Abstract
At least three domains of simian virus 40 large T antigen (TAg) participate in cellular transformation. The LXCXE motif of TAg binds to all members of the retinoblastoma protein (pRB) family of tumor suppressors. The N-terminal 70 residues of TAg have significant homology to the J domain of Hsp40/DnaJ and cooperate with the LXCXE motif to inactivate the pRB family. A bipartite C-terminal domain of TAg binds to p53 and thereby disrupts the ability of p53 to act as a sequence-specific transcription factor. The contribution of these three domains of TAg to cellular transformation was evaluated in cells that contained inactivating mutations in the pRB and p53 pathways. Cells that stably expressed wild-type or selected mutant forms of TAg were generated in mouse embryo fibroblasts (MEFs) containing homozygous deletions in the RB, INK4a, and ARF loci. It was determined that the J domain, the LXCXE motif, and the p53-binding domain of TAg were required for full transformation of wild-type and RB−/− MEFs. In contrast, INK4a−/− MEFs that lacked expression of p16INK4a and p19ARF and ARF−/− MEFs that lacked p19ARF but expressed p16INK4a acquired anchorage-independent growth when expressing wild-type TAg or mutant derivatives that disrupted either the pRB-binding or p53-binding domain. The expression and function of the pRB family members were not overly disrupted in ARF−/− MEFs expressing LXCXE mutants of TAg. These results suggest that inactivating mutations of p19ARF can relieve the requirement for the LXCXE motif in TAg-mediated transformation and that TAg may have additional functions in transformation.
Simian virus 40 (SV40) large T antigen (TAg) has been used extensively as a model system to study cellular transformation. TAg has the ability to transform a wide variety of normal cells seemingly by affecting the functions of a small number of cellular proteins. To transform wild-type (WT) mouse embryo fibroblasts (MEFs), TAg utilizes at least three domains: the J domain, the LXCXE motif that binds to the retinoblastoma protein (pRB) family of proteins (pRB, p107, and p130), and the p53-binding domain (12, 13, 17, 20, 65, 77, 81). The J domain is a highly conserved element present in all members of the DnaJ/Hsp40 family of molecular chaperones as well as all polyomavirus T antigens (39). DnaJ proteins bind specifically to hsp70 homologues to perform various chaperone activities, including the destruction of specific proteins (reviewed in reference 63). The J domain of TAg binds to hsc70 and participates in the inactivation of pRB family members (27, 56, 60, 68, 69). The J domain and the LXCXE motif of TAg cooperate to disrupt the ability of pRB family members to repress E2F-dependent transcription and to decrease the levels of hyperphosphorylated p107 and p130 (27, 60, 64, 65, 68, 69, 77). Thus, the J domain and LXCXE motif of TAg appear to induce transformation and promote cell growth by interfering with the functions of pRB, p107, and p130. The p53-binding domain of TAg binds to the specific DNA-binding domain of p53, thereby directly interfering with the ability of p53 to activate transcription (3, 21, 51). Therefore, it is believed that TAg can transform cells primarily by interfering with p53 and the pRB family.
The original observations that TAg binds to p53 and the pRB family set into motion a large field of research that has led to a more complete understanding of the role of these tumor suppressors in the development of cancer (13, 44, 46). Not only is pRB itself mutated in a wide variety of cancers, wild-type pRB can be functionally inactivated by expression of an LXCXE-containing viral oncoprotein. pRB can also be inactivated by hyperphosphorylation as a result of overexpression of cyclin D1 or loss of expression p16INK4a. p16INK4a binds to cdk4 and blocks the association of cdk4 with D-type cyclins (57). Overexpression of p16INK4a in RB+/+ cells prevents phosphorylation and subsequent inactivation of pRB by cyclin D1-cdk4 and promotes a cell cycle arrest in G1 (41, 48). In contrast, p16INK4a is unable to induce a cell cycle arrest in RB−/− cells, suggesting that the pRB pathway is required for p16INK4a-mediated cell cycle arrest (48). p16INK4a may also affect the phosphorylation status of p107 and p130, as each of these proteins migrates as a lower-phosphorylation form in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis after p16INK4a overexpression (74; J. Zalvide and J. A. DeCaprio, unpublished observations). Therefore, p16INK4a may regulate all three members of the pRB family.
In addition to p16INK4a, the INK4a locus encodes a second gene product, p19ARF (54). While each of these proteins is encoded by a unique first exon, the second and third exons encode p19ARF in an alternate reading frame than p16INK4a (54). It has been demonstrated that p19ARF binds and inactivates MDM2 (36, 52, 67, 80). Since MDM2 contributes to the degradation of p53, overexpression of p19ARF can result in an increased amount of p53, leading to an arrest in G1 (28, 80). Therefore, through the expression of p16INK4a and p19ARF, the INK4a locus apparently participates in the regulation of both the pRB and p53 pathways.
Although TAg-mediated transformation is thought to involve primarily the inactivation of pRB family members and p53, studies of mice that contain mutations in the INK4a locus would seem to indicate that the functional inactivation of pRB and p53 is not sufficient to cause cellular transformation. INK4a knockout mice lack the second and third exons of the INK4a locus and do not express p16INK4a or p19ARF, although they may express a truncated version of p19ARF that may be partially active (5, 53, 58). Despite this interference with both the p53 and pRB pathways, INK4a−/− MEFs were not transformed. Although they readily became immortalized, INK4a−/− MEFs were unable to grow in soft agar and required the expression of activated H-ras to become transformed (37, 58). INK4a−/− mice do have an increased incidence of several types of tumors including lymphomas and fibrosarcomas, which presumably occur after the mice receive additional genetic mutations (58). Curiously, the ARF−/− mice that fail to express p19ARF but retain expression of WT p16INK4a display a similar range of tumors as the INK4a−/− mice that have lost both p19ARF and p16INK4a (35, 37). In addition, the ARF−/− MEFs were readily immortalized and could be transformed by activated H-ras alone similarly to the INK4a−/− MEFs (37). Therefore, at least in the mouse model, it has been difficult to observe the specific contribution of p16INK4a to tumor suppression since the INK4a−/− and ARF−/− knockouts result in a similar phenotype.
Given that disruption of the INK4a locus was not sufficient to induce transformation of MEFs, the pRB and p53 pathways may retain some of their tumor suppressor activity in INK4a−/− cells. Indeed, it has been noted that p53 can be activated in the ARF−/− MEFs by ionizing radiation but not by adenovirus E1A (14, 37). It is thus possible that p53 may retain the ability to act as a tumor suppressor under some conditions in the ARF−/− and INK4a−/− mice. Alternatively, there may be tumor suppressor pathways other than pRB and p53 that retain activity in the INK4a−/− or ARF−/− MEFs. We wished to determine whether the loss or functional inactivation of tumor suppressor genes that are targeted by TAg would abrogate the need for the J domain, the LXCXE motif, or the p53-binding domain in TAg-mediated transformation. Using this approach, it had been previously demonstrated that the LXCXE motif was required to transform RB−/− MEFs, suggesting that p107 and p130 were also targeted by the LXCXE motif (11, 76). To identify additional pathways that may be targeted by SV40 TAg, we generated WT, RB−/−, ARF−/−, and INK4a−/− MEFs that stably expressed WT or selected mutants of TAg. We observed that TAg could fully transform MEFs from each of these genetic backgrounds. Notably, the LXCXE motif was absolutely required for transformation of WT and RB−/− MEFs but was dispensable for anchorage-independent growth of ARF−/− and INK4a−/− MEFs, suggesting that the pRB pathway was functionally inactivated by loss of p19ARF. However, pRB family members were expressed normally, and their ability to repress E2F activity was intact in the ARF−/− and INK4a−/− MEFs expressing the LXCXE mutants of TAg. These results suggest the possibility that TAg's LXCXE motif may functionally inactivate other growth suppressors in addition to the pRB family and that p19ARF may have activity beyond the regulation of MDM2 and p53.
