ABSTRACT
Although macroautophagy/autophagy has been proposed as a critical defense mechanism against HIV-1 by targeting viral components for degradation, its contribution as a catabolic process in providing optimal anti-HIV-1 immunity has never been addressed. The failure to restore proper antiviral CD8A/CD8 T-cell immunity, especially against HIV-1, is still the major limitation of current antiretroviral therapies. Consequently, it is of clinical imperative to provide new strategies to enhance the function of HIV-1-specific CD8A T-cells in patients under antiretroviral treatments (ART). Here, we investigated whether targeting autophagy activity could be an optional solution to make this possible. Our data show that, after both polyclonal and HIV-1-specific activation, CD8A T-cells from ART displayed reduced autophagy-dependent degradation of lysosomal contents when compared to naturally HIV-1 protected elite controllers (EC). We further confirmed in EC, by using specific BECN1 gene silencing and lysosomal inhibitors, the critical role of active autophagy in superior CD8A T-cell protection against HIV-1. More importantly, we found that an IL21 treatment was effective in rescuing the antiviral CD8A T-cell immunity from ART in an autophagy-dependent manner. Finally, we established that IL21-dependent rescue occurred due to the enhanced degradation of endogenous lipids via autophagy, referred to as lipophagy, which fueled the cellular rates of mitochondrial beta-oxidation. In summary, our data show that autophagy/lipophagy can be considered as a therapeutic tool to elicit functional antiviral CD8 T-cell responses. Our results also provide additional insights toward the development of improved T-cell-based prevention and cure strategies against HIV-1.
Abbreviations: ART: patients under antiretroviral therapy; BaF: bafilomycin A1; BECN1: beclin 1; CEF: cytomegalo-, Epstein-Barr- and flu-virus peptide pool; Chloro.: chloroquine; EC: elite controllers; FAO: fatty acid beta-oxidation; HIVneg: HIV-1-uninfected control donors; IFNG/IFN-γ: interferon gamma; IL21: interleukin 21; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; PBMC: peripheral blood mononuclear cells; SQSTM1: sequestosome 1; ULK1: unc-51 like autophagy activating kinase 1.
KEYWORDS: Antiretroviral therapy, elite controllers, FAO, HIV-1, IL21, lipophagy, polyfunctionality
Introduction
The maintenance of polyfunctional CD8A/CD8 T-cell responses, especially against human immunodeficiency virus type-1 (HIV-1), is associated with better viral control and slower disease progression [1]. Polyfunctional CD8A T-cells are defined by their ability, upon activation, to secrete multiple cytokines simultaneously with cytotoxic molecules such as PRF1 (perforin 1) and GZMB (granzyme B) [2]. However, despite successful viral suppression, patients under antiretroviral therapies (ART) display defective antiviral CD8A T-cells with reduced polyfunctional responses when compared to elite controllers (EC) [3–7]. EC, who are able to maintain a natural control of HIV-1 due to strong antiviral CD8A T-cell responses, can help in the design of new strategies aiming at reinforcing CD8A T-cell immunity in ART [8].
Recently, Angin M. et al. have shown that antiviral CD8A T-cells from ART display a restrictive glucose dependency, which is a distinct metabolic defect not observed in EC [4]. In this study, the authors suggest that the superior CD8A T-cell immunity found in EC might be explained by their ability to use additional metabolic resources other than glucose. This consequently allows them to sustain effective mitochondrial respiration with high ATP production. This immune advantage, referred to as metabolic plasticity, has to be achieved in ART through the design of new strategies, but the molecular mechanisms responsible for it need to first be clarified among EC. In cancer, data show that the diverse metabolic fuel sources that can be produced by macroautophagy/autophagy, a lysosomal-dependent catabolic process, provide tumors with metabolic plasticity [9–11]. Since previous observations have proposed that autophagy may contribute to limiting HIV-1 pathogenesis in EC by targeting viral components for degradation, we raised the question of whether it could rather provide their CD8A T-cells with metabolic plasticity and superior antiviral immunity [12,13]. In the present study, we showed that following polyclonal and antigen-specific cell activation, CD8A T-cells from ART displayed reduced autophagic activity when compared to EC. Using specific gene silencing of BECN1, one major regulator of autophagic activity, we confirmed the critical role of this catabolic process in highly functional CD8A T-cell responses in EC [14]. We demonstrated that IL21 production by CD4 T-cells, which was lower in ART when compared to EC, was associated with the levels of autophagic activity in CD8A T-cells [15]. Our data further confirmed that IL21 addition in ART’s cultures was effective in rescuing their antiviral CD8A T-cells in an autophagy-dependent manner. More precisely, we found that the IL21-mediated rescue was specifically provided due to increased lipophagy, which describes the autophagic degradation of endogenous lipids [16–18]. Finally, we concluded our study by confirming in CD8A T-cells from ART that the enhanced lipophagy following IL21 treatment impacted the cellular energetic balance directly through lipid breakdown and fatty acid beta-oxidation (FAO).
Altogether, this study puts lipophagy at the center of the molecular pathway by which IL21 signaling leads to enhanced mitochondrial metabolism in protective CD8A T-cell immunity during persistent HIV-1 infection. Our results also provide new information to consider for the development of effective T cell-based vaccines and cure strategies against HIV-1, where effective HIV-1-specific CD8A T-cell responses are warranted [19–22].
Results
Confirmation of reduced autophagic activity in HIV-1-specific CD8A T-cells from ART
Autophagy is a highly dynamic intracellular degradative system that involves the capture, isolation and lysosomal digestion of intracellular materials in specialized structures called autolysosomes (ALs) [23]. Although previous findings suggest that autophagy in EC may be associated with virologic benefits by targeting HIV-1 components for degradation, no information is available regarding the potential immunological advantage that autophagy may provide in EC, especially in their antiviral CD8A T-cells [12,13].
We decided to use several complementary methods to assess autophagy in CD8A T-cells from all study groups such as multi-parameter and ImageStream flow cytometry, pulse-chase assay, and electron microscopy. First and foremost, we validated all methods by using CD8A T-cells from HIVneg donors (n = 10) that have been starved for 2 h, which is the most common way to increase autophagic activity in culture (Fig. S1A-D). As expected, we found significant increases in the expression levels for the autophagy-related protein BECN1 (beclin 1) and SQSTM1 (sequestosome 1) (Fig. S1A). Our data also showed increased autophagy-dependent lytic activity in starved CD8A T-cells by using a pulse-chase assay method that has previously been developed on macrophages (Fig. S1B) [24]. Then, by using Imaging flow cytometry, we evaluated the lysosomal content of MAP1LC3/LC3 (microtubule associated protein 1 light chain 3) protein [25], which was also shown increased under starvation. This was determined by the colocalization index of fluorescently labeled LC3 and lysosomal marker Lyso-ID, which is called bright detail similarity (BDS) (Fig. S1Ci-iii). However, in agreement with the previous observations that were also generated on CD8 T-cells [25], we did not found any difference in LC3 puncta counts after cell starvation (Fig. S1Civ). This indicated that, due to the nature of lymphocytes (displaying almost no cytoplasm compartment) and the limitation of the Imaging-based assay, LC3 puncta counts could not be used as a quantitative and reproducible indicator values of CD8A T-cell-related autophagy.
Once validated, we assessed the levels of protein expression of the two key autophagy-related genes, ULK1 and BECN1, in ART and compared them to those from age-matched EC and uninfected donor controls (HIVneg). The expression levels of ULK1 and BECN1 were determined by multiparameter flow cytometry in CD8 T-cells after 6 h of polyclonal and antiviral activations. To elicit antiviral activations, we used a combined CEF peptide pool (cytomegalo-virus with Epstein-Barr and flu viruses) or HIV-1 Gag. Antigen-specific CD8A T-cells were determined by their positive staining for IFNG/IFN-γ, following the antiviral activations, as previously done [26]. Of note, we did not find any difference in the percentage of antigen-activated CD8A T-cells represented as IFNG+ cells between ART and EC, and we used HIVneg as a negative control for HIV-1 Gag stimulation (Fig. S1E and S1F). In addition to ULK1 and BECN1, we decided to investigate intracellular accumulation of the SQSTM1, when cells were cultured with BaF. Our data showed that basal levels of ULK1, BECN1 and SQSTM1, as determined by the percentages of positive cells, were similar in non-activated CD8A T-cells for all study groups (Figure 1A-C). In contrast, after both polyclonal and antiviral activations, we found that the expression levels for all molecules were significantly reduced in ART when compared to EC and HIVneg.
Figure 1.
