause of epilepsy. Curr Opin Neurol. 2003;16(2):171–176. doi: 10.1097/00019052-200304000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Oyrer J, et al. Ion channels in genetic epilepsy: from genes and mechanisms to disease-targeted therapies. Pharmacol Rev. 2018;70(1):142–173. doi: 10.1124/pr.117.014456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aartsma-Rus A, Krieg AM. FDA approves eteplirsen for duchenne muscular dystrophy: the next chapter in the eteplirsen saga. Nucleic Acid Ther. 2017;27(1):1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corey DR. Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nat Neurosci. 2017;20(4):497–499. doi: 10.1038/nn.4508. [DOI] [PubMed] [Google Scholar]
- 7.Bennett CF. Therapeutic antisense oligonucleotides are coming of age. Annu Rev Med. 2019;70:307–321. doi: 10.1146/annurev-med-041217-010829. [DOI] [PubMed] [Google Scholar]
- 8.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 9.Liao Y, et al. Molecular correlates of age-dependent seizures in an inherited neonatal-infantile epilepsy. Brain. 2010;133(pt 5):1403–1414. doi: 10.1093/brain/awq057. [DOI] [PubMed] [Google Scholar]
- 10.Gazina EV, et al. ‘Neonatal’ Nav1.2 reduces neuronal excitability and affects seizure susceptibility and behaviour. Hum Mol Genet. 2015;24(5):1457–1468. doi: 10.1093/hmg/ddu562. [DOI] [PubMed] [Google Scholar]
- 11.Wolff M, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140(5):1316–1336. doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]
- 12.Sanders SJ, et al. Progress in understanding and treating SCN2A-mediated disorders. Trends Neurosci. 2018;41(7):442–456. doi: 10.1016/j.tins.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howell KB, et al. SCN2A encephalopathy: a major cause of epilepsy of infancy with migrating focal seizures. Neurology. 2015;85(11):958–966. doi: 10.1212/WNL.0000000000001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvill GL, et al. Targeted resequencing in epileptic encephalopathies identifies de novo mutations in CHD2 and SYNGAP1. Nat Genet. 2013;45(7):825–830. doi: 10.1038/ng.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen AS, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berecki G, et al. Dynamic action potential clamp predicts functional separation in mild familial and severe de novo forms of SCN2A epilepsy. Proc Natl Acad Sci U S A. 2018;115(24):E5516–E5525. doi: 10.1073/pnas.1800077115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kordasiewicz HB, et al. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74(6):1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ihara Y, et al. Retigabine, a Kv7.2/Kv7.3-channel opener, attenuates drug-induced seizures in knock-in mice harboring Kcnq2 mutations. PLoS One. 2016;11(2):e0150095. doi: 10.1371/journal.pone.0150095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatta T, et al. Neurotoxic effects of phenytoin on postnatal mouse brain development following neonatal administration. Neurotoxicol Teratol. 1999;21(1):21–28. doi: 10.1016/S0892-0362(98)00028-2. [DOI] [PubMed] [Google Scholar]
- 20.Planells-Cases R, et al. Neuronal death and perinatal lethality in voltage-gated sodium channel alpha(II)-deficient mice. Biophys J. 2000;78(6):2878–2891. doi: 10.1016/S0006-3495(00)76829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogiwara I, et al. Nav1.2 haplodeficiency in excitatory neurons causes absence-like seizures in mice. Commun Biol. 2018;1:96. doi: 10.1038/s42003-018-0099-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenk GM, et al. Scn8a antisense oligonucleotide is protective in mouse models of SCN8A encephalopathy and dravet syndrome. Ann Neurol. 2020;87(3):339–346. doi: 10.1002/ana.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Z, et al. Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome. Sci Transl Med. 2020;12(558):eaaz6100. doi: 10.1126/scitranslmed.aaz6100. [DOI] [PubMed] [Google Scholar]
- 24.Rigo F, et al. Pharmacology of a central nervous system delivered 2’-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther. 2014;350(1):46–55. doi: 10.1124/jpet.113.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearney JA, et al. A gain-of-function mutation in the sodium channel gene Scn2a results in seizures and behavioral abnormalities. Neuroscience. 2001;102(2):307–317. doi: 10.1016/S0306-4522(00)00479-6. [DOI] [PubMed] [Google Scholar]
- 26.Schattling B, et al. Activity of NaV1.2 promotes neurodegeneration in an animal model of multiple sclerosis. JCI Insight. 2016;1(19):e89810. doi: 10.1172/jci.insight.89810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Semple BD, et al. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol. 2013;106-107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finkel RS, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388(10063):3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 29.Chiriboga CA, et al. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86(10):890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkel RS, et al. Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med. 2017;377(18):1723–1732. doi: 10.1056/NEJMoa1702752. [DOI] [PubMed] [Google Scholar]
- 31.Mousa MA, et al. A comprehensive institutional overview of intrathecal nusinersen injections for spinal muscular atrophy. Pediatr Radiol. 2018;48(12):1797–1805. doi: 10.1007/s00247-018-4206-9. [DOI] [PubMed] [Google Scholar]
- 32.Wurster CD, et al. Intrathecal administration of nusinersen in adolescent and adult SMA type 2 and 3 patients. J Neurol. 2019;266(1):183–194. doi: 10.1007/s00415-018-9124-0. [DOI] [PubMed] [Google Scholar]
- 33.Kornbrust D, et al. Oligo safety working group exaggerated pharmacology subcommittee consensus document. Nucleic Acid Ther. 2013;23(1):21–28. doi: 10.1089/nat.2012.0399. [DOI] [PubMed] [Google Scholar]
- 34.Spratt PWE, et al. The autism-associated gene Scn2a contributes to dendritic excitability and synaptic function in the prefrontal cortex. Neuron. 2019;103(4):673–685. doi: 10.1016/j.neuron.2019.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatsukawa T, et al. Scn2a haploinsufficient mice display a spectrum of phenotypes affecting anxiety, sociability, memory flexibility and ampakine CX516 rescues their hyperactivity. Mol Autism. 2019;10:15. doi: 10.1186/s13229-019-0265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walf AA, Frye CA. The use of the elevated plus maze as an assay of anxiety-related behavior in rodents. Nat Protoc. 2007;2(2):322–328. doi: 10.1038/nprot.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Shalom R, et al. Opposing effects on NaV1.2 function underlie differences between SCN2A variants observed in individuals with autism spectrum disorder or infantile seizures. Biol Psychiatry. 2017;82(3):224–232. doi: 10.1016/j.biopsych.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang XH, et al. Translation efficiency of mRNAs is increased by antisense oligonucleotides targeting upstream open reading frames. Nat Biotechnol. 2016;34(8):875–880. doi: 10.1038/nbt.3589. [DOI] [PubMed] [Google Scholar]
- 39.Liang XH, et al. Antisense oligonucleotides targeting translation inhibitory elements in 5’ UTRs can selectively increase protein levels. Nucleic Acids Res. 2017;45(16):9528–9546. doi: 10.1093/nar/gkx632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rigo F, et al. Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv Exp Med Biol. 2014;825:303–352. doi: 10.1007/978-1-4939-1221-6_9. [DOI] [PubMed] [Google Scholar]
- 41. Franklin KBJ, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. Academic Press; 2007. [Google Scholar]
- 42.Farr TD, et al. Bilateral alteration in stepping pattern after unilateral motor cortex injury: a new test strategy for analysis of skilled limb movements in neurological mouse models. J Neurosci Methods. 2006;153(1):104–113. doi: 10.1016/j.jneumeth.2005.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.