MATERIALS AND METHODS
Plasmids.
The SV40 large TAg cDNA expression plasmids pSG5-T, pSG5-H42Q, pSG5-K1, pSG5-C105G, and pSG5-PVU-1 have been previously described (69, 76). pSG5-dl434-444 was cloned by substituting the PflMI-PstI fragment from dl434-444 containing the mutant p53-binding site into pSG5-T (40). pSG5-HQ-K1 was similarly cloned by substituting the PflMI-PstI fragment of pSG5-K1 into pSG5-HQ. pSG5-T1-135 and pSG5-T1-350 were generated by PCR amplification of a TAg cDNA with the appropriate primers. DNA sequencing confirmed the identity of all constructs.
The p21 promoter-luciferase reporter, pWWP-luc, has been previously described (19), as has the 3xE2F-luciferase reporter, 3xE2F-Luc, containing three specific E2F DNA-binding sites (50). The cyclin G-luciferase reporter, pGL3-cyclin G-Luc, was obtained from Carol Prives. The β-galactosidase reporter plasmid, pCMX-β-Gal, was used as a control for transfection efficiency (23).
Cells.
INK4a−/− MEFs and ARF−/− MEFs have been previously described (37, 58). RB−/− and WT MEFs were prepared from 13.5-day embryos as previously described (32, 76). All MEFs were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% Fetal Clone Serum I (HyClone) and penicillin-streptomycin. MEFs were cotransfected with pEpuro and fivefold excess SV40 TAg-encoding plasmid by the calcium phosphate precipitation method or Fugene 6 (Boehringer Mannheim). After 16 h of exposure to the plasmid DNA, cells were refed with complete medium and grown for 24 h before splitting into medium containing puromycin (2 μg/ml). Before reaching confluence, colonies were pooled and expanded in three 100-mm-diameter plates, at which point they were considered to be established. Cells of no more than three passages after this stage were used for transformation studies and were always passaged at subconfluence.
Western blotting and antibodies.
Cells were lysed in high-salt EBC (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 0.5% Nonidet P-40) containing aprotinin (10 μg/ml), leupeptin (10 μg/ml), 0.1 mM phenylmethylsulfonyl fluoride, 4 mM sodium fluoride, and 0.1 mM sodium orthovanadate. Lysates were cleared by centrifugation at 14,000 × g, and protein concentration was determined by the Bradford assay (Bio-Rad); 100 μg of each sample was separated in SDS-polyacrylamide gels and transferred to nitrocellulose membranes (Bio-Rad). The membranes were blocked for 1 h with 5% nonfat dry milk and 1% goat serum in TBS-T (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Tween 20), before overnight incubation with the primary antibody in TBS-T containing 1% bovine serum albumin at 4°C. The following antibodies were used in this study: anti-TAg polyclonal antibodies (PAb) 101 and 419 (26) (American Type Culture Collection), anti-RB PAb 245 (Pharmingen), anti-p107 C-18 (Santa Cruz Biotechnology), anti-p130 C-20 (Santa Cruz), anti-p53 FL-393 (Santa Cruz), anti-p15INK4b Ab-3 (Neomarkers), anti-p16INK4a M-156 (Santa Cruz), and anti-p21CIP1 C-19 (Santa Cruz). The anti-p19ARF rabbit PAb was previously described (37). Detection of proteins was performed with the appropriate horseradish peroxidase-conjugated secondary rabbit or goat antibody (Pierce) at a 1:5,000 dilution in TBS-T containing 2.5% milk and 0.5% goat serum. Immunoblots were developed using enhanced chemiluminescence (Pierce) according to the manufacturer's protocol.
Transformation assays.
Soft agar assays were performed in 35-mm-diameter dishes coated with 2 ml of DMEM containing 10% fetal bovine serum (HyClone) and 0.6% agarose (Gibco-BRL). Cells were seeded on top of this layer at a density of 5 × 104 cells per plate in DMEM containing 10% fetal bovine serum and 0.3% agarose. Soft agar colony formation was evaluated 6 weeks after the initial plating. To determine growth to high density, cells were seeded at low density (5 × 104 per 60-mm-diameter dish) and fed every 3 days with DMEM containing 10% fetal bovine serum. Triplicate plates of cells were counted every 2 or 3 days.
Promoter reporter assays.
WT and ARF−/− cell lines were plated at a density of 105 cells per 35-mm-diameter dish. Cells were transfected the following day with Fugene 6 and 0.5 μg of the appropriate luciferase reporter construct and pCMX-β-Gal (23). At 48 h after transfection, cells were washed twice in phosphate-buffered saline and assayed for β-galactosidase and luciferase activities as previously described (31).
RESULTS
Immortalization of MEFs by SV40 TAg.
It has been reported that TAg-mediated transformation of WT MEFs requires the inactivation of the pRB family by the LXCXE motif and J domain and inactivation of p53 function through the p53-binding domain (10, 76, 81). It is possible that these transforming domains of TAg have other functions or affect additional cellular targets. To determine whether these TAg domains contributed to transformation in cells that had undergone inactivation of the pRB and p53 pathways, we used selected knockout mouse strains. INK4a−/− MEFs contain a deletion of the second and third exons of the INK4a locus (58). This prevents the expression of both p16INK4a and p19ARF, effectively disrupting both the p53 and the pRB pathways. ARF−/− MEFs selectively delete the first exon of the p19ARF protein and retain an intact p16INK4a (37). RB−/− MEFs lose pRB but express the pRB-related proteins p107 and p130 (76).
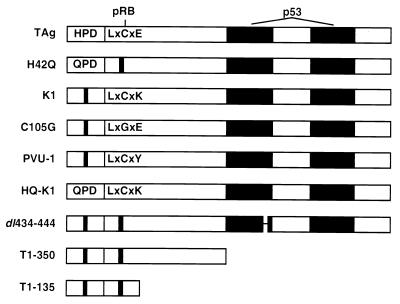
To determine which domains of TAg are required for transformation of WT, INK4a−/−, ARF−/−, and RB−/− MEFs, we established cell lines that express WT TAg or various mutants of TAg as depicted in Fig. 1. An intact HPD motif (residues 42 to 44) is essential for J domain function of TAg (38, 65, 68). The H-to-Q point substitution (H42Q) in the HPD motif has been shown to inactivate several J domain functions, including SV40 TAg-mediated ori-dependent DNA replication (6). The LXCXE motif of TAg, contained within residues 103 to 107, binds to and inactivates pRB family members (13, 16, 20, 76). The K1 (E107K), C105G, and PVU-1 substitution mutations disrupt the ability of TAg to bind to pRB, p107, and p130. The bipartite p53-binding domain of TAg is located between residues 350 and 550 (40, 51). A small in-frame deletion of residues 434 to 444 completely disrupts the ability of TAg to bind p53 (40). The truncation mutants T1-350 and T1-135 lack the entire p53-binding region but retain an intact J domain and LXCXE motif. All of the SV40 TAg cDNA constructs expressed similar levels of large TAg and did not express small t antigen (68, 76; data not shown).
FIG. 1.
SV40 TAg constructs. The J domain is contained within the first 82 residues and includes the conserved HPD (residues 42 to 44). The point substitution mutant H42Q inactivates the J domain function of TAg (6, 68). The LXCXE motif is present in residues 103 to 107. The K1, C105G, and PVU-1 mutations disrupt binding to all pRB family members (76). The HQ-K1 mutant contains inactivating mutations in the J domain and LXCXE motif. The dl434-444 mutant disrupts the ability of TAg to bind to p53 (40). T1-350 contains the first 350 amino acids of TAg. T1-135 contains the first 131 amino acids of TAg plus 4 additional residues (79). All TAg constructs were expressed as cDNAs in a pSG5 plasmid under an SV40 promoter.