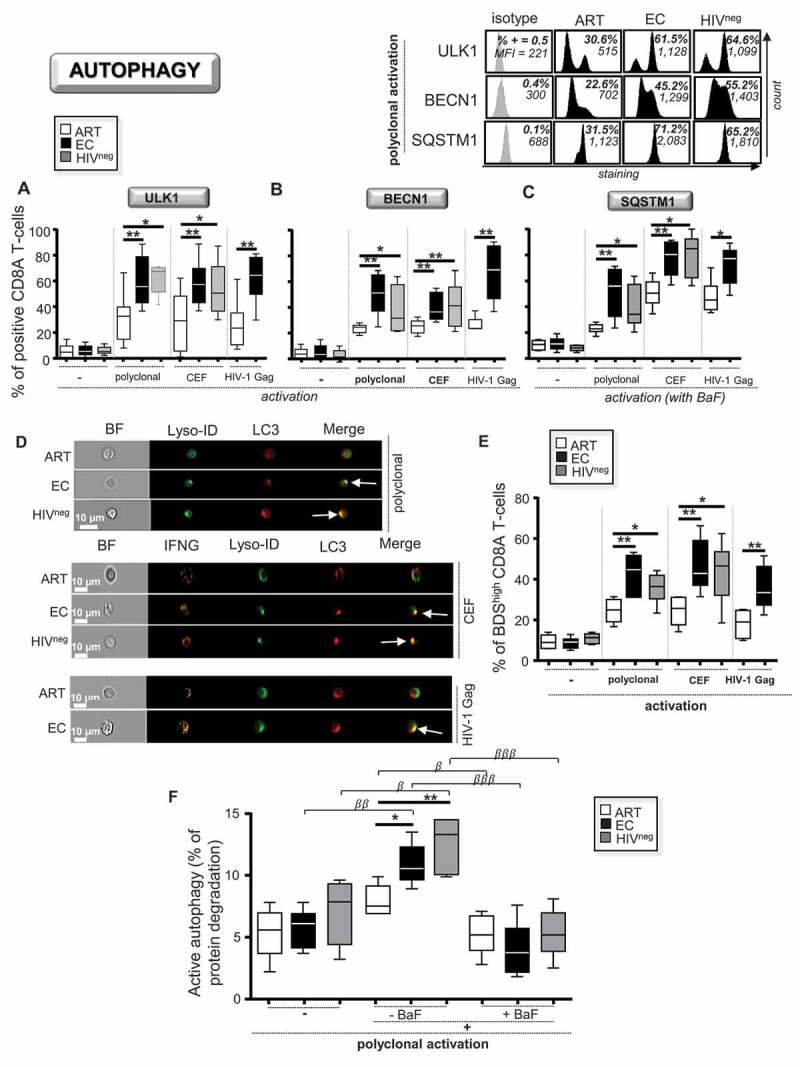
Reduced autophagic activity in ART after CD8A T-cell activation. (A-E) CD8A T-cells from ART, EC and HIVneg were either polyclonally or antigen-specifically (CEF peptides or Gag antigens) activated and cultured without activation for 6 h (n = 6). Of note, antigen-specific CD8A T-cells were identified at 6 h post-activation by positive staining for IFNG. The percentages of positive cells were then determined in CD8A T-cells for the autophagy-related players (A) ULK1 and (B) BECN1. (C) We also assessed the percentages of SQSTM1+ CD8A T-cells when lysosomal activity was blocked by BaF during the culture. Representative histograms including isotype controls after 6 h of post-polyclonal activation are also shown above. (D and E) Lysosomal content of LC3 determined by Imaging flow cytometry. (D) Representative images of single CD3W+ CD8A+ (polyclonal activation) and CD3W+ CD8A+ IFNG+ T-cells (CEF and HIV-1 activations) merged for fluorescent signals from LC3 and Lyso-ID. BF, bright field. (E) Percentages of BDShigh LC3+ Lyso-ID+ CD8A T-cells in ART, EC and HIVneg. The colocalization index BDS stands for bright detail similarity. (F) Autophagy-dependent proteolytic degradation of long-lived proteins determined in purified CD8A T-cells after polyclonal activation in the presence or absence of BaF, by using a pulse-chase approach. β, symbol used for paired t test (comparison between treated CD8A T-cells and their untreated control). *, symbol used for Mann-Whitney test (comparison between study groups). One symbol, 0.05 > P> 0.01; two symbols, 0.01 > P> 0.001; and three symbols, 0.001 > P> 0.0001
We next used an ImageStream-based autophagy assay. Our data showed reduced lysosomal content of LC3 in activated CD8A T-cells from ART (Figure 1D and Figure 1E). We further quantified the autophagy-dependent lytic activity as previously done in macrophages, but performed here in CD8A T-cells after 6 h of polyclonal activation with or without BaF [24]. Once again, our results showed that activated CD8A T-cells from ART displayed lower percentages of autophagy-dependent degradation when compared to EC and HIVneg (Figure 1F). We decided to use this pulse-chase assay for the rest of our study to appreciate autophagic activity in CD8A T-cells. Indeed, not only did we confirm the efficacy of BaF in blocking autophagic activity with the assay (Figure 1F), but we also found that the results positively correlated with the expression levels of ULK1, BECN1 and SQSTM1, as well as with the lysosomal content of LC3 (Fig. S2).
Finally, we analyzed the polyclonally-activated CD8A T-cells for all study groups at the ultrastructural level. Similarly to others, we used electron microscopy to evaluate the number of both autophagic vacuoles (AV) and ALs [12]. In contrast to AV that were identified as a double-membrane-structure containing undigested cytoplasmic material, ALs were generated by the fusion (complete or in progress) of the autophagic vacuoles with the lysosomes and were limited by a single membrane (Figure 2A). We validated the use of this method by starving CD8A T-cells from HIVneg donors (n = 3), which showed increased AL numbers (Fig. S1D). Our data showed that activated CD8A T-cells from ART displayed less AV and ALs than those of EC and HIVneg (as determined by both numbers and percentages of positivity per 40 cells) (Figure 2B–E). Altogether, our results indicate that antiviral CD8A T-cells from ART, including those specific to HIV-1, display reduced autophagic activity along with lower numbers of ALs when compared to EC.
Figure 2.
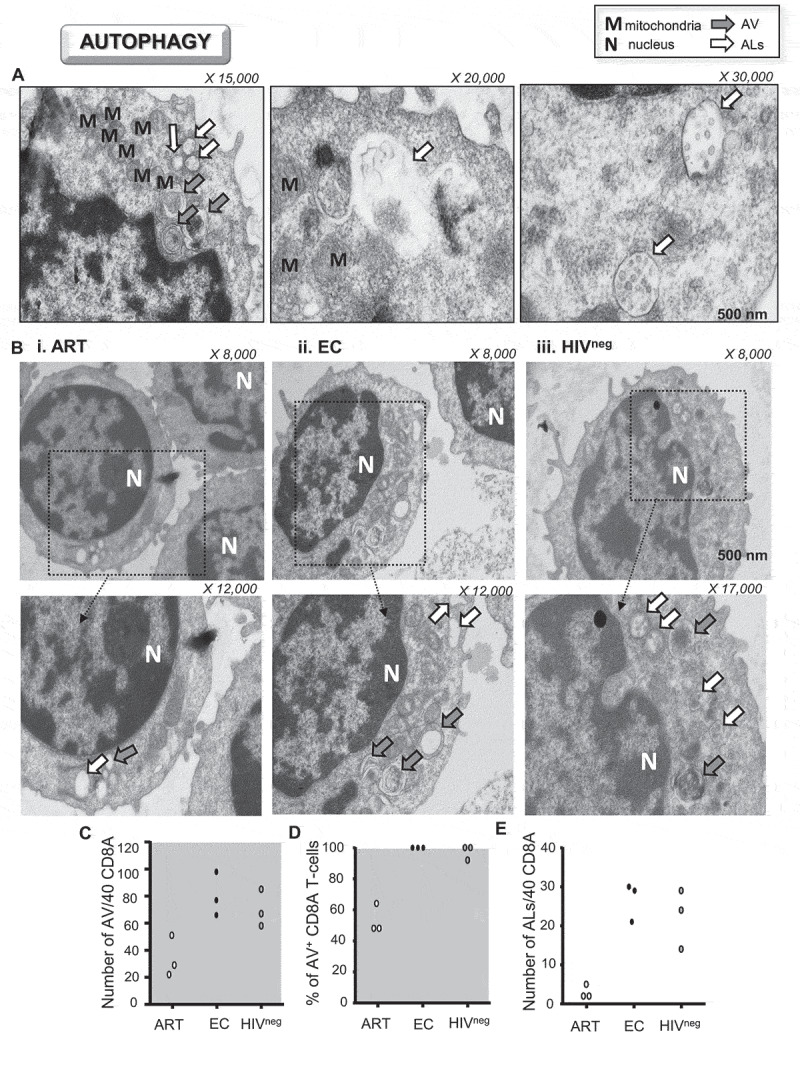
Lower amounts of ALs in activated CD8A T-cells from ART. (A-E) Ultrastructural analysis of purified CD8A T-cells from ART, EC and HIVneg after 6 h of polyclonal activation (n = 3). (A) Representative ultrastructural micrographs from uninfected controls to differentiate AV (autophagic vacuoles; gray arrows) from ALs (autolysosomes; white arrows). (B) Ultrastructural micrographs in i. ART, ii. EC and iii. HIVneg including strategic magnifications to appreciate the numbers of cellular vacuoles (X 8,000–17,000). Quantitative analysis of (C) the number and (D) percentages of positive CD8A T-cells for AV per 40 cells. (E) Number of ALs per 40 CD8A T-cells
Strong autophagy in EC drives their anti-HIV-1 CD8A T-cell polyfunctionality
Given the key role that autophagy plays in protective CD8A T-cell immunity with mouse models of viral infection, it was reasonable to believe that it may be involved in natural immune protection against persistent HIV-1 infection as well [27].
To investigate whether autophagy was responsible for enhanced antiviral CD8A T-cell responses in EC, we specifically inhibited BECN1 expression using small interfering silencing RNAs (siRNA). Briefly, purified CD8A T-cells from all study groups were either electroporated or transfected with siRNA specific for BECN1 or with negative control siRNA for 2 h. Then, cells were washed twice and cultured with their autologous CD8A-depleted PBMC (ratio CD8A:PBMC = 1:5). At 24 h post-transfection, cells were activated either polyclonally or specifically for an additional 6 h. At 6 h post-activation, we finally determined both the expression levels of BECN1 and cell apoptosis in CD8A T-cells by multi-parameter flow cytometry. Electroporation alone or transfection with negative siRNA did not affect BECN1 expression when compared to cells that were not electroporated. In contrast, CD8A T-cells that were transfected with BECN1 siRNA displayed up to 86.1% and 82.5% reductions of BECN1 expression, respectively in EC and HIVneg (Fig. 3Ai and S3A-C). In fact, BECN1 silencing in EC and HIVneg led to reductions of its protein expression to levels comparable to ART. Of note, we found similar percentages of cell apoptosis for all conditions of electroporation or transfection, regardless of the study groups (Fig. 3Aii). We further investigated whether specifically interfering with BECN1 expression in activated CD8A T-cells led to reduced autophagic activity. We also used the lysosomal inhibitors BaF and chloroquine (Chloro.), which are potent and well-acknowledged inhibitors of cellular autophagy by targeting lysosomal activity, as controls. Levels of active autophagy in activated CD8A T-cells that were electroporated or transfected with negative control siRNA were similar to those of cells that were not electroporated. Following polyclonal or antiviral activations, our data confirmed that both specific BECN1 silencing and lysosomal inactivation led to significant reductions of active autophagy in EC and HIVneg to levels comparable to ART (Figure 3B).
Figure 3.