TAg expression plasmids were cotransfected with a plasmid expressing a puromycin resistance gene (pEpuro) into WT, ARF−/−, INK4a−/−, and RB−/− MEFs. Early-passage MEFs were transfected to minimize the possibility that the cells had acquired spontaneous mutations. Pools of transfectants were selected in puromycin and tested for TAg expression. Expression of WT TAg or TAg mutants was quite high in all pools of cells and varied less than twofold (data not shown). WT and RB−/− MEFs usually senesce within 10 to 20 passages, and any puromycin-resistant clone was likely to have undergone immortalization (76). In WT MEFs, we observed that only cells that expressed TAg constructs containing an intact p53-binding domain became immortalized, as had been previously reported (40). An intact J domain or LXCXE motif was not required for immortalization. WT MEFs transfected with pEpuro only failed to produce any immortal lines. Similar results were observed with RB−/− MEFs, although a few small colonies expressing either the dl434-444 or T1-135 construct were observed (40, 71). Since the RB−/− MEFs expressing the dl434-444 construct grew more slowly than cells transfected with other TAg constructs, they were not used in transformation studies. The T1-135 RB−/− MEFs grew slightly faster and were tested in the soft agar assay.
Both ARF−/− and INK4a−/− cells immortalized readily, as determined by the generation of stable pools of puromycin-resistant cells transfected by the puromycin resistance plasmid alone. This observation is consistent with prior reports that ARF−/− and INK4a−/− MEFs were either already immortalized or underwent immortalization very readily (37, 58). Furthermore, the p53-binding domain of TAg was not required for the immortalization of INK4a−/− or ARF−/− MEFs. We were able to generate pools of cells that stably expressed WT TAg or the J domain, LXCXE motif, or p53-binding domain mutants in the ARF−/− and INK4a−/− MEFs. When these cells were examined by indirect immunofluorescence using a monoclonal antibody for TAg, more than 95% of the cells expressed detectable levels of TAg (data not shown). In contrast, when we attempted to generate ARF−/− or INK4a−/− MEFs expressing TAg with mutations in both the pRB-binding and the p53-binding domains, only about 50% of the cells expressed the TAg mutants (data not shown). We were unable to generate populations of ARF−/− or INK4a−/− MEFs that expressed in every cell the K1-dl434-444 double mutation or T1-135 or T1-350 containing a mutation in the LXCXE motif (T1-135K1 or T1-350K1). Similarly, a TAg triple mutant in which the J domain, the LXCXE motif, and the p53-binding domain were mutated (H42Q-K1-dl434-444) also did not yield clones that expressed TAg in every cell. Although each of these double- and triple-mutant transfectants formed as many colonies as WT TAg transfectants, we found that less than half of the cells lacked detectable TAg expression. Given the lack of uniform expression, INK4a−/− or ARF−/− MEFs transfected with double- or triple-mutant TAg constructs were not studied further.
Role of the LXCXE motif in anchorage-independent growth.
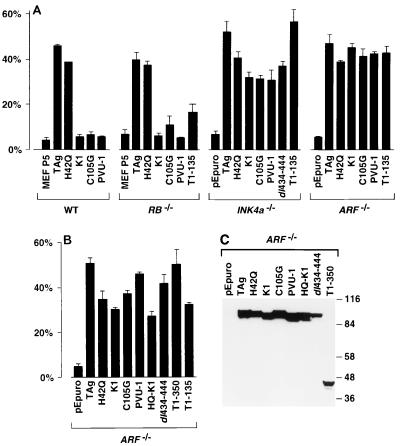
To determine the ability of TAg or mutant derivatives to confer anchorage-independent growth, the MEF cell lines were plated in soft agar. Growth was evaluated visually, and colonies containing eight or more cells were scored as transformants (Fig. 2A). WT TAg expression in WT and RB−/− MEFs was capable of inducing growth in soft agar in approximately 40 to 50% of cells seeded. In WT and RB−/− MEFs, the J domain mutant H42Q transformed almost as well as WT TAg, with nearly the same number and size of colonies, indicating that the J domain was not required for anchorage-independent growth (68). In contrast, TAgs containing mutations in the LXCXE motif were unable to transform WT or RB−/− MEFs (68, 76). Thus, an intact LXCXE motif but not the J domain was necessary for the transformation of WT and RB−/− MEFs. Similar results were observed in p107−/−, p130−/−, and p107/p130−/− MEFs (Zalvide and DeCaprio, unpublished). In each of these genetic backgrounds, the LXCXE mutants failed to induce growth in soft agar, while both WT TAg and the J domain mutant of TAg induced soft agar growth in more than 40% of the cells seeded (data not shown). The truncated T1-135, lacking the p53-binding domain, was able to induce soft agar growth of RB−/− MEFs only slightly more efficiently than the full-length LXCXE mutants, suggesting that the p53-binding domain of TAg may contribute to anchorage-independent growth of RB−/− MEFs.
FIG. 2.
Ability of TAg to form soft agar colonies in WT, RB−/−, INK4a−/−, and ARF−/− MEFs. (A) TAg-immortalized MEFs from the indicated cultures were plated in soft agar and allowed to grow for 6 weeks. Colonies containing eight or more cells were scored as positive and indicated as percentage of cells seeded. Nonimmortalized WT and RB−/− MEFs from the fifth passage (P5) were used as controls. Error bars indicate standard deviation. (B) Soft agar colony formation in an independent transformation of ARF−/− MEFs. (C) Western blot of TAg expression in ARF−/− MEFs. Equivalent amount of lysates prepared from each of the indicated MEFs were separated in an SDS–7.5% polyacrylamide gel and blotted with an anti-TAg monoclonal antibody. Sizes are indicated in kilodaltons.
Since INK4a−/− MEFs disrupt the expression of both p16INK4a and p19ARF, these cells are expected to have at least partially inactivated the pRB and p53 tumor suppressor pathways. We therefore reasoned that TAg might not require the LXCXE motif or the p53-binding region to transform INK4a−/− MEFs. As seen in Fig. 2A, WT TAg or mutants containing point substitutions in the LXCXE motif (K1, C105G, or PVU-1), a small in-frame deletion in the p53-binding domain (dl434-444), or a deletion of the entire p53-binding domain (T1-135) were able to induce soft agar colony growth in INK4a−/− MEFs. Notably, some function of TAg was required for transformation of INK4a−/−, as the pEpuro-only transfectants were unable to grow in soft agar (Fig. 2A). Thus, expression of TAg containing an intact LXCXE motif or an intact p53-binding region conferred anchorage-independent growth in the INK4a−/− cells.
Surprisingly, ARF−/− MEFs that express p16INK4a but not p19ARF were transformed by the same TAg constructs that were able to transform INK4a−/− MEFs. TAg constructs containing mutations in the J domain (H42Q), the LXCXE motif (K1, C105G, or PVU-1) or the p53-binding domain (T1-135) were able to induce ARF−/− MEFs to grow in soft agar with nearly the same efficiency as WT TAg (Fig. 2A). Since p19ARF was suspected to affect MDM2 and p53 function but not pRB function, the ability of the LXCXE mutants to form soft agar colonies in ARF−/− MEFs was unexpected. However, three different mutant constructs of the LXCXE motif, each unable to bind pRB family members, were able to induce soft agar colony growth as efficiently as WT TAg. Notably, the truncation mutant T1-135, unable to bind to p53, also induced colony formation in soft agar.
To confirm the ability of LXCXE TAg mutants to induce soft agar growth in ARF−/− MEFs, we established a second series of cells with additional TAg constructs. The level of TAg expression was similar in each pool of ARF−/− MEFs, as confirmed by Western blotting (Fig. 2C). Again, the three different LXCXE mutant constructs were each capable of conferring the ability of ARF−/− MEFs to grow in soft agar relative to the pEpuro-only pools of transfectants (Fig. 2B). Furthermore, a J domain-LXCXE double mutant, HQ-K1, was also capable of growth in soft agar. This double mutant, though expected to have completely disrupted any ability to perturb the pRB family of tumor suppressors, was nearly as effective as the K1 and H42Q single mutants in inducing soft agar growth. In addition, three different p53-binding-defective mutants of TAg, dl434-444, T1-350, and T1-135, were each able to transform ARF−/− MEFs. Thus, in both INK4a−/− and ARF−/− cells, mutants expressing either an intact p53-binding domain or LXCXE motif were able to induce transformation, as measured by the ability to induce anchorage-independent growth.
Growth to high density requires the J domain and the LXCXE motif.
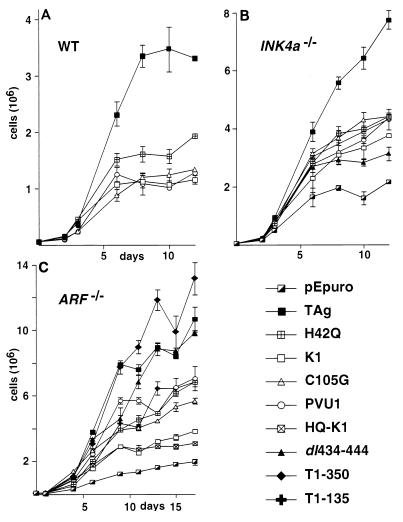
The TAg-transformed phenotype is also reflected in the ability of adherent cells to grow to high density on plastic surfaces. TAg-transformed cells can grow to a higher density than nontransformed cells. To overcome a density arrest, cells must overcome signals for growth arrest due to contact inhibition as well as growth factor depletion. TAg induces high-density growth in cultured cells (76). To determine the domains of TAg that participate in inducing high-density growth, the growth rate of TAg-immortalized MEF cell lines was determined. Cells were plated on a 10-cm-diameter plate at a low density and refed with medium containing 10% serum every 3 days. At various intervals, three replica plates of each transformant were harvested and counted. This growth experiment was repeated at least twice, and the results of a typical experiment are shown in Fig. 3.
FIG. 3.
Ability of TAg-transformed MEFs to grow to high density. Fifty thousand WT, INK4a−/−, or ARF−/− MEFs stably expressing the indicated TAg constructs were seeded on replica 100-mm-diameter plates and fed with complete medium containing 10% serum every 2 to 3 days. Triplicate plates were harvested at the indicated day after seeding, and cells were counted. Error bars indicate standard deviation.
WT TAg was able to induce growth to high density in WT MEFs, resulting in approximately a 100-fold increase in cell number after 12 days (Fig. 3A). In contrast, the J domain mutant of TAg (H42Q) grew to approximately half the density as wild-type TAg before undergoing a density arrest. This result suggests that although the J domain had only a very minor influence on the ability of MEFs to grow in soft agar, an intact J domain enhanced the ability of TAg to induce growth to high density. The LXCXE mutants, K1, C105G, and PVU-1, also underwent a density arrest, resulting in three- to fourfold fewer cells relative to WT TAg. The observed contribution of the J domain and LXCXE motif to TAg's ability to overcome density arrest in WT and RB−/− MEFs is entirely consistent with previous reports (68).
WT TAg induced growth to very high density in INK4a−/− MEFs, nearly twice as high as observed in WT MEFs (Fig. 3B). In contrast, the J domain (H42Q) and LXCXE (K1, C105G, and PVU-1) transformants grew to a density half of that observed for MEFs expressing WT TAg but nearly twice that of the pEpuro transformants. The p53-binding-defective mutant dl434-444 grew to a slightly lower density than the J domain and LXCXE mutants but still resulted in significantly more cells than the pEpuro-only MEFs. In contrast to the results observed in the anchorage-independent growth assay, loss of the J domain, the LXCXE motif, or the p53-binding domain impaired the ability of INK4a−/− MEFs to grow to high density.
WT TAg induced ARF−/− MEFs to grow to approximately twice the densities of MEFs transfected with the J domain mutant, H42Q, and the LXCXE mutants, C105G and PVU-1 (Fig. 3C). The LXCXE mutant K1 and the J domain-LXCXE double mutant HQ-K1 grew to a slightly lesser extent than the other LXCXE mutants but still grew to a higher density than the pEpuro-only transfectants. Notably, the TAg p53-binding domain mutants dl434-444 and T1-350 grew as well as WT TAg, suggesting that the p53-binding domain is not necessary for growth to high density in ARF−/− MEFs. Furthermore, ARF−/− MEFs that expressed T1-135 did not grow to as high a density as the full-length TAg, T1-350, or dl434-444 constructs, suggesting that there may be an additional transforming function within residues 135 to 350 of TAg. Evidence for an additional transforming domain within these residues has previously been proposed (15). The near-wild-type TAg ability of the dl434-444 mutant to induce growth in ARF−/− MEFs compared to INK4a−/− MEFs may be accounted for by the possibility that the INK4a−/− MEFs express an N-terminal fragment of the p19ARF protein encoded by the intact exon 1β in the INK4a locus (53). The truncated p19ARF may be able to inhibit MDM2 activity and thereby activate p53 (36).
Expression and activity of pRB in ARF−/− MEFs.
The soft agar experiments shown in Fig. 2A and B suggest that while the LXCXE motif was required for TAg-dependent anchorage-independent growth in WT and RB−/− MEFs, it was dispensable in INK4a−/− and ARF−/− MEFs. This suggests that the loss of p19ARF may have resulted in a partial inactivation of the pRB growth suppression pathway in addition to the p53 pathway. To explore this possibility, we examined the pRB and p53 pathways in WT and ARF−/− MEFs.
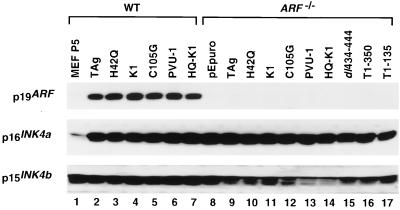
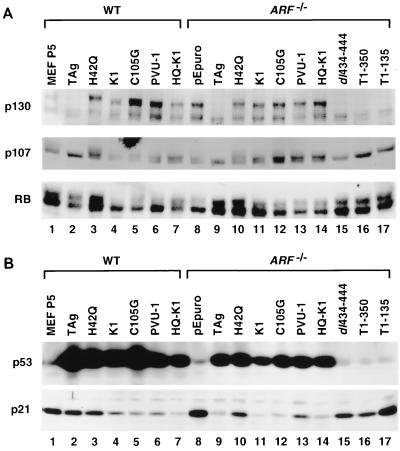
First, we considered that either the targeted disruption of the 1β exon in the ARF−/− MEFs or the stable expression of TAg may have affected expression of the nearby p15INK4b or p16INK4a genes. Loss of either p15INK4b or p16INK4a could lead to an increase in cyclin D-cdk4 activity, resulting in the hyperphosphorylation and inactivation of the pRB family members. However, as shown in Fig. 4, p16INK4a was expressed at relatively similar levels in WT and ARF−/− MEFs established by various TAg constructs, and only a slight variation of p15INK4b expression was noted in the ARF−/− MEFs. Thus, it is unlikely that the loss of p16INK4a or p15INK4b expression was responsible for the ability of the LXCXE mutant in ARF−/− MEFs to overcome the pRB growth suppression pathway in the soft agar assay. Notably, the levels of p16INK4a and p19ARF were low in the early-passage primary WT MEFs and became significantly increased upon TAg-induced immortalization. This increase in expression of p19ARF has been reported during serial passage of WT MEFs (37).