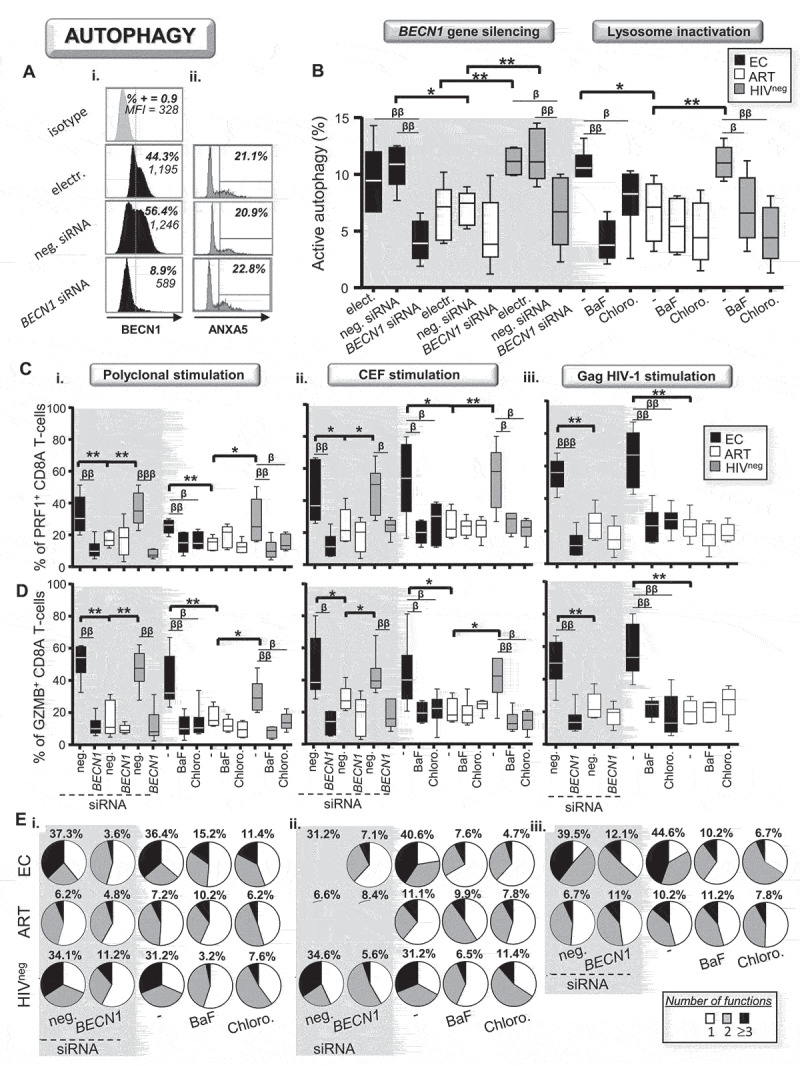
Active autophagy plays a key role in antiviral CD8A immunity during HIV-1 infection. (A-E) Active autophagy and effector function were both determined in activated CD8A T-cells under specific BECN1 gene silencing or lysosomal inactivation with Baf or Chloro (n = 6). (A) Representative histograms of i. BECN1 expression and ii. ANXA5 staining in transfected CD8A T-cells from EC at 6 h post-polyclonal activation. (B) Autophagy-dependent proteolytic degradation of long-lived proteins assessed in polyclonally activated CD8A T-cells when active autophagy was inhibited or not. Percentages of (C) PRF1+ and (D) GZMB+ CD8A T-cells in the context of autophagy blockade. Results were shown in the context of i. polyclonal, ii. CEF-specific and iii. HIV-1-specific activation. (E) Pie chart representations of CD8A T-cell polyfunctionality that were determined after i. polyclonal and ii and iii. antigen-specific activations with or without autophagy blockade. Percentages of highly functional CD8A T-cells (expressing three or more antiviral cytokines and cytotoxic molecules in addition to IFNG) were also indicated in bold for all study groups and conditions. β, symbol used for paired t test (comparison between treated CD8A T-cells and their untreated control). *, symbol used for Mann-Whitney test (comparison between study groups). One symbol, 0.05 > P> 0.01; two symbols, 0.01 > P> 0.001; and three symbols, 0.001 > P> 0.0001
Next, using multi-parameter flow cytometry, we determined in activated CD8A T-cells from EC, ART, and HIVneg whether BECN1 gene silencing and lysosomal inactivation could impact their antiviral immune responses. To this aim, we first analyzed the cells’ ability to produce antiviral cytokines (IFNG and IL2) and cytotoxic molecules (PRF1, GZMA and GZMB) with or without autophagy inhibition (Fig. S3D). When autophagy was untouched in CD8A T-cells, we found that both polyclonal and antiviral activations led to higher percentages of PRF1+ and GZMB+ cells in EC and HIVneg when compared to ART (Figure 3C and Figure 3D). In contrast, targeting active autophagy in activated CD8A T-cells from EC and HIVneg, by BECN1 gene silencing or with lysosomal inhibitors, both led to significant reductions of both PRF1 and GZMB productions to levels comparable to ART (Figure 3C and Figure 3D). Of note, we did not find any differences between EC, ART and HIVneg for the single production of IFNG, IL2 and GZMA, even when autophagy was inhibited.
Finally, we assessed CD8A T-cell polyfunctionality including in HIV-1-specific cells for all study groups and treatments. Polyfunctional CD8A T-cells, which were characterized by their ability to produce multiple cytokines and cytotoxic molecules in addition to IFNG, are highly predictive of protective immunity against pathogens including HIV-1 [2–6]. In contrast, we characterized monofunctional CD8A T-cells by their single staining for IFNG. Our data showed that, when autophagy was untouched, activated CD8A T-cells in EC and HIVneg displayed superior cell polyfunctionality. This was illustrated by higher percentages of cells producing three or more molecules along with reduced percentages of monofunctional cells, when compared to ART (Figure 3E). Once again, autophagy inhibition in EC and HIVneg, regardless of the method used, led to significant inhibition of CD8A T-cell polyfunctionality to levels that were comparable to ART. Overall, our results demonstrate a critical role of active autophagy in the superior CD8A anti-HIV-1 potential, as determined by cell polyfunctionality and production of cytotoxic molecules.
IL21 enhances antiviral CD8A T-cell responses from ART in an autophagy-dependent manner
We investigated whether autophagy in activated CD8A T-cells during HIV-1 infection were controlled by intrinsic regulation or involved in cell cooperation with other immune key players such as CD4 helpers or CD14+ monocytes. In this context, we first purified CD8A T-cells and activated them either alone or with autologous monocyte- and CD4 T-cell-depleted PBMC cultures for 6 h. Once again, we assessed both the expression levels of autophagy-related genes (ULK1, BECN1 and SQSTM1 in the context of lysosomal inactivation) and the autophagy-dependent lytic activity for all study groups and activation methods (polyclonal, CEF- and HIV-1 Gag-specific activations). Our data showed that activating CD8A T-cells alone or removing CD4 T-cells from cultures both led to significant inhibition of the expression levels of the autophagy-related genes along with reduced lytic activity in EC and HIVneg to levels comparable with ART (Figure 4). In summary, our data confirmed that the strong autophagic activity found in activated CD8A T-cells from EC and HIVneg required the presence of CD4 T-cells in cultures. In this regard, we decided to investigate the role of IL21 (interleukin-21), which is a CD4-related cytokine known to promote antiviral CD8A T-cell responses [28].
Figure 4.
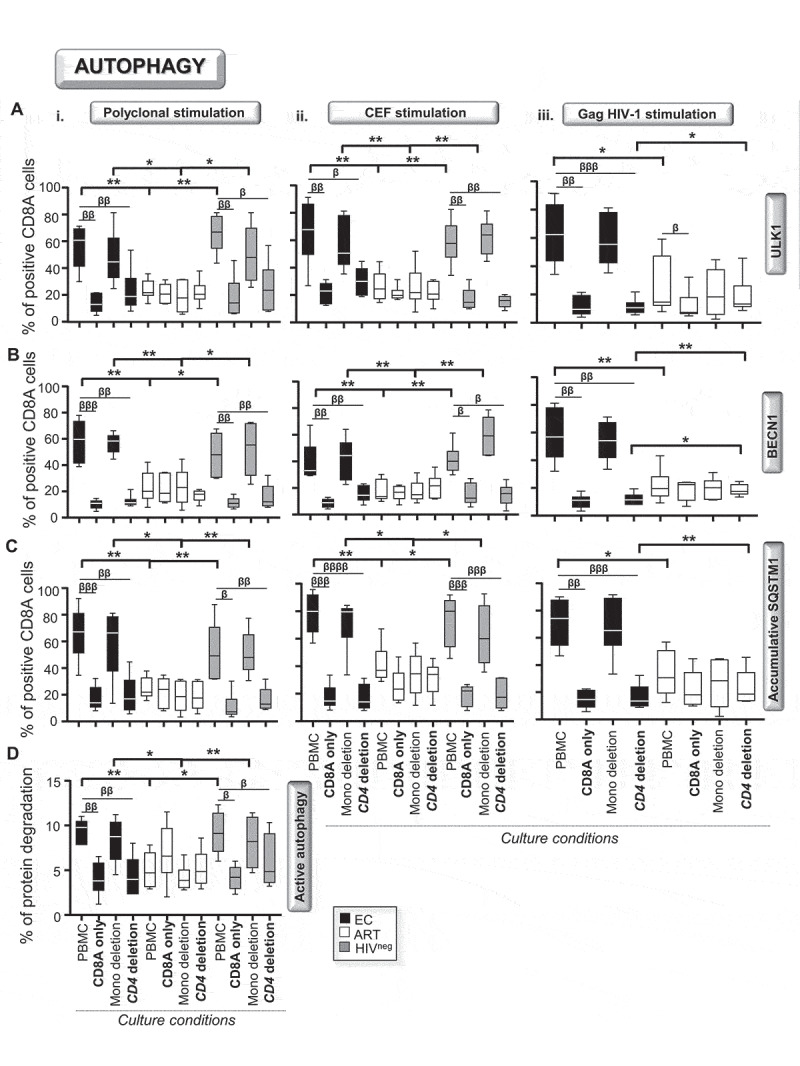
CD4 T-cell help is required to ensure high autophagic activity in EC. We first activated total PBMC, purified CD8A T-cells and PBMC whose CD4 T-cells or monocytes have been depleted either i. polyclonally, ii. CEF- or iii. HIV-1 Gag-specifically for 6 h (n = 6). Then, we assessed the percentages of (A) ULK1+, (B) BECN1+ and (C) SQSTM1+ (with BaF) in CD8A T-cells for all conditions by flow cytometry. (D) We also determined at 6 h post-polyclonal activation on purified CD8A T-cells the autophagy-dependent proteolytic degradation of long-lived proteins using the pulse-chase approach. β, symbol used for paired t test (comparison between treated CD8A T-cells and their untreated control). *, symbol used for Mann-Whitney test (comparison between study groups). One symbol, 0.05 > P> 0.01; two symbols, 0.01 > P> 0.001; and three symbols, 0.001 > P> 0.0001
First, our data showed that cultures from ART had lower percentages of IL21+ CD4 T-cells when compared to those from EC and HIVneg after 6 h of polyclonal activation (Figure 5A) [15,29]. Importantly, we found a positive and highly significant correlation between the percentages of IL21-producing CD4 T-cells and levels of autophagic activity in CD8A T-cells with all subjects (Figure 5B). Altogether, these first sets of data strongly suggested an important role of IL21 in dictating the levels of active autophagy among antiviral CD8A T-cells during HIV-1 infection.
Figure 5.