FIG. 4.
Western blot for p19ARF, p16INK4a, and p15INK4b in WT and ARF−/− MEFs expressing various TAg constructs. Equivalent amounts of lysates prepared from WT and ARF−/− MEFs stably expressing the indicated TAg constructs were separated in SDS-polyacrylamide gels and blotted with specific antibodies.
Alternatively, it was possible that loss of p19ARF in ARF−/− MEFs affected expression of pRB family members. This was a distinct possibility given the observation that p19ARF affects MDM2 function and MDM2 has been reported to bind to pRB (73). However, as shown in Fig. 5A, there was no significant difference between TAg-expressing WT and ARF−/− MEFs in the steady-state levels or phosphorylation state of pRB, p107, and p130. As noted in the introduction, the LXCXE motif and J domain of TAg cooperate to alter the phosphorylation state of p130 and p107 (68, 69). As can be seen in Fig. 5A, the hyperphosphorylated forms of p130 and p107 were not present in the WT or ARF−/− MEFs expressing WT TAg (lanes 2 and 9), dl434-444 (lane 15), T1-350 (lane 16), and T1-135 (lane 17). In contrast, the hyperphosphorylated forms of p107 and p130 were present in WT and ARF−/− MEFs established by the J domain mutant H42Q (lanes 3 and 10), the LXCXE mutant constructs K1 (lanes 4 and 11), C105G (lanes 5 and 12), and PVU-1 (lanes 6 and 13), and the double mutant HQ-K1 (lanes 7 and 14). Thus, the pRB family of proteins were expressed and normally phosphorylated in the presence of LXCXE and J domain mutants of TAg in a manner indistinguishable between WT and ARF−/− MEFs.
FIG. 5.
Expression of pRB family members, p53, and p21 in WT and ARF−/− MEFs. (A) Western blot for pRB, p107, and p130 in WT and ARF−/− MEFs expressing various TAg constructs. (B) Western blot for p53 and p21 in WT and ARF−/− MEFs expressing various TAg constructs.
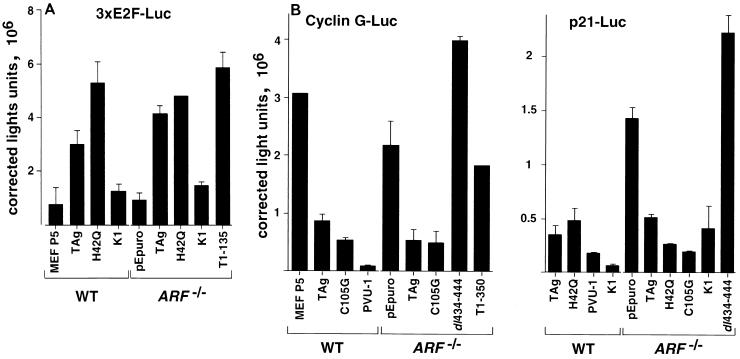
The E2F transcription factors are among the best-characterized targets of the pRB family. pRB, p107, and p130 bind to and repress the transcriptional activity of certain members of the E2F family (7, 9, 24, 30, 33, 62, 72). To test whether the pRB family retained the ability to repress E2F activity in ARF−/− MEFs that expressed TAg or an LXCXE mutant, we examined the activity of an E2F promoter reporter containing three consensus E2F sites (43). As shown in Fig. 6A, WT TAg or the J domain mutant H42Q could transactivate the 3xE2F-Luc reporter relative to non-TAg-expressing WT or ARF−/− MEFs. Similarly, the p53-binding mutant T1-135 could also transactivate the 3xE2F-Luc reporter in ARF−/− MEFs. In contrast, the activity of the E2F promoter reporter was significantly lower in MEFs established by LXCXE mutants of TAg. The LXCXE mutant K1 was unable to transactivate the 3xE2F-Luc reporter relative to controls in either WT or ARF−/− MEFs. A promoter reporter containing three mutated E2F sites was not affected by expression of TAg or any of the mutated TAg constructs (data not shown). We conclude that the ability of the pRB family to decrease transcription through the E2F site was not abrogated in the LXCXE mutant-expressing ARF−/− MEFs.
FIG. 6.
E2F and p53 promoter reporter activity in TAg-transformed WT and ARF−/− MEFs. The reporter constructs (A) 3xE2F-Luc and pCMV-β-Gal were cotransfected into the indicated stable TAg-transformed MEFs. Luciferase activity was normalized to β-galactosidase activity and expressed as relative luminometer units. The experiment was performed in triplicate; error bars indicate standard deviation. (B) Cyclin G promoter-luciferase reporter (left) and p21 promoter-luciferase promoter (right) activities were determined as described above.
Expression and activity of p53 in ARF−/− MEFs.
SV40 TAg has a well-described ability to bind to p53 and to increase its level of expression at least in part through decreasing the turnover rate (70). As shown in Fig. 5B (top panel), p53 expression was extremely low in the pEpuro ARF−/− MEFs (lane 8) as well as in early-passage cultures of WT MEFs (lane 1). In contrast, WT and ARF−/− MEFs expressing WT TAg (lanes 2 and 9), LXCXE mutants (lanes 4 to 6 and 11 to 13), or J domain mutants (lanes 3, 7, 10, and 14), which are capable of binding to p53, had significantly increased levels of p53. This observation suggests that TAg could stabilize p53 both in WT cells and in cells that have lost p19ARF. In contrast, the expression of p53-binding mutants dl434-444 (lane 15), T1-350 (lane 16), and T1-135 (lane 17) did not result in increased steady-state levels of p53.
To determine whether p53 retained the ability to transactivate certain promoters in ARF−/− MEFs, we measured the activity of two p53-responsive promoter reporters, cyclin G and p21, in WT and ARF−/− MEFs. As shown in Fig. 6B, WT TAg and C105G, an LXCXE mutant, strongly repressed the transcription of the cyclin G promoter relative to the activity observed in the pEpuro-transfected ARF−/− MEFs. In contrast, p53-binding mutants of TAg, dl434-444 and T1-350, failed to repress the activity of the cyclin G promoter. Similar results were observed with the p21CIP promoter. WT TAg, the J domain mutant H42Q, and two different LXCXE mutants repressed the activity of the p21CIP promoter, while the p53-binding mutant dl434-444 was unable to repress this promoter. The levels of p21CIP protein seen in TAg-expressing WT and ARF−/− MEFs reflected the changes seen in promoter activity (Fig. 5B, lower panel). p21CIP levels were high in WT MEFs and in the ARF−/− pEpuro-only transfectants (lanes 1 and 8). p21CIP levels were also quite high in the ARF−/− cell lines expressing any of the p53-binding mutants of TAg, dl434-444 (lane 15), T1-135 (lane 16), or T1-350 (lane 17). These results suggest that in ARF−/− MEFs stably expressing p53-binding mutants of TAg, p53 retained the ability to act as a DNA-specific transcription factor.
DISCUSSION
In this report, the contribution of the J domain, LXCXE motif, and p53-binding domain of TAg to transformation of WT, RB−/−, INK4a−/−, and ARF−/− MEFs was examined. The goal was to determine whether the loss or functional inactivation of certain tumor suppressor genes would reduce the requirement for these domains in TAg transformation. WT SV40 TAg was able to induce growth of MEFs derived from each of these genetic backgrounds to high density and in an anchorage-independent manner. In WT and RB−/− MEFs, TAg's p53-binding domain was required for efficient immortalization, the LXCXE motif was needed for anchorage-independent and high-density growth, and the J domain was required for high-density growth. In contrast, TAg had different requirements for transformation of MEFs that did not express p16INK4a or p19ARF. The p53-binding domain of TAg was not required for immortalization, high-density growth, or soft agar growth of INK4a−/− or ARF−/− MEFs. These results were not unexpected given that the loss of p19ARF expression has been shown to perturb the p53 growth suppression pathway, thereby reducing the requirement for p53 inactivation by TAg binding. The LXCXE motif and J domain were required for high-density growth of INK4a−/− MEFs, suggesting that the pRB family was functionally intact despite the loss of p16INK4a expression. The most unexpected observation was that the LXCXE motif was not required for anchorage-independent growth of either INK4a−/− or ARF−/− MEFs. This latter result would seem to suggest that the growth suppression functions of the pRB family were disrupted in ARF−/− and INK4a−/− MEFs during anchorage-independent growth. Notably, the ARF−/− MEFs which expressed p16INK4a but not p19ARF behaved similarly as INK4a−/− MEFs in response to TAg. Both cell types were induced to grow in soft agar when expressing either p53-binding domain or LXCXE motif mutants of TAg. Since p19ARF was reported to regulate the p53 pathway but not the pRB pathway, the LXCXE mutants were not expected to induce anchorage-independent growth in ARF−/− MEFs that expressed p16INK4a.
It is unlikely that INK4a−/− and ARF−/− MEFs expressing the LXCXE mutant TAg constructs have acquired additional specific genetic modifications that enabled them to grow in soft agar. All assays were performed with pools of transfected cells to reduce the possibility of clonal variation. Furthermore, similar transformation results were obtained with several independently performed transfections. INK4a−/− or ARF−/− MEFs could be readily immortalized in the absence of TAg, as evidenced by the efficient ability to select for colonies in the presence of the puromycin resistance gene alone. However, INK4a−/− or ARF−/− MEFs expressing the puromycin resistance gene alone were not transformed, as determined by their inability to grow in soft agar or to high density. Expression of TAg conferred an additional transforming activity that permitted growth under these more stringent conditions. These observations are consistent with the demonstration that expression of activated H-ras induced transformation of ARF−/− and INK4a−/− MEFs (37, 58).
Growth in soft agar measures the ability of fibroblasts to acquire anchorage-independent growth. Anchorage-independent growth has been considered a stringent in vitro method for predicting the tumor formation potential of TAg-transformed fibroblasts (55). However, it has been reported that the ability of TAg-transformed cells to form tumors in nude mice does not require an intact LXCXE motif (71). To date, there has been an absolute requirement for an intact LXCXE motif in anchorage-independent growth of TAg-transformed WT MEFs in soft agar (10, 11, 71, 76). Here, we observed that the LXCXE motif was not required for soft agar growth of INK4a−/− and ARF−/− MEFs.
Notably, expression of HQ-K1, a TAg containing inactivating mutations in both the LXCXE motif and J domain, induced INK4a−/− or ARF−/− MEFs to grow in soft agar. Despite loss of the J domain and LXCXE motif, HQ-K1 could bind to and stabilize p53. Transformation by this and other LXCXE mutants would seem to indicate that binding to p53 conferred a growth advantage and that p53 function was not completely eliminated by the loss of p19ARF. Consistent with this possibility, p53 appeared to be functional in the ARF−/− MEFs that express TAg with mutations in the p53-binding domain, as demonstrated by the increased activity of the p21CIP and cyclin G promoters (Fig. 6B) as well as by the increased expression of p21CIP (Fig. 5B). This potential for activation of p53 in ARF−/− MEFs did not seem to pose a barrier to immortalization or to the ability of the p53-binding mutant TAgs to induce growth in soft agar or to high density.
The ability of adherent cells to grow to high density may reflect the cell's response to limiting amounts of growth factors and nutrients as the cell number increases. An intact J domain and LXCXE motif were both required for TAg to induce growth to high density in WT, RB−/−, ARF−/−, and INK4a−/− MEFs (Fig. 3 and reference 68). The contribution of the J domain to high-density growth of these MEFs is consistent with previous work that suggests that the J domain cooperates with the LXCXE motif to inactivate pRB, p107, and p130 (27, 60, 68). Further evidence for the J domain activity includes the reduction in phosphorylation of p107 and p130 in WT and ARF−/− MEFs expressing TAg (Fig. 5B). We have previously reported that J domain mutant constructs were as effective as WT TAg in promoting the high-density growth of p130−/− p107−/− double-knockout MEFs, suggesting that the J domain specifically contributed to the functional inactivation of p107 and p130 (68). The observation that the J domain and the LXCXE motif were required for overcoming density arrest of INK4a−/− and ARF−/− MEFs suggests that the pRB family including p107 and p130 was functionally intact in adherent INK4a−/− and ARF−/− MEFs. Therefore, loss of p16INK4a or p19ARF expression did not perturb the pRB growth suppression activity in the density arrest assay. It will be interesting to determine whether TAgs containing J domain or LXCXE motif mutants will have the ability to induce high-density or soft agar growth in MEFs homozygously deficient in Rb, p107, and p130.
The J domain cooperates with the LXCXE motif to inactivate pRB function in a variety of assays including dissociation of pRB-E2F complexes and overriding pRB repression of E2F-dependent promoters (68, 77). Recent reports suggest that the LXCXE motif of SV40 and polyomavirus large T antigens may also inhibit certain pRB functions independently of the J domain. For example, WT TAg and a J domain mutant (H42Q) but not an LXCXE mutant (K1) could override a p53-induced growth arrest (25). The LXCXE motif but not the J domain was required for polyomavirus large T antigen-induced apoptosis of C2C12 myoblasts upon serum withdrawal (61). In this report, we observed that an intact LXCXE motif but not the J domain was required for activation of the 3xE2F promoter reporter in WT and ARF−/− MEFs (Fig. 6A). In addition, an intact J domain was not required for inducing soft agar growth of MEFs derived from any of the genetic backgrounds studied. These distinctions may reflect the ability of TAg to perturb different growth-suppressing functions of the pRB family. Alternatively, the LXCXE motif may target other cellular proteins that do not require the contribution of the J domain.
As mentioned earlier, the most unexpected observation was that the LXCXE mutants of TAg could induce anchorage-independent growth of ARF−/− MEFs. There are several models to explain how the LXCXE mutant TAgs were able to confer anchorage-independent growth of the ARF−/− and INK4a−/− MEFs. One possibility is that p19ARF is involved in the regulation of both p53 and pRB and that loss of p19ARF led to at least partial deregulation of both pathways. If this were true, it could explain why ARF−/− mice display the same tumor types as INK4a−/− mice. Several reports suggest that p19ARF may regulate pRB function. For example, the expression of an antisense construct to p19ARF could overcome a growth arrest induced by p16INK4a whereas the expression of an antisense construct to p16INK4a was unable to overcome a growth arrest induced by p19ARF, suggesting that p19ARF has functions that overlap with those of p16INK4a (8). In addition, overexpression of p19ARF was able to induce a growth arrest in p53−/− cells but not in p53−/− cells that overexpressed E2F-1, again suggesting that p19ARF may be able to suppress growth through regulation of pRB as well as p53 (8). In addition, MDM2 may be directly involved in the regulation of pRB by binding to the C-terminal region of pRB (73). Alternatively, MDM2 may promote the rapid degradation of E2F-1 in cells that lack p19ARF (4). Under such conditions, it is likely that pRB would be at least partially unable to act as a tumor suppressor gene, since pRB binding to E2F-1 is required for transcriptional repression of E2F-dependent promoters and tumor suppression (22, 75). However, our data do not support such a scenario since we were unable to detect any differences in E2F activity between WT and ARF−/− MEFs transformed by LXCXE mutants of TAg. Furthermore, the steady-state levels of E2F-1 appeared similar between the WT and ARF−/− cells as determined by Western blotting (data not shown). We have not extensively tested the ability of the pRB family to bind and inactivate other members of the E2F family in the TAg-transformed MEFs. It is possible that the activities of one or more members of the E2F family are diminished in ARF−/− MEFs. Alternatively, it is possible that the LXCXE region of TAg targets an additional growth or tumor-suppressing protein whose activity may be lacking or diminished in ARF−/− MEFs.