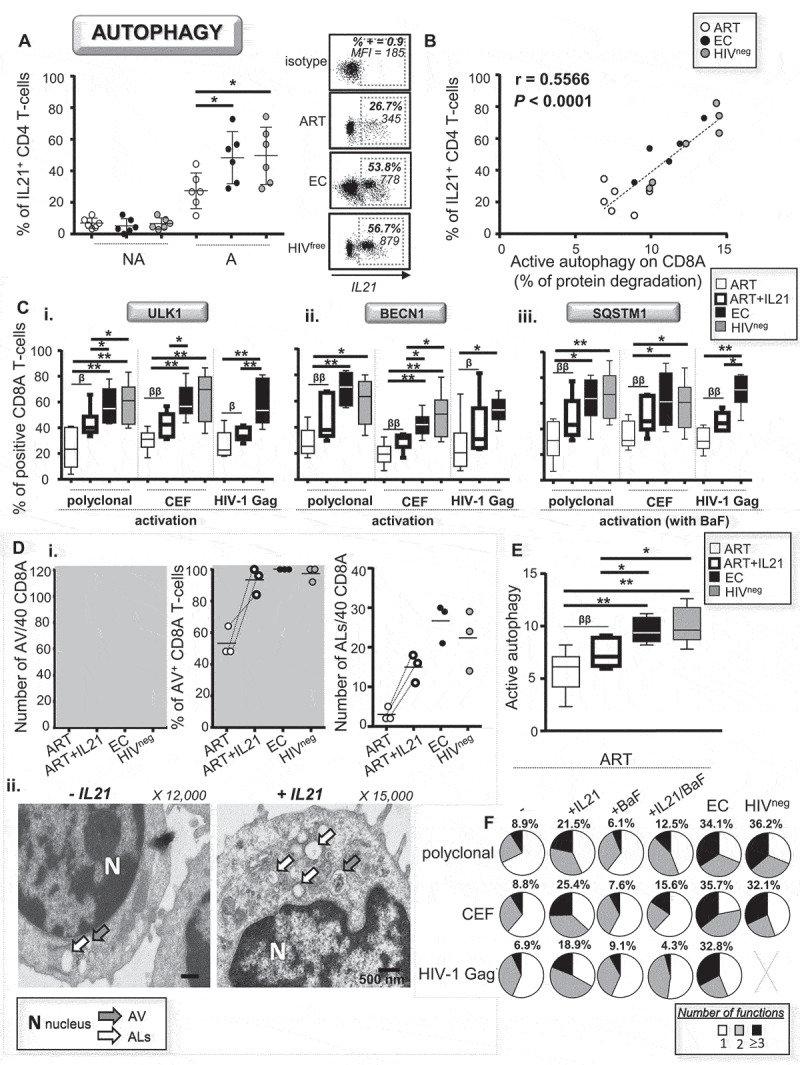
Rescue of antiviral CD8A immunity in ART by IL21 is driven by enhanced active autophagy. (A) Percentages of IL21-producing CD4 helpers from all study groups were determined after 6 h of polyclonal activation by flow cytometry. Representative dot plots are also shown including isotype control. (B) Correlation between the percentages of IL21-producing CD4 helpers and levels of active autophagy in activated CD8A T-cells for all participants. (C-F) 10 ng/mL recombinant IL21 was added in cultures form ART when their CD8A T-cells were activated either polyclonally or antigen-specifically. (C) Percentages of i. ULK1+, ii. BECN1+ and iii. SQSTM1+ CD8A T-cells were then assessed by flow cytometry. (D) Ultrastructural analysis of purified CD8A T-cells following polyclonal activation including cells from ART that have been stimulated with or without IL21. i. Number and percentage of positivity for AV and ALs per 40 CD8A T-cells. ii. Representative micrographs with and without IL21. (E) Autophagy-dependent proteolytic degradation of long-lived protein determined in purified CD8A T-cells form ART with or without IL21. (F) CD8A T-cell polyfunctionality was assessed in activated CD8A T-cells from ART (with or without IL21 and BaF treatments), EC and HIVneg. Percentages of highly functional CD8A T-cells (expressing three or more antiviral cytokines and cytotoxic molecules in addition to IFNG) were also indicated in bold. n = 6, except for (B) n = 18 and (D) n = 3. β, symbol used for paired t test (comparison between treated CD8A T-cells and their untreated control). *, symbol used for Mann-Whitney test (comparison between study groups). One symbol, 0.05 > P> 0.01; two symbols, 0.01 > P> 0.001; and three symbols, 0.001 > P> 0.0001
Therefore, we decided to add recombinant IL21 in cultures to confirm whether or not the treatment was effective in enhancing the autophagic activity among CD8A T-cells from ART. After 6 h of polyclonal and antiviral activations with or without IL21, we first assessed the levels of ULK1, BECN1 and SQSTM1 by multi-parameter flow cytometry. Our data showed that IL21 treatment with ART was indeed effective in enhancing the percentages of CD8A T-cells positive for autophagy-related genes; however, they did not fully reach the levels observed in EC and HIVneg (Figure 5C). Using electron microscopy, we also confirmed that IL21 treatment with ART increased the numbers of AVs and ALs in their activated CD8A T-cells (Figure 5D). Furthermore, treating CD8A T-cells from ART with IL21 improved, although partially when compared to EC and HIVneg controls, their autophagic activity (Figure 5E). In contrast, IL21 treatment with EC and HIVneg had no effect on their autophagy status (Fig. S4A and S4B).
Finally, we aimed to confirm that IL21 treatment in ART was effective in rescuing their antiviral CD8A T-cell responses, especially those against HIV-1, due to enhanced autophagic activity. To do so, we activated CD8A T-cells from ART (either polyclonally or antigen-specifically) with or without IL21, co-cultured them in the presence or absence of BaF, and assessed their antiviral polyfunctionality. In addition to increased expression levels of PRF1 and GZMB, IL21 treatment with ART led to significant enhancements of CD8A T-cell polyfunctionality including in cells specific for HIV-1 (Figure 5F). IL21 rescue of antiviral CD8A T-cell responses in ART was autophagy-dependent, as co-treatment with BaF abrogated the enhancement of cell polyfunctionality (Figure 5F). Once again, IL21 treatment with EC and HIVneg had no effect on their antiviral CD8A T-cell responses (Fig. S4C). In summary, our data indicate that endogenous IL21 was sufficient in eliciting optimal autophagic activity and related superior antiviral CD8A T-cell potential in EC. We further confirmed that IL21 treatment could be considered as a tool to rescue HIV-1-specific CD8A T-cell responses in ART in an autophagy-dependent manner.
IL21 promotes the autophagy-dependent degradation of endogenous lipids in CD8A T-cells from ART
Mounting an antiviral immune response against pathogens is an energetically demanding process. In this regard, autophagy can be essential for the maintenance of mitochondrial energetic function by providing metabolic substrates through lysosomal degradation. Since intrinsic lipolysis is critical for developing CD8A T-cell immunity after infection, we decided to investigate the levels of autophagy-mediated degradation of endogenous lipids, referred to as lipophagy, in our study groups [30].
By using electron microscopy with the osmium tetroxide negative-staining, we first confirmed the presence of lipid content within ALs from activated CD8A T-cells in ART (with or without IL21), EC and HIVneg (Figure 6A). However, although we were able to detect lipid content for all study groups, we found higher numbers of ALs containing lipids per 40 cells in EC, HIVfree and IL21-treated ART when compared to untreated ART (Figure 6A and Figure 6B). These data strongly indicated that IL21 treatment in ART could be associated with increased lipophagy.
Figure 6.
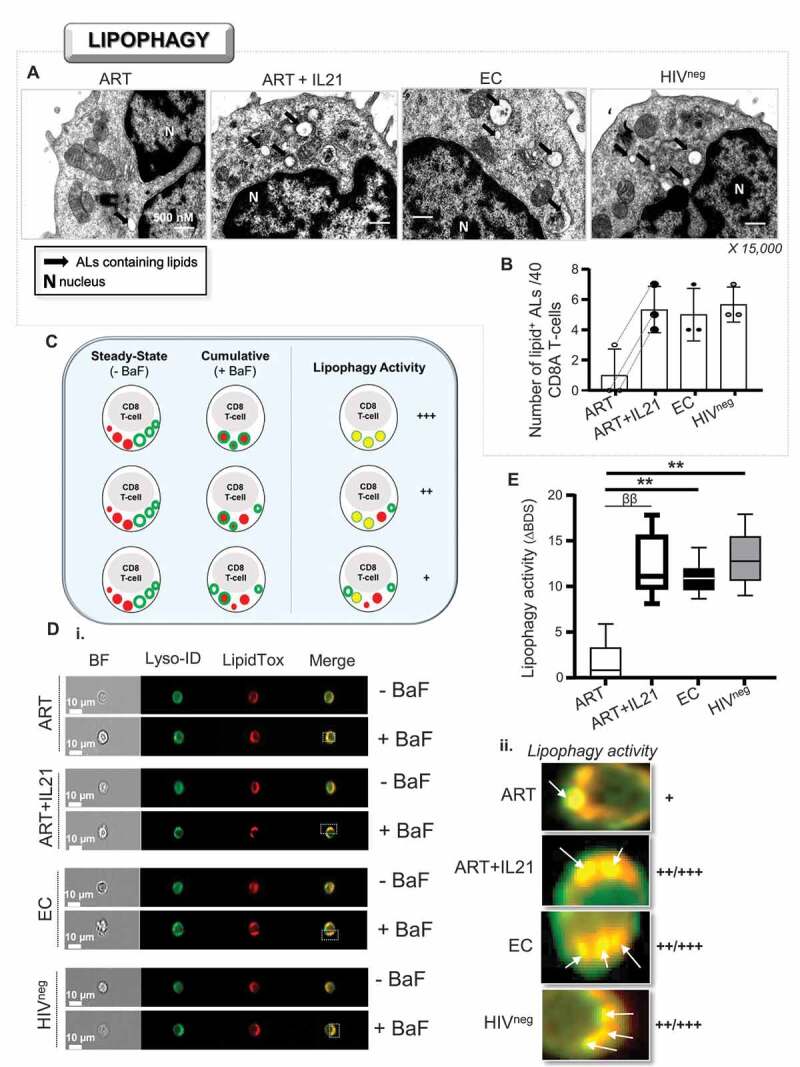
Enhanced lipophagy in ART in response to IL21 treatment. (A and B) Visualization of ALs containing endogenous lipids using osmium tetroxide negative-staining in activated CD8A T-cells (n = 3). (A) Magnified images showing ALs containing lipids (black arrows) for all study groups including ART that have been treated or not with IL21 (15,000 X). (B) Quantitative analysis of the number of ALs containing lipids per 40 cells. (C-E) Assessment of lipophagy activity by using a novel ImageStream-based lipophagy assay (n = 6). (C) Schematic representation for assessing lipophagy activity. (D) i. Representative images of single Lyso-ID+LipidTox+ CD8A T-cells for all groups after 6 h of polyclonal activation in the presence or absence of BaF (“cumulative” and “steady-state” conditions, respectively). BF, bright field. ii. Magnified images showing increased lysosomal content of endogenous lipids in BaF-treated cells, which is an indicator of lipophagy activity when compared to cells without BaF. (E) Quantification of lipophagy activity determined in Lyso-ID+LipidTox+ CD8A T-cells in ART (with or without IL-21), EC and HIVneg. Lipophagy activity was determined by the formula: ΔBDS = (% of BDShigh cells with BaF) – (% of BDShigh cells without BaF). β, symbol used for paired t test (comparison between treated CD8A T-cells and their untreated control). *, symbol used for Mann-Whitney test (comparison between study groups). One symbol, 0.05 > P> 0.01; two symbols, 0.01 > P> 0.001; and three symbols, 0.001 > P> 0.0001
Next, in order to measure the lipophagic activity among CD8A T-cells, we developed a novel ImageStream-based assay, which relies on the quantification of increased lipid content within lysosomes when cultures are conducted with BaF (Figure 6C). Briefly, we cultured cells for each treatment condition with and without BaF. Then, we collected and stained cells with Lyso-ID and LipidTox, which are lysosomal and lipid dyes respectively. Finally, we determined their lipophagic activity by the formula: ΔBDS = (BDShigh Lyso-ID+LipidTox+ cells with BaF) – (BDShigh Lyso-ID+LipidTox+ cells without BaF). First and foremost, to ascertain that this technique can measure the lipophagic activity in primary CD8A T-cells, we validated the protocol with hepatic Huh 7.5 cells which are starved for 2 h (Fig. S5). Indeed, nutrient starvation using hepatic cells is the most common way to increase lipophagy in culture [18]. Increases of lipophagy in starved Huh 7.5 cells were illustrated by higher ΔBDS in comparison to those from cells that were cultured in complete DMEM (Fig. S5A and S5B). Similarly, we found higher mean LipidTox puncta per cell with starved Huh 7.5 cells, when compared to cultures with complete media (Fig. S5C). Once validated, we assessed the lipophagic activity in activated CD8A T-cells from ART (with and without IL21), EC and HIVneg. Our data showed that, after polyclonal activation, CD8A T-cells from ART displayed less lipophagic activity than EC and HIVneg (Figure 6D and Figure 6E). We also found that IL21 treatment in ART led to increases in their lipophagic activity. Of note, although we found higher mean LipidTox puncta per cell in ART with IL21 treatment when compared to untreated ART, no significant differences were observed between study groups (Fig. S5D). Altogether, our data show that IL21 treatment of CD8A T-cells from ART enhanced their lipophagy. This indicates that metabolic reprogramming through IL21 treatment in ART is not out of reach and may abrogate their glucose dependency by providing cells with lipid substrates to fuel mitochondrial respiration.