The ability of the LXCXE mutant constructs to induce anchorage-independent growth in ARF−/− MEFs may also reflect a novel transforming function of TAg. This additional activity could overcome the growth inhibition induced by the pRB family. p300 is an especially interesting candidate for this additional activity. p300 was originally cloned as an adenovirus E1A-associated protein (18), and both p300 and the highly related CREB-binding protein (CBP) can be functionally inactivated by E1A (1, 49). E1A binds directly to p300 and CBP through a domain distinct from its pRB family-binding motif. E1A can stimulate entry into the cell cycle from quiescence using either the p300/CBP-binding region or the pRB-binding LXCXE motif (66, 78). Thus, E1A binding to p300/CBP can circumvent at least some of the growth-inhibitory functions of pRB. The mechanism of the E1A/p300 growth-promoting activity, however, is not completely understood. The effect of p300 binding to TAg also remains unknown; however, it is possible that this permits TAg to overcome the growth-suppressing function of the pRB family in ARF−/− MEFs (2, 45). The p53-binding domain of TAg can also bind to MDM2 (29). Thus, TAg could perturb the function of several growth-regulatory factors, including p53, MDM2, p300/CBP, and pRB.
In addition to the three TAg domains described here, it is possible that TAg has additional domains that contribute to transformation. The N terminus of TAg is known to bind to other cellular proteins in addition to the pRB family, most notably a 185-kDa protein that is as yet uncharacterized (42). In addition, there has been some suggestion that the DNA-binding domain of TAg (residues 131 to 280) may contribute to growth promotion and transformation. The DNA-binding region is involved in transcriptional transactivation of many cellular and viral promoters that may promote cellular growth. For example, a point substitution mutation in the DNA-binding domain reduced the ability of TAg to induce DNA synthesis in quiescent cells (15, 34). In our experiments, ARF−/− MEFs expressing T1-135 grew to half the density of those expressing full-length TAg, T1-350, or dl434-444 (Fig. 3). This may indicate that the DNA-binding region (residues 131 to 280) or perhaps the zinc finger (residues 302 to 320) can contribute to TAg-mediated transformation, though the nature and extent of this transforming activity are as yet uncharacterized.
In conclusion, these experiments have demonstrated that mutations in either the p53-binding domain or the LXCXE motif did not diminish the ability of TAg to confer anchorage-independent growth of INK4a−/− and ARF−/− MEFs. These results support the notion that the N-terminal and C-terminal regions of TAg have transforming functions beyond the disruption of the pRB and p53 tumor suppressor pathways. Furthermore, these results suggest that p19ARF may have tumor suppression functions in addition to control of the p53 tumor suppressor gene.
ACKNOWLEDGMENTS
We are grateful to Sorab Dalal and Hilde Stubdal for critical reading of the manuscript. We thank members of the DeCaprio laboratory for advice and support. We gratefully acknowledge the gift of ARF−/− MEFs and p19ARF antiserum from Martine F. Roussel (St. Jude Children's Hospital), INK4a−/− MEFs from Ronald A. DePinho (Dana-Farber Cancer Institute), the cyclin G promoter reporter plasmid pGL3-cyclin G-Luc from Carol Prives (Columbia University), pWWP-luc from Bert Vogelstein (The Johns Hopkins University School of Medicine), and dl434-444 from M. Judy Tevethia (Pennsylvania State University College of Medicine).
H.A.H.C. was supported by a fellowship from the Deutsche Forschungsgemeinschaft. A.M.B. was supported by NIH training grant 2T32CA09361 and NRSA fellowship F32CA81745. J.A.D. is a Scholar of the Leukemia and Lymphoma Society. This work was supported in part by Public Health Service grant RO1-CA63113.
REFERENCES
- 1.Arany Z, Newsome D, Olread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 2.Avantaggiati M L, Carbone M, Graessmann A, Nakatani Y, Howard B, Levine A S. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 1996;15:2236–2248. [PMC free article] [PubMed] [Google Scholar]
- 3.Bargonetti J, Reynisdottir I, Friedman P N, Prives C. Site-specific binding of wild-type p53 to cellular DNA is inhibited by SV40 T antigen and mutant p53. Genes Dev. 1992;6:1886–1898. doi: 10.1101/gad.6.10.1886. [DOI] [PubMed] [Google Scholar]
- 4.Blattner C, Sparks A, Lane D. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calabro V, Parisi T, Di Cristofano A, La Mantia G. Suppression of Ras-mediated NIH3T3 transformation by p19ARF does not involve alterations of cell growth properties. Oncogene. 1999;18:2157–2162. doi: 10.1038/sj.onc.1202532. [DOI] [PubMed] [Google Scholar]
- 6.Campbell K S, Mullane K P, Ibraham I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schauffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 7.Cao L, Faha B, Dembski M, Tsai L-H, Harlow E, Dyson N. Independent binding of the retinoblastoma protein and p107 to the transcription factor E2F. Nature. 1992;355:176–179. doi: 10.1038/355176a0. [DOI] [PubMed] [Google Scholar]
- 8.Carnero A, Hudson J D, Price C M, Beach D H. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat Cell Biol. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- 9.Chellappan S P, Hiebert S, Mudryj M, Horowitz J M, Nevins J R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Paucha E. Identification of a region of simian virus 40 large T antigen required for cell transformation. J Virol. 1990;64:3350–3357. doi: 10.1128/jvi.64.7.3350-3357.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensen J B, Imperiale M J. Inactivation of the retinoblastoma susceptibility protein is not sufficient for the transforming function of the conserved region 2-like domain of simian virus 40 large T antigen. J Virol. 1995;69:3945–3948. doi: 10.1128/jvi.69.6.3945-3948.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conzen S D, Cole C N. The three transforming regions of SV40 T antigen are required for immortalization of primary mouse embryo fibroblasts. Oncogene. 1995;11:2295–2302. [PubMed] [Google Scholar]
- 13.DeCaprio J A, Ludlow J W, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston D M. SV40 large T antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 14.de Stanchina E, McCurrach M E, Zindy F, Shieh S Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickmanns A, Zeitvogel A, Simmersbach F, Weber R, Arthur A K, Dehde S, Wideman A G, Fanning E. The kinetics of simian virus 40-induced progression of quiescent cells into S phase depend on four independent functions of large T antigen. J Virol. 1994;68:5496–5508. doi: 10.1128/jvi.68.9.5496-5508.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, Vandyke T, Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyson N, Buchkovich K, Whyte P, Harlow E. The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell. 1989;58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 18.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livinston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 19.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 20.Ewen M E, Xing Y, Lawrence J B, Livingston D M. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell. 1991;66:1155–1164. doi: 10.1016/0092-8674(91)90038-z. [DOI] [PubMed] [Google Scholar]