IL21-induced lipophagy in ART leads to a better use of their mitochondrial respiration due to FAO
The whole mitochondrial respiration, which can be enhanced in CD8A T-cells by IL15 treatment, is known to be critical to cover the energetic requirements for the effector and memory developments [4,31,32]. Therefore, we aimed to assess if IL21 treatment in polyclonally activated CD8A T-cells from ART was also associated with enhancement of their whole mitochondrial respiration. Following the manufacturer’s instruction, the respiratory kinetics that resulted from the sequential addition of pharmacological agents to the respiring cells allowed us to calculate the SRC and ATP-linked respiration. As expected, we found increases in both the whole mitochondrial SRC and ATP-linked respiration in ART when IL21 was added in culture (Fig. S6A).
Our next thought was to investigate whether IL21-induced lipophagy in ART provided free fatty acids to engage the mitochondrial fatty acid beta-oxidation (FAO). The protocol, developed to appreciate FAO, relies on the capacity of the cells to oxidize the fatty acid palmitate when other exogenous substrates are limited. Briefly, we polyclonally activated purified CD8A T-cells from ART for 6 h with or without IL21, BaF and the FAO inhibitor etomoxir. Cells were finally incubated with or without palmitate in substrate-limited media for an additional 45 min before assessing their mitochondrial respiration. Our data showed that IL21 treatment in ART led to increases of the FAO-mediated SRC and FAO ATP-linked respiration, which were both abrogated by BaF co-treatment (Figure 7A–C). Of note, cells that were treated with BaF alone had similar respiratory kinetics compared to untreated cells. As expected, IL21-induced FAO was prevented by etomoxir pre-treatment, which validated our protocol as well (Fig. 7Aii). Finally, to assess the role of induced FAO on enhanced antiviral CD8A T-cells responses when IL21 was added in ART, we activated cells either polyclonally, CEF- or HIV-1- Gag-specifically for 6 h with or without IL21 and etomoxir. We found that the enhancements of PRF1 and GZMB expressions, as well as cell polyfunctionality that were mediated by IL21 were all prevented by etomoxir co-treatment (Figure 7D and Figure 7E). In summary, our data show that IL21-mediated enhancement of antiviral CD8A T-cell responses in ART, including those against HIV-1, is driven by increased FAO.
Figure 7.
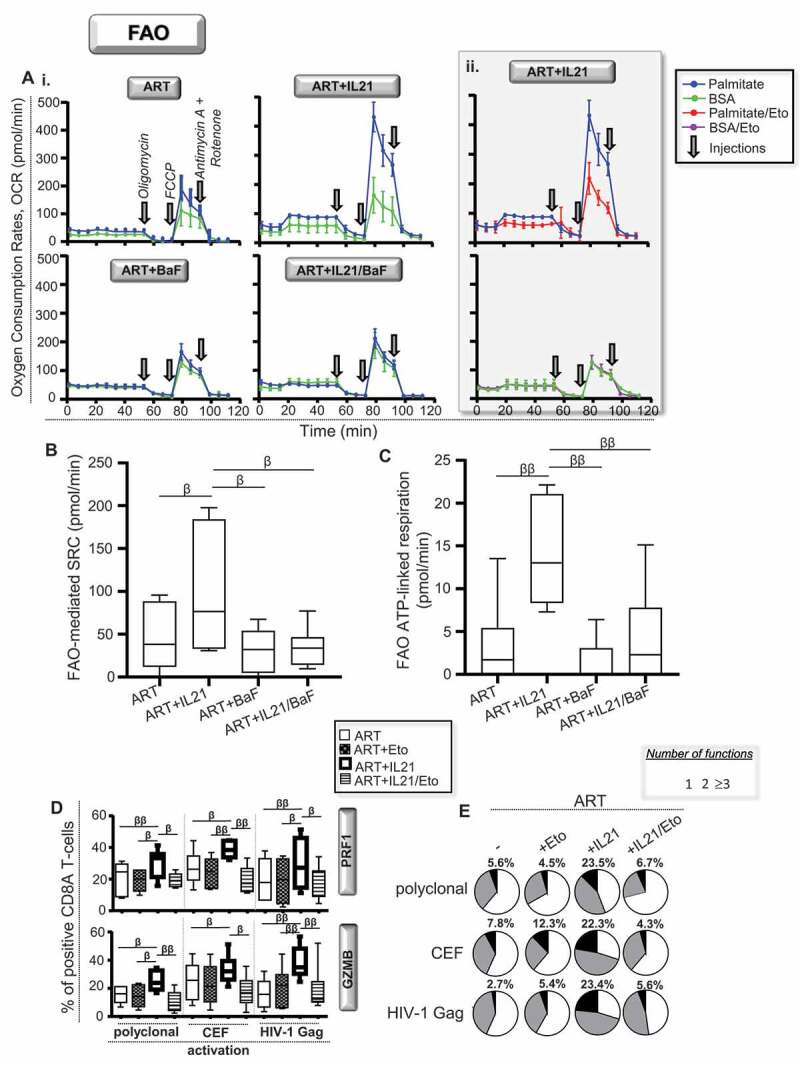
Lipophagy-induced IL21 in ART improves the cellular rates of mitochondrial beta-oxidation. (A-C) FAO determined in purified CD8A T-cells from ART at 6 h of polyclonal activation with or without IL21 and BaF (n = 6). (A) Representative graphs showing real-time oxygen consumption rates (OCR) in response to injections of mitochondrial respiration modulators, which are Oligomycin, Carbonyl cyanide 4-(trifluoromethoxy)-phenylhydrazone (FCCP) and Rotenone and Antimycin A, respectively in i. cells that have been incubated for the last 45 min with palmitate (blue kinetic) or BSA (green kinetic) and in ii. IL21-treated cells that have also been treated with the FAO inhibitor etomoxir (red and purple kinetics). Assessment of (B) FAO-mediated respiration and (C) related ATP-linked respiration for all conditions. Of note, FAO-mediated spare respiratory capacity (SRC) was determined with the formula: (SRC with palmitate) – (SRC with BSA). (D and E) CD8 T-cell polyfunctionality determined in CD8 T-cells from ART that have been polyclonally, CEF- and HIV-1 Gag- activated specifically in the presence or absence of IL21 and etomoxir (n = 6). (D) Percentages of PRF1+ and GZMB+ cells. (E) Pie chart representations of CD8A T-cell polyfunctionality. Percentages of highly functional CD8A T-cells were also indicated in bold. β, symbol used for paired t test (comparison between treated CD8 T-cells and their untreated control). β, 0.05 > P> 0.01; ββ, 0.01 > P> 0.001 and βββ, 0.001 > P> 0.0001
Discussion
Although it is well-acknowledged that antiretroviral therapies fail to restore effective anti-HIV-1 CD8A T-cell responses, strategies aiming at rescuing them are limited in numbers [3–7] EC, who control HIV-1 infection due to the maintenance of highly functional CD8A T-cells, represent a critical study group to identify which molecular mechanisms may be targeted to improve ART outcomes [3–5,8,33]. In a similar way, our previous works, which also included EC, have demonstrated the critical role of the transcription factor FOXO3/FOXO3a and its pro-apoptotic target genes on the loss of memory CD4 T- and B-cells in ART [34,35]. In the present study, we were interested in investigating the molecular mechanisms that were regulating the expression levels of both antiviral cytokines (IL2 and IFNG) and cytotoxic molecules (PRF1, GZMA and GZMB) in activated CD8A T-cells during chronic HIV-1 infection. Our data confirmed that antiviral CD8A T-cells from ART, including those specific to HIV-1, displayed both reduced expression of PRF1 and GZMB along with lower cell polyfunctionality. We also found that HIV-1-specific CD8A T-cells from ART had less autophagic activity along with lower numbers of ALs when compared to EC. We further confirmed that cytotoxic and polyfunctional HIV-1-specific CD8A T-cell responses in EC were autophagy-dependent. Altogether, the first sets of evidence pointed to a beneficial effect of stimulating autophagy-mediated lysosomal degradation in ART to potentiate their immune response to HIV-1. Of note, there is a possibility that the induction of autophagy, which is required for stimulating CD8A Tcell-related immune protection against HIV-1, might be transient and only found during the first hours of the antigen-stimulation. Indeed, although Xu X. et al. also confirmed that autophagy is essential for cytotoxic CD8A T-cell immune responses against the lymphocytic choriomeningitis virus, they showed that autophagy was finally found reduced in effector cells after 5 d of the viral infection [27]. In the context of HIV-1 infection, it will be interesting to investigate what is the status of autophagy activity in HIV-1-specific CD8A T-cells after a longer time of Gag stimulation.
Our study differs from others since autophagy has always been investigated in HIV-1 infection as a type of selective lytic process that targets the virus, referred to as xenophagy [12,36]. Another unique aspect of our study is based on the fact that the reduced autophagic activity in ART is not mediated by the productive HIV-1 infection and the expression of viral regulatory proteins such as Tat or Nef as reported in the past [13,37]. In contrast, our data showed that the reduced autophagy in ART was mediated by lower production of IL21, which is an immune defect reported after years of persistent HIV-1 infection despite antiretroviral therapy [15,29]. We showed that IL21-mediated rescue of antiviral CD8A T-cell responses in ART was driven by lipophagy, whose catabolic process of endogenous lipids has never been addressed in HIV-1 infection before. To do so, we have developed a high throughput, statistically robust method that quantifies lipophagic activity in primary human cells using the ImageStream cytometry. Several cytokines have been shown to promote autophagic activity, yet the role of IL21 in metabolic reprogramming through autophagy/lipophagy induction was unknown until now [38]. Altogether, our results showed that autophagy/lipophagy represents a new mechanism by which polyfunctional antiviral CD8A T-cells, including those specific to HIV-1, support mitochondrial respiration through FAO.
The efficiency of IL21-based therapy to promote effective HIV-1- and simian immunodeficiency virus (SIV)-specific CD8A T-cells has been repeatedly confirmed by in vitro and in vivo testing [39–42]. Besides HIV-1, IL21 can also serve as a critical factor that shapes the functional quality of antiviral CD8A T-cells against different viruses such as vaccinia virus, lymphocytic choriomeningitis virus, and hepatitis virus B [28,43,44]. Interestingly, IL21-based therapy, which has been proposed as a means of enhancing anti-tumor specific CD8A T-cell responses as well, led to a similar metabolic reprogramming toward FAO [45]. Therefore, our study, which reports how IL21 is mechanistically able to promote FAO, may provide useful information for designing therapies against viral infections as well as cancers. IL15 has also been proven effective in enhancing the antiviral capacity of HIV-1-specific CD8A T-cells in ART [4,46]. Enhancement by IL15 of HIV-1-specific CD8A T-cell responses has been associated with higher rates of oxidative metabolism as well [4]. However, recent data showed that the anti-HIV-1 capacity of IL21-stimulated CD8A T-cells exceeded that of IL15-stimulated cells [41,42]. This may be explained by the fact that, although IL15 enhanced the rates of oxidative metabolism, it failed to promote FAO in contrast to IL21 (Fig. S6B and S6C). Similar to IL15, IL21 was also found to enhance the rates of lipid uptake in ART (Fig. S6D) [4]. Although IL21-mediated rescue of antiviral CD8A T-cells in ART was driven by lipophagy, the fact remains that the rescue was only partial when compared to EC. This suggests that the highly functional CD8A T-cells from EC may not only depend on potent lipophagic activity, but also on other autophagy-dependent mechanisms. We cannot rule out the involvement of the autophagy-dependent proteolysis, referred to as proteophagy, when it comes to EC. The pulse-chased assay, which was used here to uncover the reduced autophagic activity in ART when compared to EC, was based on protein degradation. Proteophagy may provide CD8A T-cells from EC with free amino-acids such as glutamine, leucine, serine, alanine, and arginine. These amino acids are all reported to fuel mitochondrial respiration and lay the foundation for effective T-cell immune protection, even in the context of glucose restriction [47–50].
Defective HIV-1-specific CD8A T-cells in ART have been characterized by other immune defects in addition to reduced polyfunctionality when compared to EC. For example, HIV-1-specific CD8A T-cells from ART may be prone to cell death due to high levels of CASP3 activation [51]. Although we found similar levels of basal apoptosis between ART and EC regardless of autophagy blockade, we cannot exclude the possibility that HIV-1-specific CD8A T-cells from ART display increased sensitivity to Fas-induced apoptosis [52]. HIV-1-specific CD8A T-cells from ART have also been shown to have a lower capacity than EC to eliminate both productively infected and latently infected CD4 T-cells [33,53,54]. Finally, HIV-1-specific CD8A T-cells from ART may display reduced expression levels for the CCL4/MIP-1β chemokine [3,5]. It would be of clinical imperative to investigate if each of these defects in ART can be rescued by IL21 and if enhanced autophagy/lipophagy is involved in the process.
In summary, we are just beginning to appreciate how and why autophagy is a fundamental mechanism by which CD8A T-cells adapt their energetic input under specific circumstances. Up until recently, the conventional view of CD8A T-cell metabolism was that, upon antigen-specific activation, antiviral CD8A T-cells primarily used glycolysis over mitochondrial respiration to support quick bioenergetic needs [32,55]. In contrast, it was established that, once an antigen is cleared, memory CD8A T-cells relied on FAO to fuel mitochondrial respiration in order to persist and be metabolically prepared for antigen recall [30–32,56]. We show that the metabolism of antiviral CD8A T-cells, which is induced upon antigen-specific activation, is more plastic than originally thought and can be promoted by the autophagy-lysosomal pathway. To conclude, our data provide the rationale for considering the IL21/lipophagy/FAO axis as a therapeutic target for boosting specific immunity against HIV-1. Our data may also aid in the development of HIV vaccine and cure strategies in ART, whose success depends mostly on their ability to induce protective CD8A T-cell responses [19–22].
Materials and methods
HIV-1-infected patients
The experiments described in this study mainly rely on the comparison of total and HIV-1-specific CD8A T-cell features from patients under antiretroviral therapy (ART) to those from naturally protected EC. EC are unique patients displaying a long-term viral control with no treatments. All virus-infected patients were participants of the Montreal HIV infection study that received approval from the McGill University Health Center Ethical Review Board (ethic reference number SL-00.069). All subjects provided an informed and written consent for participation. The inclusion criteria of untreated ECs and ART are: middle-aged subjects (31–51 years old), presumed to have HIV-1 infection for a minimum of 3.6 years (3.6–7.5 years), no protective gene polymorphisms such as CCR5 Δ32 and B27/B57 haplotypes, with CD4 counts over 400 cells/μl in blood and undetectable viral loads (< 50 copies/ml) for 3 years or more. It is worth mentioning that ART patients in this study display years of chronic HIV- infection and antiretroviral therapy. Of note, we selected 6 age-matched and HIV-1-uninfected subjects as controls of non-infection.
Media and products
RPMI-1640 media (Wisent, 350–000-EL) and DMEM media (Wisent, 319–005-CL), FBS (Wisent, 080–450), penicillin-streptomycin antibiotics (Wisent, 450–201-EL), and PBS (Wisent, 311–010-CL) were obtained from Wisent Inc. The starvation EBSS medium was purchased from ThermoFisher Scientific (ThermoFisher Scientific, 24,010,043). All monoclonal antibodies used for multi-parameter and Imaging flow cytometry such as anti-CD3W/CD3 (BD Biosciences, 557,943) and anti-CD8A (BD Biosciences, 560,347) were purchased in BD Biosciences, except for those against ULK1 (Santa Cruz Biotechnology, sc-390,904 AF647), BECN1 (Abcam, ab246760), SQSTM1 (MBL Life Sciences, M162-A48), LC3 (MBL Life Sciences, M152-3), PRF1/perforin (Biolegend, 353,314), and GZMA/granzyme A (ThermoFisher Scientific, 12–9177-42). Recombinant IL21 (Sigma Aldrich, SRP3087) and IL15 (Sigma Aldrich, I8648), lysosomal inhibitors bafilomycin A1 (Sigma Aldrich, B1793) and chloroquine (Sigma Aldrich, C6628), and etomoxir (Sigma Aldrich, E1905) were all provided from Sigma Aldrich. Zenon labeling kits were purchased from Invitrogen (ThermoFisher Scientific, Z-25,008). Concentrations for IL21, IL15, bafilomycin A1, chloroquine, and etomoxir were 10 ng/mL, 10 ng/mL, 100 nM, 100 nM and 5 μM, respectively.
CD8A purification and cell subset removals
Some experiments required the purification of CD8A T-cells, or the removal of specific cell subsets such as CD4 T-cells and monocytes from the collected peripheral blood mononuclear cells (PBMC), before further analyses. 1. CD8A purification: CD8A T-cells were isolated by using the EasySep human CD8A T-cell enrichment kit (StemCell Technologies, 17,953). Our developed protocol allows for more than 95% purification without any cell stimulation or apoptosis. 2. Subset-depleted PBMC: We removed CD4 T-cells and CD14+ monocytes from the CD8A-depleted PBMC by using the EasySep human CD4 (StemCell Technologies, 17,852) and CD14 (StemCell Technologies, 17,858) positive selection kits II, respectively. The percentage of cell subset removal was determined at around 96.2% by using flow cytometry.
In-vitro activation assays
Total, cell subset-depleted PBMC and purified CD8A T-cells for all study groups were either polyclonally or antigen-specifically activated for 6 h in the presence of GolgiPlug (BD Biosciences, 555,029) and GolgiStop (BD Biosciences, 554,724) to assess autophagy and the intracellular production of effector molecules. For the polyclonal activation, we treated cells with 0.5 μg/mL of anti-CD3W (BioLegend, 317,326) and of 1 μg/mL anti-CD28 Abs (BD Biosciences, 555,726). To elicit the antigen-specific activations, we used either 2 μg/mL of CEF (cytomegalo- virus, with Epstein-Barr and flu viruses) peptide pool (Mabtech, 3616–1) or 5 μg/mL of HIV-1 p55 (Austral Biologicals, HI1A-730-5) and p24 Gag antigens (Austral Biologicals, HI1A-720-5) on PBMC. Of note, all cultures from ART were conducted with 10 μM of AZT (Sigma Aldrich, A2169-25 MG) to prevent any de novo viral production (confirmed by the sensitive HIV-1 p24 ELISA (Abcam, ab218268) in culture supernatants).