- 21.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Wild-type p53 activates transcription in vitro. Nature. 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 22.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W G, Jr, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 23.Forman B M, Umesono K, Chen J, Evans R M. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 24.Ginsberg D, Vairo G, Chittenden T, Xiao Z-X, Xu G, Wydner K, DeCaprio J A, Lawrence J B, Livingston D M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 25.Gjoerup O, Chao H, DeCaprio J A, Roberts T M. pRB-dependent, J domain-independent function of simian virus 40 large T antigen in override of p53 growth suppression. J Virol. 2000;74:864–874. doi: 10.1128/jvi.74.2.864-874.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris K F, Christensen J B, Radany E H, Imperiale M J. Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol Cell Biol. 1998;18:1746–1756. doi: 10.1128/mcb.18.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 29.Henning W, Rohaly G, Kolzau T, Knippschild U, Maacke H, Deppert W. MDM2 is a target of simian virus 40 in cellular transformation and during lytic infection. J Virol. 1997;71:7609–7618. doi: 10.1128/jvi.71.10.7609-7618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hijmans E M, Voorhoeve P M, Beijersbergen R L, van't Veer L J, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hofmann F, Martelli F, Livingston D M, Wang Z. The retinoblastoma gene product protects E2F-1 from degradation by the ubiquitin-proteasome pathway. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 32.Jacks T, Fazelli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 33.Kaelin W G J, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A, Livingston D M, Flemington E K. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 34.Kalderon D, Smith A E. In vitro mutagenesis of a putative DNA binding domain of SV40 large-T. Virology. 1984;139:109–137. doi: 10.1016/0042-6822(84)90334-9. [DOI] [PubMed] [Google Scholar]
- 35.Kamijo T, Bodner S, van de Kamp E, Randle D H, Sherr C J. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–2222. [PubMed] [Google Scholar]
- 36.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 38.Kelley W L, Georgopoulos C. The T/t common exon of simian virus 40, JC, and BK polyomavirus T antigens can functionally replace the J-domain of the Escherichia coli DnaJ molecular chaperone. Proc Natl Acad Sci USA. 1997;94:3674–3684. doi: 10.1073/pnas.94.8.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley W L, Landry S J. Chaperone power in a virus? Trends Biochem Sci. 1994;19:277–278. doi: 10.1016/0968-0004(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 40.Kierstead T D, Tevethia M J. Association of p53 binding and immortalization of primary C57BL/6 mouse embryo fibroblasts by using simian virus 40 T-antigen mutants bearing internal overlapping deletion mutations. J Virol. 1993;67:1817–1829. doi: 10.1128/jvi.67.4.1817-1829.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koh J, Enders G H, Dynlacht B D, Harlow E. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 42.Kohrman D C, Imperiale M J. Simian virus 40 large T antigen stably complexes with a 185-kilodalton host protein. J Virol. 1992;66:1752–1760. doi: 10.1128/jvi.66.3.1752-1760.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krek W, Livingston D M, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science. 1993;262:1557–1560. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 44.Lane D P, Crawford L V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 45.Lill N, Tevethia M J, Eckner R, Livingston D M, Modjtahedi N. p300 family members associate with the carboxyl terminus of simian virus 40 large tumor antigen. J Virol. 1997;71:129–137. doi: 10.1128/jvi.71.1.129-137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linzer D I H, Levine A J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 47.Lloyd A C, Obermuller F, Staddon S, Barth C F, McMahon M, Land H. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes Dev. 1997;11:663–677. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- 48.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 49.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 50.Neuman E, Flemington E K, Sellers W R, Kaelin W G J. Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peden K W, Srinivasan A, Farber J M, Pipas J M. Mutants with changes within or near a hydrophobic region of simian virus 40 large tumor antigen are defective for binding cellular protein p53. Virology. 1989;168:13–21. doi: 10.1016/0042-6822(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 52.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 53.Quelle D E, Cheng M, Ashmun R A, Sherr C J. Cancer-associated mutations at the INK4a locus cancel cell cycle arrest by p16INK4a but not by the alternative reading frame protein p19ARF. Proc Natl Acad Sci USA. 1997;94:669–673. doi: 10.1073/pnas.94.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 55.Risser R, Pollack R. A nonselective analysis of SV40 transformation of mouse 3T3 cells. Virology. 1974;59:477–489. doi: 10.1016/0042-6822(74)90457-7. [DOI] [PubMed] [Google Scholar]
- 56.Sawai E T, Butel J. Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J Virol. 1989;63:3961–3973. doi: 10.1128/jvi.63.9.3961-3973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Serrano M, Hannon G J, Beach D. A regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 58.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 59.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 60.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheng Q, Love T M, Schaffhausen B. J domain-independent regulation of the Rb family by polyomavirus large T antigen. J Virol. 2000;74:5280–5290. doi: 10.1128/jvi.74.11.5280-5290.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shirodkar S, Ewen M, DeCaprio J A, Morgan J, Livingston D M, Chittenden T. The transcription factor E2F interacts with the retinoblastoma product and a p107-cyclin A complex in a cell cycle-regulated manner. Cell. 1992;68:157–166. doi: 10.1016/0092-8674(92)90214-w. [DOI] [PubMed] [Google Scholar]
- 63.Silver P A, Way J C. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell. 1993;74:5–6. doi: 10.1016/0092-8674(93)90287-z. [DOI] [PubMed] [Google Scholar]
- 64.Slinskey A, Barnes D, Pipas J M. Simian virus 40 large T antigen J domain and Rb-binding motif are sufficient to block apoptosis induced by growth factor withdrawal in a neural stem cell line. J Virol. 1999;73:6791–6799. doi: 10.1128/jvi.73.8.6791-6799.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky F L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, Peters G. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stubdal H, Zalvide J, DeCaprio J A. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J Virol. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tan T H, Wallis J, Levine A J. Identification of the p53 protein domain involved in formation of the simian virus 40 large T-antigen–p53 protein complex. J Virol. 1986;59:574–583. doi: 10.1128/jvi.59.3.574-583.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson D L, Kalderon D, Smith A E, Tevethia M J. Dissociation of Rb-binding and anchorage-independent growth from immortalization and tumorigenicity using SV40 mutants producing N-terminally truncated large T antigens. Virology. 1990;178:15–34. doi: 10.1016/0042-6822(90)90375-2. [DOI] [PubMed] [Google Scholar]
- 72.Vairo G, Livingston D M, Ginsberg D. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 1995;9:869–881. doi: 10.1101/gad.9.7.869. [DOI] [PubMed] [Google Scholar]
- 73.Xiao Z X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 74.Xiao Z X, Ginsberg D, Ewen M, Livingston D M. Regulation of the retinoblastoma protein-related protein p107 by G1 cyclin-associated kinases. Proc Natl Acad Sci USA. 1996;93:4633–4637. doi: 10.1073/pnas.93.10.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 76.Zalvide J, DeCaprio J A. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zerler B, Roberts R J, Mathews M B, Moran E. Different functional domains of the adenovirus E1A gene are involved in regulation of host cell cycle products. Mol Cell Biol. 1987;7:821–829. doi: 10.1128/mcb.7.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zerrahn J, Knippschild U, Winkler T, Deppart W. Independent expression of the transforming aminoterminal domain of SV40 large T antigen from an alternatively spliced third SV40 early mRNA. EMBO J. 1993;12:4739–4746. doi: 10.1002/j.1460-2075.1993.tb06162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 81.Zhu J, Rice P W, Gorsch L, Abate M, Cole C N. Transformation of a continuous rat embryo fibroblast cell line requires three separate domains of simian virus 40 large T antigen. J Virol. 1992;66:2780–2791. doi: 10.1128/jvi.66.5.2780-2791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]