BECN1-specific gene silencing
We first purified 107 CD8A T-cells from all tested groups and electroporated them using Nucleofector II technology according to the Amaxa Biosystems manufactor’s protocol. Specific BECN1 siRNA (ThermoFisher Scientific, AM16708) and negative control siRNA (ThermoFisher Scientific, 4,390,843) were obtained from ThermoFisher Scientific. Of note, 7 μg of siRNA were either transfected for each condition for 2 h without antibiotics. Purified CD8A T-cells were washed three times thereafter to remove dead necrotic cells, counted and incubated 18 h with autologous CD8A-depleted PBMC (at ratio CD8A/PBMC = 1⁄5). Then, we activated cells for 6 h to assess the autophagy activity and effector function in transfected CD8A T-cells. At 6 h post-activation, some cells were also collected to confirm BECN1 inhibition and to assess the percentages of cell apoptosis with ANXA5/annexin-V staining by flow cytometry.
Multi-parameter flow cytometry
1. Intracellular staining: To assess the expression levels of autophagy-related genes, cytokines and cytotoxic molecules, we subjected cells to intracellular staining assays as previously described [26]. Briefly, after surface staining with specific antibodies for CD8A T-cell phenotyping, we fixed and then permeabilized cells with 0.25% saponin (Sigma Aldrich, 47,036) before the intracellular staining per se. After three washes, stained cells were finally ready for flow cytometry analyses. The viability marker 7-aminoactinomycin D or 7-AAD (ThermoFisher Scientific, 00–6993-50) was used to exclude dead cells from analyses. Of note, to assess the cumulative expression of the SQSTM1 during the 6 h-long cultures, we also cultured our cells with BaF. In order to appreciate the antiviral and cytotoxic programs on activated and viable 7-AADneg CD3W+ CD8A+ T-cell, we used the following multi-parameter antibody cocktail: anti-IL2-FITC (BD Biosciences, 561,055) and anti-IFNG/IFN-γ-AF647 (BD Biosciences, 563,495) antibodies, and GZMA/granzyme A-PE, GZMB/granzyme B-PE-CF594 (BD Biosciences, 562,462) and anti-PRF1/perforin-PerCP-Cy5.5 antibodies (Fig. S3D). 2. Palmitate uptake: At 6 h post-activation, we incubated the cells for 5 min with 5 μM of C16 BODIPYTM (ThermoFisher Scientific, D3821). After three washes, lipid uptake was determined on gated CD8A T-cells and expressed as mean intensity fluorescence (MFI). Regarding the palmitate uptake assay, we did not perform any technical replicates. In fact, we only performed the palmitate BODIPY uptake in each single tube per condition. 3. Data analyses: All samples were finally acquired using the BD LSRII Fortessa flow cytometer and analyzed with DIVA software (BD). 200,000–500,000 gated cells were analyzed for each sample.
Imaging flow cytometry
1. Lysosomal content of LC3 (autophagy): Lysosomal content of endogenous LC3 is a well-acknowledged marker for autophagy. In this context, we used a previously validated method to detect the lysosomal localization of LC3 in primary cells [25]. Briefly, after 6 h of cell activation with BaF, PBMC were first stained with the lysosomal Lyso-ID green dye according to the manufacturer’s instructions (Enzo Life Sciences, ENZ-51,005). Anti-CD3W-APC H7 (BD Biosciences, 560,176) and anti-CD8A-V450 Abs were used for surface staining. Cells were then fixed, permeabilized and finally labeled with anti-LC3-Alexa Fluor 647 Abs with or without anti-IFNG/IFN-γ-PE (BD Biosciences, 559,327) Abs. Lysosomal localization of LC3 was measured using a morphology mask to determine a similarity score, which quantifies the correlation of pixel values of the Lyso-ID and LC3 images on a per cell basis. A similarity score >1 was used as a cutoff for nuclear localization. Cells in individual bins were visually inspected to confirm subcellular localization (values < or >1). 2. Lysosomal content of endogenous lipids (lipophagy): For the purpose of this study, we have developed a high throughput and statistically robust technique that quantitates lipophagy in primary human cells. The principle of this method consists in measuring the accumulation of lipid contents in ALs when cultures were performed with the lysosomal inhibitor BaF. This culture condition was referred as the “cumulative state” and compared to the BaF-free “steady-state” condition. At 6 h post-activation with or without BaF, cells were collected and co-stained with Lyso-ID green dye and the HCS LipidTox deep red neutral lipid dye (ThermoFisher Scientific, H34477) according to the manufacturer’s instructions. Anti-CD3W-APC H7 and anti-CD8A-V450 Abs were used for surface staining. Our novel method to appreciate lipophagy activity was validated by using hepatic Huh 7.5 cell line that have been starved for 2 h (Fig. S5) [18]. 3. Data analyses: Samples were acquired using the Image Stream X MKII flow cytometer and analyzed with IDEAS software (Amnis). 200,000–500,000 gated cell singlets were analyzed for each sample. Results were expressed as co-localization index BDS (for bright detail similarity) between the lysosomal dye Lyso-ID and the other fluorescently labeled marker of interest. Of note, we determined the lipophagy activity by the formula: (% of BDShigh Lyso-ID+ LipidTox+ cells with BaF) – (% of BDShigh Lyso-ID+ LipidTox+ cells without BaF). Finally, the Spot Wizard in IDEAS was used to create an algorithm to calculate the number of LC3 and LipidTox spots (referred as puncta). Two “truth population” were manually selected with 30 events per population: a positive population of cells, which have many spots, and a negative population of cells, which have no spots. The Wizard was then used to calculate the mean number of marker spots per cells.
Autophagy-dependent lytic assays
Autophagy-dependent lytic degradation of long-lived proteins in 106 purified CD8A T-cells was quantified as previously described [24]. The method is based on a pulse-chase approach, whereby cellular proteins are radiolabeled by [14C] valine (PerkinElmer, NEC291EU050UC). Briefly, 106 purified CD8A T-cells were seeded and incubated for 18 h in complete RPMI with 0.2 µCi/ml of L-[14C] valine in order to label intracellular proteins (Pulse media). Cells were then washed three times with PBS to eliminate any unincorporated radioactivity. The short-lived proteins are allowed to be degraded for 24 h in fresh RPMI (Chase media). After that, cells were polyclonally activated for 6 h in the presence or absence of BaF, and then seeded in 10% of trichloroacetic acid (TCA) containing RPMI overnight. After centrifugation, precipitated cells were washed twice with cold 10% TCA RPMI and dissolved in 0.2 M NaOH for 2 h. Of note, supernatants contained the acid-soluble radioactivity fraction. Radioactivity was finally quantified by liquid scintillation counting. The rate of autophagy-dependent degradation of long-lived proteins was calculated from the ratio of the acid-soluble radioactivity in the medium to the one in the acid-precipitable cell fraction.
Electron microscopy analysis
Electron microscopy analysis was done as previously reported, but with a few modifications [12]. Briefly, after 6 h of polyclonal activation with or without IL21, we purified 2.106 CD8A T-cells for microscopic observation. Purified CD8A T-cells were first fixed overnight at 4°C with 2.5% glutaraldehyde (Mecalab, 1206) in 0.05 M cacodylate buffer (pH 7.4). Cells were then rinsed, post-fixed in 1.3% osmium tetroxide (Mecalab, 1605) in collidine buffer (pH 7.4), dehydrated, and embedded in two successive baths of SPURR (TedPella, 18,300–4221). Grids were rinsed in distilled water, stained with aqueous 2% uranyl acetate for 15 min and finally photographed with a Hitachi H-7100 electron microscope fitted with an AMT XR111 camera (Hitachi High-Tech America Inc.). A number of 40 cells per slide were observed. We calculated the percentage of CD8A T-cells containing autophagic vacuoles (AV) and the total numbers of AV and autolysosomes (ALs) by using different magnifications. Finally, we also used osmium tetroxide negative-staining to visualize the presence of endogenous lipids in ALs for all study groups. Of note, lipid staining with osmium is characterized by bright areas.
Metabolic flux assay
At 6 h post-polyclonal activation with or without cytokines, we assessed both the overall and FAO-mediated mitochondrial respiration in purified CD8A T-cells from ART using a Seahorse XF96 metabolic analyzer. Of note, 4.105 purified cells per condition were needed to ensure reproducible observations (in triplicate). 1. Whole mitochondrial respiration: The oxygen consumption rates (OCR) were determined in ART by using the Seahorse XF Cell Mito Stress Test kit (Agilent Technologies, 103,015–100). Briefly, 4.105 activated CD8A T-cells were seeded on XF96 well plates (Agilent Technologies, 102,601–100) in complemented Agilent RPMI with glucose 10 mM, glutamine 2 mM and pyruvate 1 mM (Agilent Technologies, 103,576–100). The XF Cell Mito Stress test kit was used according to the manufacturer protocol. OCR was determined under basal conditions and in response to modulators of respiration that were injected during the assay to reveal key parameters of mitochondrial functions. The modulators included in this assay were oligomycin (Agilent Technologies, 103,015–100), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP; Agilent Technologies, 103,015–100), rotenone (Agilent Technologies, 103,015–100) and antimycin A (Agilent Technologies, 103,015–100) (1.5 µM, 2 µM and 0.5 µM, respectively). We determined the spare respiratory capacity (SRC) and ATP-linked respiration for each condition as follows: [(maximal OCR determined after FCCP treatment) – (basal OCR determined before oligomycin treatment)] and [(basal OCR) – (minimal OCR determined after Rotenone and Antimycin A treatment)], respectively. 2. Fatty acid beta-oxidation (FAO): To determine the FAO status in ART, we used the Seahorse XF Palmitate Oxidation Stress Test kit (Agilent Technologies, 102,720–100) protocol with a minor modification. Indeed, the 4.105 activated CD8A T-cells were seeded on XF96 plates without going through the suggested nutrient restriction pre-step. This was made to keep a satisfying cell viability (superior to 90% of cell viability confirmed by flow cytometry). However, cells were still cultured under nutrient restriction, but at the end of culture and for 45 min. In this context, cells were cultured in a CO2-free incubator at 37°C with a pre-warmed XF FAO media containing only a necessary minimal content of glucose (2.5 mM) and 0.5 mM of L-carnitine (Sigma Aldrich, C0283). L-carnitine is an amino-acid derivative that transports fatty acids into cells to be processed for energy. The nutrient restriction step was also conducted in the presence of lipid substrate palmitate or bovine serum albumin (BSA; Agilent Technologies, 102,720–100). Of note, 5 μM of FAO inhibitor etomoxir was added or not at the onset of culture and during the first injection of the assay, as positive control for lipid metabolism. Once again, OCR values, that were determined under basal conditions and in response to modulators of respiration, were used to calculate SRC and ATP-linked respiration for each culture condition. FAO was determined according to the manufacturer’s instructions and by the formula: (values with palmitate) – (values with BSA). 3. Data normalization: All values were normalized to the number of viable cell events per seeded well thanks to the CytoFLEX benchtop flow cytometer (Beckman Coulter).
Statistical analysis
We used the non-parametric Mann-Whitney U test that assumes independent samples for all statistical analyses between study groups of subjects (* symbol). On the other hand, statistical analyses between two different in vitro conditions were performed using two-sided Student paired t test (β symbol). Spearman’s correlation test was used to identify association between two variables. P values of less than 0.05 were considered significant. One symbol, 0.05 > P> 0.01; two symbols, 0.01 > P> 0.001; three symbols, 0.001 > P> 0.0001; and four symbols, P< 0.0001.
Supplementary Material
Acknowledgments
We are grateful to all subjects participating in this study, their physicians, and attending staff members from the Reseau SIDA/Maladies Infectieuses of the Fonds de la Recherche Quebec-Sante (FRQ-S; Montreal, QC, Canada). We thank Danica Albert, Angie Massicotte, Natacha Cotta-Grand, and Mario Legault for technical and administrative assistance. We also thank Roxann Hétu-Arbour (M.Sc.) and Arnaldo Nakamura (M.Sc.) for technical support in Imaging flow cytometry and electron microscopy, respectively. The present study was conducted with research funds from the Fonds de Recherche du Québec-Santé (FRQ-S) and Natural Sciences and Engineering Research Council of Canada (NSERC). We also thank Fondation Armand-Frappier for providing funds requested for the purchase of the Agilent Seahorse XFe96 cell metabolic flux analyzer platform. Funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Statement
This work was supported by the Canadian Network for Research and Innovation in Machining Technology, Natural Sciences and Engineering Research Council of Canada; Fonds de Recherche du Québec - Santé.
Disclosure statement
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Graw F, Regoes RR.. Predicting the impact of CD8+ T cell polyfunctionality on HIV disease progression. J Virol. 2014;88:10134–10145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Boyd A, Almeida JR, Darrah PA, et al. Pathogen-specific T cell polyfunctionality is a correlate of T cell efficacy and immune protection. PLoS One. 2015;10:e0128714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Almeida JR, Price DA, Papagno L, et al. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J Exp Med. 2007;204:2473–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Angin M, Volant S, Passaes C, et al. Metabolic plasticity of HIV-specific CD8(+) T cells is associated with enhanced antiviral potential and natural control of HIV-1 infection. Nat Metab. 2019;1:704–716. [DOI] [PubMed] [Google Scholar]
- [5].Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Migueles SA, Weeks KA, Nou E, et al. Defective human immunodeficiency virus-specific CD8+ T-cell polyfunctionality, proliferation, and cytotoxicity are not restored by antiretroviral therapy. J Virol. 2009;83:11876–11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ndhlovu ZM, Proudfoot J, Cesa K, et al. Elite controllers with low to absent effector CD8+ T cell responses maintain highly functional, broadly directed central memory responses. J Virol. 2012;86:6959–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Loucif H, Gouard S, Dagenais-Lussier X, et al. Deciphering natural control of HIV-1: A valuable strategy to achieve antiretroviral therapy termination. Cytokine Growth Factor Rev. 2018;40:90–98. [DOI] [PubMed] [Google Scholar]
- [9].Guo JY, Teng X, Laddha SV, et al. Autophagy provides metabolic substrates to maintain energy charge and nucleotide pools in Ras-driven lung cancer cells. Genes Dev. 2016;30:1704–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Perera RM, Stoykova S, Nicolay BN, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Strohecker AM, White E.. Autophagy promotes BrafV600E-driven lung tumorigenesis by preserving mitochondrial metabolism. Autophagy. 2014;10:384–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nardacci R, Amendola A, Ciccosanti F, et al. Autophagy plays an important role in the containment of HIV-1 in nonprogressor-infected patients. Autophagy. 2014;10:1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nardacci R, Ciccosanti F, Marsella C, et al. Role of autophagy in HIV infection and pathogenesis. J Intern Med. 2017;281:422–432. [DOI] [PubMed] [Google Scholar]
- [14].Kang R, Zeh HJ, Lotze MT, et al. The beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Iannello A, Boulassel MR, Samarani S, et al. Dynamics and consequences of IL-21 production in HIV-infected individuals: a longitudinal and cross-sectional study. J Immunol. 2010;184:114–126. [DOI] [PubMed] [Google Scholar]
- [16].Kounakis K, Chaniotakis M, Markaki M, et al. Emerging roles of lipophagy in health and disease. Front Cell Dev Biol. 2019;7:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schulze RJ, Sathyanarayan A, Mashek DG. Breaking fat: the regulation and mechanisms of lipophagy. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862:1178–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Collins DR, Gaiha GD, Walker BD. CD8(+) T cells in HIV control, cure and prevention. Nat Rev Immunol. 2020;20:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Excler JL, Robb ML, Kim JH. HIV-1 vaccines: challenges and new perspectives. Hum Vaccin Immunother. 2014;10:1734–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jones RB, Walker BD. HIV-specific CD8(+) T cells and HIV eradication. J Clin Invest. 2016;126:455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Perdomo-Celis F, Taborda NA, Rugeles MT. CD8(+) T-cell response to HIV infection in the era of antiretroviral therapy. Front Immunol. 2019;10:1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014;20:460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Roberts EA, Deretic V. Autophagic proteolysis of long-lived proteins in nonliver cells. Methods Mol Biol. 2008;445:111–117. [DOI] [PubMed] [Google Scholar]
- [25].Phadwal K, Alegre-Abarrategui J, Watson AS, et al. A novel method for autophagy detection in primary cells: impaired levels of macroautophagy in immunosenescent T cells. Autophagy. 2012;8:677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dagenais-Lussier X, Loucif H, Cadorel H, et al. USP18 is a significant driver of memory CD4 T-cell reduced viability caused by type I IFN signaling during primary HIV-1 infection. PLoS Pathog. 2019;15:e1008060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xu X, Araki K, Li S, et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat Immunol. 2014;15:1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Novy P, Huang X, Leonard WJ, et al. Intrinsic IL-21 signaling is critical for CD8 T cell survival and memory formation in response to vaccinia viral infection. J Immunol. 2011;186:2729–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cubas R, van Grevenynghe J, Wills S, et al. Reversible reprogramming of circulating memory T follicular helper cell function during chronic HIV infection. J Immunol. 2015;195:5625–5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].O’Sullivan D, van der Windt GJ, Huang SC, et al. Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].van der Windt GJ, Everts B, Chang CH, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].van der Windt GJ, Pearce EL. Metabolic switching and fuel choice during T-cell differentiation and memory development. Immunol Rev. 2012;249:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Saez-Cirion A, Sinet M, Shin SY, et al. Heterogeneity in HIV suppression by CD8 T cells from HIV controllers: association with Gag-specific CD8 T cell responses. J Immunol. 2009;182:7828–7837. [DOI] [PubMed] [Google Scholar]
- [34].van Grevenynghe J, Cubas RA, Noto A, et al. Loss of memory B cells during chronic HIV infection is driven by Foxo3a- and TRAIL-mediated apoptosis. J Clin Invest. 2011;121:3877–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].van Grevenynghe J, Procopio FA, He Z, et al. Transcription factor FOXO3a controls the persistence of memory CD4(+) T cells during HIV infection. Nat Med. 2008;14:266–274. [DOI] [PubMed] [Google Scholar]
- [36].Sagnier S, Daussy CF, Borel S, et al. Autophagy restricts HIV-1 infection by selectively degrading Tat in CD4+ T lymphocytes. J Virol. 2015;89:615–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dinkins C, Pilli M, Kehrl JH. Roles of autophagy in HIV infection. Immunol Cell Biol. 2015;93:11–17. [DOI] [PubMed] [Google Scholar]
- [38].Botbol Y, Patel B, Macian F. Common gamma-chain cytokine signaling is required for macroautophagy induction during CD4+ T-cell activation. Autophagy. 2015;11:1864–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chevalier MF, Julg B, Pyo A, et al. HIV-1-specific interleukin-21+ CD4+ T cell responses contribute to durable viral control through the modulation of HIV-specific CD8+ T cell function. J Virol. 2011;85:733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mendez-Lagares G, Lu D, Merriam D, et al. IL-21 therapy controls immune activation and maintains antiviral CD8+ T cell responses in acute simian immunodeficiency virus infection. AIDS Res Hum Retroviruses. 2017;33(S1):S81–S92. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].White L, Krishnan S, Strbo N, et al. Differential effects of IL-21 and IL-15 on perforin expression, lysosomal degranulation, and proliferation in CD8 T cells of patients with human immunodeficiency virus-1 (HIV). Blood. 2007;109:3873–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wu K, Zhang S, Zhang X, et al. IL-21 expands HIV-1-specific CD8(+) T memory stem cells to suppress HIV-1 replication in vitro. J Immunol Res. 2019;2019:1801560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tang L, Chen C, Gao X, et al. Interleukin 21 reinvigorates the antiviral activity of hepatitis B virus (HBV)-specific CD8+ T cells in chronic HBV infection. J Infect Dis. 2019;219:750–759. [DOI] [PubMed] [Google Scholar]
- [44].Yi JS, Du M, Zajac AJ. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 2009;324:1572–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Loschinski R, Bottcher M, Stoll A, et al. IL-21 modulates memory and exhaustion phenotype of T-cells in a fatty acid oxidation-dependent manner. Oncotarget. 2018;9:13125–13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mueller YM, Bojczuk PM, Halstead ES, et al. IL-15 enhances survival and function of HIV-specific CD8+ T cells. Blood. 2003;101:1024–1029. [DOI] [PubMed] [Google Scholar]
- [47].Geiger R, Rieckmann JC, Wolf T, et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167:829–42e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ma EH, Bantug G, Griss T, et al. Serine is an essential metabolite for effector T cell expansion. Cell Metab. 2017;25:345–357. [DOI] [PubMed] [Google Scholar]
- [49].Ron-Harel N, Ghergurovich JM, Notarangelo G, et al. T cell activation depends on extracellular alanine. Cell Rep. 2019;28:3011–21e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ron-Harel N, Santos D, Ghergurovich JM, et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 2016;24:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yan J, Sabbaj S, Bansal A, et al. HIV-specific CD8+ T cells from elite controllers are primed for survival. J Virol. 2013;87:5170–5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mueller YM, De Rosa SC, Hutton JA, et al. Increased CD95/Fas-induced apoptosis of HIV-specific CD8(+) T cells. Immunity. 2001;15:871–882. [DOI] [PubMed] [Google Scholar]
- [53].Migueles SA, Osborne CM, Royce C, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shan L, Deng K, Shroff NS, et al. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity. 2012;36:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Buck MD, O’Sullivan D, Klein Geltink RI, et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Pearce EL, Walsh MC, Cejas PJ, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


